Abstract
Familial British dementia (FBD) and familial Danish dementia (FDD) are caused by mutations in the BRI2 gene. These diseases are characterized clinically by progressive dementia and ataxia and neuropathologically by amyloid deposits and neurofibrillary tangles. Herein, we investigate BRI2 protein accumulation in FBD, FDD, Alzheimer disease and Gerstmann-Sträussler-Scheinker disease. In FBD and FDD, we observed reduced processing of the mutant BRI2 pro-protein, which was found accumulating intracellularly in the Golgi of neurons and glial cells. In addition, we observed an accumulation of a mature form of BRI2 protein in dystrophic neurites, surrounding amyloid cores. Accumulation of BRI2 was also observed in dystrophic neurites of Alzheimer disease and Gerstmann-Sträussler-Scheinker disease cases. Although it remains to be determined whether intracellular accumulation of BRI2 may lead to cell damage in these degenerative diseases, our study provides new insights into the role of mutant BRI2 in the pathogenesis of FBD and FDD and implicates BRI2 as a potential indicator of neuritic damage in diseases characterized by cerebral amyloid deposition.
Keywords: BRI2, Amyloid, Familial British dementia, Familial Danish dementia, Alzheimer disease, Gerstmann-Sträussler-Scheinker disease
1. Introduction
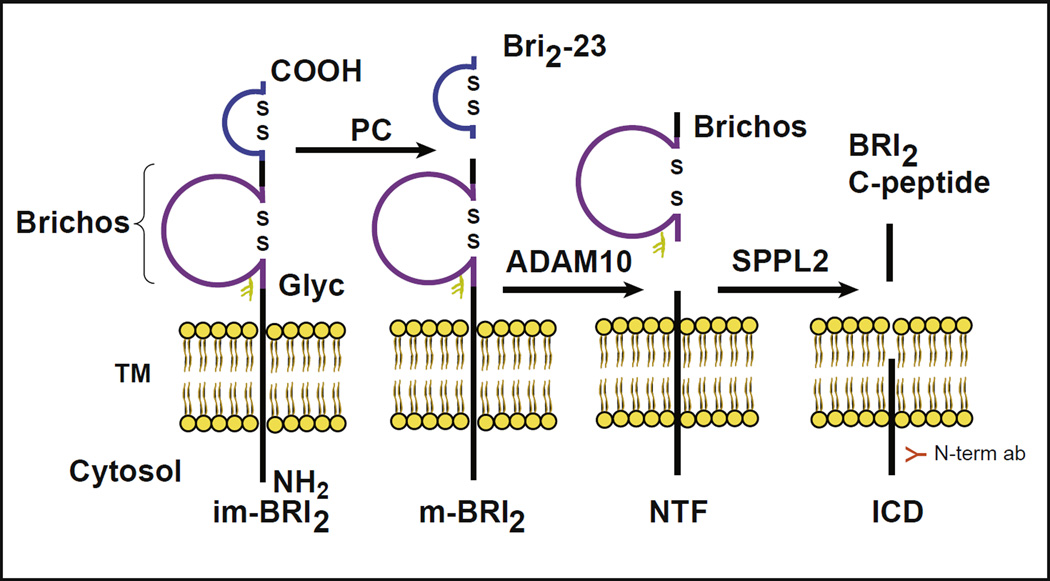
Mutations in the BRI2 gene are associated with familial British dementia (FBD) and familial Danish dementia (FDD; Vidal et al., 1999, 2000). These diseases are characterized clinically by a progressive dementia and ataxia and neuropathologically by amyloid deposition and tau aggregation (Holton et al., 2001, 2002; Vidal et al., 1999, 2000). The BRI2 protein contains 266 amino acids (aa) and belongs to a family of integral type II single trans-membrane domain proteins (Deleersnijder et al., 1996; Vidal et al., 1999, 2001). As other proteins of the regulated pathway, BRI2 is produced as a pro-protein with a pro-peptide sequence that is cleaved by pro-protein convertases (PCs). Cleavage by PCs between the BRI2 ectodomain (residue Arg243 and Glu244, KGIQKR↓EAS; Choi et al., 2004; Kim et al., 1999, 2002) produces a mature protein (mature BRI2 protein or m-BRI2) and a 23 aa C-terminal pro-peptide (Bri2-23; Fig. 1). Ectodomain cleavage also releases the ABri and ADan amyloids (last 34 aa C-terminal peptides) from the mutant BRI2 polypeptides (277 aa). These peptides are transported via regulated secretory pathway through axons for secretion (Choi et al., 2004). Processing of BRI2 is complex, involving several proteases in addition to PCs. The BRI2 pro-protein and m-BRI2 protein may also be cleaved by the disintegrin and metalloproteinase domain-containing protein 10 (ADAM10), releasing a BRICHOS domain (Sanchez-Pulido et al., 2002), and an N-terminal fragment (NTF). The NTF is also the subject of additional proteolysis by signal peptide peptidase-like 2 (SPPL2; Fluhrer et al., 2012), releasing an intracellular domain and a BRI2 C-peptide (Fig. 1; Choi et al., 2004; Garringer et al., 2010, 2013; Martin et al., 2008, 2009). The BRICHOS domain consists of ~ 100 aa and is found in several different type II membrane proteins, including BRI2, Chondromodulin-I, CA11, and surfactant protein C (Sanchez-Pulido et al., 2002; Willander et al., 2011). The domain has 2 conserved cysteine residues that form a disulfide bridge. Disulfide-bonded loops within pro-proteins and neuropeptides have been shown to be important for sorting of peptides from the trans-Golgi network to regulated secretory pathway (Glombik et al.,1999; Krömer et al.,1998). BRI2 has a single N-linked glycosylation site (Asp170) inside the BRICHOS domain that has been shown in vitro to be important for BRI2 trafficking to the cell surface and its steady state levels at the plasma membrane (Tsachaki et al., 2011).
Fig. 1.
Proteolytic processing of BRI2. Scheme showing the processing of the type-II single trans-membrane (TM) domain BRI2 protein. Processing of the pro-protein (or immature protein, im-BRI2) by pro-protein convertases (PCs) generates a 23 amino acid (aa) peptide (Bri2-23) and a mature form of BRI2 (m-BRI2). Processing of the FBD and FDD forms of BRI2 by PCs releases the 34 aa amyloid peptides (ABri and ADan). Processing by ADAM10 in the ectodomain of BRI2 releases the BRICHOS domain (~ 100 aa) and the N-terminal fragment (NTF). The NTF is also the subject of additional proteolysis by SPPL2, releasing an intracellular domain (ICD) and a C-terminal peptide fragment (BRI2 C-peptide). Disulfide-bonded loops in the BRICHOS domain and in the carboxy-terminus of BRI2 (aa 5 and 22 of the Bri2-23 peptide) are indicated as well as the single N-glycosylation site (Glyc) at position 170. The figure shows the location of the epitope recognized by the N-term abs.
The mutant BRI2 pro-protein is not efficiently processed by PCs to generate m-BRI2 and has been found to accumulate intracellularly in a transgenic mouse model of FDD (Tg-FDD; Garringer et al., 2013; Tamayev et al., 2010a,b; Vidal et al., 2009). In addition, dystrophic neurites (DNs) consisting of large globular processes surrounding ADan amyloid cores in Tg-FDD mice also show the presence of BRI2, in agreement with previous immunohistochemical studies that have shown BRI2 deposits in DNs of Alzheimer disease (AD), around ischemic lesions, in torpedoes and grumose changes in the cerebellum, as well as in Lewy neurites, ballooned neurons, and neurons undergoing hypoxia (Akiyama et al., 2004; Baron and Pytel, 2016; Del Campo et al., 2014). DNs in Tg-FDD mice are labeled by antibodies (abs) against the N-terminus of BRI2 but not by abs against the C-terminus of the mutant BRI2 protein (Garringer et al., 2013), suggesting that they contain m-BRI2 rather than immature BRI2 protein.
Herein, we studied BRI2 processing and intracellular accumulation of BRI2 in FBD, FDD, AD, and Gerstmann-Sträussler-Scheinker disease (GSS), diseases characterized neuropathologically by amyloid deposits and tau accumulation. As controls, intracellular accumulation of BRI2 was assessed in familial multiple system tauopathy with presenile dementia (MSTD) and normal individuals. Our studies show that the ectodomain processing of the mutant form of BRI2 by PCs is compromised in FBD and FDD, leading to the intracellular accumulation of immature BRI2 in the Golgi compartment of neurons and glial cells. In addition, we observe the accumulation of m-BRI2 in DNs in association with the presence of amyloid cores in FBD, FDD, AD, and GSS. Accumulation of m-BRI2 was not seen in association with neurofibrillary tangles (NFTs) of FBD, FDD, AD, and GSS, as well as in tau deposits in MSTD. Although it is not known if intracellular accumulation of BRI2 ultimately results in cell damage, these studies provide new insights into the mechanisms of neurodegeneration in brain diseases characterized by amyloid deposition and tau aggregation.
2. Methods
2.1. Generation of expression constructs
The cDNA encoding human wild-type BRI2, and the FBD and FDD mutants forms of BRI2 with the addition of a Kozak sequence and an N-terminal Myc epitope tag were PCR amplified, gel purified, and sub-cloned into pcDNA 3.1 directional TOPO vector (Invitrogen). Plasmids were transformed into DH5alpha cells (Invitrogen), and positive colonies were screened by restriction enzyme analysis and direct DNA sequencing. To introduce an Asp to Ala change at aa 170, site-directed mutagenesis (Agilent Technologies) was done according to the manufacturer’s protocol using primers forward (5′-CTA TGT GAT CCC TCT GGC CAC TTC CAT TGT TAT GC-3′) and reverse (5′-GCA TAA CAA TGG AAG TGG CCA GAG GGA TCA CAT AG-3′).
2.2. Cell culture and transfection
Mouse neuroblastoma N2a cells (ATCC) were maintained in 50% Dulbecco’s modified Eagle’s medium (Lonza) supplemented with 5% fetal bovine serum and 100 units/ml penicillin/streptomycin/amphotericin (Pen/Strep/Amp). Hek293 cells (ATCC) were maintained in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal bovine serum, 100 units/ml Pen/Strep/Amp, and 2-mM L-Glutamine. N2a cells were transiently transfected using LipofectAMINE PLUS (Invitrogen) for 5–6 hours with 2 µg of supercoiled plasmid DNA and analyzed after 48 hours. To generate stable cell lines, Hek293 and N2a cells were transfected using Fugene 6 (Roche). Twenty-four hours following transfection, the media were replaced with selection media, growth media supplemented with 750-ug/mL G418 (Mediatech). Selected expressing clones were used for further analysis.
2.3. Antibodies
For immunohistochemical and biochemical studies, we use antibodies against the ABri and ADan amyloid peptides which also recognize the C-terminus of the immature forms of mutant BRI2 (Vidal et al., 2009). We also used abs against the N-terminus of BRI2: ab14307 (Abcam) and 11–26 (Akiyama et al., 2004). Abs against c-Myc (Santa Cruz) and against beta-actin (AC-15, Sigma) were also used. Secondary abs used for Western blot ECL detection were donkey anti-rabbit IgG, HRP (NA934, GE Healthcare), anti-mouse IgG, HRP (NA931, GE Healthcare), and anti-chicken IgY, HRP (A9046, Sigma-Aldrich). For confocal studies, BRI2 proteins were detected with anti-c-Myc and secondary Alexa Fluor 594 goat anti-mouse (Invitrogen). Calnexin was detected with anti-Calnexin (Abcam) and secondary Alexa Fluor 488 goat anti-rabbit (Invitrogen). GM130 was detected with anti-GM130 (Abcam) and secondary Alexa Fluor 488 goat anti-rabbit (Invitrogen).
2.4. Sample preparation and Western blot
Transfected cells were washed in phosphate-buffered saline (PBS) at 4 °C and lysed in tris-buffered saline containing 1% sodium dodecyl sulfate and protease inhibitors (Complete, Roche). Post-nuclear supernatants were prepared by homogenizing the mouse neocortex in 3 volumes of cold Hepes-sucrose buffer (20-mM Hepes, 1-mM EDTA, 1-mM EGTA, 0.25-M sucrose, all from Sigma) containing protease inhibitors (Complete, 1-mM pepstatin, 100-mM TLCK- HCl, 200-mM TPCK, and 1-mM leupeptin; all from Roche). The homogenates were centrifuged at 1000 g for 10 minutes and the postnuclear supernatants retained for western analysis. For PNGase treatment, protein samples were treated with PNGase (New England BioLab) according to the manufacturer’s instructions. Protein concentrations were determined with the BCA Protein Assay Kit (Pierce). Forty micrograms of protein were used for Western blot analysis. Proteins were separated on Mini-Protean TGX Precast gels (Bio-Rad), run under denaturing conditions and electrotransferred to Immobilon PVDF membrane (Millipore). Membranes were blocked with 0.01% milk in PBS (10-mM phosphate, pH 7.4, 150-mM NaCl) with 0.05% Tween-20 (PBS-T, all from Sigma) using the SNAP ID protein detection system (Millipore) then incubated 2 hours or overnight with the primary ab. Following washes with PBS-T buffer, membranes were incubated 1 hour with the appropriate HRP-conjugated secondary ab and visualized using the Pierce ECL Pico kit (Pierce). To ensure equal protein loading, blots were probed with anti-β-actin. Blots were scanned and quantified using Image J software from NIH. Statistical analysis was done using GraphPad Prism (GraphPad Software).
2.5. Histology and immunohistochemistry
Mice were anesthetized and perfusion fixed with 4% paraformaldehyde in 0.1-M phosphate buffer, pH 7.2 (Sigma-Aldrich). Brains were removed, embedded in paraffin. Sections (8-µm thick) were cut and mounted on poly-l-lysine-coated slides and stained with hematoxylin and eosin. Immunohistochemical staining was performed as previously described (Garringer et al., 2013). Immunohistochemical studies were also carried out in paraffin sections from cerebral cortex, hippocampus, and cerebellum from individuals affected by sporadic AD, familial AD (FAD; PSEN1-V261I mutation; Miravalle et al., 2005), GSS (PRNP-F198S mutation; Ghetti et al., 1989, 1995), FBD, and FDD (Vidal et al., 1999, 2000), MSTD (MAPT IVS10+3 G>A; Spillantini et al., 1998), and age-matched controls.
2.6. Confocal microscopy
Glass 4-chamber slides (Nunc) were coated with 0.1-mg/ml Poly-D-Lysine (Invitrogen) diluted in sterile water. The surface of each well was covered with the poly-D-lysine solution for 5 minutes at room temperature, then rinsed with sterile water, and allowed to air dry 2 hours before the use. Stably transfected Hek293 cells were plated 2000 cells per well and cultured in growth media 24 hours. The slides were then fixed for 15 minutes at 37 °C in 4% paraformaldehyde, blocked with 2% BSA (Sigma) and 1% Triton (Acros Organics) in PBS overnight at 4 °C. The slides were incubated with primary abs and with the appropriate fluorescently labeled secondary ab for 60 minutes at 37 °C in the dark. BRI2 proteins were detected with anti-c-Myc and secondary Alexa Fluor 594 goat anti-mouse. Endoplasmic reticulum (ER) was detected with anti-Calnexin, secondary Alexa Fluor 488 goat anti-rabbit. The Golgi apparatus was detected with GM130, secondary Alexa Fluor 488 goat anti-rabbit.
3. Results
3.1. BRI2 protein, processing, and post-translational modifications
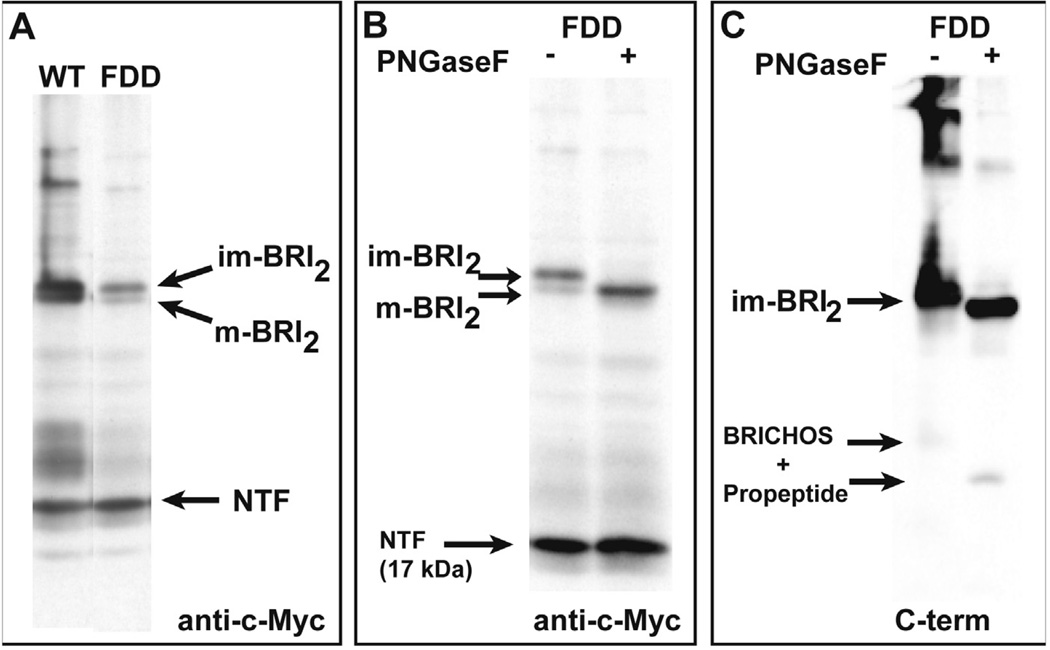
Cleavage of BRI2 pro-protein by PCs to generate m-BRI2 is more efficient for the wild-type BRI2 protein than mutant BRI2. Further processing of m-BRI2 by ADAM10 releases a large part of the ectodomain of BRI2 containing the BRICHOS domain and leaves a membrane-associated NTF of ~ 17 kDa (Fig. 2A). Deglycosylation using PNGaseF of protein extracts from transfected cell lines affects the mobility of full-length BRI2 and m-BRI2 proteins, but not that of the NTF (Fig. 2B), suggesting that ADAM10 cleavage in BRI2 occurs N-terminal from the glycosylation site at Asn 170. Treatment with PNGaseF also affects the mobility of the C-terminal fragment, which contains the pro-peptide sequence and the glycosylated BRICHOS domain, suggesting that processing by ADAM10 can occur independently from PC cleavage of BRI2 (Fig. 2C). The remaining NTF attached to the cell membrane may undergo RIP by SPPL2a or SPPL2b, releasing an intracellular domain and a C-terminal peptide fragment (BRI2 C-peptide; Fig. 1).
Fig. 2.
BRI2 proteolytic fragments after the treatment with PNGaseF. (A). Analysis of BRI2 processing in transfected N2a cells. Expression constructs encoding an N-terminal c-Myc-tagged wild-type BRI2 and the FDD mutant form of BRI2 (FDD) were transiently transfected into N2a cells. Western blot analysis of cell lysates using anti-c-Myc abs shows the ectodomain processing of BRI2 generating m-BRI2. Processing of the FDD mutant form of BRI2 by PCs is compromised, with accumulation of im-BRI2. The NTF (~ 17 kDa) released by ADAM10 processing is indicated. (B). Mobility of BRI2 fragments after the treatment with PNGaseF. Western blot analysis of N2a cells using anti-c-Myc abs shows that PNGaseF removed the oligosaccharide chains from both the full-length and mature forms of BRI2 (shown for the FDD mutant form of BRI2) without affecting the migration of the NTF since the glycosylation site is included in the domain released by ADAM10 processing. (C). Western blot analysis of N2a cells using the C-terminal ab that recognizes the C-terminus of the mutant precursor protein. Treatment with PNGaseF removed the oligosaccharide chains from both the full-length im-BRI2 and the C-terminal peptide that is released by ADAM10 cleavage from the NTF. The C-terminal peptide has the sequence of the BRICHOS domain and the pro-peptide, which contains the sequence of the ADan amyloid peptide before cleavage by PCs. m-BRI2 could not be detected since PC cleavage removes the C-terminal sequence recognized by the ab. Samples were run in triplicates. Representative samples are shown.
3.2. Intracellular BRI2 accumulation and role of glycosylation
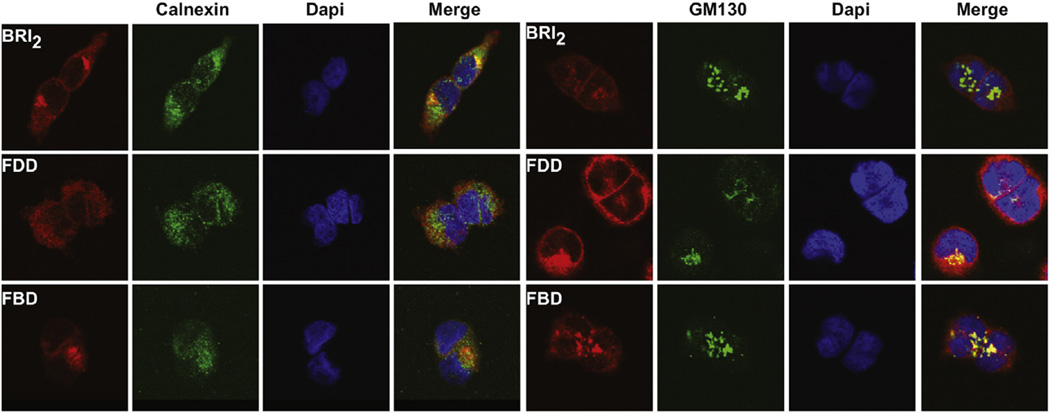
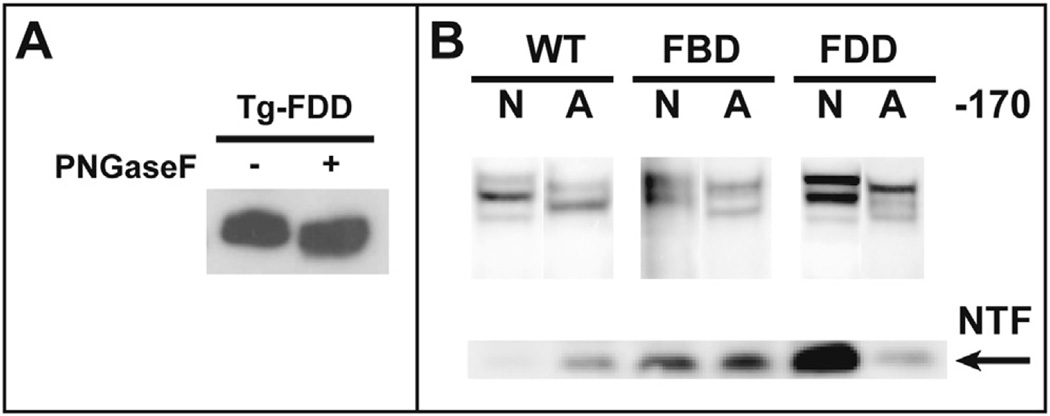
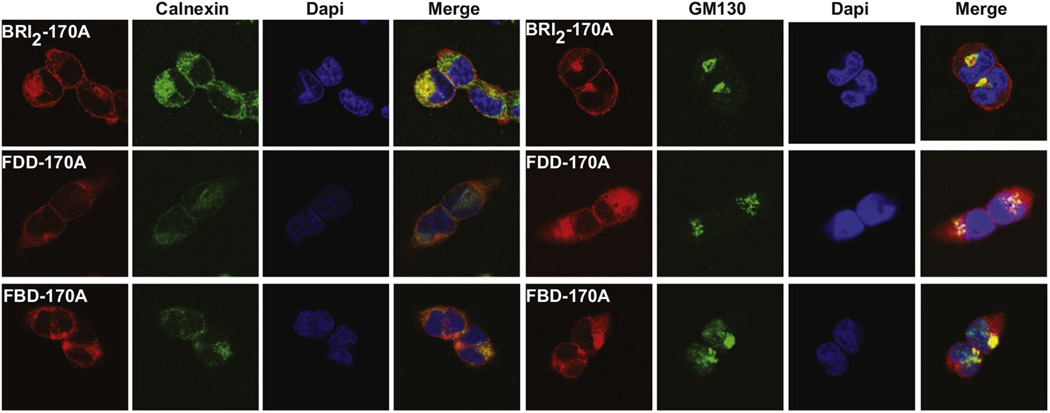
Western blot analysis shows that ectodomain processing by PCs of the FBD and FDD mutant forms of BRI2 is compromised, leading to an intracellular accumulation of mutant BRI2 pro-protein. Confocal microscopic analysis of stable transfected HEK293 cells using abs against a c-Myc tag on the N-terminus of the BRI2 pro-protein shows that the protein localizes to the cell membrane, with the wild-type protein also colocalizing with the ER marker calnexin (Fig. 3). Analysis of cells expressing the FBD and FDD mutant forms of the BRI2 protein by confocal microscopy shows that the mutant proteins accumulate and colocalize with the Golgi marker GM130 (Fig. 3). Analysis of brain protein extracts from Tg-FDD mice suggests that the glycosylated mutant form of the BRI2 protein accumulates intracellularly in vivo, whereas the treatment with PNGaseF modifies the mobility of the FDD-mutant BRI2 pro-protein and m-BRI2 protein (Fig. 4A). Since synthesis of complex type N-linked oligosaccharide chains begins with the cotranslational addition of core mannose–rich oligosaccharide chains to the protein, which are then trimmed and new sugars are added as the protein is processed through the Golgi (Kornfeld and Kornfeld, 1985), we determined whether protein accumulation had any effect on the glycosylation of BRI2. Knock-out of the glycosylation site (Asn170Ala mutation) in wild-type and mutant BRI2 led to an increase in the mobility of BRI2 pro-protein and m-BRI2 but did not change the mobility of the NTF (Fig. 4B). Lack of glycosylation did not seem to modify the processing of the BRI2 protein. Confocal analysis of stable-transfected Hek293 cells with the Asn170Ala mutated BRI2 protein showed colocalization of the protein with the ER marker calnexin, but a significant amount of the immunoreactivity was also seen to colocalize with the Golgi marker GM130 (Fig. 5). Analysis of the FBD and FDD mutant forms of BRI2 with the Asn170Ala change did not show a significant change in the localization of the mutant proteins (Fig. 5).
Fig. 3.
Subcellular localization of wild-type and mutant forms of BRI2. Confocal immunofluorescence microscopy shows that the BRI2 protein localize to the cell membrane, with the wild-type protein also colocalizing with the ER marker calnexin. Analysis of the localization of the FBD and FDD mutant precursor proteins shows a different pattern. In addition to some degree of ER co-localization, the mutant proteins colocalize with the Golgi marker GM130. Confocal immunofluorescence microscopy was performed in Hek293 cells stable transfected with wild-type (BRI2), the FDD mutant form of BRI2 (FDD), and the FBD mutant form of BRI2 (FBD) using the N-terminal Myc ab (red), an ab against Calnexin (green) and an ab against GM130 (green). Nuclei were stained with DAPI (blue). A merge of the images is shown on the right panels. Abbreviations: FBD, familial British dementia; FDD, familial Danish dementia. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
Fig. 4.
BRI2-Asn170 and BRI2 processing. (A). Glycosylation of BRI2 in Tg-FDD mice. Treatment with PNGaseF of PNS from neocortex of Tg-FDD mice affects the mobility of the mutant im-BRI2 and m-BRI2 proteins, demonstrating that the protein is glycosylated in vivo. (B). Knock-out of the glycosylation site at position 170 of BRI2. An Asn170Ala change in the BRI2 sequence leads to an increase in the mobility of the immature and mature forms of BRI2 without altering the migration of the NTF or the overall processing pattern of the protein in wild-type and the FBD and FDD mutant forms of BRI2. Analysis was performed in Hek293 cells stable transfected with wild-type BRI2 (WT), the FBD mutant form of BRI2 (FBD), and the FDD mutant form of BRI2 (FDD) using the N-terminal Myc ab. N indicates Asn at aa 170; A indicates Ala at aa 170. Samples were run in triplicates. Representative samples are shown. Abbreviations: FBD, familial British dementia; FDD, familial Danish dementia; NTF, N-terminal fragment; PNS, postnuclear supernatants.
Fig. 5.
Subcellular localization of wild-type and mutant forms of BRI2 with an Asn170Ala substitution. Confocal analysis shows that the Asn170Ala BRI2 protein still colocalizes with the ER marker calnexin, but a significant amount of the immunoreactivity now also colocalizes with the Golgi marker GM130. Analysis of the FBD and FDD mutant forms of BRI2 with the Asn170Ala change show, in addition to a small degree of ER colocalization, a strong colocalization with the Golgi marker GM130 as did the FBD and FDD mutants with Asn 170. Confocal immunofluorescence microscopy was performed in Hek293 cells stable transfected with wild-type (BRI2), the FDD mutant form of BRI2 (ADanPP), and the FBD mutant form of BRI2 (ABriPP) using the N-terminal Myc ab (red), an ab against Calnexin (green), and an ab against GM130 (green). Nuclei were stained with DAPI (blue). A merge of the images is shown on the right panels. Abbreviations: ER, endoplasmic reticulum; FBD, familial British dementia; FDD, familial Danish dementia. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
3.3. Accumulation of BRI2 in glial cell bodies in FBD and FDD
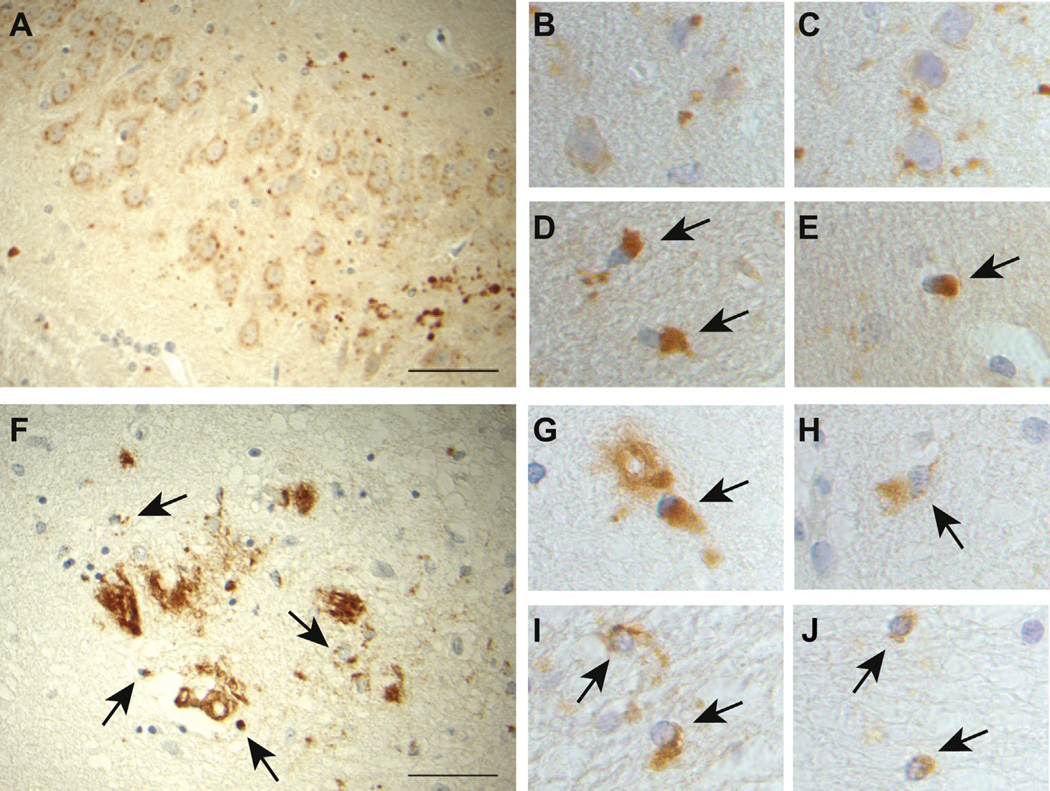
Immunohistochemical studies using abs against the N-terminus of BRI2 revealed the presence of intracellular immunopositive perinuclear accumulation of ubiquitinated (not shown) BRI2 in neurons and glial cells of Tg-FDD mice (Fig. 6A–C). Abs that recognize the C-terminus of the mutant precursor protein and the ADan amyloid peptide revealed also the presence of intracellular protein accumulation in addition to amyloid deposits (Fig. 6D and E). The finding of immunoreactivities for both N- and C-terminal epitopes suggests the accumulation of full-length mutant BRI2 pro-protein. Immunohistochemical studies using the C-terminal ab revealed the presence of intracellular perinuclear accumulation of BRI2 in a patient with FDD (Fig. 6F–H, black arrows) and a patient with FBD (Fig. 6I and J, black arrows) in addition to amyloid deposits.
Fig. 6.
Accumulation of mutant BRI2 in FBD and FDD. Immunohistochemical studies using the BRI2-N -terminal ab show intracellular deposits in neurons and glia (A–C) and globular DNs (A) in Tg-FDD mice. The ab against the C-terminus of the mutant precursor protein reveals only the presence of intracellular perinuclear aggregates in Tg-FDD mice (D, E, black arrows). Immunohistochemical studies using abs against the C-terminal sequence that recognizes the C-terminus of the mutant precursor protein in a patient with FDD reveal the presence of intracellular protein accumulation (F–H, black arrows) in addition to amyloid deposits. Immunohistochemical studies using abs that recognize the C-terminus of the mutant precursor protein and the ABri amyloid peptide in a patient with FBD show intracellular protein accumulation (I, J, black arrows) in addition to amyloid deposits. Sections were from the hippocampus (A, E) and neocortex (B–D) of a 21-month old Tg-FDD mouse, neocortex of an FDD case (F–H), and neocortex of an FBD case (I, J). Immunohistochemistry using the N-term BRI2 ab14307 (A–C), the C-term ab against the Danish mutant (D–H), and the C-term ab against the British mutant (I, J). Scale bars: A, F, 50 µm; B–E and G–J are high magnification from 100×. Abbreviations: DNs, dystrophic neurites; FBD, familial British dementia; FDD, familial Danish dementia.
3.4. Accumulation of BRI2 in DNs in AD and GSS
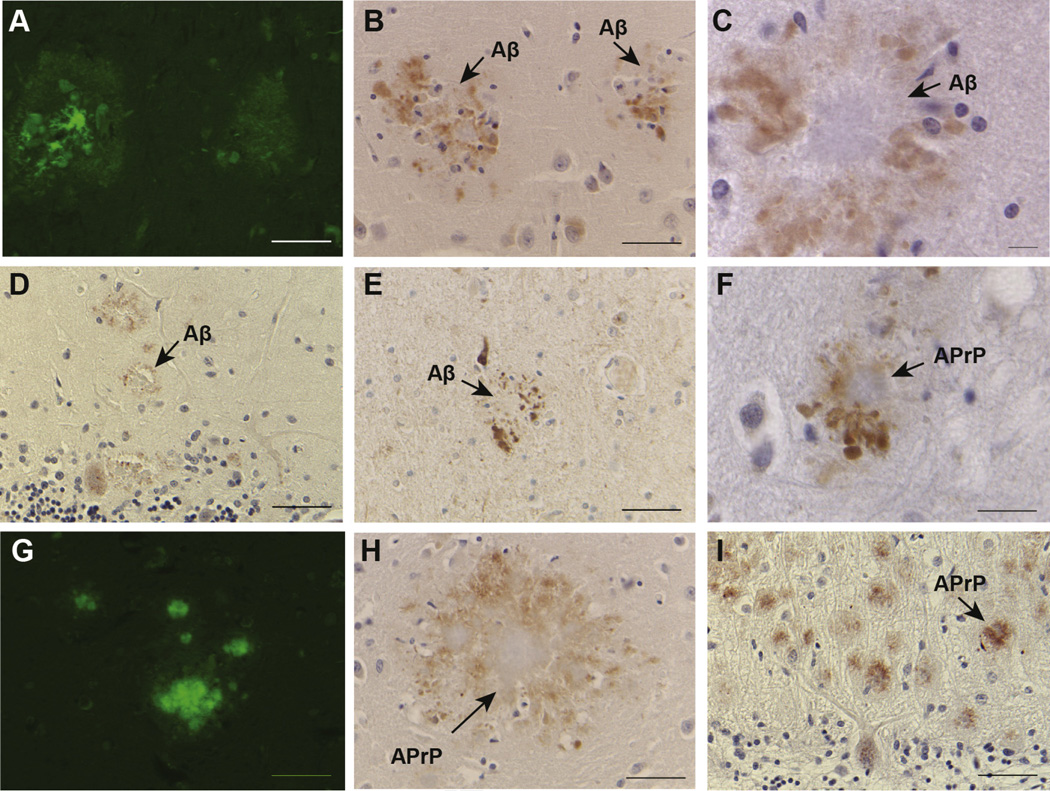
In FBD, FDD, and Tg-FDD mice, analysis of brain sections shows clusters of swollen neurites surrounding amyloid cores that are recognized by abs specific for the N-terminus of BRI2 but not by the C-terminal abs (Garringer et al., 2013; Vidal et al., 2009). Since these profiles appeared to be associated with amyloid cores, we analyzed sections from the cerebral cortex, hippocampus, and cerebellum from the cases of sporadic AD (Fig. 7A–C and E), FAD (Fig. 7D), and GSS (Fig. 7F–I), which are characterized by extracellular amyloid deposition and intracellular NFTs. In all cases, we detected N-terminal BRI2 immunoreactive DNs surrounding amyloid cores. Staining was not observed after omitting the first ab, whereas a similar immunoreactive profile was observed using 2 different primary abs against the N-terminus of BRI2 (Fig. 7E). In the cerebellum, in addition to the swollen neurites around amyloid cores, the N-terminal BRI2 ab labeled cell bodies, dendrites, and axons of Purkinje cells (Fig. 7D and I). Immunostaining of tissue from normal controls and a case of MSTD showed the previously described pattern of normal cellular staining using N-terminal BRI2 abs (Akiyama et al., 2004).
Fig. 7.
Intracellular accumulation of m-BRI2 in patients with Alzheimer disease and Gerstmann-Sträussler-Scheinker disease. The BRI2-amino-terminal ab labels globular DNs in sections from patients with AD (B, C), FAD (D), and GSS (F, H, I). These profiles are associated with amyloid cores (A, G), although some clusters of DNs may also be seen in the absence of amyloid deposits. DNs on a section from a patient with AD are also labeled using the BRI2-amino-terminal ab 11–26 (E). In the cerebellum, the N-terminal BRI2 ab labeled swollen neurites at the periphery of cerebellar amyloid cores, and cell bodies, dendrites, and axons of Purkinje cells of AD and GSS cases (D, I). Immunostaining of normal controls and a case of MSTD showed normal cellular staining. Arrows indicate the presence of Aβ and APrP amyloid cores. Sections were from the hippocampus of a sporadic AD case (A–C, E), cerebellum from a FAD case (D), and cerebellum from a GSS case (F–I). Thioflavine S (A, G). Immunohistochemistry using the BRI2 N-term ab14307 (B–D, F, H–I) and ab 11–26 (E). Scale bars: A, B, D, E, G–I, 50 µm; F, 20 µm; C, 10 µm. Abbreviations: DNs, dystrophic neurites; FBD, familial British dementia; FDD, familial Danish dementia; GSS, Gerstmann-Sträussler-Scheinker disease; MSTD, multiple system tauopathy with presenile dementia.
4. Discussion
Cleavage by PCs within the BRI2 ectodomain is the main proteolytic event that releases the mutant peptides found deposited as ABri amyloid in FBD patients and ADan amyloid in FDD patients (Vidal et al., 1999, 2000). The mutant precursor proteins are inefficiently cleaved by PCs, leading to the intracellular accumulation of immature mutant BRI2 pro-protein in neurons and glial cells in patients with FBD, FDD, and a transgenic mouse model of FDD, but the significance of this finding for our understanding of the pathologic process associated with FBD and FDD is not known. The accumulation of immature mutant BRI2 pro-protein may be caused by the mutant sequences interfering with the correct folding of BRI2 and the enzymatic activity of PCs. In addition, they may also disrupt information required for efficient Golgi exit. The molecular mechanism responsible for the abnormal processing of mutant BRI2 may involve the BRICHOS domains, which seems to have intramolecular chaperone-like function protecting and avoiding pro-peptide aggregation (Johansson et al., 2006, 2009; Peng et al., 2010). Several interstitial lung diseases have been linked to mutations in the SP-C gene, SFTPC, with the great majority of these mutations found within the BRICHOS domain of the pro-peptide where mutations are believed to affect pro-peptide folding and conformation. Mutagenesis of one or both of the cysteines in the BRICHOS domain of SP-C results in misfolding of the pro-peptide and mistargeting of the unprocessed mutant protein (Kabore et al., 2001). In addition, in vitro experiments have shown that the BRICHOS domain can bind amyloidogenic peptides and affect fibril formation (Willander et al., 2011, 2012). Thus, in FBD and FDD, the BRICHOS domain may regulate negatively the enzymatic processing of the pro-protein by interacting with the amyloidogenic sequences of ABri and ADan in the mutant precursor proteins. The interaction between the BRICHOS domain and the amyloidogenic sequences may alter the accessibility of PCs to the cleavage site, resulting in the abnormal processing of the immature proteins and the targeting of the unprocessed precursor to the Golgi. Since the glycosylation site (aa 170) is present in the BRICHOS domain of BRI2, changes in the folding of BRI2 could affect glycosylation of BRI2 and eventually the processing and the trafficking of BRI2. Vice versa, some glycosylation sites seem to be crucial for protein folding but their removal from folded proteins often does not affect the protein fold and function (Shental-Bechor and Levy, 2008). We observed that mutagenesis of Asn 170 had no remarkable effect on wild-type and mutant BRI2 processing by PCs or ADAM10 but leads to the intracellular accumulation of BRI2 as previously reported (Tsachaki et al., 2011). Wild-type BRI2 with the N170A substitution accumulated in a Golgi compartment, similarly to the FBD and FDD mutant forms of BRI2, suggesting that glycosylation plays an important role in protein trafficking of BRI2. These data highlight the importance of considering the intracellular accumulation of immature mutant BRI2 pro-protein in the pathobiology of FBD and FDD. In AD, numerous lines of evidence suggest that in addition to Aβ, some of the neurotoxicity associated with AD may be due to the accumulation of C-terminal proteolytic fragments of the amyloid β precursor protein (AβPP), which have been found in patients with the disease and in animal models (Chang and Suh, 2005; Vidal et al., 2012).
In FBD and FDD, we have also observed the presence of large and rounded processes, strongly immunolabeled by abs against the N-terminus of BRI2 (but not the C-terminus) in association with amyloid cores, although some of these processes may be also observed in the absence of amyloid cores. The finding of m-BRI2 in DNs, which symbolize neuritic damage (Masliah et al., 1993), suggest an accumulation of m-BRI2 protein that is different from the accumulation of the immature protein observed in the Golgi in FBD and FDD. The N-terminal abs also showed similar accumulation of m-BRI2 in DNs in association with Aβ in AD and APrP in GSS. No immunolabeling was observed in NFTs of AD and GSS, nor in tau deposits of MSTD. Our results are in agreement with those by Akiyama (Akiyama et al., 2004), who reported positive BRI2 immunostaining in globular DNs in senile plaques in AD using an N-terminal ab. As in our study, NFT of AD cases were negative for BRI2. Similar immunohistochemical data in AD have been recently reported, although the authors suggest accumulation of an immature form of BRI2 based on Western blot data (Del Campo et al., 2014). Accumulation of m-BRI2 in DNs may occur as a consequence of a reduction of tau binding to microtubules and resulting impairment in transport of m-BRI2 (Garringer et al., 2013), leading to m-BRI2 accumulation in DNs. They are immunoreactive for a wide variety of synaptic, cytoskeletal, and growth-related proteins, including AβPP (Blanchard et al., 2003). A potential molecular mechanism by which abnormal accumulation of m-BRI2 in DNs may be deleterious to the cell could involve the role of BRI2 in the processing of AβPP (Fotinopoulou et al., 2005; Matsuda et al., 2005). Mature BRI2 binds the C-terminus of AβPP, masking the cleavage sites of β- and α-secretase and the γ-secretase docking site on C99. Interestingly, this interaction occurs only between m-BRI2 and mature AβPP (m-AβPP), downstream of the trans-Golgi complex (Matsuda et al., 2008). Indeed, maturation of BRI2 into m-BRI2 and transport of m-BRI2 along the secretory pathway are required to generate a functional AβPP processing inhibitor (Matsuda et al., 2011a). Thus, abnormal accumulation and subcellular localization of m-BRI2 could reduce the interaction between m-BRI2 and m-AβPP in vivo leading to an increase in the production of Aβ peptides (Matsuda et al., 2009, 2011b; Tamayev and D’Adamio, 2012; Tamayev et al., 2011). The finding of Aβ codeposited with ADan in the brain of patients with FDD seems to support this hypothesis (Vidal et al., 2000). In FDD, parenchymal and vascular ADan deposition exceeds that of Aβ, with some deposits being composed of Aβ or ADan alone, and in a considerable number of cases showing colocalization of both peptides (Holton et al., 2002). Importantly, the finding of m-BRI2 accumulation in DNs in GSS suggests that m-BRI2 may be involved in a similar cellular degenerative pathway in these diseases, marked by amyloid accumulation and tau fibrillar deposition and neurodegeneration.
In summary, we showed that the extended C-terminus of BRI2 originated by mutations in the BRI2 gene in FBD and FDD reduced the processing of the protein leading to the intracellular accumulation of the immature protein in the Golgi. Future studies will determine the molecular mechanism(s) involved in the intracellular accumulation of mutant BRI2 and its potential role in the pathogenesis of FBD and FDD. We also observed a second type of BRI2 accumulation in the form of DNs associated with 4 different cerebral amyloidogenic peptides (ABri, ADan, Aβ, and APrP), likely containing m-BRI2. The study of m-BRI2 protein accumulation in DNs may provide insights into the molecular mechanism(s) underlying neuronal dysfunction in association with cerebral amyloidosis.
Acknowledgments
This study was supported by grants from the National Institute of Health NS050227, AG037338 and AG10133. Acknowledgment is made to the Ralph W. and Grace M. Showalter Research Trust and the donors of Alzheimer’s Disease Research, a program of Bright-Focus Foundation, for support of this research (A2008-304). The authors thank Dr. T. Revesz (Queen Square Brain Bank for Neurological Disorders) for FBD and FDD tissues and Dr. H. Akiyama (Tokyo Institute of Psychiatry) for antibody 11–26. They also thank Debra Lucas and Rose Richardson for the technical assistance.
Footnotes
Disclosure statement
None of the authors have any competing interest.
Uncited references
References
- Akiyama H, Kondo H, Arai T, Ikeda K, Kato M, Iseki E, Schwab C, McGeer PL. Expression of BRI, the normal precursor of the amyloid protein of familial British dementia, in human brain. Acta Neuropathol. 2004;107:53–58. doi: 10.1007/s00401-003-0783-1. [DOI] [PubMed] [Google Scholar]
- Baron BW, Pytel P. Expression pattern of the BCL6 and ITM2B proteins in normal human brains and in Alzheimer disease. Appl. Immunohistochem. Mol. Morphol. 2016 doi: 10.1097/PAI.0000000000000329. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- Blanchard V, Moussaoui S, Czech C, Touchet N, Bonici B, Planche M, Canton T, Jedidi I, Gohin M, Wirths O, Bayer TA, Langui D, Duyckaerts C, Tremp G, Pradier L. Time sequence of maturation of dystrophic neurites associated with Abeta deposits in APP/PS1 transgenic mice. Exp. Neurol. 2003;184:247–263. doi: 10.1016/s0014-4886(03)00252-8. [DOI] [PubMed] [Google Scholar]
- Chang KA, Suh YH. Pathophysiological roles of amyloidogenic carboxy-terminal fragments of the beta-amyloid precursor protein in Alzheimer’s disease. J. Pharmacol. Sci. 2005;97:461–471. doi: 10.1254/jphs.cr0050014. [DOI] [PubMed] [Google Scholar]
- Choi SI, Vidal R, Frangione B, Levy E. Axonal transport of British and Danish amyloid peptides via secretory vesicles. FASEB J. 2004;18:373–375. doi: 10.1096/fj.03-0730fje. [DOI] [PubMed] [Google Scholar]
- Del Campo M, Hoozemans JJ, Dekkers LL, Rozemuller AJ, Korth C, Müller-Schiffmann A, Scheltens P, Blankenstein MA, Jimenez CR, Veerhuis R, Teunissen CE. BRI2-BRICHOS is increased in human amyloid plaques in early stages of Alzheimer’s disease. Neurobiol. Aging. 2014;35:1596–1604. doi: 10.1016/j.neurobiolaging.2014.01.007. [DOI] [PubMed] [Google Scholar]
- Deleersnijder W, Hong G, Cortvrindt R, Poirier C, Tylzanowski P, Pittois K, Van Marck E, Merregaert J. Isolation of markers for chondro-osteogenic differentiation using cDNA library subtraction. Molecular cloning and characterization of a gene belonging to a novel multigene family of integral membrane proteins. J. Biol. Chem. 1996;271:19475–19482. doi: 10.1074/jbc.271.32.19475. [DOI] [PubMed] [Google Scholar]
- Fluhrer R, Martin L, Klier B, Haug-Kröper M, Grammer G, Nuscher B, Haass C. The α-helical content of the transmembrane domain of the British dementia protein-2 (Bri2) determines its processing by signal peptide peptidase-like 2b (SPPL2b) J. Biol. Chem. 2012;287:5156–5163. doi: 10.1074/jbc.M111.328104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fotinopoulou A, Tsachaki M, Vlavaki M, Poulopoulos A, Rostagno A, Frangione B, Ghiso J, Efthimiopoulos S. BRI2 interacts with amyloid precursor protein (APP) and regulates amyloid beta (Abeta) production. J. Biol. Chem. 2005;280:30768–30772. doi: 10.1074/jbc.C500231200. [DOI] [PubMed] [Google Scholar]
- Garringer HJ, Murrell J, D’Adamio L, Ghetti B, Vidal R. Modeling familial British and Danish dementia. Brain Struct. Funct. 2010;214:235–244. doi: 10.1007/s00429-009-0221-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garringer HJ, Murrell J, Sammeta N, Gnezda A, Ghetti B, Vidal R. Increased tau phosphorylation and tau truncation, and decreased synaptophysin levels in mutant BRI2/tau transgenic mice. PLoS One. 2013;8:e56426. doi: 10.1371/journal.pone.0056426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghetti B, Dlouhy SR, Giaccone G, Bugiani O, Frangione B, Farlow MR, Tagliavini F. Gerstmann-Sträussler-Scheinker disease and the Indiana kindred. Brain Pathol. 1995;5:61–75. doi: 10.1111/j.1750-3639.1995.tb00578.x. [DOI] [PubMed] [Google Scholar]
- Ghetti B, Tagliavini F, Masters CL, Beyreuther K, Giaccone G, Verga L, Farlow MR, Conneally PM, Dlouhy SR, Azzarelli B, Bugiani O. Gerstmann-Sträussler-Scheinker disease. II. Neurofibrillary tangles and plaques with PrP-amyloid coexist in an affected family. Neurology. 1989;39:1453–1461. doi: 10.1212/wnl.39.11.1453. [DOI] [PubMed] [Google Scholar]
- Glombik MM, Krömer A, Salm T, Huttner WB, Gerdes HH. The disulfide-bonded loop of chromogranin B mediates membrane binding and directs sorting from the trans-Golgi network to secretory granules. EMBO J. 1999;18:1059–1070. doi: 10.1093/emboj/18.4.1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardy J, Allsop D. Amyloid deposition as the central event in the aetiology of Alzheimer’s disease. Trends Pharmacol. Sci. 1991;12:383–388. doi: 10.1016/0165-6147(91)90609-v. [DOI] [PubMed] [Google Scholar]
- Hardy JA, Higgins GA. Alzheimer’s disease: the amyloid cascade hypothesis. Science. 1992;256:184–185. doi: 10.1126/science.1566067. [DOI] [PubMed] [Google Scholar]
- Holton JL, Ghiso J, Lashley T, Rostagno A, Guerin CJ, Gibb G, Houlden H, Ayling H, Martinian L, Anderton BH, Wood NW, Vidal R, Plant G, Frangione B, Revesz T. Regional distribution of amyloid-Bri deposition and its association with neurofibrillary degeneration in familial British dementia. Am. J. Pathol. 2001;158:515–526. doi: 10.1016/S0002-9440(10)63993-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holton JL, Lashley T, Ghiso J, Braendgaard H, Vidal R, Guerin CJ, Gibb G, Hanger DP, Rostagno A, Anderton BH, Strand C, Ayling H, Plant G, Frangione B, Bojsen-Moller M, Revesz T. Familial Danish dementia: a novel form of cerebral amyloidosis associated with deposition of both amyloid-Dan and amyloid-beta. J. Neuropathol. Exp. Neurol. 2002;61:254–267. doi: 10.1093/jnen/61.3.254. [DOI] [PubMed] [Google Scholar]
- Johansson H, Eriksson M, Nordling K, Presto J, Johansson J. The Brichos domain of prosurfactant protein C can hold and fold a transmembrane segment. Protein Sci. 2009;18:1175–1182. doi: 10.1002/pro.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson H, Nordling K, Weaver TE, Johansson J. The Brichos domain-containing C-terminal part of pro-surfactant protein C binds to an unfolded poly-val transmembrane segment. J. Biol. Chem. 2006;281:21032–21039. doi: 10.1074/jbc.M603001200. [DOI] [PubMed] [Google Scholar]
- Kabore AF, Wang WJ, Russo SJ, Beers MF. Biosynthesis of surfactant protein C: characterization of aggresome formation by EGFP chimeras containing propeptide mutants lacking conserved cysteine residues. J. Cell Sci. 2001;114(Pt 2):293–302. doi: 10.1242/jcs.114.2.293. [DOI] [PubMed] [Google Scholar]
- Kim SH, Creemers JW, Chu S, Thinakaran G, Sisodia SS. Proteolytic processing of familial British dementia-associated BRI variants: evidence for enhanced intracellular accumulation of amyloidogenic peptides. J. Biol. Chem. 2002;277:1872–1877. doi: 10.1074/jbc.M108739200. [DOI] [PubMed] [Google Scholar]
- Kim SH, Wang R, Gordon DJ, Bass J, Steiner DF, Lynn DG, Thinakaran G, Meredith SC, Sisodia SS. Furin mediates enhanced production of fibrillogenic ABri peptides in familial British dementia. Nat. Neurosci. 1999;2:984–988. doi: 10.1038/14783. [DOI] [PubMed] [Google Scholar]
- Kornfeld R, Kornfeld S. Assembly of asparagine-linked oligosaccharides. Annu. Rev. Biochem. 1985;54:631–664. doi: 10.1146/annurev.bi.54.070185.003215. [DOI] [PubMed] [Google Scholar]
- Krömer A, Glombik MM, Huttner WB, Gerdes HH. Essential role of the disulfide-bonded loop of chromogranin B for sorting to secretory granules is revealed by expression of a deletion mutant in the absence of endogenous granin synthesis. J. Cell Biol. 1998;140:1331–1346. doi: 10.1083/jcb.140.6.1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin L, Fluhrer R, Haass C. Substrate requirements for SPPL2b-dependent regulated intramembrane proteolysis. J. Biol. Chem. 2009;284:5662–5670. doi: 10.1074/jbc.M807485200. [DOI] [PubMed] [Google Scholar]
- Martin L, Fluhrer R, Reiss K, Kremmer E, Saftig P, Haass C. Regulated intramembrane proteolysis of Bri2 (Itm2b) by ADAM10 and SPPL2a/SPPL2b. J. Biol. Chem. 2008;283:1644–1652. doi: 10.1074/jbc.M706661200. [DOI] [PubMed] [Google Scholar]
- Masliah E, Mallory M, Deerinck T, DeTeresa R, Lamont S, Miller A, Terry RD, Carragher B, Ellisman M. Re-evaluation of the structural organization of neuritic plaques in Alzheimer’s disease. J. Neuropathol. Exp. Neurol. 1993;52:619–632. doi: 10.1097/00005072-199311000-00009. [DOI] [PubMed] [Google Scholar]
- Matsuda S, Giliberto L, Matsuda Y, Davies P, McGowan E, Pickford F, Ghiso J, Frangione B, D’Adamio L. The familial dementia BRI2 gene binds the Alzheimer gene amyloid-beta precursor protein and inhibits amyloid-beta production. J. Biol. Chem. 2005;280:28912–28916. doi: 10.1074/jbc.C500217200. [DOI] [PubMed] [Google Scholar]
- Matsuda S, Giliberto L, Matsuda Y, McGowan EM, D’Adamio L. BRI2 inhibits amyloid beta-peptide precursor protein processing by interfering with the docking of secretases to the substrate. J. Neurosci. 2008;28:8668–8676. doi: 10.1523/JNEUROSCI.2094-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda S, Matsuda Y, Snapp EL, D’Adamio L. Maturation of BRI2 generates a specific inhibitor that reduces APP processing at the plasma membrane and in endocytic vesicles. Neurobiol. Aging. 2011a;32:1400–1408. doi: 10.1016/j.neurobiolaging.2009.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda S, Tamayev R, D’Adamio L. Increased AβPP processing in familial Danish dementia patients. J. Alzheimers Dis. 2011b;27:385–391. doi: 10.3233/JAD-2011-110785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miravalle L, Calero M, Takao M, Roher AE, Ghetti B, Vidal R. Amino-terminally truncated Aβ peptide species are the main component of cotton wool plaques. Biochemistry. 2005;44:10810–10821. doi: 10.1021/bi0508237. [DOI] [PubMed] [Google Scholar]
- Peng S, Fitzen M, Jörnvall H, Johansson J. The extracellular domain of Bri2 (ITM2B) binds the ABri peptide (1–23) and amyloid beta-peptide (Abeta1–40): implications for Bri2 effects on processing of amyloid precursor protein and Abeta aggregation. Biochem. Biophys. Res. Commun. 2010;393:356–361. doi: 10.1016/j.bbrc.2009.12.122. [DOI] [PubMed] [Google Scholar]
- Sanchez-Pulido L, Devos D, Valencia A. BRICHOS: a conserved domain in proteins associated with dementia, respiratory distress and cancer. Trends Biochem. Sci. 2002;27:329–332. doi: 10.1016/s0968-0004(02)02134-5. [DOI] [PubMed] [Google Scholar]
- Shental-Bechor D, Levy Y. Effect of glycosylation on protein folding: a close look at thermodynamic stabilization. Proc. Natl. Acad. Sci. U. S. A. 2008;105:8256–8261. doi: 10.1073/pnas.0801340105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spillantini MG, Murrell JR, Goedert M, Farlow MR, Klug A, Ghetti B. Mutation in the tau gene in familial multiple system tauopathy with presenile dementia. Proc. Natl. Acad. Sci. U. S. A. 1998;95:7737–7741. doi: 10.1073/pnas.95.13.7737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamayev R, D’Adamio L. Memory deficits of British dementia knock-in mice are prevented by Aβ-precursor protein haploinsufficiency. J. Neurosci. 2012;32:5481–5485. doi: 10.1523/JNEUROSCI.5193-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamayev R, Giliberto L, Li W, d’Abramo C, Arancio O, Vidal R, D’Adamio L. Memory deficits due to familial British dementia BRI2 mutation are caused by loss of BRI2 function rather than amyloidosis. J. Neurosci. 2010b;30:14915–14924. doi: 10.1523/JNEUROSCI.3917-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamayev R, Matsuda S, Fà M, Arancio O, D’Adamio L. Danish dementia mice suggest that loss of function and not the amyloid cascade causes synaptic plasticity and memory deficits. Proc. Natl. Acad. Sci. U. S. A. 2010a;107:20822–20827. doi: 10.1073/pnas.1011689107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamayev R, Matsuda S, Giliberto L, Arancio O, D’Adamio L. APP heterozygosity averts memory deficit in knockin mice expressing the Danish dementia BRI2 mutant. EMBO J. 2011;30:2501–2509. doi: 10.1038/emboj.2011.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsachaki M, Serlidaki D, Fetani A, Zarkou V, Rozani I, Ghiso J, Efthimiopoulos S. Glycosylation of BRI2 on asparagine 170 is involved in its trafficking to the cell surface but not in its processing by furin or ADAM10. Glycobiology. 2011;21:1382–1388. doi: 10.1093/glycob/cwr097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidal R, Barbeito AG, Miravalle L, Ghetti B. Cerebral amyloid angiopathy and parenchymal amyloid deposition in transgenic mice expressing the Danish mutant form of human BRI2 . Brain Pathol. 2009;19:58–68. doi: 10.1111/j.1750-3639.2008.00164.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidal R, Calero M, Revesz T, Plant G, Ghiso J, Frangione B. Sequence, genomic structure and tissue expression of Human BRI3, a member of the BRI gene family. Gene. 2001;266:95–102. doi: 10.1016/s0378-1119(01)00374-2. [DOI] [PubMed] [Google Scholar]
- Vidal R, Frangione B, Rostagno A, Mead S, Revesz T, Plant G, Ghiso J. A stop-codon mutation in the BRI gene associated with familial British dementia. Nature. 1999;399:776–781. doi: 10.1038/21637. [DOI] [PubMed] [Google Scholar]
- Vidal R, Revesz T, Rostagno A, Kim E, Holton JL, Bek T, Bojsen-Moller M, Braendgaard H, Plant G, Ghiso J, Frangione B. A decamer duplication in the 3’ region of the BRI gene originates an amyloid peptide that is associated with dementia in a Danish kindred. Proc. Natl. Acad. Sci. U. S. A. 2000;97:4920–4925. doi: 10.1073/pnas.080076097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidal R, Sammeta N, Garringer HJ, Sambamurti K, Miravalle L, Lamb BT, Ghetti B. The Psen1-L166P-knock-in mutation leads to amyloid deposition in human wild-type amyloid precursor protein YAC transgenic mice. FASEB J. 2012;26:2899–2910. doi: 10.1096/fj.12-205542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willander H, Askarieh G, Landreh M, Westermark P, Nordling K, Keränen H, Hermansson E, Hamvas A, Nogee LM, Bergman T, Saenz A, Casals C, Åqvistg J, Jörnvall H, Berglund H, Presto J, Knight SD, Johansson J. High-resolution structure of a BRICHOS domain and its implications for anti-amyloid chaperone activity on lung surfactant protein C. Proc. Natl. Acad. Sci. U. S. A. 2012;109:2325–2329. doi: 10.1073/pnas.1114740109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willander H, Hermansson E, Johansson J, Presto J. BRICHOS domain associated with lung fibrosis, dementia and cancer—a chaperone that prevents amyloid fibril formation? FEBS J. 2011;278:3893–3904. doi: 10.1111/j.1742-4658.2011.08209.x. [DOI] [PubMed] [Google Scholar]