Abstract
The components of chloroplast translation are similar to those of prokaryotic translation but contain some additional unique features. Proteomic analysis of the Chlamydomonas reinhardtii chloroplast ribosome identified an S1-like protein, plastid-specific ribosomal protein-7 (PSRP-7), as a stoichiometric component of the 30S subunit. Here, we report that PSRP-7 is part of a polyprotein that contains PSRP-7 on its amino end and two translation elongation factor Ts (EF-Ts) domains at the carboxy end. We named this polyprotein PETs (for polyprotein of EF-Ts). Pets is a single-copy gene containing the only chloroplast PSRP-7 and EF-Ts sequences found in the C. reinhardtii genome. The pets precursor transcript undergoes alternative splicing to generate three mRNAs with open reading frames (ORFs) of 1.68, 1.8, and 3 kb. A 110-kD pro-protein is translated from the 3-kb ORF, and the majority of this protein is likely posttranslationally processed into the 65-kD protein PSRP-7 and a 55-kD EF-Ts. PETs homologs are found in Arabidopsis thaliana and rice (Oryza sativa). The conservation of the 110-kD PETs polyprotein in the plant kingdom suggests that PSRP-7 and EF-Ts function together in some aspects of chloroplast translation and that the PETs pro-protein may have a novel function as a whole.
INTRODUCTION
In the chloroplast, where proteins of the photosynthetic apparatus and the carbon-fixation enzymes are synthesized, gene expression is primarily controlled posttranscriptionally by mechanisms affecting mRNA splicing, RNA stability, and translation (Mayfield et al., 1995; Barkan and Goldschmidt-Clermont, 2000). Chloroplast translation has long been thought to be similar to translation in prokaryotic organisms, based on similarities in rRNAs and on the sensitivity of chloroplast ribosomes to bacterial antibiotics (reviewed in Sugiura et al., 1998). These similarities support the endosymbiotic theory, which states that organelles, including chloroplasts and mitochondria, originated from a photosynthetic prokaryote that was integrated into a eukaryotic host (Bhattacharya et al., 2004). It is now recognized that chloroplast translation is unique and more complex than in prokaryotic systems (Harris et al., 1994; Beligni et al., 2004). Many studies have identified nuclear factors that affect translation of specific chloroplast mRNAs, and the majority of these translational factors have no obvious orthologs in prokaryotic organisms. Fewer studies have focused on the basic machinery involved in chloroplast translation, including ribosomes and translation factors, mainly because it was assumed that most of these components would be conserved between bacteria and chloroplasts. Proteomic and biochemical analysis of chloroplast ribosomes identified several plastid-specific ribosomal proteins (PSRPs), in addition to many bacterial orthologs (Yamaguchi et al., 2002, 2003; Yamaguchi and Subramanian, 2003). It has been proposed that PSRPs may take part in unique aspects of chloroplast translation, such as regulation by light. To date, however, there is little experimental data to support this idea (Yamaguchi and Subramanian, 2003). Thus, the identification and characterization of PSRPs should help define the mechanisms of chloroplast translation and, hence, the understanding of photosystem biogenesis, plastid differentiation, and ultimately plant development and function.
Among the proteins found on the small subunit of the Chlamydomonas reinhardtii chloroplast ribosome, we identified a novel protein that contains two S1 domains, which was named PSRP-7 (Yamaguchi et al., 2002). In bacteria, binding of the 30S subunit near the initiation codon of mRNAs is facilitated by a ribosome-mRNA interaction promoted by a Shine-Dalgarno sequence and the ribosomal protein S1 (Subramanian, 1983; Jacques and Dreyfus, 1990). The Escherichia coli S1 protein has six S1 domains (Subramanian, 1983; Gribskov, 1992). In C. reinhardtii chloroplasts, the homolog of bacterial S1 is a nuclear-encoded 49-kD protein with only three S1 domains (Yamaguchi et al., 2002; Merendino et al., 2003). Because the S1 homolog is truncated in C. reinhardtii and PSRP-7 contains two S1 domains, we proposed that PSRP-7 could supplement the function of S1 in mRNA recognition and factor-mediated translation initiation in the chloroplast (Yamaguchi et al., 2002).
Here, we report that C. reinhardtii PSRP-7 is part of a novel polyprotein that contains not only PSRP-7, but also two translation elongation factor Ts (EF-Ts) domains. We named this protein PETs (for polyprotein of EF-Ts). The EF-Ts domains are near perfect homologs of Synechocystis EF-Ts, and no other chloroplast EF-Ts genes have been identified in the C. reinhardtii genome. The role of EF-Ts in translation elongation in bacteria has been widely studied (Nyborg and Kjeldgaard, 1996; Gromadski et al., 2002; Ramakrishnan, 2002). Briefly, elongation factor Tu (EF-Tu)-GTP forms a complex with aminoacyl-tRNA, brings it rapidly to the A site of the ribosome, and subsequently dissociates from the ribosome in an inactive GDP form. EF-Ts functions as a nucleotide-exchange factor by binding EF-Tu and accelerating GDP dissociation from EF-Tu.
In this work, we demonstrate that the pets precursor transcript undergoes alternative splicing to generate three mRNAs coding for proteins of 110 (PETs precursor), 79, and 65 kD (PSRP-7). The 110-kD protein is likely cleaved to form the 65-kD mature PSRP-7 and the 55-kD EF-Ts polypeptide. PETs has orthologs in Arabidopsis thaliana and rice (Oryza sativa), implying functional significance in chloroplast translation. Together, our findings suggest that the novel structure of the pets gene, its alternative splicing, and the posttranslational cleavage of the precursor PETs protein drive the coordinated expression of the chloroplast ribosomal S1-like protein and EF-Ts. We also suggest that the 110-kD PETs pro-protein may play a novel role on its own.
RESULTS
PETs Is a Novel Chloroplast Polyprotein That Contains PSRP-7 and EF-Ts
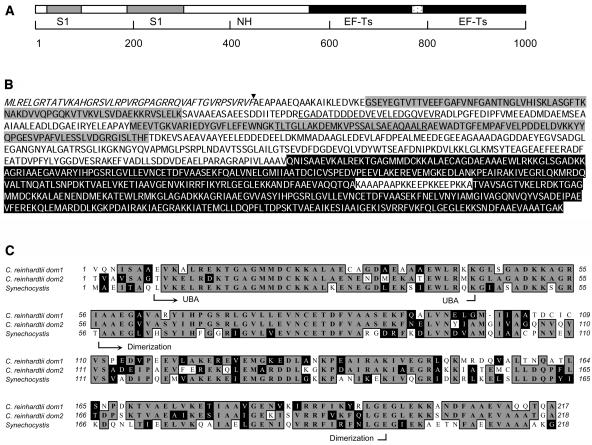
PSRP-7 was first identified as a C. reinhardtii chloroplast ribosomal protein of ∼66 kD that contained two S1 domains (Yamaguchi et al., 2002, 2003). With the purpose of characterizing the psrp-7 gene, we obtained the longest EST clone encoding PSRP-7 from the Kazusa DNA Research Institute (Asamizu et al., 2000) and completely sequenced it. Surprisingly, instead of the expected ∼1.7-kb open reading frame (ORF), the full-length cDNA contained a 3-kb ORF that was predicted to encode a 110-kD protein (Figure 1A). The encoded protein, diagrammed in Figure 1A, contained the S1 domains of PSRP-7 at the N-terminal end as well as two tandem repeats homologous to translation EF-Ts at the carboxy end. We named this novel protein PETs. Figure 1B shows the sequence of the full-length PETs protein; the amino acids shaded in gray indicate the two S1 domains and the amino acids shaded in black correspond to the two EF-Ts domains. The 226-residue middle region of PETs showed no homology to other proteins in the National Center for Biotechnology Information databanks. We have referred to this portion as the PETs nonhomologous region (NH in Figure 1A). According to PETs protein sequence, we predicted that the C terminus of the NH region was the C terminus of PSRP-7.
Figure 1.
C. reinhardtii PETs Precursor Protein Contains PSRP-7 and EF-Ts.
(A) Diagram of the PETs precursor protein. The two S1 domains (gray boxes), the NH region (white box), and the two EF-Ts domains (black boxes) are indicated. The scale in number of amino acids is shown underneath.
(B) C. reinhardtii PETs protein sequence. The domain colors are as in (A). The chloroplast transit peptide cleavage site is indicated with an arrowhead. The peptides previously identified from PSRP-7 by proteomic analysis are underlined.
(C) Alignment between EF-Ts domains 1 and 2 of C. reinhardtii PETs with Synechocystis EF-Ts. Identical amino acids are shaded in gray and similar amino acids in black. The UBA and dimerization domains are shown. Amino acid numbers are indicated on both sides.
The N-terminal sequence of the mature PSRP-7 was determined by automated Edman degradation, and an unambiguous sequence of Ala-Glu-Ala-Pro-Ala- was obtained, establishing that the transit peptide cleavage occurs between residues 43 and 44 of the cytoplasmic precursor, as indicated by the arrowhead in Figure 1B.
The first S1 domain of PETs has 55.7% identity with the S1 domain of TEX, a member of a family of transcriptional accessory proteins from Bordetella pertusis (Fuchs et al., 1996). The second S1 domain has only 20% identity to other S1 domains (Yamaguchi et al., 2002). Comparison of the EF-Ts domains of PETs with bacterial EF-Ts proteins reveals high sequence conservation. EF-Ts domain 1 of PETs has 54% identity and 81.2% similarity to Synechocystis EF-Ts (Figure 1C), whereas domain 2 has 59% identity and 83.5% similarity. The two domains have a 60% identity and 74% similarity to each other. EF-Ts monomers are composed of two domains: a 50–amino acid N-terminal domain that participates in GDP exchange called the ubiquitin-associated (UBA) domain (Kawashima et al., 1996) and a 146–amino acid carboxy domain that participates in the dimerization of EF-Ts (Jiang et al., 1996). As shown in Figure 1C, both features can be identified in each of the PETs EF-Ts domains.
The pets Gene Is Single Copy and Generates at Least Two Detectable mRNAs in C. reinhardtii
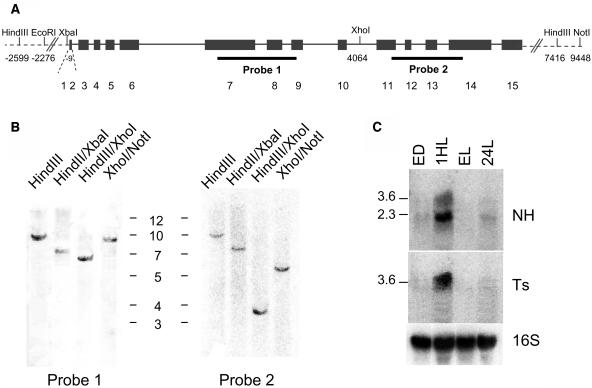
In addition to the EST sequence, database searches of the C. reinhardtii genome (http://genome.jgi-psf.org/chlre2/chlre2.home.html) identified pets as a single-copy gene containing the only PSRP-7 and EF-Ts coding regions. To confirm this, we performed DNA gel blot analyses using probes from the PSRP-7 or the EF-Ts regions. Figure 2A is a map of the C. reinhardtii pets gene, showing the restriction enzymes used for DNA gel blot analysis as well as their position relative to the translation start codon, considered as +1. Figure 2A also shows the hybridization sites for probes 1 (PSRP-7 region) and 2 (EF-Ts region). Figure 2B shows that pets is indeed a single-copy gene in C. reinhardtii and that the fragments obtained with the different restriction enzyme digests and probes 1 and 2 are consistent with the S1 and EF-Ts domains being physically linked in the same fragment of genomic DNA.
Figure 2.
More Than One mRNA Is Generated from the Single-Copy C. reinhardtii pets Gene.
(A) Map of C. reinhardtii pets gene. Exons (black boxes) are numbered underneath; introns are shown as black lines. Restriction enzymes (top), their positions on the gene (bottom), and the probes (black lines, bottom) used for DNA gel blots are indicated.
(B) DNA gel blot of C. reinhardtii pets gene. Ten micrograms of genomic DNA were digested with the restriction enzymes indicated at the top; membranes were hybridized with the probes indicated on (A). The sizes of the 1-kb ladder are indicated.
(C) Two pets mRNAs are detected by RNA gel blot analysis. Twenty micrograms of C. reinhardtii total RNAs from cells grown at 12 h light/12 h dark and harvested at the end of the dark (ED), 1 h upon illumination (1HL), and at the end of the light period (EL), or grown in 24-h light (24HL) were loaded in each lane. Membranes were hybridized with Ts, NH, and 16S rDNA probes. The sizes of the bands in kilobases are indicated on the left.
Because the 65-kD PSRP-7 protein is contained within the 110-kD ORF of the pets gene, we hypothesized that the mature protein must be generated by either posttranscriptional and/or posttranslational events. As an initial approach to determine which of these scenarios was involved, we examined pets mRNA accumulation. Figure 2C shows an RNA gel blot of C. reinhardtii total RNA from cells grown at 24-h light or under a 12-h-light/12-h-dark cycle and harvested at the end of the dark, 1 h upon illumination, and at the end of the light period. A probe corresponding to the nonhomologous region of pets (NH) hybridizes with two mRNAs of ∼3.6 and 2.6 kb. The same band was obtained when a probe against the S1 domains was used (data not shown). By contrast, using a probe that consisted of the EF-Ts domains (Ts), a single 3.6-kb mRNA was identified. The sizes of these mRNAs suggest that the small transcript could code for the mature PSRP-7 protein, whereas the longer one corresponds to the 110-kD protein. The RNA gel blot analyses suggest that the 65-kD PSRP-7 protein could be produced after alternative splicing of the pets precursor transcript but that the EF-Ts domains must arise from the longer mature transcript.
C. reinhardtii PETs Pro-Protein Is Likely Posttranslationally Processed to Form Two Mature Proteins
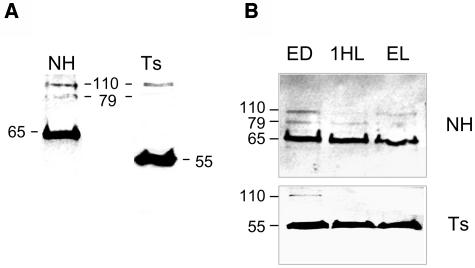
To elucidate which proteins are generated from the pets ORF, we used antisera raised against the NH region as well as a monoclonal antibody against Streptococcus EF-Ts (Ts) in protein gel blot analyses of C. reinhardtii total soluble proteins. Figure 3A shows that the anti-NH antisera recognized a major 65-kD protein and two less intense polypeptides of 110 and ∼79 kD. The anti-Ts antibody recognized a major ∼55-kD band, the expected size of a protein containing only the two EF-Ts domains as well as a less intense band at 110 kD (Figure 3A). PETs protein accumulation was also tested in cells grown under 12 h light/12 h dark. Figure 3B shows that 65-kD protein (PSRP-7, assayed with the anti-NH antibody) was always the major form. With the anti-Ts antibody, the 110-kD band was visible only at the end of the dark, and the 79-kD band was not seen in the conditions examined (Figure 3B).
Figure 3.
Four Proteins Are Generated from the pets Gene.
C. reinhardtii total soluble proteins were separated by 12% SDS-PAGE. Anti-NH or anti-Ts antibodies were used. The protein sizes in kilodaltons are shown.
(A) Protein gel blot of 20 μg of total soluble proteins from cells grown under 24-h light (24HL).
(B) Cells were grown under 12 h light/12 h dark; samples were harvested at the end of the dark (ED), 1 h upon illumination (1HL), and at the end of the light period (EL). Ten micrograms of protein were loaded in each lane.
The protein gel blot analyses suggest that PETs is translated as a 110-kD PSRP7-EF-Ts fusion pro-protein that exists as a stable entity but that a majority of the polyprotein is likely processed to yield the 65-kD PSRP-7 and the 55-kD EF-Ts polypeptide composed of two EF-Ts domains. These results also show that the 79-kD species contains at least parts of the NH region but does not contain enough Ts epitopes to be recognized by the anti-Ts antibody.
Mapping of the C. reinhardtii pets Splicing Sites Indicates That Three mRNAs Are Produced
By RNA gel blot analysis we detected two mRNAs (Figure 2C): the larger 3.6-kb mRNA, proposed to give rise to the 110-kD pro-protein, and a smaller 2.6-kb mRNA that could give rise to either the 65- or the 79-kD protein or to both. To determine precisely which mRNAs are generated from the pets gene and to clarify the origin of the 65- and 79-kD proteins, we performed RT-PCR on C. reinhardtii total RNA. For PCR, we used a forward oligo at the initiator AUG and oligo(dT) as reverse oligo. We amplified two mRNAs that were fully sequenced: (1) one with a 3-kb coding region whose sequence matched the EST previously sequenced and (2) another with a 1.68-kb coding region whose sequence revealed that intron 9 and 10 failed to be spliced. This last transcript reaches a stop codon six nucleotides downstream of the exon 9–intron 9 unspliced junction and is predicted to yield a 560–amino acid protein of ∼61 kD, a close approximate to the 65-kD mature PSRP-7. The sequence of the 3′ untranslated region (UTR) consisted in the rest of intron 9, exon 10, and 242 nucleotides of intron 10, until a 19-nucleotide poly-A tail was reached. This would create an ∼2.7 mRNA, consistent with the size of the smaller band seen in RNA gel blots. These results also confirm what we expected about the C-terminal end of PSRP-7, which is therefore located at the end of the NH region, upstream of where the EF-Ts domains start.
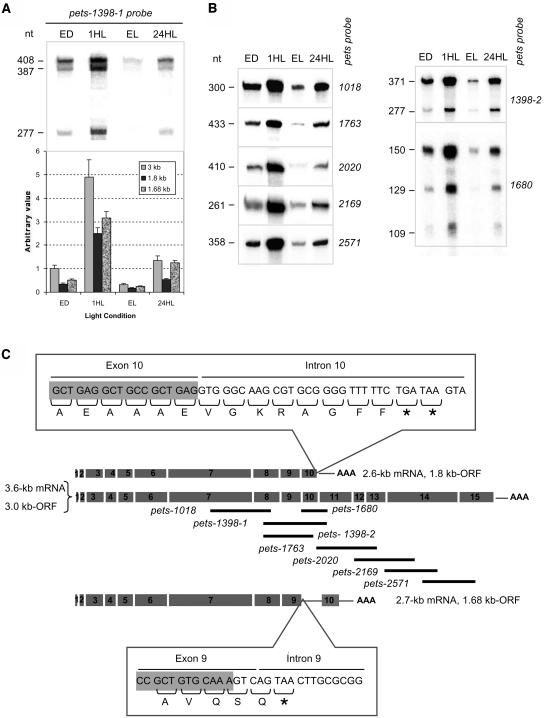
We also performed RNA protection assays with C. reinhardtii total RNA isolated from cells grown under a 12-h-light/12-h-dark regime and harvested at the end of the dark period, 1 h after shifting the cells to the light, and at the end of the light period, or from cells grown under continuous light. Figures 4A and 4B show the results of the RNase protection assays performed using several anti-sense RNA probes, which are diagrammed in Figure 4C as black lines under the scheme of the pets full-length cDNA. When using the pets-1398-1 probe, three major bands of 408, 387, and 277 nucleotides were obtained (Figure 4A). When overexposed, an extra 109-nucleotide band was observed (data not shown). The first band corresponds to the full-length protected probe. The 277-nucleotide and the minor 109-nucleotide bands are consistent with the ∼2.7-kb mRNA (1.68-kb ORF) amplified by RT-PCR. However, the 387-nucleotide band must arise from a different pets mRNA form in which exon 11 is not present downstream of exon 10. Other probes spanning other exon–intron junctions were used to identify whether other splice variants were generated (Figure 4B, left panel, see Figure 4C for probe detail). Only a single band of the size of the full-length probe was obtained in all cases. Taking the RNA gel blot, RNase protection, and RT-PCR assays into account, we proposed that three mRNA forms are produced from the pets gene: a 3.6-kb mRNA with a 3-kb ORF, a 2.7-kb mRNA with a 1.68-kb ORF, and a 2.6-kb mRNA with a 1.8-kb ORF. This last mRNA was not amplified by RT-PCR but is predicted to have the polyadenylation site in intron 10 mentioned above, which would make a 2.6-kb mRNA. The top inset in Figure 4C shows the 3′ sequence of the ORF of this mRNA, showing that a stop codon is reached 27 nucleotides downstream of the exon–intron junction, creating a 1.8-kb ORF. This mRNA would translate a protein with a predicted size of 72 kD, which most likely corresponds to the 79-kD protein detected by protein gel blot analysis (Figure 3). The bottom inset in Figure 4C shows the 3′ sequence of the 1.68-kb ORF, which would generate PSRP-7.
Figure 4.
Mapping of the Three mRNAs Produced from the C. reinhardtii pets Gene and Their Differential Accumulation during a Light/Dark Cycle.
(A) RNase protection assay using C. reinhardtii total RNA from wild-type cells grown under continuous light (24HL) or at 12 h light/12 h dark and harvested at the end of the dark period (ED), 1 h after shifting cells to light (1HL), and at the end of the light period (EL). Pets-1398-1 probe was used; the sizes of the protected bands in number of nucleotides (nt) are shown on the left. The intensity of each band (top) was quantified, corrected according to their size and normalized to the corresponding band from the end of the dark condition (bottom). Bars are the mean values of three independent experiments; error bars indicate standard deviations.
(B) RNase protection assays using the probes indicated on the right; the sizes of the protected bands (nucleotides [nt]) are shown on the left.
(C) Scheme of the three pets-derived mRNAs: exons are shown as numbered gray boxes, 3′ UTRs and poly(A) tails are also schematized. The antisense RNA probes used are indicated as black lines. The top and bottom insets show the 3′ end of the 1.8- and 1.68-kb ORFs, respectively. The corresponding amino acids are shown. The asterisks indicate the stop codons.
Two additional probes were used to confirm the presence of the 1.8- and 1.68-kb ORFs. One of them, pets-1398-2, generated two protected bands of 277 (Figure 4B) and 83 nucleotides (data not shown, only seen when markedly overexposed) corresponding to the 1.68-kb ORF and a 371-nucleotide band corresponding to both the 1.8-kb and the full-length ORF (Figure 4B). The other probe, pets-1680, gave a 129-nucleotide band from the 1.8-kb ORF, a 150-nucleotide band for the full-length ORF, and a 109-nucleotide band from the 1.68-kb ORF as expected (Figure 4B).
Taken together, the RNA and protein analyses of the pets gene indicate that the 110-, 79-, and 65-kD proteins may be explained by translation of the 3-, 1.8-, and 1.68-kb ORFs, respectively, and that the 55-kD EF-Ts polypeptide can only be generated from posttranslational processing of the 110-kD PETs precursor.
Light-Regulated Expression of PETs
To assess the effect of the transition from dark to light on pets mRNA accumulation, we quantified the intensity of the three protected bands obtained by RNase protection using the pets-1398 probe. Figure 4A shows that the accumulation of all three mRNAs greatly increases after the shift to the light and decreases thereafter. The relative amount of each band indicates that the mRNA with the 3-kb ORF is the major product under all growth conditions, whereas the mRNAs with the 1.8- and the 1.68-kb ORFs accumulate to between 30 and 90% of the 3-kb mRNA. That is in agreement with the RNA gel blot analysis (Figure 2B), considering that the two smaller 2.6- and 2.7-kb mRNAs are likely unresolved within the ∼2.6-kb band in the RNA gel blot analysis. As shown in Figure 3B, the 65- and 55-kD proteins are the major products under all light conditions tested and show no significant variation between conditions. These data indicate that, despite the variations in mRNA accumulation, the levels of the 65- and 55-kD proteins remains stable, perhaps because of unequal protein turnover under different light conditions or because of differential translation.
In Vitro Activities of the S1 and EF-Ts Domains of PETs
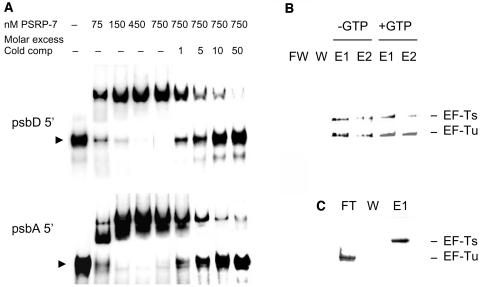
The identification of S1 domains in PSRP-7 suggests that this protein might function during translation, similar to other S1 proteins, as an RNA binding protein. Gel shift assays were used to identify whether the PSRP-7 region of PETs is capable of binding to the 5′ UTRs of C. reinhardtii chloroplast mRNAs. Figure 5A shows that, indeed, recombinant PSRP-7 binds radioactively labeled psbD (top panel) and psbA (bottom panel) 5′ UTRs at concentrations as low as 75 nM. Binding to these RNAs was saturated at protein concentrations between 450 and 750 nM. Moreover, the binding of 750 nM protein to 100 pM of radiolabeled UTRs was competed by a 10- to 50-fold molar excess of the same RNA used as cold competitor (Figure 5A).
Figure 5.
In Vitro Activities of the PSRP-7 and EF-Ts Domains of PETs.
(A) One hundred picomole [α-32P]UTP-labeled psbD (top) or psbA (bottom) 5′ UTRs from C. reinhardtii were incubated with pet19b-expressed PSRP-7 (0, 75, 150, 450, and 750 nM) in the presence of 0, onefold, fivefold, 10-fold, and 50-fold molar excess of the same unlabeled 5′ UTR. Reactions were resolved in 8% TBE-acrylamide gels. The arrowheads indicate the free probes.
(B) A binding reaction of His-tagged PETs EF-Ts (200 pmoles) with Flag-tagged EF-Tu (200 pmoles) was purified on Ni-NTA (−GTP). Alternatively, after binding to the column and a first wash, 2 mL of 1 mM GTP in binding buffer was run through the column, washed again, and then the proteins eluted in 200-μL aliquots (+GTP). FT, flow through; W, wash; E1 and E2, elutions 1 and 2, respectively.
(C) EF-Ts and EF-Tu recombinant proteins were run through the same column without a prior binding reaction. Representative silver-stained gels are shown in (B) and (C).
The identification of UBA and dimerization domains in the EF-Ts domains of PETs (Figure 1B) suggests that these are functional EF-Ts domains. To identify whether the EF-Ts 55-kD polypeptide was active in EF-Tu binding, we purified this recombinant protein fused to an N-terminal His tag and tested binding to recombinant Flag-tagged C. reinhardtii chloroplast EF-Tu in vitro. Both recombinant proteins were incubated together in binding buffer at 0°C for 90 min. After the binding reaction, the proteins were purified by nickel-nitrilotriacetic acid agarose (Ni-NTA) and the fractions separated by SDS-PAGE and either silver stained (Figures 5B and 5C) or assayed by protein gel blots with anti-His and anti-Flag antibodies (data not shown). Flag-tagged EF-Tu eluted together with the EF-Ts polypeptide (Figure 5B, −GTP). As a control, the same proteins were incubated with Ni-NTA agarose without a prior binding reaction. As shown in Figure 5C, the Flag-tagged EF-Tu came out in the flow through, whereas the His-tagged EF-Ts came out in the elution fractions, as expected. The addition of 1 mM GTP did not disrupt binding (Figure 5B, +GTP), similar to what occurs for other chloroplast and mitochondrial EF-Tu • EF-Ts complexes (Spremulli and Spremulli, 1987; Zhang and Spremulli, 1998). These data show that the EF-Ts region of PETs is capable of binding EF-Tu and hence could be the functional EF-Ts in the chloroplast.
Localization of PSRP-7 and EF-Ts in Polysomes
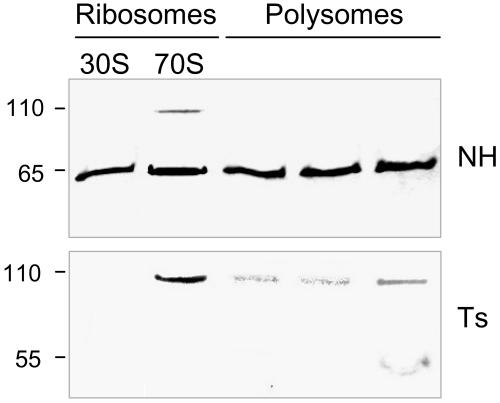
Because PSRP-7 was identified as a component of the 30S subunit, we assayed for the presence of PSRP-7 species in isolated 30S subunits, 70S ribosomes, and translating polysomes. Figure 6 shows that the 65-kD protein (PSRP-7) is present in both 30S and 70S fractions, as previously reported (Yamaguchi et al., 2002, 2003), as well as in polysomes. Interestingly, the 110-kD protein also showed association with the 70S ribosomes.
Figure 6.
Localization of PETs Protein Forms in Ribosomes and Polysomes.
C. reinhardtii isolated ribosomes and polysomes were subjected to protein gel blot analysis (1 μg/lane) using the NH (top) and Ts antibodies (bottom). Polysome fractions correspond to tetrasomes, pentasomes, and nanosomes (left to right). The protein sizes (kD) are indicated on the left side.
A functional EF-Ts would be expected to transiently associate with polysomes during translation elongation, and as shown in Figure 6, the 55-kD protein was observed in polysome fractions, but not in the 30S and 70S fractions. By contrast, the 110-kD protein was detected by the Ts antisera in the 70S fractions, whereas lower levels of this protein were also observed in polysome fractions (Figure 6). The discrepancy between anti-NH and anti-Ts in identifying the 110-kD protein in polysomes suggests that anti-Ts is probably better at recognizing epitopes on PETs precursor at the protein concentration used for polysome analysis (1 μg/lane).
The pets Gene Is Organized in a Similar Manner in Higher Plants as in C. reinhardtii
To determine whether the arrangement of the PSRP-7 and EF-Ts proteins in a single gene was unique to C. reinhardtii, we examined other plant species by database searching. In A. thaliana and O. sativa, we identified both a full-length ORF capable of producing the 110-kD PETs polyprotein and a shorter ORF very similar to the C. reinhardtii 1.68-kb ORF (PSRP-7). Database information for other plant species is not as complete as for A. thaliana and O. sativa, but ESTs corresponding to the N- and C-terminal regions of PETs suggest that PETs homologs are found in many, and perhaps all, plant species (data not shown; Benichou et al., 2003). Figure 7 shows a sequence alignment of the PETs proteins from A. thaliana, O. sativa, and C. reinhardtii and shows that the EF-Ts domains are almost identical and the S1 domains quite similar. The nonhomologous regions differ between the three species.
Figure 7.
PETs Has Counterparts in Higher Plants.
Homology comparison between PETs from C. reinhardtii, A. thaliana, and O. sativa. Identical amino acids are highlighted in gray; similar amino acids are highlighted in black. S1, NH, and EF-Ts regions are indicated. Amino acid numbers are shown on the sides. Dashes represent gaps introduced by the alignment parameters.
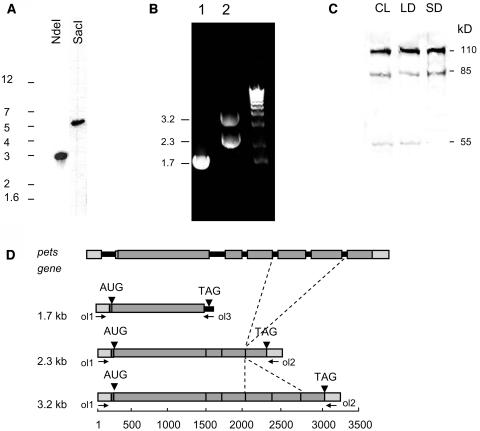
We examined pets expression in A. thaliana in greater detail. Figure 8A shows a DNA gel blot analysis of A. thaliana genomic DNA using a probe consisting of the S1 domain portion of the pets gene. Similar to C. reinhardtii, and as predicted by the genome database, pets is a single-copy gene in A. thaliana. We also performed RT-PCR reactions with oligonucleotides specific for short and long ORFs extracted from the database sequences. When oligos 1 and 2 were used (Figure 8D), we obtained an expected 3.2-kb band and a 2.3-kb band (Figure 8B, lane 2). When oligos 1 and 3 were used, a 1.7-kb band was obtained (Figure 8B, lane 1). The PCR products were subsequently cloned and sequenced. Figure 8D shows a diagram of the A. thaliana psrp-7 gene and the three products obtained by RT-PCR. The pets gene of A. thaliana contains the first exon and intron upstream of the initiator AUG; exon 2 is quite long and spans 16 nucleotides of the 5′ UTR plus 1345 nucleotides of coding region. The 3.2-kb band corresponds to an mRNA with a coding region of 2.9 kb capable of producing the full-length 110-kD PETs precursor homolog. The 1.7-band corresponds to an mRNA that correctly splices intron 1 to give the mature 5′ UTR but fails to splice intron 2 and therefore reaches a stop codon 21 nucleotides downstream of the splice site. This generates a 1.37-kb ORF capable of producing a mature PSRP-7 homolog. Unexpectedly, the 2.3-kb band corresponds to an mRNA undergoing another form of alternative splicing in which exons 5 and 6 are omitted in the mature messenger, giving rise to a 2.1-kb coding region (Figure 8D). The protein translated from this mRNA would be ∼85 kD and would contain the two S1 domains, the NH region, and just one EF-Ts domain. However, this EF-Ts domain would be constituted by the N-terminal half of EF-Ts domain 1 and the C-terminal half of EF-Ts domain 2. This 85-kD protein does not have a counterpart among the PETs-derived proteins that were detected in C. reinhardtii.
Figure 8.
The pets Gene of A. thaliana Undergoes Alternative Splicing.
(A) DNA gel blot of A. thaliana pets gene. Ten micrograms of genomic DNA were digested with NdeI or SacI. Membranes were hybridized with a probe corresponding to the A. thaliana pets S1 domain region. The sizes of the DNA markers are shown on the left.
(B) The A. thaliana pets gene generates three detectable RNAs by RT-PCR using oligos 1 and 3 (lane 1) and oligos 1 and 2 (lane 2) depicted in (D). Right lane, 1-kb ladder. The sizes of the PCR products are indicated on the left.
(C) Protein gel blot analysis of A. thaliana total soluble proteins extracted from 14-d-old seedlings grown under either continuous light (CL), long day (12 h light/12 h dark; LD), or short day (8 h light/16 h dark; SD). Ten micrograms were run in 10% SDS-PAGE. The anti-EF-Ts antibody was used. The sizes of the bands in kilodaltons are shown on the right.
(D) Scheme showing A. thaliana pets gene and its three mature mRNA products. Gray boxes indicate exons; black lines represent introns. Arrowheads indicate start and stop codons. The oligos used for PCR are shown with arrows; the sizes of the PCR products are indicated on the left. The dashed lines indicate the two exons that are spliced out in the 2.3-kb mRNA. The scale at the bottom represents the number of nucleotides.
PETs Is Processed in Higher Plants in a Manner Similar to C. reinhardtii
To ascertain whether a PSRP-7 and EF-Ts fusion protein was produced in higher plants, as in algae, we performed protein gel blot analysis of 14-d-old A. thaliana seedlings grown under different conditions with the anti-EF-Ts antibody used for C. reinhardtii. Figure 8C shows that, in all cases, the major band identified corresponded to the 110-kD PETs precursor. In addition, an 85-kD protein (expected from the 2.1-kb ORF) and the 55-kD EF-Ts protein were also identified by protein gel blot analysis, but at much lower abundance than the 110-kD species (Figure 8C). These results indicate that, despite qualitative similarities between C. reinhardtii and A. thaliana, there are quantitative differences that might be relevant to translation in both organisms.
Comparison of PETs mRNA and Protein Species between A. thaliana, O. sativa, and C. reinhardtii
Table 1 summarizes all the PETs ORFs and protein products identified in the three species. Gene names and accession numbers for the short and long cDNAs are also provided. In general terms, both PSRP-7 and the full-length PETs precursor are conserved in higher plants and algae. However, species-specific pets ORFs are found, such as the C. reinhardtii 1.8-kb ORF, which contains 44 amino acids of the EF-Ts domain region and the A. thaliana 2.1-kb ORF, which contains only one EF-Ts domain (Table 1). Note that the length of the short protein (PSRP-7) is quite different between the three species, but alignment of the full-length precursors (Figure 6) shows that their C-terminal ends always reside at the end of the nonhomologous region, just before the start of the EF-Ts domains.
Table 1.
Comparison of PETs-Derived Proteins from C. reinhardtii, A. thaliana, and O. sativa
| Short Protein (PSRP-7)
|
Full-Length Precursor (PETs)
|
Other Proteins
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Species | Gene Name | Exonsa | ORF | Accession No. | No. Amino Acids | ORF | Accession No. | No. Amino Acids | Exonsa | No. Amino Acids |
| C. reinhardtiib | C_160057 | 1–9 | 1.68 | N.Fc | 560 | 3.04 | AV626377d | 1013 | 1–10 | 604 |
| A. thalianae | At4g29060 | 1 and 2 | 1.35 | AY057550f | 455 | 2.90 | NM_119050f | 953 | 1–4, 7 | 710 |
| O. sativag | CHR12v07232003 | 1 and 2 | 2.06 | AK062564f | 687 | 3.50 | AK067367f | 1147 | N.F. | N.F. |
Exons present in mature ORF.
Database information was taken from the JGI Web page (http://genome.jgi.psf.org/chire2/chire2.home.html).
Not found in the database.
NCBI accession number of longest EST (Kazusa DNA Research Institute).
Database information was extracted from the TAIR Web page (http://www.arabidopsis.org).
NCBI accession number of full-length ORFs from databases.
Database information was obtained from the TIGR rice genome database (http://www.tigr.org/tdb/e2k1/osa1).
DISCUSSION
PSRP-7 is a novel 65-kD ribosomal protein that contains two S1 domains and accumulates in stoichiometric amounts with other ribosomal proteins in the 30S subunit of C. reinhardtii chloroplast ribosomes (Yamaguchi et al., 2002). Here, we show that the C. reinhardtii PSRP-7 protein is encoded by a gene whose complete ORF codes for a 110-kD polyprotein. This polyprotein, named PETs, contains the PSRP-7 protein on the N terminus and two tandem repeats of EF-Ts on the carboxy end. Genomic database and DNA gel blot analyses show that pets is a single-copy gene containing the only chloroplast PSRP-7 and EF-Ts homologous sequences within the C. reinhardtii genome. Another C. reinhardtii ORF (genie.2076.1 = C_4650003) encoding a 332–amino acid protein with a single EF-Ts–like sequence, showed partial and weaker similarity with Synechocystis EF-Ts. The highest overall similarity of this ORF (170/329, 51%) was seen with the EF-Ts of Rickettsia prowazekii, a close relative of the proposed ancestors of mitochondria, suggesting that C_4650003 might be the mitochondrial EF-Ts. The anti-EF-Ts monoclonal antibody used in this work did not recognize this protein (34.3 kD), probably because of the low similarity between Streptococcus EF-Ts and the putative C. reinhardtii mitochondrial EF-Ts.
RNase protection assays identified three distinct pets mRNAs: (1) a 3.6-kb mRNA with a 3-kb ORF, in which all the introns and exons are correctly spliced and which produces the 110-kD PETs polyprotein, (2) a 2.7-kb mRNA in which introns 9 and 10 are not spliced, generating a 1.68-kb ORF that yields the 65-kD PSRP-7 protein, and (3) a 2.6-kb mRNA in which intron 10 fails to be spliced, producing a 1.8-kb ORF that encodes a protein with an estimated size of 72 kD, most likely the 79-kD protein seen in protein gel blots. We did not detect any mRNA that could produce the 55-kD EF-Ts polypeptide alone, indicating that this protein must arise from the full-length mRNA. Protein gel blot analysis using antisera for PSRP-7 identifies the 110-kD polyprotein, a 79-kD protein, and the mature 65-kD PSRP-7, whereas antibodies for EF-Ts identify the 110-kD polyprotein and a 55-kD polypeptide composed of two EF-Ts domains. These results together suggest that the PETs precursor protein is likely posttranslationally cleaved into the 65-kD PSRP-7 and the 55-kD EF-Ts polypeptide.
Analysis of Chlamydomonas chloroplast ribosomes identified the 65-kD PSRP-7 as the only form of PETs associated with the 30S subunit (Figure 3C; Yamaguchi et al., 2002). We have shown that PSRP-7 is capable of binding chloroplast mRNAs in vitro (Figure 5). Although the role of the PSRP-7 protein in translation is still unknown, the RNA binding activity associated with the S1 domains may be a key feature of PSRP-7 function in chloroplast translation.
We also suggest that the 55-kD EF-Ts polypeptide could be the functional EF-Ts in the chloroplast by its ability to bind to EF-Tu in vitro (Figure 5) and by its association to chloroplast polysomes (Figure 3C). By contrast, the 110-kD PETs pro-protein was also found in translationally incompetent 70S ribosomes.
Provided that chloroplast EF-Ts is contained within PETs, the identification of two EF-Ts domains in a single polypeptide is somewhat unique compared with other EF-Ts proteins. Bacterial EF-Ts and eukaryotic EF-1 β, an analog of EF-Ts, are comprised of only one domain per polypeptide and have an approximate molecular weight of 30,000. It has been reported that dimerization of Thermus thermophilus EF-Ts is required for its nucleotide exchange function, in contrast with E. coli EF-Ts, which functions as monomers (Nesper et al., 1998). Perhaps chloroplast EF-Ts proteins also require dimerization for their function and that could be accomplished by having two EF-Ts domains in the same polypeptide. The fusion of a ribosomal protein and an elongation factor is also novel but harder to rationalize. Perhaps PSRP-7 and EF-Ts may need to function together, and physical connection may be a great advantage in this scenario. In A. thaliana, PETs accumulates mainly as the 110-kD species, suggesting that this protein is capable of functioning as the fusion protein, although some processed 55-kD protein is also observed by protein gel blot analysis of A. thaliana proteins. The polyprotein may even have a different function compared with the mature forms. As an example, several reports have shown that, besides its role in translation elongation, EF-Tu participates in translation arrest (Snyder et al., 2003) and/or ribosome rescue (Wower et al., 2000). In the second case, EF-Tu works together with the S1 protein and SmpB, a transfer-mRNA binding protein, to regulate transfer-mRNA binding to ribosomes. Because EF-Ts forms a complex with EF-Tu and PETs contains S1 and EF-Ts domains, we can speculate that the PETs precursor protein may have a role in translation arrest and/or the rescue of stalled ribosomes, especially if we consider that PETs was found associated with nontranslationally active 70S ribosomes.
Why would cells use two different mechanisms, mRNA splicing and posttranslational processing, to generate PSRP-7 is not clear at this time. It is possible that only PSRP-7 expression is required under some conditions, whereas coordinate expression with EF-Ts may be needed at other times. In the first case, the 1.68-kb ORF would be preferentially produced by splicing, and only the mature PSRP-7 protein expressed. When both proteins are required, it might be an energetic advantage for the cell to transcribe a single mRNA and translate a single protein that is imported into the chloroplast and then proteolytically cleaved. This later hypothesis is supported by the fact that the pets precursor is the major mRNA species, whereas the mature 55- and 65-kD are the main protein forms.
PETs could potentially be targeted to both chloroplasts and mitochondria, and its alternative splicing and processing could be a way of regulating this dual targeting. Both MitoProt and Predotar softwares and the C. reinhardtii Joint Genome Institute database predict PETs as a mitochondrial protein. Dual targeting has been reported for several proteins that participate in processes shared by both organelles, such as antioxidant proteins and aminoacyl-tRNA synthases (Chew et al., 2003; Goggin et al., 2003). The accuracy and efficiency of dual targeting is regulated by mechanisms such as usage of alternative start codons and alternative splicing of precursor mRNAs (Watanabe et al., 2001; De la Fuente van Bentem et al., 2003).
All together, the data presented in this work strengthen the idea that chloroplast translation is distinct from bacterial translation, both in terms of the components and how these components are related structurally and functionally. The relevance of PETs existing as a stable entity and being conserved in green algae (C. reinhardtii), dicots (A. thaliana), and monocots (O. sativa), and most likely all chloroplast-containing species, suggests that the linkage of PSRP-7 and EF-Ts in PETs plays an important functional role, although the exact nature of its role in translation has yet to be determined.
METHODS
Chlamydomonas reinhardtii Strains and Culture Conditions
C. reinhardtii wild-type strain 137c (mt+) was obtained from the Chlamydomonas Genetic Center, Durham, NC. Cells were grown in TAP media (Gorman and Levine, 1965) on a rotary shaker at 23°C at 4500 lux for RNA preparations and on TAP-agar plates for protein and genomic DNA preparations. Liquid cultures were grown in either continuous light or under a 12-h-light/12-h-dark regime. Cultures were maintained at a cell density of 1 × 106 cells per mL for at least 48 h before harvest.
Arabidopsis thaliana Growth Conditions
A. thaliana Columbia ecotype seeds were plated on half MS-agar medium supplemented with 0.5% sucrose and grown for 14 d at 20°C in continuous light, long day (12 h light/12 h dark), or short day conditions (8 h light/16 h dark) at 3000 lux.
DNA Cloning and Sequencing, DNA Gel Blots, and RNA Gel Blots
Classical molecular biology techniques were followed for all DNA and RNA manipulations as described by Sambrook et al. (1989), as well as DNA gel blots, RNA gel blots, and 32P labeling of DNA for use as probes. RNA gel blots and DNA gel blots were visualized using a Packard Cyclone storage phosphor system equipped with Optiquant software (Packard Instrument Company, Downers Grove, IL). Total digital light units were measured for each lane of the blot as well as an arbitrary space for background subtraction.
The longest pets EST was obtained from the Kazusa DNA Research Institute (http://www.kazusa.or.jp/en/plant/chlamy/EST/) (Asamizu et al., 2000). C. reinhardtii total cDNAs were made by reverse transcription of 1 μg of total RNA using Superscript II (Invitrogen, Carlsbad, CA) according to the manufacturer's instructions. Pets cDNAs were amplified by PCR and cloned into pCR 2.1 TOPO (Invitrogen). The oligos were as follows: 5′-ATGCTGCGGGAGCTCGGACGC-3′ and 5′ (T)23 3′ as reverse oligo. A. thaliana pets cDNAs were obtained by PCR using the Superscript A. thaliana cDNA library (Invitrogen) using the following oligos: oligo 1, 5′-CTCGAGGCACAGGGGAGAGACAAAATGAG-3′; oligo 2, 5′-GACTAGTCATGAACAAAAAATTACAGG-3′; oligo 3, 5′-GACTAGTGTGATTTTTAGTCAACTTACTTTTGG-3′. DNA sequencing was performed by capillary electrophoresis technology using an ABI 3700 DNA analyzer (Applied Biosystems, Foster City, CA; Nucleic Acid Core Facility, Scripps Research Institute).
RNase Protection Assays
RNA probes were made as follows: DNAs were amplified by PCR using forward oligos with a BamHI site and reverse oligos with an EcoRI site proceeded by the T7 RNA polymerase promoter to be able to make antisense RNAs as follows: for pets-1018, forward 5′-GGATCCGCTGTTGCTGCCTACG-3′ and reverse 5′-GAATTCTAATACGACTCACTATAGGGAGCACATCCTTGGGGATG-3′; for pets-1398-1 and pets-1680, forward 5′-CCGGATCCGCTGGTGGACTACTGGACC-3′ and reverse 5′-CGAATTCTAATACGACTCACTATAGGGCAGACCCTTCTTGCGCAGCC-3′; for pets-1398-2, forward 5′-CCGGATCCGCTGGTGGACTACTGGACC-3′ and reverse 5′-CCGAATTCTAATACGACTCACTATAGGGCGCACTCAGCCAGCGCCTTC-3′; for pets-1763, forward 5′-GGGGATCCGTGACGCTGAGGCTGCCGC-3′ and reverse 5′-GGAATTCTAATACGACTCACTATAGGGCTTGACCAGCTCGGCCACGG-3′; for pets-2020, forward 5′-GCGGATCCGAGGTGCTGGCAAAGG-3′ and reverse 5′-GGAATTCTAATACGACTCACTATAGGGGGTCTTGTCGCGCAGC-3′; for pets-2169, forward 5′-GGATCCAACCCCGACAAGACCGTGG-3′ and reverse 5′-GAATTCTAATACGACTCACTATAGGGGTTCACCTCCAGCAGCACACC-3′; for pets-2571, forward 5′-CGGGATCCGTCCTACATCCACCCC-3′ and reverse 5′-GGAATTCTAATACGACTCACTATAGGGTGGCGGCGATGGACTC-3′. PCR products were digested with EcoRI and BamHI and cloned into pUC18. All clones were linearized with BamHI, except for pets-1680 that was linearized with BstXI. DNA templates were in vitro transcribed using T7 RNase polymerase (Roche, Indianapolis, IN) according to the manufacturer's instructions using [α32P]UTP as the radioactive ribonucleotide. Full-length RNA probes were gel-purified from 8% acrylamide-TBE-urea gels in RNA elution buffer (0.5 M sodium acetate, 0.5% SDS, and 1 mM EDTA) and precipitated with 0.4 M ammonium acetate and two volumes of 100% ethanol.
Twenty-five micrograms of C. reinhardtii total RNAs were resuspended in 20 μL of RNase protection assay hybridization buffer (40 mM Pipes, pH 6.4, 1 mM EDTA, pH 8.0, 0.4 M NaCl, and 80% formamide) containing 1× 105 cpm of the corresponding RNA probe. The RNAs were denatured at 85°C for 10 min and then annealed at 60°C for 18 h. After cooling to room temperature, the samples were incubated in 300 μL of RNase mix (300 mM NaCl, 10 mM Tris-HCl, pH 7.4, 5 mM EDTA, pH 8, 2 μg/mL of RNase T1, and 40 μg/mL of RNase A) at 30°C for 60 min. RNases were inactivated by adding 20 μL of 10% SDS and 10 μL 10 mg/mL proteinase K at 37°C for 30 min. After phenol:chloroform:isoamyl alcohol extraction, RNA-RNA complexes were precipitated by adding 750 μL of 100% ethanol and 10 μg of tRNA, incubated at −80°C for 15 min and spun down at 12,000g for 30 min. Protected RNAs were denatured at 85°C RNA and separated in 8% TBE-urea-acrylamide gels. The intensity of the bands obtained with the pets-1398-1 probe was determined by phosphor imaging and corrected by the amount of uridines present compared with the full-length probe. Thus, the full-length protected band (408 nucleotides) was multiplied by 1, the 387-nucleotide band by 1.055, and the 277-nucleotide band by 1.6.
Ribosome and Polysome Preparation
Chlamydomonas chloroplast ribosomes and the 30S subunits were prepared as previously described (Yamaguchi et al., 2002), and the chloroplast polysomes were prepared as previously reported (Cohen et al., 1998). Polysome fraction numbers 15, 17, and 21 were used for protein gel blot analysis, corresponding approximately to tetrasomes, pentasomes, and nanosomes.
Protein Gel Blotting
Polyclonal antisera against the nonhomologous region of C. reinhardtii PSRP-7 (NH) antibodies were raised in rabbits (Scripps Research Institute, Protein and Nucleic Acid Core Facility). Anti-Streptococcus suis serotype 2 EF-Ts monoclonal antibodies were provided by M. Gottschalk. Alkaline phosphatase-labeled anti-rabbit and anti-mouse secondary antibodies were used (Sigma, St. Louis, MO). Protein gel blots were developed using nitroblue tetrazolium and 5-bromo-4-chloro-3-indolyl phosphate (Sigma).
Edman Degradation
Determination of the N-terminal sequence of the mature PSRP-7 protein was done using 100 pM of protein immobilized on a polyvinylidene difluoride membrane according to the original procedure for Edman degradation (Heirwegh and Edman, 1957).
PSRP-7 and EF-Ts in Vitro Activity Measurements
The PSRP-7 and EF-Ts regions of PETs were amplified by PCR introducing an NdeI site at the initiator AUG and a BamHI site downstream of the stop codon. Digested PCR products were cloned into pET19b (Novagen, Madison, WI) and transformed into BL21 competent cells. Protein induction and purification were done using Ni-NTA agarose according to the manufacturer's instructions (QIAexpressionist; Qiagen, Valencia, CA) and dialyzed into the corresponding binding buffers.
Binding of the EF-Ts region of PETs to chloroplast EF-Tu was tested as reported previously (Zhang and Spremulli, 1998). C. reinhardtii chloroplast EF-Tu (GenBank accession number S09153) was amplified by RT-PCR using a forward oligo that introduced an in-frame NdeI site: 5′-AACACATATGTCACGTGCTAAGTTTGAACG-3′ and a reverse oligo that created a fusion with a 1× Flag tag and introduced a BamHI site downstream of the stop codon: 5′-GGATCCTTACTTGTCGTCGTCGTCCTTGTAGTCGGTACCTTGAACAATATTAGTTACAACACC-3′, The PCR product was digested with NdeI and BamHI and cloned into pET3a vector (Novagen) and transformed into BL21 competent cells. Protein induction and purification was done using anti-Flag M2-agarose affinity gel according to the manufacturer's instructions (Sigma).
Binding of PSRP-7 to RNA was determined by gel shift assays. PET19b-expressed PSRP-7 (75, 150, 450, and 750 nM) was incubated with a 100 pM concentration [α-32P]UTP-labeled psbD or psbA 5′ UTRs from C. reinhardtii. For competition assays, unlabeled psbD or psbA 5′ UTRs were added at onefold, fivefold, 10-fold, and 50-fold molar excess. Reactions were done in 30 mM Tris-HCl, pH 7, 5 mM MgCl2, 5 mM DTT, 50 mM KCl, 3% glycerol, 20 units of SUPERASE-in RNase inhibitor (Ambion, Austin, TX), and 1 μg of tRNA. Samples were incubated on ice for 30 min and resolved in 8% acrylamide-TBE gels at 4°C. Gels were fixed in 10% methanol and 10% acetic acid for 10 min, dried, exposed to sensitive screens, and developed by phosphor imaging.
Binding of the EF-Ts region of PETs to chloroplast EF-Tu was tested as reported previously (Zhang and Spremulli, 1998). Two hundred picomoles of His-tagged PETs EF-Ts domain region (55 kD) were incubated with the same amount of Flag-tagged EF-Tu in binding buffer (50 mM Tris-HCl, pH 8, 10 mM MgCl2, 0.1 M NH4Cl, and 1 mM DTT) on ice for 90 min and purified through Ni-NTA or anti-Flag affinity columns. Two conditions were tested: in one, −GTP, the binding reaction was run through the column. Alternatively, after the binding of the EF-Ts-EF-Tu complex to the column and a first wash, 2 mL of 1 mM GTP in column binding buffer were run through, the column washed again and then eluted in 200-μL aliquots (+GTP). Fifty-microliter aliquots were run in 12% SDS-PAGE and either silver-stained or visualized by protein gel blot using anti-His and anti-Flag antibodies. Controls for both recombinant proteins without doing the binding reaction were run in both Ni-NTA and anti-Flag columns.
Computational Analyses
EST and genome information from C. reinhardtii was obtained from the Chlamydomonas Joint Genome Institute (http://genome.jgi-psf.org/chlre2/chlre2.home.html). A. thaliana information was obtained from the Arabidopsis Information Resource (http://www.arabidopsis.org). Rice (Oryza sativa) information was extracted from The Institute for Genomic Research (http://www.tigr.org/tdb/e2k1/osa1). Partial DNA sequences were assembled using CAP (http://www.infobiogen.fr/services/analyseq/cgi-bin/cap_in.pl). Homology comparison was done using BLAST 2 sequences (National Center for Biotechnology Information). UBA and dimerization domains were estimated taking into account Thermus thermophilus EF-Ts (Jiang et al., 1996). Multiple sequence alignments were performed using MacVector 7.0 software. For both softwares, BLOSUM62 matrix was used with a gap open = 11, gap extension = 1, x_dropoff = 50, expect = 10, and without filter. Prediction of subcellular localization was done using ChloroP, MitoProt, and Predotar.
Sequence data from this article have been deposited with the EMBL/GenBank data libraries under accession numbers AY661459 to AY661462.
Acknowledgments
We thank S. Franklin, A. Cohen, J. Schultz, D. Barnes, and R. Henry for critical reading of the manuscript, A. Coragliotti for the provision of RNA, M. Gottschalk for the anti-EF-Ts antibody, and J. Harper for the A. thaliana seeds. This work was supported by funds from the U.S. Department of Energy (93ER70116) and the National Institutes of Health (GM54659) to S.P.M. M.V.B. was supported by a Skaggs Postdoctoral Fellowship.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Stephen P. Mayfield (mayfield@scripps.edu).
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.104.026708.
References
- Asamizu, E., Miura, K., Kucho, K., Inoue, Y., Fukuzawa, H., Ohyama, K., Nakamura, Y., and Tabata, S. (2000). Generation of expressed sequence tags from low-CO2 and high-CO2 adapted cells of Chlamydomonas reinhardtii. DNA Res. 7, 305–307. [DOI] [PubMed] [Google Scholar]
- Barkan, A., and Goldschmidt-Clermont, M. (2000). Participation of nuclear genes in chloroplast gene expression. Biochimie 82, 559–572. [DOI] [PubMed] [Google Scholar]
- Beligni, M.V., Yamaguchi, K., and Mayfield, S.P. (2004). The translational apparatus of C. reinhardtii. Photosyn. Res., in press. [DOI] [PubMed]
- Benichou, M., Li, Z., Tournier, B., Chaves, A., Zegzouti, H., Jauneau, A., Delalande, C., Latche, A., Bouzayen, M., and Spremulli, L.L. (2003). Tomato EF-Ts(mt), a functional mitochondrial translation elongation factor from higher plants. Plant Mol. Biol. 53, 411–422. [DOI] [PubMed] [Google Scholar]
- Bhattacharya, D., Yoon, H.S., and Hackett, J.D. (2004). Photosynthetic eukaryotes unite: Endosymbiosis connects the dots. Bioessays 26, 50–60. [DOI] [PubMed] [Google Scholar]
- Chew, O., Whelan, J., and Millar, A.H. (2003). Molecular definition of the ascorbate-glutathione cycle in Arabidopsis mitochondria reveals dual targeting of antioxidant defenses in plants. J. Biol. Chem. 278, 46869–46877. [DOI] [PubMed] [Google Scholar]
- Cohen, A., Yohn, C.B., Bruick, R.K., and Mayfield, S.P. (1998). Translational regulation of chloroplast gene expression in Chlamydomonas reinhardtii. Methods Enzymol. 297, 192–209. [Google Scholar]
- De la Fuente van Bentem, S., Vossen, J.H., Vermeer, J.E., de Vroomen, M.J., Gadella, T.W. Jr., Haring, M.A., and Cornelissen, B.J. (2003). The subcellular localization of plant protein phosphatase 5 isoforms is determined by alternative splicing. Plant Physiol. 133, 702–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs, T.M., Deppisch, H., Scarlato, V., and Gross, R. (1996). A new gene locus of Bordetella pertussis defines a novel family of prokaryotic transcriptional accessory proteins. J. Bacteriol. 178, 4445–4452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goggin, D.E., Lipscombe, R., Fedorova, E., Millar, A.H., Mann, A., Atkins, C.A., and Smith, P.M. (2003). Dual intracellular localization and targeting of aminoimidazole ribonucleotide synthetase in cowpea. Plant Physiol. 131, 1033–1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorman, D.S., and Levine, R.P. (1965). Cytochrome f and plastocyanin: Their sequence in the photosynthetic electron transport chain of Chlamydomonas reinhardtii. Proc. Natl. Acad. Sci. USA 54, 1665–1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gribskov, M. (1992). Translational initiation factors IF-1 and eIF-2 alpha share an RNA-binding motif with prokaryotic ribosomal protein S1 and polynucleotide phosphorylase. Gene 119, 107–111. [DOI] [PubMed] [Google Scholar]
- Gromadski, K.B., Wieden, H.J., and Rodnina, M.V. (2002). Kinetic mechanism of elongation factor Ts-catalyzed nucleotide exchange in elongation factor Tu. Biochemistry 41, 162–169. [DOI] [PubMed] [Google Scholar]
- Harris, E.H., Boynton, J.E., and Gillham, N.W. (1994). Chloroplast ribosomes and protein synthesis. Microbiol. Rev. 58, 700–754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heirwegh, K., and Edman, P. (1957). Purification and N-terminal determination of crystalline pepsin. Biochim. Biophys. Acta 24, 219–220. [DOI] [PubMed] [Google Scholar]
- Jacques, N., and Dreyfus, M. (1990). Translation initiation in Escherichia coli: Old and new questions. Mol. Microbiol. 4, 1063–1067. [DOI] [PubMed] [Google Scholar]
- Jiang, Y., Nock, S., Nesper, M., Sprinzl, M., and Sigler, P.B. (1996). Structure and importance of the dimerization domain in elongation factor Ts from Thermus thermophilus. Biochemistry 35, 10269–10278. [DOI] [PubMed] [Google Scholar]
- Kawashima, T., Berthet-Colominas, C., Wulff, M., Cusack, S., and Leberman, R. (1996). The structure of the Escherichia coli EF-Tu.EF-Ts complex at 2.5 A resolution. Nature 379, 511–518. Erratum. Nature 381, 172. [DOI] [PubMed] [Google Scholar]
- Mayfield, S.P., Yohn, C.B., Cohen, A., and Danon, A. (1995). Regulation of chloroplast gene expression. Annu. Rev. Plant Physiol. Plant Mol. Biol. 46, 147–166. [Google Scholar]
- Merendino, L., Falciatore, A., and Rochaix, J.D. (2003). Expression and RNA binding properties of the chloroplast ribosomal protein S1 from Chlamydomonas reinhardtii. Plant Mol. Biol. 53, 371–382. [DOI] [PubMed] [Google Scholar]
- Nesper, M., Nock, S., Sedlak, E., Antalik, M., Podhradsky, D., and Sprinzl, M. (1998). Dimers of Thermus thermophilus elongation factor Ts are required for its function as a nucleotide exchange factor of elongation factor Tu. Eur. J. Biochem. 255, 81–86. [DOI] [PubMed] [Google Scholar]
- Nyborg, J., and Kjeldgaard, M. (1996). Elongation in bacterial protein biosynthesis. Curr. Opin. Biotechnol. 7, 369–375. [DOI] [PubMed] [Google Scholar]
- Ramakrishnan, V. (2002). Ribosome structure and the mechanism of translation. Cell 108, 557–572. [DOI] [PubMed] [Google Scholar]
- Sambrook, J., Fritsch, E.F., and Maniatis, T. (1989). Molecular Cloning: A Laboratory Manual. (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press).
- Snyder, L., Blight, S., and Auchtung, J. (2003). Regulation of translation of the head protein of T4 bacteriophage by specific binding of EF-Tu to a leader sequence. J. Mol. Biol. 334, 349–361. [DOI] [PubMed] [Google Scholar]
- Spremulli, G.H., and Spremulli, L.L. (1987). Effect of GDP on the interactions between chloroplast EF-Ts and chloroplast and E. coli EF-Tu. Biochem. Biophys. Res. Commun. 148, 1490–1495. [DOI] [PubMed] [Google Scholar]
- Subramanian, R. (1983). Structure and functions of ribosomal protein S1. Prog. Nucleic Acid Res. Mol. Biol. 28, 101–142. [DOI] [PubMed] [Google Scholar]
- Sugiura, M., Hirose, T., and Sugita, M. (1998). Evolution and mechanisms of translation in chloroplasts. Annu. Rev. Genet. 32, 437–459. [DOI] [PubMed] [Google Scholar]
- Watanabe, N., Che, F.S., Iwano, M., Takayama, S., Yoshida, S., and Isogai, A. (2001). Dual targeting of spinach protoporphyrinogen oxidase II to mitochondria and chloroplasts by alternative use of two in-frame initiation codons. J. Biol. Chem. 276, 20474–20481. [DOI] [PubMed] [Google Scholar]
- Wower, I.K., Zwieb, C.W., Guven, S.A., and Wower, J. (2000). Binding and cross-linking of tmRNA to ribosomal protein S1, on and off the Escherichia coli ribosome. EMBO J. 19, 6612–6621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi, K., Beligni, M.V., Prieto, S., Haynes, P.A., McDonald, W.H., Yates III, J.R., and Mayfield, S.P. (2003). Proteomic characterization of the Chlamydomonas reinhardtii chloroplast ribosome. Identification of proteins unique to the 70 S ribosome. J. Biol. Chem. 278, 33774–33785. [DOI] [PubMed] [Google Scholar]
- Yamaguchi, K., Prieto, S., Beligni, M.V., Haynes, P.A., McDonald, W.H., Yates III, J.R., and Mayfield, S.P. (2002). Proteomic characterization of the small subunit of Chlamydomonas reinhardtii chloroplast ribosome: Identification of a novel S1 domain-containing protein and unusually large orthologs of bacterial S2, S3, and S5. Plant Cell 14, 2957–2974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi, K., and Subramanian, A.R. (2003). Proteomic identification of all plastid-specific ribosomal proteins in higher plant chloroplast 30S ribosomal subunit. Eur. J. Biochem. 270, 190–205. [DOI] [PubMed] [Google Scholar]
- Zhang, Y., and Spremulli, L.L. (1998). Roles of residues in mammalian mitochondrial elongation factor Ts in the interaction with mitochondrial and bacterial elongation factor Tu. J. Biol. Chem. 273, 28142–28148. [DOI] [PubMed] [Google Scholar]