Here, Ziolkowski et al. combine high-throughput fluorescence methods to measure crossovers with natural Arabidopsis ecotypes in order to identify the first trans-acting modifier of meiotic recombination in plants. The authors found that HEI10, which encodes a conserved ubiquitin E3 ligase, naturally limits Arabidopsis crossovers and has the potential to influence the response to selection.
Keywords: meiosis, recombination, modifier, HEI10, ZMM, Arabidopsis
Abstract
During meiosis, homologous chromosomes undergo crossover recombination, which creates genetic diversity and balances homolog segregation. Despite these critical functions, crossover frequency varies extensively within and between species. Although natural crossover recombination modifier loci have been detected in plants, causal genes have remained elusive. Using natural Arabidopsis thaliana accessions, we identified two major recombination quantitative trait loci (rQTLs) that explain 56.9% of crossover variation in Col×Ler F2 populations. We mapped rQTL1 to semidominant polymorphisms in HEI10, which encodes a conserved ubiquitin E3 ligase that regulates crossovers. Null hei10 mutants are haploinsufficient, and, using genome-wide mapping and immunocytology, we show that transformation of additional HEI10 copies is sufficient to more than double euchromatic crossovers. However, heterochromatic centromeres remained recombination-suppressed. The strongest HEI10-mediated crossover increases occur in subtelomeric euchromatin, which is reminiscent of sex differences in Arabidopsis recombination. Our work reveals that HEI10 naturally limits Arabidopsis crossovers and has the potential to influence the response to selection.
The majority of eukaryotes reproduces via the meiotic cell division, where a diploid cell replicates DNA once and segregates chromosomes twice to produce tetrads of haploid gametes (Barton and Charlesworth 1998). Genetic diversity is generated between gametes due to independent chromosome segregation in addition to recombination between homologous chromosomes during meiotic prophase I (Barton and Charlesworth 1998). Despite the importance of crossovers for balanced chromosome segregation during meiosis and fertility, extensive genetic variation in recombination frequency is observed within and between species (Sanchez-Moran et al. 2002; Coop and Przeworski 2006; Fledel-Alon et al. 2011; Hinch et al. 2011; Sandor et al. 2012; Bauer et al. 2013; Rodgers-Melnick et al. 2015; Ziolkowski et al. 2015; Johnston et al. 2016). Importantly, natural variation that modifies crossover frequency has the potential to widely influence genetic adaptation and the response to selection (Hill and Robertson 1966; Feldman et al. 1996; Barton and Charlesworth 1998).
Genetic polymorphisms that modify crossover frequency can be classified as cis- or trans-acting, according to whether they control recombination on the same chromosome or throughout the genome, respectively (Coop and Przeworski 2006; Yandeau-Nelson et al. 2006; Baudat and de Massy 2007; Ziolkowski et al. 2015). Examples of human trans modifier loci include the RNF212 meiotic E3 ligase gene, which controls crossover levels (Kong et al. 2008; Fledel-Alon et al. 2011), and the PRDM9 zinc finger SET domain gene, which specifies recombination hot spot locations (Fledel-Alon et al. 2011; Hinch et al. 2011). Polymorphisms are also known to exert local cis effects, where heterozygous polymorphisms can inhibit crossover repair of interhomolog strand invasion events (Borts and Haber 1987; Baudat and de Massy 2007; Mercier et al. 2015). Structural variation (for example, insertions and deletions [indels], translocations, and inversions) are also associated with crossover suppression at larger physical scales (Fransz et al. 2016). Extensive evidence for cis and trans modification of crossover frequency exists in plants, including Arabidopsis thaliana (Timmermans et al. 1997; Barth et al. 2001; Yandeau-Nelson et al. 2006; Esch et al. 2007; McMullen et al. 2009; López et al. 2012; Salomé et al. 2012; Bauer et al. 2013; Martín et al. 2014; Rodgers-Melnick et al. 2015; Ziolkowski et al. 2015). Therefore, we sought to use high-throughput fluorescent reporter systems to measure recombination and identify trans-acting crossover modifier loci that vary between A. thaliana accessions.
Meiotic recombination initiates via formation of DNA double-strand breaks (DSBs) by SPO11 transesterases (Mercier et al. 2015). DSBs are resected to generate ssDNA, which is bound by the RecA-related recombinases RAD51 and DMC1 (Mercier et al. 2015). The resulting nucleoprotein filaments can then invade a replicated sister chromatid or a homologous DNA duplex to form a displacement loop (Mercier et al. 2015). Immunostaining of Arabidopsis meiocytes for DMC1, RAD51, or the DSB-associated histone variant γH2A.X has revealed ∼100–200 foci distributed along the paired homologous chromosomes at the leptotene stage (Girard et al. 2015; Yelina et al. 2015). Approximately 10 of these initiating meiotic DSBs mature into crossovers per Arabidopsis meiosis (Giraut et al. 2011; Salomé et al. 2012; Rowan et al. 2015; Choi et al. 2016).
Meiotic interhomolog strand invasion intermediates can follow alternative repair fates, including crossover or noncrossover, which differ in exchange of flanking markers (Mercier et al. 2015). The majority of Arabidopsis crossovers is dependent on the conserved ZMM pathway (named after the budding yeast genes Zip1, Zip2, Zip3, Zip4, Msh4, Msh5, and Mer3), which includes SHOC1, HEI10, ZIP4, MSH4, MSH5, MER3, PTD, MLH1, and MLH3 (Lynn et al. 2007; Mercier et al. 2015). Crossovers generated by the ZMM pathway show interference, where double crossovers are spaced farther apart than expected at random, which is detectable over the scale of megabases in Arabidopsis (Lynn et al. 2007; Mercier et al. 2015). The ZMM pathway is thought to stabilize interhomolog recombination intermediates, including double Holliday junctions, and promote crossover resolution (Lynn et al. 2007; Gray and Cohen 2016). A minority of Arabidopsis crossovers is generated by a noninterfering pathway that includes MUS81 (Mercier et al. 2015). Noncrossover repair of strand invasion events is promoted by multiple nonredundant pathways that include FANCONI ANEMIA COMPLEMENTATION GROUP M (FANCM), MHF1, MHF2, FIDGETIN-LIKE1 (FIGL1), RECQ4A, RECQ4B, TOPOISOMERASE3α (TOP3α), and MSH2 (Mercier et al. 2015). The combined action of these anti-crossover pathways results in repair of ∼90% of initiating meiotic DSBs as noncrossovers.
In this study, we identified an Arabidopsis natural genetic variation that acts in trans to control meiotic crossover frequency. Although A. thaliana is predominantly self-fertilizing, clear evidence for outcrossing exists. For example, Arabidopsis linkage disequilibrium decays rapidly over kilobase distances, and strong historical crossover hot spots are detectable (Kim et al. 2007; Horton et al. 2012; Choi et al. 2013). Genotyping of natural Arabidopsis populations has also revealed standing heterozygosity and evidence for local outcrossing between subpopulations (Bomblies et al. 2010). Therefore, recombination-modifying polymorphisms have had the opportunity to exert an effect on the genetic history of this species. Here we identify natural genetic polymorphisms in the HEI10 meiotic E3 ligase gene that associate with quantitative variation in crossover frequency between Arabidopsis accessions. We further show that HEI10 is highly dosage-sensitive and that transformation of additional HEI10 copies is sufficient to more than double crossover recombination throughout euchromatin. Together, this demonstrates that HEI10 is a limiting factor for interference-sensitive crossover formation in Arabidopsis.
Results
Detecting recombination modifier loci using Col/Ler chromosome substitution lines (CSLs)
Genetic segregation of linked, hemizygous T-DNAs expressing different colors of fluorescent proteins in pollen or seed (fluorescent-tagged lines [FTLs]) can be used to measure Arabidopsis crossover frequency (Fig. 1; Emmanuel et al. 2006; Berchowitz and Copenhaver 2008; Yelina et al. 2013; Ziolkowski et al. 2015). We previously analyzed crossovers in an F2 population derived from crosses between the Col-420 subtelomeric FTL and Catania-1 (Ct) parents, which did not identify significant trans-acting recombination modifier loci (Ziolkowski et al. 2015). To further screen for natural crossover modifiers, we generated a Col-420×Landsberg erecta (Ler) F2 population, which showed higher mean recombination than Col-420×Ct (20.2 cM vs. 15.0 cM) (Fig. 1A; Supplemental Tables S1, S2) and significantly greater variation in crossover frequency between individuals (Brown-Forsythe test, P = 2.91 × 10−32). This is consistent with the presence of trans modifier loci (Fig. 1A; Supplemental Tables S1, S2).
Figure 1.
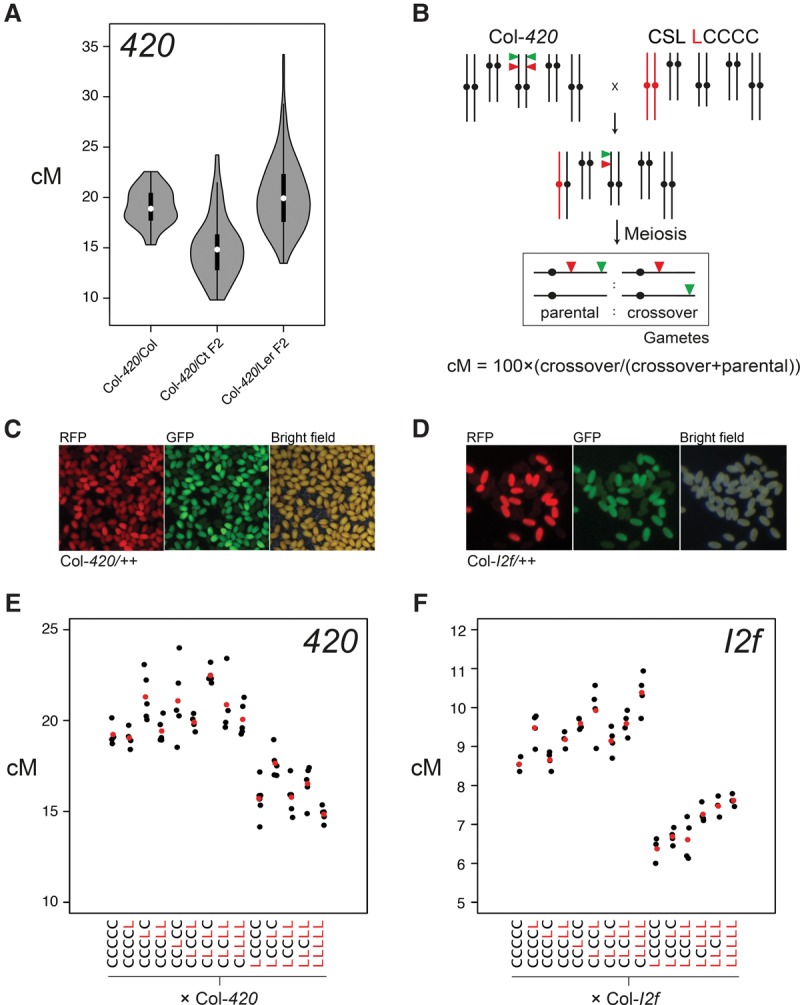
A dosage-sensitive trans-acting recombination modifier on Arabidopsis chromosome 1. (A) Col-420 FTL crossover frequency (in centimorgans) in Col-420/Col, Col-420/Ct F2 (Ziolkowski et al. 2015), or Col-420/Ler F2 populations. (B) Crossing scheme used to analyze CSLs. Arabidopsis chromosomes are colored black (Col) or red (Ler), with FTL transgenes indicated by colored triangles. Parental and crossover fluorescence ratios were used to measure genetic distance (in centimorgans). (C) Representative micrographs of seeds from a 420/++ hemizygote imaged under bright-field or showing green and red fluorescence. (D) As for C, but showing pollen from an I2f/++ hemizygote. (E) 420 crossover frequency in F1 plants derived from Col-420×CSL crosses. CSL genotypes are indicated by “C” (Col) and “L” (Ler) for each chromosome. Replicate F1 data are shown by black dots, and mean values are indicated by red dots. (F) As for E, but measuring crossover frequency (in centimorgans) within FTL interval I2f.
To identify trans recombination modifier loci, we first used Col/Ler CSLs (Fig. 1B; Wijnker et al. 2012). For example, CSL LCCCC denotes Ler (L) and Col (C) genotypes for each of the five chromosomes (Fig. 1B). Fourteen CSLs were crossed to Col-I2f and Col-420 FTLs (crossover reporters located on chromosomes 2 and 3, respectively), and replicate F1 measurements were collected (Fig. 1B–F; Supplemental Tables S3, S4). We observed that all F1 genotypes that were chromosome 1 Col/Ler heterozygous showed significantly reduced crossovers compared with control Col-420×CCCCC F1 plants, with weaker effects detected from the other chromosomes (Fig. 1E,F; Supplemental Tables S3–S5). This reveals the presence of a semidominant trans-acting recombination modifier on chromosome 1.
Genetic mapping of Arabidopsis recombination quantitative trait loci (rQTLs)
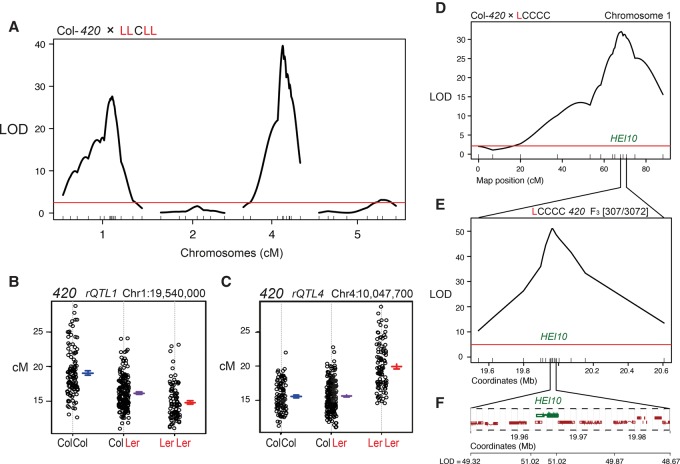
We observed previously that juxtaposition of homozygous and heterozygous regions can influence recombination in cis at the megabase scale (Ziolkowski et al. 2015). To eliminate cis effects and specifically map trans recombination modifiers, we generated an F2 population from a Col-420×LLCLL cross. In this population, chromosome 3 is Col/Col homozygous, which is where the 420 FTL interval is located, and therefore cis effects were excluded. We identified two major trans rQTLs on chromosomes 1 and 4, with logarithm of the (base 10) odds ratio (LOD) scores of 40.2 and 53.5, which explain 23.3% and 33.6% of the variance in recombination, respectively (F-test, P < 2 × 10−16) (Fig. 2A; Supplemental Table S6). rQTL1Ler genotypes associate with low recombination, with heterozygotes showing intermediate crossover frequency (Fig. 2B), consistent with the semidominant effects observed for chromosome 1 in the CSL F1 experiments (Fig. 1E,F; Supplemental Tables S3, S4). In contrast, rQTL4Ler associates with high recombination and behaves recessively, explaining why it was not detected in the CSL experiments (Fig. 2C).
Figure 2.
The rQTL1 recombination modifier locus maps to the meiotic E3 ligase gene HEI10. (A) LOD scores for genetic markers and crossover frequency from a Col-420×LLCLL F2 population. Genetic map positions (in centimorgans) of markers are indicated on the X-axis. The red line indicates the 95% significance threshold. (B) Effects plots showing 420 crossovers (in centimorgans) from Col/Col, Col/Ler, or Ler/Ler individuals for a rQTL1 maker. (C) As for B, but showing a rQTL4 marker. (D) LOD scores for genetic markers and crossover frequency in a Col-420×LCCCC F2 population. The red line indicates the 95% significance threshold. The approximate position of HEI10 is labeled. (E) As for D, but showing the marker LOD for 420 crossover frequency in a 1.7-Mb interval from a F3 Col-420×LCCCC population derived from D. (F) Marker LOD associated with 420 crossovers in proximity to HEI10 (green) and adjacent genes (red).
To investigate the influence of rQTL1 and rQTL4 on meiotic recombination elsewhere in the genome, we performed cytogenetic analysis in Col and Ler in addition to recombinants with low (rQTL1Ler rQTL4Col) or high (rQTL1Col rQTL4Ler) 420 crossovers (Supplemental Fig. S1A–E; Supplemental Tables S7–S9). MLH1 foci occurring along the meiotic synaptonemal complex (visualized by ZYP1 immunostaining) serve as a measure of total interfering crossovers per nucleus (Lambing et al. 2015). We observed significantly more MLH1 foci in rQTL1Col rQTL4Ler lines compared with the other genotypes (Mann-Whitney-Wilcoxon test, P = 0.0396) (Supplemental Fig. S1A,C; Supplemental Table S7). We confirmed the same trend via analysis of chiasmata at metaphase I (Mann-Whitney-Wilcoxon test, P = 2.20 × 10−5) (Supplemental Fig. S1B,D; Supplemental Table S8; Sanchez-Moran et al. 2002). These analyses confirm that Col and Ler polymorphisms underlying rQTL1 and rQTL4 influence crossovers not only in the 420 interval but throughout the chromosomes.
Genetic variation in the HEI10 meiotic E3 ligase gene underlies rQTL1
We sought to identify rQTL1 using an F2 population derived from a Col-420×LCCCC cross, which again revealed a major rQTL on chromosome 1 (Fig. 2D; Supplemental Table S10). We selected an F2 individual that was Col/Ler heterozygous spanning rQTL1 (19.54–21.24 Mb), which was self-fertilized to generate a large F3 population (n = 3072) (Fig. 2E; Supplemental Table S11). Genotyping identified 307 F3 plants with crossovers within the rQTL1 region, which were then measured for 420 crossover frequency and genotyped for 15 additional markers (Fig. 2E; Supplemental Table S11). This narrowed the credible rQTL1 interval to a 34-kb region containing 14 genes (Fig. 2E,F). The most strongly associated marker pair (LOD = 51.02) defined a 4.3-kb interval containing two genes: MRD1 (At1g53480) and HEI10 (At1g53490) (Fig. 2F). As HEI10 belongs to a conserved gene family, which encodes RING domain SUMO/ubiquitin E3 ligases that promote crossovers in diverse eukaryotes, this was the strongest candidate gene for rQTL1 (Supplemental Figs. S2, S3; Bhalla et al. 2008; Kong et al. 2008; Fledel-Alon et al. 2011; Chelysheva et al. 2012; Sandor et al. 2012; Serrentino et al. 2013; De Muyt et al. 2014; Qiao et al. 2014; Johnston et al. 2016; Rao et al. 2017). HEI10 family proteins possess N-terminal RING domains, central coiled-coil domains, and C-terminal regions of unknown function (Supplemental Figs. S2, S3B; Gray and Cohen 2016).
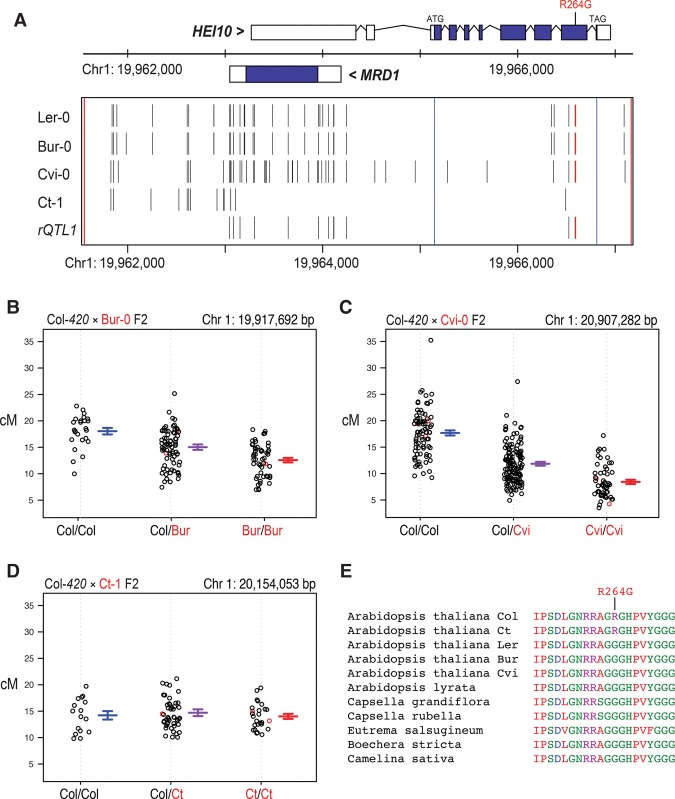
To further investigate HEI10 polymorphisms associated with rQTL1, we sequenced the Ler accession used in our experiments and identified a single nonsynonymous (R264G) substitution and three synonymous intragenic variants relative to the Col reference sequence (Fig. 3A; Supplemental Table S12), which was consistent with 1001 Genomes project data from the closely related Ler-1 and La-0 accessions (Alonso-Blanco et al. 2016). The HEI10 promoter, which overlaps the antisense gene MRD1, is also polymorphic, with 26 single-nucleotide polymorphisms (SNPs) or indels upstream of the start codon (Fig. 3A; Supplemental Table S12). We generated additional F2 populations derived from Col-420×Bur-0 or Col-420×Cvi-0 crosses and again observed significant association between the HEI10 region and crossover frequency (Bur-0 LOD = 8.30, 95% significance threshold LOD = 2.82; Cvi-0 LOD = 31.74, 95% significance threshold LOD = 2.97) (Fig. 3B,C; Supplemental Table S13). We sequenced HEI10Bur and HEI10Cvi and observed 43 and 30 polymorphisms, respectively, relative to HEI10Col (Fig. 3A; Supplemental Table S12). As we previously observed an absence of trans rQTLs in Col-420×Ct-1 populations (Fig. 3D; Ziolkowski et al. 2015), we also sequenced HEI10Ct (Fig. 3A; Supplemental Table S12). Twelve polymorphisms are shared between HEI10Ler, HEI10Cvi, and HEI10Bur but absent from HEI10Ct, including the R264 substitution, which we consider as candidates for rQTL1 causal variants (Fig. 3A; Supplemental Table S12). HEI10 transcript and protein levels measured by quantitative RT–PCR (qRT–PCR) and immunocytogenetic analysis, respectively, did not show significant differences between Col, Ler, and Col/Ler F1 (Supplemental Fig. S1E; Supplemental Tables S14–S16), consistent with the causal rQTL1 polymorphism influencing HEI10 protein function or expression timing rather than expression level. The nonsynonymous R264G polymorphism occurs in the HEI10 C-terminal region (Fig. 3E; Supplemental Figs. S2, S3), which, by analogy with other RING E3 ligases, may play a role in substrate recognition (Deshaies and Joazeiro 2009). We queried 1001 Genomes project data for the frequency and geographic distribution of the HEI10 R264G variants (Supplemental Fig. S4; Alonso-Blanco et al. 2016). Both alleles are globally distributed, with the majority of accessions (959 out of 1008) showing the Ler G264 genotype, and the Col R264 reference allele present in the remaining 123 (11.4%) (Supplemental Fig. S4). As other Brassicaceae HEI10 orthologs show glycine at position 264, this is consistent with the R264 variant observed in Col-0 and Ct-1 being more recently derived within A. thaliana (Fig. 3E).
Figure 3.
Candidate rQTL1 Col/Ler polymorphisms. (A) Plot showing the HEI10 region on chromosome 1. The positions of HEI10 (forward strand) and MRD1 (reverse strand) gene annotations are plotted as black boxes, with coding regions shown in blue. Blue vertical lines indicate HEI10 ATG and TAG codons. Black X-axis ticks show the positions of Ler-0, Bur-0, Cvi-0, and Ct-1 polymorphisms, identified by Sanger sequencing. Red ticks show the nonsynonymous HEI10 substitution R264G. (B) 420 recombination rate (in centimorgans) from individual plants in a Col-420×Bur-0 F2 population, according to the Col-0/Bur-0 genotype at marker 19,917,692 base pairs (bp). (C) As for B but plotting for marker 20,907,282 bp in a Col-420×Cvi-0 F2 population. (D) As for B but plotting for marker 20,154,053 bp in a Col-420×Ct-1 F2 population (the marker is below the 95% significance threshold LOD = 2.27). (E) A multiple sequence alignment of Brassicacea HEI10 orthologs in the region of the R264G substitution.
Arabidopsis crossover frequency is sensitive to HEI10 dosage
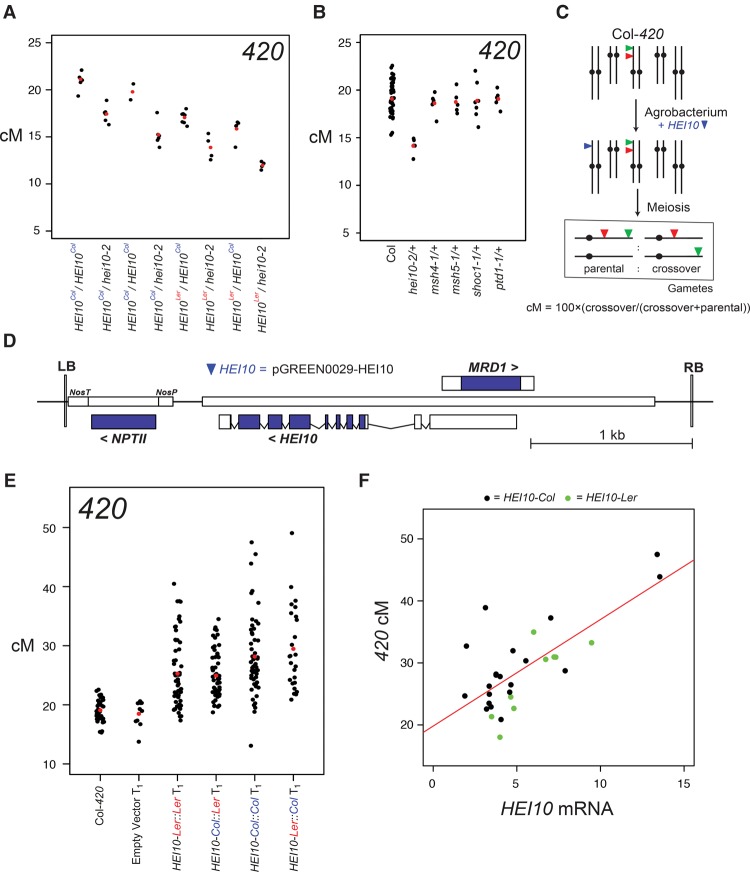
As a genetic test of rQTL1 allelism with HEI10, we crossed recombinant 420 lines with the null hei10-2 allele, which was isolated in the Col background and shows substantially reduced crossovers and fertility when homozygous (Chelysheva et al. 2012). Four independent recombinant lines were used for crosses, two of which were rQTL1Col homozygous, and two that were rQTL1Ler homozygous (Supplemental Table S17). All lines analyzed were rQTL4Ler homozygous. F1 individuals that were hei10-2 heterozygous showed significantly reduced recombination compared with wild-type crosses (Fig. 4A; Supplemental Table S17). This indicates HEI10 dosage sensitivity, which is similar to haploinsufficiency of mouse hei10 and rnf212 mutations (Reynolds et al. 2013; Qiao et al. 2014). Consistent with our previous rQTL mapping, the progeny from rQTL1Ler crosses showed significantly fewer 420 crossovers compared with rQTL1Col progeny (X2, P < 2.2 × 10−16) (Fig. 4A; Supplemental Table S17), which we interpret as reflecting the different activity of HEI10 Col/Ler variants, including R264G. To investigate whether haploinsufficiency is a general property of mutants in the Arabidopsis ZMM pathway, we compared Col-420 F1 crossover measurements using hei10-2, msh4-1, msh5-1, shoc1-1, and ptd1 heterozygotes (Fig. 4B; Supplemental Table S18). Among these mutations, only hei10-2/+ heterozygotes showed significantly reduced crossovers compared with wild type (X2, P = 1.63 × 10−38) (Fig. 4B; Supplemental Table S18), revealing that dosage sensitivity was specific to HEI10.
Figure 4.
HEI10 is a dosage-sensitive regulator of Arabidopsis crossovers. (A) 420 crossovers (in centimorgans) in F1 individuals derived from crosses between HEI10Col or HEI10Ler homozygotes and wild-type (Col) or null hei10-2 mutants. Replicate individuals are shown as black dots, and mean values are shown as red dots. (B) 420 crossovers (in centimorgans) in F1 individuals derived from crosses with interfering crossover ZMM pathway mutants. (C) Schematic showing the transformation of Col-420 with additional HEI10 copies (blue triangles). (D) Diagram illustrating the HEI10 T-DNA construct used for Arabidopsis transformation via Agrobacterium. (LB) T-DNA left border sequence; (RB) T-DNA right border sequences. (E) 420 crossover frequency (in centimorgans) in empty vector, HEI10Ler::Ler, HEI10Col::Ler, HEI10Ler::Col, and HEI10Col::Col transformants compared with untransformed Col-420/Col controls. Data from individual plants are shown as black dots, and mean values are shown in red. (F) Correlation between 420 crossovers (in centimorgans) and HEI10 transcript levels measured by qRT–PCR from HEI10Col (black) and HEI10Ler (green) transformant flowers. A regression line is plotted in red.
Increased HEI10 dosage elevates euchromatic crossovers genome-wide
Due to HEI10 dosage sensitivity, we next investigated whether increasing copy number would elevate recombination beyond wild type (Fig. 4C–E). We transformed Col-420 plants with a HEI10 transgene under the control of its endogenous promoter, amplified from either Col or Ler genomic DNA (referred to here as HEI10Col and HEI10Ler) (Fig. 4C–E). HEI10Col and HEI10Ler T1 populations, but not empty vector T1, showed significantly higher recombination than untransformed Col-420 controls (Mann-Whitney-Wilcoxon test: HEI10Col, P = 4.03 × 10−14; HEI10Ler, P = 3.64 × 10−10; empty, P = 0.474) (Fig. 4E; Supplemental Tables S2, S19). Wide variation in recombination rate was observed within T1 populations (Fig. 4E), which was likely caused by varying HEI10 transgene copy numbers and position effects that influence expression level. Indeed, qRT–PCR analysis of HEI10 expression from meiotic stage flower buds (floral stages 1–12) of T1 transformants revealed a positive correlation with 420 recombination (r = 0.727; P = 7.96 × 10−6) (Fig. 4F; Supplemental Table S20). HEI10Col transformants showed higher recombination than HEI10Ler transformants (Mann-Whitney-Wilcoxon test, P = 8.93 × 10−3) (Fig. 4E; Supplemental Table S19), which is consistent with the different recombination activity of Col/Ler HEI10 variants. To further investigate polymorphisms responsible for differences in HEI10 function, we generated Col::Ler and Ler::Col promoter swap constructs and repeated transformation (Fig. 4E; Supplemental Table S19). The HEI10-Ler::Col transformants showed significantly higher recombination than HEI10-Col::Ler transformants (Mann-Whitney-Wilcoxon test, P = 1.1 × 10−2). Furthermore, HEI10-Ler::Col were not significantly different from HEI10-Col::Col (Mann-Whitney-Wilcoxon test, P = 0.841), and HEI10-Col::Ler were not different from HEI10-Ler-Ler (Mann-Whitney-Wilcoxon test, P = 0.259). Together, this is consistent with intragenic HEI10 polymorphisms, including R264G, causing differences in recombination activity (Fig. 4E; Supplemental Table S19).
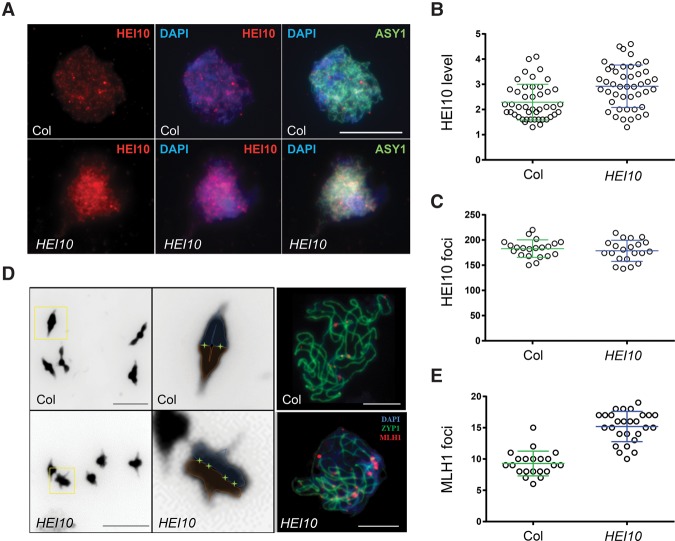
A HEI10Col T1 line showing high 420 recombination (C2; 33.74 cM) was selected for cytological investigation. Immunostaining of leptotene stage meiotic nuclei for HEI10 showed a significant increase in signal intensity (Mann-Whitney-Wilcoxon test, P = 3.90 × 10−4), although focus numbers were not changed (Mann-Whitney-Wilcoxon test, P = 0.5971) (Fig. 5A–C; Supplemental Tables S21, S22). To investigate the effect of HEI10Col transformation on crossover formation, we performed MLH1 immunostaining at the pachytene stage (Fig. 5D,E). There were close to double the number of MLH1 foci along HEI10Col chromosomes compared with wild type (mean = wild type 9.3, HEI10Col 16.2; Mann-Whitney-Wilcoxon test, P = 4.83 × 10−8) (Fig. 5D,E; Supplemental Table S23). HEI10Col also showed more compact bivalents at metaphase I, which is indicative of greater crossover numbers in the chromosome arms (Fig. 5D; Sanchez-Moran et al. 2002). This provides cytological evidence that increased HEI10 dosage and expression level elevates crossovers throughout the genome.
Figure 5.
Increased HEI10 dosage promotes formation of meiotic MLH1 foci. (A) Representative images showing leptotene stage male meiocytes from Col or HEI10Col (line C2) immunostained for HEI10 (red) and ASY1 (green) and counterstained with DAPI (blue). Bars, 10 µm. (B) Quantification of HEI10 expression level via immunostaining of Col and HEI10Col (line C2). (C) As for B but showing quantification of HEI10 foci. (D) Representative images of DAPI-stained bivalents at metaphase I in wild type (Col) (top left panel) and HEI10Col (line C2) (bottom left panel). Bars, 5 µm. (Top middle panel) A magnified view of a wild-type ring bivalent is shown with homologs outlined in red and blue. (Bottom middle panel) The inferred chiasmata sites are marked with an “X.” A magnified view of a HEI10Col ring bivalent is shown. Representative images showing leptotene stage male meiocytes from Col (top right panel) or HEI10Col (line C2) (bottom right panel) stained for MLH1 (red), ASY1 (green), and DAPI (DNA; blue). Bars, 10 μm. (E) Quantification of MLH1 foci on pachytene stage meiotic chromosomes in wild type and HEI10Col (line C2).
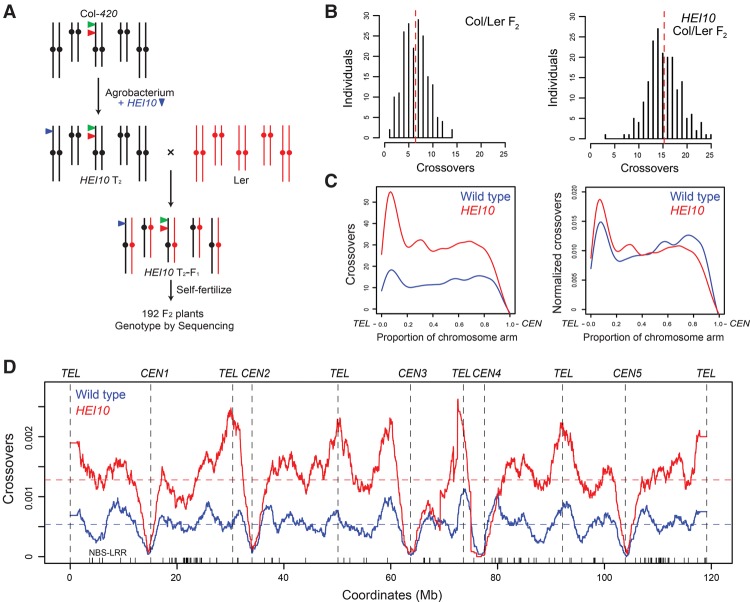
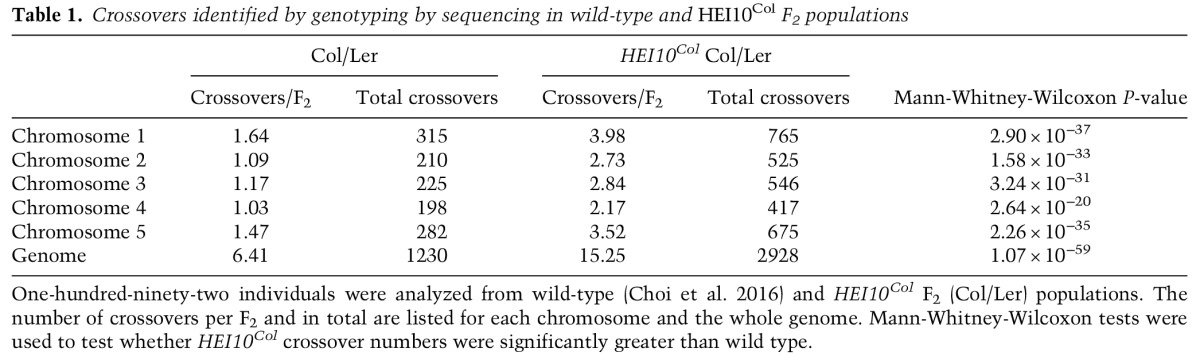
To investigate the effect of increased HEI10 dosage on crossovers at higher resolution, we used genotyping by sequencing (Fig. 6A). The HEI10Col C2 line was backcrossed to Ler alongside a wild-type Col control (Fig. 6A). HEI10Col×Ler F1 plants showed highly elevated 420 recombination compared with Col/Ler F1 (Supplemental Tables S19, S24), demonstrating that HEI10 increases crossovers in both hybrid (Col/Ler) and inbred (Col/Col) backgrounds. F2 populations (n = 192) were then generated from wild-type and HEI10Col Col/Ler F1 plants and sequenced to identify crossover locations (Fig. 6A; Supplemental Table S25; Choi et al. 2016). The HEI10Col population contained more than double the number of wild-type crossovers (1230 vs. 2928 crossovers; mean per F2, 6.41 vs. 15.25; Mann-Whitney-Wilcoxon test, P = 1.07 × 10−59) (Fig. 6B; Supplemental Fig. S5; Supplemental Table S1), consistent with our MLH1 focus analysis (Fig. 5D,E). Euchromatic chromosome arms showed the greatest increase in HEI10Col crossovers (2.6×, Mann-Whitney-Wilcoxon test, P = 1.98 × 10−8), with the largest effects in the subtelomeric regions (Fig. 6C,D; Supplemental Tables S26, S27). A lower yet significant crossover increase was observed in the pericentromeres (1.6×, Mann-Whitney-Wilcoxon test, P = 2.34 × 10−9) (Fig. 6C; Supplemental Fig. S6; Supplemental Tables S26, S27). In contrast, centromeric suppression of crossovers was observed in both populations (Fig. 6C; Supplemental Fig. S6; Supplemental Tables S26, S27; Copenhaver et al. 1999; Yelina et al. 2015).
Figure 6.
Increased HEI10 dosage elevates euchromatic crossover frequency genome-wide. (A) Diagram showing genetic mapping using the HEI10Col (line C2) following crosses to Ler (red). (B) Crossover numbers mapped by genotyping by sequencing in wild-type (Col/Ler) and HEI10Col/Ler F2 populations (Table 1). Mean values are indicated by the vertical red dotted lines. (C) Crossovers analyzed along the proportional length of chromosome arms from telomeres (TEL) to centromeres (CEN) in wild-type (blue) and HEI10 (red) populations. Plots are shown analyzing total crossovers or after normalizing by total crossover events. (D) Crossover frequency along the five chromosomes in wild-type (blue) and HEI10Col (red) populations. Mean values are shown by the dotted horizontal lines, and telomere (TEL) and centromere (CEN) positions are indicated by vertical dotted lines and labels.
Table 1.
Crossovers identified by genotyping by sequencing in wild-type and HEI10Col F2 populations
Crossovers were mapped using ∼1×–2× depth sequencing data and the TIGER analysis pipeline (Supplemental Table S25; Rowan et al. 2015), which resolved events to a mean width of 976 base pairs (bp). To analyze the fine-scale distribution of wild type versus HEI10 crossovers, we overlapped them with gene and transposon annotations and compared them with matched sets of randomly chosen intervals (Supplemental Fig. S6A; Supplemental Table S28). Both wild-type and HEI10 crossovers show increased intergenic and decreased transposon overlap compared with random (Supplemental Fig. S6A; Supplemental Table S28), which is consistent with Arabidopsis crossover hot spots associating with euchromatic gene promoters and terminators (Choi et al. 2013; Mercier et al. 2015). We also compared DNA methylation levels and observed that crossovers from both populations were hypomethylated in CG, CHG, and CHH sequence contexts compared with random (Supplemental Fig. S6B; Stroud et al. 2013). This is further consistent with both wild-type and HEI10 crossovers being enriched within euchromatic regions along the chromosome arms.
Discussion
Plants, fungi, and invertebrates possess a single HEI10/RNF212 ortholog (Bhalla et al. 2008; Chelysheva et al. 2012; Wang et al. 2012; Serrentino et al. 2013; De Muyt et al. 2014; Lake et al. 2015), whereas vertebrates encode separate RNF212 and HEI10 proteins that function as antagonistic SUMO and ubiquitin E3 ligases (Ward et al. 2007; Reynolds et al. 2013; Qiao et al. 2014; Gray and Cohen 2016; Rao et al. 2017). In mice, HEI10 and RNF212 promote stable accumulation of MSH4/MSH5 (MutSγ) heterodimers on recombining meiotic chromosomes via a SUMO–ubiquitin relay, which promotes recruitment of MLH1/MLH3 (MutLγ) and crossover formation (Ward et al. 2007; Reynolds et al. 2013; Qiao et al. 2014; Gray and Cohen 2016; Rao et al. 2017). However, it remains unclear which proteins are direct ubiquitin/SUMO targets of the HEI10/RNF212 E3 ligases, although rice HEI10 can directly interact with MSH5 (Wang et al. 2012). HEI10/RNF212 RING domains are C3HC4 zinc fingers, which generally function as protein interaction domains to recruit E2 ubiquitin-conjugating enzymes to substrates (Deshaies and Joazeiro 2009). Regions outside the RING domain are known to contribute to substrate recognition (Deshaies and Joazeiro 2009). Therefore, we postulate that the HEI10 C-terminal R264 variant may alter substrate recognition efficiency and SUMO/ubiquitin transfer during regulation of meiotic recombination.
Beyond genetic variation that alters HEI10 function, we demonstrated that Arabidopsis crossover frequency is exquisitely sensitive to HEI10 dosage. We propose that higher HEI10 concentration at meiotic repair foci quantitatively promotes crossovers via increased SUMO or ubiquitin transfer to substrate recombination factors. The dosage sensitivity of Arabidopsis HEI10 is strikingly reminiscent of rnf212 and hei10 mutations in mice, which show haploinsufficiency (Reynolds et al. 2013; Qiao et al. 2014). Furthermore, polymorphisms in RNF212 and HEI10 genes have been associated with variation in recombination rate in human, cattle, and sheep populations (Kong et al. 2008; Fledel-Alon et al. 2011; Sandor et al. 2012; Johnston et al. 2016). We propose that haploinsufficiency and dosage sensitivity of HEI10/RNF212 genes predisposes them to acting as trans recombination modifiers in diverse eukaryotic lineages. It is interesting to note that increased HEI10 dosage in Arabidopsis led to the greatest crossover increase in subtelomeric euchromatin, which is similar to the sex differences in recombination observed in both plants and mammals (Coop and Przeworski 2006; Giraut et al. 2011). For example, Arabidopsis male meiosis shows subtelomeric increases in crossover frequency (Giraut et al. 2011). Therefore, we speculate that differences in HEI10/RNF212 expression or regulation have the potential to contribute to sex differences in recombination. We also note that increasing HEI10 copy number may be an attractive mechanism to elevate crossover numbers during breeding of crop species.
Crossover modifier loci are able to alter population responses to selection (Feldman et al. 1996). For example, recombination can mitigate the effects of Hill-Robertson interference when linked loci are under selection (Hill and Robertson 1966; Barton and Charlesworth 1998). Therefore, loci that modify crossover frequency may influence genetic adaptation to diverse environments and conditions. Interestingly, total recombination levels compared across eukaryotes are generally low, typically with one or two crossovers per chromosome per meiosis, despite wide variation in physical genome size (Mercier et al. 2015). It is possible that high recombination levels might cause infertility and be selected against. However, Arabidopsis anti-crossover pathway mutants show normal fertility despite greatly elevated crossover frequency, at least in the short term (Girard et al. 2015; Mercier et al. 2015). Therefore, we propose that Arabidopsis recombination modifiers may act to maintain relatively low crossover numbers. As rQTL1Col and rQTL4Col alleles show opposite effects on crossover frequency, this example is consistent with antagonistic modifiers acting to balance recombination. It is also important to note that the effect of modifiers will be highly dependent on genome architecture and outcrossing levels. Crossover modifiers may be especially common in plants, where frequent polyploidization causes challenges for balanced meiotic genome transmission (Bomblies et al. 2016). Indeed, meiotic axis proteins (ASY1, ASY3, PDS5, ZYP1a, ZYP1b, SMC1, and REC8) have been strongly selected during polyploid evolution in Arabidopsis arenosa (Yant et al. 2013), and the Ph1 locus is required for promotion of homologous versus homeologous recombination in hexaploid bread wheat (Martín et al. 2014). Therefore, further study of plant meiotic modifier loci is likely to reveal insights into the control of recombination and how this interacts with selection during evolution.
Materials and methods
Arabidopsis strains
Crossover frequency was measured using fluorescent reporters in seeds (Col-420) and pollen (Col-I2f) (Emmanuel et al. 2006; Berchowitz and Copenhaver 2008; Yelina et al. 2013; Ziolkowski et al. 2015). In F2 populations derived from FTL hemizygotes, only a subset of progeny will contain the fluorescent protein-encoding transgenes also in a hemizygous state, which is necessary for crossover measurement. When using the seed-based 420 line, it is possible to enrich for FTL hemizygous F2 plants by examining seed under a fluorescence microscope prior to sowing and separating nonfluorescent, hemizygous, and homozygous seeds based on eGFP and dsRed fluorescence intensities. CSLs were kindly provided by Erik Wijnker, Jose van der Belt, and Joost Keurentjes (University of Wageningen) (Wijnker et al. 2012), with the exception of LCCCC, which was obtained from an esd7-1 backcross line (del Olmo et al. 2010). Mt-0, Ct-1, and Cvi-0 accessions were obtained from the Nottingham Arabidopsis Stock Centre. The ZMM mutant alleles used were hei10-2 (Salk_014624), msh4-1 (Salk_136296), msh5-1 (Salk_110240), ptd1 (Salk_ 127447), and shoc1-1 (Salk_057589). Genotyping primer sequences for these mutations used with the LBb1.3 T-DNA left border primer are in Supplemental Table S29.
rQTL mapping
Genomic DNA was extracted using CTAB and genotyped using PCR amplification of Col/Ler SSLP, CAPS, or dCAPS markers (Supplemental Tables S30, S31). We performed one- and two-dimensional QTL mapping using the R statistical package rQTL. We implemented the Haley-Knott regression algorithm using 2.5-cM steps across the genome and 0.1-cM steps for rQTL1 fine mapping. To fit models with multiple QTLs, we used the fitqtl function with Haley-Knott regression. We used 10,000 permutations for each mapping population to empirically calculate genome-wide LOD score significance thresholds.
HEI10 transformation
HEI10 was amplified from Col or Ler genomic DNA using primers HEI10-XbaI and HEI10-BamHI (Supplemental Table S29). Amplification products were cloned into the pGREEN-0029 binary vector using XbaI and BamHI restriction enzymes. Promoter swap constructs were generated using XbaI/PacI digestion and vector religation. These vectors were transformed into Col-420 FTL hemizygous plants using Agrobacterium strain GV3101 and floral dipping.
Quantitative gene expression analysis
RNA was extracted from ∼40 mg of immature flower buds (closed buds up to stage 12, which contain all meiotic stages) using TRI reagent (Sigma-Aldrich). Reverse transcription was performed with SuperScript II reverse transcriptase (ThermoFisher Scientific). Relative HEI10 expression was measured by qPCR using primers HEI10-qPCR1 and HEI10-qPCR2, and the meiosis-specific gene DMC1 was amplified using primers DMC1-qPCR1 and DMC1-qPCR2 as a control for ΔCt calculations (Supplemental Table S29). For HEI10 T1 analysis, the 2−ΔΔCt method was used to quantify relative transcript levels in comparison with untransformed plants.
Meiotic cytology and immunostaining
Chromosome spreads of Arabidopsis pollen mother cells and rDNA in situ hybridization were performed as described using fixed buds (Sanchez-Moran et al. 2007). Pachytene stage meiocytes were immunostained for ASY1, ZYP1, MLH1, and HEI10 using fresh buds as described (Armstrong et al. 2002; Sanchez-Moran et al. 2007) with the following antibodies: α-ASY1 (rabbit; 1:500 dilution), α-MLH1 (rabbit, IgG-purified; 1:200 dilution), α-ZYP1 (rat; 1:500 dilution), and α-HEI10 (rabbit; 1:200 dilution) (Armstrong et al. 2002; Sanchez-Moran et al. 2007; Chelysheva et al. 2012; Lambing et al. 2015). HEI10-immunostained slides within experiments were prepared side by side, and images were captured using the same exposure times. HEI10-immunostained cell images were acquired as Z-stacks of 10-µm × 0.4-µm optical sections, and maximum intensity projections were reconstructed using ImageJ (Lambing et al. 2015). Cell boundaries were defined manually, and total signal intensity within cells was measured. An adjacent image region was used to measure background intensity, and this value was subtracted from the cell intensity. The fluorescence signal from an adjacent Inspeck Red microsphere (ThermoFisher Scientific) was also used to normalize HEI10 signal intensity. Microscopy was conducted using a DeltaVision personal DV microscope (Applied Precision/GE Healthcare) equipped with a CDD Coolsnap HQ2 camera (Photometrics). Image capture was performed using SoftWoRx software version 5.5 (Applied precision/GE Healthcare). For MLH1 and ZYP1 coimmunostaining of pachytene nuclei, individual cell images were acquired as Z-stacks of 0.1-µm optical sections, and the maximum intensity projection for each cell was rendered using ImageJ. Numbers of MLH1 foci associated with the synaptonemal complex were scored. DAPI staining of chromosomes from metaphase I nuclei and chiasma counting were performed as described (Sanchez-Moran et al. 2002). Image capture was conducted using a Nikon 90i fluorescence microscope. Images were analyzed with NIS-Elements-F software and ImageJ.
Mapping crossovers via genotyping by sequencing
DNA was extracted using CTAB and used to generate sequencing libraries as described (Rowan et al. 2015; Yelina et al. 2015) with the following modifications. DNA was extracted from three rosette leaves of 5-wk-old plants, and 150 ng of DNA was used as input for each library. DNA was sheared for 20 min at 37°C with 0.4 U of DNA Shearase (Zymo research). Each set of 96 libraries was sequenced on one lane of an Illumina NextSeq500 instrument (300-cycle Mid Output run). FastQ sequencing data files are available from ArrayExpress accessions E-MTAB-4657 (wild type) (Choi et al. 2016) and E-MTAB-4967 (HEI10). Sequencing data were analyzed to identify crossovers as reported previously using the TIGER pipeline (Rowan et al. 2015; Yelina et al. 2015; Choi et al. 2016). Crossovers were tallied in 10-kb windows and plotted along chromosomes after smoothing using the R function “filter.” Crossovers were counted and compared between populations using 2 × 2 contingency tables and χ2 tests.
Supplementary Material
Acknowledgments
We thank Mathilde Grelon (Institut National de la Recherche Agronomique, Versailles, France) for hei10-2 seed and HEI10 antibody; Erik Wijnker, Jose van der Belt, and Joost Keurentjes (University of Wageningen) for CSLs; Gregory Copenhaver (University of North Carolina) and Avi Levy (Weizmann Institute) for FTLs; and the Gurdon Institute Imaging Facility for access to microscopes. This work was funded by a Royal Society University Research Fellowship, the Gatsby Charitable Foundation (GAT2962), Polish Mobility Plus Fellowship 605/MOB/2011/0, a William Miller Fellowship from the Watson School of Biological Sciences, ERA-CAPS/BBSRC “DeCOP” grant BB/M004937/1, Meiogenix-Biotechnology and Biological Sciences Research Council Industrial Partnership Award BB/N007557/1, and the Marie-Curie “COMREC” network FP7 ITN-606956.
Footnotes
Supplemental material is available for this article.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.295501.116.
Freely available online through the Genes & Development Open Access option.
References
- Alonso-Blanco C, Andrade J, Becker C, Bemm F, Bergelson J, Borgwardt KMM, Cao J, Chae E, Dezwaan TMM, Ding W, et al. 2016. 1,135 genomes reveal the global pattern of polymorphism in Arabidopsis thaliana. Cell 166: 481–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armstrong SJ, Caryl AP, Jones GH, Franklin FCH. 2002. Asy1, a protein required for meiotic chromosome synapsis, localizes to axis-associated chromatin in Arabidopsis and Brassica. J Cell Sci 115: 3645–3655. [DOI] [PubMed] [Google Scholar]
- Barth S, Melchinger AE, Devezi-Savula B, Lübberstedt T. 2001. Influence of genetic background and heterozygosity on meiotic recombination in Arabidopsis thaliana. Genome 44: 971–978. [DOI] [PubMed] [Google Scholar]
- Barton NH, Charlesworth B. 1998. Why sex and recombination? Science 281: 1986–1990. [PubMed] [Google Scholar]
- Baudat F, de Massy B. 2007. Cis- and trans-acting elements regulate the mouse Psmb9 meiotic recombination hotspot. PLoS Genet 3: e100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer E, Falque M, Walter H, Bauland C, Camisan C, Campo L, Meyer N, Ranc N, Rincent R, Schipprack W, et al. 2013. Intraspecific variation of recombination rate in maize. Genome Biol 14: R103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berchowitz LE, Copenhaver GP. 2008. Fluorescent Arabidopsis tetrads: a visual assay for quickly developing large crossover and crossover interference data sets. Nat Protoc 3: 41–50. [DOI] [PubMed] [Google Scholar]
- Bhalla N, Wynne DJ, Jantsch V, Dernburg AF. 2008. ZHP-3 acts at crossovers to couple meiotic recombination with synaptonemal complex disassembly and bivalent formation in C. elegans. PLoS Genet 4: e1000235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bomblies K, Yant L, Laitinen RA, Kim S-T, Hollister JD, Warthmann N, Fitz J, Weigel D. 2010. Local-scale patterns of genetic variability, outcrossing, and spatial structure in natural stands of Arabidopsis thaliana. PLoS Genet 6: e1000890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bomblies K, Jones G, Franklin C, Zickler D, Kleckner N. 2016. The challenge of evolving stable polyploidy: could an increase in ‘crossover interference distance’ play a central role? Chromosoma 125: 287–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borts RH, Haber JE. 1987. Meiotic recombination in yeast: alteration by multiple heterozygosities. Science 237: 1459–1465. [DOI] [PubMed] [Google Scholar]
- Chelysheva L, Vezon D, Chambon A, Gendrot G, Pereira L, Lemhemdi A, Vrielynck N, Le Guin S, Novatchkova M, Grelon M. 2012. The Arabidopsis HEI10 is a new ZMM protein related to Zip3. PLoS Genet 8: e1002799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi K, Zhao X, Kelly KA, Venn O, Higgins JD, Yelina NE, Hardcastle TJ, Ziolkowski PA, Copenhaver GP, Franklin FCH, et al. 2013. Arabidopsis meiotic crossover hot spots overlap with H2A.Z nucleosomes at gene promoters. Nat Genet 45: 1327–1336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi K, Reinhard C, Serra H, Ziolkowski PA, Underwood CJ, Zhao X, Hardcastle TJ, Yelina NE, Griffin C, Jackson M, et al. 2016. Recombination rate heterogeneity within Arabidopsis disease resistance genes. PLOS Genet 12: e1006179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coop G, Przeworski M. 2006. An evolutionary view of human recombination. Nat Rev Genet 8: 23–34. [DOI] [PubMed] [Google Scholar]
- Copenhaver GP, Nickel K, Kuromori T, Benito MI, Kaul S, Lin X, Bevan M, Murphy G, Harris B, Parnell LD, et al. 1999. Genetic definition and sequence analysis of Arabidopsis centromeres. Science 286: 2468–2474. [DOI] [PubMed] [Google Scholar]
- Del Olmo I, López-González L, Martín-Trillo MM, Martínez-Zapater JM, Piñeiro M, Jarillo JA. 2010. EARLY IN SHORT DAYS 7 (ESD7) encodes the catalytic subunit of DNA polymerase ε and is required for flowering repression through a mechanism involving epigenetic gene silencing. Plant J 61: 623–636. [DOI] [PubMed] [Google Scholar]
- De Muyt A, Zhang L, Piolot T, Kleckner N, Espagne E, Zickler D. 2014. E3 ligase Hei10: a multifaceted structure-based signaling molecule with roles within and beyond meiosis. Genes Dev 28: 1111–1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshaies RJ, Joazeiro CAP. 2009. RING domain E3 ubiquitin ligases. Annu Rev Biochem 78: 399–434. [DOI] [PubMed] [Google Scholar]
- Emmanuel E, Yehuda E, Melamed-Bessudo C, Avivi-Ragolsky N, Levy AA. 2006. The role of AtMSH2 in homologous recombination in Arabidopsis thaliana. EMBO Rep 7: 100–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esch E, Szymaniak JM, Yates H, Pawlowski WP, Buckler ES. 2007. Using crossover breakpoints in recombinant inbred lines to identify quantitative trait loci controlling the global recombination frequency. Genetics 177: 1851–1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldman MW, Otto SP, Christiansen FB. 1996. Population genetic perspectives on the evolution of recombination. Annu Rev Genet 30: 261–295. [DOI] [PubMed] [Google Scholar]
- Fledel-Alon A, Leffler EM, Guan Y, Stephens M, Coop G, Przeworski M. 2011. Variation in human recombination rates and its genetic determinants. PLoS One 6: e20321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fransz P, Linc G, Lee C-R, Aflitos SA, Lasky JR, Toomajian C, Hoda A, Peters J, van Dam P, Ji X, et al. 2016. Molecular, genetic and evolutionary analysis of a paracentric inversion in Arabidopsis thaliana. Plant J 88: 159–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girard C, Chelysheva L, Choinard S, Froger N, Macaisne N, Lehmemdi A, Mazel J, Crismani W, Mercier R. 2015. AAA–ATPase FIDGETIN-LIKE 1 and helicase FANCM antagonize meiotic crossovers by distinct mechanisms. PLoS Genet 11: e1005369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giraut L, Falque M, Drouaud J, Pereira L, Martin OC, Mézard C. 2011. Genome-wide crossover distribution in Arabidopsis thaliana meiosis reveals sex-specific patterns along chromosomes. PLoS Genet 7: e1002354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray S, Cohen PE. 2016. Control of meiotic crossovers: from double-strand break formation to designation. Annu Rev Genet 50: 175–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill WG, Robertson A. 1966. The effect of linkage on limits to artificial selection. Genet Res 8: 269–294. [PubMed] [Google Scholar]
- Hinch AG, Tandon A, Patterson N, Song Y, Rohland N, Palmer CD, Chen GK, Wang K, Buxbaum SG, Akylbekova EL, et al. 2011. The landscape of recombination in African Americans. Nature 476: 170–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horton MW, Hancock AM, Huang YS, Toomajian C, Atwell S, Auton A, Muliyati NW, Platt A, Sperone FG, Vilhjálmsson BJ, et al. 2012. Genome-wide patterns of genetic variation in worldwide Arabidopsis thaliana accessions from the RegMap panel. Nat Genet 44: 212–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston SE, Bérénos C, Slate J, Pemberton JM. 2016. Conserved genetic architecture underlying individual recombination rate variation in a wild population of Soay sheep (Ovis aries). Genetics 203: 583–598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S, Plagnol V, Hu TT, Toomajian C, Clark RM, Ossowski S, Ecker JR, Weigel D, Nordborg M. 2007. Recombination and linkage disequilibrium in Arabidopsis thaliana. Nat Genet 39: 1151–1155. [DOI] [PubMed] [Google Scholar]
- Kong A, Thorleifsson G, Stefansson H, Masson G, Helgason A, Gudbjartsson DF, Jonsdottir GM, Gudjonsson SA, Sverrisson S, Thorlacius T, et al. 2008. Sequence variants in the RNF212 gene associate with genome-wide recombination rate. Science 319: 1398–1401. [DOI] [PubMed] [Google Scholar]
- Lake CM, Nielsen RJ, Guo F, Unruh JR, Slaughter BD, Hawley RS. 2015. Vilya, a component of the recombination nodule, is required for meiotic double-strand break formation in Drosophila. Elife 4: e08287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambing C, Osman K, Nuntasoontorn K, West A, Higgins JD, Copenhaver GP, Yang J, Armstrong SJ, Mechtler K, Roitinger E, et al. 2015. Arabidopsis PCH2 mediates meiotic chromosome remodeling and maturation of crossovers. PLoS Genet 11: e1005372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- López E, Pradillo M, Oliver C, Romero C, Cuñado N, Santos JL. 2012. Looking for natural variation in chiasma frequency in Arabidopsis thaliana. J Exp Bot 63: 887–894. [DOI] [PubMed] [Google Scholar]
- Lynn A, Soucek R, Börner GV. 2007. ZMM proteins during meiosis: crossover artists at work. Chromosom Res 15: 591–605. [DOI] [PubMed] [Google Scholar]
- Martín AC, Shaw P, Phillips D, Reader S, Moore G. 2014. Licensing MLH1 sites for crossover during meiosis. Nat Commun 5: 4580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMullen MD, Kresovich S, Villeda HS, Bradbury P, Li H, Sun Q, Flint-Garcia S, Thornsberry J, Acharya C, Bottoms C, et al. 2009. Genetic properties of the maize nested association mapping population. Science 325: 737–740. [DOI] [PubMed] [Google Scholar]
- Mercier R, Mézard C, Jenczewski E, Macaisne N, Grelon M. 2015. The molecular biology of meiosis in plants. Annu Rev Plant Biol 66: 297–327. [DOI] [PubMed] [Google Scholar]
- Qiao H, Prasada Rao HBD, Yang Y, Fong JH, Cloutier JM, Deacon DC, Nagel KE, Swartz RK, Strong E, Holloway JK, et al. 2014. Antagonistic roles of ubiquitin ligase HEI10 and SUMO ligase RNF212 regulate meiotic recombination. Nat Genet 46: 194–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao HBDP, Qiao H, Bhatt SK, Bailey LRJ, Tran HD, Bourne SL, Qiu W, Deshpande A, Sharma AN, Beebout CJ, et al. 2017. A SUMO–ubiquitin relay recruits proteasomes to chromosome axes to regulate meiotic recombination. Science 355: 403–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds A, Qiao H, Yang Y, Chen JK, Jackson N, Biswas K, Holloway JK, Baudat F, De Massy B, Wang J, et al. 2013. RNF212 is a dosage-sensitive regulator of crossing-over during mammalian meiosis. Nat Genet 45: 269–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodgers-Melnick E, Bradbury PJ, Elshire RJ, Glaubitz JC, Acharya CB, Mitchell SE, Li C, Li Y, Buckler ES. 2015. Recombination in diverse maize is stable, predictable, and associated with genetic load. Proc Natl Acad Sci 112: 3823–3828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowan BA, Patel V, Weigel D, Schneeberger K. 2015. Rapid and inexpensive whole-genome genotyping-by-sequencing for crossover localization and fine-scale genetic mapping. G3 (Bethesda) 5: 385–398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salomé PA, Bomblies K, Fitz J, Laitinen RAE, Warthmann N, Yant L, Weigel D. 2012. The recombination landscape in Arabidopsis thaliana F2 populations. Heredity (Edinb) 108: 447–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Moran E, Armstrong SJ, Santos JL, Franklin FCH, Jones GH. 2002. Variation in chiasma frequency among eight accessions of Arabidopsis thaliana. Genetics 162: 1415–1422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Moran E, Santos J-L, Jones GH, Franklin FCH. 2007. ASY1 mediates AtDMC1-dependent interhomolog recombination during meiosis in Arabidopsis. Genes Dev 21: 2220–2233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandor C, Li W, Coppieters W, Druet T, Charlier C, Georges M. 2012. Genetic variants in REC8, RNF212, and PRDM9 influence male recombination in cattle. PLoS Genet 8: e1002854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serrentino M-E, Chaplais E, Sommermeyer V, Borde V. 2013. Differential association of the conserved SUMO ligase Zip3 with meiotic double-strand break sites reveals regional variations in the outcome of meiotic recombination. PLoS Genet 9: e1003416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stroud H, Greenberg MVC, Feng S, Bernatavichute YV, Jacobsen SE. 2013. Comprehensive analysis of silencing mutants reveals complex regulation of the Arabidopsis methylome. Cell 152: 352–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timmermans MC, Das OP, Bradeen JM, Messing J. 1997. Region-specific cis- and trans-acting factors contribute to genetic variability in meiotic recombination in maize. Genetics 146: 1101–1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K, Wang M, Tang D, Shen Y, Miao C, Hu Q, Lu T, Cheng Z. 2012. The role of rice HEI10 in the formation of meiotic crossovers. PLoS Genet 8: e1002809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward JO, Reinholdt LG, Motley WW, Niswander LM, Deacon DC, Griffin LB, Langlais KK, Backus VL, Schimenti KJ, O'Brien MJ, et al. 2007. Mutation in mouse Hei10, an E3 ubiquitin ligase, disrupts meiotic crossing over. PLoS Genet 3: 1550–1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wijnker E, van Dun K, de Snoo CB, Lelivelt CLC, Keurentjes JJB, Naharudin NS, Ravi M, Chan SWL, de Jong H, Dirks R. 2012. Reverse breeding in Arabidopsis thaliana generates homozygous parental lines from a heterozygous plant. Nat Genet 44: 467–470. [DOI] [PubMed] [Google Scholar]
- Yandeau-Nelson MD, Nikolau BJ, Schnable PS. 2006. Effects of trans-acting genetic modifiers on meiotic recombination across the a1-sh2 interval of maize. Genetics 174: 101–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yant L, Hollister JD, Wright KM, Arnold BJ, Higgins JD, Franklin FCH, Bomblies K. 2013. Meiotic adaptation to genome duplication in Arabidopsis arenosa. Curr Biol 23: 2151–2156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yelina NE, Ziolkowski PA, Miller N, Zhao X, Kelly KA, Muñoz DF, Mann DJ, Copenhaver GP, Henderson IR. 2013. High-throughput analysis of meiotic crossover frequency and interference via flow cytometry of fluorescent pollen in Arabidopsis thaliana. Nat Protoc 8: 2119–2134. [DOI] [PubMed] [Google Scholar]
- Yelina NE, Lambing C, Hardcastle TJ, Zhao X, Santos B, Henderson IR. 2015. DNA methylation epigenetically silences crossover hot spots and controls chromosomal domains of meiotic recombination in Arabidopsis. Genes Dev 29: 2183–2202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziolkowski PA, Berchowitz LE, Lambing C, Yelina NE, Zhao X, Kelly KA, Choi K, Ziolkowska L, June V, Sanchez-Moran E, et al. 2015. Juxtaposition of heterozygous and homozygous regions causes reciprocal crossover remodelling via interference during Arabidopsis meiosis. Elife 4: e03708. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.