Abstract
Whereas the presence of RNA in mature ejaculate spermatozoa is now established, its functional significance, if any, is still a matter of debate. This reflects the accepted description that spermatozoa are highly differentiated, specialized cells of minimal cytoplasm and compacted nucleus that are transcriptionally inactive. A significant proportion of the RNA required for the later, haploid stages of terminal spermatogenic differentiation (spermiogenesis) is synthesized prior to transcriptional arrest then stably stored until its translation during spermiogenesis. Spermatozoal RNAs, including messenger RNAs (mRNAs) are therefore considered to be stored remnants. Any role in fertilization and early development has, until recently, seemed unlikely, since the oocyte contains large stores of maternal mRNAs known to be required for early embryonic development prior to zygotic genome activation. Although the spermatozoon can deliver its RNA to the oocyte at fertilization, it has been generally assumed that compared to the oocyte RNA reserve, the spermatozoan payload is too small to be functional in embryo development. However, the debate continues as recent studies suggest that in specific instances sperm RNA is functional. This review presents and discusses the functional significance of spermatozoal RNA in relation to some recent advances in the field.
Introduction
Spermatogenesis ultimately yields the spermatozoa, a highly differentiated transcriptionally inactive specialized cell of minimal cytoplasm and compacted nucleus. The presence of RNA within this cell is now established, however its functional significance, if any, is still a matter of debate. The population of spermatozoal RNAs reflects the significant proportion of the RNA synthesized prior to transcriptional arrest that is stored in a stable form in preparation for its translation during the later stages of terminal spermatogenic differentiation [Steger, 2001]. Accordingly, the RNAs that are present, including messenger RNAs (mRNAs), are considered remnants.
It has been generally assumed that compared to the oocyte that contains large stores of maternal mRNAs [Stitzel and Seydoux, 2007] these remnant spermatogenic RNAs are too few in number to represent a key player in fertilization and early development even if preceding zygotic genome activation. However, spermatozoa do transmit RNA to the oocyte at fertilization [Ostermeier et al., 2004] as part of the multilayered paternal contribution that also provides essential genomic, organelle (centriole in humans and primates) and male-specific proteomic (PLCz, PT32, STAT4, ETS) components (reviewed in [Krawetz, 2005]). This has fuelled and continued the debate of the functional significance of the paternal contribution. Recent studies suggest that in specific instances spermatozoal RNAs do have a biological function [Rassoulzadegan et al., 2006]; [Gur and Breitbart, 2006]. This review presents and discusses the functional significance and proposed roles of the various types of spermatozoal RNA.
Legacy and classes of spermatozoal transcripts
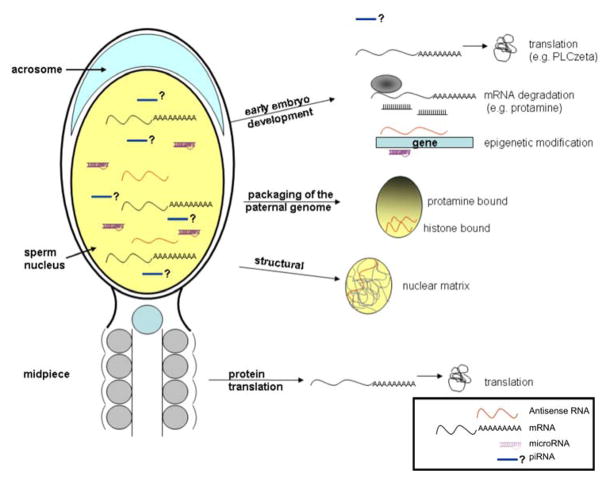
The legitimacy of spermatozoal RNAs has been hotly debated for nearly 50 years and consensus as to their authenticity has now been reached. The debate began following the initial observation that spermatozoa could incorporate radioactively labelled ribonucleotides [Abraham and Bhargava, 1963] and were therefore transcriptionally active. This was later discounted as reflecting mitochondrial associated activity [MacLaughlin and Terner, 1973]. Indeed, the absence of transcriptional activity has recently been reaffirmed [Grunewald et al., 2005]. Similarly, the presence of cytoplasmic ribosomal RNA was described [Betlach and Erickson, 1973] but never confirmed. The presence of spermatozoa RNAs was established by the initial independent studies using reverse transcription PCR (rt-PCR) [Miller et al., 1994] and in situ hybridization [Wykes et al., 1997]. The veracity of this observation is evidenced by the continued growing number of independent studies assessing the presence of specific transcripts in mammalian spermatozoa, [Cheng et al., 2006; De Ambrogi et al., 2007; Fiore et al., 2006; Teng et al., 2007; Yeung and Cooper, 2007]. This has now blossomed with the description of a myriad of transcripts contained within the spermatozoon as determined with the use of various microarray platforms [Dadoune et al., 2005; Gilbert et al., 2007; Lambard et al., 2004; Mao et al., 2004; Zhang et al., 2006; Zhao et al., 2006]. A spirited discussion of their putative roles as summarized in Figure 1, has now begun.
Figure 1.
Functions of sperm RNA. Different outcomes proposed for RNA populations, mRNA, antisense and microRNA, retained in mature spermatozoa. The RNA transferred to the oocyte at fertilization may function in early embryo development. mRNA could be translated, e.g. PLCzeta to sustain Ca2+ oscillation. Some mRNAs will be degraded, e.g. protamine. Antisense and microRNAs may epigenetically modify and modulate early embryonic gene expression. Sperm RNA may also have a structural role. For example, nuclear RNA may target chromosome regions remaining histone-bound. RNA located in the midpiece may be translated under certain conditions, e.g. capacitation.
The application and use of spermatozoa transcripts as markers of male factor fertility status continues to evolve. This arose from the initial hypothesis that the well orchestrated process of spermatogenesis would naturally lead to a well conserved set of spermatozoal transcripts in the healthy fertile male [Ostermeier et al., 2002]. The hypothesis continues to be tested with each technological advance. This has now included the use of high-resolution oligonucleotide-based microarray profiling that has allowed us to begin to dissect the pathways underlying male factor infertility [Platts et al., 2007].
Large scale microarray dataset surveys have now been set in motion to examine the degree of transcript-transcript covariance between fertile and infertile individuals [Emery et al., 2007a]. These studies have begun to reveal a series of very promising clinical markers of spermiogenic dysgenesis [Emery et al., 2007a; Emery et al., 2007b]. Evidence for large RNA networks displaying stable ratios of transcripts relative to each other in healthy individuals is accumulating. This contrasts the variable absolute signal levels that fluctuate between individuals due to downstream processes such as RNA degradation. The stable relative transcript pairs that have been identified likely reflects the well controlled processes of transcriptional regulation.
Along with carrying messenger RNA (mRNA), spermatozoa are enriched in antisense RNAs as well as microRNAs. The presence of microRNAs in spermatozoa has been independently confirmed in mouse [Amanai et al., 2006]. As with the other spermatozoal components the microRNAs are delivered to the oocyte at fertilization and can be detected up to the 2-cell stage although they are thought to serve no function [Amanai et al., 2006]. Like the corresponding mRNA population their presence has been confirmed in testis [Ro et al., 2007]. Addressing whether they indeed play a role in early embryonic development is required. The detailed characterization of the various transcripts present in the spermatozoon awaits the use of the new massively parallel sequencing technologies that has just commenced.
Functions during spermiogenesis and early embryo development
The functional significance of the transcripts that are present within the spermatozoon remains to be established. As summarized in Figure 1, one can envisage that these RNAs may play many roles. These may include maintenance and/or structural and/or support functions and perhaps are essential to early embryonic development. While evidence has been provided for each function, the role(s) of spermatozoal RNA and their mechanism of action remains to be established.
The cataloging of the nuclear encoded spermatozoal transcripts corresponding to a host of pathways has led to re-examining whether these RNAs could be translated in the absence of cytoplasmic rRNAs. Perhaps their products serve a support and/or maintenance function. Recently under capacitating conditions, the incorporation of both [35S]-methionine and [35C]-cysteine into newly synthesized spermatozoal polypeptides has been observed [Gur and Breitbart, 2006]. In the absence of a functional 80S ribosome the site of translation was localized to the midpiece. Translation was not affected by inhibitors of 80S cytoplasmic ribosomes. However, translation was completely blocked paralleling decreased sperm motility, capacitation and fertilization rate by inhibitors of mitochondrial translation and FCCP, a mitochondrial uncoupler. Whether the latter simply reflects inhibition of mitochondrial respiration remains to be established. Perhaps the nuclear encoded transcripts are translated by mitochondrial-like ribosomes [Amikura et al., 2005]. If non-mitochondrial nuclear encoded RNAs are translated, the products either lack arginine or a cytosolic tRNAArg must be available at the site of translation (D. Wallace, personal communication). Nevertheless, while questions linger, this intriguing avenue remains to be extended.
The demonstration of the delivery of spermatozoal RNA to the oocyte at fertilization was essential to developing the hypothesis that they played a critical role in normal early embryonic development [Ostermeier et al., 2004]. Their fate has been the subject of intense examination. It provides one of the first tests of the hypothesis. The profile of various paternal RNAs, protamine 1 and 2, transition protein 2, Gapd-s, and ropporin, in mouse embryos derived from ROSI (round spermatid injection) and ICSI (intracytoplasmic sperm injection) have been compared [Hayashi et al., 2003]. They observed a disappearance of the paternal transcripts during the early stages of embryogenesis, with virtually no paternal transcripts present at the 4-cell stage. This suggested that the sperm RNAs delivered to the mouse oocyte are likely discarded during the initial stages of mouse embryo development. One does not expect that all RNAs retained in the spermatozoon will have a function in early embryogenesis. As observed, some of the spermatozoal transcripts will have no effect on embryogenesis and will be degraded [Hayashi et al., 2003]. The destruction of others like the chromatin compacting protamines that complex and effectively silence nucleic acids is not unexpected and most certainly required.
Considering the quantity and the various types of transcripts that are delivered one would expect that at least some would be necessary. This is clearly exemplified by the sperm specific PLC-zeta that signals MII oocyte activation through a series of long-lived calcium oscillations following fertilization [reviewed in Swann et al., 2006]. Injection of the PLC-zeta RNAs into human, mouse, and pig oocytes elicits a similar calcium oscillatory response [Sone et al., 2005]. Interestingly, the PLC-zeta transcript has been detected in human spermatozoa [Platts et al., 2007]. Perhaps its translation at fertilization ensures this long-lived response.
Degradation or use in signalling fertilization may not be the fate of all RNAs delivered by the spermatozoon. Indeed, RNAs carried by mouse spermatozoa may epigenetically alter the phenotype of offspring while preserving a wild-type genotype through paramutation [Rassoulzadegan et al., 2006]. This occurs when two alleles of a single locus interact such that one allele influences the other to yield a heritable change. Even if the allele causing the change is not directly transmitted the paramutation can be inherited for more than one generation.
Paramutation was observed when a reporter cassette disrupted the Kit gene presenting as an unusual non-lethal white spotted fur phenotype, although the mutated allele was not detected by genotyping. This was remarkable considering the critical role of Kit in varied developmental processes. An increase in different length Kit transcripts in late spermatogenic cells including epididymal spermatozoa was observed even though expression is restricted to spermatogonia in this differentiative pathway. Injection of RNA into fertilized oocytes from tissues of mice presenting the paramutation phenotype, as well as microRNAs targeting the Kit gene, yielded the same phenotype. This data has strongly supported the view that the RNA delivered at fertilization can have a function, and in this case, act as an epigenetic modifier of early embryo development.
Sperm-Mediated Gene Transfer (SMGT) [Coward et al., 2007; Spadafora, 2008] may have uncovered another avenue whereby spermatozoa RNA can impact embryonic fate. SMGT is based on the ability of spermatozoa to take up exogenous DNA and deliver it to the oocyte upon fertilization. Through an endogenous reverse transcriptase (RT) activity [Sciamanna et al., 2003] spermatozoa can also retrotranscribe exogenous RNA. The product is maintained as an extrachromosomal structure that can then be passed via the fertilizing spermatozoon to the next generation. This or a similar system may actively engage the RNAs retained in and delivered by the spermatozoa. If correct, this provides a novel route for the introduction of non-Mendelian traits in subsequent offspring.
Localization of the RNA with the Mature Spermatozoon
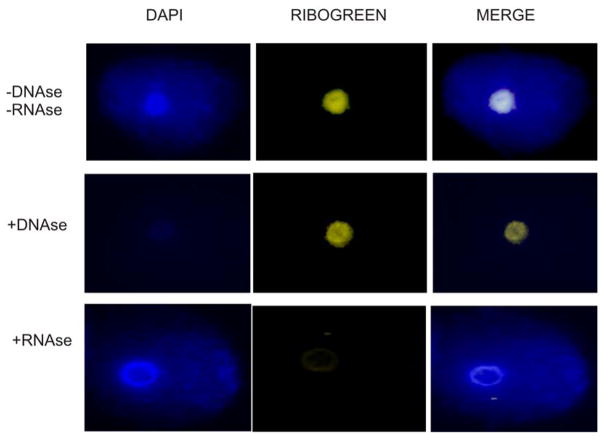
Spermatozoal RNAs are long lived, surviving in rather cramped quarters. The localization of RNA in the sperm nuclei has been reported [Dadoune et al., 2005; Pessot et al., 1989; Wykes et al., 1997] and its compartmentalization within the euchromatic and heterochromatic regions of the genome [Paul and Duerksen, 1975] described. In somatic cells, RNA is part of a proteinaceous structure interior to the nuclear envelope called the nuclear matrix. This dynamic structure is involved in the organization of the DNA into loop domains [Ward et al., 1989] through its discrete attachment to the nuclear matrix providing a platform for both transcription and replication [Tsutsui et al., 2005]. The importance of the integrity of the sperm nuclear matrix for the success of fertilization has been demonstrated in mouse [Ward et al., 1999; Ward et al., 2000], and mouse sperm nuclear halos can transform into chromosomes upon injection into oocytes [Mohar et al., 2002]. The formal conjecture that sperm RNA is part of the nuclear matrix [Miller et al., 2005] has been confirmed. Indeed, as shown in Figure 2, using an RNA-specific dye, sperm nuclear RNA is clearly observed as an integral component of the nuclear matrix. As expected, the RNA is lost when the nuclear matrix is treated with RNAse, but not with DNAse. The question remains, what is the population of RNAs that are associated with the sperm nuclear matrix?
Figure 2.
Localization of RNA to the nuclear matrix of human sperm nuclei. The histones and protamines were extracted using a solution containing 10 mM DTT and 2M NaCl. The DNA free of those proteins loops away and forms a halo around the core of DNA that remains attached to the nuclear matrix. The RiboGreen reagent was used to localize RNA to the human sperm halos. The specificity of the signal to RNA was verified by treating the halos with DNAse 1 (signal present) of RNAse (signal absent) prior to in situ hybridization.
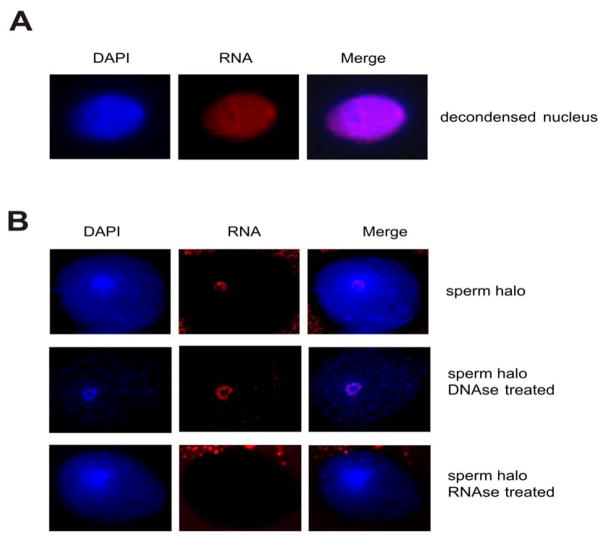
We do have some information on what these RNAs are likely to be. Spermatozoal transcripts delivered to the oocyte at fertilization that may function in early embryogenesis have been specified [Ostermeier et al., 2004]. In the initial study spermatozoal RNA profiles were compared to that of the unfertilized and fertilized oocytes. Their presence in human sperm was confirmed by RT-PCR and subsequently by RNA FISH. A typical example showing the localization of ropporin 1 that is present in the human sperm nucleus is shown in Figure 3. In partially decondensed sperm nuclei, the transcript is localized over the entire nucleus (Figure 3A). When the sperm nuclear matrix is cytologically prepared, halos of looped DNA bound by a nuclear matrix central core are resolved. Ropporin 1 RNA is localized to the nuclear matrix (Figure 3B). As expected it is resistant to DNAse 1 digestion, but rapidly degraded when treated with RNAse. At least a portion of the spermatozoal RNA that is delivered to the oocyte at fertilization is embedded within the nuclear matrix.
Figure 3.
Localization of the ropporin 1 transcript in human sperm nucleus. A) Localization in human sperm nucleus partially decondensed in the presence of DTT and Heparin. B) Localization in sperm halo preparations using 10 mM DTT and 2M NaCl to extract the protamines and histones respectively. The specificity of the signal for RNA was verified on decondensed nuclei and halo preparations treated with DNAse 1 (signal present) and RNAse (signal absent) before in situ hybridization.
Packaging the paternal genome
The retention of RNA and its association with the nuclear matrix is particularly intriguing in view of the observed differential packaging of spermatozoal DNA. During spermiogenesis, the haploid phase of spermatogenesis, most histones are progressively replaced by transition proteins and then by the smaller, more basic protamines, compacting the nucleus to 1/13th that of the oocyte [Martins and Krawetz, 2007]. During this period, transcription ceases and spermatids rely solely on stored RNAs to support protein synthesis [Hecht, 1998]. However, protamine repackaging is not complete in many species. For example, human spermatozoa, retain approximately 15% of their DNA as histone bound [Bench et al., 1996; Gatewood et al., 1987]. It is not yet known whether the histone component is passively excluded from the repackaging process or is an active consequence. The former is favored because chaperones that are known to be involved in nucleosomal deposition are not present in condensing sperm chromatin [van der Heijden et al., 2006].
Evidence from several laboratories including our own suggests that differential packaging is functionally significant. For example, the human developmental, temporally controlled and tandemly expressed β-globin gene cluster is differentially packaged. The embryonic, ε and γ globin genes ‘reside’ in the histone compartment, while the post-embryonic, β and δ globin genes are in the protamine compartment [Gardiner-Garden et al., 1998]. The differential packaging of the maternally imprinted (paternally expressed), histone bound IGF2 gene has been confirmed [Wykes and Krawetz, 2003]. Interestingly, the packaging of a DNAse 1-sensitive locus encompassing the PRM1→PRM2→TNP2 domain that are so critical to the chromatin repackaging shows no clear correspondence between DNAse 1-sensitivity and differential histone/protamine packaging. Some DNAse sensitive sequences are protamine packaged like the coding region of the PRM1 gene while the majority shows a mixture of histones and protamines.
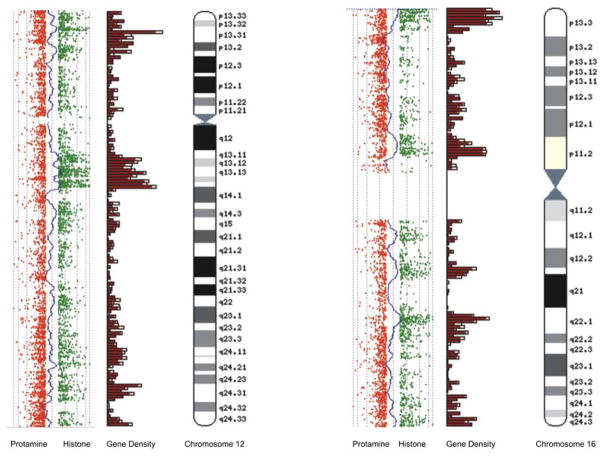
Recent genome-wide data indicates that in contrast to the mainly noncoding composition of auto-digested murine sperm DNA, the histone component from human spermatozoa is coding sequence enriched. An example of the packaging ‘preference’ for histone versus protamine bound DNA using CGH, i.e., Comparative Genomic Hybridization is shown in Figure 4. A close correspondence between the location of CCCTC factor binding sites of CTCF, a nuclear matrix associated factor and the sperm histone component has been observed. CTCF sites and their binding protein are known to be involved in mediating epigenetic effects on gene expression including the differential expression of parental alleles on imprinted genes while functioning as boundary elements preventing the spread of transcription from one gene to a closely adjacent, but ‘inappropriate’ gene [Ishihara et al., 2006; Kim et al., 2007]. They are also closely associated with gene rich rather than gene poor regions. It is tempting to speculate that in this special case of the transcriptionally silent sperm nucleus, CTCF plays a role in coordinating repackaging such that 15% of the available ‘space’ in human spermatozoa chromatin is ‘reserved’ for the coding and regulatory segments of the paternal genome. Considering the rapid displacement of all protamines by histones following fertilization, why should there be a need for spermatozoa to segregate coding from noncoding regions of the genome? One possibility is that by excluding most genes from protamine packaging, the paternal genome delivers an epigenetic ‘blueprint’ to the egg for all protein coding genes that is somehow compared with the maternal genome to ensure they are compatible. Alternatively, the paternal genome may prevent or delay the demethylation and subsequent de-repression of retroposon elements by holding them in the protamine component as post-zygotic remodeling proceeds [Bestor and Bourc’his, 2004].
Figure 4.
Comparative Genomic Hybridization of histone and protamine bound spermatozoa DNA. Alignment of CGH profiles for histone (green) and protamine (red) probe signals with Ensembl gene density profiles for chromosomes 12 and 16. Note the tendency for gene density to more closely correspond with histone rather than protamine profiles.
Perhaps the spermatozoal RNA is required to differentially package the paternal genome by shutting down the paternal genome prior to chromatin repackaging. Could the coding regions be excluded because they are marked as exempt from this mechanism? This is congruent with a role for CTCF binding where sperm RNA may act to suppress transcription from these bypassed sequences via an atypical RNA mediated down-regulatory process similar to RNAi. This could be provoked by either or both spermatozoal antisense RNAs and microRNAs perhaps employing a mechanism similar to that of XIST inactivation. Alternatively, could sense RNAs facilitate a similar shut down? If so, then one might expect spermatozoal mRNAs to mirror histone-bound sequences. Although the number of individual mRNA species in spermatozoa is not sufficient to achieve this, if flexible, a subset of mRNAs representative of all locations if not genes may be all that is required.
Conclusions
Sperm RNA has been a subject for debate for over 50 years. Its presence in the ejaculated cell has been investigated, and is it is now generally accepted that spermatozoa carry mRNAs, antisense and microRNAs into the oocyte. Whether they also carry piRNAs remains to be determined. The availability of multiple-species array platforms and the use of massive parallel sequencing will enable their detailed characterization. Undoubtedly by examining spermatozoal RNA complexity through conservation, function may then be inferred.
The recent discovery that sperm RNA can mediate epigenetic modification(s) is intriguing. Whether such RNA-mediated genetic modifications occur during “normal” fertilization remains unknown. One can easily envisage a role for these RNAs in early embryo development. “Dad having a function” warrants further investigation but this remains a direct challenge as to what may be the appropriate model of the human system. One must consider that in addition to the differential packaging of the paternal genomes, zygotic genome activation occurs at the late 1 to early 2-cell stage in the mouse embryo yet is delayed until the 4–8 cell stage embryo in humans [Braude et al., 1988]. Perhaps studies in other species with similar developmental timing, like bovine or non-human primates may better model the human system and provide the answer. The level of difficulty of this challenge is further emphasized by the small quantity of transcripts present and the somewhat heterogeneous nature of the sperm population. Rising to this challenge spermatozoa are poised to yield surprises. Our interest in this intriguing cell has yet to peak.
Acknowledgments
This work was supported in part by a National Institute of Child Health and Development Grant HD36512 to SAK and in part by a grant from the BBSRC (BBS/B/04900) to DM. CL is supported in part by the Deans Postdoctoral Recruiting Award from Wayne State School of Medicine. The authors gratefully acknowledge the use of resources at Ensembl (http://ww.ensembl.org) in the preparation of this manuscript.
LIST of ABBREVIATIONS
- SMGT
Sperm-Mediated Gene Transfer
- PRM1
protamine 1
- PRM2
protamine 2
- TNP2
transition nuclear protein 2
- CGH
Comparative Genomic Hybridization
- CTCF
CCCTC factor
- piRNA
piwi-associated RNA
References
- Abraham KA, Bhargava PM. The uptake of radioactive amino acids by spermatozoa. The intracellular site of incorporation into proteins. Biochem J. 1963;86:308–13. doi: 10.1042/bj0860308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amanai M, Brahmajosyula M, Perry AC. A restricted role for sperm-borne microRNAs in mammalian fertilization. Biol Reprod. 2006;75:877–84. doi: 10.1095/biolreprod.106.056499. [DOI] [PubMed] [Google Scholar]
- Amikura R, Sato K, Kobayashi S. Role of mitochondrial ribosome-dependent translation in germline formation in Drosophila embryos. Mech Dev. 2005;122:1087–93. doi: 10.1016/j.mod.2005.06.003. [DOI] [PubMed] [Google Scholar]
- Bench GS, Friz AM, Corzett MH, Morse DH, Balhorn R. DNA and total protamine masses in individual sperm from fertile mammalian subjects. Cytometry. 1996;23:263–71. doi: 10.1002/(SICI)1097-0320(19960401)23:4<263::AID-CYTO1>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- Bestor TH, Bourc’his D. Transposon silencing and imprint establishment in mammalian germ cells. Cold Spring Harb Symp Quant Biol. 2004;69:381–7. doi: 10.1101/sqb.2004.69.381. [DOI] [PubMed] [Google Scholar]
- Betlach CJ, Erickson RP. A unique RNA species from maturing mouse spermatozoa. Nature. 1973;242:114–5. doi: 10.1038/242114a0. [DOI] [PubMed] [Google Scholar]
- Braude P, Bolton V, Moore S. Human gene expression first occurs between the four- and eight-cell stages of preimplantation development. Nature. 1988;332:459–61. doi: 10.1038/332459a0. [DOI] [PubMed] [Google Scholar]
- Cheng YS, Kuo PL, Teng YN, Kuo TY, Chung CL, Lin YH, Liao RW, Lin JS, Lin YM. Association of spermatogenic failure with decreased CDC25A expression in infertile men. Hum Reprod. 2006;21:2346–52. doi: 10.1093/humrep/del163. [DOI] [PubMed] [Google Scholar]
- Coward K, Kubota H, Parrington J. In vivo gene transfer into testis and sperm: developments and future application. Arch Androl. 2007;53:187–97. doi: 10.1080/01485010701426455. [DOI] [PubMed] [Google Scholar]
- Dadoune JP, Pawlak A, Alfonsi MF, Siffroi JP. Identification of transcripts by macroarrays, RT-PCR and in situ hybridization in human ejaculate spermatozoa. Mol Hum Reprod. 2005;11:133–40. doi: 10.1093/molehr/gah137. [DOI] [PubMed] [Google Scholar]
- De Ambrogi M, Spinaci M, Galeati G, Tamanini C. Leptin receptor in boar spermatozoa. Int J Androl. 2007;30:458–61. doi: 10.1111/j.1365-2605.2006.00735.x. [DOI] [PubMed] [Google Scholar]
- Emery BR, Krawetz SA, Platts AE, Carrell DT. mRNA profiles in sperm of patients with protamine deregulation indicate a more generalized developmental pathology. American Society of Andrology Annual Meeting; Tampa, FL, USA. 2007a. [Google Scholar]
- Emery BR, Platts AE, Lalancette C, Krawetz SA. Protamine 1 and 2 transcript levels retained in mature sperm correlate with different cohorts of the mature sperm transcriptome. Genetics of Male Infertility Symposium; Florence, Italy. 2007b. [Google Scholar]
- Fiore C, Sticchi D, Pellati D, Forzan S, Bonanni G, Bertoldo A, Massironi M, Calo L, Fassina A, Rossi GP, Armanini D. Identification of the mineralocorticoid receptor in human spermatozoa. Int J Mol Med. 2006;18:649–52. [PubMed] [Google Scholar]
- Gardiner-Garden M, Ballesteros M, Gordon M, Tam PP. Histone- and protamine-DNA association: conservation of different patterns within the beta-globin domain in human sperm. Mol Cell Biol. 1998;18:3350–6. doi: 10.1128/mcb.18.6.3350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatewood JM, Cook GR, Balhorn R, Bradbury EM, Schmid CW. Sequence-specific packaging of DNA in human sperm chromatin. Science. 1987;236:962–4. doi: 10.1126/science.3576213. [DOI] [PubMed] [Google Scholar]
- Gilbert I, Bissonnette N, Boissonneault G, Vallee M, Robert C. A molecular analysis of the population of mRNA in bovine spermatozoa. Reproduction. 2007;133:1073–86. doi: 10.1530/REP-06-0292. [DOI] [PubMed] [Google Scholar]
- Grunewald S, Paasch U, Glander HJ, Anderegg U. Mature human spermatozoa do not transcribe novel RNA. Andrologia. 2005;37:69–71. doi: 10.1111/j.1439-0272.2005.00656.x. [DOI] [PubMed] [Google Scholar]
- Gur Y, Breitbart H. Mammalian sperm translate nuclear-encoded proteins by mitochondrial-type ribosomes. Genes Dev. 2006;20:411–6. doi: 10.1101/gad.367606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi S, Yang J, Christenson L, Yanagimachi R, Hecht NB. Mouse preimplantation embryos developed from oocytes injected with round spermatids or spermatozoa have similar but distinct patterns of early messenger RNA expression. Biol Reprod. 2003;69:1170–6. doi: 10.1095/biolreprod.103.016832. [DOI] [PubMed] [Google Scholar]
- Hecht NB. Molecular mechanisms of male germ cell differentiation. Bioessays. 1998;20:555–61. doi: 10.1002/(SICI)1521-1878(199807)20:7<555::AID-BIES6>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- Ishihara K, Oshimura M, Nakao M. CTCF-dependent chromatin insulator is linked to epigenetic remodeling. Mol Cell. 2006;23:733–42. doi: 10.1016/j.molcel.2006.08.008. [DOI] [PubMed] [Google Scholar]
- Kim TH, Abdullaev ZK, Smith AD, Ching KA, Loukinov DI, Green RD, Zhang MQ, Lobanenkov VV, Ren B. Analysis of the vertebrate insulator protein CTCF-binding sites in the human genome. Cell. 2007;128:1231–45. doi: 10.1016/j.cell.2006.12.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krawetz SA. Paternal contribution: new insights and future challenges. Nat Rev Genet. 2005;6:633–42. doi: 10.1038/nrg1654. [DOI] [PubMed] [Google Scholar]
- Lambard S, Galeraud-Denis I, Martin G, Levy R, Chocat A, Carreau S. Analysis and significance of mRNA in human ejaculated sperm from normozoospermic donors: relationship to sperm motility and capacitation. Mol Hum Reprod. 2004;10:535–41. doi: 10.1093/molehr/gah064. [DOI] [PubMed] [Google Scholar]
- MacLaughlin J, Terner C. Ribonucleic acid synthesis by spermatozoa from the rat and hamster. Biochem J. 1973;133:635–9. doi: 10.1042/bj1330635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao XM, Ma WL, Feng CQ, Zou YG, Zheng WL. An initial examination of the spermatozoal gene expression profile. Di Yi Jun Yi Da Xue Xue Bao. 2004;24:1033–6. [PubMed] [Google Scholar]
- Martins RP, Krawetz SA. Nuclear organization of the protamine locus. In: Juengel JI, Murray JF, Smith MF, editors. Reproduction in Domestic Ruminants VI. Nottingham, UK: Nottingham University Press; 2007. pp. 1–12. [DOI] [PubMed] [Google Scholar]
- Miller D, Ostermeier GC, Krawetz SA. The controversy, potential and roles of spermatozoal RNA. Trends Mol Med. 2005;11:156–63. doi: 10.1016/j.molmed.2005.02.006. [DOI] [PubMed] [Google Scholar]
- Miller D, Tang PZ, Skinner C, Lilford R. Differential RNA fingerprinting as a tool in the analysis of spermatozoal gene expression. Hum Reprod. 1994;9:864–9. doi: 10.1093/oxfordjournals.humrep.a138607. [DOI] [PubMed] [Google Scholar]
- Mohar I, Szczygiel MA, Yanagimachi R, Ward WS. Sperm nuclear halos can transform into normal chromosomes after injection into oocytes. Mol Reprod Dev. 2002;62:416–20. doi: 10.1002/mrd.10147. [DOI] [PubMed] [Google Scholar]
- Ostermeier GC, Dix DJ, Miller D, Khatri P, Krawetz SA. Spermatozoal RNA profiles of normal fertile men. Lancet. 2002;360:772–7. doi: 10.1016/S0140-6736(02)09899-9. [DOI] [PubMed] [Google Scholar]
- Ostermeier GC, Miller D, Huntriss JD, Diamond MP, Krawetz SA. Reproductive biology: delivering spermatozoan RNA to the oocyte. Nature. 2004;429:154. doi: 10.1038/429154a. [DOI] [PubMed] [Google Scholar]
- Paul J, Duerksen JD. Chromatin-associated RNA content of heterochromatin and euchromatin. Mol Cell Biochem. 1975;9:9–16. doi: 10.1007/BF01731728. [DOI] [PubMed] [Google Scholar]
- Pessot CA, Brito M, Figueroa J, Concha II, Yanez A, Burzio LO. Presence of RNA in the sperm nucleus. Biochem Biophys Res Commun. 1989;158:272–8. doi: 10.1016/s0006-291x(89)80208-6. [DOI] [PubMed] [Google Scholar]
- Platts AE, Dix DJ, Chemes HE, Thompson KE, Goodrich R, Rockett JC, Rawe VY, Quintana S, Diamond MP, Strader LF, Krawetz SA. Success and failure in human spermatogenesis as revealed by teratozoospermic RNAs. Hum Mol Genet. 2007;16:763–73. doi: 10.1093/hmg/ddm012. [DOI] [PubMed] [Google Scholar]
- Rassoulzadegan M, Grandjean V, Gounon P, Vincent S, Gillot I, Cuzin F. RNA-mediated non-mendelian inheritance of an epigenetic change in the mouse. Nature. 2006;441:469–74. doi: 10.1038/nature04674. [DOI] [PubMed] [Google Scholar]
- Ro S, Park C, Sanders KM, McCarrey JR, Yan W. Cloning and expression profiling of testis-expressed microRNAs. Dev Biol. 2007;311:592–602. doi: 10.1016/j.ydbio.2007.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sciamanna I, Barberi L, Martire A, Pittoggi C, Beraldi R, Giordano R, Magnano AR, Hogdson C, Spadafora C. Sperm endogenous reverse transcriptase as mediator of new genetic information. Biochem Biophys Res Commun. 2003;312:1039–46. doi: 10.1016/j.bbrc.2003.11.024. [DOI] [PubMed] [Google Scholar]
- Sone Y, Ito M, Shirakawa H, Shikano T, Takeuchi H, Kinoshita K, Miyazaki S. Nuclear translocation of phospholipase C-zeta, an egg-activating factor, during early embryonic development. Biochem Biophys Res Commun. 2005;330:690–4. doi: 10.1016/j.bbrc.2005.03.032. [DOI] [PubMed] [Google Scholar]
- Spadafora C. SBiRM: Systems Biology in Reproductive Medicine. 2008. A reverse transcriptase-dependent mechanism plays central roles in fundamental biological processes. [DOI] [PubMed] [Google Scholar]
- Steger K. Haploid spermatids exhibit translationally repressed mRNAs. Anat Embryol (Berl) 2001;203:323–34. doi: 10.1007/s004290100176. [DOI] [PubMed] [Google Scholar]
- Stitzel ML, Seydoux G. Regulation of the oocyte-to-zygote transition. Science. 2007;316:407–8. doi: 10.1126/science.1138236. [DOI] [PubMed] [Google Scholar]
- Swann K, Saunders CM, Rogers NT, Lai FA. PLCzeta(zeta): a sperm protein that triggers Ca2+ oscillations and egg activation in mammals. Semin Cell Dev Biol. 2006;17:264–73. doi: 10.1016/j.semcdb.2006.03.009. [DOI] [PubMed] [Google Scholar]
- Teng YN, Chung CL, Lin YM, Pan HA, Liao RW, Kuo PL. Expression of various CDC25B isoforms in human spermatozoa. Fertil Steril. 2007;88:379–82. doi: 10.1016/j.fertnstert.2006.11.186. [DOI] [PubMed] [Google Scholar]
- Tsutsui KM, Sano K, Tsutsui K. Dynamic view of the nuclear matrix. Acta Med Okayama. 2005;59:113–20. doi: 10.18926/AMO/31953. [DOI] [PubMed] [Google Scholar]
- van der Heijden GW, Derijck AA, Ramos L, Giele M, van der Vlag J, de Boer P. Transmission of modified nucleosomes from the mouse male germline to the zygote and subsequent remodeling of paternal chromatin. Dev Biol. 2006;298:458–69. doi: 10.1016/j.ydbio.2006.06.051. [DOI] [PubMed] [Google Scholar]
- Ward WS, Kimura Y, Yanagimachi R. An intact sperm nuclear matrix may be necessary for the mouse paternal genome to participate in embryonic development. Biol Reprod. 1999;60:702–6. doi: 10.1095/biolreprod60.3.702. [DOI] [PubMed] [Google Scholar]
- Ward WS, Kishikawa H, Akutsu H, Yanagimachi H, Yanagimachi R. Further evidence that sperm nuclear proteins are necessary for embryogenesis. Zygote. 2000;8:51–6. doi: 10.1017/s0967199400000824. [DOI] [PubMed] [Google Scholar]
- Ward WS, Partin AW, Coffey DS. DNA loop domains in mammalian spermatozoa. Chromosoma. 1989;98:153–9. doi: 10.1007/BF00329678. [DOI] [PubMed] [Google Scholar]
- Wykes SM, Krawetz SA. The structural organization of sperm chromatin. J Biol Chem. 2003;278:29471–7. doi: 10.1074/jbc.M304545200. [DOI] [PubMed] [Google Scholar]
- Wykes SM, Visscher DW, Krawetz SA. Haploid transcripts persist in mature human spermatozoa. Mol Hum Reprod. 1997;3:15–9. doi: 10.1093/molehr/3.1.15. [DOI] [PubMed] [Google Scholar]
- Yeung CH, Cooper TG. Potassium channels involved in human sperm volume regulation-quantitative studies at the protein and mRNA levels. Mol Reprod Dev. 2007;75:659–668. doi: 10.1002/mrd.20812. [DOI] [PubMed] [Google Scholar]
- Zhang JS, Liu Q, Li YM, Hall SH, French FS, Zhang YL. Genome-wide profiling of segmental-regulated transcriptomes in human epididymis using oligo microarray. Mol Cell Endocrinol. 2006;250:169–77. doi: 10.1016/j.mce.2005.12.041. [DOI] [PubMed] [Google Scholar]
- Zhao Y, Li Q, Yao C, Wang Z, Zhou Y, Wang Y, Liu L, Wang L, Qiao Z. Characterization and quantification of mRNA transcripts in ejaculated spermatozoa of fertile men by serial analysis of gene expression. Hum Reprod. 2006;21:1583–90. doi: 10.1093/humrep/del027. [DOI] [PubMed] [Google Scholar]