Abstract
The B3 DNA binding domain is shared amongst various plant-specific transcription factors, including factors involved in auxin-regulated and abscisic acid–regulated transcription. Herein, we report the NMR solution structure of the B3 domain of the Arabidopsis thaliana cold-responsive transcription factor RAV1. The structure consists of a seven-stranded open β-barrel and two α-helices located at the ends of the barrel and is significantly similar to the structure of the noncatalytic DNA binding domain of the restriction enzyme EcoRII. An NMR titration experiment revealed a DNA recognition interface that enabled us to propose a structural model of the protein–DNA complex. The locations of the DNA-contacting residues are also likely to be similar to those of the EcoRII DNA binding domain.
INTRODUCTION
Auxin and abscisic acid are two of the main phytohormones. The former is central to the control of growth and development of various organs (Abel and Theologis, 1996), whereas the latter plays critical roles in regulating seed maturation and germination (Leung and Giraudat, 1998). Transcription factors responding to auxin are designated as auxin response factors (ARFs). They recognize auxin responsive elements of a sequence TGTCTC existing in the promoter regions of auxin-responsive genes (Ulmasov et al., 1997, 1999; Tiwari et al., 2003). ARFs form one of the major transcription factor families containing 23 members in Arabidopsis thaliana (Riechmann et al., 2000). Several abscisic acid response factors have been identified from the analyses of abscisic acid insensitive mutants (McCarty et al., 1989; Koornneef et al., 1998). Among them maize (Zea mays) VIVIPAROUS1 (VP1) (McCarty et al., 1989, 1991) and Arabidopsis ABSCISIC ACID–INSENSITIVE3 (ABI3) (Giraudat et al., 1992) are transcription factors believed to be orthologs and recognize the Sph/RY element of sequence CATGCA (Suzuki et al., 1997; Ezcurra et al., 2000). Fourteen factors similar to ABI3 and VP1 were identified in the Arabidopsis genome (Riechmann et al., 2000). ARFs and ABI3/VP1-like factors share a homologous region corresponding to a DNA binding domain (DBD) of ∼120 amino acid residues designated B3 that was originally identified as the third basic region in the ABI3 and VP1 proteins (Giraudat et al., 1992).
RAV1 (RAV for related to ABI3/VP1) was identified as a DNA binding protein possessing an N-terminal AP2/ERF (or EREBP)-type DBD and a C-terminal B3 domain (Kagaya et al., 1999). Six proteins of this combination were found in the Arabidopsis genome and form the RAV-like family (Riechmann et al., 2000). RAV1 has the ability to bind to DNAs possessing a bipartite consensus sequence, corresponding to two separate DBDs, in which the CACCTG sequence is recognized by the B3 domain (Kagaya et al., 1999). It was revealed recently that RAV1 is a cold-responsive transcription factor that is likely to trigger another regulatory pathway than that the well investigated DREB/CBF transcription factors do (Fowler and Thomashow, 2002).
It should be noted that the recognition sequences of the B3 domains of these three families are 6 bp in length and that they share some similarity. That is, when the recognition sequences of the B3 domains of RAV1 and ARFs are compared, the consensus sequence is YRYCTS, where Y is C or T, R is A or G, and S is G or C, whereas when those of RAV1 and ABI3/VP1 are compared, the consensus is CAYSYR. Therefore, it is expected that these B3 domains share a common structural framework for sequence-specific DNA recognition, although they differ in the direct base recognition. In this study, we determined the three-dimensional structure of the B3 domain of RAV1 by heteronuclear NMR spectroscopy and proposed a DNA binding mechanism of the B3 domain.
RESULTS AND DISCUSSION
Structural Description
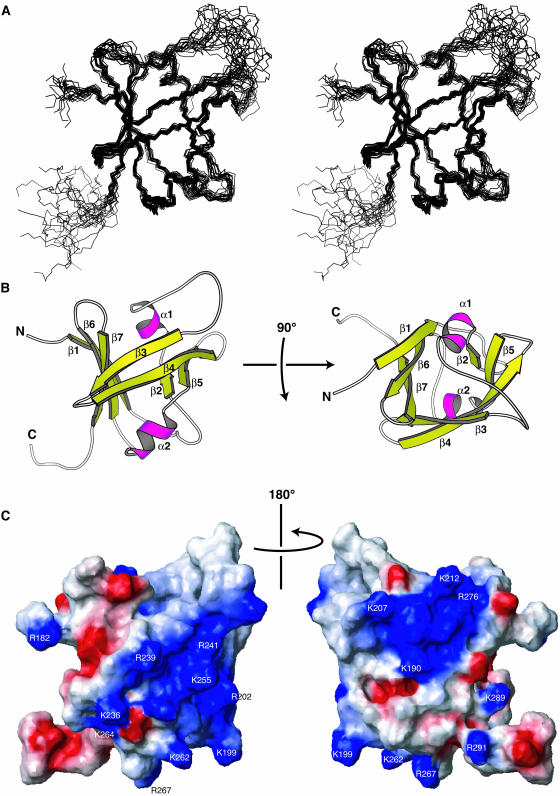
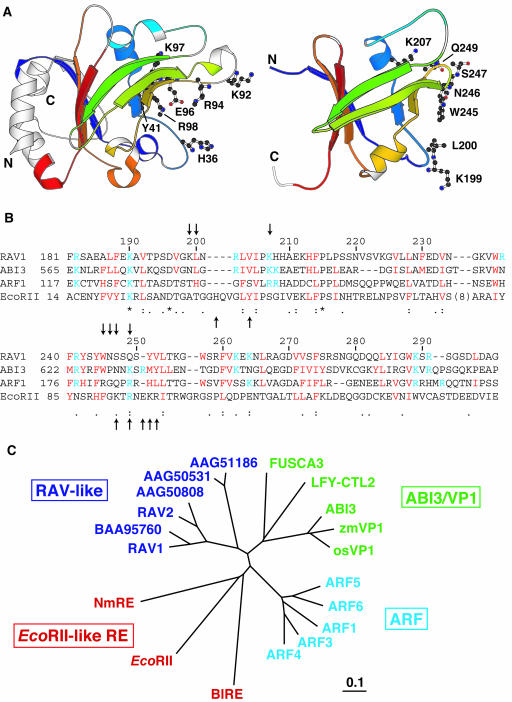
The B3 domain corresponding to Arg182–Ala298 of RAV1 was used for the structural analysis. The experimental constraints and stereochemical properties of the NMR solution structure of the B3 domain of RAV1 (RAV1-B3) are shown in Table 1. The secondary structure elements are seven β-strands (β1, Glu185–Ala191; β2, Leu203–Ile205; β3, Val226–Asp232; β4, Lys236–Trp245; β5, Ser250–Thr254; β6, Val271–Ser277; β7, Leu284–Lys289) forming a β-sheet and two α-helices (α1, Lys207–Lys212; α2, Trp257–Glu263) formed between strands β2 and β3, and β5 and β6, as defined by the program Procheck-NMR (Laskowski et al., 1996) (Figures 1 and 2B). The β-strands are connected in an antiparallel manner except for a parallel connection between β3 and β7. The angles between the strands allow the sheet to form a β-barrel–like structure, open between strands β1 and β2. The two α-helices are located at the ends of this open β-barrel.
Table 1.
Structural Statistics
| Structural Constraints | Numbers | |
|---|---|---|
| Sequential NOEsa | 378 | |
| Medium-range NOEs [2 ≤ |i-j| ≤ 4] | 173 | |
| Long-range NOEs [|i-j| > 4] | 582 | |
| Hydrogen bonds | 82 | |
| Torsion (φ) angles | 70 | |
| Torsion (χ1) angles | 31 | |
| Total | 1316 | |
| Characteristics | Ensemble of 20 Structures | Minimized Mean Structure |
| Root mean square deviation from constraints | ||
| NOEs and hydrogen bonds (Å) | 0.0025 ± 0.0010 | 0.0020 |
| Torsion angles (°) | 0.0659 ± 0.0219 | 0.169 |
| van der Waals energy (kcal/mol)b | 8.0 ± 1.1 | 7.2 |
| Root mean square deviation from the ideal geometry | ||
| Bond lengths (Å) | 0.0008 ± 0.0001 | 0.0007 |
| Bond angles (°) | 0.271 ± 0.004 | 0.267 |
| Improper angles (°) | 0.097 ± 0.006 | 0.097 |
| Average root mean square deviation from the unminimized mean structure (Å)c | ||
| N, Cα, and C | 0.49 ± 0.07 | |
| All heavy atoms | 0.99 ± 0.06 | |
| Ramachandran plot | ||
| Most favored region (%) | 75.9 | 78.6 |
| Additionally allowed region (%) | 21.3 | 19.4 |
| Generously allowed region (%) | 0.9 | 1.0 |
| Disallowed region (%) | 2.0 | 1.0 |
Nuclear Overhauser enhancements.
Values calculated with the force field for NMR structure determination in the CNS package.
Values calculated with Ala184–Val197, Arg202–Phe214, Val226–Trp245, Ser250–Ser277, and Gln283–Ser290.
Figure 1.
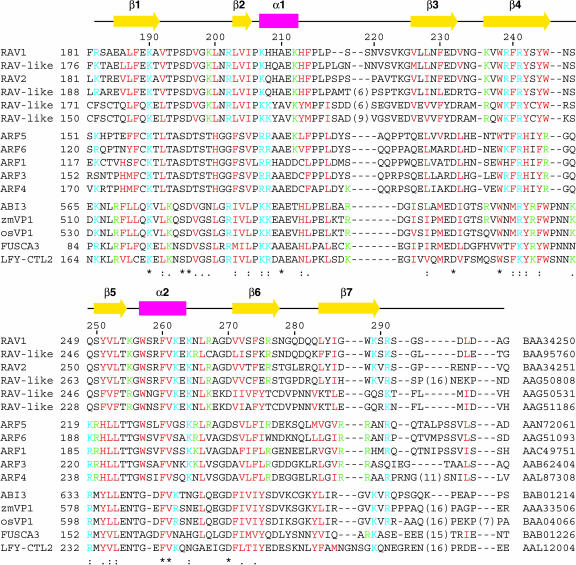
Sequence Alignment of the B3 Domains of Proteins Belonging to the RAV-Like, ARF, and ABI3/VP1 Families Produced by the ClustalX Program (Thompson et al., 1997).
Six RAV-like family proteins, five ARFs (ARF1, 3, 4, 5, and 6), and five ABI3/VP1 proteins (ABI3, 2 VP1s, FUSCA3, and leafy cotyledon 2 protein) were obtained from the National Center for Biotechnology Information (NCBI) database (http://www.ncbi.nlm.nih.gov). Entry codes are shown at the ends of the sequences. All are Arabidopsis proteins except for two VP1 proteins from maize (zmVP1) and rice (Oryza sativa) (osVP1). Numbers above the sequences are for RAV1, whereas those of the first residues of the aligned sequences of the individual proteins are indicated beside the sequences. Basic (Arg and Lys) residues conserved in more than half of the proteins presented here are cyan, whereas hydrophobic (Ile, Leu, Met, and Val) or aromatic (His, Phe, Trp, and Tyr) residues conserved in more than half of the proteins are red. Other than these residues, basic residues conserved in 80% or more of the proteins within the individual groups are green. Bold bars in magenta and arrows in yellow above the sequence alignment indicate regions of helices and strands, respectively, of RAV1-B3. Below the sequence, identical (*), conserved (:), and semiconserved residues (.) are marked as produced by the program ClustalX.
Figure 2.
Solution Structure of the RAV1-B3 Domain.
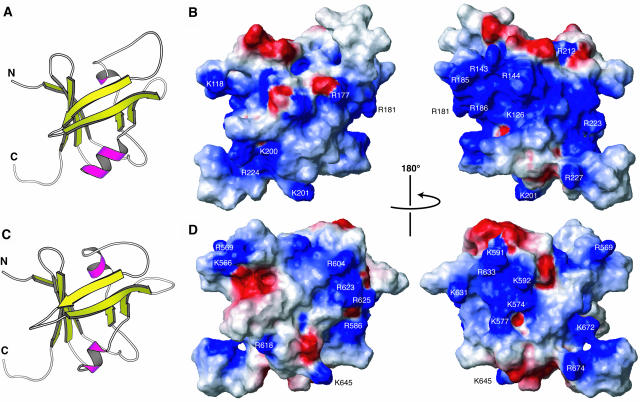
The ensemble of the selected structures in stereo view (A), ribbon diagrams of the minimized mean structure (B), and the contact surface with the presentation of the electrostatic polarization (C) (blue, positive; red, negative) are shown, where the orientation in (A) is the same as in the left panels of (B) and (C). In (B), α-helices are magenta, and β-strands are yellow. The secondary structure units are marked in (B), and the conserved basic residues colored either cyan or green in Figure 1 are indicated in (C). The figures were produced by the programs Molscript (Kraulis, 1991) and MOLMOL (Koradi et al., 1996).
The structural core formed inside of the β-barrel consists of the hydrophobic side chains of Phe188, Leu203, Ile205, Leu228, Phe230, Trp238, Phe240, Tyr242, Leu253, Val272, Phe274, Leu284, and Ile286. Aromatic residues in the two α-helices and the surrounding regions, such as His209, His213, Phe214, Trp257, and Phe260, form contacts to the core residues listed above, stabilizing the packing of the α-helices to the barrel. It should be noted that all of these hydrophobic and aromatic residues, except for His209, are highly conserved in the B3 domains of the three families (Figure 1), indicating that the structural architecture is essentially the same amongst the B3 domains of the different families.
Electrostatic interactions are observed between the side chains of Lys190–Asp196, Arg202–Asp196, Lys207–Glu211, Lys212–Glu211, Lys236–Glu263, Arg259–Glu263, Lys264–Asp232, Lys264–Glu263, Arg267–Asp270, Arg276–Asp281, Lys289–Glu189, and Lys289–Asp297, as defined by the minimal Hη (or Hζ)–Oδ (or Oɛ) distances <5.0 Å in at least two structures of the ensemble. These electrostatic interactions are also likely to contribute to the stability of the structure. It should be noted that for the electrostatic interaction of Lys190–Asp196, the two residues are conserved in all the B3 domains listed in Figure 1, and the basic or acidic residues forming the electrostatic interactions at positions 202, 207, 211, 212, 232, 236, 263, 264, 267, 270, 276, and 289 (hereafter, residues are numbered according to their positions in RAV1 in the alignment in Figure 1, unless specifically stated) are highly conserved (Figure 1).
DNA Recognition by RAV1-B3
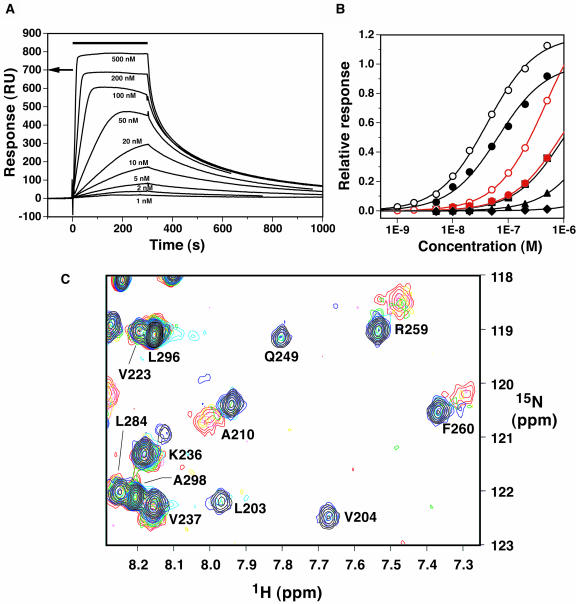
The recognition sequence of RAV1-B3 was determined as CACCTG by a random-oligonucleotide selection experiment (Kagaya et al., 1999). Using surface plasmon resonance (SPR), RAV1-B3 was demonstrated to bind significantly to a double-stranded DNA containing this consensus sequence (Figures 3A and 3B), with a binding constant of 2.0 × 107 M−1 (pH 6.0) or 1.5 × 107 M−1 (pH 7.0) at low ionic strength (100 mM KCl). The binding is sensitive to the ionic strength and was not observed at high salt concentration (500 mM KCl; Figure 3B). The response ratio of the maximal binding of the protein to the DNA immobilized on the sensor chip suggests that RAV1-B3 binds to DNA with a stoichiometry of 1:1. Nonspecific binding to a DNA with an unrelated sequence was also observed, although the apparent binding constant is smaller by a factor of ∼10 (Figure 3B). Thus, the recognition specificity to the sequence CACCTG was observed for the domain used in this study.
Figure 3.
DNA Binding of RAV1-B3 Observed by SPR and NMR.
(A) SPR difference sensorgrams (the responses in the control flow cell were subtracted) for binding of RAV1-B3 to a double-stranded 16-mer DNA (5′-CGATCACCTGAGGCTG-3′/5′-CAGCCTCAGGTGATCG-3′) (the consensus recognition sequence of RAV1-B3, as determined by Kagaya et al., 1999, is underlined), where the protein concentrations are 1, 2, 5, 10, 20, 50, 100, 200, and 500 nM. The solution is at pH 6.0 and a KCl concentration of 100 mM. The protein solutions were injected during the period indicated by the horizontal bar. The arrow indicates the maximum response value expected when the protein molecules bind to all the immobilized DNA molecules at a 1:1 stoichiometry.
(B) Equilibrium response values relative to the expected maximum values at a 1:1 stoichiometry as functions of the protein concentration. The binding profiles of RAV1-B3 to the double-stranded 16-mer DNA (as above) (black) or to a 17-mer DNA with an unrelated sequence (5′-GACGTCCGTACAACAAG-3′/5′-CTTGTTGTACGGACGTC-3′) (red) at pH 6.0 (open symbols) or pH 7.0 (closed symbols) are shown. The KCl concentration was 100 (circles), 200 (squares), 300 (triangles), or 500 (diamonds) mM. Fitting curves to the simple 1:1 binding model are shown.
(C) An overlay of a selected region of the 1H-15N HSQC spectra of RAV1-B3 in the absence (black) or the presence of 0.25 (blue), 0.5 (cyan), 0.75 (green), 1.0 (yellow), 1.25 (light magenta), and 1.5 (red) equimolar concentrations of the 16-mer DNA. The final spectrum shown in red was recorded at approximately three times larger the number of scans than the first spectrum. Spectra were recorded at the proton frequency of 750 MHz.
The binding constant of RAV1-B3 to the DNA containing the consensus sequence is not very strong (1.5 to 2.0 × 107 M−1) for a specific interaction because binding constants for specific protein–DNA interactions obtained by SPR are typically 108 to 109 M−1 (e.g., Bondeson et al., 1993; Seimiya and Kurosawa, 1996; Kamionka et al., 2004; Yamasaki et al., 2004). This weak binding, however, may be overcome by the contribution of the other DBD of the AP2/ERF-type existing in the RAV1 protein. It should be noted that ABI3/VP1 proteins form a complex with basic domain/leucine zipper-type DNA binding proteins to elongate the recognition sequence (Hobo et al., 1999; Nakamura et al., 2001). Also, most of the ARF proteins possess the ability to form dimers through their C-terminal dimerization domain and to recognize two sets of the palindromically arranged recognition sequences (Ulmasov et al., 1997, 1999). Therefore, even if the sequence-specific DNA binding by the isolated B3 domain is not very strong, binding of the full-length proteins or protein complexes could be stronger in vivo to regulate the expression of specific genes.
The regions of a protein surface responsible for its binding to DNA tend to be positively charged, simply because the DNA is negatively charged. Two significantly large, continuous areas of the surface of RAV1-B3 appear to be positively charged (Figure 2C). The larger of the areas (seen at left in Figure 2C) contains nine conserved basic residues, whereas the smaller (seen at right in Figure 2C) contains four conserved basic residues. Because the two areas cover so large a portion of the surface that some of these basic residues (e.g., Lys212 and Lys262) are located on the opposite sides of the molecule, it is unlikely that all of these residues are involved in the binding to DNA. Some of them, for example, Lys190, Lys202, Lys207, Lys212, Lys236, Lys264, Arg267, and Arg276, are involved in the intramolecular electrostatic interactions mentioned above, although this does not exclude the possibility of their involvement in DNA binding.
An NMR titration experiment was performed to elucidate the protein–DNA interface (Figure 3C). By adding increasing amounts of the DNA, chemical shift perturbations were observed in heteronuclear single-quantum coherence (HSQC) spectra as follows. The positions of some cross-peaks did not change (e.g., Val223, Leu284, Leu296, and Ala298 in Figure 3C) or shift only slightly so that the chemical shift changes were easy to follow (e.g., Lys236 and Val237 in Figure 3C). Although the changes could not be followed, some cross-peaks originating from the complex appear in a position close to that of the free protein (e.g., Ala210, Arg259, and Phe260 in Figure 3C). For others, however, the difference was very large so that the cross-peaks of the protein–DNA complex could not be assigned (e.g., Leu203, Val204, and Gln249 in Figure 3C). These chemical shift changes were completed when the concentration ratio of DNA to protein reached ∼1.0, which indicated a 1:1 binding stoichiometry of the protein–DNA complex.
It should be noted that the peak positions appear to continuously change (fast exchange in the NMR time scale) when the difference is as small as ∼0.03 ppm in 1H dimension (or ∼0.3 ppm in 15N dimension), that is, ∼20 Hz (e.g., Lys236 and Val237 in Figure 3C), whereas the change is not continuous (slower exchange) when the difference is as large as ∼0.1 ppm in 1H dimension, that is, ∼80 Hz (e.g., Ala210, Arg259, and Phe260 in Figure 3C). Therefore, if the chemical exchange reaction is a global event and common among these residues, its reaction time should be in the region of 20 to 80 Hz because the fast exchange indicates that the exchange reaction is faster than the chemical shift difference, and slow exchange indicates that the reaction is slower than the chemical shift difference. This presumable global exchange event may correspond to equilibrium between specific binding and nonspecific association but not that between the binding and free states, considering that a much slower dissociation reaction was observed in the SPR experiment (Figure 3A). It also should be noted that there is a possibility that the cross-peaks of Leu203, Val204, and Gln249 in Figure 3C are broadened by matching the chemical shift difference with the rate of the exchange reaction described above (intermediate exchange). We have recorded HSQC spectra of the complex at a different magnetic field (1H frequency of 500 MHz) and different temperatures (293, 298, and 303 K; the protein is unstable at higher temperature), which induces perturbation in the chemical shift difference or exchange rate. However, any new peaks did not appear in the vicinity of the cross-peaks of Leu203, Val204, and Gln249 of the free protein (data not shown). Therefore, it is more likely that the cross-peaks of these residues in the complex are not broadened but that they appear in significantly distant positions.
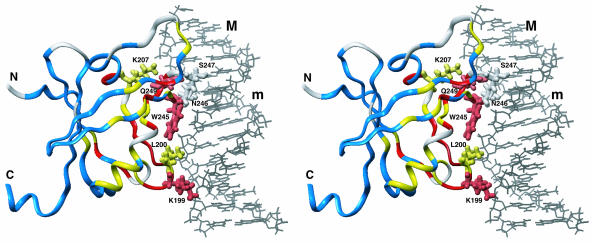
By classifying the residues according to their chemical shift differences, it became clear that one side of the structure is largely affected by the binding of DNA (Figure 4). Based on this result, we have built a structural model of the complex of RAV1-B3 and standard B-type DNA using a computational approach (Figure 4). In this model, side chains of Asn, Ser, and Gln residues, which are typical base-recognizing residues through the formation of hydrogen bonds (Pabo and Sauer, 1992), at positions 246, 247, and 249, respectively, on the loop between β-strands 4 and 5 (loop 45), deeply enter the major groove and contact bases. Also, Lys199 and Leu200 on the loop between β-strands 1 and 2, Lys207 on α-helix 1, and Trp245 on loop 45 contact the sugar–phosphate backbone, among which Lys199 and Lys207 form hydrogen bonds (and electrostatic interactions) to the phosphate groups. Thus, the protein–DNA interface in the proposed model does not correspond to either of the two positively charged areas described above (Figure 2C), but it involves parts of the both areas.
Figure 4.
A Predicted Model of the Complex of RAV1-B3 (Ribbon Representation of Different Colors Described Below) and a Standard B-Type DNA Molecule (Black Wire) in Stereo View.
The molecules are orientated as in Figure 2A. Amino acid residues with backbone chemical shifts affected by the DNA binding are red [combined chemical shift difference (ΔδH2+ (ΔδN/5.0)2)1/2 ≥ 0.2 ppm; residues 191, 193, 198, 199, 201 to 204, 208, 209, 225, 242, 245, 249 to 251, and 254) or yellow (combined chemical shift difference ≥ 0.1 ppm; residues 190, 196, 197, 200, 205, 207, 221, 222, 227, 239, 241, 244, 252, 253, 257, 259 to 263, and 272). Those only slightly affected (combined chemical shift difference < 0.1 ppm; residues 184 to 189, 192, 210 to 214, 216, 223, 224, 226, 228 to 238, 240, 243, 258, 264 to 271, 273 to 279, 282 to 284, and 286 to 298) are shown in blue. Residues for which no meaningful information, that is, those with unassigned or unobserved backbone resonances in the free protein, or Pro residues (residues 182, 183, 194, 195, 206, 215, 217 to 220, 246 to 248, 255, 256, 280, 281, and 285) are shown in white. The residues contacting DNA in this model are shown in stick representation. The major and minor grooves of DNA are indicated by M and m, respectively. It should be noted that this is a general model and does not specifically indicate detailed protein–DNA interaction, possessing uncertainty of the relative orientation of the two molecules (see supplemental data online). The figure was produced by the Insight II molecular display program (Accelrys).
It should be noted that at position 207, basic residues are completely conserved amongst the B3 domains of the three families (Figure 1). Although the Lys residues conserved at position 199 of RAV-like proteins are Thr or Asn in most of the ARFs (ARFs other than those listed in Figure 1 are also considered) and Ser or Asn in ABI3/VP1 proteins, all of these amino acids are capable of hydrogen bond formation to the DNA phosphate group. Also, at position 200, aliphatic or aromatic residues are conserved, although the residues at this position are not involved in the formation of the structural core, and may therefore be involved in DNA binding. It is more important to note that at positions 245, 246, 247, and 249, different residues are conserved for each family, as follows. At position 245, Trp is conserved for the B3 domains of RAV-like and ABI3/VP1 families, whereas Arg is conserved in ARFs (Figure 1). At position 246, Asn is conserved in RAV-like proteins (Arg and Lys for two members) and ABI3/VP1 proteins, whereas Gly is conserved in ARFs. This is likely to be consistent with the conservation pattern at position 245, considering the possibility that the Arg residue of ARFs, which is also a typical base-recognizing residue (Pabo and Sauer, 1992), at this position enters the groove instead of the Gly residue at position 246. It is also important to note that Ser and Gln are conserved at positions 247 and 249, respectively, in RAV-like proteins, whereas they are Gln/Thr and Lys/Arg/Gln in ARFs (ARFs other than those listed in Figure 1 are also considered) and Lys (except for an Asn) and Arg in ABI3/VP1 proteins, although the insertion of two residues (between position 245 and 246 in Figure 1) in ABI3/VP1 proteins makes the alignment ambiguous in this region.
These observations suggest that the DNA-recognition modes adopted by the B3 domains of the three families share a common structural framework, but, on the other hand, that the differences in the base-contacting residues are responsible for the differences in the recognition sequence. In this model, only 2 to 4 bp can be recognized by three amino acid side chains. The possibility cannot be excluded, however, that a slight kinking or untwisting of the DNA and concomitant reorientation of the amino acid side chains will make additional residues, such as Lys207 and Trp245, available to contact the bases, and thereby allow recognition of 6 bp of the consensus recognition sequence. Therefore, the detailed mechanism of the sequence-specific DNA recognition by the B3 domains should be analyzed in the future by an experimental determination of the structure of the complex.
Structures of ARF1- and ABI3-B3
By a homology modeling approach based on the RAV1-B3 structure, the structures of the B3 domains of ARF1 (ARF1-B3) and ABI3 (ABI3-B3) were predicted. The modeled structures are very similar to RAV1-B3 and are energetically favored, which confirms that the structural architecture is shared amongst these B3 domains. Slight differences, however, are observed in the length of secondary structure units. It should be noted that the length of α-helix 2 in ABI3-B3 is significantly shorter than the other two B3 domains (Figure 5). This is likely to be caused by the deletion of a residue and the replacement of a Trp residue by Gly in the region corresponding to α-helix 2 (Figure 1).
Figure 5.
Structural Models of ARF1-B3 (Top) and ABI3-B3 (Bottom).
The ribbon diagrams ([A] and [C]) and the contact surfaces with the presentation of the electrostatic polarization ([B] and [D]) (blue, positive; red, negative) are shown where the orientation in (A) and (C) and in the left panels of (B) and (D) is the same as in the left panel of Figure 2B. Positions of the conserved basic residues are shown on the surfaces.
The electrostatic potential of these two B3 domains is not strikingly similar to that of RAV1-B3 (Figures 5B and 5D). A significantly larger area is positively charged in the surface of ARF1, suggesting a stronger binding to DNA. However, the possible DNA binding regions suggested above, which include Arg143, Arg181, and Arg185 of ARF1 or Arg586, Lys591, Lys631, and Arg633 of ABI3, form continuous, positively charged patches on the surface (Figures 5B and 5D), which is consistent with the proposed common structural framework of DNA binding.
Similarity to the EcoRII N-Terminal Domain
The Protein Data Bank was searched for structures similar to that of RAV1-B3 using the program DALI (Holm and Sander, 1993). Only a single structure, that of the N-terminal effector binding domain (hereafter referred to as the N domain) of the restriction enzyme EcoRII (Zhou et al., 2004), was identified to be significantly similar to that of RAV1-B3, possessing a Z-score of 6.3 and a root mean square deviation value of 3.4 Å (Figure 6). This restriction enzyme, in addition to a catalytic domain, possesses an effector binding domain, which binds specifically to the DNA sequence CCWGG, where W is A or T, without catalyzing cleavage (Mücke et al., 2002). The structure possesses an eight-stranded open β-barrel and seven α-helices and was considered to be a novel fold (Zhou et al., 2004). A comparison of the structures (Figure 6A) makes it clear that the first seven of the eight β-strands of the EcoRII N domain are connected in exactly the same manner as the seven β-strands of RAV1-B3. In addition, some of the α-helices of the EcoRII N domain structure are located at equivalent positions to the two α-helices of RAV1-B3. The major structural difference is localized to the N and C termini; that is, the EcoRII N domain possesses an additional α-helix at its N terminus and an α-helix/β-strand/α-helix structure at its C terminus.
Figure 6.
Structural and Sequence Similarity between the N-Terminal Effector Binding Domain of the Restriction Enzyme EcoRII and the B3 Domains.
(A) Structures of the EcoRII N-terminal domain (left) and RAV1-B3 (right) are viewed from the same orientation as aligned by the program DALI (Holm and Sander, 1993). The aligned segments by DALI are shown in the same colors. The candidate residues involved in the binding to DNA, as suggested by Zhou et al. (2004), and in this study (Figure 4) are shown in the ball-and-stick representation. The figure was produced by Molscript (Kraulis, 1991).
(B) Sequence alignment of the B3 domains of RAV1, ABI3, and ARF1 and the N-terminal domain of EcoRII produced by the ClustalX program (Thompson et al., 1997). Numbers above the sequences are for RAV1, whereas those of the first residues of the aligned sequences of the individual proteins are indicated beside the sequences. Below the sequence, identical and similar residues are marked as produced by the program ClustalX. The conserved basic and hydrophobic or aromatic residues that are in cyan and red, respectively, in Figure 1 are colored in the same way. In addition, basic, hydrophobic, and aromatic residues located in the same position as the above residues in this alignment are also colored. Note that the alignment of the first three sequences is not exactly the same as that in Figure 1. The predicted DNA-contacting residues are indicated by the arrows pointing downward (RAV1) or upward (EcoRII).
(C) A phylogenetic tree of the three B3 domain families and the N-terminal domains of EcoRII-like restriction enzymes as produced by ClustalX. The same B3 domains as in Figure 1 were selected, in which some of the RAV-like family proteins are shown by their NCBI entry codes. NmRE (NCBI code AAF41665) and BlRE (AAN25267) are putative restriction enzymes of Neisseria meningitidis and Bifidobacterium longum, respectively, possessing high similarity to EcoRII.
The amino acid sequences of the three representative B3-domains and the EcoRII N domain were compared in Figure 6B, where identical and similar residues are observed throughout the region. The pairwise alignment of the respective sequences in the region shown in Figure 6B revealed identity values of 19.3 and 57.1%, where conservative changes were permitted, for RAV1-B3 and the EcoRII N domain (ratios were obtained using the amino acid length of RAV1-B3). These values are comparable to those obtained by a comparison of RAV1-B3 and ARF1-B3 (23.5% identity and 68.9% permitting conservative changes). Also, a pairwise sequence comparison of ARF1-B3 or ABI3-B3 and the EcoRII N domain revealed identity values of 16.8 or 15.0% and 58.0 or 55.0%, permitting conservative changes. A phylogenetic tree for all the B3 domains listed in Figure 1 and the N-terminal domains of EcoRII and similar putative restriction enzymes (Figure 6C) indicates that the distances between the N domains of these restriction enzymes are similar to those of the B3 domains of different families.
Residues of the EcoRII N domain involved in DNA binding were suggested by a biochemical analysis (Reuter et al., 1999) and the cleft-like shape formed by the relevant region (Zhou et al., 2004). The locations of these residues are very similar to those of RAV1-B3 that are suggested to directly contact the DNA as discussed above, in both the tertiary structure and the primary sequence (Figure 6). Therefore, the two structures are related not only in the secondary structure arrangement but probably also in the DNA binding mechanism. To our knowledge, this is the second example that the DBDs of plant-specific transcription factors and the prokaryotic endonucleases are structurally and functionally related, where the first example is that of the AtERF1 protein and Tn916 integrase (Allen et al., 1998; Wojciak et al., 1999), which use a three-stranded β-sheet for DNA binding. These two cases are similar to each other in that the prokaryotic endonucleases possess an N-terminal noncatalytic DBD and a C-terminal catalytic domain, the former being related to the DBD of a plant-specific transcription factor.
METHODS
Sample Preparation
The DNA that codes for RAV1-B3 (Arg182-Ala298) was subcloned by PCR from an Arabidopsis thaliana full-length cDNA clone (Seki et al., 2002) with the identification code RAFL06-79-P13 (Munich Information Center for Protein Sequences code At1g13260). The PCR primers used were designed so that the T7 promoter sequence, ribosome binding site, and oligohistidine tag, as well as the cleavage site for tobacco etch virus protease are attached at the 5′ end, and the T7 terminator sequence is attached at the 3′ end (T. Yabuki, Y. Motoda, M. Saito, N. Matsuda, T. Kigawa, and S. Yokoyama, unpublished results). Consequently, additional amino acids derived from the expression vectors were attached to the protein fragments, that is, GlySerSerGlySerSerGly to the N terminus and SerGlyProSerSerGly to the C terminus. The 13C-, 15N-labeled, and unlabeled proteins were expressed by a cell-free system developed in RIKEN (Kigawa et al., 1999, 2004; Yokoyama et al., 2000). The protein was purified by HiTrap chelating and HiTrap SP (Amersham, Piscataway, NJ) columns. The buffers used were 50 mM sodium phosphate, pH 8.0, 500 mM NaCl, and 20 to 500 mM imidazole for HiTrap chelating chromatography, and 20 mM sodium phosphate, pH 7.0, and 200 mM to 1 M NaCl for HiTrap SP chromatography. The apparent molecular mass of the purified protein is 17.2 kD as obtained by a gel filtration analysis (data not shown), which indicates a monomeric state considering the theoretical molecular mass of 14.3 kD. Protein concentration was determined by A280 values and molar absorption coefficients calculated from the amino acid sequences (Pace et al., 1995). For NMR measurements, ∼1.0 mM proteins were dissolved in 20 mM potassium phosphate buffer, pH 7.0, containing 500 mM KCl, 0.1 mM sodium 2,2-dimethyl-2-silapentane-5-sulfonate, and 5% D2O, unless otherwise stated.
NMR Measurements and Resonance Assignments
Typical homonuclear and heteronuclear NMR spectra (Wüthrich, 1986; Bax, 1994) were recorded on Bruker DMX-750 (750.13 MHz for 1H and 76.02 MHz for 15N) and DMX-500 (500.13 MHz for 1H, 125.76 MHz for 13C, and 50.68 MHz for 15N) spectrometers (Bruker Instruments, Billerica, MA) at 293 K, essentially as previously described (Yamasaki et al., 2004). The recorded spectra were analyzed for the backbone and side chain resonances assignments also as described previously (Yamasaki et al., 2004). HSQC spectra of samples containing 100% D2O were recorded at 293 K, by which 41 hydrogen bond donors were identified. Using the HMQC-J experiment (Kay and Bax, 1989), 70 3J HNHα coupling values were obtained. By analyzing the NOESY, TOCSY, and DQF-COSY spectra, 26 and five pairs of Hβ and Val Hγ resonances, respectively, were assigned stereospecifically (Wagner et al., 1987).
Determination of the Three-Dimensional Structure
The distance constraints derived from the NOESY spectra were classified into four categories, 1.5 to 2.8, 1.5 to 3.5, 2.0 to 4.5, and 2.5 to 6.0 Å, according to the relationship that NOE intensity is inversely proportional to the sixth power of distance, which was calibrated with the average intensities of the intraresidue NOEs of Hδ–Hɛ pairs of Phe or Tyr possessing fixed distances (2.5 Å). When a pair of NOEs from chemically equivalent protons (e.g., Hβ1 and Hβ2) without stereospecific assignment are classified into the same distance category, two separate constraints were imposed as if they were stereospecifically assigned. For other NOEs from stereospecifically unassigned protons, the constraints were imposed using the sum-averaged distances from all the equivalent protons (Nilges, 1993). For maintaining a hydrogen bond, a constraint of 1.5 to 2.5 Å was imposed on the distance between the hydrogen and acceptor oxygen, and another constraint of 2.5 to 3.5 Å was imposed on the distance between the donor nitrogen and acceptor oxygen.
The φ angle constraints were classified into two categories, −120° ± 40° and −55° ± 35°, corresponding to the 3JαN coupling values of 7.0 Hz or larger and 6.0 Hz or smaller, respectively. For stereospecifically assigned residues, χ1 torsion angle constraints classified into three categories, 60° ± 40°, 180° ± 40°, and −60° ± 40°, were imposed.
Random simulated annealing (Nilges et al., 1988) was performed using the program CNS (Brünger et al., 1998). At the initial stage of the structure calculation, NOE distance and dihedral angle constrains were imposed. Force constants of 150 kcal mol−1 Å−2 and 100 to 200 kcal mol−1 rad−2 were used for the distance and dihedral angle constraints, respectively. The hydrogen bond distance constraints were used only at the final stage of the calculation so that the hydrogen bond acceptors were determined using the structures obtained at an earlier stage. From 50 initial structures, 20 structures with no distance violations larger than 0.2 Å, no torsion angle violations larger than 2°, and the lowest total energies were selected as accepted structures. The minimized mean structures were produced by a protocol for the selection of the accepted structures in the CNS program. The average root mean square deviations of the coordinates from the unminimized mean structure of the ensemble (Table 1) were calculated by the program MOLMOL (Koradi et al., 1996). The φ and ψ dihedral angles were analyzed using the program Procheck-NMR (Laskowski et al., 1996). The secondary structure elements were also identified by Procheck-NMR.
SPR
Experiments were performed at 293 K using a Biacore X apparatus (BIACORE, Uppsala, Sweden). The running buffer was 20 mM potassium phosphate (pH 6.0 or 7.0) containing 100 to 500 mM KCl and 0.005% Tween 20. Altogether, 551 and 596 response units of two double-stranded DNAs (5′-bio-CGATCACCTGAGGCTG-3′/5′-CAGCCTCAGGTGATCG-3′) (the consensus recognition sequence of RAV1-B3, as determined by Kagaya et al., 1999, is underlined), and (5′-bio-GACGTCCGTACAACAAG-3′/5′-CTTGTTGTACGGACGTC-3′), respectively (bio indicates the 5′-biotinylated strand), were immobilized on the surfaces of Sensor Chip SAs (BIACORE). Solutions containing RAV1-B3 at concentrations of 1 to 500 nM were injected into the flow cells at 20 μL min−1 for 5 min. The equilibrium binding constants were obtained by fitting the equilibrium response values at different protein concentrations to the simple 1:1 binding model using BIAevaluation 3.0 software (BIACORE).
NMR Titration Analysis
HSQC spectra of RAV1-B3 at the initial concentration of 0.2 mM dissolved in 20 mM potassium phosphate buffer, pH 7.0, containing 300 mM KCl were recorded at 293 K and 1H frequency of 750.13 MHz, by adding increasing amounts of 6.0 mM of the double-stranded 16-mer DNA (5′-CGATCACCTGAGGCTG-3′/5′-CAGCCTCAGGTGATCG-3′) (the consensus recognition sequence of RAV1-B3, as determined by Kagaya et al., 1999, is underlined). The concentration of the double-stranded DNA was determined using an extinction coefficient calculated after digestion of the strands with phosphodiesterase I (Worthington Biochemical, Lakewood, NJ).
Structural Modeling of ARF1-B3, ABI3-B3, and the RAV1-B3/DNA Complex
The HOMOLOGY module of Insight II (Accelrys, San Diego, CA) was used for the structural modeling of the ARF1- and ABI3-B3 domains based on the sequence homology to RAV1-B3. A stepwise energy minimization procedure was performed using the cff95 force field parameter set and the DISCOVER 3 program module in Insight II package (Accelrys) as follows. Initially, to relax steric clash found in several sites of the molecules, energy minimization was performed for side chains of the amino acid residues having atoms within a cutoff distance of 3 Å from each clashing site. Then, further energy minimization was performed by changing the cutoff distance to 6 Å, and finally, 10 Å. Similar three-step calculations were performed for all of the atoms, including the main chain, of the amino acid residues within the cutoff distances. Then, the entire structures of the molecules were relaxed using the same energy minimization. In addition to above, the entire structures were refined by evaluating the solvation effects with the use of the Generalized Born method implemented in the sander module of the AMBER 7 molecular modeling program package (Case et al., 2002).
The model of the RAV1-B3/DNA complex was built essentially as described previously (Yamasaki et al., 2004).
The coordinates of the determined RAV1-B3 structure have been deposited to the Protein Data Bank under the accession identifier 1WID.
Supplementary Material
Acknowledgments
The authors thank N. Matsuda, Y. Motoda, Y. Fujikura, M. Saito, Y. Miyata, K. Hanada, A. Kobayashi, N. Sakagami, M. Ikari, F. Hiroyasu, Y. Nishimura, M. Watanabe, M. Sato, M. Hirato, and Y. Kamewari at RIKEN and H. Yamaguchi at the National Institute of Advanced Industrial Science and Technology for technical assistance, as well as P. Reay at RIKEN for critical reading of the manuscript. The molecular modeling calculations in this study were partially conducted using the resources in the Computer Center for Agriculture, Foresty, and Fisheries Research, the Ministry of Agriculture, Forestry, and Fisheries, Japan. This work was supported in part by the RIKEN Structural Genomics/Proteomics Initiative and the National Project on Protein Structural and Functional Analyses, Ministry of Education, Culture, Sports, Science, and Technology.
The authors responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) are: Kazuo Shinozaki (sinozaki@rtc.riken.go.jp) and Shigeyuki Yokoyama (yokoyama@biochem.s.u-tokyo.ac.jp).
Online version contains Web-only data.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.104.026112.
References
- Abel, S., and Theologis, A. (1996). Early genes and auxin action. Plant Physiol. 111, 9–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen, M.D., Yamasaki, K., Ohme-Takagi, M., Tateno, M., and Suzuki, M. (1998). A novel mode of DNA recognition by a β-sheet revealed by the solution structure of the GCC-box binding domain in complex with DNA. EMBO J. 17, 5484–5496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bax, A. (1994). Multidimensional nuclear magnetic resonance methods for protein studies. Curr. Opin. Struct. Biol. 4, 738–744. [Google Scholar]
- Bondeson, K., Frostell-Karlsson, A., Fagerstam, L., and Magnusson, G. (1993). Lactose repressor-operator DNA interactions: Kinetic analysis by a surface plasmon resonance biosensor. Anal. Biochem. 214, 245–251. [DOI] [PubMed] [Google Scholar]
- Brünger, A.T., et al. (1998). Crystallography and NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 54, 905–921. [DOI] [PubMed] [Google Scholar]
- Case, D.A., et al. (2002). AMBER 7. (San Francisco: University of California).
- Ezcurra, I., Wycliffe, P., Nehlin, L., Ellerström, M., and Rask, L. (2000). Transactivation of the Brassica napus napin promoter by ABI3 requires interaction of the conserved B2 and B3 domains of ABI3 with different cis-elements: B2 mediates activation through an ABRE, whereas B3 interacts with an RY/G-box. Plant J. 24, 57–66. [DOI] [PubMed] [Google Scholar]
- Fowler, S., and Thomashow, M.F. (2002). Arabidopsis transcriptome profiling indicates that multiple regulatory pathways are activated during cold acclimation in addition to the CBF cold response pathway. Plant Cell 14, 1675–1690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giraudat, J., Hauge, B.M., Valon, C., Smalle, J., Parcy, F., and Goodman, H.M. (1992). Isolation of the Arabidopsis-ABI3 gene by positional cloning. Plant Cell 4, 1251–1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobo, T., Kowyama, Y., and Hattori, T. (1999). A bZIP factor, TRAB1, interacts with VP1 and mediates abscisic acid-induced transcription. Proc. Natl. Acad. Sci. USA 96, 15348–15353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holm, L., and Sander, C. (1993). Protein structure comparison by alignment of distance matrices. J. Mol. Biol. 233, 123–138. [DOI] [PubMed] [Google Scholar]
- Kagaya, Y., Ohmiya, K., and Hattori, T. (1999). RAV1, a novel DNA-binding protein, binds to bipartite recognition sequence through two distinct DNA-binding domains uniquely found in higher plants. Nucleic Acids Res. 27, 470–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamionka, A., Bogdanska-Urbaniak, J., Scholz, O., and Hillen, W. (2004). Two mutations in the tetracycline repressor change the inducer anhydrotetracycline to a corepressor. Nucleic Acids Res. 32, 842–847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kay, L.E., and Bax, A. (1989). New methods for measurement of NH-CαH coupling constants in 15N-labelled proteins. J. Magn. Reson. 86, 110–126. [Google Scholar]
- Kigawa, T., Yabuki, T., Matsuda, N., Matsuda, T., Nakajima, R., Tanaka, A., and Yokoyama, S. (2004). Preparation of Escherichia coli cell extract for highly productive cell-free protein expression. J. Struct. Funct. Genomics 5, 63–68. [DOI] [PubMed] [Google Scholar]
- Kigawa, T., Yabuki, T., Yoshida, Y., Tsutsui, M., Ito, Y., Shibata, T., and Yokoyama, S. (1999). Cell-free production and stable-isotope labelling of milligram quantities of proteins. FEBS Lett. 442, 15–19. [DOI] [PubMed] [Google Scholar]
- Koornneef, M., Leon-Kloosterziel, K.M., Schwarts, S.H., and Zeevaart, J.A.D. (1998). The genetic and molecular dissection of abscisic acid biosynthesis and signal transduction in Arabidopsis. Plant Physiol. Biochem. 36, 83–89. [Google Scholar]
- Koradi, R., Billeter, M., and Wüthrich, K. (1996). MOLMOL: A program for display and analysis of macromolecular structures. J. Mol. Graph. 14, 51–55. [DOI] [PubMed] [Google Scholar]
- Kraulis, P.J. (1991). MOLSCRIPT: A program to produce both detailed and schematic plots of protein structures. J. Appl. Crystallogr. 24, 946–950. [Google Scholar]
- Laskowski, R.A., Rullmann, J.A., MacArthur, W.M., Kaptein, R., and Thornton, J.M. (1996). AQUA and PROCHECK-NMR: Programs for checking the quality of protein structures solved by NMR. J. Biomol. NMR 8, 477–486. [DOI] [PubMed] [Google Scholar]
- Leung, J., and Giraudat, J. (1998). Abscisic acid signal transduction. Annu. Rev. Plant Physiol. Plant Mol. Biol. 49, 199–222. [DOI] [PubMed] [Google Scholar]
- McCarty, D.R., Carson, C.B., Lazar, M., and Simonds, S.C. (1989). Transposable element induced mutations of the vivapaurous-1 gene of maize. Dev. Genet. 10, 473–481. [Google Scholar]
- McCarty, D.R., Hattori, T., Carson, C.B., Vasil, V., Lazar, M., and Vasil, I.K. (1991). The vivapaurous-1 developmental gene of maize encodes a novel transcriptional activator. Cell 66, 895–905. [DOI] [PubMed] [Google Scholar]
- Mücke, M., Grelle, G., Behlke, J., Kraft, R., Krünger, D.H., and Reuter, M. (2002). EcoRII: A restriction enzyme evolving recombination functions? EMBO J. 21, 5262–5268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura, S., Lynch, T.J., and Finkelstein, R.R. (2001). Physical interactions between ABA response loci of Arabidopsis. Plant J. 26, 627–635. [DOI] [PubMed] [Google Scholar]
- Nilges, M. (1993). A calculation strategy for the structure determination of symmetric dimers by 1H NMR. Proteins 17, 297–309. [DOI] [PubMed] [Google Scholar]
- Nilges, M., Clore, G.M., and Gronenborn, A.M. (1988). Determination of the three-dimensional structures of proteins from inter-proton distance data by dynamic simulated annealing from a random array of atoms. FEBS Lett. 239, 129–136. [DOI] [PubMed] [Google Scholar]
- Pabo, C.O., and Sauer, R.T. (1992). Transcription factors: Structural families and principles of DNA recognition. Annu. Rev. Biochem. 61, 1053–1095. [DOI] [PubMed] [Google Scholar]
- Pace, C.N., Vajdos, F., Fee, L., Grimsley, G., and Gray, T. (1995). How to measure and predict the molar absorption coefficient of a protein. Protein Sci. 4, 2411–2423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reuter, M., Shneider-Mergener, J., Kupper, D., Meisel, A., Mackeldanz, P., Krünger, D.H., and Schroeder, C. (1999). Regions of endonuclease EcoRII involved in DNA target recognition identified by membrane-bound peptide repertoires. J. Biol. Chem. 274, 5213–5221. [DOI] [PubMed] [Google Scholar]
- Riechmann, J.L., et al. (2000). Arabidopsis transcription factors: Genome-wide comparative analysis among eukaryotes. Science 290, 2105–2110. [DOI] [PubMed] [Google Scholar]
- Seimiya, M., and Kurosawa, Y. (1996). Kinetics of binding of Antp homeodomain to DNA analyzed by measurements of surface plasmon resonance. FEBS Lett. 398, 279–284. [DOI] [PubMed] [Google Scholar]
- Seki, M., et al. (2002). Functional annotation of a full-length Arabidopsis cDNA collection. Science 296, 141–145. [DOI] [PubMed] [Google Scholar]
- Suzuki, M., Kao, C.Y., and McCarty, D.R. (1997). The conserved B3 domain of VIVIPAROUS1 has a cooperative DNA binding activity. Plant Cell 9, 799–807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson, J.D., Gibson, T.J., Plewniak, F., Jeanmougin, F., and Higgins, D.G. (1997). The CLUSTAL-X windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25, 4876–4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiwari, S.B., Hagen, G., and Guilfoyle, T. (2003). The roles of auxin response factor domains in auxin-responsive transcription. Plant Cell 15, 533–543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulmasov, T., Hagen, G., and Guifoyle, T.J. (1997). ARF1, a transcription factor that binds to auxin response elements. Science 276, 1865–1868. [DOI] [PubMed] [Google Scholar]
- Ulmasov, T., Hagen, G., and Guifoyle, T.J. (1999). Dimerization and DNA binding of auxin response factors. Plant J. 19, 309–319. [DOI] [PubMed] [Google Scholar]
- Wagner, G., Braun, G., Havel, T.F., Schaumann, T., Go, N., and Wüthrich, K. (1987). Protein structures in solution by nuclear magnetic resonance and distance geometry. The polypeptide fold of the basic pancreatic trypsin inhibitor determined using two different algorithms, DISGEO and DISMAN. J. Mol. Biol. 196, 611–639. [DOI] [PubMed] [Google Scholar]
- Wojciak, J.M., Connolly, K.M., and Clubb, R.T. (1999). NMR structure of the Tn916 integrase-DNA complex. Nat. Struct. Biol. 6, 366–373. [DOI] [PubMed] [Google Scholar]
- Wüthrich, K. (1986). NMR of Proteins and Nucleic Acids. (New York: John Wiley & Sons).
- Yamasaki, K., et al. (2004). A novel zinc-binding motif revealed by solution structures of DNA-binding domains of Arabidopsis SBP-family transcription factors. J. Mol. Biol. 337, 49–63. [DOI] [PubMed] [Google Scholar]
- Yokoyama, S., et al. (2000). Structural genomics projects in Japan. Nat. Struct. Biol. 7 (suppl.), 943–945. [DOI] [PubMed] [Google Scholar]
- Zhou, X.E., Wang, Y., Reuter, M., Mücke, M., Krüger, D.H., Meehan, E.J., and Chen, L. (2004). Crystal structure of type IIE restriction endonuclease EcoRII reveals an autoinhibition mechanism by a novel effector-binding fold. J. Mol. Biol. 335, 307–319. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.