Abstract
Because of its key role in secondary phenylpropanoid metabolism, Phe ammonia-lyase is one of the most extensively studied plant enzymes. To provide a basis for detailed structure–function studies, the enzyme from parsley (Petroselinum crispum) was crystallized, and the structure was elucidated at 1.7-Å resolution. It contains the unusual electrophilic 4-methylidene-imidazole-5-one group, which is derived from a tripeptide segment in two autocatalytic dehydration reactions. The enzyme resembles His ammonia-lyase from the general His degradation pathway but contains 207 additional residues, mainly in an N-terminal extension rigidifying a domain interface and in an inserted α-helical domain restricting the access to the active center. Presumably, Phe ammonia-lyase developed from His ammonia-lyase when fungi and plants diverged from the other kingdoms. A pathway of the catalyzed reaction is proposed in agreement with established biochemical data. The inactivation of the enzyme by a nucleophile is described in detail.
INTRODUCTION
Phe ammonia-lyase (PAL; EC 4.3.1.5) is at the gateway from the primary metabolism into the important secondary phenylpropanoid metabolism in plants (Hahlbrock and Scheel, 1989). PAL catalyzes the nonoxidative elimination of ammonia from l-Phe to give trans-cinnamate (Figure 1A). Trans-cinnamate is the precursor of numerous phenylpropanoid compounds that fulfill various essential functions as mechanical supports (lignins) (Whetten and Sederoff, 1995), as protectants against biotic and abiotic stress (antipathogenic phytoalexins, antioxidants, and UV-absorbing compounds) (Dixon and Paiva, 1995), as pigments like the anthocyanins (Holton and Cornish, 1995), and in signaling as, for instance, flavonoid nodulation factors (Weisshaar and Jenkins, 1998). Besides its important role in plant development, PAL is also a key enzyme in plant stress response. Its biosynthesis is stimulated on pathogenic attack, tissue wounding, UV irradiation, low temperature, or low levels of nitrogen, phosphate, or iron (Dixon and Paiva, 1995). The enzyme is accumulated in the vicinity of the affected tissue (Mauch-Mani and Slusarenko, 1996; Ehness et al., 1997). Because PAL is ubiquitously distributed in plants and absent in animals, it is a promising target for herbicides; various inhibitors are currently being developed (Laber et al., 1986; Appert et al., 2003).
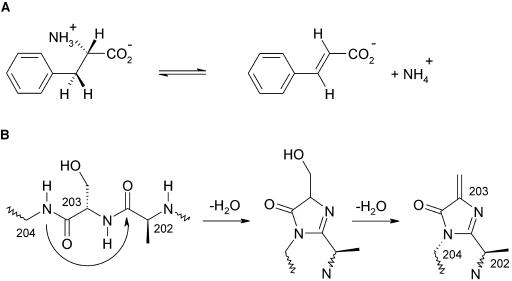
Figure 1.
Nonoxidative Deamination of Phe.
(A) Reaction catalyzed by PAL from plants and fungi resulting in trans-cinnamate and ammonia. Besides, phenylalanine is processed by other enzymes to Tyr and to phenylpyruvate.
(B) Proposed autocatalytic formation of MIO from the tripeptide 202Ala-Ser-Gly by two water elimination steps. The sp3 conformation at 204-N is taken from the wild-type structure of HAL (Schwede et al., 1999). It is probably enforced by the surrounding polypeptide, increasing the electrophilicity of the 203-Cβ atom.
The unusual nonoxidative deamination reaction of PAL requires an electrophilic group in the enzyme, which is not available among the 20 standard amino acid residues. Because PAL lacks a cofactor to provide such an electrophilic group, it was suggested that the electrophile is produced by a posttranslational dehydration of a Ser to form a dehydroalanine (Hanson and Havir, 1970). The Ser was later identified as Ser203 (Schuster and Retey, 1994). On elucidation of the crystal structure of the homologous enzyme His ammonia-lyase (HAL), however, the proposed dehydroalanine turned out to be part of a 4-methylidene-imidazole-5-one (MIO) group (Schwede et al., 1999), which is formed autocatalytically by cyclization and dehydration of an Ala-Ser-Gly tripeptide as shown in Figure 1B (Baedeker and Schulz, 2002a). In a sequence alignment between HAL and PAL, residues 202Ala-Ser-Gly of PAL were assigned to form the MIO (Schwede et al., 1999) in agreement with the identified Ser203. A closely related autocatalytic tripeptide cyclization was detected earlier with the green fluorescent protein (Ormö et al., 1996; Barondeau et al., 2003) which is, however, neither sequence- nor structure-related to HAL or PAL.
A medical application of PAL arose in context with the human genetic disease phenylketonuria that leads to severe mental retardation (Levy, 1999; Sarkissian et al., 1999). It has been shown in vivo that PAL can be used to convert poisonous excess Phe in the blood into the harmless compounds trans-cinnamate and ammonia so that PAL may become a useful palliative for phenylketonuria (Ambrus et al., 1978; Safos and Chang, 1995). In a further application, PAL is used for producing the essential amino acid l-Phe from trans-cinnamate and ammonia (Yamada et al., 1981; Evans et al., 1987) and for the synthesis of numerous l-arylalanines (Gloge et al., 2000). Here, we report the crystal structure of the key plant enzyme PAL at high resolution and suggest a reaction pathway for the catalysis.
RESULTS AND DISCUSSION
Crystal Structure
For protein production, we used a gene of PAL that had been adapted to Escherichia coli by numerous silent mutations (Baedeker and Schulz, 1999). The redesigned gene was cloned into vector pT7-7, transformed into E. coli strain BL21(DE3), and expressed in the cytosol. Narrow peaks were cut out during the chromatographic steps to obtain a pure and homogeneous enzyme. The crystals grew from an unbuffered solution within a couple of days, assuming feather-like shapes without a planar surface as depicted in the insert of Figure 2. However, these feathers were single crystals that diffracted to high resolution.
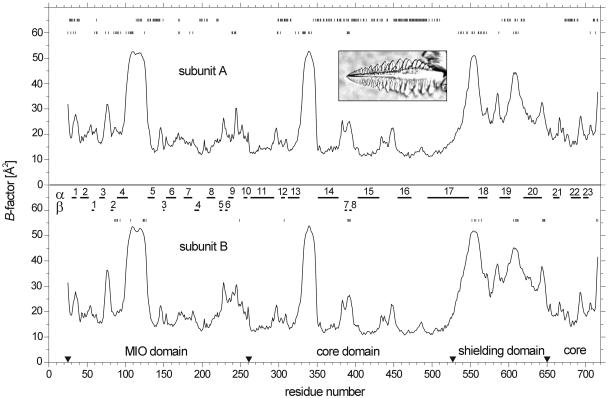
Figure 2.
B-Factor Distributions of the Two Crystallographically Independent Subunits of the Tetrameric PAL Together with the Secondary Structures.
The domain borders are marked by arrowheads. The residues in the molecular interfaces (top line) and in the crystal packing contacts involving subunit A (second line) and subunit B (third line) are marked. The exceptional habit of a typical single crystal of PAL is shown in the insert (crystal length = 400 μm).
For phasing, we produced selenium-labeled protein using a Met-auxotrophic E. coli strain. In electrospray ionization mass spectrometry, the labeled PAL showed a mass increase of 976 D with respect to a parallel measurement of wild-type PAL. This difference is close to the expected value of 985 D for the incorporation of all 21 seleno-Met. Labeled PAL yielded the same crystals as the wild type. One labeled crystal was used in a multiwavelength anomalous diffraction (MAD) experiment reaching 2.2-Å resolution (Table 1). The MAD data revealed 34 clear selenium sites, which were then used for phasing, giving rise to an electron density map that was subsequently improved by solvent flattening and histogram matching. The quality of the resulting map sufficed to build a model of two PAL subunits in the crystallographic asymmetric unit, which was confirmed by all selenium positions being at Met. The model was then transferred to the wild-type crystals and refined to 1.7-Å resolution, yielding quality indices in a favorable range (Table 1).
Table 1.
Crystallographic Data Statistics
| Data Collectiona | SeMet Labeled | Wild Type | ||
|---|---|---|---|---|
| Wavelength [Å] | Peak 0.9798 | Inflection 0.9800 | Remote 0.9824 | 0.8200 |
| Resolution [Å] | 50–2.0 (2.10–2.00) | 50–2.2 (2.33–2.20) | 50–2.2 (2.33–2.20) | 50–1.7 (1.76–1.70) |
| Observations | 755,820 | 547,977 | 550,577 | 1,113,883 |
| Unique reflections | 94,213 | 132,273 | 68,083 | 156,788 |
| Completeness (%) | 98.9 (93.9) | 98.6 (92.0) | 98.7 (92.4) | 96.8 (88.2) |
| Multiplicity | 8.0 (6.9) | 4.1 (3.7) | 8.1 (7.2) | 7.1 (6.0) |
| Rsym-I [%] | 9.2 (42) | 7.0 (34) | 8.7 (42) | 4.9 (39) |
| Average I/σI | 14.5 (5.4) | 13.4 (5.0) | 17.0 (6.9) | 21.4 (4.5) |
| Refinement of the Wild Type | ||||
| Resolution range [Å]/number of protein atoms/water molecules | 50–1.7/10,576/1,072 | |||
| Average B-factors: main chain [Å2]/all atoms [Å2] | 23.4/25.2 | |||
| Rcryst [%]/Rfree [%]/rmsd bond lengths [Å]/rmsd bond angles [°] | 16.6/19.9/0.014/1.40 | |||
| Ramachandran angles: most favored region [%]/allowed region [%] | 90.0/10.0 | |||
Values in parentheses refer to the highest resolution shells; rmsd, root mean squares deviation.
Both crystals belonged to space group C2221 with cell dimensions a = 119.8 Å, b = 160.5 Å, and c = 141.3 Å, containing two subunits of 77,818 D per asymmetric unit. The phases were determined using the MAD method. The figure of merit was 0.44 after phasing with MLPHARE and 0.90 after the refinement.
The model comprises 692 of the 716 amino acid residues in each of the two asymmetric subunits (Figure 3A). These two subunits resemble each other closely as indicated by their B-factor distributions (Figure 2). A structural comparison resulted in an rmsd of 0.51 Å for the Cα atoms, with the largest deviations at packing contacts. Because of lacking electron density, residues 1 to 24 could not be modeled. Moreover, residues 104 to 128 and 334 to 347 had only weak density support in both asymmetric units, resulting in high B-factors (Figure 2). The tetrameric structure of PAL is clearly defined in the crystal, confirming gel permeation chromatography data in solution. The tetramer shows the common D2 symmetry (Figure 3B). One of the three molecular twofold axes is crystallographic, and the others deviate by 20° from the nearest translation vector of the orthorhombic unit cell. The four subunits are tightly interconnected. Each subunit has a surface of 30,330 Å2 and buries 29% of it in contacts of 4215 Å2, 3660 Å2, and 880 Å2 with the other subunits. Consequently, a single subunit is most likely unstable in solution. The interface residues are specified in Figure 2. The packing of PAL in the crystal is very dense, giving rise to a Vm of 2.18 Å3/D, which is at the lower end of the distribution for proteins of this size (Kantardjieff and Rupp, 2003). Despite dense packing, the involved contacts bury merely 7215 Å2 or 8% of each PAL tetramer surface of 86,940 Å2. The residues in packing contacts are marked in Figure 2.
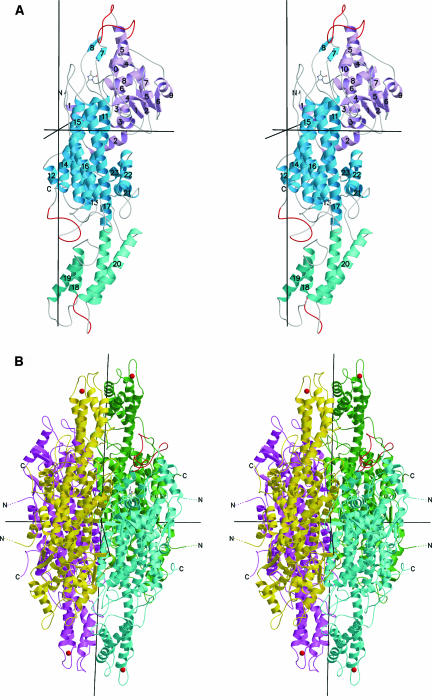
Figure 3.
Stereo Ribbon Plot of PAL.
(A) One subunit of the D2-symmetric PAL homotetramer with numbered secondary structure elements. The chain is color coded for the MIO domain (pink), the core domain (blue), and the inserted shielding domain (cyan). The highly mobile loops at residues 110, 340, and 550 are red.
(B) Full tetramer with view into the active center of the blue subunit as indicated by the ball-and-stick model of MIO. The vertical twofold axis is crystallographic. The blue active center is covered by the shielding domains of the green and yellow subunits and by the two highly mobile loops (red) at positions 110 from the blue subunit and 340 from the green subunit. Thr549 corresponding to the phosphorylated Thr of the closely homologous PAL from French beans (82% sequence identity; Allwood et al., 1999) is marked by a red ball at the Cα position.
Molecular Architecture
PAL is a predominantly α-helical protein with 52% of the residues in 23 α-helices and only 5% of the residues in eight β-strands. A chainfold superposition with HAL shows that each PAL subunit should be subdivided into four domains as indicated by arrowheads in Figure 2, by colors in Figure 3A, and in sequence detail in Figure 4. The mobile N-terminal 24-residue peptide occurs only in PAL and not in HAL. Such extensions are widespread; they may anchor the enzyme at other cell components. The MIO domain (residues 25 to 261) is also present in HAL, although HAL lacks helices α1 and α2, which fortify the interface to the core domain (residues 262 to 527 and 650 to 716). The core domain is also present in HAL, whereas the inserted shielding domain (residues 528 to 649) is a specialty of PAL. Further additions in PAL, mostly small insertions into external loops, are detailed in the structure-based sequence alignment of Figure 4. The size difference between PAL and HAL is demonstrated in Figure 5. The most prominent addition of PAL with respect to HAL is the shielding domain, two of which form an arch over the active center, restricting substrate access to a narrow channel.
Figure 4.
Structure-Based Sequence Alignment of the Reported PAL from Parsley (Top Line) with HAL from Pseudomonas putida.
Every tenth residue is marked by a dot or comma wherever possible. The secondary structure is from PAL. Lower-case letters indicate lack of structure. The underlined residues align within a 3-Å distance cutoff for Cα atoms. The asterisks denote 97 strictly conserved positions within the PAL family (above the line) and 65 within the HAL family (below). Thr549 corresponding to the phosphorylated Thr of the PAL from French beans (Allwood et al., 1999) is black/white inverted. The alignment results in 144 identical residues (not marked).
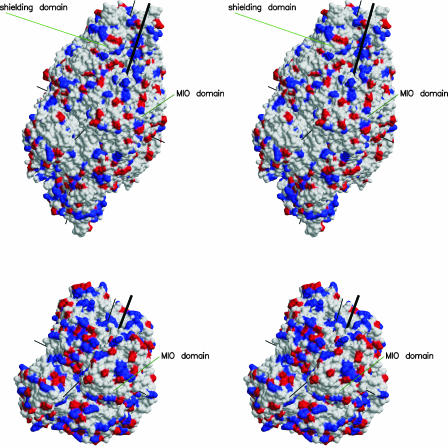
Figure 5.
Stereoview of the Surfaces of the PAL Tetramer (Top) and the HAL Tetramer in the Same Orientation, Which Is Roughly That of Figure 3.
The surfaces show negative (red) and positive (blue) electrostatic charges. Thin black lines denote the three twofold axes. The entrances to the active centers are marked by thick black lines. The MIO domain and the shielding domain are indicated.
The B-factor plot of Figure 2 reveals two highly mobile segments around positions 110 and 340. Both loops are located at the entrance of the active center. In particular, loop 110 is near the MIO of the same subunit, whereas loop 340 is near the MIO of the other subunit in the crystallographically asymmetric unit. Both are marked in Figure 3. The same highly mobile loops are known from HAL (Baedeker and Schulz, 2002b), indicating that their mobility is required for catalysis. Figure 2 shows further that the whole shielding domain is very mobile, although it is tightly connected to the core domain through the exceptionally long 55-residue helix α17. Interestingly, the detected phosphorylation site of PAL from French beans (Phaseolus vulgaris) (Allwood et al., 1999) is located at the end of this helix as marked in Figures 3B and 4. This position has the highest mobility of the shielding domain (Figure 2). It seems quite possible that the phosphorylation shifts this domain, changing the access to the active center.
Prosthetic Group MIO
The active center of PAL is defined by the location of MIO, which is depicted in Figure 6. The autocatalytic formation of MIO from the polypeptide is most likely achieved by exerting mechanical pressure during the folding process as demonstrated in HAL (Baedeker and Schulz, 2002a). When fitting MIO to the electron density, however, we found appreciable density in front of its electrophilic carbon atom 203-Cβ, which could not be explained by water molecules. Moreover, the MIO atom 204-N assumed an sp2 conformation like the corresponding atom of HAL inhibited by a Cys-MIO adduct (Baedeker and Schulz, 2002b). This sp2 conformation is quite distinct from the sp3 conformation assumed in wild-type HAL (Schwede et al., 1999).
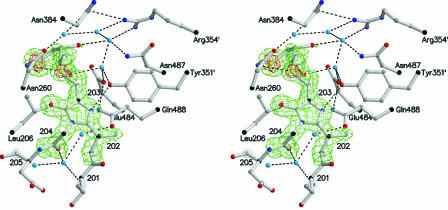
Figure 6.
Stereoview of the DTT-MIO Adduct Showing the Simulated Annealing Omit (Fo-Fc)-Electron Density Map Contoured at 3 σ (Green) and at 6 σ (Red).
Hydrogen bonds are indicated by dashed lines. The water molecules and several side chains at the active center are depicted. The three water molecules below MIO participate in the autocatalytic formation of the prosthetic group MIO (Baedeker and Schulz, 2002a).
The prepared PAL was always kept in 5 mM DTT to avoid intermolecular disulfide bonds that may have hindered crystallization. Therefore, we concluded that the analyzed crystal contained a nucleophilic DTT bound to the electrophilic MIO. This conclusion is supported by the electron density distribution in front of MIO that was modeled as a covalently bound DTT showing the high densities of the sulfur atoms (Figure 6). It is further corroborated by the sp2 conformation of the 204-N atom that corresponds to a similarly modified HAL. It agreed also with the electrospray ionization mass spectrometry value for our wild-type PAL that was 163 D above the expected value and with the electrospray ionization mass spectrometry value for selenomethionine-labeled PAL that was 152 D above expectation. Both differences fit the 154-D mass of DTT very well.
The DTT derivative was finally confirmed by enzyme kinetics. Wild-type PAL showed a specific activity of 1.9 units/mg with a Km of 68 μM, which is in the commonly observed range (Baedeker and Schulz, 1999). When 0.01 to 2.0 mM DTT was added to wild-type PAL, we observed that Vmax did not change appreciably, indicating a competitive type inhibition with a Ki of 100 μM. Furthermore, in repeated measurements over a whole day, we observed that PAL incubation with 0.5 mM DTT at activity test conditions resulted in a linear decay to 25% of the original activity, whereas a reference without DTT decreased merely to 90%. Moreover, a measurement of a dissolved crystal showed no activity above the detection level, which was 2% of the wild-type activity. These findings demonstrate that DTT binds to MIO, resulting in a reversible competitive-type inhibition over short periods. After an incubation of several hours, however, the compound is probably oxidized by dioxygen, resulting in an irreversible inhibition. A similar oxidation occurs in the green fluorescent proteins (Barondeau et al., 2003).
Therefore, we suggest that the inactivated PAL of the crystal contains an oxidized DTT-MIO adduct, which is a MIO (Figure 1B) with a sulfur-substituted 203-Cβ atom. Obviously, the sulfur substitution at 203-Cβ changes the electronic structure of MIO to an sp2 conformation at the 204-N atom, which by itself, should also prefer sp2 because it forms an amide with 203-O. Presumably, the sp3 conformation of 204-N observed in wild-type HAL and expected in wild-type PAL is probably enforced by the polypeptide as it increases the electrophilicity of the 203-Cβ atom and thus promotes the enzymatic reaction.
Reaction Pathway
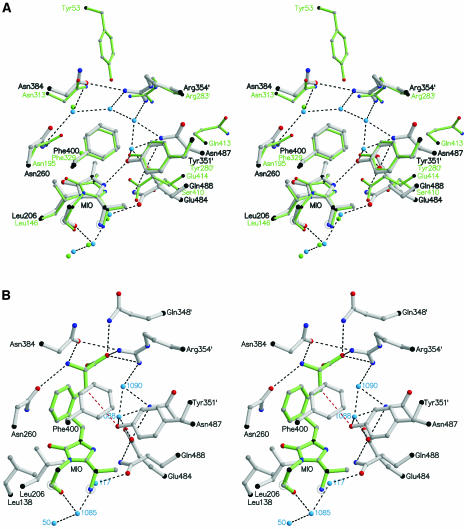
After the structure of HAL was elucidated, a sequence comparison suggested a similar active center arrangement and, therefore, a similar reaction pathway for PAL (Schwede et al., 1999). Now, a superposition of the two active centers confirms the close relationship as shown in Figure 7A. To explain the catalysis, we modeled an l-Phe adduct of PAL (Figure 7B). In this model, the amino group is bound to Asn260 and Asn384. The carboxylate is accommodated by Gln348′ and Arg354′ from the other subunit and probably by Tyr110 from the same subunit as in the modeled His-HAL adduct (Baedeker and Schulz, 2002b). However, Tyr110 is on a highly mobile loop and is displaced in the PAL crystal structure. The substrate phenyl group fits rather tightly but may be relaxed by an induced fit on substrate binding. Such an induced fit seems likely because the two highly mobile loops around positions 110 and 340 at the active center should be structured during catalysis. However, a decision about this proposal depends on further structure analyses.
Figure 7.
Stereoview of the Catalytic Center.
Hydrogen bonds are indicated by black dashed lines.
(A) Superposition of the structures of PAL and HAL (green) based on the 34 Cα atoms within a 13-Å distance of the 203-Cβ atom of MIO.
(B) Model of the reaction intermediate showing the expected position of l-Phe (green). The HS proton is abstracted by Tyr351′ (red dashed line) and released via Wat1088 and Glu484 to the bulk solvent.
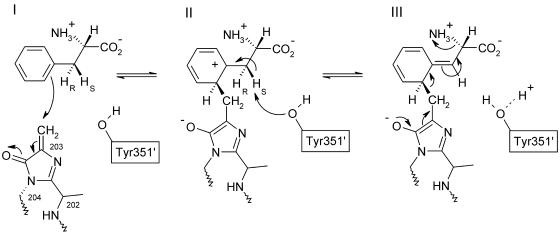
The suggested reaction pathway is shown in Figure 8. First, an electron pair of the phenyl ring attacks the 203-Cβ atom of MIO (state I). On this attack MIO becomes aromatic and the positively charged σ-complex of the substrate phenyl group is stabilized by the produced 203-O anion (state II). Moreover, the positive charge is stabilized by an interaction with the π-electrons of Phe400 (Figure 7B). The conversion of MIO to the aromatic state changes the conformation of 204-N from sp3 to sp2, causing a small peptide displacement, which was observed in HAL where it was easily accommodated. The electron-deficient phenyl ring of the substrate renders the two hydrogens at the Cβ atoms acidic, and the HS proton is abstracted by Tyr351′ from another subunit in agreement with isotope labeling experiments (Hermes et al., 1985). Tyr351′ is in turn hydrogen bonded via Wat1088 and Glu484 to the bulk solvent so that the proton is easily released. The following rearrangement of the Phe-MIO adduct in state III eliminates the amino group and regenerates MIO. The resulting ammonia is occluded by the other product trans-cinnamate and will be released into the solvent after the cinnamate has diffused away, as was suggested by kinetic data (Hermes et al., 1985).
Figure 8.
Mechanism Proposed for the Elimination of Ammonia from l-Phe as Catalyzed by PAL in Agreement with Biochemical Data (Langer et al., 1995).
Note the change of the 204-N atom geometry from an electrophilicity-enhancing sp3 conformation to sp2 in the aromatic ring. The products trans-cinnamate and ammonia are depicted in Figure 1A.
The active center residues of PAL were also confirmed by mutational studies guided by a homology model based on HAL (Röther et al., 2002). Most intriguingly, the mutation Tyr110→Phe resulted in a complete loss of activity, although this Tyr should not be that important for the reaction as it is expected to contact merely the substrate carboxylate group. However, because the introduced Phe110 is in a highly mobile loop (Figure 2), it may reach the active center to bind like the substrate and inhibit the enzyme.
The PAL and HAL Superfamily
A chainfold search in the Protein Data Bank using DALI (Holm and Sander, 1993) revealed that only HAL with a Z-score of 39.6 is significantly similar to PAL. The second best Z-score is 10.8 for an adenylosuccinate lyase, which does not indicate a significant relationship. Accordingly, PAL and HAL constitute a superfamily that is separated from all other known proteins by chainfold divergence. The PAL chainfold is essentially that of HAL supplemented by a 54-residue N-terminal extension, a 122-residue shielding domain, and some small insertions (Figures 3A and 4). A Sequence Data Bank search revealed ∼90 complete PAL sequences, ∼70 complete HAL sequences, and no other clearly sequence-related protein, confirming the delimitation of the superfamily at the sequence level.
The PAL sequences are from 46 plant and five fungi species as well as from one bacterium, whereas the HAL sequences are spread over all species. The bacterial PAL is special and will be discussed below. Using the reported PAL from parsley (Petroselinum crispum) as a reference, the most distant PAL is from a fungus and has 29% identical amino acid residues, whereas the closest HAL is from Pseudomonas syringe and shows only 25% identities. Because all HAL sequences are related to each other and have >32% identities in binary comparisons, the superfamily splits clearly into a PAL and a HAL family. The 97 and 65 residues strictly conserved within the PAL and HAL families, respectively, are marked in Figure 4. Among them, 31 are conserved in the superfamily.
The homology within the plant PAL species is above 60% identical residues. Several plants contain multiple isoenzymes. Parsley has the three isoenzymes PAL1 (reported here), PAL2, and PAL3, whereas Arabidopsis thaliana, for instance, has the four isoenzymes PAL1 to PAL4. Sequence comparisons among these seven isoenzymes showed that they cluster by species so that those within one species are more closely related to each other than to isoenzymes of the other species. Accordingly, the diversification occurred in each species separately.
PAL is known to accept Tyr as a poor substrate (Langer et al., 2001). This agrees with our model depicted in Figure 7B because there is some space left for the additional hydroxyl group. In maize (Zea mays), a PAL was found showing comparable catalytic efficiencies for Tyr and Phe (Rösler et al., 1997). This specificity shift cannot be explained easily by amino acid exchanges around the active center. Moreover, an efficient Tyr ammonia-lyase (TAL) with a low side activity for Phe was detected in Rhodobacter capsulatus, which uses the produced trans-p-hydroxycinnamic acid for chromophore synthesis (Kyndt et al., 2002). Again, the specificity change cannot be related to the residue exchanges.
Sequence alignments with the TAL from R. capsulatus indicated that it was developed from HAL rather than from PAL because it lacks the N-terminal extension and the shielding domain characteristic for the plant and fungi PAL (Figure 3A). Recently, a bacterial PAL was detected in Streptomyces maritimus, which uses the produced trans-cinnamic acid as a precursor for the antibiotic enterocin (Xiang and Moore, 2002). Again, sequence alignments revealed that the characteristic insertions were missing, indicating a closer relationship to HAL than to the plant and fungi PAL. The development of the two bacterial enzymes from HAL is further supported by the levels of residue identity because the TAL from R. capsulatus, the PAL from S. maritimus, and the HAL from P. putida (Schwede et al., 1999) show 34% identity in binary comparisons within this group and only 24% identity in comparisons with the PAL from parsley.
Taken together, HAL is the basic enzyme of the superfamily because it participates in a central metabolic pathway and is therefore ancient and distributed over numerous species. PAL and TAL were derived from HAL to fulfill special tasks. Applying merely amino acid exchanges, the bacterial PAL and TAL were developed directly from HAL and used for the syntheses of an antibiotic and a chromophore, respectively. The plant and fungi PAL was derived from HAL by inserting ∼200 residues in addition to numerous amino acid exchanges. Presumably, this happened in early evolution when fungi and plants established their secondary phenylpropanoid metabolism based on trans-cinnamic acid and diverged from the species of the other kingdoms.
CONCLUSION
The structure of PAL shows that this key plant enzyme is most likely an offspring of HAL from the central metabolic His degradation pathway. In contrast with HAL, the active center of the plant and fungi PAL is well shielded by a separate domain. The shielding domain restricts the access to the active center so that the risk of inactivation by nucleophiles in conjunction with dioxygen is minimized. This may help PAL to function, for instance, in stressed plant tissue. It should be noted that PAL forms its electrophilic prosthetic group autocatalytically from its own polypeptide, rendering it independent of any cofactor and thus facilitating its upregulation. In a medical application, the known surface of PAL can now be used for designing a recombinant nonimmunogenic enzyme for the treatment of the human genetic disease phenylketonuria.
METHODS
Enzyme Expression, Purification, Activity, and Crystallization
Because of inefficient codon usage, heterologous expression of the 2.2-kb gene of isoenzyme-1 of the PAL from parsley (Petroselinum crispum) in Escherichia coli failed to yield enough homogenous protein for a structure analysis (Schuster and Retey, 1995). Therefore, we used a gene especially designed for E. coli (Baedeker and Schulz, 1999). For expression, we took vector pT7-7-PAL in E. coli BL21(DE3) together with the Hsp60 chaperone-containing vector pREP4-groESL. The cells were grown at 37°C to OD600 0.8 using 2YT medium supplemented with 100 μg/mL of ampicillin (PAL plasmid) and 25 μg/mL of kanamycin (groESL plasmid). The temperature was then lowered to 20°C, and the expression was induced by adding 0.1 mM isopropylthio-β-d-thiogalactoside to run for 20 h. The cells were harvested, resuspended in 35 mL of buffer A (50 mM KH2PO4, pH 7.5, and 5 mM DTT) with 1 mM tosylfluorid (phenylmethylsulfonyl fluoride) and 500 units of Benzonase (Merck, Darmstadt, Germany), and sonicated for 10 min. After centrifugation (60 min at 48,000g), the supernatant was dialyzed overnight against buffer A and then applied to an anion-exchange column (Source 30Q; Pharmacia, Uppsala, Sweden), which was subsequently eluted using buffer A with a gradient of 0 to 2 M NaCl. PAL was identified by SDS-PAGE, separated, concentrated, and applied to a gel permeation column (Superdex 200-26/60; Pharmacia) equilibrated in buffer A with 600 mM NaCl and 3% (w/v) maltose. PAL-containing fractions were concentrated and dialyzed overnight against deionized water with 5 mM DTT. A typical yield was 15 to 20 mg PAL per liter of culture.
The activity of PAL was determined by monitoring the reaction product trans-cinnamate at 290 nm (ɛ290 = 10,000 M−1cm−1). The assay contained 0.1 M Tris-HCl, pH 8.8, and various concentrations of l-Phe and DTT in 1 mL of water and was run at 30°C. The reaction was started by adding 9.3 μg of PAL dissolved in 5 μL of water. The l-Phe and DTT concentrations were 0.01 to 10 mM and 0.01 to 2.0 mM, respectively.
Selenomethionine (SeMet)-labeled PAL was produced by transforming the expression vector pT7-7-PAL into the Met-auxotrophic E. coli strain B834(DE3) without cotransforming pREP4-groESL. Cells were cultured in LeMaster medium (Hendrickson et al., 1990) containing 25 mg/L of seleno-l-Met (Acros, Geel, Belgium). The cultivation and purification procedures were the same as with wild-type PAL. The preparation yielded ∼10 mg of SeMet-labeled PAL per liter of culture.
The protein was crystallized using the hanging drop vapor diffusion method at 20°C. After screening, suitable crystals were obtained from a 1:1 mixture of a 8-mg/mL protein solution with a reservoir solution containing 6 to 11% (w/v) polyethylene glycol 6000 and 30 to 60 mM MgCl2. Fewer and larger crystals grew if the reservoir solution was diluted with water to a volume of 120 to 160% after the drop had been placed. Wild-type and SeMet-labeled PAL crystallized under identical conditions and yielded similar crystals. The crystals appeared within 2 d and grew to final dimensions of 500 × 150 × 100 μm3. For data collection, the crystals were transferred in three steps to a solution containing 30% (w/v) glycerol, 18% (w/v) polyethylene glycol 6000, and 40 mM MgCl2 before they were mounted on a cryoloop and flash-frozen to 100 K in a nitrogen gas stream.
X-Ray Data Collection and Analysis
The data of one single SeMet-labeled PAL crystal at 100 K were collected at the Swiss Light Source (Villigen, Switzerland) at three different wavelengths, which were selected on the basis of an x-ray fluorescence spectrum taken from the same crystal. In addition, we collected a data set of a wild-type PAL crystal at 100 K. All data were processed with programs XDS and XSCALE (Kabsch, 1993). The initial selenium sites were determined with SHELX-D (Uson and Sheldrick, 1999). They were refined and used for initial phasing with MLPHARE (Collaborative Computational Project Number 4, 1994). Phase extension to 1.7 Å by density modification and noncrystallographic symmetry averaging and the construction of an initial model were performed with RESOLVE (Terwilliger, 2003). A second, independent model was built with ARP/wARP (Perrakis et al., 1997). The two protein models were combined and completed manually with XFIT (McRee, 1999). Water molecules were automatically introduced by the ARP/waters routine of REFMAC (Murshudov et al., 1997) and then edited using XFIT. The MIO group was generated and energy minimized using the PRODRG2 server (van Aalten et al., 1996). The protein model was refined using REFMAC without noncrystallographic symmetry restraint. Superpositions were done with LSQMAN (Kleywegt and Jones, 1995). The figures were produced with POVScript+ (Fenn et al., 2003) and Raster3D (Merritt and Bacon, 1997).
The atomic coordinates and structure factors are deposited in the Protein Data Bank under accession code 1W27. The accession codes for the sequences of Figure 4 are Y07654 (PAL) and M35140 (HAL). The accession code for the coordinates of HAL is 1B8F.
Acknowledgments
We thank the team of the Swiss Light Source (Villigen, Switzerland) for their help in data collection. The project was supported by the Deutsche Forschungsgemeinschaft under SFB-388.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Georg E. Schulz (georg.schulz@ocbc.uni-freiburg.de).
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.104.025288.
References
- Allwood, E.G., Davies, D.R., Gerrish, C., Ellis, B.E., and Bolwell, G.P. (1999). Phosphorylation of phenylalanine ammonia-lyase: Evidence for a novel protein kinase and identification of the phosphorylated residue. FEBS Lett. 457, 47–52. [DOI] [PubMed] [Google Scholar]
- Ambrus, C.M., Ambrus, J.L., Horvath, C., Pedersen, H., Sharma, S., Kant, C., Mirand, E., Guthrie, R., and Paul, T. (1978). Phenylalanine depletion for the management of phenylketonuria: Use of enzyme reactors with immobilized enzymes. Science 201, 837–839. [DOI] [PubMed] [Google Scholar]
- Appert, C., Zon, J., and Amrhein, N. (2003). Kinetic analysis of the inhibition of phenylalanine ammonia-lyase by 2-aminoindan-2-phosphonic acid and other phenylalanine analogues. Phytochemistry 62, 415–422. [DOI] [PubMed] [Google Scholar]
- Baedeker, M., and Schulz, G.E. (1999). Overexpression of a designed 2.2 kb gene of eukaryotic phenylalanine ammonia-lyase in Escherichia coli. FEBS Lett. 457, 57–60. [DOI] [PubMed] [Google Scholar]
- Baedeker, M., and Schulz, G.E. (2002. a). Autocatalytic peptide cyclization during chain folding of histidine ammonia-lyase. Structure 10, 61–67. [DOI] [PubMed] [Google Scholar]
- Baedeker, M., and Schulz, G.E. (2002. b). Structures of two histidine ammonia-lyase modifications and implications for the catalytic mechanism. Eur. J. Biochem. 269, 1790–1797. [DOI] [PubMed] [Google Scholar]
- Barondeau, D.P., Putnam, C.D., Kassmann, C.J., Tainer, J.A., and Getzoff, E.D. (2003). Mechanism and energetics of green fluorescent protein chromophore synthesis revealed by trapped intermediate structures. Proc. Natl. Acad. Sci. USA 100, 12111–12116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collaborative Computational Project Number 4 (1994). The CCP4 suite: Programs for protein crystallography. Acta Crystallogr. D 50, 760–763. [DOI] [PubMed] [Google Scholar]
- Dixon, R.A., and Paiva, N.L. (1995). Stress-induced phenylpropanoid metabolism. Plant Cell 7, 1085–1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehness, R., Ecker, M., Godt, D.E., and Roitsch, T. (1997). Glucose and stress independently regulate source and sink metabolism and defense mechanisms via signal transduction pathways involving protein phosphorylation. Plant Cell 9, 1825–1841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans, C.T., Hanna, K., Payne, C., Conrad, D., and Misawa, M. (1987). Biotransformation of trans-cinnamic acid to L-phenylalanine: Optimization of reaction conditions using whole yeast cells. Enzyme Microb. Technol. 9, 417–421. [Google Scholar]
- Fenn, T.D., Ringe, D., and Petsko, G.A. (2003). POVScript+: A program for model and data visualization using persistence of vision ray-tracing. J. Appl. Crystallogr. 36, 944–947. [Google Scholar]
- Gloge, A., Zon, J., Kövari, A., Poppe, L., and Retey, J. (2000). Phenylalanine ammonia-lyase: The use of its broad substrate specificity for mechanistic investigations and biocatalysis—synthesis of L-arylalanines. Chemistry 6, 3386–3390. [DOI] [PubMed] [Google Scholar]
- Hahlbrock, K., and Scheel, D. (1989). Physiology and molecular biology of phenylpropanoid metabolism. Annu. Rev. Plant Physiol. Plant Mol. Biol. 40, 347–369. [Google Scholar]
- Hanson, K.R., and Havir, E.A. (1970). L-phenylalanine ammonia-lyase. IV. Evidence that the prosthetic group contains a dehydroalanyl residue and mechanism of action. Arch. Biochem. Biophys. 141, 1–17. [DOI] [PubMed] [Google Scholar]
- Hendrickson, W.A., Horton, J.R., and LeMaster, D.M. (1990). Selenomethionyl proteins produced for analysis by multiwavelength anomalous diffraction (MAD): A vehicle for determination of three-dimensional structure. EMBO J. 9, 1665–1672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermes, J.D., Weiss, P.M., and Cleland, W.W. (1985). Use of nitrogen-15 and deuterium isotope effects to determine the chemical mechanism of phenylalanine ammonia-lyase. Biochemistry 24, 2959–2967. [DOI] [PubMed] [Google Scholar]
- Holm, L., and Sander, C. (1993). Protein structure comparison by alignment of distance matrices. J. Mol. Biol. 233, 123–138. [DOI] [PubMed] [Google Scholar]
- Holton, T.A., and Cornish, E.C. (1995). Genetics and biochemistry of anthocyanin biosynthesis. Plant Cell 7, 1071–1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabsch, W. (1993). Automatic processing of rotation diffraction data from crystals of initially unknown symmetry and cell constants. J. Appl. Crystallogr. 26, 795–800. [Google Scholar]
- Kantardjieff, K.A., and Rupp, B. (2003). Matthews coefficient probabilities: Improved estimates for unit cell contents of proteins, DNA, and protein-nucleic acid complex crystals. Protein Sci. 12, 1865–1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleywegt, G.T., and Jones, T.A. (1995). Where freedom is given, liberties are taken. Structure 3, 535–540. [DOI] [PubMed] [Google Scholar]
- Kyndt, J.A., Meyer, T.E., Cusanovich, M.A., and Van Beeumen, J.J. (2002). Characterization of a bacterial tyrosine ammonia lyase, a biosynthetic enzyme for the photoactive yellow protein. FEBS Lett. 512, 240–244. [DOI] [PubMed] [Google Scholar]
- Laber, B., Kiltz, H.-H., and Amrhein, N. (1986). Inhibition of phenylalanine ammonia-lyase in vitro and in vivo by (1-amino-2-phenylethyl)phosponic acid, the phosphonic analog of phenylalanine. Z. Naturforsch. C 41, 49–55. [Google Scholar]
- Langer, M., Pauling, A., and Rétey, J. (1995). The role of dehydroalanine in catalysis by histidine ammonia-lyase. Angew. Chem. Int. Ed. Engl. 34, 1464–1465. [Google Scholar]
- Langer, B., Langer, M., and Rétey, J. (2001). Methylidene-imidazolone (MIO) from histidine and phenylalanine ammonia-lyase. Adv. Protein Chem. 58, 175–214. [DOI] [PubMed] [Google Scholar]
- Levy, H.L. (1999). Phenylketonuria: Old disease, new approach to treatment. Proc. Natl. Acad. Sci. USA 96, 1811–1813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mauch-Mani, B., and Slusarenko, A.J. (1996). Production of salicylic acid precursors is a major function of phenylalanine ammonia-lyase in the resistance of arabidopsis to Peronospora parasitica. Plant Cell 8, 203–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McRee, D.E. (1999). XtalView/Xfit—A versatile program for manipulating atomic coordinates and electron density. J. Struct. Biol. 125, 156–165. [DOI] [PubMed] [Google Scholar]
- Merritt, E.A., and Bacon, D.J. (1997). Raster3d. Photorealistic molecular graphics. Methods Enzymol. 277, 505–524. [DOI] [PubMed] [Google Scholar]
- Murshudov, G.N., Vagin, A.A., and Dodson, E.J. (1997). Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D 53, 240–255. [DOI] [PubMed] [Google Scholar]
- Ormö, M., Cubitt, A.B., Kallio, K., Gross, L.A., Tsien, R.Y., and Remington, S.J. (1996). Crystal structure of the Aequorea victoria green fluorescent protein. Science 273, 1392–1395. [DOI] [PubMed] [Google Scholar]
- Perrakis, A., Sixma, T.K., Wilson, K.S., and Lamzin, V.S. (1997). wARP: Improvement and extension of crystallographic phases by weighted averaging of multiple refined dummy atomic models. Acta Crystallogr. D 53, 448–455. [DOI] [PubMed] [Google Scholar]
- Rösler, J., Krekel, F., Amrhein, N., and Schmid, J. (1997). Maize phenylalanine ammonia-lyase has tyrosine ammonia-lyase activity. Plant Physiol. 113, 175–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Röther, D., Poppe, L., Morlock, G., Viergutz, S., and Retey, J. (2002). An active site homology model of phenylalanine ammonia-lyase from Petroselinum crispum. Eur. J. Biochem. 269, 3065–3075. [DOI] [PubMed] [Google Scholar]
- Safos, S., and Chang, T.M. (1995). Enzyme replacement therapy in ENU2 phenylketonuric mice using oral microencapsulated phenylalanine ammonia-lyase: A preliminary report. Artif. Cells Blood Substit. Immobil. Biotechnol. 23, 681–692. [DOI] [PubMed] [Google Scholar]
- Sarkissian, C.N., Shao, Z., Blain, F., Peevers, R., Su, H., Heft, R., Chang, T.M.S., and Scriver, C.R. (1999). A different approach to treatment of phenylketonuria: Phenylalanine degradation with recombinant phenylalanine ammonia lyase. Proc. Natl. Acad. Sci. USA 96, 2339–2344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuster, B., and Retey, J. (1994). Serine-202 is the putative precursor of the active site dehydroalanine of phenylalanine ammonia lyase. Site-directed mutagenesis studies on the enzyme from parsley (Petroselinum crispum L.). FEBS Lett. 349, 252–254. [DOI] [PubMed] [Google Scholar]
- Schuster, B., and Retey, J. (1995). The mechanism of action of phenylalanine ammonia-lyase: The role of prosthetic dehydroalanine. Proc. Natl. Acad. Sci. USA 92, 8433–8437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwede, T.F., Retey, J., and Schulz, G.E. (1999). Crystal structure of histidine ammonia-lyase revealing a novel polypeptide modification as the catalytic electrophile. Biochemistry 38, 5355–5361. [DOI] [PubMed] [Google Scholar]
- Terwilliger, T.C. (2003). Automated main-chain model building by template matching and iterative fragment extension. Acta Crystallogr. D 59, 38–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uson, I., and Sheldrick, G.M. (1999). Advances in direct methods for protein crystallography. Curr. Opin. Struct. Biol. 9, 643–648. [DOI] [PubMed] [Google Scholar]
- van Aalten, D.M.F., Bywater, R., Findlay, J.B.C., Hendlich, M., Hooft, R.W.W., and Vriend, G. (1996). PRODRG, a program for generating molecular topologies and unique molecular descriptors from coordinates of small molecules. J. Comput. Aided Mol. Des. 10, 255–262. [DOI] [PubMed] [Google Scholar]
- Weisshaar, B., and Jenkins, G.I. (1998). Phenylpropanoid biosynthesis and its regulation. Curr. Opin. Plant Biol. 1, 251–257. [DOI] [PubMed] [Google Scholar]
- Whetten, R., and Sederoff, R. (1995). Lignin biosynthesis. Plant Cell 7, 1001–1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang, L., and Moore, B.S. (2002). Inactivation, complementation, and heterologous expression of encP, a novel bacterial phenylalanine ammonia-lyase gene. J. Biol. Chem. 277, 32505–32509. [DOI] [PubMed] [Google Scholar]
- Yamada, S., Nabe, K., Izuo, N., Nakamichi, K., and Chibata, I. (1981). Production of L-phenylalanine from trans-cinnamic acid with Rhodotorula glutinis containing L-phenylalanine ammonia-lyase activity. Appl. Environ. Microbiol. 42, 773–778. [DOI] [PMC free article] [PubMed] [Google Scholar]