Abstract
Development of winter hardiness in trees is a two-stage process involving sequential perception of distinct environmental cues, short-day (SD) photoperiod and low temperature (LT). We have shown that both SD and LT are recognized by leaves of silver birch (Betula pendula cv Roth) leading to increased freezing tolerance, and thus leaves can be used as an experimental model to study the physiological and molecular events taking place during cold acclimation. To obtain a molecular marker for the acclimation process in birch we cloned a gene, designated Bplti36, encoding a 36-kD acidic SK2 type of dehydrin. The gene was responsive to LT, drought, salt, and exogenous abscisic acid. This responsiveness to abiotic stresses and abscisic acid was retained when Bplti36 was introduced to Arabidopsis (Arabidopsis thaliana). The LT induction of the gene appeared to be under the control of the C-repeat-binding factor pathway as suggested by the presence of several C-repeat/dehydration-responsive element/LT-responsive elements in the Bplti36 promoter and its constitutive expression in C-repeat-binding factor overproducing Arabidopsis. In birch SD photoperiod at normal-growth temperature did not result in significant induction of Bplti36. However, preexposure to SD followed by LT treatment resulted in a remarkable increase in Bplti36 transcript accumulation as compared to LT-treated plants grown at long-day photoperiod. This suggests that SD photoperiod potentiates the LT response by conditioning the leaf tissue to be more responsive to the LT stimulus.
Woody plants in temperate climates are regularly exposed to freezing temperatures during the winter months. Their ability to survive is based on adaptive mechanisms by which plants enter a state of dormancy and develop freezing tolerance. As proposed by Weiser (1970), this acclimation process includes two distinct stages. First, a decrease in day length triggers growth cessation and dormancy development as well as a moderate increase in freezing tolerance. In the second stage, freezing tolerance is further promoted by low temperature (LT), enabling the overwintering parts of the plants to survive the winter temperatures (Weiser, 1970). LT alone will also increase frost hardiness of woody species, although the combination of short-day (SD) photoperiod and LT result in the greatest hardiness level (Christersson, 1978; Li et al., 2002). In contrast, the length of the photoperiod does not seem to play a significant role in development of freezing tolerance in annual herbaceous plants, but the cold acclimation process is stimulated primarily by low nonfreezing temperatures (Sakai and Larcher, 1987). However, a more complex regulation of freezing tolerance appears to be operational in winter annuals involving both developmental and environmental signals. In winter cereals a vernalization requirement postponing transition to reproductive phase and photoperiod sensitivity allows plants to maintain freezing tolerance for a longer period of time under SD compared with long-day (LD) environments (Fowler et al., 2001; Danyluk et al., 2003).
Ability to cold acclimate is a quantitative trait manifested in LT-induced transcriptional reprogramming and leading to several biochemical and physiological alterations in the plant (Hughes and Dunn, 1996; Palva and Heino, 1998; Thomashow, 1999; Heino and Palva, 2003). Genes induced by cold fall into two main categories. First, genes encoding enzymes or structural components believed to participate in direct protection of cells against freezing damage. This group includes genes encoding e.g. late embryogenesis-abundant (LEA) proteins, enzymes required for osmolyte biosynthesis, antifreeze proteins, chaperones, and detoxification enzymes (for review, see Bray, 1993; Ingram and Bartels, 1996; Palva and Heino, 1998; Thomashow, 1999). The second category includes genes encoding transcription factors and other regulatory proteins controlling the LT responses either transcriptionally or posttranscriptionally (Thomashow, 1998, 1999; Heino and Palva, 2003).
Accumulation of dehydrins (LEA D-II proteins; for review, see Close, 1996, 1997; Svensson et al., 2002) is frequently associated with development of freezing tolerance in plants. Dehydrins are also induced by other abiotic stresses leading to water deficit, such as drought and salinity, as well as by the phytohormone abscisic acid (ABA; Bray, 1993; Close, 1996, 1997; Campbell and Close, 1997). Although dehydrin genes have mostly been studied in herbaceous plants, dehydrins have also been characterized in several woody species, including Poncrinus trifoliate (Cai et al., 1995), Pseudotsuga menziesii (Jarvis et al., 1996), Prunus persica (Arora and Wisniewski, 1994; Artlip et al., 1997), white spruce (Picea glauca; Richard et al., 2000), Prunus amyggdalus (Campalans et al., 2000), and Populus euramericana (accession no. Q9AR85). In woody plants, expression of dehydrin genes and accumulation of dehydrins appear to correlate with the seasonal variation in freezing tolerance and/or dormancy (Wisniewski et al., 1996; Arora et al., 1997; Artlip et al., 1997; Welling et al., 1997, 2004; Rinne et al., 1998; Lim et al., 1999). In particular, there appears to be a link between dehydrin accumulation and development of freezing tolerance (Arora et al., 1992, 1996, 1997; Arora and Wisniewski, 1994, 1996; Cai et al., 1995; Wisniewski et al., 1996; Welling et al., 1997).
The LT-induced expression of cold-responsive genes, e.g. DHNs, is mediated by several distinct signal transduction pathways (Ishitani et al., 1997; Shinozaki and Yamaguchi-Shinozaki, 2000; Fowler and Thomashow, 2002). One of the most intensively studied cold-response pathways is the C-repeat-binding factor (CBF) pathway, which has been established as an integral component of the cold-acclimation response (Shinozaki and Yamaguchi-Shinozaki, 2000; Thomashow, 2001). Central to the CBF pathway are the transcriptional activators, named CBF1/DREB1b, CBF2/DREB1c, and CBF3/DREB1a (Stockinger et al., 1997; Liu et al., 1998; Gilmour et al., 1998; Shinwari et al., 1998), that bind to the C-repeat (CRT)/dehydration responsive element (DRE)/LT-responsive element (LTRE) elements present in the promoters of several cold- and drought-inducible genes. The CBF/DREB1 genes themselves are transiently induced by cold stress, and this induction precedes that of the downstream target genes with the CRT/DRE/LTRE cis-elements (Thomashow, 1999). Constitutive overexpression of CBF1/DREB1b (Jaglo-Ottosen et al., 2000) or CBF3/DREB1a (Liu et al., 1998; Kasuga et al., 1999) in transgenic Arabidopsis (Arabidopsis thaliana) plants results in accumulation of transcripts of cold-responsive CRT/DRE/LTRE-containing genes without an LT stimulus, indicating that temperature-dependent posttranslational modifications are not needed for the activity of the corresponding proteins. Components of the CBF pathway have been identified from several other plant species, and the pathway seems to be conserved in flowering plants (Jaglo et al., 2001). However, no information is currently available of the function or components of the CBF pathway in woody plants.
We are studying the molecular mechanism of cold acclimation in woody plants by using silver birch (Betula pendula) as a model. We have previously established that both SD photoperiod and LT can induce development of freezing tolerance in birch leaves in addition to the overwintering parts of the plant. Consequently, leaves offer a convenient model system for elucidating the early events in woody plant cold acclimation (Li et al., 2002). To obtain a molecular marker for these studies we cloned a birch dehydrin gene. The expression of this gene followed acclimation in birch leaves and appeared to be under CBF control in Arabidopsis. Interestingly, preexposure of birch seedlings to SD photoperiod was needed for the maximal induction of the gene by LT, suggesting that SD sensitizes the plants to the LT stimulus.
RESULTS
Structural Characterization of a Birch Dehydrin Gene
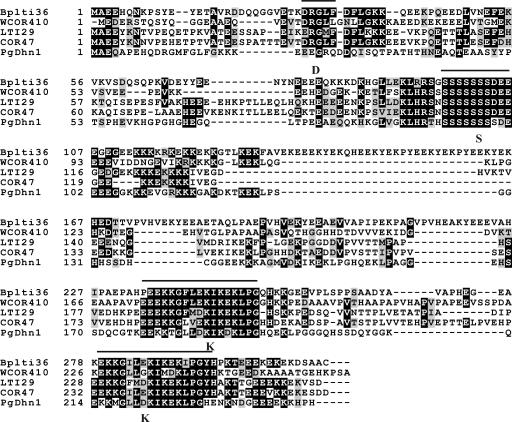
Increase in freezing tolerance during cold acclimation in herbaceous and woody plants has been correlated with activation of specific genes, in particular those for dehydrins (Palva and Heino, 1998; Thomashow, 1999; Puhakainen et al., 2004). To obtain a marker gene for molecular studies of cold acclimation in birch, we isolated cDNA and genomic clones encoding birch dehydrins. Degenerated primers corresponding to the conserved DRGVFD and IKEKLP segments found in different dehydrins were used for PCR amplification of dehydrin sequences from birch-genomic DNA. The obtained PCR fragment (BpDHN1-fragment) was sequenced and found to exhibit homology to previously characterized dehydrin genes from other plant species (data not shown). A full-length dehydrin cDNA was isolated from a χ-FIX cDNA library by using the BpDHN1-fragment as a probe. The BpDHN1-fragment was also used as a probe to screen a birch-genomic library, and a 13-kb genomic clone was isolated and characterized by restriction mapping and Southern-blotting analysis. A 3.2-kb fragment was subsequently sequenced and found to contain the complete coding sequence of the corresponding gene named Bplti36, interrupted with one 169-bp intron. Comparison with previously described dehydrin sequences revealed that the intron was located at a conserved position in the middle of the Ser repeat. The polypeptide encoded by Bplti36 contains the two conserved amino acid motifs typical to dehydrins, the Ser-rich repeat, followed by four acidic amino acids (S-segment) in the middle of the polypeptide and two Lys-rich repeats (K-segments) in the C-terminal part of the polypeptide (Fig. 1). Between the conserved S- and K-segments, the Bplti36 polypeptide has seven unique Glu-rich repeats. In general, Bplti36 is Glu (23.8%) and Lys (17.4%) rich and its total content of charged amino acids is 46.3%. It has high homology with acidic dehydrins (classified by Danyluk et al., 1994) found in Arabidopsis (Welin et al., 1994), wheat (Triticum aestivum; Danyluk et al., 1994), and white spruce (Richard et al., 2000; Fig. 1). Similar to previously characterized dehydrins, Bplti36 lacks Trp, but contains one Cys, which is uncharacteristic to dehydrins.
Figure 1.
Comparison of deduced amino acid sequence of Bplti36 and dehydrins from Arabidopsis (LTI29, COR47), wheat (WCOR410), and P. glauca (PgDhn1). The Lys-rich repeat (K-segment), the Ser-repeat (S-segment), and the D/ERGL/MF/L area (D) common to acidic dehydrins presented here are overlined and marked with bold letters K, S, and D, respectively. Identical and similar amino acids are shown in black and grey boxes, respectively.
The 3.2-kb genomic clone of Bplti36 also included 2 kb DNA upstream of the coding sequence. The transcription start site of the Bplti36 gene was determined by primer extension and it was found to be 311 bp from the translation initiation codon (Supplemental Fig. 1, available at www.plantphysiol.org). No consensus TATA-box motifs close to the transcription start site were evident. Promoter analysis made by the PLACE program (Prestridge, 1991; Higo et al., 1999) suggested that the promoter area contained several sequence motifs showing homology to cis-acting elements conferring stress responsiveness to previously characterized plant genes. These included one ABA response element (Marcotte et al., 1989) and five CRT/DRE/LTRE elements (Nordin et al., 1993; Baker et al., 1994; Yamaguchi-Shinozaki and Shinozaki, 1994; Supplemental Fig. 1).
Bplti36 Is Induced by Abiotic Stresses That Lead to Cellular Water Deficit and by ABA
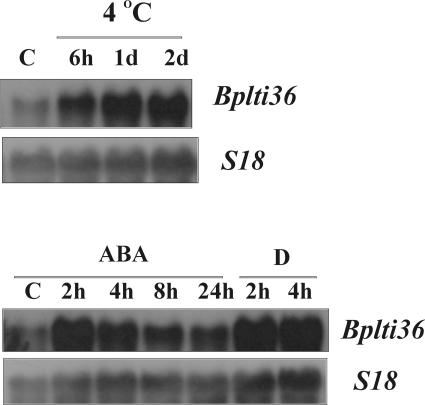
Cellular dehydration is a common denominator for abiotic stresses like freezing, drought, or high salt, and many of the cold-responsive target genes are also induced by these other stresses in addition to LT (Seki et al., 2002a). The phytohormone ABA is known to mediate responses to these stresses, and many of the stress-induced genes are also responsive to ABA (Chen et al., 1983; Lång et al., 1994; Seki et al., 2002b). The responsiveness of Bplti36 to cold, drought, high salt, and exogenous ABA in birch leaves was characterized by RNA gel-blot hybridizations. Bplti36 was shown to be highly responsive to LT, drought, and ABA (Fig. 2) and to a lesser extend to salt but only marginally to wounding and salicylic acid (data not shown). Transcript level of Bplti36 increased steadily during exposure to 4°C for 2 d or by drought treatment. In contrast, ABA application caused an early peak in Bplti36 transcript accumulation, followed by a decrease to a new steady-state level, which was higher than the level in the controls.
Figure 2.
The accumulation of Bplti36 mRNA in leaves of birch seedlings exposed to cold (4°C) for 6 h, 1 d, and 2 d and in leaf discs either infiltrated with exogenous ABA (100 μm; 2, 4, and 8 h) or left to dry on filter paper for 2 and 4 h.
Bplti36 Is Controlled by the CBF Pathway in Arabidopsis
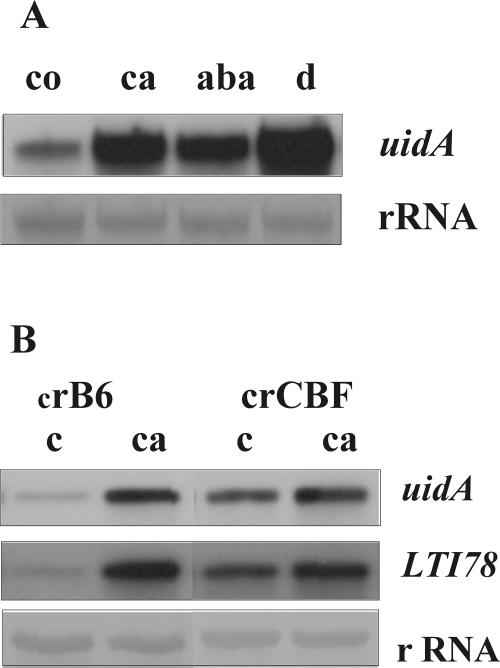
Bplti36 promoter was predicted to contain several elements previously demonstrated to confer stress responsiveness to genes in other plant species. To elucidate the functionality of these cis-elements, transgenic Arabidopsis plants were generated carrying Bplti36 promoter fused to the uidA reporter gene. The transgenic plants were exposed to cold, drought, and exogenous ABA, and the expression of the fusion construct was quantified by RNA gel-blot hybridization. Both stress treatments as well as ABA application resulted in considerable expression of uidA in the transgenic plants (Fig. 3A), suggesting that the cis-elements in the Bplti36 promoter are indeed operational in Arabidopsis. The Bplti36 promoter contains five putative CRT/DRE/LTRE elements (Supplemental Fig. 1) acting as possible binding sites for the CBF/DREB1 transcription factors in Arabidopsis. To verify the functionality of these elements a transgenic Arabidopsis line carrying Bplti36 promoter-uidA fusion was crossed both with an Arabidopsis line overproducing CBF3 and a corresponding vector control line B6 (Gilmour et al., 2000). The F1 plants from the cross with the CBF3 overproducer exhibited constitutive expression of uidA at normal-growth temperature and a further enhanced expression when cold acclimated (Fig. 3B). The cross with B6 did not lead to expression of uidA in control condition, but considerable enhancement in uidA expression was found after exposure to LT (Fig. 3B). The expression pattern of LTI78/RD29A (Nordin et al., 1993), known CBF-target gene in Arabidopsis, was similar to that of uidA expression in the F1 progeny of both crosses (Fig. 3B).
Figure 3.
A, Activation of the Bplti36 promoter in response to LT (ca), exogenous ABA (aba), and drought (d) in transgenic Arabidopsis carrying the Bplti36 promoter fused to the β-glucuronidase reporter gene (uidA). Plants were transferred from 20°C to 4°C, treated by 60 μm ABA for 6 h, or exposed to drought for overnight. The effects of different treatments on the activity of Bplti36 promoter were determined by using uidA as a probe. co, Control plants grown at normal growth conditions. B, Activation of the Bplti36 promoter in CBF3 overproducers (crCBF3) and corresponding vector controls (crB6) in normal-growth conditions (c) and after exposure to cold (ca; 4°C) for 6 h. Transgenic Arabidopsis carrying Bplti36 promoter fused to uidA-reporter gene were crossed with CBF3 overproducer (crCBF) and corresponding-vector control B6 (crB6). The activity of Bplti36 promoter in crosses were determined by using uidA as a probe. The efficacy of the cold treatment and equal RNA loading were controlled with an LTI78 probe and total RNA staining with methyl blue staining, respectively.
SD Potentiates the LT-Induced Expression of Bplti36 in Birch
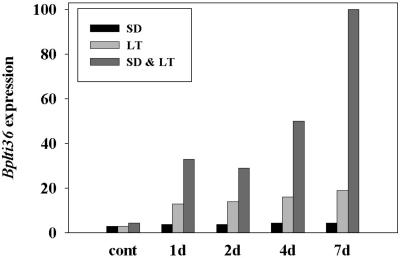
In woody plants acclimation and consequently development of freezing tolerance proceeds sequentially in response to SD and LT. The synergistic effect of these two factors manifested in enhanced freezing tolerance of buds and stem tissue is also evident in birch leaves (Table I; Li et al., 2002). To elucidate the response of Bplti36 expression to the SD signal, birch seedlings were exposed to a short two-stage acclimation process and the accumulation of Bplti36 mRNA was followed. Birch seedlings were exposed to either SD, LT, or SD followed by LT. SD exposure led to a slight increase in the accumulation of Bplti36 transcripts but not to the level observed in LT-treated plants (Fig. 4). LT alone triggered a moderate accumulation of Bplti36 transcripts (Fig. 4). In contrast to this modest induction of Bplti36, exposure to SD followed by LT treatment resulted in a substantial increase in Bplti36 level compared to LT treatment alone (Fig. 4). These data indicate that although SD exposure hardly effects Bplti36 expression by itself, it strongly enhances the effect of LT exposure on Bplti36 expression.
Table I.
The effect of photoperiod and temperature on freezing tolerance of birch leaves
| Condition | Cont | 1 d | 2 d | 4 d | 7 d | 14 d |
|---|---|---|---|---|---|---|
| LT50 | ||||||
| SD | −2.5 ± 0.21 | −3.4 ± 0.25 | −4.5 ± 0.31 | −5.9 ± 0.28 | −6.4 ± 0.35 | −7.1 ± 0.6 |
| LT | −2.5 ± 0.21 | −4.7 ± 0.28 | −6.8 ± 0.54 | −10.5 ± 0.47 | −11.6 ± 0.53 | |
| SD + LT | −6.4 ± 0.35 | −9.6 ± 0.65 | −12.0 ± 0.73 | −14.8 ± 0.66 | −15.8 ± 0.84 |
Birch seedlings were exposed to SD length (12 h light) for 7 d; LT in LD condition (4°C, 24 h light) for 7 d and SD for 7 d followed by LT (4°C) for 7 d. The freezing tolerance (LT50) of leaves (means ± se) was measured by ion-leakage test (Sukumaran and Weiser, 1972).
Figure 4.
The accumulation of Bplti36 mRNA in birch leaves under SD (12 h light, 18°C), LT (24 h light, 4°C), and SD plus LT (12 h light, 4°C, LT preceded by 7 d in SD). Histograms show the normalized values (after standardization to ribosomal signal intensities) presented as percentage of the highest value (7 d in SD plus LT).
DISCUSSION
Shortening of day length initiates the seasonal acclimation process in woody plants, triggering growth cessation and dormancy development as well as a moderate increase in freezing tolerance (Weiser, 1970; Welling et al., 1997; Li et al., 2002). Full freezing tolerance is then developed by a subsequent exposure to LT (Weiser, 1970). Our previous results have demonstrated that the signal transduction pathways starting from the perception of either SD or LT signals and leading to cold acclimation are operational in birch leaves (Li et al., 2002). The synergistic effect of SD and LT on development of freezing tolerance in buds and stems is also evident in birch leaves. Consequently, leaves provide a convenient model system to elucidate the molecular processes that are taking place during the early phases of seasonal cold acclimation in birch.
To obtain a marker gene for the acclimation process a dehydrin gene was cloned from birch. Dehydrins (LEA D-II proteins) have been associated to the development of freezing tolerance both in herbaceous and woody plants. Accumulation of dehydrin-like proteins follows the development of freezing tolerance in both herbaceous (Danyluk et al., 1998) and woody plants (Arora et al., 1992, 1996, 1997; Arora and Wisniewski, 1994, 1996; Cai et al., 1995; Wisniewski et al., 1996; Welling et al., 1997; Lim et al., 1999). Furthermore, in woody plants dehydrin levels have been shown to exhibit seasonal variation, which is correlating to the level of freezing tolerance (Arora and Wisniewski, 1994; Wisniewski et al., 1996). The expression of Bplti36 shows similar seasonal variation in stems of birch being highest during midwinter and decreasing to undetectable level in late spring (data not shown).
The cloned birch dehydrin gene Bplti36 encoded a polypeptide belonging to the SK2 type of acidic dehydrins (Danyluk et al., 1994; Close, 1996). Analogously with previously characterized dehydrins associated with cold acclimation, the expression pattern of Bplti36 showed specificity for stresses that lead to cellular-water deficit, e.g. the accumulation of the Bplti36 transcripts in birch leaves was induced in response to LT, drought, high salinity, and exogenous ABA. This pattern of expression indicates that Bplti36 provides a good molecular marker for the cold-acclimation process. However, we cannot linearly correlate the accumulation of Bplti36 mRNA with the level of freezing tolerance. Firstly, we have measured only transcript accumulation, which might not directly reflect Bplti36 protein levels. Secondly, other components might limit the development of tolerance in leaves. Studies conducted with woody plants have mainly been focused on analysis of dehydrin expression/accumulation in overwintering parts/tissues, such as buds and bark. However, in overwintering parts the development of freezing tolerance and dormancy overlap, and it is difficult to distinguish these two phenomena from each other. Only a few studies have been made to analyze dehydrin accumulation/gene expression in leaves. Arora et al. (1996) have compared development of freezing tolerance and accumulation of a 60-kD dehydrin-like protein in leaves of sibling deciduous and evergreen peach and shown that these two phenomena are related to each other in the more freezing-tolerant deciduous species, but not in less freezing-tolerant evergreen species.
In accordance with the observed stress-induced expression of Bplti36, analysis of the promoter sequence of this gene revealed several cis-elements previously demonstrated to mediate stress responsiveness in herbaceous plants. These include five putative CRT/DRE/LTRE elements previously shown to be binding sites of CBF/DREB transcription factors. This suggested that the LT activation of cold-responsive genes in woody plants could employ similar mechanisms as in herbaceous plants. This is supported by the results showing that the pBplti36-uidA construct when transferred to Arabidopsis is responsive to cold. The functionality of the putative CRT/DRE/LTRE elements in the Bplti36 promoter is further indicated by two lines of evidence: (1) Introduction of the pBplti36-uidA construct into a CBF3 overproducer in Arabidopsis results in constitutive expression of Bplti36 (Fig. 3B), and (2) deletion of CRT/DRE/LTRE elements abolishes the LT induction of the Bplti36 gene in Arabidopsis (data not shown). These results also suggest that the CBF pathway mediating LT responses in Arabidopsis and other herbaceous plants (Stockinger et al., 1997; Liu et al., 1998; Jaglo et al., 2001) is conserved in trees. This notion is supported by the presence of putative CBF orthologues in a birch expressed sequence tag database recently generated in our laboratory (A. Tuikkala, J. Ulvila, S. Mallick, K. Ojala, M. Aalto, P. Heino, R. Ruonala, L. Kauppinen, J. Immanen, Y. Helariutta, L. Holm, J. Kangasjärvi, L. Paulin, S. Rudd, and E.T. Palva, unpublished data).
Our results indicate that although the two environmental cues, SD photoperiod and LT, involved in cold acclimation in woody plants individually have an effect on development of freezing tolerance and expression of LT-responsive genes, their effect is somewhat limited. However, the SD stimulus appears to sensitize the birch cells to subsequent LT exposure. This effect can be seen both in the increased freezing tolerance and in particular in the dramatic increase in Bplti36 mRNA levels in plants exposed to both SD and LT. This potentiation seems to be specific for birch/trees, because no potentiation could be seen in the expression of Bplti36 when transgenic Arabidopsis carrying a pBplti36-uidA construct were exposed to corresponding SD and LT (data not shown). However, we cannot rule out the possibility that the increased level of Bplti36 mRNA observed in birch is not due to increased stability of the mRNA in SD + LT treated plants. Weiser (1970) originally proposed that SD will result in the formation of translocatable hardening-promoting factor in leaves, which could sensitize cells to perceive the LT signal. Our results are in agreement with and support this proposition. The potentiation of LT-induced gene expression by photoperiod appears to be distinct from the complex developmental regulation seen in overwintering cereals in which photoperiod sensitivity allows plants to maintain LT-responsive genes in an up-regulated state for a longer period of time under SD compared to LD environments (Fowler et al., 2001).
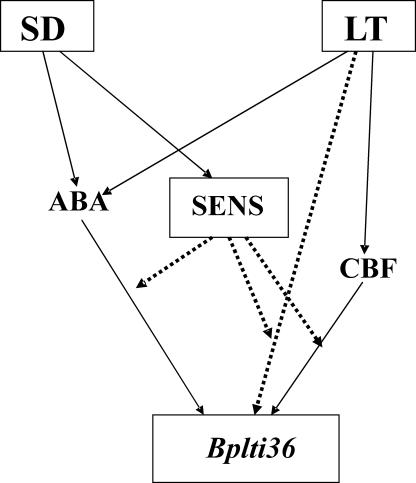
The interaction between SD and LT responses in woody plants is presumably involving phytochrome A, as overproduction of oat phytochrome A has been shown to impair cold-acclimation responses in hybrid aspen (Olsen et al., 1997; Welling et al., 2002). Thus, it is possible that SD signal in woody plants is initially perceived by phytochrome A and then transduced to responses including increase in ABA and sensitization to LT, as seen in enhanced expression of the Bplti36 (Fig. 5). In birch both SD photoperiod and LT exposure result in an increase in ABA content (Li et al., 2002). In this scenario, the accumulation of ABA could be responsible for the modest activation of Bplti36 in response to SD photoperiod only. LT exposure would in turn activate other pathway(s), including that mediated by CBF. However, the potentiation of Bplti36 expression appears not to be due to enhanced expression of CBFs, as SD did not enhance the LT-induced expression of the CBF orthologues in birch leaves (data not shown). This indicates that the effect of SD is either downstream from CBF expression or through some other pathway, which could still cooperate with the CBF pathway (Fig. 5). Sensitization to LT would be either through ABA or another, uncharacterized factor in LT pathway(s) (Fig. 5).
Figure 5.
A working model for the signaling pathways leading to Bplti36 expression in birch leaves by SD length and LT. Dotted lines represent hypothetical pathways. SENS, sensitization.
MATERIALS AND METHODS
Plant Material and Growth Conditions
Silver birch (Betula pendula cv Roth.) seeds were germinated and sown in peat:vermiculite (2:1) mixture and grown in a greenhouse under a long photoperiod, (24 h, 90–100 μmol m−2 s−1 photosynthetically active radiation [PAR], +18°C). The 24-h day length was used to avoid any interference from natural light conditions, which are highly variable at this latitude. The plants were fertilized with slow-release fertilizer (7% nitrogen, 5% phosphorus, 26% potassium), which was added to each pot (5g) at the beginning of the experiment. Three-month-old seedlings were used in all experiments.
Arabidopsis (Arabidopsis thaliana) plants were germinated and grown for 6 d in 12-h light, and 20°C on 1/2× Murashige and Skoog, 0.1% MES, pH 5.7, agar plates and thereafter transferred into 24-well plates containing the same medium. Two-week-old plants were used in all experiments.
PCR Amplification of Genomic DNA with Degenerated Primers
Genomic DNA from birch leaves was isolated according to Lodhi et al. (1994). The degenerated PCR primers were designed by using two conserved amino acid-sequences (DRGVFD and IKEKLP) of known dehydrins in Arabidopsis. The 5′-primer was 5′-GAT/C AGA/G GGI GTA/CGT TTT/C GAT/C TT-3′ and the 3′-primer was 5′-CC IGG IAG C/T TT T/CTC C/T TT T/GC AT-3′. Two hundred nanograms of birch genomic DNA was used as a template in the reaction. The PCR was performed with Peltier Thermal Cycler (PTC-200; MJ Research, Watertown, MA) with 35 cycles: denaturation, 94°C 1 min; annealing, 55°C 1 min; and elongation, 72°C 30 s. A 7-min denaturation at 96°C and 30-min elongation at 72°C was done before the first and after the last cycle, respectively. The PCR products were cloned into a pGEM-T vector (pGEM-T Vector System I; Promega, Madison, WI) and sequenced.
Cold and Photoperiod Treatments
Three-month-old birch seedlings were exposed to cold in a controlled-environmental chamber at 4°C constant temperature under LD conditions (24-h photoperiod; 90–100 μmol m−2 s−1 PAR). For short two-stage cold acclimation 3-month-old seedlings were divided into three lots for separate experimental treatments in growth chambers as follows: One-third of the plants were moved from LD to SD (12-h photoperiod; 18°C and 90–100 μmol m−2 s−1 PAR) for 1 week, then to SD + LT (12-h photoperiod; 4°C and 90–100 μmol m−2 s−1 PAR) for 1 week. Two-thirds of the plants were kept under LD conditions for 1 week, then one-half of the plants were moved to LT (24-h photoperiod; 4°C and 90–100 μmol m−2 s−1PAR) for one week, and the other half was moved to SD (12-h photoperiod, 18°C and 90–100 μmol m−2 s−1 PAR) for 2 weeks. The 12-h photoperiod was chosen since the critical day length for the ecotype used (Viitasaari 63°14′N; 26°07′E, Central Finland) is 14 h. For northern analysis fully expanded leaves were used.
Determination of Freezing Tolerance
Freezing tolerance was determined from five replicate samples. Leaf pieces cut from fully expanded leaves were wrapped in Miracloth (Calbiochem, La Jolla, CA) and placed in test tubes in a controlled-freezing bath. Extracellular ice formation was initiated at −1.5°C and after a 1-h equilibration period the bath temperature was decreased by 2°C/h. Samples were withdrawn, thawed on ice overnight, and the extent of freezing injury was determined by the ion-leakage method (Sukumaran and Weiser, 1972), and LT50 was calculated to correspond to the temperature giving 50% ion leakage.
Drought, ABA, and Salt Treatments
Drought, ABA, and salt treatments were done with leaf discs from fully expanded leaves of 3-month-old seedlings. Leaf discs were air dried for 2 and 4 h. ABA application was done by incubating leaf discs in 100 μm ABA for 2, 4, 8, and 24 h. In salt treatments leaf discs were incubated in 100 mm NaCl for 6 h. Leaf discs incubated in distilled water were used as controls.
Screening of cDNA and Genomic Library and Sequence Analysis
Screening of a λFIX II cDNA library (DIFCO Laboratories, Detroit) constructed from poly(A)+ RNA isolated from leaves of birch (Kiiskinen et al., 1997) and genomic library (λFIX II (Stratagene, La Jolla, CA) was carried out according to standard laboratory procedures (Sambrook et al., 1989) by using the isolated dehydrin PCR fragment as a probe. The cDNA and genomic clones were subcloned into pSport vector (Life Technologies/Gibco-BRL, Cleveland) and pBluescript KS II (+/−) phagemid vector, respectively, and sequenced.
DNA Sequence Analysis
The nucleotide sequence analysis was done by using the Applied Biosystems automated sequencer (PE-Applied Biosystems, Foster City, CA).
Promoter Analysis
Promoter sequence analysis was done with PLACE Signal Scan Search program (Prestridge, 1991; Higo et al., 1999). Primer extension was done according to Sambrook et al. (1989) with poly(A)+ RNA from 2-d cold-acclimated leaves of birch. An 18-mer (5′ATC ATC CCG AAC AGC GGT 3′) oligonucleotide corresponding to the DNA sequence from position −60 to −42 downstream the translation initiation codon was used as a primer. The extension products were analyzed on 6% polyacrylamide gel and compared to a sequence produced from the 5′-untranslated region of the Bplti36 gene.
RNA Isolation and Northern Hybridization
RNA was isolated from birch leaves according to Chang et al. (1993). For northern blotting total RNA (7.5 μg/sample) was separated in 1.5% formaldehyde gel and capillary blotted onto positively charged nylon membrane (Boehringer-Mannheim/Roche, Basel) essentially as described by Sambrook et al. (1989). The membranes were stained with methylene blue to confirm the RNA quality by ribosomal RNA integrity. The methylene blue was removed from the membranes by washing them in 0.1× SSC, 0.5% SDS at 68°C for 15 min. PCR-amplified cDNA insert of Bplti36 cDNA (50 ng) was labeled either with DIG Easy Hyb–UTP (Boehringer Mannheim) or α32dCTP, using Boehringer Mannheim High Prime kit (catalog no. 1 585 592) and purified with ProbeQuant G-50 Micro Columns (catalog no. 27 5335 01; Amersham Biosciences, Piscataway, NJ) before northern hybridizations. RNA hybridizations were done as described in DIG manual (Boehringer Mannheim) or according to Church and Gilbert (1984). The probe was removed from the membrane with 0.1% SDS at 95°C for 30 min and rehybridized with a birch 18S ribosomal RNA probe. Hybridization signals were quantified by Phospho-Image scanner (BAS 1500; Fujifilm, Tokyo) and quantified by Phosphoimager analyzer software (TINA version 2.09c). Results were normalized for an equal loading by the ribosomal-RNA signal.
Transformation of Arabidopsis
The 2-kb promoter fragment of Bplti36 was isolated by PCR by using primers including HindIII and NcoI restriction sites; 5′ggg gga agc tta cgt aaa tgt tgg ctt 3′and 5′ctc cgc cat ggt caa aca aat gg 3′, respectively. The promoter fragment was then cut with HindIII and NcoI and cloned in front of the uidA cDNA previously cloned into pBlueScript KS (+/−). Full-length promoter plus uidA was then isolated by restriction with HindIII and PstI (blunted) and inserted into pDE1001 (Denecke et al., 1992) that was opened with HindIII and NcoI (blunted). The vector was introduced into an Agrobacterium tumefaciens strain carrying the plasmid pGV2260 and used to transform Arabidopsis Columbia ecotype with in planta transformation method (Bechtold et al., 1993). Transgenic plants were selected by germinating T1 seeds on one-half Murashige and Skoog medium, 0.1% MES, pH 5.7, plates containing 100 μm kanamycin. The presence of transgenes in kanamycin-resistant plants was verified by Southern blot (data not shown) and lines with one insert were selected for further studies.
Arabidopsis Crossings
Transgenic Arabidopsis, line tp27/5, carrying Bplti36 promoter connected to the uidA reporter gene, was crossed both with a transgenic Arabidopsis line overproducing CBF3 (Gilmour et al., 2000) and the corresponding vector control line B6 (Gilmour et al., 2000). CBF3 overproducers and B6 line were used as acceptor plants in crossings. T1 plants from the crossings were used in experiments described below.
Expression Studies with Transgenic Arabidopsis
Arabidopsis seedlings were exposed to LT (+4°C) and 60 μm ABA for 6 h and to drought by removing the lid of 24-well tissue culture plates for overnight. RNA was extracted both from control plants (grown in 12 h light, 20°C) and treated plants by using RNAeasy Plant Mini kit (Qiagen USA, Valencia, CA). Northern hybridization was done as described above by using the uidA or LTI78/RD29A cDNAs as probes.
Sequence data from this article have been deposited with the EMBL/GenBank data libraries under accession number Q9AR85.
Supplementary Material
Acknowledgments
Prof. Michael Thomashow is thanked for the use of CBF3 overproducing and corresponding B6 lines. Arja Ikävalko and Mirva Tirkkonen are thanked for excellent technical assistance.
This work was supported by the Academy of Finland (grant nos. 38034, 44252, 44883, and 49952), by the Finnish Centre of Excellence Program 2000–2005, and by Biocentrum Helsinki.
The online version of this article contains Web-only data.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.104.047258.
References
- Arora R, Rowland LJ, Panta GR (1997) Chill-responsive dehydrins in blueberry: are they associated with cold hardiness or dormancy transitions? Physiol Plant 101: 8–16 [Google Scholar]
- Arora R, Wisniewski ME (1994) Cold acclimation in genetically related (sibling) deciduous and evergreen peach (Prunus persica (L.) Batsch): a 60-kilodalton bark protein in cold-acclimated tissues of peach is heat stable and related to the dehydrin family of proteins. Plant Physiol 105: 95–101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arora R, Wisniewski ME (1996) Accumulation of a 60-kD dehydrin protein in peach xylem tissues and its relationship to cold acclimation. HortScience 3: 923–925 [Google Scholar]
- Arora R, Wisniewski ME, Rowland LJ (1996) Cold acclimation and alterations in dehydrin-like and bark storage proteins in the leaves of sibling deciduous and evergreen peach. J Am Soc Hortic Sci 121: 915–919 [Google Scholar]
- Arora R, Wisniewski ME, Scorza R (1992) Cold acclimation in genetically related (sibling) deciduous and evergreen peach (Prunus persica (L.) Batsch): seasonal changes in cold hardiness and polypeptides of bark and xylem tissues. Plant Physiol 99: 1562–1568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Artlip TS, Callahan AM, Bassett CL, Wisniewski ME (1997) Seasonal expression of a dehydrin gene in sibling deciduous and evergreen genotypes of peach (Prunus persica (L.) Batsch). Plant Mol Biol 33: 61–70 [DOI] [PubMed] [Google Scholar]
- Baker SS, Wilhelm KS, Thomashow MF (1994) The 5′-region of Arabidopsis thaliana cor 15a has cis-acting elements that confer cold-, drought- and ABA-regulated gene expression. Plant Mol Biol 24: 701–713 [DOI] [PubMed] [Google Scholar]
- Bechtold N, Ellis J, Pelletier G (1993) In-planta Agrobacterium-mediated gene-transfer by infiltration of adult Arabidopsis thaliana plants. C R Acad Sci Ser III Life Sci 316: 1194–1199 [Google Scholar]
- Bray EA (1993) Molecular responses to water deficit. Plant Physiol 103: 1035–1040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai Q, Moore GA, Guy CL (1995) An unusual group 2 LEA gene family in citrus responsive to low temperature. Plant Mol Biol 29: 11–23 [DOI] [PubMed] [Google Scholar]
- Campalans A, Pages M, Messeguer R (2000) Protein analysis during almond embryo development: identification and characterization of a late embryogenesis abundant protein. Plant Physiol Biochem 38: 449–457 [Google Scholar]
- Campbell SA, Close TJ (1997) Dehydrins: genes, proteins, and associations with phenotypic traits. New Phytol 137: 61–74 [Google Scholar]
- Chang S, Puryear J, Cairney J (1993) A simple and efficient method for isolating RNA from pine trees. Plant Mol Biol Rep 11: 113–116 [Google Scholar]
- Chen HH, Li PH, Brenner ML (1983) Involvement of abscisic acid in potato cold acclimation. Plant Physiol 71: 362–365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christersson L (1978) The influence of photoperiod and temperature on the development of frost hardiness in seedlings of Pinus silvestris and Picea abies. Physiol Plant 44: 288–294 [Google Scholar]
- Church GM, Gilbert W (1984) Genomic sequencing. Proc Natl Acad Sci USA 81: 1991–1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Close TJ (1996) Dehydrins: emergence of a biochemical role of a family of plant dehydration proteins. Physiol Plant 97: 795–803 [Google Scholar]
- Close TJ (1997) Dehydrins: a commonalty in the response of plants to dehydration and low temperature. Physiol Plant 100: 291–296 [Google Scholar]
- Danyluk J, Houde M, Rassart E, Sarhan F (1994) Differential expression of a gene encoding an acidic dehydrin in chilling sensitive and freezing tolerant graminae species. FEBS Lett 344: 20–24 [DOI] [PubMed] [Google Scholar]
- Danyluk J, Kane NA, Breton G, Limin AE, Fowler B, Sarhan F (2003) TaVRT-1, a putative transcription factor associated with vegetative to reproductive transition in cereals. Plant Physiol 132: 1849–1860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danyluk J, Perron A, Houde M, Limin A, Fowler B, Benhamou N, Sarhan F (1998) Accumulation of an acidic dehydrin in the vicinity of the plasma membrane during cold acclimation of wheat. Plant Cell 10: 623–638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denecke J, Derycke R, Botterman J (1992) Plant and mammalian sorting signals for protein retention in the endoplasmic-reticulum contain a conserved epitope. EMBO J 11: 2345–2355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fowler DB, Breton G, Limin AE, Mahfoozi S, Sarhan F (2001) Photoperiod and temperature interactions regulate low-temperature-induced gene expression in barley. Plant Physiol 127: 1676–1681 [PMC free article] [PubMed] [Google Scholar]
- Fowler S, Thomashow MF (2002) Arabidopsis transcriptome profiling indicates that multiple regulatory pathways are activated during cold acclimation in addition to the CBF cold response pathway. Plant Cell 14: 1675–1690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilmour SJ, Sebolt AM, Salazar MP, Everard JD, Thomashow MF (2000) Overexpression of the Arabidopsis CBF3 transcriptional activator mimics multiple biochemical changes associated with cold acclimation. Plant Physiol 124: 1854–1865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilmour SJ, Zarka DG, Stockinger EJ, Salazar MP, Houghton JM, Thomashow MF (1998) Low temperature regulation of the Arabidopsis CBF family of AP2 transcriptional activators as an early step in cold-induced COR gene expression. Plant J 16: 433–442 [DOI] [PubMed] [Google Scholar]
- Heino P, Palva ET (2003) Signal transduction in plant cold acclimation. In H Hirt, K Shinozaki, eds, Topics in Current Genetics 4. Springer-Verlag, Berlin, pp 151–186
- Higo K, Ugawa Y, Iwamoto M, Korenaga T (1999) Plant cis-acting regulatory DNA elements (PLACE) database: 1999. Nucleic Acids Res 27: 297–300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes MA, Dunn MA (1996) The molecular biology of plant acclimation to low temperature. J Exp Bot 296: 291–305 [Google Scholar]
- Ingram J, Bartels D (1996) The molecular basis of dehydration tolerance in plants. Annu Rev Plant Physiol Plant Mol Biol 47: 377–403 [DOI] [PubMed] [Google Scholar]
- Ishitani M, Xiong L, Stevenson B, Zhu J-K (1997) Genetic analysis of osmotic and cold stress signal transduction in Arabidopsis thaliana: interactions and convergence of abscisic acid-dependent and abscisic acid-independent pathways. Plant Cell 9: 1935–1949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaglo KR, Kleff S, Amundsen KL, Zhang X, Haake V, Zhang JZ, Deits T, Thomashow MF (2001) Components of the Arabidopsis C-repeat/dehydration-responsive element binding factor cold-response pathway are conserved in Brassica napus and other plant species. Plant Physiol 127: 910–917 [PMC free article] [PubMed] [Google Scholar]
- Jaglo-Ottosen KR, Gilmour SJ, Zarka DG, Schabenberger O, Thomashow MF (2000) Arabidopsis CBF overexpression induces COR genes and enhances freezing tolerance. Science 280: 104–106 [DOI] [PubMed] [Google Scholar]
- Jarvis SB, Taylor MA, MacLeod MR, Davies HV (1996) Cloning and characterization of the cDNA clones of three genes that are differentially expressed during dormancy-breakage in the seeds of Douglas fir (Pseudotsuga menziesii). J Plant Physiol 147: 559–566 [Google Scholar]
- Kasuga M, Liu Q, Miura S, Yamaguchi-Shinozaki K, Shinozaki K (1999) Improving plant drought, salt, and freezing tolerance by gene transfer of a single stress-inducible transcription factor. Nat Biotechnol 17: 287–291 [DOI] [PubMed] [Google Scholar]
- Kiiskinen M, Korhonen M, Kangasjärvi J (1997) Isolation and characterization of cDNA for a plant mitochondrial phosphate translocator (Mpt1) mRNA accumulation in birch (Betula pendula Roth). Plant Mol Biol 35: 271–279 [DOI] [PubMed] [Google Scholar]
- Lång V, Mäntylä E, Welin B, Sundberg B, Palva ET (1994) Alterations in water status, endogenous abscisic acid content, and expression of rab18 gene during the development of freezing tolerance in Arabidopsis thaliana. Plant Physiol 104: 1341–1349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Puhakainen T, Welling A, Viherä-Aarnio A, Ernstsen A, Junttila O, Heino P, Palva ET (2002) Cold acclimation in silver birch (Betula pendula Roth): development of freezing tolerance in different tissues and climatic ecotypes. Physiol Plant 116: 478–488 [Google Scholar]
- Lim CC, Krebs SL, Arora R (1999) A 25-kDa dehydrin associated with genotype and age-dependent leaf freezing-tolerance in Rhododendron: a genetic marker for cold hardiness? Theor Appl Genet 99: 912–920 [Google Scholar]
- Liu Q, Kasuga M, Sakuma Y, Abe H, Miura S, Yamaguchi-Shinozaki K, Shinozaki K (1998) Two transcription factors, DREB1 and DREB2, with an EREB/AP2 DNA binding domain separate two cellular signal transduction pathways in drought- and low-temperature-responsive gene expression, respectively, in Arabidopsis. Plant Cell 10: 1391–1406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lodhi MA, Ye G-N, Weeden NF, Reisch BI (1994) A simple and efficient method for DNA extraction from grapevine cultivars and vitis species. Plant Mol Biol Rep 12: 6–13 [Google Scholar]
- Marcotte WR, Russell SH, Quatrano RS (1989) Abscisic acid-responsive sequences from the Em gene of wheat. Plant Cell 1: 969–976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordin K, Vahala T, Palva ET (1993) Differential expression of two related, low-temperature-induced genes in Arabidopsis thaliana (L.) Heynh. Plant Mol Biol 21: 641–653 [DOI] [PubMed] [Google Scholar]
- Olsen JE, Junttila O, Nilsen J, Eriksson ME, Martinussen I, Olsson O, Sandberg G, Moritz T (1997) Ectopic expression of oat phytochrome A in hybrid aspen changes critical daylength for growth and prevents cold acclimatization. Plant J 12: 1339–1350 [Google Scholar]
- Palva ET, Heino P (1998) Molecular mechanism of plant cold acclimation and freezing tolerance. In P Li, T Chen, eds, Plant Cold Hardiness. Plenum Press, New York, pp 3–14
- Puhakainen T, Hess M, Mäkelä P, Svensson J, Heino P, Palva ET (2004) Overexpression of multiple dehydrin genes enhances tolerance to freezing stress in Arabidopsis. Plant Mol Biol 54: 743–753 [DOI] [PubMed] [Google Scholar]
- Prestridge DS (1991) SIGNAL SCAN: a computer program that scans DNA sequences for eukaryotic transcriptional elements. CABIOS 7: 203–206 [DOI] [PubMed] [Google Scholar]
- Richard S, Morency MJ, Drevet C, Jouanin L, Séguin A (2000) Isolation and characterization of a dehydrin gene from white spruce induced upon wounding, drought and cold stresses. Plant Mol Biol 43: 1–10 [DOI] [PubMed] [Google Scholar]
- Rinne P, Welling A, Kaikuranta P (1998) Onset of freezing tolerance in birch (Betula pubescens Ehrh.) involves LEA proteins and osmoregulation and is impaired in an ABA-deficient genotype. Plant Cell Environ 21: 601–611 [Google Scholar]
- Sakai A, Larcher W, editors (1987) Frost Survival of Plants: Responses and Adaptation to Freezing Stress. Springer-Verlag, Berlin
- Sambrook J, Fritsch EF, Maniatis T (1989) Molecular Cloning: A Laboratory Manual, Ed 2. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY
- Seki M, Ishida J, Narusaka M, Fujita M, Nanjo T, Umezawa T, Kamiya A, Nakajima M, Enju A, Sakurai T, et al (2002. b) Monitoring the expression pattern of around 7000 Arabidopsis genes under ABA treatments using a full-length cDNA microarray. Funct Integr Genomics 2: 282–291 [DOI] [PubMed] [Google Scholar]
- Seki M, Narusaka M, Ishida J, Nanjo T, Fujita M, Oono Y, Kamiya A, Nakajima M, Enju A, Sakurai T, et al (2002. a) Monitoring the expression profiles of 7000 Arabidopsis genes under drought, cold and high-salinity stresses using a full-length cDNA microarray. Plant J 31: 279–292 [DOI] [PubMed] [Google Scholar]
- Shinozaki K, Yamaguchi-Shinozaki K (2000) Molecular responses to dehydration and low-temperature: differences and cross-talk between two stress signaling pathways. Curr Opin Plant Biol 3: 217–223 [PubMed] [Google Scholar]
- Shinwari ZK, Nakashima K, Miura S, Kasuga M, Seki M, Yamaguchi-Shinozaki K, Shinozaki K (1998) An Arabidopsis gene family encoding DRE/CRT binding proteins involved in low-temperature-responsive gene expression. Biochem Biophys Res Commun 250: 161–170 [DOI] [PubMed] [Google Scholar]
- Stockinger EJ, Gilmour SJ, Thomashow MF (1997) Arabidopsis thaliana CBF1 encodes an AP2 domain-containing transcriptional activator that binds to the C-repeat/DRE, a cis-acting DNA regulatory element that stimulates transcription in response to low temperature and water deficit. Proc Natl Acad Sci USA 94: 1035–1040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sukumaran NP, Weiser CJ (1972) An excised leaflet test for evaluation potato frost tolerance. HortScience 7: 467–468 [Google Scholar]
- Svensson J, Ismail AM, Palva ET, Close TJ (2002) Dehydrins. In KB Storey, JM Storey, eds, Sensing, Signaling and Cell Adaptation. Elsevier, Amsterdam, pp 155–171
- Thomashow MF (1998) Role of cold-responsive genes in plant freezing tolerance. Plant Physiol 118: 1–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomashow MF (1999) Plant cold acclimation: freezing tolerance genes and regulatory mechanisms. Annu Rev Plant Physiol Plant Mol Biol 50: 571–599 [DOI] [PubMed] [Google Scholar]
- Thomashow MF (2001) So what's new in the field of cold acclimation? Lots! Plant Physiol 125: 89–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiser CJ (1970) Cold resistance and injury in woody plants. Science 169: 1269–1278 [DOI] [PubMed] [Google Scholar]
- Welin B, Olson Å, Nylander M, Palva ET (1994) Characterization and differential expression of dhn/lea/rab-like genes during cold acclimation and drought stress in Arabidopsis thaliana. Plant Mol Biol 26: 131–144 [DOI] [PubMed] [Google Scholar]
- Welling A, Kaikuranta P, Rinne P (1997) Photoperiodic induction of dormancy and freezing tolerance in Betula pubescens: involvement of ABA and dehydrins. Physiol Plant 100: 119–125 [Google Scholar]
- Welling A, Moritz T, Palva ET, Junttila O (2002) Independent activation of cold acclimation by low temperature and short photoperiod in hybrid aspen. Plant Physiol 129: 1633–1641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welling A, Rinne P, Viherä-Aarnio A, Kontunen-Soppela S, Heino P, Palva ET (2004) Photoperiod and temperature differentially regulate the expression of two dehydrin genes during overwintering of birch (Betula pubescens Ehrh.). J Exp Bot 55: 507–516 [DOI] [PubMed] [Google Scholar]
- Wisniewski M, Close TJ, Artlip T, Arora R (1996) Seasonal patterns of dehydrins and 70-kDa heat-shock proteins in bark tissues of eight species of woody plants. Physiol Plant 96: 496–505 [Google Scholar]
- Yamaguchi-Shinozaki K, Shinozaki K (1994) A novel cis-acting element in an Arabidopsis gene is involved in responsiveness to drought, low-temperature, or high-salt-stress. Plant Cell 6: 251–264 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.