Preface
Most people think of human development only in terms of ‘human’ cells and organs. Here, we discuss another facet involving human-associated microbial communities. A microbial perspective of human development provides opportunities to refine our definitions of healthy pre- and postnatal growth and to develop new strategies for disease prevention and treatment. Considering the dramatic changes in lifestyles and disease patterns that are occurring with globalization, we issue a call for human microbial observatory programs designed to examine microbial community development in birth cohorts representing populations with diverse anthropologic characteristics, including those undergoing rapid change.
Introduction
A consideration of the biological landscape that encompasses human development should consider all facets of what it means to be ‘human’. At least as many of the cells, and the vast majority of unique genes, in the human body are microbial1–3. As such, we can view ourselves as holobionts4. The dynamic microbe-microbe and microbe-host interactions that allow our microbial communities to assemble and endure are as yet largely uncharacterized. Our relationships with microbes begin before birth, represent potentially modifiable features of postnatal development, and likely contribute to intra- and interpersonal variations in many aspects of our normal human physiology, metabolism, immunity, and neurology, as well as to our disease predispositions.
The past ten years have produced a magnificent and still rapidly evolving toolbox of experimental and computational techniques for culture-independent identification of the microorganisms that comprise our body habitat-associated microbial communities (microbiota), as well as their microbial genes (microbiome) and gene products. These tools allow a number of hypotheses about microbial contributions to human development to be tested. One hypothesis is that maternal microbial ecology impacts pregnancy, fetal development, pregnancy outcomes, and the future health of offspring. If true, this hypothesis suggests the possibility of prenatal prognostic and diagnostic measurements and therapeutic interventions targeting the maternal microbiota that might guide healthy fetal development and avoid premature birth or other untoward outcomes. Another hypothesis is that following birth, there are microbial taxa whose changing pattern of representation can be used to define ‘normal’ programs of development of microbial communities occupying a given body habitat in biologically unrelated individuals with healthy growth phenotypes (defined by anthropometric indices; http://www.who.int/childgrowth/mgrs/en/). Corollaries to this hypothesis are that these developmental programs provide an additional microbial definition of healthy growth and that deviations from these normal programs of community assembly represent a way to characterize abnormal development, including states of immaturity or precocious maturation. Establishing a causal relationship between the state of microbial community development and healthy growth would allow deviations from normal microbiota development to be employed as a parameter for risk assessment or classification of a number of diseases that manifest themselves early or later in life, yield new insights about disease pathogenesis, and provide a starting point for developing microbiota-directed therapeutic interventions or new approaches for disease prevention.
In this Perspective, we discuss evolving concepts about (i) the relationship between maternal microbial ecology (before, during, and after pregnancy) and pregnancy outcomes, (ii) the relationship between human breast milk oligosaccharides, the establishment and expressed functions of the gut microbiota, and healthy postnatal growth, and (iii) the need for long-term birth cohort studies to identify shared as well as distinctive features of microbial community development, within and across different populations, and delineate how normal execution (and perturbations) of this facet of human developmental biology is related to health status.
Maternal microbial ecology
The structure and function of maternal microbial communities, and the impact of these communities on maternal and infant health outcomes has been considered in several body habitats, including the vagina, the distal gut, and the mouth.
Vaginal microbiota
Culture-based studies have suggested for decades that lactobacilli are the most prevalent constituents of the vaginal microbiota in non-pregnant and pregnant women 5. More recently, culture-independent studies demonstrating that vaginal communities frequently are highly uneven and dominated numerically by a single Lactobacillus species have prompted some investigators to assign vaginal communities into a relatively limited number of discrete ‘community state types’ (CSTs). These state types are classified by either the dominant Lactobacillus species or the presence of a relatively diverse, lactobacillus-poor community (CST IV)6. The resolution and veracity of the vaginal CST model remains unsettled: some investigators have proposed additional stable or transitional states beyond the five CSTs described initially7. Others have highlighted potential pitfalls, including the extent to which the detection of ‘state types’ is dependent upon the analytic workflow8. Regardless of the ultimate usefulness of the CST model, the relatively limited diversity of abundant taxa in vaginal communities suggests that a deterministic process of community assembly, such as habitat filtering, governs the overall structure of the adult vaginal microbiota.
CST IV is similar to the microbiota structure encountered in bacterial vaginosis, a dysbiosis associated with adverse health outcomes, including preterm birth9,10. In non-pregnant North American women, CST prevalence varies with self-reported race/ethnicity. CST IV is observed in ~40% of African-American and Hispanic women, but in only ~20% of Asian-Americans and ~10% of Caucasians6. This skewed distribution suggests that a diverse non-Lactobacillus-dominated community (CST IV) might represent a normal variant in a subset of women and argues for an expanded assessment of what comprises a ‘healthy’ vaginal microbiota.
Relatively little is known about the development of the vaginal microbiota prior to and following puberty, and how different vaginal community ‘fates’ (structural and functional states) in adulthood are determined. One area that should be investigated is the relationship between the glycan content of the vaginal mucosa and community state (CST), including the biogeographical features of each state. In addition, much remains to be learned about the effects of bacterial and eukaryotic taxa (and their viruses) on vaginal epithelial cell differentiation programs, vaginal mucosal metabolism, and the activities of components of the innate and adaptive arms of the immune system represented in this body habitat. The development of microarrays composed of purified microbial glycans11 provides one way to characterize immunological responses to bacterial antigens represented in a vaginal microbiota and thus creates another approach to community state classification. Representative preclinical models are needed for testing whether causal relationships exist between these and other environmental factors and community states, and for characterizing the mechanisms that shape community assembly, determine community responses to various perturbations, underlie community resiliency, and mediate the effects of CSTs on host biology.
A compelling question is whether there is a discernable ‘program’ of change in the properties of the vaginal microbiota before, during, and after pregnancy, and the degree to which such changes recapitulate features of the ‘original’ developmental biology of the community. A related question is whether and how functional alterations in vaginal microbial community states and the other body site microbiota during pregnancy impact intrauterine growth (see Text Box). Recent work has focused on bacterial taxonomic composition, rather than on functional features expressed by these microbial communities. To date, studies suggest that during pregnancy the bacterial composition of the microbiota is more stable than at other times during adulthood12–15. The diverse CST IV of the vagina appears to be the least stable during pregnancy, exhibiting a substantially higher rate of transition to alternate CSTs on a week-to-week timescale, compared to the four Lactobacillus-dominated CSTs14. A cautionary note is that community composition has not been defined in time-series studies where individuals are sampled prior to conception, following pregnancy, and during subsequent pregnancies. In addition, little is known about the non-bacterial membership of the pregnancy-associated microbiota.
Text Box. Concepts and hypotheses about the evolving maternal-fetal landscape and microbial ecosystem.
Beneficial activities of maternal microbiota may impact fetal nutrition and development
Altered compositions and expressed activities of maternal microbiota may contribute to gestational outcome, including adverse outcomes such as premature labor and birth
Microbes transferred to offspring at, or prior to delivery, reflect prepartum environmental exposures of the mother (e.g., diet)
Persisting post-partum disturbances in vaginal microbiota may pose risk for preterm birth in subsequent pregnancies
Variations in maternal transfer of microbes to infants may affect early postnatal development
Some factors that promote vaginal microbiota structural stability during pregnancy are recognized, such as lack of menses. However, many factors remain unknown, along with the degree to which structural stability is accompanied by functional stability and how this relates to the initial transfer of taxa from mothers to infants during the immediate peripartum period. From an anthropologic perspective, it is interesting that prescribed diets during pregnancy are an important part of the cultural traditions of some human populations16. It is unclear how these treatments affect vaginal (and gut) microbial community structure, function, and stability during pregnancy and in the immediate peripartum period. The answers to these questions may yield new agents for deliberate microbiota manipulation.
In contrast to the structural stability of the vaginal microbiota during pregnancy, studies of women in the USA, Europe, Africa, and Asia have shown that upon delivery, the vaginal microbiota commonly undergoes an abrupt and striking alteration in its taxonomic composition14,17–19. This alteration is characterized by significantly increased alpha-diversity and is driven by a decrease in the abundance of Lactobacillus spp. and a commensurate increase in diverse anaerobic species. Although many features of altered post-partum communities remain to be elucidated (e.g., time to return to ‘baseline’), it appears that in many women, they can persist for at least one year14. A short interpregnancy interval (<12 mo) is associated with an increased risk of preterm birth; whether a persisting altered post-partum vaginal community plays a contributing role warrants further study.
Gut microbiota
Much more information is needed about whether the structural and functional properties of the gut microbiota of women change as a function of pregnancy and, if so, whether and how such change relates to maternal and fetal health, as well as the subsequent health status of infants and children. The relationship of maternal nutritional status at the time of conception to the health of the newborn is well-established20. One study of pregnant Finnish women reported a significant increase in fecal energy content, as determined by bomb calorimetry, between the first and third trimesters despite stable diets and energy intake. This change correlated with compositional shifts in the microbiota21. However, studies of women residing in the USA14 and in Tanzania17, conducted at higher temporal resolution, found that their fecal microbiota manifested compositional stability throughout pregnancy (as measured by trends of alpha diversity, week-to-week variation within subjects, and beta diversity across gestational time), though the reasons for these divergent findings are unclear. Maternal microbiota and diet have the potential to influence both fetal and maternal epigenomes as well, although a discussion of this topic is beyond the scope of this Perspective.
Oral microbiota
Mothers harbor complex microbial communities in their mouths whose compositional states22 and transcriptional activities23 are altered in the setting of periodontitis, a condition associated with adverse health outcomes, including intra-uterine growth restriction, preterm birth, and low birth weight24. A study of the taxonomic composition of the oral microbiota of women living in the USA and Africa indicated that it remains stable during pregnancy14,17. However, comparisons with pre-conception data obtained from the same women were lacking. Microbial taxa that likely originate from the mouth have been detected in amniotic fluid25–28 and in the placenta29, particularly in association with unhealthy states and/or adverse outcomes such as preterm labor with intact fetal (chorioamniotic) membranes and with premature rupture of these membranes. Disentangling adverse effects on pregnancy originating from the oral microbiota is challenging, especially if disease results from a perturbation among relatively minor constituents30.
Development of the oral microbiota has not been comprehensively defined through time series studies of healthy infants and children. Therefore, the effects of maternal prenatal history, gestational age, route of delivery, and milk feeding history remain to be characterized. One approach to defining ‘normal’ is illustrated by a recent report of 50 children who were studied from 4 to 6 years of age31. The authors observed a strong effect of chronological age on the taxonomic composition of the oral microbiota. This effect was more pronounced for the composition of supragingival plaque bacterial communities than it was for those in saliva, which suggests habitat site-specific differences in community assembly programs. In this study, deviations from early, normal community compositions were predictive of subsequent development of dental caries31.
Fetal microbial exposure and preterm delivery
The sterility of the fetal environment has been pondered from the ‘birth’ of microbiology32. Early studies suggested universal sterility of the amniotic cavity prior to labor33, though subsequent indirect evidence challenged that assumption34. More recent PCR-based studies indicate that microbial invasion of the amniotic cavity (MIAC) occurs more frequently and involves a greater diversity of microbes than presumed historically25,26. Endometrial sampling of the intra-uterine cavity in non-pregnant women has yielded widely varying rates of microbial recovery across culture-based studies (0–89%)35. Recent molecular-based studies suggest that the uterus of most women harbors microbes, with Lactobacillus, Prevotella, and Bacteroides among the genera encountered most commonly35,36. However, data obtained during pregnancy are lacking.
The placental basal plate contains intracellular bacteria in a minority of women (~27%), but in about half of those who deliver spontaneously before 28 weeks37. More recently, the placenta has been reported to harbor a complex set of microbes based on detection of bacterial DNA sequences38. However, unlike densely colonized body sites such as the gut and mouth, samples of placenta are overwhelmingly culture-negative39. Assessments of potential microbes in the placenta, and other low microbial biomass sites, are particularly prone to confounding findings from ‘background’ DNA40,41 and should be interpreted with caution in the absence of appropriate controls. The degree to which the fetal-placental environment has evolved to serve as a venue for programmed engagement of diverse microbes, as opposed to a site that simply tolerates stochastic low-level microbial exposures, remains unclear and merits further study.
A recent report suggested that among women with spontaneous preterm birth, those with histologic evidence of severe chorioamnionitis had fewer bacterial species in the placental membrane (adjacent to the fetal side) than did those without severe chorioamnionitis42. This difference may have been driven largely by a high abundance of a limited number of clonal pathogens (as is typical of many clinical infections) in women with severe chorioamnionitis. Additional studies with appropriate negative controls are needed to corroborate these findings and to resolve unanswered questions, such as the primary provenance (i.e., body site of origin) of detected microbes, as well as the directionality and timing of microbial translocation across adjacent tissues43.
Microbes have been detected in first-pass meconium samples from approximately two-thirds of healthy, vaginally delivered, breastfed term infants but at very low levels44. Detection is more common in meconium from neonates born before 33 gestational weeks, with considerable taxonomic overlap with microbes reported in amniotic fluid25,45. Molecular evidence of MIAC has provided associations (space, time, and ‘dose’) that support a causal relationship with preterm birth25. Microbial taxa associated with preterm birth most frequently originate from the mother and exploit one of three natural routes for invading the amniotic cavity46: ascending from the vagina and cervix, transfer via the fallopian tubes, or translocation from more distant colonized body sites, presumably via the bloodstream27. The majority of invading microbes appear to be derived from the vagina25,28,46, though microbes from other body habitats, most notably the oral cavity27,47 and gut26, likely play a role in some cases. In particular, taxa associated with CST IV communities, such as Ureaplasma and Prevotella species, are among the more common invaders. By contrast, Lactobacillus species are very rarely encountered in amniotic fluid, even after membrane rupture26, despite the relatively high prevalence and abundance of lactobacilli in vaginal communities. This suggests that features of specific microbial taxa, or groups of co-occurring taxa, found in CST IV communities underpin factors that promote amniotic cavity invasion (e.g., virulence genes and divergent host immune responses48). Whether particular vaginal CSTs, or the presence/abundance of individual taxa, are associated with preterm birth is an unresolved question of great interest. Studies have produced conflicting results12,14. If vaginal CST IV communities are indeed associated with preterm birth in some women, this would be broadly consistent with epidemiologic evidence linking bacterial vaginosis, which shares taxonomic similarity with CST IV communities, to an increased risk of preterm birth9.
The impact of preterm delivery on the development of microbial communities in premature infants has been examined principally from the perspective of the infant. Characterizing development of their microbial communities versus those in full term infants could amend current, and yield new, definitions of biological immaturity, although such definitions are confounded by the frequent practice of preemptive antibiotic administration to prematurely-born individuals. On the other hand, maternal microbial communities also exert significant influence. A recent study demonstrated that transient microbial colonization of pregnant germ-free mice is sufficient to modulate the function of small intestinal innate immune cell populations in their germ-free offspring49. Microbial products were detectable in both the dam’s milk and the placenta, suggesting that ‘indirect’ exposure through the mother is sufficient to shape neonatal development. Findings of this type suggest that systematically characterizing multiple body habitat-associated microbial communities in mothers with preterm versus full term pregnancies creates opportunities to examine whether there are identifiable programs of change in maternal microbial ecology during pregnancy, and to determine how disruption of these programs impact initial transfer of microbes to their offspring (and subsequent development of their microbiota). This knowledge could change clinical practice so that more attention is placed on careful stewardship of maternal microbial resources in those at risk for preterm delivery50 and so that deliberate efforts are made to transfer these microbes to their offspring (with potential supplementation of key taxa that are not present).
Breast milk and the infant gut microbiota
Recent work has begun to uncover the how breast milk changes over time from parturition and how it shapes the structural and functional maturation of infant-associated microbial communities.
Breast milk-associated microbiota
While neonates acquire bacteria from various sources (the delivery process, physical interactions with mother and siblings, etc.), a relatively ignored and poorly understood source is breast milk. Studies of milk-associated microbiota reveal highly individualized assemblages51. These assemblages are routinely dominated by skin-associated bacteria, such as staphylococci and streptococci, which generally do not persist in the infant gut in significant numbers beyond a few weeks52. Some anaerobic species, such as Bifidobacterium, have been isolated from breast milk, suggesting a route for transit of specific strains that eventually colonize the infant colon. However, the factors that contribute to the strain specific composition in breast milk are not yet clear and are the subject of debate.
Human milk oligosaccharides (HMOs)
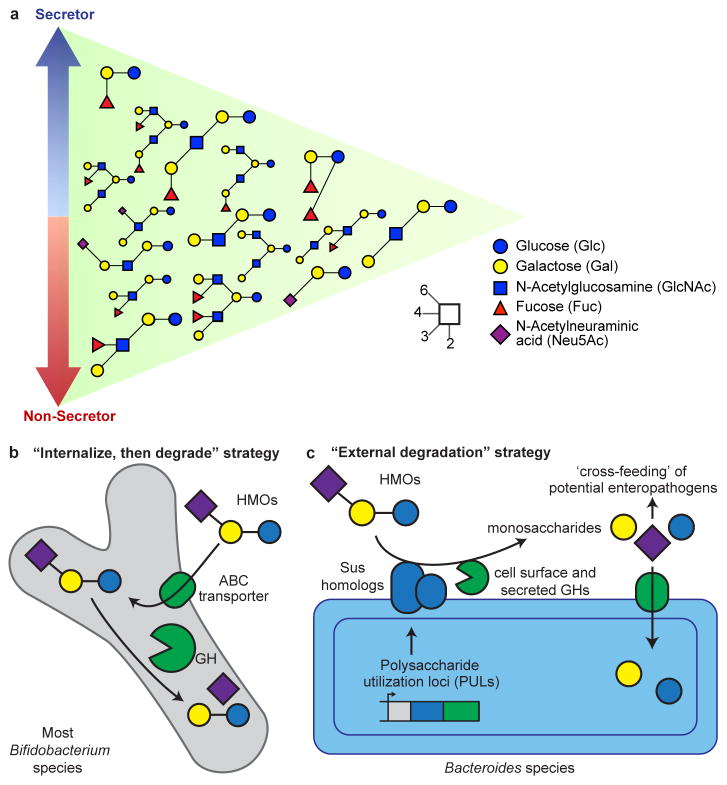
From a molecular perspective, breast milk is the best characterized food we consume. The most abundant dry component of milk is lactose, which provides nutrition for the infant, though many bacterial taxa can also digest this disaccharide. Lactose is made available specifically to bacterial colonizers of the infant gut by extending it with 3 to 20 monosaccharide units to yield oligosaccharide structures, known collectively as human milk oligosaccharides (HMOs)53,54. HMOs are all constructed on a lactose core together with combinations of glucose, galactose, N-acetyl galactosamine, fucose, and sialic acid (N-acetylneuraminic acid or Neu5Ac)55 residues. HMOs are often terminated by fucose or sialic acid (Figure 1A). The fraction of fucosylated HMO structures out of the total HMO pool is approximately 60%, while sialylated structures range from 5 to 20%56,57.
Figure 1. Representation of abundant HMOs present in milk and strategies employed by infant gut microbiota for their degradation.
(A) Secretor status and HMO composition. HMO structures that are most abundant in secretors are indicated by the blue arrow, while those that are most abundant in the breast milk of non-secretors are indicated by the red arrow. Structures at the intersection are found in both secretor and non-secretor mothers in similar abundances. The structures of these HMOs, along with their glycosidic linkages, are described by the inset key. (B) Most strains of Bifidobacterium use an ‘internalize, then degrade’ strategy where HMO structures are first imported using ABC transporters and degraded by intracellular glycoside hydrolases (GH). (C) Strains of Bacteroides typically employ an ‘external degradation’ strategy for HMO structures that involves cell surface associated carbohydrate binding proteins and secreted glycoside hydrolases encoded by polysaccharide utilization loci (PULs) that have features similar the prototypic starch utilization system (Sus) of Bacteroides thetaiotaomicron. This external degradation can result in ‘cross-feeding’ of secondary consumers in the infant gut microbiota.
The role of HMOs has become more apparent with the application of nanoflow liquid chromatography mass spectrometry (nanoLC-MS). This method has detected >300 structures in breast milk samples pooled from several mothers, with concentrations spanning four orders of magnitude. For a given mother, the number of HMO structures is often >100, while the profile of HMO structures varies between mothers53,54. Some HMOs contain the Lewis blood group antigens Lea, Leb, Lex, and Ley.58 Individuals who produce Leb epitopes [α(1,2)-fucosylated structures] in their secretions, due to the presence of an active fucosyltransferase 2 (FUT2) gene, are known as secretors59. Secretors tend to have higher amounts of HMOs compared to non-secretors (as much as 20% more). They also contain higher levels of fucosylated structures (nearly two-fold more). However, non-secretors often contain higher levels of sialylated HMO structures57 (Figure 1A). The percentage of non-secretors varies geographically: they comprise about 20% of the population in Europe and up to 40% in West Africa60.
A key question is how the HMO profiles of breast milk change as a function of time after delivery, and how compositional differences relate to the development of the microbiota and healthy growth of the infant. Because of the relatively recent application of nanoLC-MS for HMO profiling and constraints imposed by assay throughput, there is limited information regarding the temporal patterns of change in specific HMO structures in healthy mothers, and whether consistent differences exist across groups of women representing different ages, parity, geographic locales, nutritional states, culinary traditions, and socioeconomic status. The general trend over the lactation period is a decrease in total HMO from colostrum through mature milk, with the largest drop occurring in the first month postpartum61. However, the total amount of milk delivered as colostrum is quite small compared to mature milk (matching the size of the infant stomach and intestine). Thus, throughout lactation, the relative abundances of each class and even each compound remain relatively constant61–63.
Premature birth can significantly affect the structural profile of the mother’s HMOs64. HMOs in these mothers are as yet unpredictable. Many mothers delivering preterm have fucosylated HMOs that are as low as 20–40% of total HMOs while other mothers have levels greater than 60%. This discrepancy is not corrected over time.
A recent study revealed that HMO content in Malawian mothers correlates with infant growth outcomes65. Breast milk samples collected from Malawian mothers at 6 months postpartum were divided into two groups: samples from mothers whose infants exhibited healthy growth at the time of collection (as defined by anthropometry), and those from mothers whose offspring exhibited severe stunting. Liquid chromatography-time-of-flight mass spectrometry (LC-TOF MS) revealed that mothers of stunted infants had significantly lower concentrations of total, sialylated, and fucosylated HMOs, with the most growth discriminatory sialylated HMO being sialyllacto-N-tetraose b (LSTb), and most discriminatory fucosylated HMOs being 2′-fucosyllactose (2′FL) and lacto-N-fucopentaose I (LNFP I).
Sialic acids constitute a group of nine-carbon monosaccharides, derived from neuraminic acid, and include N-acetylneuraminic acid (Neu5Ac). UDP-N-acetylglucosamine-2-epimerase, the rate-limiting enzyme in sialic acid biosynthesis is produced at low levels in the livers of infants66. Thus, breast milk represents an important source of this sugar. The availability of sialic acid impacts many organs, including the brain, where Neu5Ac is a component of gangliosides and is covalently linked to neural cell adhesion molecules (NCAMs) that mediate cell-cell interactions involved in synaptogenesis and memory67,68. Dietary supplementation with sialylated glycoproteins and sialyllactose increases polysialylation of NCAM and sialyated gangliosides, with some reports showing improved memory in animal models69. In addition, a preclinical model has shown that 6′-sialyllactose increases muscle mass and contractility70.
Several HMO structures have been produced chemically and enzymatically71. However, producing the wide array of structures encountered in human milk is not yet commercially feasible. There is a ~25% overlap between bovine milk oligosaccharide (BMO) and HMO structures. Sialylated milk oligosaccharides are present in mature human milk at concentrations up to 20-fold greater than in mature bovine milk72,73. Therefore, bovine milk-derived infant formulas and complementary or therapeutic foods used to treat children with undernutrition are deficient in these compounds. However, BMOs with structural similarity to HMOs are present in the by-products of dairy processing, presenting an opportunity to purify them at a scale sufficient for preclinical and clinical studies, and potentially for wider spread distribution should such studies show safety, efficacy, and yield an understanding of their mechanism of action.
Preclinical studies have provided direct evidence that sialylated milk oligosaccharides are causally related to growth65. Young germ-free mice and newborn germ-free piglets were colonized with members of the gut microbiota of a stunted Malawian infant. Recipient animals were fed a Malawian diet representative of foods consumed after weaning by this human population, with or without supplementation with sialylated BMOs (S-BMO) purified from a whey waste stream generated during the manufacture of cheese. Feeding these animals this Malawian diet, or an isocaloric Malawian diet supplemented with S-BMO revealed that S-BMO promoted lean body mass gain and impacted metabolism in ways indicative of improved metabolic flexibility (the capacity to rapidly shift from anabolism in the fed state to lipid oxidation in the fasted state)74. Bone growth was also positively impacted in gnotobiotic mice. These effects were not ascribable to differences in food consumption and were microbiota-dependent, as they were not observed in germ-free animals. Moreover, when animals were provided an isocaloric Malawian diet supplemented with a mixture of fructo-oligosaccharides, a component of some infant formulas, growth promotion was not observed.
Development of a Milk Oriented Microbiota (MOM)
The initial microbiota of nursing infants is an assemblage of microbes derived from mother’s fecal, vaginal, and skin microbiota52. Within weeks, pro- and antimicrobial agents in breast milk guide development of a MOM. A common enrichment involves members of the Actinobacteria, chiefly Bifidobacterium species that frequently dominate the gut microbiota of breastfed infants, in some cases representing 70–90% of the fecal community75. Intriguingly, this enrichment is less pronounced in more industrialized countries75–78. Bifidobacterial enrichment is linked to maternal genotype; secretor mothers’ milk appear to enrich bifidobacteria more rapidly76.
Several beneficial functions have been attributed to a bifidobacteria-dominated MOM. For example, lactate and acetate, the primary end products of bifidobacterial fermentation, are known to be an important energy source for colonocytes, to lower intestinal pH, and to contribute to gut barrier function79. Robust colonization by a single bifidobacterial subspecies, Bifidobacterium longum subsp. infantis, correlates with improved vaccine responses during the first year of life77. Moreover, intestinal bifidobacteria produce essential nutrients, including folate and riboflavin80.
Two dominant bifidobacterial species, Bifidobacterium longum and B. breve, routinely colonize breastfed infants throughout the world, although other species, including B. bifidum, B. catenulatum, and B. pseudocatenulatum are also commonly observed. In general, bifidobacterial species are prolific consumers of HMOs and possess an array of glycoside hydrolases (notably fucosidases and sialidases81) that catalyze cleavage of key glycosidic linkages, permitting metabolism of some or all of the sugar monomers embedded in HMOs. The mechanisms for consumption of HMOs by these organisms illustrate two different strategies82. B. longum subsp. infantis (and to a lesser extent B. longum subsp. longum, B. breve, and B. pseudocatenulatum) transport HMOs directly into the cell via ATP binding cassette (ABC) transporters and cleave these oligosaccharides with intracellular glycoside hydrolases (Figure 1B)83. In contrast, B. bifidum deploys some glycoside hydrolases to the cell wall for extracellular cleavage of HMOs prior to importing select degradation products. Similarly, Bacteroides species, another important set of HMO consumers (and frequent MOM-associated species), also deploy external glycoside hydrolases to degrade these structures prior to their internalization (Figure 1C)83.
The “internalize, then degrade” approach for HMO consumption adopted by the majority of infant-borne bifidobacteria can be viewed as an ingenious strategy for protecting the neonate. These bacteria prevent growth of competitor strains by simple sequestration of available sugar substrates in the colon, a concept consistent with the inverse correlation observed between fecal HMO concentrations and the level of bifidobacterial colonization76,84. An important consideration is whether there are deleterious consequences associated with harboring a MOM dominated by bacteria that degrade HMO externally. An antibiotic-treated mouse model was used to show that mucin, a glycan that contains structures similar to HMOs, can be externally degraded by Bacteroides spp. to release fucose and sialic acid monomers that ‘cross-feed’ various pathogens85. Could external degradation of HMOs lead to growth of pathogens or pathobionts in the low diversity neonatal gut microbiota? Three recent studies point to this potential risk. In gnotobiotic mice that harbored a stunted Malawian infant’s microbiota, external degradation of S-BMO by Bacteroides fragilis released its constituent monosaccharides, including sialic acid, that cross-fed Escherichia coli populations65. Others have observed Bacteroides cross-feeding Enterobacteriaceae in mice fed sialyllactose (an oligosaccharide common to mammalian milks) and in nursing piglets86,87. Enterobacteriaceae are considered by some to be harbinger of dysbiosis88.
Together, these findings suggest that the potential for bacterial cross-feeding on HMOs may be a risk factor for neonates. They also illustrate the extreme caution that should be afforded when composing diets for neonates that harbor low diversity gut microbiota during early stages of community development. In cases where a single oligosaccharide prebiotic is being considered, such as the use of fucosyllactose or sialyllactose for infant formula, it would help to know the composition of the infant MOM to avoid potential cross-feeding of problematic bacterial clades and/or enteropathogens. Alternatively, this problem may be alleviated by use of synbiotic applications (combinations of pre- and probiotics) where the probiotic component is known to readily consume the oligosaccharides provided and/or derived monomers.
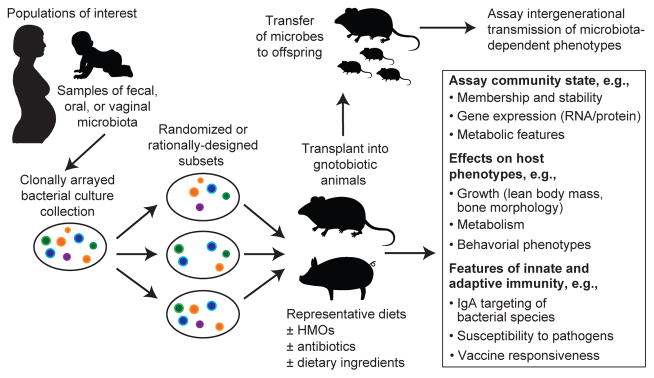
Several challenging questions need to be addressed. First, we know very little about the functions of various HMO structures. Why has mammalian evolution produced such a diverse consortium of structures? Even more diversity is possible given the number of possible glycosidic linkages, suggesting that observed HMOs structures were selected by evolution. Second, we need to better characterize the interactions and relative effect sizes of the ‘anti-microbial’ and ‘pro-microbial’ components of milk on MOM development. One approach for addressing these questions is to use gnotobiotic animals colonized with MOMs from infants representing different gestational ages, milk feeding histories, and growth phenotypes. Alternatively, these animals can be colonized with defined collections of cultured bacterial strains, generated from a given donor’s microbiota; these clonally arrayed collections can be manipulated so that all members, or subsets, are added, with or without pathogens/pathobionts, to recipient animals (Figure 3). Gnotobiotic animals colonized with these communities can be fed breast milk or infant formula supplemented with defined milk oligosaccharide structures (many of the antimicrobial elements of breast milk, including antibodies, lactoferrin, and lysozyme, are absent from these formulas). These models represent one way for determining the “rules” that govern early phases of human gut microbiota development.
Moving from MOM to a weaning-oriented microbiota and beyond
Recent work provides an approach for identifying a program of gut microbial community development that is executed during the first 2–3 years of postnatal life as infants move from a diet dominated by milk through a period of complementary feeding to a fully weaned state. Monthly collection of fecal samples from members of a Bangladeshi birth cohort with healthy growth phenotypes (defined by anthropometry) allowed bacterial 16S rRNA-sequence based datasets to be generated that described the bacterial composition of their developing gut communities78. Random Forests-based regression identified a group of 24 of the most age-discriminatory bacterial strains; their relative abundances in these biologically unrelated individuals were used as a microbial signature to describe the state of development (‘age’) of their microbiota. This approach allows the age or state of maturation of any child’s microbiota in the population to be computed using the Random Forests-derived model and compared to the reference healthy cohort (whose microbiota age and chronologic age are highly correlated). Deviations from normal are expressed in the form of a Microbiota for Age Z-score (MAZ). A similar approach was used to characterize normal gut microbiota development in Malawian birth cohorts. Remarkably, the resulting Malawian Random Forests-generated model shared many of the age-discriminatory strains that were represented in the Bangladeshi-derived model89.
Calculating MAZ scores disclosed that microbiota development was impaired in Malawian and Bangladeshi children presenting with moderate acute malnutrition (MAM) or severe acute malnutrition (SAM)78,89. Their microbiota appeared “younger” than would be expected based on their chronologic age, with the severity of immaturity being greater in those with SAM. Moreover, studies of children with SAM indicated that their microbiota immaturity was not durably repaired after treatment with either of two ready-to-use therapeutic foods. In other words, these children had a persistent defect affecting their gut microbial communities78,89. Transplanting immature microbiota from stunted/underweight Malawian children, or microbiota from chronologically age-matched donors with healthy growth phenotypes, to young germ-free mice fed a diet resembling that consumed by the microbiota donors disclosed that immature microbiota transmitted impaired growth phenotypes89.
These and other studies provide preclinical proof-of-concept that gut microbiota development is causally related to healthy growth89,90. They also provide a microbial measure of normal as well as perturbed postnatal development. Knowledge of a subset of age-discriminatory bacterial strains that promote growth in these gnotobiotic animal models89 allows attention to be focused on factors that may support their establishment, adequate representation, and/or expressed functions. These factors include gestational age, delivery mode, milk feeding history, exposures to antibiotics, and enteropathogen burden (Figure 3).
A related question is how microbiota development affects development of the immune system. This issue can be addressed in part by defining (gut) mucosal IgA responses to members of the microbiota91,92, using fecal samples serially collected from members of birth cohort studies. This approach provides one way to identify interrelationships between community development, development of the immune system, breast milk HMO content, and host growth phenotypes.
A call for human microbial community observatories
Characterizing normal gut microbiota development and the development of other body habitat-associated microbial communities in members of birth cohorts provides a framework for exploring the degree to which these processes vary across populations of infants and children with healthy growth phenotypes, as well as whether and how perturbations of these programs are related to growth faltering and the risk for and pathogenesis of various diseases. These studies should include an examination of the host ecological landscape that gives rise to the newborn child (i.e., the mother and her microbial communities starting at the time of conception). The results could yield insights about as yet unappreciated microbial contributions to a wide range of disorders that are overtly manifest, or foreshadowed, by changes microbial community structure and function in infancy or childhood (e.g., obesity93–95, immunologic disorders that include atopic states96, and neurodevelopmental disorders97).
Given the dramatic, myriad, and rapid changes wrought by globalization in our lifestyles, and the vast differences in sanitation and hygiene experienced by different populations, we propose that a series of ‘human microbial observatories’ be established whose purpose is to characterize the evolution of microbial communities in mothers before, during, and following their pregnancies and the development of microbial communities in their offspring (and perhaps in the future, the subsequent pregnancies and offspring of these children). We propose that the populations selected for study should not only illustrate currently distinct lifestyles and geographies, but also contain segments that are likely to undergo lifestyle changes within a generation. Entities, both private and public, that are committed to addressing global health challenges have already made investments that have established durable, trusting relationships between health care providers and such populations, and the infrastructure required to obtain informed consent and apply validated procedures for collecting and archiving biospecimens and associated metadata [e.g., the Global Enteric Multicenter Study (GEMS)98, the MAL-ED network99, and WASH100]. These investments should be leveraged for the proposed human microbial observatory programs, which will require sustained support given the duration of the required period of observation. Developing effective and innovative strategies for achieving such durable support is a subject that requires expertise from multiple disciplines, and in our opinion, is a compelling challenge whose solution(s) have broad implications for obtaining answers to this biological question as well as myriad others related to the promotion of human flourishing (eudaimonia) in the broadest sense.
Wise and effective stewardship of our human microbial resources is a responsibility that extends across generations and national boundaries. Knowledge of how our microbial communities evolve in health and how their development is jeopardized or overtly disrupted provides an opportunity to discover strategies and tools for timely repair. However, understanding how such repair can be achieved brings great responsibility. The immediate as well as long term consequences of such interventions applied early in the course of a human life need to be determined. Rigorous preclinical tests of safety and efficacy have to be designed and applied in representative animal models when available. Very thoughtful consideration must be given to the ethical, regulatory, and societal issues and consequences that could arise from early interventions that shape the composition and function of our microbial communities. This is a time for inspiration and awe as we gain insights about how we function as holobionts, and a time for mindfulness and sobriety as we consider how to deliberately shape facets of our own developmental biology to improve wellness during our human lifecycle.
Figure 2. Discovery pipeline for characterizing the functional properties of developing human microbial communities.
Gnotobiotic animals can be employed as preclinical models for determining the effects of various states of microbial community development on host developmental biology.
Acknowledgments
Work cited from the authors’ labs was supported in part by grants from the National Institutes of Health (DK30292, HD061923, AT007079, AT008759), the Bill & Melinda Gates Foundation, the March of Dimes Foundation, and the Thomas C. and Joan M. Merigan Endowment at Stanford University.
Footnotes
Author Contributions – Each author contributed to the writing of this paper.
Competing financial interests J.I.G. is co-founder of Matatu, Inc., a company characterizing the role of diet-by-microbiota interactions in animal health. Upon completion of his PhD studies, M.R.C. has joined Matatu, Inc. as a research scientist. C.B.L and D.A.M., are co-founders of Evolve Biosystems, Inc., a company focused on diet-based manipulation of the gut microbiota. D.A.R. is a member of the Scientific Advisory Board of Seres Health. The other authors declare that they have no competing interests.
References
- 1.Sender R, Fuchs S, Milo R. Are we really vastly outnumbered? Revisiting the ratio of bacterial to host cells in humans. Cell. 2016;164:337–340. doi: 10.1016/j.cell.2016.01.013. [DOI] [PubMed] [Google Scholar]
- 2.Qin J, et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010;464:59–65. doi: 10.1038/nature08821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.The Human Microbiome Project Consortium. Structure, function and diversity of the healthy human microbiome. Nature. 2012;486:207–214. doi: 10.1038/nature11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gordon JI, Knowlton N, Relman DA, Rohwer F, Youle M. Superorganisms and Holobionts. Microbe. 2013;8:152–153. [Google Scholar]
- 5.Levison M, Corman L, Carrington E, Kaye D. Quantitative microflora of the vagina. Am J Obstet Gynecol. 1977;127:80–85. doi: 10.1016/0002-9378(77)90318-0. [DOI] [PubMed] [Google Scholar]
- 6.Ravel J, et al. Vaginal microbiome of reproductive-age women. Proc Natl Acad Sci U S A. 2011;108(Suppl 1):4680–4687. doi: 10.1073/pnas.1002611107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dareng EO, et al. Prevalent high-risk HPV infection and vaginal microbiota in Nigerian women. Epidemiol Infect. 2016;144:123–137. doi: 10.1017/S0950268815000965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Koren O, et al. A guide to enterotypes across the human body: meta-analysis of microbial community structures in human microbiome datasets. PLoS Comput Biol. 2013;9:e1002863. doi: 10.1371/journal.pcbi.1002863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hillier SL, et al. Association between bacterial vaginosis and preterm delivery of a low-birth-weight infant. The vaginal infections and prematurity study group. N Engl J Med. 1995;333:1737–1742. doi: 10.1056/NEJM199512283332604. [DOI] [PubMed] [Google Scholar]
- 10.Horner-Devine MC, Bohannan BJ. Phylogenetic clustering and overdispersion in bacterial communities. Ecology. 2006;87:S100–108. doi: 10.1890/0012-9658(2006)87[100:pcaoib]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 11.Stowell SR, et al. Microbial glycan microarrays define key features of host-microbial interactions. Nat Chem Biol. 2014;10:470–476. doi: 10.1038/nchembio.1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Romero R, et al. The vaginal microbiota of pregnant women who subsequently have spontaneous preterm labor and delivery and those with a normal delivery at term. Microbiome. 2014;2:18. doi: 10.1186/2049-2618-2-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Romero R, et al. The composition and stability of the vaginal microbiota of normal pregnant women is different from that of non-pregnant women. Microbiome. 2014;2:4. doi: 10.1186/2049-2618-2-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.DiGiulio DB, et al. Temporal and spatial variation of the human microbiota during pregnancy. Proc Natl Acad Sci U S A. 2015;112:11060–11065. doi: 10.1073/pnas.1502875112. This study of 40 women with weekly sampling of their vaginal, gut, and oral microbiota during pregnancy, and with monthly sampling postpartum, disclosed that (i) the bacterial composition of these communities remains relatively stable during pregnancy, followed by a marked postpartum change in the vaginal community than can persist for ≥12 months, and (ii) the prevalence of a Lactobacillus-poor vaginal community state type (CST 4) is inversely correlated with gestational age at delivery. These findings raise a series of questions for future research, including those related to how the configurations of maternal microbial communities function to facilitate transmission of microbes to infants, and the how the vaginal microbiota operates to shape pregnancy outcomes/maternal health, including in situations where the inter-pregnancy interval is short. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Aagaard K, et al. A metagenomic approach to characterization of the vaginal microbiome signature in pregnancy. PLoS ONE. 2012;7 doi: 10.1371/journal.pone.0036466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wilson CS. Nutritionally beneficial cultural practices. World Rev Nutr Diet. 1985;45:68–95. doi: 10.1159/000410264. [DOI] [PubMed] [Google Scholar]
- 17.Bisanz JE, et al. Microbiota at multiple body sites during pregnancy in a rural Tanzanian population and effects of moringa-supplemented probiotic yogurt. Appl Environ Microbiol. 2015;81:4965–4975. doi: 10.1128/AEM.00780-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.MacIntyre DA, et al. The vaginal microbiome during pregnancy and the postpartum period in a European population. Sci Rep. 2015;5:8988. doi: 10.1038/srep08988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huang YE, et al. Homogeneity of the vaginal microbiome at the cervix, posterior fornix, and vaginal canal in pregnant Chinese women. Microb Ecol. 2015;69:407–414. doi: 10.1007/s00248-014-0487-1. [DOI] [PubMed] [Google Scholar]
- 20.Burke BS, Stevenson SS. Nutrition studies during pregnancy; relation of maternal nutrition to condition of infant at birth; study of siblings. J Nutr. 1949;38:453–467. doi: 10.1093/jn/38.4.453. [DOI] [PubMed] [Google Scholar]
- 21.Koren O, et al. Host remodeling of the gut microbiome and metabolic changes during pregnancy. Cell. 2012;150:470–480. doi: 10.1016/j.cell.2012.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu B, et al. Deep sequencing of the oral microbiome reveals signatures of periodontal disease. PLoS ONE. 2012;7:e37919. doi: 10.1371/journal.pone.0037919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Duran-Pinedo AE, et al. Community-wide transcriptome of the oral microbiome in subjects with and without periodontitis. ISME J. 2014;8:1659–1672. doi: 10.1038/ismej.2014.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Siqueira FM, et al. Intrauterine growth restriction, low birth weight, and preterm birth: adverse pregnancy outcomes and their association with maternal periodontitis. J Periodontol. 2007;78:2266–2276. doi: 10.1902/jop.2007.070196. [DOI] [PubMed] [Google Scholar]
- 25.DiGiulio DB, et al. Microbial prevalence, diversity and abundance in amniotic fluid during preterm labor: a molecular and culture-based investigation. PLoS ONE. 2008;3:e3056. doi: 10.1371/journal.pone.0003056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.DiGiulio DB, et al. Prevalence and diversity of microbes in the amniotic fluid, the fetal inflammatory response, and pregnancy outcome in women with preterm pre-labor rupture of membranes. Am J Reprod Immunol. 2010;64:38–57. doi: 10.1111/j.1600-0897.2010.00830.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Han YW, et al. Transmission of an uncultivated Bergeyella strain from the oral cavity to amniotic fluid in a case of preterm birth. J Clin Microbiol. 2006;44:1475–1483. doi: 10.1128/JCM.44.4.1475-1483.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Han YW, Shen T, Chung P, Buhimschi IA, Buhimschi CS. Uncultivated bacteria as etiologic agents of intra-amniotic inflammation leading to preterm birth. J Clin Microbiol. 2009;47:38–47. doi: 10.1128/JCM.01206-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Swati P, Thomas B, Vahab SA, Kapaettu S, Kushtagi P. Simultaneous detection of periodontal pathogens in subgingival plaque and placenta of women with hypertension in pregnancy. Arch Gynecol Obstet. 2012;285:613–619. doi: 10.1007/s00404-011-2012-9. [DOI] [PubMed] [Google Scholar]
- 30.Costalonga M, Herzberg MC. The oral microbiome and the immunobiology of periodontal disease and caries. Immunol Lett. 2014;162:22–38. doi: 10.1016/j.imlet.2014.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Teng F, et al. Prediction of early childhood caries via spatial-temporal variations of oral microbiota. Cell Host Microbe. 2015;18:296–306. doi: 10.1016/j.chom.2015.08.005. [DOI] [PubMed] [Google Scholar]
- 32.Kustner O. Beitrag zur Lehre von der puerperalen Infection der Neugeborenen. Arch Gynak. 1877;11:256. [Google Scholar]
- 33.Harris JW, Brown H. Bacterial content of the uterus at cesarean section. Am J Obstet Gynecol. 1927;13:133. [Google Scholar]
- 34.Benirschke K. Routes and types of infection in the fetus and the newborn. AMA J Dis Child. 1960;99:714–721. doi: 10.1001/archpedi.1960.02070030716003. [DOI] [PubMed] [Google Scholar]
- 35.Verstraelen H, et al. Characterisation of the human uterine microbiome in non-pregnant women through deep sequencing of the V1-2 region of the 16S rRNA gene. Peer J. 2016;4:e1602. doi: 10.7717/peerj.1602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mitchell CM, et al. Colonization of the upper genital tract by vaginal bacterial species in nonpregnant women. Am J Obstet Gynecol. 2015;212:611.e1–9. doi: 10.1016/j.ajog.2014.11.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stout MJ, et al. Identification of intracellular bacteria in the basal plate of the human placenta in term and preterm gestations. Am J Obstet Gynecol. 2013;208:226 e1–7. doi: 10.1016/j.ajog.2013.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Aagaard K, et al. The placenta harbors a unique microbiome. Sci Transl Med. 2014;6:237ra65. doi: 10.1126/scitranslmed.3008599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bhola K, et al. Placental cultures in the era of peripartum antibiotic use. Aust N Z J Obstet Gynaecol. 2008;48:179–184. doi: 10.1111/j.1479-828X.2008.00833.x. [DOI] [PubMed] [Google Scholar]
- 40.Kliman HJ. Comment on ‘The placenta harbors a unique microbiome’. Sci Transl Med. 2014;6:254le4. doi: 10.1126/scitranslmed.3009864. [DOI] [PubMed] [Google Scholar]
- 41.Salter SJ, et al. Reagent and laboratory contamination can critically impact sequence-based microbiome analyses. BMC Biol. 2014;12:87. doi: 10.1186/s12915-014-0087-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Prince AL, et al. The placental microbiome is altered among subjects with spontaneous preterm birth with and without chorioamnionitis. Am J Obstet Gynecol. 2016:305–307. doi: 10.1016/j.ajog.2016.01.193. S0002-9378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kim MJ, et al. Widespread microbial invasion of the chorioamniotic membranes is a consequence and not a cause of intraamniotic infection. Lab Invest. 2009;89:924–936. doi: 10.1038/labinvest.2009.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hansen R, et al. First-pass meconium samples from healthy term vaginally-delivered neonates: an analysis of the microbiota. PLoS ONE. 2015;10:e0133320. doi: 10.1371/journal.pone.0133320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ardissone AN, et al. Meconium microbiome analysis identifies bacteria correlated with premature birth. PLoS ONE. 2014;9:e90784. doi: 10.1371/journal.pone.0090784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Romero R, et al. The role of infection in preterm labour and delivery. Paediatr Perinat Epidemiol. 2001;15(Suppl 2):41–56. doi: 10.1046/j.1365-3016.2001.00007.x. [DOI] [PubMed] [Google Scholar]
- 47.Bearfield C, Davenport ES, Sivapathasundaram V, Allaker RP. Possible association between amniotic fluid micro-organism infection and microflora in the mouth. BJOG. 2002;109:527–533. doi: 10.1111/j.1471-0528.2002.01349.x. [DOI] [PubMed] [Google Scholar]
- 48.Menon R, Peltier MR, Eckardt J, Fortunato SJ. Diversity in cytokine response to bacteria associated with preterm birth by fetal membranes. Am J Obstet Gynecol. 2009;201:306.e1–6. doi: 10.1016/j.ajog.2009.06.027. [DOI] [PubMed] [Google Scholar]
- 49.Gomez de Aguero M, et al. The maternal microbiota drives early postnatal innate immune development. Science. 2016;351:1296–1302. doi: 10.1126/science.aad2571. An elegant gnotobiotic mouse model of transient microbial colonization demonstrates that maternal exposure to microbes during pregnancy shapes immunological development and function in the neonate. [DOI] [PubMed] [Google Scholar]
- 50.Muglia LJ, Katz M. The enigma of spontaneous preterm birth. N Engl J Med. 2010;362:529–535. doi: 10.1056/NEJMra0904308. [DOI] [PubMed] [Google Scholar]
- 51.McGuire MK, McGuire MA. Human milk: mother nature’s prototypical probiotic food? Adv Nutr. 2015;6:112–123. doi: 10.3945/an.114.007435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Palmer C, Bik EM, DBD, Relman DA, Brown PO. Development of the human infant intestinal microbiota. PLoS Biol. 2007;5:e177. doi: 10.1371/journal.pbio.0050177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wu S, Tao N, German JB, Grimm R, Lebrilla CB. Development of an annotated library of neutral human milk oligosaccharides. J Proteome Res. 2010;9:4138–4151. doi: 10.1021/pr100362f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wu S, Grimm R, German JB, Lebrilla CB. Annotation and structural analysis of sialylated human milk oligosaccharides. J Proteome Res. 2011;10:856–868. doi: 10.1021/pr101006u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kunz C, Rudloff S. Biological functions of oligosaccharides in human milk. Acta Paediatr. 1993;82:903–912. doi: 10.1111/j.1651-2227.1993.tb12597.x. [DOI] [PubMed] [Google Scholar]
- 56.Ninonuevo MR, et al. A strategy for annotating the human milk glycome. J Agric Food Chem. 2006;54:7471–7480. doi: 10.1021/jf0615810. [DOI] [PubMed] [Google Scholar]
- 57.Totten SM, et al. Comprehensive profiles of human milk oligosaccharides yield highly sensitive and specific markers for determining secretor status in lactating mothers. J Proteome Res. 2012;11:6124–6133. doi: 10.1021/pr300769g. [DOI] [PubMed] [Google Scholar]
- 58.Bode L. Human milk oligosaccharides: every baby needs a sugar mama. Glycobiology. 2012;22:1147–1162. doi: 10.1093/glycob/cws074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Thurl S, et al. Variation of human milk oligosaccharides in relation to milk groups and lactational periods. Br J Nutr. 2010;104:1261–1271. doi: 10.1017/S0007114510002072. [DOI] [PubMed] [Google Scholar]
- 60.Totten SM, et al. Rapid-throughput glycomics applied to human milk oligosaccharide profiling for large human studies. Anal Bioanal Chem. 2014;406:7925–7935. doi: 10.1007/s00216-014-8261-2. Recently developed nanoLC-MS methods allow rapid and reproducible detection of HMO structures in small volumes of biological samples, enabling large-scale clinical studies. [DOI] [PubMed] [Google Scholar]
- 61.Coppa GV, et al. Changes in carbohydrate composition in human milk over 4 months of lactation. Pediatrics. 1993;91:637–641. [PubMed] [Google Scholar]
- 62.Ninonuevo MR, et al. Daily variations in oligosaccharides of human milk determined by microfluidic chips and mass spectrometry. J Agr Food Chem. 2008;56:618–626. doi: 10.1021/jf071972u. [DOI] [PubMed] [Google Scholar]
- 63.Chaturvedi P, et al. Fucosylated human milk oligosaccharides vary between individuals and over the course of lactation. Glycobiology. 2001;11:365–372. doi: 10.1093/glycob/11.5.365. [DOI] [PubMed] [Google Scholar]
- 64.De Leoz MLA, et al. Lacto-N-tetraose, fucosylation, and secretor status are highly variable in human milk oligosaccharides from women delivering preterm. J Proteome Res. 2012;11:662–4672. doi: 10.1021/pr3004979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Charbonneau MR, et al. Sialylated milk glycans promote growth in gnotobiotic mice and pigs with a stunted Malawian infant gut microbiota. Cell. 2016;164:859–871. doi: 10.1016/j.cell.2016.01.024. A mass spectrometry-based study of Malawian mothers’ breast milk samples obtained at 6 months postpartum revealed that those whose infants were stunted/underweight had significant reductions in levels of a number of HMO structures, including those that were sialylated, compared to mothers of infants with healthy growth phenotypes. Follow-up characterization of gnotobiotic mice and piglets, colonized with consortia of cultured bacterial strains from a severely stunted/underweight 6-month old Malawian infant and fed a macro- and micronutrient deficient Malawian diet, provided preclinical evidence that sialylated milk oligosaccharides play a causal, microbiota-dependent role in promoting lean body mass gain and bone growth, and in enhancing host metabolism. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gal B, et al. Development changes in UDP-N-acetylglucosamine 2-epimerase activity of rat and guinea-pig liver. Comp Biochem Physiol B. 1997;108:13–15. doi: 10.1016/s0305-0491(97)00016-3. [DOI] [PubMed] [Google Scholar]
- 67.Wang B. Sialic acid is an essential nutrient for brain development and cognition. Annu Rev Nutr. 2009;29:177–222. doi: 10.1146/annurev.nutr.28.061807.155515. [DOI] [PubMed] [Google Scholar]
- 68.Wang B, Brand-Miller J. The role and potential of sialic acid in human nutrition. Eur J Clin Nutr. 2003;57:1351–1369. doi: 10.1038/sj.ejcn.1601704. [DOI] [PubMed] [Google Scholar]
- 69.Wang B, et al. Dietary sialic acid supplementation improves learning and memory in piglets. Am J Clin Nutr. 2007;85:561–569. doi: 10.1093/ajcn/85.2.561. [DOI] [PubMed] [Google Scholar]
- 70.Yonekawa T, et al. Sialyllactose ameliorates myopathic phenotypes in symptomatic GNE myopathy model mice. Brain. 2014;137:2670–2679. doi: 10.1093/brain/awu210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Chen X. Advances in Carbohydrate Chemistry and Biochemistry. 2015;72 doi: 10.1016/bs.accb.2015.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Aldredge DL, et al. Annotation and structural elucidation of bovine milk oligosaccharides and determination of novel fucosylated structures. Glycobiology. 2013;23:664–676. doi: 10.1093/glycob/cwt007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sundekilde UK, et al. Natural variability in bovine milk oligosaccharides from Danish Jersey and Holstein-Friesian breeds. J Agr Food Chem. 2012;60:6188–6196. doi: 10.1021/jf300015j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Muoio DM. Metabolic inflexibility: when mitochondrial indecision leads to metabolic gridlock. Cell. 2014;159:1253–1262. doi: 10.1016/j.cell.2014.11.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Mueller NT, Bakacs E, Combellick J, Grigoryan Z, Dominguez-Bello MG. The infant microbiome development: mom matters. Trends Mol Med. 2015;21:109–117. doi: 10.1016/j.molmed.2014.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lewis ZT, et al. Maternal fucosyltransferase 2 status affects the gut bifidobacterial communities of breastfed infants. Microbiome. 2015;3:13. doi: 10.1186/s40168-015-0071-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Huda MN, et al. Stool microbiota and vaccine responses of infants. Pediatrics. 2014;134:e362–72. doi: 10.1542/peds.2013-3937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Subramanian S, et al. Persistent gut microbiota immaturity in malnourished Bangladeshi children. Nature. 2014;510:417–421. doi: 10.1038/nature13421. A machine learning approach was used to identify a group of bacterial strains that together define a program of ‘normal’ microbiota development share by biologically unrelated individuals who were members of birth cohort of healthy Bangladeshi infants/children. This program is perturbed in undernourished children in ways that yield microbial communities that appear significantly younger (more immature) than those of chronologically age-matched healthy individuals. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Fukuda S, et al. Bifidobacteria can protect from enteropathogenic infection through production of acetate. Nature. 2011;469:543–547. doi: 10.1038/nature09646. [DOI] [PubMed] [Google Scholar]
- 80.Sugahara H, Odamaki T, Hashikura N, Abe F, Xiao JZ. Differences in folate production by bifidobacteria of different origins. Biosci Microbiota Food Health. 2015;34:87–93. doi: 10.12938/bmfh.2015-003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Garrido D, Dallas DC, Mills DA. Consumption of human milk glycoconjugates by infant-associated bifidobacteria: mechanisms and implications. Microbiology. 2013;159:649–664. doi: 10.1099/mic.0.064113-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Garrido D, et al. Comparative transcriptomics reveals key differences in the response to milk oligosaccharides of infant gut-associated bifidobacteria. Sci Rep. 2015;5:13517. doi: 10.1038/srep13517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Marcobal A, et al. Bacteroides in the infant gut consume milk oligosaccharides via mucus-utilization pathways. Cell Host Microbe. 2011;10:507–514. doi: 10.1016/j.chom.2011.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.De Leoz ML, et al. Human milk glycomics and gut microbial genomics in infant feces show a correlation between human milk oligosaccharides and gut microbiota: a proof-of-concept study. J Proteome Res. 2015;14:491–502. doi: 10.1021/pr500759e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Ng KM, et al. Microbiota-liberated host sugars facilitate post-antibiotic expansion of enteric pathogens. Nature. 2013;502:96–99. doi: 10.1038/nature12503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Frese SA, Mills DA. Should infants cry over spilled milk? Fecal glycomics as an indicator of a healthy infant gut microbiome. J Pediatr Gastroenterol Nutr. 2015;60:695. doi: 10.1097/MPG.0000000000000798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Frese SA, Parker K, Calvert CC, Mills DA. Diet shapes the gut microbiome of pigs during nursing and weaning. Microbiome. 2015;3:28. doi: 10.1186/s40168-015-0091-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Shin NR, Whon TW, Bae JW. Proteobacteria: microbial signature of dysbiosis in gut microbiota. Trends Biotechnol. 2015;33:496–503. doi: 10.1016/j.tibtech.2015.06.011. [DOI] [PubMed] [Google Scholar]
- 89.Blanton LV, et al. Gut bacteria that prevent growth impairments transmitted by microbiota from malnourished children. Science. 2016;351:aad3311. doi: 10.1126/science.aad3311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Schwarzer M, et al. Lactobacillus plantarum strain maintains growth of infant mice during chronic undernutrition. Science. 2016;351:854–857. doi: 10.1126/science.aad8588. [DOI] [PubMed] [Google Scholar]
- 91.Kau AL, et al. Functional characterization of IgA-targeted bacterial taxa from undernourished Malawian children that produce diet-dependent enteropathy. Sci Transl Med. 2015;7:276ra24. doi: 10.1126/scitranslmed.aaa4877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Palm NW, et al. Immunoglobulin A coating identifies colitogenic bacteria in inflammatory bowel disease. Cell. 2014;158:1000–1010. doi: 10.1016/j.cell.2014.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Cox LM, et al. Altering the intestinal microbiota during a critical developmental window has lasting metabolic consequences. Cell. 2014;158:705–721. doi: 10.1016/j.cell.2014.05.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Dogra S, et al. Dynamics of infant gut microbiota are influenced by delivery mode and gestational duration and are associated with subsequent adiposity. mBio. 2015;6:e02419–14. doi: 10.1128/mBio.02419-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Cho I, et al. Antibiotics in early life alter the murine colonic microbiome and adiposity. Nature. 2012;488:621–626. doi: 10.1038/nature11400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Arrieta M, et al. Early infancy microbial and metabolic alterations affect risk of childhood asthma. Science Translational Medicine. 2015;7:307ra152–307ra152. doi: 10.1126/scitranslmed.aab2271. [DOI] [PubMed] [Google Scholar]
- 97.Goyal MS, Venkatesh S, Milbrandt J, Gordon JI, Raichle ME. Feeding the brain and nurturing the mind: Linking nutrition and the gut microbiota to brain development. Proc Natl Acad Sci U S A. 2015;112:14105–14112. doi: 10.1073/pnas.1511465112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Levine MM, Kotloff KL, Nataro JP, Muhsen K. The Global Enteric Multicenter Study (GEMS): impetus, rationale, and genesis. Clin Infect Dis. 2012;55(Suppl 4):S215–S224. doi: 10.1093/cid/cis761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.MAL-ED Network Investigators. The MAL-ED study: A multinational and multidisciplinary approach to understand the relationship between enteric pathogens, malnutrition, gut physiology, physical growth, cognitive development, and immune responses in infants and children up to 2 years of age in resource-poor environments. Clin Infect Dis. 2014;59(Suppl 4):S193–S206. doi: 10.1093/cid/ciu653. A large multi-site birth cohort study that includes an effort to serially sample microbial communities in infants/children in order to identify correlations between microbiota composition/development, postnatal growth phenotypes and other facets of health status. [DOI] [PubMed] [Google Scholar]
- 100.Ngure FM, et al. Water, sanitation, and hygiene (WASH), environmental enteropathy, nutrition, and early child development: making the links. Ann NY Acad Sci. 2014;1308:118–128. doi: 10.1111/nyas.12330. [DOI] [PubMed] [Google Scholar]