ABSTRACT
Enterococci are ancient commensal bacteria that recently emerged as leading causes of antibiotic-resistant, hospital-acquired infection. Vancomycin-resistant enterococci (VRE) epitomize why drug-resistant enterococcal infections are a problem: VRE readily colonize the antibiotic-perturbed gastrointestinal (GI) tract where they amplify to large numbers, and from there, they infect other body sites, including the bloodstream, urinary tract, and surgical wounds. VRE are resistant to many antimicrobials and host defenses, which facilitates establishment at the site of infection and confounds therapeutic clearance. Having evolved to colonize the GI tract, VRE are comparatively ill adapted to the human bloodstream. A recent study by Honsa and colleagues (E. S. Honsa et al., mBio 8:e02124-16, 2017, https://doi.org/10.1128/mBio.02124-16) found that a strain of vancomycin-resistant Enterococcus faecium evolved antibiotic tolerance within the bloodstream of an immunocompromised host by activating the stringent response through mutation of relA. Precisely how VRE colonize and infect and the selective pressures that led to the outgrowth of relA mutants are the subjects of ongoing research.
INTRODUCTION
Bacteria in the genus Enterococcus occur among the intestinal consortia of hosts that span the animal kingdom (1). These intrinsically rugged bacteria have become leading multidrug-resistant hospital pathogens. The enterococci are among the vanguard of antibiotic-resistant bacteria for at least two reasons: they evolved in the gastrointestinal (GI) tracts of insects and other invertebrates that naturally live and feed on organic matter that contains antibiotics and antibiotic-producing microorganisms (2), and they are naturally hardy and are able to withstand starvation, desiccation, and other stresses better than most other microbes. Enterococci are intrinsically resistant to many antibiotics, and they readily mutate or acquire resistance genes to others as needed. In addition to being leading multidrug-resistant hospital pathogens, they also serve as reservoirs and transmit resistance genes to other bacteria. Enterococcus faecium and Enterococcus faecalis are the enterococcal species most often associated with multidrug-resistant nosocomial infection, and about 30 years ago, both species acquired resistance to the important last-line bactericidal drug, vancomycin.
Vancomycin-resistant enterococci (VRE) pose a special threat to immunocompromised patients, who often undergo antibiotic treatment or prophylaxis in hospitals. The study by Honsa and colleagues (3) describes the evolution of a vancomycin-resistant E. faecium strain within such a patient. How did this pediatric patient become infected with VRE? Nearly all VRE infections begin with colonization of the GI tract by bacteria that are ingested from the hospital environment (Fig. 1). Enterococci are resistant to desiccation and starvation, and they are difficult to eradicate from the hospital with disinfectants (1). From contaminated patient-proximal surfaces, including bed rails and thermometer handles, enterococci survive transit through the stomach and small intestine, and then they replicate in the large intestine where they may reach very high densities due to limited competition because of prior antibiotic treatment. Dense intestinal colonization with VRE recontaminates the patient-proximal environment, perpetuating its presence within modern hospitals (4). The risk of developing a VRE infection is very likely directly proportional to the number of drug-resistant enterococci in the GI tract; more colonization equals greater risk of infection.
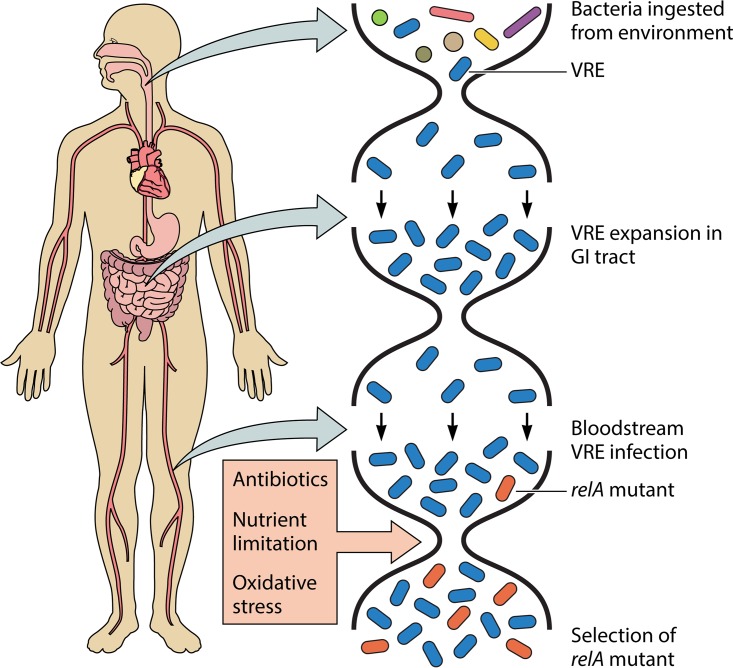
FIG 1 .
Simplified model of bacterial population dynamics during VRE colonization, infection, and selection of relA mutants in an immunocompromised patient. Bacteria, including VRE, are first ingested from the patient-proximal hospital environment. Transit through the upper GI tract coupled with broad-spectrum antibiotic treatment kill off nearly all other microbes, allowing VRE to grow to very high densities in the lower GI tract. A small number of bacteria from the GI tract population seed the bloodstream, where the population expands again. Once a relA mutant bacterium occurs in the bloodstream VRE population, it is able to grow to a high enough density to be detected, because it provides a survival advantage against antibiotic treatment, nutrient limitation, and/or oxidative stress.
The patient described in this study (3) was uniquely susceptible to vancomycin-resistant E. faecium colonization and infection. Only 6 weeks old, she had an immature GI tract microbiome, which was further confounded by chemotherapy to treat acute myeloid leukemia, causing impaired host defenses, accompanied by antibiotic treatment. Among the agents used was a broad-spectrum cephalosporin, to which enterococci are intrinsically resistant, and which prior work has shown limits the occurrence of competing commensal anaerobes (5–7). The VRE that colonized the patient’s GI tract most likely seeded her bloodstream, but whether that occurred through translocation through the gut wall, which often becomes inflamed during chemotherapy (8) or from contaminants that made their way to the skin and were introduced into the bloodstream through the central venous catheter is unclear.
It was recently shown that hospital-adapted, multidrug-resistant strains of E. faecium belong to a distinct lineage (clade A) that diverged from human commensal strains (clade B) of the species approximately 3,000 years ago (9). The lineage of hospital-adapted E. faecium, known as clade A1 or clonal cluster 17 (CC17), is so different from commensal strains that an argument can be made that they should be considered a different species entirely (9). The VRE strain studied by Honsa et al. (3), in fact belongs to clade A1/CC17, which is globally disseminated. Genetic factors enriched in this hospital-adapted E. faecium clade include the following: (i) altered cell wall polysaccharide and capsule biosynthesis genes that appear to enhance its fitness for the hospital environment (10); (ii) an accumulation of mobile elements likely due to a missing clustered regularly interspaced short palindromic repeat (CRISPR)-Cas system (9, 11); and (iii) a unique phosphotransferase system (PTS) that allows for the utilization of amino sugars, which occur on epithelial cell surfaces and mucin. This PTS has been shown to enhance the ability of E. faecium to colonize the antibiotic-perturbed GI tract (12), and suggests that hospital-adapted VRE strains adopt a parasitic lifestyle, feeding on host-derived secretions, such as mucin, when other dietary carbohydrates are not available.
By sequencing the genomes of 22 consecutive bloodstream VRE isolates from the same patient, Honsa and colleagues (3) were able to identify mutations that occurred during infection and growth in blood, an environment to which enterococci are not naturally adapted. A mutation in relA was repeatedly seen in 8 of the 22 isolates, so the authors investigated possible biological consequences of this mutation. They found that relA mutant strains constitutively activated the stringent response through elevated baseline levels of the alarmone (p)ppGpp. The relA mutant strains were not more drug resistant as defined by the ability to grow planktonically in higher levels of drug, but in biofilms, the mutant strains showed greater tolerance to lethal doses of both linezolid and daptomycin. Enigmatically, biofilms formed by the relA mutants were less adherent and more sparsely populated under the conditions studied.
The authors suggest that antibiotic use may have provided the selective pressure that led to the outgrowth of relA mutant strains. The adaptation of enterococci to growth in the bloodstream is complex, and the selective pressure or pressures may be more complex than initially thought. Daptomycin has been shown to select for relA mutations in vitro (13), and daptomycin treatment in this patient may have helped sustain the relA mutants at a high frequency in the population. Nevertheless, daptomycin could not have been the original selective pressure that drove the mutation to a high enough level to be detected in the first place, because the initial relA mutant was isolated a week prior to daptomycin therapy. Other antibiotics given to the patient preceding the appearance of the relA mutation were cefepime, vancomycin, meropenem, and linezolid. Of these antibiotics, vancomycin is known to have the ability to induce the stringent response (14); however, this strain was already highly vancomycin resistant and was likely inert to the effects of the drug. Use of beta-lactams and linezolid have been shown to be associated with relA mutations in Staphylococcus aureus (15, 16), but these drugs do not appear to directly induce the stringent response. Indeed, Honsa and colleagues found that linezolid exposure did not increase (p)ppGpp levels in the wild-type strain (3).
What besides antibiotic pressure may have selected for activation of the stringent response in this patient? First, nutrient limitation is well-known to activate the stringent response (17). While blood may not seem like a nutrient-limited environment, nutrient withholding is a key component of innate immunity (18), and the nutrient profile of blood differs substantially from that of the GI tract. Enterococci therefore need to alter their metabolism and physiology considerably to grow efficiently in the bloodstream. Second, exposure to reactive oxygen species during bacterial engulfment by immune cells of an immunologically normal host, and possibly with residual function in this infant receiving chemotherapy, is a potent activator of the stringent response (17). Could one or more of these mechanisms have selected for constitutive stringent response expression by the VRE isolated from the pediatric patient studied by Honsa and colleagues (3)? Despite severe neutropenia and bone marrow that was not producing functional immune cells due to chemotherapy, tissue-resident macrophages may still have been present and functioning (19). Overall, our understanding of how VRE adapt to nutrient limitation, antibiotic pressure, and oxidative stress in bloodstream infections is far from complete, and which of these may have selected for the outgrowth of bacteria with relA mutations in this case remains highly speculative.
Regardless of the pressure or pressures that selected for them, the relA mutants were unable to completely displace the wild-type strain within the bloodstream of this patient. Only 8 of the 22 isolates collected possessed the relA mutation, and wild-type strains were isolated throughout the course of the infection. Honsa and colleagues (3) showed that the relA mutants were compromised in their ability to form biofilms; this finding demonstrates how context matters when assessing whether a mutation is beneficial or not. In the face of cellular stresses, such as those described above, mutating relA in order to accumulate (p)ppGpp appears to be beneficial. However, in other situations, such as needing to compete for resources in order to grow and persist in the bloodstream, the same mutation is likely detrimental. For the population as a whole, it seems that retaining a mixture of wild-type and relA mutant bacteria might have offered the best of both worlds.
Ultimately, bloodstream infection with VRE is likely an evolutionary dead end for the bacteria. Because VRE are transmitted between patients by the fecal-oral route, bacteria growing in the bloodstream are highly unlikely to find their way into a subsequent patient. Thus, bacteria must “relearn” how to grow within the bloodstream of each new patient that they infect. Honsa and colleagues (3) show that activation of the stringent response is an advantageous adaptation in the bloodstream of an immunocompromised host, and this is the first study to document an E. faecium strain evolving a relA mutation in vivo. Future surveillance will determine whether this adaptation is generally encountered in enterococcal bacteremia or whether the outgrowth of these mutants was unique to the ecological conditions present in the bloodstream of this infant.
ACKNOWLEDGMENTS
We thank the members of the Gilmore lab, and in particular Anthony O. Gaca, for helpful discussions and feedback during the preparation of this article.
This work is supported by PHS grant AI083214 (Harvard-wide Program on Antibiotic Resistance), as well as grants AI072360 and AI108710 from the National Institutes of Health.
The views expressed in this Commentary do not necessarily reflect the views of this journal or of ASM.
Footnotes
For the article discussed, see https://doi.org/10.1128/mBio.02124-16.
Citation Van Tyne D, Gilmore MS. 2017. Raising the alarmone: within-host evolution of antibiotic-tolerant Enterococcus faecium. mBio 8:e00066-17. https://doi.org/10.1128/mBio.00066-17.
REFERENCES
- 1.Van Tyne D, Gilmore MS. 2014. Friend turned foe: evolution of enterococcal virulence and antibiotic resistance. Annu Rev Microbiol 68:337–356. doi: 10.1146/annurev-micro-091213-113003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gilmore MS, Lebreton F, van Schaik W. 2013. Genomic transition of enterococci from gut commensals to leading causes of multidrug-resistant hospital infection in the antibiotic era. Curr Opin Microbiol 16:10–16. doi: 10.1016/j.mib.2013.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Honsa ES, Cooper VS, Mhaissen MN, Frank M, Shaker J, Iverson A, Rubnitz J, Hayden RT, Lee RE, Rock CO, Tuomanen EI, Wolf J, Rosch JW. 2017. RelA mutant Enterococcus faecium with multiantibiotic tolerance arising in an immunocompromised host. mBio 8:e02124-16. doi: 10.1128/mBio.02124-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Arias CA, Murray BE. 2012. The rise of the Enterococcus: beyond vancomycin resistance. Nat Rev Microbiol 10:266–278. doi: 10.1038/nrmicro2761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brandl K, Plitas G, Mihu CN, Ubeda C, Jia T, Fleisher M, Schnabl B, DeMatteo RP, Pamer EG. 2008. Vancomycin-resistant enterococci exploit antibiotic-induced innate immune deficits. Nature 455:804–807. doi: 10.1038/nature07250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Donskey CJ, Chowdhry TK, Hecker MT, Hoyen CK, Hanrahan JA, Hujer AM, Hutton-Thomas RA, Whalen CC, Bonomo RA, Rice LB. 2000. Effect of antibiotic therapy on the density of vancomycin-resistant enterococci in the stool of colonized patients. N Engl J Med 343:1925–1932. doi: 10.1056/NEJM200012283432604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rice LB. 2001. Emergence of vancomycin-resistant enterococci. Emerg Infect Dis 7:183–187. doi: 10.3201/eid0702.700183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ubeda C, Taur Y, Jenq RR, Equinda MJ, Son T, Samstein M, Viale A, Socci ND, van den Brink MR, Kamboj M, Pamer EG. 2010. Vancomycin-resistant Enterococcus domination of intestinal microbiota is enabled by antibiotic treatment in mice and precedes bloodstream invasion in humans. J Clin Invest 120:4332–4341. doi: 10.1172/JCI43918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lebreton F, van Schaik W, McGuire AM, Godfrey P, Griggs A, Mazumdar V, Corander J, Cheng L, Saif S, Young S, Zeng Q, Wortman J, Birren B, Willems RJ, Earl AM, Gilmore MS. 2013. Emergence of epidemic multidrug-resistant Enterococcus faecium from animal and commensal strains. mBio 4:e00534-13. doi: 10.1128/mBio.00534-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Palmer KL, Godfrey P, Griggs A, Kos VN, Zucker J, Desjardins C, Cerqueira G, Gevers D, Walker S, Wortman J, Feldgarden M, Haas B, Birren B, Gilmore MS. 2012. Comparative genomics of enterococci: variation in Enterococcus faecalis, clade structure in E. faecium, and defining characteristics of E. gallinarum and E. casseliflavus. mBio 3:e00318-11. doi: 10.1128/mBio.00318-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Palmer KL, Gilmore MS. 2010. Multidrug-resistant enterococci lack CRISPR-cas. mBio 1:e00227-10. doi: 10.1128/mBio.00227-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang X, Top J, de Been M, Bierschenk D, Rogers M, Leendertse M, Bonten MJ, van der Poll T, Willems RJ, van Schaik W. 2013. Identification of a genetic determinant in clinical Enterococcus faecium strains that contributes to intestinal colonization during antibiotic treatment. J Infect Dis 207:1780–1786. doi: 10.1093/infdis/jit076. [DOI] [PubMed] [Google Scholar]
- 13.Hachmann AB, Sevim E, Gaballa A, Popham DL, Antelmann H, Helmann JD. 2011. Reduction in membrane phosphatidylglycerol content leads to daptomycin resistance in Bacillus subtilis. Antimicrob Agents Chemother 55:4326–4337. doi: 10.1128/AAC.01819-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Abranches J, Martinez AR, Kajfasz JK, Chávez V, Garsin DA, Lemos JA. 2009. The molecular alarmone (p)ppGpp mediates stress responses, vancomycin tolerance, and virulence in Enterococcus faecalis. J Bacteriol 191:2248–2256. doi: 10.1128/JB.01726-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gao W, Chua K, Davies JK, Newton HJ, Seemann T, Harrison PF, Holmes NE, Rhee HW, Hong JI, Hartland EL, Stinear TP, Howden BP. 2010. Two novel point mutations in clinical Staphylococcus aureus reduce linezolid susceptibility and switch on the stringent response to promote persistent infection. PLoS Pathog 6:e1000944. doi: 10.1371/journal.ppat.1000944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mwangi MM, Kim C, Chung M, Tsai J, Vijayadamodar G, Benitez M, Jarvie TP, Du L, Tomasz A. 2013. Whole-genome sequencing reveals a link between β-lactam resistance and synthetases of the alarmone (p)ppGpp in Staphylococcus aureus. Microb Drug Resist 19:153–159. doi: 10.1089/mdr.2013.0053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Poole K. 2012. Bacterial stress responses as determinants of antimicrobial resistance. J Antimicrob Chemother 67:2069–2089. doi: 10.1093/jac/dks196. [DOI] [PubMed] [Google Scholar]
- 18.Cassat JE, Skaar EP. 2013. Iron in infection and immunity. Cell Host Microbe 13:509–519. doi: 10.1016/j.chom.2013.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Davies LC, Jenkins SJ, Allen JE, Taylor PR. 2013. Tissue-resident macrophages. Nat Immunol 14:986–995. doi: 10.1038/ni.2705. [DOI] [PMC free article] [PubMed] [Google Scholar]