Abstract
Rice (Oryza sativa) produces ent-copalyl diphosphate for both gibberellin (GA) phytohormone and defensive phytoalexin biosynthesis, raising the question of how this initial biosynthetic step is carried out for these distinct metabolic processes. Here, a functional genomics approach has been utilized to identify two disparate ent-copalyl diphosphate synthases from rice (OsCPS1ent and OsCPS2ent). Notably, it was very recently demonstrated that only one of these (OsCPS1ent) normally operates in GA biosynthesis as mutations in this gene result in severely impaired growth. Evidence is presented here strongly indicating that the other (OsCPS2ent) is involved in related secondary metabolism producing defensive phytochemicals. In particular, under appropriate conditions, OsCPS2ent mRNA is specifically induced in leaves prior to production of the corresponding phytoalexins. Thus, transcriptional control of OsCPS2ent seems to be an important means of regulating defensive phytochemical biosynthesis. Finally, OsCPS1ent is significantly more similar to the likewise GA-specific gene An1/ZmCPS1ent in maize (Zea mays) than its class II terpene synthase paralogs involved in rice secondary metabolism. Hence, we speculate that this cross-species conservation by biosynthetic process reflects derivation of related secondary metabolism from the GA primary biosynthetic pathway prior to the early divergence between the separate lineages within the cereal/grass family (Poaceae) resulting in modern rice and maize.
Plants produce a vast and diverse array of low-Mr organic compounds. A small number of these are primary metabolites, which are common to all plant species as they are directly required for growth and development. The remaining, overwhelming majority of these natural products are considered secondary metabolites, although many have important ecological roles, particularly in plant defense (Dixon, 2001). Terpenoids, which comprise the largest class of natural products, are particularly abundant in plants and can be found in both primary and secondary metabolism (Croteau et al., 2000). Notably, a substantial fraction of the known terpenoids can be classified as labdane-related diterpenoids (20 carbon). These are defined here as minimally containing the bicyclic hydrocarbon structure found in the labdane class of diterpenoids, although this core structure can be further cyclized and/or rearranged, as in the related/derived structural classes (e.g. kauranes, abietanes, and [iso] pimaranes). Significantly, this includes the GA growth hormones as primary metabolites. Nevertheless, the vast majority of the >5,000 labdane-related diterpenoids are secondary metabolites.
Biosynthesis of labdane-related diterpenoids is initiated by class II terpene synthases, which catalyze formation of the characteristic bicyclic backbone in producing specific stereoisomers of labdadienyl/copalyl diphosphate (CPP) from the universal diterpenoid precursor (E,E,E)-geranylgeranyl diphosphate (GGPP). This protonation-initiated cyclization is fundamentally different than the diphosphate ionization initiated reactions catalyzed by the more common class I terpene synthases. Nevertheless, the class II enzymes clearly fall within the terpene synthase gene family (Bohlmann et al., 1998). However, rather than the DDXXD metal-binding motif functionally associated with class I activity (Davis and Croteau, 2000), class II terpene synthases contain a distinct DXDD motif (Sun and Kamiya, 1994), which has been functionally associated with terpene synthase class II cyclization (Peters et al., 2001). Significantly, action of the class II terpene synthase commits, or at least severely restricts, the metabolic fate of the cyclized product molecule. Thus, terpenoid biosynthesis is often controlled, in part, by regulating class II terpene synthase activity (e.g. GA biosynthesis in Arabidopsis [Arabidopsis thaliana]; Silverstone et al., 1997).
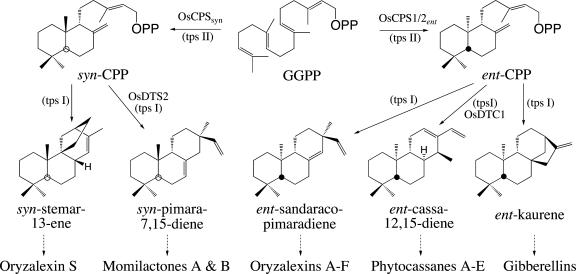
Rice (Oryza sativa) produces a number of labdane-related diterpenoids beyond the ubiquitous GAs (Fig. 1). These are momilactones A and B (Kato et al., 1973; Cartwright et al., 1981), oryzalexins A to F (Akatsuka et al., 1985; Sekido et al., 1986; Kato et al., 1993, 1994), oryzalexin S (Kodama et al., 1992), and phytocassanes A to E (Koga et al., 1995, 1997). All of these natural products are produced in leaves in response to infection with the blast pathogenic fungus Magneportha grisea and exhibit antimicrobial properties, thus qualifying as phytoalexins (VanEtten et al., 1994). Notably, rice leaves produce all of these secondary metabolites after UV irradiation as well as blast fungal infection (Kodama et al., 1988), providing a convenient method for inducing biosynthesis of these natural products and, presumably, transcription of the corresponding enzymatic machinery. In particular, UV irradiation induces biosynthesis of ent-sandaracopimaradiene, syn-pimara-7,15-diene, and syn-stemar-13-ene, the putative precursors to oryzalexins A to F, momilactones A and B, and oryzalexin S, respectively (Wickham and West, 1992). These polycyclic diterpene hydrocarbons have been shown to be selectively produced via CPP of the corresponding stereochemistry (i.e. ent or syn; Mohan et al., 1996). Recent work has identified the class I diterpene synthase producing ent-cassa-12,15-diene, the putative precursor to phytocassanes A to E (Yajima et al., 2004), stereoselectively from ent-CPP (Cho et al., 2004). Thus, the phytoalexins phytocassanes A to E and oryzalexins A to F share a common biosynthetic step, the production of ent-CPP from GGPP, with GA anabolism, bringing into question the relationship between the enzyme(s) catalyzing this step in primary versus secondary metabolism.
Figure 1.
Known cyclization steps in labdane-related diterpenoid biosynthesis in rice. Reactions catalyzed by class II (tpsII) or class I (tpsI) terpene synthases are indicated, along with the corresponding classes of natural products derived from each of the named polycyclic hydrocarbon structures (dashed arrows indicate multiple enzymatic steps).
In the only case investigated to date where reactions from GA biosynthesis have been recruited to secondary metabolism, the production of stevioside in Stevia rebaudiana, a shared class II terpene cyclase, ent-CPP synthase (CPS), was found to be common to both of these processes (Richman et al., 1999). In the case of rice labdane-related diterpenoid biosynthesis, it was very recently reported that only a single CPP synthase gene (OsCPS1) is involved in GA biosynthesis (Sakamoto et al., 2004). This was demonstrated by the severe growth defect (i.e. dwarf phenotype) of the corresponding mutant plant, along with its rescue by exogenous application of GA3. Although two other class II terpene synthase genes can be found in the rice genome, gene isolation and biochemical characterization were not reported, leaving in question the role and specific activity of these additional cyclases. We have previously demonstrated that one of these is the syn-CPP synthase involved in phytoalexin/allelochemical biosynthesis (Xu et al., 2004). Here we report that rice contains two disparate ent-CPP synthases. One of these (OsCPS1ent) corresponds to the GA metabolism-specific gene identified by Sakamoto et al. (2004), while evidence is provided strongly indicating that the other (OsCPS2ent) is involved in secondary labdane-related diterpenoid biosynthesis. Further, the sequence conservation pattern for class II terpene synthases from rice and maize (Zea mays) is used to suggest an ancient origin for the derivation of secondary (defensive phytochemical) metabolism from primary (GA phytohormone) biosynthesis in the cereal crop plant family.
RESULTS
Functional Identification of Two ent-CPP Synthases from Rice
Due to our interest in the functionally distinct class II terpene synthases as potentially significant targets for metabolic engineering and biochemical analysis, we identified three putative class II terpene synthases from the extensive sequence information available for rice (Goff et al., 2002; Yu et al., 2002; Kikuchi et al., 2003). One of these was readily amplified from mRNA prepared from UV-irradiated rice leaves and found to produce syn-CPP for phytoalexin/allelochemical biosynthesis (accession AY530101; Xu et al., 2004). The second class II terpene synthase gene was not as easily isolated. Nevertheless, we have now cloned the corresponding full-length cDNA and deposited the associated sequence into the various nucleotide databases as accession AY602991. The third gene was available from the rice full-length cDNA sequencing project (accession AK100333; Kikuchi et al., 2003) and obtained from the Rice Genome Resource Center (www.rgrc.dna.affrc.go.jp). Notably, this last gene sequence contains EXDD in place of the otherwise conserved DXDD motif functionally associated with class II cyclization. This was confirmed by independently cloning a gene fragment covering this region.
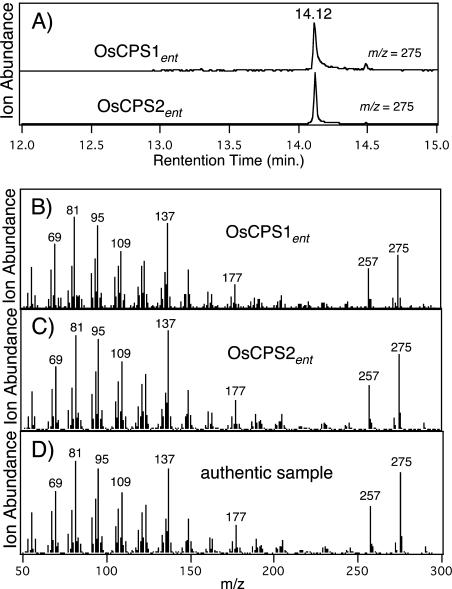
Work with OsCPSsyn suggested that removal of the N-terminal transit peptide, which is only required for plastid import of diterpene synthases in planta, to form a pseudomature construct, is required for functional expression (Xu et al., 2004). Thus, for each of the two unidentified class II terpene synthases, truncated versions were constructed for recombinant expression (see Fig. 4). These were based on truncation analysis of the Arabidopsis CPSent (S. Prisic and R.J. Peters, unpublished data). Enzymatic assays were carried out with partially purified recombinant preparations and GGPP as substrate. Phosphatase treatment was then employed to remove the pyrophosphate from GGPP and any enzymatically formed derivative to enable straightforward extraction of the resulting alcohol into organic solvent. Enzymatic conversion of GGPP was analyzed by gas chromatography-mass spectrometry (GC-MS) of the resulting extracts, demonstrating production of an altered prenyl diphosphate structure (this is not observed with extracts from untransformed bacteria). Comparison of the enzymatically formed compound to similarly dephosphorylated authentic samples of ent- and syn-CPP demonstrated that both enzymes produce ent-CPP (Fig. 2). Therefore, we have functionally identified two rice ent-CPP synthases (OsCPS1ent and OsCPS2ent).
Figure 4.
Amino acid alignment of class II terpene synthases from Poaceae. Truncations for recombinant pseudomature constructs are indicated by the arrowhead and the (D,E)XD(D,N) motif by an underline.
Figure 2.
Production of ent-CPP. A, GC-MS analysis (275 m/z extracted ion chromatographs) of the dephosphorylated reaction products from GGPP produced by the indicated enzyme. B, Mass spectrum of the GC-MS 275 m/z chromatograph peak for OsCPS1ent (RT = 14.12 min); C, that for OsCPS2ent (RT = 14.12); and D, that for authentic dephosphorylated ent-CPP (i.e. ent-copalol), which also exhibits RT = 14.12 min and is clearly separable from dephosphorylated syn-CPP (i.e. syn-copalol; RT = 13.96 min) and GGPP (i.e. geranylgeraniol; RT = 13.87 min).
OsCPS1ent and OsCPS2ent Have Distinct Metabolic Functions
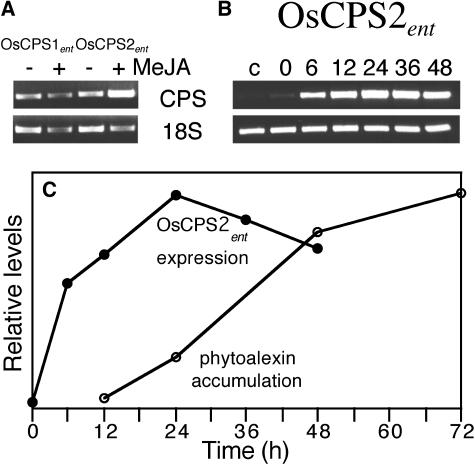
The recent publication by Sakamoto et al. (2004) clearly demonstrated that only a single class II terpene synthase, which they termed OsCPS1, is normally involved in GA metabolism. As expected, one of the two ent-CPP synthases identified here corresponds to OsCPS1, specifically, the gene derived from the large-scale cDNA project (accession AK10033) that we have designated OsCPS1ent to more precisely reflect its function. Because the other functional CPS identified here (accession AY602991) does not compensate for mutations in OsCPS1ent and, therefore, is not expressed in conjunction with GA biosynthesis, we hypothesized that this gene (designated OsCPS2ent as it corresponds to the OsCPS2 of Sakamoto et al., 2004) might be alternatively expressed for production of the known ent-labdane-related diterpenoid defensive secondary metabolites. This was initially examined by characterization of gene transcription in response to methyl jasmonate (MeJA). Application of this important plant defense signaling molecule previously has been shown to induce phytoalexin biosynthesis in rice cell culture (Nojiri et al., 1996). Further, we have recently demonstrated that transcription of OsCPSsyn (which corresponds to OsCPS4 from Sakamoto et al., 2004) that can only be involved in defensive secondary metabolism (see Fig. 1) is up-regulated in response to MeJA (Xu et al., 2004). While transcription of the GA-specific OsCPS1ent gene is not significantly altered, OsCPS2ent mRNA levels were consistently increased approximately 5-fold by MeJA treatment (Fig. 3A), thus indicating a role for OsCPS2ent in defensive secondary metabolism. Also consistent with a role in defense, 16 of the 21 expressed sequence tags currently associated with OsCPS2ent in the TIGR Gene Index are from blast pathogen infected rice expressed sequence tag projects (www.tigr.org; rice-TC187448).
Figure 3.
Expression analysis. Semiquantitative RT-PCR analysis of mRNA expression levels for the indicated genes. Specific bands corresponding to CPS or the 18S rRNA control are indicated. A, Expression in germinated seedlings in response to application of 0.5 mm MeJA. B, Expression of OsCPS2ent in detached leaves from 4-week-old plants in response to UV irradiation. Time (hours) after exposure is indicated (c = control leaves after approximately 18 h). C, Relative graphical comparison of OsCPS2ent mRNA levels (black circles) and ent-labdane-related diterpenoid phytoalexin accumulation (white circles; as described by Kodama et al., 1988) in UV-irradiated detached leaves.
Previous review of the relevant literature has been used to suggest that plant secondary metabolism is most often regulated at the level of transcription (Peters and Croteau, 2004). Transcriptional control is manifested by up-regulation of enzymatic mRNA levels, with subsequent phytochemical accumulation, in response to the appropriate environmental conditions. This was investigated through use of UV irradiation, which has long been appreciated to induce phytoalexin biosynthesis in rice (Cartwright et al., 1977). In addition, quantitative analysis of phytochemical accumulation for the detached leaf UV-irradiation method used here has previously been reported (Kodama et al., 1988). Consistent with the hypothesized role in phytoalexin biosynthesis, transcription of OsCPS2ent is dramatically increased in UV-irradiated rice leaves prior to the reported phytochemical accumulation (Kodama et al., 1988; Fig. 3, B and C). Similar results have been reported for other terpene synthases that function in phytoalexin biosynthesis, including the ent-CPP specific (i.e. downstream) class I terpene synthase OsDTC1 (Cho et al., 2004; Sakamoto et al., 2004; Wilderman et al., 2004; Xu et al., 2004). Therefore, these results indicate that biosynthesis of the ent-labdane-related diterpenoid phytoalexins also may be regulated, at least to some extent, through transcriptional control of the associated class II terpene synthase OsCPS2ent.
Conservation of Class II Terpene Synthases within Poaceae
Not surprisingly, given their common origin from within the cereal and grass plant family (Poaceae), the rice class II terpene synthases are most similar to An1/ZmCPS1ent from maize (Bensen et al., 1995; Fig. 4). Outside of Poaceae, amino acid sequence alignment of OsCPS1ent and OsCPS2ent with known class II terpene synthases also demonstrates the expected homology. Specifically, OsCPS1ent shares 42% to 45% identity with the other known GA-specific CPSent, all of which are from various dicot species (Sun and Kamiya, 1994; Ait-Ali et al., 1997; Smith et al., 1998; Rebers et al., 1999; Richman et al., 1999), and approximately 30% identity with the known gymnosperm bifunctional class II/I terpene synthases (Stofer Vogel et al., 1996; Schepmann et al., 2001; Martin et al., 2004). Somewhat lower homology is found for OsCPS2ent, which shares 37% to 40% identity with CPSent from dicots and approximately 27% identity with the gymnosperm bifunctional terpene synthases. This is similar to the homology levels observed with OsCPSsyn, which shares 38% to 42% identity with dicot CPSent and approximately 28% identity with the bifunctional gymnosperm enzymes. Significantly, OsCPS1ent is much more similar to An1/ZmCPS1ent (64% identity) than either of its paralogs, OsCPS2ent or OsCPSsyn, which are each only 44% identical to OsCPS1ent. OsCPS2ent and OsCPSsyn are also only marginally more similar to An1/ZmCPS1ent (42% and 44% identity, respectively) than to CPSent from dicots. However, despite the difference in product stereochemistry, OsCPS2ent and OsCPSsyn are relatively closely related, exhibiting 53% identity. This striking conservation pattern of class II terpene synthases within Poaceae is also evident at the evolutionarily more relevant nucleotide level (Table I).
Table I.
Cereal CPS nucleotide identity
| Nucleotide Identity | OsCPS1ent | OsCPS2ent | OsCPSsyn |
|---|---|---|---|
| An1/ZmCPS1ent | 74% | 57% | 56% |
| OsCPS1ent | – | 57% | 56% |
| OsCPS2ent | – | – | 65% |
Percentage identity over aligned region of nucleotide sequence is shown. The significant identity between functionally related CPS is indicated by the bold numbers (dash indicates redundant data).
DISCUSSION
Terpene synthases seem to be conserved by taxonomic origin rather than mechanistic considerations (Bohlmann et al., 1998). Therefore, to provide a model system for investigation of structure-function relationships in class II terpene synthases, we have functionally characterized the corresponding genes from rice, which is known to produce two stereochemically varied class II cyclization products (i.e. ent- and syn-CPP). In combination with the previously described syn-CPP synthase (Xu et al., 2004), the two genes reported here to encode ent-CPP synthases provide an ideal model system for biochemical study. In particular, OsCPS2ent and OsCPSsyn are approximately 53% identical at the amino acid level, and investigation of the differential determinants for the observed change in product stereochemical outcome is expected to be informative. More immediately, the ability of OsCPS1ent to carry out class II cyclization, despite containing an alternative EXDD rather than DXDD motif, demonstrates the ability of Glu to functionally substitute for the first Asp. This is consistent with previous mutational analysis of this motif in the bifunctional class II/I terpene cyclase abietadiene synthase from grand fir (Abies grandis), where only the first Asp could be mutated to Glu with retention of class II activity (Peters and Croteau, 2002). In addition, this same mutational analysis also found that Asn could only functionally substitute for the last Asp, and we speculate that the corresponding alternative DXDN motif may also be naturally found. Accordingly, the catalytically requisite class II cyclization DXDD motif might be more correctly defined as (D,E)XD(D,N), which should be useful in guiding functional annotation of terpene synthases.
Significantly, the two ent-CPP synthases identified here are not functionally equivalent, but operate in different metabolic processes in planta. In particular, the recent report by Sakamoto et al. (2004) identifying rice GA biosynthetic genes by mutant plant phenotype (i.e. dwarfism) demonstrated that only OsCPS1ent operates in GA biosynthesis. Hence, the fact that OsCPS2ent does not compensate for loss of OsCPS1ent in GA biosynthesis, despite its biochemically identical activity, clearly demonstrates that these class II terpene synthases have distinct expression patterns. Accordingly, the expression analysis presented here (Fig. 3) strongly indicates that OsCPS2ent is involved in biosynthesis of the known ent-CPP derived labdane-related diterpenoid rice phytoalexins (Fig. 1). More specifically, OsCPS2ent mRNA levels are increased prior to phytochemical production, suggesting that transcriptional control of OsCPS2ent represents a regulatory point for phytoalexin biosynthesis. Notably, OsCPS2ent resides in a terpene synthase gene cluster in the rice genome along with the previously identified class I terpene synthase OsDTC1 that acts specifically on ent-CPP for phytoalexin biosynthesis (Cho et al., 2004). This is also consistent with a role in secondary metabolism for OsCPS2ent as the likewise phytoalexin-specific and similarly consecutively acting OsCPSsyn and class I syn-CPP specific 9β-pimara-7,15-diene synthase OsDTS2 are also clustered together, suggesting that this provides an evolutionary mechanism linking their functionally coupled, sequential biosynthetic activity (Wilderman et al., 2004).
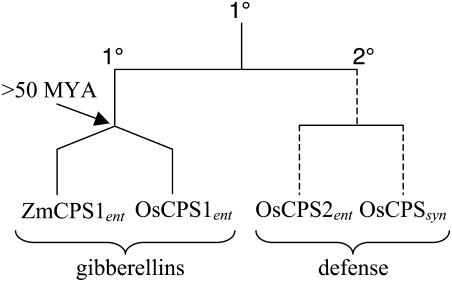
Interestingly, OsCPS1ent is significantly more similar to the maize CPS An1/ZmCPS1ent that also functions in GA metabolism (Bensen et al., 1995), rather than OsCPS2ent. In contrast, OsCPS2ent is most similar to OsCPSsyn, which is also involved in phytoalexin anabolism (Xu et al., 2004), despite their stereochemical difference in product outcome. Thus, we speculate that OsCPS2ent and OsCPSsyn share a common ancestral gene that was similarly involved in secondary (defensive phytochemical), rather than primary (GA phytohormone), metabolism. Such evolution of syn-CPP synthase activity by duplication and divergence of an already appropriately controlled (i.e. secondary metabolism specific) ancestral ent-CPP specific gene would be consistent with the suggestion that secondary metabolism might most readily evolve as stepwise changes first in regulation and then specificity (Pichersky and Gang, 2000). Further, the observed conservation pattern also suggests that gene duplication and specialization for primary (GA phytohormone) or secondary (defensive phytochemical) metabolism occurred prior to the speciation event leading to the split between rice and maize. Notably, extensive phylogenetic analysis of the large grass plant family indicates that rice and maize represent the two major lineages within Poaceae, which together constitute over 95% of the constituent genera and particularly include all cereal crops (Grass Phylogeny Working Group, 2001). Evidence from the fossil record (Stebbins, 1981) has been used to suggest that these lineages diverged over 50 million years ago (Wolfe et al., 1989). Hence, this split between primary and secondary labdane-related diterpenoid metabolism appears to be quite ancient (Fig. 5). Intriguingly, maize has also been shown to produce labdane-related diterpenes in response to fungal infection (Mellon and West, 1979) and must contain a second CPS that can partially compensate for loss of An1/ZmCPS1ent in GA biosynthesis (i.e. inactivating insertional mutagenesis of An1/ZmCPS1ent only results in a semidwarf phenotype; Bensen et al., 1995). This is consistent with early derivation (i.e. evolution) of related secondary metabolism from GA biosynthesis and may further indicate broad retention of this type of secondary metabolism for defensive purposes in the important cereal crop plant family.
Figure 5.
Putative phylogenetic relationships between Poaceae class II terpene synthases. Approximate cladogram indicating initial gene duplication and specialization for primary (1°) and secondary (2°) metabolism, followed by gene duplication resulting in OsCPS2ent and OsCPSsyn, and ancient divergence between rice and maize lineages that produced the GA-specific OsCPS1ent and An1/ZmCPS1ent. Horizontal lines indicate gene duplication events, while diagonal lines indicate species divergence (>50 MYA = over 50 million years ago). Note that the dashed lines indicate the uncertainty inherent in positioning the split of OsCPSsyn from OsCPS2ent given the need for duplicated genes to assume different activities (e.g. stereochemically distinct products) to provide evolutionary pressure for retention of both genes.
As a final note, while this manuscript was under review, essentially identical data and results were reported (Otomo et al., 2004), although the alternative EXDD motif, functional clustering of terpene synthases, and evolutionary implications were not discussed. While some significant differences can be observed between our amino acid sequences and those originally reported by Otomo et al. (2004), these have apparently been resolved (T. Toyomasu, personal communication), and the remaining relatively minor differences presumably reflect allelic differences between the subsp. japonica rice used by Otomo et al. (2004) and the subsp. indica used here and in our previous study (Xu et al., 2004). In particular, their OsCyc1 (AB066270) corresponds to our OsCPSsyn (Xu et al., 2004), their OsCyc2 (AB066271) to the OsCPS2ent reported here, and their OsCPS1 (AB126932) to the OsCPS1ent reported here.
CONCLUSION
In summary, we have used the extensive sequence information available for rice to functionally identify two novel, disparate ent-CPP synthases (OsCPS1ent and OsCPS2ent). In combination with the previously identified syn-CPP synthase (Xu et al., 2004), these offer an ideal model system for future structure-function investigations of class II terpene synthases. Notably, these two biochemically identical genes are not functionally equivalent, rather being separately responsible for primary or secondary metabolic processes. Specifically, GA biosynthesis is dependent on OsCPS1ent, while OsCPS2ent almost certainly acts in defensive secondary metabolism. Further, as expected for a role in initiating labdane-related diterpenoid biosynthesis, transcription of OsCPS2ent is induced by conditions that stimulate phytoalexin biosynthesis, indicating a regulatory control point for this important metabolic process. In addition, the conservation of class II terpene synthases within the cereal/grass plant family may indicate an ancient split of labdane-related diterpenoid metabolism into separate GA and defensive phytochemical biosynthetic pathways in Poaceae.
MATERIALS AND METHODS
Chemicals
GGPP and ent- and syn-CPP were synthesized as previously described (Mohan et al., 1996). Unless otherwise noted, all other chemicals were purchased from Fisher Scientific (Loughborough, Leicestershire, UK).
Plant Material
Rice plants (Oryza sativa L. subsp. indica cv IR24) and seedlings (subsp. japonica cv Nipponbare) were those previously described (Xu et al., 2004). Briefly, detached leaves from 4-week-old greenhouse grown plants were UV irradiated at 254 nm from 15-cm distance for 25 min and then incubated for the indicated period of time under dark humid conditions at 30°C. Seeds were surface sterilized and germinated for 1 week under sterile humid conditions at 30°C in the dark. After germination, the seedlings underwent MeJA treatment, being sprayed with approximately 2 mL 0.1% Tween 20 ± 0.5 mm MeJA/g of plant weight, and the seedlings were then incubated for two more days under the same conditions. RNA was isolated using Concert Plant Reagent (Invitrogen, Carlsbad, CA). Semiquantitative reverse transcription (RT)-PCR expression analysis, using 0.5 μg total RNA and the gene-specific primers described below or QuantumRNA 18S standard primers (Ambion, Austin, TX), was also carried out as described by Xu et al. (2004). In particular, appropriate control experiments were performed to ensure that the final amplification conditions were in the linear response range (CPS expression was quantified by normalization against the corresponding 18S band) and each set of expression conditions (seedlings ± MeJA and detached plant leaves ± UV irradiation) was analyzed in two independent experiments, yielding essentially identical results.
Cloning
Three putative class II terpene synthases were identified by BLAST queries of GenBank (www.ncbi.nlm.nih.gov/BLAST/), The Institute for Genomic Research (TIGR; tigrblast.tigr.org/tgi/), and the rice cDNA database at KOME (cdna01.dna.affrc.go.jp/cDNA/) with the nucleotide sequence of CPS from Arabidopsis (Arabidopsis thaliana; Sun and Kamiya, 1994). RT-PCR reactions were performed to verify expression of the predicted genes in UV-irradiated leaves by generating fragments of the corresponding sequences. After our initial success with OsCPSsyn (Xu et al., 2004), we continued our efforts to verify expression for the remaining rice class II terpene synthases, which required trials with a number of different primer pairs. The ultimately successful primer pairs were OsCPS1-F (GAAAAGGATGGAGCGTTTCAT) and OsCPS1-R (TCAAATAACTTGCTCAAAAATCACTTCAGAAATGTGTTGGTCAA), and OsCPS2-F (CCTACCGCGCCTCGCAG) and OsCPS2-R (GTCGATGAGCTCATCAAGTG). All fragments were cloned into pCR-Zero-Blunt (Invitrogen) and verified by complete sequencing. Notably, the OsCPS1ent fragment covers the region containing the alternative EXDD motif. Thus, OsCPS1ent from both the subsp. indica used here and the originally reported subsp. japonica (Kikuchi et al., 2003) contain this alternative motif. The full-length cDNA for OsCPS1ent was obtained from the rice full-length cDNA sequencing project (www.rgrc.dna.affrc.go.jp). To obtain the open reading frame for OsCPS2ent, 3′ RACE (GeneRacer; Invitrogen) was performed using the 5′ end primer ATGCAGATGCAGGTGCTCACC. The 5′ untranslated region was obtained by 5′ RACE using the OsCPS2-R primer. Both RACE products were cloned into pCR-Zero-Blunt and verified by complete sequencing of two independent isolates, demonstrating identical sequence in the 1,932-bp overlap. Truncated versions of both OsCPS1ent and OsCPS2ent were then cloned into pENTR/SD/D-TOPO via PCR amplification for directional topoisomerization and verified by complete sequencing. These were then transferred by directional recombination to the T7 based expression vector pDEST14 (Gateway system; Invitrogen).
Recombinant Expression and Functional Characterization
Expression was carried out with the BL21 derived C41 strain (Miroux and Walker, 1996), as described for OsCPSsyn by Xu et al. (2004). Briefly, cells were grown to midlog phase at 37°C then shifted to 16°C for 1 to 2 h prior to induction (1 mm isopropylthio-β-galactoside) and overnight expression. Lysates were prepared by sonication and the recombinant protein partially purified by batch binding to ceramic hydroxyapatite type II (Bio-Rad, Hercules, CA) and elution into 0.2 M sodium phosphate, pH 6.8. Enzymatic assays were performed with these preparations, also as described by Xu et al. (2004). Briefly, reactions with 5 μm GGPP were carried out for 1 h at 30°C prior to enzymatic dephosphorylation (calf intestinal phosphatase; New England Biolabs, Beverly, MA) to allow extraction into organic solvent (hexanes) and GC-MS analysis. This was performed using an HP-5 column on an Agilent (Palo Alto, CA) 6890N GC instrument with 5973N mass selective detector. Samples (5 μL) were injected at 40°C in the splitless mode. After holding 3 min at 40°C, the temperature was increased at 20°C/min to 300°C, where it was held for 3 min. MS data was collected from 50 to 500 m/z during the temperature ramp.
Sequence Analysis and Alignments
Sequence alignments, identity calculations, and phylogenetic analysis were performed with the AlignX program in the Vector NTI software package (Invitrogen) using standard parameters. An1/ZmCPS1ent (L37750; Bensen et al., 1995) was the reference sequence for alignment and phylogenetic analysis of Poaceae class II terpene synthases. For individual comparisons with dicot CPSent the rice genes were defined as the reference sequence. The CPSent sequences were those from Arabidopsis (U11034; Sun and Kamiya, 1994), Pisum sativum (U63652; Ait-Ali et al., 1997), Curcurbita maxima (AF049905 and AF049906; Smith et al., 1998), and Stevia rebaudiana (AF034545; Richman et al., 1999). The gymnosperm bifunctional class II/I terpene synthase sequences were those from Abies grandis (U50765; Stofer Vogel et al., 1996), Ginkgo biloba (AF331704; Schepmann et al., 2001), and Picea abies (AY473620 and AY473621; Martin et al., 2004).
Sequence data from this article have been deposited with the EMBL/GenBank data libraries under accession number AY602991.
Acknowledgments
The authors thank Dr. Adam Bogdanove and his laboratory for supplying rice plants and seeds, Dr. Robert Thornburg for MeJA, Dr. Robert M. Coates and his laboratory for the ent- and syn-CPP samples used in this study, Dana J. Morrone for critically reading the manuscript, and Dr. Eran Pichersky for helpful feedback regarding gene evolution.
This work was supported by funds from Iowa State University and the Roy J. Carver Charitable Trust (grant no. 03–190).
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.104.050567.
References
- Ait-Ali T, Swain SM, Reid JB, Sun T, Kamiya Y (1997) The LS locus of pea encodes the gibberellin biosynthesis enzyme ent-kaurene synthase A. Plant J 11: 443–454 [DOI] [PubMed] [Google Scholar]
- Akatsuka T, Kodama O, Sekido H, Kono Y, Takeuchi S (1985) Novel phytoalexins (oryzalexins a, b, and c) isolated from rice blast leaves infected with Pyricularia oryzae. Part I: isolation, characterization and biological activities of oryzalexins. Agric Biol Chem 49: 1689–1694 [Google Scholar]
- Bensen RJ, Johal GS, Crane VC, Tossberg JT, Schnable PS, Meeley RB, Briggs SP (1995) Cloning and characterization of the maize An1 gene. Plant Cell 7: 75–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohlmann J, Meyer-Gauen G, Croteau R (1998) Plant terpenoid synthases: molecular biology and phylogenetic analysis. Proc Natl Acad Sci USA 95: 4126–4133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cartwright DW, Langcake P, Pryce RJ, Leworthy DP, Ride JP (1977) Chemical activation of host defence mechanisms as a basis for crop protection. Nature 267: 511–513 [Google Scholar]
- Cartwright DW, Langcake P, Pryce RJ, Leworthy DP, Ride JP (1981) Isolation and characterization of two phytoalexins from rice as momilactones A and B. Phytochemistry 20: 535–537 [Google Scholar]
- Cho E-M, Okada A, Kenmoku H, Otomo K, Toyomasu T, Mitsuhashi W, Sassa T, Yajima A, Yabuta G, Mori K, et al (2004) Molecular cloning and characterization of a cDNA encoding ent-cassa-12,15-diene synthase, a putative diterpenoid phytoalexin biosynthetic enzyme, from suspension-cultured rice cells treated with a chitin elicitor. Plant J 37: 1–8 [DOI] [PubMed] [Google Scholar]
- Croteau R, Kutchan TM, Lewis NG (2000) Natural products (secondary metabolites). In B Buchanan, W Gruissem, R Jones, eds, Biochemistry and Molecular Biology of Plants. American Society of Plant Biologists, Rockville, MD, pp 1250–1318
- Davis EM, Croteau R (2000) Cyclization enzymes in the biosynthesis of monoterpenes, sesquiterpenes, and diterpenes. Top Curr Chem 209: 53–95 [Google Scholar]
- Dixon RA (2001) Natural products and plant disease resistance. Nature 411: 843–847 [DOI] [PubMed] [Google Scholar]
- Goff SA, Ricke D, Lan TH, Presting G, Wang R, Dunn M, Glazebrook J, Sessions A, Oeller P, Varma H, et al (2002) A draft sequence of the rice genome (Oryza sativa L. ssp. japonica). Science 296: 92–100 [DOI] [PubMed] [Google Scholar]
- Grass Phylogeny Working Group (2001) Phylogeny and subfamilial classification of the grasses (Poaceae). Ann Mo Bot Gard 88: 373–457 [Google Scholar]
- Kato H, Kodama O, Akatsuka T (1993) Oryzalexin E, a diterpene phytoalexin from UV-irradiated rice leaves. Phytochemistry 33: 79–81 [Google Scholar]
- Kato H, Kodama O, Akatsuka T (1994) Oryzalexin F, a diterpene phytoalexin from UV-irradiated rice leaves. Phytochemistry 36: 299–301 [Google Scholar]
- Kato T, Kabuto C, Sasaki N, Tsunagawa M, Aizawa H, Fujita K, Kato Y, Kitahara Y, Takahashi N (1973) Momilactones, growth inhibitors from rice, Oryza sativa L. Tetrahedron Lett 14: 3861–3864 [Google Scholar]
- Kikuchi S, Satoh K, Nagata T, Kawagashira N, Doi K, Kishimoto N, Yazaki J, Ishikawa M, Yamada H, Ooka H, et al (2003) Collection, mapping, and annotation of over 28,000 cDNA clones from japonica rice. Science 301: 376–379 [DOI] [PubMed] [Google Scholar]
- Kodama O, Li WX, Tamogami S, Akatsuka T (1992) Oryzalexin-S, a novel stemarane-type diterpene rice phytoalexin. Biosci Biotechnol Biochem 56: 1002–1003 [DOI] [PubMed] [Google Scholar]
- Kodama O, Suzuki T, Miyakawa J, Akatsuka T (1988) Ultraviolet-induced accumulation of phytoalexins in rice leaves. Agric Biol Chem 52: 2469–2473 [Google Scholar]
- Koga J, Ogawa N, Yamauchi T, Kikuchi N, Ogasawara N, Shimura M (1997) Functional moiety for the antifungal activity of phytocassane E, a diterpene phytoalexin from rice. Phytochemistry 44: 249–253 [Google Scholar]
- Koga J, Shimura M, Oshima K, Ogawa N, Yamauchi T, Ogasawara N (1995) Phytocassanes A, B, C, and D, novel diterpene phytoalexins from rice, Oryza sativa L. Tetrahedron 51: 7907–7918 [Google Scholar]
- Martin DM, Faldt J, Bohlmann J (2004) Functional characterization of nine norway spruce TPS genes and evolution of gymnosperm terpene synthases of the TPS-d subfamily. Plant Physiol 135: 1908–1927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellon JE, West CA (1979) Diterpene biosynthesis in maize seedlings in response to fungal infection. Plant Physiol 64: 406–410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miroux B, Walker JE (1996) Over-production of proteins in Escherichia coli: mutant hosts that allow synthesis of some membrane proteins and globular proteins at high levels. J Mol Biol 260: 289–298 [DOI] [PubMed] [Google Scholar]
- Mohan RS, Yee NKN, Coates RM, Ren YY, Stamenkovic P, Mendez I, West CA (1996) Biosynthesis of cyclic diterpene hydrocarbons in rice cell suspensions: conversion of 9,10-syn-labda-8(17),13-dienyl diphosphate to 9β-pimara-7,15-diene and stemar-13-ene. Arch Biochem Biophys 330: 33–47 [DOI] [PubMed] [Google Scholar]
- Nojiri H, Sugimora M, Yamane H, Nishimura Y, Yamada A, Shibuya N, Kodama O, Murofushi N, Omori T (1996) Involvement of jasmonic acid in elicitor-induced phytoalexin production in suspension-cultured rice cells. Plant Physiol 110: 387–392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otomo K, Kenmoku H, Oikawa H, Konig WA, Toshima H, Mitsuhashi W, Yamane H, Sassa T, Toyomasu T (2004) Biological functions of ent- and syn-copalyl diphosphate synthases in rice: key enzymes for the branch point of gibberellin and phytoalexin biosynthesis. Plant J 39: 886–893 [DOI] [PubMed] [Google Scholar]
- Peters RJ, Croteau RB (2002) abietadiene synthase catalysis: conserved residues involved in protonation-initated cyclization of geranylgeranyl diphosphate to (+)-copalyl diphosphate. Biochemistry 41: 1836–1842 [DOI] [PubMed] [Google Scholar]
- Peters RJ, Croteau RB (2004) Metabolic engineering of plant secondary metabolism. In G Kishore, ed, Handbook of Plant Biotechnology: Applications of Plant Biotechnology in Agriculture, the Pharmaceutical Industry, and Other Industries, Vol 2. John Wiley & Sons, London, pp 609–628
- Peters RJ, Ravn MM, Coates RM, Croteau RB (2001) Bifunctional abietadiene synthase: free diffusive transfer of the (+)-copalyl diphosphate intermediate between two distinct active sites. J Am Chem Soc 123: 8974–8978 [DOI] [PubMed] [Google Scholar]
- Pichersky E, Gang DR (2000) Genetics and biochemistry of secondary metabolites in plants: an evolutionary perspective. Trends Plant Sci 5: 439–445 [DOI] [PubMed] [Google Scholar]
- Rebers M, Kaneta T, Kawaide H, Yamaguchi S, Yang YY, Imai R, Sekimoto H, Kamiya Y (1999) Regulation of gibberellin biosynthesis genes during flower and early fruit development of tomato. Plant J 17: 241–250 [DOI] [PubMed] [Google Scholar]
- Richman AS, Gijzen M, Starratt AN, Yang Z, Brandle JE (1999) Diterpene synthesis in Stevia rebaudiana: recruitment and up-regulation of key enzymes from the gibberellin biosynthetic pathway. Plant J 19: 411–421 [DOI] [PubMed] [Google Scholar]
- Sakamoto T, Miura K, Itoh H, Tatsumi T, Ueguchi-Tanaka M, Ishiyama K, Kobayashi M, Agrawal GK, Takeda S, Abe K, et al (2004) An overview of gibberellin metabolism enzyme genes and their related mutants in rice. Plant Physiol 134: 1642–1653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schepmann HG, Pang J, Matsuda SP (2001) Cloning and characterization of Ginkgo biloba levopimaradiene synthase, which catalyzes the first committed step in ginkolide biosynthesis. Arch Biochem Biophys 392: 263–269 [DOI] [PubMed] [Google Scholar]
- Sekido H, Endo T, Suga R, Kodama O, Akatsuka T, Kono Y, Takeuchi S (1986) Oryzalexin D (3,7-dihydroxy-(+)-sandaracopimaradiene), a new phytoalexin isolated from blast-infected rice leaves. J Pestic Sci 11: 369–372 [Google Scholar]
- Silverstone AL, Chang C-W, Krol E, Sun T-P (1997) Developmental regulation of the gibberellin biosynthetic gene GA1 in Arabidopsis thaliana. Plant J 12: 9–19 [DOI] [PubMed] [Google Scholar]
- Smith MW, Yamaguchi S, Ait-Ali T, Kamiya Y (1998) The first step of gibberellin biosynthesis in pumpkin is catalyzed by at least two copalyl diphosphate synthases encoded by differentially regulated genes. Plant Physiol 118: 1411–1419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stebbins GL (1981) Coevolution of grasses and herbivores. Ann Mo Bot Gard 68: 75–86 [Google Scholar]
- Stofer Vogel B, Wildung MR, Vogel G, Croteau R (1996) Abietadiene synthase from grand fir (Abies grandis). J Biol Chem 271: 23262–23268 [DOI] [PubMed] [Google Scholar]
- Sun T-P, Kamiya Y (1994) The Arabidopsis GA1 locus encodes the cyclase ent-kaurene synthetase A of gibberellin biosynthesis. Plant Cell 6: 1509–1518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- VanEtten HD, Mansfield JW, Bailey JA, Farmer EE (1994) Two classes of plant antibiotics: phytoalexins versus ‘phytoanticipins’. Plant Cell 6: 1191–1192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickham KA, West CA (1992) Biosynthesis of rice phytoalexins: identification of putative diterpene hydrocarbon precursors. Arch Biochem Biophys 293: 320–332 [DOI] [PubMed] [Google Scholar]
- Wilderman PR, Xu M, Jin Y, Coates RM, Peters RJ (2004) Identification of syn-pimara-7,15-diene synthase reveals functional clustering of terpene synthases involved in rice phytoalexin/allelochemical biosynthesis. Plant Physiol 135: 2098–2105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfe KH, Gouy M, Yang Y-W, Sharp PM, Li W-H (1989) Date of the monocot-dicot divergence estimated from chloroplast DNA sequence data. Proc Natl Acad Sci USA 86: 6201–6205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu M, Hillwig ML, Prisic S, Coates RM, Peters RJ (2004) Functional identification of rice syn-copalyl diphosphate synthase and its role in initiating biosynthesis of diterpenoid phytoalexin/allelopathic natural products. Plant J 39: 309–318 [DOI] [PubMed] [Google Scholar]
- Yajima A, Mori K, Yabuta G (2004) Total synthesis of ent-cassa-12,15-diene, a putative precursor of rice phytoalexins, phytocassanes A-E. Tetrahedron Lett 45: 167–169 [Google Scholar]
- Yu J, Hu S, Wang J, Wong GK, Li S, Liu B, Deng Y, Dai L, Zhou Y, Zhang X, et al (2002) A draft sequence of the rice genome (Oryza sativa L. ssp. indica). Science 296: 79–92 [DOI] [PubMed] [Google Scholar]