Abstract
Pectin methylesterase (PME) and invertase are key enzymes in plant carbohydrate metabolism. Inhibitors of both enzymes constitute a sequence family of extracellular proteins. Members of this family are selectively targeted toward either PME or invertase. In a comparative structural approach we have studied how this target specificity is implemented on homologous sequences. By extending crystallographic work on the invertase inhibitor Nt-CIF to a pectin methylesterase inhibitor (PMEI) from Arabidopsis thaliana, we show an α-helical hairpin motif to be an independent and mobile structural entity in PMEI. Removal of this hairpin fully inactivates the inhibitor. A chimera composed of the α-hairpin of PMEI and the four-helix bundle of Nt-CIF is still active against PME. By contrast, combining the corresponding segment of Nt-CIF with the four-helix bundle of PMEI renders the protein inactive toward either PME or invertase. Our experiments provide insight in how these homologous inhibitors can make differential use of similar structural modules to achieve distinct functions. Integrating our results with previous findings, we present a model for the PME-PMEI complex with important implications.
INTRODUCTION
At the posttranslational level, the activity of enzymes is commonly regulated by various mechanisms, including residue-directed protein modifications such as phosphorylation, glycosylation, and interaction with specific inhibitors. The nature of these inhibitors may range from small molecules to entire proteins, as found with the well-studied inhibitors of proteases (Bode and Huber, 1992). In plants, inhibitory proteins are often targeted toward sugar-modifying enzymes that escape cellular control mechanisms upon secretion into the plant cell wall or the vacuole (Juge et al., 2004). We are focusing on the structure–function relationship of an inhibitor family that regulates the activity of plant acid invertase and pectin methylesterase (PME).
Invertases convert the transport sugar sucrose into its building blocks, fructose and glucose. In higher plants, invertases exist in compartment specific isoforms, with only extracytosolic species being sensitive to inhibitory proteins. Altered activity of extracellular invertase has been shown to have dramatic effects on growth and development (Cheng et al., 1996; Tang et al., 1999; Goetz et al., 2001). This is consistent with roles of invertase activity in vital cellular processes such as carbohydrate transport (Roitsch et al., 2003), sugar signaling (Koch, 1996, 2004; Smeekens, 1998; Wobus and Weber, 1999), and stress response (Ehness et al., 1997; Roitsch et al., 2003). Protein inhibitors of invertase (Greiner et al., 1998) affect enzyme activity in a strictly pH-dependent manner (Rausch and Greiner, 2004) and have been proposed as transgenic tools to engineer post-harvest sucrose metabolism in crop plants (Greiner et al., 1999).
PMEs catalyze the demethylesterification of the homogalacturonan component of pectins, highly heterogeneous polymers (Vorwerk et al., 2004) that represent a major constituent of the plant primary cell wall. As the degree of demethylesterification determines the solidity of the wall, physiological processes requiring rearrangement of the cell wall architecture are affected by PME activity (Micheli, 2001). These include root development (Wen et al., 1999), stem elongation, and fruit ripening (Frenkel et al., 1998; Pilling et al., 2000; Brummell and Harpster, 2001). PME appears to be also involved in plant–pathogen interaction by serving as a host receptor for Tobacco mosaic virus (Chen et al., 2000; Chen and Citovsky, 2003). A protein inhibitor of plant PME (Giovane et al., 2004) has first been purified directly from kiwi fruit (Actinidia deliciosa; Camardella et al., 2000). Recently, two homologous species from Arabidopsis thaliana were recombinantly expressed and identified as PME inhibitors (PMEI; Wolf et al., 2003; Raiola et al., 2004).
PME and invertase inhibitors form a large plant sequence family named PMEI-related proteins (PMEI-RP). Family members share moderate sequence homology, and are selectively targeted toward apparently unrelated enzymes. Nothing is known about the molecular basis for the target specificity. As a first step to investigate this issue, we have previously determined the structure of the invertase inhibitor Nt-CIF from tobacco, CIF hereafter. The structural analysis revealed a four-helix bundle, preceded by an uncommon N-terminal extension (Hothorn et al., 2004). We suspected this small helical motif to play an important role in the inhibitory mechanism but were unable to test this hypothesis because truncated forms of the inhibitor were insoluble and thus not suitable for biochemical analysis.
In this work we have extended our studies to the PMEI, the second representative of the protein family. We report the three-dimensional structure of At-PMEI1 from Arabidopsis, PMEI hereafter. Comparative structural analysis of the two inhibitors inspired us to engineer protein chimera and investigate their interaction with PME and invertase. By crystallographic analysis and functional characterization of mutants, we are now able to define major determinants of target specificity for both functional classes of inhibitors.
RESULTS AND DISCUSSION
Overall Structure of PMEI
PMEI has been expressed, purified, and crystallized as described in the Methods section. Despite the moderate sequence identity between PMEI and CIF (∼20%), we could solve the structure by molecular replacement using the coordinates of CIF as search model in calculations with the program EPMR (Kissinger et al., 1999). The final model of the asymmetric unit, refined at 2.86-Å resolution, comprises three almost-complete chains of PMEI and 22 water molecules.
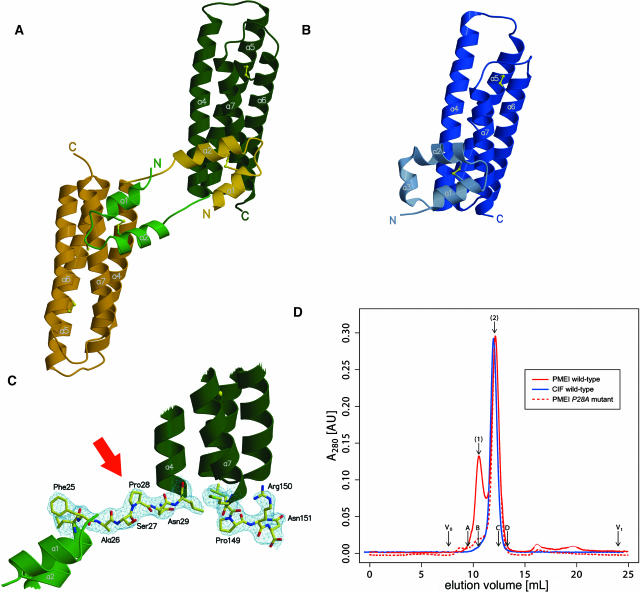
PMEI is composed of a four-helix bundle (residues 29 to 153) that arranges the helical components (helices α4 to α7) in an up-down–up-down topology, thereby creating an arrangement highly similar to that seen in CIF (root-mean-square deviation [RMSD] between 114 bundle Cα atoms is lower than 1.5 Å). The structural similarity between the bundles is considerably higher than expected from the degree of sequence conservation (Chothia and Lesk, 1986), possibly attributable to the presence of a conserved disulfide bridge (Cys-71PMEI/Cys-111PMEI, Cys-73CIF/Cys-114CIF) linking helix α5 to α6 (Figures 1A and 1B). The bundle core of the inhibitor is preceded by a 28-residue extension, basically resembling the molecular architecture already observed with the invertase inhibitor CIF (Hothorn et al., 2004). The extension in PMEI can be superimposed well with the corresponding segment in CIF (RMSD between 24 corresponding Cα atoms is <0.7 Å) but is radically reoriented with respect to the bundle core. This results in extensive contacts with the bundle of a neighboring molecule (Figures 1A and 1B). The arrangement resembles a molecular handshake of the two α-hairpins, forming a dimer that may also be present in solution (see below). The third molecule in the asymmetric unit is involved in lattice contacts essentially similar to those observed in the dimer.
Figure 1.
Structure of PMEI and Comparison with the Invertase Inhibitor CIF.
(A) Ribbon representation of the PMEI dimer with the respective molecules shown in green and yellow.
(B) CIF shown in the same orientation as the green molecule in (A).
(C) The linker region (residues 25PMEI to 29PMEI) interconnecting the dimer as well as a C-terminal extension shown in bonds representation and including the final 2 |Fobs-Fcalc| electron density map (contoured at 1.2 σ).
(D) A 280-nm absorbance trace of an analytical size-exclusion chromatography reveals the presence of PMEI (shown in red) dimers (peak 1) and monomers (peak 2). The invertase inhibitor CIF (shown in blue) appears to be exclusively monomeric. PMEI mutant P28A (dashed red line) does not resemble the dimeric state. Void (V0) and total (Vt) volume are shown for the column together with the elution volumes of molecular weight standards (A, BSA; B, ovalbumin; C, chymotrypsinogen A; D, ribonuclease A). The estimated molecular weight values of the At-PMEI1 monomer and dimer are 19,600 and 37,000, respectively. The calculated monomer molecular weight is 16,400.
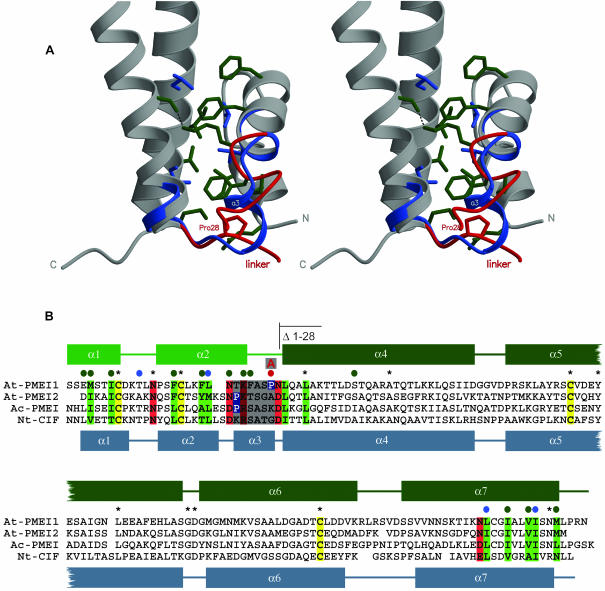
The helical extensions of CIF and PMEI participate in remarkably similar and mostly hydrophobic interfaces with the helix bundle that is contacted in cis in the former and in trans in the latter case (Figures 2A and 2B). In PMEI, this results in a completely unwound conformation of the linker (Figure 2B, highlighted in gray) between the helical hairpin and the bundle (Figure 1C).
Figure 2.
The α-Helical Hairpin Module in PMEI and CIF.
(A) Stereo close-up view of the bundle-hairpin interface in PMEI with invariant (blue) and conserved residues (green) contributing to interface stabilization included. The small helix-α3 connecting hairpin and bundle in CIF (blue) is unwound in PMEI (red).
(B) Sequence comparison of representative inhibitors with secondary structure assignment according to DSSP (Kabsch and Sander, 1983) and invariant Cys residues shown in yellow. Residues contributing to the bundle-hairpin interface are highlighted, dependent on their properties, in green and red. Conserved residues shown in (A) are denoted with a colored dot. The linker region discussed in the text is highlighted in gray; the linker Pro in PMEI is shown in blue.
Size-exclusion chromatography (see Methods) indicates a mixture of PMEI monomers and dimers in solution, compatible with the presence of a dimer in our crystals. The stability of this dimer is not affected by buffer variations in a range tested between pH 6.0 and 8.0. However, substantial reduction of ionic strength and protein concentration indicates monomer-dimer equilibrium and, as seen in the structural model, mainly hydrophobic stabilization of the dimeric state. In contrast with PMEI, CIF elutes exclusively as monomer in experiments performed under identical conditions (Figure 1D). We discuss functional aspects of this behavior below.
Truncation of the N-Terminal Extension Inactivates PMEI
We previously suspected the N-terminal α-hairpin to play a role in the inhibitory mechanism of CIF but were unable to test this hypothesis because truncated forms of the inhibitor were insoluble and thus not suitable for biochemical analysis (Hothorn et al., 2004).
Given the high overall structural similarity between CIF and PMEI, the large conformational differences in this segment prompted us to create a truncated version of the latter, deleting the entire α-hairpin (Δ1-28). The resulting construct could be purified to homogeneity using similar protocols as established for the wild-type inhibitor (Wolf et al., 2003). From size-exclusion chromatography and circular-dichroism experiments we conclude the remaining part of the inhibitor to be folded (data not shown).
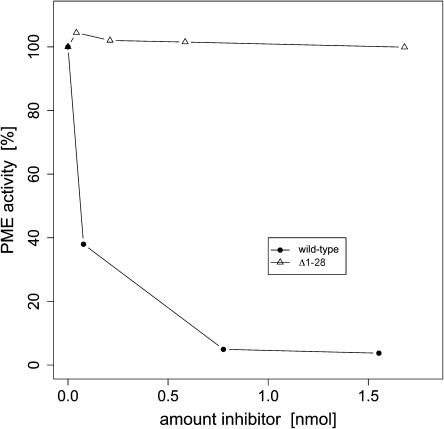
Truncated PMEI is inactive in dose-dependent inhibition assays (Grsic-Rausch and Rausch, 2004), monitoring the inactivation of PME preparations from Arabidopsis flowers (Figure 3), as well as preparations from other sources. Only dramatically increased inhibitor concentrations (∼5000-fold) show a mild inhibitory effect on the enzyme (data not shown). Our observations identify the N-terminal extension as a crucial determinant of PMEI activity.
Figure 3.
Removal of the N-terminal α-Helical Hairpin Inactivates PMEI.
Dose-dependent inhibition effect of the wild-type inhibitor (solid circle) and the Δ1-28 truncation on a preparation of PME from Arabidopsis flowers, prepared as described (Wolf et al., 2003).
Structural Determinants of the N-Terminal Flexibility
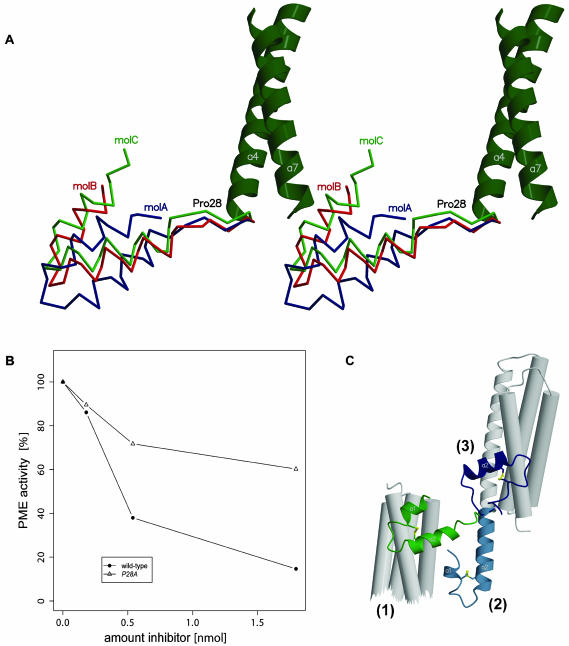
To further analyze the role of the N-terminal extension, we investigated whether alterations in the linker between hairpin and bundle can modulate structural and functional properties of PMEI. Conformational flexibility of this linker is already apparent by the different orientations of the helix hairpin in lattice dimers as observed in the wild-type inhibitor crystal (Figure 4A, shown in blue).
Figure 4.
Structural Determinants of Flexibility within the N-terminal Hairpin Module.
(A) Stereo view of the three PMEI molecules in the asymmetric unit, superimposed with respect to the four-helix bundle. The relative displacement indicates conformational variability. Note that the difference in orientation between molA and molC is almost 90°.
(B) Decreased inhibitory power of the PMEI P28A mutant in comparison with the wild-type inhibitor (solid circle) in plant PME inhibition assays.
(C) Structural superposition of the wild-type PMEI dimer (green) and two P28A mutant structures shown in dark (form A) and light blue (form B) highlight conformational flexibility of the PMEI hairpin. The flipped-out state (2) might resemble an intermediate in transition between dimer (1) and monomer (3).
Considering the frequently observed role of Pro in structural rearrangements we replaced Pro-28 by Ala (Figure 1C; P28A mutant), hoping to induce a conformation similar to that seen in CIF. In contrast with the wild-type inhibitor, the mutant protein elutes as a monomer in size-exclusion chromatography (Figure 1D). Moreover, we observed reduced inhibitory power in activity assays against plant PME (Figure 4B).
The significant conformational alterations in solution prompted us to explore structural effects of the mutation in detail. We have determined the structure of the P28A mutant in two crystal forms (Table 1; see Methods). Remarkably, crystalline P28A mutants present the N-terminal extension in two different orientations. As anticipated from the mutant design strategy one of these conformations (form A) is strikingly similar to Nt-CIF (Hothorn et al., 2004, shown in dark blue in Figure 4C). Form A and CIF superimpose well within the bundle region (RMSD of 114 Cα atoms <1.5 Å) whereas the α-hairpin is slightly displaced. Moreover, in the bundle-hairpin interface we find the same residues involved in structural stabilization as for the wild-type dimer (Figure 2A; see above) with the exception of Phe-25 in the linker peptide that is now flipped outwards. In the second crystal form (form B) we find the linker wound up, integrating helices α2 and α4 into a single long α-helix (Figure 4C, shown in light blue). The mutant structures demonstrate both an open and a closed conformation of PMEI. By structural analogy with CIF we hypothesize that the closed form (form A) would represent a stable monomer in solution that can also be attained by the wild-type protein (Figure 1D). The open form suggests an intermediate configuration in monomer-dimer transition (see states [1] to [3] in Figure 4C) although crystal-packing effects cannot be ruled out.
Table 1.
Summary of the Crystallographic Analysis
| Data Collection | PMEI Wild Type | PMEI P28A Mutant (form A) | PMEI P28A Mutant (form B) |
|---|---|---|---|
| Space group; unit cell (Å, °) | I222; a = 60.77, b = 106.19, c = 186.2, α = β = γ = 90 | C2; a = 108.40, b = 62.44, c = 22.34, α = γ = 90, β = 94.05 | H3; a = b = 82.41, 105.41 α = β = 90, γ = 120 |
| Wavelength (Å) | 1.0 | 0.933 | 0.8166 |
| Resolution (Å) | 2.86 | 1.50 | 2.68 |
| Highest shell (Å) | 3.04–2.86 | 1.50–1.54 | 2.85–2.68 |
| No. of unique reflections | 14,136 (2,307) | 24,884 (1,833) | 7,391 (1,184) |
| Multiplicity | 3.5 (3.6) | 3.6 (3.3) | 5.14 (3.8) |
| I/σa | 16.9 (5.4) | 15.4 (3.6) | 19.8 (6.56) |
| Rsym (%)a | 5.9 (27.4) | 5.3 (35.8) | 6.1 (22.1) |
| Completeness (%) | 98.4 (98.1) | 99.8 (98.7) | 98.5 (93.3) |
| Refinement | |||
| Resolution range (Å) | 29.06–2.86 | 54.23–1.50 | 19.46–2.68 |
| No. of reflections | 14,129 | 24,884 | 7,391 |
| Rwork (%)b | 21.8 | 18.3 | 21.5 |
| Rfree (%)b | 28.0 | 20.7 | 25.6 |
| No. of atoms | |||
| Protein | 3,355 | 1,166 | 2,198 |
| Solvent | 22 | 104 | 44 |
| RMS deviations | |||
| Bond length | 0.007 | 0.017 | 0.010 |
| Angles | 1.2 | 1.5 | 1.4 |
As defined in XDS (Kabsch, 1993).
As defined in CNS (Brunger et al., 1998) or REFMAC5 (Collaborative Computational Project Number 4, 1994).
Taken together, we have used the P28A mutant to visualize the different conformational states of PMEI; configurations that we believe can be adopted by the wild-type inhibitor (Figure 1D) but may not be favored at protein concentrations typically used in crystallization. Our results clearly demonstrate flexibility of the N-terminal extension. Although the effect of the mutation on the inhibitory activity of PMEI is moderate, we believe that flexibility of the N-terminal module is relevant for inhibitor function.
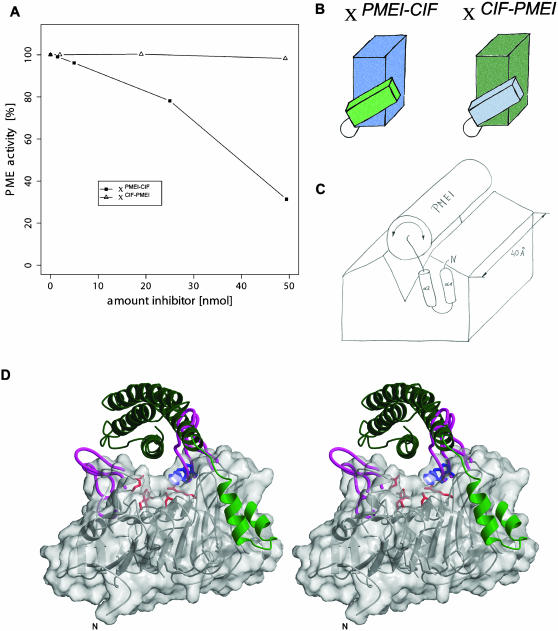
Protein Chimera Shed Light on Target Specificity
The presence of a structurally distinct and flexible motif on an apparently rigid inhibitor core prompted us to investigate whether this module represents a determinant of target specificity. We therefore engineered chimera that combine the α-hairpin module of PMEI with the four-helix bundle of CIF (XPMEI-CIF) and vice versa (XCIF-PMEI; Figure 5B; see Methods). Biophysical analyses of the purified proteins indicated the engineered inhibitors were folded.
Figure 5.
Determinants of Target Specificity.
(A) Dose-dependent inhibition of Arabidopsis PME by protein chimera between PMEI and the invertase inhibitor CIF.
(B) Schematic representation of protein chimera. In XPMEI-CIF, the α-hairpin of PMEI has been connected to the bundle core of CIF. XCIF-PMEI combines the corresponding hairpin segment of CIF with the four-helix bundle of PMEI. The color coding follows the structural representations in Figure 1.
(C) Proposed model for the inactivation of plant PME by PMEI. Schematic view, in which the four-helix bundle covers the enzyme's pectin binding cleft, which measures ∼40 Å in length. The helical hairpin anchors the inhibitor to its target enzyme and mediates specificity.
(D) Manual docking of PMEI onto the pectin binding cleft of plant PME. Carrot PME (PDB ID 1gq8) is shown in both ribbon and molecular surface representation in a view along the cleft region. The inhibitor (in green) covers the entire cleft. Active-site residues and residues W227/252 discussed in the text are highlighted in red and blue, respectively. The longer loop regions of the bacterial enzyme (PDB ID 1qjv) are indicated in magenta.
Strikingly, XPMEI-CIF is able to inactivate plant PME in dose-dependent inhibition assays (Figure 5A). By sharp contrast, XCIF-PMEI did not show inhibitory activity toward either PME or invertase under similar conditions (Figure 5A; data not shown). These results clearly indicate that the PMEI-hairpin module is necessary and sufficient to inhibit PME when attached to a four-helix bundle common to the sequence family (XPMEI-CIF in Figure 5A).
In this respect, it is noteworthy that PMEI and CIF share only ∼17% of their residues within the bundle region (Figure 2B), most of which are located in the interior of the protein (Hothorn et al., 2004). Therefore, the surface-charge distribution is unlikely to play an important role in PME inactivation, although the inhibitory activity of XPMEI-CIF is decreased with respect to the wild-type inhibitor.
The inability of XCIF-PMEI to inactivate invertase (data not shown) suggests that the hairpin motif is not sufficient for the invertase inhibitory function. Asking whether the CIF bundle would represent the major functional module instead, we attempted to test XPMEI-CIF in invertase inhibition assays. Unfortunately, the protein precipitated at buffer conditions established for invertase inhibition assays (see Methods). By contrast, the XCIF-PMEI chimera is stable even at acidic pH and could therefore be used in invertase assays (see above).
Our observations provide compelling evidence that invertase and PMEIs have established distinct target inactivation mechanisms on virtually identical structural scaffolds. In the case of PMEI, specificity is apparently encoded in the α-hairpin, whereas the entire inhibitor and/or the four-helix bundle of CIF may contain the major determinants for specificity toward invertase.
A Model for the PME-PMEI Complex
Our analysis of PMEI has interesting implications for the interaction of the inhibitor with its target enzyme. To illustrate this, we have manually docked the inhibitor (see Methods) to the crystal structure of carrot PME (Johansson et al., 2002), an enzyme that can be inactivated by PMEI (S. Wolf, unpublished data). In the resulting model, the α-helical bundle covers the pectin binding cleft of the enzyme that harbors the substrate and active-site residues (Jenkins et al., 2001; Johansson et al., 2002; see also Figures 5C and 5D). PMEI would bind in an open conformation exposing the N-terminal α-hairpin to interact with a C-terminal helix at the surface of PME. For the following reasons we find this model, which uses large areas of the inhibitor for interaction (∼1500 Å2 buried surface), particularly attractive. (1) PMEI almost completely covers the pectin binding cleft (40 Å in length; Figure 5C) that contains the active site of the enzyme (Johansson et al., 2002). Trp residues located in this cleft become shielded upon inhibitor binding as concluded from fluorescence studies (D'Avino et al., 2003). (2) Biochemical studies indicated a 1:1 complex between PME and PMEI (D'Avino et al., 2003), consistent with our model. (3) Our docking model nicely explains why the homologous bacterial PME is not sensitive to PMEI (Wolf et al., 2003): several loop regions protruding from areas flanking the cleft (Jenkins et al., 2001; Wolf et al., 2003) would interfere with the binding of the PMEI bundle, a selection mechanism proposed earlier (D'Avino et al., 2003). (4) Our model is consistent with a fully automated docking approach employing the program FTDOCK (Gabb et al., 1997). Using surface complementarity as quality criterion, FTDOCK presents a docking solution close to our model based on biochemical data among the top 10 out of 10,000 trials (see Methods). In addition, most of the 100 top-ranked solutions cluster around the pectin binding cleft.
It is also noteworthy that our complex model brings the C terminus of PMEI in close proximity to the N terminus of PME. Such a scenario would allow convenient binding of PMEI homologous Pro-regions in type-I PMEs (Micheli, 2001) in a similar mode, although the functional role of these portions is yet unclear.
In the suggested complex model, the α-hairpin would serve as an anchor, which is essential for the positioning of the four-helix bundle that then mediates the inhibitory activity. This view is consistent with our observations that the bundle alone has no activity and that replacement of the bundle by its CIF counterpart is still functional.
Taken together, our docking model of the interaction between PME and PMEI represents a valuable hypothesis that can now be tested in biochemical studies using site-directed mutagenesis and, finally, by structure determination of the PME-PMEI complex.
Concluding Remarks
Our work reveals plant invertase inhibitors and PMEIs to represent a protein family that has implemented inhibitory activity toward different target enzymes on similar structural scaffolds. By structural comparison of PMEI with its counterpart CIF (Hothorn et al., 2004) and protein-engineering approaches we have identified determinants of target specificity within this class of proteins. It is known that sequence comparison alone does not allow predicting whether family members inhibit one or the other target. Our analysis has narrowed the major determinants of specificity to a set of ∼28 residues that can now be analyzed in detail for their role in the inhibition process.
The presented protein chimeras suggest different mechanisms of enzyme inhibition by PMEI and CIF. Understanding these mechanisms in detail will require the structure of the cognate inhibitor-enzyme complexes. Invertase/PMEIs have been used to silence post-translationally their target enzymes in transgenic plants (Greiner et al., 1999; Balibrea Lara et al., 2004). From the presented protein chimera we conclude that protein engineering may represent a useful tool to gain further insights into specificity toward PME and invertase isoenzymes. These studies will allow a more specific interference with key enzymes in plant carbohydrate metabolisms and may inspire novel biotechnological applications.
METHODS
Expression, Crystallization, and Data Collection
Wild-type PMEI and mutant forms have been expressed and purified as described (Wolf et al., 2003) following protocols established for the related invertase inhibitor Nt-CIF (Hothorn et al., 2003). Before crystallization, samples were concentrated to ∼5 mg/mL using a Vivapore 10/20 mL concentrator (7.5 kD MWCO; Vivascience, Hannover, Germany) and dialyzed against 100 mM NaCl, 10 mM Hepes, pH 7.0. In the case of the wild-type protein, orthorhombic crystals were grown at room temperature by vapor diffusion from hanging drops composed of equal volumes (2 + 2 μL) of protein solution and crystallization buffer (10% [v/v] PEG 8000, 0.3 M NaCl, 0.1 M Na+/K+ Pi, pH 6.2) suspended over 1.0 mL of the latter as reservoir solution. Thin plates of ∼200 × 80 × 20 μm were transferred into reservoir solution containing 10% (v/v) glycerol and flash-frozen in liquid nitrogen. A data set at 2.86-Å resolution has been collected at beam line PX06 (Swiss Light Source, Villigen, Switzerland). Monoclinic crystals (form A; see Table 1) of the P28A mutant grew in 0.2 M (NH4)2SO4, 0.3 M Na+/K+ tartrate, 0.1 M sodium citrate, pH 5.6, and were cryoprotected by addition of 10% (v/v) glycerol. A data set at 1.5-Å resolution has been recorded at beam line ID14-2 (European Synchrotron Radiation Facility, Grenoble, France). Rhombohedral crystals (form B in Table 1) of the P28A mutant developed in 2.5 M (NH4)2SO4 and 4% (v/v) isopropanol and were cryoprotected by addition of 20% (v/v) glycerol. Data collection at beam line X11 (EMBL/DESY, Hamburg) yielded a data set at 2.68-Å resolution. Data processing and scaling was performed with XDS (Kabsch, 1993; December 2003 version).
Structure Determination and Refinement
The structure of PMEI has been determined by molecular replacement in six-dimensional searches with the program EPMR (Kissinger et al., 1999) using a polyalanine model of the previously determined invertase inhibitor Nt-CIF (PDB accession code 1RJ1; Hothorn et al., 2004). The solution comprises a trimer in the asymmetric unit, accounting for a solvent content of ∼60%. The small helix-α3 connecting hairpin motif and bundle in Nt-CIF produced clashes in the crystal packing. Removal of this helix, rigid-body refinement, and the use of strict noncrystallographic symmetry allowed the calculation of an initial electron density map at 3.4 Å, in which a disulfide bridge and two Tyr residues could be located. During refinement, strong maxima in the |Fobs-Fcalc| and 2 |Fobs-Fcalc| electron density maps indicated the unfolding of helix α3 into a linker region connecting the N-terminal α-hairpin motif with the bundle of a neighboring molecule. The structure was completed in alternating cycles of model correction using the program O (Jones et al., 1991) and restrained refinement as implemented in CNS (Brunger et al., 1998). The structure of the rhombohedral P28A crystal form has been solved by molecular replacement using CNS. The solution comprises a dimer in the asymmetric unit (starting Rfree 0.41). Finally, the monoclinic crystal form of the mutant protein, comprising a monomer in the asymmetric unit, has been built using Arp/Warp 6.0 (Lamzin and Wilson, 1993) starting from a molecular replacement solution calculated with CNS. In this case, Refmac5 (Collaborative Computational Project Number, 1994) has been used for the final rounds of refinement. A summary of the crystallographic analysis is shown in Table 1.
Inspection of the refined models with PROCHECK (Laskowski et al., 1993) revealed a good stereochemistry except for some residues in poorly defined loop regions. Structure visualization was done with POVSCRIPT (Fenn et al., 2003) and RASTER3D (Merritt and Bacon, 1997).
Size-exclusion chromatography was performed using an analytical grade Superdex 75 HR 10/30 column (Amersham Biosciences, Piscataway, NJ) preequilibrated in 0.3 M NaCl, 0.1 M Na+/K+ Pi, pH 6.2. Fifty microliters of the sample (10.0 mg/mL) were loaded onto the column and elution at 0.8 mL/min was monitored by ultraviolet absorbance at 280 nm.
Site-Directed Mutagenesis
Site-specific mutations were introduced with the QuickChange mutagenesis kit (Stratagene, La Jolla, CA) following the manufacturer's instructions and verified by DNA sequencing.
A truncated version of PMEI lacking 28 N-terminal residues was PCR amplified using sense primer 5′-ATAGCTAAATCCATGGACTCGCCTAATCTTCAAGCCTTG-3′, antisense primer 3′-AAATTGTCAAGGTACCTTAATTACGTGGTAACATGTTAG-5′, and a pQE30 vector (Qiagen USA, Valencia, CA) containing At-PMEI1 (at1g48020) as the template. The NcoI/KpnI-restricted fragment was cloned into pETM20, a modified pET21d vector (Novagen, Madison, WI) providing thioredoxin A (trxA) followed by a 6×His tag (amplified from pET32a [Novagen]) and a tobacco etch virus (Tev) protease cleavage site as an N-terminal fusion partner (Hothorn et al., 2003).
Protein Chimera
XPMEI-CIF has been constructed by BamHI/EcoRV digestion of wild-type Nt-CIF in pQE30 (Greiner et al., 1998) producing a fragment that encodes the CIF bundle starting at residue Ile-32. The PMEI hairpin was PCR amplified with sense primer 5′-ATAGCTAAATCCATGGACAGTTCAGAAATGAGCACAATC-3′ and antisense primer 5′-AAATTGTCAAGATATCAGGCGATGCGAATTTCGTATTG-3′ containing Asp-29 and an EcoRV site. After ligation, XPMEI-CIF was amplified and ligated into expression vector pETM20. XCIF-PMEI has been constructed by simultaneously ligating PCR fragments encoding the CIF hairpin and PMEI bundle into a pETM-20 expression vector. The CIF bundle was amplified using sense primer 5′-ATCGGACTGCAGCCATGGCAAATAATCTAGTAGAAACTAC-3′, antisense primer 5′-CGATACGGCATCGCTAGCGCTTGAAGATTCCCTGTTGCACTTCGTTTGTC-3′, and Nt-CIF in pQE30 as the template. The PMEI bundle was amplified with sense primer 5′-ATCGGACTGCAGGCTAGCAAAAACCACACTTGATTCTAC-3′, antisense primer 5′-AAATTGTCAAGGTACCTTAATTACGTGGTAACATGTTAG-3′, and At-PMEI1 in pQE30 as the template. Subsequently, the CIF hairpin fragment was restricted with NcoI and NheI, whereas the PMEI bundle was restricted with NheI and KpnI. Both fragments were ligated into the NcoI/KpnI-restricted pETM-20 vector. Constructs were verified by DNA sequencing.
Activity Assay for Inhibitor Function
PME preparations from a mixture of Arabidopsis flowers and siliques were obtained by homogenizing the tissue in 2 mL/g extraction buffer (25 mM maleic acid, 75 mM Tris base, pH 7.0, 1 M NaCl, complemented with a complete mini EDTA–free protease inhibitor tablet (Roche, Mannheim, Germany). After incubation on ice for 30 min with gentle agitation, the homogenate was centrifuged twice at 11,000g for 10 min and the supernatant kept to perform inhibition assays. PME activity was determined by a coupled enzymatic assay as described (Grsic-Rausch and Rausch, 2004). In brief, the assay was performed in 50 mM phosphate buffer, pH 7.5, in the presence of 0.4 mM NAD. PME activity using commercially available pectin (Sigma, St. Louis, MO) as substrate was measured by the amount of the produced methanol, which was first oxidized to formaldehyde by alcohol oxidase (1 unit; Sigma), followed by oxidation to formate via formaldehyde dehydrogenase (0.35 unit; Sigma). The produced NADH was measured at OD340nm in a spectrophotometer.
Acid invertase activity (assay buffer: 30 mM sucrose, 20 mM triethanol amine, 7 mM citric acid, 1 mM phenyl methyl sulfonyl fluoride, pH 4.6) was measured by enzymatic determination of released glucose in a coupled assay with hexokinase and glucose-6-phosphate dehydrogenase as described (Weil and Rausch, 1994).
Rigid-Body Protein Docking
A molecular surface of carrot PME (PDB ID 1gq8) calculated with VOIDOO (Kleywegt and Jones, 1994) has been used to manually navigate the inhibitor onto the PME pectin binding cleft using the program O (Jones et al., 1991). Furthermore, we used the program FTDock (Gabb et al., 1997) to perform a rigid-body docking of the PME to its inhibitor. The algorithm discretizes the surfaces of the two interacting molecules and performs a global scan of the translational and rotational space. FTDock evaluates millions of possible relative orientations between the two molecules and keeps the 10,000 solutions that show the best surface complementarity. To speed up the calculations, the PME (larger molecule) was kept static and its inhibitor was translated and rotated to explore the six degrees of associational freedom. Finally, we rescored the list of 10,000 solutions and ranked them according to electrostatics complementarity between the two interacting interfaces and empirical pair potentials (Moont et al., 1999).
Atomic coordinates and structure factors have been submitted to the Protein Data Bank (http://www.rcsb.org) with codes 1X8Z (wild type), 1X91 (P28A mutant form A), and 1X90 (P28A mutant form B).
Acknowledgments
We are grateful to Thomas Rausch for continuous support and discussions on PMEI-RP physiology during the preparation of the manuscript. We thank the staff at the European Synchrotron Radiation Facility, Grenoble, France, the Deutsches Elektronensynchrotron, Hamburg, Germany, and the Swiss Light Source, Villigen, Switzerland for technical support during data collection. We acknowledge financial support from the Suedzucker AG, Mannheim, Germany and the KWS Saat AG, Einbeck, Germany (grants to S.G.). M.H. gratefully acknowledges financial support from the Peter and Traudl Engelhorn Stiftung Penzberg, Germany.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Klaus Scheffzek (scheffzek@embl.de).
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.104.025684.
References
- Balibrea Lara, M.E., Gonzalez Garcia, M.C., Fatima, T., Ehness, R., Lee, T.K., Proels, R., Tanner, W., and Roitsch, T. (2004). Extracellular invertase is an essential component of cytokinin-mediated delay of senescence. Plant Cell 16, 1276–1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bode, W., and Huber, R. (1992). Natural protein proteinase inhibitors and their interaction with proteinases. Eur. J. Biochem. 204, 433–451. [DOI] [PubMed] [Google Scholar]
- Brummell, D.A., and Harpster, M.H. (2001). Cell wall metabolism in fruit softening and quality and its manipulation in transgenic plants. Plant Mol. Biol. 47, 311–340. [PubMed] [Google Scholar]
- Brunger, A.T., et al. (1998). Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 54, 905–921. [DOI] [PubMed] [Google Scholar]
- Camardella, L., Carratore, V., Ciardiello, M.A., Servillo, L., Balestrieri, C., and Giovane, A. (2000). Kiwi protein inhibitor of pectin methylesterase amino-acid sequence and structural importance of two disulfide bridges. Eur. J. Biochem. 267, 4561–4565. [DOI] [PubMed] [Google Scholar]
- Chen, M.H., and Citovsky, V. (2003). Systemic movement of a tobamovirus requires host cell pectin methylesterase. Plant J. 35, 386–392. [DOI] [PubMed] [Google Scholar]
- Chen, M.H., Sheng, J., Hind, G., Handa, A.K., and Citovsky, V. (2000). Interaction between the tobacco mosaic virus movement protein and host cell pectin methylesterases is required for viral cell-to-cell movement. EMBO J. 19, 913–920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng, W.-H., Taliercio Earl, W., and Chourey Prem, S. (1996). The Miniature1 seed locus of maize encodes a cell wall invertase required for normal development of endosperm and maternal cells in the pedicel. Plant Cell 8, 971–983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chothia, C., and Lesk, A.M. (1986). The relation between the divergence of sequence and structure in proteins. EMBO J. 5, 823–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collaborative Computational Project Number 4. (1994). The CCP4 suite: Programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 50, 760–763. [DOI] [PubMed] [Google Scholar]
- D'Avino, R., Camardella, L., Christensen, T.M., Giovane, A., and Servillo, L. (2003). Tomato pectin methylesterase: Modeling, fluorescence, and inhibitor interaction studies-comparison with the bacterial (Erwinia chrysanthemi) enzyme. Proteins 53, 830–839. [DOI] [PubMed] [Google Scholar]
- Ehness, R., Ecker, M., Godt Dietmute, E., and Roitsch, T. (1997). Glucose and stress independently regulate source and sink metabolism and defense mechanisms via signal transduction pathways involving protein phosphorylation. Plant Cell 9, 1825–1841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fenn, T.D., Ringe, D., and Petsko, G.A. (2003). POVScript+: A program for model and data visualization using persistence of vision ray-tracing. J. Appl. Crystallogr. 36, 944–947. [Google Scholar]
- Frenkel, C., Peters, J.S., Tieman, D.M., Tiznado, M.E., and Handa, A.K. (1998). Pectin methylesterase regulates methanol and ethanol accumulation in ripening tomato (Lycopersicon esculentum) fruit. J. Biol. Chem. 273, 4293–4295. [DOI] [PubMed] [Google Scholar]
- Gabb, H.A., Jackson, R.M., and Sternberg, M.J. (1997). Modelling protein docking using shape complementarity, electrostatics and biochemical information. J. Mol. Biol. 272, 106–120. [DOI] [PubMed] [Google Scholar]
- Giovane, A., Servillo, L., Balestrieri, C., Raiola, A., D'Avino, R., Tamburrini, M., Ciardiello, M.A., and Camardella, L. (2004). Pectin methylesterase inhibitor. Biochim. Biophys. Acta 1696, 245–252. [DOI] [PubMed] [Google Scholar]
- Goetz, M., Godt, D.E., Guivarc'h, A., Kahmann, U., Chriqui, D., and Roitsch, T. (2001). Induction of male sterility in plants by metabolic engineering of the carbohydrate supply. Proc. Natl. Acad. Sci. USA 98, 6522–6527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greiner, S., Krausgrill, S., and Rausch, T. (1998). Cloning of a tobacco apoplasmic invertase inhibitor. Proof of function of the recombinant protein and expression analysis during plant development. Plant Physiol. 116, 733–742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greiner, S., Rausch, T., Sonnewald, U., and Herbers, K. (1999). Ectopic expression of a tobacco invertase inhibitor homolog prevents cold-induced sweetening of potato tubers. Nat. Biotechnol. 17, 708–711. [DOI] [PubMed] [Google Scholar]
- Grsic-Rausch, S., and Rausch, T. (2004). A coupled spectrophotometric enzyme assay for the determination of pectin methylesterase activity and its inhibition by proteinaceous inhibitors. Anal. Biochem. 133, 14–18. [DOI] [PubMed] [Google Scholar]
- Hothorn, M., Bonneau, F., Stier, G., Greiner, S., and Scheffzek, K. (2003). Bacterial expression, purification and preliminary X-ray crystallographic characterization of the invertase inhibitor Nt-CIF from tobacco. Acta Crystallogr. D Biol. Crystallogr. 59, 2279–2282. [DOI] [PubMed] [Google Scholar]
- Hothorn, M., D'Angelo, I., Marquez, J.A., Greiner, S., and Scheffzek, K. (2004). The invertase inhibitor Nt-CIF from tobacco: A highly thermostable four-helix bundle with an unusual N-terminal extension. J. Mol. Biol. 335, 987–995. [DOI] [PubMed] [Google Scholar]
- Jenkins, J., Mayans, O., Smith, D., Worboys, K., and Pickersgill, R.W. (2001). Three-dimensional structure of Erwinia chrysanthemi pectin methylesterase reveals a novel esterase active site. J. Mol. Biol. 305, 951–960. [DOI] [PubMed] [Google Scholar]
- Johansson, K., El-Ahmad, M., Friemann, R., Jornvall, H., Markovic, O., and Eklund, H. (2002). Crystal structure of plant pectin methylesterase. FEBS Lett. 514, 243–249. [DOI] [PubMed] [Google Scholar]
- Jones, T.A., Zou, J.-Y., Cowan, S.W., and Kjeldgaard, M. (1991). Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A 47, 110–119. [DOI] [PubMed] [Google Scholar]
- Juge, N., Svensson, B., Henrissat, B., and Williamson, G. (2004). Plant proteinaceous inhibitors of carbohydrate-active enzymes. Biochim. Biophys. Acta 1696, 141. [Google Scholar]
- Kabsch, W. (1993). Automatic processing of rotation diffraction data from crystals of initially unknown symmetry and cell constants. J. Appl. Crystallogr. 26, 795–800. [Google Scholar]
- Kabsch, W., and Sander, C. (1983). Dictionary of protein secondary structure: Pattern recognition of hydrogen-bonded and geometrical features. Biopolymers 22, 2577–2637. [DOI] [PubMed] [Google Scholar]
- Kissinger, C.R., Gehlhaar, D.K., and Fogel, D.B. (1999). Rapid automated molecular replacement by evolutionary search. Acta Crystallogr. D Biol. Crystallogr. 55, 484–491. [DOI] [PubMed] [Google Scholar]
- Kleywegt, G.J., and Jones, T.A. (1994). Detection, delineation, measurement and display of cavities in macromolecular structures. Acta Crystallogr. D Biol. Crystallogr. 50, 178–185. [DOI] [PubMed] [Google Scholar]
- Koch, K. (2004). Sucrose metabolism: Regulatory mechanisms and pivotal roles in sugar sensing and plant development. Curr. Opin. Plant Biol. 7, 235–246. [DOI] [PubMed] [Google Scholar]
- Koch, K.E. (1996). Carbohydrate-modulated gene expression in plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 47, 509–540. [DOI] [PubMed] [Google Scholar]
- Lamzin, V.S., and Wilson, K.S. (1993). Automated refinement of protein models. Acta Crystallogr. D Biol. Crystallogr. 49, 129–147. [DOI] [PubMed] [Google Scholar]
- Laskowski, R.A., MacArthur, M.W., Moss, D.S., and Thornton, J.M. (1993). PROCHECK: A program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 26, 283–291. [Google Scholar]
- Merritt, E.A., and Bacon, D.J. (1997). Raster3D: Photorealistic molecular graphics. Methods Enzymol. 277, 505–524. [DOI] [PubMed] [Google Scholar]
- Micheli, F. (2001). Pectin methylesterases: Cell wall enzymes with important roles in plant physiology. Trends Plant Sci. 6, 414–419. [DOI] [PubMed] [Google Scholar]
- Moont, G., Gabb, H.A., and Sternberg, M.J. (1999). Use of pair potentials across protein interfaces in screening predicted docked complexes. Proteins 35, 364–373. [PubMed] [Google Scholar]
- Pilling, J., Willmitzer, L., and Fisahn, J. (2000). Expression of a Petunia inflata pectin methyl esterase in Solanum tuberosum L. enhances stem elongation and modifies cation distribution. Planta 210, 391–399. [DOI] [PubMed] [Google Scholar]
- Raiola, A., Camardella, L., Giovane, A., Mattei, B., De Lorenzo, G., Cervone, F., and Bellincampi, D. (2004). Two Arabidopsis thaliana genes encode functional pectin methylesterase inhibitors. FEBS Lett. 557, 199–203. [DOI] [PubMed] [Google Scholar]
- Rausch, T., and Greiner, S. (2004). Plant protein inhibitors of invertases. Biochim. Biophys. Acta 1696, 253–261. [DOI] [PubMed] [Google Scholar]
- Roitsch, T., Balibrea, M.E., Hofmann, M., Proels, R., and Sinha, A.K. (2003). Extracellular invertase: Key metabolic enzyme and PR protein. J. Exp. Bot. 54, 513–524. [DOI] [PubMed] [Google Scholar]
- Smeekens, S. (1998). Sugar regulation of gene expression in plants. Curr. Opin. Plant Biol. 1, 230–234. [DOI] [PubMed] [Google Scholar]
- Tang, G.Q., Luscher, M., and Sturm, A. (1999). Antisense repression of vacuolar and cell wall invertase in transgenic carrot alters early plant development and sucrose partitioning. Plant Cell 11, 177–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vorwerk, S., Somerville, S., and Somerville, C. (2004). The role of plant cell wall polysaccharide composition in disease resistance. Trends Plant Sci. 9, 203–209. [DOI] [PubMed] [Google Scholar]
- Weil, M., and Rausch, T. (1994). Acid invertase in Nicotiana tabacum crown-gall cells: Molecular properties of the cell-wall isoform. Planta 193, 430–437. [DOI] [PubMed] [Google Scholar]
- Wen, F., Zhu, Y., and Hawes, M.C. (1999). Effect of pectin methylesterase gene expression on pea root development. Plant Cell 11, 1129–1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wobus, U., and Weber, H. (1999). Sugars as signal molecules in plant seed development. Biol. Chem. 380, 937–944. [DOI] [PubMed] [Google Scholar]
- Wolf, S., Grsic-Rausch, S., Rausch, T., and Greiner, S. (2003). Identification of pollen-expressed pectin methylesterase inhibitors in Arabidopsis. FEBS Lett. 555, 551–555. [DOI] [PubMed] [Google Scholar]