Abstract
Meat adulteration is a worldwide concern. In this paper, a new droplet digital PCR (ddPCR) method was developed for the quantitative determination of the presence of chicken in sheep and goat meat products. Meanwhile, a constant (multiplication factor) was introduced to transform the ratio of copy numbers to the proportion of meats. The presented ddPCR method was also proved to be more accurate (showing bias of less than 9% in the range from 5% to 80%) than real-time PCR, which has been widely used in this determination. The method exhibited good repeatability and stability in different thermal treatments and at ultra-high pressure. The relative standard deviation (RSD) values of 5% chicken content was less than 5.4% for ultra-high pressure or heat treatment. Moreover, we confirmed that different parts of meat had no effect on quantification accuracy of the ddPCR method. In contrast to real-time PCR, we examined the performance of ddPCR as a more precise, sensitive and stable analytical strategy to overcome potential problems of discrepancies in amplification efficiency discrepancy and to obtain the copy numbers directly without standard curves. The method and strategy developed in this study can be applied to quantify the presence and to confirm the absence of adulterants not only to sheep but also to other kinds of meat and meat products.
Introduction
Adulteration of meat products occurs frequently worldwide [1–3]. The condition occurs such as deliberately adding cheaper derivatives to bulk out minced meat products, unwitting adulteration by poor manufacturing methods, or mislabeling (deliberate or unwitting) of one meat as another. In China, the sheep and goat meats are often adulterated with chicken because chicken is much less expensive than sheep or goat meat [3–5]. To protect the interests of consumers, many countries and regions have issued regulations regarding the labeling of meat products [6–8], and several methods for the identification of animal species based on DNA [9–12] or protein (peptides) [13–16] have been developed to coordinate with the implementation of those regulations. However, determining of the ratio of adulteration remains a critical issue because it is difficult to discriminate whether the adulterants were included deliberately or inadvertently.
Numerous papers have demonstrated the applicability of real-time PCR for meat quantification [17–21]. However, the copy numbers calculated using the Ct value from real-time PCR that can be affected by amplification efficiency and impurities in the DNA solution. Discrepancies from the actual copy numbers would result in greater deviation in the quantification of the weight proportion of species. Digital PCR (dPCR) is a new technique for precise quantification that can eliminate the effects of matrixes, improve the sensitivity and precision, and provide an absolute measurement of nucleic acid concentration without the use of standard curves [22]. This technique has been extensively used in several areas, including quantitative gene expression analysis [23], bacterial abundance [24], identification of genetically modified organisms [25] and food authentication [26]. In the quantification of DNA from transgenic soy, dPCR may be more suitable for quantitative analysis, as it exhibits a measurement uncertainty of only 17% or below for a single reaction [27]. However, compared with other methods, there have been fewer reports about digital PCR for the quantitative analysis of meat species.
The difficulty in the quantification of meat is transforming copy numbers to the weight proportion of meat. The DNA yields (the copies of each gene) vary, even with equal weights of different types of meat, because the cell density, genome size and the copy numbers of target genes in the genomic DNA vary among different animal species [28]. Recent research reported that there was a close linear relationship between the raw meat weight and DNA content and gene copy numbers [29]. Such an approach requires two calibration curves, which would increase the experimental complexity and the bias of the results. Additionally, the adulterant often comes from various parts of the animal, and meat products are usually treated with high temperatures or high hydrostatic pressure. To the best of our knowledge, very little work has been performed to study the robustness of quantification by ddPCR when using meat from different parts or using processed meat.
Hence, in this study, a droplet digital PCR (ddPCR) assay for the accurate and precise quantification of the chicken weight fraction in sheep or goat was developed and validated. To address the difficulty of using the copy numbers calculated by ddPCR for quantification of the weight proportions of meats, a constant (multiplication factor) was introduced. We compared the established method with real-time PCR, which is widely used in the detection of meat adulteration. Additionally, to verify the repeatability and accuracy of this method, we analyzed the influence of different parts of the chicken and different processing conditions, such as thermal and ultra-high pressure treatments. The goals of this study were to determine the fractions of chicken in sheep or goat, whether added unwittingly or deliberately, and to provide a precise method to aid law enforcement agencies in the control of food adulteration.
Materials and methods
Preparation of reference mixed meat samples
Authentic fresh meat (skeletal muscle) samples from pork (Sus scrofa), beef (Bostaurus), horse (Equus caballus), rabbit (Oryctolagus cuniculus), donkey (Equus asinus), sheep (Ovis aries), goat (Capra hircus), dog (Canis lupus), chicken (Gallus gallus), duck (Anas platyrhynchos), pigeon (Columba livia), goose (Anser anser), and turkey (Meleagris gallopavo), along with commercial sheep and goat samples, including minced mutton, meat rolls, kebabs, dumplings, lamb sausage and cumin lamb, were purchased from a local market in Beijing, China. To determine the dynamic range, repeatability and limits of quantification of the methods, different proportions of fresh chicken and sheep meat were prepared to cover a range of 1–80% chicken/sheep (160 mg chicken/40 mg sheep, 100 mg chicken/100 mg sheep, 40 mg chicken/160 mgsheep, 20 mg chicken/180 mg sheep, 10 mg chicken/190 mg sheep, 2 mg chicken/198 mg sheep).
To ensure that the extracted DNA accurately represented the proportions of different meats, 200 mg samples of the tissue mixtures described above were frozen in liquid nitrogen and ground at 25 times/s for 60 s using a TissueLyserⅡ (QIAGEN, Germany) until mixed evenly.
DNA extraction
DNA were extracted from 200 mg of each meat sample by using a modified cetyltrimethyl ammonium bromide (CTAB) protocol (ISO 21571:2005) [30]. Following DNA extraction, the purity and concentration of the DNA solutions were measured by UV photometry with NanoDrop 1000 (Germany).
Primers and probes
To detect sheep, goat and chicken, the housekeeping gene replication protein A1 (RPA1) was selected as the target detection sequence. The RPA1 sequences of sheep, goat, and chicken were BLASTed against representative species, including Sus scrofa (NC 010454.3), Bos Taurus (AC 000176.1), Gallus gallus (NC 006106.3), Anas platyrhynchos (NW 004677611.1), Equus caballus (NC 009154.2), Meleagris gallopavo (NC 015031.2) and other distant taxa. The primers and probes for digital PCR were designed using Primer Premier 5.0, and target sequences were aligned using DNAMAN. TaqMan probes were labeled with 6-carboxyfluorescein (FAM) at the 5’ end and black hole quencher (BHQ I) at the 3’ end (Table 1).
Table 1. Primer and probe sequences for quantitative PCR.
| Primer/Probe | Sequences (5’-3’) | NCBI Reference Sequence |
|---|---|---|
| Sheep-F (Goat-F) | CTGACACACGGGACACMTCTCC | NC_019468.1 |
| Sheep-R (Goat-R) | AAGCTAAACATGGACCCACATG | |
| Sheep-P (Goat-P) | FAM-TAAGCCAGCCTTGTGCGTGTGGTCC-BHQ1 | |
| Chicken-F | CAGAACCACACTCAACCTGTCTGA | NC_006106.3 |
| Chicken-R | TCGGGGAAATGTCTTACTGCAAG | |
| Chicken-P | FAM-CTCCTAGCAGCCTGTGCCAAGGCCA-BHQ1 |
Droplet digital PCR assay and specificity detection
The ddPCR assays were carried out in a total volume of 20 μL containing 50 ng of DNA template, 10 μL 2 ×ddPCR Master Mix (Bio-Rad, USA), 1 μL of 10 μM primer-F, 1 μL of 10 μM primer-R, and 0.5 μL of 10 μM probes. A Bio-Rad QX200 ddPCR droplet generator (Bio-Rad, USA) was used to divide the 20 μL mixture into approximately 20,000 droplets, with the target DNA segments and PCR reagents being randomly distributed into the droplets. The primer and probe sequences are shown in Table 1. The thermal parameters were as follows: 10 min at 95°C, 30 s at 94°C, followed by 50 cycles of 30 s at 94°C and 1 min at 60°C, followed by enzyme inactivation at 98°C for 10 min and holding at 4°C. Finally, the amplified products were analyzed using a QX200 droplet reader (Bio-Rad, USA).
After the PCR assays were finished, the copy numbers were automatically analyzed using QuantaSoft Version 1.6.6 (Bio-Rad, USA). The absolute copy numbers per panel were estimated according to the number of positive wells and the total number of partitions. According to the Poisson distribution, the copy number per droplet was calculated using the following equation:
| (1) |
Where the R value was the total number of separated droplets and P was the number of wells positive for the RPA1 gene [31].
To test the specificity of the sheep, goat and chicken ddPCR reactions, 200 mg of each of 13 types of fresh meat (sheep, goat, pork, chicken, duck, goose, pigeon, beef, horse, dog, rabbit, turkey and donkey) was frozen and ground. DNA was extracted using the CTAB protocol mentioned above. Approximately 50 ng of DNA was used for each ddPCR assay.
Quantification strategy based on multiplication factor
Because there is a discrepancy between the DNA fraction and the mass proportion of meats, to avoid deviation, it is necessary to correct the concentration of species-specific genes quantified by digital PCR by using multiplication factors. The copy numbers of chicken and sheep in the DNA solution were related to the weight of mixture used for DNA extraction and the copy numbers of the chicken and sheep per unit of mass. Hence, to yield a sample with the specific type of meat, Eq 2 was used:
| (2) |
where M is the mass in weight of specific meat in the sample, Q is the copy number of the RPA1 gene in the DNA solution as calculated by ddPCR measurement, and C is the RPA1 gene copy number of unit mass.
Since the ratios of mass of chicken and sheep could help us obtained the chicken proportion, we should calculate the ratios of the mass of chicken and sheep. So we deduced Eq 3:
| (3) |
where Mc and Ms are the masses of chicken and sheep, respectively; Qc and Qs are the copy numbers of chicken and sheep in the mixtures, respectively; and Cc and Cs represent the copy numbers per unit mass of chicken and sheep, respectively.
Under a fixed procedure for the extraction of DNA (grinding, protease and denaturant), for a mixed meat product with a type of processing and tissue composition, the multiplication factor k (Cs/Cc) could be considered invariable. Therefore,
| (4) |
To determine the k value, five mixtures of different proportions of sheep and chicken (1%, 10%, 20%, 50% and 80% chicken (w/w)) underwent ddPCR quantification of the concentration of the species-specific RPA1 gene. Each sample was tested three times in parallel.
To verify the accuracy and stability under a fixed level of quality, six parallel mixtures of 50% (w/w) sheep and chicken were prepared in order to quantify the copy numbers of the RPA1 genes from chicken and sheep. The k value and RSD were calculated.
Real-time PCR quantification approaches
Real-time PCR was performed using an Applied Biosystems 7500 real-time PCR system (Applied Biosystems, USA) in 25 μL reaction mixtures, which consisted of 12.5 μL of 2× TaqMan Gene Expression Master Mix (Applied Biosystems, USA), 1.0 μL of each primer (10 μM), 0.5 μL of probe (10 μM), 5 μL of template DNA (20 ng/μL and 5 μL of ddH2O. PCR was initiated at 50°C for 2 min, 95°C for 10 min, followed by 40 cycles at 95°C for 15 s and 60°C for 60 s. Every round of the experiment was performed on a single 96-well reaction plate to minimize variability due to thermal cycling.
To simplify the experimental procedure and reduce the complexity of the conversion from copy numbers to the weight of meat, equal weights of sheep and chicken were mixed as a standard sample. To correct the discrepancy of copy numbers of equal weights of chicken and sheep, the equal virtual concentration, 1×105 copies/μL, was assumed to represent the copies of RPA1 for sheep and chicken in the standard sample DNA solution. Six 4-fold dilutions of a standard sample DNA solution were measured in triplicate. By plotting mean CT vs. log(DNA copy numbers), standard curves for both species were constructed.
For unbiased comparisons between the two quantification methods, mixtures with chicken levels of 80%, 50%, 20%, 10%, 5%, and 1% were selected for real-time PCR assay and ddPCR. The CT values recorded by sheep and chicken detectors were directly interpolated on the standard curve to yield the concentration of species-specific genes. The proportion of chicken meat in the sample was expressed as:
| (5) |
where Cchicken and Csheep were the copy numbers obtained from the standard curve of sheep and chicken detectors, respectively.
Dynamic range, limit of quantification (LOQ), and limit of detection (LOD)
The squared regression coefficient (R2) of the linear regression line between the actual content of adulterant (x-axis) and the measured values of the meat fraction (y-axis) was used to evaluate the dynamic range of the quantification of mixed meat.
The LOQ was determined by evaluating the relative standard deviation (RSD) and bias of meat mixtures ranging from 1%-10%. Three replicates of ddPCR tests were run for each level.
The chicken in the binary (refer the mixture used) was accordingly diluted to 1%, 0.8%, 0.5%, 0.2% and 0.1% for sensitivity detection. Each level of concentration was analyzed in three replicates.
Repeatability and accuracy
To determine the repeatability and accuracy of the method for detection of adulterant ingredients, six replicates of ddPCR tests were run for each mixture, with chicken levels of 10%, 5%, 4%, 3%, 2% and 1% (w/w), by different operators in our lab. In order to minimize the impact of other factors, the operators started with extracted DNA.
Robustness of the method
The texture, processing (heat treatment or ripening) and tissue ratios of the meat fractions used for the production of mixed meat products may influence the results produced by methods that measure the content of DNA, such as quantitative PCR [32]. To evaluate the effect of different body parts of chicken on quantification, the meat from chicken legs, breasts and wings were mixed with sheep in two proportions (5% and 50%), treated with liquid nitrogen, and ground in a TissueLyser II (QIAGEN). The measured values were corrected by a k value, and the deviation of two levels (5% and 50%) of mixtures tested by ddPCR was calculated.
The effects of thermal treatment with different temperatures and times on the quantitative method were investigated. The binary mixtures of chicken and sheep in two proportions (5% and 50%) were treated in a refrigerator at -18°C for 24 h or at 4°C for 12 h, respectively. In addition, binary mixtures were steamed at 50°C, 80°C, or 100°C in a digital circulation water bath (Memmert), and thermally processed at 120°C in an autoclave (Tuttnauer 3850 EL) for 10 minutes and 20 minutes, respectively.
Further, the influence of different intensities of ultra-high pressure on the accuracy of quantification was also considered. Both of the two proportions (5% and 50%) of meat mixtures were processed at 200 MPa, 300 MPa, 400 MPa, 500 MPa and 600 MPa for 10 minutes and 20 minutes by 650-5L HHP (high hydrostatic pressure, HHP, KeFa). The processed mixtures were ground, and the DNA was extracted and tested in the same way as the fresh samples. Then, the copy numbers measured by ddPCR were converted to the proportion of meat by the k value, which were deduced theoretically and verified by experiments as described in the “Quantification strategy based on multiplication factor” section.
Results and discussion
Determination and verification of multiplication factors
The ratios of the copy numbers of unit mass of chicken and sheep in five proportions (1%, 10%, 20%, 50% and 80% chicken) are shown in Table 2. The average value of the k value for five different meat fractions was 0.8, and the relative standard deviation (RSD) was 9.9%, demonstrating that the k values were stable against any mass ratio of chicken and sheep.
Table 2. The ratios of copy numbers of unit mass with different proportions of chicken.
| Chicken proportion | Three parallel tests of the concentration of chicken (copies/μL) | Three parallel tests of the concentration of sheep(copies/μL) | K | Means | RSD | ||||
|---|---|---|---|---|---|---|---|---|---|
| 80% | 770 | 769 | 768 | 160 | 159 | 160 | 0.8 | 0.8 | 9.9% |
| 50% | 375 | 378 | 376 | 292 | 295 | 305 | 0.8 | ||
| 20% | 163 | 157 | 161 | 484 | 472 | 476 | 0.7 | ||
| 10% | 103 | 100 | 99 | 714 | 718 | 721 | 0.8 | ||
| 1% | 9.7 | 9.1 | 7.6 | 556 | 536 | 547 | 0.8 | ||
To verify the accuracy and stability of the k value, equal masses of muscle were used to obtain the multiplication factor, which depicted the difference in target copy numbers between the two species. To determine the k value, six parallel references mixtures (50chicken/50sheep) were measured by ddPCR. The average copy numbers for sheep in the mixture were 265 copies/μL, 336 copies/μL, 324 copies/μL, 373.3 copies/μL, 379.3 copies/μL, and 327 copies/μL, while the average copy numbers for chicken in the mixture were 335.33 copies/μL, 406 copies/μL, 395.3 copies/μL, 471.7 copies/μL, 480 copies/μL, and 384.7 copies/μL. The k value was approximately 0.8 (RSD = 3.1%), which could be used to convert copies of target DNA extracted from lean mixtures to the weight proportions of each species (S1 Table).
These results suggest that the k value is always approximately 0.8, being independent of the relative content of chicken and sheep. Therefore, the k value can be applied to correct for the proportion of meat. Compared to previously reported ddPCR methods, which require the production of a matrix-adapted series of proportional materials encompassing the whole range of expected meat proportions [29], the use of a single-point equal mass proportion material in ddPCR-based food assays in the present work simplifies the experimental step for species quantification without producing standard curves.
Comparison of ddPCR and RT-PCR for quantification
The following formula (4) was used: Mc/Ms = k×(Qc/Qs), k = 0.8, Mc/Ms = 0.8×(Qc/Qs).The measured chicken fractions in the meat mixtures (80%, 50%, 20%, 10%, 5%, and 1% w/w) were 79.1% (RSD = 0.4%), 49.8% (RSD = 1.3%), 20.8% (RSD = 1.9%), 9.9% (RSD = 2.0%), 4.8% (RSD = 0.5%), and 1.3% (RSD = 13.8%), respectively (Table 3).
Table 3. Comparison of the limit of quantification for sheep and chicken mixtures (weight/weight) by ddPCR and real-time PCR.
| Actual value (%) | Measured value (%) | RSD (%) | Bias (%) | |||
|---|---|---|---|---|---|---|
| ddPCR | qPCR | ddPCR | qPCR | ddPCR | qPCR | |
| 80 | 79.1±0.3 | 68.1±0.1 | 0.4 | 2.5 | -1.2 | -14.9 |
| 50 | 49.8±0.7 | 34.9±0.1 | 1.3 | 4.8 | -0.4 | -30.2 |
| 20 | 20.8±0.4 | 12.7±0.1 | 1.9 | 1.9 | 4.2 | -36.5 |
| 10 | 9.9±0.2 | 5.4±0.1 | 2.0 | 9.1 | -1.0 | -45.8 |
| 5 | 4.8±0.1 | 2.9±0.1 | 0.5 | 8.7 | -3.4 | -41.1 |
| 1 | 1.3±0.2 | 0.6±0.1 | 13.8 | 8.6 | 24.8 | -35.8 |
Actual value (%): Chicken content is expressed as the actual percentage.
Measured value (%): The average content of chicken as measured by ddPCR and real-time PCR.
Bias (%): Bias of the average content of chicken measured by ddPCR and real-time PCR compared with the actual value.
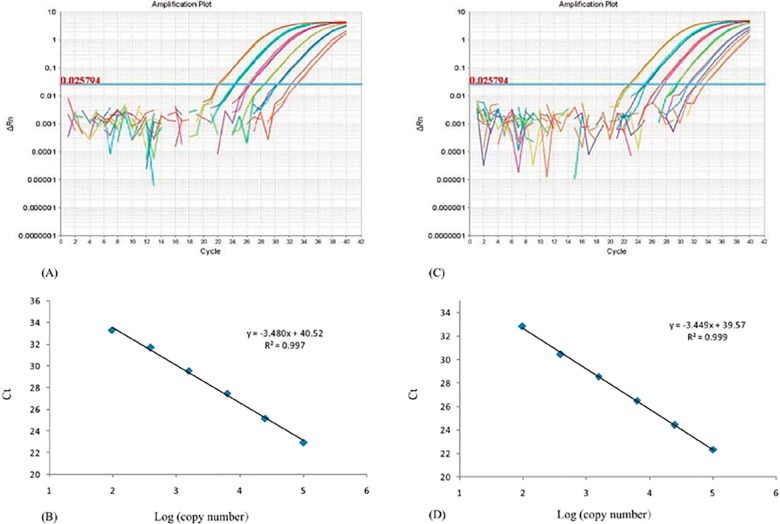
The real-time PCR method utilizes a linear relationship between the Ct value and the logarithm of initial template copy numbers in reactions, which is assumed to be proportional to the weight of chicken and sheep in the mixtures. In this work, two standard curves were established for the quantification of chicken and sheep. The amplification efficiencies for chicken and sheep in meat mixtures were 94.96% and 93.80%, respectively. In addition, from the standard curve generated with Ct values against log (copy numbers of chicken) or log (copy numbers of sheep), the equations were established as follows: Ct = - 3.449log (copy numbers of chicken) + 39.57 (R2 = 0.999); Ct = - 3.480 log (copy numbers of sheep) + 40.52 (R2 = 0.997) (Fig 1). These two equations could be used to estimate the chicken and sheep contents in a mixture; thus, the fraction of chicken in sheep (w/w) could be determined based on the copy ratio of chicken and sheep.
Fig 1. Amplification plots and standard curves for the RPA1 genes of chicken and sheep.
(A) Amplification plots of 4-fold dilution series of binary mixture DNA of chicken (from 1×105 to 9.7×101 copies/μL). (B) Linearity test, regression line parameters of 4-fold dilution series of chicken DNA (from 1×105 to 9.7×101 copies/μL) as standards. (C) Amplification plots of 4-fold dilution series of binary mixture DNA of sheep (from 1×105 to 9.7×101 copies/μL). (D) Linearity test, regression line parameters of 4-fold dilution series of binary mixture DNA of sheep (from 1×105 to 9.7×101 copies/μL) as standards. Each data point represents the mean of three replicates.
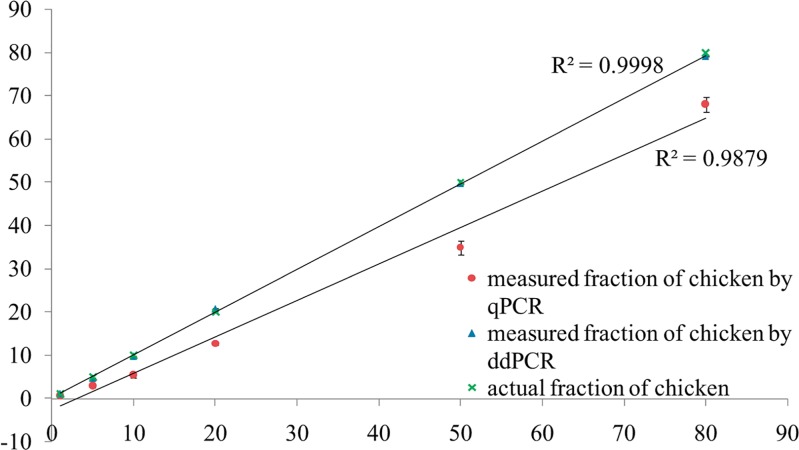
According to the results obtained from the same mixtures analyzed with the two methods shown in Table 3, both the ddPCR and qPCR methods had a relative standard diversion (RSD) ≤ 25% for quantification and showed good repeatability. However, the bias of qPCR was greater than 25%, while that of ddPCR was less than 5% for chicken mixtures ranging from 5% to 50%. The deviation of 80% chicken was -14.94% with real-time PCR, whereas the bias was -1.15% with ddPCR. The results showed that the difference reached 44.80% for the 10% chicken fraction. According to Fig 2, the correlation coefficient (R2) for the actual chicken fraction in a mixture of chicken and sheep was 0.9879 based on real-time PCR. The accuracy of qPCR quantification was lower than that of ddPCR.
Fig 2. Dynamic range of the ddPCR assay for quantification of chicken and sheep fractions.
The vertical axis represents the measured fraction of chicken in sheep (w/w) by ddPCR and qPCR. The horizontal axis shows the actual fraction of chicken in sheep (w/w). Three replicates for each data point were analyzed. The points indicate the concentrations of 80%, 50%, 20%, 10%, 5% and 1% (w/w) of chicken in sheep. Linearity between the actual chicken fraction (w/w) and the measured chicken fraction (w/w) by ddPCR. The correlation coefficient (R2) for the weight of chicken was 0.9998 for ddPCR and 0.9878 for real-time PCR.
Digital PCR is an absolute quantification technique used to calculate the copy number without a standard curve and Ct, thereby avoiding the discrepancy produced by differing amplification efficiencies of different samples. The bias of 1% chicken samples measured by real-time PCR was -35.8%, whereas it was 24.8% when measured using ddPCR. Compared to a previous study, the bias of real-time PCR for 1% sample is 65%. Overall, the ddPCR method is more accurate than real-time PCR in meat adulteration quantification [32].
Real-time PCR has been widely used in the quantification of nucleic acids in various species, as demonstrated with a large range of adulteration concentrations (experimentally from 1% to 80%). Compared to real-time PCR, ddPCR is much more accurate and easier to perform; accordingly, it requires much more costly equipment and consumables [33].
Dynamic range, limit of quantification (LOQ) and limit of detection (LOD)
The measurement of the proportion of chicken in sheep, the slope of the standard curve, and the correlation coefficient (R2) of the respective species in meat mixtures are summarized in Fig 2. The lower amounts that decreased to 1% w/w still showed a good recovery. The results showed that the deviation was less than 2% when the chicken concentration ranged from 5% to 80%, and the quantification method presented in our study could provide an accurate analysis with low RSD (Table 3). The chicken proportion of the dynamic range of sheep was between 1% and 80%.
The LOQ was defined as 1% chicken in sheep, which could be precisely quantified with an RSD less than 25% following the FAO Guidelines on the performance criteria and validation of methods for the detection, identification and quantification of specific DNA sequences and specific proteins in food [34] (Table 4). In contrast, in another quantification study based on the ddPCR technique, which used a two-step conversion that increased the bias of quantification, the LOQ was 10 mg (10%) for pork, and the bias was approximately 17% [29]. In our study, the deviation of 10% (w/w) was approximately less than ±8.17%, and the LOQ was defined as 1%. Because digital PCR is an extremely precise quantification technique [31,35,36], it can be used to help law enforcement agencies and testing organizations to ascertain whether or not adulteration is intentional. The 1% lowest level of quantification of adulterant is achieved with routine testing and food quality control.
Table 4. LOQ, repeatability and accuracy test.
| Meat mixtures2 | A1 | B | The RSD of different operators | ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Six parallels of quantification | Mean Value (%) | RSD (%) | Bias (%) | Six parallels of quantification | Mean Value (%) | RSD (%) | Bias (%) | ||||||||||||
| 10% | 9.22 | 9.26 | 9.03 | 9.19 | 9.39 | 9.01 | 9.18±0.14 | 1.56 | -8.17 | 9.24 | 9.49 | 9.01 | 9.14 | 9.25 | 9.84 | 9.33±0.30 | 3.18 | -6.72 | 2.54% |
| 5% | 5.08 | 4.75 | 4.74 | 5.03 | 4.87 | 5.31 | 4.97±0.22 | 4.45 | -0.70 | 5.19 | 5.20 | 4.89 | 4.80 | 4.77 | 5.04 | 4.98±0.19 | 3.85 | -0.35 | 3.97% |
| 4% | 4.24 | 3.72 | 3.85 | 4.1 | 3.80 | 4.23 | 3.99±0.23 | 5.75 | -0.24 | 4.23 | 4.32 | 3.87 | 3.75 | 4.10 | 4.11 | 4.06±0.21 | 5.34 | 1.55 | 5.37% |
| 3% | 2.72 | 3.02 | 2.89 | 2.70 | 2.82 | 3.12 | 2.88±0.17 | 5.74 | -4.08 | 2.79 | 2.91 | 3.07 | 2.72 | 2.86 | 2.80 | 2.86±0.12 | 4.28 | -4.71 | 4.85% |
| 2% | 2.14 | 2.22 | 2.10 | 2.13 | 2.33 | 2.11 | 2.17±0.09 | 4.08 | 8.60 | 2.34 | 2.11 | 2.28 | 2.12 | 2.29 | 2.27 | 2.24±0.10 | 4.26 | 11.81 | 4.26% |
| 1% | 1.30 | 1.32 | 1.14 | 1.13 | 1.08 | 1.36 | 1.22±0.11 | 9.71 | 22.10 | 1.15 | 1.34 | 1.28 | 1.12 | 1.23 | 1.20 | 1.21±0.08 | 7.23 | 21.04 | 8.19% |
1A and B represents the different operators in our lab. 2Meat mixtures indicate the chicken weight percentage of the sheep content.
The limit of detection (LOD) was defined as the lowest percentage of chicken content that could be reliably detected. To determine the LOD of our method, mixtures containing various percentages, ranging from 0.1% to 1% were prepared. The sensitivity levels observed are shown in S2 Table. The results showed that the positive wells could be stably seen even in the 0.1% group. Thus the LOD of the system reached 0.1% chicken content in sheep.
Specificity, repeatability and accuracy
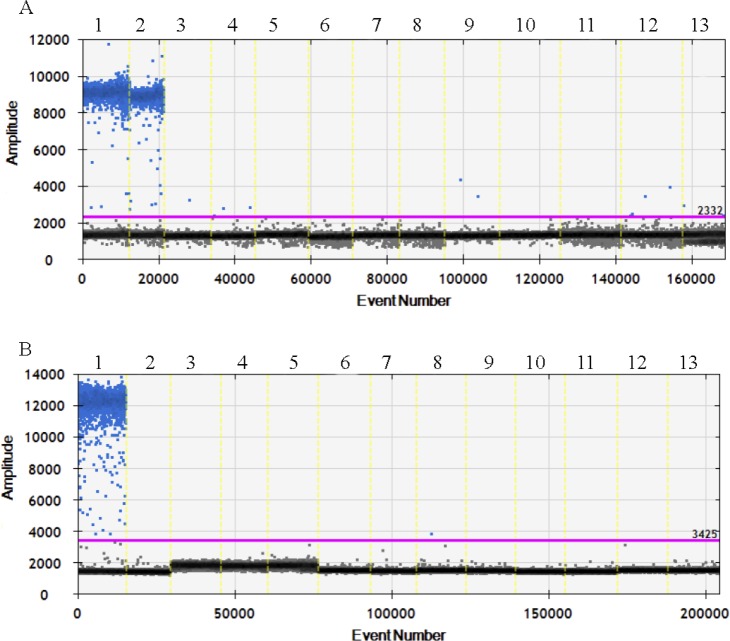
Specificity is the crucial precondition for a successful qualitative and quantitative method in food authentication. To determine the specificity of the sheep and chicken ddPCR reaction, meats from 13 animal species were tested: sheep, goat, chicken, pork, duck, goose, pigeon, beef, horse, dog, rabbit, turkey and donkey. Six varieties of chicken, five varieties of sheep and three varieties of goat were amplified to sequencing, the results showed that the amplified region is conservation for our research (S1 Fig). The two pairs of species-specific primers and probes only amplified target sequences from sheep, goat and chicken, respectively, and they showed no cross-reaction with any non-target species. These results confirmed that the method was capable of identifying chicken mixed with or substituted for sheep and goat specifically (Fig 3).
Fig 3. The specificity results of primers and probes of chicken, sheep and goat.
The horizontal axis indicates the event number of 13 kinds of meats. The vertical axis indicates the amplitude of samples. (A) Upper frame: droplet cluster positive for FAM (sheep, goat and other meats). Lower frame: droplet cluster negative for FAM (sheep, goat and any other meats) negative droplet cluster. Lanes: 1, sheep; 2, goat; 3, pork; 4, beef; 5, horse; 6, rabbit; 7, donkey; 8, dog; 9, chicken; 10, duck; 11, pigeon; 12, goose; 13, turkey. (B) Upper region: droplet cluster positive for FAM (chicken and any other meats). Lower region: droplet cluster negative for FAM (chicken and any other meats) negative droplet cluster. Lanes: 1, chicken; 2, sheep; 3, goat; 4, beef; 5, horse; 6, rabbit; 7, donkey; 8, dog; 9, chicken; 10, duck; 11, pigeon; 12, goose; 13, turkey.
Intra-laboratory repeatability validation was performed by two different experienced operators to determine the chicken content of the same sample on two different days. Variability of≤10% was observed in each of the six cartridges for the determination of the chicken and sheep fractions by two operators (Table 4). The RSD values in the 1% to 10% chicken mixtures were less than 9.71%. The deviations in 2% and 10% chicken mixtures were less than 12%, and that from the 1% chicken mixtures was less than 22.10%. These results demonstrate that meat fractions can be quantitated repeatedly and accurately with this assay.
Robustness of the method
Effects of different parts of the animal species
To verify whether the repeatability and accuracy of quantification in the adulteration assay change with the body location, we selected three different parts of chicken (legs, breasts and wings) to make a meat mixture for testing. The results showed that the bias of the measured value in three locations of chicken mixed in sheep is less than ±9%, and the RSD values were all under 6% (S3 Table). We concluded that meats located in different parts of the body did not significantly affect the quantitative results in this method.
Mitochondrial DNA has been extensively used for PCR templates in species determination [37–40] due to its high copy numbers. However, nuclear genomic DNA is better for meat quantification because its constant haploid copy number makes the concentration of genes per gram of meat relevant to the quantity of meat tested. Thus, it is more predictive than mitochondrial markers, which vary sharply in the copy numbers in different organs and individuals [41,42]. According to a previous report, there is a five-fold inter-tissue variation in mtDNA content per cell that results in either an underestimation (-70%) or overestimation (+160%) of DNA content [26]. In the present study, the nuclear genomic gene RPA1 was used for meat quantification, it is not necessary to determine which parts of meat came from.
Effect of the treatment temperature
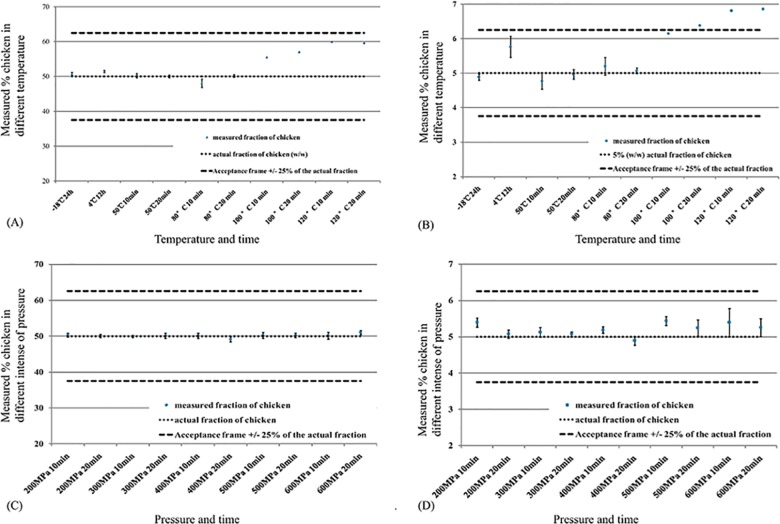
To demonstrate the robustness of the ddPCR assay, different temperature treatments were implemented, including -18°C for 24 h; 4°C for 12 h; 50°C, 80°C, 100°C and 120°C for 10 min and 20 min (S4 Table). Using the precision analysis, the results showed that the RSD values of all samples, with the temperature ranging from -18°C to 120°C, were less than 10%. This illustrated that the method in the present study maintained a very high precision and stability. Bias in the measurement of 50% and 5% chicken fraction samples processed under 100°C varied in the ranges of ±5% and ±15%, respectively. The method maintained a high level of accuracy for samples treated at -18°C to 80°C. Bias in the measurement of mixtures containing 50% and 5% chicken became larger with higher temperatures and longer treatments. When the temperature increased from 100°C to 120°C, for 50% mixtures, the errors increased from +10% to +20% (Fig 4A), and for 5% chicken, the errors increased from +22% to +37% (Fig 4B), respectively. This suggested that 100°C is the turning point for accuracy in the measurement of heat-treated samples. The ddPCR quantification method established here is more accurate when mixtures were treated at temperatures less than 100°C, and the accuracy decreases significantly with the length of processing. In addition, we found that the deviation had a positive relationship with the extension of time and the increase of temperature when treated above 100°C.
Fig 4. The repeatability results of 50% and 5% chicken in sheep processed under different temperature and pressure conditions for different durations.
The target chicken fraction is indicated by a dotted line. The acceptance criterion for repeatability is ±25% of the target proportion, represented by the dashed lines. Error bars represent the standard deviation between the replicates for each treated condition. (A) The 50% chicken fraction processed at different temperatures for different durations and measured by ddPCR (three replicates in ten conditions). (B) The 5% chicken fraction processed at different temperatures for different durations and measured by ddPCR (three replicates in ten conditions). (C) The 50% chicken fraction processed at different pressure intensities and for different durations and measured by ddPCR (three replicates in ten conditions). (D) The 5% chicken fraction processed at different pressure intensities and for different durations and measured by ddPCR (three replicates in ten conditions).
It has been suggested that the loss of accuracy is probably due to the different degradation rates of DNA from different species, and it will increase with the temperature of heat processing [43,44]. The loss of accuracy may also be caused by the recovery rate of genomic DNA in extractions, since protein-protein and protein-DNA (polysaccharides) interactions that occur during heat treatment lead to discrepancies in DNA release in extraction [45]. Both events induced variations of the k value, which may explain the positive increase in deviation in synchronization with the increase of temperature from 100°C to 120°C. Therefore, it is recommended that the reference meat mixture be treated at the same treatment for calculating the multiplication factor.
Effect of ultra-high pressure (UHP) processing
In the meat industry, ultra-high pressure is mainly used to increase shelf life and improve the food safety of ready-to-eat meat products as a novel post-packaging non-thermal decontamination technology. Therefore, UHP has been employed in the meat industry for the past two decades [46]. The effect of ultra-high pressure on the quantification of meat mixtures is summarized in S5 Table. The RSD values and bias were less than 25% in 50% and 5% chicken fraction mixtures subjected to treatment with pressure from 200 MPa to 600 MPa (Fig 4C and 4D). High pressure can destroy the tertiary structure and quaternary structure of proteins but has little effect on covalent bonds. Heating causes DNA degradation and may influence the efficiency of PCR; in contrast, treatment with high pressure at room temperature does not damage the primary structure of DNA. The precision and accuracy of quantitative determination are not correlated with treatment with high hydrostatic pressure in the range of 200–600 MPa. In general, high pressure has no significant effect on the quantification accuracy of meat mixtures.
Testing of commercial samples
To demonstrate the applicability of the ddPCR assay, 14 commercial sheep and goat samples from local supermarkets were analyzed by ddPCR and real-time PCR. Table 5 indicated that 5 samples were positive for chicken DNA by ddPCR, and one brand of lamb sausage was positive for ddPCR, but negative for real-time PCR. These results demonstrated that the sensitivity of the developed ddPCR method was better than that of real-time PCR. Dumplings, the other lamb sausage and a brand of kebab were positive for chicken according to both ddPCR and real-time PCR, showing adulteration with chicken meat at concentrations from 30% to 75%. However, the RSD values of ddPCR method were lower than those of real-time PCR, indicating that ddPCR showed higher precision and accuracy. The percentage of chicken content in cumin lamb was lower than the LOQ (1%); thus, we presumed that the sample might have been contaminated unintentionally. These results showed that this developed technology can be utilized for commercially available meat products and that it shows better sensitivity, precision and accuracy than real-time PCR.
Table 5. Identification and quantification of chicken in commercial goat/sheep products.
| Sample | Identification | Quantification | |||
|---|---|---|---|---|---|
| ddPCR | Real-time PCR | ddPCR | Real-time PCR | ||
| Fresh meat | Minced mutton | Negative | Negative | 0.0 | 0.0 |
| Meat rolls 1 | Negative | Negative | 0.0 | 0.0 | |
| Meat rolls 2 | Negative | Negative | 0.0 | 0.0 | |
| Meat rolls 3 | Negative | Negative | 0.0 | 0.0 | |
| Meat rolls 4 | Negative | Negative | 0.0 | 0.0 | |
| Kebab 1 | Positive | Positive | 39.04±0.59 | 40±5.38 | |
| Kebab 2 | Negative | Negative | 0.0 | 0.0 | |
| Kebab 3 | Negative | Negative | 0.0 | 0.0 | |
| Kebab 4 | Negative | Negative | 0.0 | 0.0 | |
| Kebab 5 | Negative | Negative | 0.0 | 0.0 | |
| Dumplings | Positive | Positive | 58.57±1.20 | 55.47±5.56 | |
| Meat products | Lamb sausage 1 | Positive | Positive | 73.92±1.86 | 76.64±7.29 |
| Lamb sausage 2 | Positive | Negative | 0.12±5.44 | — | |
| Cumin lamb 1 | Positive | Positive | 0.65±2.98 | 1.23±8.98 | |
Meat rolls 1–4 represent four brands of sheep meat rolls, respectively.
Kebabs 1–5 represent five different brands of kebab, respectively.
Lamb sausages 1, 2 represent two different brands of lamb sausage, respectively.
Conclusion
In this work, we showed that the ddPCR method is a novel, accurate, reliable and easy quantification strategy for the determination of animal species in fresh and processed meat products. Normally, the copy numbers cannot be converted to proportions directly because the quantities of the haploid genome per gram of meat and the densities of cells are different in meats from different species. The strategy that we tested was to introduce a constant (multiplication factor) deduced theoretically to transform the ratio of copy numbers to the proportion of meats. This quantification method can be reliably used down to the 1% (w/w) level for chicken and sheep mixtures, and the LOD is 0.1% (w/w). Compared to real-time PCR, the bias of the chicken proportion in the range from 5% to 80% was less than 9% according to this assay. Moreover, it was concluded that the part of chicken meat had no effect on the accuracy of quantification. Further, this research also showed that a high level of accuracy was maintained under different pressure and temperature conditions. This method, which introduces a constant to transform the ratio of copy numbers to the proportion of meats, has the potential to be applied in routine assays for quantification of food adulteration, not only in raw or processed meat products, but also in many other applied fields in which this information is needed.
Supporting information
The multi-alignments of (A) RPA1 gene target sequences from Beijing fatty chicken and other varieties, (B) RPA1 gene target sequences from sheep, goat and other varieties.
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
Acknowledgments
We would like to express our deepest gratitude to all the members of the China Agricultural University and Chinese Academy of Inspection and Quarantine who contributed to our study. We would also like to thank the company BIO-RAD for offering their guidance of operation.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
The authors received no specific funding for this work.
References
- 1.O’MAHONY P J. Finding horse meat in beef products: a global problem. QJM: An International Journal of Medicine. 2013;106(6):595–597. [DOI] [PubMed] [Google Scholar]
- 2.Cawthorn D M, Steinman H A, Hoffman L C. A high incidence of species substitution and mislabelling detected in meat products sold in South Africa. Food Control. 2013;32(2),440–449. [Google Scholar]
- 3.Suiping Tang, Yan Zhang, Jinghui Huang. Analysis of beef and lamp products adulteration in Guangdong province. Journal of food safety and quality. 2016;7:1882–1886. [Google Scholar]
- 4.Nan Li, Jiahui Wang, Qing Shen, Fengqin Li, Jin Xu, Tao Jiang. Survey of duck, chicken and pork component in lamb or beef slices sold in Beijing. Chineses journal of food hygiene. 2016;26: 227–232. [Google Scholar]
- 5.Ping Jin, Hongliu Ding, Pei Li, Lili Fan, Chunling Fu. Analysis of meat products adulterated in Suzhou area in 2013. Chineses journal of food hygiene. 2014;26: 168–172. [Google Scholar]
- 6.European Commission. Commission Directive 2002/86/EC. Official Journal ofthe European Communities, L 305/19, Brussels.
- 7.Food and Drug Administration. (2011). FDA Food Safety Modernization Act (FSMA).
- 8.Zukál E, Körmendy L. On calculation of ‘meat content’according to the quantitative ingredient declarations (QUID). Journal of Food Engineering.2007;78: 614–621. [Google Scholar]
- 9.Amaral JS, Santos CG, Melo VS, Oliveira MBP, Mafra I. Authentication of a traditional game meat sausage (Alheira) by species-specific PCR assays to detect hare, rabbit, red deer, pork and cow meats. Food Research International.2014;60:140–145. [Google Scholar]
- 10.Karabasanavar NS, Singh SP, Kumar D, Shebannavar SN. Detection of pork adulteration by highly-specific PCR assay of mitochondrial D-loop. Food Chemistry.2014;145: 530–534. 10.1016/j.foodchem.2013.08.084 [DOI] [PubMed] [Google Scholar]
- 11.Druml B, Hochegger R, Cichna-Markl M. Duplex real-time PCR assay for the simultaneous determination of the roe deer (Capreolus capreolus) and deer (sum of fallow deer, red deer and sika deer) content in game meat products. Food Control.2015;57: 370–376. [Google Scholar]
- 12.Dooley JJ, Paine KE, Garrett SD, Brown HM.Detection of meat species using TaqMan real-time PCR assays. Meat Science. 2004;68: 431–438. 10.1016/j.meatsci.2004.04.010 [DOI] [PubMed] [Google Scholar]
- 13.Montowska M, Pospiech E. Species-specific expression of various proteins in meat tissue: Proteomic analysis of raw and cooked meat and meat products made from beef, pork and selected poultry species. Food chemistry. 2013;136:1461–1469. 10.1016/j.foodchem.2012.09.072 [DOI] [PubMed] [Google Scholar]
- 14.Cai Q F, Wang XC, Liu GM, Zhang L, Ruan M M, Liu Y, et al. Development of a monoclonal antibody-based competitive enzyme linked-immunosorbent assay (c-ELISA) for quantification of silver carp parvalbumin. Food Control.2013;29:241–247. [Google Scholar]
- 15.Liu L, Chen FC, Dorsey JL, Hsieh Y HP.Sensitive Monoclonal Antibody-based Sandwich ELISA for the Detection of Porcine Skeletal Muscle in Meat and Feed Products. Journal of food science.2006;71:M1–M6. [Google Scholar]
- 16.Leitner A, Castro-Rubio F, Marina M L, Lindner W.Identification ofmarker proteins for the adulteration of meat products with soybean proteins bymultidimensional liquid chromatography–tandem mass spectrometry. Journal of Proteome Research.2006;5: 2424–2430. 10.1021/pr060145q [DOI] [PubMed] [Google Scholar]
- 17.López-Andreo M, Lugo L, Garrido-Pertierra A, Prieto M I, Puyet A.Identification and quantitation of species in complex DNA mixtures by real-time polymerase chain reaction. Analytical Biochemistry.2005;339: 73–82. 10.1016/j.ab.2004.11.045 [DOI] [PubMed] [Google Scholar]
- 18.Cheng X, He W, Huang F, Huang M, Zhou G.Multiplex real-time PCR for the identification and quantification of DNA from duck, pig and chicken in Chinese blood curds. Food Research International.2014;60: 30–37. [Google Scholar]
- 19.Köppel R, Daniels M, Felderer N, Brünen-Nieweler C.Multiplex real-time PCR for the detection and quantification of DNA from duck, goose, chicken, turkey and pork. European Food Research and Technology.2013;236: 1093–1098. [Google Scholar]
- 20.Iwobi A, Sebah D, Kraemer I, Losher C, Fischer G, Busch U, et al. A multiplex real-time PCR method for the quantification of beef and pork fractions in minced meat. Food chemistry. 2015;169: 305–313. 10.1016/j.foodchem.2014.07.139 [DOI] [PubMed] [Google Scholar]
- 21.López-Andreo M, Aldeguer M, Guillén I, Gabaldón J A, Puyet A. Detection and quantification of meat species by qPCR in heat-processed food containing highly fragmented DNA. Food Chemistry. 2012;134(1): 518–523. [Google Scholar]
- 22.Pinheiro LB, Coleman V A, Hindson C M, Herrmann J, Hindson B J, Bhat S, et al. Evaluation of a droplet digital polymerase chain reaction format for DNA copy number quantification. Analytical Chemistry.2011;84: 1003–1011. 10.1021/ac202578x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guo G, Huss M, Tong G Q, Wang C, Sun L L, Clarke N D, et al. Resolution of cell fate decisions revealed by single-cell gene expression analysis from zygote to blastocyst. Development cell. 2010;18: 675–685. [DOI] [PubMed] [Google Scholar]
- 24.Dreo T, Pirc M, Ramšak Ž, Pavšič J, Milavec M, Žel J, et al. Optimising droplet digital PCR analysis approaches for detection and quantification of bacteria: a case study of fire blight and potato brown rot. Analytical and bioanalytical chemistry.2014;406:6513–6528. 10.1007/s00216-014-8084-1 [DOI] [PubMed] [Google Scholar]
- 25.Morisset D, Štebih D, Milavec M, Gruden K, Žel J.Quantitative analysis of food and feed samples with droplet digital PCR. Plos One.2013;8: e62583 10.1371/journal.pone.0062583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Floren C, Wiedemann I, Brenig B, Schütz E, Beck J.Species identification and quantification in meat and meat products using droplet digital PCR (ddPCR). Food chemistry. 2015;173: 1054–1058. 10.1016/j.foodchem.2014.10.138 [DOI] [PubMed] [Google Scholar]
- 27.Köppel R, Bucher T, Frei A, Waiblinger HU. Droplet digital PCR versus multiplex real-time PCR method for the detection and quantification of DNA from the four transgenic soy traits MON87769, MON87708, MON87705 and FG72, and lectin. European Food Research Technology.2015;241: 521–527. [Google Scholar]
- 28.Ballin NZ, Vogensen F K, Karlsson A H.Species determination–Can we detect and quantify meat adulteration? Meat Science.2009;83: 165–174. 10.1016/j.meatsci.2009.06.003 [DOI] [PubMed] [Google Scholar]
- 29.Cai Y, Li X, Lv R, Yang J, Li J, He Y, et al. Quantitative analysis of pork and chicken products by droplet digital PCR. Biomed Research International.2014;810209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Scaravelli E, Brohée M, Marchelli R, van Hengel AJ.Development of three real-time PCR assays to detect peanut allergen residue in processed food products. European Food Research Technology.2008;227: 857–869. [Google Scholar]
- 31.Hindson BJ, Ness KD, Masquelier DA, Belgrader P, Heredia N J, Makarewicz AJ, et al. High-throughput droplet digital PCR system for absolute quantitation of DNA copy number. Analytical Chemistry.2011;83: 8604–8610. 10.1021/ac202028g [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Köppel R, Ruf J,Rentsch J. Multiplex real-time PCR for the detection and quantification of DNA from beef, pork, horse and sheep. European Food Research Technology.2011;232: 151–155. [Google Scholar]
- 33.Wee E J H, Wang Y, Chang-Hao T S, et al. Simple, Sensitive and Accurate Multiplex Detection of Clinically Important Melanoma DNA Mutations in Circulating Tumour DNA with SERS Nanotags. Theranostics. 2016,6(10):1506–1513. 10.7150/thno.15871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Codex Committee on Methods of Analysis and Sampling. Guidelines on performance criteria and validation of methods for detection, identification and quantification of specific DNA sequences and specific proteins in foods. Rome: Codex Alimentarius-FAO.2010:CAC/GL 7402010.
- 35.Baker M. Digital PCR hits its stride.NatureMethods.2012;9: 541–544. [Google Scholar]
- 36.Day E, Dear P H, McCaughan F. Digital PCR strategies in the development and analysis of molecular biomarkers for personalized medicine. Methods.2013;59: 101–107. 10.1016/j.ymeth.2012.08.001 [DOI] [PubMed] [Google Scholar]
- 37.Cammà C, Di Domenico M, Monaco F.Development and validation of fast Real-Time PCR assays for species identification in raw and cooked meat mixtures. Food Control.2012;23: 400–404. [Google Scholar]
- 38.Wang P, Hu Y, Yang H, Han J, Zhao Y, Chen Y. DNA-based Authentication Method for Detection of Yak (Bos grunniens) in Meat Products. Journal of AOAC International.2013;96: 142–146. [DOI] [PubMed] [Google Scholar]
- 39.Hou B, Meng X, Zhang L, Guo J, Li S, Jin H. Development of a sensitive and specific multiplex PCR method for the simultaneous detection of chicken, duck and goose DNA in meat products. Meat Science.2015;101: 90–94. 10.1016/j.meatsci.2014.11.007 [DOI] [PubMed] [Google Scholar]
- 40.Kesmen Z, Gulluce A, Sahin F, Yetim H.Identification of meat species by TaqMan-based real-time PCR assay. Meat Science.2009;82: 444–449. 10.1016/j.meatsci.2009.02.019 [DOI] [PubMed] [Google Scholar]
- 41.Alonso A, Martí P, Albarrán C, Garcí P, Garcí O, de Simón L F, et al. Real-time PCR designs to estimate nuclear and mitochondrial DNA copy number in forensic and ancient DNA studies. Forensic Science International.2004;139: 141–149. 10.1016/j.forsciint.2003.10.008 [DOI] [PubMed] [Google Scholar]
- 42.Ng J, Satkoski J, Premasuthan A, Kanthaswamy S.A nuclear DNA-based species determination and DNA quantification assay for common poultry species. Journal of Food Science andTechnology.2014;51: 4060–4065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hird H, Chisholm J, Sánchez A, Hernandez M, Goodier R, Schneede K, et al. Effect of heat and pressure processing on DNA fragmentation and implications for the detection of meat using a real-time polymerase chain reaction. Food additives and contaminants. 2006;23: 645–650. 10.1080/02652030600603041 [DOI] [PubMed] [Google Scholar]
- 44.Kingombe C I B, Lüthi E, Schlosser H, Howald D, Kuhn M, Jemmi T. A PCR-based test for species-specific determination of heat treatment conditions of animal meals as an effective prophylactic method for bovine spongiform encephalopathy. Meat Science.2001;57: 35–41. [DOI] [PubMed] [Google Scholar]
- 45.Tornberg E. Effects of heat on meat proteins–Implications on structure and quality of meat products. Meat Science.2005;70: 493–508. 10.1016/j.meatsci.2004.11.021 [DOI] [PubMed] [Google Scholar]
- 46.Bajovic B, Bolumar T, Heinz V.Quality considerations with high pressure processing of fresh and value added meat products. Meat science.2012;92: 280–289. 10.1016/j.meatsci.2012.04.024 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The multi-alignments of (A) RPA1 gene target sequences from Beijing fatty chicken and other varieties, (B) RPA1 gene target sequences from sheep, goat and other varieties.
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.