Here, Dewar et al. use a proteomic screen in Xenopus egg extracts to identify factors that are enriched on chromatin when CMG unloading from chromatin, which is a key event during eukaryotic replication termination, is blocked. Their results show that CRL2Lrr1 is a master regulator of replisome disassembly during vertebrate DNA replication termination.
Keywords: DNA replication, replication termination, p97, CMG, ubiquitin
Abstract
A key event during eukaryotic replication termination is the removal of the CMG helicase from chromatin. CMG unloading involves ubiquitylation of its Mcm7 subunit and the action of the p97 ATPase. Using a proteomic screen in Xenopus egg extracts, we identified factors that are enriched on chromatin when CMG unloading is blocked. This approach identified the E3 ubiquitin ligase CRL2Lrr1, a specific p97 complex, other potential regulators of termination, and many replisome components. We show that Mcm7 ubiquitylation and CRL2Lrr1 binding to chromatin are temporally linked and occur only during replication termination. In the absence of CRL2Lrr1, Mcm7 is not ubiquitylated, CMG unloading is inhibited, and a large subcomplex of the vertebrate replisome that includes DNA Pol ε is retained on DNA. Our data identify CRL2Lrr1 as a master regulator of replisome disassembly during vertebrate DNA replication termination.
In eukaryotic cells, genomic DNA replication can be divided into four phases: licensing, initiation, elongation, and termination (Siddiqui et al. 2013; Bell and Labib 2016). During licensing, which occurs in the G1 phase of the cell cycle, ORC, Cdc6, and Cdt1 load pairs of Mcm2–7 ATPases at each origin of replication. In vertebrates, ∼50,000 origins are licensed genome-wide (Prioleau and MacAlpine 2016). Initiation occurs at the G1/S transition when Cdc45 and the GINS complex (consisting of Sld5, Psf1, Psf2, and Psf3) associate with Mcm2–7 to form active CMG helicases. CMG encircles and travels along the leading strand template (Fu et al. 2011; Kang et al. 2012). During elongation, the leading strand is copied by DNA polymerase ε (Pol ε). The lagging strand is synthesized as ∼150-nucleotide Okazaki fragments (Burgers 2009). Each Okazaki fragment is initiated by the DNA Pol α/primase complex, which makes a short RNA–DNA primer that is extended by DNA Pol δ. DNA Pol δ acquires processivity by interacting with the sliding clamp PCNA, which is loaded onto DNA by the five-subunit RFC complex. When the 3′ end of one Okazaki fragment encounters the 5′ end of another, DNA Pol δ displaces the RNA–DNA primer, and the resulting 5′ flap is removed by FEN1 followed by ligation by DNA ligase I (Burgers 2009). Long flaps with secondary structure are removed by the helicase/nuclease Dna2. During termination, replisomes converge, DNA synthesis is completed, the replisome is disassembled, and daughter duplexes are decatenated. Importantly, a single defective termination event has the potential to disrupt chromosome segregation and cause genome instability, whereas failure to initiate DNA synthesis at one or a few origins is readily mitigated by firing of other origins. Despite the importance of replication termination, this process has received little attention compared with other stages of the replication cycle.
Recently, considerable progress has been made in understanding the events underlying replication termination in eukaryotic cells. Use of a vertebrate cell-free system that supports site-specific and synchronous termination revealed that the leading strands of converging replication forks pass each other without stalling and that they are rapidly ligated to downstream Okazaki fragments (Dewar et al. 2015). These observations are in contrast to termination of SV40 DNA tumor virus replication, where forks stall (Levine et al. 1970; Tapper and DePamphilis 1978; Seidman and Salzman 1979). Moreover, converging CMG helicases are not unloaded until after the leading strand of one fork has been ligated to the downstream Okazaki fragment of the converging fork, implying that CMG is unloaded from dsDNA (Dewar et al. 2015). Most likely, converging CMGs pass each other and continue translocating until they reach a downstream Okazaki fragment. They then appear to pass over the ssDNA–dsDNA junction and probably translocate along dsDNA, as shown previously for purified Mcm2–7 and CMG complexes (Kaplan et al. 2003; Kang et al. 2012), before being unloaded.
Important aspects of CMG unloading have been revealed. Late in S phase, the Mcm7 subunit of CMG undergoes K48-linked polyubiquitylation by a Cullin RING E3 ubiquitin ligase (Maric et al. 2014; Moreno et al. 2014). The ligase corresponds to SCFDia2 in yeast but remains unknown in higher eukaryotes. In the absence of SCFDia2, CMG is retained on chromatin along with Ctf4/And-1 (also known as Wdhd1) and Csm3/Timeless (vertebrate names after slash), which stably interact with CMG as part of a “replisome progression complex” (RPC) that also includes Tof1/Tipin, Pol α, Mcm10, Mrc1/Claspin, FACT, and Topoisomerase I. These observations suggest that Mcm7 ubiquitylation is the trigger for unloading and disassembly of CMG and other RPC proteins. Yeast mutants lacking Dia2 (dia2Δ) show signs of genomic instability, suggesting that CMG unloading is essential to preserve genomic stability (Blake et al. 2006; Koepp et al. 2006). CMG unloading also depends on the p97 ATPase (Franz et al. 2011; Maric et al. 2014; Moreno et al. 2014), a “segregase” that extracts polyubiquitylated proteins from their local environment in collaboration with different adaptor proteins (Meyer et al. 2012; Buchberger et al. 2015). Unloading may also require Ufd1 and Npl4 (Franz et al. 2011), which form a heterodimer that binds p97 as well as the p97 adaptor Faf1 (Franz et al. 2016). Despite this progress, critical questions remain unanswered: Which E3 ubiquitin ligase ubiquitylates Mcm7 in higher eukaryotic cells? When is Mcm7 ubiquitylated and what triggers this event? How is Mcm7 ubiquitylation coordinated with replisome disassembly? What is the nature of the p97 complex that promotes unloading?
To identify the vertebrate Mcm7 ubiquitin ligase, we performed a mass spectrometry (MS) screen in Xenopus egg extracts for proteins that remain on chromatin when CMG unloading is blocked. This approach identified CRL2Lrr1 (a Cul2-based RING E3 ubiquitin ligase), several p97 adaptor proteins, other potential regulators of replication termination, and many components of the replisome. In the absence of CRL2Lrr1, Mcm7 ubiquitylation and CMG unloading are inhibited, as is the unloading of DNA Pol ε and numerous other replisome components. CRL2Lrr1 chromatin loading, Mcm7 ubiquitylation, and p97 recruitment occur only when forks terminate. Our data suggest that convergence of CMGs during replication termination triggers de novo CRL2Lrr1 recruitment, Mcm7 ubiquitylation, and coordinated unloading of a large replisome subcomplex by p97.
Results
Mcm7 ubiquitylation and p97 chromatin recruitment are termination-specific events
To study the mechanism of CMG unloading during replication termination, we replicated a plasmid in Xenopus egg extracts and analyzed binding of replisome proteins by plasmid pull-down (PPD). To this end, magnetic beads were coated with lac repressor (LacR) and incubated with the chromatinized plasmid DNA in ice-cold buffer. The LacR-coated beads efficiently bound DNA in a sequence-independent manner (Budzowska et al. 2015), which allowed us to isolate the plasmid and examine chromatin-bound proteins by Western blotting (Supplemental Fig. S1A). As shown previously for sperm chromatin replicating in egg extracts (Moreno et al. 2014), CMG unloading from plasmid was blocked by the inhibitor MLN4924 (“Cul-i”) (Supplemental Fig. S1A, cf. lanes 2 and 4), which inhibits the activity of Cullin E3 ubiquitin ligases by preventing their neddylation (Supplemental Fig. S1B, cf. lanes 2 and 3, Cul2 blot). CMG unloading was also blocked by the p97 inhibitor NMS-873 (“p97-i”) (Supplemental Fig. S1A, lane 3). In contrast, bortezomib had no effect (Supplemental Fig. S1A, lane 5), indicating that proteasome activity is not required. In the presence of p97-i but not Cul-i, Mcm7 appeared as a series of high-molecular-weight species (Supplemental Fig. S1A, lanes 3,4), which represent polyubiquitin chains (Supplemental Fig. S1B) that were primarily K48-linked (Supplemental Fig. S1C). The polyubiquitin chains observed in the presence of p97-i were abolished upon coaddition of Cul-i, indicating that they represent Cullin-dependent ubiquitylation (Supplemental Fig. S1A, lane 10). Importantly, when CMG unloading was blocked by p97-i, fork convergence, the completion of DNA synthesis, and decatenation were all unaffected (Supplemental Fig. S2A–J). Our results show that plasmid replication recapitulates Cullin RING ligase-dependent and p97-dependent CMG unloading as seen on sperm chromatin (Moreno et al. 2014) and in cells (Maric et al. 2014) and support our previous conclusion that CMG removal is not required for the DNA transactions underlying termination (Dewar et al. 2015).
Mcm7 ubiquitylation occurs late in S phase (Maric et al. 2014; Moreno et al. 2014), but whether this event is coupled to termination remains unknown. To address this, we took advantage of a reversible LacR-based replication fork barrier to induce synchronous and localized termination (Dewar et al. 2015). Plasmid DNA harboring 32 tandem lacO sequences (p[lacOx32]) was preincubated with LacR and replicated for 18 min in egg extract, leading to fork arrest on either side of the LacR array (Fig. 1A; Supplemental Fig. S2K; Dewar et al. 2015). We then added buffer (which maintains the arrest) or IPTG (which disrupts the LacR array and induces termination) and analyzed chromatin-bound proteins by PPD (which was not affected by the presence of IPTG) (data not shown) and Western blotting. In the presence of IPTG, CMG dissociated by 30 min, whereas, in the presence of buffer, CMG was retained (Fig. 1A, Sld5 blot, cf. lanes 8 and 10). When p97-i was added with IPTG, CMG unloading was blocked, and all of the chromatin-associated Mcm7 became polyubiquitylated (Fig. 1A, lane 9), whereas, when p97-i was added with buffer, there was no Mcm7 ubiquitylation (Fig. 1A, lane 11). Importantly, at an earlier time point before CMG unloading occurred, Mcm7 ubiquitylation was detected whether or not p97-i was added but only in the presence of IPTG (Fig. 1A, lanes 4–7). These results show that Mcm7 ubiquitylation requires disruption of the LacR array with IPTG.
Figure 1.
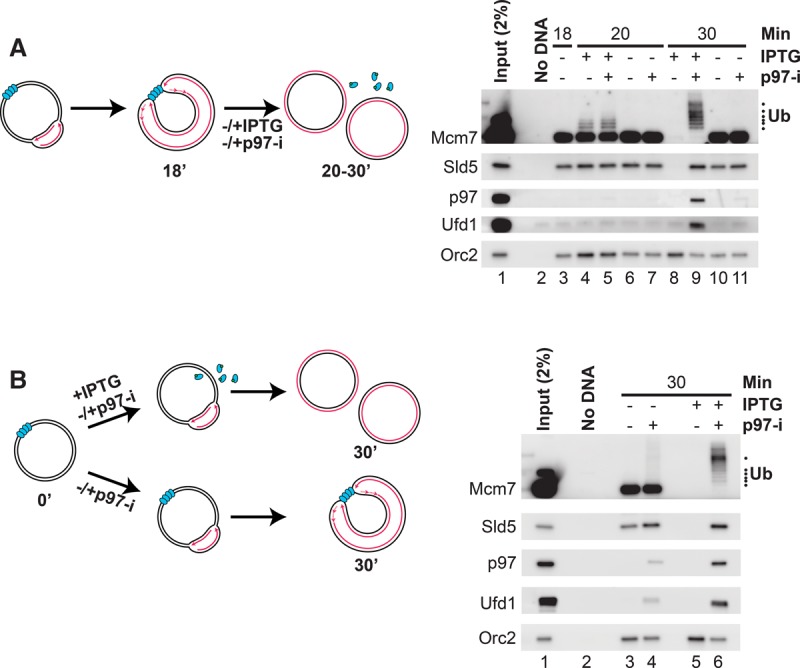
Mcm7 ubiquitylation and p97 recruitment are termination-specific. (A) p[lacOx32] was incubated with LacR and then replicated until forks converged on the LacR array. At 18 min, IPTG and/or p97-i was added, as indicated. Chromatin-bound proteins were recovered by PPD at 18, 20, or 30 min and analyzed by Western blotting to detect Mcm7, Sld5, p97, Ufd1, and Orc2 (loading control). In parallel, reactions including radionucleotides were assembled to assess DNA replication (Supplemental Fig. S2K). (B) p[lacOx32] was incubated with LacR and then replicated in the presence or absence of p97-i and/or IPTG, as indicated. Chromatin-bound proteins were recovered by PPD at 30 min and blotted for Mcm7, Sld5, p97, Ufd1, and Orc2 (loading control). In parallel, reactions including radionucleotides were assembled to assess DNA replication (Supplemental Fig. S2L). The same protein samples were also analyzed in Figure 4B.
The IPTG-dependent polyubiquitylation of Mcm7 could be caused by termination or the resumption of fork progression after IPTG addition. To distinguish between these possibilities, we compared two conditions. In one, fork progression and termination occurred, whereas, in the other, only fork progression was allowed. To create the first condition, we replicated LacR-bound p[lacOx32] and added IPTG from the start of the reaction to immediately disrupt the LacR array and allow fork progression and termination (Fig. 1B, top scheme; Supplemental Fig. S2L). To create the second condition, we added buffer instead of IPTG, in which case forks progressed to the array, but no termination occurred (Fig. 1B, bottom scheme; Supplemental Fig. S2L). We also included p97-i throughout both reactions to trap ubiquitylated Mcm7 that arose at any stage of the procedure. As shown in Figure 1B, efficient Mcm7 ubiquitylation was detected only in the presence of IPTG, when termination was allowed (cf. lanes 4 and 6). The small amount of Mcm7 ubiquitylation seen in the absence of IPTG (Fig. 1B, lane 4) likely reflects a small proportion of plasmids that undergo more than one initiation event (data not shown), leading to a low level of termination events in the presence of a LacR array (depicted in Supplemental Fig. S2M). Control reactions lacking p97-i demonstrated that IPTG induced efficient CMG unloading (Fig. 1B, cf. lanes 3 and 5), and replication showed the expected efficiency in the two conditions (Supplemental Fig. S2N). Our data strongly argue that ubiquitylation of Mcm7 is a termination-specific event.
We also addressed when p97 associates with chromatin. Strikingly, p97 was detected only in the presence of p97-i (Supplemental Fig. S1A, lane 3), suggesting that p97 binds stably to chromatin only when its catalytic activity is inhibited. Like Mcm7 ubiquitylation, efficient p97 binding occurred only when replication termination was allowed (Fig. 1A, lane 9, B, lane 6). Furthermore, p97 binding in the presence of p97-i was inhibited by coaddition of Cul-i, which blocks Mcm7 ubiquitylation (Supplemental Fig. S1A, lane 8 vs. 10), or Geminin, which blocks replication initiation (Supplemental Fig. S1A, lane 8 vs. 9). The p97 cofactor Ufd1, which heterodimerizes with Npl4, exhibited behavior similar to that of p97 (Fig. 1A,B). Together, the data indicate that when replisomes terminate, they undergo de novo ubiquitylation by a Cullin RING ligase followed by recruitment of p97–Ufd1–Npl4, which extracts CMG from chromatin.
A proteomic screen for proteins associated with terminated CMGs
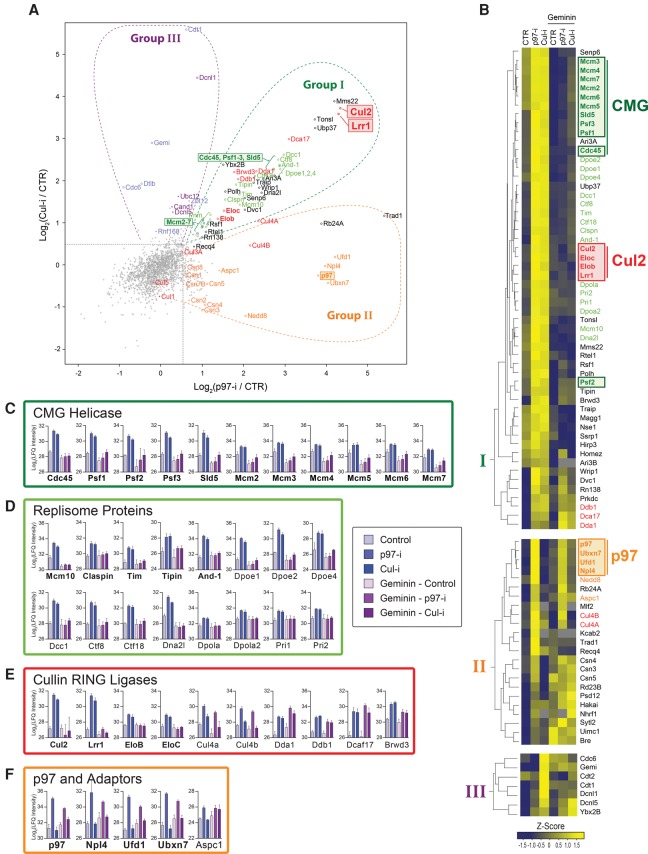
To understand how replication termination is regulated, we wanted to identify the vertebrate Cullin E3 ligase that ubiquitylates Mcm7. We reasoned that, in the presence of Cul-i, the E3 ligase might bind to terminating CMGs but fail to promote Mcm7 ubiquitylation (due to the absence of Cullin neddylation) and thus become trapped on chromatin. Similarly, p97-i might stabilize the E3 ligase on chromatin because p97-i traps CMG on DNA while allowing extensive Cullin-dependent polyubiquitylation of Mcm7 (Supplemental Fig. S1A). Furthermore, these inhibitors should identify any proteins that are retained with terminated CMGs on chromatin. As such, this approach might identify other regulators of termination as well as replisome components that are coordinately unloaded with CMG. To implement this strategy, we used chromatin MS (“CHROMASS”) (Räschle et al. 2015), which involves isolation of sperm chromatin by centrifugation followed by MS analysis of the recovered proteins. Sperm chromatin was replicated to completion in the presence of vehicle, p97-i, or Cul-i. To identify proteins whose enrichment in the presence of inhibitors was independent of replication, a parallel set of samples was pretreated with the replication licensing inhibitor Geminin. Four biochemical replicates of each condition were analyzed by single-shot MS and subjected to label-free quantification (LFQ), which provides a relative measure of protein abundance. Using a nonparametric test with a permutation-based false discovery rate (FDR) cutoff, we found that only 84 of the 2581 identified proteins significantly changed between the vehicle control sample and samples treated with either p97-i or Cul-i (FDR < 0.05, S0 = 0.1) (see Supplemental Fig. S3A,B; Supplemental Table S1). The majority of these hits accumulated on chromatin in response to both inhibitors (Fig. 2A,B, group I), while fewer proteins were enriched only in the presence of p97-i (Fig. 2A,B, group II) or Cul-i (Fig. 2A,B, group III).
Figure 2.
CHROMASS analysis of proteins enriched on DNA in the presence of p97-i or Cul-i. (A) Sperm chromatin was replicated to completion in the presence of vehicle, p97-i, or Cul-i. Chromatin-bound proteins were recovered by chromatin spin-downs and analyzed by MS. For each protein detected in the screen, fold enrichment relative to the vehicle control was calculated for p97-i or Cul-i treatment. Significant hits are indicated with colored markers and grouped according to whether they were a hit in both conditions (group I), only in the presence of p97-i (group II), or only in the presence of Cul-i (group III). In parallel, a set of control experiments was performed in which replication was blocked by Geminin treatment (Supplemental Fig. S3C,D). (B) Heat map showing the mean of the z-scored log2 LFQ intensity from four independent replicates described in A. LFQ values are arbitrary units, which reflect the abundance of different proteins recovered from chromatin in each condition. All proteins that were significantly up-regulated with p97-i and/or Cul-i (84 in total) (see the Materials and Methods) were subjected to nonsupervised hierarchical clustering. The main clusters corresponding to group I (enhanced binding with p97-i or Cul-i), group II (enhanced binding with p97-i only), or group III (enhanced binding with Cul-i only) were extracted and reclustered. (C) Mean log2 LFQ intensity across all four replicates is shown for Cdc45, GINS (Psf1, Psf2, Psf3, and Sld5), and Mcm2–7 across all conditions assessed by MS, including Geminin treatment. Error bars represent standard deviation. (D) As in C but for Mcm10, Claspin, Timeless–Tipin (Tim and Tipin), And-1, DNA Pol ε (Dpoe1, Dpoe2, and Dpoe4), Ctf18–Ctf8–Dcc1, Dna2 (Dna2l), and DNA Pol α/primase complex (Dpola, Dpola2, Pri1, and Pri2). (E) As in C but for components of potential Cullin ligases, the ligase CRL2Lrr1 (Cul2, Lrr1, EloB, and EloC), and Cul4a and Cul4b scaffolds and interacting proteins (Cul4a, Cul4b, Dda1, and Ddb1) as well as putative DCAFs (Dcaf17 and Brwd3) that are expected to form CRL4Dcaf17 and CRL4Brwd3. (F) As in C but for p97 and known substrate adapters: Ufd1–Npl4, Ubxn7, and Aspc1.
As expected, both p97-i and Cul-i led to robust enrichment of the CMG complex compared with a control reaction without inhibitor (Fig. 2C [blue bars], see also A,B). Mcm2–7 subunits were less enriched than Cdc45 or GINS subunits, probably due to the presence of some chromatin-bound Mcm2–7 that had not undergone replication initiation. The enrichment of CMG components in the presence of p97-i and Cul-i was replication-dependent (Fig. 2C [blue vs. magenta bars], cf. also B; Supplemental Fig. S3C,D; Supplemental Table S1). Strikingly, we detected numerous replication proteins that, like CMG, exhibited inhibitor- and replication-dependent enrichment (Fig. 2D). Many of these proteins directly interact with CMG, including Mcm10 (Merchant et al. 1997), Claspin (Lee et al. 2005), Timeless–Tipin (Gotter et al. 2007), And-1 (also known as Wdhd1 or Ctf4 in yeast) (Gambus et al. 2009; Simon et al. 2014; Villa et al. 2016), and DNA Pol ε (Langston et al. 2014). We note that all of these, apart from DNA Pol ε, are components of the RPC that assembles around CMG in yeast (Fig. 2D, bold protein names; Gambus et al. 2006). Several of the other enriched proteins interact indirectly with CMG, including Ctf18–Ctf8–Dcc1 (which binds DNA Pol ε) (Garcia-Rodriguez et al. 2015) as well as Dna2 and Pol α/primase (which are probably tethered to CMG by And-1) (Gambus et al. 2009; Villa et al. 2016). In contrast, replication proteins that do not interact with CMG (PCNA, Fen1, DNA ligase I, and DNA Pol δ) were not significantly enriched (Supplemental Fig. S4A). Notably, RPA was not a hit in our screen (Rfa1–3) (Supplemental Fig. S4A), consistent with our model that impaired replisome unloading does not block ligation of leading strands to the Okazaki fragments of the converging fork (Dewar et al. 2015). These data suggest that a large replisome complex comprising CMG and numerous proteins involved in the synthesis of the leading strand is unloaded from DNA in a Cullin- and p97-dependent manner.
Two of the top hits in group I were Cul2 and Lrr1, both of which were enriched ∼16-fold in the presence of both Cul-i and p97-i compared with the control reactions (Fig. 2A,E). Cul2 and Lrr1 are components of the Cullin RING ligase CRL2Lrr1, which also includes the adapters Elongin B (EloB), EloC, and the RING subunit Rbx1 (Kamura et al. 2004). EloB and EloC were also enriched in our analysis (Fig. 2E), whereas the small protein Rbx1 was not detected with our CHROMASS approach. Enrichment of Cul2, Lrr1, EloB, and EloC was blocked by Geminin, demonstrating that CRL2Lrr1 recruitment was replication-dependent (Fig. 2E). We confirmed that Cul2, Lrr1, and EloC form a complex in egg extracts (Supplemental Fig. S4B). The only other Cullin ligases identified in our analysis were CRL4Dcaf17 (represented by Cul4, the Ddb1 adaptor, and the Dcaf17 substrate receptor) (Jin et al. 2006) and CRL4Brwd3 (represented by Cul4, Ddb1, and the Brwd3 substrate receptor) (Ozturk et al. 2013). We confirmed the existence of CRL4Dcaf17 and CRL4Brwd3 complexes in egg extracts (Supplemental Fig. S4B). However, Dcaf17 enrichment was replication-independent, whereas Brwd3 enrichment was modest compared with Lrr1 and only partially replication-dependent (Fig. 2A,E). Taken together, our results single out Cul2Lrr1 as the top candidate for an E3 ligase that ubiquitylates Mcm7 during replication termination (as examined further below). Other than the above Cullin RING ubiquitin ligases, group I includes many other proteins that are potential regulators of, or participants in, termination (Discussion; Supplemental Fig. S4D).
Within group II, p97 and a specific set of p97 adaptor proteins (Ufd1, Npl4, and Ubxn7) were enriched ∼16-fold in the presence of p97-i but not with Cul-i (Fig. 2A,B,F), suggesting that these adaptors could mediate recognition of the polyubiquitin chain that forms on Mcm7. Recruitment was diminished substantially by Geminin, indicating that a fraction of p97, Ufd1, Npl4, and Ubxn7 bound chromatin in a replication-coupled manner (Fig. 2F). All four proteins behaved very similarly across all six conditions, consistent with p97, Ufd1, Npl4, and Ubxn7 forming a complex (Fig. 2A,B; Alexandru et al. 2008; den Besten et al. 2012; Raman et al. 2015). Aspc1, which is thought to promote disassembly of p97 complexes (Orme and Bogan 2012; Cloutier et al. 2013), was also enriched in the presence of p97-i and not Cul-i (Fig. 2A,B, F) but less so than p97, Ufd1, Npl4, and Ubxn7. Only two other proteins (Rb24A and Trad1) showed similarly high and inhibitor-specific enrichment (Fig. 2A,B; Supplemental Fig. S4D), suggesting that they may also be part of the p97 machinery. We also detected modest enrichment of proteins that regulate Cullin RING ligases, including Nedd8 and subunits of the COP9 signalosome (Csn1–5, Csn7b, and Csn8) (Fig. 2A,B; Supplemental Fig. S4D).
Within group III, we detected the licensing factors Cdt1 and Cdc6 (Fig. 2A,B; Supplemental Fig. S4D). Consistent with this, replication-dependent Cdt1 destruction is prevented by Cul-i (Moreno et al. 2014), likely leading to its retention on chromatin. Consistent with an accumulation of inactive Cullin E3 ligases, several regulators of the neddylation cascade (Ubc12, Dcnl1, and Dcnl5) as well as the inhibitor Cand1, which binds only to nonneddylated cullins, accumulated specifically in the presence of Cul-i but not p97-i (Fig. 2A,B; Supplemental Fig. S4D).
In summary, our proteomic analysis identified a large group of replication factors that are retained with CMG in a p97-i-dependent and Cul-i-dependent manner as well as potential new regulators of termination. Among these, Cul2Lrr1 is the lead candidate for a Mcm7 ubiquitin ligase.
CRL2Lrr1 depletion inhibits Mcm7 ubiquitylation and CMG unloading
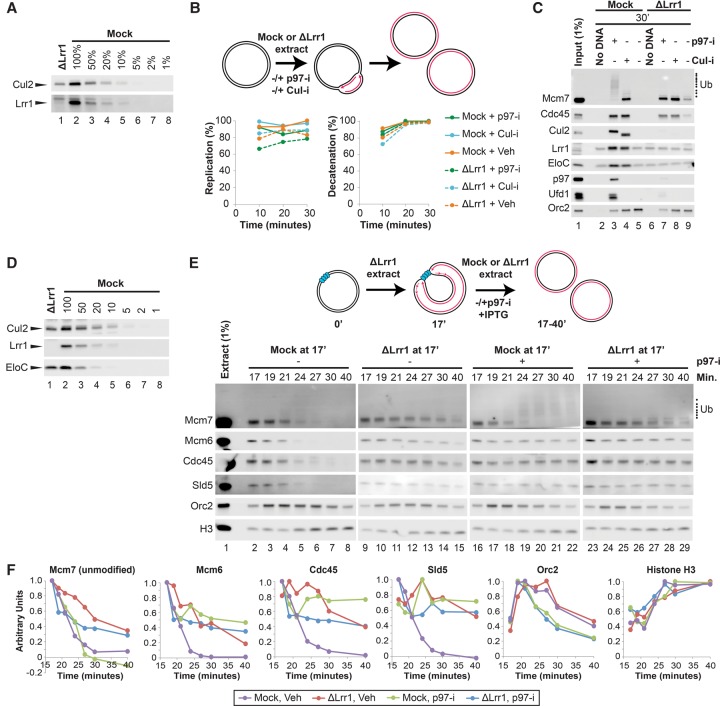
We next addressed whether CRL2Lrr1 plays a role in termination. Immunodepletion of Lrr1 from egg extracts partially codepleted Cul2 (Fig. 3A) and did not affect plasmid replication efficiency or decatenation (Fig. 3B), the final DNA transaction of termination (Dewar et al. 2015). In contrast, CMG unloading was impaired in the absence of Lrr1 (Fig. 3C, lanes 5,9), and Mcm7 polyubiquitylation was undetectable (Fig. 3C, lanes 3,7). Moreover, the p97-i- and Cul-i-dependent binding of Cul2, EloC, p97, and Ufd1 was greatly reduced in the absence of Lrr1 (Fig. 3C, cf. lanes 3 and 7, note that Cul-i increases the mobility of Cul2 due to deneddylation). To carefully quantify the CMG unloading defect in the absence of CRL2Lrr1, we first allowed forks to converge on a LacR array in Lrr1-depleted extracts (Fig. 3D). After 17 min, we split the reaction and added back mock- or Lrr1-depleted extracts containing IPTG and examined the kinetics of CMG unloading at closely spaced intervals (Fig. 3E,F). Whereas CMG unloading was largely complete 24 min after addition of mock-depleted extract, CMG was still bound after 40 min in the presence of Lrr1-depleted extract (Fig. 3E,F, cf. purple and red graphs; see also Supplemental Fig. S5C). A recombinant complex of Xenopus Lrr1, EloB, and EloC did not restore Mcm7 ubiquitylation or CMG unloading (data not shown), perhaps because it lacked Cul2, and the endogenous Cul2 remaining in Lrr1-depleted extracts (Fig. 3A) was bound to other substrate receptors. Together with data presented below, our results strongly suggest that CRL2Lrr1 is the primary E3 ligase responsible for Mcm7 polyubiquitylation and CMG unloading in Xenopus egg extracts.
Figure 3.
Depletion of Lrr1 eliminates Mcm7 polyubiquitylation and inhibits CMG unloading. (A) Mock-depleted (mock) and Lrr1-depleted (ΔLrr1) extracts were blotted with Cul2 and Lrr1 antibodies. The bands corresponding to Cul2 and Lrr1 are marked with arrowheads. Over 90% of Lrr1 was depleted. (B) The extracts from A were used to replicate plasmids in the presence or absence of p97-i and/or Cul-i. Radionucleotides were included, and the relative extent of replication and decatenation was quantified at different time points (as in Supplemental Fig. S2H–J). (C) In parallel to B, reactions lacking radionucleotides were assembled. At 30 min, chromatin-bound proteins were recovered by PPD and blotted for Mcm7, Cdc45, Cul2, Lrr1, EloC, p97, Ufd1, and Orc2 (loading control). The same samples were also used in Supplemental Figure S7F. (D) Immunodepleted extracts were analyzed as in A except that EloC was also analyzed. Over 90% of Lrr1 was depleted. (E) p[lacOx32] was incubated with LacR and then replicated in Lrr1-depleted (ΔLrr1) extracts from D until forks converged on the LacR array. At 17 min, mock- or Lrr1-depleted extract was added along with IPTG to induce termination in the presence or absence of p97-i. Chromatin-bound proteins were recovered by PPD at time points between 17 and 40 min and then analyzed by Western blotting using antibodies against Mcm7, Mcm6, Cdc45, Sld5, Orc2, and Histone H3 (H3). Orc2 served as a loading control for the Mcm7 blot, and Histone H3 served as the loading control for the other blots. (F) Quantification of Mcm7, Mcm6, Cdc45, Sld5, Orc2, and Histone H3 protein recovery from E (see the Materials and Methods). As expected, in the presence p97-i, the unmodified form of Mcm7 disappeared in the presence of mock-depleted extract due to ubiquitylation (green graph) but not in LRR1-depleted extract, where ubiquitylation is inhibited (blue graphs).
CRL2Lrr1 associates with chromatin during termination
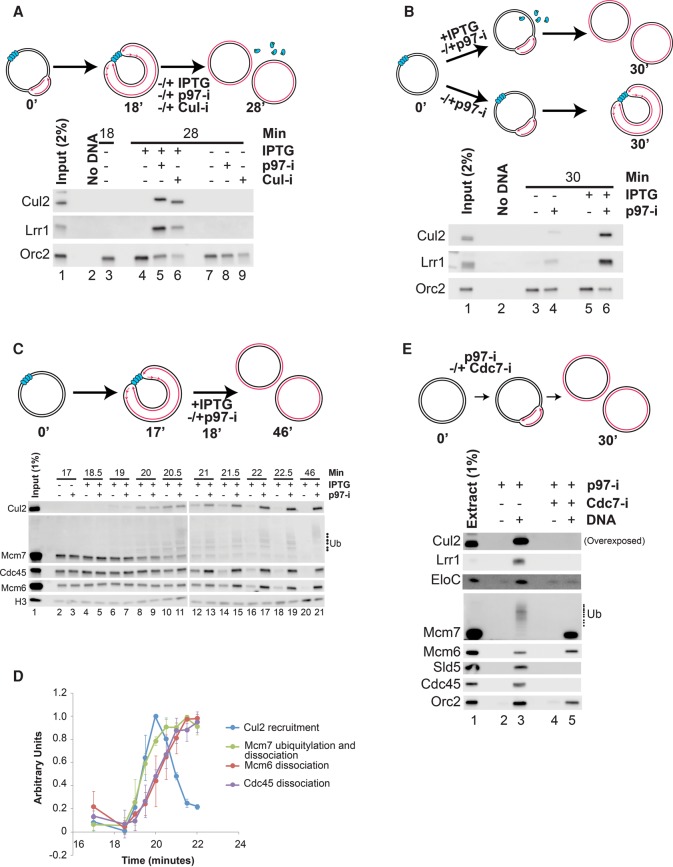
To understand the regulation of termination, we asked under what conditions CRL2Lrr1 binds to chromatin. Analogous to Mcm7 ubiquitylation (Fig. 1), we found that, in the presence of p97-i or Cul-i, Cul2 and Lrr1 did not bind efficiently to forks arrested at a LacR array (Fig. 4A, lanes 8,9) and associated with chromatin only upon the addition of IPTG, which induces termination (Fig. 4A, lanes 5,6). Moreover, when fork progression but not termination was allowed in the continued presence of p97-i, binding was poor (Fig. 4B, lane 4), whereas, when termination was also allowed, binding was efficient (Fig. 4B, lane 6). Therefore, like p97 binding and Mcm7 ubiquitylation (Fig. 1), CRL2Lrr1 binding to chromatin in the presence of inhibitors is termination-specific.
Figure 4.
CRL2Lrr1 is specifically recruited during termination. (A) p[lacOx32] was incubated with LacR and then replicated until forks converged on the LacR array. At 18 min, IPTG was added to induce termination or withheld to maintain the block along with either p97-i or Cul-i to block CMG unloading or vehicle control. Chromatin-bound proteins were recovered by PPD at 18 or 28 min and analyzed by Western blotting to detect Cul2, Lrr1, and Orc2 (loading control). In parallel, reactions including radionucleotides were assembled and analyzed to assess replication (Supplemental Fig. S7A). Samples are the same as those used in Figure 5, A and B. (B) The same samples described in Figure 1B were blotted for Cul2, Lrr1, and Orc2 (loading control). (C) Termination was induced and monitored as in A except that, after IPTG addition, chromatin was recovered by PPD at 17 or 18.5 min and every 30 sec thereafter and blotted for Cul2, Mcm7, Cdc45, Mcm6, and Histone H3 (H3; loading control). (D) Quantification of Cul2 binding, Mcm6 and Cdc45 dissociation, and Mcm7 ubiquitylation/dissociation from the Western blots shown in C and two additional experimental replicates (see the Materials and Methods). The mean is plotted, and error bars represent the standard deviation. Only samples lacking p97-i (even-numbered lanes in C) are quantified. (E) Plasmid DNA was incubated in egg extracts with p97-i and either vehicle or Cdc7-i. Chromatin-bound proteins were recovered by PPD after 30 min and blotted for Cul2, Lrr1, EloC, Mcm7, Mcm6, Sld5, Cdc45, and Orc2 (loading control).
We next addressed whether CRL2Lrr1 is recruited to chromatin in the absence of inhibitors. To this end, LacR-bound p[lacOx32] was replicated in egg extract to arrest forks at the array (Fig. 4C). At 18 min, IPTG was added along with vehicle or p97-i, and, every 30 sec thereafter, plasmid was recovered and analyzed. In the absence of p97-i, CMG unloading (exemplified by Mcm6 and Cdc45) began shortly after IPTG addition and was largely complete by 22.5 min (Fig. 4C [even numbered lanes], D [red and purple lines for quantification]). Importantly, in the presence or absence of p97-i, Cul2 accumulated on chromatin at similar levels between 19 and 20.5 min (Fig. 4C, lanes 6–11), suggesting that p97-i does not affect the level or kinetics of Cul2 binding. In the absence of p97-i, Cul2 subsequently dissociated and was no longer bound by 22.5 min, while, in the presence of the inhibitor, Cul2 continued to accumulate before reaching a peak at 22.5 min. These data indicate that Cul2 is normally recruited to chromatin during unperturbed termination and lost during CMG unloading.
We wanted to know when Cul2 associates with chromatin relative to Mcm7 ubiquitylation and CMG dissociation. To quantify the kinetics of Mcm7 ubiquitylation, we measured the disappearance of the unmodified Mcm7 band (Fig. 4C), as this was more accurate than quantifying the multiple ubiquitylated Mcm7 species. We validated this strategy by demonstrating that treatment of the recovered plasmid with Usp21 (Ye et al. 2011), a nonspecific deubiquitylating enzyme, abolished the slow-mobility Mcm7 species while increasing the level of unmodified Mcm7 (Supplemental Fig. S6A,B). Using this approach, we found that Mcm7 ubiquitylation and Cul2 recruitment were closely correlated (Fig. 4D, green and blue traces), whereas these events preceded CMG dissociation by ∼45 sec (Fig. 4D, cf. green and blue vs. red and purple traces). Together, the data argue that, during termination, CRL2Lrr1 is recruited to chromatin, whereupon Mcm7 is rapidly ubiquitylated and then extracted by p97.
CRL2Lrr1 does not bind to licensed Mcm2–7 complexes
In the presence of Cul-i or p97-i, CRL2Lrr1 was bound to chromatin even when the vast majority of replicated plasmid DNA was in the closed circular duplex form (Figs. 3C [lanes 3,4], B, 4A [lanes 5,6]; Supplemental Fig. S2K; Fig. 4B, lane 6; Supplemental Fig. S2L), implying that CRL2Lrr1 recognizes CMG that encircles dsDNA. If this model is correct, it raises the possibility that CRL2Lrr1 might bind to uninitiated Mcm2–7 complexes, which also encircle dsDNA (Evrin et al. 2009; Remus et al. 2009). To address this possibility, we measured CRL2Lrr1 binding to chromatin in the presence of the Cdc7 inhibitor PHA-767491 (“Cdc7-i”), which blocks the conversion of licensed Mcm2–7 into active replication forks (Montagnoli et al. 2008). p97-i was included to stabilize any CRL2Lrr1 complexes on chromatin. As expected, Cdc7-i eliminated DNA replication (Supplemental Fig. S6C) and CMG assembly (Fig. 4E, lane 5, Sld5 and Cdc45 blots). Importantly, Cdc7-i also eliminated CRL2Lrr1 chromatin binding and Mcm7 ubiquitylation (Fig. 4E, cf. lanes 3 and 5). Therefore, while CRL2Lrr1 interacts efficiently with dsDNA that contains CMG complexes, it does not associate with dsDNA containing Mcm2–7 complexes. This discrimination insures that CRL2Lrr1 does not reverse the licensing process.
CRL2Lrr1-dependent unloading of a replisome subcomplex
Our proteomic analysis showed that when CMG unloading was blocked by Cul-i or p97-i, numerous other replication proteins were retained (Fig. 2D). We wanted to test whether these proteins are associated with forks and whether they are coordinately unloaded during termination. To this end, LacR-bound p[lacOx32] was replicated in egg extracts for 18 min to arrest forks at the array (Fig. 5A; Supplemental Fig. S7A). IPTG was then added to induce termination or omitted to maintain the block, and we also included vehicle, p97-i, or Cul-i. As expected, the CMG complex (represented by Mcm7, Cdc45, and Sld5) was associated with arrested forks (Fig. 5A, lanes 3,7–9), lost when termination was induced with IPTG (Fig. 5A, lane 4), and retained when Cul-i or p97-i was added with IPTG (Fig. 5A, lanes 5,6). Claspin, Mcm10, DNA Pol ε, Ctf18, and Dna2 exhibited similar behavior, as did the ubiquitin ligase Traip (Fig. 5A). Timeless and Tipin, which form a complex, also behaved similarly, although they associated at reduced levels with terminated compared with arrested replisomes (Fig. 5A, cf. lanes 5,6 and 8,9). DNA Pol α was present at arrested forks, as expected (Fig. 5B, lanes 3,7–9), but was not detectable upon termination even in the presence of Cul-i or p97-i (Fig. 5B, lanes 4–6) despite being detected in our proteomic analysis (Fig. 2D). This difference is likely due to the greater stringency of the PPD procedure compared with the gentle sperm chromatin isolation method used in the proteomic analysis. Surprisingly, even though we detected DNA Pol α at arrested forks, we could not detect And-1 (Fig. 5B, lanes 3,7–9), which suggests that, unlike Ctf4 in yeast (Gambus et al. 2009; Simon et al. 2014), And-1 does not tether DNA Pol α to replication forks. Importantly, we were able to detect And-1 at earlier time points (Supplemental Fig. S7B, lanes 2,3), which supports its previously described role in replication initiation (Zhu et al. 2007) and suggests that And-1 recruits DNA Pol α during initiation but not at elongating forks. Remarkably, binding of And-1 was greatly enhanced upon termination (Fig. 5B, lanes 5,6; Supplemental Fig. S7B,D), suggesting that And-1 binds stably to terminated but not elongating replisomes.
Figure 5.
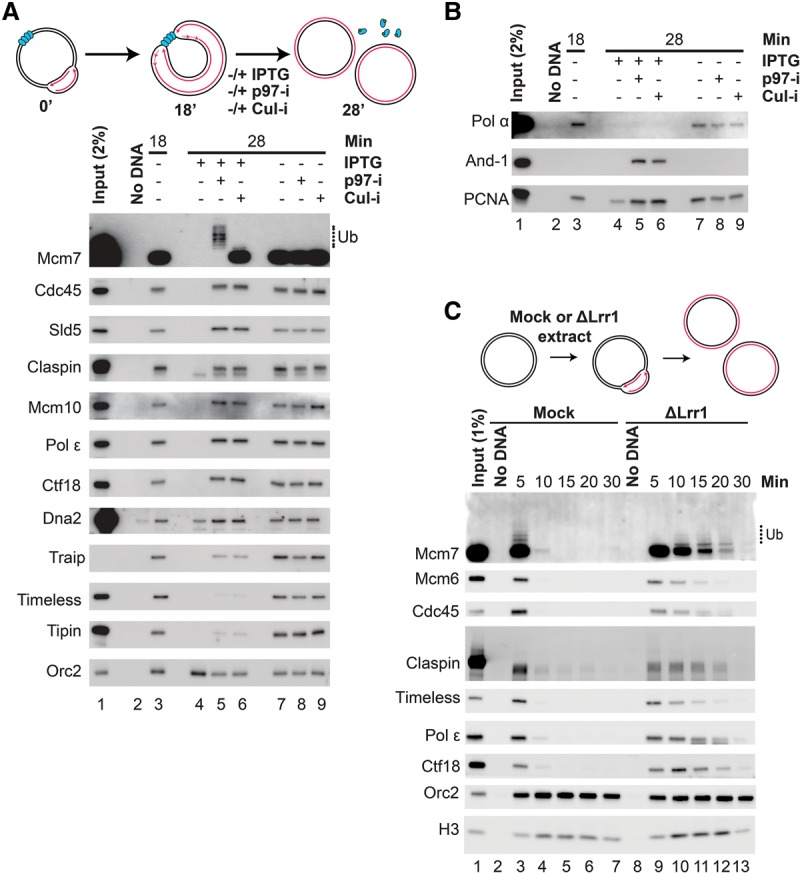
CRL2Lrr1 is involved in disassembly of a vertebrate RPC. (A) The samples in Figure 4A were blotted for Mcm7, Cdc45, Sld5, Claspin, Mcm10, DNA Pol ε, Ctf18, Dna2, Traip, Timeless, Tipin, and Orc2 (loading control). In parallel, reactions including radionucleotides were assembled and analyzed to assess replication (Supplemental Fig. S7A). Traip was not detected in the input, but the band shown was depleted from extracts by immunoprecipitation using the Traip antibody (data not shown). (B) The same samples analyzed in A were also analyzed by Western blotting to detect DNA Pol α, And-1, and PCNA. (C) Plasmids were replicated in Lrr1-depleted extracts. At the indicated times, chromatin-bound proteins were recovered by PPD and blotted for Mcm7, Mcm6, Cdc45, Claspin, Timeless, Pol ε, Ctf18, Orc2, and Histone H3 (H3). Orc2 served as a loading control for Mcm7, Mcm6, and Cdc45, whereas Histone H3 served as a loading control for Claspin, Timeless, Pol ε, and Ctf18.
In contrast to the proteins discussed above, DNA Pol δ and RPA were not associated with terminated replisomes (Supplemental Fig. S7E). The loss of RPA may explain the reduced binding of Timeless and Tipin to terminated replisomes (Fig. 5A), as these proteins also interact with RPA. PCNA unloading was largely termination-independent (Supplemental Fig. S7B, cf. lanes 2,8 vs. 10) as shown previously (Moreno et al. 2014; Dungrawala et al. 2015). However, we clearly detected retention of some PCNA when CMG unloading was blocked (Fig. 5B, lane 4 vs. 5,6; Supplemental Fig. S7B, lane 10 vs. 11), which probably reflects PCNA associated with DNA Pol ε. Our results indicate that the vertebrate replisome contains a large, highly stable subassembly containing CMG, Claspin, Mcm10, DNA Pol ε, Ctf18–Ctf8–Dcc1, Dna2, Traip, and PCNA along with more weakly associated components such as Timeless–Tipin and DNA Pol α. When forks terminate but CMG unloading is blocked, this subassembly is retained on chromatin and appears to recruit additional factors such as And-1.
To further test the idea that this replisome subassembly is coordinately unloaded from chromatin, we examined its fate in Lrr1-depleted egg extracts. As shown in Figure 5C and Supplemental Figures S5 and S7F, Claspin, Timeless, DNA Pol ε, and Ctf18 all persisted on chromatin in the absence of Lrr1. These data suggest that CRL2Lrr1 triggers the coordinated removal of CMG and numerous other replication proteins from chromatin during replication termination.
Discussion
We performed a proteomic screen for factors that are retained on chromatin with terminated CMG complexes and identified the E3 ubiquitin ligase CRL2Lrr1, many CMG-associated proteins, and potential new regulators of termination. Immunodepletion of Lrr1 from egg extracts greatly impaired the unloading of CMG and associated factors. Although we were unable to rescue these defects with recombinant Lrr1 (see the Results), the following evidence supports a central and direct role for CRL2Lrr1 in replisome disassembly. First, CMG unloading requires a Cullin RING ligase (Supplemental Fig. S1A; Moreno et al. 2014), and CRL2Lrr1 is the only CRL whose binding to chromatin is strictly replication-dependent. Second, CRL2Lrr1 recruitment to chromatin is absolutely dependent on replication termination, and the kinetics of Mcm7 ubiquitylation and Cul2 recruitment are highly correlated. Third, when we depleted Lrr1, Mcm7 ubiquitylation was virtually undetectable, and unloading of CMG and CMG-associated proteins was greatly delayed. Together with the observation that p97 recruitment to chromatin is termination-dependent and inhibited in Lrr1-depleted extract, the data support the following mechanism: Once CMGs have terminated, CRL2Lrr1 is recruited de novo and ubiquitylates Mcm7, whereupon CMG is extracted from chromatin by p97, leading to the dissociation of CMG-associated proteins (Fig. 6). Consistent with our work, others recently discovered that CUL-2LRR−1 promotes CMG unloading in worms and Xenopus egg extracts (Sonneville et al. 2017). In possible further agreement with our model, genetic interactions between LRR-1 and various replisome components, including PSF-2 and PSF-3, have been detected in worms (Ossareh-Nazari et al. 2016).
Figure 6.
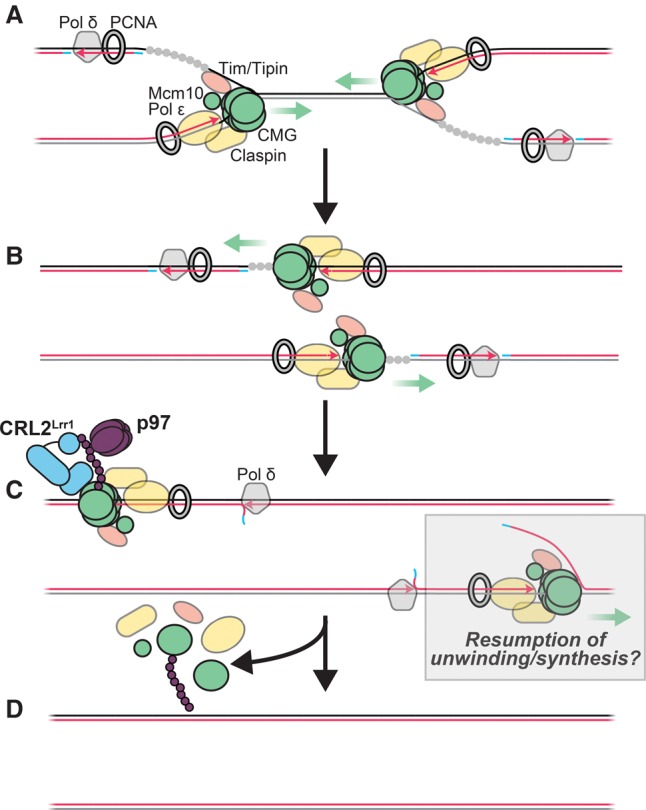
Model for unloading of a vertebrate RPC by CRL2Lrr1. (A) During replication, replication forks are bound by a vertebrate RPC (vRPC) that includes CMG, Mcm10, DNA Pol ε, Timeless/Tipin (Tim/Tipin), Claspin, Ctf18–RLC (not shown), Dna2 (not shown), Traip (not shown), and PCNA. (B) vRPCs pass each other and remain associated with CMGs as they translocate along ssDNA. (C) vRPCs translocate over the downstream lagging strands and encircle dsDNA. This prevents CMG and DNA Pol ε from blocking Okazaki fragment processing and allows binding by DNA Pol δ, which performs strand displacement synthesis. The presence of vRPCs on dsDNA leads to binding of And-1 and CRL2Lrr1, which ubiquitylates Mcm7. If the replisome encounters a long downstream 5′ flap at the penultimate Okazaki fragment prior to unloading, reinitiation and unwinding by CMG and DNA synthesis by DNA Pol ε may be triggered. (D) vRPCs are extracted from chromatin by p97.
What recruits CRL2Lrr1 to chromatin? Remarkably, when CMG unloading is blocked in the presence of p97-i or Cul-i, CRL2Lrr1 binds stably to chromatin even after all of the DNA is fully replicated and ligated. We proposed previously that, as the helicase passes over a downstream Okazaki fragment, the replacement of ssDNA with dsDNA in CMG's central channel triggers CMG unloading (Fig. 6; Dewar et al. 2015). We now speculate that the presence of dsDNA in CMG's central channel leads to CRL2Lrr1 recruitment, possibly through a conformational change in CMG. Given that CRL2Lrr1 does not interact with licensed Mcm2–7 complexes, which also encircle dsDNA, recognition of terminated replisomes must also involve Cdc45, GINS, or other CMG-associated factors. However, we cannot rule out mechanisms that are independent of dsDNA. For example, the juxtaposition of converged CMGs or their interaction with PCNA molecules of the converging fork might stimulate CRL2Lrr1 binding. In yeast, SCFDia2 coimmunoprecipitates with CMG after nuclease digestion of chromatin (Gambus et al. 2006), suggesting that SCFDia2 may bind replisomes constitutively, but it cannot be ruled out that this association occurs after preparation of cell lysates. Lrr1 shares 15% sequence identity with Dia2 and, like Dia2, contains leucine-rich repeats. However, Lrr1 lacks a tetratricopeptide repeat (TPR) domain, which mediates SCFDia2 binding to the replisome (Maculins et al. 2015). This may explain why CRL2Lrr1 does not bind stably to replisomes in the absence of replication termination. Further work will be required to determine how replication termination stimulates the de novo recruitment of CRL2Lrr1, whether recruitment is sufficient for Mcm7 ubiquitylation, and whether this mechanism is conserved in other organisms.
Our proteomic screen identified four p97 adaptor proteins (Ufd1, Npl4, Ubxn7, and Aspc1). Like p97, the Ufd1–Npl4 heterodimer (as measured by Ufd1 Western blotting) bound chromatin in a manner dependent on CRL2Lrr1, consistent with a role in termination. Similarly, Ubxn7 chromatin binding was at least partially replication-dependent in the MS screen (Fig. 2F). The Ufd1–Npl4 heterodimer interacts with p97 in numerous chromatin-associated processes, including eviction of RNA Pol II at sites of DNA damage (Verma et al. 2011). Ubxn7 contains a ubiquitin-interacting motif (UIM) that binds neddylated Cullins and thereby mediates interactions between p97 and active Cullin RING ligases (Alexandru et al. 2008; den Besten et al. 2012; Raman et al. 2015). Importantly, the interaction of Ubxn7 with p97 depends on Ufd1 and Npl4 (Hanzelmann et al. 2011). Together, the data indicate that a p97–Ufd1–Npl4–Ubxn7 complex exists in egg extracts, and we propose that it is involved in CMG unloading. Aspc1, a negative regulator of p97 (Orme and Bogan 2012; Cloutier et al. 2013), may antagonize the p97–Ufd1–Npl4–Ubxn7 complex in CMG unloading. Previous work suggests that UFD-1/NPL-4 promote CMG unloading in worms (Franz et al. 2011), and recent work indicates that the same is true in budding yeast (K. Labib, pers. comm.), highlighting the similarities in CMG disassembly in diverse eukaryotes.
Our data provide new insights into replisome architecture. We identified many proteins that are present at forks and retained when CMG unloading is blocked by Cul-i, p97-i, or Lrr1 depletion. These include Claspin (Mrc1), Mcm10, Pol ε, and Timeless–Tipin (Csm3–Tof1), all of which bind stably to CMG. A subpopulation of PCNA molecules is also retained, which probably reflects the Pol ε-bound pool of PCNA described previously (Dungrawala et al. 2015). We also identified other proteins that interact indirectly with CMG: Ctf18–Ctf8–Dcc1 binds Pol ε (Garcia-Rodriguez et al. 2015), while Traip binds PCNA (Hoffmann et al. 2016). Additionally, we identified Dna2 and DNA Pol α but can provide no obvious explanation for how they are connected to CMG given that they are thought to be recruited by And-1 (Gambus et al. 2006; Langston et al. 2014), which does not appear to bind stably to forks (see below). In sum, we propose the existence of a stable vertebrate replisome subassembly consisting of CMG, Claspin, Mcm10, DNA Pol ε, Timeless–Tipin, PCNA, Ctf18–Ctf8–Dcc1, Traip, Dna2, and DNA Pol α. Given its similarity to the yeast RPC (which contains CMG, Claspin/Mrc1, Mcm10, and Timeless–Tipin/Csm3–Tof1), we call this complex the vertebrate RPC (vRPC). The simplest interpretation of our data is that unloading of the entire vRPC is triggered by CRL2Lrr1-dependent Mcm7 ubiquitylation, but we cannot rule out the possibility that CRL2Lrr1 or other ubiquitin ligases target additional vRPC components.
The absence of several proteins from the replication fork merits discussion. In particular, And-1 (Ctf4) was barely detectable at arrested forks but bound robustly to terminated replisomes (Fig. 5B). This observation is consistent with recent findings that Ctf4 is dispensable for synthesis of leading and lagging strands in vitro (Devbhandari et al. 2017; Yeeles et al. 2017). Since And-1 can be detected at replication forks when cross-linking is used (Dungrawala et al. 2015), we propose that And-1 interacts dynamically with elongating replisomes and more stably with terminated replisomes. Additionally, our proteomic analysis did not detect robust enrichment of Topoisomerase I or FACT (Spt16 and Ssrp1), which are components of the yeast RPC. Rfc2, Rfc3, Rfc4, and Rfc5, which form an alternative RFC complex with Ctf18–Ctf8–Dcc1, were also not enriched. Failure to observe enrichment of these proteins in the presence of p97-i or Cul-i was probably due to their high nonspecific backgrounds in CHROMASS (see Supplemental Table S1). In the future, it will be important to clarify how these complexes bind to replicating and terminated replisomes.
Our results have interesting implications for Okazaki fragment processing and maintenance of genome stability. Unlike DNA Pol δ, DNA Pol ε has limited strand displacement synthesis activity (Garg et al. 2004; Georgescu et al. 2014) and cannot support maturation of Okazaki fragments, which requires the formation of 5′ flaps that are removed by Fen1. Therefore, when a terminating leading strand meets the 5′ end of a downstream Okazaki fragment from the converged fork, DNA Pol ε must be replaced with DNA Pol δ (Fig. 6C). Our model that CMG continues moving on dsDNA at the downstream Okazaki fragment not only eliminates interference of Okazaki fragment processing by the helicase but also drags DNA Pol ε away from the 3′ end of the leading strand. This allows Pol ε to be replaced with Pol δ without prior unloading of CMG or PCNA, both of which probably contact Pol ε. This mechanism, although attractive, has a potential disadvantage. If the CMG–DNA Pol ε complex is not unloaded rapidly once it passes onto dsDNA, it would promote rereplication if it encounters a long flap at another Okazaki fragment (Fig. 6C). Consistent with this idea, mutation of lrr-1 in worms causes DNA damage, checkpoint activation, and evidence of rereplication (Merlet et al. 2010). It will be important to test whether termination ever stimulates rereplication and what the consequences are for chromosome stability.
Several other proteins identified in our proteomic analysis (Supplemental Fig. S4D, group I) may represent novel vRPC components or regulators of termination. As Mms22l–Tonsl is reported to interact with CMG (O'Connell et al. 2010; O'Donnell et al. 2010; Piwko et al. 2010), we speculate that it may be a vRPC component. The same may be true of DNA Pol η, which interacts with the Ctf18–RLC complex (Shiomi et al. 2007). Unlike CRL4Dcaf17, CRL4Brwd3 exhibited some replication-dependent chromatin binding, suggesting a possible role in termination. We speculate that Usp37, which is a deubiquitylating enzyme, could function as a negative regulator of Mcm7 ubiquitylation. Consistent with this notion, overexpression of Usp37 increases levels of Mcm7 on chromatin, although this effect has been attributed to deregulation of Cdt1 (Hernandez-Perez et al. 2016). It will be interesting to test these hypotheses and determine whether the other hits, such as Wrip1, Ari3A, Dvc1, Rnf138, Rd23b, and Senp6, are vRPC replisome components or whether they cooperate in some way with CRL2Lrr1 to extract CMG from chromatin.
Materials and methods
Protein purification
Biotinylated LacR was purified as reported previously (Dewar et al. 2015).
Plasmids
p[lacOx32] was prepared as described previously (Dewar et al. 2015).
Xenopus egg extracts, sperm chromatin, and DNA replication
Xenopus egg extracts were prepared essentially as described (Lebofsky et al. 2009). Briefly, licensing mixes were prepared by mixing high-speed supernatant (HSS) of egg cytoplasm with either plasmid DNA at a final concentration of 7.5–15 ng/µL or demembranated sperm chromatin at a final concentration of 10,000 sperm per microliter. Licensing mixes were incubated for 30 min at room temperature to assemble prereplicative complexes (pre-RCs). To block pre-RC formation, Geminin was added to HSS at a final concentration of 10 µM and incubated for 10 min at room temperature prior to addition of plasmid DNA or sperm chromatin. To initiate replication, 1 vol of licensing mix was mixed with 2 vol of nucleoplasmic extract (NPE). For experiments containing radiolabeled DNA, [α-32P]dATP was added to NPE prior to replication initiation. Reactions were typically stopped in 5–10 vol of replication stop buffer (8 mM EDTA, 0.13% phosphoric acid, 10% ficoll, 5% SDS, 0.2% bromophenol blue, 80 mM Tris at pH 8), treated with 1 µg/µL Proteinase K, and then directly analyzed by gel electrophoresis. Where dissolution and ligation were monitored, reactions were stopped in 10 vol of stop solution (0.5% SDS, 25 mM EDTA, 50 mM Tris-HCl at pH 7.5) and treated with 190 ng/µL RNase A and 909 ng/µL Proteinase K, and termination assays were performed as described previously (Dewar et al. 2015). Where p97-i and Cul-i were used, 0.05 vol in DMSO was added to yield the following working concentrations: 200 µM NMS-873 (Sigma) and 200 µM MLN4924 (Active Biochem). For sperm chromatin assays, 5 µM MLN4924 was used, as described previously (Moreno et al. 2014). Where Cdc7-i was used, 0.2 vol in MilliQ water was added to yield a final working concentration of 100 µM PHA-767491 (Sigma). Ubiquitin mutants (Boston Biochem) were added to a final concentration of 100 µM. Samples were separated by agarose gel electrophoresis. Gels were exposed to phosphorscreens and imaged on the Typhoon FLA 7000 phosphorimager (GE Healthcare). Band or total lane intensities were quantified using Multi-Gauge software (Fujifilm) with subtraction of an appropriate background. To determine relative replication efficiency (e.g., Fig. 3B), the most intense lane was set to 100.
Western blotting
Protein samples were run on Mini-PROTEAN TGX precast protein gels (Bio-Rad) and transferred to PVDF membranes (Perkin Elmer). Membranes were blocked in a solution of 5% dry milk dissolved in 1× PBST for 30–60 min and then incubated with primary antibody at a dilution of 1:500 to 1:20,000 in 1× PBST containing 1% BSA for 12–16 h at 4°C. After extensive washing in 1× PBST, membranes were incubated with goat anti-rabbit horseradish peroxidase (HRP)-conjugated secondary antibodies (Jackson ImmunoResearch) at a dilution of 1:20,000 to 1:30,000 in 5% dry milk and 1× PBST for 1 h at room temperature. Membranes were washed extensively and briefly incubated in HyGLO chemiluminescent HRP antibody detection reagent (Denville) or SuperSignal West Femto maximum sensitivity substrate (ThermoFisher) and developed using the chemiluminescence function on the Amersham Imager 600 (GE).
Induction of termination
LacR-bound p[lacOx32] was used to induce termination in Xenopus egg extracts, as described previously (Dewar et al. 2015).
PPDs
PPDs were performed as described previously (Budzowska et al. 2015). At least two biological replicates were performed for all PPDs. Proteins associated with the chromatin fraction were visualized by Western blotting as described above. Using ImageJ, we measured the intensity of each band and an appropriate background and subtracted the latter from the former. For each protein examined, the highest-intensity band in each blot was assigned a value of 1. In Figure 4D, to quantify Mcm7 ubiquitylation/dissociation, Mcm6 dissociation, and Cdc45 dissociation, the band intensity obtained above was subtracted from 1 and plotted. In Figure 4D and Supplemental Figure S6B, the mean and standard deviation of three biological replicates were graphed.
CHROMASS
Sperm chromatin was replicated for 60 min with or without Geminin pretreatment in the presence of DMSO, NMS-873, or MLN4924. Chromatin and associated proteins were isolated by centrifugation through a sucrose cushion and then processed and analyzed by CHROMASS as described previously (Räschle et al. 2015). Two biological replicates of the experiment were performed, and two parallel reactions were assembled for each condition during each replicate, yielding four biochemical replicates.
Liquid chromatography-tandem MS (LC-MS/MS) analysis
Peptides were separated on reversed-phase columns (50 cm, 75-µm inner diameter, packed in house with ReproSil-Pur C18-AQ 1.8 µm resin [Dr. Maisch GmbH]) and directly injected into the mass spectrometer (Q Exactive HF, Thermo Scientific). Using a Nanoflow HPLC (Thermo Scientific), peptides were loaded in buffer A (0.5% acetic acid) and eluted with a 4-h gradient (5%–95% buffer B [80% acetonitrile, 0.5% acetic acid]) at a constant flow rate of 250 nL/min. For the reversed-phase separation, the column was maintained at a constant temperature of 60°C. The mass spectrometer was operated in a data-dependent fashion using a top N method for peptide sequencing.
Data processing in MaxQuant
Raw data were analyzed with MaxQuant version 1.5.3.31 using a label-free algorithm (Cox et al. 2014). A FDR of 0.01 for proteins and peptides and a minimum peptide length of 7 amino acids were required. MS/MS spectra were searched against a nonredundant Xenopus database (available on request). For the Andromeda search, trypsin was chosen for enzyme specificity, allowing for cleavage N-terminal to proline. Cysteine carbamidomethylation was selected as a fixed modification, while protein N-terminal acetylation and methionine oxidation were selected as variable modifications. Maximally, two missed cleavages were allowed. Initial mass deviation of precursor ions was up to 7 ppm, and mass deviation for fragment ions was 0.5 Da. Quantification in MaxQuant was performed using the built-in LFQ algorithm. “Match between run” was used to transfer identities between all samples. Raw files and MaxQuant output tables are available at ProteomeXchange (http://proteomecentral.proteomexchange.org; ID = PXD004828).
Statistical analysis
All statistical analysis was performed using Perseus software (Tyanova et al. 2016). All calculations were based on the LFQ intensities reported by MaxQuant. A modified t-test with permutation-based FDR statistics was used to identify proteins that were significantly up-regulated with p97-i or Cul-i compared with control reactions (two-sample test implemented in Perseus version 1.5.5.5 with the following parameters: FDR < 0.05, S0 = 0.1, minimal number of valid values: four in at least one replicate group). Missing values were imputed into control samples only (CTR and CTR + Geminin), with numbers from a normal distribution with a mean and standard deviation chosen to best simulate low abundance values close to the detection limit. All test results are reported in Supplemental Table S1 (column B–M). Proteins that were significantly up-regulated in the presence of p97-i or Cul-i compared with a control reaction without drug were further analyzed by nonsupervised hierarchical clustering. Log2 LFQ intensities were z-scored, and the mean of the four replicates was used for clustering. Main clusters were extracted and subjectively assigned to group I (up with p97-i and Cul-i), group II (up with p97-i only), or group III (up with Cul-i only). Each group was reclustered and is represented as a heat map (see Fig. 2B).
Coimmunoprecipitations
Coimmunoprecipitations were performed with antibodies raised against Lrr1, Dcaf17, and Brwd3 that were affinity-purified to 1 mg mL−1. For mock immunoprecipitations, nonspecific IgGs were used at 1 mg mL−1. Three volumes of antibody was incubated with 1 vol of Protein A Sepharose Fast Flow resin (PAS) (GE Healthcare) overnight at 4°C. Five volumes of 20% NPE containing 0.25% NP-40 was added to the conjugated resin and incubated for 2 h at 4°C. The supernatant was harvested and mixed with an equal volume of 2× SDS sample buffer (100 mM Tris at pH 6.8, 4% SDS, 0.2% bromophenol blue, 20% glycerol, 10% β-mercaptoethanol) for Western blot analysis. The resin was washed five times with phospho-buffered saline (PBS) + 0.25% NP-40 followed by elution of immunoprecipitated proteins with 1× SDS sample buffer (50 mM Tris at pH 6.8, 2% SDS, 0.1% bromophenol blue, 10% glycerol, 5% β-mercaptoethanol) for analysis by Western blotting.
Immunodepletions
For Lrr1 depletion, 3 vol of Lrr1 antibody at 1 mg mL−1 was incubated with 1 vol of Protein A Sepharose Fast Flow (PAS) (GE Healthcare) overnight at 4°C. For mock depletion, 3 vol of nonspecific IgGs, concentrated to 1 mg mL−1, was used. One volume of antibody-conjugated Sepharose was then added to 5 vol of precleared HSS or NPE and incubated for 1 h at 4°C. The HSS or NPE was collected and incubated two more times with antibody-conjugated sepharose for a total of three rounds of depletion. The depleted HSS or NPE was collected and used immediately for DNA replication, as described above.
Usp21 treatment
PPDs were performed as described above, except that proteins associated with 20–30 ng of plasmid were washed additionally with Usp21 buffer (150 mM NaCl, 10mM DTT, 50 mM Tris at pH 7.5) to remove any detergent, resuspended in 20 µL of Usp21 buffer rather than 2× sample buffer, and then split into two 10-µL samples. One microliter of Usp21 (gift of D. Finley) or buffer was added, and the sample was incubated for 1 h at 37°C to allow for deubiquitylation. The reaction was stopped by addition of 2× SDS sample buffer (100 mM Tris at pH 6.8, 4% SDS, 0.2% bromophenol blue, 20% glycerol, 10% β-mercaptoethanol).
Antibodies
Cul2 antibodies were raised by New England Peptide against a synthetic 16-amino-acid peptide from the N terminus of Xenopus laevis Cul2 (H2N-MSLKPRVVDFDETWNKC-amide). Lrr1 antibodies used for immunodepletions and immunoprecipitations were raised by New England Peptide against a synthetic 16-amino-acid peptide from the C terminus of X. laevis Lrr1 (Ac-CYSQFLDKYLQSTRV-OH). Lrr1 antibodies used for Western blotting were raised by Abgent against a bacterially expressed 180-amino-acid fragment from the N terminus of X. laevis Lrr1. EloC antibodies were raised by New England Peptide against a synthetic 11-amino-acid peptide from the N terminus of X. laevis EloC (H2N-MDGEEKTYGGC-amide). Traip antibodies were raised by New England Peptide against a synthetic 16-amino-acid peptide from the C terminus of X. laevis Traip (Ac-CTSSLANQPRLEDFLK-OH). Cul4b antibodies were raised by New England Peptide against a synthetic 15-amino-acid peptide from the C terminus of X. laevis Cul4b (Ac-CERDKENPNQYNYVA-OH). Ddb1 antibodies were raised by New England Peptide against a synthetic 14-amino-acid peptide from the C terminus of X. laevis Ddb1 (Ac-CDDLIKVVEELTRIH-OH). Dcaf17 antibodies were raised by New England Peptide against a synthetic 14-amino-acid peptide from the C terminus of X. laevis Dcaf17 (Ac-CRLVKKRFALLDDDP-OH). Brwd3 antibodies were raised by New England Peptide against a synthetic 14-amino-acid peptide from the C terminus of X. laevis Brwd3 (Ac-CTEKARVSHLMGWNG-OH). Histone H3 antibodies were obtained from Cell Signaling (no. 9715S). Timeless antibodies were obtained from Abcam (ab50943). Antibodies against Ctf18 were a generous gift of T. Takahashi. Antibodies against Tipin were a generous gift from V. Costanzo (London Research Instituite). Antibodies against And-1 were a generous gift from A. Dutta (University of Virginia). The following antibodies used in this study were described previously: Mcm7 and Orc2 (Fang and Newport 1993), Cdc45 (Mimura and Takisawa 1998); RPA (Walter and Newport 2000), DNA Pol ε (p60 subunit) and DNA Pol δ (Pacek et al. 2006), Claspin (Byun et al. 2005); Mcm10 (Wohlschlegel et al. 2002), PCNA (Kochaniak et al. 2009), DNA Pol α (Arias and Walter 2005), and p97, Ufd1, and Npl4 (Raman et al. 2011). Antibodies against Mcm6, Sld5, and Dna2 will be described elsewhere.
Supplementary Material
Acknowledgments
We thank Wade Harper, Daniel Semlow, and Benjamin Stinson for feedback on the manuscript, and members of the Walter laboratory for helpful discussions. We thank Jürgen Cox, Tikira Temu, Korbinian Mayr, Igor Paron, and Gabi Sowa for bioinformatics and technical support. We thank Vincenzo Costanzo for the Tipin antibody, Anindya Dutta for the And-1 antibody, and Tatsuro Takahashi for the Ctf18 antibody. We thank Daniel Finley for the Usp21 protein. We thank K. Labib and A. Gambus for discussing unpublished results. This work was supported by National Institutes of Health grant GM80676 to J.C.W., and the Center for Integrated Protein Science Munich (CIPSM). J.C.W. is an investigator of the Howard Hughes Medical Institute. J.M.D. performed the experiments described in Figures 1, 2, 4 (A and B), and 5 (A and B) and Supplemental Figures S1 (A and C), S2, S3, S4 (A, C, and D), and S7 (A–E). E.L. performed the experiments described in Figures 3, 4 (C–E), and 5C and Supplemental Figures S1B, S4B, S5, S6, and S7F. M.R. performed the MS and analysis described in Figure 2 and Supplemental Figures S3 and S4. J.C.W. and J.M.D. conceived the study. J.C.W., J.M.D., E.L., M.M., and M.R. designed the experiments. J.C.W., J.M.D., E.L., and M.R. wrote the manuscript.
Footnotes
Supplemental material is available for this article.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.291799.116.
References
- Alexandru G, Graumann J, Smith GT, Kolawa NJ, Fang R, Deshaies RJ. 2008. UBXD7 binds multiple ubiquitin ligases and implicates p97 in HIF1α turnover. Cell 134: 804–816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arias EE, Walter JC. 2005. Replication-dependent destruction of Cdt1 limits DNA replication to a single round per cell cycle in Xenopus egg extracts. Genes Dev 19: 114–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell SP, Labib K. 2016. Chromosome duplication in Saccharomyces cerevisiae. Genetics 203: 1027–1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blake D, Luke B, Kanellis P, Jorgensen P, Goh T, Penfold S, Breitkreutz BJ, Durocher D, Peter M, Tyers M. 2006. The F-box protein Dia2 overcomes replication impedance to promote genome stability in Saccharomyces cerevisiae. Genetics 174: 1709–1727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchberger A, Schindelin H, Hanzelmann P. 2015. Control of p97 function by cofactor binding. FEBS Lett 589: 2578–2589. [DOI] [PubMed] [Google Scholar]
- Budzowska M, Graham TG, Sobeck A, Waga S, Walter JC. 2015. Regulation of the Rev1-pol ς complex during bypass of a DNA interstrand cross-link. EMBO J 34: 1971–1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgers PMJ. 2009. Polymerase dynamics at the eukaryotic DNA replication fork. J Biol Chem 284: 4041–4045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byun TS, Pacek M, Yee MC, Walter JC, Cimprich KA. 2005. Functional uncoupling of MCM helicase and DNA polymerase activities activates the ATR-dependent checkpoint. Genes Dev 19: 1040–1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cloutier P, Lavallee-Adam M, Faubert D, Blanchette M, Coulombe B. 2013. A newly uncovered group of distantly related lysine methyltransferases preferentially interact with molecular chaperones to regulate their activity. PLoS Genet 9: e1003210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox J, Hein MY, Luber CA, Paron I, Nagaraj N, Mann M. 2014. Accurate proteome-wide label-free quantification by delayed normalization and maximal peptide ratio extraction, termed MaxLFQ. Mol Cell Proteomics 13: 2513–2526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- den Besten W, Verma R, Kleiger G, Oania RS, Deshaies RJ. 2012. NEDD8 links cullin-RING ubiquitin ligase function to the p97 pathway. Nat Struct Mol Biol 19: 511–516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devbhandari S, Jiang J, Kumar C, Whitehouse I, Remus D. 2017. Chromatin constrains the initiation and elongation of DNA replication. Mol Cell 65: 131–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewar JM, Budzowska M, Walter JC. 2015. The mechanism of DNA replication termination in vertebrates. Nature 525: 345–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dungrawala H, Rose KL, Bhat KP, Mohni KN, Glick GG, Couch FB, Cortez D. 2015. The replication checkpoint prevents two types of fork collapse without regulating replisome stability. Mol Cell 59: 998–1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evrin C, Clarke P, Zech J, Lurz R, Sun J, Uhle S, Li H, Stillman B, Speck C. 2009. A double-hexameric MCM2–7 complex is loaded onto origin DNA during licensing of eukaryotic DNA replication. Proc Natl Acad Sci 106: 20240–20245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang F, Newport JW. 1993. Distinct roles of cdk2 and cdc2 in RP-A phosphorylation during the cell cycle. J Cell Sci 106: 983–994. [DOI] [PubMed] [Google Scholar]
- Franz A, Orth M, Pirson PA, Sonneville R, Blow JJ, Gartner A, Stemmann O, Hoppe T. 2011. CDC-48/p97 coordinates CDT-1 degradation with GINS chromatin dissociation to ensure faithful DNA replication. Mol Cell 44: 85–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franz A, Pirson PA, Pilger D, Halder S, Achuthankutty D, Kashkar H, Ramadan K, Hoppe T. 2016. Chromatin-associated degradation is defined by UBXN-3/FAF1 to safeguard DNA replication fork progression. Nat Commun 7: 10612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu YV, Yardimci H, Long DT, Guainazzi A, Bermudez VP, Hurwitz J, van Oijen A, Scharer OD, Walter JC. 2011. Selective bypass of a lagging strand roadblock by the eukaryotic replicative DNA helicase. Cell 146: 931–941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gambus A, Jones RC, Sanchez-Diaz A, Kanemaki M, van Deursen F, Edmondson RD, Labib K. 2006. GINS maintains association of Cdc45 with MCM in replisome progression complexes at eukaryotic DNA replication forks. Nat Cell Biol 8: 358–366. [DOI] [PubMed] [Google Scholar]
- Gambus A, van Deursen F, Polychronopoulos D, Foltman M, Jones RC, Edmondson RD, Calzada A, Labib K. 2009. A key role for Ctf4 in coupling the MCM2–7 helicase to DNA polymerase α within the eukaryotic replisome. EMBO J 28: 2992–3004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Rodriguez LJ, De Piccoli G, Marchesi V, Jones RC, Edmondson RD, Labib K. 2015. A conserved Pol binding module in Ctf18–RFC is required for S-phase checkpoint activation downstream of Mec1. Nucleic Acids Res 43: 8830–8838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garg P, Stith CM, Sabouri N, Johansson E, Burgers PM. 2004. Idling by DNA polymerase δ maintains a ligatable nick during lagging-strand DNA replication. Genes Dev 18: 2764–2773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgescu RE, Langston L, Yao NY, Yurieva O, Zhang D, Finkelstein J, Agarwal T, O'Donnell ME. 2014. Mechanism of asymmetric polymerase assembly at the eukaryotic replication fork. Nat Struct Mol Biol 21: 664–670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gotter AL, Suppa C, Emanuel BS. 2007. Mammalian TIMELESS and Tipin are evolutionarily conserved replication fork-associated factors. J Mol Biol 366: 36–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanzelmann P, Buchberger A, Schindelin H. 2011. Hierarchical binding of cofactors to the AAA ATPase p97. Structure 19: 833–843. [DOI] [PubMed] [Google Scholar]
- Hernandez-Perez S, Cabrera E, Amoedo H, Rodriguez-Acebes S, Koundrioukoff S, Debatisse M, Mendez J, Freire R. 2016. USP37 deubiquitinates Cdt1 and contributes to regulate DNA replication. Mol Oncol 10: 1196–1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann S, Smedegaard S, Nakamura K, Mortuza GB, Räschle M, Ibanez de Opakua A, Oka Y, Feng Y, Blanco FJ, Mann M, et al. 2016. TRAIP is a PCNA-binding ubiquitin ligase that protects genome stability after replication stress. J Cell Biol 212: 63–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin J, Arias EE, Chen J, Harper JW, Walter JC. 2006. A family of diverse Cul4–Ddb1-interacting proteins includes Cdt2, which is required for S Phase destruction of the replication factor Cdt1. Mol Cell 23: 709–721. [DOI] [PubMed] [Google Scholar]
- Kamura T, Maenaka K, Kotoshiba S, Matsumoto M, Kohda D, Conaway RC, Conaway JW, Nakayama KI. 2004. VHL-box and SOCS-box domains determine binding specificity for Cul2–Rbx1 and Cul5–Rbx2 modules of ubiquitin ligases. Gene Dev 18: 3055–3065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang YH, Galal WC, Farina A, Tappin I, Hurwitz J. 2012. Properties of the human Cdc45/Mcm2–7/GINS helicase complex and its action with DNA polymerase ε in rolling circle DNA synthesis. Proc Natl Acad Sci 109: 6042–6047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan DL, Davey MJ, O'Donnell M. 2003. Mcm4,6,7 uses a ‘pump in ring’ mechanism to unwind DNA by steric exclusion and actively translocate along a duplex. J Biol Chem 278: 49171–49182. [DOI] [PubMed] [Google Scholar]
- Kochaniak AB, Habuchi S, Loparo JJ, Chang DJ, Cimprich KA, Walter JC, van Oijen AM. 2009. Proliferating cell nuclear antigen uses two distinct modes to move along DNA. J Biol Chem 284: 17700–17710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koepp DM, Kile AC, Swaminathan S, Rodriguez-Rivera V. 2006. The F-box protein Dia2 regulates DNA replication. Mol Biol Cell 17: 1540–1548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langston LD, Zhang D, Yurieva O, Georgescu RE, Finkelstein J, Yao NY, Indiani C, O'Donnell ME. 2014. CMG helicase and DNA polymerase ε form a functional 15-subunit holoenzyme for eukaryotic leading-strand DNA replication. Proc Natl Acad Sci 111: 15390–15395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebofsky R, Takahashi T, Walter JC. 2009. DNA replication in nucleus-free Xenopus egg extracts. Methods Mol Biol 521: 229–252. [DOI] [PubMed] [Google Scholar]
- Lee J, Gold DA, Shevchenko A, Dunphy WG. 2005. Roles of replication fork-interacting and Chk1-activating domains from Claspin in a DNA replication checkpoint response. Mol Biol Cell 16: 5269–5282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine AJ, Kang HS, Billheimer FE. 1970. DNA replication in SV40 infected cells. I. Analysis of replicating SV40 DNA. J Mol Biol 50: 549–568. [DOI] [PubMed] [Google Scholar]
- Maculins T, Nkosi PJ, Nishikawa H, Labib K. 2015. Tethering of SCF(Dia2) to the replisome promotes efficient ubiquitylation and disassembly of the CMG helicase. Curr Biol 25: 2254–2259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maric M, Maculins T, De Piccoli G, Labib K. 2014. Cdc48 and a ubiquitin ligase drive disassembly of the CMG helicase at the end of DNA replication. Science 346: 1253596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merchant AM, Kawasaki Y, Chen Y, Lei M, Tye BK. 1997. A lesion in the DNA replication initiation factor Mcm10 induces pausing of elongation forks through chromosomal replication origins in Saccharomyces cerevisiae. Mol Cell Biol 17: 3261–3271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merlet J, Burger J, Tavernier N, Richaudeau B, Gomes JE, Pintard L. 2010. The CRL2LRR-1 ubiquitin ligase regulates cell cycle progression during C. elegans development. Development 137: 3857–3866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer H, Bug M, Bremer S. 2012. Emerging functions of the VCP/p97 AAA-ATPase in the ubiquitin system. Nat Cell Biol 14: 117–123. [DOI] [PubMed] [Google Scholar]
- Mimura S, Takisawa H. 1998. Xenopus Cdc45-dependent loading of DNA polymerase α onto chromatin under the control of S-phase Cdk. EMBO J 17: 5699–5707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montagnoli A, Valsasina B, Croci V, Menichincheri M, Rainoldi S, Marchesi V, Tibolla M, Tenca P, Brotherton D, Albanese C, et al. 2008. A Cdc7 kinase inhibitor restricts initiation of DNA replication and has antitumor activity. Nat Chem Biol 4: 357–365. [DOI] [PubMed] [Google Scholar]
- Moreno SP, Bailey R, Campion N, Herron S, Gambus A. 2014. Polyubiquitylation drives replisome disassembly at the termination of DNA replication. Science 346: 477–481. [DOI] [PubMed] [Google Scholar]
- O'Connell BC, Adamson B, Lydeard JR, Sowa ME, Ciccia A, Bredemeyer AL, Schlabach M, Gygi SP, Elledge SJ, Harper JW. 2010. A genome-wide camptothecin sensitivity screen identifies a mammalian MMS22L–NFKBIL2 complex required for genomic stability. Mol Cell 40: 645–657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Donnell L, Panier S, Wildenhain J, Tkach JM, Al-Hakim A, Landry MC, Escribano-Diaz C, Szilard RK, Young JTF, Munro M, et al. 2010. The MMS22L–TONSL complex mediates recovery from replication stress and homologous recombination. Mol Cell 40: 619–631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orme CM, Bogan JS. 2012. The ubiquitin regulatory X (UBX) domain-containing protein TUG regulates the p97 ATPase and resides at the endoplasmic reticulum-golgi intermediate compartment. J Biol Chem 287: 6679–6692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ossareh-Nazari B, Katsiarimpa A, Merlet J, Pintard L. 2016. RNAi-based suppressor screens reveal genetic interactions between the CRL2LRR-1 E3-ligase and the DNA replication machinery in Caenorhabditis elegans. G3 (Bethesda) 6: 3431–3442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozturk N, VanVickle-Chavez SJ, Akileswaran L, Van Gelderb RN, Sancar A. 2013. Ramshackle (Brwd3) promotes light-induced ubiquitylation of Drosophila cryptochrome by DDB1–CUL4–ROC1 E3 ligase complex. Proc Natl Acad Sci 110: 4980–4985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pacek M, Tutter AV, Kubota Y, Takisawa H, Walter JC. 2006. Localization of MCM2–7, Cdc45, and GINS to the site of DNA unwinding during eukaryotic DNA replication. Mol Cell 21: 581–587. [DOI] [PubMed] [Google Scholar]
- Piwko W, Olma MH, Held M, Bianco JN, Pedrioli PG, Hofmann K, Pasero P, Gerlich DW, Peter M. 2010. RNAi-based screening identifies the Mms22L–Nfkbil2 complex as a novel regulator of DNA replication in human cells. EMBO J 29: 4210–4222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prioleau MN, MacAlpine DM. 2016. DNA replication origins-where do we begin? Genes Dev 30: 1683–1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raman M, Havens CG, Walter JC, Harper JW. 2011. A genome wide screen identifies p97 as an essential regulator of DNA damage-dependent CDT1 destruction. Mol Cell 44: 72–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raman M, Sergeev M, Garnaas M, Lydeard JR, Huttlin EL, Goessling W, Shah JV, Harper JW. 2015. Systematic proteomics of the VCP-UBXD adaptor network identifies a role for UBXN10 in regulating ciliogenesis. Nat Cell Biol 17: 1356–1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Räschle M, Smeenk G, Hansen RK, Temu T, Oka Y, Hein MY, Nagaraj N, Long DT, Walter JC, Hofmann K, et al. 2015. DNA repair. Proteomics reveals dynamic assembly of repair complexes during bypass of DNA cross-links. Science 348: 1253671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remus D, Beuron F, Tolun G, Griffith JD, Morris EP, Diffley JF. 2009. Concerted loading of Mcm2–7 double hexamers around DNA during DNA replication origin licensing. Cell 139: 719–730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seidman MM, Salzman NP. 1979. Late replicative intermediates are accumulated during simian virus 40 DNA replication in vivo and in vitro. J Virol 30: 600–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiomi Y, Masutani C, Hanaoka F, Kimura H, Tsurimoto T. 2007. A second proliferating cell nuclear antigen loader complex, Ctf18–replication factor C, stimulates DNA polymerase η activity. J Biol Chem 282: 20906–20914. [DOI] [PubMed] [Google Scholar]
- Siddiqui K, On KF, Diffley JF. 2013. Regulating DNA replication in eukarya. Cold Spring Harb Perspect Biol 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon AC, Zhou JC, Perera RL, van Deursen F, Evrin C, Ivanova ME, Kilkenny ML, Renault L, Kjaer S, Matak-Vinkovic D, et al. 2014. A Ctf4 trimer couples the CMG helicase to DNA polymerase α in the eukaryotic replisome. Nature 510: 293–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonneville R, Priego-Moreno S, Knebel A, Johnson C, Hastie J, Gartner A, Gambus A, Labib K. 2017. CUL-2LRR-1 and UBXN-3/FAF1 drive replisome disassembly during DNA replication termination and mitosis. Nat Cell Biol 10.1038/ncb3500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tapper DP, DePamphilis ML. 1978. Discontinuous DNA replication: accumulation of Simian virus 40 DNA at specific stages in its replication. J Mol Biol 120: 401–422. [DOI] [PubMed] [Google Scholar]
- Tyanova S, Temu T, Sinitcyn P, Carlson A, Hein MY, Geiger T, Mann M, Cox J. 2016. The Perseus computational platform for comprehensive analysis of (prote)omics data. Nat Methods 13: 731–740. [DOI] [PubMed] [Google Scholar]
- Verma R, Oania R, Fang R, Smith GT, Deshaies RJ. 2011. Cdc48/p97 mediates UV-dependent turnover of RNA Pol II. Mol Cell 41: 82–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villa F, Simon AC, Ortiz Bazan MA, Kilkenny ML, Wirthensohn D, Wightman M, Matak-Vinkovic D, Pellegrini L, Labib K. 2016. Ctf4 Is a hub in the eukaryotic replisome that links multiple CIP-box proteins to the CMG helicase. Mol Cell 63: 385–396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter J, Newport J. 2000. Initiation of eukaryotic DNA replication: origin unwinding and sequential chromatin association of Cdc45, RPA, and DNA polymerase α. Mol Cell 5: 617–627. [DOI] [PubMed] [Google Scholar]
- Wohlschlegel JA, Dhar SK, Prokhorova TA, Dutta A, Walter JC. 2002. Xenopus mcm10 binds to origins of DNA replication after mcm2–7 and stimulates origin binding of cdc45. Mol Cell 9: 233–240. [DOI] [PubMed] [Google Scholar]
- Ye Y, Akutsu M, Reyes-Turcu F, Enchev RI, Wilkinson KD, Komander D. 2011. Polyubiquitin binding and cross-reactivity in the USP domain deubiquitinase USP21. EMBO Rep 12: 350–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeeles JT, Janska A, Early A, Diffley JF. 2017. How the eukaryotic replisome achieves rapid and efficient DNA replication. Mol Cell 65: 105–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu W, Ukomadu C, Jha S, Senga T, Dhar SK, Wohlschlegel JA, Nutt LK, Kornbluth S, Dutta A. 2007. Mcm10 and And-1/CTF4 recruit DNA polymerase α to chromatin for initiation of DNA replication. Genes Dev 21: 2288–2299. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.