Abstract
Hexadeca 7,10,13-trienoic acid (16:3Δ7,10,13) is one of the most abundant fatty acids in Arabidopsis (Arabidopsis thaliana) and a functional component of thylakoid membranes, where it is found as an sn-2 ester of monogalactosyldiacylglycerol. The Arabidopsis fad5 mutant lacks activity of the plastidial palmitoyl-monogalactosyldiacylglycerol Δ7-desaturase FAD5, and is characterized biochemically by the absence of 16:3Δ7,10,13 and physiologically by reduced chlorophyll content and a reduced recovery rate after photoinhibition. While the fad5 mutation has been mapped, the FAD5 gene was not unambiguously identified, and a formal functional characterization by complementation of fad5 mutant phenotypes has not been reported. Two candidate genes (At3g15850 and At3g15870) predicted to encode plastid-targeted desaturases at the fad5 chromosomal locus were cloned from fad5 plants and sequenced. A nonsense mutation changing codon TGG (Trp-98) into TGA (stop) was identified in At3g15850 (ADS3), whereas the fad5 At3g15870 allele was identical to wild type (after correction of a sequencing error in the published wild-type genomic At3g15870 sequence). Expression of a genomic clone or cDNA for wild-type At3g15850 conferred on fad5 plants the ability to synthesize 16:3Δ7,10,13 and restored leaf chlorophyll content. Arabidopsis carrying a T-DNA insertion in At3g15870 had wild-type levels of both 16:3Δ7,10,13 and chlorophyll. Together, these data formally prove that At3g15850 is FAD5. Interestingly, the fad5 phenotype was partially complemented when extraplastidial Δ9-desaturases of the Arabidopsis desaturase (ADS) family were expressed as fusions with a plastidial transit peptide. Tight correlation between leaf 16:3Δ7,10,13 levels and chlorophyll content suggests a role for plastidial fatty acid desaturases in thylakoid formation.
Plants that accumulate hexadeca 7,10,13-trienoic acid (16:3Δ7,10,13) are termed 16:3 plants and include rape (Brassica napus), spinach (Spinacia oleracea), and Arabidopsis (Arabidopsis thaliana). In these plants, 16:3Δ7,10,13 can comprise up to 30% of the total fatty acid (Browse et al., 1986), making 16:3Δ7,10,13 the second-most-abundant fatty acid after α-linolenic acid (18:3Δ9,12,15). Pioneering work by Siebertz and Heinz established that 16:3Δ7,10,13 is confined to the sn-2 position of monogalactosyldiacylglycerol (MGDG) and results from a sequence of desaturation steps starting with 16:0 (Siebertz and Heinz, 1977). In Arabidopsis, a three-step desaturation pathway is defined by mutants in FAD5 (Kunst et al., 1989), FAD6 (Falcone et al., 1994), and FAD7 (Iba et al., 1993) or FAD8 (McConn et al., 1994), genes that encode desaturases that sequentially introduce the Δ7, Δ10, and Δ13 double bonds, respectively. Plants in which the initial FAD5 Δ7-desaturation step was compromised and that were devoid of unsaturated 16-carbon fatty acids on MGDG were identified in an ethyl methanesulfonate mutant screen (Kunst et al., 1989). Aside from the absence of 16:3Δ7,10,13 from their membrane lipids and a moderate reduction in chlorophyll content (Kunst et al., 1989), fad5 Arabidopsis plants exhibit a growth and chlorosis phenotype at low temperature (Hugly and Somerville, 1992) and delayed recovery of PSII after photoinhibition by high-light stress, a process that has been extensively studied (Vijayan and Browse, 2002). In addition, fad5 plants are incapable of producing the oxylipin dinor-oxo-phytodienoic acid implicated in wound signaling (Weber et al., 1997).
Although 16:3Δ7,10,13 is a prominent plant fatty acid, the FAD5 gene encoding the enzyme catalyzing the first committed step in its synthesis has not been unambiguously identified. The fad5 mutation was mapped on Arabidopsis chromosome 3 (Hugly et al., 1991), and, based on sequence analysis, Mekhedov et al. (2000) proposed that a mutation in one of two desaturase-like genes (At3g15850, At3g15870) in the vicinity of the fad5 locus was responsible for the loss of FAD5 activity. Both these genes are members of the Arabidopsis desaturase (ADS) multigene family, which comprises a total of nine members and is the largest family of fatty acid desaturase-like genes in the Arabidopsis genome (Fig. 1). The functional annotation of ADS enzymes is largely based on characterizations performed in heterologous systems or after ectopic expression in Arabidopsis, and the physiological roles of the majority of ADS enzymes are not clear. A white spruce (Picea glauca) homolog of At3g15850 has previously been characterized as encoding an 18:0 Δ9-desaturase by heterologous expression in yeast (Saccharomyces cerevisiae; Marillia et al., 2002). Similarly, two cytoplasmic ADS enzymes from Arabidopsis, ADS1 and ADS2, formed exclusively Δ9-desaturated fatty acids when expressed in yeast (Heilmann et al., 2004), or predominantly Δ9 when overexpressed using a seed-specific promoter in Arabidopsis (Heilmann et al., 2004) or in Brassica juncea (ADS1 only; Yao et al., 2003). To date, no formal study of the function of ADS enzymes in the Arabidopsis fad5 mutant has been reported, nor has the restoration of physiological phenotypes associated with this lesion been investigated. Here, we identify the mutation in the fad5 mutant allele of At3g15850 and demonstrate complementation of the fad5 biochemical phenotype by the wild-type allele of At3g15850 (ADS3), including accumulation of 16:3Δ7,10,13, and the restoration of the physiological phenotype of chlorophyll contents to wild-type levels. Interestingly, the expression of in-frame fusions between cDNAs encoding the ADS3 plastidial transit peptide (ADS31–71) and the cytoplasmic Δ9-desaturases ADS1 or ADS2 (corresponding to genes At1g06080 and At2g31360, respectively) also resulted in complementation of the fad5 phenotype, consistent with previous ectopic expression studies on ADS enzymes (Heilmann et al., 2004). Complementation of the fad5 phenotype with cDNA constructs encoding engineered plastid-targeted ADS enzymes was only partial, and our characterization of the resulting intermediate phenotypes indicates a tight correlation between leaf 16:3Δ7,10,13 levels and leaf chlorophyll content.
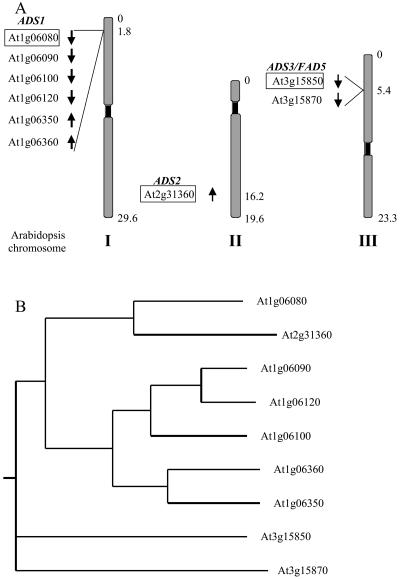
Figure 1.
The ADS multigene family. A, Comprising nine members, the ADS family is the largest family of desaturase-like genes in the Arabidopsis genome. ADS1 and ADS2 have previously been identified by sequence homology to cyanobacterial acyl-lipid desaturases and mammalian acyl-CoA desaturases (Fukuchi-Mizutani et al., 1998). Boxes, ADS genes studied here; numbers denote approximate chromosomal location in mega base pairs. B, ADS cDNA sequences were clustered using ClustalW (http://www.ebi.ac.uk/clustalw/), and a phylogram was created using PHYLIP (http://evolution.genetics.washington.edu/phylip.html). Chromosomal loci as indicated.
RESULTS
In a separate study, we tested the heterologous expression of ADS1, ADS2, and ADS3 in yeast and ectopic expression in seeds of fab1 fae1 double mutant Arabidopsis plants, which are deficient in both plastidial and extraplastidial fatty acid elongation, respectively. The results of these experiments showed that several ADS gene products, including ADS3, are capable of converting 16:0 to 16:1Δ7 in the plastid and that this desaturation step occurs on fatty acids esterified to MGDG rather than on other lipid classes (Heilmann et al., 2004). In this study, we identify the genetic lesion underlying the fad5 phenotype and demonstrate, by complementation, that At3g15850 is the FAD5 locus, based on its ability to restore the fad5 biochemical and physiological phenotypes.
The fad5 Allele of At3g15850 (ADS3) Contains a Nonsense Mutation
The closely linked genes At3g15850 (ADS3) and At3g15870 (ADS3.2) have previously been proposed as candidates for FAD5 (Mekhedov et al., 2000). This prediction was based on similarity to acyl-CoA-dependent desaturases of animal and fungal origin, apparent presence of plastidial transit peptides in the deduced amino acid sequences, and proximity to the mapped location of the fad5 mutation (Hugly et al., 1991; Hugly and Somerville, 1992). To substantiate the prediction experimentally and to unambiguously identify the FAD5 gene, genomic DNA for ADS3 and ADS3.2 was amplified from fad5 plants and sequenced. We identified two differences compared to the sequences of the corresponding wild-type genes in GenBank file 30698537: a nonsense mutation in At3g15850 changing codon 98 from TGG (W) into TGA (stop), and a frameshift in At3g15870, the latter representing a sequencing error in the published GenBank file (Fig. 2). The polypeptide predicted from the fad5 allele of At3g15850 carrying the nonsense mutation is severely truncated and likely biochemically inactive because a polypeptide containing residues 1 to 97 lacks the three His boxes shown to be necessary for catalytic activity (Fig. 2; Shanklin et al., 1994).
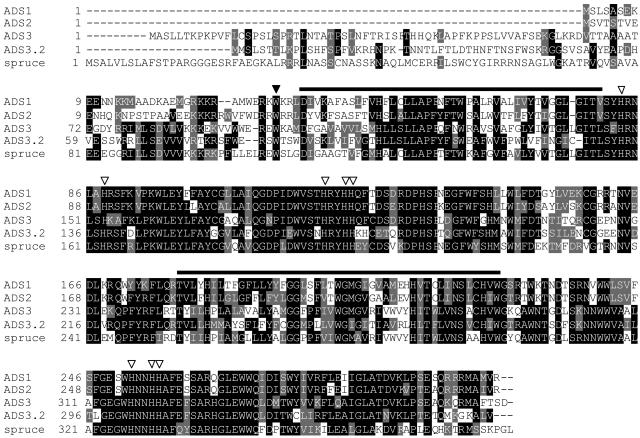
Figure 2.
Alignment of deduced amino acid sequences of ADS enzymes. ADS1 to 3, Deduced sequences of the enzymatically active desaturases encoded by At1g06080, At2g31360, and At3g15850, respectively. ADS3.2, Polypeptide predicted from the corrected sequence of At3g15870; spruce, ADS-related polypeptide from white spruce encoded by AF438199. White triangles, His residues conserved in membrane-bound desaturases; black triangle, position of the nonsense mutation (W98stop) in the ADS3 polypeptide deduced from the fad5 allele of At3g15850; bars, transmembrane domains predicted by TMPRED (http://www.ch.embnet.org/software/TMPRED_form.html).
Expression of a Genomic DNA Fragment Containing At3g15850 (ADS3) Restores the Ability to Synthesize 16:3Δ7,10,13 to Arabidopsis fad5 Mutant Plants
A genomic DNA fragment amplified from wild-type plants and containing At3g15850 under the control of its own promoter was expressed in Arabidopsis fad5 mutant plants. The fatty acid composition of leaf samples from third-generation homozygous transgenic plants derived from transformation event 5-3 (lines 5–3 C1, 5–3 C10, and 5–3 C T3) was analyzed by gas chromatography/mass spectrometry (Fig. 3; Table I). While 16:3Δ7,10,13 was absent from fad5 plants, the compound accumulated in leaves of fad5 plants expressing the genomic fragment, and fatty acid patterns from the complemented plants were indistinguishable from those of wild-type plants. The 16:3 formed in wild type and in complemented fad5 mutants coeluted with an authentic 16:3Δ7,10,13 standard and had a mass spectral fragmentation pattern indistinguishable from that of the standard (data not shown). Functional complementation of the fad5 mutant by At3g15850 proves that this gene encodes a palmitoyl-MGDG Δ7-desaturase.
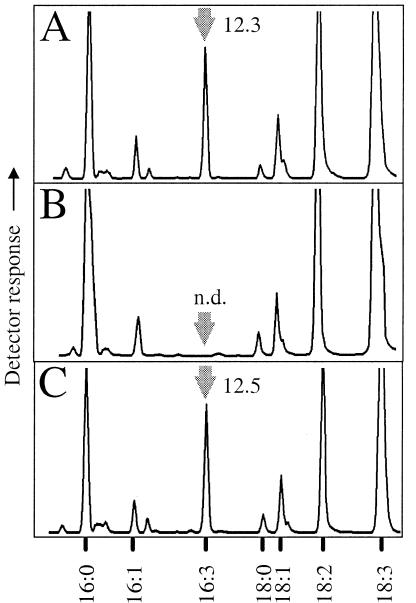
Figure 3.
16:3Δ7,10,13 content in leaves of Arabidopsis fad5 mutant plants expressing genomic DNA containing At3g15850. A genomic ADS3 clone was expressed in fad5 mutant plants, and fatty acid patterns from 1-week-old second-generation leaves were analyzed by gas chromatography/mass spectrometry. A, Wild type; B, fad5; C, fad5 line expressing the genomic ADS3 fragment under its endogenous promoter. Arrows, Elution of 16:3Δ7,10,13. Numbers denote 16:3Δ7,10,13 quantifications as mol% of total fatty acids. Elution of other fatty acids as indicated.
Table I.
Fatty acid composition in leaves of wild-type Arabidopsis, fad5 mutants, fad5 mutants expressing either genomic DNA containing ADS3 or ADS cDNA constructs, and SALK_142406, a T-DNA insertion mutant of ADS3.2
| Fatty Acid
|
Wild Type
|
fad5
|
Genomic ADS3
|
cDNA
|
SALK_142406
|
||||
|---|---|---|---|---|---|---|---|---|---|
| Line C1 | Line C10 | Line C T3 | ADS31-71-ADS1 | ADS31-71-ADS2 | ADS3 | ||||
| 16:0 | 14.1 ± 2.2 | 19.6 ± 1.8 | 15.2 ± 2.0 | 14.2 ± 1.5 | 14.6 ± 1.8 | 16.6 ± 2.1 | 17.0 ± 2.1 | 16.4 ± 2.4 | 13.8 ± 2.0 |
| 16:1 | 2.7 ± 0.5 | 3.1 ± 0.8 | 2.9 ± 0.5 | 3.1 ± 0.6 | 2.6 ± 0.5 | 2.1 ± 0.3 | 2.0 ± 1.0 | 2.8 ± 1.3 | 2.9 ± 0.5 |
| 16:3 | 12.1 ± 1.6 | n.d. | 11.2 ± 1.1 | 10.0 ± 1.2 | 12.4 ± 1.1 | 4.5 ± 1.2 | 3.5 ± 0.9 | 6.6 ± 2.3 | 12.3 ± 1.7 |
| 18:0 | 1.5 ± 0.2 | 2.0 ± 0.4 | 2.8 ± 0.3 | 3.1 ± 0.4 | 1.8 ± 0.6 | 6.3 ± 0.9 | 4.1 ± 0.5 | 3.3 ± 1.0 | 1.8 ± 0.5 |
| 18:1 | 5.6 ± 0.2 | 6.2 ± 0.4 | 5.9 ± 0.4 | 4.9 ± 0.3 | 3.7 ± 0.7 | 6.6 ± 0.2 | 5.5 ± 1.2 | 5.8 ± 0.9 | 4.9 ± 0.6 |
| 18:2 | 17.0 ± 2.0 | 20.3 ± 1.6 | 17.5 ± 1.3 | 16.3 ± 2.1 | 15.8 ± 1.5 | 15.2 ± 2.2 | 17.3 ± 3.5 | 17.7 ± 2.9 | 16.2 ± 2.5 |
| 18:3 | 46.4 ± 3.4 | 49.2 ± 2.6 | 43.2 ± 2.1 | 47.8 ± 2.5 | 48.1 ± 2.6 | 47.3 ± 2.5 | 48.6 ± 2.7 | 46.8 ± 2.2 | 47.6 ± 3.6 |
Data as mol% of total fatty acids ± sd; n.d., not detectable; C1, C10, and C T3, transgenic fad5 lines expressing genomic ADS3; SALK_142406, ADS3.2 T-DNA insertion mutant.
Correction of the At3g15870 GenBank Sequence Reveals a Second ADS Gene Encoding a Potential Plastidial Targeting Peptide
According to current GenBank annotations, only one ADS gene, At3g15850, encodes an N-terminal chloroplast transit peptide and would thus be the only candidate for FAD5. However, detailed reanalysis of the published genomic sequence for At3g15870, located on chromosome 3 in close proximity to At3g15850, revealed a sequencing error in GenBank files 30698537 and 18400932. The sequencing error represents a frameshift in the N-terminal region of the deduced ADS3.2 polypeptide. The corrected genomic sequence (accession no. AY734685) reveals an open reading frame with a longer predicted N terminus for ADS3.2 that includes an extension recognized by TargetP software (http://www.cbs.dtu.dk/services/TargetP/) as a plastidial targeting peptide. The new deduced amino acid sequence substantially improves the similarity between the N-terminal portion of the deduced ADS3.2 polypeptide and the other ADS enzymes. Figure 2 shows an alignment of the amino acid sequences of the enzymatically active ADS1, ADS2, and ADS3 desaturases with that deduced from the corrected ADS3.2 sequence and with that deduced from a gene from white spruce with sequence similarity to Arabidopsis ADS3 (Marillia et al., 2002).
Analysis of ADS-related sequences in publicly available databases reveals that most members of the ADS multigene family, including ADS3, are represented by numerous expressed sequence tags (ESTs) and therefore likely well expressed. Northern-blot analysis indicated that ADS3 is highly expressed in young leaves of wild-type Arabidopsis and that expression in roots is less than 10% of that in leaves (data not shown), an expression pattern consistent with At3g15850 encoding a plastidial desaturase. By contrast, ADS3.2 is the only ADS gene for which no corresponding ESTs can be found in public databases, indicating low transcript abundance in the tissues studied. The sequence of a partial ADS3.2 cDNA isolated from flowers (GenBank file 42463933) is consistent with our corrected genomic sequence. However, the partial ADS3.2 cDNA sequence appears to contain several introns that have not been excised, raising the possibility that ADS3.2 may be nonfunctional, and the cDNA may represent the consequence of expression of a pseudogene. A Brassica rapa EST (L38104) similar to ADS3.2 has been reported that aligns well over the regions encoding the targeting peptide, suggesting that B. rapa may contain a functional homolog to ADS3.2. Because the fad5 mutation had been mapped to a chromosomal locus containing both ADS3 and ADS3.2 (Hugly et al., 1991), and because our sequence analysis suggested the presence of a transit peptide in ADS3.2, we tested the possibility that the ADS3.2 gene contributes to the FAD5 phenotype in Arabidopsis.
At3g15870 (ADS3.2) Is Not Related to the Arabidopsis FAD5 Phenotype
From the complete loss of FAD5 activity in fad5 mutant plants, which carry the nonsense mutation in ADS3 but contain the wild-type allele of ADS3.2, it must be concluded that ADS3.2 does not even partially compensate for the disruption of ADS3. In order to directly verify that ADS3.2 does not contribute to the FAD5 phenotype, we characterized an Arabidopsis T-DNA insertion mutant (SALK_142406), which contains a T-DNA insertion in exon 2 of the corrected ADS3.2 open reading frame. Figure 4 illustrates that the T-DNA is present in the ADS3.2 gene and that the T-DNA-disrupted allele is homozygous because the wild-type allele is not present in the SALK_142406 line used for analysis. Fatty acid patterns from leaves from this line did not resemble those of fad5 leaves but showed wild-type levels for both 16:3Δ7,10,13 (compare Figs. 4C and 3A; Table I) and leaf chlorophyll content (Table II). The phenotype of the homozygous SALK_142406 proves that ADS3.2 does not contribute to the FAD5 phenotype.
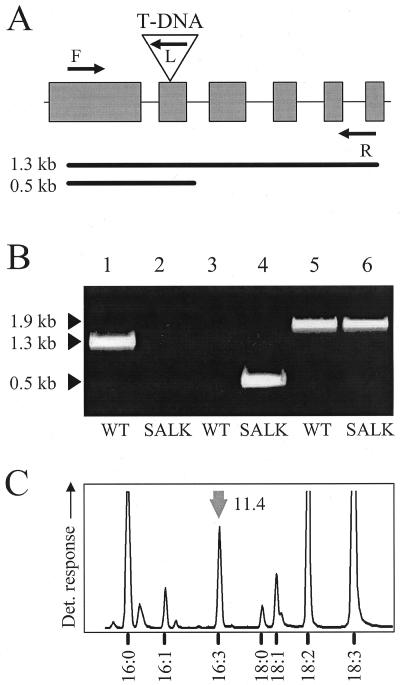
Figure 4.
Identification of a T-DNA insertion in locus At3g15870 encoding ADS3.2. Genomic DNA from wild type (WT) and an individual of line SALK_142406 (SALK) were analyzed by PCR for the presence or absence of wild-type and T-DNA-disrupted alleles of At3g15870. A, Predicted intron/exon structure of the corrected At3g15870 gene. Triangle, Position of the T-DNA insertion in SALK_142406; arrows, primers used for amplification (F, forward primer; L, T-DNA left-border primer; R, reverse primer); bars, amplification products for wild-type allele (top) and T-DNA-disrupted allele (bottom). B, Two primer pairs were used to screen genomic DNA from wild type (lanes 1, 3, and 5) and a line of SALK_142406 (lanes 2, 4, and 6) for the presence of wild-type (F, R) and/or a T-DNA-disrupted (F, L) allele of At3g15870. Lanes 5 and 6 show the amplification of wild-type At3g15850 in both samples. C, Leaf fatty acid pattern of homozygous SALK_142406 line. Arrow, Elution position of 16:3Δ7,10,13. Number denotes 16:3Δ7,10,13 quantification as mol% of total fatty acids. Elution positions of other fatty acids are indicated.
Table II.
Total chlorophyll content in leaves of wild-type Arabidopsis, fad5 mutants, fad5 mutants expressing either genomic DNA containing ADS3 or ADS cDNA constructs, and in a T-DNA insertion mutant of ADS3.2
| Chlorophyll Content
|
Wild Type
|
fad5
|
Genomic ADS3
|
cDNA
|
SALK_142406
|
||||
|---|---|---|---|---|---|---|---|---|---|
| Line C1 | Line C10 | Line C T3 | ADS31-71-ADS1 | ADS31-71-ADS2 | ADS3 | ||||
| mg g−1 fresh weight ± sd | 1.34±0.06 | 0.95±0.04 | 1.31±0.04 | 1.29±0.04 | 1.45±0.04 | 1.11±0.02 | 1.05±0.06 | 1.23±0.04 | 1.38±0.08 |
| Relative (% of wild type) | 100% | 70.5% | 98% | 96% | 108% | 83% | 78% | 92% | 103% |
C1, C10, and C T3, Transgenic fad5 lines expressing genomic ADS3; SALK_142406, ADS3.2 T-DNA insertion mutant.
Leaf 16:3Δ7,10,13 Accumulation Is Partially Restored in fad5 Plants Constitutively Expressing cDNAs Encoding Plastid-Targeted ADS1 or ADS2, or an ADS3 cDNA
In a separate study, the cDNA sequence encoding the ADS3 N-terminal transit peptide (ADS31–71) was fused in frame to cDNAs encoding the extraplastidial ADS1 or ADS2 to engineer plastid-targeted ADS31–71-ADS1 and ADS31–71-ADS2 (Heilmann et al., 2004). Each of these constructs or a cDNA encoding ADS3 was individually expressed in fad5 mutant plants under the control of the constitutive cassava vein mosaic virus (CVMV) promoter. The standardized expression conditions allowed us to compare the relative effectiveness of the redirected ADS1 and ADS2 desaturases with that of the plastidial ADS3 desaturase in their ability to synthesize 16:3Δ7,10,13 and to affect the associated physiological marker, chlorophyll content.
While 16:3Δ7,10,13 was absent from fad5 plants (Fig. 3B), it accumulated in leaves of fad5 plants constitutively expressing the plastid-targeted ADS1 and ADS2 enzymes as well as in those expressing ADS3 (Table I). These results are consistent with our earlier finding that when directed to the plastid, ADS1 and ADS2 can act as 16:1Δ7 desaturases on palmitoyl-MGDG in the pathway leading to the formation of 16:3Δ7,10,13 (Heilmann et al., 2004). While 16:3Δ7,10,13 was clearly synthesized with ADS expression driven by the CVMV promoter, 16:3Δ7,10,13 levels did not reach those of wild-type plants or those of fad5 plants complemented with the genomic clone of ADS3. The average accumulation of 16:3Δ7,10,13 with each expressed ADS enzyme can serve as a relative measure of palmitoyl-MGDG Δ7-desaturase activity in the plastid. The data indicate that ADS3, an enzyme naturally residing in the plastid, performed significantly better than the cytosolic ADS1 or ADS2 retargeted to the plastid (Table I): Leaf 16:3Δ7,10,13 accumulation with expression of plastid-targeted ADS1 and ADS2 was 53% and 68%, respectively, of the levels observed with constitutive expression of ADS3.
Partial Complementation of the fad5 Mutant Reveals a Tight Correlation between Leaf 16:3Δ7,10,13 Levels and Chlorophyll Content
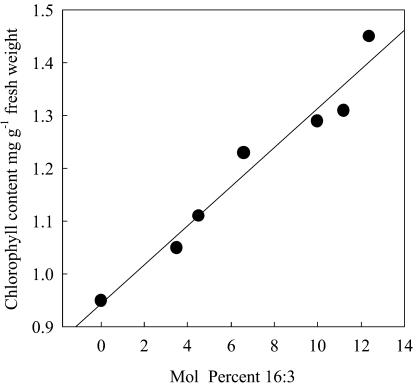
As a physiological marker for fad5 complementation, we investigated leaf chlorophyll content in 1-week-old leaves from wild-type Arabidopsis, from fad5 plants, and from fad5 plants expressing the genomic At3g15850 fragment or cDNAs encoding plastid-targeted ADS enzymes (Table II). Whereas the fad5 mutant plants were characterized by an approximately 30% reduction in chlorophyll content compared to wild-type plants, expression of the genomic ADS3 fragment or of cDNAs encoding plastid-targeted ADS enzymes significantly increased the leaf chlorophyll content (Table II) over that of the fad5 mutant. Interestingly, plants with intermediate leaf 16:3Δ7,10,13 accumulation, such as fad5 plants expressing cDNAs encoding plastid-targeted ADS enzymes, also showed intermediate leaf chlorophyll levels (compare Tables I and II). The correlation between accumulation of 16:3Δ7,10,13 and leaf chlorophyll content in partially or fully complemented fad5 plants from the same accession (Fig. 5) has a correlation coefficient (r2) of 0.96, indicating a significant linear correlation.
Figure 5.
Correlation between leaf 16:3Δ7,10,13 levels and leaf chlorophyll content in fad5 Arabidopsis mutants and in partially or fully complemented fad5 plants.
DISCUSSION
A nonsense mutation was identified in the fad5 allele of At3g15850 (ADS3). The expression of genomic DNA containing the wild-type allele of this gene under its endogenous promoter resulted in complementation of the Arabidopsis fad5 phenotype, restoring both 16:3Δ7,10,13 levels and chlorophyll content in leaves to wild-type levels. Together, these data prove that At3g15850 encodes a plastidial palmitoyl-MGDG Δ7-desaturase that acts in the 16:3Δ7,10,13 synthetic pathway. The data presented are consistent with 16:1Δ7 accumulation observed in Arabidopsis seeds with ectopic, seed-specific expression of plastid-targeted ADS enzymes, including ADS3 (Heilmann et al., 2004).
While data from this study suggest that At3g15870, a gene located in close proximity to ADS3, may also encode an ADS enzyme with a plastidial targeting peptide, a contribution of At3g15870 to the FAD5 phenotype can be ruled out because the sequence of the fad5 allele of At3g15870 was found identical to its wild-type allele, and because a T-DNA disruption of At3g15870 displayed a wild-type rather than a fad5 phenotype.
A link between a plant's ability to generate 16:3Δ7,10,13 and the expression of ADS-like genes has previously been reported based on relative EST abundance in several plant species (Mekhedov et al., 2000). Analysis of the complete genomic sequence of rice (Oryza sativa), an 18:3 plant that does not accumulate 16:3Δ7,10,13, confirms this correlation—the rice genome does not contain homologs of Arabidopsis ADS3 or ADS3.2 or, in fact, of any other ADS gene (Goff et al., 2002; Yu et al., 2002; ftp://ftp.tigr.org/pub/data/Eukaryotic_Projects/o_sativa/annotation_dbs/pseudomolecules/version_2.0/all_chrs/all.pep). GenBank files 32992879 and 32994161 contain cDNA clones from rice with similarity to ADS3 that have subsequently been attributed to fungal contamination (Kikuchi et al., 2003). The absence of the ADS multigene family in rice may not only mirror differences in membrane lipid content between Arabidopsis and rice but may also indicate differences in the mechanisms of membrane temperature acclimation between 16:3 and 18:3 plants, reviewed by Falcone et al. (2004).
The physiological functions of ADS enzymes other than FAD5, including those of ADS1 and ADS2, remain to be determined. ADS1 and ADS2 have been demonstrated to be functional fatty acid desaturases (Yao et al., 2003; Heilmann et al., 2004) that lack transit peptides and likely reside in the endoplasmic reticulum. ADS1 and ADS2 gene expression levels are differentially regulated in response to changes in ambient temperature. ADS1 and ADS2 transcripts have been reported only in flowers (Fukuchi-Mizutani et al., 1998); however, analysis of massively parallel signature sequencing data suggests that ADS2 transcripts are highly abundant in flowers, inflorescences, leaves, roots, and siliques of Arabidopsis (data not shown). The degree of membrane lipid unsaturation influences the biophysical properties of membranes at high or low temperatures (Nishida and Murata, 1996), suggesting a role for ADS1 and ADS2 in temperature acclimation of Arabidopsis reproductive organs (Nishida and Murata, 1996). Although characterized as Δ9-desaturases in yeast and in seeds of Arabidopsis (Heilmann et al., 2004) and B. juncea (ADS1 only; Yao et al., 2003), the data presented here show that, when retargeted to the plastid, ADS1 and ADS2 can act as palmitoyl-MGDG Δ7-desaturases and functionally complement the fad5 mutation. The molecular mechanism underlying this apparent switch in regiospecificity has recently been shown to involve interactions between desaturases and different head groups of the lipid substrates found in different subcellular compartments (Heilmann et al., 2004). The characterization of a white spruce gene with similarity to Arabidopsis ADS3, encoding an 18:0 Δ9-desaturase (Marillia et al., 2002), by heterologous expression in yeast is consistent with our data, and it is possible that in planta this enzyme may exhibit FAD5 functionality.
The fad5 mutant Arabidopsis plants used in this study showed leaf chlorophyll contents of approximately 70% of wild-type levels. The observed reduction in chlorophyll content was thus more pronounced than that previously reported for fad5 plants, with approximately 85% of wild-type levels (Kunst et al., 1989), likely reflecting differences in culture conditions or the age of the plants used. The partial complementation of the fad5 phenotype observed in this study with constitutive expression of cDNAs encoding plastid-targeted ADS1, ADS2, or ADS3 may be explained by lower promoter strength of the constitutive CVMV promoter, in comparison to the endogenous FAD5 promoter driving expression of the genomic DNA fragment, which resulted in full complementation. In all partially or fully complemented fad5 plants, the resulting accumulation of 16:3Δ7,10,13 was tightly correlated with increases in chlorophyll content. This correlation may be explained by the increased availability of sn-1/sn-2 polyunsaturated MGDG in the complemented plants versus sn-2 saturated (palmitoyl-) MGDG in the mutant plants (Kunst et al., 1989). While the role of MGDG in thylakoid assembly is not understood in detail, MGDG has been demonstrated to mediate the formation of extensive lamellar organization of pea (Pisum sativum) PSII chlorophyll a/b light-harvesting antenna protein (LHCII) in vitro, resembling thylakoid assembly (Simidjiev et al., 2000). MGDG used for the studies on pea LHCII was present in an inverted hexagonal (HII) phase, the formation of which requires a high degree of polyunsaturation on MGDG (for review, see Bruce, 1998). Thus, a high degree of fatty acid unsaturation associated with MGDG would favor thylakoid assembly and represents a possible explanation for the observed fad5 physiological phenotype of reduced leaf chlorophyll content.
In summary, we report several lines of evidence for the assignment of At3g15850 (ADS3) as the gene encoding the plastidial palmitoyl-MGDG Δ7-desaturase FAD5, including the identification of a mutation in the fad5 allele of ADS3, and the functional complementation of the biochemical and physiological fad5 phenotypes, i.e. the accumulation of 16:3Δ7,10,13 and the restoration of leaf chlorophyll content to wild-type levels. These data are consistent with previous seed overexpression of ADS enzymes in that the cytoplasmic fatty acid Δ9-desaturases, ADS1 and ADS2, can partially complement the fad5 phenotype when retargeted to the plastid. In addition to identifying the fad5 lesion, our data indicate that the reduction in leaf chlorophyll content in fad5 plants may be due to a reduced level of unsaturated MGDG adversely affecting photosystem assembly or stability, an observation that invites more detailed investigation.
MATERIAL AND METHODS
Genomic Constructs
DNA fragments of At3g15850 and At3g15870 were amplified from genomic DNA extracted from fad5 plants with four pairs of primers designed based on the sequence of wild-type Arabidopsis (Arabidopsis thaliana; GenBank file 30698537). Restriction endonuclease sites suitable for cloning into conventional vectors were added to 5′ ends of the primers. The At3g15850 promoter and coding regions were amplified separately, using the primer combinations 5′-CGCGAATTCGAGAGGTGCAAGAGTCGTGG-3′/5′-AATGGATCCCTCCGTCGCTGCAGCAGCT-3′ and 5′-CGCGAATTCTCTTCCTTCTTTCTCTTAGCCAT-3′ (Primer 1)/5′-AATGGATCCAGTTGAGTATCTAGAATTGCCGT-3′ (Primer 2), respectively.
The At3g15870 promoter and coding region were amplified separately, using the primer combinations 5′-AATGGATTCTGTGCAAGAGATTGTATGTGACAA-3′/5′-CGCGAATTCTGCCTTTACATGCATATTCAATGTGCT-3′ and 5′-AATGGATCCGCATGCATTAATGCATATCGAATGT-3′ (Primer 3)/5′-CGCGAATTCCATATGGTTTCATTTGAGATTCCCT-3′ (Primer 4), respectively. The amplification protocol included an initial 10-min denaturation step at 94°C, 30 cycles of 30 s of denaturation at 94°C, 30 s of annealing at 60°C, and 3 min of extension at 72°C, followed by 15 min of extension at 72°C. To minimize PCR artifacts, Pfu polymerase was used. DNA fragments amplified from genomic DNA in three independent PCR reactions were cloned into pBlueScript(SK+) vectors (Stratagene, La Jolla, CA) and were sequenced individually using universal and custom sequencing primers. After identification of the fad5 nonsense mutation in At3g15850 and the potential sequencing error causing the frameshift in At3g15870, the corresponding fragments from two more initial PCR reactions were sequenced to confirm the observed changes. In order to rule out PCR contamination by cloned DNA and to get additional proof of the nonsense mutation, a DNA fragment of At3g15850 spanning the mutation site was amplified from fad5 plants with one end extending beyond the sequence amplified and cloned in the first amplification. The following primer combination was used for this step: 5′-CGCGAATTCTAGTACTACTAATTGTGAAGCATAC-3′/5′-AATGGATCCAGGCTTTATGAGAAAGATTCCTATG-3′. The PCR protocol was as above with the exception of annealing at 65°C. Cloning of the resulting PCR fragment and sequencing confirmed the nonsense mutation in At3g15850 of fad5 plants.
To generate a construct containing wild-type At3g15850 with its own promoter, a fragment spanning the At3g15850 coding region was amplified with primers 1 and 2 from genomic DNA obtained from wild-type plants. The promoter fragment amplified from fad5 plants in the first experiment had wild-type sequence and was fused to the wild-type coding region using the naturally occurring HindIII site (codons 42 and 43 of the At3g15850 reading frame). The resulting fragment was inserted into pBI101.1 vector (Jefferson et al., 1987) and its sequence was confirmed. This construct was used for plant transformation. A genomic fragment corresponding to the coding region of wild-type At3g15870 was amplified with primers 3 and 4. It was used for sequencing to demonstrate that the observed frameshift was not associated with fad5 mutation.
cDNA Constructs
ADS3 including the sequence encoding the transit peptide was amplified from Arabidopsis flower cDNA using the primer combination 5′-GGGGACAAGTTTGTACAAAAAAGCAGGCTATGGCTTCTCTTCTAACA-3′ (Primer 5)/5′-GGGGACCACTTTGTACAAGAAAGCTGGGTTCAGTCGGTGAATGC-3′ (Primer 6). The sequence encoding the ADS3 transit peptide was fused to ADS1 and ADS2 cDNA fragments by overlap extension PCR, creating ADS31–71-ADS1 and ADS31–71-ADS2. ADS1 and ADS2 cDNA fragments were amplified from the respective pYES2 yeast expression constructs described previously (Heilmann et al., 2004) using the primer combinations 5′-GCTGCTGCAGCGACGTTGTCATTGTCAGCC-3′/5′-GGGGACCACTTTGTACAAGAAAGCTGGGTTCCGAGACGTCGTTCCATATC-3′ (Primer 7) and 5′-GCTGCTGCAGCGACGTTGTCGGTGACATCAACGG-3′/5′-GGGGACCACTTTGTACAAGAAAGCTGGGTTCAACGAACTATAGCCAT-3′ (Primer 8), respectively. The amplification protocol for ADS genes included an initial 2-min denaturation step at 94°C, 25 cycles of 30 s of denaturation at 94°C, 30 s of annealing at 57°C, and 2 min of extension at 72°C, followed by 15 min of extension at 72°C. The cDNA encoding the transit peptide ADS31–71 was amplified from ADS3 cDNA using the primer combinations 5′-GGGGACAAGTTTGTACAAAAAAGCAGGCTATGGCTTCTCTTCTAACA-3′/5′-GGCTGACAATGACAACGTCGCTGCAGCAGC-3′, or 5′-GGGGACAAGTTTGTACAAAAAAGCAGGCTATGGCTTCTCTTCTAACA-3′/5′-CCGTTGATGTCACCGACAACGTCGCTGC-3′, respectively, for fusion to ADS1 or ADS2 cDNA fragments. The amplification protocol included 2 min of denaturation at 94°C, 10 min of annealing at 45°C, and 10 min of extension at 72°C, followed by 29 cycles of 1 min of denaturation at 94°C, 1 min of annealing at 50°C, and 1 min of extension at 72°C, followed by 15 min of extension at 72°C. Subsequently, the amplification products were purified, and the fusion product assembled using the primer combinations 5/7 and 5/8, respectively. Amplification was carried out as described above for amplification of the ADS genes. The resulting cDNA fragments were moved into the binary plasmid pGATE-CVMV for constitutive expression under the CVMV promoter in Arabidopsis, using GATEWAY technology (Invitrogen, Carlsbad, CA) according to the manufacturer's instructions.
Identification of a T-DNA Insertion in At3g15870
Individual plants from a mixed population of SALK_142406 were screened by PCR using genomic DNA and the gene-specific primer 5′-ATGGAGAAGACTCTTGTCGG-3′ (Primer F) in combination with 5′-TGGTTCACGTAGTGGGCCATCG-3′ (Primer L) to screen for T-DNA inserts (Alonso et al., 2003) or with 5′-TCAAACCAAAGCCTTTCCCTTCA-3′ (Primer R) to amplify the wild-type allele of At3g15870. Genomic fragments of At3g15850 were amplified as controls using the primer combination 5/6.
All amplifications were carried out using Advantage HF2 Taq-polymerase (CLONTECH, Palo Alto, CA). All restriction endonucleases and T4 DNA ligase were from New England Biolabs (Beverly, MA). Sequences of all constructs were verified prior to transformation.
Plant Growth and Transformation
Arabidopsis fad5 plants containing only trace amounts of desaturated 16-carbon fatty acids in MGDG (Kunst et al., 1989) were grown in soil in controlled-environment growth chambers at 22°C under continuous illumination of approximately 300 μmol photons m−2 s−1. Agrobacterium tumefaciens cells were transformed with the pGATE-CVMV constructs by electroporation. Bacterial cultures were grown in Luria-Bertani medium containing 35 μg/L each of Rifampicin, Gentamicin, and Spectinomycin overnight at 30°C in 300-mL cultures shaking at 200 rpm. Cells were harvested by centrifugation, and cell pellets were resuspended in 300 mL of 5% (w/w) Suc containing 0.5% Silwet L-77 (OSi Specialties, Sistersville, WV). Seven-week-old Arabidopsis plants were transformed using the floral dip method (Clough and Bent, 1998) by submerging developing flowers for 5 s in A. tumefaciens suspension 4 times over a 1-week period. The resulting transgenic plants were selected in soil for resistance to aerosolic ammonium glufosinate (BASTA).
Biochemical Analysis
Fatty acids were extracted and methylated by adding 200 μL of boron trichloride (BCl3) directly to young leaves and incubating for 30 min at 80°C. Fatty acid methyl esters were reextracted with 2 mL of hexane and dried under N2 flow. Fatty acid methyl esters were analyzed using a HP5890 gas chromatograph (Hewlett-Packard, Palo Alto, CA) fitted with a 30 m × 250-μm InnoWax capillary column (Hewlett-Packard). The oven temperature was raised from 100°C to 240°C at a rate of 15°C min−1 with a flow rate of 2.0 mL min−1. An authentic 16:3Δ7,10,13 standard was obtained from Larodan Lipids (Malmö, Sweden) and used for identification. Leaf chlorophyll was quantified in 80% (v/v) acetone extracts by measuring the A663 and 645 nm and using the Arnon equations for total chlorophyll content (Arnon, 1949).
Distribution of Materials
Upon request, all novel materials described in this publication will be made available in a timely manner for noncommercial research purposes, subject to receiving the requisite permission from any third-party owners of all or parts of the material. Obtaining any permission will be the responsibility of the requestor.
The updated annotation for At3g15850 and the corrected sequence for At3g15870 were deposited in GenBank under the accession numbers AY734684 and AY734685, respectively.
Acknowledgments
We thank Dr. Mark S. Pidkowich for constructing the pGATE-CVMV plasmid.
This work was supported by the Office of Basic Energy Sciences of the U.S. Department of Energy, the Oilseed Engineering Alliance of The Dow Chemical Company and Dow Agrosciences, the National Science Foundation (grant no. IBN–0084329), and an Emmy Noether fellowship (German Science Foundation; to I.H.).
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.104.052951.
References
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, et al (2003) Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657 [DOI] [PubMed] [Google Scholar]
- Arnon D (1949) Copper enzymes in isolated chloroplasts. Polyphenoloxidase in Beta vulgaris. Plant Physiol 24: 1–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browse J, Warwick N, Somerville CR, Slack CR (1986) Fluxes through the prokaryotic and eukaryotic pathways of lipid synthesis in the ‘16:3’ plant Arabidopsis thaliana. Biochem J 235: 25–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruce BD (1998) The role of lipids in plastid protein transport. Plant Mol Biol 38: 223–246 [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Falcone DL, Gibson S, Lemieux B, Somerville C (1994) Identification of a gene that complements an Arabidopsis mutant deficient in chloroplast omega 6 desaturase activity. Plant Physiol 106: 1453–1459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falcone DL, Ogas JP, Somerville CR (2004) Regulation of membrane fatty acid composition by temperature in mutants of Arabidopsis with alterations in membrane lipid composition. BMC Plant Biol 4: 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuchi-Mizutani M, Tasaka Y, Tanaka Y, Ashikari T, Kusumi T, Murata N (1998) Characterization of Δ9 acyl-lipid desaturase homologues from Arabidopsis thaliana. Plant Cell Physiol 39: 247–253 [DOI] [PubMed] [Google Scholar]
- Goff SA, Ricke D, Lan TH, Presting G, Wang R, Dunn M, Glazebrook J, Sessions A, Oeller P, Varma H, et al (2002) A draft sequence of the rice genome (Oryza sativa L. ssp. japonica). Science 296: 92–100 [DOI] [PubMed] [Google Scholar]
- Heilmann I, Pidkowich MS, Girke T, Shanklin J (2004) Switching desaturase enzyme specificity by alternate subcellular targeting. Proc Natl Acad Sci USA 101: 10266–10271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hugly S, Kunst L, Somerville C (1991) Linkage relationships of mutations that affect fatty-acid composition in Arabidopsis. J Hered 82: 484–488 [Google Scholar]
- Hugly S, Somerville C (1992) A role for membrane lipid polyunsaturation in chloroplast biogenesis at low temperature. Plant Physiol 99: 197–202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iba K, Gibson S, Nishiuchi T, Fuse T, Nishimura M, Arondel V, Hugly S, Somerville C (1993) A gene encoding a chloroplast omega-3 fatty acid desaturase complements alterations in fatty acid desaturation and chloroplast copy number of the fad7 mutant of Arabidopsis thaliana. J Biol Chem 268: 24099–24105 [PubMed] [Google Scholar]
- Jefferson RA, Kavanagh TA, Bevan MW (1987) GUS fusions: beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J 6: 3901–3907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikuchi S, Satoh K, Nagata T, Kawagashira N, Doi K, Kishimoto N, Yazaki J, Ishikawa M, Yamada H, Ooka H, et al (2003) Collection, mapping, and annotation of over 28,000 cDNA clones from japonica rice. Science 301: 376–379 [DOI] [PubMed] [Google Scholar]
- Kunst L, Browse J, Somerville C (1989) A mutant of Arabidopsis deficient in desaturation of palmitic acid in leaf lipids. Plant Physiol 90: 943–947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marillia EF, Giblin EM, Covello PS, Taylor DC (2002) A desaturase-like protein from white spruce is a Delta(9) desaturase. FEBS Lett 526: 49–52 [DOI] [PubMed] [Google Scholar]
- McConn M, Hugly S, Browse J, Somerville C (1994) A mutation at the fad8 locus of Arabidopsis identifies a second chloroplast ω-3 desaturase. Plant Physiol 106: 1609–1614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mekhedov S, de Ilarduya OM, Ohlrogge J (2000) Toward a functional catalog of the plant genome. A survey of genes for lipid biosynthesis. Plant Physiol 122: 389–402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishida I, Murata N (1996) Chilling sensitivity in plant and cyanobacteria: the crucial contribution of membrane lipids. Annu Rev Plant Physiol Plant Mol Biol 47: 541–568 [DOI] [PubMed] [Google Scholar]
- Shanklin J, Whittle E, Fox BG (1994) Eight histidine residues are catalytically essential in a membrane-associated iron enzyme, stearoyl-CoA desaturase, and are conserved in alkane hydroxylase and xylene monooxygenase. Biochemistry 33: 12787–12794 [DOI] [PubMed] [Google Scholar]
- Siebertz HP, Heinz E (1977) Labelling experiments on the origin of hexa- and octa-decatrienoic acids in galactolipids from leaves. Z Naturforsch 32: 193–205 [Google Scholar]
- Simidjiev I, Stoylova S, Amenitsch H, Javorfi T, Mustardy L, Laggner P, Holzenburg A, Garab G (2000) Self-assembly of large, ordered lamellae from non-bilayer lipids and integral membrane proteins in vitro. Proc Natl Acad Sci USA 97: 1473–1476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vijayan P, Browse J (2002) Photoinhibition in mutants of Arabidopsis deficient in thylakoid unsaturation. Plant Physiol 129: 876–885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber H, Vick BA, Farmer EE (1997) Dinor-oxo-phytodienoic acid: a new hexadecanoid signal in the jasmonate family. Proc Natl Acad Sci USA 94: 10473–10478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao K, Bacchetto RG, Lockhart KM, Friesen LJ, Potts DA, Covello PS, Taylor DC (2003) Expression of the Arabidopsis ADS1 gene in Brassica juncea results in a decreased level of total saturated fatty acids. Plant Biotechnol 1: 221–229 [DOI] [PubMed] [Google Scholar]
- Yu J, Hu S, Wang J, Wong GK, Li S, Liu B, Deng Y, Dai L, Zhou Y, Zhang X, et al (2002) A draft sequence of the rice genome (Oryza sativa L. ssp. indica). Science 296: 79–92 [DOI] [PubMed] [Google Scholar]