Abstract
Human retinal ganglion cells (RGCs) derived from pluripotent stem cells (PSCs) have anticipated value for human disease study, drug screening, and therapeutic applications; however, their full potential remains underdeveloped. To characterize RGCs in human embryonic stem cell (hESC) derived retinal organoids we examined RGC markers and surface antigen expression and made comparisons to human fetal retina. RGCs in both tissues exhibited CD184 and CD171 expression and distinct expression patterns of the RGC markers BRN3 and RBPMS. The retinal progenitor cells (RPCs) of retinal organoids expressed CD184, consistent with its expression in the neuroblastic layer in fetal retina. In retinal organoids CD184 expression was enhanced in RGC competent RPCs and high CD184 expression was retained on post-mitotic RGC precursors; CD171 was detected on maturing RGCs. The differential expression timing of CD184 and CD171 permits identification and enrichment of RGCs from retinal organoids at differing maturation states from committed progenitors to differentiating neurons. These observations will facilitate molecular characterization of PSC-derived RGCs during differentiation, critical knowledge for establishing the veracity of these in vitro produced cells. Furthermore, observations made in the retinal organoid model closely parallel those in human fetal retina further validating use of retinal organoid to model early retinal development.
Keywords: retinal organoid, retinal ganglion cell, stem cell derived retina, CD184(CXCR4), CD171 (L1CAM), retina development, RBPMS
1. Introduction
RGCs are neurons that indirectly receive visual information from photoreceptors in the retina. Their axons form the optic nerve that relays this information to the brain’s primary vision centers. RGC death is the cause of blindness in two prominent diseases, glaucoma, and optic nerve hypoplasia. Optic nerve hypoplasia is the most common cause of congenital blindness, the etiology of which is unknown and therapy non-existent (Borchert, 2012; Garcia-Filion and Borchert, 2013). Glaucoma is the leading cause of irreversible blindness worldwide, a disease about which much is known but that still lacks a cure (Almasieh et al., 2012; Weinreb et al., 2014).
Whereas animal models for study of RGC biology and disease have great value, they have limitations and cannot accurately reproduce distinctly human biology (Bouhenni et al., 2012). Human RGCs derived from PSCs may provide an alternative model to study RGC development and disease. In addition to being human cells, they can be produced with the genetic background of afflicted patients, which can be a uniquely powerful tool for studying human cell biology, disease phenotype, and drug or environmental responses. Additionally the prospect of using PSC derived RGCs for cell replacement therapy is a component of an ultimate curative goal for advanced disease (Cooke and Meyer, 2015; Jayaram et al., 2011; Pearson and Martin, 2015).
While goals for PSC derived RGCs are lofty, there has been little characterization of the in vitro produced cells. There is a need for rigorous study of molecular and cellular properties of these cells to establish the degree to which they mimic bona fide human embryonic RGCs including their competence to produce the multitude of RGC types observed in adult retina (Sanes and Masland, 2015). This will define their utility and/or limitations. In this work, we produce RGCs from hESCs in the context of a developing embryonic retinal tissue or retinal organoid. This culture methodology reproducibly produces a significant number of embryonic RGCs in a cellular environment created within the context of a self-forming three-dimensional tissue which replicates many intercellular cell signaling and cell-cell interactions of tissue developing in vivo (Hynds and Giangreco, 2013; Nakano et al., 2012). We provide an initial characterization of the RGCs produced as well as establish the utility of the surface antigens, CD184 (CXCR4) and CD171 (L1CAM), for RGC isolation.
2. Materials and Methods
2.1 hESC culture and differentiation to retinal organoids
All work was performed with Children’s Hospital Los Angeles (CHLA) Stem Cell Research Oversight Committee approval. WA09 (WiCell) hESCs were maintained on mouse embryonic fibroblasts in ESCM (DMEM/F12, 20% KSR, 2 mM glutamine, 100 μM NEAA, 100 μM BME, and 4 ng/ml bFGF) and passaged manually following collagenase treatment every four or five days. For production of retinal organoids, WA09 cells were differentiated to neural retina (not optic cup) in three dimensional culture as described (Nakano et al., 2012) except hESC colonies were dissociated with Accutase (Thermo Fisher Scientific) and reaggregated in Lipidure-coated U well 96-well plates (Nof America) using 5000 cells per well in 30 μl Aggrewell media (Stem Cell Technologies) + 10 μM Y27632 (Tocris). Media was changed to retinal differentiation media (Nakano et al., 2012) on day 1 and Growth Factor Reduced (GFR) Matrigel (Corning) plus 3 μM IWR1 (Cayman Chemical) were added on day 2. Matrigel was added to 0.5%; the concentration varied by lot, but this was generally about 50 μg/ml. Tissue was incubated at 37°C, 5% CO2, 20% oxygen until use. Usually the entire aggregate formed retinal tissue with the apical surface on the exterior (Figure 1A), so an excision step described was unnecessary (Nakano et al., 2012). We observed some variation in appearance of the differentiated retinal tissue, but all studies were performed with tissue selected for the bright appearance of retinal epithelium (Figure 1A).
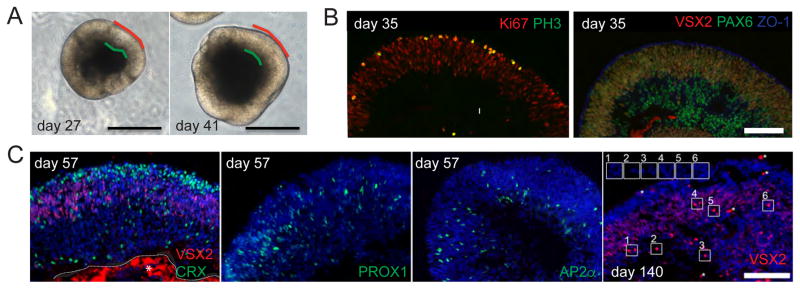
Figure 1. Production of retinal organoid tissue.
A) Typical in vitro differentiated retinal organoids with phase bright tissue. Apical surface is marked in red while green demarcates the basal side of the retinal epithelium. Scale bar: 500 μm. B) Immunostaining of cryosectioned retinal organoids demonstrating that the RPCs (VSX2+ PAX6+ Ki67+) differentiated from hESCs undergo mitosis (PH3) at the apical surface (ZO-1) scale bar: 100 μm. C) RPCs give rise to photoreceptors (CRX), horizontal (PROX1), amacrine (AP2α), and bipolar (bright VSX2 day 140, but not day 57) cells. Insets show bright VSX2 signal that co-stains with DAPI while asterisks (*) indicate bright artifacts. Non-specific red (VSX2 day 57) staining is outlined and marked with an asterisk. Scale bar: 100 μm.
2.2 Fixation and embedding
Retinal organoids were fixed in 4% paraformaldehyde (PFA) in phosphate buffered saline (PBS) at room temperature (RT) for 12–15 min and cryoprotected in 30% sucrose in PBS overnight at 4°C. For embedding, 100 μl OCT Tissue-Tek, spiked with Trypan Blue, was added to the organoids and frozen on dry ice within a larger volume of OCT, the Trypan enabling visualization of the organoids during sectioning. Cryosections (10 μm) were placed on positively charged slides. Blocks and slides were stored at −80°C. Slides were heated at 37°C for 15 min prior to staining, post fixed 5 min in 4% PFA, and rinsed 3 x 5 min in TBS + 0.05% Triton X-100 (TBST).
Fetal retina was obtained with approval from the CHLA Institutional Review Board. Eyes were collected in IMDM media and transported on ice. Cornea and lens were removed under a dissecting microscope, with the globe submerged in chilled PBS. The dissected globe was fixed in 4% PFA in PBS for 40 min (pf week 11.5), 25 min (pf week 13), 50 min (pf week 18) at RT followed by 3 PBS washes. Fixed eyes were placed in 30% sucrose in PBS for 15 min on ice and embedded on dry ice in OCT:sucrose (1:2). Cryosectioning and storage were as above for retinal organoids. For staining, slides were thawed at RT for few minutes, post-fixed in freshly prepared 4% PFA for 5 min and rinsed 3 x 5 min with TBST.
2.3 Immunofluorescence
Tissue sections were blocked for 1 hr with blocking buffer (5% normal donkey serum and 3% bovine serum albumin (BSA) in TBS with 0.2% Triton X-100) at RT. Sections were incubated with primary antibodies (see Table S1) diluted in blocking buffer for 2 hr at RT or overnight at 4°C, washed 3 X 5 min in TBST, incubated with secon dary antibodies diluted in blocking buffer for 1 hr at RT in the dark, washed 3 X 5 min in TBST with DAPI (1:1000) included in the last wash, and cover-slipped using Mowial mounting medium plus DABCO or ProLong Diamond (Thermo Fisher Scientific). For staining of frozen sections with surface antigens CD24-PE, CD90-FITC, CD184-PE or CD171-PE no post-fixing was performed, BD block solution (5% FBS, PBS, 0.1% NaN3, 0.05 mM EDTA) was substituted for blocking buffer, PBS substituted for TBST in washes, and TrueBlack (Biotium) used as per instructions to reduce autofluorescence. CD90-FITC staining of retinal organoids was enhanced using an Alexa Fluor 488 Tyramide Signal Amplification Kit (Thermo Fisher Scientific).
For immunofluorescent analysis of sorted cells, 5000 cells were centrifuged 400g 6 min, suspended in 10 μl RDM3-S (DMEM/F12, 10% FBS, 1X N2, 0.5X Glutamax, 0.5 μM retinoic acid, and penn-strep) transferred to poly-L-lysine (SIGMA), GFR Matrigel coated coverslips, allowed to attach at 37°C for 1 hour before fixing for 5 min at RT in 4% PFA. Cells were washed 3X with PBS for storage at 4°C and stained as above. Post staining, cells were treated with TrueBlack (Biotium) as per manufacturer’s guidelines, and stained with DAPI (1:1000 in PBS) for 5 min after the TrueBlack. For experiments involving EdU, the Click-It kit (Life Technologies- Thermo) staining protocol was followed as specified by the manufacturer.
Fluorescence images were acquired with the Nikon Eclipse Ti microscope at 20X and 10X magnification. Image intensity was adjusted to present the signal that exceeded most of the background signal in routinely performed IgG controls, except when rabbit antibodies were used, in which case a no primary antibody control was used for background signal adjustment.
2.4 Quantitation of RGCs in retinal organoids
Stained sections to analyze were selected based on the morphology (basal border below the RGC layer and lack of tears or folding of tissue). Single channel images of BRN3 and DAPI were manually globally thresholded in ImageJ to include all positive cells and minimal noise and background. To avoid inaccuracies associated with identifying closely packed individual cells the total thresholded area was recorded as a measurement proportional to cell number. The ratios of the BRN3 area (RGCs) to DAPI area (total cells) were computed for normalization.
2.5 Axon outgrowth
GFR Matrigel was diluted 3-fold with RDM3-S and 1 ml used to coat a 35 mm tissue culture dish which was incubated at 37°C for 30 min to create a thin, firm gel. Three retinal organoids cultured to the indicated day and 1 ml of RDM3-S were added on top of each gel. The retinal organoids were allowed to adhere to the gel and project neurites. Phase-contrast images were taken after 5 days of growth at 37°C, 5% CO2.
2.6 Flow cytometry
Retinal spheres of specified age were washed with HBSS and dissociated for 30 min at 37°C with Accutase (200 μl/sphere) with intermittent trituration. Cells were pelleted at 400g 6 min, resuspended in RDM3-S media, treated with DNase (100U/ml) for 15 min at RT, pelleted again and resuspended in BD block solution such that after subsequent antibody addition there were 10^4 cells/μl. Antibodies were used at recommended dilutions (Table S1) and incubated for 45 min to one hour at 4°C in the dark. After washing with BD block solution, the cells were resuspended in 5% FBS, PBS containing DAPI, placed on ice, and analyzed on a BD FACSCanto II or BD LSR II, or sorted on a BD FACSARIA into RDM3-S on ice. FACs DIVA software analysis of single color labeled beads was used to set compensation for all flow experiments except no compensation was performed for those using only PE, APC and DAPI. Doublets were eliminated on FCS-H vs FCS-W and SSC-H vs SSC-W plots and DAPI positive cells were excluded from all data shown or prior to sorting. “All cells” or “total cells” refers to the population of DAPI negative cells collected for analysis independent of surface marker characteristics. For experiments analyzing fixed cells by flow cytometry, cells were processed and stained with surface antigens as above, fixed in 4% PFA in PBS for 15 min RT, washed 2X with PBS, permeabilized with 90% methanol for 30 min at 4°C in the dark, washed 2X with 1% BSA in PBS (BSA/PBS), incubated with primary antibody diluted in BSA/PBS for 30 min RT in the dark, washed 2X in BSA/PBS, incubated in secondary antibody 30 min RT in the dark, washed 2X in BSA/PBS, and suspended in BSA/PBS for flow cytometry analysis. After the fixation step, cells were pelleted at 3000g for 5 min. When EdU detection was required the cells were processed as above except that 0.5% Triton in PBS for 15 min at RT was used for permeabilization and the Click-IT reaction was carried out as specified by instructions (Thermo Fisher Scientific) following permeabilization. Where noted, Zombie NIR (Biolegend) was used to eliminate dead cells and used according to the manufacturer’s instructions prior to surface antigen antibody incubation.
2.7 Quantification of sorted single cells
Single cell images had their channels separated and converted into .tiff files. Using ImageJ, background subtraction with a rolling ball radius of 50 was performed on all channels. Then the DAPI channel was manually thresholded so that only the nuclei were highlighted. Channels other than DAPI were specified to be measured in the analysis. ImageJ’s Analyze Particle function was then used to measure mean grey values (MGVs) of the specified channels for particles larger than 25 μm2 highlighted by the DAPI threshold. The tabulated MGVs for each condition were binned, and a cutoff determined as one greater than the highest MGV positive bin in the experiment controls. The percentage of cells above the MGV cutoff was used as an indicator for percentage of positive signal. For goat and mouse antibodies a concentration matched species specific IgG was used for the control. For rabbit antibodies no primary antibody was used as the control.
2.8 RT-qPCR
For RT-qPCR, RNA was extracted from 5000 or 10,000 sorted cells using a PicoPure RNA Isolation kit (Thermo Fisher Scientific) -including the DNase digestion step- and cDNA generated with Superscript III (Thermo Fisher Scientific) according to instructions. Realtime PCR reactions utilized primers and hydrolysis probes (Table S2), Taqman Universal Mastermix II (Thermo Fisher Scientific) and an Applied Biosystems 7900HT realtime PCR machine. Non template controls were performed in parallel. 10 μl assays were performed in triplicate and ΔΔCt calculated relative to the total population of live cells using GAPDH as a reference gene, except for Figure S2E where UBC served as the reference. Error is the propagated standard deviation of triplicate measures.
3. Results
3.1 Basic properties of RGCs produced in retinal organoid culture
Retinal organoids were produced from hESCs using a differentiation protocol similar to that developed in the Sasai laboratory (Nakano et al., 2012). RPCs developed in a polarized epithelium within two and a half to three weeks of differentiation giving rise to RGCs beginning week three to four, followed by horizontal, photoreceptor, amacrine, and bipolar cells in subsequent weeks (Figures 1 and 2).
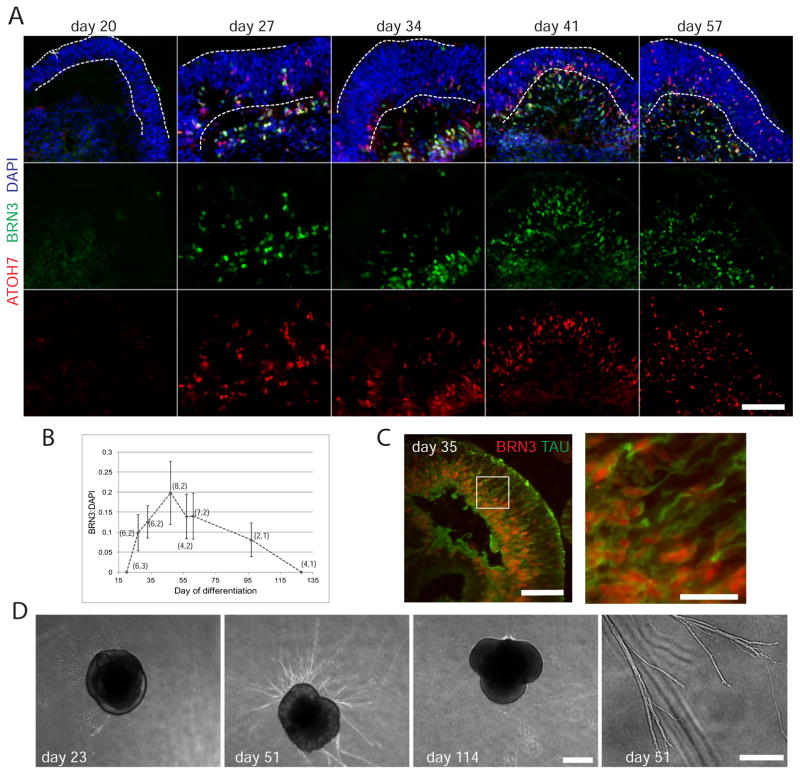
Figure 2. Characterization of RGCs in retinal organoids.
A) Immunostaining time course demonstrating RGC production in retinal organoid culture at the indicated day following initiation of differentiation from hESCs. The NBL resides roughly between the dotted lines and the basal layer is below. Scale bar: 100 μm. B) Quantitation of BRN3 immunofluorescent signal relative to DAPI signal from images similar to those shown in A. Data is the mean signal (see methods) from n images from m distinct preps. (n,m) is shown next for each point. Error is standard deviation. C) Tau immunostain demarcates axon outgrowth. Inset is magnified to the right. Scale bars: 100 μm, 25 um. D) Phase contrast images demonstrating axon outgrowth from a day 51 retinal organoid placed on Matrigel, but not from day 23 or 114 retinal organoids, prior to RGC genesis or after RGC death, respectively. Scale bar: 500 μm. Axon branching and growth cones at day 51 are shown at higher magnification. Scale bar: 100 μm.
To characterize RGCs in the retinal organoids we evaluated expression of the common molecular markers ATOH7 (MATH5), which identifies RGC competent RPCs as well as early post-mitotic RGCs, and BRN3 (BRN3a-c, POU4f1-3), which is a family of transcription factors required for RGC differentiation (Figure 2A) (Brown et al., 2001; Mu et al., 2005; Wang et al., 2001; Xiang et al., 2011; Yang et al., 2003). In retinal organoids ATOH7+ cells were first detected by day 27 and observed interspersed in the neuroblastic layer (NBL) as retinal organoids mature. They were also apparent in an emerging basal layer. The ATOH7+ cell population overlaps with BRN3+ cells that were also primarily observed in the emergent basal layer. This pattern is expected of ATOH7+ RPCs that mature first to BRN3+ ATOH7+ RGC precursors, and then to BRN3+, ATOH7− RGCs (Fu et al., 2009; Xiang et al., 2011).
The number of BRN3+ RGCs increased until roughly day 50 and then exhibited a slow decline (Figure 2A and B). Axonogenesis (TAU staining) from BRN3+ cells was observed in retinal organoids, and when provided a Matrigel substrate retinal organoid derived axons exhibited competence for robust extension (Figure 2C and D). No axon outgrowth was observed at ages prior to RGC production (day 23) or after RGC death (day 114) consistent with a RGC origin of these axons (Figure 2D).
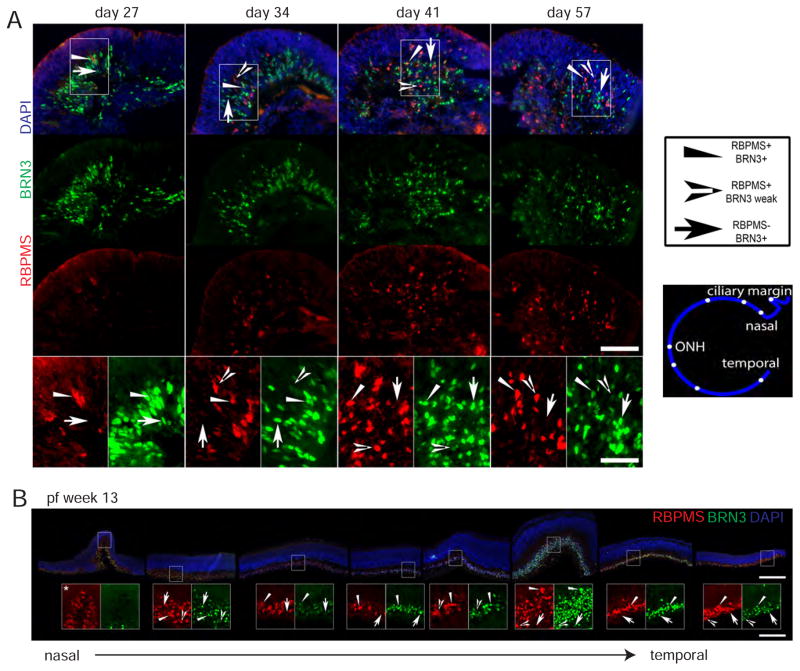
We also examined expression of RBPMS, an RNA binding protein thought to be expressed in all RGCs of vertebrates (Hörnberg et al., 2013; Kwong et al., 2010; Rodriguez et al., 2014). In retinal organoids BRN3+ cells were abundant at day 27 while RBPMS+ cells were rare; however, RBPMS cells became abundant at later ages, consistent with BRN3 being a transcription factor required early to specify RGC development and RBPMS functioning in a differentiating cell (Hörnberg et al., 2013; Mu et al., 2005; Xiang et al., 2011) (Figure 3A). Interestingly, in retinal organoids relatively few cells appeared to co-stain for BRN3 and RBPMS; however, upon close inspection in all RBPMS+ cells BRN3 signal was detected, albeit weakly compared to many BRN3+ cells. The opposite was not true in that not all BRN3+ cells expressed a detectable level of RBPMS (Figure 3A). This pattern of RBPMS− BRN3+, RBPMS+ BRN3+, or RBPMS+ BRN3 weak was observed in human fetal retina at post-fertilization (pf) week 13 as well (Figure 3B). Also apparent in fetal retina is that the relative expression patterns of RBPMS and BRN3 were not uniform across the developing retina, with BRN3 predominant in central, more mature, cone-rich retina and RBPMS more dominant in the less mature, rod-rich periphery.
Figure 3. RBPMS and BRN3 expression in embryonic human RGCs.
A) Immunostaining of retinal organoids. Insets show RBPMS− BRN3+, RBPMS+ BRN3+, and RBPMS+ BRN3 weak cells as indicated by the different arrows. The exposure is enhanced 2X in these images compared to above for clarity. Scale bar: 100 μm, 50 μm. B) Immunostaining of fetal retina with insets (enlarged 3X) demonstrating RBPMS− BRN+, RBPMS+ BRN3+, and RBPMS+ BRN3 weak RGCs as indicated by the type of arrowhead. Also enlarged is an inset of the ciliary margin where BRN3 appears absent from RBPMS+ cells at pf week 13. From left to right noncontiguous pictures (rotated and cropped) are organized to show the progression from nasal to temporal retina with cartoon showing approximate picture location in the retina. All pictures are from the same section. Scale bars: 250μm, 83 μm.
The results show that RGCs in retinal organoids are derived from ATOH7 competent progenitors, migrate to the basal layer of the retina, form distinct BRN3 and RBPMS expressing embryonic populations, and exhibit a robust competency to produce and extend axons; thus demonstrating that RGCs in hESC-derived retinal tissue share basic characteristics with RGCs in vitro.
3.2 CD184 and CD171 identify RGCs at differing stages of maturation in retinal organoid culture
An ability to isolate retinal organoid RGCs facilitates a more rigorous molecular characterization. Accordingly, we examined expression of CD90 (Thy-1), which has proven useful for RGC purification in rodent retina (Barnstable and Dräger, 1984; Barres et al., 1988). In human fetal retina, CD90 was detected in a pattern consistent with expression in maturing RGC processes: strong signal in the RGC nerve fiber layer (NFL) and weaker signal in the inner plexiform layer (IPL) (Figure S1A). However, immunostaining and flow cytometry of CD90 expression in retinal organoids was not consistent with expression solely on RGCs since CD90 expression was fluid through time, and persisted after the time when most RGCs have died (day 93) (Figure S1B, Figure 2B). A subset of cells, however, exhibited relatively high CD90 expression at days 34, 42 and 48, when RGCs were prevalent (Figure S1B). A small subset of these expressed BRN3B as expected of RGCs, but the majority expressed VSX2 and NES suggesting that they may be a subset of RPCs (Figure S2). Thus, CD90 expression appears to be confined to RGCs in fetal retina, but not in retinal organoids.
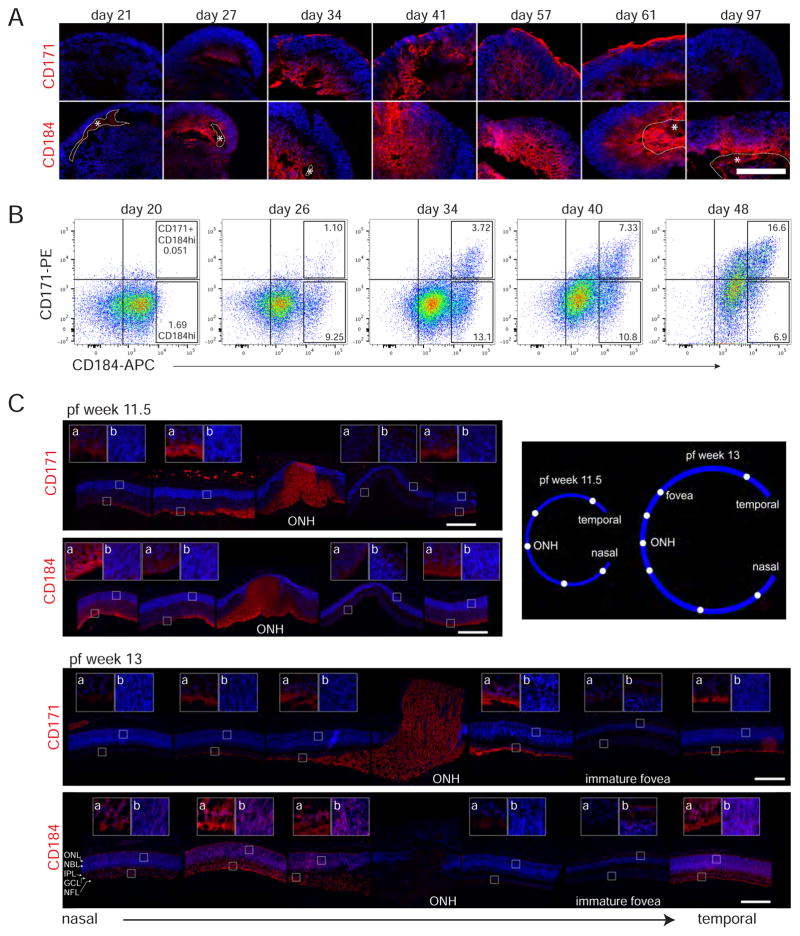
As CD90 did not clearly identify RGCs in hESC-derived tissue, the utility of other surface proteins that have reported expression in RGCs in mouse, chick, or zebrafish, namely CD171 (L1CAM), and CD184 (CXCR4), was investigated (Bartsch et al., 1989; Chalasani et al., 2003; Demyanenko and Maness, 2003; Q. Li et al., 2005; Lyckman et al., 2000; Pujic et al., 2006). The patterns of both CD171 and CD184 staining in retinal organoids were consistent with RGC expression (Figures 4 and S4). The onset of signal coincided roughly with the onset of RGC production, day 27, and signal was observed in the basal layer where RGCs were located (Figure 4A compared to Figure 2A). While both antigens were detected at day 27, CD171 appeared weak compared to CD184, while by day 34 the two surface markers had a similar intensity. This suggests that CD184 expression precedes that of CD171. CD171, similar to TAU, was detected on processes traversing the basal to apical dimension of the retinal organoids (for example Figure 2C, day 57 Figure 4A and days 36 and 41 Figure S4). In addition, both CD171 and TAU were detected at the apical surface of retinal organoids (Figure 2C, Figure 4A day 34–61 and Figure S4 d41–76). This detection of CD171 expression on the apical surface of the retinal organoids, is thus consistent with its well-documented expression on RGC axons (Demyanenko and Maness, 2003; Lyckman et al., 2000). Notably the CD171 surface signal diminished along with the basal signal with timing consistent with rapid death of RGCs (day 97). In contrast, the CD184 staining in retinal organoids persisted through day 97 (Figure 4A); however, detection of CD184 at this age varied which may be due to the presence or absence of RGCs at a culture period when they were rapidly dying (Figure 2B, data not shown).
Figure 4. Expression of surface markers CD184 and CD171 in hESC-derived and human fetal retina.
A) Developmental time course of hESC-derived retina immunostained with anti-CD171-PE (Life Technologies) and anti-CD184-PE. DAPI is blue. With the CD184 antibody areas of non-specific staining or probable non specificity are outlined and marked with an asterisk. An independent CD171 antibody showed similar staining pattern (Figure S4). Scale bar: 100 μm. B) Flow cytometry plots of dissociated retinal organoid cells of the indicated age labeled with anti-CD184-APC and anti-CD171-PE. Each plot shows data comprised of cells from six dissociated retinal organoids. Quadrant lines drawn based on unstained cell or IgG control signal (both exhibited equivalent signal), while boxes gate the CD184hi or CD184hi CD171+ populations with percent of total cells shown within. CD184hi is discriminated from CD184+ based on a visual assessment of the minor population exhibiting higher levels of CD184. C) Immunostaining of pf weeks 11.5 and 13 human fetal retina. Noncontiguous pictures show the progression from nasal to temporal retina with cartoon showing approximate picture location in the retina. Insets show either cells from the GCL(a) or NBL(b) at 3.5X ONH (optic nerve head). At pf 13 inset b includes part of the emerging ONL as well. Staining (including that in Figure S5 which shows BRN3 detection for reference) was performed on adjacent retina sections, each stained with different antibodies. For each antibody all pictures were taken from the same section with identical exposure times. The only exception is the pf 11.5 central picture (temporal to the ONH) because the ONH was not found in the same section as the most central retina stained. Scale bar: 250 μm.
Examination of CD184 and CD171 expression in retinal organoids in flow cytometry time courses suggest that most cells acquire a weak CD184 expression around day 20 since in one experiment the majority of cells are unlabeled (Figure S3) and in a second experiment the majority of cells are labeled (Figure 4B). If one assumes different preps exhibit some variability in maturity, then this is consistent with day 20 being an approximate time when the cell population is in transition from non-expressing to weakly expressing CD184. A subset of the retinal organoid cells acquired stronger CD184 (CD184hi) immunoreactivity by day 26, a population of cells which peaked at day 34 to 40 and then declined (Figures 4B and S3). Few cells exhibited CD171 expression on day 26, but that number continued to increase throughout the time course and the proportion of cells exhibiting a CD184hi CD171+ phenotype increased as retinal organoids mature (Figure 4B and S3). Both the flow cytometry and immunostaining data suggests that CD184 expression precedes that of CD171.
Expression of CD184 and CD171 was also examined in human fetal retina. Both CD184 and CD171 were clearly seen in the NFL on the axons below the ganglion cell layer (GCL) at pf weeks 11.5 and 13 although not in precisely the same pattern (Figures 4C and S5). CD171 exhibited strong expression on axons in the relatively mature central retina and those exiting the optic nerve head, with somewhat lower expression in the less mature nasal peripheral region at both ages. Also there was a notable absence of or weak CD171 expression in ganglion cells axons in the central/foveal region, yet there were clearly BRN3+ cells present (Figure S5). This suggests that central/foveal RGCs either do not express CD171 or do so with a differential timing.
CD184 was detected in pf weeks 11.5 and 13 as well. At pf week 11.5 CD184 was primarily detected in the NFL; however, in the nasal peripheral region expression appeared to pervade the emerging RGC layer consistent with localization to migrating cell bodies and neurites of migrating RGCs (Figure 4C). At the later developmental stage of pf week 13, CD184 expression was more ubiquitous, observed in the NFL, the RGC cell bodies, emerging IPL, as well as in the NBL and emerging ONL (Figure 4C). Like CD171, expression appeared absent from the central/foveal retina at these ages. In addition to being detected on cell types other than RGCs, CD184 expression appeared dynamic in RGC axons, very bright on GC axons at exiting the optic nerve at pf week 11.5, but largely disappearing by pf week 13, suggesting transient CD184 expression in RGC axons.
Comparing the most peripheral expression at pf week 11.5, CD184 appears to precede CD171 expression (Figure 4). Overall, subtleties in the expression patterns of CD171 and CD184 suggest that surface marker expression exhibits a complexity in the developing fetal retina that will require a more extensive analysis to document precisely, but indicate a specificity to RGC neurites for CD171 and suggests a transient expression in RGCs for CD184.
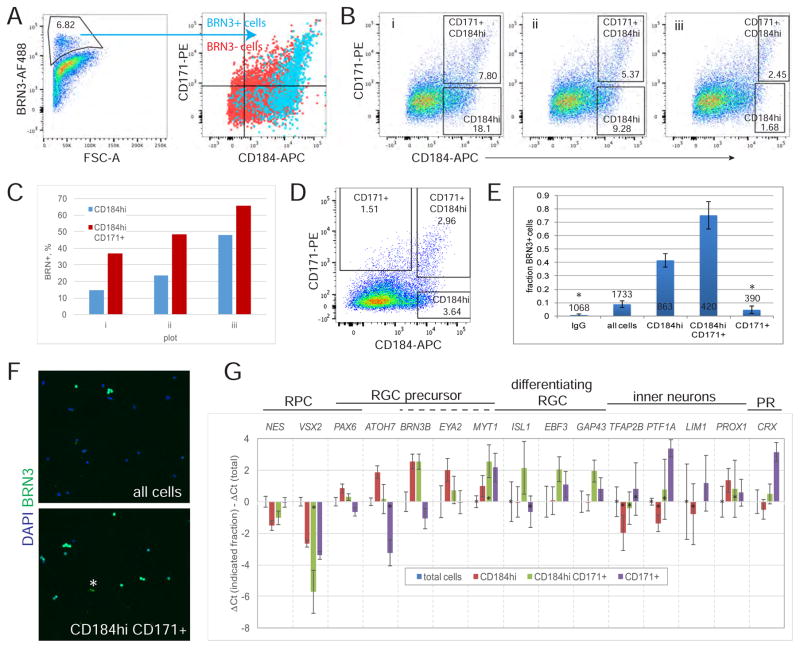
The expression in the RGC layer of both fetal and hESC-derived retina suggested that sorting of the CD184hi cells and/or the CD184hi CD171+ cells could be useful for RGC isolation. To test this idea dissociated retinal organoid cells were fixed, co-immunolabeled for CD171, CD184 and BRN3 and analyzed by flow cytometry. BRN+ cells comprised about 7% of the total cells after 35 days of differentiation and most expressed CD184 or CD184 and CD171 (Figure 5A). BRN3+ cells were most enriched when limiting selection to the highest CD184 expression (Figure 5B and C). Sorting of live cells from the hESC derived retinal tissue at differentiation day 33 demonstrated that about 75% of cells exhibited BRN3 expression in a CD184hi CD171+ fraction and about 40% in a CD184hi fraction (Figure 5D, E and F). Since BRN3− cells were present in a CD184hi CD171+ fraction we addressed the possibility that these cells expressed BRN3 weakly but might have strong RBPMS expression as was observed for a subset of RGCs by immunostaining (Figure 2A). Therefore, in a similar cell sorting experiment, cells were stained for RBPMS in addition to BRN3. Most RBPMS+ cells were also BRN3+, but RBPMS+ cells BRN3− cells comprised only an additional 5% of the cells, meaning that the precise nature of the remaining BRN3− RPBMS− cells remains unclear (data not shown).
Figure 5. Sorting retinal organoid cells using CD184 and CD171 enriches for RGCs.
A–C) At day 35 retinal organoids were dissociated to single cells, labeled with anti-CD184-APC and anti-CD171-PE, subsequently fixed and immunostained for BRN3, then analyzed by flow cytometry. A) The gate used to define BRN3+ cells is shown, as is the distribution of these cells on a CD171 versus CD184 plot. No cells segregated from the main population in IgG controls. B) Plots of the data in A shown with gates drawn to include increasingly higher levels of CD184. Within each gate is the % total cells. C) Data derived from the plots shown in B is plotted. Percent of BRN3+ cells within each gate was determined by applying the BRN3+ gate in shown in A to cells in the gated population. D–G) At day 33 of differentiation 30 retinal organoids were dissociated to single cells, labeled with anti-CD184-APC and anti-CD171-PE, and live cells sorted using the gates indicated in D. Gates were selected to maximize the percent of RGCs based on data in B and C. E) Immunodetection of BRN3+ cells in sorted cell population is shown as average +/− SD from 3 (or 2*) coverslips. The total number of cells analyzed to generate the data is indicated. F) Representative images used to generate the graphed data in E. * artifact excluded from analysis. G) RT-qPCR analysis of sorted cells relative to total cells. Those samples where average Ct values were greater than 37 are marked (*) since this is near the limit of detection and contributes to large errors for some samples, especially when the gene is weakly expressed in the total cell population. Qualitatively similar data was obtained in four similar experiments, two of which are shown in Figure S6.
Enrichment for RGCs was also seen by RT-qPCR since BRN3B transcripts were enhanced in both CD184hi and CD184hi CD171+ fractions, but not CD171+ fractions (Figures 5G and S6). Genes identifying RPCs (VSX2 and NES) were relatively depleted in the RGC enriched fractions (CD184hi and CD184hi CD171+) suggesting a reduction in the percent of RPCs, the cells that dominate the retinal organoids at this age. ATOH7, required for RGC formation but not expressed in mature RGCs, was enriched primarily in the CD184hi fraction. EYA2, which is thought, along with BRN3B, to be a direct target of ATOH7, was detected in both CD184hi and CD184hi CD171+ fractions (Gao et al., 2014; Hutcheson and Vetter, 2001). Other genes expressed in RGCs including ISL1, EBF3, MYT1 and GAP43 were concentrated primarily in the CD184hi CD171+ fraction. EBF3 and GAP43 are expressed in a more mature, differentiating RGCs and, in the human retina, ISL1 expression is detected subsequent to BRN3 (Pacal and Bremner 2014). Thus, the pattern of enrichment of all of these genes suggests that the CD184hi CD171+ fraction contains more mature BRN3+ RGCs than the CD184hi fraction.
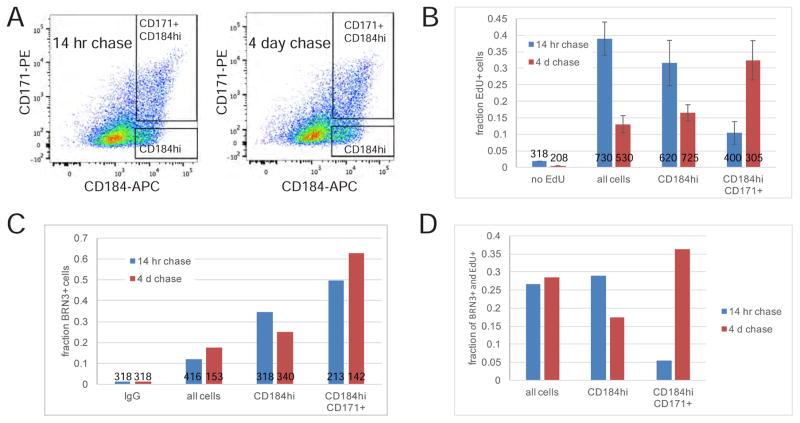
To further evaluate whether CD184hi cells are less mature than the RGCs in the CD184hi CD171+ fraction, we performed EdU pulse-chase experiments designed to observe progression of immature, but post-mitotic RGCs to more differentiated RGCs (Figure 6). Intact retinal organoids were labeled with a 10 hour EdU pulse in order to label a large number of proliferating RPCs, followed by a relatively short (14 hour) and long (4 day) label-free chase. Some of the RPCs (EdU+ CD184+) were assumed to exit the cell cycle during the chase period and enhance expression of CD184 and CD171 to become EdU+ CD184hi or EdU+ CD184hi CD171+ cells. If after leaving the cell cycle, EdU+ cells first express high CD184 (but not CD171), and subsequently, as cells mature, CD171 is expressed, then after the “short” chase (14 hours) more EdU+ cells would be present in the CD184hi fraction than the CD184hi CD171+ fraction. However, after the “long” chase (4 days), the EdU+ cells would be expected to distribute in both fractions with fewer EdU+ cells in the CD184hi fraction compared to CD184hi CD171+ fraction, as they would have matured to populate the CD184hi CD171+ fraction. Alternatively, if there were no difference in expression timing of CD184 and CD171 then similar numbers of EdU+ cells in the different sorted fractions would be expected. The results showed that after a 14 hour chase about 30% of BRN3+ cells in the CD184hi population were EdU+, whereas only 5% of BRN3+ cells were EdU+ in the CD184hi CD171+ population (Figure 6D). After a 4 day chase the percent of BRN3+ and EdU+ cells in the CD184 hi fraction had dropped to 17% while in the CD184hi CD171+ population the percent of BRN3+ and EdU+ cells increased to 36% (Figure 6D). This data is consistent with newly post mitotic CD184hi BRN3+ RGCs maturing to express CD171.
Figure 6. RGC precursors express CD184 prior to CD171.
A) On day 31 and day 34 of differentiation 15 retinal organoids were labeled with 2 μM EdU for 10 hours followed by a 4 day or 14 hour chase, respectively. On day 35, following dissociation, retinal organoid cells were stained with anti-CD184-APC and anti-CD171-PE then live cells sorted using the gates shown. B) EdU+ cells in sorted populations detected by Click-iT chemistry are shown as the average +/− SD EdU+ cells on 2 coverslips. The total number of cells analyzed to generate data is indicated in the respective columns. C) Immunodetection of BRN3+ cells in the sorted populations. The number of cells analyzed to generate data in columns from left to right are indicated. D) The EdU+ and BRN3+ cell data (B and C) was used to calculate the fraction of BRN3+ cells labeled by EdU.
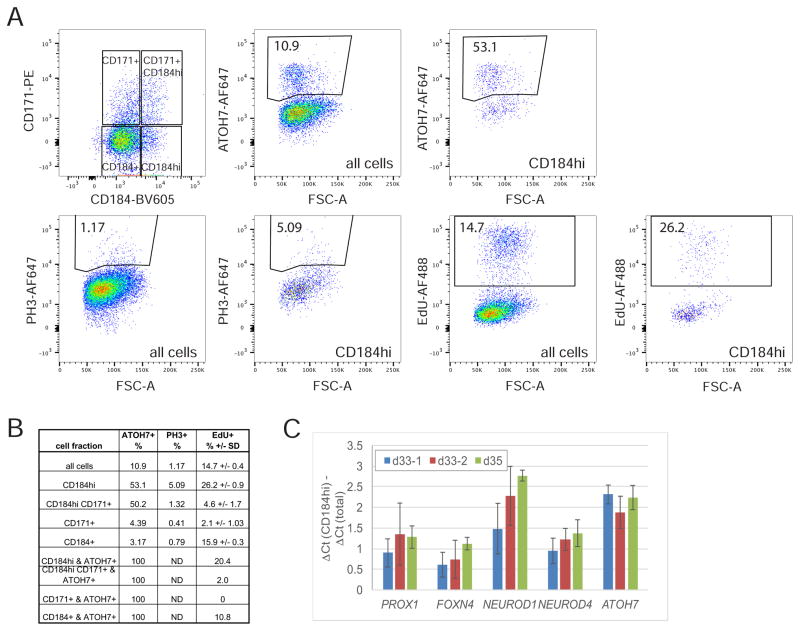
RPCs in retinal organoids were expressing CD184 around day 20 in culture (Figure 4B and S3), and by day 26 some cells expressed a higher level of CD184 (Figure 4B and S3). We wondered whether the high CD184 expression is a property only of post-mitotic cells or alternatively, whether at least some of these cells remain in the cell cycle. To address this, retinal organoids were subjected to a 1 hour EdU pulse to label only cells in S-phase, then co-labeled with CD184, CD171, and ATOH7 or PH3 and analyzed by flow cytometry. About 26% of the CD184hi cells were labeled by EdU, and hence in S-phase, and about 5% were in mitosis (PH3+) (Figure 7A and B), demonstrating that a significant fraction of CD184hi cells had not yet exited the cell cycle. In addition, 20% of the CD184hi ATOH7+ cells were EdU+, similar to the 25% percent Atoh7 expressing RPCs reported to be actively proliferating in the mouse retina (Figure 7B) (Kiyama et al., 2011). This data indicates that many RPCs expressing ATOH7, or those RPCs competent to become RGCs, also expressed the higher level of CD184. Also, RT-qPCR analysis of the CD184hi fraction of sorted cells showed an enrichment for several factors, FOXN4, PROX1, NEUROD1, NEUROD4(MATH3), in addition to ATOH7, that are known to be expressed as RPCs acquire competence for generating amacrine, horizontal, and photoreceptor cells, and transition toward cell cycle exit (Akagi et al., 2004; Boije et al., 2013; Dyer et al., 2003; Fujitani et al., 2006; Inoue et al., 2002; S. Li et al., 2004; Mao et al., 2008; 2013; Xiang and S. Li, 2013) (Figure 7C). Together these data raise the possibility that as some RPCs move toward cell cycle exit or neurogenic divisions to generate RGCs, and possibly other cell types, they boost CD184 expression to higher levels. This is consistent with the observed CD184 staining of the NBL in fetal retina, although the fetal staining varies with intensity depending upon age or location within the developing retina suggestive of an unresolved complexity (Figure 4). Overall, the data suggests that RPCs in retinal organoids acquire a low level CD184 expression around day 20, then roughly coincident with an RPCs’ acquisition of ATOH7 expression, or competence to differentiate to RGCs, it enhances CD184 expression prior to exiting the cell cycle.
Figure 7. CD184hi cells include progenitors that have not exited the cell cycle.
A) Retinal organoids day 41 were incubated for 1 hour with 10 μM EdU then immediately dissociated, labeled with Zombie NIR, anti-CD184-BV-605, anti-CD171-PE, anti ATOH7-AF647 or anti PH3-AF647, and EdU Click-IT linked to AF488. The total population, as well as four gates in the CD171 versus CD184 plot, were analyzed for the %EdU+ cells or %ATOH7 or PH3+ as shown for CD184hi fraction. Gates were drawn by eye to include distinct populations, not detected in controls, of positive cells. The % positive cells for CD184+, CD184hi CD171+ and CD171+ are shown in the table in B. C) RT-qPCR analysis of several competence conferring transcription factors in CD184hi sorted cells relative to total cells from three independent sort experiments. The gates used to sort the cells analyzed are shown in Figure 5D for d33-1 and Figure S5 for d33-2 and d35.
4. Discussion
In this work we characterized basic properties of hESC-derived RGCs as well as demonstrated that antibodies to CD184 and CD171 could be used to identify maturation stages of hESC-derived RGCs from committed progenitors to post-mitotic maturing RGC precursors.
4.1 Human retinal ganglion cell characterization
RGCs have been identified in retinal organoids by expression of BRN3 (Nakano et al., 2012; Phillips et al., 2012; Zhong et al., 2014), and axons attributed to organoid RGCs are capable of producing sodium-dependent action potentials as well as microtubule dependent antero- and retrograde transport (Maekawa et al., 2015; Tanaka et al., 2015). However, RGCs in this system otherwise remain uncharacterized. Here we demonstrated that several properties of RGCs in organoids are similar to RGCs in vivo. 1) As anticipated of bona fide RGCs, the data suggested that retinal organoid RGCs were generated from ATOH7+ RPCs, that mature into BRN3+ATOH7− RGC cell precursors. 2) In human RGCs, BRN3 expression preceded that of ISL (Pacal and Bremner, 2014), whereas in the mouse where these genes were expressed co-temporally (Mu et al., 2008; Rachel et al., 2002). The human organoid RGCs reproduced the human pattern. 3) hESC-derived RGCs, like human fetal RGCs expressed RBPMS in a subset of BRN3+ cells, and many of the RBPMS+ cells expressed BRN3 relatively weakly. 4) Developing human RGCs in both retinal organoid and fetal tissue expressed CD184 and CD171. 5) CD184 was expressed on organoid RGCs with a timing that temporarily preceded CD171, an observation made in the retinal organoids, but consistent with the staining pattern observed in fetal retina. 6) BRN3 enriched populations in retinal organoids were shown to express genes previously identified in mammalian RGC precursors (PAX6, ATOH7, EYA2, MYT1) and differentiating RGCs (EBF3, GAP43) (Gao et al., 2014; Mu et al., 2005). 7) The production of axons from retinal organoid retinal ganglion cells reported here and by others (Maekawa et al., 2015; Tanaka et al., 2015) suggests that the retinal organoid environment is providing appropriate factors to induce axon outgrowth, a process which does not occur by default but rather requires trophic signaling and is enhanced by electrical activity (Goldberg et al., 2002). These data, and the fact that some of our observations (numbers 3, 4, and 5 above) were first made in in vitro in retinal organoids and subsequently confirmed in fetal retina, serves to extend the current morphological and cell marker evidence (Meyer et al., 2011; Nakano et al., 2012; Zhong et al., 2014) documenting the validity of employing retinal organoids as an experimental model to study early human retinal development.
This work however also served to uncover some deviations from normal developmental processes. Extension of nascent axons to the apical surface (Figure 2C) suggests RGC polarize improperly since axons normally extend basally to the inner limiting membrane. In zebrafish, RGC polarization occurs shortly after cell cycle exit and depends on extrinsic signals, such as laminin in the inner limiting membrane (Randlett et al., 2011; Zolessi et al., 2006). That the lens and ciliary body synthesize proteins comprising this basement membrane (Halfter et al., 2008; 2005) but these structures are absent in the organoid cultures perhaps offers an explanation for this result. The death of all RGCs within a couple months also does not normally occur in vivo. The organoid RGC axons lack not only a proper inner limiting membrane, but also the optic nerve head, optic track and appropriate neuronal targets. This raises the possibility that the observed decrease in RGC number may be analogous to RGC apoptosis that occurs in vivo when RGCs are unable to innervate appropriate targets (Bähr, 2000; Isenmann et al., 2003; Provis, 1987; Provis and Penfold, 1988). We also observed aberrant CD90 expression as it was not restricted to RGCs as it largely appears to be in the human fetal retina. This may be an artifact of in vitro culture. These observations define some limitations of the organoid system which could be targeted with culture refinements to enhance the value as a model system.
This work revealed several other potentially interesting observations of developing human RGCs that will require further study to validate functional relevance. The expression patterns of BRN3 and RBPMS in pf week13 fetal retina elucidated distinct RGC populations during development: RBPMS− BRN3+, RBPMS+ BRN3+, or RBPMS+ BRN3 weak cells. This could be representative of different states of maturation, which based on expression timing observed in organoids, is predicted to progress from immature RBPMS− BRN3+ cells to more mature RBPMS+ BRN3 weak cells. However, the distribution in the fetal retina, where the less mature retina in the rod rich periphery is heavily populated by the RBPMS+ BRN3 weak cells, whereas the more mature cone rich central retina is enriched in the RBPMS− BRN3+ cells, argues against a developmental progression and raises the possibility that these markers distinguish distinct developmental paths that early human RGCs follow en route to diversification to their many types. In contrast, in adult rodent retina RBPMS is thought to be universally detected in RGCs (Piri et al., 2006; Rodriguez et al., 2014). This difference could be due to the maturity of RGCs (embryonic versus adult) or species differences.
Additionally, CD171 was readily detected in the human fetal retina on RGC neurites at pf weeks 11.5 and 13 as was expected given that CD171 (L1CAM) is implicated in axon guidance or targeting functions of at least some RGCs (Buhusi et al., 2008; Demyanenko and Maness, 2003; Feldheim and O’Leary, 2010). However, CD171 staining was absent in the central/foveal region implying that these RGCs do not utilize CD171 for axon biology, at least at these developmental stages.
CD184 staining of the fetal retina is also interesting since it’s expression appears transient in RGC axons as it is readily detected on fibers exiting the optic nerve head at pf 11.5, but only a very modest expression is seen at pf 13 on optic nerve head fibers. This may be analogous to a brief period of CD184 expression during RGC differentiation that has been described in zebrafish (Pujic et al., 2006). Transient expression may explain the absence in the relatively mature central/foveal at the ages examined.
4.2 Expression of CD184 and CD171 on human retinal organoid RGCs
This work demonstrated that the timing of CD184 and CD171 expression in differentiating retinal organoids was concurrent with RGC production and maturation and showed that high levels of CD184 expression were characteristic of RGC competent progenitors and early post-mitotic cells, while maturing RGCs maintained CD184 expression but also expressed CD171. This conclusion is supported by the detection of ATOH7+ cells in S-phase in CD184hi fractions, the appearance of CD184hi expression chronologically prior to CD171, the disappearance of CD184hi cells and appearance of greater number of dual positive cells through time, and the detection of pulsed-chased EdU in the CD184hi population prior to the CD184hi CD171+ cells. There also was a lack of enrichment of genes associated with RGC differentiation in the CD184hi cells, whereas these cells showed greater enrichment for ATOH7 which is only transiently expressed in developing RGCs.
The data discussed above demonstrated that CD184 and CD171 were expressed on RGCs in retinal organoids and thus enables employment of these surface markers to obtain a cell population enriched for RGCs or RGC precursors. However, there were BRN3− and RBPMS− immunonegative cells present in sorted fractions. This suggests that CD184hi and CD184hi CD171+ sorted fractions may also contain other cell types. As suggested by expression of FOXN4, PROX1, NEUROD1, and NEUROD4 in Figure 7C, the CD184hi fraction, in addition to RGC competent progenitors and RGC precursors, may contain RPCs competent to become amacrine, horizontal, and/or photoreceptor cells. Furthermore ATOH7 expressing progenitors do not exclusively develop into RGCs, as lineage tracing of ATOH7+ cells revealed that many ATOH7+ cells become amacrine cells, horizontal cells, and rod and cone photoreceptors (Brzezinski et al., 2012; Feng et al., 2010). Thus, it will be interesting in future studies to understand the nature of all the cells in the CD184hi population especially given the heterogeneous character of RPCs and probabilistic decisions associated with RPC cell fate commitment (Boije et al., 2014; Cepko, 2014; Kiyama et al., 2011; Trimarchi et al., 2008). The CD184hi CD171+ fraction showed variable enrichment of transcripts for inner neurons (TFAB2B, PTF1A, LIM1, PROX1) and photoreceptors (CRX) suggesting that photoreceptors, horizontal and amacrine cells can be present (Figures 5G and S5). Notably the relative level of these transcripts was reduced when the gate used for sorting was more restricted to include only the highest CD184 expressing cells (compare Figure S6A to Figures 4D and G and S6B), consistent with the data showing BRN3+ cells were most enriched when limiting selection to the highest CD184 expression (Figure 5B and C). The data is consistent with a fraction of the non BRN3+ cells being photoreceptors, amacrine cells, or horizontal cells. Thus, this work demonstrates that CD171 and CD184 permit significant enrichment of differentiating RGCs from retinal organoids, but not isolation to purity unless detection of other cell type transcripts is not associated with translation into proteins or is a reflection of plasticity and heterogeneity at this early stage in development (Trimarchi et al., 2008; 2007).
In conclusion, CD184 and CCD171, by enabling enrichment of RGCs at different stages of maturation, should be useful tools for further characterization of human retinal organoid RGCs.
Supplementary Material
Appendix A: Supplementary data
Appendix B: Supplementary tables
Highlights.
Retinal ganglion cells in 3D culture and fetal retina exhibit similar properties.
ATOH7+ progenitors and differentiating retinal ganglion cells express CD184.
CD171 is expressed on differentiating retinal ganglion cells.
CD184 and CD171 identify different differentiation stages of retinal ganglion cells.
Acknowledgments
We thank Esteban Fernandez for imaging assistance and ImageJ scripts for imaging analysis, Michael Sheard, Jackie Lin, Ann George, and Jonathon Kirzner for flow cytometry support, Hardeep Singh for assistance with fetal retina, Matthew Thornton and Brendan Grubbs for fetal retina procurement, and David Cobrinik, Oscar Aparicio, and Mark Borchert for comments on the manuscript. This work was supported by the AB Reins Foundation, the Neonatal Blindness Fund, the Shavelle Foundation, the National Institutes of Health grant R21EY025419, a BD Biosciences Stem Cell Research Grant (JGA), a Caltech Summer Undergraduate Research Fellowship (JMC), and the Children’s Hospital Los Angeles Stem Cell Core Facility, a CIRM funded shared laboratory.
Abbreviations
- GCL
ganglion cell layer
- hESC
human embryonic stem cells
- IPL
inner plexiform layer
- NBL
neuroblastic layer
- NFL
nerve fiber layer
- ONH
optic nerve head
- ONL
outer nuclear layer
- pf
post fertilization
- PSC
pluripotent stem cell
- RPC
retinal progenitor cell
- RGC
retinal ganglion cell
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Akagi T, Inoue T, Miyoshi G, Bessho Y, Takahashi M, Lee JE, Guillemot F, Kageyama R. Requirement of multiple basic helix-loop-helix genes for retinal neuronal subtype specification. J Biol Chem. 2004;279:28492–28498. doi: 10.1074/jbc.M400871200. [DOI] [PubMed] [Google Scholar]
- Almasieh M, Wilson AM, Morquette B, Cueva Vargas JL, Di Polo A. The molecular basis of retinal ganglion cell death in glaucoma. Progress in Retinal and Eye Research. 2012;31:152–181. doi: 10.1016/j.preteyeres.2011.11.002. [DOI] [PubMed] [Google Scholar]
- Barnstable CJ, Dräger UC. Thy-1 antigen: a ganglion cell specific marker in rodent retina. Neuroscience. 1984;11:847–855. doi: 10.1016/0306-4522(84)90195-7. [DOI] [PubMed] [Google Scholar]
- Barres BA, Silverstein BE, Corey DP, Chun LL. Immunological, morphological, and electrophysiological variation among retinal ganglion cells purified by panning. Neuron. 1988;1:791–803. doi: 10.1016/0896-6273(88)90127-4. [DOI] [PubMed] [Google Scholar]
- Bartsch U, Kirchhoff F, Schachner M. Immunohistological localization of the adhesion molecules L1, N-CAM, and MAG in the developing and adult optic nerve of mice. J Comp Neurol. 1989;284:451–462. doi: 10.1002/cne.902840310. [DOI] [PubMed] [Google Scholar]
- Bähr M. Live or let die - retinal ganglion cell death and survival during development and in the lesioned adult CNS. Trends in Neurosciences. 2000;23:483–490. doi: 10.1016/s0166-2236(00)01637-4. [DOI] [PubMed] [Google Scholar]
- Boije H, MacDonald RB, Harris WA. Reconciling competence and transcriptional hierarchies with stochasticity in retinal lineages. Curr Opin Neurobiol. 2014;27:68–74. doi: 10.1016/j.conb.2014.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boije H, Shirazi Fard S, Ring H, Hallböök F. Forkheadbox N4 (FoxN4) triggers context-dependent differentiation in the developing chick retina and neural tube. Differentiation. 2013;85:11–19. doi: 10.1016/j.diff.2012.12.002. [DOI] [PubMed] [Google Scholar]
- Borchert M. Reappraisal of the optic nerve hypoplasia syndrome. J Neuroophthalmol. 2012;32:58–67. doi: 10.1097/WNO.0b013e31824442b8. [DOI] [PubMed] [Google Scholar]
- Bouhenni RA, Dunmire J, Sewell A, Edward DP. Animal models of glaucoma. J Biomed Biotechnol. 2012;2012:692609. doi: 10.1155/2012/692609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown NL, Patel S, Brzezinski J, Glaser T. Math5 is required for retinal ganglion cell and optic nerve formation. Development. 2001;128:2497–2508. doi: 10.1242/dev.128.13.2497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brzezinski JA, Prasov L, Glaser T. Math5 defines the ganglion cell competence state in a subpopulation of retinal progenitor cells exiting the cell cycle. Developmental Biology. 2012;365:395–413. doi: 10.1016/j.ydbio.2012.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buhusi M, Schlatter MC, Demyanenko GP, Thresher R, Maness PF. L1 interaction with ankyrin regulates mediolateral topography in the retinocollicular projection. Journal of Neuroscience. 2008;28:177–188. doi: 10.1523/JNEUROSCI.3573-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cepko C. Intrinsically different retinal progenitor cells produce specific types of progeny. Nature Reviews Neuroscience. 2014;15:615–627. doi: 10.1038/nrn3767. [DOI] [PubMed] [Google Scholar]
- Chalasani SH, Baribaud F, Coughlan CM, Sunshine MJ, Lee VMY, Doms RW, Littman DR, Raper JA. The chemokine stromal cell-derived factor-1 promotes the survival of embryonic retinal ganglion cells. Journal of Neuroscience. 2003;23:4601–4612. doi: 10.1523/JNEUROSCI.23-11-04601.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooke JA, Meyer JS. Human Pluripotent Stem Cell-Derived Retinal Ganglion Cells: Applications for the Study and Treatment of Optic Neuropathies. Curr Ophthalmol Rep. 2015;3:200–206. doi: 10.1007/s40135-015-0081-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demyanenko GP, Maness PF. The L1 cell adhesion molecule is essential for topographic mapping of retinal axons. Journal of Neuroscience. 2003;23:530–538. doi: 10.1523/JNEUROSCI.23-02-00530.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyer MA, Livesey FJ, Cepko CL, Oliver G. Prox1 function controls progenitor cell proliferation and horizontal cell genesis in the mammalian retina. Nat Genet. 2003;34:53–58. doi: 10.1038/ng1144. [DOI] [PubMed] [Google Scholar]
- Feldheim DA, O’Leary DDM. Visual map development: bidirectional signaling, bifunctional guidance molecules, and competition. Cold Spring Harbor Perspectives in Biology. 2010;2:a001768. doi: 10.1101/cshperspect.a001768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng L, Xie ZH, Ding Q, Xie X, Libby RT, Gan L. MATH5 controls the acquisition of multiple retinal cell fates. Mol Brain. 2010;3:36. doi: 10.1186/1756-6606-3-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu X, Kiyama T, Li R, Russell M, Klein WH, Mu X. Epitope-tagging Math5 and Pou4f2: new tools to study retinal ganglion cell development in the mouse. Dev Dyn. 2009;238:2309–2317. doi: 10.1002/dvdy.21974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujitani Y, Fujitani S, Luo H, Qiu F, Burlison J, Long Q, Kawaguchi Y, Edlund H, Macdonald RJ, Furukawa T, Fujikado T, Magnuson MA, Xiang M, Wright CVE. Ptf1a determines horizontal and amacrine cell fates during mouse retinal development. Development. 2006;133:4439–4450. doi: 10.1242/dev.02598. [DOI] [PubMed] [Google Scholar]
- Gao Z, Mao CA, Pan P, Mu X, Klein WH. Transcriptome of Atoh7 retinal progenitor cells identifies new Atoh7-dependent regulatory genes for retinal ganglion cell formation. Dev Neurobiol. 2014;74:1123–1140. doi: 10.1002/dneu.22188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Filion P, Borchert M. Optic nerve hypoplasia syndrome: a review of the epidemiology and clinical associations. Curr Treat Options Neurol. 2013;15:78–89. doi: 10.1007/s11940-012-0209-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg JL, Espinosa JS, Xu Y, Davidson N, Kovacs GTA, Barres BA. Retinal ganglion cells do not extend axons by default: promotion by neurotrophic signaling and electrical activity. Neuron. 2002;33:689–702. doi: 10.1016/s0896-6273(02)00602-5. [DOI] [PubMed] [Google Scholar]
- Halfter W, Dong S, Dong A, Eller AW, Nischt R. Origin and turnover of ECM proteins from the inner limiting membrane and vitreous body. Eye. 2008;22:1207–1213. doi: 10.1038/eye.2008.19. [DOI] [PubMed] [Google Scholar]
- Halfter W, Dong S, Schurer B, Ring C, Cole GJ, Eller A. Embryonic synthesis of the inner limiting membrane and vitreous body. Investigative Ophthalmology & Visual Science. 2005;46:2202–2209. doi: 10.1167/iovs.04-1419. [DOI] [PubMed] [Google Scholar]
- Hörnberg H, Wollerton-van Horck F, Maurus D, Zwart M, Svoboda H, Harris WA, Holt CE. RNA-binding protein Hermes/RBPMS inversely affects synapse density and axon arbor formation in retinal ganglion cells in vivo. Journal of Neuroscience. 2013;33:10384–10395. doi: 10.1523/JNEUROSCI.5858-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutcheson DA, Vetter ML. The bHLH factors Xath5 and XNeuroD can upregulate the expression of XBrn3d, a POU-homeodomain transcription factor. Developmental Biology. 2001;232:327–338. doi: 10.1006/dbio.2001.0178. [DOI] [PubMed] [Google Scholar]
- Hynds RE, Giangreco A. Concise review: the relevance of human stem cell-derived organoid models for epithelial translational medicine. Stem Cells. 2013;31:417–422. doi: 10.1002/stem.1290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue T, Hojo M, Bessho Y, Tano Y, Lee JE, Kageyama R. Math3 and NeuroD regulate amacrine cell fate specification in the retina. Development. 2002;129:831–842. doi: 10.1242/dev.129.4.831. [DOI] [PubMed] [Google Scholar]
- Isenmann S, Kretz A, Cellerino A. Molecular determinants of retinal ganglion cell development, survival, and regeneration. Progress in Retinal and Eye Research. 2003;22:483–543. doi: 10.1016/S1350-9462(03)00027-2. [DOI] [PubMed] [Google Scholar]
- Jayaram H, Limb GA, Becker S. Stem Cell Based Therapies for Glaucoma. 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiyama T, Mao CA, Cho JH, Fu X, Pan P, Mu X, Klein WH. Overlapping spatiotemporal patterns of regulatory gene expression are required for neuronal progenitors to specify retinal ganglion cell fate. Vision Research. 2011;51:251–259. doi: 10.1016/j.visres.2010.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwong JMK, Caprioli J, Piri N. RNA binding protein with multiple splicing: a new marker for retinal ganglion cells. Investigative Ophthalmology & Visual Science. 2010;51:1052–1058. doi: 10.1167/iovs.09-4098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q, Shirabe K, Thisse C, Thisse B, Okamoto H, Masai I, Kuwada JY. Chemokine signaling guides axons within the retina in zebrafish. Journal of Neuroscience. 2005;25:1711–1717. doi: 10.1523/JNEUROSCI.4393-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Mo Z, Yang X, Price SM, Shen MM, Xiang M. Foxn4 controls the genesis of amacrine and horizontal cells by retinal progenitors. Neuron. 2004 doi: 10.1016/j.neuron.2004.08.041. [DOI] [PubMed] [Google Scholar]
- Lyckman AW, Moya KL, Confaloni A, Jhaveri S. Early postnatal expression of L1 by retinal fibers in the optic tract and synaptic targets of the Syrian hamster. J Comp Neurol. 2000;423:40–51. [PubMed] [Google Scholar]
- Maekawa Y, Onishi A, Matsushita K, Koide N, Mandai M, Suzuma K, Kitaoka T, Kuwahara A, Ozone C, Nakano T, Eiraku M, Takahashi M. Optimized Culture System to Induce Neurite Outgrowth From Retinal Ganglion Cells in Three-Dimensional Retinal Aggregates Differentiated From Mouse and Human Embryonic Stem Cells. Curr Eye Res. 2015:1–11. doi: 10.3109/02713683.2015.1038359. [DOI] [PubMed] [Google Scholar]
- Mao CA, Wang SW, Pan P, Klein WH. Rewiring the retinal ganglion cell gene regulatory network: Neurod1 promotes retinal ganglion cell fate in the absence of Math5. Development. 2008;135:3379–3388. doi: 10.1242/dev.024612. [DOI] [PubMed] [Google Scholar]
- Mao CA, Mao C-A, Cho JH, Cho J-H, Wang J, Wang J, Gao Z, Gao Z, Pan P, Pan P, Tsai W-W, Tsai WW, Frishman LJ, Klein WH. Reprogramming amacrine and photoreceptor progenitors into retinal ganglion cells by replacing Neurod1 with Atoh7. 2013 doi: 10.1242/dev.099549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer JS, Howden SE, Wallace KA, Verhoeven AD, Wright LS, Capowski EE, Pinilla I, Martin JM, Tian S, Stewart R, Pattnaik B, Thomson JA, Gamm DM. Optic vesicle-like structures derived from human pluripotent stem cells facilitate a customized approach to retinal disease treatment. Stem Cells. 2011;29:1206–1218. doi: 10.1002/stem.674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mu X, Fu X, Beremand PD, Thomas TL, Klein WH. Gene regulation logic in retinal ganglion cell development: Isl1 defines a critical branch distinct from but overlapping with Pou4f2. Proceedings of the National Academy of Sciences. 2008;105:6942–6947. doi: 10.1073/pnas.0802627105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mu X, Fu X, Sun H, Beremand PD, Thomas TL, Klein WH. A gene network downstream of transcription factor Math5 regulates retinal progenitor cell competence and ganglion cell fate. Developmental Biology. 2005;280:467–481. doi: 10.1016/j.ydbio.2005.01.028. [DOI] [PubMed] [Google Scholar]
- Nakano T, Ando S, Takata N, Kawada M, Muguruma K, Sekiguchi K, Saito K, Yonemura S, Eiraku M, Sasai Y. Self-formation of optic cups and storable stratified neural retina from human ESCs. Cell Stem Cell. 2012;10:771–785. doi: 10.1016/j.stem.2012.05.009. [DOI] [PubMed] [Google Scholar]
- Pacal M, Bremner R. Induction of the ganglion cell differentiation program in human retinal progenitors before cell cycle exit. Dev Dyn. 2014;243:712–729. doi: 10.1002/dvdy.24103. [DOI] [PubMed] [Google Scholar]
- Pearson C, Martin K. Stem cell approaches to glaucoma: from aqueous outflow modulation to retinal neuroprotection. Prog Brain Res. 2015 doi: 10.1016/bs.pbr.2015.04.005. [DOI] [PubMed] [Google Scholar]
- Phillips MJ, Wallace KA, Dickerson SJ, Miller MJ, Verhoeven AD, Martin JM, Wright LS, Shen W, Capowski EE, Percin EF, Perez ET, Zhong X, Canto-Soler MV, Gamm DM. Blood-derived human iPS cells generate optic vesicle-like structures with the capacity to form retinal laminae and develop synapses. Investigative Ophthalmology & Visual Science. 2012;53:2007–2019. doi: 10.1167/iovs.11-9313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piri N, Kwong JMK, Song M, Caprioli J. Expression of hermes gene is restricted to the ganglion cells in the retina. Neurosci Lett. 2006;405:40–45. doi: 10.1016/j.neulet.2006.06.049. [DOI] [PubMed] [Google Scholar]
- Provis JM. Patterns of cell death in the ganglion cell layer of the human fetal retina. J Comp Neurol. 1987;259:237–246. doi: 10.1002/cne.902590205. [DOI] [PubMed] [Google Scholar]
- Provis JM, Penfold PL. Cell death and the elimination of retinal axons during development. Progress in Neurobiology. 1988;31:331–347. doi: 10.1016/0301-0082(88)90018-4. [DOI] [PubMed] [Google Scholar]
- Pujic Z, Omori Y, Tsujikawa M, Thisse B, Thisse C, Malicki J. Reverse genetic analysis of neurogenesis in the zebrafish retina. Developmental Biology. 2006;293:330–347. doi: 10.1016/j.ydbio.2005.12.056. [DOI] [PubMed] [Google Scholar]
- Rachel RA, Dolen G, Hayes NL, Lu A, Erskine L, Nowakowski RS, Mason CA. Spatiotemporal features of early neuronogenesis differ in wild-type and albino mouse retina. Journal of Neuroscience. 2002;22:4249–4263. doi: 10.1523/JNEUROSCI.22-11-04249.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randlett O, Poggi L, Zolessi FR, Harris WA. The oriented emergence of axons from retinal ganglion cells is directed by laminin contact in vivo. Neuron. 2011;70:266–280. doi: 10.1016/j.neuron.2011.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez AR, de Sevilla Müller LP, Brecha NC. The RNA binding protein RBPMS is a selective marker of ganglion cells in the mammalian retina. J Comp Neurol. 2014;522:1411–1443. doi: 10.1002/cne.23521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanes JR, Masland RH. The Types of Retinal Ganglion Cells: Current Status and Implications for Neuronal Classification. Annu Rev Neurosci. 2015 doi: 10.1146/annurev-neuro-071714-034120. [DOI] [PubMed] [Google Scholar]
- Tanaka T, Yokoi T, Tamalu F, Watanabe SI, Nishina S, Azuma N. Generation of retinal ganglion cells with functional axons from human induced pluripotent stem cells. Sci Rep. 2015;5:8344. doi: 10.1038/srep08344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trimarchi JM, Stadler MB, Cepko CL. Individual Retinal Progenitor Cells Display Extensive Heterogeneity of Gene Expression. PLoS ONE. 2008;3:e1588. doi: 10.1371/journal.pone.0001588.s013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trimarchi JM, Stadler MB, Roska B, Billings N, Sun B, Bartch B, Cepko CL. Molecular heterogeneity of developing retinal ganglion and amacrine cells revealed through single cell gene expression profiling. J Comp Neurol. 2007;502:1047–1065. doi: 10.1002/cne.21368. [DOI] [PubMed] [Google Scholar]
- Wang SW, Kim BS, Ding K, Wang H, Sun D, Johnson RL, Klein WH, Gan L. Requirement for math5 in the development of retinal ganglion cells. Genes & Development. 2001;15:24–29. doi: 10.1101/gad.855301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinreb RN, Aung T, Medeiros FA. The pathophysiology and treatment of glaucoma: a review. JAMA. 2014;311:1901–1911. doi: 10.1001/jama.2014.3192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang M, Li S. Foxn4: a multi-faceted transcriptional regulator of cell fates in vertebrate development. Sci China Life Sci. 2013;56:985–993. doi: 10.1007/s11427-013-4543-8. [DOI] [PubMed] [Google Scholar]
- Xiang M, Qiu F, Jiang H, Jin K. Molecular Control of Retinal Ganglion Cell Specification and Differentiation 2011 [Google Scholar]
- Yang Z, Ding K, Pan L, Deng M, Gan L. Math5 determines the competence state of retinal ganglion cell progenitors. Developmental Biology. 2003;264:240–254. doi: 10.1016/j.ydbio.2003.08.005. [DOI] [PubMed] [Google Scholar]
- Zhong X, Gutierrez C, Xue T, Hampton C, Vergara MN, Cao LH, Peters A, Park TS, Zambidis ET, Meyer JS, Gamm DM, Yau KW, Canto-Soler MV. Generation of three-dimensional retinal tissue with functional photoreceptors from human iPSCs. Nature Communications. 2014;5:4047. doi: 10.1038/ncomms5047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zolessi FR, Poggi L, Wilkinson CJ, Chien CB, Harris WA. Polarization and orientation of retinal ganglion cells in vivo. Neural Dev. 2006;1:2. doi: 10.1186/1749-8104-1-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix A: Supplementary data
Appendix B: Supplementary tables