Abstract
Type II inositol polyphosphate 5-phosphatases (5PTases) in yeast and animals have been known to regulate the level of phosphoinositides and thereby influence various cellular activities, such as vesicle trafficking and actin organization. In plants, little is known about the phosphatases involved in hydrolysis of phosphoinositides, and roles of type II 5PTases in plant cellular functions have not yet been characterized. In this study, we demonstrate that the FRAGILE FIBER3 (FRA3) gene of Arabidopsis thaliana, which encodes a type II 5PTase, plays an essential role in the secondary wall synthesis in fiber cells and xylem vessels. The fra3 mutations caused a dramatic reduction in secondary wall thickness and a concomitant decrease in stem strength. These phenotypes were associated with an alteration in actin organization in fiber cells. Consistent with the defective fiber and vessel phenotypes, the FRA3 gene was found to be highly expressed in fiber cells and vascular tissues in stems. The FRA3 protein is composed of two domains, an N-terminal localized WD-repeat domain and a C-terminal localized 5PTase catalytic domain. In vitro activity assay demonstrated that recombinant FRA3 exhibited phosphatase activity toward PtdIns(4,5)P2, PtdIns(3,4,5)P3, and Ins(1,4,5)P3, with the highest substrate affinity toward PtdIns(4,5)P2. The fra3 missense mutation, which caused an amino acid substitution in the conserved motif II of the 5PTase catalytic domain, completely abolished the FRA3 phosphatase activity. Moreover, the endogenous levels of PtdIns(4,5)2 and Ins(1,4,5)P3 were found to be elevated in fra3 stems. Together, our findings suggest that the FRA3 type II 5PTase is involved in phosphoinositide metabolism and influences secondary wall synthesis and actin organization.
INTRODUCTION
Phosphoinositides are phospholipids present ubiquitously in all eukaryotic cell membranes. They are generated by sequential phosphorylation of phosphatidylinositol by specific kinases. Studies in yeast and animals have documented that phosphoinositides are important signaling molecules that regulate a variety of cellular functions, including cell differentiation and proliferation, cell death, vesicle trafficking, and actin cytoskeleton organization (Takenawa and Itoh, 2001). One of the best-studied phosphoinositides is phosphatidylinositol 4,5-bisphosphate [PtdIns(4,5)P2]. PtdIns(4,5)P2 not only serves as a precursor for the production of signaling molecules diacylglycerol and inositol 1,4,5-triphosphate [Ins(1,4,5)P3], but also by itself regulates vesicle trafficking and actin organization by binding to and modulating the activity of actin-associated proteins, including profilin, α-actinin, vinculin, and gelsolin (Takenawa and Itoh, 2001). Termination of phosphoinositide signaling is mediated by inositol polyphosphate phosphatases (PTases) that hydrolyze the 3-, 4-, or 5-position phosphate from the inositol ring of phosphoinositides. In plants, all forms of phosphoinositides except PtdIns(3,4,5)P3 have been detected (Mueller-Roeber and Pical, 2002), and PtdIns(4,5)P2 has been shown to be important for the polar growth of pollen tubes (Kost et al., 1999). However, little is known about the phosphatases that remove phosphate from phosphoinositides in plants (Mueller-Roeber and Pical, 2002).
PTases are traditionally classified into four groups based on the position of phosphate they hydrolyze, 1-, 3-, 4-, or 5-phosphatases. The best characterized group is inositol polyphosphate 5-phosphatases (5PTases), which is further divided into four types according to their distinct substrate specificities for inositol phosphates and/or phosphoinositides (Majerus et al., 1999). Type I 5PTases hydrolyze only water-soluble substrates Ins(1,4,5)P3 and Ins (1,3,4,5)P4. Type II 5PTases hydrolyze not only water-soluble Ins(1,4,5)P3 and Ins (1,3,4,5)P4 but also phosphoinositides PtdIns(4,5)P2 and PtdIns(3,4,5)P3.
In plants, several genes encoding inositol polyphosphate 1- or 5-phosphatases and a group of putative SAC domain phosphoinositide phosphatases have been identified (Berdy et al., 2001; Sanchez and Chua, 2001; Xiong et al., 2001; Despres et al., 2003; Zhong and Ye, 2003). It has been shown that mutation of a bifunctional enzyme with 3′(2′),5′-bisphosphate nucleotidase and inositol polyphosphate 1-phosphatase activities in the Arabidopsis thaliana fiery1 mutant causes altered sensitivities to abscisic acid (ABA) induction (Quintero et al., 1996; Xiong et al., 2001). Eleven putative type I 5PTase genes have been identified in the Arabidopsis genome (Berdy et al., 2001), and two of them, At5PTase1 and AtIP5P2, exhibit typical type I 5PTase substrate specificity [i.e., they hydrolyze only water-soluble inositol phosphates Ins(1,4,5)P3 and Ins(1,3,4,5)P4] (Berdy et al., 2001; Sanchez and Chua, 2001). Recently, it was shown that another 5PTase, AtPTase11, displays phosphatase activity toward phospholipids (Ercetin and Gillaspy, 2004). Overexpression of At5PTase1 or AtIP5P2 in transgenic Arabidopsis plants has been demonstrated to reduce the Ins(1,4,5)P3 level and alter the ABA-induced plant responses (Sanchez and Chua, 2001; Burnette et al., 2003). Mutation of the COTYLEDON VASCULAR PATTERN2 (CVP2) gene, which encodes a putative type I 5PTase, has been shown to alter leaf venation pattern, Ins(1,4,5)P3 level, and ABA sensitivity (Carland and Nelson, 2004). In addition, overexpression of the human type I 5PTase in tobacco (Nicotiana tabacum) cells causes a significant reduction in the Ins(1,4,5)P3 level but no apparent effects on cell growth and morphology (Perera et al., 2002).
Although several type I 5PTases have been studied in plants, the type II 5PTases have not been characterized except for a report of the presence of four putative type II 5PTase genes in the Arabidopsis genome (Berdy et al., 2001). Mutations of several human and yeast type II 5PTases have been shown to cause dramatic effects on development or stress response. Mutations of human OCRL1 have been linked to oculocerebrorenal syndrome of Lowe, a rare X-linked developmental disorder. In Lowe syndrome, it was found that the 5PTase activity of OCRL1 is abolished, and concomitantly there is an increase in the level of PtdIns(4,5)P2, which is thought to be the cause of the syndrome (Zhang et al., 1998). Mutation of the type II 5PTase domain in the yeast Inp51p has been shown to cause an elevation of PtdIns(4,5)P2 and confer cold tolerance (Stolz et al., 1998). However, the biochemical activities and cellular functions of type II 5PTases in plants remain unknown.
In this study, we demonstrate that the FRAGILE FIBER3 (FRA3) gene, which is required for the secondary wall formation and mechanical strength of fiber cells, encodes a type II 5PTase. We show that the fra3 mutation dramatically reduces secondary wall thickening, and this alteration is associated with abnormal actin organization in fiber cells. The FRA3 protein consists of an N-terminal localized WD-repeat domain and a C-terminal localized 5PTase catalytic domain. We demonstrate that FRA3 exhibits type II 5PTase activity in vitro and shows the highest substrate affinity toward PtdIns(4,5)P2. Moreover, we have found that the endogenous levels of PtdIns(4,5)2 and Ins(1,4,5)P3 are elevated in fra3 stems. Together, our findings provide genetic evidence suggesting that the FRA3 type II 5PTase plays an essential role in cell wall synthesis and actin organization.
RESULTS
The fra3 Mutations Cause Defects in Secondary Wall Thickening in Fiber Cells and Xylem Vessels
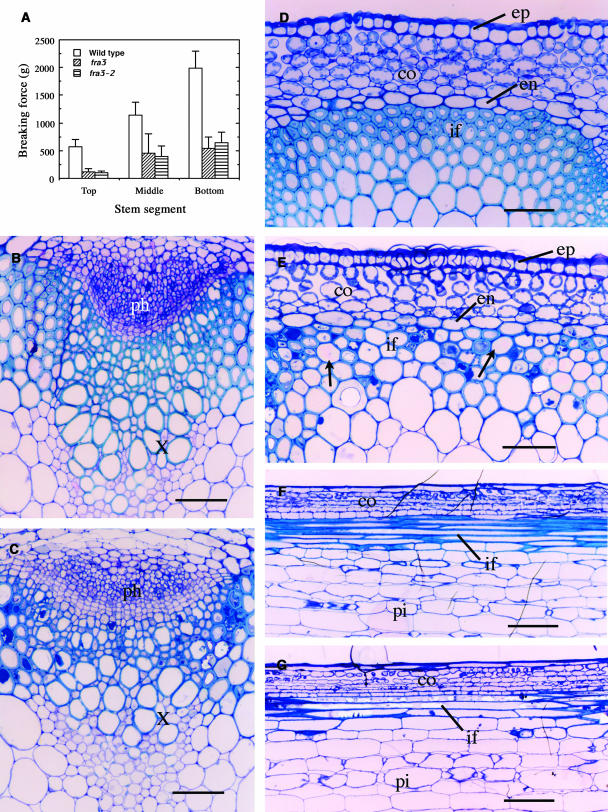
Arabidopsis inflorescence stems develop three to four layers of fiber cells between vascular bundles, and the mechanical strength of mature stems is largely attributable to these interfascicular fibers (Zhong et al., 1997, 2001). To investigate the molecular mechanisms underlying fiber cell formation, we isolated two allelic fra3 mutants defective in stem strength. Quantitative analysis showed that the breaking strength of fra3 and fra3-2 stems was reduced by approximately threefold compared with that of the wild type (Figure 1A), indicating that the fra3 mutations cause a dramatic reduction in the mechanical strength of stems.
Figure 1.
Alterations of Stem Strength and Fiber Secondary Wall Thickness by the fra3 Mutation.
Inflorescence stems of 8-week-old plants were used for breaking force measurement, and their bottom internodes were examined for fiber cell morphology. co, cortex; en, endodermis; ep, epidermis; if, interfascicular fiber; ph, phloem; pi, pith; x, xylem. Bars = 60 μm in (B) to (E) and 190 μm in (F) and (G).
(A) Breaking force measurement showing that the force to break stems apart was about three times lower in fra3 and fra3-2 than in the wild type.
(B) and (C) Cross sections of stems showing vascular bundles in the wild type (B) and fra3 (C).
(D) Cross section of an interfascicular region of the wild type showing fiber cells with thick walls.
(E) Cross section of an interfascicular region of fra3 showing fiber cells with thin walls. Note that some fiber cells (arrows) had extremely thinner walls.
(F) and (G) Longitudinal sections of interfascicular regions showing long fiber cells in both the wild type (F) and fra3 (G).
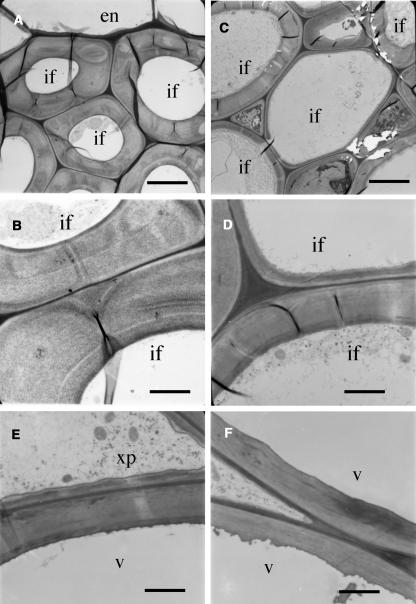
Because interfascicular fibers contribute largely to the mechanical strength of stems, we investigated the fiber morphology in the fra3 mutants. Both fra3 and fra3-2 mutants exhibited the same cellular phenotypes; therefore, only the data from the fra3 mutant are presented in this study. Cross-section examinations revealed that although the interfascicular fiber cells in the wild type had relatively uniform and thick secondary walls (Figure 1D), those in the fra3 mutant showed an apparent reduction in wall thickness (Figure 1E). It was noted that the wall thickness appeared not to be uniform among fra3 fiber cells, and some of the fiber cells with extremely thin walls looked larger in diameter than wild-type ones (Figure 1E). Transmission electron microscopy further confirmed that the fra3 mutation caused a dramatic reduction in the wall thickness among fiber cells (Figures 2A to 2D, Table 1). It was also observed that the wall thickness of xylem vessels was slightly reduced in the fra3 mutant compared with the wild type (Figures 1B, 1C, 2E, and 2F, Table 1). Quantitative analysis of cell wall composition demonstrated that the fra3 mutation resulted in a 37% reduction in glucose but no major effects on other sugar content (Table 2). This suggests that the fra3 mutation mainly affects cellulose content, which is consistent with the dramatic decrease in secondary wall thickness in fiber cells. Longitudinal sections showed no apparent differences in the length of fibers and pith cells between the wild type and the fra3 mutant (Figures 1F and 1G). No anatomical changes were observed in other cell types in the fra3 mutant. These results indicate that the stem strength defect caused by the fra3 mutation is a result of reduced fiber cell wall thickness.
Figure 2.
Nonuniform Reduction of Secondary Wall Thickness in Fiber Cells of the fra3 Mutant.
Fibers and vessels from the bottom internodes of 8-week-old plants were sectioned and examined for their wall thickness under a transmission electron microscope. en, endodermis; if, interfascicular fiber; v, vessel; xp, xylem parenchyma. Bars = 5.3 μm in (A) and (C) and 2.4 μm in (B), (D), (E), and (F).
(A) and (B) Wild-type interfascicular fibers showing uniformly thick walls. High magnification of the walls is shown in (B).
(C) and (D) fra3 interfascicular fibers showing nonuniform reduction of secondary wall thickness. High magnification of the walls is shown in (D).
(E) and (F) Vessel walls were slightly thinner in fra3 (F) than in the wild type (E).
Table 1.
Wall Thickness of Fibers and Vessels in the Stems of Wild Type and fra3 Mutant (μm)
| Sample | Fiber Cells | Vessels |
|---|---|---|
| Wild type | 3.54 ± 0.25 | 2.23 ± 0.18 |
| fra3 | 2.11 ± 0.45 | 1.64 ± 0.21 |
The wall thickness was determined from transmission electron microscope micrographs of fibers and vessels. Data are means ± se from 25 cells.
Table 2.
Cell Wall Sugar Composition of the Stems of the Wild Type and fra3 Mutant (mg/g Dry Cell Wall)
| Sample | Glucose | Xylose | Mannose | Galactose | Arabinose | Rhamnose |
|---|---|---|---|---|---|---|
| Wild type | 277 ± 0.7 | 144 ± 4.2 | 17.4 ± 0 | 15.7 ± 0.4 | 13.5 ± 0.5 | 4.1 ± 0.6 |
| fra3 | 174 ± 16 | 136 ± 4.9 | 17.8 ± 0.7 | 13.2 ± 0 | 12.1 ± 0.6 | 3.9 ± 0.2 |
Cell wall materials were prepared from mature stems of 10-week-old plants. Data are means ± se of two independent assays.
Except for the defective secondary wall thickening and stem strength phenotypes, the fra3 mutant did not exhibit any abnormality in plant growth and development. Both dark- and light-grown fra3 seedlings were indistinguishable from the wild type. Mature fra3 plants had similar height as the wild type and underwent reproductive growth as normally as the wild type (data not shown).
The fra3 Mutation Alters Actin Organization in Fiber Cells
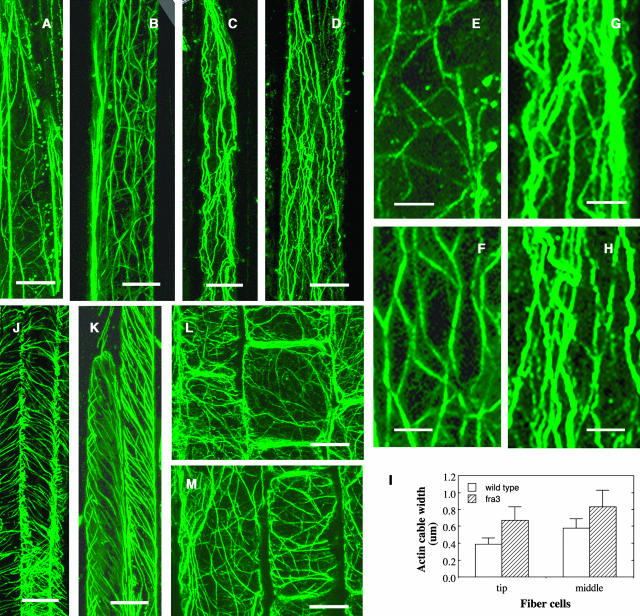
Because cell wall synthesis is influenced by cytoskeletons, we next investigated whether the fra3 mutation caused any defects in cytoskeleton organization (Figure 3). Immunolabeling experiments showed that the organization of cortical microtubules in developing fiber cells did not exhibit any noticeable differences between the wild type and fra3 (Figures 3J and 3K), indicating that the fra3 mutation did not affect cortical microtubule organization. Examination of the organization of filamentous actin (F-actin) showed that although the F-actin in developing wild-type fiber cells formed a fine network (Figures 3A and 3B), the F-actin in developing fra3 fiber cells was apparently disrupted (Figures 3C and 3D). The F-actin cables in fra3 fiber cells appeared to lose their fine network pattern and became much thicker compared with the wild type (Figures 3E to 3H). Some of the thick F-actin cables appeared to be caused by bundling, which is a reminiscence of F-actin bundling caused by the actin drug latrunculin B (Mathur et al., 2003). Quantitative measurement of the width of actin cables based on their fluorescent signals showed that compared with the wild type, the width of F-actin cables in the tip and middle regions of fra3 fiber cells increased by 72 and 43%, respectively (Figure 3I). Examination of the cytoskeleton in elongating pith cells of the fra3 mutant showed no apparent alterations in the organization of F-actin (Figures 3L and 3M) and cortical microtubules (data not shown) compared with the wild type. These results demonstrated that the fra3 mutation caused an alteration in F-actin organization in fiber cells, which is consistent with the defective fiber phenotype observed in the fra3 mutant.
Figure 3.
Alteration of F-Actin Organization in Developing Fiber Cells of the fra3 Mutant.
Longitudinal sections of stems were immunolabeled for F-actin and microtubules with monoclonal antibodies against actin or α-tubulin and fluorescein isothiocyanate–conjugated secondary antibodies. Fluorescence-labeled F-actin and cortical microtubules were visualized under a confocal microscope. Bars = 18 μm in (A) to (D), 6 μm in (E) to (H), 11 μm in (J) and (K), and 12 μm in (L) and (M).
(A) and (B) The tip (A) and middle (B) regions of a wild-type fiber cell showing fine F-actin network.
(C) and (D) The tip (C) and middle (D) regions of a fra3 fiber cell showing thick F-actin cables.
(E) and (F) High magnification of areas from (A) and (B), respectively, showing fine F-actin cables.
(G) and (H) High magnification of areas from (C) and (D), respectively, showing thick F-actin cables.
(I) Quantitative measurement of the width of fluorescence-labeled F-actin cables in the tip and middle regions of fiber cells.
(J) and (K) Fiber cells showing cortical microtubules aligned in parallel in both the wild type (J) and fra3 (K).
(L) and (M) Pith cells showing the fine F-actin network in both the wild type (L) and fra3 (M).
Map-Based Cloning of the FRA3 Gene
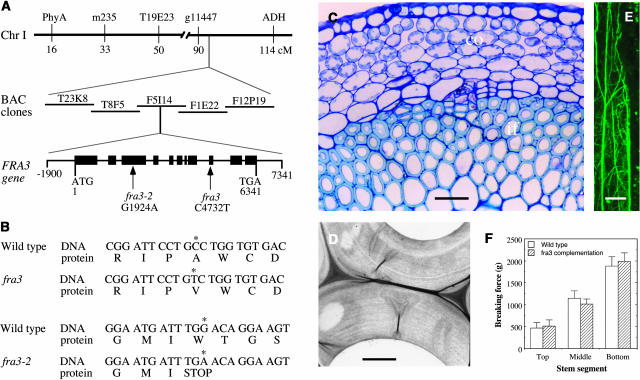
To investigate the molecular nature that was responsible for the defective fiber phenotype, we pursued the map-based cloning of the FRA3 gene. Using codominant amplified polymorphic sequences (CAPS) markers, we mapped the fra3 locus near the g11447 marker on chromosome 1. Fine mapping with additional CAPS markers revealed that the fra3 locus was located within a 27-kb region in the BAC clone F5I14 (Figure 4A). Based on the gene annotations of F5I14 from the Arabidopsis genome database, this 27-kb region contains six genes. Sequencing of these genes from the fra3 and fra3-2 mutants revealed point mutations in one of these genes, F5I14.11 (At1g65580) (Figure 4A). The fra3 mutant carries a C-to-T mutation, and the fra3-2 mutant contains a G-to-A mutation. The fra3 mutation causes loss of a BstNI site in the gene, which was confirmed by digestion of PCR-amplified DNA fragment with BstNI (data not shown). Transformation of the wild-type F5I14.11 gene into the fra3 and fra3-2 mutants completely restored the fiber wall thickness, F-actin organization, and stem strength to the wild-type level (Figures 4C to 4F). Together, these results unequivocally demonstrated that the point mutations in the F5I14.11 gene are responsible for the fra3 mutant phenotypes; therefore, the F5I14.11 gene represents the FRA3 gene.
Figure 4.
Map-Based Cloning of the FRA3 Gene and Complementation of the fra3 Mutant.
(A) Map-based cloning of the FRA3 gene. The fra3 locus was mapped to a 27-kb region located in the BAC clone F5I14 on chromosome 1. The FRA3 gene consists of 11 exons and 10 introns. The fra3 mutation causes a C-to-T transition in the 9th exon, and the fra3-2 mutation results in a G-to-A transition in the 3rd exon. Black boxes represent exons and lines between exons denote introns in the FRA3 gene diagram.
(B) Nucleotide and amino acid sequences around the fra3 mutation sites. The fra3 mutation changes a wild-type codon encoding Ala into a codon encoding Val. The fra3-2 mutation changes a wild-type codon encoding Trp into a stop codon.
(C) Cross section of an interfascicular region of fra3 complemented with the wild-type FRA3 gene. Note the fiber cells with thick walls. The bottom internode of 8-week-old plants was used for sectioning. co, ortex; if, interfascicular fiber. Bar = 75 μm.
(D) Thick walls of interfascicular fiber cells of fra3 complemented with the wild-type FRA3 gene. Bar = 2.2 μm.
(E) Fine F-actin network in an interfascicular fiber cell of fra3 complemented with the wild-type FRA3 gene. F-actin was immunolabeled with a monoclonal antibody against actin and fluorescein isothiocyanate–conjugated secondary antibodies. Bar = 18 μm.
(F) Breaking force measurement showing that the force to break stems apart was similar between the wild type and fra3 complemented with the wild-type FRA3 gene.
Sequence Analysis of the FRA3 Gene and Its Encoded Protein
To perform molecular characterization of the FRA3 gene, we isolated the full-length FRA3 cDNA from an Arabidopsis stem cDNA library. Comparison of the sequences of FRA3 gene and its cDNA revealed that the FRA3 gene consists of 11 exons and 10 introns with 6341 bp in length from the start codon to the stop codon (Figure 4A). The longest open reading frame in the FRA3 cDNA is 3306 bp long, and it encodes a protein of 1101 amino acids with a predicted molecular mass of 121.6 kD and a predicted pI of 5.9. The fra3 mutation occurred in the ninth exon and fra3-2 in the third exon of the FRA3 gene (Figure 4A). Comparison of the wild-type and fra3 cDNAs and their deduced amino acid sequences showed that the fra3 missense mutation changed an Ala codon GCC to a Val codon GTC (Figure 4B). The fra3-2 mutation changed a Trp codon TGG to a stop codon TGA (Figure 4B), which results in a truncated protein with deletion of 681 amino acid residues.
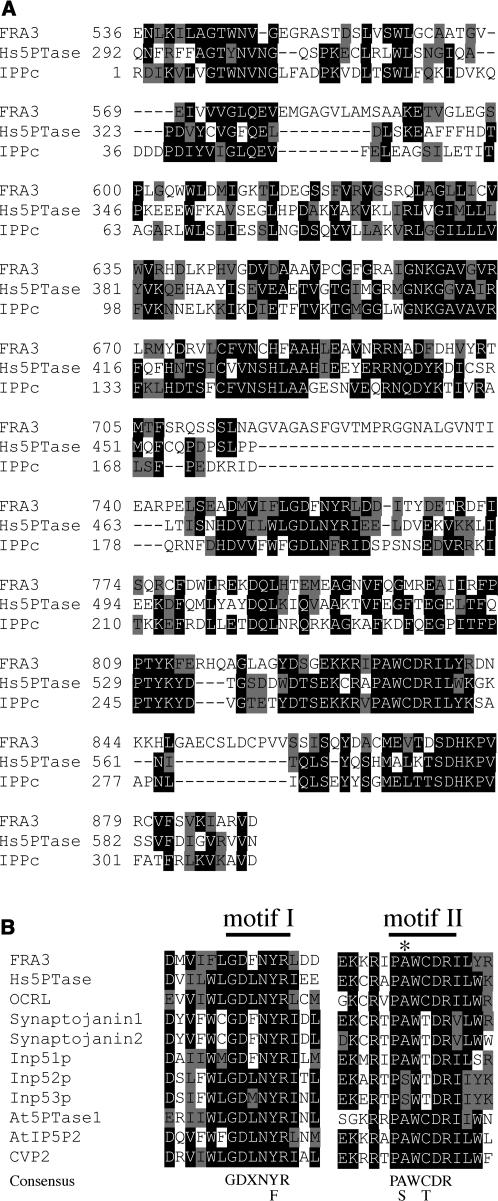
A search of the GenBank database showed that the deduced FRA3 protein contains a domain that shares high sequence similarity to a group of type II 5PTases from yeast and animals. All known 5PTases contain a conserved catalytic domain that is ∼350 amino acid residues in length. Within this domain, two highly conserved motifs have been shown to be important for the 5-phosphatase activity (Majerus et al., 1999). The putative 5-phosphatase catalytic domain of FRA3 protein shares 28% identity and 48% similarity with the catalytic domain of human type II 5PTase (Figure 5A). In particular, the two conserved 5PTase catalytic motifs (GDXNY/FR and PA/SWC/TDR) are completely conserved in the FRA3 5PTase domain (Figure 5B). It was noted that the fra3 missense mutation occurred at the Ala residue in motif II (Figure 5B), suggesting that the Ala residue is important for FRA3 function. The fra3-2 non-sense mutation caused deletion of the whole 5PTase domain (Figure 6C), indicating that fra3-2 is devoid of the 5PTase function.
Figure 5.
Sequence Analysis of the 5PTase Domain of FRA3.
(A) Alignment of the 5-phosphatase domain of FRA3 with that of human type II 5PTase (Hs5PTase) and the conserved inositol polyphosphate phosphatase catalytic domain (IPPc, smart00128) from the GenBank conserved domain database. The numbers shown at the left of each sequence are the positions of amino acid residues in the corresponding proteins. Gaps (marked with dashes) were introduced to maximize the sequence alignment. Identical and similar amino acid residues are shaded with black and gray, respectively.
(B) Sequence alignment of the two conserved 5-phosphatase catalytic motifs of FRA3 and other 5-phosphatases. The consensus sequences for the two motifs are shown below the aligned sequences. The fra3 mutation changes an Ala (marked with an asterisk) into Val in motif II. Shown in the alignment are motifs from type II 5PTases (Hs5PTase, OCRL, synaptojanin1, synaptojanin2, Inp51p, Inp52p, and Inp53p) and Arabidopsis 5PTases (At5PTase1, AtIP5P2, and CVP2).
Figure 6.
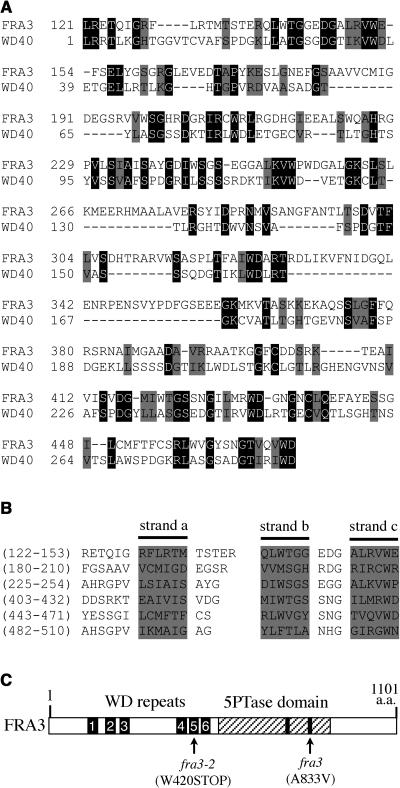
Sequence Analysis of the WD Repeats of FRA3.
(A) Sequence alignment of the N-terminal region of FRA3 with the conserved WD-repeat domain (WD40 domain, cd00200) from the GenBank conserved domain database. Gaps (marked with dashes) were introduced to maximize the sequence alignment. Identical and similar amino acid residues are shaded with black and gray, respectively.
(B) Alignment of the six WD repeats in FRA3. The WD repeats were predicted using a program that identifies protein repeats. The shaded sequences shown in each repeat represent three of the four putative β strands (a, b, and c) as predicted with the secondary structure prediction program.
(C) Diagram of the FRA3 protein showing the organization of the WD repeats and the 5-phosphatase domain. The numbered bars represent the six WD repeats. The hatched region represents the 5-phosphatase domain in which the two conserved motifs are marked with black bars. The fra3 and fra3-2 mutation sites are marked.
In addition to the putative 5PTase domain located at the C-terminal region, FRA3 contains an N-terminal region with ∼500 amino acids in length. A search of the GenBank conserved domain database revealed the presence of a WD-repeat domain in the FRA3 N-terminal region (Figure 6A). The putative FRA3 WD-repeat domain shows 23% identity and 35% similarity to the conserved WD-repeat domain. The WD repeat is a structurally conserved motif that comprises a 44– to 60–amino acid sequence with a GH dipeptide 11 to 24 residues from its N terminus and a WD dipeptide at the C terminus. It has been shown that neither the GH dipeptide nor the WD dipeptide is completely conserved among WD repeats. Instead, the WD repeat is a conserved structural motif that forms a four-stranded antiparallel β sheet (Smith et al., 1999). WD-repeat proteins are defined as having at least four or more WD repeats that are required to form a β propeller structure (Smith et al., 1999). Further analysis of the FRA3 amino acid sequence using a program that identifies protein repeats revealed that FRA3 contains six WD repeats (Figure 6B). Each WD repeat in FRA3 was predicted to form four β strands by the secondary structure prediction program. Because FRA3 contains more than four WD repeats that are required for formation of a β propeller structure, FRA3 is by definition also a WD-repeat protein. Together, these results show that FRA3 is composed of two conserved domains, a WD-repeat domain and a 5PTase domain (Figure 6C).
Expression Pattern of the FRA3 Gene
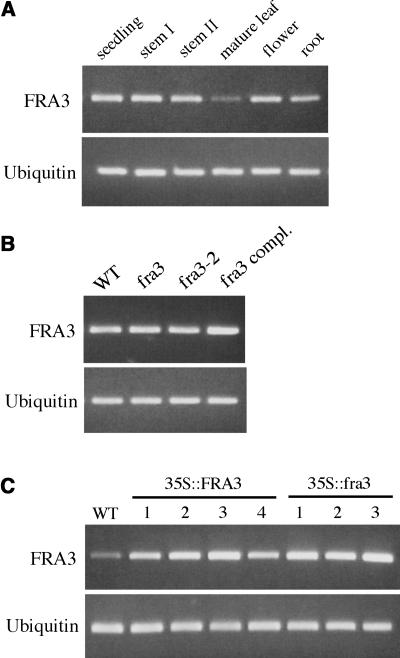
To investigate the expression pattern of the FRA3 gene, we first used the semiquantitative RT-PCR method to detect its expression in various organs. The FRA3 gene was found to be expressed at similar levels in seedlings, stems, roots, and flowers, but a much lower expression level was detected in mature leaves (Figure 7A). This indicates that the FRA3 gene is expressed ubiquitously in various Arabidopsis organs. The fra3 and fra3-2 point mutations did not affect the accumulation of FRA3 mRNA (Figure 7B).
Figure 7.
Gene Expression Analysis of FRA3 by Semiquantitative RT-PCR.
(A) Expression of the FRA3 gene in various Arabidopsis organs. The expression level of a ubiquitin gene was used as an internal control. The seedlings used were 2 weeks old. Mature leaves were from 6-week-old plants. Flowers and mature roots were from 8-week-old plants. Stems I and II were from 4- and 8-week-old plants, respectively.
(B) Expression of the FRA3 gene in seedlings of the wild type, fra3 mutants, and a representive fra3 complementation line (fra3 compl.).
(C) Overexpression of wild-type FRA3 cDNA (35S::FRA3) or mutant fra3 cDNA (35S::fra3) in wild-type seedlings under the control of the 35S promoter of Cauliflower mosaic virus. Shown are four representative lines of 35S::FRA3 and three representative lines of 35S::fra3.
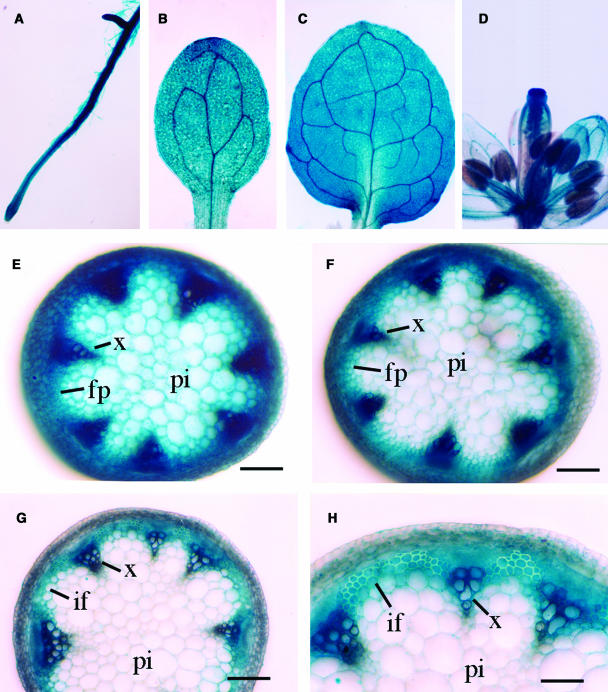
We next used the β-glucuronidase (GUS) reporter gene to further study the FRA3 gene expression pattern. A 2-kb 5′ upstream sequence and the entire exon and intron regions of the FRA3 gene were ligated in frame with the GUS reporter gene in the binary vector pBI101. The construct was introduced into both wild-type and fra3 plants. Because the construct completely rescued the fra3 mutant phenotypes (data not shown), it indicates that the region of the FRA3 gene used in the GUS reporter construct contains all elements responsible for the expression of the endogenous FRA3 gene. Examination of GUS activity in the transgenic plants showed GUS expression in young roots (Figure 8A), cotyledons (Figure 8B), leaves (Figure 8C), and flowers (Figure 8D). In rapidly elongating inflorescence stems, the GUS signals were present in nearly all tissues (Figure 8E). However, the signals gradually became more concentrated in vascular bundles and interfascicular fiber cells when the internodes were near cessation of elongation (Figure 8F). In nonelongating internodes in which fiber cells undergo massive secondary wall deposition (Ye et al., 2002), the GUS staining was predominantly seen in interfascicular fibers and vascular bundles (Figures 8G and 8H). The predominant expression of the FRA3 gene in developing fibers and vascular cells is consistent with the defective fiber and vessel phenotypes seen in the fra3 mutant.
Figure 8.
Gene Expression Analysis of FRA3 Using the GUS Reporter Gene.
The FRA3 gene, including a 2-kb 5′ upstream sequence and the entire exon and intron region, was ligated in frame with the GUS reporter gene and transformed into wild-type Arabidopsis. Various organs from the transgenic plants were stained for GUS activity. fp, fiber precursor; if, interfascicular fiber; pi, pith; x, xylem. Bars = 165 μm in (E) to (G) and 330 μm in (H).
(A) and (B) Primary root (A) and cotyledon (B) from 3-d-old seedlings showing the GUS staining in all tissues.
(C) Leaf from 10-d-old seedlings showing the GUS staining in mesophyll cells and veins.
(D) Flower showing the GUS staining in various floral parts.
(E) Section from young elongating internodes showing GUS staining in all tissues.
(F) Section from internodes at the stage near the cessation of elongation showing intensive GUS staining in vascular bundles and interfascicular regions.
(G) Section from nonelongating internodes showing the GUS staining predominantly in vascular bundles and interfascicular fibers.
(H) High magnification of (G) showing fiber cells with thick secondary walls and strong GUS staining.
FRA3 Possesses Phosphatase Activity toward PtdIns(4,5)P2, PtdIns(3,4,5)P3, and Ins(1,4,5)P3
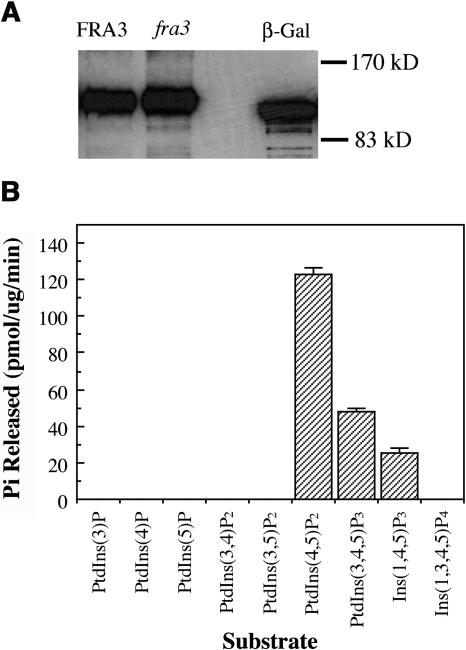
The finding that FRA3 contains a putative type II 5PTase domain prompted us to investigate its phosphatase activity. We expressed His-tagged full-length FRA3 protein in yeast and purified the recombinant FRA3 protein on a nickel-nitrilotriacetic acid agarose (Ni-NTA) column (Figure 9A). The purified protein was assayed for its phosphatase activity using various phospholipids and water-soluble inositol phosphates as substrates. It was found that FRA3 hydrolyzed phosphate from both PtdIns(4,5)P2 and PtdIns(3,4,5)P3 but not from other phospholipids, including PtdIns(3)P, PtdIns(4)P, PtdIns(5)P, PtdIns(3,4)P2, and PtdIns(3,5)P2 (Figure 9B). FRA3 was also capable of hydrolyzing phosphate from Ins(1,4,5)P3 but not Ins(1,3,4,5)P4. The fra3 mutant protein, which contains a missense mutation in the catalytic motif II, did not show any detectable phosphatase activity toward any substrates, indicating that the mutation completely abolishes its enzymatic activity. These results demonstrate that FRA3 is indeed a 5PTase. Because it hydrolyzes both phospholipids and water-soluble inositol phosphate, FRA3 belongs to the type II 5PTase group.
Figure 9.
FRA3 Exhibits Phosphatase Activity toward PtdIns(4,5)P2, PtdIns(3,4,5)P3, and Ins(1,4,5)P3.
His-tagged full-length FRA3 protein was expressed in yeast, purified on a Ni-NTA column, and used for assay of its phosphatase activity.
(A) Detection of recombinant FRA3 protein expressed in yeast. Shown are the His-tagged wild-type FRA3 protein (FRA3), fra3 mutant protein (fra3), and β-galactosidase (β-Gal). The recombinant proteins were detected with a monoclonal antibody against the Xpress epitope and horseradish peroxidase–labeled secondary antibodies.
(B) Assay of the FRA3 phosphatase activity toward various phospholipids and two water-soluble inositol polyphosphates. FRA3 exhibits an apparent phosphatase activity toward PtdIns(4,5)P2, PtdIns(3,4,5)P3, and Ins(1,4,5)P3. No phosphatase activity was detected for the fra3 mutant protein. The recombinant β-galactosidase was used as a control in the assay.
FRA3 Exhibits Differential Magnesium Sensitivity toward Different Substrates
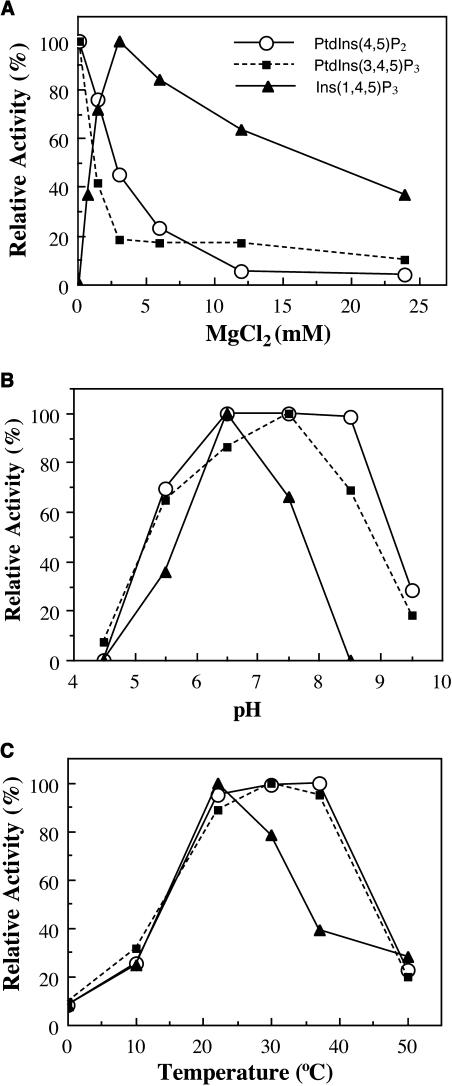
Because the enzymatic activity of 5PTases in yeast and animals requires divalent cations, such as Mg2+ (Majerus et al., 1999), we investigated whether FRA3 exhibited similar biochemical properties as its yeast and animal counterparts. It was found that in the presence of the divalent cation chelator EDTA, FRA3 did not exhibit any phosphatase activity (data not shown). Addition of Mg2+ dramatically stimulated the FRA3 phosphatase activity, indicating that FRA3 requires Mg2+ for its enzymatic activity (Figure 10A). It was interesting to find that the FRA3 phosphatase activity toward different substrates exhibited differential Mg2+ sensitivity (Figure 10A). The FRA3 phosphatase activity toward phospholipids exhibits its maximum in the presence of 0.1 mM Mg2+. A concentration of 1.5 mM Mg2+ or higher dramatically lowered its activity toward phospholipids. By contrast, FRA3 did not show any detectable activity toward Ins(1,4,5)P3 in the presence of 0.1 mM Mg2+. However, its activity toward Ins(1,4,5)P3 gradually increased with increasing concentrations of Mg2+ up to 3 mM, at which concentration the activity reached its maximum (Figure 10A). These results demonstrate that the FRA3 phosphatase activity toward phospholipids and water-soluble Ins(1,4,5)P3 can be differentially influenced by Mg2+ concentration.
Figure 10.
Effects of Magnesium, pH, and Temperature on the FRA3 Phosphatase Activity.
The FRA3 phosphatase activity was assayed in the presence of 50 μM substrates under various conditions as shown, and the data are presented as percentages of the highest value. The assays were run in duplicates and repeated twice.
(A) Effects of Mg2+ concentration on the FRA3 phosphatase activity. Various concentrations of Mg2+ were included in the assay buffer. The lowest Mg2+ used for the activity assay was 0.1 mM as shown in the figure.
(B) Effects of pH on the FRA3 phosphatase activity toward its substrates.
(C) Effects of temperature on the FRA3 phosphatase activity toward its substrates.
The finding that the FRA3 activity can be regulated by Mg2+ concentration indicates that the in vivo physiological conditions could affect its phosphatase activity. This finding prompted us to investigate the effects of different pH and temperature conditions on the FRA3 phosphatase activity. It was found that although FRA3 was active toward PtdIns(4,5)P2 and PtdIns(3,4,5)P3 between pH 5.5 and 8.5, its activity toward Ins(1,4,5)P3 was more sensitive to pH (Figure 10B). The optimal temperature for the FRA3 activity toward PtdIns(4,5)P2 and PtdIns(3,4,5)P3 was between 22 and 37°C, whereas that toward Ins(1,4,5)P3 was around 22°C (Figure 10C). These results indicate that the FRA3 phosphatase activity toward Ins(1,4,5)P3 is more sensitive to pH and temperature compared with the phospholipid substrates.
FRA3 Exhibits the Highest Substrate Affinity toward PtdIns(4,5)P2
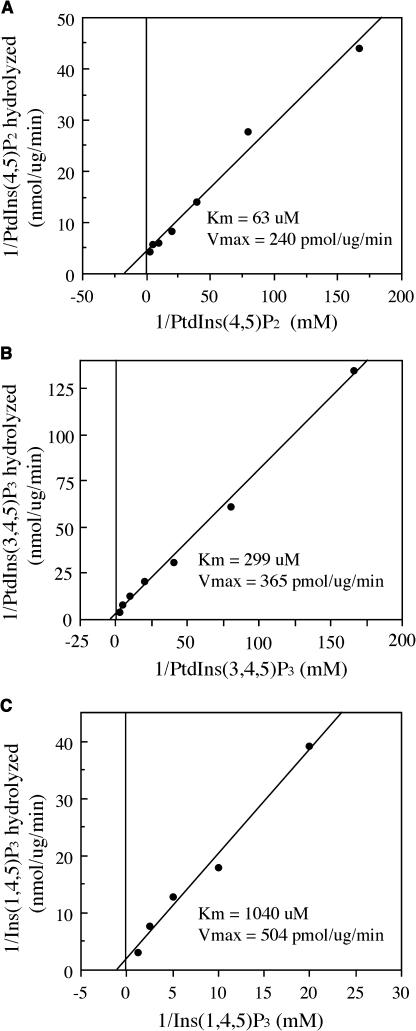
To test the substrate affinity of FRA3, we examined the kinetic properties of FRA3. The FRA3 phosphatase activity was assayed using various concentrations of PtdIns(4,5)P2, PtdIns(3,4,5)P3, and Ins(1,4,5)P3. The Km and Vmax values were calculated by the Lineweaver-Burk analysis. It was found that the Km values of FRA3 phosphatase activity toward PtdIns(4,5)P2, PtdIns(3,4,5)P3, and Ins(1,4,5)P3 were 63, 299, and 1040 μM, respectively (Figures 11A to 11C). The Km values of FRA3 were comparable with those of human type II 5PTase [180 μM for PtdIns(4,5)P2 and 1150 μM for Ins(1,4,5)P3] (Ijuin et al., 2000). The kinetic analysis demonstrates that FRA3 has a much higher substrate affinity toward PtdIns(4,5)P2 than PtdIns(3,4,5)P3 and Ins(1,4,5)P3, suggesting that FRA3 might preferentially hydrolyze PtdIns(4,5)P2 in vivo.
Figure 11.
Substrate Affinity of the FRA3 Phosphatase.
The kinetic activity of FRA3 phosphatase was assayed in the presence of various concentrations of substrates. The results were analyzed by Lineweaver-Burk plots to determine the Km and Vmax values.
(A) The kinetic property of FRA3 toward PtdIns(4,5)P2. PtdIns(4,5)P2 (6 to 400 μM) was incubated with 0.25 μg of recombinant FRA3 protein.
(B) The kinetic property of FRA3 toward PtdIns(3,4,5)P3. PtdIns(3,4,5)P3 (6 to 400 μM) was incubated with 0.5 μg of recombinant FRA3 protein.
(C) The kinetic property of FRA3 toward Ins(1,4,5)P3. Ins(1,4,5)P3 (50 to 800 μM) was incubated with 0.5 μg of recombinant FRA3 protein.
The fra3 Mutation Causes an Elevation in the Endogenous Levels of PtdIns(4,5)P2 and Ins(1,4,5)P3
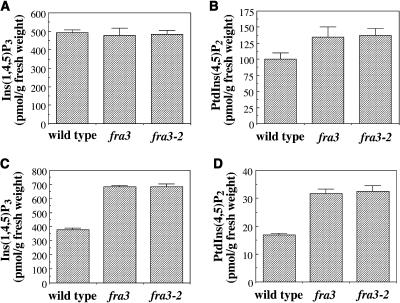
To investigate whether abolishment of the phosphatase activity by the fra3 mutation caused any accumulation of its substrates in vivo, we measured the amount of endogenous Ins(1,4,5)P3 and PtdIns(4,5)P2 in wild-type and fra3 plants. In fra3 seedlings, although the level of Ins(1,4,5)P3 was not altered (Figure 12A), the level of PtdIns(4,5)P2 slightly increased compared with the wild type (Figure 12B). In fra3 stems, the levels of both Ins(1,4,5)P3 (Figure 12C) and PtdIns(4,5)P2 (Figures 12D) were elevated compared with the wild type. These results demonstrate that the fra3 mutations cause an accumulation of PtdIns(4,5)P2 and Ins(1,4,5)P3 in the inflorescence stems, which is consistent with the defective fiber and vessel phenotypes in the fra3 mutant.
Figure 12.
Measurement of the Endogenous Levels of Ins(1,4,5)P3 and PtdIns(4,5)P2 in the Wild Type and the fra3 Mutant.
The seedlings used were 10 d old, and the inflorescence stems were from 8-week-old plants. The levels of Ins(1,4,5)P3 and PtdIns(4,5)P2 are expressed as picomoles per gram of fresh weight.
(A) The level of Ins(1,4,5)P3 in wild-type and fra3 seedlings.
(B) The level of PtdIns(4,5)P2 in wild-type and fra3 seedlings.
(C) The level of Ins(1,4,5)P3 in wild-type and fra3 stems.
(D) The level of PtdIns(4,5)P2 in wild-type and fra3 stems.
It has been shown previously that the sensitivity of seed germination to ABA treatment could be altered by a change in Ins(1,4,5)P3 level (Sanchez and Chua, 2001; Burnette et al., 2003; Carland and Nelson, 2004). Comparison of the seed germination of fra3 with that of the wild-type revealed no changes in the sensitivity to ABA inhibition (data not shown), which was in agreement with the unaltered level of Ins(1,4,5)P3 in seedlings.
Overexpression of the FRA3 5PTase Does Not Alter Plant Growth and Development
To study whether overexpression of the FRA3 5PTase affects plant growth and development, we introduced the wild-type FRA3 cDNA driven by the 35S promoter of Cauliflower mosaic virus into wild-type plants. A total of 72 transgenic plants were obtained, and none of them showed any abnormality in plant growth and development. RT-PCR analysis confirmed that 25 of the transgenic lines accumulated a much higher level of FRA3 mRNA than the wild type (four representatives are shown in Figure 7C). It is worth noting that the mutant fra3 cDNA was overexpressed as a control (Figure 7C) in the overexpression study, and no abnormality in plant growth and development was observed. It has been reported that overexpression of several type I 5PTases did not cause any altered phenotypes in normal plant growth and development (Sanchez and Chua, 2001; Perera et al., 2002; Burnette et al., 2003; Carland and Nelson, 2004).
DISCUSSION
Type II 5PTases have been known in yeast and animals to regulate the level of phosphoinositides and thereby influence various cellular activities such as actin cytoskeleton and vesicle trafficking (Takenawa and Itoh, 2001). In plants, several genes encoding putative 5PTases have been identified, and overexpression of two type I 5PTase genes has been shown to affect ABA sensitivity (Sanchez and Chua, 2001; Burnette et al., 2003). However, the putative type II 5PTase genes have not been characterized, and their roles in plant cellular functions are not known. Our finding that FRA3 encodes a type II 5PTase that is essential for actin organization and secondary wall synthesis in fiber cells provides a functional study of a type II 5PTase in plants.
FRA3 Is a Type II 5PTase
Two lines of evidence demonstrate that the FRA3 gene encodes a type II 5PTase. First, FRA3 possesses a domain that shows the highest sequence similarity to the catalytic domains of type II 5PTases from yeast and animals. The two conserved motifs that are known to be essential for the 5-phosphatase activity are completely conserved in FRA3. The importance of these two catalytic motifs was further evidenced in the fra3 mutant, in which a missense mutation of the Ala residue in motif II completely abolishes the FRA3 phosphatase activity. The Ala/Ser residue in motif II is conserved in all known 5PTases (Majerus et al., 1999). Our finding that the fra3 mutation completely abolishes its phosphatase activity provides biochemical evidence for the functional importance of the Ala/Ser residue in motif II of 5PTases. Secondly, recombinant FRA3 protein is able to hydrolyze phosphate from both phospholipids [PtdIns(4,5)P2 and PtdIns(3,4,5)P3] and water-soluble inositol polyphosphate [Ins(1,4,5)P3], which is a feature of type II 5PTases from yeast and animals (Majerus et al., 1999). The available evidence suggests that the FRA3 type II 5PTase shares both sequence and catalytic activity conservations with those from yeast and animals.
It is of interest to find that in addition to fibers and vascular tissues, the FRA3 gene is also expressed in other tissues. However, the defects caused by the fra3 mutations are only seen in fiber cells and xylem vessels. This fact suggests that other FRA3-like 5PTases might compensate for the loss of FRA3 in other cell types. The Arabidopsis genome contains three additional genes that encode proteins with high sequence similarity to FRA3. One of these FRA3-like proteins, At5PTase14, exhibits the same phosphatase activity as FRA3 [i.e., it shows activity toward PtdIns(4,5)P2, PtdIns(3,4,5)P3, and Ins(1,4,5)P3] (Zhong and Ye, 2004). In addition, the lack of alteration of Ins(1,4,5)P3 level in the fra3 seedlings might be because of compensation by type I 5PTases that are known to regulate the Ins(1,4,5)P3 level (Sanchez and Chua, 2001; Burnette et al., 2003; Carland and Nelson, 2004).
It is intriguing to note that FRA3 is capable of hydrolyzing PtdIns(3,4,5)P3, although PtdIns(3,4,5)P3 has not yet been detected in plants. A putative Tyr phosphatase from Arabidopsis has also been shown to be able to hydrolyze PtdIns(3,4,5)P3 (Gupta et al., 2002). Because of the lack of evidence for the existence of PtdIns(3,4,5)P3 in plants, the functional importance of FRA3 phosphatase toward PtdIns(3,4,5)P3 is currently unknown.
FRA3 Contains a WD-Repeat Domain
In addition to the phosphatase catalytic domain, some type II 5PTases in yeast and animals possess additional modules, such as SAC domain, Rho GTPase activating protein domain, and Pro-rich domain (Whisstock et al., 2002). The SAC domain in synaptojanins has been shown to hydrolyze 3- or 4-position phosphate from PtdIns(3)P, PtdIns(4)P, or PtdIns(3,5)P2, and together with the activity of the type II 5PTase domain, it has been thought to be able to completely terminate the phosphoinositide signaling molecules (Hughes et al., 2000). The Rho GTPase activating protein domain in OCRL1 has been demonstrated to interact with Rac GTPase, which was proposed to play a role in localizing OCRL1 to the trans-Golgi network (Faucherre et al., 2003).
It is extremely interesting to find that FRA3 contains a WD-repeat domain, which is not found in any known 5PTases from yeast and animals. WD-repeat proteins have been found in all eukaryotes, and some of them have been shown to be involved in many important biological processes, including signal transduction, RNA synthesis/processing, chromatin assembly, vesicle trafficking, cytoskeletal assembly, cell cycle control, and apoptosis (Li and Roberts, 2001). It has been proposed that the WD repeats in these proteins create a stable platform for protein–protein interactions (Smith et al., 1999). Indeed, some of the WD-repeat proteins have been shown to be involved in the assembly of multiprotein complexes, such as the β subunit of the G protein, the TAFII transcription factor, and the E3 ubiquitin ligase complexes (Li and Roberts, 2001). In plants, many WD-repeat proteins have been identified (van Nocker and Ludwig, 2003), and the WD-repeat domain in COP1 has been shown to interact with other effectors (Holm et al., 2001). The possible role of the WD-repeat domain in the FRA3 protein remains to be investigated.
Magnesium Concentration Differentially Regulates the FRA3 Phosphatase Activity toward Phosphoinositides and Ins(1,4,5)P3
Biochemical studies of 5PTases in yeast and animals have shown that 5PTases require Mg2+ for their catalytic activity. It was proposed that the 5PTases share a common mechanism for catalysis with a family of Mg2+-dependent nucleases, including DNase I and the apurinic/apyrimidinic base excision repair endonucleases (Whisstock et al., 2002). Because FRA3 shows high sequence similarity to 5PTases from yeast and animals, it was not surprising to find that FRA3 also requires Mg2+ for its activity. However, it was intriguing to find that the optimal Mg2+ concentration required for the FRA3 phosphatase activity toward Ins(1,4,5)P3 and phosphoinositides differs dramatically. This finding suggests that in plant cells, the free Mg2+ concentration, which may fluctuate depending on the physiological status of the cell, might influence the FRA3 phosphatase activity toward phosphoinositides and Ins(1,4,5)P3.
FRA3 Is Required for Normal Actin Organization in Fiber Cells
The fact that the fra3 mutation alters the normal actin organization in fiber cells indicates that regulation of the phosphoinositide metabolism by the FRA3 phosphatase is essential for actin organization. In agreement with this finding, it has been shown that mutations of OCRL1, a human type II 5PTase, cause a decrease in long actin stress fibers and an increase in punctate F-actin staining (Suchy and Nussbaum, 2002). Both human synaptojanins and yeast synaptojanin homologs, which contain type II 5PTase domains, are known to be important for actin organization (Mitchell et al., 2002).
It has been shown in yeast and animals that PtdIns(4,5)P2 regulates actin polymerization by modulating actin regulatory proteins, such as cofilin, profilin, and capping proteins CapZ and gelsolin (Takenawa and Itoh, 2001). In plants, PtdIns(4,5)P2 has also been shown to be able to bind profilin (Kovar et al., 2001). Because FRA3 is able to hydrolyze PtdIns(4,5)P2 in vitro and its mutation causes an accumulation of PtdIns(4,5)P2 in vivo, it is reasonable to propose that the abnormal actin organization seen in the fra3 mutant is a result of the alteration in PtdIns(4,5)P2 level.
FRA3 Influences Secondary Wall Synthesis in Fiber Cells
The fra3 mutant was isolated based on its reduced fiber strength phenotype. Anatomical examination revealed that the fra3 mutation dramatically reduced the secondary wall thickness in fiber cells, which likely accounts for the weak fiber strength phenotype. A prominent phenotype in fra3 is that the reduction in wall thickness is not uniform among fiber cells. Occasionally, some fiber cells are nearly devoid of secondary walls. This phenotype is reminiscent of that caused by mutation of the ROOT HAIR DEFECTIVE3 gene in the fra4 mutant. The fra4 mutant exhibited aggregation of actin cables in fiber cells, which was proposed to partially account for the defective secondary wall thickening phenotype (Hu et al., 2003). Because mutation of the FRA3 gene also resulted in altered actin organization, it is possible that the defective secondary wall thickening seen in the fra3 mutant is caused partially by the altered actin organization. This is in agreement with the known functions of F-actin in facilitating the transport of vesicles carrying cell wall polysaccharides and vesicles containing cellulose-synthesizing enzymes. An alteration in actin organization might impede the transport of these vesicles, thereby causing a reduction in the overall cell wall synthesis. The possibility that the signaling molecules PtdIns(4,5)P2 and Ins(1,4,5)P3, which are substrates for FRA3, might directly affect secondary wall synthesis could not be excluded.
In summary, we have demonstrated that FRA3 belongs to the type II 5PTase group and exhibits the highest substrate affinity toward PtdIns(4,5)P2. Mutation of the FRA3 gene was found to cause a dramatic reduction in secondary wall thickness, an alteration in actin organization, and an increase in PtdIns(4,5)P2 and Ins(1,4,5)P3 levels. We propose that FRA3 type II 5PTase regulates the metabolism of phosphoinositides and that this regulation is important for normal actin organization and secondary wall synthesis.
METHODS
Mutant Isolation and Map-Based Cloning
Ethyl methanesulfonate–mutagenized M2 Arabidopsis thaliana (ecotype Columbia) plants were grown in a greenhouse, and their inflorescence stems were screened for mutants with reduced stem strength. The stem breaking strength was measured as described previously (Zhong et al., 1997). The mutants were backcrossed with Columbia three times to eliminate any background mutations. The fra3 and fra3-2 mutants were crossed with each other for allelic analysis. The mutants were crossed with wild-type Arabidopsis ecotype Landsberg erecta to produce 1400 F2 mapping plants. The fra3 locus was mapped using CAPS markers (Konieczny and Ausubel, 1993). CAPS markers were developed by PCR amplification of genomic DNA fragments from ecotypes Columbia and Landsberg and digestion of the DNA fragments with various restriction enzymes.
For complementation analysis, the wild-type FRA3 gene, including a 2-kb 5′ upstream sequence, the entire exon and intron regions, and a 1-kb sequence 3′ downstream sequence, was PCR amplified by high-fidelity DNA polymerase with gene-specific primers (5′-TGGAGGAGAATCTTAGAAGTCTC-3′ and 5′-TGTATTGATTGTCTTGTTGTTG-3′), confirmed by sequencing, and cloned into the binary vector pBI101. The construct was introduced into the fra3 mutants by Agrobacterium tumefaciens–mediated transformation (Bechtold and Bouchez, 1994). Transgenic plants were selected on MS medium (Sigma-Aldrich, St. Louis, MO) containing 50 μg/mL of kanamycin and grown into adult plants for examination of rescued phenotypes. FRA3 genomic DNA and cDNA were sequenced using a dye-base cycle sequencing kit (Applied Biosystems, Foster City, CA).
Microscopy
The basal internodes of inflorescence stems of 8-week-old plants were fixed in 2% (v/v) glutaraldehyde in PEMT buffer (50 mM Pipes, 2 mM EGTA, 2 mM MgSO4, and 0.05% [v/v] Triton X-100, pH 7.2), post-fixed in 2% (v/v) OsO4, and embedded in Araldite/Embed 812 resin (Electron Microscopy Sciences, Fort Washington, PA). One-micrometer-thick sections were cut and stained with toluidine blue for light microscopy. For transmission electron microscopy, ultrathin sections (90 nm) were cut, post-stained with uranyl acetate and lead citrate, and observed under a Zeiss EM 902A electron microscope (Zeiss, Jena, Germany).
Cell Wall Analysis
Mature inflorescence stems of 10-week-old plants were collected for cell wall preparation and cell wall composition analysis. Stems were ground into fine powder in liquid nitrogen and further homogenized in a polytron. After extraction in ethanol, the cell wall residues were dried and used for analysis. Cell wall sugars (as alditol acetates) were measured according to Hoebler et al. (1989).
Immunolabeling of Actin Filaments and Microtubules
Stem segments, in which developing fiber cells just began to deposit lignin as stained with phloroglucinol HCl, were used for immunolabeling of actin filaments and cortical microtubules in fiber cells. Elongating stems were used for immunolabeling of actin filaments and cortical microtubules in pith cells. Samples were fixed in PEMT buffer containing 1.5% (v/v) formaldehyde and 0.5% (v/v) gutaraldehyde. Aldehyde fixation has been shown to preserve actin filaments well in plant cells (Vitha et al., 2000), and the F-actin patterns revealed by histolocalization and green fluorescent protein–tagged talin are comparable (Frank et al., 2003). After fixation, the stem segments were sectioned longitudinally with a vibratome into 100-μm-thick sections that were then processed for immunolabeling. Sections were incubated with monoclonal antibodies against chicken actin (ICN, Aurora, OH) or chicken α-tubulin (Sigma-Aldrich) and subsequently with fluorescein isothiocyanate-conjugated secondary antibodies. The fluorescence-labeled F-actin and microtubules were observed under a Leica TCs SP2 spectral confocal microscope (Leica Microsystems, Heidelberg, Germany). Images were saved and processed with Adobe Photoshop version 7.0 (Adobe Systems, San Jose, CA). The width of the fluorescence-labeled actin cables was measured using the ruler tool of Adobe Photoshop. A total of 125 actin cables were measured for calculation of the averaged width and standard errors.
Expression Analysis
Total RNA was isolated from leaves, stems, roots, flowers, and seedlings using a Qiagen RNA isolation kit (Qiagen, Valencia, CA). One microgram of the purified RNA was treated with DNase I to remove any potential genomic DNA contamination and then used for first strand cDNA synthesis. One-twentieth of the first strand cDNA was used for PCR amplification of FRA3 with gene-specific primers (5′-AGACATGACCTTAAACCTCACGTG-3′ and 5′-AAGAGCCTCTTGCTCGGTGGTCGT-3′). The primers used for RT-PCR span six introns so that any potential amplification from genomic DNA could be easily identified based on its larger size. No genomic DNA was amplified in the RT-PCR reactions. The PCR was performed for variable cycles to determine the logarithmic phase of amplifications for the samples. The RT-PCR reactions were repeated three times, and identical results were obtained. The expression of a ubiquitin gene was used as an internal control for determining the RT-PCR amplification efficiency among different samples.
For the GUS reporter gene study, the FRA3 gene containing a 2-kb 5′ upstream sequence and the entire exon and intron regions was PCR amplified by high-fidelity DNA polymerase with primers 5′-TGGAGGAGAATCTTAGAAGTCTC-3′ and 5′-TGGACTGTGCAAGTTCTTTAGCTG-3′, confirmed by sequencing, and ligated in frame with the GUS reporter gene in the pBI101 vector. The construct was introduced into wild-type and fra3 mutant plants by Agrobacterium-mediated transformation. Transgenic plants were selected on MS medium containing 50 μg/mL of kanamycin. For analysis of the GUS activity, seedlings or stem segments were first immersed in 90% ice-cold acetone for 20 min and then incubated in the GUS staining solution (100 mM sodium phosphate, pH 7.0, 10 mM EDTA, 0.5 mM ferricyanide, 0.5 mM ferrocyanide, and 1 mM 5-bromo-4-chloro-3-indolyl β-d-glucuronic acid) at 37°C. The tissues were cleared in 70% ethanol and then used directly for observation or hand sectioned before observation.
Generation of FRA3-Overexpressing Plants
The full-length wild-type or mutant FRA3 cDNA was PCR amplified using high-fidelity DNA polymerase from cDNAs derived from wild-type or fra3 mutant stems, respectively. The amplified cDNA was confirmed by sequencing and ligated downstream of the 35S promoter of Cauliflower mosaic virus in pBI121. The constructs were transformed into wild-type Arabidopsis plants using the Agrobacterium-mediated transformation procedure. Transgenic plants were selected on kanamycin, and the T2 generation was used for expression analysis and examination of any potential phenotypes caused by FRA3 overexpression.
Sequence Analysis
The WD-repeat domain and 5PTase catalytic domain in FRA3 were identified by searching the public database at the National Center for Biotechnology Information (http:://www.ncbi.nlm.nih.gov/BLAST). The domain sequences were aligned using the ClustalW 1.8 program (http://searchlauncher.bcm.tmc.edu/multi-align/multi-align.html). The WD repeats were identified using a protein repeats searching program (http://www.embl-heidelberg.de/∼andrade/papers/rep/search.html) and further analyzed using the secondary structure prediction program (http://www.predictprotein.org/) to determine the four β strands that potentially form the antiparallel β sheet.
Expression of Recombinant Proteins in Yeast
Full-length cDNAs of wild-type FRA3 and mutant fra3 were PCR amplified using specific primers (5′-ATGGAAGATCGTCAAAACGATCAAAAC-3′ and 5′-CTATGGACTGTGCAAGTTCTTTAG-3′) and confirmed by sequencing. The cDNAs were ligated in frame into the yeast expression vector pYES2/NT, which is tagged with six His and the Xpress epitope (DLYDDDDK) at the N terminus (Invitrogen, Carlsbad, CA). The constructs were transformed into the yeast strain INVSc1 (Invitrogen). The expression of recombinant proteins was induced in the presence of 2% galactose for 24 h. After induction, yeast cells were broken using glass beads, and the crude protein extracts were passed through an Ni-NTA column for purification of recombinant proteins.
The expression of recombinant proteins was confirmed by immunoblot analysis. Two hundred nanograms of purified recombinant protein was separated on a 4 to 20% SDS gel and transferred onto a nitrocellulose membrane. The recombinant proteins were detected by incubation with the monoclonal antibody against the N-terminal tagged Xpress epitope (Invitrogen) and horseradish peroxidase–conjugated secondary antibodies. The control protein used was β-galactosidase expressed under the same conditions. Both recombinant wild-type FRA3 and mutant fra3 proteins were confirmed to be expressed in yeast cells.
Phosphatase Activity Assay
Purified recombinant proteins were used for inositol polyphosphate phosphatase activity assay. Phospholipids were dissolved in the reaction buffer (50 mM Tris-HCl, pH 7.0, 0.25% β-d-octylglucoside, and 1 mM phenylmethylsulfonyl fluoride) with the aid of sonication. Phosphatase activity was measured by mixing 0.5 μg of recombinant proteins and 50 μM of substrates in the reaction buffer and incubated at 22°C. The enzyme activity was linear for 30 min; thus, all assays described in this study were run for 15 min. The FRA3 activity toward Ins(1,4,5)P3 was assayed in the presence of 3 mM Mg2+ in the assay buffer, and the FRA3 activity toward PtdIns(4,5)P2 and PtdIns(3,4,5)P3 was assayed in the presence of 0.1 mM Mg2+. The free phosphate released from the substrates was detected by the malachite green method (Kodama et al., 1986). The control protein used for the assays was β-galactosidase in replace of FRA3 in the reaction mixtures. All assays were run in duplicates and repeated at least twice, and identical results were obtained.
To test the effects of Mg2+ concentration on the FRA3 activity, different concentrations of MgCl2 were added in the reaction buffer. For analysis of the effects of pH and temperature, the FRA3 phosphatase activity was assayed under the same conditions as described above except for various pH or temperatures. For the kinetic analysis, the FRA3 activity toward a range of concentrations of substrates was assayed under the same conditions as described above except that pH 6.5 was used for Ins(1,4,5)P3.
Measurement of the Endogenous Levels of Ins(1,4,5)P3 and PtdIns(4,5)P2
Ten-day-old seedlings grown on agar containing MS medium and inflorescence stems of 8-week-old plants grown in a greenhouse were collected for measurement of Ins(1,4,5)P3 and PtdIns(4,5)P2. Tissues were ground into a fine powder in liquid nitrogen with a mortar and pestle, and the fresh weight of the ground tissues was determined in preweighed tubes. For extraction of Ins(1,4,5)P3, 1 g of freshly ground tissues were mixed with 0.8 mL of 20% ice-cold perchloric acid and incubated on ice for 20 min. The mixture was centrifuged at 4,000g for 20 min, and the supernatant was saved and neutralized to pH 7.5 with 1.5 M KOH containing 60 mM Hepes and 5% universal pH indicator (Fisher Scientific, Pittsburgh, PA). The neutralized solution was centrifuged at 2,000g for 15 min, and the supernatant was saved for Ins(1,4,5)P3 assay.
Ins(1,4,5)P3 was measured using the Ins(1,4,5)P3 [3H] Biotrak Assay System (Amersham Biosciences, Piscataway, NJ) according to the manufacturer's protocol. Serial dilutions of the samples and all the controls were included in the assay according to the manufacturer's recommendation. The amount of Ins(1,4,5)P3 in the samples was interpolated from a standard curve generated with known amount of Ins(1,4,5)P3. The assays were performed in duplicates and the experiments were repeated twice.
For extraction of PtdIns(4,5)P2, 1 g of freshly ground tissues was mixed with 2.1 mL of methanol:chloroform:12 M HCl (80:20:1, v/v/v) for 15 min. After further extraction by addition of 0.7 mL of chloroform and 1.25 mL of 0.1 M HCl, the chloroform phase was separated from the aqueous phase by centrifugation at 1,000g for 15 min. The lipid-containing chloroform phase was saved and dried in a stream of nitrogen. PtdIns(4,5)P2 in the dried sample was converted to Ins(1,4,5)P3 by addition of 250 μL KOH and heating at 100°C for 15 min. After cooling on ice, the solution was neutralized to pH 7.5 with 20% of perchloric acid containing 5% universal pH indicator. The neutralized solution was extracted with 1-butanol:light peteroleum ether (5:1, v/v), and the lower aqueous phase containing Ins(1,4,5)P3 was saved for Ins(1,4,5)P3 assay. The samples were spiked with [3H] PtdIns(4,5)P2 to determine the efficiency of conversion of PtdIns(4,5)P2 to Ins(1,4,5)P3. The amount of PtdIns(4,5)P2 was calculated based on the conversion efficiency.
Accession Numbers
Sequence data for the DNA and protein sequences from this article have been deposited with the EMBL/GenBank data libraries under accession numbers AY761186 (FRA3), M74161 (Hs5PTase), M88162 (OCRL), U45479 (synaptojanin1), U90312 (synaptojanin2), NP_012264 (Inp51p), NP_014293 (Inp52p), NP_014752 (Inp53p), AAG17824 (At5PTase1), AAG17825 (AtIP5P2), and At1g05470 (CVP2).
Acknowledgments
We thank Beth Richardson and John Shields for their help with microscopy and the editor and reviewers for their constructive comments. This work was supported by grants from the USDA and the Department of Energy.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instruction for Authors (www.plantcell.org) is: Zheng-Hua Ye (zhye@plantbio.uga.edu).
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.104.027466.
References
- Bechtold, N., and Bouchez, D. (1994). In planta Agrobacterium-mediated transformation of adult Arabidopsis thaliana plants by vacuum infiltration. In Gene Transfer to Plants, I. Potrykus, and G. Spangenberg, eds (Berlin: Springer-Verlag), pp. 19–23.
- Berdy, S.E., Kudla, J., Gruissem, W., and Gillaspy, G.E. (2001). Molecular characterization of At5Ptase1, an inositol phosphatase capable of terminating inositol triphosphate signaling. Plant Physiol. 126, 801–810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burnette, R.N., Gunesekera, B.M., and Gillaspy, G.E. (2003). An Arabidopsis inositol 5-phosphatase gain-of-function alters abscisic acid signaling. Plant Physiol. 132, 1011–1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carland, F.M., and Nelson, T. (2004). COTYLEDON VASCULAR PATTERN2-mediated inositol (1,4,5) triphosphate signal transduction is essential for closed venation patterns of Arabidopsis floral organs. Plant Cell 16, 1263–1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Despres, B., Bouissonnié, F., Wu, H.-J., Gomord, V., Guilleminot, J., Grellet, F., Berger, F., Delseny, M., and Devic, M. (2003). Three SAC1-like genes show overlapping patterns of expression in Arabidopsis but are remarkably silent during embryo development. Plant J. 34, 293–306. [DOI] [PubMed] [Google Scholar]
- Ercetin, M.E., and Gillaspy, G.E. (2004). Molecular characterization of an Arabidopsis gene encoding a phospholipid-specific inositol polyphosphate 5-phosphatase. Plant Physiol. 135, 938–946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faucherre, A., Desbois, P., Satre, V., Lunardi, J., Dorseuil, O., and Gacon, G. (2003). Lowe syndrome protein OCRL1 interacts with Rac GTPase in the trans-Golgi network. Hum. Mol. Genet. 12, 2449–2456. [DOI] [PubMed] [Google Scholar]
- Frank, M.J., Cartwright, H.N., and Smith, L.G. (2003). Three brick genes have distinct functions in a common pathway promoting polarized cell division and cell morphogenesis in the maize leaf epidermis. Development 130, 753–762. [DOI] [PubMed] [Google Scholar]
- Gupta, R., Ting, J.T.L., Sokolov, L.N., Johnson, S.A., and Luan, S. (2002). A tumor suppressor homolog, AtPTEN1, is essential for pollen development in Arabidopsis. Plant Cell 14, 2495–2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoebler, C., Barry, J.L., David, A., and Delort-Laval, J. (1989). Rapid acid-hydrolysis of plant cell wall polysaccharides and simplified quantitative determination of their neutral monosaccharides by gas-liquid chromatography. J. Agric. Food Chem. 37, 360–367. [Google Scholar]
- Holm, M., Hardtke, C.S., Gaudet, R., and Deng, X.-W. (2001). Identification of a structural motif that confers specific interaction with the WD40 repeat domain of Arabidopsis COP1. EMBO J. 20, 118–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu, Y., Zhong, R., Morrison, W.H., and Ye, Z.-H. (2003). The Arabidopsis RHD3 gene is required for cell wall biosynthesis and actin organization. Planta 217, 912–921. [DOI] [PubMed] [Google Scholar]
- Hughes, W.E., Cooke, F.T., and Parker, P.J. (2000). Sac phosphatase domain proteins. Biochem. J. 350, 337–352. [PMC free article] [PubMed] [Google Scholar]
- Ijuin, T., Mochizuki, Y., Fukami, K., Funaki, M., Asano, T., and Takenawa, T. (2000). Identification and characterization of a novel inositol polyphosphate 5-phosphatase. J. Biol. Chem. 275, 10870–10875. [DOI] [PubMed] [Google Scholar]
- Kodama, T., Fukui, K., and Kometani, K. (1986). The initial phosphate burst in ATP hydrolysis by Myosin and subfragment-1 as studied by a modified malachite green method for determination of inorganic phosphate. J. Biochem. 99, 1465–1472. [DOI] [PubMed] [Google Scholar]
- Konieczny, A., and Ausubel, F.M. (1993). A procedure for mapping Arabidopsis mutations using co-dominant ecotype-specific PCR-based markers. Plant J. 4, 403–410. [DOI] [PubMed] [Google Scholar]
- Kost, B., Lemichez, E., Spielhofer, P., Hong, Y., Tolias, K., Carpenter, C., and Chua, N.-H. (1999). Rac homologues and compartmentalized phosphatidylinositol 4,5-bisphosphate act in a common pathway to regulate polar pollen tube growth. J. Cell Biol. 145, 317–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovar, D.R., Drobak, B.K., Collings, D.A., and Staiger, C.J. (2001). The characterization of ligand-specific maize (Zea mays) profilin mutants. Biochem. J. 358, 49–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, D., and Roberts, R. (2001). WD-repeat proteins: Structure characteristics, biological function, and their involvement in human diseases. Cell. Mol. Life Sci. 58, 2085–2097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majerus, P.W., Kisseleva, M.V., and Norris, F.A. (1999). The role of phosphatases in inositol signaling reactions. J. Biol. Chem. 274, 10669–10672. [DOI] [PubMed] [Google Scholar]
- Mathur, J., Mathur, N., Kernebeck, B., and Hülskamp, M. (2003). Mutations in actin-related proteins 2 and 3 affect cell shape development in Arabidopsis. Plant Cell 15, 1632–1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell, C.A., Gurung, R., Kong, A.M., Dyson, J.M., Tan, A., and Ooms, L.M. (2002). Inositol polyphosphate 5-phosphatases: Lipid phosphatases with flair. IUBMB Life 53, 25–36. [DOI] [PubMed] [Google Scholar]
- Mueller-Roeber, B., and Pical, C. (2002). Inositol phospholipid metabolism in Arabidopsis. Characterization and putative isoforms of inositol phospholipid kinase and phosphoinositide-specific phospholipase C. Plant Physiol. 130, 22–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perera, I.Y., Love, J., Heilmann, I., Thompson, W.F., and Boss, W.F. (2002). Up-regulation of phosphoinositide metabolism in tobacco cells constitutively expressing the human type I inositol polyphosphate 5-phosphatase. Plant Physiol. 129, 1795–1806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quintero, F.J., Garciadeblas, B., and Rodriguez-Navarro, A. (1996). The SAL1 gene of Arabidopsis, encoding an enzyme with 3′(2′),5′-bisphosphate nucleotidase and inositol polyphosphate 1-phosphatase activities, increases salt tolerance in yeast. Plant Cell 8, 529–537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez, J.-P., and Chua, N.-H. (2001). Arabidopsis PLC1 is required for secondary responses to abscisic acid signals. Plant Cell 13, 1143–1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, T.F., Gaitatzes, C., Saxena, K., and Neer, E.J. (1999). The WD repeat: A common architecture for diverse functions. Trends Biochem. Sci. 24, 181–185. [DOI] [PubMed] [Google Scholar]
- Stolz, L.E., Kuo, W.J., Longchamps, J., Sekhon, M.K., and York, J.D. (1998). INP51, a yeast inositol polyphosphate 5-phosphatase required for phosphatidylinositol-4,5-bisphosphate homeostasis and whose absence confers a cold-resistant phenotype. J. Biol. Chem. 273, 11852–11861. [DOI] [PubMed] [Google Scholar]
- Suchy, S.F., and Nussbaum, R.L. (2002). The deficiency of piP2 5-phosphatase in Lowe syndrome affects actin polymerization. Am. J. Hum. Genet. 71, 1420–1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takenawa, T., and Itoh, T. (2001). Phosphoinositides, key molecules for regulation of actin cytoskeletal organization and membrane traffic from the plasma membrane. Biochim. Biophys. Acta 1533, 190–206. [DOI] [PubMed] [Google Scholar]
- van Nocker, S., and Ludwig, P. (2003). The WD-repeat protein superfamily in Arabidopsis: Conservation and divergence in structure and function. BMC Genomics 4, 50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitha, S., Baluska, F., Braun, M., Samaj, J., Volkmann, D., and Barlow, P.W. (2000). Comparison of cryofixation and aldehyde fixation for plant actin immunocytochemistry: Aldehydes do not destroy F-actin. Histochem. J. 32, 457–466. [DOI] [PubMed] [Google Scholar]
- Whisstock, J.C., Wiradjaja, F., Waters, J.E., and Gurung, R. (2002). The structure and function of catalytic domains within inositol polyphosphate 5-phosphatases. IUBMB Life 53, 15–23. [DOI] [PubMed] [Google Scholar]
- Xiong, L., Lee, B., Ishitani, M., Lee, H., Zhang, C., and Zhu, J.-K. (2001). FIERY1 encoding an inositol polyphosphate 1-phosphatase is a negative regulator of abscisic acid and stress signaling in Arabidopsis. Genes Dev. 15, 1971–1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye, Z.-H., Freshour, G., Hahn, M.G., Burk, D.H., and Zhong, R. (2002). Vascular development in Arabidopsis. Int. Rev. Cytol. 220, 225–256. [DOI] [PubMed] [Google Scholar]
- Zhang, X., Hartz, P.A., Philip, E., Racusen, L.C., and Majerus, P.W. (1998). Cell lines from kidney proximal tubules of a patient with Lowe syndrome lack OCRL inositol polyphosphate 5-phosphatase and accumulate phosphatidylinositol 4,5-bisphosphate. J. Biol. Chem. 273, 1574–1582. [DOI] [PubMed] [Google Scholar]
- Zhong, R., Burk, D.H., and Ye, Z.-H. (2001). Fibers. A model for studying cell differentiation, cell elongation, and cell wall biosynthesis. Plant Physiol. 126, 477–479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong, R., Taylor, J.J., and Ye, Z.-H. (1997). Disruption of interfascicular fiber differentiation in an Arabidopsis mutant. Plant Cell 9, 2159–2170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong, R., and Ye, Z.-H. (2003). The SAC domain-containing protein gene family in Arabidopsis. Plant Physiol. 132, 544–555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong, R., and Ye, Z.-H. (2004). Molecular and biochemical characterization of three WD-repeat domain-containing inositol polyphosphate 5-phosphatases in Arabidopsis thaliana. Plant Cell Physiol., in press. [DOI] [PubMed]