Abstract
In Arabidopsis thaliana brassinosteroid (BR), perception is mediated by two Leu-rich repeat receptor-like kinases, BRASSINOSTEROID INSENSITIVE1 (BRI1) and BRI1-ASSOCIATED RECEPTOR KINASE1 (BAK1) (Arabidopsis SOMATIC EMBRYOGENESIS RECEPTOR-like KINASE3 [AtSERK3]). Genetic, biochemical, and yeast (Saccharomyces cerevisiae) interaction studies suggested that the BRI1-BAK1 receptor complex initiates BR signaling, but the role of the BAK1 receptor is still not clear. Using transient expression in protoplasts of BRI1 and AtSERK3 fused to cyan and yellow fluorescent green fluorescent protein variants allowed us to localize each receptor independently in vivo. We show that BRI1, but not AtSERK3, homodimerizes in the plasma membrane, whereas BRI1 and AtSERK3 preferentially heterodimerize in the endosomes. Coexpression of BRI1 and AtSERK3 results in a change of the steady state distribution of both receptors because of accelerated endocytosis. Endocytic vesicles contain either BRI1 or AtSERK3 alone or both. We propose that the AtSERK3 protein is involved in changing the equilibrium between plasma membrane–located BRI1 homodimers and endocytosed BRI1-AtSERK3 heterodimers.
INTRODUCTION
Brassinosteroid (BR) signaling is one of the best-studied signal transduction pathways in plants. In contrast with animals where steroid hormones are perceived by nuclear receptors, plants employ plasma membrane receptors that include the BRASSINOSTEROID INSENSITIVE1 (BRI1) protein. The Leu-rich repeat (LRR) receptor-like kinase (RLK) BRI1 was identified as a loss-of-function BR-insensitive Arabidopsis thaliana mutant that cannot be rescued by exogenous application of BRs (Li and Chory, 1997). The BRI1 protein consists of an extracellular domain, a single transmembrane domain, and a cytoplasmic Ser/Thr kinase. The extracellular domain contains 25 LRRs and a 70–amino acid island domain between the 21st and the 22nd LRR that was found essential for BR binding (He et al., 2000; Wang et al., 2001). A second LRR RLK, BRI1-ASSOCIATED RECEPTOR KINASE1 (BAK1), was identified in an activation-tagging screen for bri1 suppressors (Li et al., 2002) and in a yeast (Saccharomyces cerevisiae) two-hybrid screen for BRI1 kinase domain interacting proteins (Nam and Li, 2002). BAK1 has a shorter extracellular domain, with only five LRRs, and it lacks the 70–amino acid island domain. BAK1 is identical to the previously described Arabidopsis SOMATIC EMBRYOGENESIS RECEPTOR-like KINASE3 (AtSERK3), a member of a small family of related RLKs (Hecht et al., 2001). Genetic and molecular data support the notion that BRI1 and BAK1 (AtSERK3) are part of the same BR receptor complex, although two other BRI1 homologs have been reported to function as BR receptors (Yin et al., 2002b). First, bak1 null alleles are semidwarfs with reduced BR insensitive phenotypes. Second, overexpression of BAK1 (AtSERK3) suppresses weak bri1 alleles with mutations in the extracellular and in the kinase domain and gives rise to a BRI1-overexpression phenotype in wild-type plants (reviewed in Clouse, 2002; Li, 2003). Several downstream components in BR signaling have also been identified. Negative and positive regulators of BR signaling were identified as a GSK-3/Shaggy-like kinase called Brassinosteroid Insensitive 2 (Li and Nam, 2002) and the nuclear-localized Ser/Thr phosphatase bri1 Suppressor 1 family (Mora-García et al., 2004), respectively. Their potential substrates are the nuclear proteins BRASSINAZOLE RESISTANT1 (Wang et al., 2002) and bri1-EMS-SUPPRESSOR1 (BES1) (Yin et al., 2002a). Neither of these proteins has been shown to be a direct target of the BRI1/BAK1 (AtSERK3) heterodimer. BRI1 and BAK1 (AtSERK3) localize to the plasma membrane (Friedrichsen et al., 2000; Li et al., 2002), whereas BRASSINAZOLE RESISTANT1 and BES1 proteins accumulate in the nucleus after BR application (Wang et al., 2002; Yin et al., 2002a). How the BRI1-BAK1 (AtSERK3) receptor complex transmits the BR signal is not known, although two recent models suggest similarities to either animal Try kinase or transforming growth factor-β (TGF-β) cell surface receptor activation (reviewed in Clouse, 2002; Li, 2003).
Signaling events in animal cells require endocytosis, and endosomes are considered to function as signaling compartments in addition to a more general role in receptor recycling (reviewed in Gonzáles-Gaitán, 2003). During endocytosis, several intermediate compartments are distinguished. In the early sorting endosomes, a decision is made to direct endocytosed receptors either toward late endosomes and to degradation in lysosomes or to recycle the receptors back to the plasma membrane via recycling endosomes (reviewed in Gruenberg, 2001). In plants, endocytosis is poorly understood, but similar mechanisms are proposed (Ueda et al., 2001). Recently, studies of the GDP/GTP exchange factor for small G-proteins of the auxin responsive factor class (ARF-GEF), GNOM (Geldner et al., 2003), and of sterol trafficking in Arabidopsis (Grebe et al., 2003) demonstrate the importance of endocytosis in plant development. Although previously demonstrated in plants (Horn et al., 1989), virtually nothing is known about the role of endocytosis in plant receptor-mediated signaling. Previously, we showed that the AtSERK1 protein internalizes into early endosomes when coexpressed with the PP2C type KINASE ASSOCIATED PROTEIN PHOSPHATASE (KAPP) in cowpea (Vigna unguiculata) protoplasts (Shah et al., 2002). It was proposed that the function of KAPP was tightly coupled to a mechanism of plant receptor internalization.
To study the dynamics of the BRI1/AtSERK3 interaction in living cells and to determine whether endocytosis of the two receptors is taking place, we tagged both BRI1 and AtSERK3 proteins with the green fluorescent protein (GFP) variants exhibiting either yellow or cyan fluorescent proteins (YFP or CFP, respectively). This allowed us to follow both receptors simultaneously in vivo and to apply imaging techniques, such as fluorescence lifetime imaging microscopy (FLIM) to determine Förster resonance energy transfer (FRET), indicative of receptor heterodimerization. These techniques are essential for imaging protein interactions in living plant (Shah et al., 2001, 2002; Immink et al., 2002) and animal cells (Sorkin et al., 2000; Haj et al., 2002).
Our results show that BRI1 and AtSERK3 are constantly recycled via endosomes. When coexpressed, the two receptors are sorted into different endosomal compartments containing either BRI1 or AtSERK3 alone, or both. Interestingly, BRI1 and AtSERK3 do not constitutively interact because they show interaction mainly in the endosomes and in restricted areas on the plasma membrane. This resembles the endocytic pathway of internalization of animal receptors and suggests a function of the AtSERK3 protein in redistributing the BRI1 receptor.
RESULTS
AtSERK3 Accelerates BRI1 Endocytosis in Protoplasts
To determine the subcellular localization of BRI1 and AtSERK3 proteins in plant cells, we performed in vivo targeting experiments in cowpea and Arabidopsis protoplasts derived from leaf tissue. BRI1 and AtSERK3 proteins were tagged at the C terminus with either CFP or YFP and then transiently expressed in protoplasts under control of the constitutive 35S promoter of Cauliflower mosaic virus. The localization of each fusion protein was examined using confocal laser scanning microscopy (CLSM). To show the morphology of the protoplast and the localization of the fluorescent proteins for each confocal optical section of a cell, CFP or YFP fluorescence is presented in a separate channel (cyan and yellow, respectively), followed by the chlorophyll autofluorescence in the red channel and by an overlay image combining both.
The transient protein expression system in protoplasts has been successfully used to study gene regulation, signal transduction (reviewed in Sheen, 2001), protein targeting, and trafficking in plant cells (Jin et al., 2001, 2003; Kim et al., 2001; Ueda et al., 2001; Sohn et al., 2003; Park et al., 2004). In spite of many advantages, protoplasts also have limitations. First, localization of the transiently expressed proteins may not always reflect that of the endogenous proteins in intact plant cells because of overexpression. Second, protoplasts are devoid of cell walls and plasmodesmata and lack normal cell–cell communication and polarity cues, so the transiently expressed proteins may behave differently from the endogenous proteins in intact plant cells. Thus, the results obtained using transient expression in protoplasts should be considered with those limitations in mind.
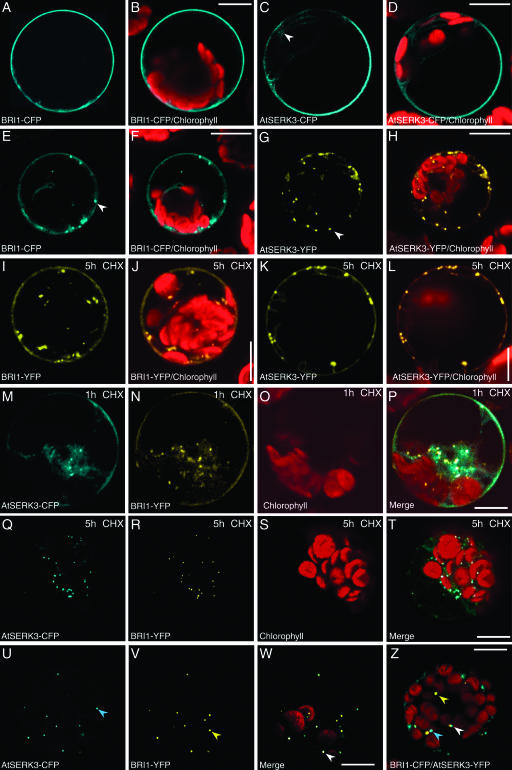
In single transfections, BRI1-CFP and AtSERK3-CFP fusion proteins are localized to the plasma membrane as early as 3 h after transfection, and this pattern was unchanged up to 16 h (overnight) incubation of the protoplasts. In Figures 1A to 1D, representative images are shown after 8 h of incubation. Residual AtSERK3-CFP fluorescence was also seen in the cytoplasm, similar to what was observed for AtSERK1 (Shah et al., 2002; indicated by an arrowhead in Figure 1C), the previously described close homolog of AtSERK3. When protoplasts were optically cross-sectioned close to the periphery of the cell, multiple fluorescently labeled vesicle-like compartments were observed (examples are shown in Figures 1E and 1F for BRI1-CFP and in Figures 1G and 1H for AtSERK3-YFP). Often those organelles appeared to be at a site near the plasma membrane (indicated by arrowheads in Figures 1E and 1G). The protein synthesis inhibitor cycloheximide (CHX) was added to protoplasts expressing either BRI1-CFP or AtSERK3-YFP 3 h after transfection, and the protoplasts were examined up to 5 h after the treatment (Figures 1I and 1J and Figures 1K and 1L, respectively). In CHX-treated protoplasts, no change in the distribution of either BRI1-CFP or AtSERK3-YFP fluorescence between the plasma membrane and the vesicle-like compartments was seen when compared with untreated protoplasts (data not shown) or to protoplasts expressing BRI1-YFP and AtSERK3-YFP overnight. In general, the expression was higher when the longer incubation time was used (cf. Figures 1I and 1K with Figures 1E and 1G). The constant level of membrane-localized receptors in the absence of protein synthesis suggests that neither undergoes rapid internalization when expressed alone.
Figure 1.
Localization of BRI1 and AtSERK3 Proteins in Cowpea and in Arabidopsis Protoplasts.
(A) to (H) Confocal images of cowpea protoplast transfected with single constructs, BRI1-CFP ([A] and [E]), AtSERK3-CFP, and AtSERK3-YFP in (C) and (G), respectively, and incubated in protoplast medium for 8 h. Because no differences in the localization and the expression levels between the CFP- and the YFP-tagged versions of the proteins were observed, we present the data with either the CFP or the YFP construct. BRI1 and AtSERK3 localized to the plasma membrane as shown in (A) for BRI1-CFP and in (C) for AtSERK3-CFP (cyan). An arrowhead in (C) indicates some residual AtSERK3-CFP fluorescence in the cytoplasm. BRI1 and AtSERK3 also localized to the multiple vesicles when protoplast transiently expressing BRI1-CFP ([E], cyan) and AtSERK3-YFP ([G], yellow) were optically cross-sectioned through the periphery of the cell. Note the arrowheads pointing to vesicles budding from the plasma membrane. The combined images of (A), (C), (E), and (G) with the chlorophyll autofluorescence (red) are shown in (B), (D), (F), and (H), respectively.
(I) to (L) Confocal images of cowpea protoplasts transfected with BRI1-YFP ([I] and [J]) and AtSERK3-YFP ([K] and [L]) incubated for 3 h in protoplast medium and then for 5 h in the presence of 50 μM CHX. The images combined with the chlorophyll autofluorescence (red) images are shown in (J) and (L), respectively.
(M) to (T) Confocal images of cowpea protoplast cotransfected with AtSERK3-CFP and BRI1-YFP incubated for 3 h in protoplast medium and then for 1 h ([M] to [P]) and for 5 h ([Q] to [T]) in the presence of 50 μM CHX. The AtSERK3-CFP fluorescence is shown in (M) and (Q). The BRI1-YFP fluorescence is shown in (N) and (R). The chlorophyll autofluorescence is shown in (O) and (S) and the combined images in (P) and (T), respectively.
(U) to (W) Confocal images of cowpea protoplast cotransfected with AtSERK3-CFP and BRI1-YFP recorded 3 h after transfection. The AtSERK3-CFP and the BRI1-YFP fluorescence localized in the endosomes is shown in (U) and (V), respectively, and the combined image in (W). Note the arrowheads pointing to the three different types of endosomes present in the cell.
(Z) A confocal image of Arabidopsis protoplast cotransfected with BRI1-CFP and AtSERK3- YFP recorded 16 h after transfection. Bar = 10 μM.
When BRI1 and AtSERK3 were coexpressed for 3 h as CFP and YFP fusions, respectively, followed by 5 h of CHX treatment, there was almost compete depletion of both BRI1 and AtSERK3 fluorescent proteins from the plasma membrane (cf. Figures 1M to 1P with Figures 1Q to 1T). This suggested that the presence of AtSERK3 altered the membrane location of BRI1. This could be because of accelerated endocytosis and/or inefficient recycling back to the plasma membrane. An interesting observation in the BRI1/AtSERK3 coexpression experiments was the lack of complete colocalization between the two fluorescently tagged proteins in the endosomes (Figures 1U to 1W). Surprisingly, we distinguished three different types of vesicle-like compartments, compartments that contained BRI1 and AtSERK3 together (indicated by an arrowhead in Figure 1W) and compartments that contained either AtSERK3 (cyan arrowhead in Figure 1U) or BRI1 alone (yellow arrowhead in Figure 1V). The ratio between the numbers of each type of compartment differed among different protoplasts. Similar observations were made when Arabidopsis protoplasts were cotransfected with BRI1 and AtSERK3 fluorescent proteins (Figure 1Z). Because an endocytic compartment will contain more than a single molecule of the fluorescently labeled receptor, this excludes the possibility of a random event. Thus, we conclude that the endosomal-sorting mechanism in plant cells can distinguish among different plasma membrane receptors.
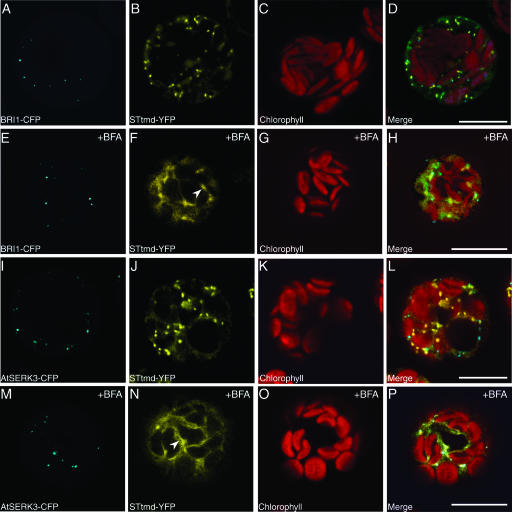
To verify that the vesicular origin of the BRI1 and AtSERK3 fluorescent proteins is the plasma membrane and does not represent proteins in transit from the Golgi membranes, we used rat sialyltransferase fused to YFP (STtmd-YFP) as a Golgi marker for colocalization experiments with either BRI1-CFP (Figures 2A to 2D) or AtSERK3-CFP–tagged proteins (Figures 2I to 2L). It was previously shown that the STtmd-YFP localizes to the trans side of the Golgi compartments in tobacco (Nicotiana tabacum) cells (Boevink et al., 1998), cowpea protoplasts (Carette et al., 2000), and in Arabidopsis (Grebe et al., 2003). No colocalization between STtmd-YFP and either BRI1-CFP (Figure 2D) or AtSERK3-CFP fluorescence (Figure 2L) was observed.
Figure 2.
Colocalization of BRI1 and AtSERK3 Proteins with the Golgi STtmd-YFP Marker in Cowpea Protoplasts.
Confocal images of protoplast cotransfected with BRI1-CFP/STtmd-YFP ([A] to [D]) and with AtSERK3-CFP/STtmd-YFP ([I] to [L]) recorded 3 h after transfection. The BRI1-CFP and AtSERK3-CFP fluorescence localized to the endosomes is shown in (A) and (I) (cyan). The STtmd-YFP fluorescence localized to the Golgi stacks is shown in (B) and (J) (yellow). The chlorophyll autofluorescence (red) is shown in (C) and (K) and the combined images in (D) and (L). Protoplast cotransfected with BRI1-CFP/STtmd-YFP ([E] to [H]) and with AtSERK3-CFP/STtmd-YFP ([M] to [P]) were allowed to express the proteins for 3 h and were then incubated in the presence of 20 μg/mL BFA for 30 min. The BRI1-CFP and AtSERK3-CFP fluorescence is shown in (E) and (M), respectively. The STtmd-YFP fluorescence is shown in (F) and (N). Membranes resembling the endoplasmic reticulum are indicated by arrowheads in (F) and (N). The chlorophyll autofluorescence is shown in (G) and (O) and the combined images in (H) and (P). Bars = 10 μM.
In Arabidopsis roots, Brefeldin A (BFA) inhibits post-Golgi vesicle trafficking by targeting large ARF-GEFs, resulting in rapid internalization of plasma membrane proteins in so-called BFA compartments (Geldner et al., 2001) containing fused endosomes (Geldner et al., 2003). BFA added to the cowpea protoplasts coexpressing STtmd-YFP and either BRI1-CFP or AtSERK3-CFP, destroyed the integrity of the Golgi vesicles and causing accumulation of the STtmd-YFP fluorescence on the Golgi membrane as early as 30 min after application (indicated by arrowheads in Figures 2F and 2N). Similar effects of BFA in cowpea protoplasts were previously observed using the Golgi marker ERD2-YFP (Pouwels et al., 2002). No change in the morphology and localization of BRI1 and AtSERK3 fluorescently labeled vesicles was observed after BFA application (cf. Figures 2E and 2M with Figures 2A and 2I). This suggested that in cowpea protoplasts derived from a mesophyll tissue, the internalization and recycling of BRI1 and AtSERK3 proteins are BFA insensitive.
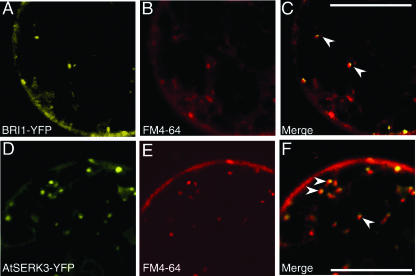
To confirm the endosomal localization of the BRI1 and AtSERK3 proteins, we examined the localization of BRI1-YFP and AtSERK3-YFP fusion proteins simultaneously with the lipophilic dye FM4-64 (Figure 3). FM4-64 is routinely used to follow the endocytic pathway in yeast and in plant cells (Vida and Emr, 1995; Ueda et al., 2001). As indicated by arrowheads in Figures 3C and 3F, most of the BRI1- and AtSERK3-containing vesicles are stained with FM4-64, suggesting that BRI1 and AtSERK3 proteins are indeed recycled and internalized via an endocytic mechanism.
Figure 3.
FM4-64 Labeling.
Confocal images of cowpea protoplast expressing BRI1-YFP (A) and AtSERK3-YFP ([D], yellow) were labeled with FM4-64 (red) and viewed after 3 h for BRI1-YFP (B) and for AtSERK3-YFP (E). The superimposed images of (A) and (B) resulted in (C), and the superimposed images of (D) and (E) resulted in (F). Arrowheads point to the overlapping yellow and red dots representing endosomes in (C) and (F). Bars = 10 μM.
BRI1 and AtSERK3 Heterodimerize in the Plasma Membrane and in Endocytic Compartments
To determine where BRI1 and AtSERK3 receptor homodimerization or heterodimerization occurs in cells, we applied FLIM. This technique captures FRET occurring between the CFP donor and the YFP acceptor molecules when both are in close proximity of ∼5 nm. This distance is regarded to be indicative of direct protein–protein interaction (Bastiaens and Pepperkok, 2000; Hink et al., 2002). Using a combination of a two-photon laser-scanning microscope and a time-correlated single photon counting acquisition card, a fluorescence intensity image of the tagged receptors is obtained. Subsequently, the fluorescence lifetime (τ) of the CFP donor molecule is calculated for every pixel in the image according to a double exponential decay model. Upon FRET, the τ of the CFP donor molecules will decrease and is visualized as a false color-coded image superimposed over the visible light image. The mean fluorescence lifetime ranges from dark blue (τ = 2.5 ns; no interaction) to dark orange or red (τ = 1.9 ns; interaction) (Figure 4). FLIM measurements performed with all receptor combinations used in this analysis are summarized in Table 1.
Figure 4.
FRET between BRI1 and AtSERK3 Imaged by FLIM.
(A) to (D) FLIM on cowpea protoplast transiently expressing AtSERK3-CFP/AtSERK3-YFP ([A] and [B]) and BRI1-CFP/BRI1-YFP ([C] and [D]) for 16 h. This time point was selected because a sufficient amount of fluorescence is required to measure FRET. Intensity images representing a steady state of the donor CFP fluorescence are presented in (A) for AtSERK3-CFP and in (C) for BRI1-CFP, respectively. The mean fluorescence lifetime values (τ) were calculated as described in Methods, and the lifetime distribution of the outlined regions in (A) and (C) are presented as enlarged pseudocolor images in (B) and (D). Arrowheads point to the areas with a significant reduction of the lifetime (dark orange to green, τ = 1.9 to 2.0 ns). Note the color bar where dark blue is used to display τ = 2.5 ns (no interaction) and the red to dark orange to display τ = 1.9 ns (interaction).
(E) to (J) FLIM on cowpea protoplast transiently coexpressing AtSERK3-CFP and BRI1-YFP proteins for 16 h. The intensity images of the donor CFP fluorescence are presented in (E), (G), and (I). Enlargements of the outlined regions in (E), (G), and (I) are presented as pseudocolor images in (F), (H), and (J), respectively. The arrowheads point to the areas with short lifetime, indicative for FRET.
Table 1.
Fluorescence Lifetime Analysis and FRET Characterization in Cowpea Protoplasts
| N
|
||||||
|---|---|---|---|---|---|---|
| Proteins | Cellular Localization | Lifetime τ (ns) ± sd | FRET Efficiency (E) (%) | FRET+ | FRET− | n |
| CFP | Cytoplasm | 2.57 ± 0.03 | – | 3 | 1 | |
| BRI1-CFP | PM | 2.47 ± 0.03 | – | 5 | 3 | |
| AtSERK3-CFP | PM | 2.53 ± 0.04 | – | 6 | 3 | |
| AtSERK1-CFP | PM | 2.51 ± 0.02 | – | 6 | 3 | |
| BRI1-CFP/AtSERK3-YFP& | PM | 2.00 ± 0.11 | 20 | 9 | 6 | 3 |
| BRI1-YFP/AtSERK3-CFP | Endosomes | 2.00 ± 0.09 | 20 | 6 | 0 | 1 |
| BRI1-CFP/AtSERK3-YFP& | PM | 2.01 ± 0.10 | 20 | 3 | 1 | 1 |
| BRI1-YFP/AtSERK3-CFP+BL | ||||||
| BRI1-CFP/BRI1-YFP | PM | 1.90 ± 0.11 | 23 | 7 | 0 | 2 |
| AtSERK3-CFP/AtSERK3-YFP | PM | 2.49 ± 0.02 | – | 2 | 12 | 2 |
| AtSERK1-CFP/AtSERK1-YFP | PM | 1.96 ± 0.11 | 23 | 7 | 0 | 1 |
FRET efficiency (E) is determined as described in Methods. N, total number of protoplasts analyzed; FRET+, number of protoplasts showing FRET; FRET−, number of protoplasts not showing FRET; n, number of independent transfection experiments; PM, plasma membrane; –, FRET efficiency is zero (nonexistent).
We first investigated whether AtSERK3 and BRI1 were able fto form homodimers (Figures 4A and 4B and Figures 4C and 4D, respectively). In protoplasts expressing AtSERK3-CFP/AtSERK3-YFP constructs, the lifetime of AtSERK3-CFP at the plasma membrane remained 2.49 ns (displayed as a homogeneous dark blue membrane in Figure 4B and corresponding τ values in Table 1). Occasionally, single small areas showing lifetime reduction in the plasma membrane were found in a few of the AtSERK3-CFP/AtSERK3-YFP–expressing protoplasts (Table 1). By contrast, in BRI1-CFP/BRI1-YFP–expressing protoplasts, a reduction in the fluorescence lifetime of the donor CFP molecules (τ = 2.5 to 1.9 ns) was detected, suggesting that BRI1 was able to form homodimers in the plasma membrane (FRET efficiency 23%). The reduction in the fluorescence lifetime of BRI1-CFP in the presence of BRI1-YFP was comparable to the reduction of the fluorescence lifetime of AtSERK1-CFP in the presence of AtSERK1-YFP (Table 1), which was previously shown to form homodimers in the plasma membrane (Shah et al., 2001; M.A. Hink, K. Shah, E. Russinova, S.C. de Vries, and A.J.W.G. Visser, unpublished results). The areas where BRI1/BRI1 homodimers in the plasma membrane occur do not appear to be organized but are fairly frequent (note the dark orange areas indicated by arrowheads in Figure 4D). We conclude that the BRI1 receptor exists as a homodimer in the plasma membrane of living plant cells. We cannot exclude the possibility that in some cases the lack of FRET is because of a competitive interaction between endogenous and transiently expressed BR receptors. This depends on the abundance of the endogenous receptors in the respective tissues. Although the BRI1 receptor is expressed relatively high in mesophyll cells (Friedrichsen et al., 2000), FRET was always observed between transiently coexpressed BRI1-CFP and BRI1-YFP receptors. The abundance of AtSERK3 transcripts in mesophyll cells was low as predicted by RT-PCR (C. Albrecht and S.C. de Vries, unpublished results), suggesting that the lack of interaction between transiently coexpressed AtSERK3-CFP and AtSERK3-YFP receptors is unlikely to be caused by interaction with its native counterpart and most likely reflects the monomeric state of AtSERK3.
To determine whether BRI1/AtSERK3 heterodimerization occurred in vivo, we transfected protoplasts with either BRI1-CFP and AtSERK3-YFP or AtSERK3-CFP and BRI1-YFP constructs. Because the plasma membrane of the cells cotransfected with BRI1 and AtSERK3 is rapidly depleted of the receptors, we first analyzed those parts where the fluorescent proteins were still present (Figures 4E and 4F). In such areas, the fluorescence intensity image of a protoplast cotransfected with, for example, AtSERK3-CFP and BRI1-YFP showed a uniform distribution of the CFP fluorescence (the boxed plasma membrane area in Figure 4E). In most parts of the plasma membrane, no (τ = 2.5 ns) or very little reduction in the fluorescence lifetime (τ = 2.47 ns) was observed (visualized by the dark blue color in Figure 4F). Those values corresponded to the values of the fluorescence lifetimes determined for protoplasts transfected with either BRI1-CFP (τ = 2.47 ns) or AtSERK3-CFP (τ = 2.53 ns) constructs alone (Table 1) and indicated no interaction occurring between AtSERK3 and BRI1 in those areas. In addition, multiple small patches that showed a significant reduction in τ from 2.5 to 2.0 ns (Table 1) of the donor (BRI1-CFP) molecules (visualized as green to orange patches, and indicated by arrowheads, in Figure 4F) were observed. The FRET efficiency in these areas was ∼20% (Table 1). The size and the frequency of these discrete areas varied among different protoplasts.
We next determined the lifetime of the AtSERK3-CFP donor molecules in the presence of BRI1-YFP in the endocytic compartments at a site near the plasma membrane (Figures 4G and 4H) or located in the cytoplasm (Figures 4I and 4J). Twin-like organelles next to the plasma membrane showed a reduction in τ of the AtSERK3-CFP from 2.5 to 2.0 ns (arrowheads in Figure 4H; FRET efficiency 20%). Comparable reductions in the fluorescence lifetime of the AtSERK3-CFP were observed in most of the BRI1 and AtSERK3 containing endocytic compartments located in the cytoplasm (see Figure 4J where spotted areas with different fluorescence lifetimes are visualized in green and orange and indicated by arrowheads). We conclude that BRI1-AtSERK3 heterodimerization is nonuniformly distributed in the plasma membrane and appears to coincide with developing endocytic compartments that contain both receptors.
AtSERK3-Accelerated Endocytosis of BRI1 in Protoplasts Represents an Activated BR Signaling
To determine if brassinolide (BL) treatment of the cowpea protoplasts coexpressing BRI1 and AtSERK3 fluorescently tagged proteins would alter the amount or the size of the membrane areas in which FRET occurred, we performed FLIM experiments on protoplasts coexpressing BRI1 and AtSERK3 in the presence of 1 μM BL. No changes in the τ values of the CFP donor molecules (Table 1) or in the number and distribution of the membrane areas showing FRET were observed (data not shown). Because BL has been shown to specifically bind to BRI1 and increase BRI1 phosphorylation (Wang et al., 2001), one possibility could be that BRI1 and AtSERK3 show ligand-independent endocytosis. Alternatively, the protoplasts could contain sufficient endogenous amounts of BL to activate the introduced fluorescent receptors.
To determine whether our FLIM data represent elements of an activated BR signaling pathway, we made use of the BES1 protein, one of the two proposed nuclear targets of BR signaling. Unphosphorylated BES1 protein was reported to accumulate in the nucleus in response to BL and to promote cell elongation in dark-grown Arabidopsis hypocotyls (Yin et al., 2002a). We transiently expressed the BES1-GFP fusion protein in protoplasts and examined the cells before and after BL application. The GFP fusion with the gain-of-function mutant bes1-D, which was previously shown to accumulate in the nucleus without BL application (Yin et al., 2002a), was used as a positive control. In cowpea protoplasts, both the BES1-GFP and the mutant bes1-GFP fusions were observed in the nucleus as early as 3 h after transfection (arrowheads in Figures 5A and 5B). BES1-GFP and bes1-GFP fluorescent proteins were also seen in the cytoplasm and in the plasma membrane (Figures 5A and 5B). Overnight expression of either BES1-GFP or mutant bes1-GFP proteins induced cell elongation and eventually the rupture of the protoplasts, indicating the presence of functional BES1 proteins in the protoplast system (data not shown). Application of BL had no effect on the localization of either BES1 or bes1-D fluorescent proteins (data not shown). We used Arabidopsis protoplasts isolated from mutants defective in the biosynthesis of BRs, such as cbb1, det2-1, and dwf4 to transiently express BES1-GFP protein (Figures 5C to 5E, respectively). In all mutant backgrounds, BES1 protein was localized in the nucleus without BL treatment. We conclude that contrary to the previous observation made by Yin et al. (2002a), in mesophyll cells BES1 nuclear localization is not dependent on the BR application. Similar observations were reported previously when the nuclear localization of BES1 was examined in light-grown root tips (Zhao et al., 2002). We next checked the phosphorylation state of BES1 protein without and with BL treatment in Arabidopsis protoplasts. In the absence of ligand, BES1 was found in phosphorylated form after BL treatment was shifted into a dephosphorylated form of faster electrophoretic mobility. It was proposed that the dephosphorylated form of BES1 modulates the transcription of BL-regulated genes (Yin et al., 2002a). As shown in Figure 5F, BES1-GFP was detected as a fast and a slow migrating band after SDS-PAGE and immunoblotting with anti-BES1 antibody or anti-GFP antibody (data not shown) representing phosphorylated and unphosphorylated forms (Yin et al., 2002a). The presence of the two forms in our system was not dependent on BL application (Figure 5F, cf. lane 1 with lane 2). Based on the presence of unphosphorylated BES1 protein in the nucleus, we conclude that in protoplasts the BR signaling is active. Therefore, our observations concerning BRI1/AtSERK3 endocytosis appear to represent active BR signaling.
Figure 5.
BES1 Expression in Protoplast.
(A) and (B) Confocal images of cowpea protoplasts transiently expressing BES1-GFP (A) and bes1-GFP ([B], green) 3 h after transfection. The nuclear-localized fluorescent protein is indicated by arrowheads.
(C) to (E) Confocal images of Arabidopsis protoplasts derived from different BR biosynthetic mutants, cbb1 (C), det2-1 (D), and dwf4 (E), and transiently expressing BES1-GFP. In all images, the chlorophyll autofluorescence is shown in red.
(F) Immunoblot analysis of the transiently expressed BES1-GFP protein in Arabidopsis protoplasts detected with anti-BES1 antibody. The blot shows that fast migrating band of ∼65 kD representing the unphosphorylated BES1-GFP is present in Arabidopsis protoplasts without and after BL treatment (lanes 1 and 2, respectively). Lane 3 contains protein extract from protoplasts cotransfected with BRI1-CFP and AtSERK3-YFP. An asterisk indicates additional bands that correspond in size to the endogenous BES1 proteins.
BRI1 Is Endocytosed in Roots in Arabidopsis Plants
RNA gel blot experiments (Li and Chory, 1997) and real-time quantitative RT-PCR (Shimada et al., 2003) have shown that in Arabidopsis the BRI1 gene is expressed ubiquitously and displays very little organ specificity. BRI1-GFP fluorescence was observed in cells of the hypocotyls, roots, and cotyledons of seedlings stably expressing BRI1-GFP fusion protein from the BRI1 promoter (Friedrichsen et al., 2000). Although not organ specific, BRI1-GFP expression was temporally regulated, and adult tissues always displayed low levels of BRI1-GFP fluorescence. In previous studies, BRI1-GFP and BAK1 (AtSERK3)-GFP fluorescence was reported to be exclusively plasma membrane localized (Friedrichsen et al., 2000; Li et al., 2002; Nam and Li, 2002). Based on our observation that in protoplasts coexpression of BRI1 and AtSERK3 resulted in accelerated endocytosis of the complex, we further explored the possibility that a similar phenomenon could be observed in planta. We therefore introduced the BRI1-GFP fusion under the control of the BRI1 endogenous promoter in wild-type Arabidopsis plants. The same construct was shown to rescue the bri1-104 mutation (Friedrichsen et al., 2000). Two independent lines containing a single copy of the BRI1-GFP construct were selected and analyzed further. Using CLSM, we looked at whether BRI1-GFP was internalized in roots of 7-d-old light-grown seedlings. As previously described, BRI1-GFP fluorescence was detected on the plasma membrane in roots but surprisingly also in endocytic compartments (Figure 6B). This occurred first at the apical part of the root meristem immediately adjacent to the quiescent center and to a lesser extent in the lateral and the columella root cap cells (Figure 6A) and in the elongation zone. In the mature parts of the root, BRI1-GFP fluorescence was reduced but also seen in the plasma membrane and in the endosomes (data not shown). This observation supports the hypothesis that the increased rate of BRI1 endocytosis is correlated with the places where active BR signaling is occurring in roots.
Figure 6.
BRI1 Endocytosis in Arabidopsis Roots.
(A) and (B) Confocal images of root from 7-d-old ProBRI1-BRI1-GFP transgenic seedling. BRI1-GFP signal is detected internalized in the cells immediately adjacent to the quiescent center ([A], arrowhead) and in the meristem zone as shown in (B).
(C) A confocal image of root meristem from bak1/serk3 mutant seedlings expressing ProBRI1-BRI1-GFP construct.
(D) and (E) Confocal images of wild-type BRI1-GFP expressing roots after BFA application in the presence of CHX (D) followed by 2 h washing out in (E). Note the BFA-induced accumulation of BRI1-GFP indicated by an arrowhead.
(F) to (H) Confocal images of FM4-64–stained root meristem area. BRI1-GFP fluorescence (green) is shown in (F). FM4-64–stained cells (red) are shown in (G). The combined images of (F) and (G) resulted in (H). Arrowheads mark vesicles colabeled by both BRI1-GFP and FM4-64.
(I) to (K) Confocal images of protoplasts derived from 7-d-old ProBRI1-BRI1-GFP transgenic seedling.
(I) Root protoplasts expressing BRI1-GFP.
(J) Protoplasts from the aerial part of the seedling expressing BRI1-GFP.
(K) Mixture of root and shoot protoplasts expressing BRI-GFP after BFA application. Note the formation of BFA compartments only in the chlorophyll-free cells (arrowhead). Bar = 10 μM.
FM4-64 staining was used to colabel the vesicles expressing BRI1-GFP. As shown in Figures 6F to 6H, FM4-64 colocalizes with most of the BRI1-GFP containing vesicles. We next analyzed the effect of BFA on BRI1-GFP endocytosis in Arabidopsis roots. BFA application in the presence of CHX caused reversible aggregation of the BRI1-GFP fluorescence in compartments similar in morphology and localization to the BFA-induced compartments of the pin-formed (PIN) proteins (Geldner et al., 2001, 2003) (Figures 6D and 6E). These results suggest that endosome-mediated BRI1 internalization as observed in cowpea protoplasts also occurs in Arabidopsis root cells. To show that the observed difference in BFA sensitivity between cowpea mesophyll cells and Arabidopsis root cells is not an artifact of the cowpea protoplasts, we generated protoplasts from 5- to 7-d-old BRI1-GFP transgenic seedlings (Figures 6I and 6J). BFA application of the protoplasts caused formation of BFA bodies only in the chlorophyll-free cells derived from either root or hypocotyl parts of the plant (arrowhead in Figure 6K). We never observed formation of BFA compartments in the chlorophyll-containing cells under the same conditions (Figure 6K), although some peripheral aggregation of BRI1 fluorescent protein was occasionally observed. This observation also suggests there are differences in the response of different plant cells to application of BFA.
To determine whether the rate of BRI1 endocytosis in Arabidopsis root cells is dependent on or reduced in the absence of the AtSERK3 protein, the BRI1-GFP transgenic line was crossed to a serk3/bak1 null mutant. No change in the rate of BRI1-GFP endocytosis in the serk3/bak1 background compared with the wild type was observed (cf. Figure 6B with 6C). We conclude that the observed BFA-sensitive trafficking of BRI1 in Arabidopsis roots reflects general recycling as was also observed for the PIN proteins.
DISCUSSION
BRI1 and AtSERK3 Dynamics in Protoplasts
When transiently expressed as translational CFP or YFP fusions in cowpea protoplasts, our results indicate that BRI1 and AtSERK3 are localized to the plasma membrane, similar to what was observed in Arabidopsis roots for BRI1 (Friedrichsen et al., 2000) and BAK1 (Li et al., 2002; Nam and Li, 2002). We also detected both proteins in small vesicle-like compartments in the cytoplasm close to the plasma membrane. Results of several experiments led to the conclusion that these vesicles represent endosomes. First, the vesicles containing BRI1 and AtSERK3 colocalize with the fluorescent endocytic tracer FM4-64 that is used to colabel early endosomes in yeast (Vida and Emr, 1995) and in plant cells (Ueda et al., 2001). Second, the BRI1 and AtSERK3 containing vesicles do not contain a trans-Golgi marker. Third, in contrast with the trans-Golgi compartments that were larger, less mobile, and highly sensitive to BFA, the BRI1 and AtSERK3 containing vesicles were smaller, highly motile, and BFA insensitive.
The turnover of BRI1 and AtSERK3 proteins in protoplasts expressing either BRI1 or AtSERK3 alone was relatively slow, but when coexpressed in protoplasts, BRI1 and AtSERK3 first colocalized at the plasma membrane and then are rapidly internalized into endosomes. Most of the internalized BRI1 and AtSERK3 proteins were not recycled back to the membrane and were possibly targeted for degradation, resulting in a nearly complete depletion of BRI1 and AtSERK3 fluorescent proteins from the plasma membrane. We believe that the rapid internalization of the coexpressed BRI1 and AtSERK3 proteins is not a result of overexpression or heterodimerization with cowpea orthologs because neither BRI1-CFP/YFP nor AtSERK3-CFP/YFP coexpression resulted in an increased rate of protein internalization. Because no differences between cowpea and Arabidopsis expression systems were observed, we conclude that receptor internalization via endocytosis generally occurs in plant cells.
Receptor recycling is well documented in animal cells. For instance, all members of the epidermal growth factor receptor (EGFR) family are predominantly localized in the plasma membrane and even in the absence of ligands are slowly internalized yet quickly recycled back to the membrane through endosomes. Epidermal growth factor ligand binding and activation of the EGFR results in an accelerated endocytosis eventually leading to receptor downregulation in the lysosomes (reviewed in Wiley, 2003). Evidence that similar mechanisms operate in plants came from recent studies showing that membrane proteins, such as the auxin efflux carrier PIN1 (Geldner et al., 2001), cell wall pectins (Baluška et al., 2002), and sterols (Grebe et al., 2003), are actively recycled through endosomes in either Arabidopsis or maize (Zea mays) roots. PIN1 recycling and proper targeting required the BFA sensitive ARF-GEF, GNOM, because the BFA-induced rapid internalization of PIN1 in BFA compartments was because of blocking of the resecretion of the protein via GNOM from the endosomes to the plasma membrane (Geldner et al., 2003). We have observed differences between mesophyll and root cells in the BFA sensitivity of the endosomal compartments containing BRI1 and AtSERK3 proteins. So far the assembly of the BFA compartments enriched in plasma membrane proteins, sterols, or pectins was mainly investigated in roots. Although highly speculative, the different BFA sensitivity of the endocytic compartments in different cell types might reflect differences in the actin cytoskeleton (reviewed in Geldner, 2004; Šamaj et al., 2004). Differences in the distribution of the Golgi markers in responses to BFA were previously reported between maize and onion (Allium cepa) root cells (Satiat-Jeunemaitre and Haves, 1992) and tobacco mesophyll cells (Boevink et al., 1998), BY-2 cells (Ritzenthaler et al., 2002), or Arabidopsis mesophyll protoplasts (Kim et al., 2001). Ritzenthaler et al. (2002) proposed a model in which the early effects of BFA depends on the physiological status of the Golgi and can result in either fusion of the Golgi with the endoplasmic reticulum or clustering of the Golgi around perinuclear BFA compartments as observed in root cells (Satiat-Jeunemaitre and Haves, 1992; Geldner et al., 2003). Because BR signaling is active in young tissues of shoots and roots (Shimada et al., 2003), it is very unlikely that differences in the BFA sensitivity reflect differences in the BR signaling mechanism.
The mechanism of animal receptor internalization is well documented but not completely understood. It is proposed that EGFR internalization is mediated mainly by clathrin-coated pits (Galperin and Sorkin, 2003), whereas an additional raft-caveolar internalization pathway was recently demonstrated for the TGF-β receptor (Di Guglielmo et al., 2003). When internalized, the ligand-receptor complexes pass through a series of endosomal compartments where they are sorted to different intracellular destinations. It is generally accepted that the early endosomes are the first sorting station of the internalized receptors (reviewed in Gruenberg, 2001). From that point, some receptors are recycled to the plasma membrane through the recycling endosomes. Others are sent to the late endosomal compartments and lysosomes for degradation, which is facilitated by ubiquitination of the receptors (reviewed in Katzmann et al., 2002; Sorkin and Von Zastrow, 2002).
When BRI1 and AtSERK3 were coexpressed together, three different endosomal compartments were observed. Surprisingly, endosomes could contain either BRI1 or AtSERK3 alone, or BRI1 and AtSERK3 together. This intriguing observation suggests that BRI1 and AtSERK3 might undergo receptor-specific sorting. It is not known whether this occurs before or after heterodimerization or how the observed proportions among endosomes containing individual monomeric, homodimeric, or heterodimeric receptors were accomplished. It seems likely that the three different endosomes arise directly from the plasma membrane. Studies of endocytosis in plants are fairly limited, but the analysis of the Arabidopsis genome point to conservation in most of the endocytic machinery (reviewed in Jürgens and Geldner, 2002). The high number of putative endosomal Rab GTPases and the fact that protein recycling through the ARF-GEF GNOM is not exclusive in Arabidopsis suggests the existence of a rather complex endosomal system in plants. This may consist of several functionally independent endosomes involved in distinct recycling pathways (Rutherford and Moore, 2002; Ueda and Nakano, 2002; Geldner et al., 2003).
Previously, it was shown that BRI1 and BAK1 proteins could phosphorylate each other in vitro and in vivo and that bak1 mutants phenotypically resemble weak bri alleles (Li et al., 2002; Nam and Li, 2002). In our protoplast system, we observed that the interaction between BRI1 and AtSERK3 was restricted to only a few small parts of the plasma membrane. This was surprising because BRI1 and AtSERK3 were colocalized in the entire plasma membrane. BRI1-AtSERK3 heterodimerization was also consistently observed in vesicles that were at the site near the plasma membrane and vesicles that were internalized and located in the cytoplasm. We therefore propose that heterodimerization between both receptors mainly occurs upon the onset of endocytosis. Whether this can be equated with transphosphorylation events between BRI1 and AtSERK3 is at present not known. For instance, for the EGF receptor ErbB1 it has been shown that intermolecular phosphorylation can occur between individual receptor molecules in the absence of a ligand (Verveer et al., 2000).
We observed that in protoplasts, exogenous BRs were not required for BRI1 and AtSERK3 interaction. This could mean that the observed endocytosis is a ligand-independent process. In that case, it apparently does not require BL-dependent BRI1 activation via phosphorylation (He et al., 2000; Wang et al., 2001). However, the presence of unphosphorylated BES1-GFP protein in protoplast nuclei similar to the BL-dependent system, described by Yin et al. (2002a), suggests that the observed endocytosis of BRI1 and AtSERK3 takes place in cells that show active BL signaling responses.
In protoplasts, a part of the BRI1 receptor molecules are in the homodimeric state. AtSERK1, a close homolog of AtSERK3, is similarly able to homodimerize when expressed in protoplasts (Shah et al., 2001). By contrast, AtSERK3 was completely unable to homodimerize in protoplasts. AtSERK3 lacks the second Cys pair flanking the LRR in the extracellular domain, and this pair might be essential for intermolecular interactions and receptor homodimerizaton (Diévart and Clark, 2003).
Although not excluded, BRI1 homodimerization has so far not been demonstrated in plants, and bri1 mutant analysis suggested that BRI1 might function in a heterodimer (Li et al., 2002; Nam and Li, 2002). If a situation such as observed in our protoplasts reflects the situation in plants, a mosaic of homodimeric and heterodimeric BRI receptors could be envisaged. In that scenario, BRI1 can transduce a signal through either BRI1/BRI1 homodimers or through heterodimers with AtSERK3 (BAK1). The weak phenotype of the serk3/bak1 null allele suggests that the BRI11/AtSERK3 (BAK1) pair is also not exclusive, and BRI1 can form heterodimers with other AtSERK3-like proteins. We can propose that one of the roles of AtSERK3 (BAK1) would be to fine tune BR signaling by shifting the equilibrium of the membrane-localized BRI1 receptors toward the endosomes.
BRI1 Dynamics in Planta
Our work demonstrates that BRI1 receptor internalization occurs in planta. In Arabidopsis roots, BRI1 undergoes endocytosis in the primary meristem zone and thus correlates with an area where BR-controlled cell growth occurs. BRI1 recycling is BFA sensitive so, at least in part, resembles the ARF-GEF GNOM-mediated endocytic pathway also used by the PIN1 protein. Therefore, this pathway may function to maintain the required level of several membrane proteins reminiscent of clathrin-mediated receptor recycling in animal cells. Our observation that BRI1 receptor internalization in Arabidopsis roots was not affected in bak1/serk3 mutant plants is likely because of functional redundancy. AtSERK3 belongs to a small family of at least five closely related homologs in Arabidopsis (Hecht et al., 2001; Peng and Li, 2003). Whereas a direct link between BR signaling and other members of the SERK family has not yet been demonstrated, FRET experiments showed that at least two other SERK homologs are able to form heterodimers with BRI1 in protoplasts (E. Russinova, J.W. Borst, and S.C. de Vries, unpublished results).
Recently, endocytosis has been demonstrated to be not only a mechanism for receptor downregulation but also a prerequisite for signaling through animal receptor Try kinases and TGF-β receptors (reviewed in Gonzáles-Gaitán, 2003). In animal TGF-β receptor signaling, it was recently demonstrated that ligand-independent constitutive trafficking occurs via a different endocytic pathway. One of these pathways follows the classic clathrin-dependent pathway, while simultaneously a second one is dependent on lipid rafts (Di Guglielmo et al., 2003). Of particular interest in the TGF-β model is that ligands do not regulate receptor trafficking but rather stabilize the heteromeric-receptor complex involved in active signaling during subsequent intracellular trafficking. If such a scenario would apply to the BRI1-AtSERK3 heterodimer, the endosomes containing both receptors as we observed in this work would be candidates for endosomes directly involved in BR signaling.
METHODS
Plant Material and Growth Conditions
Arabidopsis thaliana ecotype Columbia (Col) was used as the wild type. Seeds were germinated either on half-strength MS medium (Duchefa, Haarlem, The Netherlands) plus 1% sucrose or directly in soil. Plants were grown under fluorescent light (16-h-light/8-h-dark cycles). Arabidopsis BRI1-GFP–expressing plants were generated by introducing the ProBRI1-BRI1-GFP construct (Friedrichsen et al., 2000) using the floral dip method (Clough and Bent, 1998). Transgenic seedlings were selected on half-strength MS medium containing 50 mg/L kanamycin. Seven-day-old Arabidopsis seedlings from two independent homozygous ProBRI1-BRI1-GFP lines (T3 generation) were used to determine the BRI1-GFP localization in roots using CLSM. A ProBRI1-BRI1-GFP transgenic line was crossed with a bak1/serk3 null mutant (SALK_034523.56.00.x) generated by the SALK Institute (http://signal.salk.edu). Seeds from cbb1, det2-1, and dwf4 Arabidopsis mutants were obtained from the the Arabidopsis Stock Center.
Construction of the CFP/YFP-Tagged Proteins
The full-length cDNA of BRI1 was PCR amplified from an EST (Asamizu et al., 2000) obtained from Kazusa DNA Research Institute (Kisarazu City, Japan) with primers BRI1-F, 5′-CATGCCATGGATGAAGACTTTTTCAAGC-3′, and BRI1-R, 5′-CATGCCATGGCTAATTTTCCTTCAGGAA-3′. The BRI1 cDNA was then inserted in the NcoI site upstream of the CFP/YFP tags of the pMON999 (Monsanto, St. Louis, MO) vectors to generate BRI1-CFP and BRI1-YFP fusions, respectively. The AtSERK3 cDNA was PCR amplified with primers S3-F, 5′-CATGCCATGGAACGAAGATTAATGATC-3′, and S3-R, 5′-CATGCCATGGCTCTTGGACCCGAGGG-3′, and subcloned in the NcoI site of the vectors pMON999-CFP/YFP. All constructs were verified by sequencing. The binary constructs Pro35S-BES1-GFP and Pro35S-bes1-GFP used for transient assays in protoplasts were the same as described by Yin et al. (2002a). The full-length AtSERK1-CFP and YFP fusions were described before (Shah et al., 2001).
Transient Expression in Cowpea and Arabidopsis Protoplasts, Protoplast Treatments, and FM4-64 Staining
Cowpea and Arabidopsis mesophyll protoplasts were prepared and transfected as described previously by Shah et al. (2002), Sheen (2001) (http://genetics.mgh.harvard.edu/sheenweb), and Birnbaum et al. (2003). After transfection, the protoplasts were incubated in protoplast medium either overnight or for 3 h followed by addition of CHX (Sigma, St. Louis, MO) in concentration 50 μM from 50 mM stock. The protoplasts were used for observations until 5 h after adding the CHX. BFA (Sigma) was added to the protoplasts or applied to BRI1-GFP–expressing Arabidopsis seedlings in concentration 20 μg/mL from 5 mg/mL in DMSO. The protoplasts or the Arabidopsis roots were observed with CLSM in intervals of 30 min after the BFA application. The BFA wash-off experiments were performed as described by Geldner et al. (2001), and 24-epibrassinolide (Sigma) was applied to the protoplasts in concentration 1 μM from 0.5 mM stock in 80% ethanol. The protoplasts were taken for observation or FRET measurements after overnight or 1 to 2 h incubation with the 24-epibrassinolide. FM4-64 staining of the protoplasts and the Arabidopsis roots was performed exactly as described by Shah et al. (2002). The protoplasts were washed from the dye and incubated in protoplast medium for 0.5, 1, 2, and 3 h.
Immunoblotting
Arabidopsis protoplasts transiently expressing BES1-GFP protein for 3 to 4 h were used for immunoblotting analysis as previously described by Yin et al. (2002a) using anti-BES1 antibody.
Fluorescence Microscopy
The CFP and YFP fluorescence and the FM4-64 in protoplasts and in Arabidopsis seedlings were analyzed with the Confocal Laser Scanning Microscope 510 (Carl Zeiss, Jena, Germany) as described before by Shah et al. (2002). In addition, the GFP was excited by the 488-nm laser line in combination with the main dichroic 488, and GFP fluorescence was detected by a band-pass 505- to 550-nm filter. A 40× oil immersion objective (numerical aperture 1.3) was used for scanning. The pinhole setting was 60 μm, which yielded a theoretical thickness (full width at half-maximum) of 1 μm. Images and data captures were analyzed with Zeiss LSM510 software (version 3.2).
FLIM
FLIM was performed using a Bio-Rad Radiance 2100 MP system (Hercules, CA) in combination with a Nikon TE 300 inverted microscope (Tokyo, Japan). Two-photon excitation pulses were generated by a Ti:Sapphire laser (Coherent Mira; Santa Clara, CA) that was pumped by a 5-W Coherent Verdi laser. Pulse trains of 76 MHz (150 fs pulse duration, 860 nm center wavelength) were produced. The excitation light was directly coupled into the microscope and focused into the sample using a CFI Plan Apochromat 60× water immersion objective lens (numerical aperture 1.2). Fluorescent light was detected using the nondescanned single photon counting detection, which is the most sensitive solution for two-photon imaging. For the FLIM experiment, the Hamamatsu R3809U MCP PMT (Hamamatsu City, Japan) was used, which has a typical time resolution ∼50 ps. CFP emission was selected using a 480DF30 band-pass filter. Images with a frame size of 64 × 64 pixels were acquired, and the average count rate was 2 × 104 photons/s for an acquisition time of 90 s (Borst et al., 2003; Chen et al., 2003; Becker et al., 2004; Chen and Periasamy, 2004). From the intensity images obtained, complete fluorescence lifetime decays were calculated per pixel and fitted using a double exponential decay model. The fluorescence lifetime of one component was fixed to the value found for AtSERK1-CFP (2.5 ns) (Table 1). The FRET efficiency (E) was determined by E = 1−τDA/τD, where τD is the fluorescence lifetime of the donor in the absence of acceptor and τDA that of the donor in the presence of acceptor at a distance (R). The distance between the donor and the acceptor was determined from the relation τDA = τD/(1 + (R0/R)6, where R0 is the Förster radius, the distance between the donor and acceptor at which 50% energy transfer takes place (Elangovan et al., 2002).
Acknowledgments
We thank Joan Wellink and Jeroen Pouwels for providing us with the STtmd-YFP Golgi marker, for growing the cowpea plants, and for protoplast transfections; Antonie Visser and Mark Hink for help with FRET measurements and during data analysis; and Niko Geldner and Jiří Friml for helpful discussions and the critical reading of the manuscript. J.C. is an investigator of the Howard Hughes Medical Institute. This work was supported by Grant ERBIO4-CT96-0689 from the European Union Biotechnology program, Grant QLG2-2000-00602 from the European Union Quality of Life and Management of Living Recourses program, and Wageningen University, Department of Agrotechnology and Food Sciences to E.R., M.K., and S.C.D.; by the Human Frontier Science Program Organization long-term fellowship to A.C-D.; and grants from the USDA and the Human Frontier Science Program to J.C.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Sacco C. de Vries (sacco.devries@wur.nl).
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.104.025387.
References
- Asamizu, E., Nakamura, Y., Sato, S., and Tabata, S. (2000). A large scale analysis of cDNA in Arabidopsis thaliana: Generation of 12,028 non-redundant expressed sequence tags from normalized and size-selected cDNA libraries. DNA Res. 7, 175–180. [DOI] [PubMed] [Google Scholar]
- Baluška, F., Hlavacka, A., Šamaj, J., Palme, K., Robinson, D.G., Matoh, T., McCurdy, D.W., Menzel, D., and Volkmann, D. (2002). F-actin-dependent endocytosis of cell wall pectins in meristematic root cells. Insights from Brefeldin A-induced compartments. Plant Physiol. 130, 422–431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bastiaens, P.I.H., and Pepperkok, R. (2000). Observing proteins in their natural habitat: The living cell. Trends Biochem. Sci. 25, 631–637. [DOI] [PubMed] [Google Scholar]
- Becker, W., Bergmann, A., Hink, M.A., Konig, K., Benndorf, K., and Biscup, C. (2004). Fluorescence lifetime imaging by time-correlated single-photon counting. Microsc. Res. Tech. 63, 58–66. [DOI] [PubMed] [Google Scholar]
- Birnbaum, K., Shasha, D.E., Wang, J.Y., Jung, J.W., Lambert, G.M., Galbraith, D.W., and Benfey, P.N. (2003). A gene expression map of the Arabidopsis root. Science 302, 1956–1960. [DOI] [PubMed] [Google Scholar]
- Boevink, P., Oparka, K., Santa Cruz, S., Martin, B., Betteridge, A., and Hawes, C. (1998). Stacks on tracks: The plant Golgi apparatus traffics on an actin/ER network. Plant J. 15, 441–447. [DOI] [PubMed] [Google Scholar]
- Borst, J.W., Hink, M., van Hoek, A., and Visser, A.J.W.G. (2003). Multiphoton microspectroscopy in living plant cells. In Proceedings of SPIE, Vol. 4963: Multiphoton Microscopy in the Biomedical Sciences III, A. Periasamy and P.T. So, eds (Bellingham, WA: SPIE), pp. 231–238.
- Carette, J.E., Stuiver, M., Van Lent, J., Wellink, J., and Van Kammen, A. (2000). Cowpea mosaic virus infection induces a massive proliferation of endoplasmic reticulum but not Golgi membranes and is dependent on de novo membrane synthesis. J. Virol. 74, 6556–6563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, Y., Mills, J.D., and Periasamy, A. (2003). Protein localization in living cells and tissues using FRET and FLIM. Differentiation 71, 528–541. [DOI] [PubMed] [Google Scholar]
- Chen, Y., and Periasamy, A. (2004). Characterization of two-photon excitation fluorescence lifetime imaging microscopy for protein localization. Microsc. Res. Tech. 63, 72–80. [DOI] [PubMed] [Google Scholar]
- Clough, S.J., and Bent, A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743. [DOI] [PubMed] [Google Scholar]
- Clouse, S.D. (2002). Brassinosteroid signal transduction: Clarifying the pathway from ligand perception to gene expression. Mol. Cell 10, 973–982. [DOI] [PubMed] [Google Scholar]
- Diévart, A., and Clark, S.E. (2003). Using mutant alleles to determine the structure and function of leucine-rich repeat receptor-like kinases. Curr. Opin. Plant Biol. 6, 507–516. [DOI] [PubMed] [Google Scholar]
- Di Guglielmo, G.M., Le Roy, C., Goodfellow, A.F., and Wrana, J.L. (2003). Distinct endocytic pathways regulate TGF-β receptor signalling and turnover. Nat. Cell Biol. 5, 410–421. [DOI] [PubMed] [Google Scholar]
- Elangovan, M., Day, R.N., and Periasamy, A. (2002). Nanosecond fluorescence resonance energy transfer-fluorescence lifetime imaging microscopy to localize the protein interactions in a single living cell. J. Microsc. 205, 3–14. [DOI] [PubMed] [Google Scholar]
- Friedrichsen, D.M., Joazeiro, C.A.P., Li, J., Hunter, T., and Chory, J. (2000). Brassinosteroid-insensitive-1 is ubiquitously expressed leucine-rich receptor serine/threonine kinase. Plant Physiol. 123, 1247–1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galperin, E., and Sorkin, A. (2003). Visualization of Rab5 activity in living cells by FRET microscopy and influence of plasma-membrane-targeted Rab5 on clathrin-dependent endocytosis. J. Cell Sci. 116, 4799–4810. [DOI] [PubMed] [Google Scholar]
- Geldner, N. (2004). The plant endosomal system—Its structure and role in signal transduction and plant development. Planta 219, 547–560. [DOI] [PubMed] [Google Scholar]
- Geldner, N., Anders, N., Wolters, H., Keicher, J., Kornberger, W., Muller, P., Delbarre, A., Ueda, T., Nakano, A., and Jürgens, G. (2003). The Arabidopsis GNOM AFR-GEF mediates endosomal recycling, auxin transport, and auxin-dependent plant growth. Cell 112, 219–230. [DOI] [PubMed] [Google Scholar]
- Geldner, N., Friml, J., Stierhof, Y.-D., Jurgens, G., and Palme, K. (2001). Auxin transport inhibitors block PIN1 cycling and vesicle trafficking. Nature 413, 425–428. [DOI] [PubMed] [Google Scholar]
- Gonzáles-Gaitán, M. (2003). Signal dispersal and transduction through the endocytic pathway. Nat. Rev. Mol. Cell Biol. 4, 213–224. [DOI] [PubMed] [Google Scholar]
- Grebe, M., Xu, J., Möbius, W., Ueda, T., Nakano, A., Geuze, H.J., Rook, M.B., and Scheres, B. (2003). Arabidopsis sterol endocytosis involves actin-mediated trafficking via ARA6-positive early endosomes. Curr. Biol. 13, 1378–1387. [DOI] [PubMed] [Google Scholar]
- Gruenberg, J. (2001). The endocytic pathway: A mosaic of domains. Nat. Rev. Mol. Cell Biol. 2, 721–730. [DOI] [PubMed] [Google Scholar]
- Haj, F.G., Verveer, P.J., Squire, A., Neel, B.G., and Bastiaens, P.I.H. (2002). Imaging sites of receptor dephosphorylation by PTP1B on the surface of the endoplasmic reticulum. Science 295, 1708–1711. [DOI] [PubMed] [Google Scholar]
- He, Z., Wang, Z.-Y., Li, J., Zhu, Q., Lamb, C., Ronald, P., and Chory, J. (2000). Perception of brassinosteroids by the extracellular domain of the receptor kinase BRI1. Science 288, 2360–2363. [DOI] [PubMed] [Google Scholar]
- Hecht, V., Velle-Calzada, J.-P., Hartog, M.V., Schmidt, E.D.L., Boutilier, K., Grossniklaus, U., and de Vries, S.C. (2001). The Arabidopsis SOMATIC EMBRYOGENESIS RECEPTOR KINASE 1 gene is expressed in developing ovules and embryos and enhances embryogenic competence in culture. Plant Physiol. 127, 803–816. [PMC free article] [PubMed] [Google Scholar]
- Hink, M.A., Bisseling, T., and Visser, A.J.W.G. (2002). Imaging protein-protein interactions in living cells. Plant Mol. Biol. 50, 871–883. [DOI] [PubMed] [Google Scholar]
- Horn, M.A., Heinstein, P.F., and Low, P.S. (1989). Receptor-mediated endocytosis in plant cells. Plant Cell 1, 1003–1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Immink, R.G.H., Gadella, T.W.J., Ferrario, S., Busscher, M., and Angenent, G.C. (2002). Analysis of MADS box protein-protein interactions in living plant cells. Proc. Natl. Acad. Sci. USA 99, 2416–2421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin, J.B., Bae, H., Kim, S.J., Jin, Y.H., Goh, C.-H., Kim, D.H., Lee, Y.J., Tse, Y.C., Jiang, L., and Hwang, I. (2003). The Arabidopsis dynamin-like proteins ADL1C and ADL1E play a critical role in mitochondria morphogenesis. Plant Cell 15, 2357–2369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin, J.B., Kim, Y.A., Kim, S.J., Lee, S.H., Kim, D.H., Cheong, G.-W., and Hwang, I. (2001). A new dynamin-like protein, ADL6, is involved in trafficking from the trans-Golgi network to the central vacuole in Arabidopsis. Plant Cell 13, 1511–1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jürgens, G., and Geldner, N. (2002). Protein secretion in plants: From the trans-Golgi network to the outer space. Traffic 3, 605–613. [DOI] [PubMed] [Google Scholar]
- Katzmann, D.J., Odorizzi, G., and Emr, S.D. (2002). Receptor downregulation and multivesicular-body sorting. Nat. Rev. Mol. Cell Biol. 3, 893–905. [DOI] [PubMed] [Google Scholar]
- Kim, D.H., Eu, Y.-J., Yoo, C.M., Kim, Y.-W., Pih, K.T., Jin, J.B., Kim, S.J., Stenmark, H., and Hwang, I. (2001). Trafficking of phosphatidylinositol 3-phosphate from the trans-Golgi network to the lumen of the central vacuole in plant cells. Plant Cell 13, 287–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, J. (2003). Brassinosteroids signal through two receptor-like kinases. Curr. Opin. Plant Biol. 6, 494–499. [DOI] [PubMed] [Google Scholar]
- Li, J., and Chory, J. (1997). A putative leucine-rich repeat receptor kinase involved in brassinosteroid signal transduction. Cell 90, 929–938. [DOI] [PubMed] [Google Scholar]
- Li, J., and Nam, K.H. (2002). Regulation of brassinosteroid singling by a GSK3/SHAGGY-like kinase. Science 295, 1299–1301. [DOI] [PubMed] [Google Scholar]
- Li, J., Wen, J., Lease, K.A., Doke, J.T., Tax, F.E., and Walker, J.C. (2002). BAK1, an Arabidopsis LRR receptor-like protein kinase, interacts with BRI1 and modulates brassinosteroid signaling. Cell 110, 213–222. [DOI] [PubMed] [Google Scholar]
- Mora-García, S., Vert, G., Yin, Y., Caño-Delgado, A., Cheong, H., and Chory, J. (2004). Nuclear protein phosphatases with Kelch-repeat domains modulate the response to brassinosteroids in Arabidopsis. Genes Dev. 18, 448–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nam, K.H., and Li, J. (2002). BRI1/BAK1, a receptor kinase pair mediating brassinosteroid signaling. Cell 110, 203–212. [DOI] [PubMed] [Google Scholar]
- Park, M., Kim, S.J., Vitale, A., and Hwang, I. (2004). Identification of the protein storage vacuole and protein targeting to the vacuole in leaf cells of three plant species. Plant Physiol. 134, 625–639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng, P., and Li, J. (2003). Brassinosteroid signal transduction: A mix of conservation and novelty. J. Plant Growth Regul. 22, 298–312. [DOI] [PubMed] [Google Scholar]
- Pouwels, J., Van Der Krogt, G.N.M., Van Lent, J., Bisseling, T., and Wellink, J. (2002). The cytoskeleton and the secretory pathway are not involved in targeting the cowpea mosaic virus movement protein to the cell periphery. Virology 297, 48–56. [DOI] [PubMed] [Google Scholar]
- Ritzenthaler, C., Nebenführ, A., Movafedhi, A., Stussi-Garaud, C., Behnia, L., Pimpl, P., Staehelin, L.A., and Robinson, D.G. (2002). Reevaluation of the effects of Brefeldin A on plant cells using tobacco Bright Yellow 2 cells expressing Golgi-targeted green fluorescent protein and COPI antisera. Plant Cell 14, 237–261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutherford, S., and Moore, I. (2002). The Arabidopsis Rab GTPase family: Another enigma variation. Curr. Opin. Plant Biol. 5, 518–528. [DOI] [PubMed] [Google Scholar]
- Šamaj, J., Baluška, F., Voigt, B., Schlicht, M., Volkmann, D., and Menzel, D. (2004). Endocytosis, actin cytoskeleton, and signaling. Plant Physiol. 135, 1150–1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satiat-Jeunemaitre, B., and Haves, C. (1992). Redistribution of a Golgi gycoprotein in plant cells treated with Brefeldin A. J. Cell Sci. 103, 1153–1166. [Google Scholar]
- Shah, K., Gadella, T.W.J., van Erp, H., Hecht, V., and de Vries, S.C. (2001). Subcellular localization and dimerization of the A. thaliana somatic embryogenesis receptor kinase 1 protein. J. Mol. Biol. 309, 641–655. [DOI] [PubMed] [Google Scholar]
- Shah, K., Russinova, E., Gadella, T.W.J., Jr., Willemse, J., and de Vries, S.C. (2002). The Arabidopsis kinase-associated protein phosphatase controls internalization of the somatic embryogenesis receptor kinase 1. Genes Dev. 16, 1707–1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheen, J. (2001). Signal transduction in maize and Arabidopsis mesophyll protoplasts. Plant Physiol. 127, 1466–1475. [PMC free article] [PubMed] [Google Scholar]
- Shimada, Y., Goda, H., Nakamura, A., Takatsuto, S., Fujioka, S., and Yoshida, S. (2003). Organ-specific expression of brassinosteroid-biosynthetic genes and distribution of endogenous brassinosteroids in Arabidopsis. Plant Physiol. 131, 287–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sohn, E.J., Kim, E.S., Zhao, M., Kim, S.J., Kim, H., Kim, Y.-W., Lee, Y.J., Hillmer, S., Sohn, U., Jiang, L., and Hwang, I. (2003). Rha1, an Arabidopsis Rab5 homolog, plays a critical role in the vacuolar trafficking of soluble cargo proteins. Plant Cell 15, 1057–1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorkin, A., McClure, M., Huang, F., and Carter, R. (2000). Interaction of EGF receptor and grb2 in living cells visualized by fluorescence resonance energy transfer (FRET) microscopy. Curr. Biol. 10, 1395–1398. [DOI] [PubMed] [Google Scholar]
- Sorkin, A., and Von Zastrow, M. (2002). Signal transduction and endocytosis: Close encounters of many kinds. Nat. Rev. Mol. Cell Biol. 3, 600–614. [DOI] [PubMed] [Google Scholar]
- Ueda, T., and Nakano, A. (2002). Vesicular traffic: An integral part of plant life. Curr. Opin. Plant Biol. 5, 513–517. [DOI] [PubMed] [Google Scholar]
- Ueda, T., Yamaguchi, M., Uchimiya, H., and Nakano, A. (2001). Ara6, a plant-unique novel type Rab GTPase, functions in the endocytic pathway of Arabidopsis thaliana. EMBO J. 20, 4730–4741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verveer, P.J., Wouters, F.S., Reynolds, A.R., and Bastiaens, P.I.H. (2000). Quantitative imaging of lateral ErbB1 receptor signal propagation in the plasma membrane. Science 290, 1567–1570. [DOI] [PubMed] [Google Scholar]
- Vida, T.A., and Emr, S.D. (1995). A new vital stain for visualizing vacuolar membrane dynamics and endocytosis in yeast. J. Cell Biol. 128, 779–792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Z.-Y., Nakano, T., Gendron, J., He, J., Chen, M., Vafeados, D., Yang, Y., Fujioka, S., Yoshida, S., Asami, T., and Chory, J. (2002). Nuclear-localized BZR1 mediates brassinosteroid-induced growth and feedback suppression of brassinosteroid biosynthesis. Dev. Cell 2, 505–513. [DOI] [PubMed] [Google Scholar]
- Wang, Z.-Y., Seto, H., Fujioka, S., Yoshida, S., and Chory, J. (2001). BRI1 is a critical component of a plasma-membrane receptor for plant steroids. Nature 410, 380–383. [DOI] [PubMed] [Google Scholar]
- Wiley, H.S. (2003). Trafficking of the ErbB receptors and its influence on signaling. Exp. Cell Res. 284, 78–88. [DOI] [PubMed] [Google Scholar]
- Yin, Y., Wang, Z.-Y., Mora-Garcia, S., Li, J., Yoshida, S., Asami, T., and Chory, J. (2002. a). BES1 accumulates in the nucleus in response to brassinosteroids to regulate gene expression and promote stem elongation. Cell 109, 181–191. [DOI] [PubMed] [Google Scholar]
- Yin, Y., Wu, D., and Chory, J. (2002. b). Plant receptor kinases: Systemin receptor identified. Proc. Natl. Acad. Sci. USA 99, 9090–9092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, J., Peng, P., Schmitz, R.J., Decker, A.D., Tax, F.E., and Li, J. (2002). Two putative BIN2 substrates are nuclear components of brassinosteroids signaling. Plant Physiol. 130, 1221–1229. [DOI] [PMC free article] [PubMed] [Google Scholar]