Here, Petrenko et al. present the first integrative analysis of the molecular properties of circadian clocks in α and β pancreatic cells and provide new insights into the complex regulation of islet cell physiology at transcriptional and functional levels.
Keywords: mouse α cells and β cells, circadian clock, insulin/glucagon secretion, RNA sequencing, single-cell bioluminescence–fluorescence time-lapse microscopy
Abstract
A critical role of circadian oscillators in orchestrating insulin secretion and islet gene transcription has been demonstrated recently. However, these studies focused on whole islets and did not explore the interplay between α-cell and β-cell clocks. We performed a parallel analysis of the molecular properties of α-cell and β-cell oscillators using a mouse model expressing three reporter genes: one labeling α cells, one specific for β cells, and a third monitoring circadian gene expression. Thus, phase entrainment properties, gene expression, and functional outputs of the α-cell and β-cell clockworks could be assessed in vivo and in vitro at the population and single-cell level. These experiments showed that α-cellular and β-cellular clocks are oscillating with distinct phases in vivo and in vitro. Diurnal transcriptome analysis in separated α and β cells revealed that a high number of genes with key roles in islet physiology, including regulators of glucose sensing and hormone secretion, are differentially expressed in these cell types. Moreover, temporal insulin and glucagon secretion exhibited distinct oscillatory profiles both in vivo and in vitro. Altogether, our data indicate that differential entrainment characteristics of circadian α-cell and β-cell clocks are an important feature in the temporal coordination of endocrine function and gene expression.
The circadian timing system is a mechanism developed in virtually all organisms from bacteria to humans that allows one to anticipate the changes of geophysical time (Partch et al. 2014). The mammalian circadian clock is organized in a hierarchical structure, with a master pacemaker residing in the suprachiasmatic nuclei (SCNs) of the hypothalamus that establishes phase coherence in the body by synchronizing every day the slave oscillators residing in the periphery, encompassing billions of individual cellular clocks (Partch et al. 2014). While light input represents the dominant timing cue (Zeitgeber) for the oscillators of SCN neurons, feeding cycles driven by rest–activity rhythms have a major impact on the entrainment of peripheral clocks (Dibner and Schibler 2015a). Metabolism and circadian rhythms are therefore tightly linked through complex behavioral and molecular pathways (Perelis et al. 2015b). Moreover, cell-autonomous self-sustained peripheral clocks operative in nearly every cell in the body are connected to the metabolic state of the cell (Asher and Schibler 2011; Asher and Sassone-Corsi 2015; Perrin et al. 2015).
Circadian oscillators in the endocrine pancreas have been demonstrated recently to play an essential role in driving transcription and function of the pancreatic islet in mice and humans (Marcheva et al. 2010; Sadacca et al. 2011; Pulimeno et al. 2013; Qian et al. 2013; Perelis et al. 2015a; Saini et al. 2016). Moreover, ablation of the islet clock in mouse models, including islet clock perturbation induced at an adult age, directly triggers the onset of type 2 diabetes (T2D) (Marcheva et al. 2010; Sadacca et al. 2011; Perelis et al. 2015a). These findings provide a direct functional link between the molecular clock operative in pancreatic islets, its function under physiological conditions, and the etiology of T2D.
Pancreatic islets represent an intricate model comprising α and β cells as main endocrine cell types, secreting the counterregulatory hormones glucagon and insulin, respectively. Beyond the primordial role of β-cell dysfunction in T2D development, alterations in α-cell function and resulting hyperglucagonemia are important components of the metabolic aberrations associated with T2D (Unger and Orci 1975). Therefore, studies of α-cell physiology and regulation of glucagon secretion have caught more attention, particularly with respect to potential T2D treatment (Gromada et al. 2007; Thorel et al. 2010; Zaret and White 2010).
In the pancreas, circadian regulation of its endocrine function has been studied at the level of whole islets, therefore mostly unravelling clock regulations occurring in β cells, representing >80% of the mouse islet cell content. Consequently, the circadian physiology of α cells, representing ∼10%–15% of islet cells in mice, stayed largely unexplored. We therefore focused on the physiological importance of the α-cell clock and its impact on transcription and glucagon secretion. Furthermore, we assessed α-cell and β-cell circadian properties in parallel by developing experimental settings that allowed us to simultaneously monitor the molecular makeup of the clock and the circadian physiology of separated α and β cells. Diurnal transcriptome analysis in separated populations of α and β cells, assessed through next-generation RNA sequencing (RNA-seq), revealed that a high number of key functional genes in both islet cell types exhibited rhythmic expression patterns with either common or distinct properties. Interestingly, the phase of key core clock components was shifted between α-cell and β-cell clocks in vivo. Furthermore, a similar effect was observed by in vitro population analysis of separated α and β cells synchronized with physiologically relevant cues and by high-resolution single-cell bioluminescence–fluorescence time-lapse microscopy (Pulimeno et al. 2013). The distinct coordination between α-cellular and β-cellular clocks may account for the regulation of insulin and glucagon secretion profiles, exhibiting oscillatory profiles in vivo in the blood and in vitro as recorded by cell perifusion (Saini et al. 2016). This work is the first integrative analysis on the molecular properties of circadian clocks operative in α and β cells that brings new insights into the complex regulation of islet cell physiology at the transcriptional and functional level.
Results
RNA-seq analysis reveals distinct expression patterns in α and β cells
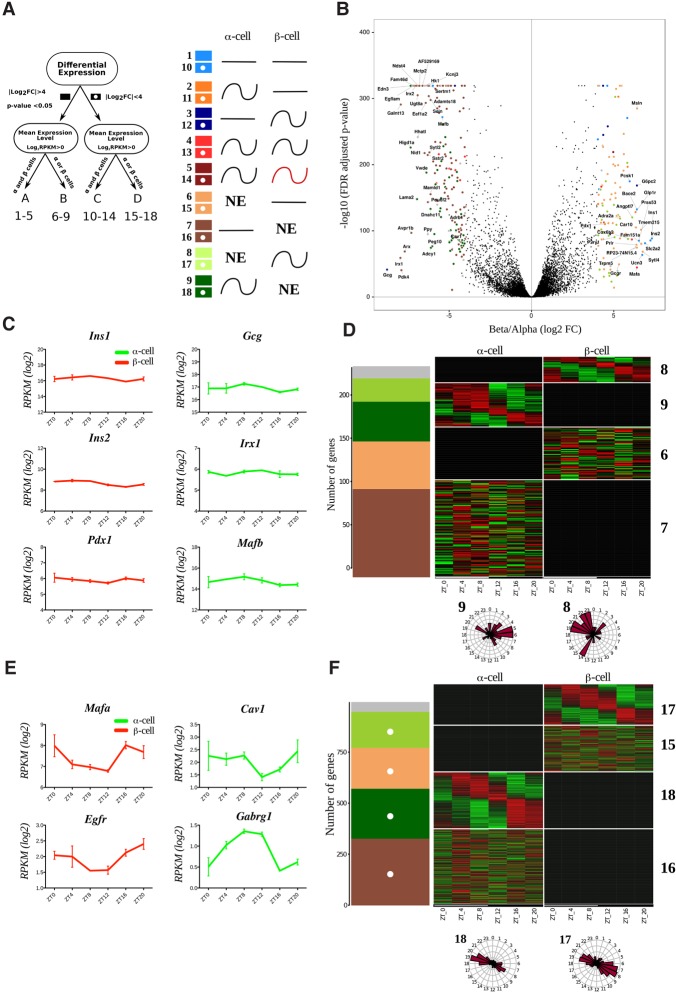
To analyze the α-cell and β-cell transcriptome and function in parallel, we combined the proglucagon (Gcg)-Venus reporter mouse (Reimann et al. 2008) with the β-cell-specific rat insulin2 promoter (RIP)-Cherry reporter mouse for specific labeling of α cells (Zhu et al. 2015) and with the Period2::Luciferase (Per2::Luc) knock-in mouse (Yoo et al. 2004) for assessment of circadian clock properties (Supplemental Fig. S1). To identify the transcripts expressed in a rhythmic manner in α and β cells, separated populations of these two cell types were isolated from triple reporter islets at six time points over 24 h and subjected to genome-wide transcriptome analysis by RNA-seq (data available at Gene Expression Omnibus [http://www.ncbi.nlm.nih.gov/geo] through accession number GSE95156) (Materials and Methods; Supplemental Figs. S1, S2). A total of 12,452 transcripts expressed with log2 reads per kilobase per million mapped reads (RPKM) > 0 in at least one cell type were identified (Supplemental Data Set 1). Two types of analyses were applied to each identified transcript: (1) differential expression level analysis between α and β cells and (2) assessment of 24-h period rhythmicity of the transcript (Fig. 1A). Based on the average expression of six time points, we considered genes exhibiting an expression difference of >16 (absolute log2 fold change >4, false discovery rate [FDR]-adjusted P-value < 0.05) between the two cell types as differentially expressed. Such differential expression analysis, illustrated in Figure 1B, identified 284 transcripts that were highly enriched in α or β cells and detected in either both cell types (40 genes; group A) (Supplemental Data Set 1) or one cell type only (244 genes; group B). An additional group of 997 genes demonstrated a lower cell-specific differential expression (absolute log2 fold change <4; group D). These transcripts were expressed in one cell type only at a globally lower level (maximum log2 RPKM < 4) (Supplemental Fig. S3A). Finally, the remaining ∼90% of all identified transcripts were expressed in both cell types with nondifferential (<16-fold difference) absolute levels (11,171 genes; group C).
Figure 1.
Temporal pattern of transcripts differentially expressed in α and β cells. (A) Groups and models assigned to transcripts with respect to their differential expression and rhythmic pattern. Transcripts with expression levels of log2 RPKM > 0 in at least one of the cell types were considered as expressed in this cell type and nonexpressed (NE) in the other cell type. Genes with expression differences >16-fold (absolute log2 fold change >4; FDR-adjusted P-value < 0.05) between two cell types were considered as differentially expressed (groups A and B; solid-colored squares), while those with expression level differences <16-fold were considered as nondifferentially expressed (groups C and D; squares with white dots). We therefore obtained four independent groups of genes: group A (differentially expressed α-cell- and β-cell-specific transcripts detectable in both cell types), group B (differentially expressed α-cell- and β-cell-specific transcripts expressed in one single cell type), group C (genes expressed in α and β cells with lower fold change), and group D (genes in one single cell type with lower fold change). Based on harmonic regression with a period of 24 h, model selection to assess rhythmicity was applied to these four groups. An arbitrary threshold of 0.4 was set on BIC weight (BICW). Genes were assigned to one of the nine pairs of models: groups A and C: genes defined as nonrhythmic (models 1 and 10), genes defined as rhythmic in α cells (models 2 and 11), genes defined as rhythmic in β cells (models 3 and 12), genes defined as rhythmic in both cell types with similar parameters (models 4 and 13), and genes defined as rhythmic in both cell types with different parameters (models 5 and 14); and groups B and D: genes expressed in β cells only and defined as nonrhythmic (models 6 and 15), genes expressed in α cells only and defined as nonrhythmic (models 7 and 16), genes expressed in β cells only and defined as rhythmic (models 8 and 17), and genes expressed in α cells only and defined as rhythmic (models 9 and 18). (NE) Gene is not expressed. Genes with lower BICWs were considered as undefined (model 0) and are represented as a gray bar in D and F. (B) Volcano plot presenting the transcripts differentially expressed in α cells (log2 fold change less than −4) or β cells (log2 fold change >4). Differentially expressed genes are identified by colored dots corresponding to their respective model. (C,E) Temporal expression profiles for selected nonrhythmic (C) and rhythmic (E) transcripts differentially expressed in β cells (left) and α cells (right). Only the profile on the higher-expression cell type is shown. Data represent the mean of two biological replicates per time point (each replicate is a mix of cells from six mice). Error bars express the SD of two independent experiments. (D,F) Rhythmic expression heat maps for transcripts of groups B and D. The number of genes distributed by models (described in A) is depicted in the left panel. (Right panels) Corresponding heat maps showing relative expression indicated in green (low) and red (high). Phase distribution of rhythmic genes is presented in the adjacent polar histograms.
The temporal patterns of all analyzed transcripts were divided into 18 models according to their rhythmic properties in one or both cell types and with respect to their differential expression (Fig. 1A and legend). Importantly, along with the high number of key functional transcripts exhibiting a similar rhythmic pattern in both cell types (models 4 and 13) (Fig. 1A), numerous transcripts harboured distinct temporal profiles in α and β cells.
Temporal patterns of functional genes differentially expressed in α and β cells
Most of the classical α-cell- and β-cell-specific transcripts were differentially expressed (group A and B) (Supplemental Data Set 1), confirming the accuracy of our cell separation approach. As expected, hormone transcripts Ins1 and Ins2 and β-cell-specific transcription factors (Mafa, Ucn3, Nkx6-1, Mnx1, and Pdx1) were expressed in β cells with highest significance, while Gcg and α-cell-specific transcription factors (Mafb, Irx1, Irx2, and Arx) were significantly differentially expressed in α cells, validating the specificity of the cell populations in our experimental conditions. Most of the above-mentioned cell-specific transcription factors as well as genes encoding for insulin and glucagon were nonrhythmic (Fig. 1C), except for Mafa, which was oscillating (Fig. 1E). The low-level expression of classical α-cell- and β-cell-specific transcripts in the opposite cell type might be attributed to the actual expression in this cell type or minor cross-contamination between two cell populations during the fluorescence-activated cell sorting (FACS) procedure.
With respect to the assessment of rhythmic expression patterns (groups A and B), 34 transcripts (seven in group A and 27 in group B; models 3 and 8, respectively) were qualified as rhythmic in β cells, and 51 (five in group A and 46 in group B; models 2 and 9, respectively) were qualified as rhythmic in α cells (Fig. 1D,E; Supplemental Fig. S3B; Supplemental Data Set 1). According to gene ontology (GO) term analysis, the rhythmic β-cell-specific genes were enriched in biological processes such as cell adhesion, protein and hormone transport and secretion, and neuroactive ligand–receptor interaction. Rhythmic genes in α cells were enriched in processes such as cell signalling, development, and synaptic transmission and for pathways of the complement and coagulation cascade (Supplemental Data Sets 2, 3; Supplemental Table 1). Of note, the peak phase for the expression of α-cell-specific transcripts was mainly during the day, whereas the peak phase for β-cell genes occurred mainly during the end of the night (Fig. 1D; Supplemental Fig. S3B; Supplemental Data Set 1).
Among the genes defined as group D (Fig. 1A), 245 transcripts exhibited rhythmic expression in β cells (model 17), whereas 177 transcripts exhibited rhythmic expression in α cells (model 18) (Fig. 1F; Supplemental Data Set 1). The oscillatory profile of these transcripts showed two principal peaks at Zeitgeber time 8 (ZT8) and ZT20 in both cell types (Fig. 1F). β-Cell-specific transcripts from this group of genes were involved mainly in ion transport, while α-cell-specific transcripts were associated with intracellular component movement and morphology, cell signalling, and developmental processes (Supplemental Data Set 5).
Temporal patterns of genes nondifferentially expressed in α and β cells
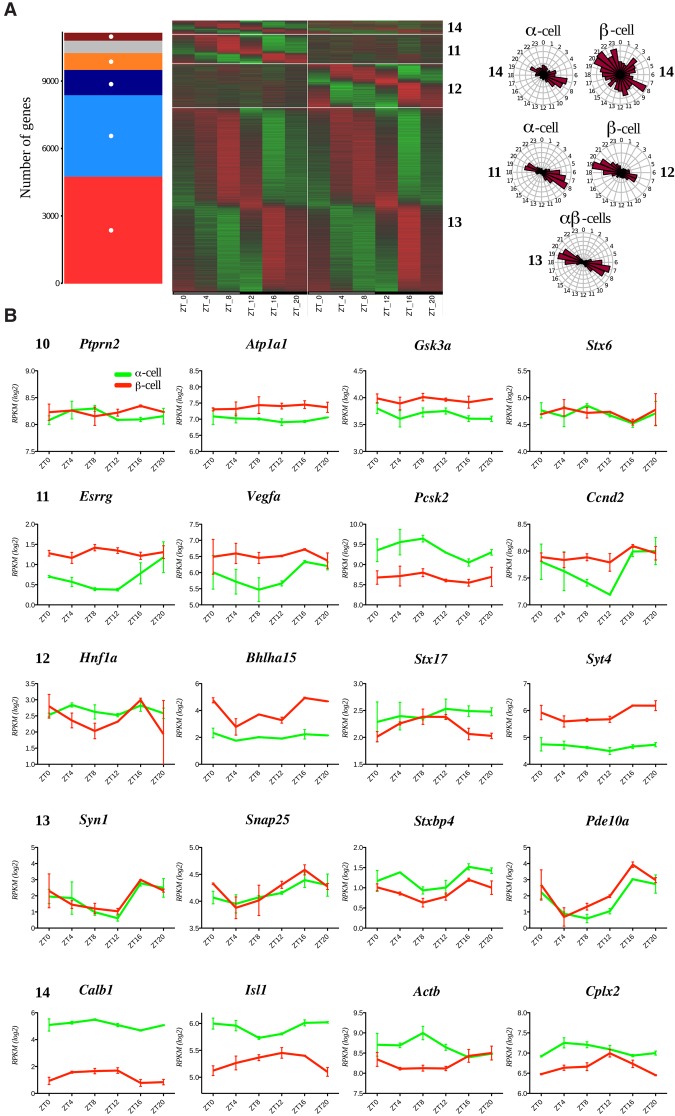
Most transcripts with nondifferential levels of expression in the two cell types, classified as group C, were distributed between five models (models 10–14) according to their temporal expression pattern (Figs. 1A, 2A). Genes comprising this group were involved in a high number of different basal metabolic processes (Supplemental Data Set 4). Interestingly, most of the genes involved in hormone secretion were rhythmic, with similar circadian parameters in both cell types (Supplemental Data Set 4, model 13). While no significant GO term enrichment could be identified for this group due to the large number of genes with a wide range of different functions (Supplemental Data Set 1, 4), many key genes involved in intracellular signalling, granule trafficking and exocytosis, mitochondrial ATP production, and ion channels were present in this group (Fig. 2, model 13; Supplemental Data Set 1; Supplemental Table 1). Of note, a group of 751 genes exhibited a rhythmic pattern in α cells only (model 11), comprising Pcsk2, Vegfa, and Ccnd3 (Fig. 2B). On the other hand, 1126 genes were rhythmic in β cells only (model 12), comprising those involved in endoplasmic reticulum-associated functions and glucose metabolism (Fig. 2A,B; Supplemental Data Set 1, 4; Supplemental Table 1). Finally, 352 genes were rhythmic in both cell types, with distinct oscillatory parameters in α and β cells (Fig. 2A,B, model 14; Supplemental Data Set 1, 4).
Figure 2.
Comparative analysis of temporal expression patterns of the transcripts expressed in α and β cells (group C in Fig. 1A). A total of 11,171 transcripts expressed in both α and β cells (log2 RPKM > 0) and exhibiting the expression level differences absolute log2 fold change <4 between the two cell types were assigned to one of the five models (models 10–14 in Fig. 1A). (A) The number of genes assigned in models and corresponding heat maps showing relative expression indicated in green (low) and red (high). The gray bar represents genes not classified in any models. Phase distribution of rhythmic genes is presented on the adjacent polar histograms for rhythmic genes in (1) one cell type (models 11 and 12), (2) both cell types with the same parameters (model 13), or (3) both cell types with different parameters (model 14). (B) Temporal profiles for selected transcripts expressed in both cell types delineated to one of the rhythmicity models. Data are expressed as mean ± SD of two independent experiments.
RNA-seq reveals distinct rhythmic phases for the expression profiles of core clock transcripts in α and β cells
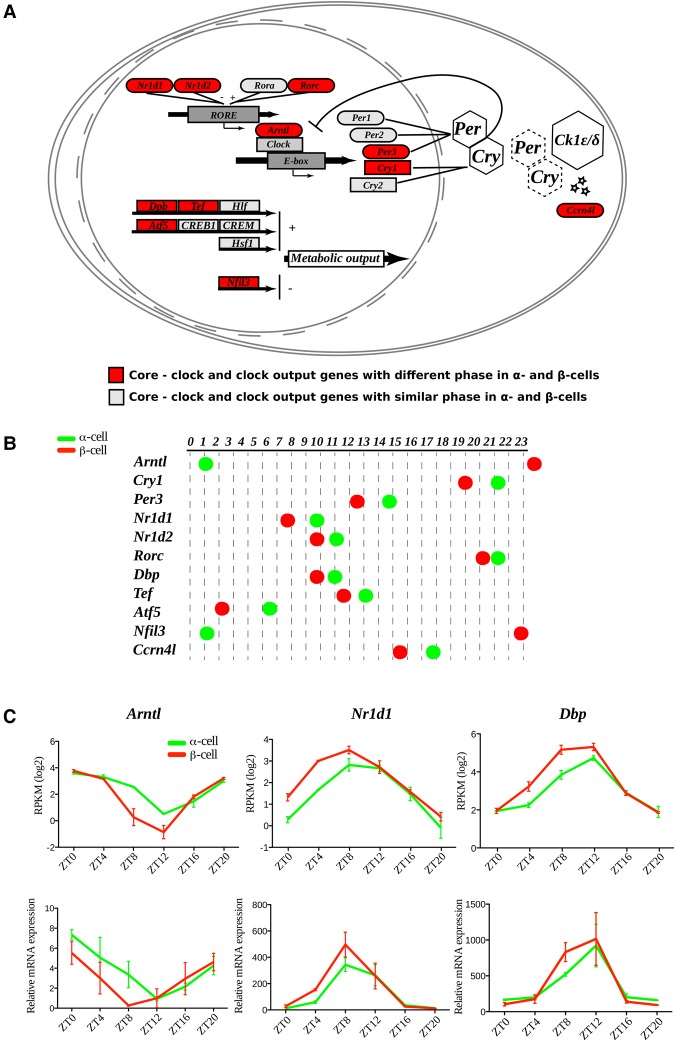
Surprisingly, the most enriched biological function in the above-mentioned group of genes with distinct oscillatory parameters (group C model 14) was the GO term “circadian rhythm” (Fig. 3A,B; Supplemental Fig. S4; Supplemental Data Set 4). Indeed, several core clock transcripts, including Bmal1 (Arntl), Reverbα (Nr1d1), and Cry1, exhibited distinct rhythmic patterns of expression in α and β cells, with the β-cell core clock genes being phase-advanced ∼4 h compared with their α-cell counterparts (Fig. 3C; Supplemental Fig. S4). The phase coherence between the rhythmic profiles of these transcripts expressed in α and β cells was further confirmed by quantitative RT–PCR (qRT–PCR) (Fig. 3C). Collectively, these data suggest that α-cellular and β-cellular clocks exhibit comparable rhythmic amplitudes but distinct phases in vivo, with β-cell clocks being phase-advanced compared with α-cell clocks.
Figure 3.
Core clock transcripts expressed in α and β cells exhibit distinct rhythmic phases in vivo. (A) Mapping of identified α-cell and β-cell molecular clock and clock-controlled transcripts into modified “circadian rhythm” KEGG (Kyoto Encyclopedia of Genes and Genomes) pathways. Transcripts exhibiting distinct rhythmic phases in α and β cells are marked in red, and those with similar phases are marked in gray. (B) Rhythmic phases of core clock and clock-controlled transcripts in α and β cells. (C) Temporal profiles of selected core clock transcripts with distinct rhythmic phases in α and β cells assessed by RNA-seq data (top panels) and qRT–PCR analysis (bottom panels). Data represent the mean of two biological replicates per time point (each replicate is a mix of cells from six mice). Error bars express the SD of two independent experiments.
α-Cellular and β-cellular clocks synchronized in vitro by forskolin exhibit distinct circadian profiles at the population and single-cell levels
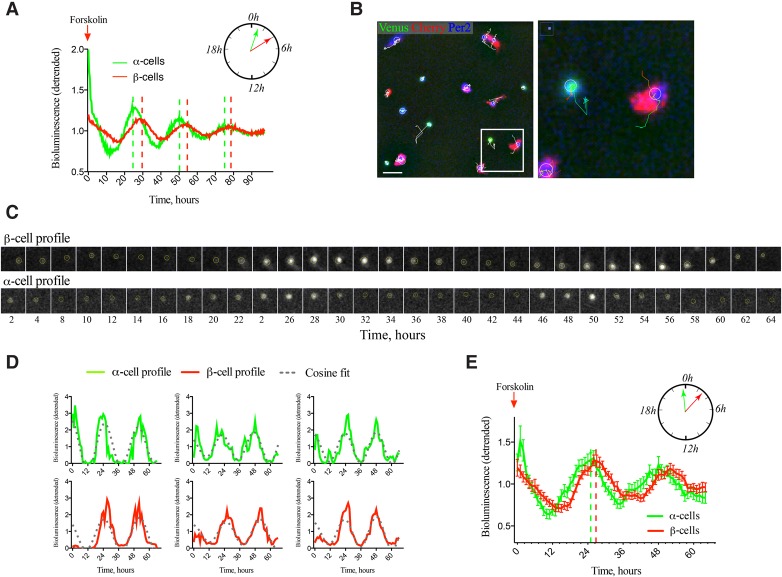
Since different rhythmic phases observed for core clock transcripts in vivo might be attributed to distinct entrainment properties of α and β cells, analysis of circadian properties of α-cell and β-cell populations was further undertaken in vitro. PER2::Luc profiles were recorded in intact islets and for dispersed islet cells synchronized with forskolin, which has been shown previously to be an efficient synchronizer for intact mouse and human islets (Perelis et al. 2015a; Saini et al. 2016). No difference was observed between the circadian profiles of whole islets and a mixed islet cell population (Supplemental Fig. S5A). Next, we conducted population analysis of separated pure α and β cells synchronized with forskolin (Fig. 4A). Forskolin induced high-amplitude oscillations in both cell types compared with the respective controls stimulated with medium change only (Supplemental Fig. S5B). Of note, separated α cells exhibited significantly earlier circadian phase in comparison with β cells, with no significant change in the circadian period length (Fig. 4A; Supplemental Table 2).
Figure 4.
α-Cell and β-cell clocks synchronized in vitro by forskolin exhibit distinct circadian phases at the population (A) and single-cell (B–E) levels. (A) Average PER::Luc oscillation profiles of forskolin-synchronized FACS separated α-cell and β-cell populations (50,000 cells per well) in n = 4 and n = 5 experiments (with an average of five animals used per experiment), respectively. Data are presented as detrended values (Pulimeno et al. 2013). Significant phase shift between the two cell types (see Supplemental Table 2) is presented schematically in the polar diagram. (B) Representative full bioluminescence–fluorescence image (512 × 512 pixels) of proGcg-Venus/RIP-Cherry/PER2::Luc-dissociated mouse islet cells subjected to time-lapse bioluminescence–fluorescence microscopy (Supplemental Movies S1–2). α Cells are Venus-positive (green labeling), and β cells are Cherry-positive (red labeling). Bioluminescence signal (blue in the image) was quantified for each cell over the nucleus within the circled area. Representative trajectories are overplayed in white over the image. (Right panel) The cropped image presents two traced cells. Bar, 40 µm. (C) Time-lapse microscopy of circadian PER2::Luc bioluminescence of a representative β cell (top row) and α cell (bottom row). Images were taken every 2 h during 64 h. (D) Analyzed bioluminescence tracks (detrended) for three representative α cells (green) and three β cells (red) with corresponding fitted cosine curves (gray) obtained using the circadian gene expression (CGE) plug-in (Sage et al. 2010). (E) Average PER2::Luc bioluminescence expression profiles (detrended) of n = 49 Venus-positive α cells and n = 55 Cherry-positive β cells after forskolin shock recorded in n = 8 independent time-lapse movies. Note the significant phase shift between two cell types (23.98 h ± 0.65 h in α cells and 26.71 h ± 0.58 h in β cells). P = 0.0023, two-tailed paired t-test. Data are expressed as mean ± SEM.
To explore individual cellular rhythms, we performed single-cell analysis of mouse α and β cells using a high-resolution combined bioluminescence–fluorescence time-lapse microscopy approach (Pulimeno et al. 2013). Dispersed triple transgenic islet cells synchronized with forskolin were subjected to time-lapse microscopy (Fig. 4B,C; Supplemental Movies S1, S2). The PER2::Luc bioluminescence profiles of α cells (Venus-positive) and β cells (Cherry-positive) were analyzed (Fig. 4D). Based on the analysis of 49 α cells and 55 β cells in eight time-lapse microscopy experiments, the circadian phase of PER2::Luc bioluminescence profile was significantly advanced in α cells when compared with β cells (Fig. 4E), in agreement with the population analysis (cf. Fig. 4A). Of note, we observed an opposite tendency for the phase shift between α-cellular and β-cellular clocks in vivo (Fig. 3B,C) compared with in vitro following forskolin synchronization (Fig. 4A,E).
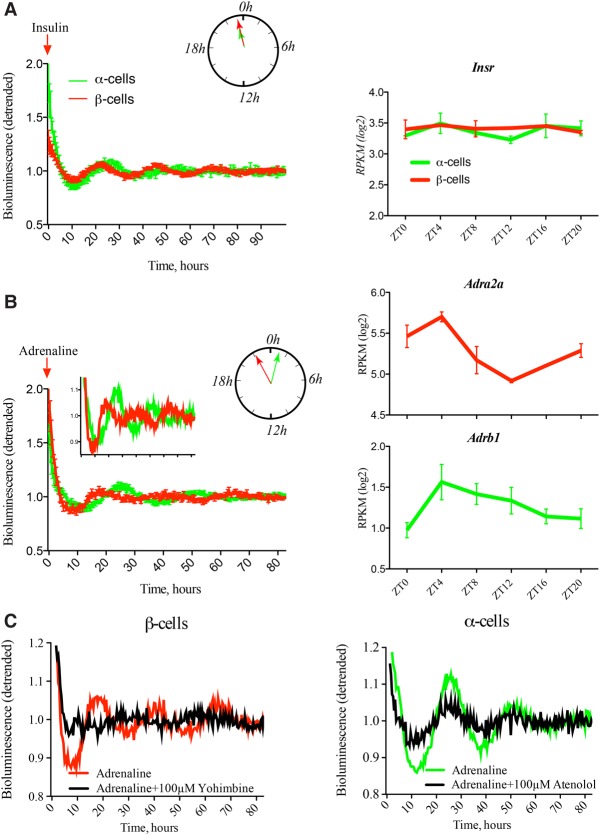
Adrenaline, but not insulin, exerts differential synchronizing effects on α-cellular and β-cellular clocks in vitro
To decipher potential differences in the α-cellular and β-cellular clock entrainment properties in vitro, the physiologically relevant synchronizers insulin and adrenaline were applied to the islet cells. In vitro synchronization with insulin and adrenaline pulses proved to be efficient for intact pancreatic islets (Supplemental Fig. S5C). When applied to isolated α and β cells, insulin induced similar profiles of circadian bioluminescence in both cell types (Fig. 5A; Supplemental Table 2). In contrast, separated α cells synchronized by adrenaline exhibited distinct oscillation properties when compared with β cells, with α cells being phase-delayed behind their β-cell counterparts (Fig. 5B; Supplemental Table 2). Thus, the phase shift between α and β cells induced by adrenaline (Fig. 5B) was in the opposite direction when compared with the one observed for forskolin synchronization in vitro (Fig. 4A,E) and coherent with the phases of core clock transcripts expressed in α and β cells in vivo (Fig. 3C).
Figure 5.
α-Cell and β-cell clocks synchronized in vitro by adrenaline, but not by insulin, exhibit distinct circadian phases. (Left panels) Average detrended PER2::Luc bioluminescence profiles for separated α-cell and β-cell populations synchronized in vitro with a 1-h pulse of 100 nM insulin (A) or 5 µM adrenaline (B). Circadian phases of the PER2::Luc bioluminescence profiles recorded from α-cell and β-cell populations, presented in the polar diagrams, were similar (A) or delayed for α-cell population (B). Mean circadian parameters for these experiments are in Supplemental Table 2. (Right panels) In vivo temporal expression transcript profiles over 24 h obtained by RNA-seq (Supplemental Data Set 1) for insulin receptor (Insr) and adrenergic receptors (Adra2a and Adrb1) in α and β cells. Data are expressed as mean ± SEM for the left panels. (A) n = 5 experiments for α cells; n = 6 experiments for β cells. (B) n = 6 experiments for α cells; n = 3 experiments for β cells. An average of five mice was used per experiment. Mean ± SD for the right panels. n = 2 independent experiments. (C) Average detrended PER2::Luc bioluminescence profiles for separated α-cell and β-cell populations synchronized with adrenaline alone or in the presence of adrenergic receptor antagonists. n = 6 experiments for α cells with adrenaline alone; n = 3 experiments for β cells with adrenaline alone; n = 3 independent experiments with antagonists for each cell type. An average of five mice was used per experiment. Coincubation of α and β cells with antagonists of β1 adrenergic receptor (100 µM atenolol) and α2 adrenergic receptor (100 µM yohimbine), respectively, attenuated the synchronizing effect of adrenaline. Mean circadian parameters for these experiments are in Supplemental Table 3.
To gain mechanistic insights into the observed differences, we compared expression profiles of the adrenaline hormone receptors in α and β cells. Importantly, while Insr exhibited similar levels and temporal expression profiles in both cell types (Fig. 5A), adrenergic receptors were differentially expressed in α and β cells (Adrb1 in α cells and Adra2a in β cells) (Fig. 5B). Therefore, the observed differences in the circadian properties of α-cellular and β-cellular clocks might be attributed to the distinct repertoire of these hormone receptors expressed on α and β cells and possibly due to their distinct temporal profiles, as demonstrated by RNA-seq analysis (Fig. 5B; Supplemental Data Set 1). To further explore this hypothesis, α2 adrenergic receptor (ADRA2) antagonist yohimbine was applied to β cells prior to adrenaline synchronization, resulting in a perturbed cellular circadian rhythm compared with adrenaline synchronization alone (Fig. 5C, left panel; Supplemental Table 3). Moreover, application of the selective β1 adrenergic receptor antagonist atenolol to α cells prior to adrenaline synchronization led to a significant attenuation of the circadian profile (Fig. 5C right panel; Supplemental Table 3).
Blood levels of the islet hormones are oscillating over 24 h
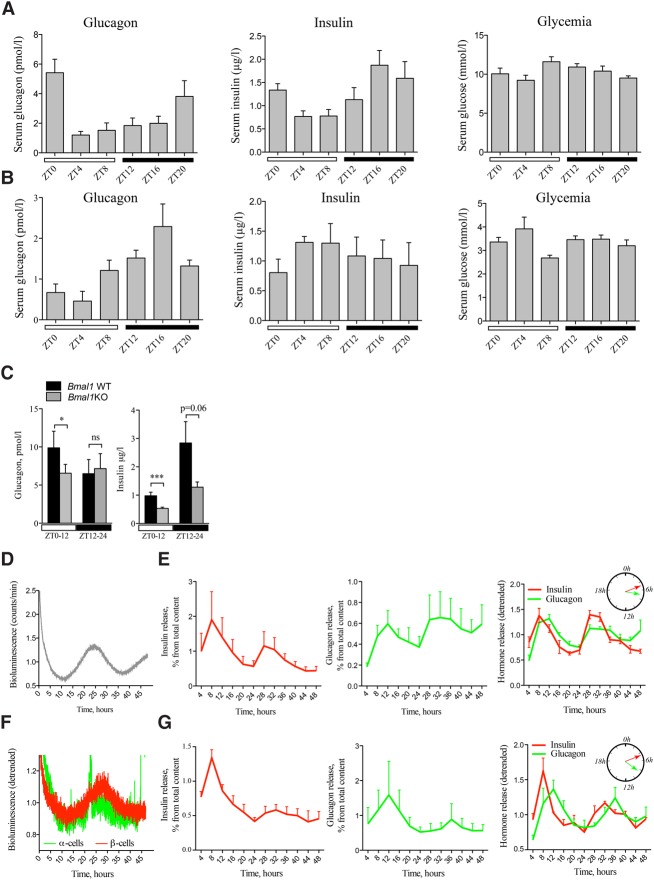
To unravel the impact of the islet cellular clocks on physiology, the temporal pattern of islet hormones was assessed around the clock. Blood levels of glucose, insulin, and glucagon were measured in night-fed animals every 4 h during 24 h (Fig. 6A) following the same design developed for islet cell transcript analysis (Supplemental Fig. S1). In agreement with previous publications, insulin levels exhibited a rhythmic profile with a peak in the middle of night (ZT16; Fig. 6A). On the other hand, glucagon levels were not considered circadian according to CosinorJ analysis, whereas a clear peak at ZT20–ZT0 following the peak of insulin was observed (Fig. 6A). Glucose levels did not show a clear rhythmic pattern in the same serum samples.
Figure 6.
Secretion of insulin and glucagon in vivo and in vitro exhibits rhythmic profiles altered in circadian mutant mice. In vivo insulin, glucagon, and glucose levels were assessed in the sera collected from night-fed animals (A) or animals fasted for 12 h prior to the experiment and during the entire period of serum collection (B). Obtained profiles were analyzed by CosinorJ and qualified as 24-h rhythmic for χ2 < 0.5 within the period length range of 18–30 h and nonrhythmic if χ2 ≥ 0.5 or outside this period length range. (A) Mouse sera were collected around the clock every 4 h (see Supplemental Fig. S1 for the design). n = 6–15 animals for each time point. Average serum insulin levels were χ2 = 0.0069 for period 22.05 h ± 2.2 h and phase 18.83 h ± 6.45 h (rhythmic), χ2 = 3.39 (nonrhythmic) for glucagon, and χ2 = 0.73 (nonrhythmic) for glucose. (B) Mouse sera were collected as described in A from n = 3–5 mice per time point. CosinorJ analysis results for average serum insulin levels were χ2 = 0.055 for period 20.1 h ± 6.04 h (rhythmic), χ2 = 0.21 for glucagon for period 24.0 h ± 1.59 h (rhythmic), and χ2 = 0.16 for glucose for period 10.5 h ± 2.17 h (nonrhythmic). (C) In vivo insulin and glucagon levels were assessed in mouse serum samples collected during the light phase (ZT0–ZT12; white square) and dark phase (ZT12–ZT24; black square) in night-fed Bmal1 knockout animals and their Bmal1 wild-type littermates. Light-phase insulin was assessed in n = 27 paired samples from Bmal1 knockout and Bmal1 wild-type mice (total of 54 animals). Dark phase insulin was assessed in 12 paired animal samples. Light-phase glucagon was measured in the blood samples from n = 16 pairs of animals (n = 12 pairs for the dark phase). Data are expressed as mean ± SEM. (***) P = 0.0006 for light-phase insulin; (*) P = 0.034 for light-phase glucagon, two-tailed paired t-test. (D,E) Circadian bioluminescence recording (D) with parallel assessment of hormone secretion (E) in perifused mixed islet cell populations after forskolin synchronization. n = 4 independent experiments with an average of four mice used per experiment. The perifusion medium contained 5.5 mM glucose and was collected every 4 h during 48 h. Hormone concentrations in the outflow medium were normalized to the total hormone content in the cell lysate at the end of each experiment and are expressed as the percentage of total content (mean ± SEM) (left and middle panels) or as superimposed detrended values (mean ± SEM) (right panel). CosinorJ analysis results for insulin levels were χ2 = 0.22 for period 19.49 h ± 1.17 h and phase 5.13 h ± 0.37 h (rhythmic) and χ2 = 0.064 for glucagon level for period 25.79 h ± 2.19 h and phase 7.05 h ± 0.75 h (rhythmic). (F,G) Circadian bioluminescence recording (F) with parallel assessment of insulin secretion in a separated β-cell population (G, left panel) and glucagon secretion in a separated α-cell population (G, middle panel). n = 3 independent experiments for α and for β cells, with and average of five mice used per experiment. α-Cell and β-cell populations were separated by FACS, plated, synchronized with forskolin pulse, and continuously perifused with culture medium containing 5.5 mM glucose for 48 h following synchronization. The outflow medium was collected every 4 h. Hormone concentrations in the outflow medium samples were normalized to the total hormone content in the cell lysate at the end of each experiment and are expressed as the percentage of total content (mean ± SEM) (left and middle panels) or as superimposed detrended values (mean ± SEM) (right panel). CosinorJ analysis results were χ2 = 0.19 for insulin level for period 23.17 h ± 0.56 h and phase 4.94 h ± 0.88 h (rhythmic) and χ2 = 0.04 for glucagon level for period 23.03 ± 1.14 h and phase 8.4 h ± 0.93 h (rhythmic).
To distinguish between the clock-driven and food-driven origins of the observed rhythms, islet hormone blood levels were assessed in fasted animals (Fig. 6B). The rhythmic profiles of glucagon and insulin persisted in the absence of feeding, although the secretion peak for both hormones was advanced ∼8 h in fasted animals as compared with night-fed mice. Thus, the phase coherence between the peaks of insulin and glucagon levels was maintained in the night-fed and fasted mice (Fig. 6A,B). As expected, the blood glucose levels were strongly diminished in fasted animals without any clear diurnal rhythmicity pattern (Fig. 6A,B, cf. right panels).
To assess the impact of a functional clock on insulin and glucagon blood levels, the islet hormone measurements were performed in clock-deficient Bmal1 knockout mouse serum samples. During the light phase, both insulin and glucagon secretion levels were significantly diminished in Bmal1 knockout mice compared with the Bmal1 wild-type littermates (Fig. 6C). During the dark phase, only insulin exhibited a tendency for dampened secretion levels in Bmal1 knockout mice, which did not reach statistical significance (P = 0.06, paired t-test), while glucagon levels remained unchanged (Fig. 6C).
Rhythmic basal secretion of glucagon and insulin by isolated α and β cells synchronized in vitro
To understand whether the oscillatory profiles of islet hormones observed in the blood might be regulated to some extent by cell-autonomous clocks, we used an in-house-developed perifusion system connected to a LumiCycle chamber (Saini et al. 2016). Basal insulin and glucagon secretion was monitored around the clock under a constant flow of culture medium in a dispersed mixed islet cell population (Fig. 6E) and FACS-sorted α and β cells (Fig. 6G) synchronized in vitro by a forskolin pulse. In parallel, PER::Luc bioluminescence was recorded from the same cells (Fig. 6D,F). These perifusion experiments suggested that both basal insulin secretion by synchronized β cells and basal glucagon secretion by synchronized α cells are circadian. Of note, the peak of glucagon secretion was delayed compared with that of insulin secretion by ∼2 h in the mixed cell population and ∼4 h in FACS-sorted α and β cells (Fig. 6E,G, cf. right panels).
Discussion
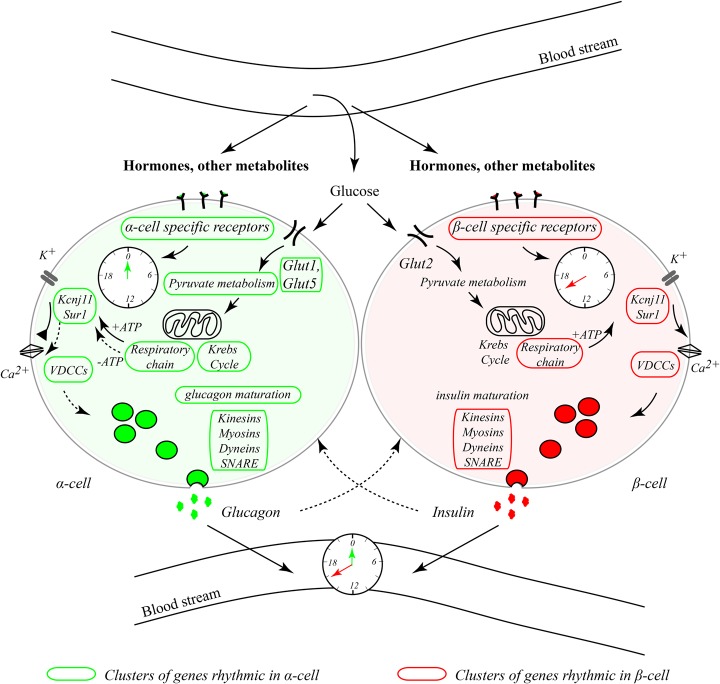
This study represents the first parallel molecular analysis of α-cell and β-cell oscillators and their impact on the gene expression and functional regulation of islet cells. We demonstrate that α-cell and β-cell clocks harbor different circadian properties in vivo and in vitro in response to physiologically relevant stimuli such as adrenaline (Fig. 7). Parallel large-scale in vivo transcriptome analysis in separated α and β cells showed oscillatory profiles for a high number of key islet genes in either one or both cell types (Figs. 1–3; Supplemental Data Set 1). Rhythmically expressed transcripts with similar or distinct characteristics in α and β cells comprised those encoding for glucose transporters, glucose metabolism enzymes, and regulators of granule trafficking and exocytosis (Fig. 7; Supplemental Data Set 1; Supplemental Table 1). We propose that, along with feeding–fasting cycles, the described distinct properties of α-cellular and β-cellular clocks (Figs. 3–5) might contribute to orchestrating temporal secretion patterns of glucagon and insulin (Fig. 6), possibly due to differential circadian expression of functional genes in α and β cells (Figs. 1, 2, 7).
Figure 7.
Inputs and outputs of α-cellular and β-cellular clocks. α-Cellular and β-cellular oscillators exhibit different circadian phases in vivo and in vitro in response to physiologically relevant stimuli, such as adrenaline, possibly due to a distinct repertoire of surface receptors and signal transduction molecules specific for each cell type. Key functional genes exhibit similar or distinct temporal patterns in α and β cells, comprising those encoding for glucose transporters, enzymes catalyzing glucose metabolism reactions (glycolysis, pyruvate metabolism, and Krebs cycle), KATP channels, voltage-dependent calcium channels (VDCCs), genes involved in glucagon maturation (but not insulin maturation), and genes responsible for granule trafficking and exocytosis. Clusters of rhythmic genes in β cells are highlighted in red, and those rhythmic in α cells are highlighted in green. We hypothesize that the distinct properties of α-cellular and β-cellular clocks, along with feeding–fasting cycles, might contribute to orchestrating different oscillating secretory patterns of glucagon and insulin stemming from the differential temporal orchestration of the functional gene transcription in α and β cells.
Pancreatic α-cellular and β-cellular clocks exhibit distinct molecular properties in vivo and in vitro
Parallel RNA-seq analysis of the temporal pattern of transcripts expressed in separated α and β cells suggested important phase differences between the two cell types (Fig. 3). Opposite phase relationships were found in isolated α and β cells synchronized by forskolin in vitro (Fig. 4A; Supplemental Table 2). These data were supported by single-cell analysis of α and β cells in mixed populations (Fig. 4B–E), raising the question of which endocrine and paracrine regulators might be relevant for islet cell clock synchronization in vivo. Synchronization of islet cellular clocks is a highly complex and dynamic process comprising feeding–fasting cycles, neural regulation, and endocrine and paracrine stimuli (Dibner and Schibler 2015b; Perelis et al. 2015a,b). Since insulin and catecholamines are key regulators of islet physiology during feeding and fasting, respectively, their synchronization properties on islet cell clocks were assessed. Indeed, these hormones had pronounced synchronizing capacity on α-cell and β-cell clocks (Fig. 5; Supplemental Fig. S5C). Insulin, which has been suggested to have a weak synchronizing effect on cultured rat fibroblasts through the transcriptional induction of core clock genes Per1 and Per2 (Balsalobre et al. 2000), is acting via its specific receptors. Insulin receptor exhibited similar expression levels and temporal patterns in both cell types according to our data (Fig. 5; Supplemental Data Set 1) and previously published data sets (Benner et al. 2014; Adriaenssens et al. 2016; DiGruccio et al. 2016). In response to a high dose of insulin, α-cellular and β-cellular clocks exhibit a similar circadian phase (Fig. 5; Supplemental Table 2).
In contrast, the adrenaline pulse generates a significant phase shift between α and β cells, similar to the phase shift observed in vivo (cf. Figs. 5B and 3C). Responses of the α-cell and β-cell clocks to adrenaline synchronization are likely to be mediated through adrenergic receptors, which are expressed differentially in α and β cells according to our RNA-seq data set (Fig. 5B, right panels). Importantly, inhibition of the β-cell-specific ADRA2A receptor with the α2 adrenergic antagonist yohimbine or of the α-cell-enriched ADRB1 receptor with the selective β1 adrenergic antagonist atenolol altered the synchronizing effect of adrenaline in these cells, respectively (Fig. 5C), further supporting a receptor-specific effect of adrenaline on islet cells. The intracellular effect of ADRB1 is mediated via activation of adenylyl cyclase (Vieira et al. 2004), while ADRA2A inhibits this enzyme (Rosengren et al. 2010). According to our RNA-seq analysis, different isoforms of adenylyl cyclase (Adcy3, Adcy1, and Adcy9) are rhythmic only in α cells and are either nonrhythmic or nonexpressed in β cells (Supplemental Data Set 1). Additionally, distinct isoforms of protein kinase C (Prkcb and Prkce) and phospholipase C (Plcb1, Plcb4, and Plcg1), second messengers of the GNAQ-activated pathway downstream from ADRA2A, exhibited differential expression levels and temporal patterns in α and β cells (Supplemental Data Set 1). Collectively, the data presented in Figure 5C together with the RNA-seq analysis imply that the distinct circadian response of α-cell and β-cell clocks to adrenaline might be mediated by the cell-specific expression of adrenergic receptors and their second messengers in these cells. This islet cell-specific synchronizing effect of adrenaline observed in vitro may also account for the phase difference of α-cellular and β-cellular clocks demonstrated in vivo (Fig. 3) due to the essential role of the sympathetic innervation for islet function (Thorens 2014).
Additionally, oscillatory patterns of blood insulin (Fig. 6A) and adrenaline (De Boer and Van der Gugten 1987) may feed back on islet clocks by influencing their circadian phase. Transient changes of these hormones levels—for instance, increase of adrenaline concentration upon stress or increase of insulin after food ingestion—may contribute to the regulation of islet cell oscillators. In summary, our experiments suggest that adrenaline and insulin may coordinate the cell-specific resetting of α-cellular and β-cellular clocks, resulting in their phase difference in vivo (Figs. 3, 5, 7).
Of note, Per2 expression exhibited a similar rhythmic expression profile in α and β cells in vivo but not in vitro, as reflected by the circadian PER2::Luc reporter (see Figs. 3B, 4, 5B). It has been demonstrated that, unlike other core clock genes, Per2 expression might be uncoupled from the core clock and driven by systemic cues independently of the local oscillator (Kornmann et al. 2007). Therefore, the observed discrepancy might be explained by the dominant effect of food entrainment on Per2, overriding the input of local oscillators in vivo and its absence in vitro under constant medium conditions.
Differential rhythmic expression pattern of the α-cell and β-cell transcriptome
Computational analysis of large-scale temporal gene expression profiles with high resolution in separated α-cell and β-cell populations resulted in establishing a unique database (Supplemental Data Set 1) comprising α-cell- and β-cell-specific genes and those expressed in both cell types and the assessment of their rhythmicity. In total, ∼60% of all detected transcripts exhibited rhythmicity in one or both cell types (Figs. 1D,F, 2A; Supplemental Fig. S3), which is higher in comparison with the recently reported fraction in whole islets (Perelis et al. 2015a). This difference might be explained by transcripts exhibiting distinct phases in two cell types (models 5 and 14), which may not be detectable as rhythmic in the whole islets. Previous studies suggested that the ratio of cycling and noncycling genes is tissue-specific, with metabolically active tissues exhibiting a greater number of rhythmic transcripts (Zhang et al. 2014). Playing a key role in glucose metabolism, α and β cells represent populations with high metabolic activities closely associated with feeding behavior. Most of the observed rhythmic transcripts exhibited peak expression levels in the middle of the light or dark phase (Figs. 1, 2), which might be attributed to the strong synchronizing effect exerted by rhythmic feeding on α-cell and β-cell clocks, as was demonstrated for other metabolically active tissues (Damiola et al. 2000; Atger et al. 2015). Among the rhythmically expressed transcripts, we found genes required for granule biogenesis, trafficking, and exocytosis (Fig. 7; Supplemental Data Set 1; Supplemental Table 1).
Rhythmic transcriptional regulation of islet hormone biogenesis
Biogenesis of insulin and glucagon shares similarities at principal steps, including hormone production, maturation, and secretory granule formation, trafficking, and exocytosis. The expression levels of insulin and glucagon were constant throughout the day (Fig. 1C), in line with previous reports on mouse and human insulin gene expression (Marcheva et al. 2010; Pulimeno et al. 2013). While the profile of the β-cell-specific proprotein convertase Pcsk1 (determining the first step of insulin maturation) (Orci et al. 1987) was nonrhythmic, expression of Pcsk2 (which regulates proinsulin processing in β cells) and the first steps of Gcg maturation in α cells (Furuta et al. 2001) were rhythmic only in α cells. Its expression was in opposite phase to the maximum of glucagon secretion, possibly to allow refilling of glucagon stores (Fig. 2B). Genes encoding for COPII proteins as well as members of common vesicle trafficking pathways and SNARE proteins—regulating immediate and delayed granule exocytosis (Regazzi et al. 1995) and their accessory factors—were rhythmically expressed, peaking in the middle of the night (Fig. 7; Supplemental Table 1; Supplemental Data Set 1). In line with our data, previous studies suggested circadian profiles for granule assembly and exocytosis regulators in in vitro synchronized islets, which were affected by clock disruption in transgenic mice (Perelis et al. 2015a) and human islet cells (Saini et al. 2016).
Rhythmic regulation of stimulus–secretion-coupling pathways in α and β cells
Furthermore, differential rhythmic control was exerted over transcripts encoding for proteins involved in basal and stimulated insulin and glucagon secretion (Fig. 7). Indeed, high glucose uptake in β cells is mediated by the nonrhythmic expression of the Glut2 transporter (Slc2a2 transcript) (Orci et al. 1989), while, in α cells, the principal glucose transporters Glut1 and Glut5 (Slc2a1 and Slc2a5 transcripts) were rhythmic. Glucokinase (Gck transcript) exhibited a rhythmic expression pattern in both cell types, simultaneously peaking at night. Genes responsible for the initial steps in glucose metabolism (pyruvate metabolism [Pdha1 and Pcx] and Krebs cycle [Aco2]) were rhythmic in α cells but not β cells. In contrast, the expression of respiratory chain components (complex V transcripts) was similarly rhythmic in both cell types, peaking at daytime (Supplemental Data Set 1; Supplemental Table 1). These data suggest a rhythmic transcriptional regulation of energy metabolism processes in mouse islet cells (in line with previous findings in clock-disrupted mice) (Lee et al. 2011) and human islet cells (Saini et al. 2016). In the liver, most of these rate-limiting mitochondrial enzymes are oscillating at the protein level (Neufeld-Cohen et al. 2016). Mitochondrial glucose metabolism differentially raises the ATP/ADP ratio in α and β cells, regulating the function of ion channels responsible for triggering insulin secretion and inhibiting glucagon release (Gromada et al. 2007). Elevated levels of ATP lead to the closure of ATP-sensitive K+ (KATP) channels (complex of KCNJ11 and sulfonylurea receptor SUR-1 encoded by Abcc8), resulting in cell depolarization and opposite regulation of glucose-induced insulin and glucagon secretion (Gromada et al. 2004). Here, we report that the rhythmic expression peaks for Kcnj11 and Abcc8 in α and β cells are temporally separated: ZT6 for Kcnj11 and ZT20 for Abcc8. Interestingly, in human islet cells, the KCNJ11 transcript was up-regulated upon siRNA-mediated CLOCK disruption (Saini et al. 2016), suggesting an inhibitory effect of the BMAL1/CLOCK complex on KCNJ11 expression. Activation of KATP upon high glucose concentration opens voltage-dependent calcium channels (VDCCs) in β cells, resulting in calcium influx, SNARE-mediated insulin granule release, and closure of these channels in α cells, inhibiting glucagon secretion (Gromada et al. 2004; Gustavsson et al. 2009). Low glucose concentrations result in low ATP levels in α cells and moderately activate KATP channels, allowing the opening of VDCCs (Fig. 7, dotted arrow), leading to Ca2+ influx in α cells and SNARE-mediated release of glucagon granules. Interestingly, multiple genes encoding for VDCC members (Supplemental Table 1) exhibited rhythmic expression, in line with the previously reported circadian regulation of VDCC expression in the SCNs (Nahm et al. 2005). Hence, the fact that the activity of VDCCs in islet cells is regulated by glucose makes them good candidates for coupling feeding regimen to intracellular molecular clocks.
Rhythmic profiles of insulin and glucagon are not aligned in vivo and in vitro
Turning to the functional output of α and β cells, plasma insulin levels exhibited significant circadian oscillation, peaking in the middle of the dark phase in night-fed mice (Fig. 6A), in agreement with previous studies reporting higher plasma insulin levels during the activity (night) phase (Marcheva et al. 2010; Perelis et al. 2015a). Parallel assessment of plasma glucagon content in the same samples suggested the oscillatory fluctuation over 24 h with the peak of secretion in the morning hours (ZT0), lagging ∼8 h after the insulin peak (Figs. 6A, 7). These data are consistent with previous work detecting weak daily rhythms in plasma glucagon levels, peaking at the end of the night in ad libitum fed rats (Ruiter et al. 2003). It is noteworthy that rhythmic profiles of both insulin and glucagon secretion were preserved in fasted animals, with peaks strongly and similarly advanced for both hormones as compared with night-fed animals (Fig. 6A,B), suggesting that the circadian clock impacts on islet hormone secretion also in the absence of rhythmic feeding. In line with a previous report (Ruiter et al. 2003), introducing a fasting period prior to the blood collection influenced the daily glucagon secretion pattern by significantly enhancing its amplitude and advancing the phase (Fig. 6A,B, cf. left panels). Interestingly, the opposite tendency has been observed for the circadian amplitude of insulin secretion, which was lower in fasted animals compared with their night-fed counterparts (Fig. 6A,B, cf. middle panels). This finding is in line with human data indicating that the circadian amplitude of blood insulin is positively correlated with blood glucose levels (Boden et al. 1996). Of note, previous experiments with SCN lesioned animals (Yamamoto et al. 1987) demonstrated a role of the central clock in generating daily rhythms of glucagon secretion. Moreover, the studies by Bass and colleagues (Marcheva et al. 2010; Perelis et al. 2015a) provided compelling evidence that insulin secretion is strongly compromised in clock-deficient mice and that this effect is cell-autonomous. Similarly, in human islets, clock disruption reduced the amount of insulin at both basal and high-glucose conditions (Saini et al. 2016). In strong agreement with these studies, our data imply that the presence of a functional clock is essential for proper islet hormone secretion, as clock-disrupted Bmal1 knockout mice had significantly altered blood levels of both insulin and glucagon (Fig. 6C), further strengthening the importance of a functional clock in regulating glucose homeostasis.
In line with in vivo data, our perifusion experiments demonstrate that mixed islet cells and pure α cells synchronized in vitro by forskolin secrete basal glucagon in a circadian manner (Fig. 6E,G, middle panels). The suggested role of the α-cell clock in regulating glucagon secretion is supported by a previous study demonstrating an inhibitory effect of the core clock component Reverba on glucagon secretion in αTC1–9 cells (Vieira et al. 2013).
A rhythmic pattern of basal insulin secretion was obtained from mixed cells and pure β cells exhibiting similar circadian phases in this case (Fig. 6E,G), consistent with earlier in vitro studies in rodent and human islet cells, suggesting a circadian profile of insulin secretion by isolated islets or islet cells in rodents and humans (Peschke and Peschke 1998; Perelis et al. 2015a; Saini et al. 2016). Remarkably, this temporal separation of glucagon and insulin secretion observed in vivo persists also in vitro, with the glucagon profile being phase-delayed compared with insulin (Fig. 6E,G, right panels). This phase shift was particularly pronounced in separated α-cell and β-cell populations, suggesting differential autonomous circadian regulation of hormone secretion in α and β cells (Fig. 7).
While the dietary glucose intake has an acute effect on regulating insulin and glucagon secretion, cell-autonomous islet cellular clocks are likely to modulate this process in a temporal manner through the diurnal regulation of genes involved in secretion (Dibner and Schibler 2015b). Similar to rodents, endocrine body rhythms in humans, including those of insulin, are tightly regulated by the circadian system (Philippe and Dibner 2014). Such oscillatory profiles of metabolically active hormones may represent an important anticipatory mechanism, allowing for the optimal metabolic orchestration at the organism level.
The incidence of metabolic diseases, including T2D, is growing exponentially in modern societies (Krug 2016). This tendency might be attributed to sleep perturbations related to the increasing rate of acute and social jet lag and the rise of shifted work schedules (Maury et al. 2014; Saini et al. 2015). Accumulating data suggest a key role of α-cell dysfunction in the development of T2D (D'Alessio 2011; Lee et al. 2016). Our data will help to dissect the contribution of the circadian clock in α-cell and β-cell physiology and pave the way for future studies aiming at deciphering α-cell and β-cell dysfunction in the etiology of metabolic diseases.
Materials and methods
Animal care and reporter mouse strain
Animal studies were performed according to the regulations of the veterinary office of the State of Geneva. The triple reporter mouse strain was established by crossing Gcg-Venus reporter (Reimann et al. 2008), RIP-Cherry (Zhu et al. 2015), and Per2::Luc (Yoo et al. 2004) mice (Supplemental Fig. S1). ProGcg-Venus and RIP-Cherry reporters exhibited very high specificity and expression levels in α and β cells, respectively (Supplemental Fig. S1; Zhu et al. 2015; Dusaulcy et al. 2016). Bmal1 knockout mice have been described previously by Jouffe et al. (2013). All experiments were done in mice between 7 and 16 wk of age under standard animal housing conditions with free access to food and water and in 12-h light/12-h dark cycles (LD). For the in vivo sample collection, animals were subjected to night-restricted feeding (Supplemental Fig. S1) 2 wk prior to the experiments and during the entire period of sample collection, allowing us to reduce the effect of individual feeding rhythms (Atger et al. 2015). For sample collection covering the 24-h period, half of the animals were entrained by inverted LD and feeding cycles during the 3 wk preceding the experiments. Fasted blood collection around the clock was performed in the absence of feeding and following 12 h of fasting prior to the first time point. For Bmal1 knockout mice and their control littermates, serum samples were collected during the light phase (ZT0–ZT12) and dark phase (ZT12–ZT24).
Pancreatic islet isolation and separation of α and β cells
Islets of Langerhans were isolated by standard procedure based on collagenase (type XI; Sigma) digestion of the pancreas followed by Ficoll purification (Wojtusciszyn et al. 2009). Islet cells were gently dissociated by trypsin (GIBCO) resuspended in KRB solution (pH 7.4) supplemented with 0.3 % free fatty acid bovine serum albumin (BSA) (Sigma), 1.4 mM glucose, and 0.5 mM EDTA). α-Cell and β-cell populations were separated by flow cytometry FACS (Astrios sorter, Beckman Coulter) based on fluorescence wavelength and intensity and cell singlet nature, size, and viability (Supplemental Fig. S2).
In vitro islets/islet cell culture, synchronization, and bioluminescence monitoring
For in vitro culture, the intact islets or dissociated or sorted cells were recovered in RPMI 1640 complete medium (11.2 mM glucose, 110 µg/mL sodium pyruvate) supplemented with 10% fetal calf serum, 110 U/mL penicillin, 110 µg/mL streptomycin, and 50 µg/mL gentamycin and attached to 35-mm dishes or multiwell plates precoated with a laminin-5-rich extracellular matrix (Parnaud et al. 2008). Adherent islets/cells were synchronized by a 1-h pulse of 10 µM forskolin (Sigma), 100 nM insulin (NovoRapid), or 5 µM adrenaline (Geneva Hospital Pharmacy) prior to continuous bioluminescence recording in RPMI supplemented with 100 µM luciferin (NanoLight Technology) (Saini et al. 2016). For detrended time series, raw luminescence signals were processed by a moving average with a window of 24 h (Saini et al. 2016). β1 adrenergic receptor antagonist atenolol (100 µM), and 100 µM α2 adrenergic receptor antagonist yohimbine were applied to α and β cells, respectively, 15 min prior to adding adrenaline for the synchronization, and cells were kept in the medium during a 1-h synchronization period.
Combined bioluminescence–fluorescence time-lapse microscopy and data analysis
Dispersed islet cells attached to glass-bottomed dishes (WillCo Wells BV) were synchronized by forskolin and subjected to combined bioluminescence–fluorescence imaging (Pulimeno et al. 2013). An Olympus LV200 workstation equipped with a 63× UPLSAPO objective and EM CCD camera (Image EM C9100-13, Hamamatsu) was used. The recorded time-lapsed images were analyzed on ImageJ 1.50 (Schneider et al. 2012), with individual cells tracked in the bioluminescence and fluorescence channels using a modified version of ImageJ plug-in CGE (Sage et al. 2010). Measuring of expression levels was performed on the labeled and tracked cells in the bioluminescence images over time. To assess the circadian characteristics of single-cell profiles, a Cosine fitting method (CosinorJ) was applied (Mannic et al. 2013).
RNA-seq
Total RNA was prepared from FACS-sorted α and β cells collected every 4 h around the clock in duplicates (total of 24 samples representing an RNA pool of six to 12 mice each) using RNeasy Plus Micro Kit (Qiagen). RNA-seq was performed on the Institute of Genetics and Genomics in Geneva genomics platform (University of Geneva, Switzerland). The TruSeq stranded total RNA with Ribo-Zero Gold kit (Illumina) was used for library preparation with 100 ng of total RNA as input. Library molarity and quality were assessed with Qubit (Life Technologies) and Tapestation using a DNA high-sensitivity chip (Agilent Technologies). Paired-end reads of 50 bases were generated using TruSeq SBS HS version 3 chemistry on an Illumina HiSeq 2500 sequencer. RNA-seq mapping and quantification and model selection and rhythmicity assessment are described in the Supplemental Material.
Measurements of insulin, glucagon, and glucose levels in the blood serum
Mice were sacrificed by decapitation at ZT0, ZT4, ZT8, ZT12, ZT16, and ZT20 with subsequent blood collection and serum preparation by immediate centrifugation at 3000 rpm for 15 min at 4°C (Supplemental Fig. S1) and storage at −80°C. Protease inhibitors PMSF (Axon), aprotinin (Sigma), and DPP4 (Millipore) were added to the samples to preserve the hormones from degradation. Insulin and glucagon concentrations were assessed by ultrasensitive mouse insulin and glucagon ELISA kits (Mercodia), and serum glucose was assessed by Accu-Chek glucometer (Roche).
Islet cell continuous perifusion
Dispersed islet cells or FACS-separated α and β cells were attached and forskolin-synchronized as described above, placed into an in-house-developed two-well horizontal perifusion chamber connected to a LumiCycle, and continuously perifused with RPMI (without sodium pyruvate) containing 5.5 mM glucose and 100 µM luciferin as described (Saini et al. 2016). Bioluminescence recordings were performed in parallel to the 4-h interval automated collection of the outflow medium. Basal insulin and glucagon levels were quantified in the outflow medium by mouse insulin and glucagon ELISA kits (Mercodia) and normalized to the total cellular content with subsequent moving average transformation. Circadian parameters of the secreted profiles were evaluated by the CosinorJ algorithm (Mannic et al. 2013).
Supplementary Material
Acknowledgments
We thank Ueli Schibler, Claes Wollheim, Eric Feraille, Hans Reinke, and Ursula Loizides-Mangold for constructive discussions; Pedro Luis Herrera and his colleagues—Simona Chera, Luiza Ghila, Fabirizio Thorel, and Olivier Fazio—for generous help with the reporter mice; Yvan Gosmain and Christian Vesin for helping with islet isolation; Andre Liani and George Severi for helping with perifusion experiments; Mylene Docquier and Brice Petit for performing RNA-seq experiments; Jean-Pierre Aubry-Lachainaye for his exceptional help with FACS experiments; and Christoph Bauer and Jerome Bosset for helping with bioimaging experiments. This work was funded by Swiss National Science Foundation grant 31003A_166700/1, the Novartis Consumer Health Foundation, the Fondation Romande pour la Recherche sur le Diabète, the European Foundation for the Study of Diabetes/Boehringer Ingelheim Basic Research Programme, the Fondation Privée des Hôpitaux Universitaires de Genève, the Bo Hjelt Foundation, and the Olga Mayenfisch Foundation (C.D.).
Footnotes
Supplemental material is available for this article.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.290379.116.
References
- Adriaenssens AE, Svendsen B, Lam BY, Yeo GS, Holst JJ, Reimann F, Gribble FM. 2016. Transcriptomic profiling of pancreatic α, β and δ cell populations identifies δ cells as a principal target for ghrelin in mouse islets. Diabetologia 59: 2156–2165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asher G, Sassone-Corsi P. 2015. Time for food: the intimate interplay between nutrition, metabolism, and the circadian clock. Cell 161: 84–92. [DOI] [PubMed] [Google Scholar]
- Asher G, Schibler U. 2011. Crosstalk between components of circadian and metabolic cycles in mammals. Cell Metab 13: 125–137. [DOI] [PubMed] [Google Scholar]
- Atger F, Gobet C, Marquis J, Martin E, Wang J, Weger B, Lefebvre G, Descombes P, Naef F, Gachon F. 2015. Circadian and feeding rhythms differentially affect rhythmic mRNA transcription and translation in mouse liver. Proc Natl Acad Sci 112: E6579–E6588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balsalobre A, Marcacci L, Schibler U. 2000. Multiple signaling pathways elicit circadian gene expression in cultured Rat-1 fibroblasts. Curr Biol 10: 1291–1294. [DOI] [PubMed] [Google Scholar]
- Benner C, van der Meulen T, Caceres E, Tigyi K, Donaldson CJ, Huising MO. 2014. The transcriptional landscape of mouse β cells compared to human β cells reveals notable species differences in long non-coding RNA and protein-coding gene expression. BMC Genomics 15: 620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boden G, Ruiz J, Urbain JL, Chen X. 1996. Evidence for a circadian rhythm of insulin secretion. Am J Physiol 271: E246–E252. [DOI] [PubMed] [Google Scholar]
- D'Alessio D. 2011. The role of dysregulated glucagon secretion in type 2 diabetes. Diabetes Obesity Metab 13: 126–132. [DOI] [PubMed] [Google Scholar]
- Damiola F, Le Minh N, Preitner N, Kornmann B, Fleury-Olela F, Schibler U. 2000. Restricted feeding uncouples circadian oscillators in peripheral tissues from the central pacemaker in the suprachiasmatic nucleus. Genes Dev 14: 2950–2961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Boer SF, Van der Gugten J. 1987. Daily variations in plasma noradrenaline, adrenaline and corticosterone concentrations in rats. Physiol Behav 40: 323–328. [DOI] [PubMed] [Google Scholar]
- Dibner C, Schibler U. 2015a. Circadian timing of metabolism in animal models and humans. J Intern Med 277: 513–527. [DOI] [PubMed] [Google Scholar]
- Dibner C, Schibler U. 2015b. Metabolism. A pancreatic clock times insulin release. Science 350: 628–629. [DOI] [PubMed] [Google Scholar]
- DiGruccio MR, Mawla AM, Donaldson CJ, Noguchi GM, Vaughan J, Cowing-Zitron C, van der Meulen T, Huising MO. 2016. Comprehensive α, β and δ cell transcriptomes reveal that ghrelin selectively activates δ cells and promotes somatostatin release from pancreatic islets. Mol Metab 5: 449–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dusaulcy R, Handgraaf S, Heddad-Masson M, Visentin F, Vesin C, Reimann F, Gribble F, Philippe J, Gosmain Y. 2016. α-Cell dysfunctions and molecular alterations in male insulinopenic diabetic mice are not completely corrected by insulin. Endocrinology 157: 536–547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuta M, Zhou A, Webb G, Carroll R, Ravazzola M, Orci L, Steiner DF. 2001. Severe defect in proglucagon processing in islet A-cells of prohormone convertase 2 null mice. J Biol Chem 276: 27197–27202. [DOI] [PubMed] [Google Scholar]
- Gromada J, Ma X, Hoy M, Bokvist K, Salehi A, Berggren PO, Rorsman P. 2004. ATP-sensitive K+ channel-dependent regulation of glucagon release and electrical activity by glucose in wild-type and SUR1−/− mouse α-cells. Diabetes 53: S181–S189. [DOI] [PubMed] [Google Scholar]
- Gromada J, Franklin I, Wollheim CB. 2007. α-Cells of the endocrine pancreas: 35 years of research but the enigma remains. Endocr Rev 28: 84–116. [DOI] [PubMed] [Google Scholar]
- Gustavsson N, Wei SH, Hoang DN, Lao Y, Zhang Q, Radda GK, Rorsman P, Sudhof TC, Han W. 2009. Synaptotagmin-7 is a principal Ca2+ sensor for Ca2+ -induced glucagon exocytosis in pancreas. J Physiol 587: 1169–1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jouffe C, Cretenet G, Symul L, Martin E, Atger F, Naef F, Gachon F. 2013. The circadian clock coordinates ribosome biogenesis. PLoS Biol 11: e1001455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornmann B, Schaad O, Bujard H, Takahashi JS, Schibler U. 2007. System-driven and oscillator-dependent circadian transcription in mice with a conditionally active liver clock. PLoS Biol 5: e34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krug EG. 2016. Trends in diabetes: sounding the alarm. Lancet 387: 1485–1486. [DOI] [PubMed] [Google Scholar]
- Lee J, Kim MS, Li R, Liu VY, Fu L, Moore DD, Ma K, Yechoor VK. 2011. Loss of Bmal1 leads to uncoupling and impaired glucose-stimulated insulin secretion in β-cells. Islets 3: 381–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YH, Wang MY, Yu XX, Unger RH. 2016. Glucagon is the key factor in the development of diabetes. Diabetologia 59: 1372–1375. [DOI] [PubMed] [Google Scholar]
- Mannic T, Meyer P, Triponez F, Pusztaszeri M, Le Martelot G, Mariani O, Schmitter D, Sage D, Philippe J, Dibner C. 2013. Circadian clock characteristics are altered in human thyroid malignant nodules. J Clin Endocrinol Metab 98: 4446–4456. [DOI] [PubMed] [Google Scholar]
- Marcheva B, Ramsey KM, Buhr ED, Kobayashi Y, Su H, Ko CH, Ivanova G, Omura C, Mo S, Vitaterna MH, et al. 2010. Disruption of the clock components CLOCK and BMAL1 leads to hypoinsulinaemia and diabetes. Nature 466: 627–631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maury E, Hong HK, Bass J. 2014. Circadian disruption in the pathogenesis of metabolic syndrome. Diabetes Metab 40: 338–346. [DOI] [PubMed] [Google Scholar]
- Nahm SS, Farnell YZ, Griffith W, Earnest DJ. 2005. Circadian regulation and function of voltage-dependent calcium channels in the suprachiasmatic nucleus. J Neurosci 25: 9304–9308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neufeld-Cohen A, Robles MS, Aviram R, Manella G, Adamovich Y, Ladeuix B, Nir D, Rousso-Noori L, Kuperman Y, Golik M, et al. 2016. Circadian control of oscillations in mitochondrial rate-limiting enzymes and nutrient utilization by PERIOD proteins. Proc Natl Acad Sci 113: E1673–E1682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orci L, Ravazzola M, Storch MJ, Anderson RG, Vassalli JD, Perrelet A. 1987. Proteolytic maturation of insulin is a post-Golgi event which occurs in acidifying clathrin-coated secretory vesicles. Cell 49: 865–868. [DOI] [PubMed] [Google Scholar]
- Orci L, Thorens B, Ravazzola M, Lodish HF. 1989. Localization of the pancreatic β cell glucose transporter to specific plasma membrane domains. Science 245: 295–297. [DOI] [PubMed] [Google Scholar]
- Parnaud G, Bosco D, Berney T, Pattou F, Kerr-Conte J, Donath MY, Bruun C, Mandrup-Poulsen T, Billestrup N, Halban PA. 2008. Proliferation of sorted human and rat β cells. Diabetologia 51: 91–100. [DOI] [PubMed] [Google Scholar]
- Partch CL, Green CB, Takahashi JS. 2014. Molecular architecture of the mammalian circadian clock. Trends Cell Biol 24: 90–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perelis M, Marcheva B, Ramsey KM, Schipma MJ, Hutchison AL, Taguchi A, Peek CB, Hong H, Huang W, Omura C, et al. 2015a. Pancreatic β cell enhancers regulate rhythmic transcription of genes controlling insulin secretion. Science 350: aac4250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perelis M, Ramsey KM, Bass J. 2015b. The molecular clock as a metabolic rheostat. Diabetes Obesity Metab 17: 99–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrin L, Loizides-Mangold U, Skarupelova S, Pulimeno P, Chanon S, Robert M, Bouzakri K, Modoux C, Roux-Lombard P, Vidal H, et al. 2015. Human skeletal myotubes display a cell-autonomous circadian clock implicated in basal myokine secretion. Mol Metab 4: 834–845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peschke E, Peschke D. 1998. Evidence for a circadian rhythm of insulin release from perifused rat pancreatic islets. Diabetologia 41: 1085–1092. [DOI] [PubMed] [Google Scholar]
- Philippe J, Dibner C. 2014. Thyroid circadian timing: roles in physiology and thyroid malignancies. J Biol Rhythms 2: 76–83. [DOI] [PubMed] [Google Scholar]
- Pulimeno P, Mannic T, Sage D, Giovannoni L, Salmon P, Lemeille S, Giry-Laterriere M, Unser M, Bosco D, Bauer C, et al. 2013. Autonomous and self-sustained circadian oscillators displayed in human islet cells. Diabetologia 56: 497–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian J, Block GD, Colwell CS, Matveyenko AV. 2013. Consequences of exposure to light at night on the pancreatic islet circadian clock and function in rats. Diabetes 62: 3469–3478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regazzi R, Wollheim CB, Lang J, Theler JM, Rossetto O, Montecucco C, Sadoul K, Weller U, Palmer M, Thorens B. 1995. VAMP-2 and cellubrevin are expressed in pancreatic β-cells and are essential for Ca2+-but not for GTPγ S-induced insulin secretion. EMBO J 14: 2723–2730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reimann F, Habib AM, Tolhurst G, Parker HE, Rogers GJ, Gribble FM. 2008. Glucose sensing in L cells: a primary cell study. Cell Metab 8: 532–539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosengren AH, Jokubka R, Tojjar D, Granhall C, Hansson O, Li DQ, Nagaraj V, Reinbothe TM, Tuncel J, Eliasson L, et al. 2010. Overexpression of α2A-adrenergic receptors contributes to type 2 diabetes. Science 327: 217–220. [DOI] [PubMed] [Google Scholar]
- Ruiter M, La Fleur SE, van Heijningen C, van der Vliet J, Kalsbeek A, Buijs RM. 2003. The daily rhythm in plasma glucagon concentrations in the rat is modulated by the biological clock and by feeding behavior. Diabetes 52: 1709–1715. [DOI] [PubMed] [Google Scholar]
- Sadacca LA, Lamia KA, deLemos AS, Blum B, Weitz CJ. 2011. An intrinsic circadian clock of the pancreas is required for normal insulin release and glucose homeostasis in mice. Diabetologia 54: 120–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sage D, Unser M, Salmon P, Dibner C. 2010. A software solution for recording circadian oscillator features in time-lapse live cell microscopy. Cell Div 5: 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saini C, Brown SA, Dibner C. 2015. Human peripheral clocks: applications for studying circadian phenotypes in physiology and pathophysiology. Front Neurol 6: 95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saini C, Petrenko V, Pulimeno P, Giovannoni L, Berney T, Hebrok M, Howald C, Dermitzakis ET, Dibner C. 2016. A functional circadian clock is required for proper insulin secretion by human pancreatic islet cells. Diabetes Obesity Metab 18: 355–365. [DOI] [PubMed] [Google Scholar]
- Schneider CA, Rasband WS, Eliceiri KW. 2012. NIH Image to ImageJ: 25 years of image analysis. Nat Methods 9: 671–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorel F, Nepote V, Avril I, Kohno K, Desgraz R, Chera S, Herrera PL. 2010. Conversion of adult pancreatic α-cells to β-cells after extreme β-cell loss. Nature 464: 1149–1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorens B. 2014. Neural regulation of pancreatic islet cell mass and function. Diabetes Obesity Metab 16(Suppl 1): 87–95. [DOI] [PubMed] [Google Scholar]
- Unger RH, Orci L. 1975. The essential role of glucagon in the pathogenesis of diabetes mellitus. Lancet 1: 14–16. [DOI] [PubMed] [Google Scholar]
- Vieira E, Liu YJ, Gylfe E. 2004. Involvement of α1 and β-adrenoceptors in adrenaline stimulation of the glucagon-secreting mouse α-cell. Naunyn Schmiedebergs Arch Pharmacol 369: 179–183. [DOI] [PubMed] [Google Scholar]
- Vieira E, Marroqui L, Figueroa AL, Merino B, Fernandez-Ruiz R, Nadal A, Burris TP, Gomis R, Quesada I. 2013. Involvement of the clock gene Rev–erbα in the regulation of glucagon secretion in pancreatic α-cells. PLoS One 8: e69939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wojtusciszyn A, Andres A, Morel P, Charvier S, Armanet M, Toso C, Choi Y, Bosco D, Berney T. 2009. Immunomodulation by blockade of the TRANCE co-stimulatory pathway in murine allogeneic islet transplantation. Transpl Int 22: 931–939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto H, Nagai K, Nakagawa H. 1987. Role of SCN in daily rhythms of plasma glucose, FFA, insulin and glucagon. Chronobiol Int 4: 483–491. [DOI] [PubMed] [Google Scholar]
- Yoo SH, Yamazaki S, Lowrey PL, Shimomura K, Ko CH, Buhr ED, Siepka SM, Hong HK, Oh WJ, Yoo OJ, et al. 2004. PERIOD2::LUCIFERASE real-time reporting of circadian dynamics reveals persistent circadian oscillations in mouse peripheral tissues. Proc Natl Acad Sci 101: 5339–5346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaret KS, White MF. 2010. Diabetes forum: extreme makeover of pancreatic α-cells. Nature 464: 1132–1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang R, Lahens NF, Ballance HI, Hughes ME, Hogenesch JB. 2014. A circadian gene expression atlas in mammals: implications for biology and medicine. Proc Natl Acad Sci 111: 16219–16224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu X, Hu R, Brissova M, Stein RW, Powers AC, Gu G, Kaverina I. 2015. Microtubules negatively regulate insulin secretion in pancreatic β cells. Dev Cell 34: 656–668. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.