Abstract
The nuclear envelope (NE) acts as a selective barrier to macromolecule trafficking between the nucleus and the cytoplasm and undergoes a complex reorganization during mitosis. Different eukaryotic kingdoms show specializations in NE function and composition. In contrast with vertebrates, the protein composition of the NE and the function of NE proteins are barely understood in plants. MFP1 attachment factor 1 (MAF1) is a plant-specific NE-associated protein first identified in tomato (Lycopersicon esculentum). Here, we demonstrate that two Arabidopsis thaliana MAF1 homologs, WPP1 and WPP2, are associated with the NE specifically in undifferentiated cells of the root tip. Reentry into cell cycle after callus induction from differentiated root segments reprograms their NE association. Based on green fluorescent protein fusions and immunogold labeling data, the proteins are associated with the outer NE and the nuclear pores in interphase cells and with the immature cell plate during cytokinesis. RNA interference–based suppression of the Arabidopsis WPP family causes shorter primary roots, a reduced number of lateral roots, and reduced mitotic activity of the root meristem. Together, these data demonstrate the existence of regulated NE targeting in plants and identify a class of plant-specific NE proteins involved in mitotic activity.
INTRODUCTION
The nucleus, a hallmark of all eukaryotic cells, is separated from the cytoplasm by a double membrane system, the nuclear envelope (NE). Increasing evidence indicates differences between plant and metazoan NE composition and function, suggesting a kingdom-specific specialization of the NE. Whereas the outer nuclear membrane (ONM) is continuous with the endoplasmic reticulum (Mattaj, 2004), the inner nuclear membrane (INM) has a different protein composition. In metazoans, the INM contains several specific integral membrane proteins, which are connected through protein–protein interactions with chromatin and with the nuclear lamina (Mattout-Drubezki and Gruenbaum, 2003).
Embedded in the NE are the nuclear pore complexes (NPCs), large multiprotein complexes that form selective channels for nucleocytoplasmic transport (Fahrenkrog and Aebi, 2003). Significant progress has been made in the identification of yeast and mammalian nucleoporins (Nups) by proteomics approaches (Allen et al., 2001; Vasu and Forbes, 2001; Cronshaw et al., 2002). In addition to forming gateways for the exchange of molecules across the NE during interphase, NPCs interact with the spindle assembly checkpoint components Mad1 and Mad2 during mitosis in human cells and yeast (Campbell et al., 2001; Iouk et al., 2002). Moreover, several Nups, such as Nup170 and RanBP2/Nup358, associate with kinetochores and play a role in microtubule organization and chromosome segregation (Kerscher et al., 2001; Joseph et al., 2002, 2004; Salina et al., 2003). Mislocalized Nups have been implicated as causes for human diseases, such as cancer and triple A syndrome (Cronshaw and Matunis, 2003; Cronshaw and Matunis, 2004).
The progress in the molecular characterization of NE and NPC components in metazoans cells, however, cannot be applied by homology to plants. The plant NE appears to differ from the metazoan NE in both function and composition. Higher plant cells lack centrosomal structures and instead use the NE as the major site of microtubule nucleation during mitosis (Canaday et al., 2000; Schmit, 2002). No bona fide lamina appears to exist in yeast and plants, and the fully sequenced Arabidopsis thaliana genome does not contain identifiable homologs of genes coding for metazoan lamins, lamina-associated integral INM proteins, or many of the 30 Nups identified from yeast and mammals (Rose et al., 2004). The molecular composition of the NE and NPC in plants therefore must differ from that of the animal kingdom. To date, only a handful of plant NE-associated proteins have been cloned and characterized, including nuclear matrix constituent protein 1 (NMCP1), MFP1 attachment factor 1 (MAF1), Ran GTPase activating protein (RanGAP), and the microtubule nucleation factor Spc98p (Masuda et al., 1997; Gindullis et al., 1999; Rose and Meier, 2001; Erhardt et al., 2002; Pay et al., 2002; Brandizzi et al., 2004). NMCP1, MAF1, and a fragment of the human INM protein lamin B receptor have been used as NE markers to study NE dynamics in plant cells (Masuda et al., 1999; Dixit and Cyr, 2002a; Irons et al., 2003).
RanGAP is an accessory protein of the small GTPase Ran, which is involved in nuclear import and export, and is targeted to the NE through protein–protein interaction. The domain structure of plant and mammalian RanGAPs and their distinct targeting mechanisms indicate that their receptors at the NE might differ (Rose and Meier, 2001). Mammalian RanGAP has a unique C-terminal domain not present in yeast or plant RanGAP, which is required for its targeting to the nuclear pore and kinetochore. When modified by SUMOylation, the SUMOylated C terminus binds to the Nup RanBP2/Nup358 (Matunis et al., 1998). By contrast, all known plant RanGAPs possess a unique N-terminal domain, which in turn is not conserved in mammalian or yeast RanGAP (Meier, 2000). This domain, called the WPP domain after a highly conserved Trp-Pro-Pro motif, is necessary and sufficient for NE targeting of the Arabidopsis RanGAP1 (Rose and Meier, 2001).
MAF1 is a 16-kD protein, which consists mainly of a domain homologous to the WPP domain of plant RanGAP. It was first identified in tomato (Lycopersicon esculentum) and was shown to concentrate at the NE of tobacco (Nicotiana tabacum) suspension culture cells. MAF1 is widely conserved in land plants but has no homolog outside the plant kingdom, consistent with a plant-specific role at the NE. Like RanGAP, it does not have a transmembrane domain and is probably associated with the NE by protein–protein interactions (Gindullis et al., 1999).
The absence of plant homologs of the animal NE proteins, the unique NE targeting mechanisms of higher plants versus metazoan RanGAP, and the presence of plant-specific NE-associated proteins such as MAF1 suggest that plants and animals have evolved unique NE protein compositions. Here, we show that two members of the MAF-homologous Arabidopsis WPP domain protein (WPP) family are associated with the NE in a developmentally regulated fashion. WPP domain protein 1 (WPP1) localizes preferentially to the ONM in the vicinity of NPCs and is redistributed to the cell plate during cytokinesis. Intriguingly, RNA interference (RNAi)-based depletion of the WPP family leads to a developmental defect in Arabidopsis roots. This phenotype is caused by a reduced number of cells entering mitosis, providing evidence for a functional link between a NE protein and cell division in plants.
RESULTS
The Arabidopsis Genome Encodes Five Proteins Containing WPP Domains
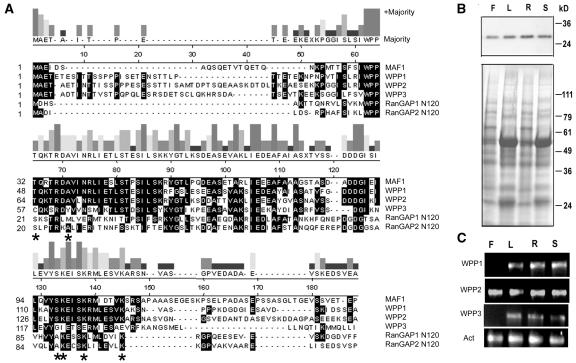
Plant RanGAP has been shown to contain an N-terminal NE targeting domain that appears to be unique to plants and is not shared with yeast or mammalian RanGAP (Meier, 2000; Rose and Meier, 2001). This domain, called the WPP domain, is similar to the NE-associated protein MAF1 from tomato (Gindullis et al., 1999). We used the tomato MAF1 sequence to identify proteins encoded in the Arabidopsis genome similar to the WPP domain. In addition to the two members of the RanGAP family, RanGAP1 and RanGAP2, there are three open reading frames encoding small WPP domain–containing proteins similar to MAF1 in Arabidopsis, termed WPP domain proteins 1, 2, and 3 (WPP1, WPP2, and WPP3). Figure 1A shows the alignment of MAF1, WPP1, WPP2, and WPP3 and the N-terminal domains of RanGAP1 and RanGAP2. Among the Arabidopsis proteins, MAF1 is most closely related to WPP1 and WPP2 (45.4 and 46.7% amino acid identity, respectively, compared with 28.9% identity with WPP3, 27.5% with RanGAP1, and 28.3% with RanGAP2). WPP1 and WPP2 are more closely related to each other (56.1% amino acid identity) than to WPP3 (34.8 and 43.9% amino acid identity, respectively), and all three family members are more closely related to each other than they are to the RanGAP WPP domains.
Figure 1.
Arabidopsis WPPs Have a Central Conserved WPP Domain and Are Expressed in Most Organs.
(A) Alignment of tomato MAF1 and the five Arabidopsis proteins containing WPP domains, WPP1, WPP2, WPP3, and the N-terminal 120 amino acids of RanGAP1 and RanGAP2. Majority, consensus strength (height of bars) and sequence based on majority (at least three of six). Black shading indicates amino acids that match the majority. Asterisks indicate otherwise conserved residues different in the WPP3 sequence (see text).
(B) The top panel shows an immunoblot analysis with OSU132 on total protein extracts from different adult plant tissues: F, flower; L, leaf; R, root; S, stem. The bottom panel shows a Coomassie blue–stained replica gel as loading control.
(C) RT-PCR analysis on total RNA from adult plant tissues (same as [B]) with primers specific for the genes indicated on the left. Act, Actin-related protein 6 (At3g33520) gene.
Expression Pattern of Arabidopsis WPP Family Members
A polyclonal rabbit antiserum was raised against recombinant WPP1. The antiserum OSU132 recognized recombinant WPP1, WPP2, WPP3, and RanGAP1 with decreasing affinity (data not shown). The top panel in Figure 1B shows an immunoblot analysis performed on total protein extracts from different Arabidopsis organs with the OSU132 antibody. A single band of ∼25 kD was detected in flowers, leaves, roots, and stems. Although the predicted size of WPP proteins is between 17 and 19 kD, all three recombinant proteins also have an apparent molecular weight in SDS-PAGE of 25 to 30 kD (data not shown). A similar observation is obtained with tomato MAF1 (Gindullis et al., 1999). In addition, a band was detected at 59 kD, which is the predicted size of RanGAP1 (data not shown). To investigate the expression pattern of each gene separately, RT-PCR with gene-specific primers was performed. Figure 1C shows that WPP1 and WPP3 expression was detected in leaves, roots, and stems but not flowers, whereas WPP2 expression was detected in all four organs.
WPP1 and WPP2 Accumulate at the NE in Arabidopsis Protoplasts
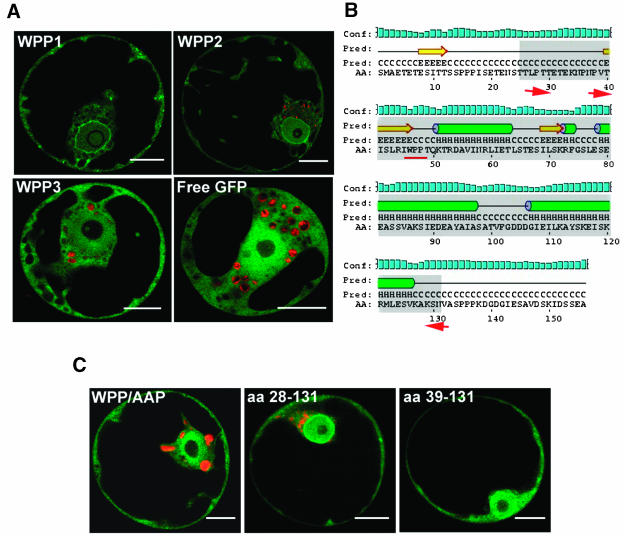
Endogenous tomato MAF1 and MAF1–green fluorescent protein (GFP) are localized at the NE of suspension-cultured cells (Gindullis et al., 1999). The subcellular localization of the Arabidopsis WPP family was investigated by expressing WPP1, WPP2, and WPP3 as GFP fusion proteins under the control of the 35S promoter in transiently transformed Arabidopsis protoplasts (Figure 2A). WPP1-GFP and WPP2-GFP accumulated at the NE and showed an additional diffuse staining in the cytoplasm and nucleus, similar to the localization pattern of MAF1 (Gindullis et al., 1999; Rose and Meier, 2001). By contrast, WPP3-GFP is present in the nucleus and in the cytoplasm but does not accumulate at the NE, resembling the distribution of free GFP. Inspection of the sequence alignment (Figure 1A) revealed that WPP3 is missing several amino acid residues conserved among the other WPP domain proteins (Figure 1A; data not shown). Therefore, a possible reason for its different behavior is that some of these conserved residues are necessary for NE targeting.
Figure 2.
An Intact WPP Domain Is Necessary to Localize WPP1 to the NE in Arabidopsis Protoplasts.
(A) GFP fluorescence of Arabidopsis protoplasts transiently expressing WPP1-GFP, WPP2-GFP, or WPP3-GFP.
(B) WPP1 secondary structure prediction with core WPP domain indicated as a gray box. Green cylinders, α-helices; yellow arrows, β-strands; black line, random coils; conf, confidence of prediction; pred, predicted secondary structure (H, helix; C, coil; E, strand). AA, amino acid sequence; red arrows, deletion end points of the constructs shown in (C); red underline, site of point mutations shown in (C).
(C) GFP fluorescence of Arabidopsis protoplasts transiently expressing WPP/AAP-mutated WPP1 (left panel), amino acids (aa) 28 to 131 of WPP1 (middle panel), and amino acids 39 to 131 of WPP1 (right panel). Bars in (A) and (C) = 10 μm.
Defining a Minimal WPP Domain
Figure 2B shows the secondary structure prediction of WPP1 using methods described by Jones (1999) and McGuffin et al. (2000). The underlined WPP motif is conserved in all known higher and lower plant WPP domain proteins (data not shown) and was shown to be crucial for RanGAP1 targeting to the NE (Rose and Meier, 2001). To determine whether the same motif is important for WPP1 NE localization, WPP was mutated to AAP. This mutation disrupts the NE targeting of WPP1 as shown in Figure 2C. To define the minimal length of the WPP domain, we took advantage of the extension of the most conserved domain in an alignment of all known WPP domain–containing proteins (data not shown), which coincides with the domain predicted to consist of a β strand and three α helices in Figure 2B. Two deletion constructs, indicated by the arrows in Figure 2B, were expressed as GFP fusions and analyzed for accumulation at the NE level. Figure 2C shows that residues 28 to 131 are sufficient for NE targeting, whereas residues 39 to 131 are not. Together, these data suggest that the central, most conserved domain of WPP1 is sufficient for NE targeting and that the WP motif is required.
WPP1-GFP and WPP2-GFP NE Targeting Is Developmentally Regulated in Arabidopsis Plants
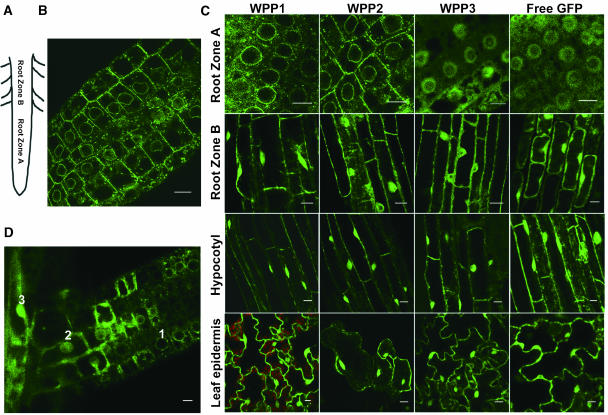
To investigate the localization pattern of WPP1-GFP, WPP2-GFP, and WPP3-GFP in different cell types, Arabidopsis transgenic lines expressing the fusion proteins under the control of the 35S promoter were created. Seven-day-old light-grown seedlings were imaged for GFP localization by confocal microscopy. Figure 3A shows the definition of zones of the primary root used in Figure 3C and the text. Root zone A includes the division and elongation zone. Root zone B includes the maturation zone, defined by the appearance of root hairs and indicative of differentiated cells. Root zone A spans ∼1 mm from the tip of a primary root in Arabidopsis seedlings. Figure 3B shows an overview of root zone A cells of a WPP2-GFP transgenic line, indicating the accumulation of the fusion protein at the NE in the small, undifferentiated cells in this zone. Figure 3C shows the localization pattern of WPP1-GFP, WPP2-GFP, WPP3-GFP, and free GFP in cells of root zone A, root zone B, hypocotyl cells, and leaf epidermis cells. As seen in Arabidopsis protoplasts (Figure 2A), the distribution of WPP3-GFP is indistinguishable from free GFP in all cell types imaged, indicating that this protein does not have a functional NE-targeting domain. WPP1-GFP and WPP2-GFP are targeted to the NE in root zone A cells. In contrast with this specific localization pattern, the distribution of GFP fluorescence in these lines resembled that of free GFP in the other three cell types. Root zone A is composed of cells that have recently divided or are primed for cell division. All other cell types shown are differentiated and have exited the cell cycle. This indicates that NE targeting of WPP1 and WPP2 is developmentally regulated and likely corresponds to the undifferentiated, cycling status of the cells.
Figure 3.
WPP1 and WPP2 Localize to the NE in Specific Cell Types.
(A) Depiction of zones A and B in a root.
(B) GFP fluorescence of transgenic Arabidopsis root tip containing WPP2-GFP.
(C) GFP fluorescence of transgenic Arabidopsis tissues (indicated to the left of each row) expressing various fusions (indicated on the top of each column).
(D) GFP fluorescence of a lateral root emerging from the primary root of a WPP1-GFP transgenic Arabidopsis plant. Bars in (B) to (D) = 10 μm.
As evident from Figures 3B and 3C, a less intense labeling of the cell cortex and of cytoplasmic speckles was also observed. Whereas NE association was consistently observed, labeling of the cell cortex and the speckles was only found in some cells (cf. e.g., Figures 3B to 3D and 5A). There was no difference between WPP1 and WPP2 with respect to this pattern. Localization in cytoplasmic speckles had also been observed for MAF1 (Gindullis et al., 1999). To investigate their identity, we performed a dual labeling experiment with transiently transformed MAF1-GFP and BIODIPY TR C5-ceramide (Molecular Probes, Eugene, OR) to label the Golgi complex and/or its derivatives in BY-2 cells. We found colocalization of some, but not all, GFP-labeled speckles with the Golgi marker, indicating that some of the speckles are related to elements in the Golgi pathway (data not shown).
Figure 5.
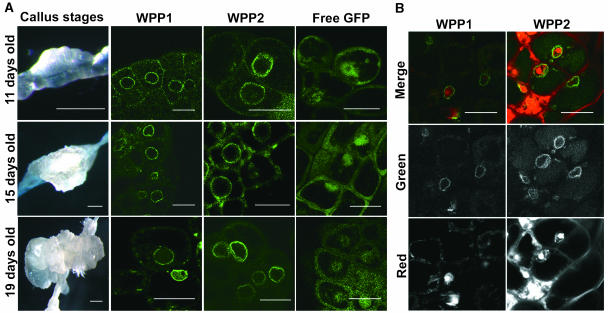
WPP1 and WPP2 Relocalize to the NE When Cell Division Is Induced.
(A) The first column (bars = 1 mm) shows calli formed 11, 15, and 19 d after placing root zone B tissue on callus-inducing medium. The second, third, and fourth columns (bars = 10 μm) show GFP fluorescence of transgenic calli expressing the fusion proteins indicated at the top of each column.
(B) Propidium iodide counterstaining of calli. Top, GFP and red propidium iodide fluorescence of callus cells originated from WPP1-GFP (left column) and WPP2-GFP (right column) transgenic zone B root tissue. Middle, GFP fluorescence; bottom, red propidium iodide fluorescence. Bars = 10 μm.
Secondary meristems are responsible for the emergence of lateral roots from root zone B. Figure 3D shows a section of a lateral root emerged from the main root. Cells at the tip of the lateral root (the secondary meristem) show WPP1-GFP at the NE, indistinguishable from the pattern at the primary meristem (cell 1 in Figure 3D). By contrast, differentiated cells in zone B of the primary root as well as older cells of the lateral root (cells 3 and 2, respectively) show diffuse GFP fluorescence in the nucleus and cytoplasm. To verify that the different localization patterns of WPP1-GFP and WPP2-GFP are not simply because of an increasing accumulation of the fusion protein, which would eventually obscure the signal at the nuclear rim, we compared the GFP-fusion protein accumulation in zone A and zone B by immunoblot analysis. Figure 4 shows that equivalent amounts of GFP-fusion protein, per total cellular protein, are present in both zones.
Figure 4.
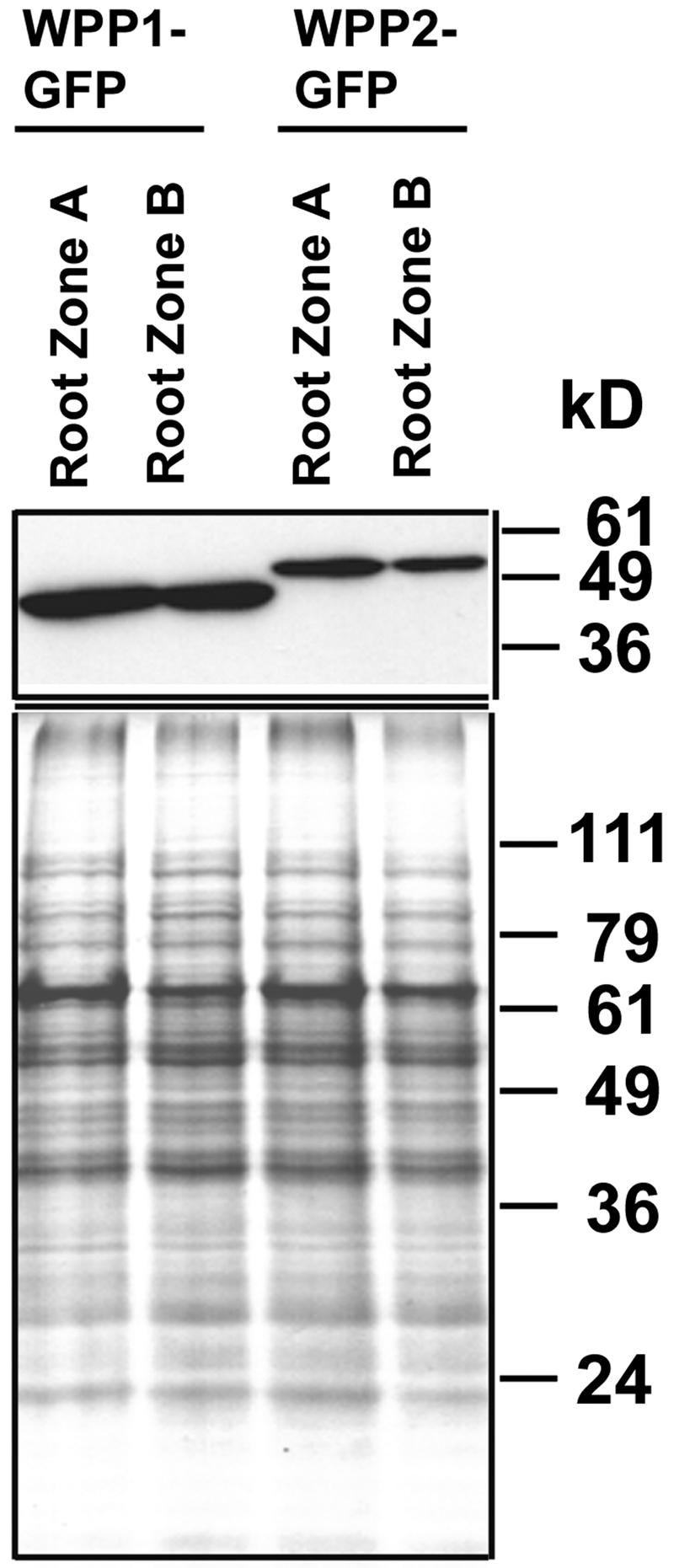
Equivalent Levels of WPP1-GFP and WPP2-GFP Accumulate in the Different Root Zones.
Top panel, immunoblot analysis with anti-GFP on total protein extracts from the different root zones; bottom panel, Coomassie blue–stained replica gel as loading control.
WPP1 and WPP2 Appear at the NE as Cells Reenter the Cell Cycle
Differentiated plant cells can be induced to dedifferentiate and reenter the cell cycle by hormone treatments. Root explants from zone B of 14-d-old WPP1-GFP, WPP2-GFP, and free GFP expressing plants were placed on callus-induction medium. Cells in developing calli were imaged 11, 15, and 19 d after transfer to callus-induction medium (Figure 5A). At all three stages, WPP1-GFP and WPP2-GFP associated with the NE in callus cells, again correlating with the dividing, dedifferentiated state of the cells. No change occurred in the localization pattern of free GFP (Figure 5A, right column). Figure 5B shows cells counterstained with propidium iodide to confirm the location of the nuclei.
WPP1-GFP Is Primarily Located at the Outer Surface of the NE
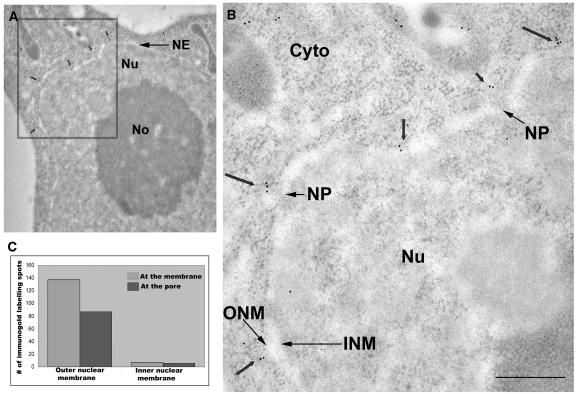
The previously described localization of tomato MAF1 in tobacco BY-2 cells was only investigated at the light microscopy level (Gindullis et al., 1999). Therefore, it was not known if MAF1 is associated with the inner or outer surface of the NE, or both. Here, we have investigated by transmission electron microscopy immunolocalizations the precise subcellular localization of WPP1. Antisera against GFP were used to localize WPP1-GFP in 15-d-old callus tissue induced from transgenic WPP1-GFP Arabidopsis roots (Figure 5). Figures 6A and 6B (expanded rectangle from A) show an example of an immunogold labeled section with the nucleus containing the nucleolus and the NE. In WPP1-GFP nuclei, clusters of two or more gold particles were found primarily at the nuclear rim and predominantly associated with the cytoplasmic side of the NE in the vicinity of nuclear pores. The bar graph in Figure 6C quantifies the distribution of gold particles at the NE level, which was calculated by counting gold particles from nuclei of three independent experiments. Few lone gold particles or doublets were found at the level of the nuclear rim in control GFP-expressing callus cells, but they were distributed equally at both sides of the NE (data not shown), and single gold particles and doublets were also found in the cytoplasm of both WPP1 and GFP expressing cells. Samples treated with preimmune rabbit serum did not show any labeling (data not shown).
Figure 6.
Subcellular Localization of WPP1-GFP in Transgenic Arabidopsis Calli.
(A) Electron micrograph showing a section of a callus cell. Nu, nucleus; NE, nuclear envelope; No, nucleolus.
(B) Expanded section of the rectangle enclosed area in (A). Arrows denote the positions of gold particles at the outer side of the NE. Cyto, cytoplasm; Nu, nucleus; ONM, location of the outer nuclear membrane; INM, location of the inner nuclear membrane; NP, nuclear pore. Bar = 400 nm.
(C) Bar graph illustrating the distribution of gold particles at the two nuclear membranes: gray bars indicate the number of gold particles on the inner and outer sides of the NE; black bars indicate the number of NE-associated particles located at the nuclear pores.
Tomato MAF1 Is Localized at the Immature Cell Plate during Cell Division
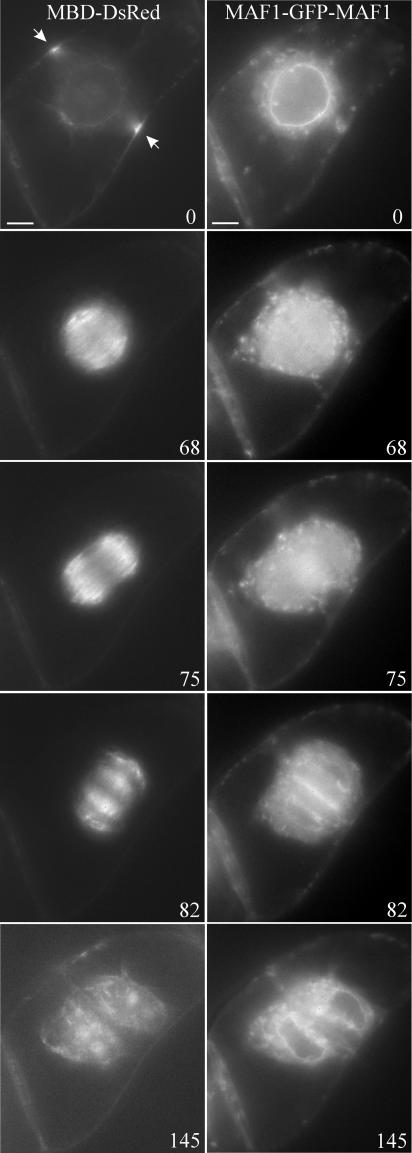
A MAF1-GFP transgenic tobacco BY-2 cell line was created to visualize MAF1 localization during cell division. This transgenic line also contained a microtubule binding domain (MBD)-DsRed marker to visualize microtubules during cell division. Figure 7 shows images collected at different time points during cell division in this cell line (for a time-lapse movie, see supplemental data online). The MAF1-GFP marker accumulates at the NE during preprophase (time = 0) and also shows a faint but consistent localization at the preprophase band site. Upon NE breakdown and the onset of metaphase, MAF1-GFP intrudes into the spindle apparatus and the surrounding cytoplasm (time = 68). Later during anaphase, this marker shows preferential localization to the midzone (time = 75) and is incorporated to those regions of the nascent cell plate where vesicle activity (i.e., vesicle fusion and maturation) is highest (time = 82). This accumulation is transient and is not seen in more developed areas of the cell plate (time = 145). MAF1-GFP repartitions to the reforming NEs during telophase (time = 82), where it persists at the end of cytokinesis (time = 145).
Figure 7.
MAF1 Localization during Cell Division.
See supplemental data online for a time-lapse movie. The MBD-DsRed+MAF1-GFP dual-marker BY-2 cell line was used to visualize the localization pattern of MAF1 during mitosis. Images were collected using 0.7-s exposure times, for both the red and the green channels, at 1-min intervals. The MBD-DsRed microtubule marker was used to identify the different stages of mitosis. The numbers represent time in minutes. Bars for all images = 10 μm.
WPP Protein Family Underexpression Slows Root Growth
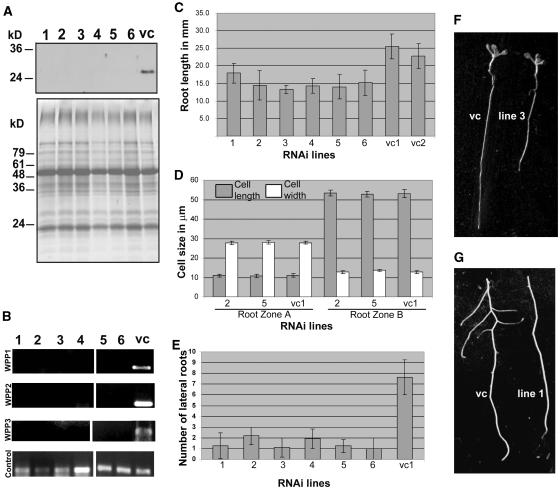
To investigate the loss-of-function phenotype of WPP family members, RNAi-expressing transgenic Arabidopsis plants were created (see Methods). Several lines were recovered that showed no detectable protein accumulation (Figure 8A). Separate lines were constructed with WPP1-RNAi, WPP2-RNAi, and WPP3-RNAi constructs. However, subsequent analysis of the lines by gene-specific RT-PCR showed that all three constructs were capable of silencing the expression of all three genes (Figure 8B).
Figure 8.
WPP Family Underexpression Slows Root Growth.
(A) The top panel shows immunoblot analysis with OSU132 antibody on total seedling protein extracts from the different RNAi lines (1 to 6) and the vector control (vc). Bottom panel, Coomassie stain of a replica gel as loading control.
(B) RT-PCR analysis on total RNA from seedling tissue on the different RNAi lines (1 to 6) and the vector control (vc) with specific primers for the genes indicated on the left. Control, 300-bp fragment from the 5′-end of the RanGAP1 mRNA.
(C) Root length of WPP RNAi lines (1 to 6) and two vector control lines (vc1 and vc2).
(D) Cell length and width of RNAi lines 2 and 5 and vector control (vc1).
(E) Number of lateral roots of WPP RNAi lines and vc1.
(F) Root length comparison between RNAi line 3 and vector control (vc) at 7 d after germination.
(G) Comparison of the number of lateral roots present between RNAi line 1 and vector control at 10 d after germination.
Plants grown on vertical MS agar plates developed a significantly shorter primary root compared with plants transformed with the empty vector. Figure 8C shows the average length of primary roots from 7-d-old light-grown seedlings. Figure 8F shows a typical seedling from line 3, compared with the vector control. No other features of the plant, such as hypocotyl length, shoot size, leaf number, leaf size, or flowering time, were significantly affected in the RNAi lines. To test whether the short root phenotype was caused by reduced cell elongation, length and width of root cells were measured. No noticeable cell size differences were detected between the vector control and RNAi lines in root zones A and B (Figure 8D). Figure 8E shows that the number of lateral roots was also reduced in 10-d-old light-grown seedlings. This is evident in Figure 8G, which shows the shortest vector-control root placed next to the longest root from RNAi line 1. It is apparent that the RNAi line 1 has fewer lateral roots compared with the vector-control root of a comparable developmental stage. Similarly, shorter roots and fewer lateral roots were also observed in WPP2 antisense lines (data not shown).
Mitotic Activity in RNAi Lines Is Reduced
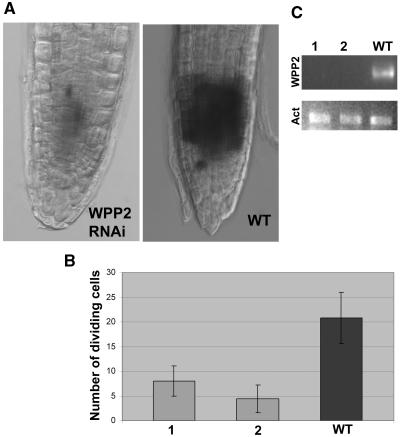
To examine mitotic activity in RNAi lines, we transformed transgenic Arabidopsis plants containing a PCYCB1;1:β-glucuronidase (GUS) fusion gene with the WPP2 RNAi construct. Because of a mitotic degradation signal in the protein, the reporter gene activity is seen only in actively dividing cells (Donnelly et al., 1999).
The number of cells undergoing mitosis, as visualized by the GUS staining, is reduced from 20.8 ± 5.2 in wild-type roots to 8 ± 3.0 in RNAi line 1 and 4.4 ± 2.7 in RNAi line 2 (Figure 9B). Figure 9A shows representative images of a wild-type root tip next to a root tip of RNAi line 1.
Figure 9.
Decreased Mitotic Activity in RNAi Lines.
(A) PCYCB1;1:GUS expression in mitotic cells of WPP2 RNAi and wild-type roots.
(B) Bar graph depicting number of dividing cells in RNAi lines (1 and 2) and wild-type roots.
(C) RT-PCR depicting loss of WPP2 expression in PCYCB1;1:GUS RNAi lines 1 and 2. Act, Actin-related protein 6 (At3g33520) gene was used as control.
DISCUSSION
In recent years, several developmental and cytokinetic defects in animals have been traced back to mutations in NE and NPC proteins. In contrast with metazoans, little is known about NE composition and NE protein function in plant cells. Our data for the plant-specific WPP family provides evidence for a functional link between plant NE proteins and cell division, indicating an analogous important role for the NE in plant cell division.
The NE Targeting Mechanism of WPP1 and RanGAP1 Share the Requirement for a Minimal Conserved WPP Domain
MAF-like proteins in higher plants are highly conserved and similar to the N terminus of plant RanGAP (Meier, 2000). Tomato MAF1, Arabidopsis RanGAP1, and Arabidopsis RanGAP2 are all targeted to the NE (Gindullis et al., 1999; Rose and Meier, 2001; Dixit and Cyr, 2002a; Pay et al., 2002). Tomato MAF1 was originally identified in a yeast two-hybrid screen with the DNA binding coiled-coil protein MFP1 (Gindullis et al., 1999). It has been subsequently established that MFP1 is a predominantly plastidic (not nuclear) protein, making it unlikely that the affinity of MAF1 for MFP1 is related to its NE targeting (Jeong et al., 2003). The MAF-like WPP domain of RanGAP1 is necessary and sufficient for this targeting, and a highly conserved WPP signature motif within the targeting domain is required for NE targeting (Rose and Meier, 2001). In this study, we demonstrated that the conserved core WPP domain of WPP1 is also sufficient to target GFP to the NE. In analogy to RanGAP1, mutation of WPP to AAP in WPP1-GFP also causes loss of NE localization, indicating a similar targeting mechanism for both proteins. It is therefore likely that plant RanGAP and WPP proteins bind to the same acceptor protein(s) at the NE.
Despite Strong Sequence Conservation, Arabidopsis WPP Proteins Show Differential NE Targeting
The high sequence conservation and similar NE targeting of the WPP domains of plant RanGAP and WPPs invite the assumption that all WPP domain–containing proteins might localize to the NE. However, we show here that members of the Arabidopsis WPP protein family differentially localize to the NE in cell culture and in planta. Only WPP1-GFP and WPP2-GFP concentrate at the NE in transiently transformed protoplasts as well as root tip cells of stably transformed plants. By contrast, WPP3-GFP is not concentrated at the NE in these cell types and instead shows a distribution similar to free GFP. Therefore, the presence of a WPP domain is not sufficient for NE targeting. WPP3 is missing several otherwise highly conserved amino acid residues. Therefore, our current working model is that these residues are important for interaction with an acceptor protein at the NE.
WPP1 and WPP2 Associate with the NE in a Developmentally Regulated Manner Linked to Mitotic Activity
In addition to the difference in NE targeting of Arabidopsis WPP1, WPP2, and WPP3 in the same cell type, differential patterns of targeting could be observed for stably expressed WPP1-GFP and WPP2-GFP in planta. Strikingly, the localization of both proteins changes from a predominantly NE accumulation to a diffuse cytoplasmic and nuclear labeling in differentiated cells of the root (zone B), hypocotyl, and leaf epidermis. This observation indicates that WPP1 and WPP2 may play a specialized role in undifferentiated cells. This localization pattern was not observed for RanGAP1, which concentrates at the NE in differentiated cells (Pay et al., 2002). A role of WPP1 and WPP2 at the NE in dividing cells would predict a reestablishment of NE targeting in cells reentering mitosis. Indeed, when root zone B cells were induced to dedifferentiate and reenter the cell cycle, WPP1-GFP and WPP2-GFP were again concentrated at the NE. This supports the hypothesis that NE localization occurs specifically in cells that are either primed for cell division or have recently divided. This developmental regulation of targeting provides an intriguing link between plant NE proteins and mitotic activity unique to the WPP protein family.
WPP1-GFP Is Associated with the Outer NE in Interphase
The NE—comprised of the ONM, INM, and NPCs—disassembles and reassembles during mitosis. The localization of WPPs at the NE as well as their dynamics during the NE-free stages of the cell cycle might pinpoint the site of WPP function. The immunogold localization data for WPP1 are consistent with those obtained by confocal microscopy and support the notion that WPP1 is primarily associated with the ONM in the vicinity of the nuclear pores, making it an unlikely candidate for a plant INM or nuclear lamina component. Instead, a cytoplasmic NPC-specific function seems more likely. Whereas the localization of plant RanGAP is presently not resolved at the ultrastructural level, the localization of WPP1 is in good correlation with electron microscopy images obtained from animal cells showing that metazoan RanGAP and its interaction partner RanBP2/Nup358 are associated with the outer surface of the nuclear pores (Wu et al., 1995; Matunis et al., 1996).
Tomato MAF1 Is Redirected to Regions of Active Vesicle Trafficking during Cytokinesis
After NE breakdown, MAF1-GFP distributes to the cytoplasm surrounding the spindle in tobacco BY-2 cells. Intriguingly, MAF1-GFP concentrates at the immature cell plate during the early stages of phragmoplast formation and is associated with the growing edge of the cell plate as the phragmoplast expands. This localization pattern differs from the localization pattern of Golgi, endoplasmic reticulum, and lamin-B receptor markers during cytokinesis, which label all stages of the developing cell plate and do not show specific localization to the immature or growing edges of the developing cell plate (Nebenführ et al., 2000; Dixit and Cyr, 2002b; Brandizzi et al., 2004). Notably, the cell plate localization pattern of the MAF1-GFP marker correlates to the sites where vesicle trafficking and vesicle fusion are concentrated in a developing phragmoplast (Staehelin and Hepler, 1996; Smith, 2002; Mayer and Jürgens, 2004; Seguí-Simarro et al., 2004). An antiserum against carrot (Daucus carota) NMCP-1, the only other plant NE-associated protein studied during mitosis, also labels the cytoplasm during metaphase but does not decorate the cell plate during cytokinesis in carrot or celery (Apium graveolens) cells (Masuda et al., 1999). Our finding that MAF1 is associated with two sites of vesicle delivery and membrane fusion events during plant cell division, the reforming NE as well as the newly forming cell plate, might indicate a function of WPP proteins in membrane trafficking.
Reduced Expression of the WPP Protein Family in Arabidopsis Delays Root Development through Reduction of Mitotic Activity
To characterize the function of the WPP protein family in Arabidopsis, RNAi was used to downregulate its expression. RNAi constructs of individual members of the WPP gene family proved to reduce RNA and protein expression level of all family members; therefore, no data are currently available for the effect of individual knockdowns. Global reduction in WPP expression caused significantly delayed root growth and development in otherwise normally developing Arabidopsis seedlings. Reduced root length is not caused by changes in cell size, indicating that WPP underexpression does not affect root cell elongation. Consistent with these data, no changes in auxin sensitivity were detected in roots from WPP RNAi lines (data not shown). Instead, a clear reduction of the mitotic activity of the root meristem was observed, indicating a cytokinesis-specific function of WPP proteins. Intriguingly, the mitosis-promoting activity of WPPs correlates well with the specific NE localization patterns of WPP1 and WPP2 observed in meristematic root cells and rapidly cycling cells in callus and liquid cell culture.
Possible Modes of WPP Action during Cell Division
Based on the data presented here, several modes of action can be proposed for WPPs. It is possible that WPPs act as regulators at the NPC, being either involved in NPC assembly and composition or functions of the NPC, such as nucleocytoplasmic transport. In yeasts, M phase–specific rearrangements of the NPCs inhibit nucleocytoplasmic transport (Makhnevych et al., 2003), linking NPC composition and transport with cell division. The regulation of nucleocytoplasmic transport is emerging as an important factor in plant environmental and developmental signaling, such as light, phytohormone, and stress signal transduction (reviewed in Merkle, 2004). The observed effect on root growth and cell division in WPP underexpressing plants might point at a possible involvement in the signaling pathway for the plant hormone cytokinin (Werner et al., 2003). The Ran cycle plays a major role in nucleocytoplasmic transport and NE dynamics. It is therefore tempting to speculate that competition of WPPs with RanGAP for shared binding sites regulates RanGAP function at the NE during cell division in plant cells.
The finding that MAF1 targets GFP to two sites of membrane fusion events is intriguing. Both Golgi vesicles and tubular endoplasmic reticulum components are recruited to the division plane during plant mitosis and cytokinesis (Cutler and Ehrhardt, 2002; Seguí-Simarro et al., 2004). Our results add to the body of knowledge regarding elements that contribute to the formation of the cell plate (Nacry et al., 2000; Waizenegger et al., 2000; Bednarek and Falbel, 2002; Hepler et al., 2002; Nishihama et al., 2002; Smith, 2002; Strompen et al., 2002; Mayer and Jürgens, 2004; Müller et al., 2004; Pan et al., 2004). WPPs might function as a marker for sorting vesicles designated to fuse either at the NE or the cell plate. Interestingly, mutations in components of the Ran cycle lead to defects in NPC assembly as well as accumulation of Nup-containing vesicles in yeast, connecting NPC assembly with components of the nucleocytoplasmic transport machinery as well as vesicle trafficking (Ryan et al., 2003). Therefore, another attractive hypothesis is that WPPs could act as regulators of these newly emerging functions by affecting either RanGAP interactions or NPC composition in plant cells.
In summary, the WPP family in Arabidopsis exhibits a unique developmentally regulated NE-association pattern and influences the mitotic activity of root meristems. Consistent with the observed data, several intriguing hypotheses are now open for investigation to identify the mechanism of WPP action.
METHODS
Sequence Analysis
WPP1 (At5g43070), WPP2 (At1g47200), and WPP3 (At5g27940) were identified by sequence similarity searches using BLAST (Altschul et al., 1997) performed with the previously described tomato (Lycopersicon esculentum) MAF1 (Gindullis et al., 1999). MEGALIGN protein alignment software (DNASTAR, Madison, WI) was used for multiple sequence alignments using the Clustal algorithm. The secondary structure was predicted using the PSIPRED protein structure prediction server (Jones, 1999; McGuffin et al., 2000).
Cloning
WPP1, WPP2, and WPP3 are intronless genes and could therefore be PCR amplified from genomic DNA isolated from Arabidopsis thaliana leaves. The WPP1 PCR product was isolated using the primers 5′-TCCATGGCCGAAACCGAA-3′ and 5′-CTAAGTTCACTTCGAACTGCTC-3′. The primers 5′-TTGAATTCATGGCAGAAACCGCCGAGACA-3′ and 5′-TTGAATTCTCAAGCCTCACTCTTCTC-3′ were used for WPP2, and primers 5′-TTGAATTCATGGCAGAAACCGCC-3′ and 5′-TTGAATTCCTAAATCAGCAACTGCAT-3′ were used for WPP3 gene amplification. The WPP1 PCR product was cloned into the TOPO-TA vector using the TOPO-TA cloning kit from Invitrogen (Carlsbad, CA) and subsequently subcloned into the expression vector pRSET B using the NcoI and EcoRI restriction sites. The PCR products of WPP2 and WPP3 were directly cloned into pRSET B using the internal EcoRI sites of the PCR primers.
GFP fusions for transient expression were created by cloning PCR-amplified, TOPO-cloned, and NcoI-digested fragments of the three genes into the NcoI site of pRTL2-mGFPS65T (von Arnim et al., 1998). Primers were designed to eliminate stop codons and fuse the coding sequences to the 5′ end of the GFP gene. For WPP1, the same 5′ primer as above was used with the 3′ primer 5′-CCATGGAAGCTTCACTTGAATC-3′. For WPP2, the 5′ primer 5′-GCCATGGCAGAAACCGC-3′ and 3′ primer 5′-GCCATGGAAGCCTCACTCTTC-3′ were used. WPP3 was amplified using the same 5′ primer as above with the 3′ primer 5′-GCCATGGAAATCAGCAACTGC-3′. GFP fusions for Arabidopsis transformation were created by subcloning the WPP1, WPP2, and WPP3 open reading frames from pRTL2 into the binary vector pFGC5941 (http://www.chromdb.org/plasmids/table1.html) distributed through the Arabidopsis Biological Resource Center (http://www.biosci.ohio-state.edu/∼plantbio/Facilities/abrc/abrchome.htm) using NcoI restriction sites with the GFP gene at the C terminus of each gene.
For RNAi lines, WPP1, WPP2, and WPP3 PCR products were cloned into the RNAi vector pFGC1008 (http://www.chromdb.org/plasmids/table1.html) using restriction sites AscI/SwaI and BamHI/SpeI for the first and second cloning directions, respectively. Primers 5′-TTACTAGTGGCGCGCCATGGCCGAAACCGAAACG-3′ and 5′-TTGGATCCATTTAAATCTCGTCAAGCTTCACTT-3′ were used for WPP1 PCR amplification. Primers 5′-TTACTAGTGGCGCGCCATGGCAGAAACCGCCGAG-3′ and 5′-TTGGATCCATTTAAATCAAGCCTCACTCT-3′ were used for WPP2 PCR amplification. Primers 5′-TTACTAGTGGCGCGCCATGGCAGAAACCGCCGAT-3′ and 5′-TTGGATCCATTTAAATTCAGCAACTGCATCATC-3′ were used for WPP3 PCR amplification.
Site-Directed Mutagenesis
The conserved WPP motif in WPP1 was mutated to AAP using the mutagenic primer 5′-CATTTCCCTCAGAATCGCGGCACCGACTCAAAAAACTC-3′ and its complementary primer with pRTL2-mGFPS65T containing the WPP1 gene as template, as described previously for RanGAP1 (Rose and Meier, 2001).
Antibody Production
The WPP1 open reading frame cloned in pRSET B was expressed as a recombinant protein with His and Xpress epitope tags in BL21 DE3 cells. Protein purification was performed using nickel-nitrilotriacetic acid agarose superflow column and the protocol given by Qiagen (Valencia, CA). Rabbit antiserum against WPP1 (OSU132) was produced by Cocalico Biologicals (Reamstown, PA).
Plant Material
Arabidopsis ecotype Columbia was used for all analyses. Seedlings were grown on MS medium (Caisson Laboratories, Rexburg, ID) supplemented with B5 vitamins, 2% sucrose, and 0.8% agar at 22°C in continuous white light. Plants were grown on soil (BFG Supply Company, Xenia, OH) under 16-h-light and 8-h-dark conditions. For callus induction, 1× Gamborg's B5 medium (Caisson Laboratories), 2% (w/v) glucose, 1.7 mM Mes, pH 5.7, 0.8% (w/v) agar, 0.5 mg/L 2,4-D, and 0.05 mg/L of kinetin was used.
Plant Cell Cultures
Green Arabidopsis suspension culture cells were cultured in Gamborg's B5 basal medium with minimal organics (Caisson Laboratories, Rexburg, ID) supplemented with 2% (w/v) sucrose, 5 μM 2,4-D, and 1.7 mM Mes, pH 5.7. Cultures were maintained by shaking at 200 rpm in constant light at 24°C and subcultured weekly by 1:10 dilution with fresh medium.
Generation of Transgenic MAF1-GFP BY-2 Culture
The tomato MAF1-GFP transgene (Gindullis et al., 1999) was introduced into the pCAMBIA1300 vector (CAMBIA, Canberra, Australia) between a 35S promoter and Nos terminator. This construct was electroporated into C58C1 Agrobacterium tumefaciens cells, which were used to transform the MBD-DsRed tobacco (Nicotiana tabacum) cell line (Dixit and Cyr, 2003). Stably transformed tobacco cells were selected using 100 mg/L of kanamycin and 20 mg/L of hygromycin, and individual calli were used to establish liquid cultures.
Arabidopsis Protein Extracts
Total protein extracts from different tissues were prepared by heating liquid N2 ground tissue in 62.5 mM Tris-HCl, pH 6.8, 20% glycerol, 4% SDS, and 1.4 M 2-mercaptoethanol at 70°C for 10 min and recovering the supernatant after centrifugation at 12,000 rpm.
Immunoblot Analysis
Dilutions of 1:2000 and 1:1000 were used for the OSU132 and anti-GFP (Molecular Probes) primary antibodies, respectively. A dilution of 1:20,000 was used for the horseradish peroxidase–coupled donkey anti-rabbit secondary antibody (Amersham Pharmacia Biotech, Uppsala, Sweden). Immunoblot analysis was performed as described by Sambrook et al. (1989). Enhanced chemiluminescence detection was performed as described by Amersham Pharmacia Biotech.
RT-PCR Analysis
The Arabidopsis cDNAs for WPP1, WPP2, WPP3, and the control genes (Actin and 300-bp fragment from the N terminus of RanGAP1) were synthesized and amplified by RT-PCR using specific primers for each gene. Primers 5′-TTACTAGTGGCGCGCCATGGCCGAAACCGAAACG-3′ and 5′-GAACAACACCATATTCAAGCCTCG-3′ were used for WPP1, 5′-TTACTAGTGGCGCGCCATGGCAGAAACCGCCGAG-3′ and 5′-AATAAGGAAGTAAAACCAAATTTC-3′ were used for WPP2, and 5′-TTGAATTCCTAAATCAGCAACTGCAT-3′ and 5′-TAACGCTCAAGCTTCTCCTCTAAA-3′ were used for WPP3. For Actin-related protein 6 (At3g33520) gene as control, primers 5′-AAAACCACTTACAGAGTTCGTTCG-3′ and 5′-GTTGAACGGAAGGGATTGAGAGT-3′ were used. For the RanGAP1 N terminus, the primers 5′-TTGAATTCATGGATCATTCAGCGAAA-3′ and 5′-TTGAATTCCTCAACCTCGGATTCTTC-3′ were used. Total RNA was prepared from flowers, leaves, roots, and stems of 30-d-old plants using the RNeasy plant mini kit (Qiagen). RT-PCR was performed using the ProSTAR HF single tube RT-PCR system from Stratagene (La Jolla, CA) using 1 μL of RNA, 100 ng of each primer, and 47°C annealing temperature.
Protoplast Transformation
Four-day-old Arabidopsis suspension cells were harvested and washed with 0.4 M mannitol and 20 mM Mes, pH 5.5, and incubated for 2 to 3 h at room temperature on a platform shaker at 180 rpm in 1% cellulase and 0.1% pectolyase (Karlan, Santa Rosa, CA). The protoplasts were washed once with W5 medium (154 mM NaCl, 5 mM KCl, 125 mM CaCl2, and 5 mM glucose, pH 6.0), resuspended in W5 medium at a concentration of 3 to 5 × 106 cells/mL, and incubated on ice for 2 h. For transformation, the protoplasts were resuspended in ice-cold 0.4 M mannitol, 15 mM MgCl2, and 5 mM Mes, pH 5.6, solution. Twenty micrograms of DNA, 300 μL of protoplast suspension, and 300 μL of 40% polyethylene glycol, 0.1 M Ca(NO3)2, and 0.4 M mannitol, pH 8.0, were mixed by gentle inversion at room temperature for 30 min. After washing with W5 medium, the transformed protoplasts were resuspended in 4 mL of Gamborg's medium with 0.4 M mannitol and incubated in the dark for 48 h before microscopy.
Fluorescence Microscopy
Confocal microscopy images were obtained as described by Rose and Meier (2001). For further processing, Adobe Photoshop software (Adobe Systems, Mountain View, CA) was used. For the time-course experiment, observations were conducted on MBD-DsRed+MAF1-GFP cells, 2 to 3 d after subculture. The cells were immobilized on poly-l-Lys coated cover slips in a humid chamber, and images were collected using a Plan-Neofluar 40× (numerical aperture 1.3) oil-immersion objective (Zeiss, Thornwood, NY). Wide-field fluorescence microscopy was conducted using a shutter-equipped Zeiss Axiovert S100 TV microscope, and images were captured with a CoolSNAP HQ camera (Roper Scientific, Tucson, AZ) controlled by ESee software (Inovision, Durham, NC). DsRed (515 to 560 nm excitation, long-pass 590 nm emission) and GFP (460 to 500 nm excitation and 510 to 560 nm emission) filter sets were used to visualize and discriminate between these fluorophores. Images were collected using 30% excitation light intensity from a mercury short-arc lamp. For propidium iodide staining, callus cells were incubated at room temperature with 0.1 mg/mL of propidium iodide for 5 to 10 min. After five washes with growth medium, cells were used for microscopy.
Sample Preparation for Electron Microscopy
Samples from callus tissue from roots of transgenic GFP and WPP1-GFP Arabidopsis lines were fixed with 4% paraformaldehyde and 0.2% glutaraldehyde in 0.1 M potassium phosphate buffer, pH 7.4, at room temperature for 2 h. After three washes with potassium phosphate buffer and quenching with the same buffer containing 50 mM Gly for 20 min at room temperature, samples were dehydrated in a graded ethanol series. They were then transferred to −20°C and infiltrated in a graded ethanol/LR Gold resin (Electron Microscopy Sciences, Washington, PA) series and embedded in 100% LR Gold resin containing 0.5% benzil activator in gelatin capsules. Samples were polymerized over night by placing the capsules 15 cm above a UV light box at −20°C.
Immunogold Labeling
Eighty- to ninety-nanometer thin sections were collected on formvar and carbon coated nickel grids. Grids were floated first on a drop of distilled water and then on a drop of PBS (10 mM potassium phosphate buffer, pH 7.4, containing 150 mM NaCl) for a few minutes. After blocking with Aurion goat blocking solution (Electron Microscopy Sciences, Washington, PA) for 30 min, grids were incubated overnight with a dilution of the primary antibody in the incubation buffer (10 mM potassium phosphate buffer, pH 7.4, containing 150 mM NaCl, 10 mM NaAzide, and 0.2% BSA) at 4°C. Two separate primary antibodies (Chemicon International, Temecula, CA) were used: the affinity purified rabbit polyclonal anti-GFP diluted to 1/50 and the mouse monoclonal anti-GFP at a 1/200 dilution. After six washes with PBS, grids were incubated with the 10-nm gold particle conjugated goat anti-rabbit or anti-mouse secondary antibody (Electron Microscopy Sciences) in incubation buffer for 5 h at room temperature. After six washes with PBS, followed by two washes in distilled water, grids were stained 5 min with a 2% aqueous solution of uranyl acetate and 2 min with a 0.5% solution of lead citrate. Samples were observed and photographed on a Hitachi H-7500 transmission electron microscope (Tokyo, Japan). The distribution of the immunogold labeling was evaluated by counting gold particles at the NE level in the GFP and GFP-WPP1 transformed callus tissue, from 24 separate grids (four grids for each transformant in three separate experiments).
Agrobacterium and Arabidopsis Transformation
The T-DNA constructs were electroporated into Agrobacterium strains LBA 4404 (for RNAi plants) and GV3101 (for GFP plants), and the transformed Agrobacteria were used to transform Arabidopsis plants according to Clough and Bent (1998). For RNAi lines, hygromycin (25 μg/mL) resistant plants were selected. Four WPP1 (lines 1 to 4) and two WPP2 (lines 5 and 6) RNAi lines were used for further analysis because they showed absence of the WPP protein by immunoblot analysis. For RNAi/PCYCB1;1:GUS lines, kanamycin (50 μg/mL) and hygromycin (25 μg/mL) double resistant lines were used. For GFP lines, BASTA (10 μg/mL) resistant lines expressing GFP were used.
GUS Staining
Histochemical staining of GUS expression was performed as described by Jefferson et al. (1987).
Supplementary Material
Acknowledgments
We would like to thank Maureen Petersen and Desouky Ammar for their technical help and advice in electron microscopy, Biao Ding for generous user time of his confocal microscope, Desh Pal Verma for the Arabidopsis cell suspension culture, and Jyan-Chyun Jang and John L. Celenza for the PCYCB1;1:GUS line. We thank Diane Furtney for expert manuscript editing. Financial support by the National Science Foundation (MCB-0079577, MCB-209339, and MCB-0343167) and the USDA (Plant Growth and Development 2001-01901) to I.M. is greatly acknowledged.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Iris Meier (meier.56@osu.edu).
Online version contains Web-only data.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.104.026740.
References
- Allen, N.P., Huang, L., Burlingame, A., and Rexach, M. (2001). Proteomic analysis of nucleoporin interacting proteins. J. Mol. Biol. 276, 29268–29274. [DOI] [PubMed] [Google Scholar]
- Altschul, S.F., Madden, T.L., Schaffer, A.A., Zhang, J., Zhang, Z., Miller, W., and Lipman, D.J. (1997). Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bednarek, S.Y., and Falbel, T.G. (2002). Membrane trafficking during plant cytokinesis. Traffic 3, 621–629. [DOI] [PubMed] [Google Scholar]
- Brandizzi, F., Irons, S.L., and Evans, D.E. (2004). The plant nuclear envelope: New prospects for a poorly understood structure. New Phytol. 163, 227–246. [DOI] [PubMed] [Google Scholar]
- Campbell, M.S., Chan, G.K.T., and Yan, T.J. (2001). Mitotic checkpoint protein HsMAD1 and HsMAD2 are associated with nuclear pore complexes in interphase. J. Cell Sci. 114, 953–963. [DOI] [PubMed] [Google Scholar]
- Canaday, J., Stoppin-Mellet, V., Mutterer, J., Lambert, A.M., and Schmit, A.C. (2000). Higher plant cells: Gamma-tubulin and microtubule nucleation in the absence of centrosomes. Microsc. Res. Tech. 49, 487–495. [DOI] [PubMed] [Google Scholar]
- Clough, S.J., and Bent, A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743. [DOI] [PubMed] [Google Scholar]
- Cronshaw, J.M., Krutchinsky, A.N., Zhang, W., Chait, B.T., and Matunis, M.J. (2002). Proteomic analysis of the mammalian nuclear pore complex. J. Cell Biol. 5, 915–927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cronshaw, J.M., and Matunis, M.J. (2003). The nuclear pore complex protein ALADIN is mislocalized in triple A syndrome. Proc. Natl. Acad. Sci. USA 100, 5823–5827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cronshaw, J.M., and Matunis, M.J. (2004). The nuclear pore complex: Disease associations and functional correlations. Trends Endocrinol. Metab. 15, 34–39. [DOI] [PubMed] [Google Scholar]
- Cutler, S.R., and Ehrhardt, D.W. (2002). Polarized cytokinesis in vacuolate cells of Arabidopsis. Proc. Natl. Acad. Sci. USA 99, 2812–2817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixit, R., and Cyr, R.J. (2002. a). Spatio-temporal relationship between nuclear-envelope breakdown and preprophase band disappearance in cultured tobacco cells. Protoplasma 219, 116–121. [DOI] [PubMed] [Google Scholar]
- Dixit, R., and Cyr, R.J. (2002. b). Golgi secretion is not required for marking the preprophase band site in cultured tobacco cells. Plant J. 29, 99–108. [DOI] [PubMed] [Google Scholar]
- Dixit, R., and Cyr, R.J. (2003). Cell damage and reactive oxygen species production induced by fluorescence microscopy: Effect on mitosis and guidelines for non-invasive fluorescence microscopy. Plant J. 36, 280–290. [DOI] [PubMed] [Google Scholar]
- Donnelly, P.M., Bonetta, D., Tsukaya, H., Dengler, R.E., and Dengler, N.G. (1999). Cell cycling and cell enlargement in developing leaves of Arabidopsis. Dev. Biol. 215, 407–419. [DOI] [PubMed] [Google Scholar]
- Erhardt, M., Stoppin-Mellet, V., Campagne, S., Canaday, J., Mutterer, J., Fabian, T., Sauter, M., Muller, T., Peter, C., Lambert, A.M., and Schmit, A.C. (2002). The plant Spc98p homologue colocalizes with gamma-tubulin at microtubule nucleation sites and is required for microtubule nucleation. J. Cell Sci. 115, 2423–2431. [DOI] [PubMed] [Google Scholar]
- Fahrenkrog, B., and Aebi, U. (2003). The nuclear pore complex: Nucleocytoplasmic transport and beyond. Nat. Rev. Mol. Cell Biol. 4, 757–766. [DOI] [PubMed] [Google Scholar]
- Gindullis, F., Peffer, N.J., and Meier, I. (1999). MAF1, a novel plant protein interacting with matrix attachment region binding protein MFP1, is located at the nuclear envelope. Plant Cell 11, 1755–1768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hepler, P.K., Valster, A., Molchan, T., and Vos, J.W. (2002). Roles for kinesin and myosin during cytokinesis. Philos. Trans. R. Soc. Lond. B Biol. Sci. 357, 761–766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iouk, T., Kerscher, O., Scott, R.J., Basrai, M.A., and Wozniak, R.W. (2002). The yeast nuclear pore complex functionally interacts with components of the spindle assembly checkpoint. J. Cell Biol. 159, 807–819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irons, S.L., Evans, D.E., and Brandizzi, F. (2003). The first 238 amino acids of the human lamin B receptor are targeted to the nuclear envelope in plants. J. Exp. Bot. 54, 943–950. [DOI] [PubMed] [Google Scholar]
- Jefferson, R.A., Kavanagh, T.A., and Bevan, M.W. (1987). GUS fusions: β-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 6, 3901–3907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong, S.Y., Rose, A., and Meier, I. (2003). MFP1 is a thylakoid-associated, nucleoid-binding protein with a coiled-coil structure. Nucleic Acids Res. 31, 5175–5185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, D.T. (1999). Protein secondary structure prediction based on position-specific scoring matrices. J. Mol. Biol. 292, 195–202. [DOI] [PubMed] [Google Scholar]
- Joseph, J., Liu, S.T., Jablonski, S.A., Yen, T.J., and Dasso, M. (2004). The RanGAP1-RanBP2 complex is essential for microtubule-kinetochore interactions in vivo. Curr. Biol. 14, 611–617. [DOI] [PubMed] [Google Scholar]
- Joseph, J., Tan, S.H., Karpova, T.S., McNally, J.C., and Dasso, M. (2002). SUMO-1 targets RanGAP1 to kinetochores and mitotic spindles. J. Cell Biol. 156, 595–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerscher, O., Hieter, P., Winey, M., and Basrai, M.A. (2001). Novel role for a Saccharomyces cerevisiae nucleoporin, Nup170p, in chromosome segregation. Genetics 157, 1543–1553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makhnevych, T., Lusk, C.P., Anderson, A.M., Aitchison, J.D., and Wozniak, R.W. (2003). Cell cycle regulated transport controlled by alterations in the nuclear pore complex. Cell 115, 813–823. [DOI] [PubMed] [Google Scholar]
- Masuda, K., Haruyama, S., and Fujino, K. (1999). Assembly and disassembly of the peripheral architecture of the plant cell nucleus during mitosis. Planta 210, 165–167. [DOI] [PubMed] [Google Scholar]
- Masuda, K., Xu, Z.-J., Takahashi, S., Ito, A., Ono, M., Nomura, K., and Inoue, M. (1997). Peripheral framework of carrot cell nucleus contains a novel protein predicted to exhibit a long alpha-helical domain. Exp. Cell Res. 232, 173–181. [DOI] [PubMed] [Google Scholar]
- Mattaj, I.W. (2004). Sorting out the nuclear envelope from the endoplasmic reticulum. Nat. Rev. Mol. Cell Biol. 5, 65–69. [DOI] [PubMed] [Google Scholar]
- Mattout-Drubezki, A., and Gruenbaum, Y. (2003). Dynamic interactions of nuclear lamina proteins with chromatin and transcriptional machinery. Cell. Mol. Life Sci. 60, 2052–2063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matunis, M.J., Coutavas, E., and Blobel, G. (1996). A novel ubiquitin-like modification modulates the partitioning of the Ran-GTPase-activating protein RanGAP1 between the cytosol and the nuclear pore complex. J. Cell Biol. 135, 1457–1470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matunis, M.J., Wu, J., and Blobel, G. (1998). SUMO-1 modification and its role in targeting the Ran GTPase-activating protein, RanGAP1, to the nuclear pore complex. J. Cell Biol. 140, 499–509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer, U., and Jürgens, G. (2004). Cytokinesis: Lines of division taking shape. Curr. Opin. Plant Biol. 7, 599–604. [DOI] [PubMed] [Google Scholar]
- McGuffin, L.J., Bryson, K., and Jones, D.T. (2000). The PSIPRED protein structure prediction server. Bioinformatics 16, 404–405. [DOI] [PubMed] [Google Scholar]
- Meier, I. (2000). A novel link between Ran signal transduction and nuclear envelope proteins in plants. Plant Physiol. 124, 1507–1510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merkle, T. (2004). Nucleo-cytoplasmic partitioning of proteins in plants: Implications for the regulation of environmental and developmental signaling. Curr. Genet. 44, 231–260. [DOI] [PubMed] [Google Scholar]
- Müller, S., Smertenko, A., Wagner, V., Heinrich, M., Hussey, P.J., and Hauser, M.T. (2004). The plant microtubule-associated protein AtMAP65-3/PLE is essential for cytokinetic phragmoplast function. Curr. Biol. 14, 412–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nacry, P., Mayer, U., and Jürgens, G. (2000). Genetic dissection of cytokinesis. Plant Mol. Biol. 43, 719–733. [DOI] [PubMed] [Google Scholar]
- Nebenführ, A., Frohlick, J.A., and Staehelin, L.A. (2000). Redistribution of Golgi stacks and other organelles during mitosis and cytokinesis in plant cells. Plant Physiol. 124, 135–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishihama, R., Soyano, T., Ishikawa, M., Araki, S., Tanaka, H., Asada, T., Irie, K., Ito, M., Terada, M., Banno, H., Yamazaki, Y., and Machida, Y. (2002). Expansion of the cell plate in plant cytokinesis requires a kinesin-like protein/MAPKKK complex. Cell 109, 87–99. [DOI] [PubMed] [Google Scholar]
- Pan, R., Lee, Y.R., and Liu, B. (2004). Localization of two homologous Arabidopsis kinesin-related proteins in the phragmoplast. Planta 10.1007/s00425–004–1324–4. [DOI] [PubMed]
- Pay, A., Resch, K., Frohnmeyer, H., Fejes, E., Nagy, F., and Nick, P. (2002). Plant RanGAPs are localized at the nuclear envelope in interphase and associated with microtubules in mitotic cells. Plant J. 30, 699–709. [DOI] [PubMed] [Google Scholar]
- Rose, A., and Meier, I. (2001). A domain unique to plant RanGAP is responsible for its targeting to the plant nuclear rim. Proc. Natl. Acad. Sci. USA 98, 15377–15382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose, A., Patel, S., and Meier, I. (2004). The plant nuclear envelope. Planta 218, 327–336. [DOI] [PubMed] [Google Scholar]
- Ryan, K.J., McCaffery, M., and Wente, S.R. (2003). The Ran GTPase cycle is required for yeast nuclear pore complex assembly. J. Cell Biol. 160, 1041–1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salina, D., Enarson, P., Rattner, J.B., and Burke, B. (2003). Nup358 integrates nuclear envelope breakdown with kinetochore assembly. J. Cell Biol. 162, 991–1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook, J., Fritsch, E., and Maniatis, T. (1989). Molecular Cloning: A Laboratory Manual. (Cold Spring Harbor, NY: Cold Spring Harbor Press).
- Schmit, A.C. (2002). Acentrosomal microtubule nucleation in higher plants. Int. Rev. Cytol. 220, 257–289. [DOI] [PubMed] [Google Scholar]
- Seguí-Simarro, J.M., Austin II, J.R., White, E.A., and Staehelin, L.A. (2004). Electron tomographic analysis of somatic cell plate formation in meristematic cells of Arabidopsis preserved by high-pressure freezing. Plant Cell 16, 836–856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, L. (2002). Plant cytokinesis: Motoring to the finish. Curr. Biol. 12, R206–R208. [DOI] [PubMed] [Google Scholar]
- Staehelin, L.A., and Hepler, P.K. (1996). Cytokinesis in higher plants. Cell 84, 821–824. [DOI] [PubMed] [Google Scholar]
- Strompen, G., El Kasmi, F., Richter, S., Lukowitz, W., Assaad, F.F., Jurgens, G., and Mayer, U. (2002). The Arabidopsis HINKEL gene encodes a kinesin-related protein involved in cytokinesis and is expressed in a cell cycle-dependent manner. Curr. Biol. 12, 153–158. [DOI] [PubMed] [Google Scholar]
- Vasu, S.K., and Forbes, D.J. (2001). Nuclear pores and nuclear assembly. Curr. Opin. Cell Biol. 13, 363–375. [DOI] [PubMed] [Google Scholar]
- von Arnim, A., Deng, X., and Stacey, M. (1998). Cloning vectors for the expression of green fluorescent protein fusion proteins in transgenic plants. Gene 221, 35–43. [DOI] [PubMed] [Google Scholar]
- Waizenegger, I., Lukowitz, W., Assaad, F., Schwarz, H., Jürgens, G., and Mayer, U. (2000). The Arabidopsis KNOLLE and KEULE genes interact to promote vesicle fusion during cytokinesis. Curr. Biol. 10, 1371–1374. [DOI] [PubMed] [Google Scholar]
- Werner, T., Motyka, V., Laucou, V., Smets, R., Van Onckelen, H., and Schmülling, T. (2003). Cytokinin-deficient transgenic Arabidopsis plants show multiple developmental alterations indicating opposite functions of cytokinins in the regulation of shoot and root meristem activity. Plant Cell 15, 2532–2550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, J., Matunis, M.J., Kraemer, D., Blobel, G., and Coutavas, E. (1995). Nup358, a cytoplasmically exposed nucleoporin with peptide repeats, Ran-GTP binding sites, zinc fingers, a cyclophilin A homologous domain, and a leucine-rich region. J. Biol. Chem. 9, 14209–14213. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.