Abstract
The plant hormones abscisic acid (ABA), jasmonic acid (JA), and ethylene are involved in diverse plant processes, including the regulation of gene expression during adaptive responses to abiotic and biotic stresses. Previously, ABA has been implicated in enhancing disease susceptibility in various plant species, but currently very little is known about the molecular mechanisms underlying this phenomenon. In this study, we obtained evidence that a complex interplay between ABA and JA-ethylene signaling pathways regulate plant defense gene expression and disease resistance. First, we showed that exogenous ABA suppressed both basal and JA-ethylene–activated transcription from defense genes. By contrast, ABA deficiency as conditioned by the mutations in the ABA1 and ABA2 genes, which encode enzymes involved in ABA biosynthesis, resulted in upregulation of basal and induced transcription from JA-ethylene responsive defense genes. Second, we found that disruption of AtMYC2 (allelic to JASMONATE INSENSITIVE1 [JIN1]), encoding a basic helix-loop-helix Leu zipper transcription factor, which is a positive regulator of ABA signaling, results in elevated levels of basal and activated transcription from JA-ethylene responsive defense genes. Furthermore, the jin1/myc2 and aba2-1 mutants showed increased resistance to the necrotrophic fungal pathogen Fusarium oxysporum. Finally, using ethylene and ABA signaling mutants, we showed that interaction between ABA and ethylene signaling is mutually antagonistic in vegetative tissues. Collectively, our results indicate that the antagonistic interactions between multiple components of ABA and the JA-ethylene signaling pathways modulate defense and stress responsive gene expression in response to biotic and abiotic stresses.
INTRODUCTION
In their natural environment, plants are continuously threatened by various biotic and abiotic stresses. Their survival under such conditions is dependent on the ability to perceive external signals and respond in a timely manner. Our current understanding of plant signaling pathways involved in biotic and abiotic stresses is still rudimentary. However, the emerging picture from several studies supports the notion that plant signaling pathways are composed of intricate networks with frequent cross talk that allow the plant to activate an appropriate spectrum of responses depending on the type of stimuli present. This leads to the production of proteins with direct roles in alleviating the damaging effects of stressful conditions.
The plant hormones salicylic acid (SA), jasmonic acid (JA), and ethylene are the major endogenous low molecular weight signal molecules involved in regulating defense responses in plants. In Arabidopsis thaliana, an intact JA-ethylene signaling pathway is thought to be necessary for resistance to necrotrophic pathogens, such as Botrytis cinerea and Erwinia carotovora. By contrast, the SA signaling pathway is believed to mediate the resistance to biotrophic pathogens, such as Erysiphe orontii, Peronospora parasitica, and Pseudomonas syringae (Thomma et al., 2001; Rojo et al., 2003). Thus, it is conceivable that to mount an effective defense response, the plant activates the particular signaling pathways that are most likely to enhance resistance to a class of invader. Simultaneously, other signaling pathways with minimal or no significant effects on the invading pathogen may be suppressed to avoid depletion of valuable physiological resources. An example of such antagonistic interaction occurs between the SA and the JA signaling pathways in Arabidopsis. Mutations that impair SA signaling or biosynthesis result in elevated expression of the JA-ethylene responsive antifungal defensin PDF1.2 (Spoel et al., 2003). By contrast, mutations that constitutively activate the SA-signaling pathway suppress PDF1.2 expression and render the plants susceptible to pathogens (Petersen et al., 2000). However, antagonism between the SA and JA signaling pathways does not apply in all cases, and some defense response genes require intact JA, ethylene, and SA signaling pathways after pathogen challenge (Campbell et al., 2003).
The plant hormone abscisic acid (ABA) regulates interacting signaling pathways involved in plant responses to several abiotic stresses, such as drought, salt, and cold, as well as plant growth and development. The ABA-dependent signaling pathway regulates stress-inducible gene expression through several positive and negative regulators (reviewed in Shinozaki et al., 2003). Genetic analysis of Arabidopsis mutants compromised in ABA biosynthesis or signaling has identified a complex interplay between ABA and various other phytohormone signaling pathways. One of the relatively better characterized genetic interactions occurs among ABA, ethylene, and the sugar signaling pathways (Gazzarrini and McCourt, 2001; Finkelstein and Gibson, 2002; Leon and Sheen, 2003). Genetic analyses have also demonstrated that the ABA signaling pathway interacts antagonistically with the ethylene signaling pathway and vice versa to modulate plant development (Beaudoin et al., 2000; Ghassemian et al., 2000). First, it was suggested that one of the functions of ABA is to inhibit overproduction of ethylene as ABA-deficient maize (Zea mays) (Spollen et al., 2000) and tomato (Lycopersicon esculentum) (Sharp et al., 2000) mutants overaccumulate ethylene and display stunted growth phenotypes. In addition, the maintenance of shoot growth by ABA involves, in part, suppression of ethylene synthesis (LeNoble et al., 2004). By contrast, ethylene negatively regulates ABA signaling in controlling seed dormancy (Beaudoin et al., 2000). For instance, the ethylene insensitive mutant ein2 (allelic to the enhanced response to ABA3 or era3) shows enhanced ABA sensitivity during seed germination (Ghassemian et al., 2000). Mutations in the genes (e.g., ETHYLENE RECEPTOR1 [ETR1], EIN2, and EIN3) involved in positive regulation of ethylene signaling result in a glucose (glo) and an ABA-oversensitivity phenotype with greater growth inhibition in response to exogenous glucose and ABA than in wild-type plants, whereas mutations affecting negative regulators of ethylene signaling (e.g., CONSTITUTIVE TRIPLE RESPONSE1 [CTR1]) result in reduced ABA sensitivity and a glucose insensitivity (gin) phenotype (Zhou et al., 1998; Ghassemian et al., 2000; Cheng et al., 2002). An antagonistic interaction between the ABA and the JA signaling pathways has also been observed in the jasmonic acid resistant1 (jar1) and jasmonic acid insensitive4 (jin4) mutants, which show hypersensitivity to ABA inhibition of germination (Staswick et al., 1992; Berger et al., 1996). Another study found that ABA and JA antagonistically regulate the expression of salt stress–inducible transcripts in rice (Oryza sativa) (Moons et al., 1997).
Although this seemingly antagonistic interaction between the ABA-sugar and the ethylene signaling pathways is relatively well known (reviewed in Finkelstein and Gibson, 2002; Finkelstein et al., 2002; Leon and Sheen, 2003), our current understanding on how this interaction influences other ethylene-mediated plant responses, such as defense gene expression and pathogen resistance, is very limited. Also largely unknown is how the ABA signaling pathway interacts with the JA signaling pathway that in itself partially overlaps with the ethylene signaling pathway in regulating defense gene expression and resistance to necrotrophic pathogens. An increased understanding of the potential interactions between ABA and biotic stress responses is of vital importance for engineering plants for disease resistance without compromising ABA-regulated abiotic stress response pathways.
Here, using various ABA biosynthesis or signaling mutants, we first demonstrated that ABA signaling antagonizes the JA-ethylene responsive defense gene expression and fungal disease resistance in Arabidopsis. Furthermore, using ethylene signaling mutants we demonstrated a mutually antagonistic interaction between ABA and ethylene signaling that modulates stress responsive gene expression during vegetative growth and propose potential points for such interaction. Collectively, our results suggest that the complex interplay between biotic and abiotic stress pathways provides a means of managing and prioritizing diverse stress responses and could have important implications on engineering of biotic and abiotic stress resistance in plants.
RESULTS
ABA Antagonizes JA-Ethylene Responsive Defense Gene Expression in Arabidopsis
Recent results from several studies showed that ABA negatively influences disease resistance phenotypes in various plant species (Rezzonico et al., 1998; McDonald and Cahill, 1999; Audenaert et al., 2002; Mohr and Cahill, 2003). Similarly, water stress reduces disease tolerance to some pathogens (Wildermuth and Morgan, 2004). We hypothesized that the enhanced susceptibility phenotype could be attributable to ABA's suppression of JA-ethylene responsive defense gene expression. To test this hypothesis, we first studied PDF1.2 expression, a known marker gene positively regulated by the JA-ethylene signaling pathway, in plants treated with methyl jasmonate (MJ), ethylene, ABA, or a combination of MJ and ABA or ethylene and ABA. To specifically quantify and compare the changes in defense gene transcript levels after various treatments and/or in mutant backgrounds, we used real-time quantitative RT-PCR (RT-Q-PCR) in all gene expression studies reported in this article. The use of the highly sensitive method of RT-Q-PCR permitted the measurement of basal (uninduced) transcript levels of the genes studied here as well as their induction or suppression by external stimuli. In particular, it should be noted that the quantification of reductions in transcript levels observed after treatments are not readily measured by traditional RNA gel blotting. In these analyses, we quantified target gene expression relative to the expression measured from the reference genes (e.g., three β-actin genes or β-tubulin) in the same sample. The transcript abundance of the reference genes measured as cycle threshold value did not show any significant change after treatments/inoculations (see Methods for details and also Hoth et al., 2002; Campbell et al., 2003; Schenk et al., 2003). Normalization of gene expression using three β-actin genes or β-tubulin also showed a strong correlation with expression normalized relative to that of the 25S rRNA. The high abundance of the 25S rRNA transcripts requires severalfold dilution of cDNAs before use in RT-Q-PCR reactions and therefore cannot be conveniently assayed on the same sample as the assay for the target gene (J. Anderson and K. Kazan, unpublished data).
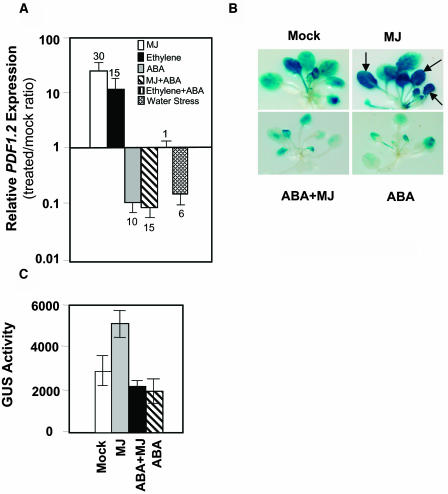
As expected, treatments with MJ and ethylene significantly induced PDF1.2 (30-fold and 15-fold, respectively) in treated wild-type plants relative to the mock-treated plants, whereas ABA treatment caused 10-fold reduction at the basal transcript levels of PDF1.2 relative to those in mock-treated wild-type plants (Figure 1A). In addition, we noted that, in the presence of ABA, neither ethylene nor MJ were able to induce PDF1.2 expression in wild-type plants (Figure 1A). Interestingly, water stress also significantly reduced (sixfold) PDF1.2 transcript levels over those measured in plants not exposed to water stress (Figure 1A).
Figure 1.
Exogenous ABA Suppresses PDF1.2 Expression in Arabidopsis.
(A) Fold changes in relative transcript abundance of the PDF1.2 in MJ-, ethylene-, ABA-, MJ+ABA-, ethylene+ABA- (second to last column), and water stress–treated wild-type (Columbia-0 [Col-0]) plants. Total RNA was isolated from plants 48 h after each treatment, converted to cDNA, and used as template in RT-Q-PCR assays. Transcript levels of PDF1.2 were normalized to the expression of β-actin genes measured in the same samples and expressed logarithmically relative to the normalized transcript levels in mock-treated wild-type plants. Average data with error bars from two independent experiments are presented. The numbers on each bar show fold increase or fold decrease caused by each treatment in PDF1.2 transcript levels relative to those in mock-treated plants.
(B) and (C) Three- to four-week-old homozygous plants transformed with the PDF1.2promoter:GUS construct were either mock treated or treated with ABA, MJ, or combination of ABA and MJ for 48 h. Histochemical GUS staining was performed overnight on 10 seedlings for each experiment (B). The leaves that display strong GUS activity after MJ treatment as evidenced by saturated blue color are indicated by arrows. GUS activity was also measured fluorometrically using at least 10 seedlings for each treatment (C). Average data from two separate experiments are shown. Error bars indicate standard deviation.
We next examined the effect of ABA on MJ-responsive expression of PDF1.2 using Arabidopsis plants transformed with the PDF1.2promoter:GUS construct (Manners et al., 1998). The expression of PDF1.2 expression is known to increase with plant age (He et al., 2002), and this explains the relatively high background GUS expression observed in mock-treated PDF1.2promoter:GUS plants. However, this attribute proved to be useful for testing whether ABA treatment could reduce background GUS activity in these plants. Indeed, both histochemical staining and enzyme activity assays showed significantly reduced GUS expression after ABA treatment as well as MJ-ABA cotreatment in these plants, whereas as reported previously by Manners et al. (1998), the MJ-treatment significantly induced GUS activity in PDF1.2promoter:GUS plants as evidenced by saturated dark blue color on the leaves (Figures 1B and 1C). Collectively, these results suggested that ABA acts antagonistically to MJ and ethylene in suppressing PDF1.2 expression.
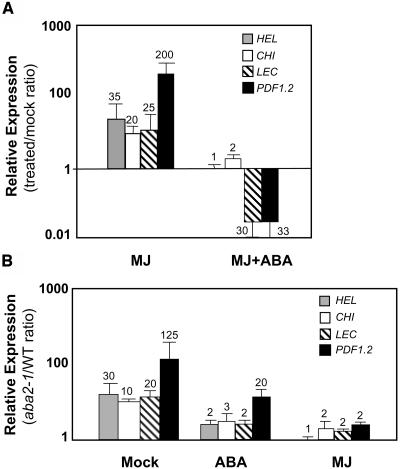
To test whether the suppression caused by ABA was specific to PDF1.2 or whether other JA-ethylene responsive defense genes also show a similar response to ABA, we quantified the transcript levels of three other MJ-ethylene responsive defense genes, namely CHI (basic chitinase), HEL (hevein-like protein or PR4), and LEC, an MJ- and ethylene-responsive lectin-like protein (Schenk et al., 2000) after MJ or MJ-ABA cotreatment. As expected, MJ treatment strongly induced the transcript levels of PDF1.2, CHI, HEL, and LEC in wild-type plants relative to the expression of these genes in mock-treated plants (Figure 2A). We next examined the effect of MJ-ABA cotreatment on defense gene induction. These experiments again showed that MJ, when applied together with ABA, was not able to fully induce the expression of these defense genes (Figure 2A). Consistent with the known ethylene inducibility of these genes, treatments of wild-type plants with ethylene produced results that are similar to those by MJ (data not shown).
Figure 2.
ABA Suppresses JA-Ethylene Responsive Defense Gene Expression in Arabidopsis.
(A) Fold changes (induction or suppression) in relative transcript abundance of PDF1.2, CHI, HEL, and LEC in MJ- and MJ+ABA-treated wild-type (Col-0) plants relative to the mock-treated wild-type plants.
(B) Fold changes in relative transcript abundance of PDF1.2, CHI, HEL, and LEC in mock-, MJ-, or ABA-treated aba2-1 mutant relative to the expression of the same genes in similarly treated wild-type plants. Total RNA was isolated from plants 48 h after each treatment, converted to cDNA, and used as template in RT-Q-PCR assays. Transcript levels in treated wild-type plants were normalized to the expression of β-actin genes measured in the same samples and expressed logarithmically relative to the similarly normalized transcript levels in mock-treated wild-type plants. The numbers on each bar show fold increase or fold decrease caused by each treatment in the transcript levels of genes relative to those in mock-treated plants (A). Transcript levels in treated aba2-1 plants were normalized relative to the expression of β-actin genes measured in the same samples and expressed logarithmically relative to the similarly normalized transcript levels in treated wild-type plants. The numbers on each bar show fold increase or fold decrease caused by each treatment in the transcript levels of genes in the aba2-1 background relative to those in similarly treated wild-type plants (B). Average data with error bars from two independent experiments are shown in both (A) and (B).
It can be argued, however, that endogenous ABA levels attained in the plant after ABA treatment do not represent physiologically relevant ABA concentrations; thus, the observed reductions in the transcript levels of defense genes after ABA treatment could be misleading. We therefore studied expression of these defense genes in the abscisic acid deficient2 (aba2-1) and aba1-2 mutants (Koornneef et al., 1982). The aba2-1 mutant contains significantly reduced levels of endogenous ABA because of a mutation in the SDR1 gene, which encodes a short-chain dehydrogenase/reductase involved in ABA biosynthesis (Cheng et al., 2002; González-Guzmán et al., 2002). We observed 125-, 10-, 30-, and 20-fold higher basal transcript levels of PDF1.2, CHI, HEL, and LEC, respectively, in untreated aba2-1 plants than in untreated wild-type plants (Figure 2B). ABA treatment reduced the transcript levels of these defense genes in both the wild type and the aba2-1 mutant. However, the aba2-1 mutant had significantly higher transcript levels of all four genes examined when compared with those in ABA-treated wild-type plants as demonstrated by the aba2-1/wild type normalized expression ratios (Figure 2B). In addition, MJ treatment induced the transcript levels of most of these defense genes to slightly higher levels (approximately twofold) in the aba2-1 mutant background than in wild-type plants (Figure 2B). We further confirmed that basal and MJ-activated PDF1.2 transcript levels in the aba1-2 (in Landsberg erecta background) mutant (Koornneef et al., 1982), which also shows an ABA-deficient phenotype because of a mutation in the ZEP (zeaxanthin epoxidase) gene (Marin et al., 1996), were higher than those in wild-type plants (data not shown). Overall, these results indicated that ABA, whether endogenously synthesized or exogenously applied, could play a role in antagonizing JA-ethylene responsive defense gene expression in Arabidopsis.
A Positive Regulator of ABA Signaling Negatively Regulates JA-Ethylene Responsive Defense Gene Expression
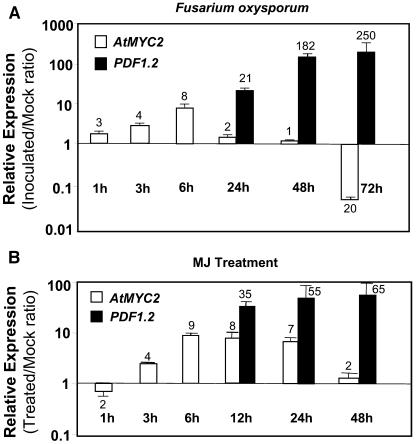
We next examined whether a positive regulator of ABA signaling pathway could have a negative effect on JA-ethylene responsive defense gene expression. The role of AtMYC2 as a positive regulator of ABA signaling has been previously established (Abe et al., 1997, 2003). To examine the potential role of this gene in plant defense, we first studied the regulation of AtMYC2 during plant defense responses. We conducted time-course inoculation experiments with the soil-borne pathogenic fungus Fusarium oxysporum and quantified the transcript abundance of AtMYC2 by RT-Q-PCR. These experiments showed that AtMYC2 is induced at early time points after inoculation with F. oxysporum relative to the expression measured in mock-inoculated plants (Figure 3A). Similarly, MJ treatment significantly induced the expression from the AtMYC2 gene relative to the expression detected in mock-treated plants (Figure 3B). As a positive control for pathogen and MJ treatments, we also measured expression from the PDF1.2 gene in the same samples. Consistent with the previous reports (Schenk et al., 2003), a significant PDF1.2 induction was detected at 12 and 24 h onwards after treatment with MJ or inoculation with F. oxysporum, respectively (Figures 3A and 3B). Interestingly, ethylene treatment significantly suppressed AtMYC2 expression relative to the untreated plants at 6 h (2 ± 0.5-fold) and 24 h (5 ± 0.2-fold) after treatment. Altered expression of AtMYC2 transcripts during plant defense suggests a role for this gene in biotic stress in addition to its known role in ABA-mediated responses to abiotic stress.
Figure 3.
AtMYC2 Is Induced during Plant Defense Responses in Arabidopsis.
AtMYC2 and PDF1.2 expression in wild-type (Col-0) plants were examined in time-course studies after inoculation with F. oxysporum (A) and after treatment with MJ (B). Total RNA was extracted from leaf tissue of 3- to 4-week-old plants (8 to 10 leaf stage) for each time point, converted to cDNA, and subjected to RT-Q-PCR analysis. The AtMYC2 and PDF1.2 transcript levels in treated/inoculated plants were normalized to the expression of β-actin measured in the same samples and expressed logarithmically relative to the similarly normalized expression levels in mock-inoculated/mock-treated plants. Each bar represents average data with error bars from two independent experiments. The numbers on each bar show fold increase or fold decrease caused by each treatment at transcript levels of AtMYC2 and PDF1.2 relative to those in mock-treated/inoculated plants.
To determine potential function(s) of AtMYC2 in plant defense, we characterized two Arabidopsis lines carrying independent T-DNA insertions in the AtMYC2 gene (Alonso et al., 2003a). Importantly, while this manuscript was in preparation, two other articles describing a series of AtMYC2 mutant alleles were concomitantly published. Of these, Lorenzo et al. (2004) reported the map-based cloning of the JASMONATE INSENSITIVE1 (JIN1/JAI1) locus, which is found to be identical to the AtMYC2 locus. Lorenzo et al. (2004) have characterized eight other mutant alleles of the JIN1/JAI1/AtMYC2 locus generated either by ethyl methanesulfonate mutagenesis (jin1-1 to jin1-6) or T-DNA insertions (jin1-7 to jin1-8). A second article by Boter et al. (2004) also described two insertional AtMYC2 mutants. One of the T-DNA mutants (SALK_040500) characterized by Boter et al. (2004) corresponds to jin1-7 of Lorenzo et al. (2004), and the second one (SALK_083483) is common between Boter et al. (2004) and this study. Following these studies, we renamed the two-AtMYC2 mutant alleles that we have characterized in this study as jin1-9 (SALK_017005) and jin1-10 (SALK_083483).
PCR amplification followed by DNA sequencing of the T-DNA–plant genomic DNA junction in one of such lines (SALK_017005) confirmed that the T-DNA is inserted into the 320th codon of AtMYC2 that encodes the amino acid Ile. Thus, the T-DNA insertion in this line truncates the basic helix-loop-helix domain necessary for dimerization and DNA binding and NH2-terminal domain necessary for either trans-activation or -repression. Given the importance of the major domains disrupted in AtMYC2, the abnormal transcript resulting from the T-DNA insertion, if expressed stably, is unlikely to be functional. Indeed, the basal transcript abundance of the AtMYC2 sequence in homozygous plants of the myc2 mutant was strongly suppressed in untreated and MJ-treated plants compared with the AtMYC2 levels in similarly treated wild-type plants (data not shown). This result provides further evidence that the T-DNA insertion interferes with the transcript accumulation as well as disrupting the coding sequence of the AtMYC2 gene.
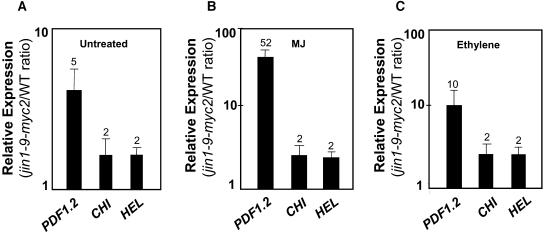
There was no detectable effect of the insertion on normal plant growth and development in the jin1-9/myc2 mutant. However, we found significantly higher basal transcript levels of the JA-ethylene responsive defense genes in untreated jin1-9/myc2 plants than untreated wild-type plants grown and sampled at the same time. Basal transcript levels of PDF1.2, CHI, and HEL were fivefold, twofold, and twofold higher, respectively, in the untreated jin1-9/myc2 mutant than in untreated wild-type plants (Figure 4A). To further confirm that the enhanced defense gene expression phenotype observed in this line was because of the disruption of AtMYC2, we examined the basal transcript level of PDF1.2 in homozygous plants of jin1-10/myc2 mutant. The T-DNA in this line is inserted into the coding region of AtMYC2, similarly truncating the AtMYC2 protein between N-terminal and basic helix-loop-helix domains (data not shown). Similarly to jin1-9/myc2, untreated homozygous plants of the jin1-9/myc2 mutant showed threefold higher basal transcription from the PDF1.2 gene; thus, all subsequent experiments were conducted on homozygous plants of the jin1-9/myc2 mutant. The molecular phenotypes of jin1/myc2 mutants reported in this article were consistent with those reported by Lorenzo et al. (2004) and Boter et al. (2004) (see also below).
Figure 4.
JA-Ethylene Responsive Defense Genes Show Elevated Levels of Basal and Activated Transcription in the jin1-9/myc2 Mutant.
Relative expression ratios of PDF1.2, CHI, and HEL transcripts in untreated (A), MJ- (B), and ethylene-treated (C) plants of the wild type (Col-0) and the homozygous plants of the jin1-9/myc2 mutant. Total RNA was extracted from leaf tissue of 3- to 4-week-old plants 24 h after MJ and ethylene treatments, converted to cDNA, and subjected to RT-Q-PCR analysis. The transcript levels in the jin1-9/myc2 mutant were normalized to the expression of β-tubulin measured in the same samples and expressed logarithmically relative to the normalized expression levels in similarly treated wild-type plants. Each bar represents average data with error bars from two independent experiments. The numbers on each bar show fold increase in defense gene transcript levels in the jin1-9/myc2 mutant relative to those in mock-treated plants.
Disruption of AtMYC2 Increases Defense Gene Induction by MJ and Ethylene
To determine whether JA-ethylene responsive defense genes are hypersensitive to induction in the jin1/myc2 mutant, we treated the wild-type and the jin1-9/myc2 mutant with MJ and ethylene and examined the transcript levels of selected JA-ethylene responsive genes in treated plants 24 h after treatment. MJ and ethylene treatments of the jin1-9/myc2 mutant resulted in 52- and 10-fold higher induction of PDF1.2 in the myc2 mutant background than in wild-type plants treated similarly (Figures 4B and 4C). CHI and HEL transcript levels were also higher in MJ- and ethylene-treated jin1-9/myc2 plants than in similarly treated wild-type plants (Figures 4B and 4C). Although increases in transcript levels of CHI and HEL in the jin1-9/myc2 mutant after MJ and ethylene treatments were relatively small (approximately twofold increase over that measured in similarly treated wild-type plants), the changes observed were highly reproducible in independent experiments. The enhanced responsiveness observed in defense gene induction in the jin1-9/myc2 mutant after MJ-ethylene treatment suggests that AtMYC2, as a positive regulator of ABA signaling, might play a role in negative regulation of JA-ethylene responsive defense genes in Arabidopsis.
AtMYC2 Overexpression in Arabidopsis
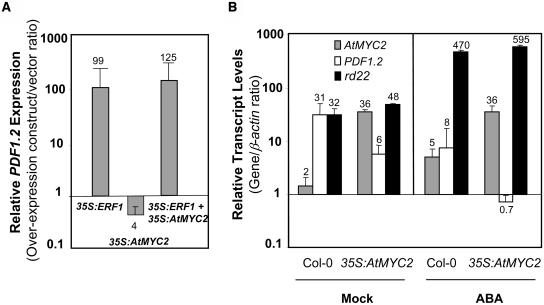
To determine whether the overexpression of AtMYC2 could inhibit defense gene expression, which would further support its putative function as a negative regulator, we first used transient protoplast transformation assays. Such assays have been extensively used to dissect plant signaling pathways (Hwang and Sheen, 2001) and similarly allowed us to test the individual and combinatorial expression of various gene constructs in a convenient way. As explained in Methods, all transformations included the 35S:GFP construct for constitutive expression of the green fluorescent protein (GFP), which was used for normalization of transformation efficiency. As a positive control, we first used the 35S:ERF1 construct, which resulted in a 10- and 99-fold increase in ERF1 (data not shown) and PDF1.2 (Figure 5A) transcript levels, respectively, relative to the expression detected in control protoplasts transformed with the vector only. The increase observed in PDF1.2 transcript levels is consistent with previous evidence that ERF1 is a positive regulator of the PDF1.2 gene (Solano et al., 1998). Overexpression of AtMYC2 resulted in an eightfold increase and a fourfold reduction in the transcript levels of AtMYC2 (data not shown) and PDF1.2 (Figure 5A), respectively, relative to the transcript levels of these genes measured in vector-transformed protoplasts. However, when both AtMYC2 and ERF1 are simultaneously overexpressed by the 35S promoter of Cauliflower mosaic virus in Arabidopsis protoplasts, no suppression of PDF1.2 transcripts was evident (Figure 5A). Taken together, these results suggest that the negative regulatory effect of AtMYC2 on PDF1.2 occurs mainly upstream of ERF1 and possibly not directly via an interaction of AtMYC2 and the PDF1.2 promoter.
Figure 5.
Transient or Stable Overexpression of AtMYC2 in Arabidopsis Suppresses PDF1.2 Expression.
(A) Transient overexpression of ERF1 in Arabidopsis protoplasts activates expression from the PDF1.2, whereas transient overexpression of AtMYC2 in Arabidopsis protoplasts suppresses the basal transcription from PDF1.2 as detected by RT-Q-PCR 48 h after transformation. AtMYC2 overexpression is not sufficient in suppressing PDF1.2 transcript levels when coexpressed with ERF1 in Arabidopsis protoplasts. Total RNA was isolated from the 35S:ERF1, 35S:AtMYC2, and 35S:ERF1+35S:AtMYC2 transformed protoplasts, converted to cDNA, and subjected to RT-Q-PCR analysis. The PDF1.2 transcript levels in the transformed samples were normalized to the expression of β-actin measured in the same samples and expressed logarithmically relative to the normalized expression levels measured in vector-only transformed protoplasts. Each bar represents average data with error bars from two independent experiments. The numbers on each bar show fold increase or fold decrease in PDF1.2 transcript levels caused by overexpression relative to the PDF1.2 levels in vector-transformed protoplasts.
(B) The stable overexpression of AtMYC2 in Arabidopsis activates rd22 expression while suppressing PDF1.2 in plants either mock-treated or treated with ABA for 11 h. Total RNA was isolated from transformed protoplasts or treated plants, converted to cDNA, and subjected to RT-Q-PCR analysis. The AtMYC2, rd22, and PDF1.2 transcript levels in untreated and ABA-treated wild type and the 35S:AtMYC2 plants were normalized to the expression of β-actin (multiplied by 1000 for clarity) measured in the same sample and expressed logarithmically. Each bar represents average data with error bars from two independent experiments. The numbers on each bar show relative transcript abundance of AtMYC2, PDF1.2, and rd22, relative to the β-actin transcript levels measured in the same samples.
Subsequently, we generated transgenic Arabidopsis plants containing the 35S:AtMYC2 construct. After screening 13 independently generated transgenic lines for elevated AtMYC2 expression, we identified two transgenic lines that contained increased transcript levels of AtMYC2 as high as 18-fold over those found in wild-type plants. Because AtMYC2 is a positive regulator of ABA responsive rd22 (Yamaguchi-Shinozaki and Shinozaki, 1993; Abe et al., 2003), we first confirmed that AtMYC2 overexpression could indeed induce rd22. The expression data, which show transcript levels of AtMYC2, rd22, and PDF1.2 relative to those of β-actin measured in the samples, are presented in Figure 5B. The rd22 transcript abundance in untreated 35S:AtMYC2 plants was indeed slightly higher (1.5-fold) than that in untreated wild-type plants. Furthermore, we found that induction of rd22 by ABA was also stronger in ABA-treated 35S:AtMYC2 plants than in ABA-treated wild-type plants (Figure 5B), confirming previous results by Abe et al. (2003). In contrast with rd22, quantification of PDF1.2 transcript abundance showed approximately fivefold lower PDF1.2 expression in untreated 35S:AtMYC2 plants than in untreated wild-type plants (Figure 5B). Furthermore, although ABA treatment reduced the transcript levels of PDF1.2 in both wild-type and the 35S:AtMYC2 plants, the reduction observed in PDF1.2 transcript levels in the 35S:AtMYC2 plants was significantly greater than that in ABA-treated wild-type plants. Overall, these results are consistent with a positive and negative regulatory role of AtMYC2 on rd22 and PDF1.2, respectively. The differences observed between the 35S:AtMYC2 and wild-type plants in the transcript levels of PDF1.2 and rd22 at 11 h after ABA treatment were no longer detectable at 24 and 48 h after ABA treatment possibly because of response saturation at later time points. Similarly, no reduction was evident in the PDF1.2 transcript levels after MJ treatment in the 35S:AtMYC2 plants relative to the treated wild-type plants (data not shown).
AtMYC2 Function Is Dispensable for Suppression of PDF1.2 by Exogenous ABA
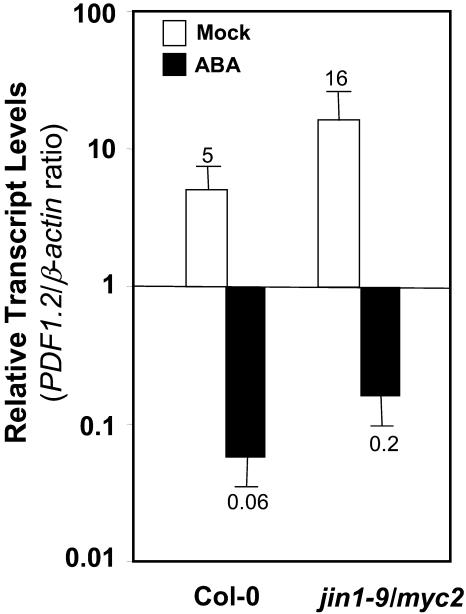
Although the data presented here strongly suggest that AtMYC2 is a negative regulator of plant defense gene expression, the exact location where AtMYC2 mediates an interaction between ABA and JA-ethylene signaling pathways is not clear. It is possible that AtMYC2, acting downstream from ABA, directly mediates the antagonistic interaction between these two pathways. If this were the case, then one would expect that PDF1.2 could not be suppressed or attenuated by exogenous ABA in the jin1-9/myc2 mutant. To test this possibility, we treated the wild type and the jin1-9/myc2 mutant with ABA and measured the transcript levels of AtMYC2 and PDF1.2 in treated plants. These experiments showed that exogenous ABA could still suppress the PDF1.2 transcript accumulation in the jin1-9/myc2 mutant plants (Figure 6). The extent of suppression (fold reduction in basal transcript abundance) of PDF1.2 in wild-type plants 24 h after treatment was not significantly different from that in jin1-9/myc2. This result suggests that AtMYC2 function is dispensable for the antagonistic effect of ABA on the JA-ethylene defense pathway and that multiple control points may exist for cross talk between these pathways.
Figure 6.
AtMYC2 Function Is Dispensable for Suppression of PDF1.2 by Exogenous ABA.
Total RNA was isolated from mock- or ABA-treated wild-type and jin1-9/myc2 plants 24 h after treatment, converted to cDNA, and subjected to RT-Q-PCR analysis. The PDF1.2 transcript levels in mock- and ABA-treated wild-type and the jin1-9/myc2 mutant were normalized to the expression of β-actin genes measured in the same samples (multiplied by 1000 for clarity). Each bar represents average data with error bars from two independent experiments. The numbers on each bar show relative transcript abundance of PDF1.2 relative to the β-actin transcript levels measured in the same samples.
ABA Signaling Pathway Negatively Regulates Resistance to F. oxysporum
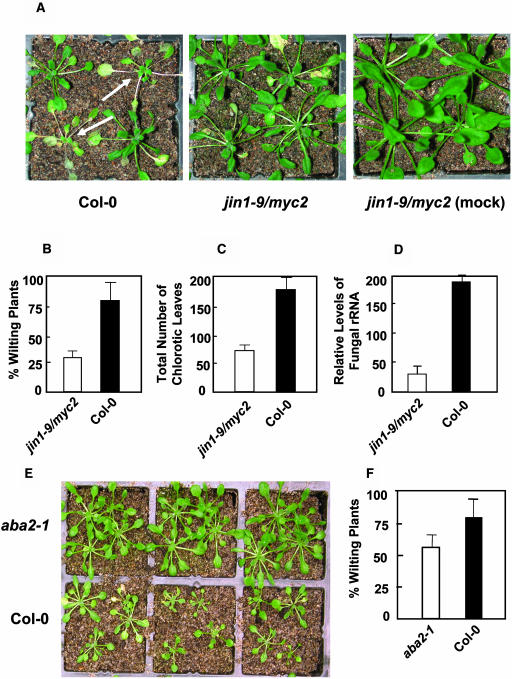
The antagonistic interaction between ABA and JA-ethylene signaling pathways could also influence resistance to the necrotrophic pathogens. To determine whether the increased levels of the JA-ethylene responsive defense gene expression in jin1-9/myc2 could provide enhanced pathogen resistance, we conducted inoculation experiments using the necrotrophic fungal pathogen F. oxysporum. This pathogen was chosen because it is known to influence the expression of AtMYC2 and PDF1.2 (this study) and is thought to be sensitive to JA-ethylene mediated defense (Berrocal-Lobo and Molina, 2004). The root dip inoculation assay in soil-grown plants by F. oxysporum is the preferred method to study infection by this soil-borne pathogen (Narasimhan et al., 2003). In inoculated plants, F. oxysporum penetrates through the roots and moves upward into the shoot via the vascular system. In three separate inoculation experiments, we inoculated the roots of 140 plants for each of the jin1-9/myc2 and wild-type plants. As shown in Figure 7A, infection by this pathogen caused stunting of shoots and chlorosis, especially in the lower leaves of infected plants. Ten days after inoculation, the number of plants showing severe wilting symptoms and the total number of chlorotic leaves were counted to assess the extent of disease severity. The number of plants that showed a strong wilting symptom was significantly lower in the jin1-9/myc2 mutant (29%) than in wild type-plants (81%; P < 0.01) (Figure 7B). We also noted that the total number of leaves that displayed chlorosis after inoculation was significantly higher in wild-type plants (181 leaves out of 140 plants) than in the myc2 mutant (68 leaves out of 140 plants), which translated to ∼2.7-fold reduction in disease development (P < 0.01) (Figure 7C). To further compare the level of disease tolerance between the wild type and the jin1-9/myc2 mutant, we estimated fungal biomass by measuring the amount of fungal RNA in plant tissue by combining all 30 plants from the wild type and the jin1-9/myc2 mutant from the inoculation experiment 3 by RT-Q-PCR, using primers specific for a transcribed spacer region of F. oxysporum rRNA. These analyses showed the presence of eightfold more fungal rRNA in wild-type plants than in the jin1-9/myc2 mutant, further supporting the conclusion that the jin1-9/myc2 mutant sustained significantly less fungal growth than the wild-type (Figure 7D).
Figure 7.
The jin1-9/myc2 and aba2-1 Mutants Show Enhanced Resistance to the Root-Infecting Fungal Pathogen F. oxysporum.
(A) to (D) In three separate experiments, 140 each of the 3-week-old wild-type (Col-0) and myc2 plants were inoculated with F. oxysporum (A). The percentage of plants showing strong wilting phenotype (arrows) (B), and the total numbers of necrotic leaves (C) were scored 10 d after inoculation. The amount of fungal RNA present in the inoculated tissue is estimated (D) using all 30 plants of each of the inoculated wild-type and myc2 plants in experiment 3 (10 d after inoculation) by RT-Q-PCR with primers specific to F. oxysporum rRNA.
(E) and (F) In two separate experiments, 60 plants of each of the wild-type (Col-0) and aba2-1 plants were inoculated with root-infecting fungal pathogen F. oxysporum (E). The plants showing strong wilting symptoms were counted 10 d after inoculation to assess the disease severity (F).
We next tested whether enhanced defense gene expression observed in the aba2-1 mutant leads to increased disease resistance; we inoculated wild-type and aba2-1 plants with F. oxysporum. These inoculation experiments showed that the number of plants showing wilt symptoms at 10 d after inoculation was significantly reduced in aba2-1 (58%) plants as compared with wild-type plants (80%; P < 0.05) (Figures 7E and 7F). This result further suggested that ABA deficiency could indeed positively affect disease resistance against F. oxysporum.
The Effect of abi1-1 and abi2-1 Mutations on PDF1.2
Because ABA itself and a positive regulator of ABA signaling AtMYC2, both appear to antagonize JA-ethylene responsive expression of defense genes and disease resistance, we explored other known mutations in ABA signaling for effects on PDF1.2 expression. The ABI1 and ABI2 genes encoding homologous Ser/Thr protein phosphatases 2C act in a negative regulatory loop of the ABA signaling pathway (Merlot et al., 2001). ABA signaling mutants, aba insensitive1 (abi1-1) and abi2-1, show a dominant-negative phenotype on ABA inhibition of seed germination (Gosti et al., 1999). Studies of genetic revertants of these mutant alleles, which show ABA hypersensitivity in seed germination assays, have lead to a proposed function for the ABI1 and ABI2 genes as negative regulators of ABA signaling (Gosti et al., 1999). Previous studies also showed that ABI1 inhibits both ABA-inducible and ABA-repressible gene expression when overexpressed transiently in maize mesophyll protoplasts (Sheen, 1996, 1998).
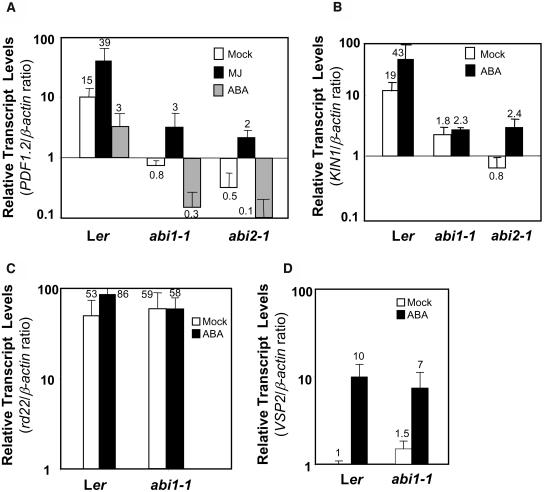
Therefore, we studied the PDF1.2 transcript levels in mock- or MJ-treated ABA signaling mutants abi1-1 and abi2-1. We observed a strong reduction at the PDF1.2 transcript levels in untreated abi1-1 and abi2-1 mutants relative to the corresponding wild-type Landsberg erecta (Ler) plants grown and sampled simultaneously. However, the response to MJ measured as the fold induction in the abi1-1 and abi2-1 mutants and the wild type were identical (Figure 8A). The regulation of PDF1.2 expression observed in these mutants confirms that there is an interaction between ABA signaling and PDF1.2 transcript accumulation. However, the effects observed on PDF1.2 expression were not consistent with the expectation that the abi1-1 and abi2-1 mutants would have a dominant negative effect on ABA signaling and therefore may abolish ABA's inhibition of PDF1.2 expression. Indeed, increased PDF1.2 transcript levels were found in the abi8 mutant (Brocard-Gifford et al., 2004). However, we found that ABA treatment could still suppress PDF1.2 expression in both Ler and abi1-1 and abi2-1 mutants, indicating that ABA antagonism on PDF1.2 was still functional in these mutants (Figure 8A). ABA is known to act through multiple signaling pathways, and these may interact differentially with the JA and ethylene pathways. For example, AtMYC2, a key regulator of the JA response, and ABI1 are thought to be on separate branches (Lorenzo et al., 2004). Our results are consistent with a dominant role for AtMYC2 in cross talk with the JA pathway when compared with that for ABI1, which appears to be more involved in regulating net expression levels of PDF1.2 rather than MJ induction.
Figure 8.
Analysis of Expression from ABA-Regulated Genes in abi1-1 and abi2-1 Mutants.
Quantification of the relative abundance of the PDF1.2, KIN1, rd22, and VSP2 transcripts in mock- or MJ- and/or ABA-treated wild-type (Ler) and the abi1-1 and abi2-1 mutants. Total RNA was isolated from plants 48 h after each treatment, converted to cDNA, and used as template in RT-Q-PCR assays. The PDF1.2, KIN1, rd22, and VSP2 transcript levels were normalized to the expression of β-actin (multiplied by 1000 for clarity) measured in the same samples and expressed logarithmically. Each bar represents average data with error bars from two independent experiments. The numbers on each bar show transcript levels of PDF1.2, KIN1, VSP2, and rd22 relative to the β-actin transcript levels measured in the same samples.
To further explore the pathways that abi1-1 and abi2-1 mutations affect, we measured the transcript levels of three ABA-responsive genes, KIN1, rd22, and VEGETATIVE STORAGE PROTEIN2 (VSP2), in the same cDNA samples used to measure PDF1.2 expression. We included KIN1 encoding a cold- and ABA-inducible protein in these studies because ABI1 and ABI2 are proposed to be the regulators of the ABA-responsive element–dependent ABA signaling branch, which positively regulates KIN1 expression (Goh et al., 2003; Chini et al., 2004). This branch of ABA signaling is thought to act independently from the ABA signaling pathway regulated by AtMYC2 (Shinozaki et al., 2003). Indeed, consistent with the dominant-negative phenotype predicted for abi1-1 and abi2-1, KIN1 transcript levels were dramatically reduced in mock-treated abi1-1 and abi2-1 plants relative to those in wild-type plants (Figure 8B). Although exogenous ABA treatment slightly induced KIN1 transcript levels in the abi2-1 mutant, the KIN1 transcript levels were still significantly lower after ABA treatment than those in untreated wild-type plants and thus could have been undetectable using traditional RNA gel blot assays.
No significant differences were observed in rd22 transcript levels between control plants of wild-type Ler and abi1-1 (Figure 8C) and abi2-1 (data not shown) mutants. In addition, in contrast with the wild type, no significant increases were detected in rd22 transcript levels in the abi1-1 (Figure 8C) and abi2-1 (data not shown) mutants treated with ABA. Overexpression of AtMYC2 increases rd22 expression and ABA induction (Figure 5B; Abe et al., 2003), and Lorenzo et al. (2004) reported that AtMYC2 expression was inducible by ABA in the abi1-1 mutant. Therefore, it is likely that abi1-1 mutation acts in a branch downstream from AtMYC2 in influencing the ABA-insensitivity phenotype of rd22 in this mutant.
In contrast with the effects on KIN1 and rd22 gene expression, VSP2 transcript levels did not show any significant change in untreated and ABA-treated plants of Ler and the abi1-1 mutant, suggesting that expression of VSP2, which encodes an ABA- and MJ-inducible vegetative storage protein (Berger et al., 1996), is independent from abi1-1 (Figure 8D). Taken together, these results are consistent with the earlier studies that multiple ABA signaling pathways may be operating in vegetative tissues of Arabidopsis and only some of these pathways are affected by the abi1 and abi2 mutations (Finkelstein, 1993).
Finally, to determine whether the reduced PDF1.2 expression observed in abi1-1 and abi2-1 mutants might increase susceptibility to F. oxysporum, we conducted pathogen inoculation experiments on wild-type Ler and abi1-1 and abi2-1 plants. However, inoculation of the abi1-1 and abi2-1 mutants with F. oxysporum did not reveal any altered (e.g., enhanced susceptibility) disease resistance as compared with inoculated wild-type plants. However, we noted that the Ler ecotype is much more susceptible to F. oxysporum than Col-0, with plants dying more rapidly and at lower spore concentrations (data not shown); therefore, it may be difficult to evaluate an increased susceptibility phenotype in this genetic background.
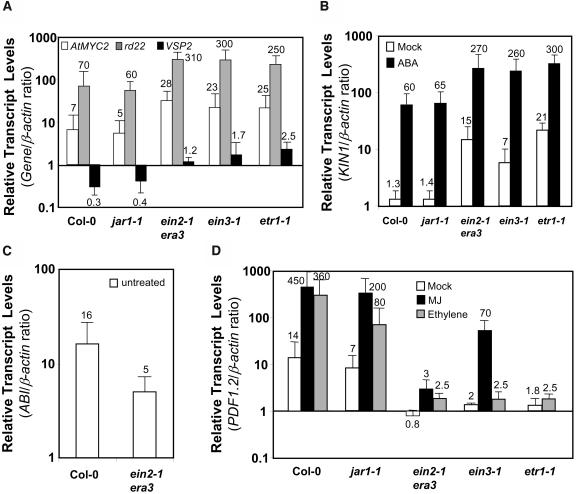
Ethylene Signaling Antagonizes ABA Responsive Gene Expression in Vegetative Tissues
The interaction among JA, ethylene, and ABA signaling is just emerging from this and other recent studies (Boter et al., 2004; Lorenzo et al., 2004). However, there is a well established interaction between ethylene and ABA signaling in relation to the regulation of seed germination (Beaudoin et al., 2000; Ghassemian et al., 2000). The ethylene insensitive mutants etr1-1, ein2-1/era3, and ein3 show increased sensitivity to ABA in germination assays (Beaudoin et al., 2000; Ghassemian et al., 2000; Yanagisawa et al., 2003) as well as reduced PDF1.2 transcript levels (Penninckx et al., 1996) and increased pathogen susceptibility (Thomma et al., 1999; Berrocal-Lobo et al., 2002; Geraats et al., 2002). By contrast, the mutations in the CTR1 gene, a negative regulator of ethylene signaling, constitutively activate the ethylene pathway while making seed germination less sensitive to ABA (Beaudoin et al., 2000; Ghassemian et al., 2000). However, it is possible that the antagonistic relationship between ethylene and ABA signaling may be different in vegetative tissues than in seeds (Fedoroff, 2002). To study the nature of interactions between JA-ethylene and ABA signaling pathways in vegetative tissues, we measured KIN1, VSP2, rd22, and AtMYC2 transcript levels in mock- and ABA-treated wild-type, JA signaling mutant jar1-1, and ethylene signaling mutants etr1-1, ein2-1/era3, and ein3-1. The transcript levels of all four genes in mock-treated etr1-1, ein2-1/era3, and ein3-1 mutants were significantly higher than those in wild-type plants (Figure 9A). In addition, we observed threefold to 4.5-fold higher expression from all four genes in the ethylene mutants after ABA treatment than in similarly treated wild-type plants. A typical result from these experiments is presented for KIN1 in Figure 9B. In contrast with results obtained with ethylene-signaling mutants, the reduced JA sensitivity as conditioned by the jar1-1 mutation did not show any effect on the ABA-responsive gene expression in these experiments (Figures 9A and 9B). Taken together, these results suggested that ethylene signaling acts antagonistically to ABA signaling in vegetative tissues.
Figure 9.
Ethylene Insensitivity Enhances ABA-Responsive Gene Expression in Vegetative Tissues.
Relative transcript levels of KIN1, rd22, VSP2, ABI1, and PDF1.2 in mock- and/or ABA-, MJ-, and/or ethylene-treated wild-type and jar1-1, ein2-1, etr1-1, and ein3-1 mutant plants. Total RNA was isolated from plants 48 h after each treatment, converted to cDNA, and used as template in RT-Q-PCR assays. The KIN1, rd22, VSP2, PDF1.2, and ABI1 transcript levels were normalized to the expression of β-actin (multiplied by 1000 for clarity) measured in the same samples and expressed logarithmically. Each bar represents average data with error bars from two independent experiments. The numbers on each bar show relative transcript levels of each gene relative to the β-actin transcript levels measured in the same samples.
To further test the interaction between ethylene and ABA signaling, we measured ABI1 transcript levels in the ein2-1/era3 mutant. The average results from three independent experiments showed that the ABI1 transcript levels in the untreated ein2-1/era3 mutant were approximately threefold lower than that observed in untreated wild-type plants (Figure 9C). This result suggests a putative link between reduced levels of negative regulators of ABA signaling and previously reported features of the ein2-1/era3 mutant, namely, increased sensitivity to ABA (Beaudoin et al., 2000; Ghassemian et al., 2000), reduced PDF1.2 expression (Penninckx et al., 1996; see also Figure 9D), and enhanced disease susceptibility (Thomma et al., 1999). This result is also consistent with the view that the interaction between the ABA and ethylene signaling pathway is mutually antagonistic in vegetative tissues in Arabidopsis.
The PDF1.2 transcript levels measured in mock-, MJ-, and ethylene-treated wild-type and mutant plants were consistent with the known features of these mutants (Figure 9D), such that we observed slightly higher PDF1.2 transcript levels in the MJ-treated ein3-1 and jar1-1 mutants than those in ein2-1/era3, but all were lower than those in wild-type plants. The partial MJ induction of PDF1.2 in these mutants detected by RT-Q-PCR is possibly because of the leaky nature of these mutants (e.g., Alonso et al., 2003b).
A Complex Interplay among ABA, JA, and Ethylene Signaling Pathways Regulates the Expression of Diverse Stress Response Gene Classes
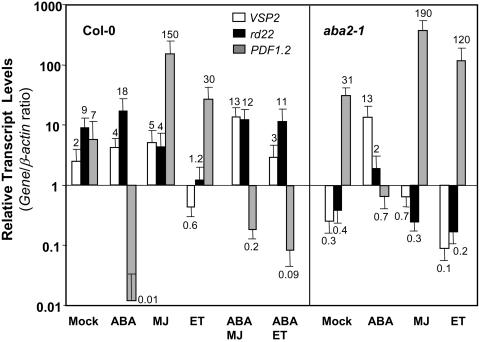
It is evident from the evidence presented so far that ABA, JA, and ethylene signaling pathways interact to regulate diverse stress responses. The nature of the interaction between these pathways appears to depend on the type of stress experienced by the plant. To further resolve these signaling interactions, we conducted independent experiments to examine transcript levels of the VSP2, rd22, and PDF1.2 genes as markers for wound, drought, and biotic stress responses, respectively, after treatments with either MJ-, ABA-, ethylene-, ABA and MJ-, or ABA and ethylene-treated wild-type plants. Overall, expression detected from PDF1.2 was consistent with the known MJ, ethylene, and ABA regulated expression patterns of this gene (Figure 10). However, as reported previously (Manners et al., 1998), the MJ-induced expression of PDF1.2 was significantly stronger than ethylene induced expression of PDF1.2. rd22 was induced by ABA but strongly suppressed by ethylene (Figure 10). Also, as expected, MJ and ABA induced VSP2, whereas ethylene suppressed expression of this gene relative to that in a mock-treated plant (Figure 10).
Figure 10.
ABA, JA, and Ethylene Signaling Pathways Interact for Regulation of Stress Responsive Gene Expression in Arabidopsis.
Relative transcript levels of VSP2, rd22, and PDF1.2 in mock-, ABA-, MJ-, ethylene-, or combination of ABA-MJ and ABA-ethylene (Col-0 only)-treated wild-type and aba2-1 mutant plants. Total RNA was isolated from plants 48 h after each treatment, converted to cDNA, and used as template in RT-Q-PCR assays. The VSP2, rd22, and PDF1.2 transcript levels were normalized to the expression of β-actin (multiplied by 1000 for clarity) measured in the same samples and expressed logarithmically. Average data from two independent experiments with error bars are presented. The numbers on each bar show transcript levels of each gene relative to the β-actin transcript levels measured in the same samples.
We also examined the expressions from these genes in wild-type plants treated with either ABA and MJ, or ABA and ethylene. As expected, neither MJ nor ethylene was able to induce PDF1.2 when applied together with ABA. In addition, ABA was also the dominant signal for the induction of rd22 as this gene was still slightly inducible by ABA in the presence of MJ or ethylene. Furthermore, the combined treatment of MJ with ABA induced VSP2 expression more strongly than either of the individual MJ or ABA treatments. The induction of VSP2 after ABA-ethylene cotreatment was lower than that after ABA treatment but higher than that in mock-treated plants (Figure 10).
Finally, we measured the expression of PDF1.2, rd22, and VSP2 in mock-, ABA-, MJ-, and ethylene-treated aba2-1 mutant. In these independent experiments, the expression of PDF1.2 was consistent with those reported earlier in Figure 2B, although differences in the actual magnitude of aba2-1/wild-type expression ratios of PDF1.2 were observed. These could be because of differences in plant growth and development between the experiments and also sensitivity of the RT-Q-PCR technique used to measure transcript levels. As expected, in mock-treated aba2-1 plants, the rd22 transcript levels were lower than those in mock-treated wild-type plants. In addition, induction of rd22 in this mutant by ABA was less, whereas suppression by MJ and ethylene were only slightly more than those in mock-treated aba2-1 plants. Analysis of the VSP2 transcript levels in the mock-treated aba2-1 mutant showed reduced VSP2 transcript levels relative to the mock-treated wild-type plants. Similarly, the VSP2 transcript levels were lower in the MJ-treated aba2-1 mutant than that in MJ-treated wild-type plants. Furthermore, the suppression of VSP2 by ethylene was stronger in the aba2-1 mutant than in wild-type plants. This suggests that ABA is required for full induction of VSP2 by MJ and also attenuation of suppression of this gene by ethylene. Here, we did not examine the antagonistic interaction between JA and ethylene signaling pathways in regulating VSP2 because expression of this gene was previously reported to be higher in untreated and MJ-treated ethylene signaling mutants than similarly treated wild-type plants (Rojo et al., 1999).
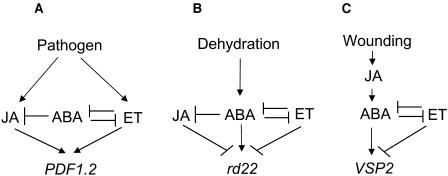
Collectively, the results from treated wild-type as well as those from treated and untreated ethylene, JA, and ABA signaling mutants suggest the presence of a complex interplay among these three signaling pathways. The simple models presented in Figure 11 summarize the signaling interactions among JA, ethylene, and ABA pathways in regulating the wound-, dehydration-, and pathogen-responsive gene expression in Arabidopsis.
Figure 11.
Proposed Model of Genetic Interactions among ABA, JA, and Ethylene Signaling Pathways for Modulation of Stress-Responsive Gene Expression in Arabidopsis.
Arrows indicate positive regulation, and blunt ends indicate negative regulation.
DISCUSSION
ABA Antagonizes Defense Gene Expression and Disease Resistance in Arabidopsis
In this article, we studied the effect of ABA and various components of ABA signaling pathway on JA-ethylene responsive defense gene expression and disease resistance in Arabidopsis. We first demonstrated that endogenous and exogenous ABA strongly reduced the transcript levels of JA-ethylene responsive defense genes. In addition, exogenous MJ or ethylene was not able to reverse the suppression caused by the ABA treatment in wild-type plants. Second, we showed that AtMYC2, a positive regulator of ABA signaling, negatively regulated expression from PDF1.2. Finally, we observed that both jin1-9/myc2 and aba2-1 mutants with substantially higher transcript levels of JA-ethylene regulated defense genes showed enhanced resistance to the necrotrophic fungal pathogen F. oxysporum. Taken together, these data strongly suggest a novel role for ABA in modulating defense gene expression and disease resistance in Arabidopsis.
Previously, there have been several indirect observations linking ABA with disease resistance in plants. For instance, exogenously supplied ABA increases the susceptibility of various plant species to fungal pathogens (Henfling et al., 1980; Ward et al., 1989; McDonald and Cahill, 1999). ABA suppresses Phe-ammonia-lyase transcript accumulation after inoculation of soybean (Glycine max) with the incompatible fungal pathogen Phytophthora megasperma f. sp glycinea (Ward et al., 1989). ABA also downregulates the β-1,3-glucanase transcript levels in tobacco (Nicotiana tabacum) cell cultures (Rezzonico et al., 1998). One recent study indicated that a tomato mutant with reduced ABA levels showed enhanced resistance to the necrotrophic pathogen B. cinerea, whereas exogenous application of ABA restored the susceptibility to this pathogen in the mutant plants (Audenaert et al., 2002). Another recent study in tomato showed that ABA-deficient plants were more resistant to infection by the bacterial pathogen P. syringae pv tomato (Pst) (Thaler and Bostock, 2004). Finally, Mohr and Cahill (2003) demonstrated that the treatment of wild-type Arabidopsis plants with ABA or water stress increases the susceptibility to Pst and P. parasitica, whereas the ABA-deficient Arabidopsis mutant aba1-1 showed reduced susceptibility to virulent isolates of P. parasitica.
The antagonistic interaction between biotic and abiotic stress responses has also been observed in other plant species, such as rice (Xiong and Yang, 2003). By contrast, other recent studies (Park et al., 2001; Mengiste et al., 2003; Chini et al., 2004) suggest that biotic and abiotic stress responses might also share common components. This further indicates the complexity of interplay among various signaling pathways during stress adaptation. The antagonistic interaction between the ABA and the JA-ethylene signaling pathways in regulating defense gene expression might be a strategy that plants employ to avoid simultaneous production of abiotic stress-related and biotic defensive proteins. Interestingly, our data also show that defense gene suppression mediated by ABA cannot be reversed by exogenous application of MJ and ethylene. This is consistent with the view that ABA action is a dominant process. One of the possible reasons for this may be that water stress affects plant survival in a more systemic and dramatic way than localized pathogen stresses, and plants have developed strategies to prioritize between these two stress responses. One would think that the antagonistic interaction between these two signaling pathways would compromise a plant's ability to tolerate both stresses should they occur simultaneously. However, simultaneous drought and necrotrophic pathogen attack may not happen very frequently in nature because these pathogens require relatively humid conditions for successful infection and under such conditions, water stress would not pose a significant threat.
ABA's involvement in plant disease resistance, however, seems to be complex and dependent on the type of the pathogen used. More recently, β-amino-butryic acid–induced resistance against leaf infecting necrotrophic pathogens Alternaria brassicicola and Plectosphaerella cucumerina was found to be compromised in the ABA-deficient mutant aba1-5 and ABA-insensitive mutant abi4-1. Furthermore, exogenous application of ABA induced callose accumulation and resistance to these two leaf-infecting pathogens in wild-type plants (Ton and Mauch-Mani, 2004). Callose accumulation, which has recently been shown to be associated with increased disease susceptibility in Arabidopsis (Nishimura et al., 2003), and to our knowledge, there has been no evidence that callose confers enhanced resistance to the soil-borne fungal pathogen F. oxysporum. By contrast, antimicrobial proteins, such as defensins, chitinases, and lectins, upregulated in the jin1/myc2 and aba2-1 mutants show strong inhibitory activities against F. oxysporum (Lay et al., 2003; Di Pietro et al., 2003; Madrid et al., 2003 and references therein). In addition, the signaling pathways conferring resistance to F. oxysporum, A. brassicicola, and P. cucumerina appear to be different. Resistance to F. oxysporum is mediated by JA-ethylene–dependent defense responses (Berrocal-Lobo and Molina, 2004), whereas these responses were found not to be effective against A. brassicicola and P. cucumerina (Ton and Mauch-Mani, 2004).
Negative Regulation of Defense Gene Expression and Disease Resistance by ABA Signaling
The data presented in this article showed that AtMYC2, a positive regulator of ABA signaling has a negative regulatory effect on defense gene expression in Arabidopsis. Importantly, while this manuscript was in preparation, Lorenzo et al. (2004) have reported the map-based cloning of the JAI1/JIN1 locus, which turned out to be identical to the AtMYC2 gene characterized here. The root elongation in the jin1/myc2 mutant shows a JA-insensitive phenotype (Berger et al., 1996; Lorenzo et al., 2004), whereas defense gene expression shows hypersensitivity to induction by JA in this mutant (Boter et al., 2004; Lorenzo et al., 2004; this study), suggesting that AtMYC2 differentially regulates different stress responses. Indeed, AtMYC2 also positively regulates the wound responsive gene VSP2 (Boter et al., 2004; Lorenzo et al., 2004). However, consistent with its negative regulatory role on defense gene expression, we showed here that the inactivation of AtMYC2 leads to heightened resistance to the necrotrophic fungal pathogen F. oxysporum. Similarly, Lorenzo et al. (2004) have reported that mutations in the JAI1/JIN1/AtMYC2 locus resulted in enhanced disease resistance against two other necrotrophic pathogens, further supporting the notion that AtMYC2 is a negative regulator of plant defense in Arabidopsis.
Currently, the exact mechanism by which AtMYC2 regulates defense gene expression is not known. Our results clearly showed that transient or stable overexpression of AtMYC2 leads to the suppression of transcription from PDF1.2. In addition, suppression on PDF1.2 was abolished when AtMYC2 and ERF1 were coexpressed in Arabidopsis protoplasts. Although these data may suggest that AtMYC2 possibly acts upstream from ERF1 in regulating defense gene expression, inactivation of AtMYC2 in the jin1/myc2 mutants does not lead to constitutive upregulation of ERF1 (Lorenzo et al., 2004). Although more experimental evidence is needed to determine the relative positions of ERF1 and AtMYC2 in defense signaling pathways, we speculate that AtMYC2's effect on defense gene suppression might not involve direct promoter binding for the following reasons. First of all, constitutive increases in PDF1.2 expression (e.g., in the cev1 mutants) do not necessarily accompany increases in ERF1 transcript levels (Brown et al., 2003), suggesting that PDF1.2 regulation may also be controlled by posttranscriptional or posttranslational processes. Secondly, the ABA-mediated suppression of PDF1.2 was not compromised in the jin1-9/myc2 mutant. This is in contrast with the role expected from a direct transcriptional repressor. Third, it is well established that the suppressive effects of c-MYC as negative regulators in animals involve interaction with other positive regulators of the target gene expression, whereas transcriptional activation by c-MYC occurs through direct DNA binding (reviewed in Wanzel et al., 2003). The same may well be the case for AtMYC2 in positively and negatively regulating different JA-ethylene and ABA-dependent responses, respectively.
It is possible that ABA and its positive regulators, such as AtMYC2, interfere with the signaling pathway regulating defense gene expression at a point upstream from ERF1, which integrates signals from both JA and ethylene pathways (Lorenzo et al., 2003). Because both JA and ethylene pathways are concomitantly required for expression of PDF1.2 (Penninckx et al., 1998), interference with either pathway would result in altered PDF1.2 expression. Our results, which showed that exogenous ABA could still suppress PDF1.2 expression in the jin1-9/myc2 mutant, suggest that ABA possibly interacts with the JA-ethylene pathway at multiple nodes, at least one of which is AtMYC2 independent. In fact, the jin1/myc2 mutant plants have reduced sensitivity to JA (i.e., less suppression of root elongation in response to JA) but do not display any altered growth in response to ethylene (Lorenzo et al., 2004). It is therefore likely that ABA interacts with JA and ethylene pathways separately and that AtMYC2 is required for interaction with the JA pathway only.
ABA's antagonistic effect on ethylene signaling may also occur upstream from ERF1 but independently from AtMYC2. EIN3, a positive regulator of ethylene signaling acting upstream from ERF1, might be a likely target for ABA in suppressing the ethylene-responsive defense gene expression. Indeed, recent evidence has suggested that glucose interferes with ethylene signaling by destabilizing the EIN3 protein, whereas ethylene enhances the stability of EIN3 (Yanagisawa et al., 2003). Glucose is also known to suppress PDF1.2 expression (Cheng et al., 2002). Interestingly, suppression of PDF1.2 transcription by glucose is abolished in the ABA-deficient aba2-1 mutant, suggesting that ABA is required for suppression of PDF1.2 expression by glucose (Cheng et al., 2002). It is therefore possible that ABA antagonizes ethylene signaling by interfering with the EIN3 function possibly through protein degradation. Indeed, ABA treatment induces expression from several F-box genes (Hoth et al., 2002), and recent studies showed that EIN3 is degraded, in the absence of ethylene, by protein ubiquitination (Guo and Ecker, 2003; Potuschak et al., 2003). Further research is needed to elucidate the actual mechanisms involved in antagonistic effect of ethylene on ABA signaling pathway.
ABA and Ethylene Signaling Are Mutually Antagonistic in Vegetative Tissues
An antagonistic effect of ethylene on ABA signaling has been previously shown during seed germination as etr1-1, ein2-1, and ein3-1 mutants compromised in reception or positive regulation of ethylene signaling show hypersensitivity to ABA for inhibition of germination (Beaudoin et al., 2000; Ghassemian et al., 2000; Yanagisawa et al., 2003). Our results presented here further extend these previous findings by showing that ABA-ethylene interaction is mutually antagonistic in vegetative tissues. First, we found higher transcript levels of ABA-responsive genes VSP2, rd22, and KIN1 as well as those of the positive regulator of ABA signaling AtMYC2 in mutants compromised in positive regulation of ethylene signaling. In addition, transcript levels of the negative regulator ABI1 were significantly reduced in the ein2/era3 mutant. Interestingly, mutations in the CTR1 gene encoding a negative regulator of ethylene signaling is known to reduce ABA sensitivity during germination, possibly because of constitutive activation of ethylene signaling (Beaudoin et al., 2000; Ghassemian et al., 2000). Consistent with this, AtMYC2 and VSP2 expression were found to be reduced in the ctr1-1 mutant (Van Zhong and Burns, 2003). Therefore, the wild-type CTR1 allele, while negatively regulating ethylene signaling, positively influences ABA signaling. Similarly, it is possible that ABI1 and ABI2, as negative regulators of ABA signaling, may act to reduce ethylene sensitivity, a function that is analogous to that of CTR1 on the ABA pathway. Our results also showed that the ein2-1/era3 mutant shows reduced transcript levels of ABI1 and PDF1.2 and increased transcript levels of ABA-responsive genes. It is therefore possible that the antagonistic effect of ethylene on the ABA pathway may be exerted partly through the activation of negative regulators of ABA signaling, such as ABI1 and ABI2. Indeed, a recent study showed that ethylene strongly induces ABI1 and ABI2 within 1 to 2 h of treatment (De Paepe et al., 2004), supporting the view that the antagonistic effect of ethylene on ABA signaling may involve negative regulators of ABA signaling.
Antagonistic effects of ethylene on ABA signaling may also require suppression of positive regulators of ABA signaling, such as AtMYC2. Our results presented here as well as those from Van Zhong and Burns (2003) showed that both endogenous ethylene itself and the ethylene signaling pathway suppress AtMYC2 and the AtMYC2-regulated genes VSP2 and rd22. Therefore, ethylene signaling most likely regulates VSP2, rd22, and PDF1.2 expression through concerted action of the positive regulator ERF1 and negative regulator AtMYC2. However, further research is required to determine the relative positions of ERF1 and AtMYC2 in these signaling pathways.
The reduced ABI1 expression and increased levels of ABA-inducible gene expression observed in this study in the ethylene signaling mutants suggest that ethylene insensitivity has the potential to increase abiotic stress tolerance in plants. Interestingly, antisense inhibition of the AtPP2CA gene, encoding an ABI1-related negative regulator of ABA signaling, was known to accelerate cold acclimation in Arabidopsis by increasing expression from cold- and ABA-responsive genes (Tahtiharju and Palva, 2001). The mutually antagonistic interactions observed between ethylene and ABA pathways might also better explain the enhanced disease tolerance observed in the ABA mutants studied here. In light of our results, it is tempting to speculate that the increased disease susceptibility observed in ethylene signaling mutants (e.g., ein2 and ein3) may be resulted from not only disruption of the ethylene pathway but simultaneous activation of antagonistic ABA pathways in these mutants.
Finally, although exogenous MJ appeared to have some inhibitory effect on rd22, no antagonism of the JA pathway on the ABA pathway was evident in the jar1-1 mutant. In fact, JA and ABA are known to act synergistically in a COI1-dependent but JAR1-independent manner for inhibition of seed germination (Ellis and Turner, 2002).
In Figure 11, we present three simple models to explain the signaling interactions among ABA, JA, and ethylene signaling pathways in regulating pathogen-, wound-, and dehydration-responsive gene expression. These models propose that in the absence of stress, the antagonistic interactions among signaling pathways help maintain low levels of expression from stress-responsive genes. Depending on the stress conditions experienced by the plant, however, one signaling pathway may become dominant over others. Consequently, a specific subset of stress-responsive genes is induced through activation of positive regulators of gene expression but also simultaneous suppression of negative regulators. For instance, induction of rd22 and suppression of PDF1.2 by ABA may require simultaneous activation of positive regulators of rd22 and suppression of the ethylene signaling pathway that positively and negatively regulates PDF1.2 and rd22, respectively. Similarly, induction of PDF1.2 by ethylene may require coordinated activation of positive regulators of PDF1.2 (e.g., ERF1) as well as suppression of negative regulation (e.g., AtMYC2) from the ABA pathway. Such cross-communication among plant hormone signaling pathways is probably achieved in a remarkably coordinated manner during adaptation to stress and would certainly enhance the plant's ability to respond to the stress factors in the most appropriate manner. Future research may reveal additional points of interactions between these signaling pathways.
METHODS
Plant Inoculations and Treatments
Arabidopsis thaliana plants were grown in plastic trays containing 30 (5 × 5 cm) cells at 24/18°C day and night temperatures, respectively, under an 8-h photoperiod (180 μmol·m−2·s−1) in a controlled growth facility. Seeds sown directly in soil were stratified at 4°C for 3 d before transfer to a growth chamber. Inoculation or treatment of plants was conducted at the 8 to 10 rosette leaf stage. Inoculations with Fusarium oxysporum were done as described before (Schenk et al., 2000, 2003; Campbell et al., 2003). All inoculation experiments were arranged in a completely randomized split-plot design on trays containing 30 (5 × 5 cm) cells (one to three plants in each cell). The disease symptoms were scored 10 d after inoculation either by counting the number of chlorotic leaves per plant, total numbers of plants showing strong wilt-symptoms, or both. Furthermore, to determine the amount of fungal mass in inoculated plant tissue, total RNA from the inoculated wild type and the myc2 mutant (30 plants of each) was isolated, converted to cDNA using random hexamers, and analyzed by RT-Q-PCR analysis using a primer set (5′-CGCCAGAGGACCCCTAAAC-3′ and 5′-ATCGATGCCAGAACCAAGAGA-3′) specific to the 18S transcribed internal spacer of F. oxysporum (National Center for Biotechnology Information accession no. AY237110) that demonstrated little or no sequence similarity in Arabidopsis. This relative expression was normalized to the levels of plant β-actin mix (see the next section), which did not show any significant difference between the RNA samples isolated from inoculated wild-type or insertion line plants.
For chemical treatments of plants, a solution containing 5% MJ (Aldrich, Milwaukee, WI) dissolved in 100% ethanol was prepared. Then, 200 μL of this was applied to a cotton ball and enclosed with each tray to be treated, to give a final concentration of 0.1 μM of MJ per liter of air. The whole tray was then sealed by two layers of opaque plastic bags and secured with masking tape. The control solution consisted of 0.1% (w/v) ethanol only and was applied to the plants (8 to 10 leaf stage) in the same manner as the MJ solution. SA (Sigma, St. Louis, MO) was dissolved in 100% ethanol, and this stock was then diluted with water to a final concentration of 5 mM. Plants were evenly sprayed with this solution before sealing as described for the MJ treatment. Control plants were sprayed evenly with 0.1% ethanol solution only. ABA (cis-trans isomer; Sigma) was dissolved to 20 mM in 100% ethanol. This stock solution was then diluted with water to a final concentration of 100 μM and sprayed evenly over the plants before sealing as described for the MJ treatment. Control plants were sprayed evenly with 0.1% (w/v) ethanol solution. Ethylene was applied to a final concentration of 200 ppm, and the plants were sealed as explained above. Untreated controls were subjected to the same conditions but without addition of the inducers. For water stress treatment, plants were left without watering for 24 h while controls grown simultaneously were watered as normally. No wilting symptom was apparent in plants exposed to water stress by the time of sampling for RNA isolations.
RT-Q-PCR
Leaf samples taken from ∼30 to 50 plants at time points after inoculation or treatments were ground to powder under liquid nitrogen for extraction of total RNA using an SV RNA isolation kit (Promega, Madison, WI) according to the manufacturer's instructions. For reverse transcription, total RNA (5 μg) was added to 1 μL of oligo(dT)23, gene specific or both primers (10 μM), and the volume adjusted to 9.25 μL with sterile Milli-Q (Millipore, Billerica, MA) water. The solution was incubated at 70°C for 5 min then immediately transferred to ice before addition of 5.25 μL of reverse transcriptase master mix containing 3 μL 5× buffer, 1.5 μL 0.1 M DTT, and 0.75 μL 10 mM dNTPs and 0.5 μL (200 units/μL) Superscript RT II reverse transcriptase (Invitrogen, Carlsbad, CA). The reaction was incubated at 42°C for 90 min before heat inactivation of reverse transcriptase by incubation at 70°C for 15 min. The cDNA was then diluted in a total volume of 300 μL with sterile Milli-Q water. A second aliquot of total RNA was treated similarly using water in place of reverse transcriptase and these samples used as minus reverse transcriptase (−RT) controls for quantification of any genomic DNA contamination or nonspecific DNA amplification. Amplification of specific regions of targeted genes and real-time detection of amplicon production were undertaken in an ABI model 7700 sequence detection system (Perkin-Elmer Applied Biosystems, Foster City, CA). RT-Q-PCR reactions contained 2.8 μL of primer mix (containing 1.5 μM of each forward and reverse primer), 2 μL cDNA template (derived from 6.6 ng/μL total RNA), 10 μL 2× SYBR-green master PCR mix (Perkin-Elmer Applied Biosystems), and 5.2 μL water to make a total volume of 20 μL. The following RT-Q-PCR primer sequences were used: for β-actin mix (β-actin-2 At3a18780, β-actin-7 At5g09810, and β-actin-8 At1g49240), a universal forward 5′-AGTGGTCGTACAACCGGTATTGT-3′ and individual reverse primers 5′-GATGGCATGAGGAAGAGAGAAAC-3′, 5′-GAGGAAGAGCATACCCCTCGTA-3′, and 5′-GAGGATAGCATGTGGAAGTGAGAA-3′ for β-actin-2, β-actin-7, and β-actin-8, respectively; for PDF1.2 (At5g44420), forward 5′-TTTGCTGCTTTCGACGCAC-3′ and reverse 5′-CGCAAACCCCTGACCATG-3′; for PR4 (At3g04720), forward 5′-TGCTACATCCAAATCCAAGCCT-3′ and reverse 5′-CGGCAAGTGTTTAAGGGTGAAG-3′; for AtMYC2 (At1g32640), forward 5′-TCATACGACGGTTGCCAGAA-3′ and reverse 5′-AGCAACGTTTACAAGCTTTGATTG-3′; for ERF1 (At1g27730), forward 5′-CGAGAAGCTCGGGTGGTAGT-3′ and reverse 5′-GCCGTGCATCCTTTTCC-3′; for LEC (At3g15356), forward 5′-GTTTCGTCTCTGGGTCATGGA-3′ and reverse 5′-GCAGCAACTTGTTATTCCTTGGA-3′; for CHI (At3g12500), forward 5′-ATCAGCGCTGCAAAGTCCTTC-3′ and reverse 5′-GTGCTGTAGCCCATCCACCTG-3′; rd22 (At5g25610), forward 5′-CTGTTTCCACTGAGGTGGCTAAG-3′ and reverse 5′-TGGCAGTAGAACACCGCGA-3′; for VSP2 (At5g24770), forward 5′-TCAGTGACCGTTGGAAGTTGTG-3′ and reverse 5′-GTTCGAACCATTAGGCTTCAATATG-3′; for ABI1 (At4g26080), forward 5′-CGGCAAAACTGCACTTCCAT-3′ and reverse 5′-CACGAGCTCCATTCCACTGAA-3′; for KIN1 (At5g15960), forward 5′-GCTGGCAAAGCTGAGGAGAA-3′ and reverse 5′-TTCCCGCCTGTTGTGCTC-3′; for β-tubulin, forward 5′-CGATTCCGTTCTCGATGTTGT-3′ and reverse 5′-AATGAGTGACACACTTGGAATCCTT-3′. Duplicate or triplicate assays were performed on all occasions using cDNA samples derived from two sets of independently grown plants for each experiment. Thermal cycling conditions consisted of 10 min at 95°C and 45 cycles of 15 s at 95°C and 1 min at 60°C before 2 min at 25°C. RT-Q-PCR reactions with minus RT controls as template were performed to correct for DNA contamination or amplification of nonspecific products. Controls with no added template were conducted for each primer pair to ensure primer dimer was not interfering with amplification detection. RT-Q-PCR results were captured and analyzed using the sequence detection software SDS version 1.7 (Perkin-Elmer Applied Biosystems). The SYBR green fluorescent signal was standardized to a passive reference dye (ROX) included in the SYBR green PCR master mix. The software generated plots of SYBR fluorescence (ΔRn) after successive cycles. The fluorescence threshold was set midway within the phase showing exponential rate of amplicon production. The cycle number at which the fluorescence passed the cycle threshold (CT) for each reaction was calculated from this graph and exported to Microsoft Excel for further analysis. Amplification from a cDNA template was used for data analysis when fluorescence from plus RT reactions crossed the threshold level at or greater than five cycles before the minus RT (i.e., −RT is less than 1/32 of +RT). Relative expression levels in each cDNA sample were obtained by normalization to the reference gene either β-tubulin or β-actin, which produced similar results (Campbell et al., 2003; Schenk et al., 2003; J. Anderson and K. Kazan, unpublished data). The level of transcript abundance relative to reference gene (termed ΔCT) was determined by subtraction of the CT for reference gene from the candidate gene CT according to the function ΔCT = CT(test gene) − CT(reference gene). To compare untreated and treated expression levels, the function ΔΔCT was first determined using the equation ΔΔCT = ΔCT(treatment) − ΔCT(control) where control represents mock-treated plants. The induction ratio of treated/control was then calculated by the formula 2−ΔΔCt in accordance with ABI Model 7700 sequence detection system user bulletin 2. After real-time PCR, amplification products for each primer set were subjected to melt-curve analysis to ensure that the fluorescence resulted from a single PCR product and did not represent primer dimer or nonspecific products. This was conducted by measuring fluorescence over a 20-min thermal gradient from 60 to 95°C and data analysis using the Dissociation Curves 1.0 software (Perkin-Elmer Applied Biosystems).
Plasmid Construction and Protoplast Transformation
Arabidopsis ecotype Col-0 plants were grown in tissue culture on MS medium for 2 weeks after germination under constant diffuse light. Leaves from 30 plants were cut into segments ∼2 mm2 and suspended in 500 mM mannitol. Protoplasts were produced from leaf segments and transformations performed according to Weigel and Glazebrook (2001). Briefly, protoplasts were assessed for viability by staining with fluorescein diacetate (0.5% FDA in W5) and subsequent observation using an ultraviolet microscope (Leica MZ6 stereomicroscope; Leica Microscopy and Scientific Instruments, Heerbrug, Switzerland). Viable protoplast concentration was calculated using a haemocytometer. Approximately 1.4 × 106 protoplasts were used for each transformation. All transformations used 20 μg of the 35S:GFP construct as an indicator of transformation efficiency. The construct for overexpression of AtMYC2 was prepared by NotI digestion of the pJIT163 vector (Hellens et al., 2000) and ligation of the band corresponding to the double 35S promoter/terminator cassette into the pGreenII vector (Hellens et al., 2000) previously linearized by digestion with HindIII, blunt ended using Klenow enzyme, and dephosphorylated using shrimp alkaline phosphatase (Roche, Nutley, NJ). The genomic sequence of AtMYC2 was PCR amplified using the primers 5′-ATGACTGATTACCGGCTACA-3′ and 5′-GACCCCATAACTTTCTAAACTTC-3′ and ligated into the HindIII-digested and blunt-ended overexpression plasmid pKEN (Brown et al., 2003). The construct for overexpression of ERF1 was based on the pBI221 vector (Clontech, Palo Alto, CA) for the 35S promoter of Cauliflower mosaic virus driven constitutive expression of the inserted gene and were individually cotransfected with the 35S:GFP vector using 20 μg of each plasmid. Control transformations were conducted using the pBI221 and 35S:GFP constructs together. All transformations were replicated twice with the control transformation replicated three times. After transformations, the cells were incubated for 24 h at room temperature and samples examined using a haemocytometer and an ultraviolet microscope (Leica MZ6 stereomicroscope) for the presence of GFP fluorescence. The percentage of cells transformed with GFP was calculated for each transformation and the remaining cell pellet used for RNA extraction using the SV RNA extraction kit (Promega). The 35S:AtMYC2 construct was also introduced into Arabidopsis plants using the floral dip transformation procedure.
GUS Activity Assays
Promoter activity was assessed by quantitative GUS assays of the uidA gene using 4-methyl-umbelliferyl-β-d-glucuronide (Sigma) as a substrate. The assay was performed according to the procedure given by Brown et al. (2003) adapted for use with microtitre plates and the Fluoroskan Ascent fluorometer (Labsystems, Helsinki, Finland).
Mutants and T-DNA Insertion Lines
All the mutant lines (i.e., aba2-1 [CS156], abi1-1 [CS22], and abi2-1 [CS23] and ein2-1 [CS8844], ein3-1 [CS8052], etr1-1 [CS8058], and jar1-1 [CS8072]) were obtained from the ABRC (The Ohio State University, Columbus, OH). All the mutant and wild-type plants used were in Arabidopsis Col-0 background except abi1-1 and abi2-1 mutants, and the corresponding wild-types which were in Ler background. The PDF1.2promoter-GUS transformed plants were in C24 background. The locations of the T-DNA insertion in the jin1/myc2 (SALK_017005 and SALK_083483) mutants were verified by PCR amplification of genomic DNA with a forward primer (5′-GCGTGGACCGCTTGCTGCAACT-3′) specific to the T-DNA insertion and a reverse primer specific to the AtMYC2 gene (5′-GATCTGATTCTCCGGCGGTTT-3′ or 5′-CGGCGAGCTCGAGTTTCACTT-3′ for SALK_017005 and SALK_083483, respectively). The amplified fragment was then sequenced to identify the exact location of the insert within the gene. To identify lines that were homozygous for the insertion, a second PCR was done using two gene-specific primers (5′-TGGCGCTCGAGGCTCTTACATC-3′ and 5′-GAAAGTCAAACCGAGGCTTCTTCG-3′ or 5′-GATCTGATTCTCCGGCGGTTT-3′ and 5′-AATTATCCGGGTCGGGTTGTG-3′). Only the wild-type allele was amplified by this PCR, whereas no amplification product was detectable from plants homozygous for the insertion. Kanamycin segregation ratios of plants identified as either wild-type or heterozygous for the T-DNA insertion at the AtMYC2 locus suggested the presence of single T-DNA insertions in both T-DNA lines.
Sequence data from this article have been deposited with the EMBL/GenBank data libraries under accession number AY237110.
Acknowledgments
We thank E. Campbell for excellent technical assistance in RT-Q-PCR analyses and pathogen inoculation assays, A. Rusu for assistance in PCR analysis of T-DNA insertion lines and GUS activity assays, B. Simpson for advice on RT-Q-PCR analyses, C. Stephens and R. Angus for advice on protoplast transformation assays, J. Botella for the 35S:GFP plasmid, R. Shivas for F. oxysporum strain, L. Forsyth for advice on pathogen inoculations; the Arabidopsis Stock Centre at The Ohio State University for seeds of mutant lines of Arabidopsis plants, the Salk Institute Genomic Analysis Laboratory for the sequence indexed Arabidopsis T-DNA insertion mutants, G.-P. Xue, R. Dolferus, I. Wilson, and L. Onate-Sanchez for critical reading of the manuscript, B. Dombrecht for useful discussions, and anonymous referees for useful comments on the manuscript. J.P.A and E.B. were recipients of an Australian postgraduate award and an Australian Agency for International Development postgraduate award, respectively.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Kemal Kazan (kemal.kazan@csiro.au).
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.104.025833.
References
- Abe, H., Urao, T., Ito, T., Seki, M., Shinozaki, K., and Yamaguchi-Shinozaki, Y. (2003). Arabidopsis AtMYC2 (bHLH) and AtMYB2 (MYB) function as transcriptional activators in abscisic acid signaling. Plant Cell 15, 63–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abe, H., Yamaguchi-Shinozaki, K., Urao, T., Iwasaki, T., Hosokawa, D., and Shinozaki, K. (1997). Role of Arabidopsis MYC and MYB homologs in drought-and abscisic acid-regulated gene expression. Plant Cell 9, 1859–1868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso, J.M., et al. (2003. a). Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301, 653–657. [DOI] [PubMed] [Google Scholar]
- Alonso, J.M., Stepanova, A.N., Solano, R., Wisman, E., Ferrari, S., Ausubel, F.M., and Ecker, J.R. (2003. b). Five components of the ethylene-response pathway identified in a screen for weak ethylene-insensitive mutants in Arabidopsis. Proc. Natl. Acad. Sci. USA 100, 2992–2997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Audenaert, K., De Meyer, G.B., and Hofte, M.M. (2002). Abscisic acid determines basal susceptibility of tomato to Botrytis cinerea and suppresses salicylic acid-dependent signaling mechanisms. Plant Physiol. 128, 491–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaudoin, N., Serizet, C., Gosti, F., and Giraudat, J. (2000). Interactions between abscisic acid and ethylene signaling cascades. Plant Cell 12, 1103–1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger, S., Bell, E., and Mullet, J.E. (1996). Two methyl jasmonate-insensitive mutants show altered expression of AtVsp in response to methyl jasmonate and wounding. Plant Physiol. 111, 525–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berrocal-Lobo, M., and Molina, A. (2004). Ethylene response factor 1 mediates Arabidopsis resistance to the soilborne fungus Fusarium oxysporum. Mol. Plant-Microbe Interact. 17, 763–770. [DOI] [PubMed] [Google Scholar]
- Berrocal-Lobo, M., Molina, A., and Solano, R. (2002). Constitutive expression of ETHYLENE-RESPONSE-FACTOR1 in Arabidopsis confers resistance to several necrotrophic fungi. Plant J. 29, 23–32. [DOI] [PubMed] [Google Scholar]
- Boter, M., Ruiz-Rivero, O., Abdeen, A., and Prat, S. (2004). Conserved MYC transcription factors play a key role in jasmonate signaling both in tomato and Arabidopsis. Genes Dev. 18, 1577–1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brocard-Gifford, I., Lynch, T.J., Garcia, M.E., Malhotra, B., and Finkelstein, R.R. (2004). The Arabidopsis thaliana ABSCISIC ACID-INSENSITIVE8 locus encodes a novel protein mediating abscisic acid and sugar responses essential for growth. Plant Cell 16, 406–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown, R.L., Kazan, K., McGrath, K.C., Maclean, D.J., and Manners, J.M. (2003). A role for the GCC-box in jasmonate-mediated activation of the PDF1.2 gene of Arabidopsis. Plant Physiol. 132, 1020–1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell, E.J., Schenk, P.M., Kazan, K., Penninckx, I.A., Anderson, J.P., Maclean, D.J., Cammue, B.P., Ebert, P.R., and Manners, J.M. (2003). Pathogen-responsive expression of a putative ATP-binding cassette transporter gene conferring resistance to the diterpenoid sclareol is regulated by multiple defense signaling pathways in Arabidopsis. Plant Physiol. 133, 1272–1284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng, W.H., Endo, A., Zhou, L., Penney, J., Chen, H.-C., Arroyo, A., Leon, P., Nambara, E., Asami, T., Seo, M., Koshiba, T., and Sheen, J. (2002). A unique short-chain dehydrogenase/reductase in Arabidopsis glucose signaling and abscisic acid biosynthesis and functions. Plant Cell 14, 2723–2743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chini, A., Grant, J.J., Seki, M., Shinozaki, K., and Loake, G.J. (2004). Drought tolerance established by enhanced expression of the CC-NBS-LRR gene, ADR1, requires salicylic acid, EDS1 and ABI1. Plant J. 38, 810–822. [DOI] [PubMed] [Google Scholar]
- De Paepe, A., Vuylsteke, M., Van Hummelen, P., Zabeau, M., and Van Der Straeten, D. (2004). Transcriptional profiling by cDNA-AFLP and microarray analysis reveals novel insights into the early response to ethylene in Arabidopsis. Plant J. 39, 537–559. [DOI] [PubMed] [Google Scholar]
- Di Pietro, A., Madrid, M.P., Caracuel, Z., Delgado-Jarana, J., and Roncero, M.I.G. (2003). Fusarium oxysporum: Exploring the molecular arsenal of a vascular wilt fungus. Mol. Plant Pathol. 4, 315–325. [DOI] [PubMed] [Google Scholar]
- Ellis, C., and Turner, J.G. (2002). A conditionally fertile coi1 allele indicates cross-talk between plant hormone signaling pathways in Arabidopsis thaliana seeds and young seedlings. Planta 215, 549–556. [DOI] [PubMed] [Google Scholar]
- Fedoroff, N.V. (2002). Cross-talk in abscisic acid signaling. Sci. STKE 140, RE10. [DOI] [PubMed] [Google Scholar]
- Finkelstein, R.R. (1993). Abscisic acid-insensitive mutations provide evidence for stage-specific signal pathways regulating expression of an Arabidopsis late embryogenesis-abundant (lea) gene. Mol. Gen. Genet. 238, 401–408. [DOI] [PubMed] [Google Scholar]
- Finkelstein, R.R., Gampala, S.S., and Rock, C.D. (2002). Abscisic acid signaling in seeds and seedlings. Plant Cell 14 (suppl.), S15–S45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkelstein, R.R., and Gibson, S.I. (2002). ABA and sugar interactions regulating development: Cross-talk or voices in a crowd? Curr. Opin. Plant Biol. 5, 26–32. [DOI] [PubMed] [Google Scholar]
- Gazzarrini, S., and McCourt, P. (2001). Genetic interactions between ABA, ethylene and sugar signaling pathways. Curr. Opin. Plant Biol. 4, 387–391. [DOI] [PubMed] [Google Scholar]
- Geraats, B.P.J., Bakker, P.A.H.M., and van Loon, L.C. (2002). Ethylene insensitivity impairs resistance to soilborne pathogens in tobacco and Arabidopsis thaliana. Mol. Plant-Microbe Interact. 15, 1078–1085. [DOI] [PubMed] [Google Scholar]
- Ghassemian, M., Nambara, E., Cutler, S., Kawaide, H., Kamiya, Y., and McCourt, P. (2000). Regulation of abscisic acid signaling by the ethylene response pathway in Arabidopsis. Plant Cell 12, 1117–1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goh, C.H., Nam, H.G., and Park, Y.S. (2003). Stress memory in plants: A negative regulation of stomatal response and transient induction of rd22 gene to light in abscisic acid-entrained Arabidopsis plants. Plant J. 36, 240–255. [DOI] [PubMed] [Google Scholar]
- González-Guzmán, M., Apostolova, N., Belles, J.M., Barrero, J.M., Piqueras, P., Ponce, M.R., Micol, J.L., Serrano, R., and Rodriguez, P.L. (2002). The short-chain alcohol dehydrogenase ABA2 catalyzes the conversion of xanthoxin to abscisic aldehyde. Plant Cell 14, 1833–1846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gosti, F., Beaudoin, N., Serizet, C., Webb, A.A., Vartanian, N., and Giraudat, J. (1999). ABI1 protein phosphatase 2C is a negative regulator of abscisic acid signaling. Plant Cell 11, 1897–1910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo, H., and Ecker, J.R. (2003). Plant responses to ethylene gas are mediated by SCFEBF1/EBF2-dependent proteolysis of EIN3 transcription factor. Cell 115, 667–677. [DOI] [PubMed] [Google Scholar]
- He, Y., Fukushige, H., Hildebrand, D.F., and Gan, S. (2002). Evidence supporting a role of jasmonic acid in Arabidopsis leaf senescence. Plant Physiol. 128, 876–884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellens, R.P., Edwards, E.A., Leyland, N.R., Bean, S., and Mullineaux, P.M. (2000). pGreen: A versatile and flexible binary Ti vector for Agrobacterium-mediated plant transformation. Plant Mol. Biol. 42, 819–832. [DOI] [PubMed] [Google Scholar]
- Henfling, J.W.D.M., Bostock, R., and Kuc, J. (1980). Effect of abscisic acid on rishitin and lubimin accumulation and resistance to Phythopthora infestans and Cladosporium cucumerinum in potato tissue slices. Phytopathology 70, 1074–1078. [Google Scholar]
- Hoth, S., Morgante, M., Sanchez, J.P., Hanafey, M.K., Tingey, S.V., and Chua, N.H. (2002). Genome-wide gene expression profiling in Arabidopsis thaliana reveals new targets of abscisic acid and largely impaired gene regulation in the abi1-1 mutant. J. Cell Sci. 15, 4891–4900. [DOI] [PubMed] [Google Scholar]
- Hwang, I., and Sheen, J. (2001). Two-component circuitry in Arabidopsis cytokinin signal transduction. Nature 413, 383–389. [DOI] [PubMed] [Google Scholar]
- Koornneef, M., Jorna, M.L., Brinkhorst-van der Swan, D.L.C., and Karssen, C.M. (1982). The isolation of abscisic acid (ABA)-deficient mutants by selection of induced revertants in non-germinating gibberellin sensitive lines of Arabidopsis thaliana (L.) Heynh. Theor. Appl. Genet. 61, 385–393. [DOI] [PubMed] [Google Scholar]
- Lay, F.T., Brugliera, F., and Anderson, M.A. (2003). Isolation and properties of floral defensins from ornamental tobacco and petunia. Plant Physiol. 131, 1283–1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeNoble, M.E., Spollen, W.G., and Sharp, R.E. (2004). Maintenance of shoot growth by endogenous ABA: Genetic assessment of the involvement of ethylene suppression. J. Exp. Bot. 55, 237–245. [DOI] [PubMed] [Google Scholar]
- Leon, P., and Sheen, J. (2003). Sugar and hormone connections. Trends Plant Sci. 8, 110–116. [DOI] [PubMed] [Google Scholar]
- Lorenzo, O., Chico, J.M., Sanchez-Serrano, J.J., and Solano, R. (2004). JASMONATE-INSENSITIVE1 encodes a MYC transcription factor essential to discriminate between different jasmonate-regulated defense responses in Arabidopsis. Plant Cell 16, 1938–1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenzo, O., Piqueras, R., Sanchez-Serrano, J.J., and Solano, R. (2003). ETHYLENE RESPONSE FACTOR1 integrates signals from ethylene and jasmonate pathways in plant defense. Plant Cell 15, 165–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madrid, M.P., Di Pietro, A., and Roncero, M.I. (2003). Class V chitin synthase determines pathogenesis in the vascular wilt fungus Fusarium oxysporum and mediates resistance to plant defence compounds. Mol. Microbiol. 47, 257–266. [DOI] [PubMed] [Google Scholar]
- Manners, J.M., Penninckx, I.A.M.A., Vermaere, K., Kazan, K., Brown, R., Morgan, A., Maclean, D.J., Curtis, M.D., Cammue, B.P.A., and Broekaert, W.F. (1998). The promoter of the plant defensin gene PDF1.2 from Arabidopsis is systemically activated by fungal pathogens and responds to methyl jasmonate but not to salicylic acid. Plant Mol. Biol. 38, 1071–1080. [DOI] [PubMed] [Google Scholar]
- Marin, E., Nussaume, L., Quesada, A., Gonneau, M., Sotta, B., Hugueney, P., Frey, A., and Marion-Poll, A. (1996). Molecular identification of zeaxanthin epoxidase of Nicotiana plumbaginifolia, a gene involved in abscisic acid biosynthesis and corresponding to the ABA locus of Arabidopsis thaliana. EMBO J. 15, 2331–2342. [PMC free article] [PubMed] [Google Scholar]
- McDonald, K.L., and Cahill, D.M. (1999). Influence of abscisic acid and the abscisic acid biosynthesis inhibitor, norflurazon, on interactions between Phytophthora sojae and soybean (Glycine max). Eur. J. Plant Pathol. 105, 651–658. [Google Scholar]
- Mengiste, T., Chen, X., Salmeron, J., and Dietrich, R. (2003). The BOS1 gene encodes an R2R3MYB transcription factor protein that is required for biotic and abiotic stress responses in Arabidopsis. Plant Cell 15, 2551–2565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merlot, S., Gosti, F., Guerrier, D., Vavasseur, A., and Giraudat, J. (2001). The ABI1 and ABI2 protein phosphatases 2C act in a negative feedback regulatory loop of the abscisic acid signaling pathway. Plant J. 25, 295–303. [DOI] [PubMed] [Google Scholar]
- Mohr, P.G., and Cahill, D.M. (2003). Abscisic acid influences the susceptibility of Arabidopsis thaliana to Pseudomonas syringae pv. tomato and Peronospora parasitica. Funct. Plant Biol. 30, 461–469. [DOI] [PubMed] [Google Scholar]
- Moons, A., Prinsen, E., Bauw, G., and Van Montagu, M. (1997). Antagonistic effects of abscisic acid and jasmonates on salt stress-inducible transcripts in rice roots. Plant Cell 9, 2243–2259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narasimhan, M.L., Lee, H., Damsz, B., Singh, N.K., Ibeas, J.I., Matsumoto, T.K., Woloshuk, C.P., and Bressan, R.A. (2003). Overexpression of a cell wall glycoprotein in Fusarium oxysporum increases virulence and resistance to a plant PR-5 protein. Plant J. 36, 390–400. [DOI] [PubMed] [Google Scholar]
- Nishimura, M.T., Stein, M., Hou, B.H., Vogel, J.P., Edwards, H., and Somerville, S.C. (2003). Loss of a callose synthase results in salicylic acid-dependent disease resistance. Science 301, 969–972. [DOI] [PubMed] [Google Scholar]
- Park, J.M., Park, C.J., Lee, S.B., Ham, B.K., Shin, R., and Paek, K.H. (2001). Overexpression of the tobacco Tsi1 gene encoding an EREBP/AP2-type transcription factor enhances resistance against pathogen attack and osmotic stress in tobacco. Plant Cell 13, 1035–1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penninckx, I.A., Eggermont, K., Terras, F.R., Thomma, B.P., De Samblanx, G.W., Buchala, A., Metraux, J.P., Manners, J.M., and Broekaert, W.F. (1996). Pathogen-induced systemic activation of a plant defensin gene in Arabidopsis follows a salicylic acid-independent pathway. Plant Cell 8, 2309–2323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penninckx, I.A., Thomma, B.P., Buchala, A., Metraux, J.P., and Broekaert, W.F. (1998). Concomitant activation of jasmonate and ethylene response pathways is required for induction of a plant defensin gene in Arabidopsis. Plant Cell 10, 2103–2113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen, M., et al. (2000). Arabidopsis map kinase 4 negatively regulates systemic acquired resistance. Cell 103, 1111–1120. [DOI] [PubMed] [Google Scholar]
- Potuschak, T., Lechner, E., Parmentier, Y., Yanagisawa, S., Grava, S., Koncz, C., and Genschik, P. (2003). EIN-3 dependent regulation of plant ethylene hormone signaling by two Arabidopsis F box proteins: EBF1 and EBF2. Cell 115, 679–689. [DOI] [PubMed] [Google Scholar]
- Rezzonico, E., Flury, N., Meins, F., Jr., and Beffa, R. (1998). Transcriptional down-regulation by abscisic acid of pathogenesis-related beta-1,3-glucanase genes in tobacco cell cultures. Plant Physiol. 117, 585–592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rojo, E., Leon, J., and Sanchez-Serrano, J.J. (1999). Cross-talk between wound signalling pathways determines local versus systemic gene expression in Arabidopsis thaliana. Plant J. 20, 135–142. [DOI] [PubMed] [Google Scholar]
- Rojo, E., Solano, R., and Sánchez-Serrano, J.J. (2003). Interactions between signaling compounds involved in plant defense. J. Plant Growth Regul. 22, 82–98. [Google Scholar]
- Schenk, P.M., Kazan, K., Manners, J.M., Anderson, J.P., Simpson, R.S., Wilson, I.W., Somerville, S.C., and Maclean, D.J. (2003). Systemic gene expression in Arabidopsis during an incompatible interaction with Alternaria brassicicola. Plant Physiol. 32, 999–1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schenk, P.M., Kazan, K., Wilson, I., Anderson, J.P., Richmond, T., Somerville, S.C., and Manners, J.M. (2000). Coordinated plant defense responses in Arabidopsis revealed by microarray analysis. Proc. Natl. Acad. Sci. USA 97, 11655–11660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp, R.E., LeNoble, M.E., Else, M.A., Thorne, E.T., and Gherardi, F. (2000). Endogenous ABA maintains shoot growth in tomato independently of effects on plant water balance: Evidence for an interaction with ethylene. J. Exp. Bot. 51, 1575–1584. [DOI] [PubMed] [Google Scholar]
- Sheen, J. (1996). Ca2+-dependent protein kinases and stress signal transduction in plants. Science 274, 1900–1902. [DOI] [PubMed] [Google Scholar]
- Sheen, J. (1998). Mutational analysis of protein phosphatase 2C involved in abscisic acid signal transduction in higher plants. Proc. Natl. Acad. Sci. USA 95, 975–980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinozaki, K., Yamaguchi-Shinozaki, K., and Seki, M. (2003). Regulatory network of gene expression in the drought and cold stress responses. Curr. Opin. Plant Biol. 6, 410–417. [DOI] [PubMed] [Google Scholar]
- Solano, R., Stepanova, A., Chao, Q., and Ecker, J.R. (1998). Nuclear events in ethylene signaling: A transcriptional cascade mediated by ETHYLENE-INSENSITIVE3 and ETHYLENE-RESPONSE-FACTOR1. Genes Dev. 12, 3703–3714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spoel, S.H., et al. (2003). NPR1 modulates cross-talk between salicylate- and jasmonate-dependent defense pathways through a novel function in the cytosol. Plant Cell 15, 760–770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spollen, W.G., LeNoble, M.E., Samuels, T.D., Bernstein, N., and Sharp, R.E. (2000). Abscisic acid accumulation maintains maize primary root elongation at low water potentials by restricting ethylene production. Plant Physiol. 122, 967–976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staswick, P.E., Su, W., and Howell, S.H. (1992). Methyl jasmonate inhibition of root growth and induction of a leaf protein are decreased in an Arabidopsis thaliana mutant. Proc. Natl. Acad. Sci. USA 89, 6837–6840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tahtiharju, S., and Palva, T. (2001). Antisense inhibition of protein phosphatase 2C accelerates cold acclimation in Arabidopsis thaliana. Plant J. 26, 461–470. [DOI] [PubMed] [Google Scholar]
- Thaler, J.S., and Bostock, R.M. (2004). Interactions between abscisic-acid-mediated responses and plant resistance to pathogens and insects. Ecology 85, 48–58. [Google Scholar]
- Thomma, B.P., Eggermont, K., Tierens, K.F., and Broekaert, W.F. (1999). Requirement of functional ethylene-insensitive 2 gene for efficient resistance of Arabidopsis to infection by Botrytis cinerea. Plant Physiol. 121, 1093–1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomma, B.P., Penninckx, I.A., Broekaert, W.F., and Cammue, B.P. (2001). The complexity of disease signaling in Arabidopsis. Curr. Opin. Immunol. 13, 63–68. [DOI] [PubMed] [Google Scholar]
- Ton, J., and Mauch-Mani, B. (2004). β-Amino-butyric acid-induced resistance against necrotrophic pathogens is based on ABA-dependent priming for callose. Plant J. 38, 119–130. [DOI] [PubMed] [Google Scholar]
- Van Zhong, G., and Burns, J.K. (2003). Profiling ethylene-regulated gene expression in Arabidopsis thaliana by microarray analysis. Plant Mol. Biol. 53, 117–131. [DOI] [PubMed] [Google Scholar]
- Wanzel, M., Herold, S., and Eilers, M. (2003). Transcriptional repression by Myc. Trends Cell Biol. 13, 146–150. [DOI] [PubMed] [Google Scholar]
- Ward, E.W.B., Cahill, D.M., and Bhattacharyya, M.K. (1989). Abscisic acid suppression of phenylalanine ammonia-lyase activity and mRNA, and resistance of soybeans to Phytophthora megasperma f. sp. glycinea. Plant Physiol. 91, 23–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weigel, D., and Glazebrook, J. (2001). Arabidopsis: A Laboratory Manual. (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press), p. 448.
- Wildermuth, G.B., and Morgan, J.M. (2004). Genotypic differences in partial resistance to crown rot caused by Fusarium pseudograminearum in relation to an osmoregulation gene in wheat. Australas. Plant Pathol. 33, 121–123. [Google Scholar]
- Xiong, L., and Yang, Y. (2003). Disease resistance and abiotic stress tolerance in rice are inversely modulated by an abscisic acid-inducible mitogen-activated protein kinase. Plant Cell 15, 745–759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi-Shinozaki, K., and Shinozaki, K. (1993). The plant hormone abscisic acid mediates the drought-induced expression but not the seed-specific expression of rd22, a gene responsive to dehydration stress in Arabidopsis thaliana. Mol. Gen. Genet. 238, 17–25. [DOI] [PubMed] [Google Scholar]
- Yanagisawa, S., Yoo, S.D., and Sheen, J. (2003). Differential regulation of EIN3 stability by glucose and ethylene signaling in plants. Nature 425, 521–525. [DOI] [PubMed] [Google Scholar]
- Zhou, L., Jang, J.C., Jones, T.L., and Sheen, J. (1998). Glucose and ethylene signal transduction crosstalk revealed by an Arabidopsis glucose-insensitive mutant. Proc. Natl. Acad. Sci. USA 95, 10294–10299. [DOI] [PMC free article] [PubMed] [Google Scholar]