In this study, McJunkin et al. show a new role for the C. elegans mir-35 family of maternally contributed essential microRNAs. They demonstrate that this microRNA family regulates the sex determination pathway at multiple levels, acting both upstream of and downstream from her-1 to prevent aberrantly activated male developmental programs in hermaphrodite embryos.
Keywords: microRNAs, embryonic development, maternal control, mir-35–41, mir-35–42, sex determination
Abstract
Gene expression in early animal embryogenesis is in large part controlled post-transcriptionally. Maternally contributed microRNAs may therefore play important roles in early development. We elucidated a major biological role of the nematode mir-35 family of maternally contributed essential microRNAs. We show that this microRNA family regulates the sex determination pathway at multiple levels, acting both upstream of and downstream from her-1 to prevent aberrantly activated male developmental programs in hermaphrodite embryos. Both of the predicted target genes that act downstream from the mir-35 family in this process, suppressor-26 (sup-26) and NHL (NCL-1, HT2A, and LIN-41 repeat) domain-containing-2 (nhl-2), encode RNA-binding proteins, thus delineating a previously unknown post-transcriptional regulatory subnetwork within the well-studied sex determination pathway of Caenorhabditis elegans. Repression of nhl-2 by the mir-35 family is required for not only proper sex determination but also viability, showing that a single microRNA target site can be essential. Since sex determination in C. elegans requires zygotic gene expression to read the sex chromosome karyotype, early embryos must remain gender-naïve; our findings show that the mir-35 family microRNAs act in the early embryo to function as a developmental timer that preserves naïveté and prevents premature deleterious developmental decisions.
MicroRNAs (miRNAs) are endogenously encoded small RNAs that counteract the translation and stability of complementary mRNA targets (Ketting 2011). Canonical miRNAs are processed from long primary transcripts by a series of nucleolytic cleavages and ultimately are loaded into Argonaute proteins to form a functional RNA-induced silencing complex (RISC) (Ha and Kim 2014).
Nucleotides 2–8 at the 5′ end of the miRNA are known as the seed sequence because these positions are the most important for target recognition. MiRNAs sharing an identical seed sequence are grouped together as a “family” because they are predicted to redundantly repress a set of common targets. Phenotypic analysis in Caenorhabditis elegans and other organisms supports this notion; mutation of all members of a miRNA family generally exhibits a more severe phenotype than deletion of a single miRNA (Ventura et al. 2008; Alvarez-Saavedra and Horvitz 2010; Parchem et al. 2015).
The roles played by miRNAs in embryonic development are incompletely understood. Notable exceptions are the mammalian miR-290 and miR-302 clusters and orthologous miR-430 (zebrafish), which promote pluripotency after the oocyte-to-embryo transition (OET) (Giraldez 2010; Greve et al. 2013). Prior to the OET, gene expression is regulated post-transcriptionally by maternally contributed factors, and many miRNA families are present during this developmental period (Chiang et al. 2010). However, for most of these maternally contributed miRNAs, their roles in early development are unknown.
Many prototypical zygotically expressed miRNAs (e.g., miR-430, lin-4, and let-7) act to control developmental timing, promoting progression to a later developmental stage when their expression is developmentally up-regulated. In contrast, maternally contributed miRNAs are present at the beginning of development and are down-regulated as development progresses. Maternally contributed miRNAs may therefore act in early embryos to enforce a naïve state, preventing developmental decisions from being executed prematurely; for example, by dampening gene expression noise and/or exerting transient repression of maternal transcripts whose expression is critical later in development. This latter hypothesis is consistent with recent findings that the mechanism of miRNA-mediated target repression in early embryogenesis differs from that observed in later embryos or differentiated tissues by favoring translational repression (which, in principle, should be reversible) rather than irreversible mRNA decay (Bazzini et al. 2012; Subtelny et al. 2014).
To better understand the biological roles that maternally contributed miRNAs play in embryonic development, we sought to dissect the genetic network controlled by the nematode-specific mir-35 family. The mir-35 family is maternally contributed as well as zygotically expressed in early embryos, suggesting that it acts at an earlier developmental stage than previously studied zygotically expressed animal miRNAs (Stoeckius et al. 2009; Alvarez-Saavedra and Horvitz 2010; Wu et al. 2010). This family is essential for embryonic development in C. elegans, but the precise nature of its essential functions are unknown (Alvarez-Saavedra and Horvitz 2010). Besides its essential functions, the mir-35 family also exhibits pleiotropic effects on diverse embryonic and post-embryonic processes, indicating that these miRNAs functionally engage with diverse gene regulatory networks in the embryo (Liu et al. 2011; Massirer et al. 2012; McJunkin and Ambros 2014; Kagias and Pocock 2015).
Here we provide evidence that one of the major roles of the mir-35 family is to act as a developmental timer, preventing premature and aberrant decision-making in the sex determination pathway. Through a partially maternal effect, the mir-35 family acts on multiple levels of the sex determination pathway to prevent the spurious activation of male developmental programs. At least two predicted mir-35 family target genes, suppressor-26 (sup-26) and NHL (NCL-1, HT2A, and LIN-41 repeat) domain-containing-2 (nhl-2), both of which encode RNA-binding proteins (RBPs), act downstream from the mir-35 family in this pathway. SUP-26 binds a broad range of targets, many of which are unrelated to sex determination, and NHL-2 is known to function in conjunction with diverse miRNAs (Hammell et al. 2009), so these findings widen the scope of pathways potentially regulated by the mir-35 family through the SUP-26 and NHL-2 axes. Furthermore, we established that one of the essential functions of the mir-35 family is repression of nhl-2. We thus defined a novel regulatory subnetwork consisting of multiple layers of post-transcriptional control that contribute to the timing and fidelity of C. elegans sex determination and other embryonic developmental processes.
Results
Most transcripts derepressed in mir-35–41(nDf50) mutant embryos are not direct targets of mir-35 family miRNAs
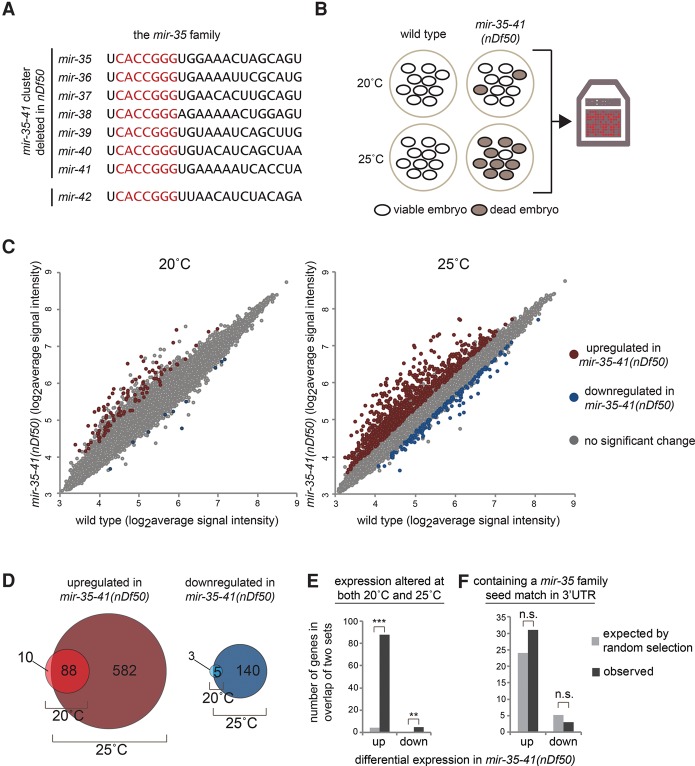
The mir-35 family consists of eight members, mirs-35–42, encoded at two loci (Alvarez-Saavedra and Horvitz 2010). The mir-35–41 cluster encodes seven members of the family on a single transcript, whereas mir-42 is encoded separately as part of the mir-42–45 cluster (Fig. 1A). Deletion of all eight mir-35–42 family members results in completely penetrant embryonic lethality with arrest at various stages of embryogenesis (Alvarez-Saavedra and Horvitz 2010). Forward genetic screens identified no suppressors of mir-35 family mutant lethality, suggesting that perturbation of multiple unknown pathways contributes to this phenotype (Alvarez-Saavedra and Horvitz 2010).
Figure 1.
Many genes are up-regulated in mir-35–41(nDf50) mutants but are not predicted mir-35 family targets. (A) Sequences of mature miRNAs comprising the mir-35 family. mir-35–41 are processed from a single transcript, while mir-42 is located in a separate cluster. Red text indicates the seed sequence. (B) mir-35–41(nDf50) is a temperature-sensitive mutant in which embryonic lethality is low at 20°C and highly penetrant at 25°C. Gene expression in embryos raised at each temperature was profiled using microarrays. (C) Many more genes are differentially expressed in mir-35–41(nDf50) embryos at 25°C than at 20°C. More genes are up-regulated than are down-regulated. (D) The genes that are differentially expressed at 20°C are largely a subset of those observed at 25°C. (E) The sets of differentially expressed genes at 20°C and at 25°C overlap much more than expected by chance. (***) P-value < 1 × 10−100; (**) P-value < 1 × 10−5, hypergeometric test. (F) The genes differentially expressed in mir-35–41(nDf50) embryos do not contain more mir-35 family seed matches than expected in a randomly chosen gene set.
Deletion of the mir-35–41 cluster alone (leaving mir-42 intact) yields a hypomorphic and temperature-sensitive phenotype (Alvarez-Saavedra and Horvitz 2010). mir-35–41(nDf50) mutants exhibit incompletely penetrant embryonic lethality, the penetrance of which increases with temperature. This malleable genetic setting has enabled the characterization of numerous post-embryonic and pleiotropic phenotypes of mir-35 family loss of function (Liu et al. 2011; Massirer et al. 2012; McJunkin and Ambros 2014; Kagias and Pocock 2015). Still, a clear molecular basis for the phenotypes of mir-35 family mutants has remained elusive. We took advantage of this temperature-sensitive allele [mir-35–41(nDf50)] to characterize the molecular phenotype associated with mir-35 family loss of function and identify downstream target genes responsible for the phenotype.
To gain insight into the molecular phenotype caused by mir-35 family loss of function, we performed gene expression analysis of mir-35–41(nDf50) mutant embryos. Since the nDf50 deletion allele acts as a temperature-sensitive hypomorphic allele of mir-35 family function, we profiled gene expression changes at both the semipermissive (20°C) and restrictive (25°C) temperatures (Fig. 1B).
Consistent with the increased severity of phenotypes at restrictive temperature, many more gene expression changes were observed in embryos raised at 25°C (670 genes) than in those raised at 20°C (98 genes; cutoff >1.5-fold, P-value < 0.05) (Fig. 1C; Supplemental Table S1). The small set of deregulated genes observed at 20°C was nearly an exact subset of the differentially expressed genes at 25°C (Fig. 1D,E). This supports the notion that the phenotypes observed at restrictive temperature are a more severe manifestation of the mild phenotypes observed at permissive temperature.
At both 20°C and 25°C, the genes up-regulated in mir-35–41(nDf50) mutants compared with wild type greatly outnumbered those down-regulated in the mutants (98 vs. eight at 20°C and 670 vs. 145 at 25°C) (Fig. 1C,D). We postulated that the large number of up-regulated genes might represent direct targets of the mir-35 family miRNAs, which are derepressed in the miRNA mutants. This was not the case. The set of up-regulated genes was not enriched for genes containing mir-35 family seed matches in their 3′ untranslated regions (UTRs) (Fig. 1F). Instead, the number of genes predicted to be targeted by the mir-35 family in this set was similar to the number expected by chance. The genes down-regulated in mir-35–41(nDf50) mutant embryos were likewise not enriched for mir-35 family seed matches. Therefore, the majority of the gene expression changes observed in mir-35–41(nDf50) mutant embryos represents downstream indirect effects of mir-35 family loss of function.
mir-35–41 prevents masculine development at multiple levels in the sex determination pathway
Previously, we demonstrated that a predicted mir-35 family target gene implicated in the sex determination pathway, sup-26, is epistatic to fecundity phenotypes observed in mir-35–41(nDf50) (McJunkin and Ambros 2014). We wondered whether mir-35–41(nDf50) mutants might have a previously undetected sex determination phenotype. Therefore, we asked whether the molecular phenotype (i.e., gene expression changes) of mir-35–41(nDf50) mimicked those of sex determination mutants by examining published data sets.
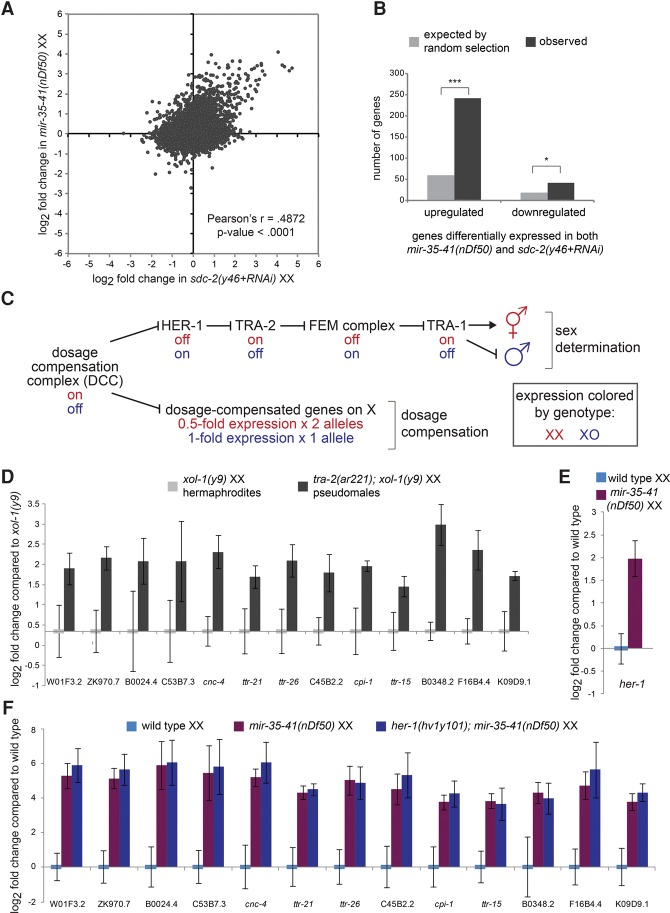
We observed a striking correlation between the gene expression changes in mir-35–41(nDf50) mutants and those in loss-of-function mutants of sdc-2 (sex determination and dosage compensation defect 2) (r = 0.49, P-value < 0.0001) (Fig. 2A; Jans et al. 2009). In particular, the derepressed genes in each genotype were highly overlapping and strongly correlated (Fig. 2A,B).
Figure 2.
Gene expression changes in mir-35–41(nDf50) mutants are similar to those in sex determination mutants. (A) Comparison of gene expression changes in mir-35–41(nDf50) embryos and sdc-2(lf) embryos, each normalized to wild type. (B) The set of genes differentially regulated in mir-35–41(nDf50) and sdc-2(lf) overlaps more than expected by chance. (***) P-value < 1 × 10−80; (*) P-value < 1 × 10−5, hypergeometric test. (C) Genetic model of C. elegans sex determination and dosage compensation. (D–F) Quantitative RT–PCR (qRT–PCR) of transcripts indicated on the X-axis in embryos of the genotypes indicated above the graph. The mean and SEM of three biological replicates are shown. (D) Both genotypes were normalized to xol-1(y9). (E,F) All genotypes were normalized to wild-type XX embryos.
Sdc-2 encodes a core component of the dosage compensation complex (DCC), a condensin-like transcriptional repressor that carries out two functions. One function of the DCC is to bind along the X chromosome to down-regulate gene expression by twofold (dosage compensation [DC]). Since the DCC is active only in XX animals after the 40-cell embryo stage due to karyotype-specific expression of sdc-2, this function equalizes X-linked gene dosage between XX hermaphrodites and XO males. The second function of the DCC is to control sex determination by repressing the master driver of male sexual fate, her-1 (hermaphroditization-1, an autosomal gene) by 40-fold, leading to hermaphroditic sexual development in XX animals (Fig. 2C; Meyer 2005). Because of the DCC's role in both processes, XX animals in which DCC function is impaired display lethality due to loss of DC but also aberrantly masculinized development (Tra [transformer] phenotype) due to derepression of her-1.
Many of the up-regulated genes common to mir-35–41(nDf50) and sdc-2(lf) were not X-linked and were up-regulated greater than twofold, suggesting that these observed gene expression changes are reflective of perturbed sex determination (a molecular Tra phenotype) rather than derepressed targets of DC. In further support of this notion, mir-35–41(nDf50) mutant gene expression changes did not significantly overlap with those in a mutant that disrupts only DC but not sex determination, dpy-27(y57) (Jans et al. 2009).
To further test whether the gene expression changes in mir-35–41(nDf50) were due to an aberrantly activated male gene expression program, we generated masculinized (Tra) embryos using an orthogonal genetic means: a mutation in the sex determination pathway. Mutant tra-2(ar221ts);xol-1(y9) XX animals are hermaphrodites when grown at 20°C but fertile pseudomales when grown at 25°C. If the high-amplitude gene expression changes observed in mir-35–41(nDf50) and sdc-2(lf) are symptomatic of masculinization, then similar changes in gene expression should be observed in the pseudomale embryos generated by perturbing the sex determination pathway directly. This was indeed the result: The 13 genes that were most highly up-regulated in mir-35–41(nDf50) embryos were similarly up-regulated in tra-2(ar221);xol-1(y9) grown at 25°C (Fig. 2D). Therefore, many of the measurable gene expression changes in mir-35–41(nDf50) embryos also occur in pseudomale embryos generated by mutation in the sex determination pathway.
Although only one (ttr-26) of the 13 genes that we examined was identified previously as showing male-biased expression, this is likely because previous genome-wide studies of sex-biased gene expression analyzed male larvae and adults, not embryos (Reinke et al. 2004; Thoemke et al. 2005). Interestingly, most of the genes identified as up-regulated in both mir-35–41(nDf50) and tra-2(ar221);xol-1(y9) embryos are of unknown function, many of which encode small secreted proteins. The diffusible nature of these proteins could play a role in coordinating sex-specific development throughout the embryo similar to HER-1, which also encodes a small secreted protein. Because these genes are up-regulated in pseudomale embryos of two genotypes [sdc-2(lf) and tra-2(ar221ts);xol-1(y9)], we propose that they are symptomatic of aberrantly activated masculine development. We term this aspect of the mir-35–41(nDf50) phenotype pseudomale gene expression (ΨGE) and use this panel of 13 genes as a readout of the ΨGE phenotype.
To determine the level at which the sex determination pathway is perturbed in mir-35–41(nDf50) mutants, we examined her-1 transcript levels by quantitative PCR (qPCR). Elevation of her-1 was observed in mir-35–41(nDf50) embryos, indicating that the sex determination pathway is deregulated upstream of her-1 (Fig. 2C,E). Surprisingly, the broader ΨGE program in mir-35–41(nDf50) mutant XX embryos is not a direct consequence of up-regulated her-1: ΨGE was still observed in mir-35–41(nDf50);her-1(hv1y101null) double mutants (Fig. 2F). This indicates that signaling in the sex determination pathway is aberrantly masculinized both upstream of and downstream from her-1 in mir-35–41(nDf50). Therefore, the mir-35–41 family acts both upstream of her-1 to indirectly repress her-1 transcription and downstream from her-1 to prevent the activation of a broader male-like gene expression program (ΨGE).
mir-35–41(nDf50) mutants are cryptically masculinized
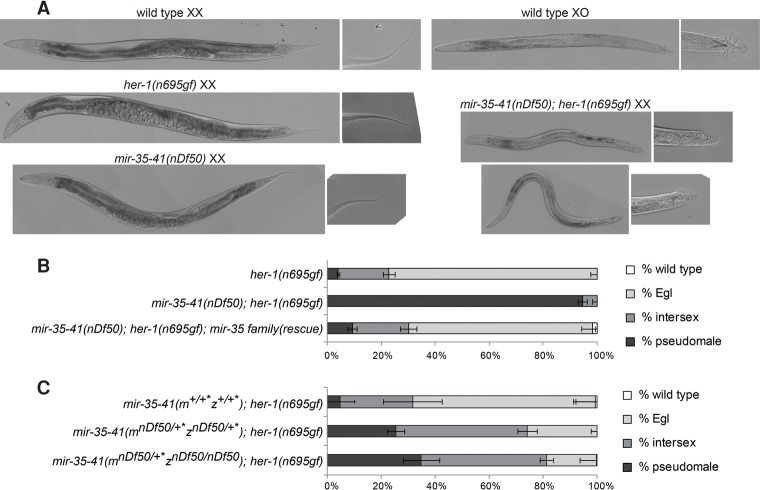
Despite the activation of her-1 transcription and the ΨGE program, sex determination of mir-35–41(nDf50) XX hermaphrodites appears superficially normal. We hypothesized that mir-35–41(nDf50) XX embryos might be cryptically masculinized and that this might be revealed by the appearance of overt masculinization in a sensitized genetic background. Accordingly, we used the her-1(n695gf) allele, which causes a weak derepression of her-1 transcription in XX animals, leading to mild masculinization in hermaphrodites, as evidenced by an egg-laying defective (Egl) phenotype. We crossed mir-35–41(nDf50) into her-1(gf) and observed that mir-35–41(nDf50);her-1(gf) double mutants exhibit dramatically enhanced synthetic masculinization (synTra phenotype); nearly all doubly mutant XX animals are self-sterile pseudomales (Fig. 3A,B). This synTra phenotype is rescued by expression of mir-35–45 from an extrachromosomal array (Fig. 3B). Thus, deletion of mir-35–41 strongly enhances masculinization of her-1(gf), and mir-35–41 therefore promotes hermaphroditic sex determination in wild-type XX animals.
Figure 3.
mir-35–41 are required for proper sex determination in hermaphrodites. (A) Differential interference contrast (DIC) micrographs of wild-type and her-1(gf) and mir-35–41(nDf50) single- and double-mutant adults on the second day of adulthood. (B) Quantification of sex determination phenotypes; darker-colored bars indicate more severe masculinization. (C) The requirement for mir-35–41 in sex determination is dose-dependent, since heterozygotes display enhanced masculinization. Removing both copies of zygotic mir-35–41 does not strongly further enhance masculinization, indicating a strong requirement for the maternal contribution of mir-35–41. (+*) mIn1 balancer chromosome.
Interestingly, sex determination is highly sensitive to the dosage of mir-35–41. Heterozygotes for mir-35–41(nDf50) also show significant enhancement of her-1(gf) masculinization (Fig. 3C). Furthermore, the maternal contribution of mir-35–41 is crucial for sex determination; animals whose mothers contain one copy of the cluster show a similar sex phenotype regardless of whether they inherit a zygotic copy of mir-35–41 (Fig. 3C, bottom two bars). Moreover, in animals lacking zygotic mir-35–41, a single maternal copy of mir-35–41 rescues pseudomale penetrance from 94% to 35% (Fig. 3, cf. middle bar of B and bottom bar of C). This strong maternal effect suggests that the mir-35 family can function at early stages of embryonic development to prevent male development of XX animals.
The predicted mir-35 family target genes sup-26 and nhl-2 are required for cryptic masculinization of mir-35–41(nDf50)
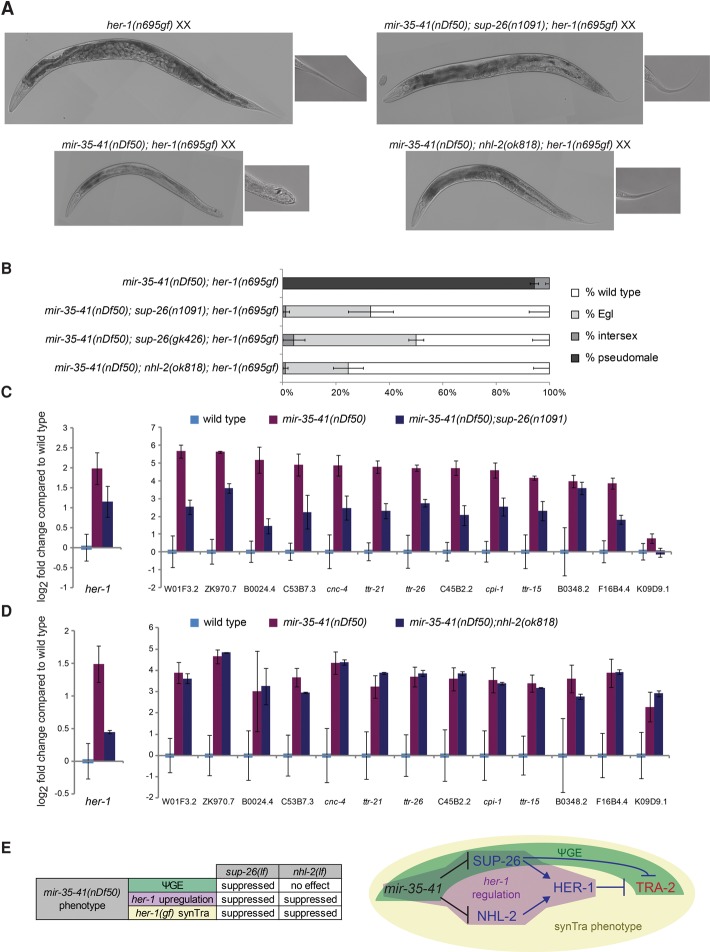
Neither her-1 nor other genes in the core sex determination pathway are predicted targets of the mir-35 family, based on searches for mir-35 seed matches in 3′ UTRs. Therefore, we sought to determine which direct mir-35 family target genes are responsible for the masculinized gene expression signature and cryptic masculinization in mir-35–41(nDf50). To this end, we screened 72 predicted mir-35 family target genes by RNAi to assess which genes were required for the synTra phenotype of mir-35–41(nDf50);her-1(gf) animals (Supplemental Table S2). Of these genes, two were required for the synTra phenotype: sup-26 and nhl-2. The effects of RNAi were recapitulated using mutant alleles of each gene. While mir-35–41(nDf50);her-1(gf) animals develop as pseudomales, mir-35–41(nDf50);her-1(gf);sup-26(lf) or mir-35–41(nDf50);her-1(gf);nhl-2(lf) animals develop as wild-type or Egl hermaphrodites (Fig, 4A,B). Thus, the predicted target genes sup-26 and nhl-2 are epistatic to the enhanced masculinization caused by mir-35–41(nDf50) and likely act downstream from mir-35–41.
Figure 4.
The mir-35 family target genes sup-26 and nhl-2 are required for the sex determination phenotypes of mir-35–41(nDf50). (A) Representative DIC micrographs of adults on the second day of adulthood. mir-35–41(nDf50);sup-26(n1091);her-1(n695gf) and mir-35–41(nDf50);nhl-2(ok818);her-1(n695gf) adults are slightly smaller than age-matched her-1(n695gf) or wild type (not shown). (B) Quantification of sex determination phenotypes; darker-colored bars indicate more severe masculinization. (C,D) qRT–PCR in embryos raised at 20°C. The mean and SEM of three biological replicates are shown. All genotypes were normalized to wild-type XX embryos. (E, left) Summary of genetic interactions of mir-35–41(nDf50) with sup-26(lf) or nhl-2(lf). (Right) Genetic model highlighting pathways whose disruption in mir-35–41(nDf50) mutants underlies each aspect of the mutant sex determination phenotype. Whether the ΨGE phenotype contributes to the synTra phenotype is yet to be determined.
sup-26 encodes an RBP containing a Q-rich low-complexity domain and two RNA recognition motifs. sup-26 was isolated previously in a screen for suppressors of the Egl phenotype of her-1(gf) (Manser et al. 2002), and we confirmed this genetic interaction (Supplemental Fig. S1). We also demonstrated previously that sup-26 is required for another aspect of the mir-35–41(nDf50) mutant phenotype: temperature-sensitive loss of fecundity (McJunkin and Ambros 2014). Previous studies have supported a role for SUP-26 in repressing translation of transformer-2 (tra-2), which encodes the target receptor for the HER-1 ligand (Fig. 2C; Mapes et al. 2010). If the function of SUP-26 downstream from mir-35–41 was entirely at the level of tra-2, then the mir-35–41(nDf50) ΨGE phenotype should be normalized in mir-35–41(nDf50);sup-26(lf) embryos, but her-1 transcription (which is largely controlled upstream of tra-2) should remain elevated. Instead sup-26(lf) alleviates the up-regulation of both the ΨGE and her-1 (Fig. 4C). Therefore, sup-26 may act on the sex determination pathway at multiple levels downstream from mir-35–41 or via a feedback loop through which sex determination can impact the DCC and hence her-1 expression (Hargitai et al. 2009).
nhl-2 encodes a protein of the TRIM (tripartite-containing motif; RING, B-box, coiled-coil domain)–NHL family homologous to Mei-P26 and Brat in Drosophila melanogaster (Hyenne et al. 2008; Hammell et al. 2009). NHL-2 plays a role in the polarity of the early C. elegans embryo, similar to Brat, which also contributes to cell polarity (Hyenne et al. 2008). NHL-2 promotes the activity of miRNAs in post-embryonic development, while Drosophila Mei-P26 represses miRNA abundance (Neumüller et al. 2008; Hammell et al. 2009). Our data, particularly the epistasis of nhl-2 to mir-35–41 and the presence of a conserved mir-35 family seed match in the nhl-2 3′ UTR, support a novel role for nhl-2 downstream from mir-35–41 as a target mRNA.
NHL-2 has not been implicated previously in the DC or sex determination pathways. Like sup-26(lf), nhl-2(lf) also suppresses the Egl phenotype of her-1(gf) and the derepression of the her-1 transcript in mir-35–41(nDf50) embryos (Fig. 4D; Supplemental Fig. S1). The suppression of her-1(gf) is particularly surprising, since nhl-2 was not isolated in mutagenesis screens for such suppressors. Unlike sup-26(lf), nhl-2(lf) did not suppress the mir-35–41(nDf50) ΨGE phenotype (Fig. 4D). Therefore, nhl-2 acts upstream of her-1 in modulating sex-specific development downstream from mir-35–41.
Interestingly, these data indicate that the synTra phenotype of mir-35–41(nDf50);her-1(gf) requires the elevation of her-1 itself and not the ΨGE program alone. Since none of the mutants examined here disrupts only the ΨGE but not her-1 up-regulation, we cannot conclude that the ΨGE is also required for the synTra phenotype. A deeper understanding of the mechanism by which sup-26 promotes the ΨGE will be necessary to dissect this genetic pathway and test whether the ΨGE does contribute to the synTra phenotype along with her-1 up-regulation (Fig. 4E).
While both sup-26 and nhl-2 are required for the sex determination phenotypes of mir-35–41(nDf50) mutants, neither gene is required for the embryonic lethality phenotype of these mutants. On the contrary, deletion of either sup-26, nhl-2, or both in combination paradoxically enhances the lethality of mir-35–41(nDf50) through a poorly understood mechanism (Supplemental Fig. S2A). Enhanced lethality in double mutants of mir-35–41(nDf50) with either sup-26 or nhl-2 is characterized by earlier developmental arrest than in mir-35–41(nDf50) or mir-35–42(nDf49 nDf50) single mutants (Supplemental Fig. S2B; Alvarez-Saavedra and Horvitz 2010), suggesting that the lethality may be altered in character rather than simply increased in penetrance. While the enhancement of lethality by sup-26(lf) or nhl-2(lf) is counterintuitive given our model, the failure to suppress lethality is consistent with the notion that perturbation of multiple pathways contributes to mir-35 family mutant lethality (since no suppressors were identified in forward genetic screens) (Alvarez-Saavedra and Horvitz 2010). Thus, since no single target gene is predicted to suppress mir-35–41(nDf50) lethality, the failure of sup-26(lf) or nhl-2(lf) to do so does not support or refute a role for either gene in contributing to this phenotype.
SUP-26 is a direct mir-35 family target gene
Our model is that translational derepression or stabilization of both sup-26 and nhl-2 together contributes to the sex determination phenotypes observed in mir-35-41(nDf50) mutant embryos. Both sup-26 and nhl-2 have strikingly long 3′ UTRs, 1163 nucleotides (nt) and 1151 nt, respectively, compared with the median 3′ UTR length of 130–140 nt in C. elegans (Mangone et al. 2010; Jan et al. 2011), suggesting that these transcripts are subject to post-transcriptional regulation. Both sup-26 (previously known as tag-310) and nhl-2 are within the top six predicted targets of the mir-35 family on Targetscan due to a well-conserved perfect 8-mer seed match to the mir-35 family located in each of their 3′ UTRs (Jan et al. 2011). Both mRNAs are also high-stringency predicted targets on mirWIP and were identified in sequencing of RNAs physically associated with RISC (Hammell et al. 2008).
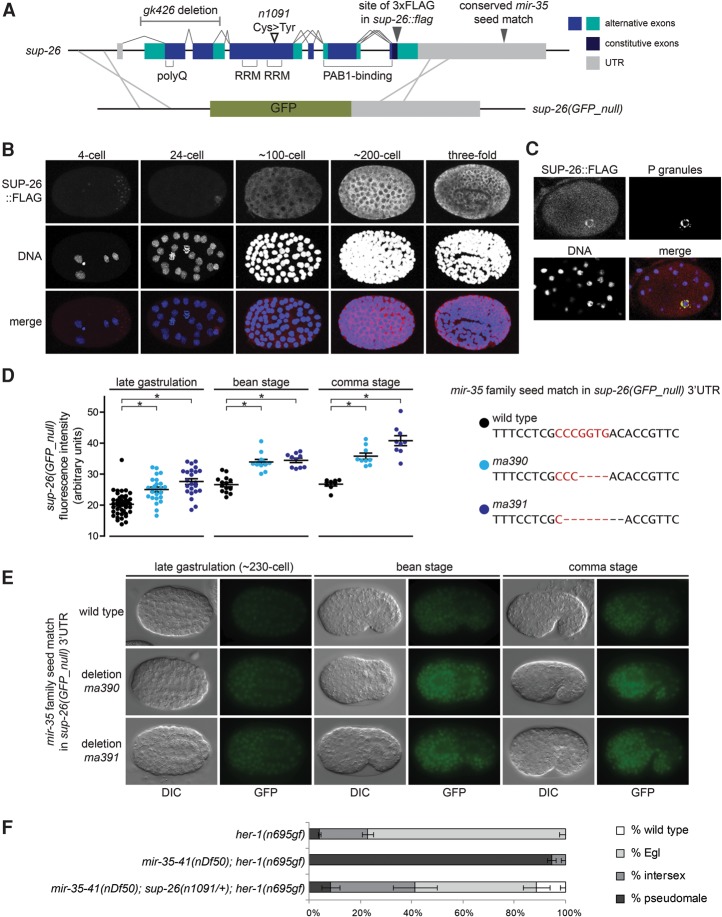
We sought to further validate the role of sup-26 as a direct mir-35 family target gene and assess the contribution of sup-26 to the mir-35–41(nDf50) sex determination phenotype. To this end, we generated new alleles of the sup-26 locus using CRISPR/Cas9 genome editing. An epitope-tagged allele was generated by fusing three Flag tags to the C terminus of SUP-26 (SUP-26::Flag), and a null allele was generated by replacing the entire SUP-26-coding sequence with the GFP-coding sequence [sup-26(GFP_null)] (Fig. 5A). The sup-26(GFP_null) allele behaved similarly to other sup-26 loss-of-function alleles with respect to her-1(gf) and mir-35–41(nDf50);her-1(gf) phenotypes (Supplemental Fig. S3B), although the latter could be tested only in mir-35–41(nDf50) heterozygotes due to the aforementioned synthetic lethality of sup-26(lf);mir-35–41(nDf50) homozygotes. Sup-26(GFP_null) males exhibited a mating defect, consistent with the role of sup-26 in promoting male sex determination (Supplemental Fig. S3C).
Figure 5.
Sup-26 is a direct target gene of the mir-35 family. (A, top) Gene model of sup-26 depicting exon structure, protein domains, mutant alleles, and CRISPR–Cas9 editing sites. (Bottom) Schematic of homologous recombination strategy to generate sup-26(GFP_null). (B) Maximum projection of confocal stacks showing immunofluorescence of SUP-26::Flag. (C) Single confocal image of SUP-26::Flag costaining with PGL-1, a core component of P granules, in an ∼48-cell stage embryo (prior to the division of the germline primordium from a single cell [P4] into two cells [Z2 and Z3]). (D) Quantification of fluorescence in embryos containing the indicated mutations at the mir-35 family seed match in the 3′ UTR of the sup-26(GFP_null) locus. (*) P < 0.0001, Student's t-test. (E) Representative epifluorescence images of embryos with wild-type or mutated mir-35 family seed match in the sup-26(GFP_null) 3′ UTR. (F) The role of sup-26 in sex determination is dose-dependent. sup-26(n1091) heterozygosity strongly suppresses the mir-35–41(nDf50);her-1(gf) synTra phenotype. (+) qC1 balancer chromosome.
The temporal expression pattern of SUP-26::Flag overlaps with that of the mir-35 family. SUP-26::Flag is expressed in the maternal germline and is maternally contributed to early embryos (Fig. 5B; Supplemental Fig. S3D). SUP-26::Flag preferentially localizes to perinuclear bodies by the 24-cell stage; these were confirmed to be germline P granules by costaining with P-granule markers (Fig. 5C). SUP-26::Flag is ubiquitously expressed and cytoplasmically localized in later stages of embryonic development (Fig. 5B). The sup-26(GFP_null) allele shows a similar expression pattern, although it is not expressed in the germline (likely due to transgene silencing of the long exogenous GFP sequence) or enriched in P granules (data not shown). This indicates that the P-granule enrichment of SUP-26::Flag could be attributable to the maternal load of SUP-26::Flag [which is silenced in sup-26(GFP_null)] and/or may be mediated by SUP-26-coding sequences that were deleted in generating sup-26(GFP_null).
Our model predicts that SUP-26 reporter abundance would be increased in the absence of mir-35–41. Surprisingly, we did not observe a robust up-regulation of SUP-26::Flag or sup-26(GFP_null) signals in the mir-35–41(nDf50) mutant background (when examined via Western blot and immunofluorescence or epifluorescence, respectively) (data not shown). qPCR of the sup-26 transcript also showed no change in abundance in mir-35–41(nDf50) mixed-stage embryo samples compared with wild type (data not shown). These results are especially surprising due to the predicted high stability of mir-35 family seed pairing and previous reports of seed match-dependent silencing of a high-copy heterologous reporter by the sup-26 3′ UTR (Garcia et al. 2011; Kagias and Pocock 2015).
We reasoned that the large changes in gene expression and physiology in mir-35–41(nDf50) embryos might confound or dampen the expression changes of a single gene. To study the loss of mir-35 targeting on sup-26 in isolation, we used CRISPR/Cas9 to delete the mir-35 family seed match in the 3′ UTR of the sup-26 reporter alleles. Two alleles of sup-26(GFP_null) in which the mir-35 family seed match is deleted showed increased GFP signal compared with the unedited sup-26(GFP_null) allele (Fig. 5D,E). Thus, the mir-35 family seed match is responsible for partial repression of transcripts bearing the sup-26 3′ UTR, validating this as a bona fide miRNA–target interaction.
SUP-26::Flag alleles in which the mir-35 family seed match was deleted did not show detectable up-regulation compared with the unedited allele (Supplemental Fig. S3E; data not shown). We envision two possible reasons for this discrepancy from sup-26(GFP_null). First, SUP-26::Flag was examined via indirect immunofluorescence on fixed and permeabilized embryos and Western blot on protein samples from populations of mixed-stage embryos. Both of those preparations are subject to caveats that can compromise quantitative measurements. In contrast, sup-26(GFP_null) was examined via direct epifluorescence of living embryos, which is likely more reproducible and more responsive to detection of small differences in protein abundance. Second, SUP-26 is an RBP, which may feed back on its own transcript and thus buffer changes in its abundance. Such a feedback loop might dampen derepression in the SUP-26::Flag context but not in the sup-26(GFP_null) context, where SUP-26 is not expressed.
The repression of sup-26 by mir-35–41 is not absolute, since sup-26(GFP_null) is still expressed when its 3′ UTR contains the wild-type mir-35 family seed match (Fig. 5D,E). Furthermore, mir-35–41 has a strongly dose-dependent effect on sex determination phenotypes (Fig. 3C). Together, these data suggest that the mir-35 family may regulate its target gene network in a highly dose-dependent manner in the endogenous setting. If this is the case, then SUP-26 may contribute to the mir-35–41(nDf50) sex determination phenotypes through relatively small changes in abundance. Supporting this notion, the effect of sup-26 on the synTra phenotype of mir-35–41(nDf50);her-1(gf) is strongly dose-dependent; heterozygosity of sup-26(n1091lf) is sufficient to suppress the phenotype (Fig. 5F). Thus, the magnitude of SUP-26 derepression necessary for the sex determination phenotypes of mir-35–41(nDf50) is on the order of twofold, indicating that the incomplete repression conferred by the mir-35 family seed match is likely biologically relevant.
Our model predicts that ablation of the mir-35 family seed matches in sup-26 and nhl-2 should recapitulate the mutant sex determination phenotypes of mir-35–41(nDf50). Deletion of the mir-35 family seed match in the 3′ UTR of sup-26::flag alleles alone did not cause lethality and had minimal or no effect on sex determination in the her-1(gf) context (Supplemental Fig. S3E,F). This is consistent with our finding that both sup-26 and nhl-2 are required for the effect of mir-35–41(nDf50) on her-1(gf), and so the simultaneous disruption of seed matches in both genes may be necessary to recapitulate the synTra phenotype. Surprisingly, the seed deletion alleles also did not recapitulate the ΨGE phenotype (Fig. 4E; data not shown), indicating that other mir-35 family target genes may contribute to this phenotype. This is plausible, since we systematically screened mir-35 target genes for suppression of the synTra phenotype but not the ΨGE phenotype.
SUP-26 is a promiscuous 3′ UTR-binding protein whose targets include nhl-2
We set out to define the complement of SUP-26-binding targets for multiple reasons. First, previous genetic and biochemical data support a role for SUP-26 in repressing tra-2 translation to regulate sex determination (Manser et al. 2002; Mapes et al. 2010). However, we observed that sup-26 was required for the change in her-1 abundance in mir-35–41 mutant embryos, suggesting that SUP-26 may regulate additional targets upstream of tra-2. Second, feedback by SUP-26 on its own transcript or cross-talk with other mir-35 family targets could confer network architecture with emergent properties for regulating development. Third, any additional targets of SUP-26 binding might also be regulated indirectly by the mir-35 family and contribute to other aspects of the mir-35 family mutant phenotype.
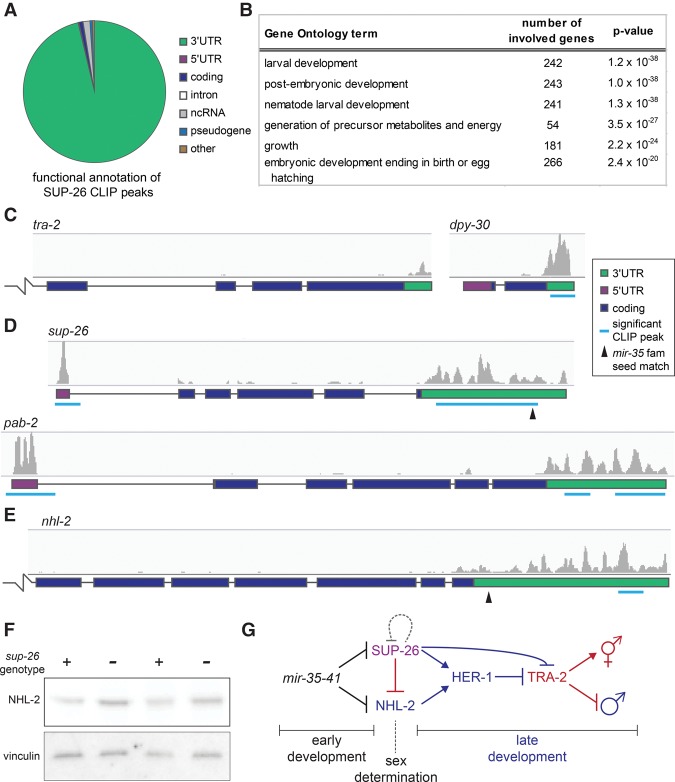
To this end, we confirmed that the Flag tags in SUP-26::Flag did not disrupt the wild-type function of SUP-26 and performed high-throughput sequencing coupled with cross-linking immunoprecipitation (HITS-CLIP) using the SUP-26::Flag strain (Supplemental Fig. S4A,B). Radiolabeling of RNA in CLIP samples showed a strong signal in cross-linked SUP-26::Flag samples but no signal in samples lacking SUP-26::Flag or in noncross-linked SUP-26::Flag samples (Supplemental Fig. S4C). This indicates that the cloned RNA was cross-linked directly to SUP-26::Flag.
SUP-26::Flag HITS-CLIP yielded 857 peaks that were detected in at least three out of four biological replicates (Supplemental Table S3). Analysis of the peak locations revealed that >95% of peaks overlapped with the 3′ UTR of 775 protein-coding genes (Fig. 6A). Thus, SUP-26 is clearly a 3′ UTR-binding protein with a large repertoire of targets. Functional annotation of transcripts bound by SUP-26::Flag revealed that the targets are enriched in genes involved in embryonic and larval development, consistent with the strong expression of SUP-26 during these stages (Fig. 6B). A significant sequence motif was not identified in SUP-26 CLIP peaks, suggesting that the specificity of SUP-26 binding may be largely determined by factors other than primary sequence.
Figure 6.
SUP-26 binds numerous 3′ UTRs, including those of sup-26 and nhl-2. (A) Functional annotation of significant SUP-26::Flag CLIP peaks. (B) Top significantly enriched gene ontology terms among transcripts bound by SUP-26::Flag. (C–E) Example CLIP traces showing SUP-26::Flag binding to the 3′ UTR of transcripts implicated in sex determination and DC (tra-2 and dpy-30) (C), the 5′ UTR and 3′ UTR of sup-26 and the poly(A)-binding protein 2 (pab-2) (D), and the 3′ UTR of nhl-2 (E). The broken line indicates that the gene model continues outside the frame. (F) Western blot showing up-regulation of NHL-2 in sup-26(GFP_null) mutants. Two biological replicates are shown. (G) Simplified working model of the mir-35–41 regulatory network. mir-35–41 likely acts in early development in both XX and XO embryos to repress sup-26 and nhl-2 prior to sex determination. sup-26 and nhl-2 promote male development and may indirectly regulate HER-1 by affecting DCC activity (not shown).
To determine whether SUP-26 binding alters mRNA abundance, we profiled gene expression in sup-26(GFP_null) and wild-type XX embryos (Supplemental Fig. S4D). No gene expression changes were observed in the whole transcriptome of sup-26(GFP_null) embryos (except for sup-26 itself, whose signal decreased due to deletion of the coding sequence). This result supports the model that SUP-26 regulates the translation but not the stability of bound mRNAs and/or may function redundantly with other RBPs.
A CLIP signal corresponding to SUP-26::Flag binding was observed in the 3′ UTR of SUP-26's previously described target, tra-2, although the signal was not identified as a significant peak (Fig. 6C). Another gene that could also contribute to the role of SUP-26 in sex determination, dpy-30, displayed stronger SUP-26::Flag binding (Fig. 6C); dpy-30 encodes a core member of the DCC. Many other genes that are not implicated in sex determination or DC also displayed strong SUP-26::Flag binding in their 3′ UTRs (Supplemental Fig. S4E).
SUP-26 binds to the 3′ UTR of its own transcript, consistent with a feedback loop buffering changes in SUP-26 abundance (Fig. 6D). Interestingly, SUP-26 also binds its own 5′ UTR and that of the poly(A)-binding protein 2 (pab-2) mRNA (Fig. 6D). This broad repertoire of binding to 3′ UTRs (775 genes) but highly specific 5′ UTR binding (two genes) is reminiscent of cytoplasmic poly(A)-binding proteins (pab-1 and pab-2 in C. elegans), which regulate the translation of their own transcripts via 5′ UTR binding (Mangus et al. 2003). While SUP-26 does not appear to bind poly(A) per se, it was shown previously to exist in a complex with PAB-1 (Fig. 5A; Mapes et al. 2010). We propose that SUP-26 may act with PAB-1 and PAB-2 to regulate translation and feed back on transcripts encoding members of this complex to tightly regulate the complex's abundance.
The newly defined targets of SUP-26::Flag binding include nhl-2. A significant CLIP peak was called in the distal region of the nhl-2 3′ UTR, nonoverlapping with the mir-35 family seed match (Fig. 6E). Western blots in mixed-stage embryo samples show that NHL-2 is up-regulated in sup-26(GFP_null) embryos, suggesting that SUP-26 binding of nhl-2 may repress its translation (Fig. 6F). The proportion of SUP-26::Flag target transcripts that contain a mir-35 family seed match is only slightly higher than in the genome at large (5.9% vs. 3.2%). However, the two targets that we identified as biologically relevant in this study are bound by SUP-26::Flag, potentially creating negative feedback on sup-26 and incoherent feed-forward on nhl-2 (Fig. 6G). Thus, upon decay of mir-35–41 during the course of development, SUP-26 derepression may buffer derepression of mir-35 family target genes (including itself and nhl-2) or act to allow only a short temporal pulse of target gene derepression. How the interconnected nature of this network affects its dynamics and robustness during development will be an important avenue for future study.
The mir-35 family seed match in the nhl-2 3′ UTR is essential
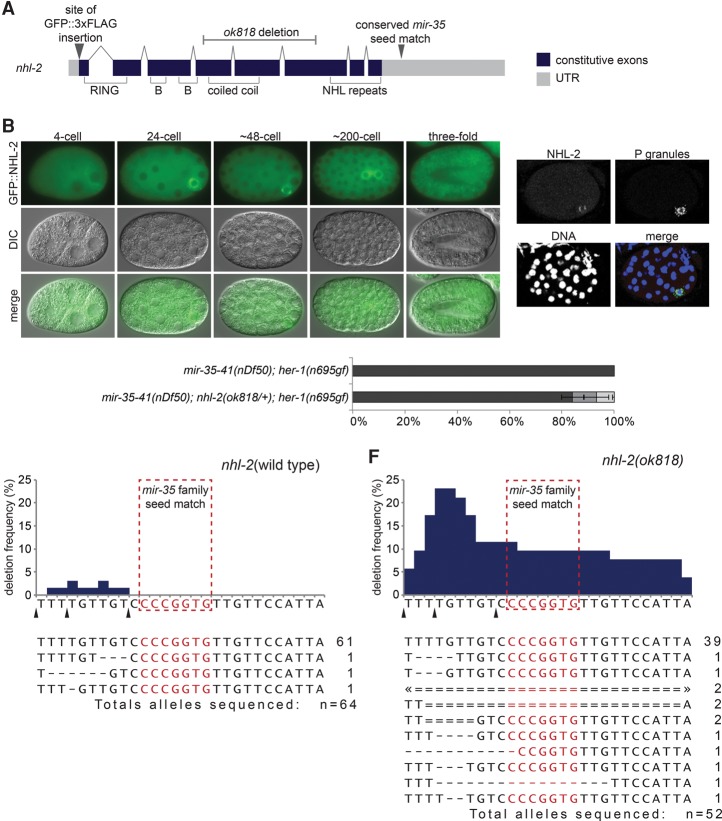
Next, we sought to characterize the nature of nhl-2 repression by the mir-35 family and the contribution of nhl-2 to the mir-35–41(nDf50) sex determination phenotype. To examine nhl-2 expression, we used CRISPR/Cas-9-mediated genome editing to tag NHL-2 at its endogenous N terminus with GFP and three Flag tags (GFP::NHL-2) (Fig. 7A). Consistent with previous reports of NHL-2 expression, GFP::NHL-2 was expressed in the maternal germline and maternally contributed to early embryos (Fig. 7B; Supplemental Fig. S5A; Hyenne et al. 2008). Like SUP-26::Flag, GFP::NHL-2 localizes to P granules by the 24-cell stage (Fig. 7C) and is ubiquitously expressed in later stages of embryonic development, although it remains P-lineage-enriched (Fig. 7B). GFP::NHL-2 P-granule enrichment is post-transcriptionally or post-translationally controlled, since reporters expressing GFP under the control of nhl-2 upstream elements but with a heterologous (let-858) 3′ UTR do not show such a bias (Supplemental Fig. S5B).
Figure 7.
The mir-35 family seed match in nhl-2 cannot be mutated in a wild-type nhl-2 background. (A) Gene model of nhl-2 depicting exon structure, protein domains, mutant alleles, and CRISPR–Cas9 editing sites. (B) Epifluorescence and corresponding DIC images of embryos expressing GFP::NHL-2. (C) Maximum projection of a confocal stack of NHL-2 costaining with a P-granule-specific antibody (K76) in a ∼48-cell stage embryo (when the germline primordium is a single cell [P4]). (D) nhl-2(ok818) heterozygosity weakly suppresses the synTra phenotype. All animals of both genotypes contain one copy of the qC1 balancer chromosome. (E,F) Representation of CRISPR/Cas9 alleles generated when targeting the mir-35 family seed match in the nhl-2 3′ UTR in a nhl-2 wild-type background (E) or nhl-2(null) background (F). (Top) Graphs indicate the frequency of genomic mutation at each nucleotide in and around the mir-35 family seed match in lines selected for visible co-CRISPR editing events. Arrowheads denote potential sites of Cas9 cleavage corresponding to injected single-guide RNAs (sgRNAs). (Bottom) The number at the right of the sequence indicates the number of alleles isolated bearing the indicated mutation. Events marked with double lines and “2” were homozygous. Arrowheads indicate that deletion extends out of frame.
Despite our model that nhl-2 is a mir-35 family target, no changes in GFP::NHL-2 abundance were observed in the mir-35–41(nDf50) mutant background (when examined via epifluorescence or Western blot) (data not shown), similar to SUP-26::Flag. Also similar to sup-26, qPCR of the nhl-2 transcript showed no change in mir-35–41(nDf50) embryos compared with wild type (data not shown). nhl-2 is weakly haploinsufficient for the mir-35–41(nDf50) sex determination phenotype, indicating that, like sup-26 and mir-35–41, nhl-2 acts in a dose-dependent manner (Fig. 7D). Thus, the amplitude of nhl-2 derepression in mir-35–41(nDf50) mutants may be on the order of twofold and thus difficult to detect in mixed-stage embryo samples.
The highly interconnected regulatory network downstream from mir-35–41 might dampen changes in expression of nhl-2 in mir-35–41(nDf50) mutants. For instance, derepression of sup-26 from mir-35 family repression may simultaneously diminish derepression of nhl-2 (Fig. 6G). Therefore, we sought to examine the derepression of nhl-2 in isolation. However, attempts to delete the mir-35 family seed match in the nhl-2 3′ UTR by genome editing were unsuccessful. The rate of genome-editing events recovered was unusually low, and none of the recovered nhl-2 3′ UTR deletions affected the mir-35 family seed match (Fig. 7E). Thirty-two genome-edited lines were selected for co-CRISPR mutations at visible marker loci (see the Supplemental Material). Of these, only three out of 64 sequenced alleles bore mutations, all just 5′ to the mir-35 family seed match (between the cleavage sites of the three injected single-guide RNAs [sgRNAs]).
We hypothesized that the deletion of the mir-35 family seed match might have a dominant lethal effect and hence be counterselected in our genome-editing experiments. If this lethality is due to elevated levels of NHL-2, then it would not occur in an nhl-2(lf) background that retains the genomic sequence corresponding to the nhl-2 3′ UTR. We therefore attempted to delete the mir-35 family seed match in the nhl-2(null) background using the same genome-editing reagents. This context yielded a much higher rate and diversity of recovered genome-editing events, and, importantly, many of the recovered deletions spanned the mir-35 family seed match (Fig. 7F). From 26 edited lines selected for co-CRISPR events, 13 out of 52 alleles bore deletions, and six of these alleles were deletions affecting the mir-35 family seed match. This result demonstrates that loss of mir-35 family targeting of NHL-2-coding transcripts is dominant-lethal, strongly supporting the notion that mir-35 targeting of nhl-2 is required for viability. By highlighting the specific requirement for the nucleotides comprising the seed match, this experiment provides strong evidence that this site mediates a direct miRNA–target interaction. This result suggests that a single miRNA target site can be essential for viability.
Because of the lethality of mutations affecting the mir-35 family seed match in nhl-2, we have been unable as yet to assess the effects of such alleles on the sex determination phenotype of her-1(n695gf) alone or in combination with sup-26 alleles also lacking mir-35 family repression. Nonetheless, the apparent negative selection of mir-35 family seed match deletions in nhl-2 seems to phenocopy another aspect of mir-35 family mutants: lethality. Nhl-2 is unlikely to be the only target gene whose mir-35 family seed match is essential, and similar genome-editing experiments at other loci will provide a new approach toward identifying essential mir-35 family target sites.
Discussion
Here we show that a major function of the maternally contributed mir-35 family is to contribute to the timing and fidelity of sex determination in C. elegans. This is the first demonstration of a role for miRNAs in the C. elegans sex determination pathway. Our data show that the predicted mir-35 family target genes sup-26 and nhl-2 act downstream from the mir-35 family in controlling this process. Taken together, our study of this miRNA family and its targets has delineated multiple new layers of post-transcriptional control (via both miRNAs and RBPs) over the sex determination pathway.
In contrast to most players in the sex determination pathway, the mir-35 family has a strong maternal effect. Most genes in sex determination act zygotically—only after the embryo's X to autosome ratio is assessed—via a process of monitoring zygotic transcription of competing X-linked signal elements (XSEs) and autosomal signal elements (ASEs; around the 40-cell embryo stage) (Meyer 2005). The maternal effect of the mir-35 family suggests that these miRNAs function prior to the onset of sex determination, since the maternal load of miRNA is established in the oocyte. Therefore, the mir-35 family may not function simply to promote hermaphroditic fates in XX zygotes after sex determination has been established. Instead, we propose that expression of mir-35 family miRNAs in the oocyte enables the mir-35 family to function prior to the onset of sex determination to prevent the deleterious premature expression of sex-specific genes. The mir-35 family is proposed to act upstream of its target genes, sup-26 and nhl-2, in this process to prevent premature commitment to male programs. Accordingly, sup-26 and nhl-2 are proposed to become relieved of mir-35 family repression in XO embryos after the onset of sex determination, allowing them to promote the proper expression of male developmental programs (Fig. 6G). This model of mir-35 family function is consistent with the temporal expression pattern of the mir-35 family, whose levels sharply decay in late stage embryos (Stoeckius et al. 2009; Wu et al. 2010). Thus, our model holds that a maternally contributed miRNA family acts as a developmental timer, promoting naïveté in early embryos and preventing the deleterious premature action of target RNAs involved in zygotic developmental decisions. This work therefore expands our understanding of miRNA functions and their contributions to developmental timing and highlights the emergence of the mir-35 family in particular as a model for understanding miRNA biology and mechanism prior to the OET.
The maternal effect of the mir-35 family also implies that these miRNAs could function in both XX and XO embryos, since the maternal load of miRNA is established in the oocyte prior to the time that the zygotic karyotype is established (at fertilization). Mutant mir-35–41(nDf50) XO animals display morphological defects in male-specific organs, which may be a result of the mistiming of sex-specific gene expression (McJunkin and Ambros 2014); that is, while activation of male-specific gene expression per se would not be expected to perturb XO male development, the premature activation of these genes might disrupt the order of developmental events and thus their fidelity. Like XX embryos, XO embryos are affected by mir-35 family mutant lethality (Supplemental Fig. S6). Whether the mistiming of sex-specific gene expression or other developmental decisions is one of the molecular bases of this lethality is yet to be determined.
SUP-26 was shown previously to modulate sex determination via translational control of tra-2. By generating a sup-26(GFP_null) allele, we demonstrated that SUP-26 not only is a modulator of the pathway but is in fact required for efficient male mating. In addition, the set of target genes bound by SUP-26 comprises a repertoire of many hundreds of mRNAs besides tra-2. In particular, SUP-26 binding of dpy-30 could contribute to SUP-26's regulation of her-1 via the DCC. Furthermore, we found that SUP-26 feeds back on its own transcript and on nhl-2 mRNA, conferring network connectivity that may buffer the effects of mir-35 family decay or temporally limit derepressed target gene expression. Future studies should examine how SUP-26 exerts translational control on its many targets and whether its feedback on mir-35 family targets contributes to the proper timing of developmental events in sex determination and other processes.
Our work is the first indication that NHL-2 regulates sex determination. NHL-2 apparently acts upstream of her-1 to promote male development through an unknown mechanism. A striking and unexpected insight arising from this work is that mir-35 family-mediated repression of nhl-2 is required for viability. This is the first identified genetic pathway that contributes to mir-35 family essentiality. The perfect conservation of the mir-35 family seed match in all nematodes in which the nhl-2 3′ UTR has been annotated indicates that this may be an ancient function of the mir-35 family. Previous studies in which nhl-2 was overexpressed from a high-copy transgene array did not report lethality (Hammell et al. 2009), suggesting that the tissue in which overexpression is lethal may be the germline, where high-copy arrays are silenced. Generating an inducible allele in which nhl-2 is derepressed from mir-35 control is a high priority and should allow the study of this axis of the mir-35 family lethality phenotype in isolation. In particular, XX specificity of the lethality would strongly support a role for nhl-2 in antagonizing the DCC rather than simply affecting sex determination downstream from the DCC. NHL domains were shown recently to bind directly to RNA (Loedige et al. 2015); identifying all of the RNA targets of NHL-2 will be critical in understanding its biological function in sex determination. In addition, the dramatic mir-35–41(nDf50);her-1(gf) synTra phenotype described here should enable forward genetic screens to identify downstream targets of NHL-2 and SUP-26 regulation.
The mir-35–41(nDf50) mutant gene expression pattern resembles that of the DCC component sdc-2 that controls DC and sex determination. The majority of the similarities between these two mutants is attributable to expression changes related to perturbed sex determination, not direct effects of DCC(lf). Nonetheless, we observed that the most upstream regulator of sex determination, her-1, is derepressed in the mir-35–41(nDf50) mutant, implying that mir-35–41 acts upstream of her-1 and possibly impacts the only known regulator of her-1 transcription, the DCC itself. However, a primary hallmark of DCC(lf) would be XX-specific lethality due to impaired DC in somatic cells (Meyer 2005), yet the lethality of mir-35 family mutants is not XX-specific (Supplemental Fig. S6). One possibility is that multiple pathways contribute to mir-35 family mutant lethality and that a form of karyotype-neutral lethality is epistatic to an underlying XX-specific lethality. It is also possible that sup-26 and nhl-2 may control the abundance of her-1 in a DCC-independent manner.
The DCC is regulated by the competing zygotic expression of autosomal and X-linked genes, known as ASEs and XSEs, respectively. These signals converge to act on the master regulator XOL-1 (XO-lethal 1), ensuring its low expression in XX embryos and high expression in XO embryos (Meyer 2005). XOL-1 prevents the expression of sdc-2 and thus DCC loading onto its target gene loci in XOs. The exquisite gene dose dependence of mir-35–41, sup-26, and nhl-2 in sex determination is consistent with these elements being involved in a counting mechanism such as the competition between ASEs and XSEs. The mir-35 family bears one characteristic of an XSE in that it may promote DCC function. However, the mir-35 family is not X-linked and has a strong maternal effect, both of which preclude its classification as an XSE. Intriguingly, in all other studied species of the Caenorhabditis clade, the mir-35–41 cluster has undergone duplication events resulting in approximately twice as many copies of mir-35 family miRNAs on X as on autosomes (Caenorhabditis brenneri: 23 on X vs. nine on autosomes; Caenorhabditis remanei: 12 on X vs. five on autosomes; and Caenorhabditis briggsae: 16 on X vs. nine on autosomes). Given the dosage-sensitive nature of this regulatory module, zygotic expression of the mir-35 family members in other Caenorhabditis species may act as a counting mechanism to promote DCC loading or hermaphrodite sex determination specifically in XX embryos.
Like the mir-35 family, small RNAs have been shown to play nonconserved roles in sex determination in Bombyx mori, Drosophila melanogaster, and Zea mays (Chuck et al. 2007; Fagegaltier et al. 2014; Kiuchi et al. 2014). The mechanism of action of small RNAs may suit them particularly well for involvement in sex determination pathways, since sex determination is a rapidly evolving process, and the target specificity of small RNAs is also extremely fluid, likely due to the small genomic space that encodes the interaction interface: the 6- to 7-nt seed (Shi et al. 2013). The mir-35 family appears unique among these known mechanisms of sex determination by small RNAs because of its maternal effect on this zygotic developmental decision.
The mir-35 family's role in developmental timing is analogous to previously characterized post-embryonically expressed miRNAs such as let-7 and lin-4. However, unlike let-7 and lin-4, whose induction drives forward developmental transitions, the decay of the mir-35 family members is likely to relieve repression of sup-26 and nhl-2, allowing for the full onset of sex determination. For this reason, one might expect the timing of the decay of mir-35 family members to be under precise developmental control. The sex determination pathway itself could potentially feed back on mir-35–41—possibly enforcing rapid decay in XO embryos—to permit the expression of male programs. How miRNAs are targeted for decay and turned over in the context of development is very poorly understood, and the newly delineated mir-35 network has the potential to illuminate such a process.
Whether repressed mRNAs can be re-expressed after decay of their cognate miRNA is another open question. In the embryo, miRNAs regulate the poly(A) tail length and translation of their targets (Wu et al. 2010; Bazzini et al. 2012; Subtelny et al. 2014). Since these mechanisms are theoretically reversible, transient miRNA-mediated repression followed by re-expression of the same mRNA molecule has the potential to occur in embryogenesis. This mode of regulation would be useful for storing maternally contributed mRNAs in an inactive state for use later in development. Taken together, our work has an immediate impact on our understanding of the biology of miRNA function and sex determination and also lays the groundwork to address many important mechanistic questions regarding miRNA activity and the regulation of miRNAs.
Materials and methods
C. elegans culture and RNA preparation
C. elegans were cultured on HB101 Escherichia coli by standard procedures at 20°C unless otherwise specified. For 25°C microarray samples and tra-2(ar221);xol-1(y9) and xol-1(y9) samples, plates were upshifted to 25°C 24 h prior to harvest. The samples for microarray analysis were grown on NGM supplemented with chicken egg yolk. Embryos were isolated by hypochlorite treatment. All samples for RNA preparation were made by resuspending the sample pellet in 4 vol of Trizol, snap freezing, and shaking for 15 min at room temperature after thawing, prior to purification according to the Trizol manufacturer's specifications (ThermoFisher). qRT–PCR was performed using Quantifast SYBR RT–PCR kit (Qiagen). See the Supplemental Material for primer sequences, list of strains used, and details on vectors used for CRISPR.
Microarrays and HITS-CLIP
Biotinylated antisense RNA was prepared using the Single-Round RNA Amplification and Biotin Labeling system (Enzo Life Sciences) and hybridized to Affymetrix 3′ IVT C. elegans microarrays. Expression data for sdc-2(lf) was acquired from NCBI Gene Expression Omnibus (GEO) series GSE14649 (Jans et al. 2009). All data were analyzed using Genespring GX software, excluding probes whose values were in the bottom quintile in every sample. CLIP was performed as in Zisoulis et al. (2012) with modifications as detailed in the Supplemental Material. All microarray and HITS-CLIP data are accessible at NCBI GEO, accession number GSE94704.
Immunofluorescence and immunoblot
Embryos were permeabilized by “freeze-cracking” (Duerr 2013) and fixed for 2 min in methanol at −20°C and then 30 min at 25°C in 4% paraformaldehyde in PBS for staining with 2 µg/mL anti-Flag antibody (M2; Sigma Aldrich) or for 10 min in methanol and then 10 min in acetone, both at −20°C, for staining with 0.2 µg/mL anti-NHL-2 polyclonal. Antibody staining was overnight at 4°C in 3% BSA with 0.1% Tween 20 and 0.1% Triton X-100 in PBS. The P-granule antibodies used were anti-PGL-1 diluted 1:500 (gift from C. Mello), and the K76 monoclonal diluted 1:100 (Strome and Wood 1983). Germlines were dissected and stained according to the Schedl laboratory protocols (http://genetics.wustl.edu/tslab/protocols).
Supplementary Material
Acknowledgments
We thank the Walker laboratory for sharing their equipment, members of the Mello laboratory for helpful discussion and the anti-PGL-1 antibody, and C. Hammell for anti-NHL-2. The K76 P-granule-specific antibody developed by S. Strome and W.B. Wood was obtained from the Developmental Studies Hybridoma Bank, created by the National Institute of Child Health and Human Development of the National Institutes of Health, and maintained at The Department of Biology at the University of Iowa. Orkan Ilbay designed and cloned some of the vector variants used for genome editing. We thank J. Calarco for sharing modified CLIP protocols, B. Meyer and R. Ketting for discussing unpublished data, and A. Sitaram for critical reading of the manuscript. Some strains were provided by the Caenorhabditis Genetics Center, which is funded by National Institutes of Health Office of Research Infrastructure Programs (P40 OD010440). This work was funded by F32 GM097895, the Charles King Trust Post-doctoral Research Fellowship, K99 GM113063-01 (to K.M.), and R01 GM34028 (to V.A.). K.M. conducted the experiments. K.M. and V.A. designed experiments, interpreted results, and wrote the paper.
Footnotes
Supplemental material is available for this article.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.290155.116.
References
- Alvarez-Saavedra E, Horvitz HR. 2010. Many families of C. elegans microRNAs are not essential for development or viability. Curr Biol 20: 367–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bazzini AA, Lee MT, Giraldez AJ. 2012. Ribosome profiling shows that miR-430 reduces translation before causing mRNA decay in zebrafish. Science 336: 233–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang HR, Schoenfeld LW, Ruby JG, Auyeung VC, Spies N, Baek D, Johnston WK, Russ C, Luo S, Babiarz JE, et al. 2010. Mammalian microRNAs: experimental evaluation of novel and previously annotated genes. Genes Dev 24: 992–1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuck G, Meeley R, Irish E, Sakai H, Hake S. 2007. The maize tasselseed4 microRNA controls sex determination and meristem cell fate by targeting Tasselseed6/indeterminate spikelet1. Nat Genet 39: 1517–1521. [DOI] [PubMed] [Google Scholar]
- Duerr JS. 2013. Antibody staining in C. elegans using ‘freeze-cracking’. J Vis Exp 10.3791/50664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fagegaltier D, König A, Gordon A, Lai EC, Gingeras TR, Hannon GJ, Shcherbata HR. 2014. A genome-wide survey of sexually dimorphic expression of Drosophila miRNAs identifies the steroid hormone-induced miRNA let-7 as a regulator of sexual identity. Genetics 198: 647–668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia DM, Baek D, Shin C, Bell GW, Grimson A, Bartel DP. 2011. Weak seed-pairing stability and high target-site abundance decrease the proficiency of lsy-6 and other microRNAs. Nat Struct Mol Biol 18: 1139–1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giraldez AJ. 2010. microRNAs, the cell's nepenthe: clearing the past during the maternal-to-zygotic transition and cellular reprogramming. Curr Opin Genet Dev 20: 369–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greve TS, Judson RL, Blelloch R. 2013. microRNA control of mouse and human pluripotent stem cell behavior. Annu Rev Cell Dev Biol 29: 213–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha M, Kim VN. 2014. Regulation of microRNA biogenesis. Nat Rev Mol Cell Biol 15: 509–524. [DOI] [PubMed] [Google Scholar]
- Hammell M, Long D, Zhang L, Lee A, Carmack CS, Han M, Ding Y, Ambros V. 2008. mirWIP: microRNA target prediction based on microRNA-containing ribonucleoprotein-enriched transcripts. Nat Methods 5: 813–819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammell CM, Lubin I, Boag PR, Blackwell TK, Ambros V. 2009. nhl-2 modulates microRNA activity in Caenorhabditis elegans. Cell 136: 926–938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hargitai B, Kutnyánszky V, Blauwkamp TA, Steták A, Csankovszki G, Takács-Vellai K, Vellai T. 2009. xol-1, the master sex-switch gene in C. elegans, is a transcriptional target of the terminal sex-determining factor TRA-1. Development 136: 3881–3887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyenne V, Desrosiers M, Labbé J-C. 2008. C. elegans Brat homologs regulate PAR protein-dependent polarity and asymmetric cell division. Dev Biol 321: 368–378. [DOI] [PubMed] [Google Scholar]
- Jan CH, Friedman RC, Ruby JG, Bartel DP. 2011. Formation, regulation and evolution of Caenorhabditis elegans 3′UTRs. Nature 469: 97–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jans J, Gladden JM, Ralston EJ, Pickle CS, Michel AH, Pferdehirt RR, Eisen MB, Meyer BJ. 2009. A condensin-like dosage compensation complex acts at a distance to control expression throughout the genome. Genes Dev 23: 602–618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagias K, Pocock R. 2015. microRNA regulation of the embryonic hypoxic response in Caenorhabditis elegans. Sci Rep 5: 11284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ketting RF. 2011. microRNA biogenesis and function: an overview. Adv Exp Med Biol 700: 1–14. [DOI] [PubMed] [Google Scholar]
- Kiuchi T, Koga H, Kawamoto M, Shoji K, Sakai H, Arai Y, Ishihara G, Kawaoka S, Sugano S, Shimada T, et al. 2014. A single female-specific piRNA is the primary determiner of sex in the silkworm. Nature 509: 633–636. [DOI] [PubMed] [Google Scholar]
- Liu M, Liu P, Zhang L, Cai Q, Gao G, Zhang W, Zhu Z, Liu D, Fan Q. 2011. mir-35 is involved in intestine cell G1/S transition and germ cell proliferation in C. elegans. Cell Res 21: 1605–1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loedige I, Jakob L, Treiber T, Ray D, Stotz M, Treiber N, Hennig J, Cook KB, Morris Q, Hughes TR, et al. 2015. The crystal structure of the NHL domain in complex with RNA reveals the molecular basis of Drosophila brain-tumor-mediated gene regulation. Cell Rep 13: 1206–1220. [DOI] [PubMed] [Google Scholar]
- Mangone M, Manoharan AP, Thierry-Mieg D, Thierry-Mieg J, Han T, Mackowiak SD, Mis E, Zegar C, Gutwein MR, Khivansara V, et al. 2010. The landscape of C. elegans 3′UTRs. Science 329: 432–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangus DA, Evans MC, Jacobson A. 2003. Poly(A)-binding proteins: multifunctional scaffolds for the post-transcriptional control of gene expression. Genome Biol 4: 223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manser J, Wood WB, Perry MD. 2002. Extragenic suppressors of a dominant masculinizing her-1 mutation in C. elegans identify two new genes that affect sex determination in different ways. Genesis 34: 184–195. [DOI] [PubMed] [Google Scholar]
- Mapes J, Chen J-T, Yu J-S, Xue D. 2010. Somatic sex determination in Caenorhabditis elegans is modulated by SUP-26 repression of tra-2 translation. Proc Natl Acad Sci 107: 18022–18027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massirer KB, Perez SG, Mondol V, Pasquinelli AE. 2012. The miR-35–41 family of microRNAs regulates RNAi sensitivity in Caenorhabditis elegans. PLoS Genet 8: e1002536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McJunkin K, Ambros V. 2014. The embryonic mir-35 family of microRNAs promotes multiple aspects of fecundity in Caenorhabditis elegans. G3 (Bethesda) 4: 1747–1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer BJ. 2005. X-chromosome dosage compensation. WormBook (ed. The C. elegans Research Community), WormBook, 10.1895/wormbook.1.8.1, http://www.wormbook.org. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumüller RA, Betschinger J, Fischer A, Bushati N, Poernbacher I, Mechtler K, Cohen SM, Knoblich JA. 2008. Mei-P26 regulates microRNAs and cell growth in the Drosophila ovarian stem cell lineage. Nature 454: 241–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parchem RJ, Moore N, Fish JL, Parchem JG, Braga TT, Shenoy A, Oldham MC, Rubenstein JLR, Schneider RA, Blelloch R. 2015. miR-302 is required for timing of neural differentiation, neural tube closure, and embryonic viability. Cell Rep 12: 760–773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinke V, Gil IS, Ward S, Kazmer K. 2004. Genome-wide germline-enriched and sex-biased expression profiles in Caenorhabditis elegans. Development 131: 311–323. [DOI] [PubMed] [Google Scholar]
- Shi Z, Montgomery TA, Qi Y, Ruvkun G. 2013. High-throughput sequencing reveals extraordinary fluidity of miRNA, piRNA, and siRNA pathways in nematodes. Genome Res 23: 497–508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoeckius M, Maaskola J, Colombo T, Rahn H-P, Friedländer MR, Li N, Chen W, Piano F, Rajewsky N. 2009. Large-scale sorting of C. elegans embryos reveals the dynamics of small RNA expression. Nat Methods 6: 745–751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strome S, Wood WB. 1983. Generation of asymmetry and segregation of germ-line granules in early C. elegans embryos. Cell 35: 15–25. [DOI] [PubMed] [Google Scholar]
- Subtelny AO, Eichhorn SW, Chen GR, Sive H, Bartel DP. 2014. Poly(A)-tail profiling reveals an embryonic switch in translational control. Nature 508: 66–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thoemke K, Yi W, Ross JM, Kim S, Reinke V, Zarkower D. 2005. Genome-wide analysis of sex-enriched gene expression during C. elegans larval development. Dev Biol 284: 500–508. [DOI] [PubMed] [Google Scholar]
- Ventura A, Young AG, Winslow MM, Lintault L, Meissner A, Erkeland SJ, Newman J, Bronson RT, Crowley D, Stone JR, et al. 2008. Targeted deletion reveals essential and overlapping functions of the miR-17∼92 family of miRNA clusters. Cell 132: 875–886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu E, Thivierge C, Flamand M, Mathonnet G, Vashisht AA, Wohlschlegel J, Fabian MR, Sonenberg N, Duchaine TF. 2010. Pervasive and cooperative deadenylation of 3′UTRs by embryonic microRNA families. Mol Cell 40: 558–570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zisoulis DG, Kai ZS, Chang RK, Pasquinelli AE. 2012. Autoregulation of microRNA biogenesis by let-7 and Argonaute. Nature 486: 541–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.