In this study, Huang et al. investigated the mechanisms by which chromobox homolog 3 (Cbx3) stimulates cell differentiation. They found that Cbx3 binds to gene promoters upon differentiation of murine ESCs to neural progenitors and recruits the Mediator subunit Med26, thus demonstrating that Cbx3 and Med26 together ensure the fidelity of lineage specification.
Keywords: Cbx3, Med26, embryonic stem cell, neural precursor, preinitiation complex, mesoderm
Abstract
Chromobox homolog 3 (Cbx3/heterochromatin protein 1γ [HP1γ]) stimulates cell differentiation, but its mechanism is unknown. We found that Cbx3 binds to gene promoters upon differentiation of murine embryonic stem cells (ESCs) to neural progenitor cells (NPCs) and recruits the Mediator subunit Med26. RNAi knockdown of either Cbx3 or Med26 inhibits neural differentiation while up-regulating genes involved in mesodermal lineage decisions. Thus, Cbx3 and Med26 together ensure the fidelity of lineage specification by enhancing the expression of neural genes and down-regulating genes specific to alternative fates.
Chromobox homolog 3 (Cbx3) is a member of the heterochromatin protein 1 (HP1) family that binds to dimethylated and trimethylated histone 3 Lys9 (H3K9) via a conserved chromodomain. Cbx3 (HP1γ) and its homologs, Cbx5 (HP1α) and Cbx1 (HP1β), have been implicated in both the repression and activation of gene transcription (Kwon and Workman 2011; Canzio et al. 2014). In Drosophila melanogaster, Cbx3 associates with both the histone chaperone FACT and RNA polymerase II (Pol II) (Kwon et al. 2010). Cbx3 occupies the transcribed regions of genes in embryonic stem cells (ESCs) and cancer cell lines, where it influences splicing and correlates with H3K9 methylation (Smallwood et al. 2012; Sridharan et al. 2013). However, Cbx3 is also enriched at promoters of mouse embryonic fibroblast (MEF)-derived pre-iPSCs (preinduced pluripotent stem cells), where it does not correlate with H3K9 methylation (Sridharan et al. 2013). RNAi knockdown of Cbx3 promotes reprogramming of fibroblasts to iPSCs (Sridharan et al. 2013).
Cbx3 plays important roles in developmental processes (Morikawa et al. 2013). In model systems, Cbx3 promotes neuronal maturation, kidney development, differentiation of ESCs to smooth muscle in culture, and embryonic arteriogenesis (Xiao et al. 2011; Dihazi et al. 2015; Oshiro et al. 2015). Little is known of how Cbx3 functions in differentiation, but its promoter localization suggested that it might directly affect transcription through the preinitiation complex (PIC) (Grunberg and Hahn 2013; Allen and Taatjes 2015). The PIC is assembled in response to activators and requires the Mediator coactivator complex to recruit the general transcription factors and Pol II (Chen et al. 2012). Core Mediator contains 25 subunits constituting three structurally distinct modules distributed into two major complexes: One bears Med26 and Pol II, and the other bears a four-subunit Cdk8 module (Malik and Roeder 2010; Conaway and Conaway 2013; Tsai et al. 2014; Allen and Taatjes 2015). The specific function of Med26 is not fully established, although it recruits the Pol II superelongation complex containing several ELL/EAF family members, whose misregulation has been associated with cancer (Takahashi et al. 2011; Zhou et al. 2012). D. melanogaster HP1 interacts directly with Med26 (Marr et al. 2014), and Cbx3 is enriched in Mediator preparations from differentiated neural precursors (Sridharan et al. 2013).
The observation that Cbx3 enriches in ESC gene bodies and pre-iPSC promoters suggested that the switch in distribution could influence gene regulation during differentiation. To determine whether this is Cbx3's mechanism of action, we used a model system in which ESCs were efficiently induced to differentiate into neural precursor cells (NPCs) (Brustle et al. 1997). The positioning and role of Cbx3 were interrogated by chromatin immunoprecipitation (ChIP) and RNAi. Our analysis revealed that Cbx3 and Med26 colocalize to promoters of both neural and mesodermal genes during differentiation and have complementary roles in stabilizing NPC fate. Decreasing Med26 or Cbx3 levels early in differentiation led to the down-regulation of neural lineage genes and up-regulation of mesodermal lineage genes. Increased binding of Cdk8 correlated with decreased mesodermal gene expression during normal differentiation and decreased neural gene expression upon Cbx3 depletion.
Results and Discussion
Cbx3 binds to gene promoters during differentiation
To address whether Cbx3 becomes enriched at promoters upon differentiation, we used ITSF medium to convert ESCs to NPCs and performed genome-wide ChIP-seq (ChIP combined with high-throughput sequencing) of Cbx3 in NPCs. Differentiation was validated by immunostaining for Oct4 in the ESCs and Nestin in the ESC-derived NPCs. Nestin-positive NPCs were able to differentiate into Tuj1+ neurons upon EGF and FGF2 withdrawal (Supplemental Fig. S1A).
Statistical analyses of Cbx3 ChIP-seq peaks showed that it reproducibly enriches at the promoters of genes in NPCs (Fig. 1A) and less so in gene bodies (Fig. 1B; Supplemental Fig. S1B). This binding pattern is distinct from ESCs, where Cbx3 bound primarily within gene bodies (Sridharan et al. 2013). Although enrichment of Pol II at the promoter correlates roughly with that of Cbx3 in the center of the distribution (Fig. 1A), there was only a modest correlation with gene expression when considering the top, middle, and bottom Cbx3-bound genes (Fig. 1C). Therefore, Cbx3 enrichment at the promoter during differentiation does not correlate with increased gene expression.
Figure 1.
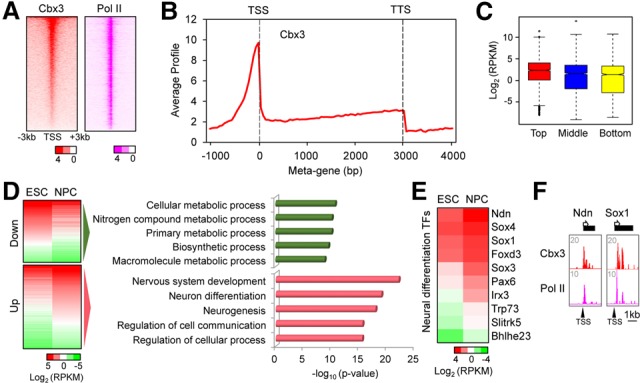
Association of Cbx3 with promoters during neural differentiation. (A) Heat map of Cbx3 and Pol II enrichment at NPC promoters. P-values of enrichment were plotted and ranked by Cbx3. (B) Metagene profile of Cbx3 enrichment in NPCs. The Y-axis is the average signal from 50-base-pair windows. (C) Box plot of gene expression level in high (red), middle (blue), and bottom (yellow) Cbx3-bound genes. Log2 expression levels are reads per kilobase per million mapped reads (RPKM) from NPC RNA sequencing (RNA-seq) data. Bound genes contained Cbx3 within a ±1-kb region flanking the transcription start site (TSS). (D, left) Gene expression heat maps of Cbx3-bound and either down-regulated or up-regulated genes during differentiation from ESCs to NPCs. (Right) Gene ontology analysis for Cbx3-bound and down genes or up genes. Regulated genes displayed a >1.5-fold change in NPCs versus ESCs. (E) The RNA-seq heat map shows Cbx3-bound up-regulated neural differentiation transcription factors (TFs) from the term “nervous system development” in D. (F) Browser track examples of Cbx3 and Pol II cobound transcription factor promoters in NPCs.
We then investigated differentiation-regulated genes bound by Cbx3 at the promoter. Gene ontology (GO) analysis revealed that genes down-regulated during differentiation were required for cellular metabolism, a characteristic of rapidly growing self-renewing ESCs (Fig. 1D, top panels). In contrast, genes involved in nervous system development and neural differentiation were up-regulated (Fig. 1D, bottom panels), including signaling molecules and transcription factors; i.e., Sox1 and Foxd3 (Fig. 1E). Browser plots of select neural fate genes showed colocalization of Pol II and Cbx3 at the promoter (Fig. 1F; Supplemental Fig. S1C). We conclude that Cbx3 localizes to promoters of differentiation-regulated genes and associates with gene activity, while its binding negatively correlates with expression of genes involved in ESC growth.
Cbx3 knockdown decreases neural differentiation and up-regulates mesodermal genes
To further explore the role of Cbx3 during differentiation, we used RNAi knockdown (Cbx3 knockdown) in either monolayer culture (Fig. 2A,B; Supplemental Fig. S2A) or embryoid body (EB) culture during neural differentiation (Fig. 2C,D). In monolayer culture, Sox1, an early marker of neural fate, appeared in most cells by day 8 (Fig. 2A; Pevny et al. 1998). Although the total cell number did not change significantly upon Cbx3 knockdown, the expression of Sox1 significantly decreased (Fig. 2A,B). Analysis of an earlier time point, day 7, revealed little Sox1 expression in the control and virtually no effect of Cbx3 knockdown (Supplemental Fig. S2B,C). To confirm the function of Cbx3, we used serum-free EBs (SFEBs), an alternate system to analyze ESC neural differentiation, and found a significant decrease in SFEBs upon Cbx3 knockdown (Fig. 2C,D; Watanabe et al. 2005; Huang et al. 2010). Collectively, these data suggest that Cbx3 positively regulates neural fate determination.
Figure 2.
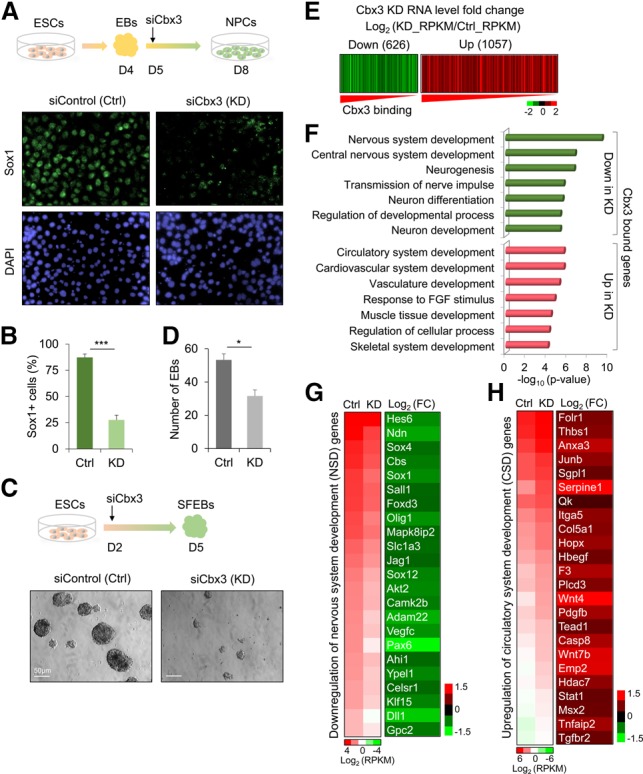
Cbx3 knockdown decreases neural and up-regulates mesodermal gene expression. (A) Cbx3 knockdown (Cbx3 KD) inhibits Sox1+ NPC generation. (Top panel) EBs formed from ESCs in medium without leukemia-inhibiting factor (−LIF). On day 4, EBs were dissociated and plated with ITSF medium for monolayer NPC generation. One day after initiating monolayer differentiation, control siRNA or Cbx3 siRNAs were transfected. (Bottom panel) Sox1 immunostaining (green) in control or Cbx3 knockdown. DAPI (blue) marks nuclei. (B) Percentage of Sox1+ versus DAPI+ cells 72 h after siRNA. (***) P < 0.005, Student's t-test. (C) SFEB formation assay for ESC neural differentiation in control (Ctrl) and Cbx3 knockdown. EBs were formed in neural induction medium. On day 2 after induction, siControl or siCbx3 RNA was transfected; total EBs were determined on day 5. (D) Quantification of EBs 3 d after siRNA transfection. (*) P < 0.05, Student's t-test. (E) Heat map of Cbx3-bound and down-regulated (green; <1.5-fold) or up-regulated (red; >1.5-fold) genes in Cbx3 knockdown versus control cells. Log2 (KD_RPKM/Ctrl_RPKM) for RNA level fold changes was plotted and ranked by Cbx3 enrichment. (F) GO analysis for 626 down-regulated genes and 1057 up-regulated genes from E. (G) Heat maps display RNA level decreases and fold change (FC) for the term “nervous system development” in F. (H) Heat maps show RNA level increase and fold change for the term “circulatory system development” in F.
To investigate the regulatory mechanisms of Cbx3 in differentiation, we performed genome-wide RNA sequencing (RNA-seq) analysis on NPCs 3 d after siControl or siCbx3 treatment (Fig. 2A). We used monolayer NPCs because the differentiation status in culture was more uniform versus SFEBs. Figure 2E is an expression heat map that plots Cbx3-bound genes down-regulated or up-regulated >1.5-fold upon siCbx3 treatment versus cells treated with the siControl. GO analysis of these Cbx3 target genes revealed that down-regulated genes were largely enriched in nervous system development and function (Fig. 2F) and included key neural lineage genes such as Hes6 and Olig1 (Fig. 2G). Remarkably, up-regulated genes represented mainly mesodermal lineage specification categories, including development of the circulatory system, vasculature, muscle, and skeleton. Figure 2H is a heat map of up-regulated genes with known roles in circulatory system development. A time course of the Cbx3 knockdown effect on the expression of select differentiation-affected genes from Figure 2, G and H, revealed altered expression in monolayer cells by 72 h but not earlier (Supplemental Fig. S2D).
Cbx3 knockdown in pluripotent ESCs versus differentiated cells demonstrated that disruption of neural fate determination specifically led to the appearance of mesodermal lineage gene expression. Supplemental Figure S2E shows that the profiles of genes up-regulated and down-regulated in ESCs upon Cbx3 knockdown are distinct and have lower significance than the mesodermal genes appearing during differentiation into NPCs. The Cbx3-bound up-regulated genes in NPCs, in addition to circulatory system development, included canonical mesodermal lineage genes such as Tnnt3, Tbx20, Tbx3, Hand1, and Pdgfra involved in cardiac lineage development and Hoxa5, Runx1, and Hoxb5 involved in hematopoietic lineage development (Supplemental Fig. S2F,G). In conclusion, the gene expression data show that Cbx3 knockdown has a distinct effect on NPC differentiation, favoring down-regulation of neural genes and up-regulation of mesodermal genes, although we have no evidence that this would eventually lead to functional mesoderm.
Cbx3 recruits Med26 to the PIC in vitro
The enrichment of Cbx3 at the promoter in NPCs suggested that it might associate with the PIC to regulate differentiation. We used the immobilized template assay to capture PICs assembled from ESC and NPC nuclear extracts using the Gal4-VP16 model activator (Fig. 3A; Chen et al. 2012; Lin and Carey 2012). We focused on Cbx3, Med26, and Cdk8, the latter two representing distinct forms of Mediator. The data revealed that Cbx3 was indeed recruited to a promoter in vitro in a Gal4-VP16-dependent manner with a strong enrichment in NPC versus ESC extracts (Fig. 3B). Med26 was enriched slightly in NPC PICs relative to input, but Cdk8 levels were low in both input and NPC PICs. We then assembled PICs from ESC extracts in the presence of increasing amounts of recombinant Cbx3 (Fig. 3C). As the concentration of Cbx3 increased, a decrease in Cdk8 was observed (Smallwood et al. 2008) but was accompanied by an increase in binding of Med26. A control Mediator subunit (Med6) and another PIC component (TBP) were unaltered. In sum, Cbx3 altered PIC composition to favor Med26 binding.
Figure 3.
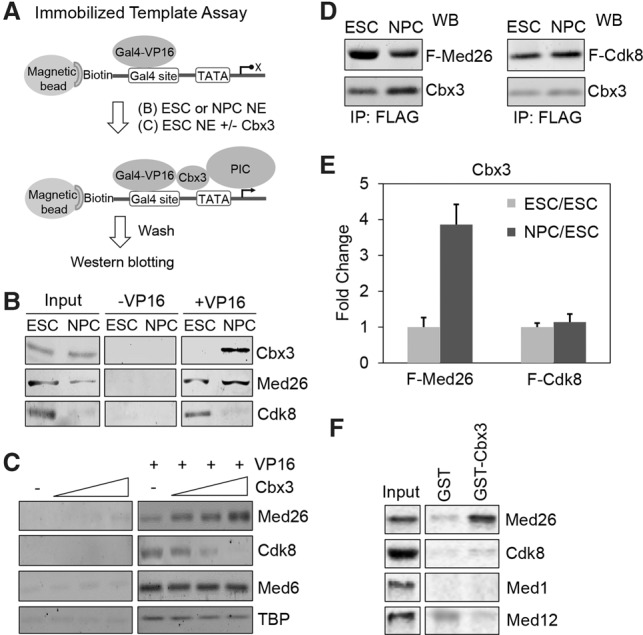
Cbx3 regulates recruitment of Med26 to the PIC in vitro. (A) Schematic of the immobilized template PIC capture assay using ESC or NPC nuclear extracts (NE) plus or minus Gal4-VP16 activator (see the Materials and Methods). (B) Recruitment of Cbx3, Med26, and Cdk8 in the ESC PIC or NPC PIC by Western blotting. (C). ESC nuclear extract was incubated with increasing amounts of purified Cbx3 in the immobilized template PIC capture assay. Western blots show Med26, Cdk8, Med6, and TBP recruitment. (D) Western blots (WB) comparing the amounts of Cbx3 from purified Flag-tagged Med26 (F-Med26) and Flag-tagged Cdk8 (F-Cdk8) in ESCs and NPCs. (E) Quantification of fold change of Cbx3 amounts in F-Med26 Mediator and F-Cdk8 Mediator during differentiation from ESCs to NPCs, respectively. (F) Binding of in vitro transcribed and translated 35S-labeled Med26, Cdk8, Med1, and Med12 to glutathione-sepharose bound GST-Cbx3 or GST.
Cbx3 was preferentially enriched in the Med26 Mediator. Flag-tagged Med26 (N-Flag) and Cdk8 (C-Flag) were expressed in doxycycline-inducible ESC lines and isolated by immunoaffinity purification (Supplemental Fig. S3A). MuDPIT (multidimensional protein identification technology) analysis of the ESC Mediators showed that their subunit composition was similar except for an abundance of the tagged subunit and relative depletion of the Cdk8 module in Flag-Med26 and vice versa (Supplemental Fig. S3B). A similar purification was then performed on ESC-derived NPCs. The immunoblots in Figure 3D and graphs in Figure 3E show that the relative amount of Cbx3 increased fourfold in NPC versus ESC Flag-Med26 Mediator, whereas the ratio of Cbx3 to Flag-Cdk8 Mediator remained the same. 35S-labeled Med26 generated by in vitro transcription and translation bound preferentially to GST-Cbx3 beads (Fig. 3F), whereas Cdk8, Med1, and Med12 did not. Collectively, we found that Cbx3 is localized at the promoter after differentiation and is enriched in a Med26 form of Mediator. We conclude that Cbx3 associates with Med26 in vitro.
Cbx3 stabilizes Med26 binding to the PIC in vivo
To determine whether Cbx3 affected Med26 recruitment in vivo, we performed ChIP-seq of Med26, Cdk8, and Pol II in the siControl-treated and Cbx3 knockdown NPCs. Cbx3 knockdown significantly decreased Med26 enrichment at promoters (−3 to +3 kb surrounding the transcription start site [TSS]) genome-wide when visualizing the data as either heat maps (Fig. 4A), the average profile (Fig. 4B), or individual browser plots (Fig. 4C). The decrease in Med26 recruitment was not an indirect effect on Med26 gene expression or protein levels (Supplemental Fig. S4A,B). Furthermore, there was little global effect of Cbx3 knockdown on Cdk8 enrichment at the promoter, albeit there was a modest decrease in Pol II (Fig. 4A,B). Collectively, the data suggest that Med26 but not Cdk8 binds cooperatively with Cbx3 at target genes in vivo.
Figure 4.
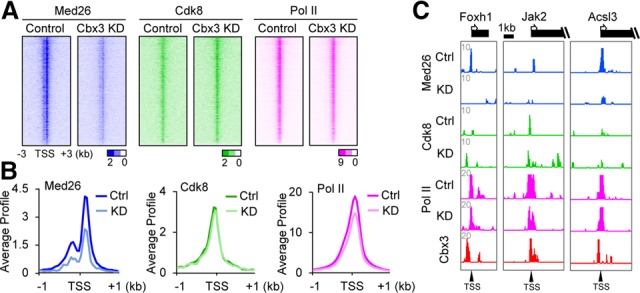
Genome-wide analysis of Cbx3 knockdown on Med26 promoter binding. (A) Heat maps of Med26, Cdk8, and Pol II at promoters in control or Cbx3 knockdown cells. P-values of enrichment were plotted and ranked by Cbx3 enrichment near the TSS. (B) Average gene profiles of Med26, Cdk8, and Pol II in control or Cbx3 knockdown NPCs. ChIP-seq signals for average distributions were plotted by significant peaks ±1 kb around the TSS. P-values for differences between control and Cbx3 knockdown were P < 2.2 × 10−16, P = 4.67 × 10−05, and P < 2.2 × 10−16, calculated by Wilcoxon rank-sum test. (C) Browser tracks show examples of Med26-, Cdk8-, and Pol II-binding changes around TSSs of neural lineage genes.
Given that the Med26 and Cdk8 forms of Mediator have different properties in vivo and in vitro (Conaway and Conaway 2011), we explored Cdk8 binding in more detail. Med26 and Cdk8 appear to display differential positioning around the TSS (Supplemental Fig. S4C). Moreover, although the average profiles of Cbx3 and Med26 in siControl-treated NPCs remained similar between promoters of Cbx3-activated and Cbx3-repressed genes, Cdk8 was enriched at promoters of Cbx3-repressed genes; i.e., those up-regulated upon Cbx3 knockdown (Supplemental Fig. S4D). These Cdk8-enriched genes were typically less active than those positively affected by Cbx3 (Supplemental Fig. S4E).
Interestingly, while the loss of Cbx3 did not change the average genomic profile of Cdk8, in many instances, it did lead to its redistribution. Med26 binding at the promoters of select nervous system development genes was typically decreased upon Cbx3 knockdown (Supplemental Fig. S4F). When specific groups of genes were surveyed, such as nervous system development genes that become deactivated upon Cbx3 knockdown (Fig. 2G), we found that decreased Med26 levels correlated with enrichment of Cdk8 (Supplemental Fig. S4G, NSD). This enrichment was clear in browser tracks of select nervous system development genes; i.e., Olig1 and Jag1 (Supplemental Fig. S4H). Conversely, when we considered the circulatory system development genes (Fig. 2H) that become derepressed upon Cbx3 knockdown, we observed decreased Cdk8 recruitment after knockdown (Supplemental Fig. S4G, CSD). The redistribution of Cdk8 is clearly evident in browser tracks of select circulatory system development genes; i.e., Itga5 and Thbs1 (Supplemental Fig. S4H). Collectively, these results suggest that Cdk8 binding correlates with decreased transcription.
Med26 is involved in Cbx3-regulated neural differentiation
Based on the above results, we hypothesized that coordination between Med26 and Cbx3 modulates gene expression during neural differentiation. To test this, we performed RNA-seq analysis in monolayer-differentiated cells 72 h after siMed26 RNA transfection (Supplemental Fig. S5). GO analysis of genes down-regulated by both Cbx3 and Med26 included genes involved in neurogenesis, as would be predicted if Med26 was necessary for transcription during neural differentiation (Fig. 5A). GO analysis of the overlapping up-regulated genes revealed that many of these genes were associated with mesodermal lineage development, including key genes controlling cardiac and hematopoietic specification (Fig. 5B). To confirm the cellular function of Med26 in differentiation, we analyzed either EB formation in SFEB culture (Fig. 5C,D) or the generation of Sox1+ NPCs in monolayer culture after Med26 knockdown (Fig. 5E,F). The data from two different cellular assays showed that Med26 knockdown impaired neural differentiation of ESCs (Fig. 5D,F). The mechanistic and functional data lead us to conclude that Med26 contributes to Cbx3-regulated gene transcription programs and positively regulates neural lineage specification.
Figure 5.
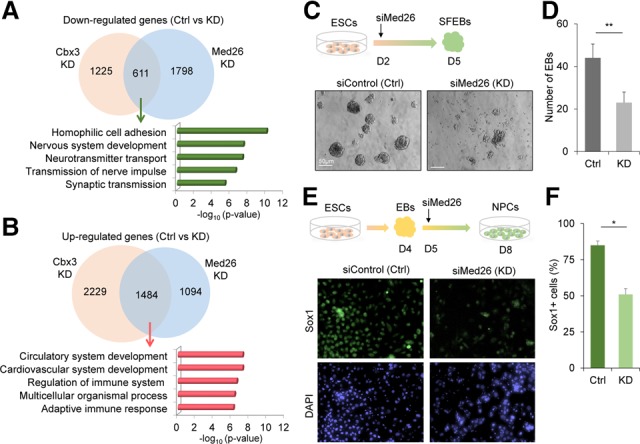
Med26 is involved in Cbx3-regulated neural differentiation. (A,B) Venn diagrams of overlap between down-regulated (A) or up-regulated (B) genes upon Cbx3 or Med26 knockdown shown above GO analysis for overlapping genes. (C) EB formation assay for neural differentiation analysis upon Med26 knockdown. ESCs (1 × 105 cells) were dissociated into single cells to form EBs in neural induction medium. On day 2 after induction, siRNA control or Med26 siRNA (Med26 knockdown) were transfected. The total number of EBs was determined on day 5. (D) Quantification of the EB number 72 h after siRNA treatments. (**) P < 0.01, Student's t-test. (E) Med26 knockdown inhibits Sox1+ NPC generation. On day 5 of differentiation, siControl or siMed26 was transfected into the cells. Immunostaining for Sox1+ cells was performed on day 8. (F) Quantification of percentage of Sox1-positive cells versus DAPI-positive cells for the immunostaining. (*) P < 0.05, Student's t-test.
Our findings suggest that Cbx3 is necessary for differentiation of ESCs to NPCs. Cbx3 binding to the promoters of differentiation-related genes regulates the neural and mesodermal gene expression programs during cell fate determination. Neuromesodermal progenitors that behave as multipotent cells or axial stem cells give rise to both neuroectoderm and mesoderm during mouse embryo development (Wilson et al. 2009). Cell signaling pathways, including Wnt, regulate the differentiation of neuromesodermal progenitors (Gouti et al. 2014; Garriock et al. 2015). Transcription factors, including Tbx6, positively regulate mesodermal fate determination in axial stem cells by inhibiting neural differentiation (Takemoto et al. 2011). As such, Cbx3 binding to promoters may be a means of maintaining lineage specificity during differentiation while also conferring plasticity to transition to the next developmental stage. The discovery of complementary functions of Cbx3 and Med26 in the activation of neural differentiation-related genes and repression of genes involved in mesodermal lineage development reveals an intriguing transcriptional mechanism for such stem cell fate decisions. The Med26 Mediator coactivator complex appears to be present on genes of both lineages to create poised complexes, but specific transcription activators and repressors likely modulate the final composition of the PIC to favor activation of one lineage and repression of the other. Recruitment of other PIC components such as Cdk8 help enforce lineage commitment, possibly by contributing to down-regulation of genes specific for the alternative fate.
Materials and methods
Cell culture and neural differentiation
V6.5 ESCs were cultured on irradiated MEFs in ESC medium (knockout DMEM [Thermo Fisher] containing 15% FBS, 2000 U/mL leukemia-inhibiting factor [LIF], 2 mM L-glutamine, 0.1 mM nonessential amino acids, 0.1 mM β-mercaptoethanol, penicillin/streptomycin). MEFs were cultured as described previously (Sridharan et al. 2013). ESCs were differentiated into NPCs through EB formation for 4 d. EBs were dissociated into single cells and plated in tissue culture dishes with ITSF medium for monolayer NPC generation (Okabe et al. 1996) and maintained in chemically defined medium containing FGF2 and EGF as described (Conti et al. 2005). EB formation assay for ESC neural differentiation analysis was as described (Huang et al. 2010). On day 2 after induction, siRNAs were transfected. Total EBs were determined on day 5.
Cell immunostaining
Cells were fixed in 4% formaldehyde for 10 min at room temperature, permeabilized in 0.1% Triton X-100 PBS buffer for 5 min, and stained with Sox1 (1:200; Abcam), Oct4 (1:200; Santa Cruz Biotechnology), Nestin (1:200; Abcam), or Tuj1 (1:200; Abcam). Secondary antibodies were goat anti-mouse IgG Alexa 488 (1:500; Abcam) and goat anti-rabbit IgG Alexa 594 (1:500; Abcam). DAPI was used for cell nucleus detection. Image quantification of Sox1 or DAPI fluorescence was performed using ImageJ (National Institutes of Health). Quantifications are a mean percentage calculated by dividing the Sox1-positive cell number by the DAPI-positive cell number in three replicates.
ChIP-seq
V6.5 ESCs and ESC-derived NPCs were subjected to ChIP-seq as described (Ferrari et al. 2012; Xue et al. 2015) using antibodies against Cbx3 (Millipore, 05-690; clone 42s2), Med26 (sc-48776), Cdk8 (sc-1521), or Pol II (sc-56767). All sequenced reads were mapped to mouse mm9 genome using Bowtie 0.12.9 (Langmead et al. 2009). Cbx3, Pol II, Med26, and Cdk8 were normalized to input using a custom script. The genome was divided into 50-base-pair windows, and significant windows with a P-value of <0.001 were selected as described (Ferrari et al. 2012). In Cbx3 knockdown ChIP-seq experiments, two biological replicates were analyzed to confirm the ChIP quality of Med26 (data not shown).
siRNA transfection
siRNAs (Dharmacon) were used at a final concentration of 20 µM and transfected using Lipofectamine RNAiMAX reagent (Thermo Fisher): Cbx3, MU-044218-01 #2; Med26, J-064504-12; and siControl, control D-001210-02.
RNA-seq
RNA libraries were prepared and processed as described previously (Xue et al. 2015). Alignment of RNA-seq reads to the mouse genome (mm9) was performed using TopHat (Trapnell et al. 2009). SAMMate was used for the reads per kilobase per million mapped reads (RPKM) (Xu et al. 2011). The average RPKM of two biological replicates was used for gene expression analysis. Quantitative RT–PCR of select genes was used with primers in the Supplemental table to validate the Cbx3 knockdown RNA-seq (Supplemental Fig. S2D).
Immobilized template assay
Immobilized template experiments were performed as described (Lin and Carey 2012). Antibodies included Cbx3 (Millipore, 05-690; clone 42s2), Med26 (Santa Cruz Biotechnology, sc-48776), Cdk8 (Santa Cruz Biotechnology, sc-1521), Med6 (Santa Cruz Biotechnology, sc366562), and TBP (Santa Cruz Biotechnology, sc-204). Blots were quantified using the LiCor Odyssey platform.
Analysis of purified proteins
Flag-Med26 or Cdk8 was obtained by targeting the ColA1 locus in V6.5 ESCs (Beard et al. 2006). Purification of protein complexes was as described previously (Smallwood et al. 2008; Chen et al. 2012).
MuDPIT analysis
The purified protein samples were processed and tandem mass spectrometry spectra were collected and analyzed as described (Law et al. 2010).
Accession numbers
The Gene Expression Omnibus accession ID for aligned and raw data is GSE89575.
Supplementary Material
Acknowledgments
We thank Hanna Mikkola for advice, and Fei Sun for technical assistance. This work was supported by National Institutes of Health (NIH) grant R01 GM074701 and funding from the University of California at Los Angeles (UCLA) Broad Stem Cell Center to M.C.; NIH R01 CA178415 to S.K.K., R01 NS072804 and R01 NS089817 to B.G.N., and P01 GM099134 to K.P.; and National Natural Science Foundation of China grant 81671396 to C.H. F.D.L. acknowledges support from a UCLA Quantitative and Computational Biosciences (QCB) Collaboratory Post-doctoral Fellowship and the QCB Collaboratory Community directed by Matteo Pellegrini.
Footnotes
Supplemental material is available for this article.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.292169.116.
References
- Allen BL, Taatjes DJ. 2015. The Mediator complex: a central integrator of transcription. Nat Rev Mol Cell Biol 16: 155–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beard C, Hochedlinger K, Plath K, Wutz A, Jaenisch R. 2006. Efficient method to generate single-copy transgenic mice by site-specific integration in embryonic stem cells. Genesis 44: 23–28. [DOI] [PubMed] [Google Scholar]
- Brustle O, Spiro AC, Karram K, Choudhary K, Okabe S, McKay RD. 1997. In vitro-generated neural precursors participate in mammalian brain development. Proc Natl Acad Sci 94: 14809–14814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canzio D, Larson A, Narlikar GJ. 2014. Mechanisms of functional promiscuity by HP1 proteins. Trends Cell Biol 24: 377–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen XF, Lehmann L, Lin JJ, Vashisht A, Schmidt R, Ferrari R, Huang C, McKee R, Mosley A, Plath K, et al. 2012. Mediator and SAGA have distinct roles in Pol II preinitiation complex assembly and function. Cell Rep 2: 1061–1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conaway RC, Conaway JW. 2011. Function and regulation of the Mediator complex. Curr Opin Genet Dev 21: 225–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conaway RC, Conaway JW. 2013. The Mediator complex and transcription elongation. Biochim Biophys Acta 1829: 69–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conti L, Pollard SM, Gorba T, Reitano E, Toselli M, Biella G, Sun Y, Sanzone S, Ying QL, Cattaneo E, et al. 2005. Niche-independent symmetrical self-renewal of a mammalian tissue stem cell. PLoS Biol 3: e283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dihazi GH, Jahn O, Tampe B, Zeisberg M, Muller C, Muller GA, Dihazi H. 2015. Proteomic analysis of embryonic kidney development: heterochromatin proteins as epigenetic regulators of nephrogenesis. Sci Rep 5: 13951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrari R, Su T, Li B, Bonora G, Oberai A, Chan Y, Sasidharan R, Berk AJ, Pellegrini M, Kurdistani SK. 2012. Reorganization of the host epigenome by a viral oncogene. Genome Res 22: 1212–1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garriock RJ, Chalamalasetty RB, Kennedy MW, Canizales LC, Lewandoski M, Yamaguchi TP. 2015. Lineage tracing of neuromesodermal progenitors reveals novel Wnt-dependent roles in trunk progenitor cell maintenance and differentiation. Development 142: 1628–1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gouti M, Tsakiridis A, Wymeersch FJ, Huang Y, Kleinjung J, Wilson V, Briscoe J. 2014. In vitro generation of neuromesodermal progenitors reveals distinct roles for wnt signalling in the specification of spinal cord and paraxial mesoderm identity. PLoS Biol 12: e1001937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grunberg S, Hahn S. 2013. Structural insights into transcription initiation by RNA polymerase II. Trends Biochem Sci 38: 603–611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C, Xiang Y, Wang Y, Li X, Xu L, Zhu Z, Zhang T, Zhu Q, Zhang K, Jing N, et al. 2010. Dual-specificity histone demethylase KIAA1718 (KDM7A) regulates neural differentiation through FGF4. Cell Res 20: 154–165. [DOI] [PubMed] [Google Scholar]
- Kwon SH, Workman JL. 2011. The changing faces of HP1: from heterochromatin formation and gene silencing to euchromatic gene expression: HP1 acts as a positive regulator of transcription. Bioessays 33: 280–289. [DOI] [PubMed] [Google Scholar]
- Kwon SH, Florens L, Swanson SK, Washburn MP, Abmayr SM, Workman JL. 2010. Heterochromatin protein 1 (HP1) connects the FACT histone chaperone complex to the phosphorylated CTD of RNA polymerase II. Genes Dev 24: 2133–2145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL. 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10: R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law JA, Ausin I, Johnson LM, Vashisht AA, Zhu JK, Wohlschlegel JA, Jacobsen SE. 2010. A protein complex required for polymerase V transcripts and RNA-directed DNA methylation in Arabidopsis. Curr Biol 20: 951–956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin JJ, Carey M. 2012. In vitro transcription and immobilized template analysis of preinitiation complexes. Curr Protoc Mol Biol 97: 12.14.1–12.14.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malik S, Roeder RG. 2010. The metazoan Mediator co-activator complex as an integrative hub for transcriptional regulation. Nat Rev Genet 11: 761–772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marr SK, Lis JT, Treisman JE, Marr MT II. 2014. The metazoan-specific mediator subunit 26 (Med26) is essential for viability and is found at both active genes and pericentric heterochromatin in Drosophila melanogaster. Mol Cell Biol 34: 2710–2720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morikawa K, Ikeda N, Hisatome I, Shirayoshi Y. 2013. Heterochromatin protein 1γ overexpression in P19 embryonal carcinoma cells elicits spontaneous differentiation into the three germ layers. Biochem Biophys Res Commun 431: 225–231. [DOI] [PubMed] [Google Scholar]
- Okabe S, Forsberg-Nilsson K, Spiro AC, Segal M, McKay RD. 1996. Development of neuronal precursor cells and functional postmitotic neurons from embryonic stem cells in vitro. Mech Dev 59: 89–102. [DOI] [PubMed] [Google Scholar]
- Oshiro H, Hirabayashi Y, Furuta Y, Okabe S, Gotoh Y. 2015. Up-regulation of HP1γ expression during neuronal maturation promotes axonal and dendritic development in mouse embryonic neocortex. Genes Cells 20: 108–120. [DOI] [PubMed] [Google Scholar]
- Pevny LH, Sockanathan S, Placzek M, Lovell-Badge R. 1998. A role for SOX1 in neural determination. Development 125: 1967–1978. [DOI] [PubMed] [Google Scholar]
- Smallwood A, Black JC, Tanese N, Pradhan S, Carey M. 2008. HP1-mediated silencing targets Pol II coactivator complexes. Nat Struct Mol Biol 15: 318–320. [DOI] [PubMed] [Google Scholar]
- Smallwood A, Hon GC, Jin F, Henry RE, Espinosa JM, Ren B. 2012. CBX3 regulates efficient RNA processing genome-wide. Genome Res 22: 1426–1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sridharan R, Gonzales-Cope M, Chronis C, Bonora G, McKee R, Huang C, Patel S, Lopez D, Mishra N, Pellegrini M, et al. 2013. Proteomic and genomic approaches reveal critical functions of H3K9 methylation and heterochromatin protein-1γ in reprogramming to pluripotency. Nat Cell Biol 15: 872–882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi H, Parmely TJ, Sato S, Tomomori-Sato C, Banks CA, Kong SE, Szutorisz H, Swanson SK, Martin-Brown S, Washburn MP, et al. 2011. Human mediator subunit MED26 functions as a docking site for transcription elongation factors. Cell 146: 92–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takemoto T, Uchikawa M, Yoshida M, Bell DM, Lovell-Badge R, Papaioannou VE, Kondoh H. 2011. Tbx6-dependent Sox2 regulation determines neural or mesodermal fate in axial stem cells. Nature 470: 394–398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Pachter L, Salzberg SL. 2009. TopHat: discovering splice junctions with RNA-seq. Bioinformatics 25: 1105–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai KL, Tomomori-Sato C, Sato S, Conaway RC, Conaway JW, Asturias FJ. 2014. Subunit architecture and functional modular rearrangements of the transcriptional mediator complex. Cell 157: 1430–1444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe K, Kamiya D, Nishiyama A, Katayama T, Nozaki S, Kawasaki H, Watanabe Y, Mizuseki K, Sasai Y. 2005. Directed differentiation of telencephalic precursors from embryonic stem cells. Nat Neurosci 8: 288–296. [DOI] [PubMed] [Google Scholar]
- Wilson V, Olivera-Martinez I, Storey KG. 2009. Stem cells, signals and vertebrate body axis extension. Development 136: 1591–1604. [DOI] [PubMed] [Google Scholar]
- Xiao Q, Wang G, Yin X, Luo Z, Margariti A, Zeng L, Mayr M, Ye S, Xu Q. 2011. Chromobox protein homolog 3 is essential for stem cell differentiation to smooth muscles in vitro and in embryonic arteriogenesis. Arterioscler Thromb Vasc Biol 31: 1842–1852. [DOI] [PubMed] [Google Scholar]
- Xu G, Deng N, Zhao Z, Judeh T, Flemington E, Zhu D. 2011. SAMMate: a GUI tool for processing short read alignments in SAM/BAM format. Source Code Biol Med 6: 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue Y, Van C, Pradhan SK, Su T, Gehrke J, Kuryan BG, Kitada T, Vashisht A, Tran N, Wohlschlegel J, et al. 2015. The Ino80 complex prevents invasion of euchromatin into silent chromatin. Genes Dev 29: 350–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Q, Li T, Price DH. 2012. RNA polymerase II elongation control. Annu Rev Biochem 81: 119–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


