Abstract
Metabolomics aims at a complete characterization of all metabolites in biological samples both in terms of their identities and concentrations. Because changes of metabolites and their concentrations are a direct reflection of cellular activity, it allows a better understanding of cellular processes and function. Although NMR spectroscopy is routinely applied to complex biological mixtures without purification, overlapping NMR peaks often pose a challenge for the comprehensive and accurate identification of the underlying metabolites. To address this problem, we present a novel nanoparticle-based strategy that differentiates between metabolites based on their electric charge. By adding electrically charged silica nanoparticles to the solution NMR sample, metabolites of opposite charge bind to the nanoparticles and their NMR signals are weakened or entirely suppressed due to peak broadening caused by the slow rotational tumbling of the nanometer sized nanoparticles. Comparison of the edited with the original spectrum facilitates analysis significantly and reduces ambiguities in the identification of metabolites. This method makes NMR directly sensitive to the detection of molecular charges at constant pH as is demonstrated here both for model mixtures and human urine. The simplicity of the approach should make it useful for a wide range of metabolomics applications.
Graphical Abstract
Introduction
Over the past decade, metabolomics has made great progress in its quest to comprehensively study complex biological systems from a small molecule perspective.1–3 Metabolomics focuses on the systematic characterization of individual components of the metabolome in biological systems at the level of biofluids, cells, tissues, and whole organisms.4–6 Identification and quantification the metabolites are important, since they reflect a real-time snapshot of many cellular activities.7
NMR spectroscopy is one of two key experimental analysis methods in metabolomics due to its high spectral resolution, its global detection capability, excellent reproducibility, and ease of sample preparation. However, depending on the complexity of the mixture, ambiguities in peak assignments and metabolite identities can exist in 1D NMR and 2D NMR experiments.8 Presently, one of the most powerful approaches for resolving such ambiguities relies on 2D TOCSY-type experiments,9 which provide chemical connectivity information between multiple nuclear spins. This information can be used to deconvolute the NMR signals of individual mixture components in complex mixtures.10 Moreover, once individual mixture components have been deconvoluted, TOCSY spectra provide useful topology information toward de novo structure elucidation of mixture components.11
Targeted sample preparation approaches have been proposed to resolve peak ambiguities and to simplify the spectra in metabolomics applications. For example, the addition of paramagnetic ions, such as gadolinium and other lanthanides, can cause differential line broadening or the disappearance of peaks of certain metabolites through paramagnetic relaxation, but such effects depend on a number of factors that are not easily predictable.12 Another approach specifically targets metabolites that contain carboxyl groups by having them react with 15N cholamine isotope tags. The modified metabolites, which are detectable by 15N NMR, can be identified provided that a customized NMR library of tagged metabolite products is available.13
Here, we present a strategy that monitors the modulation and disappearance of NMR signals of metabolites that strongly interact with electrically charged silica nanoparticles (SNPs). SNP colloid is directly mixed to the solution NMR metabolite sample forming a stable, solution-like dispersion. Nanoparticles with diameters in the tens of nanometer range have very slow rotational tumbling correlation times in the microsecond range.14 As a result, the tumbling rates of those metabolites that interact with the nanoparticles are slowed down by several orders of magnitude in a solution. This causes rapid transverse spin relaxation of these metabolites, which is manifested in dramatic line broadening and diminished peak amplitudes or complete peak disappearance. Comparison of the NMR spectra of nanoparticle-free and nanoparticle-containing metabolite samples allows the straightforward identification of NMR signals that belong to the subset of metabolites with opposite charge to the charge of the SNPs facilitating the analysis of both 1D and 2D NMR spectra. We demonstrate the SNP-editing approach with both anionic and cationic SNPs.
EXPERIMENTAL SECTION
Chemicals and Materials
A pooled urine sample from healthy humans was obtained from (Innovative Research, Inc., Novi, MI). The following metabolites were obtained for the preparation of model mixtures: citric acid monohydrate (C6H8O7•H2O, 99%), N,N-dimethylglycine (C4H9O2N, 99%), D-glucose (C6H12O6, 99.5%), L-glutamic acid (C5H9NO4, 98.5%), L-histidine (C6H9N3O2, 98.5%), sodium lactate (C3H5NaO3, 98%), sodium phosphate dibasic (Na2HPO4, 99.0%), sodium phosphate monobasic (NaH2PO4, 99.0%), and 3-(Trimethylsilyl)-1-propanesulfonic acid sodium salt (DSS) were obtained from Sigma-Aldrich. L-alanine (C3H7NO2, 98.5%), L-arginine (C6H14N4O2, 98.5%), L-asparagine monohydrate (C4H8N2O3•H2O, 98.5%), L-glutamine (C5H10N2O3, 98.5%), L-lysine hydrochloride (C6H14N2O2•HCl, 98.5%), and L-valine (C5H11NO2, 98.5%) were purchased from Fisher BioReagents. Deuterium oxide (D2O, 99.0%) was purchased from Cambridge Isotope Laboratory, Inc., Andover, MA. Amicon Ultra-15® Centrifugal Filter Units (3k and 10k NMWL) was purchased from Merck Millipore Ltd., Darmstadt, Germany.
Nanoparticle Preparation and Characterization
Bindzil® 2040 silica nanoparticles (anionic SNPs) were obtained from Eka Chemicals. The surface of these SNPs is terminated by silanol (Si-OH) groups, which experience partial deprotonation leading to a negative charge at neutral pH condition. Buffer exchange using a 10k NMWL Amicon filter unit was done for 1 mL SNP colloid by repeatedly (at least 5 times) adding 10 mL 20 mM pH 7.0 sodium phosphate buffer dissolved in D2O followed by centrifugation at 4300 rpm for 30 min to increase concentration. Finally, the volume of concentrated SNPs was adjusted back to 1 mL using the same phosphate buffer, with a recovery rate of 70%, quantified by gravimetric analysis after completely solvent evaporation. The particle concentration was estimated to be 50 μM. LUDOX® CL silica nanoparticles (cationic SNPs) were purchased from Sigma-Aldrich. According to the product description, the surface of the particles is covered with a monolayer of alumina that results in the positive surface charge. Cationic SNP stock colloid (pH 4.0) was extensively dialyzed into 100 mM NaCl in H2O and the pH was adjusted to 5.5 using 0.5 M NaOH. The particle concentration was estimated to be 28 μM. The ζ-potentials of anionic and cationic SNPs were measured to be -23.0 ± 7.4 mV and +35.7 ± 6.9 mV (average ± standard deviation), respectively, using a Malvern Zetasizer Nano ZS at 25 °C (see Supporting Information).
Model Mixture Preparation
A model mixture of common metabolites consisting of unlabeled alanine, arginine, citric acid, dimethylglycine, glucose, histidine, lactic acid, lysine, shikimic acid, and valine with a uniform concentration of 2 mM was prepared in 20 mM sodium phosphate buffer at pH 7.0 in D2O for experiments involving anionic SNPs. A two-fold diluted model mixture (1 mM) in D2O at pH 5.0 was used for cationic SNPs to maintain nanoparticle stability and prevent agglomeration.
Protein removal of Human Urine
1 mL urine sample was thawed at 4 °C and filtrated using a 3k NMWL Amicon filter unit at 4300 rpm for 20 min at 4 °C. The filter was washed twice with 2 mL nanopure water each time and all of the filtrates were pooled together before they were lyophilized and stored as a powder at −80 °C. The powder sample was dissolved in 1100 μL 20 mM sodium phosphate buffer at pH 7.0. The nanoparticle-free sample was made of 550 μL urine sample with 50 μL phosphate buffer. The SNP-containing sample was made of 550 μL urine sample with 50 μL anionic SNPs. Both samples also contained 100 μM DSS.
NMR Sample preparation
Anionic and cationic SNPs (or pure buffer for control samples) were gently mixed with the reconstituted model mixture and/or urine metabolomics samples to a final volume of 600 μL before being placed in a 5 mm NMR tube for NMR data collection. The final particle concentration of anionic SNPs was estimated to be 8 μM for the model mixture and 4 μM for the urine sample, while approximately 4 μM cationic SNPs was used in the model mixture.
NMR data collection
All NMR spectra were collected on a Bruker AVANCE 850 MHz spectrometer equipped with a cryogenically cooled probe at 298 K. 1D 1H NMR spectra of the model mixture were collected with 32,768 complex points, 16 ppm spectral width with 64 scans (for anionic SNPs) to 256 scans (for cationic SNPs). 2D 13C-1H heteronuclear single quantum coherence (HSQC) spectra of the model mixture were collected with 512x1500 (N1xN2) complex points, 8012.820 Hz in the 1H dimension and 32193.432 in the 13C dimension with 16 scans (for anionic SNPs) or 32 scans (for cationic SNPs). 1024 and 3072 data points (N1xN2) were collected with 32 scans for the urine sample. The transmitter frequency offsets were 4.70 ppm and 80 ppm in the 1H and 13C dimension, respectively.
NMR data analysis
All NMR data were zero-filled, Fourier transformed, and phase- and baseline-corrected. 1D 1H NMR spectra were processed using the ACD/1D NMR manager version 12.0 (Advanced Chemistry Development, Inc.). 2D NMR spectra were processed using NMRPipe.15 To identify metabolites in the 1D and 2D NMR spectra the COLMAR16,17 databases were queried.
Results
NMR spectral data of the model mixture
Ten compounds were chosen as a representative set of metabolites with different electrical charge properties (see Experimental Section). Figure 1 shows the 1D 1H NMR and 2D 13C-1H HSQC spectra of the model mixture in the absence (left) and the presence (right) of anionic SNPs. In the absence of SNPs, the 1D spectrum display a certain level of peak overlap, which becomes resolved by the addition of SNPs. For example, arginine overlaps with other metabolites including lysine, histidine, glucose, and dimethyglycine. The addition of anionic SNPs leads to the removal of all arginine peaks, which simplifies the analysis and quantitation of the other metabolites. To quantify the effect of SNP addition, the average signal-to-noise ratios of all unique peaks of each metabolite were measured with and without SNPs, and their ratios (SNP-free:SNP-containing) were converted into a suppression factor (Table S1). The only positively charged metabolites arginine, lysine, and histidine rank as the top 3 compounds in terms of peak suppression. Since histidine has a side chain pKa of 6.0 with a pI value of 7.59, at pH 7.0 it is only partially charged and hence it interacts with the SNPs not as strongly as arginine or lysine, which is consistent with the results in Table S1. The model mixture also demonstrates that other compounds, such as valine, with local charges, but zero net charge, can also be partially suppressed. In addition, we observed that anionic SNPs have a tendency to interact with compounds containing clusters of hydrophobic methyl groups (dimethylglycine and valine). None of the negatively charged metabolites were perturbed by the addition of SNPs. The spectral simplification induced by the nanoparticles is also obvious in the 2D 13C-1H HSQC spectrum with the suppressed peaks marked by small rectangles (Figure 1). All cross-peaks of the positively charged metabolites are suppressed entirely beyond detection limit, while the cross-peaks of the partially charged metabolites remain observable. The latter resonances do not show any noticeable chemical shift changes due to the addition the nanoparticles provided that the pH is not changed. Only very weak line broadening is observed for the non-interacting metabolites, which might be due to a slight increase in the overall viscosity of the mixture leading to somewhat slower rotational tumbling behavior.
Figure 1.
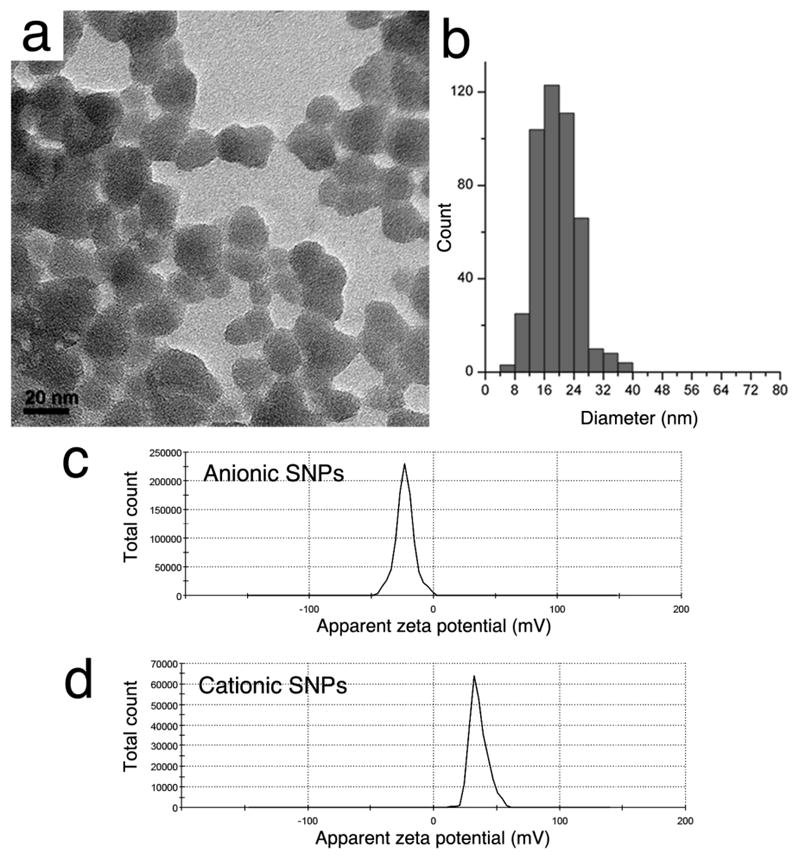
(a) TEM image and (b) size distribution of near-spherical anionic silica nanoparticles (SNPs) after being vacuum dried and concentrated on a copper grid. Their size distribution was determined to be 19.5 ± 5.3 nm. ζ-potentials of (c) anionic SNPs at pH 5.5 and (d) cationic SNPs at pH 7.0 were measured to be −23.0 ± 7.4 mV and +35.7 ± 6.9 mV, respectively.
NNR spectral data of the urine sample
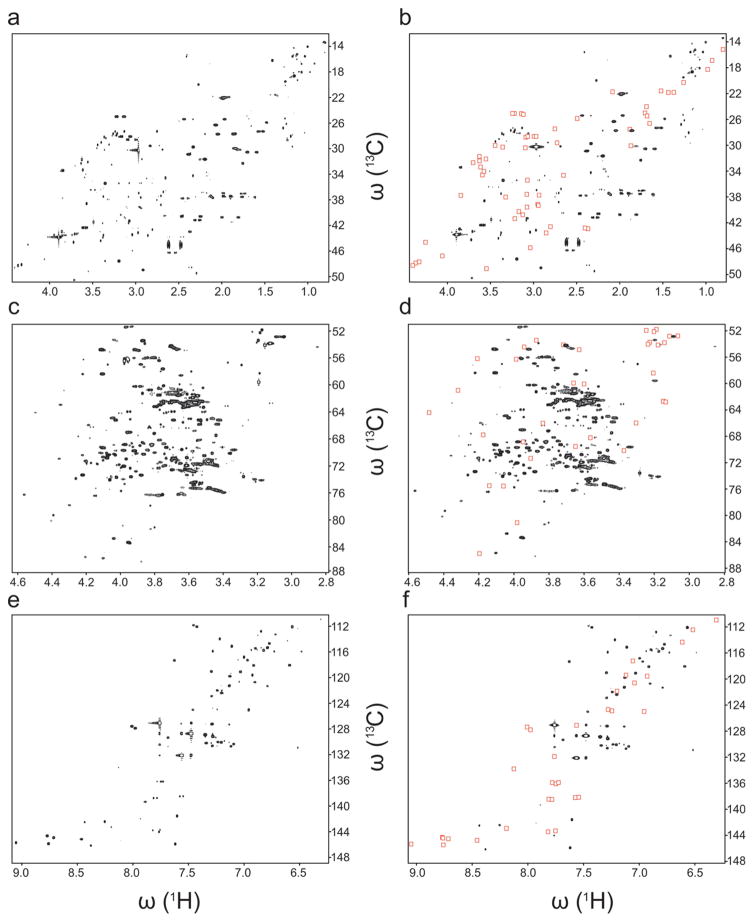
Human urine has served in metabolomics studies as an archetype among biofluids. It has been used for biomarker identification,18 disease classification19 and drug toxicity evaluation.20 It was chosen here because of its challenging metabolome displaying a high level of complexity. Our COLMAR 13C-1H HSQC web server21 was used for automatic peak assignment.16 The spectrum of SNP-free urine sample was compared to the one with anionic SNPs added, highlighting those cross-peaks stemming from compounds that were suppressed by the presence of nanoparticles (Table 1). Three representative spectral regions that show the spectral changes induced by the SNPs are depicted in Figure 3. The peak assignments of these metabolites were independently verified using 13C-1H TOCSY-HSQC and 1H-1H TOCSY NMR spectra.22 Three of these metabolites are positively charged, three of them have no net charge and possess tertiary amine groups and the other four metabolites possess a quaternary amine group. Negatively charged acidic metabolites, such as lactic acid, citric acid, and acetic acid were not perturbed. Zwitterions, such as amino acids, were only suppressed when they have a net positive charge. Many of the metabolites observed in the human urine 2D 13C-1H HSQC spectrum were observed in previous urine NMR-metabolomics studies.23 Metabolites that have only one or two cross-peaks, such as methylguanidine or dimethylglycine can be easily misassigned due to peak overlaps. Therefore, their SNP-based identification on the basis of their positive charge helps their unambiguous identification. As for the model mixture, no noticeable chemical shift changes were observed when the SNPs were added and some very weak line broadening is possibly due to overall increased viscosity.
Table 1.
List of metabolites with attenuated peaks in anionic SNP-containing urine sample.
| NP suppressed | Charge | Nitrogen number |
|---|---|---|
| 1-Methylhistidine | 0 | 1 |
| 1-Methylguanidine | 1 | 3 |
| Betaine | 0 | 1 |
| Carnitine | 0 | 1 |
| Creatinine | 0 | 3 |
| Dimethylamine | 1 | 1 |
| Dimethylglycine | 0 | 1 |
| Lysine | 1 | 2 |
| Trigonelline | 0 | 1 |
| Trimethylamine N-oxide | 0 | 1 |
Figure 3.
Effect of anionic SNPs on 2D 13C-1H HSQC NMR spectrum of human urine sample. Three representative spectral regions demonstrate the cross-peak suppression by SNP addition. Red boxes indicate missing cross-peaks in the spectrum without SNPs (a, c, e) and with SNPs (b, d, f).
Discussion
A new direction to probe metabolite properties using nanoparticles
It has been reported that uncharged hydrophobic metabolites have a lower concentration in living cells due to their lower solubility and higher chance of leakage out of the membrane. Studying charged metabolites can potentially contribute to a better understanding of metabolic homeostasis.24 NMR spectroscopy is exquisitely sensitive to local magnetic fields reflected in chemical shifts, but it is much less sensitive to electric charges (pH titration experiments represent a notable exception).25 An electrophoretic NMR technique has been proposed that focuses on the charge properties of molecules using a specially designed NMR apparatus.26,27 However, for complex biological mixtures with hundreds of small molecules, this approach does not provide the necessary resolution. Therefore, existing charge information about molecules of interest cannot be easily used for the interpretation of NMR spectra of complex mixtures without change of pH.
With the introduction of charged nanoparticles described in this work, the net charge of metabolites is directly reflected in the NMR spectra through the appearance and disappearance of NMR cross-peaks. Differentiation between metabolites based on their electric charge is possible, if the following two conditions are fulfilled: 1) SNP-bound metabolites are restricted in their rotational Brownian diffusion causing the quenching of their NMR signals; 2) the nanoparticles neither contribute to the NMR spectrum nor perturb the NMR peaks of molecules that do not interact with them. The SNP-based strategy presented here does not require new or modified hardware.
Interactions between nanoparticles and metabolites in a biological system
Progress in nanoparticle research shows promise in many different areas, including biomedical imaging, disease diagnosis and treatment, such as targeted magnetic resonance imaging (MRI),28 photoacoustic imaging,29 point-of-care diagnosis using a “lab-on-the-chip”,30 drug delivery,31 and photo-based physical therapy.32 However, the toxicity of nanoparticles has always been a practical concern and its impact on nucleic acids,33,34 proteins35,36 or even intact cells37,38 have been discussed. Yet, very little is known about the effects of metabolome-nanoparticle interactions. In this work, the presence of specific interactions between nanoparticles and certain types of metabolites was established for human urine by NMR. Considering the abundance of many of these metabolites in other biofluids and biological systems, it is likely that in these systems SNPs interact with metabolites in the same way. The fact that changes in the pattern of the metabolome can modulate and disrupt biological pathways and biomolecular activities suggests that such effects might be relevant for toxicity evaluation of biomedical applications of nanomaterials. Conversely, the interaction of nanoparticles with charged small molecules could be used to detoxify and purify biological as well as artificial chemical mixtures.
In the present work, anionic SNPs were used whose silanol (Si-OH) groups on the surface are partially deprotonated at neutral pH leading to a negative net charge,39 which is stabilized by the presence of counterions. Two categories of metabolites were found here to have a strong tendency to bind to anionic SNPs. The first category consists of metabolites with a net positive charge experiencing an attractive electrostatic interaction to the negatively charged SNPs. The positively charged metabolites often contain a neutral basic group, which after proton acceptance becomes positively charged. Examples include lysine, arginine and histidines whose NMR peaks became suppressed after binding to the much larger nanoparticles. In addition, metabolites with a quaternary nitrogen that has a positive charge also belong to this category, such as choline, betaines, and trigonelline. The second category consists of metabolites that are neutral, but which have multiple methyl groups. Especially those molecules with multiple methyl groups that are connected to tertiary amine group have a relatively strong tendency to bind to SNPs. Similar observations were made for single-stranded DNA bound to negatively charged nanoparticles where the nitrogen atoms of each nucleotide were proposed as the binding sites.40 The addition of charged nanoparticles to an NMR sample can be likened to the conversion of the NMR sample into an affinity column analogous to solid-phase extraction whereby the resins are micro- or nano-sized silica particles. We focused on silica nanoparticles in this work, but quantum dots or other nanoparticles, such as gold or silver based nanoparticles, are likely to work as well.
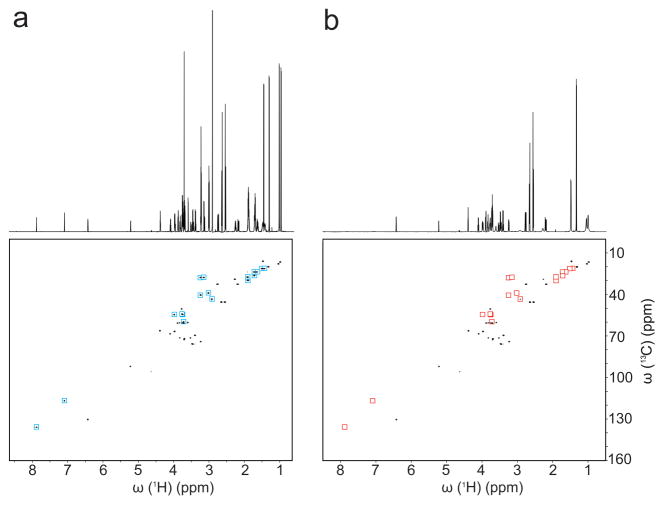
To test whether our approach also works with an opposite role of charges, namely with cationic SNPs interacting with anionic metabolites, cationic SNPs were added to the model mixture. As can be seen in Figure 4, the NMR signals of the negatively charged metabolites of the mixture, namely citric acid, lactic acid, and shikimic acid were either entirely suppressed or strongly weakened (less than 5% peak intensity left). Non-charged metabolites showed some line-broadening effects, similar to the anionic SNP case, which might be due a moderate increase in overall viscosity of the SNP solution.
Figure 4.
2D 13C-1H HSQC spectra of ten-compound model mixture a) without and b) with cationic SNPs. Red and blue squares highlight the cross-peaks that are suppressed by the presence of SNPs.
The challenge of spectral interpretation in NMR-based metabolomics
NMR-based metabolomics studies benefit from the possibility to do conduct metabolomics studies with little or no separation prior to doing the NMR experiments. Such an untargeted approach allows one to keep the metabolome as intact as possible, which is important for many studies. On the downside, the resulting NMR spectra, especially 1D spectra, can be overcrowded so that peak identification becomes notably hard. The 2D 1H-13C HSQC experiment belongs to the most popular approaches in 2D NMR-based metabolomics, because the cross-peaks are greatly dispersed along the second dimension. Nevertheless, spectral regions can still exist with significant spectral overlap. For example, in the region c) of Figure 3 many peaks are so close to each other that they cannot be discriminated. However, the presence of cationic SNPs causes the disappearance of a subset of cross-peaks significantly facilitating the analysis of the remaining ones. Since the 13C-1H HSQC spectrum does not provide information which cross-peaks belong to the same molecule, it is possible that two cross-peaks that belong to two different metabolites, not contained in NMR databases, are assigned to a single metabolite in the databases that happens to have two cross-peaks at approximately the same positions. Since the interaction of a metabolite with SNPs leads to the removal of all cross-peaks belonging to the metabolite, disappearance of only one of the two cross-peaks implies that the two cross-peaks cannot belong to the same metabolite. In this way, the addition of SNPs can disambiguate tentative peak assignments both in 1D and 2D NMR spectra. For example, the NMR peaks of arginine are often very close to or overlap with those of other metabolites of a complex mixture. When querying the spectrum against the database, arginine could not be identified. By contrast, when querying only the subgroup of peaks that in the presence of the SNPs were suppressed, arginine could be identified with a matching ratio of 0.8 and the uniqueness of 3/5 (one of the broad multiplets of a CH2 group was too weak to observe). On the other hand, metabolites that were identified based on the complete cross-peak list of the SNP-free sample, but not based on the two complementary groups of cross-peaks (suppressed vs. unaltered) are likely false positive hits requiring further analysis.
Conclusion
Metabolomics is a research field where nanomaterials have yet to make a significant impact. Here, we introduced a strategy for NMR-based metabolomics by the selective, charge-based editing of the NMR spectra of the metabolites through the addition of charged nanoparticles. This represents a fast, easy, and cost-effective alternative to other metabolite separation approaches. It allows one to highlight subset of metabolites in a complex mixture depending on molecular properties including their net charge. Specifically, combining the NMR results from a nanoparticle-free sample, a sample with anionic, and one with cationic nanoparticles permits the unambiguous distinction and identification of neutral, negatively charged, and positively charged metabolites.
The approach presented here bridging metabolomics with nanoscience provides benefits on different levels. NMR has traditionally not been considered as a sensitive detector of molecular electric charges, but the present application demonstrates how such information can be readily obtained. Additionally, NMR-based metabolomics has been limited by ambiguous peak assignments. With the help of charged nanoparticles, complicated NMR spectra, such as ones from urine samples, become simpler promising their more accurate and more efficient interpretation. Last but not the least, this study also casts light on the possibility of heterogeneous interactions between metabolites and nanoparticles in living organism by changing the composition of the metabolome with potential implications for biological function. We expect that the nanoparticle-based editing approach will prove useful for the analysis of a wide range of complex mixtures in metabolomics and elsewhere.
Supplementary Material
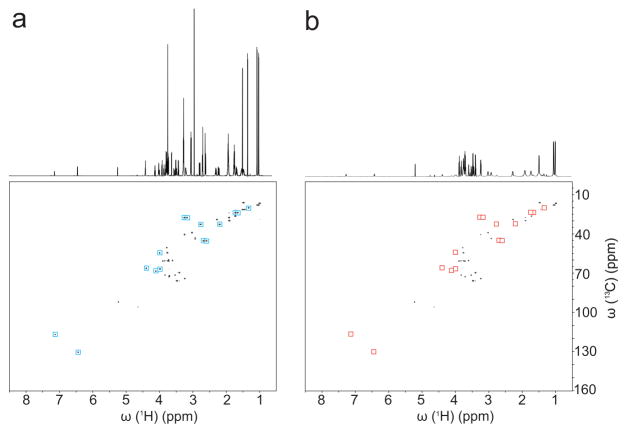
Figure 2.
1D 1H and 2D 13C-1H HSQC spectra of ten-compound model mixture a) without and b) with anionic SNPs. Red and blue squares highlight the cross-peaks that are suppressed by the presence of SNPs.
Table 2.
Effect of anionic SNPs on detection of amino acids in human urine sample.a
| Amino acid | Charge | Suppressed |
|---|---|---|
| Alanine | 0 | No |
| Arginine | +1 | Yes |
| Asparagine | 0 | No |
| Aspartic acid | −1 | No |
| Glutamic acid | −1 | No |
| Glutamine | 0 | No |
| Glycine | 0 | No |
| Lysine | +1 | Yes |
| Serine | 0 | No |
| Threonine | 0 | No |
| Tyrosine | 0 | No |
| Valine | 0 | No |
Metabolites whose NMR spectrum was suppressed by SNPs in the urine sample. Other amino acids were either not present or below the detection limit.
Acknowledgments
We thank Eka Chemicals for a generous gift of the anionic silica nanoparticles. This work was supported by the National Institutes of Health (grant R01 GM 066041 and SECIM grant U24 DK097209-01A1).
Footnotes
Supporting Information Available
Additional information about SNP measurements and properties (TEM, ζ-potential, DSL); 1D 1H NMR spectra of independently prepared replicate samples with quantitative analysis. This information is available free of charge via the Internet at http://pubs.acs.org/.
References
- 1.Nicholson JK, Lindon JC. Nature. 2008;455:1054–1056. doi: 10.1038/4551054a. [DOI] [PubMed] [Google Scholar]
- 2.Fiehn O. Plant Mol Biol. 2002;48:155–171. [PubMed] [Google Scholar]
- 3.Bingol K, Brüschweiler R. Anal Chem. 2014;86:47–57. doi: 10.1021/ac403520j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tang J. Curr Genomics. 2011;12:391–403. doi: 10.2174/138920211797248619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang A, Sun H, Wang P, Han Y, Wang X. J Proteomics. 2012;75:1079–1088. doi: 10.1016/j.jprot.2011.10.027. [DOI] [PubMed] [Google Scholar]
- 6.Wu H, Southam AD, Hines A, Viant MR. Anal Biochem. 2008;372:204–212. doi: 10.1016/j.ab.2007.10.002. [DOI] [PubMed] [Google Scholar]
- 7.Halouska S, Fenton RJ, Barletta RG, Powers R. ACS Chem Biol. 2012;7:166–171. doi: 10.1021/cb200348m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bingol K, Bruschweiler-Li L, Li DW, Brüschweiler R. Anal Chem. 2014;86:5494–5501. doi: 10.1021/ac500979g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Braunschweiler L, Ernst RR. J Magn Reson. 1983;53:521–528. [Google Scholar]
- 10.Bingol K, Brüschweiler R. Anal Chem. 2011;83:7412–7417. doi: 10.1021/ac201464y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bingol K, Zhang F, Bruschweiler-Li L, Brüschweiler R. J Am Chem Soc. 2012;134:9006–9011. doi: 10.1021/ja3033058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Correa J, Pinto LF, Riguera R, Fernandez-Megia E. Anal Chem. 2015;87:760–767. doi: 10.1021/ac5037186. [DOI] [PubMed] [Google Scholar]
- 13.Tayyari F, Gowda GA, Gu H, Raftery D. Anal Chem. 2013;85:8715–8721. doi: 10.1021/ac401712a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bloembergen N, Purcell EM, Pound RV. Phys Rev. 1948;73:679–712. [Google Scholar]
- 15.Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. J Biomol NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 16.Zhang F, Robinette SL, Bruschweiler-Li L, Brüschweiler R. Magn Reson Chem. 2009;47:S118–122. doi: 10.1002/mrc.2486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bingol K, Zhang F, Bruschweiler-Li L, Brüschweiler R. Anal Chem. 2012;84:9395–9401. doi: 10.1021/ac302197e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ganti S, Weiss RH. Urol Oncol. 2011;29:551–557. doi: 10.1016/j.urolonc.2011.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim Y, Koo I, Jung BH, Chung BC, Lee D. BMC Bioinf. 2010;11:S4. doi: 10.1186/1471-2105-11-S2-S4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang Y, Zhao F, Deng Y, Zhao Y, Ren H. J Proteome Res. 2015;14:1752–1761. doi: 10.1021/pr5011263. [DOI] [PubMed] [Google Scholar]
- 21.Bingol K, Li DW, Bruschweiler-Li L, Cabrera OA, Megraw T, Zhang F, Brüschweiler R. ACS Chem Biol. 2015;10:452–459. doi: 10.1021/cb5006382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bingol K, Brüschweiler R. [accessed April 17, 2015];J Proteome Res [online early access] doi: 10.1021/acs.jproteome.5b00184. Published Online: April 17, 2015. http://pubs.acs.org/doi/abs/10.1021/acs.jproteome.5b00184?src=recsys. [DOI]
- 23.Bouatra S, Aziat F, Mandal R, Guo AC, Wilson MR, Knox C, Bjorndahl TC, Krishnamurthy R, Saleem F, Liu P, Dame ZT, Poelzer J, Huynh J, Yallou FS, Psychogios N, Dong E, Bogumil R, Roehring C, Wishart DS. PloS One. 2013;8:e73076. doi: 10.1371/journal.pone.0073076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bar-Even A, Noor E, Flamholz A, Buescher JM, Milo R. PLoS Comput Biol. 2011;7:e1002166. doi: 10.1371/journal.pcbi.1002166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tataurova Y, Sealy MJ, Larsen RG, Larsen SC. J Phys Chem Lett. 2012;3:425–429. doi: 10.1021/jz201582d. [DOI] [PubMed] [Google Scholar]
- 26.Griffiths PC, Paul A, Hirst N. Chem Soc Rev. 2006;35:134–145. doi: 10.1039/b501286b. [DOI] [PubMed] [Google Scholar]
- 27.He QH, Liu YM, Nixon T. J Am Chem Soc. 1998;120:1341–1342. [Google Scholar]
- 28.Na HB, Song IC, Hyeon T. Adv Mater. 2009;21:2133–2148. [Google Scholar]
- 29.Yang X, Stein EW, Ashkenazi S, Wang LV. Wiley Interdiscip Rev.: Nanomed Nanobiotechnol. 2009;1:360–368. doi: 10.1002/wnan.42. [DOI] [PubMed] [Google Scholar]
- 30.Bellan LM, Wu D, Langer RS. Wiley Interdiscip Rev.: Nanomed Nanobiotechnol. 2011;3:229–246. doi: 10.1002/wnan.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.De Jong WH, Borm PJ. Int J Nanomed. 2008;3:133–149. doi: 10.2147/ijn.s596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Menon JU, Jadeja P, Tambe P, Vu K, Yuan B, Nguyen KT. Theranostics. 2013;3:152–166. doi: 10.7150/thno.5327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.An H, Jin B. Biotechnol Adv. 2012;30:1721–1732. doi: 10.1016/j.biotechadv.2012.03.007. [DOI] [PubMed] [Google Scholar]
- 34.Melzak KA, Sherwood CS, Turner RFB, Haynes CA. J Colloid Interface Sci. 1996;181:635–644. [Google Scholar]
- 35.Arvizo RR, Giri K, Moyano D, Miranda OR, Madden B, McCormick DJ, Bhattacharya R, Rotello VM, Kocher JP, Mukherjee P. PloS One. 2012;7:e33650. doi: 10.1371/journal.pone.0033650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pino Pd, Pelaz B, Zhang Q, Maffre P, Nienhaus GU, Parak WJ. Mater Horiz. 2014;1:301–313. [Google Scholar]
- 37.Cheng LC, Jiang X, Wang J, Chen C, Liu RS. Nanoscale. 2013;5:3547–3569. doi: 10.1039/c3nr34276j. [DOI] [PubMed] [Google Scholar]
- 38.Dowding JM, Das S, Kumar A, Dosani T, McCormack R, Gupta A, Sayle TX, Sayle DC, von Kalm L, Seal S, Self WT. ACS Nano. 2013;7:4855–4868. doi: 10.1021/nn305872d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rimola A, Costa D, Sodupe M, Lambert JF, Ugliengo P. Chem Rev. 2013;113:4216–4313. doi: 10.1021/cr3003054. [DOI] [PubMed] [Google Scholar]
- 40.Wishart DS, Knox C, Guo AC, Eisner R, Young N, Gautam B, Hau DD, Psychogios N, Dong E, Bouatra S, Mandal R, Sinelnikov I, Xia J, Jia L, Cruz JA, Lim E, Sobsey CA, Shrivastava S, Huang P, Liu P, Fang L, Peng J, Fradette R, Cheng D, Tzur D, Clements M, Lewis A, De Souza A, Zuniga A, Dawe M, Xiong Y, Clive D, Greiner R, Nazyrova A, Shaykhutdinov R, Li L, Vogel HJ, Forsythe I. Nucleic Acids Res. 2009;37:D603–610. doi: 10.1093/nar/gkn810. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.