Abstract
Background
The chronically instrumented fetal sheep is a widely used animal model to study fetal brain development in health and disease, but no methods exist yet to interrogate dedicated brain cell populations to identify their molecular and genomic phenotype. For example, the molecular mechanisms whereby microglia or astrocytes contribute to inflammation in the brain remain incompletely understood.
New method
Here we present a protocol to derive primary pure microglial or astrocyte cultures from near-term fetal sheep brain, after the animals have been chronically instrumented and studied in vivo. Next, we present the implementation of whole transcriptome sequencing (RNAseq) pipeline to deeper elucidate the phenotype of such primary sheep brain glial cultures.
Results
We validate the new primary cultures method for cell purity and test the function of the glial cells on protein (IL-1β) and transcriptome (RNAseq) levels in response to a lipopolysaccharide (LPS) challenge in vitro.
Comparison with existing methods
This method represents the first implementation of pure microglial or astrocytes cultures in fetal sheep brain.
Conclusions
The presented approach opens new possibilities for testing not only supernatant protein levels in response to an in vitro challenge, but also to evaluate changes in the transcriptome of glial cells derived from a large mammalian brain bearing high resemblance to the human brain. Moreover, the presented approach lends itself to modeling the complex multi-hit paradigms of antenatal and perinatal cerebral insults in vivo and in vitro.
Keywords: Fetus, Brain, Microglia, Astrocytes, LPS, in vitro, in vivo, Primary culture, RNAseq
1. Introduction
Brain injury acquired antenatally remains a major cause of long-term neurodevelopmental sequelae (Saigal and Doyle, 2008). There is growing clinical and experimental evidence for maternal and fetal infection acting via systemic and neuroinflammation to cause fetal brain injury or contributing to in utero asphyxial brain injury with consequences for postnatal health (Rees and Inder, 2005; Murthy and Kennea, 2007; Hagberg et al., 2002; Wang et al., 2006; Gotsch et al., 2007; Fahey, 2008). However, little is known about the roles of brain’s glial cells, microglia and astrocytes, in the initiation and maintenance of neuroinflammation in utero and postnatally.
We developed a protocol to culture pure fetal sheep microglia and astrocytes and expose them under in vitro conditions to a stimulus of choice, such as lipopolysaccharide (LPS), allowing a more mechanistic study of their function.
2. Protocol
2.1. Ethics statement
This study was carried out in strict accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health. The respective in vivo and in vitro protocols were approved by the Committee on the Ethics of Animal Experiments of the Université de Montréal (Permit Number: 10-Rech-1560).
2.2. Anesthesia and surgical procedure
We refer the reader to our previous report (Burns et al., 2015).
2.3. In vivo experimental protocol
The in vivo protocol has been reported elsewhere (Burns et al., 2015). Briefly, postoperatively, all animals were allowed 3 days to recover before starting the experiments. On these 3 days, at 9.00 am 9:00 am 3 ml arterial plasma sample were taken for blood gases and cytokine analysis. The in vivo protocol can vary depending on the experimental design. Therefore, it is not presented here.
2.4. Fetal sheep brain tissue dissection and cells isolation
Fetal sheep brain tissues were obtained during sheep autopsy after completion of the experiment for in vitro study (Fig. 1 a). The non-instrumented, untreated twins were designated “naïve” (NC, no exposure, such as LPS, or drug treatment in vivo) and NL when exposed to LPS in vitro for the first time. Fetal sheep microglia culture protocol was adapted from an established human adult and fetal microglia culture protocol that was modified to include a myelin removal step following high-speed centrifugation (Durafourt et al., 2013).
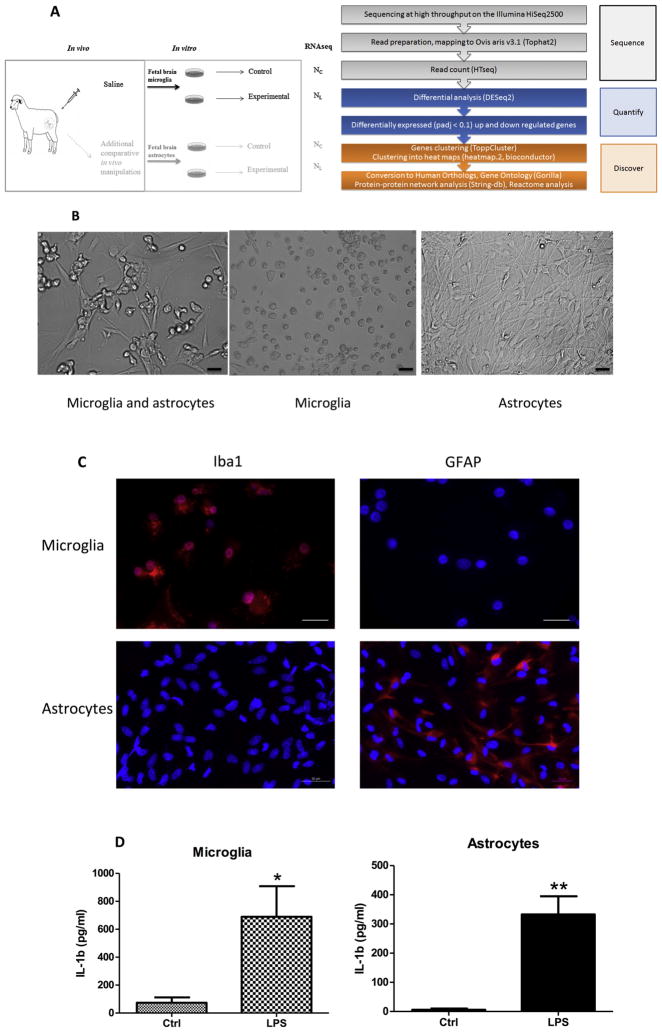
Fig. 1.
(a) In vivo–in vitro experimental design (LEFT) and RNAseq pipeline (RIGHT). In vivo, Control (saline) group is shown; in vitro, cultured cells are derived from the in vivo Control animal, named as Naïve; this simplified design renders two experimental groups: naïve Control (NC) and naïve LPS-exposed (NL). For RNAseq data comparisons, we performed differential expression analysis of NC (n = 3) and NL (n = 3). Cf. (Cao et al., 2015) where a more sophisticated experimental design is presented. (b) Cell collection and purification. Heterogeneous cells were obtained from twin fetal sheep brains and cultured in PLL-coated T75 tissue culture flask at a concentration of 2× 106 cells/ml in DMEM with 5% heat-inactivated fetal bovine serum (FBS) for 1 week; the majority of this cell population were microglia and astrocytes (Left panel). Floating microglial cells were then collected by centrifugation and seeded into 24-well plate at 1 × 105 cells/ml with 5% DMEM for another 7 days for treatment (middle panel). The adherent astrocytes were passaged 4–5 times once a week for further purification; those astrocytes were cultured in new T75 flasks every passage at a concentration of 1–2 × 106 cells/ml in DMEM with 10% FBS; the pure astrocytes were plated into 24-well tissue culture plate at 1 × 105 cells/ml with 10% DMEM for another 7 days for treatment (Right panel). Note cell morphological difference between round microglia (middle) versus elongated astrocytes (right). Images were taken with an inverted light microscope, magnification 20× for all 3 images, scale bar = 50 μm. (c) Purity validation of fetal sheep brain primary microglia and astrocytes cultures. Photomicrographs (ICC) confirming cell purity. Iba1+ staining in microglia vs. undetectable GFAP signal in microglia indicating no contamination with astrocytes in the culture (upper panel). The reverse is seen for the primary astrocyte cultures (lower panel). Scale bar = 50 μm. Magnification 40× for all images. (d) Effect of in vitro LPS treatment on microglial and astrocyte IL-1β production in conditioned media indicating cells were responding adequately to the LPS stimulus. Cultured cells were derived from in vivo Control or twin (Naïve), cell culture condition and IL-1b ELISA assay as described in the text. Results are expressed as mean ± SEM (n = 6 for microglia, n = 3 for astrocyte); data were analyzed by t-test (*P < 0.05, ** P < 0.01 compared with Control group).
-
2.4.1)
In the autopsy room, extract the entire brain from fetal skull. Weigh the brain
-
2.4.2)
Immerse brain with 70% ethanol in a plastic container for about 1 min, then rinse with cold saline 3 times to remove ethanol
-
2.4.3)
Transport brain with fresh saline in a container on ice to tissue culture (TC) hood that has been thoroughly sprayed with 70% ethanol
-
2.4.4)
In TC hood, cut brain in two hemispheres. Put brain onto a large Petri dish (150 mm × 15 mm) to process, place another hemisphere in cold saline in container. Keep on ice
-
2.4.5)
Remove all meninges quickly and efficiently to avoid fibroblast contamination, remove brainstem and white matter, only use cortex
-
2.4.6)
Transfer the cleaned brain cortex on a new Petri dish. Cut brain into <1 mm pieces (as pulp), using scalpels
-
2.4.7)
Transfer brain tissue to a 100 ml glass bottle, add 1× PBS to a final volume of about 60 ml
-
2.4.8)
Add 8 ml of 2.5% trypsin (10×, Life Technologies, Cat. No. 15090-046) and 8 ml of DNase (1 mg/ml, Roche Diagnostics, Cat. No. 10104159001) into the brain tissue (10× Trypsin and DNase aliquots in 4 ml each tube stored at −20 °C).
-
2.4.9)
Place bottle in shaking water bath and incubate at 37 °C, 180 rpm for 30 min.
-
2.4.10)
Add 10%FCS after digestion to stop tissue digestion (add 8 ml FCS in about 80 ml).
-
2.4.11)
Prepare the mesh filter (140 μm, autoclaved), place it over a 500 ml bottle to collect tissue suspension.
-
2.4.12)
Pour a small proportion of tissue mixture onto filter every time, use a 20 ml syringe to push down and let the mix tissue pass through the filter.
-
2.4.13)
Rinse mesh filter with PBS every time, until all the tissue passed through.
-
2.4.14)
Pour filtrate into 50 ml Falcon tubes (maximal 14 tubes, limited by centrifuge model), top each tube to 50 ml with PBS. Centrifuge at 1000 rpm for 15 min.
-
2.4.15)
While centrifuging, prepare same numbers of high-speed centrifuge glass tubes (round-bottom screw-cap, Nalgene, VWR 21009-681) by adding 9 ml of Percoll (VWR, CA95021-202L) each.
-
2.4.16)
After centrifugation, the supernatant should be clear and the pellet should not be loose; if they are loose, re-centrifuge.
-
2.4.17)
Carefully discard the supernatant. Top up to 20 ml with PBS in 50 ml Falcon tubes (maximum volume in a round-bottom screw-cap glass tube is 30 ml, considering 9 ml Percoll already in the glass tube; adjust volume with PBS when balancing tubes.)
-
2.4.18)
Use a serological pipette to re-suspend the pellet. Use the same pipette to transfer 10 ml and then 11 ml of tissue suspension (21 ml total) to the glass tube with 9 ml Percoll; cap the tubes.
-
2.4.19)
Weigh tubes (outside TC hood). Weigh by placing the tubes in a beaker on a scale.
-
2.4.20)
Balance tubes (inside TC hood). Pair up tubes based on weights so that both sides of the centrifuge rotor are equivalent; balance to a maximum difference of 0.05 g. If the weights are too different, top up to a maximum of 1 ml PBS, or transfer some suspension from one tube to another.
-
2.4.21)
Prepare a high-speed centrifuge (Beckman J2-21 M/E). Spray centrifuge rotor with 70% ethanol; put balanced tubes in the rotor.
-
2.4.22)
Centrifuge at 15,000 rpm (31,000g), 4 °C for 30 min, actually take about 45 min due to brake off.
-
2.4.23)
During centrifugation, prepare PLL-coated flask (poly-lysine coated, use 1:100 dilution PLL, 200 μl PLL stock in 20 ml dH2O at 37 °C for 1 h, dry for 1–2 h).
-
2.4.24)
After centrifugation, take tubes back to TC hood. Prepare as many new 50 ml Falcon tubes as there are round-bottom glass tubes (ideally 14 tubes).
-
2.4.25)
Aspirate myelin (fatty white top layer and cloudy layer under fatty layer), using a glass Pasteur pipette attached to a vacuum setting in the TC hood.
-
2.4.26)
Use a pipette to transfer as much of the clear middle layer as possible into new 50 ml Falcon tubes, without taking any of the red blood cells that settled at the tube bottom (reduce pipette speed from F to M).
-
2.4.27)
Once the clear layer has been transferred, add up to 50 ml PBS; invert tubes few times to mix.
-
2.4.28)
Centrifuge at 1200 rpm for 10 min on high brakes to remove Percoll.
-
2.4.29)
In the mean time, warm up 5% DMEM bottle at 37 °C water bath (for 500 ml all-dressed DMEM, adding 25 ml FCS (5% for microglia, *10% for astrocyte*), 5 ml glutamine (1%), 100 μl Penicillin/Streptomycin antibiotics, 10,000 U/ml).
-
2.4.30)
After centrifugation, the pellet will be extremely small. Pour off all the supernatant carefully, although the pellet is a lot more stable at this point.
-
2.4.31)
Add approximately 5 ml of PBS to the 1st tube, re-suspend the pellets one by one, and rinse each tube very well with PBS before combining them. Combine all into one tube, top up to 50 ml with PBS.
-
2.4.32)
Re-centrifuge the tube at 1200 rpm for 15 min.
-
2.4.33)
Remove as much of the supernatant as possible and add 2 ml DMEM to re-suspend the cell pellet.
-
2.4.34)
Cell counting: To estimate the cell numbers, pipette 10 μl cell suspension and mix with 10 μl of Trypan Blue dye in well (e.g., 96-well plate), take 10 μl of the mixture onto the chamber (between groove) of the haemocytometer. Under microscope, count cell numbers in the middle grid (25 small grids) and calculate the average number: Total cell numbers = (cell counted) × 2 × 104 × 2 ml (Note: *2 is dilution factor*. For example, cell count of 300 yields your total cell number in this prep to be 12 million.)
-
2.4.35)
Re-suspend cells to a concentration of 2 million cells per ml with all-dressed DMEM media.
-
2.4.36)
Plate cells in the pre-coated tissue culture flask (T75 or T175). Label flask with sheep ID number, date and your name. Put the flask into a 37 °C, 5% CO2 tissue culture incubator.
-
2.4.37)
Clean and sanitize TC hood with 70% ethanol. Bleach all used glass tubes, mesh filter and glassware overnight and then wash, rinse, and bake them for re-use.
2.5. In vitro microglia culture protocol
-
2.5.1)
late fetal sheep brain mixed cells on poly-L-lysine (PLL)-coated tissue culture T75 flasks at a concentration of 2 × 106 cells/ml in DMEM with 5% heat-inactivated fetal bovine serum (Gibco, Canada Origin), 1% penicillin/streptomycin, and 1% glutamine (5% DMEM), in which microglia are preferable to grow (Durafourt et al., 2013).
-
2.5.2)
Incubate cells for seven days at 37 °C, 5% CO2, followed by media change by centrifugation and addition of re-suspended cells back to the culture flask (Fig. 1b, Left panel)
-
2.5.3)
Continue to incubate cells for seven more days with 5% DMEM at 37 °C, 5% CO2, before floating cells were collected
-
2.5.4)
Carefully collect the floating microglia to avoid contamination with astrocytes and oligodendrocytes, incubate the cells in 24-well plate at 1 × 105 cells/ml with 5% DMEM for another 7 days (Fig. 1b, Middle panel), and then treat with or without LPS (100 ng/ml, Sigma L5024, from E. coli O127, B8) for 6 h. We chose LPS here as an example of an in vitro stimulus; this can be changed depending on your research question.
-
2.5.5)
Collect cell conditioned media for cytokine analysis; use Qiagen RNeasy Mini Kit (Cat. No. 74104) for RNA extraction
-
2.5.6)
Document cell morphology with light microscopy
-
2.5.7)
To verify microglia purity, plate another portion of floating cells into Lab-Tek 8 well chamber glass slide (Thermo Scientific) for immunocytochemistry (ICC) analysis, use Iba1 as microglia cell marker, counterstain cells with Hoechst.
2.6. In vitro astrocyte culture protocol
-
2.6.1)
Astrocytes are the major adherent cell population in flask as described above in list section 2.5.4. Purify astrocytes by passage into a new T75 flask (no PLL-coated is required) for 4–5 times before any manipulations and treatments.
-
2.6.2)
After floating microglia collection, detach the adherent cells by trypsinization (Trypsine 0.25% + EDTA 0.1%, Wisent Cat. No 325-043-EL) and re-plate into a new flask; culture for another 7 days with 10% all-dressed DMEM (see list section 2.4.29).
-
2.6.3)
Repeat step 2.6.2 for 4–5 times; the cells passage procedure takes 4–5 weeks until purified astrocytes can be used for treatment.
-
2.6.4)
Plate pure astrocytes into 24-well plate at 1 × 105 cells/ml with 10% DMEM for another 7 days (Fig. 1b, Right panel), and then treat with or without LPS for 6 h. We chose LPS here as an example of an in vitro stimulus; this can be changed depending on your research question.
-
2.6.5)
Similar to microglia culture, collect cell-conditioned media for cytokine analysis; use Qiagen RNeasy Mini Kit (Cat no 74104) for RNA extraction.
-
2.6.6)
To verify astrocytes purity, plate a portion of cells into Lab-Tek 8 well chamber glass slide (Thermo Scientific) for ICC analysis. Use GFAP as an astrocytes maker, counterstain cells with Hoechst.
2.7. Measurement of cytokines in cell culture media
-
2.7.1)
Determine cytokine concentrations (IL-1β) in cell culture media using an ovine-specific sandwich ELISA. Pre-coat mouse anti-sheep IL-1β monoclonal antibody (capture antibody, MCA1658, Bio Rad AbD Serotec at a concentration 4 μg/ml on 96-well ELISA plate (Nunc maxisorp, high capacity microtitre wells, Fisher Cat. No. 439454)) at 4 °C overnight.
-
2.7.2)
Next day, wash 3 times with washing buffer (0.05% Tween 20 in PBS, PBST)
-
2.7.3)
Block the plates for 1 h at room temperature with 10% FCS in PBST followed by 3 times washing
-
2.7.4)
Use recombinant sheep IL-1β protein (Protein Express Cat. no 968-405) as ELISA standard
-
2.7.5)
Load 50 μl of serial diluted protein standards and samples per well and incubate for 2 h at room temperature; wash the Plates 3 times. Run all standards and samples in duplicates
-
2.7.6)
Apply 50 μl of rabbit anti-sheep IL-1β polyclonal antibody (detection antibody, AHP423, Bio Rad AbD Serotec) at a dilution of 1:250 in wells and incubate for 30 min at room temperature. Wash the plates with washing buffer 5 times.
-
2.7.7)
Add 50 μl of the goat anti-rabbit IgG-HRP conjugated (dilution 1:5000, Jackson ImmunoResearch, Cat. No. 111-035-144) for 30 min
-
2.7.8)
Incubate with 50 μl of TMB substrate solution per well (BD OptEIA TMB substrate Reagent Set, BD Biosciences Cat. No. 555214)
-
2.7.9)
Stop colour development reaction at desired time with 25 μl of 2N sulphuric acid
-
2.7.10
Read the plates on ELISA plate reader at 450 nm, with 570 nm wavelength correction (EnVision 2104 Multilabel Reader, Perkin Elmer)
-
2.7.11
In this assay, the sensitivity of IL-1β ELISA was 41.3 pg/ml, the intra-assay and inter-assay coefficients of variance was <5% and <10%, respectively
2.8. Immunocytochemistry (ICC) analysis
-
2.8.1)
Culture and treat cells in chamber slides (microglia see 2.5.7 or astrocyte see 2.6.6); after media removal, fix with 1% formaldehyde for 10 min
-
2.8.2)
Gently wash 3 times for 5 min each with PBS, then block with PBST (PBS with 0.3% Triton-100) for 20 min
-
2.8.3)
For microglia, incubate cells for 1 h in dark with rabbit anti-human Iba1 (a microglia marker, Wako Cat. No. 019-19741) antibody at a dilution of 1: 250, together with Hoechst (Invitrogen H3569, dilution 1:5000), wash 3 times for 5 min each.
-
2.8.4)
Incubate cells with Alexa 568 (goat anti-rabbit, Life Tech Cat. No. A11036, dilution 1: 400) for 1 h
-
2.8.5)
After washing, mount with Fluoromount-G (Southern-Biotech, Cat. No. 0100-01) before imaging
-
2.8.6)
For microglia purity test, in control wells incubate astrocytes with GFAP-Cy3 (Sigma C9205) at a dilution of 1:500, as negative control, whereas for astrocytes purity test, those astrocytes wells are positive (Fig. 1c).
-
2.8.7)
Capture images with an immunofluorescence inverted microscope (Carl Zeiss Axiovert 200)
2.9. RNAseq approach
To demonstrate the RNAseq approach, a total of 6 samples from 3 sets of replicates were selected for RNA sequencing at high throughput on next generation sequencer, of which 3 samples were controls and 3 were exposed to LPS in vitro.
-
2.9.1)
To extract and quantify RNA, extract the total RNA from cultured microglia using TRIzol Reagent (Life Technologies)
-
2.9.2)
Establish RNA quantity and quality (RNA integrity number, RIN) using a RNA Nano Chips (Agilent RNA 6000 Nano Chips) with Agilent 2100 BioAnalyzer. For quality control, all samples should have a RIN value above 6. In our experiment all samples had a RIN value above 6 except for one sample having RIN = 5.5 but an acceptable 84% of transcripts mapped which did not affect the read count for this sample.
-
2.9.3)
Prepare RNAseq libraries using Illumina TruSeq RNA Sample Preparation v2 kit (Illumina) and perform quality control on the BioAnalyzer
-
2.9.4)
Perform single-end 100-bp sequencing on an Illumina HiSeq2500 or higher (Fig. 1a, Right panel)
2.10. RNAseq data analysis
-
2.10.1)
Reads alignment to the reference genome
-
2.10.1.1)
To maximize the amount of genes covered, map raw data to the reference genome of the sheep Ovis aris v3.1 from NCBI and Ensembl (GCA 000298735.1) as transcriptome reference, or newer version if available.
-
2.10.1.2)
Build the index of the reference fasta file with Bowtie2 (Langmead and Salzberg, 2012), then trim the adaptors of the fastQ files with TrimGalore, and map the reads to the reference genome with Tophat2 (Kim et al., 2013).
-
2.10.1.3)
From the aligned reads from Tophat2, count the number of reads per gene with HTseq and assemble into a matrix containing the read count of each gene per sample for further processing in R (Anders et al., 2014).
-
2.10.2)
Normalization and transcriptome analysis.
-
2.10.2.1)
Use DESeq2 to normalize the dataset, generate log2-fold changes and adjacent P values (padj) (Love et al., 2014). In order to minimize the false discovery rate to less than 10%, threshold the dataset of differentially expressed (DE) genes to padj < 0.1. The dataset can also be filtered for more stringent Log2 fold change of, for example, greater than |2| (Fig. 2). In our experiment, in order to find DE gene in the 3 in vitro LPS challenged samples versus 3 controls (Fig. 1a), a gene was considered DE if its adjacent p-value was strictly lower than 0.1; no criteria selection was applied to Log2 fold change. We then use the current list of DE genes for downstream analysis.
-
2.10.2.2)
Cluster the pools of DE up and down regulated genes and visualize them in heat maps generated in R using the log2 normalized counts and the heatmap.2 method of the gplots library (Warnes, 2008).
-
2.10.3)
Gene selection and Gene Ontology (GO): The sheep genome is not yet supported by most GO platforms, therefore, we recommend to perform the downstream analyses with orthologs in the human genome Homo sapiens.
-
2.10.3.1)
To select relevant genes among up and down regulated DE genes, perform enrichment analysis with ToppGenes and ToppCluster (Chen et al., 2009; Kaimal et al., 2010) for functional annotation enrichment pathways, biologic process and pathway with FDR p < 0.05.
-
2.10.3.2)
Generate protein–protein interaction networks with the STRING database; in our experiment, disconnected nodes were not represented (Franceschini et al., 2013).
-
2.10.3.3)
Perform GO analysis with Gorilla; select significant networks (P < 0.03) for further analysis (Bauer et al., 2008; Eden et al., 2009). Functional classification of selected genes can be viewed in pie chart with Panther (Mi et al., 2016).
-
2.10.3.4)
Validate selected DE genes by qRT-PCR as shown in our previous publication (Cao et al., 2015). We suggest to perform qRT-PCR validation against GADPH for significant DE genes, however potential causal network and interaction relationships should be validated by additional experiments.
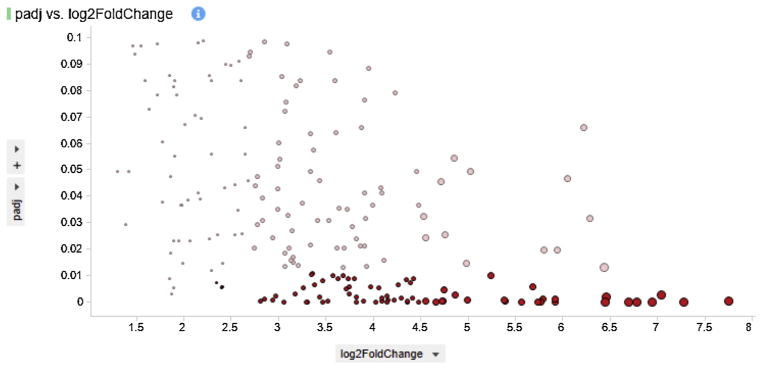
Fig. 2.
Visualization of dataset size variation with threshold variation of Log2 fold change and adjacent p-value. The analysis of 205 differentially expressed (padj < 0.1) up regulated genes can be narrowed down by two-fold selection and higher FDR value (pad < 0.01), as shown here with significant genes highlighted in red.
2.11. Statistical analyses
Cytokine responses in cell culture supernatant were analyzed using GraphPad Prism 5.0 (GraphPad Software, Inc, San Diego, CA). The results were presented as means ± SEM of at least 3 independent replicates as detailed in the figure legend. Data were analyzed by t-test within the same cell type, statistical significance was defined at p < 0.05 (*).
3. Results
3.1. Primary fetal sheep microglia and astrocytes culture
In vitro studies were conducted in primary cultures derived from six controls (naïve) animals. We were able to perform 1–2 in vitro replicates per each animal depending on cell numbers obtained. The total cell numbers per preparation varied from 24 to 65 million cells per brain and were heterogeneous, with the majority of microglial cells and astrocytes (Fig. 1b, Left panel), but also including oligodendrocytes and neurons (data not shown) (Cao et al., 2015).
To enrich for microglia or astrocytes we subjected the cells to a second step as detailed in Methods. Primary microglia were round in shape, whereas astrocytes were elongated in morphology (Fig. 1b, Middle and Right panels). To verify cell culture purity, we performed ICC with microglia and astrocyte markers confirming that the isolated primary microglia and astrocytes were high (Fig. 1c).
Next, we investigated cytokine secretion properties of these cells in the absence or presence of LPS. We found that in vitro LPS administration resulted in increased IL-1β compared to control in both cultured microglia (p < 0.05) and astrocytes (p < 0.01, Fig. 1d), indicating cells were responding adequately to the stimulus.
3.2. RNAseq approach
We focused the transcriptome analysis on microglia only leaving astrocytes RNAseq for future studies. We identified a large population of genes as differentially expressed. We considered a gene to be DE, if its adjacent p-value was lower than 0.1, equivalent to a false discovery rate of 10%. Also, among the DE genes identified, positive and negative Log2 fold changes were interpreted as up and downregulation, respectively. Applying more stringent conditions such as lowering the adjacent p-value and, or, increasing the threshold of Log2 fold change can decrease the population size of DE genes for further downstream analysis (Fig. 2).
3.3. General overview of the whole transcriptome sequencing
We sequenced the transcriptome of naïve microglia and, as a quality control, we tested the expression levels of GFAP and TNFα across our comparisons. We confirmed that all cells in our platform shared the same gene expression characteristics of microglia (Table 1). As a control measure for cell purity, our data confirmed the presence of TGF-μ1 in each sample, as previously reported (Butovsky et al., 2014).
Table 1.
Gene expression summary in naïve microglia. Differential analysis of the count data was done with the DESeq2 package; up regulated genes are highlighted in red and down regulated genes are highlighted in brown. Values in the column “naïve cells” correspond to fold change from the NC to NL (Cao et al., 2015).
| Relevance | Gene name (Human Orthologs) | Naïve cells |
|---|---|---|
| AMPK signaling pathway | PRKAA1 | 0.169 |
| PRKAA2 | 2.776 | |
| PRKAB1 | −0.189 | |
| PRKAB2 | 0.861 | |
| PRKAG1 | −0.379 | |
| PRKAG2 | −0.405 | |
| PRKAG3 | −1.329 | |
| Leptin | LEP | 5.033 |
| Adrenoceptor alpha 1A | ADRA1A | 2.572 |
| Adipocytokine signaling pathway | CAMKK1 | −1.110 |
| CAMKK2 | 0.660 | |
| Adiponectin | ADIPOQ | 2.422 |
| Calcium binding protein 39 | CAB39 | 0.279 |
| CAB39L | −0.076 | |
| Serine/threonine kinase 11 | STK11 | −0.178 |
| STE20-related kinase adaptor alpha | STRADA | −0.536 |
| STRADB | −0.506 | |
| Tak1 protein | MAP3K7 | −0.057 |
| Insulin signaling pathway | IGF1 | 1.601 |
| IGF1R | −0.125 | |
| IRS1 | 1.241 | |
| IRS4 | 4.420 | |
| PIK3-Akt signaling pathway | PIK3CA | 0.018 |
| PIK3CB | 1.793 | |
| PIK3CG | 1.950 | |
| PIK3R1 | 1.330 | |
| PIK3R3 | 1.083 | |
| PIK3R5 | 4.343 | |
| PDPK1 | 0.915 | |
| AKT2 | −0.335 | |
| AKT3 | 0.350 | |
| TSC1 | 1.099 | |
| TSC2 | 0.219 | |
| mTOR signaling pathway | RHEB | −0.257 |
| AKT1S1 | 0.908 | |
| MTOR | −0.107 | |
| RPTOR | 0.298 | |
| Inhibit cell growth and protein synthesis | RPS6KB1 | −0.078 |
| RPS6KB2 | −0.569 | |
| EIF4EBP1 | −0.822 | |
| PPARG2 | −2.093 | |
| Inhibit protein synthesis | EEF2 | −0.005 |
| EEF2K | −0.394 | |
| Growth arrest | ELAVL1 | −0.252 |
| CCNA1 | 2.574 | |
| CCNA2 | 0.143 | |
| Activated mitochondrial biogenesis | SLC2A4 | 6.386 |
| PPARGC1A | 4.842 | |
| Increased FFA oxidation | CPT1A | −0.260 |
| CPT1B | 0.496 | |
| CPT1C | 0.608 | |
| Gluconeogenesis and glucolysis | ALDOA | −0.106 |
| ALDOB | −1.659 | |
| ALDOC | −0.266 | |
| PFKP | −0.461 | |
| GPI | −0.940 | |
| Fructose-1,6-Biphosphate | FBP | −0.792 |
| NO production | NOS1AP | 0.386 |
| JNK/P38 MAPK | MAPK8 (JNK) | 0.544 |
| MAPK9 (JNK) | −0.266 | |
| MAPK10 (JNK) | 1.492 | |
| MAPK12 (P38) | −0.905 | |
| MAPK13 (P38) | 3.294 | |
| MAPK14 (P38) | −0.848 | |
| Transcription factors | C-JUN (AP1) | 0.795 |
| NFKB | 2.676 | |
| CREB1 | 0.782 | |
| ATF4 (creb TF) | 0.968 | |
| CEBPB (CEBP) | 0.586 | |
| Iron metabolism and/or anti-inflammatory | HMOX1 | −2.686 |
| NRF-2 | 0.855 | |
| P53 | TP53 | −0.554 |
| MDM4 | 3.489 | |
| Toll-like receptor 4 | TLR4 | 0.807 |
| LY96 (MD-2) | 1.213 | |
| LBP | 4.472 | |
| NF-kB signaling and inflammation | RELB | 1.503 |
| NFKB | 2.676 | |
| NFKBIA | 2.578 | |
| NFKB1 (p50) | 2.569 | |
| RELA (p65) | −0.031 | |
| PTGS2 | 5.166 | |
| TNF | 4.990 | |
| PTGS2 | 5.166 | |
| IL8 | 4.779 | |
| IL1B | 7.578 | |
| TNFAIP3 | 2.628 | |
| Lymphoid-tissue homing | CCL21 | 18.917 |
| CCL19 | 5.439 | |
| Myeloiesis and B-cell lymphopoiesis | CXCL12 (SDF-1alpha) | −0.545 |
| B-cell development and survival | TNFSF13B (BAFF) | 2.562 |
| Lymphocyte adhesion, T-cell costimulation | ICAM-1 | 4.055 |
| Initiators of the JAK-Stat pathway (JAK) | JAK1 | 0.213 |
| JAK2 | 2.289 | |
| JAK3 | 2.965 | |
| TYK2 | 0.948 | |
| STAT genes | STAT1 | −0.136 |
| STAT2 | 1.276 | |
| STAT3 | 0.660 | |
| STAT5A | 3.365 | |
| STAT5B | 1.554 | |
| Growth, proliferation, fate determination, development, immunity | IRF9 (p48) | 0.407 |
| PIM1 | 3.108 | |
| EP300 (CBP) | 0.784 | |
| CREBBP (CBP) | 0.881 | |
| CISH (CIS) | 7.170 | |
| Fractalkine/CX3CR1 axis and biological signature of microglial cells | CX3CR1 | 2.017 |
| CX3CL1 | 0.618 | |
| ITGAM (CD11b) | 0.13 | |
| IL1B | 7.578 | |
| Potential epigenetic regulator | HDAC1 | 2.271 |
| LRP phagocytosis signalling | LRP1B | 4.522 |
| LRP2 | 4.860 | |
| LRP6 | 1.052 | |
| Quality Control | TGFBR1 | 0.327 |
| TGFβ | −0.419 | |
| GFAP | −1.044 | |
| ITGAM (CD11b) | 0.130 | |
| CD40 | 4.656 | |
| IBA1 (AIF1) | 0.060 |
Bold values correspond to significant log2 fold change (padj < 0.1).
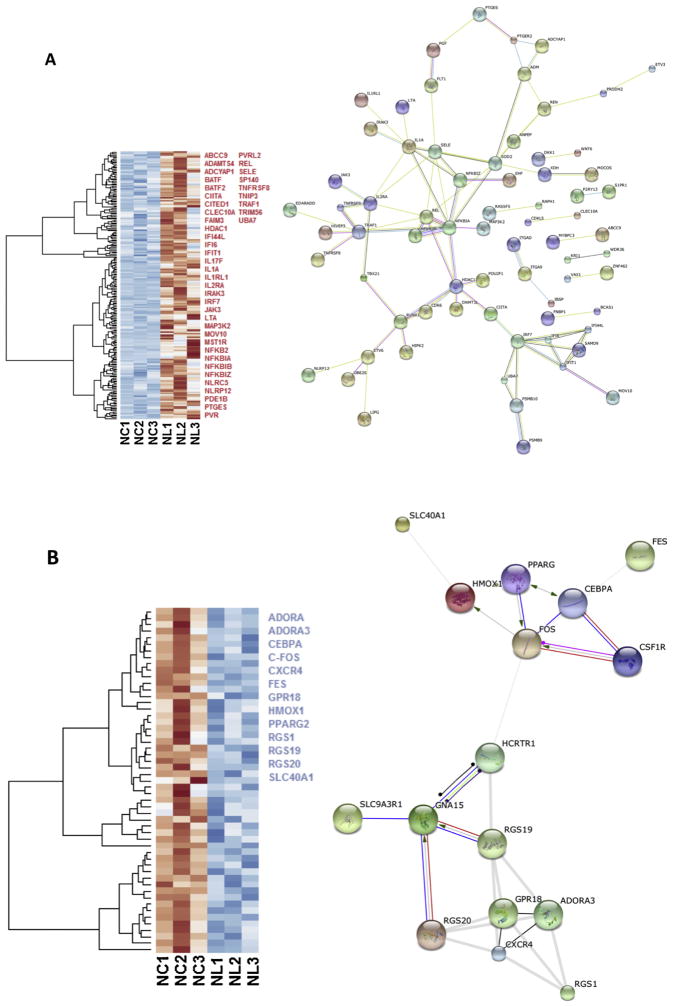
We compared gene expression between the naïve controls and naïve LPS-exposed microglial cells and found 258 differentially expressed genes (padj < 0.1), among which, 205 genes were up regulated and 53 were down regulated. We selected relevant differentially expressed genes with ToppCluster (LogP > 4.00) based on their role in the immune response (Fig. 3a, b).
Fig. 3.
Heat maps of the gene expression in microglia cells exposed to LPS. Selected up regulated (red) and down regulated (blue) genes are listed; genes were selected with ToppCluster based on their significance in the immune response (logP > 4.00) (a) Heat map of 205 differentially expressed (padj < 0.1) up regulated genes (red) in NL microglia. Interaction network highlights the position of NFKB and JAK3 genes. Protein-protein interaction networks were built with the STRING database on human orthologs and disconnected nodes were not representend. (b) Heat map of 53 differentially expressed down regulated genes in NL microglia, among selected genes indicated in blue, HMOX1 was strongly differentially expressed (Log2 = −2.686 and padj = 3.09 × 10−8). In both up and down regulated genes, we observed a different behavior for NL3 that did not affect our differential analysis. On the right, interaction network shows how HMOX1 and FOS are connected. NC = Naïve control microglia, NL = Naïve LPS-exposed microglia (Cao et al., 2015). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
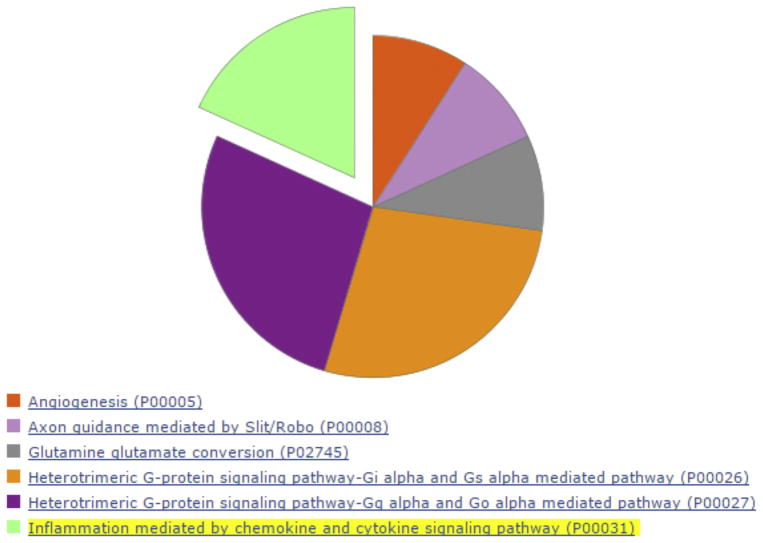
We queried the list of DE genes in the STRING database containing known and predicted protein–protein interactions. Protein-protein network interaction is a statistical, not a causal, representation of a high likelihood that these networks and pathways are either active or inactive together, thus data should be interpreted with care and validated by other means. Our analysis of protein–protein interaction suggests that among DE down regulated genes in naïve microglia, HMOX1 and FOS are likely directly interacting with each other (Fig. 3b). Further downstream analysis through the Reactome (Croft et al., 2014) suggests that the pathway G alpha (i) signaling events (p-Value 9.37 × 10−3) is potentially affected by DE down regulated genes and thus inactive. We also enriched this dataset for Biological process and Molecular function with Gorilla. The immune system process (GO:0002376, p-Value 5.8 × 10−5) seemed likely affected by the down regulation of genes: CSF1R, FOS, CD300E, RGS1, CXCR4, HMOX1, SLC40A1, DOX2, MERTK, PPARG. However further biochemical studies will be necessary to better understand how the downregulation of these genes is important in the immune system homeostasis. Enrichment for pathways with Panther revealed that inflammation mediated by chemokine and cytokine signaling pathway is affected by DE down regulated genes (Fig. 4).
Fig. 4.
DE down regulated genes in naïve microglia are enriched with Panther. Inflammation mediated by chemokine and cytokine signaling pathway is represented in light green. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
3.4. Upregulation of inflammatory and energy-sensing pathways JAK-STAT, NFKB and AMPK
Our GO and protein–protein interaction network analyses of DE genes in naïve microglia also revealed that statistically significant up regulation of inflammatory pathways, NFκB, PIK3-Akt and Jak-STAT was accompanied by a down regulation of metabolic pathways in LPS-induced inflammatory response (Fig. 3a and Table 1).
4. Discussion
We established for the first time an in vivo-in vitro (multi-hit) model of LPS exposure in mammalian microglia and astrocytes to mimic antenatal neuroinflammation. The LPS-induced inflammation is presented as an example of a possible in vivo–in vitro intervention and can be substituted by any other manipulation, such as a viral agent, a drug or an environmental stressor. The isolation of viable and highly purified microglia and astrocytes populations from in vivo exposed or non-exposed brain allowed an in vitro characterization of those cell types.
4.1. In vivo–in vitro model of perinatal inflammation
Experimentally induced inflammation in chronically instrumented non-anesthetized fetal sheep is a well-established and highly translational in vivo model of fetal physiology (Prout et al., 2010, 2012) Primary microglia cultures in different species have been reported for decades (Stansley et al., 2012). We integrated both in vivo and in vitro models into a new, hybrid system adding the layer of the whole genome analysis using RNAseq. The chief advantage of the new in vivo − in vitro model presented here is that it allows us to examine microglia and astrocyte responses to LPS-induced double-hit inflammation in situ and in vitro on integrative physiological, protein and genomic levels and in a physiologically and clinically meaningful context. This approach has the potential to uncover hitherto unseen relationships between brain and immune system on different scales of organization in the perinatal stage of development, which might accelerate discovery of new treatment strategies.
In vitro, we developed a new microglia and astrocyte isolation protocol that combines the human adult and fetal brain microglia isolation protocols (Durafourt et al., 2013) and successfully collected a highly enriched microglia and astrocytes population. The use of a modified cell isolation approach from fetal brain tissue is mainly due to the higher degree of myelination in the adult human brain compared to that of a near-term fetus (Durafourt et al., 2013). We were able to attain high purity of microglia and astrocytes, which we validated by immunocytochemistry. Moreover, RNAseq analysis of microglia showed a consistent and constant low level of expression of the astrocyte marker GFAP further confirming cell purity. We already deployed this approach on microglia (Cao et al., 2015). Here, we present this microglia paradigm in protocol-level detail and, for the first time, report on the methodology required to derive pure astrocyte cultures as well.
4.2. Methodological considerations
Microglia in vitro have been shown to secrete IL-1β preferentially when challenged with LPS, while IL-6 secretion is a hallmark of cultured astrocytes in rat (Gottschall et al., 1994). Our findings are consistent with literature and further support the cell culture purity. Aside of being efficient at achieving cell purity, the physicochemical cell purification method we present helps avoid any spurious alteration of cell properties in vitro that may occur with immunological separation techniques such as beads. Nevertheless, caution must be exerted when translating findings made in primary in vitro cultures onto in vivo systems.
In parallel to our team, the feasibility of creating a mixed primary fetal ovine brain culture has been recently demonstrated (Weaver-Mikaere et al., 2012). We have advanced this work by focusing on late rather than mid-gestation fetuses and creating primary pure microglial culture rather than mixed culture. This allowed us to then study the microglia-specific effects on the secretion profile of the inflammatory cytokine IL-1β and the sequencing at high-throughput of microglia transcriptome as a proof-of-principle.
In our approach, we used DESeq2 to normalize read counts and identify differentially expressed genes. DESeq2 was specifically designed to normalize and estimate the dispersion and differential expression in a dataset containing replicates for both control and treatment samples. Despite quality control measures prior to sequencing, the sample NL3 had a different expression pattern than the two other NL samples. We believe this is not related to RNA quality, and may have been due to environmental or other physiological conditions of the animal that we were not aware of at the time of the experiment. In interrogating the differential gene expression, we have ensured that the partially deviating pattern observed in sample NL3 did not confound our findings (Fig. 3a,b).
5. Conclusions
The presented multi-hit fetal sheep model and the genomic analysis workflow allow studying mechanisms of fetal neuroinflammation or other multi-hit fetal injury paradigms in vivo and in vitro to identify potential biomarkers, mechanisms and therapeutic targets for early postnatal intervention to prevent lasting brain injury.
HIGHLIGHTS.
After decades of relative neglect, microglia and astrocytes (also known as glia) are emerging as key players in understanding the brain function and pathophysiology, well beyond their originally conceived “neuronal support” role. Glia shape neuronal development and function in health and disease.
In fetal brain, so far methods were limited to study glial function.
Here we present a new protocol and paradigm to derive pure primary microglia and astrocytes cultures from a mature fetal sheep brain, a powerful and widely used animal model of human brain development bearing high resemblance to human brain.
This technique allows versatile studies of glia function including assessing the genomic properties using next-generation sequencing approaches.
With such model we aim to provide a useful novel platform to tackle the challenge of understanding the diversity in glial phenotype and function in a large developing brain.
Acknowledgments
The authors thank Dora Siontas, Manon Blain for cell culture and ICC, Lamia Naouel Hachehouche for help with setting up the sheep specific cytokine ELISA assay, Vania Yotova for RNAseq library preparation, Jean-Christopher Grenier for alignment to the reference genome and read count, St-Hyacinthe CHUV team and M. Michel-Robinson for technical assistance.
Funding
Supported by grants from the Canadian Institute of Health Research (CIHR) (MGF); Fonds de la recherche en santé du Québec (FRSQ) (MGF) and Molly Towell Perinatal Research Foundation (MGF).
Footnotes
Competing interests
None of the authors has any conflicts of interests.
References
- Anders S, Pyl TP, Huber W. HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer S, Grossmann S, Vingron M, Robinson PN. Ontologizer 2.0—a multifunctional tool for GO term enrichment analysis and data exploration. Bioinformatics. 2008;24:1650–1651. doi: 10.1093/bioinformatics/btn250. [DOI] [PubMed] [Google Scholar]
- Burns P, Liu HL, Kuthiala S, Fecteau G, Desrochers A, Durosier LD, Cao M, Frasch MG. Instrumentation of near-term fetal sheep for multivariate chronic non-anesthetized recordings. J Vis Exp. 2015;105:e52581. doi: 10.3791/52581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butovsky O, Jedrychowski MP, Moore CS, Cialic R, Lanser AJ, Gabriely G, Koeglsperger T, Dake B, Wu PM, Doykan CE, Fanek Z, Liu L, Chen Z, Rothstein JD, Ransohoff RM, Gygi SP, Antel JP, Weiner HL. Identification of a unique TGF-beta-dependent molecular and functional signature in microglia. Nat Neurosci. 2014;17:131–143. doi: 10.1038/nn.3599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao M, Cortes M, Moore CS, Leong SY, Durosier LD, Burns P, Fecteau G, Desrochers A, Auer RN, Barreiro LB, Antel JP, Frasch MG. Fetal microglial phenotype in vitro carries memory of prior in vivo exposure to inflammation. Front Cell Neurosci. 2015;9:294. doi: 10.3389/fncel.2015.00294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Bardes EE, Aronow BJ, Jegga AG. ToppGene Suite for gene list enrichment analysis and candidate gene prioritization. Nucleic Acids Res. 2009;37:W305–311. doi: 10.1093/nar/gkp427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croft D, Mundo AF, Haw R, Milacic M, Weiser J, Wu G, Caudy M, Garapati P, Gillespie M, Kamdar MR, Jassal B, Jupe S, Matthews L, May B, Palatnik S, Rothfels K, Shamovsky V, Song H, Williams M, Birney E, Hermjakob H, Stein L, D’Eustachio P. The reactome pathway knowledgebase. Nucleic Acids Res. 2014;42:D472–477. doi: 10.1093/nar/gkt1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durafourt BA, Moore CS, Blain M, Antel JP. Isolating culturing, and polarizing primary human adult and fetal microglia. Methods Mol Biol. 2013;1041:199–211. doi: 10.1007/978-1-62703-520-0_19. [DOI] [PubMed] [Google Scholar]
- Eden E, Navon R, Steinfeld I, Lipson D, Yakhini Z. GOrilla: a tool for discovery and visualization of enriched GO terms in ranked gene lists. BMC Bioinform. 2009;10:48. doi: 10.1186/1471-2105-10-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fahey JO. Clinical management of intra-amniotic infection and chorioamnionitis: a review of the literature. J Midwifery Women’s Health. 2008;53:227–235. doi: 10.1016/j.jmwh.2008.01.001. [DOI] [PubMed] [Google Scholar]
- Franceschini A, Szklarczyk D, Frankild S, Kuhn M, Simonovic M, Roth A, Lin J, Minguez P, Bork P, von Mering C, Jensen LJ. STRING v9.1: protein–protein interaction networks, with increased coverage and integration. Nucleic Acids Res. 2013;41:D808–D815. doi: 10.1093/nar/gks1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gotsch F, Romero R, Kusanovic JP, Mazaki-Tovi S, Pineles BL, Erez O, Espinoza J, Hassan SS. The fetal inflammatory response syndrome. Clin Obstet Gynecol. 2007;50:652–683. doi: 10.1097/GRF.0b013e31811ebef6. [DOI] [PubMed] [Google Scholar]
- Gottschall PE, Tatsuno I, Arimura A. Regulation of interleukin-6 (IL-6) secretion in primary cultured rat astrocytes: synergism of interleukin-1 (IL-1) and pituitary adenylate cyclase activating polypeptide (PACAP) Brain Res. 1994;637:197–203. doi: 10.1016/0006-8993(94)91233-5. [DOI] [PubMed] [Google Scholar]
- Hagberg H, Peebles D, Mallard C. Models of white matter injury: comparison of infectious hypoxic-ischemic, and excitotoxic insults. Ment Retard Dev Disabil Res Rev. 2002;8:30–38. doi: 10.1002/mrdd.10007. [DOI] [PubMed] [Google Scholar]
- Kaimal V, Bardes EE, Tabar SC, Jegga AG, Aronow BJ. ToppCluster: a multiple gene list feature analyzer for comparative enrichment clustering and network-based dissection of biological systems. Nucleic Acids Res. 2010;38:W96–102. doi: 10.1093/nar/gkq418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013;14:R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL. Fast gapped-read alignment with bowtie 2. Nat Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-Seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi H, Poudel S, Muruganujan A, Casagrande JT, Thomas PD. PANTHER version 10: expanded protein families and functions, and analysis tools. Nucleic Acids Res. 2016;44:D336–D342. doi: 10.1093/nar/gkv1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murthy V, Kennea NL. Antenatal infection/inflammation and fetal tissue injury. Best Pract Res Clin Obstetr Gynaecol. 2007;21:479–489. doi: 10.1016/j.bpobgyn.2007.01.010. [DOI] [PubMed] [Google Scholar]
- Prout AP, Frasch MG, Veldhuizen RA, Hammond R, Ross MG, Richardson BS. Systemic and cerebral inflammatory response to umbilical cord occlusions with worsening acidosis in the ovine fetus. Am J Obstet Gynecol. 2010;202(82):e81–89. doi: 10.1016/j.ajog.2009.08.020. [DOI] [PubMed] [Google Scholar]
- Prout AP, Frasch MG, Veldhuizen R, Hammond R, Matushewski B, Richardson BS. The impact of intermittent umbilical cord occlusions on the inflammatory response in pre-term fetal sheep. PLoS One. 2012;7:e39043. doi: 10.1371/journal.pone.0039043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rees S, Inder T. Fetal and neonatal origins of altered brain development. Early Hum Dev. 2005;81:753–761. doi: 10.1016/j.earlhumdev.2005.07.004. [DOI] [PubMed] [Google Scholar]
- Saigal S, Doyle LW. An overview of mortality and sequelae of preterm birth from infancy to adulthood. Lancet. 2008;371:261–269. doi: 10.1016/S0140-6736(08)60136-1. [DOI] [PubMed] [Google Scholar]
- Stansley B, Post J, Hensley K. A comparative review of cell culture systems for the study of microglial biology in Alzheimer’s disease. J Neuroinflamm. 2012;9:115. doi: 10.1186/1742-2094-9-115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Rousset CI, Hagberg H, Mallard C. Lipopolysaccharide-induced inflammation and perinatal brain injury. Sem Fetal Neonatal Med. 2006;11:343–353. doi: 10.1016/j.siny.2006.04.002. [DOI] [PubMed] [Google Scholar]
- Warnes GR. Gplots: Various R programming tools for plotting data. 2008. [Google Scholar]
- Weaver-Mikaere L, Gibbons HM, De Silva D, Fraser M. Primary mixed glial cultures from fetal ovine forebrain are a valid model of inflammation-mediated white matter injury. Dev Neurosci. 2012;34:30–42. doi: 10.1159/000338039. [DOI] [PubMed] [Google Scholar]