Abstract
Colorectal cancer (CRC) has become a major public health problem, ranking as the third most common type of cancer. Our previous study has revealed that TCF21 is frequently silenced by promoter hypermethylation in both CRC cell lines and primary CRC, with TCF21 methylation being significantly correlated with lymph node invasion. In this study, we further analyze the expression of TCF21 in CRC tissues and investigate the role of TCF21 in CRC in vitro and in vivo. We also explore the possible pathway regulated by TCF21. We thus demonstrate that decreased levels of TCF21 are associated with the pathological stage, clinical stage and lymph node metastasis, indicating a poor prognosis in CRC patients; overexpression of TCF21 inhibits cell proliferation, migration and invasion in the colorectal cell lines HCT116 and HT29. Furthermore, TCF21 functions as a tumor suppressor probably through inactivation of PI3K/AKT signaling and inhibition of MMPs. Our results suggest that enhancement of TCF21 levels may be a potential strategy to facilitate the prevention and treatment of CRC in the clinic.
Keywords: colorectal cancer, TCF21, prognosis, PI3K/AKT signaling, MMPs
Introduction
Colorectal cancer (CRC) has become a major public health problem, ranking as the third most common among all cancer types, which is responsible for 8%–9% of cancer-related deaths in the world.1 TCF21, located in 6q23, is known to function as a tumor suppressor and is deregulated in several types of cancers, including breast cancer, gastric cancer and clear cell renal cell carcinoma (ccRCC).2–5 The loss of function of TCF21 is associated with epigenetic alterations in different types of human cancers. For example, the methylation level of TCF21 in gastric cancer samples is much higher than that in samples of normal adjacent tissue; moreover, treatment with DNA methyltransferase can upregulate the expression of TCF21 in gastric cancer cells.5 TCF21 genetic polymorphisms are associated with the risk of breast cancer in Chinese women.6 Our previous study has revealed that TCF21 is frequently silenced by promoter hypermethylation in both CRC cell lines and primary CRC and that TCF21 methylation is significantly correlated with lymph node invasion.7 However, the role of TCF21 in CRC and the underlying mechanism remain unclear.
In this study, we analyze the expression of TCF21 in CRC tissues and investigate the role of TCF21 CRC in vitro and in vivo. We also explore the possible pathway regulated by TCF21. We demonstrate that decreased levels of TCF21 indicate a poor prognosis in CRC patients and that TCF21 functions as a tumor suppressor probably through inactivation of PI3K/AKT signaling and inhibition of MMPs. Our results suggest that enhancement of TCF21 may be a potential strategy to facilitate the prevention and treatment of CRC in the clinic.
Materials and methods
Tissue samples
All patients signed an informed consent, approved by the Independent Ethical Committee of Central South University (number 2010-S124). Tissues used for Western blot, namely, the CRC and the adjacent normal colorectal epithelial tissues used in the study, were collected from 4 patients. All samples were stored in liquid nitrogen until tissue section. These tissues were also used for immunohistochemical staining. In total, 82 samples of CRC cancer tissues (47 males and 35 females; average age: 60 years) and paired adjacent normal colorectal mucosa used in the study were collected from January 2011 to January 2014 in Xiangya Hospital of Central South University, Hunan, People’s Republic of China, and the tumor stages were classified according to National Comprehensive Cancer Network (NCCN 2009) staging criteria.8 Information related to the specimens was obtained from surgical and pathological records, and use of these samples was approved by the ethics committee of Xiangya Hospital of Central South University. The clinical features of patients are shown in Table 1.
Table 1.
Association between TCF21 expression and clinicopathologic characteristics in colorectal cancer
| Features | Number of cases | TCF21 expression
|
P-valuea | |
|---|---|---|---|---|
| Low | High | |||
| Age, years | 0.508 | |||
| >60 | 45 | 23 | 22 | |
| ≤60 | 37 | 22 | 15 | |
| Sex | 0.112 | |||
| Female | 35 | 16 | 19 | |
| Male | 47 | 29 | 18 | |
| Pathological stage | 0.033* | |||
| T1 + T2 | 57 | 27 | 30 | |
| T3 + T4 | 25 | 18 | 7 | |
| Clinical stage | 0.020* | |||
| I+II | 61 | 29 | 32 | |
| III+IV | 21 | 16 | 5 | |
| Lymph node status | 0.008** | |||
| Metastasis | 44 | 30 | 14 | |
| No metastasis | 38 | 15 | 23 | |
Notes:
χ2 test. Significant values:
P<0.05;
P<0.01. T1, the tumor has grown through the muscularis mucosa into the submucosa; T2, the tumor has grown into the muscularis propria; T3, the tumor has grown into the outermost layers of the colon or rectum but has not gone through them; T4, the tumor has invaded lumen surrounding tissues and organs.
Cell culture
Human CRC cell lines HCT116 and HT29 were obtained from the American Type Culture Collection (ATCC). All the cells were cultured in Dulbecco’s Modified Eagle’s Medium (DMEM) (Invitrogen Life Technologies, Carlsbad, CA, USA) supplemented with 10% (v/v) fetal bovine serum (FBS) and 100 U/mL penicillin/streptomycin (Invitrogen Life Technologies) at 37°C in a humidified 5% CO2 incubator.
Cell transfection
Cell transfection was conducted using Lipofectamine 3000 (Invitrogen Life Technologies), according to the manufacturer’s instruction. The lentivirus (Lv) used to overexpress TCF21 was designed and purchased from RiboBio (Guangzhou RiboBio Co, Ltd, Guangzhou, People’s Republic of China). Lv-TCF21 and Lv-NC were added into cell culture medium at MOI (multiplicity of infection) =40 for along with 5 µg/mL polybrene. Twelve hours after infection, culture medium was refreshed and cells were cultured for another 24 hours before ready for further treatments. Subsequently, the mixtures were added into the cell suspensions. After incubation for 6 h, the medium was replaced by DMEM with 10% FBS. After transfection for 48 h, the cells were used for further analysis.
Western blot analysis
The protein was extracted using radioimmunoprecipitation assay (RIPA) lysis buffer (Auragene, Changsha, People’s Republic of China) from the indicated cells, and the Bradford Protein Assay Kit (Beyotime Biotechnology, Shanghai, People’s Republic of China) was used to measure the protein concentration. After separation on 10% sodium dodecyl sulfate polyacrylamide gels by electrophoresis (SDS-PAGE), the protein was transferred to nitrocellulose membranes. Membranes were then blocked with 5% bovine serum albumin, incubated overnight at 4°C with primary antibody. The membranes were washed with Tris-buffered saline plus Tween (TBST) and then incubated with appropriate horseradish peroxidase (HRP)-conjugated secondary antibodies. Enhanced chemiluminescence (ECL) reagent was used to detect the signal on the membrane.
Immunohistochemical staining
The human cancer tissues and matched adjacent normal tissues were used for immunohistochemistry analysis, as described previously.9 Tissues were subjected to citrate-based antigen retrieval using 10 mM sodium citrate buffer (pH 6.0). The tissues were then stained with TCF21 antibody (ImmunoWay Biotechnology, Plano, TX, USA) and visualized using a 3,3′-diaminobenzidine (DAB) staining kit (Abcam, New Territories, Hong Kong, China), followed by counterstaining with hematoxylin. The staining was scored by 2 independent pathologists who were blinded to clinicopathologic features. TCF21 staining intensity was scored as 0 (negative, −), 1 (weak, +), 2 (moderate, ++) and 3 (strong, +++). The extent of staining was scored as 0–1.0 (0%–100%). The final staining score (0–3) was calculated as the multiplication product of the intensity score and the extent score. A final score ≥1 was defined as indicating high expression; otherwise, the score was said to be indicative of low expression.
3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay
Cell growth and inhibition of viability were measured by the MTT assay. Thus, 3,000 indicated cells were seeded in each 96-well plate for 6 h and further incubated for 0, 24, 48 and 72 h. Then, 1 h before the end of incubation, 100 µL MTT was added into each well at given time points followed by an additional 4 h incubation at 37°C. And then the supernatant was removed and replaced with 150 µL dimethyl sulfoxide (DMSO) in each well for 15 min. Subsequently, the optical density (OD) at 570 nm for each well was determined by an enzyme immunoassay analyzer.
Scratch assay
Cells in each group were collected and resuspended in DMEM. Each well of a 6-well plate was seeded with 1×105 cells and cultured for 24 h to 100% confluence. The cells were scratched with the head of a 20 µL tip and washed with serum-free medium. These cells were further cultured for 24 h in DMEM containing 3% FBS; then, the cells in each group were photographed for analysis.
Transwell assay
The indicated cells were starved for 12 h, resuspended in serum-free medium and added to the upper chamber of the transwell device. The lower chamber was filled with completed medium containing 10% FBS. Following 48 h culture, cells attached to the bottom were fixed and stained with crystal violet for 30 min.
Tumorigenicity assay in nude mice
The protocol of this study was approved by the Committee on the Ethics of Animal Experiments of Xiangya Hospital of Central South University. All animal experiments were performed in accordance with the guidelines of the Animal Care Committee, Xiangya Hospital, Central South University. A total of 9 male BALB/c nude mice (3 weeks old) were purchased from the Animal Center of the Chinese Academy of Sciences (Shanghai, People’s Republic of China) and randomized into 3 groups, including mock, Lv-NC (negative control) and Lv-TCF21. After 7 days’ habitation, mice were injected subcutaneously with 2×106 indicated cells into the flanks. Tumor volumes were monitored once every 5 days. Tumor volume was calculated according to the following formula: tumor volume = length × (width)2/2. After 30 days, the mice were sacrificed, and the tumor tissues were retrieved for further analysis.
Statistical analysis
Statistical analyses were performed using GraphPad Prism 5 software (Graphpad Software, Inc, La Jolla, CA, USA), and the data are presented as the mean ± standard deviation. Kaplan–Meier analysis with log-rank test was used to analyze the overall survival rate. Multivariate prognostic factors were examined using Cox’s proportional-hazards model. An unpaired 2-tailed Student’s t-test or 1-way analysis of variance (ANOVA) was used to analyze the in vitro and in vivo data. P<0.05 was considered to be statistically significant.
Results
Downregulated TCF21 predicts a poor prognosis in CRC patients
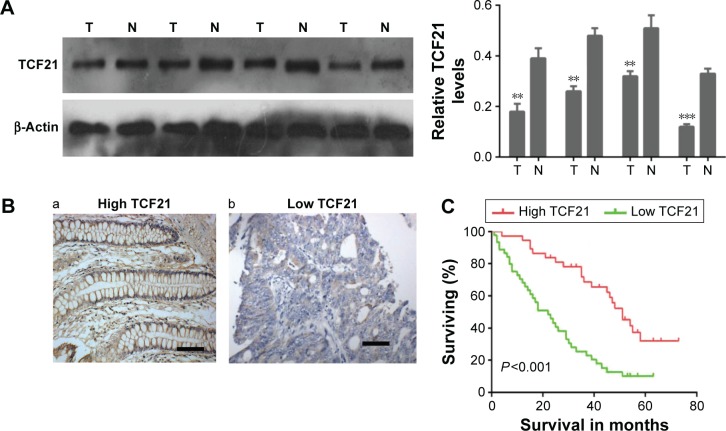
To investigate the role of TCF21 in the development of CRC, we analyzed the expression of TCF21 in CRC tissues as well as the association between its expression and overall survival. As shown in Figure 1A and B, we found that the protein levels of TCF21 were significantly downregulated in CRC compared with the levels in the paired adjacent tissues. Among 82 cases of adjacent tissues, 76 cases showed positive expression of TCF21, with a 92.7% (76/82) positive expression rate. However, among 82 cases of CRC, only 59 cases showed positive expression of TCF21, with a 72.0% (59/82) positive expression rate, which was significantly lower than in adjacent tissues. The association between TCF21 protein expression and clinicopathologic features in patients with CRC was analyzed using the χ2 test. As shown in Table 1, the TCF21 protein expression had no association with the patient’s age and sex (P>0.05). However, its protein expression showed association with the pathological stage, clinical stage and lymph node metastasis (P<0.05).
Figure 1.
Downregulation of TCF21 is associated with poor prognosis in patients with colorectal cancer.
Notes: (A) Western blot analysis for TCF21 in colorectal tumor tissues and their paired adjacent tissues (left panel), as well as the quantification (right panel). Data are presented as mean ± SD. **P<0.01, ***P<0.001. (B) Representative immunohistochemical staining for high and low TCF21 expression levels in colorectal tumor tissues. Scale bar =100 µm. (C) Kaplan–Meier curves showed worse overall survival rates for colorectal cancer patients with low TCF21 expression, compared with that in patients with high TCF21 expression.
Abbreviations: N, normal adjacent tissue; T, tumor tissue; SD, standard deviation.
Furthermore, we also analyzed the association between TCF21 expression and overall survival in patients with CRC. The Kaplan–Meier curves showed a worse overall survival rate for CRC patients with low TCF21 expression compared with the survival rate for patients with high TCF21 expression (Figure 1C).
In addition, we performed multivariate Cox model analysis, and our data suggested that the depth of invasion, the lymph node metastasis, the tumor-node-metastasis (TNM) stage and the TCF21 protein expression were independent prognostic factors for patients with colorectal carcinomas (Table 2). These results indicate that TCF21 may play an important role in the development of CRC.
Table 2.
Multivariate Cox model analysis
| Variables | RR | 95% CI | P-value |
|---|---|---|---|
| Depth of invasion (T1 + T2 vs T3 + T4) | 1.529 | 0.634–3.637 | 0.039 |
| TNM stage (I+II vs III+IV) | 2.330 | 1.189–4.565 | 0.014 |
| Lymph node metastasis (yes vs no) | 5.247 | 2.791–9.866 | <0.001 |
| TCF21 protein expression (low vs high) | 0.271 | 0.142–0.517 | <0.001 |
Abbreviations: CI, confidence interval; RR, relative risk; TNM, tumor-node-metastasis.
Upregulation of TCF21 inhibits cell proliferation and represses the ability of migration and invasion in HCT116 and HT29 cells
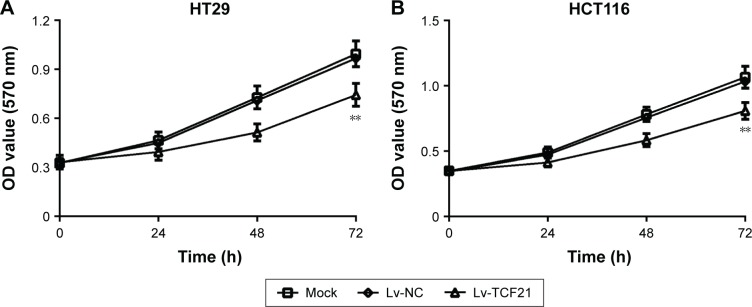
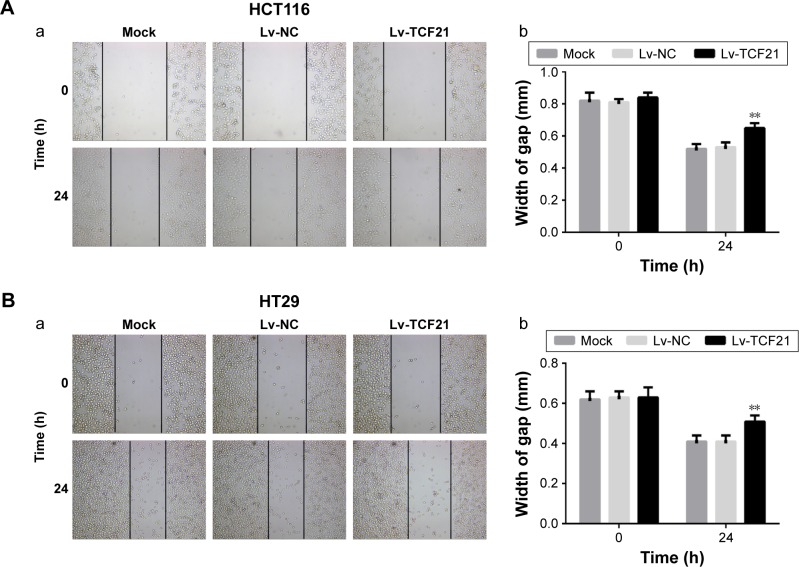
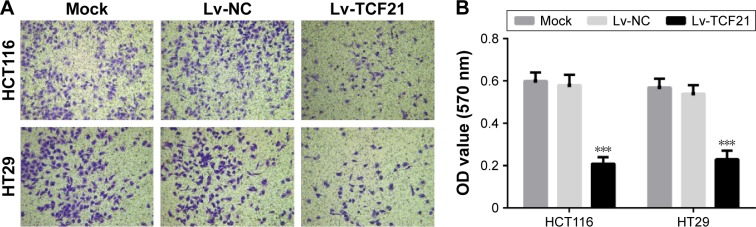
To further investigate the role of TCF21 in CRC cells, we overexpressed the expression of TCF21 in HCT116 and HT29 cells by infecting the cells with Lv. We found that overexpression of TCF21 inhibited cell proliferation in HCT116 and HT29 cells (Figure 2). In addition, we also assessed the role of TCF21 in cell migration and invasion. Upregulation of TCF21 significantly reduced the migration and invasion ability, compared with the negative control, in HCT116 and HT29 cells (Figures 3 and 4), indicating that TCF21 was able to inhibit the movement of CRC cells. Thus, TCF21 is able to repress the growth and ability of invasion in CRC cells.
Figure 2.
Overexpression of TCF21 inhibits cell proliferation in HCT116 and HT29 cells.
Notes: MTT assay was used to measured cell proliferation in (A) HT29 and (B) HCT116 cells after transfection with TCF21-expressing lentivirus. The cells without any treatment were used as mock control, and the cells treated with empty lentivirus were used as negative control. Data are presented as mean ± SD. **P<0.01. HCT116 and HT29, human colorectal cancer cell lines.
Abbreviations: Lv, lentivirus; MTT, 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide; NC, negative control; OD, optical density.
Figure 3.
Upregulation of TCF21 represses the ability of migration of HCT116 and HT29 cells.
Notes: Scratch assay was used to analyze cell migration in (A) HCT116 and (B) HT29 cells after transfection with TCF21-expressing lentivirus. (a) scratch assay; (b) width of gap for (a). The cells without any treatment were used as mock control, and the cells treated with empty lentivirus were used as negative control. Data are presented as mean ± SD. **P<0.01. HCT116 and HT29, human colorectal cancer cell lines.
Abbreviations: Lv, lentivirus; NC, negative control.
Figure 4.
Overexpression of TCF21 represses the ability of invasion of HCT116 and HT29 cells.
Notes: Transwell assay was used to analyze cell invasion in HT29 and HCT116 cells after transfection with TCF21-expressing lentivirus. The cells without any treatment were used as mock control, and the cells treated with empty lentivirus were used as negative control. (A) transwell assay with Matrigel. Magnification ×100. (B) data histogram of transwell assay. Data are presented as mean ± SD. ***P<0.001. HCT116 and HT29, human colorectal cancer cell lines.
Abbreviations: Lv, lentivirus; NC, negative control; OD, optical density.
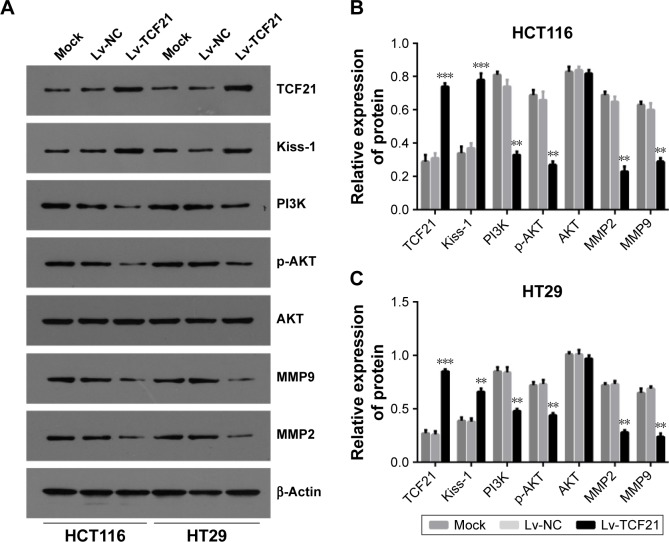
TCF21 inactivates PI3K/AKT signaling and inhibits the expression of genes encoding MMPs
Furthermore, we explored the possible downstream molecules of TCF21. As shown in Figure 5, we found that upregulation of TCF21 significantly induced the expression of Kiss-1 while decreasing the expression of PI3K and p-AKT, indicating an inactivation of PI3K/AKT signaling. In addition, overexpression of TCF21 also inhibited the expression of MMP9 and MMP2, which are involved in cancer cell invasion.
Figure 5.
TCF21 regulates key molecules of PI3K signaling and MMPs.
Notes: (A) Western blot analysis for TCF21, Kiss-1, PI3K, AKT, p-AKT, MMP2 and MMP9 in HT29 and HCT116 cells after transfection with TCF21, and the results of quantification in (B) HCT116 and (C) HT29 cells. The cells without any treatment were used as mock control, and the cells treated with empty lentivirus were used as negative control. Data are presented as mean ± SD. **P<0.01, ***P<0.001. HCT116 and HT29, human colorectal cancer cell lines.
Abbreviations: Lv, lentivirus; NC, negative control.
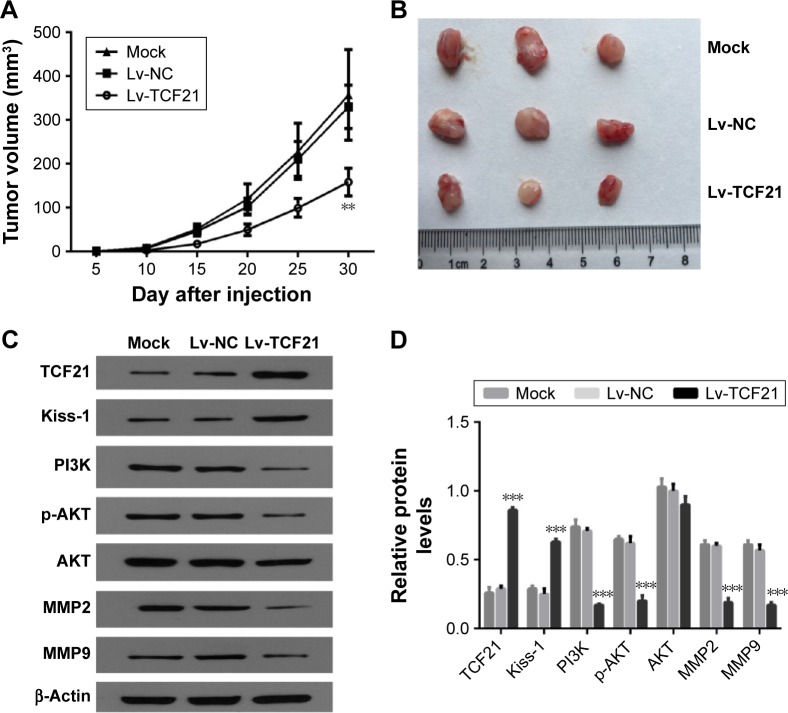
TCF21 inhibits growth of human CRC cells in nude mice
We further validated the effects of upregulated TCF21 of HCT116 cells on the growth of human CRC xenograft in nude mice. Nude mice were subcutaneously inoculated with human CRC HCT116 cells infected with blank Lv and TCF21-expressing Lv, respectively. The cells without any treatment were used as mock control. The volumes of the tumors were monitored every 5 days. After 30 days, the mice were sacrificed to remove the tumors. The average tumor volume in the Lv-NC group was significantly higher than that in the Lv-TCF21 group (Figure 6A and B). These results of the in vivo experiments suggest that TCF21 could significantly inhibit human CRC xenograft growth in nude mice. Furthermore, we also analyzed the expression of downstream molecules of TCF21 in xenograft tumor tissues. We confirmed that upregulation of TCF21 significantly increased the expression of Kiss-1 while decreasing the expression of PI3K and p-AKT, indicating an inactivation of PI3K/AKT signaling (Figure 6C). In addition, overexpression of TCF21 also inhibited the expression of MMP9 and MMP2, which are involved in cancer cell invasion (Figure 6C).
Figure 6.
TCF21 suppresses tumorigenicity of human colorectal cancer cells in vivo.
Notes: The HCT116 cells that were transfected with TCF21-expressing lentivirus were subcutaneously injected into nude mice. The cells without any treatment were used as mock control, and the cells treated with empty lentivirus were used as negative control. (A) The tumor volume was monitored every 2 days. (B) The tumors were obtained from the mice. (C) Western blot analysis for TCF21, Kiss-1, PI3K, p-AKT, AKT, MMP2 and MMP9 in the xenografted tumor tissues, as well as (D) the quantification results of the Western blot. Data are presented as mean ± SD. **P<0.01, ***P<0.001. HCT116, human colorectal cancer cell line.
Abbreviations: Lv, lentivirus; NC, negative control.
Discussion
In this study, we find that TCF21 is significantly decreased in CRC tissues, which is associated with the pathological stage, clinical stage and lymph node metastasis; furthermore, downregulated levels of TCF21 predict a poor prognosis in CRC patients. In addition, we also suggest that TCF21 functions as a tumor suppressor in vitro and in vivo, in addition to being involved in PI3K/AKT signaling.
TCF21 is located at chromosome location 6q23, a region that frequently loses heterozygosity either genetically or through epigenetic events.10 TCF21 is frequently lost in human malignancies. It has been demonstrated that TCF21 was aberrantly methylated and silenced in the majority of head-and-neck squamous cell carcinomas and non-small-cell lung cancers (NSCLCs).11 And restoration of exogenous expression of TCF21 in cells that have silenced the endogenous TCF21 results in a reduction of tumor properties both in vitro and in vivo.11 By using the DNA sequencing technique and quantitative methylation-specific polymerase chain reaction (PCR) assays, Shivapurkar et al12 found that the CpG island within exon 1 of the TCF21 gene was unmethylated in normal lung epithelial cells but predominantly methylated in lung cancer cell lines, which has been found in multiple other malignancies. Hypermethylation and decreased expression of TCF21 may be tumor specific and very frequent in all NSCLCs, even early-stage disease.13 It is important for the early detection and surveillance of lung cancer as well as multiple other malignancies. TCF21 methylation levels have been reported to provide a sensitivity rate of 92% for bladder cancer, 67% for renal cell tumors and 96% for prostate cancer.14 TCF21 methylation level is significantly increased in ccRCC. Moreover, the Kaplan–Meier survival analysis demonstrates that low expression of TCF21 is an independent prognostic factor for poor survival in patients with ccRCC.4,15 Emerging evidence also demonstrates that hypermethylation of TCF21 is observed in breast cancer, gastric cancer and head-and-neck squamous cell carcinoma,2,5,16,17 which is in line with our previous results on CRC,7 indicating that aberrant methylation is an important reason for the downregulation of the expression of TCF21 and may be associated with tumorigenesis in multiple types of cancer.
We herein confirm that upregulation of TCF21 greatly inhibits CRC cell proliferation and cell invasion in vitro, as well as repressing tumorigenesis in nude mice. Our results further indicate that the inhibitory effects of TCF21 on CRC cancer are routed through Kiss-1, PI3K/AKT signaling and MMPs.
The KiSS-1 gene was initially described as a suppressor of metastasis in malignant melanoma. Subsequent research has revealed that KiSS-1 is the strongest independent prognostic factor among the conventional prognosticators for gastric cancer, ovarian cancer, breast cancer, hepatocellular carcinoma and pancreatic cancer.18–21 Furthermore, KiSS-1 has an antiproliferative effect on cancer cells and could be responsible for decreased tumor growth and increased cancer patient survival.18
MMPs can degrade the extracellular matrix and thus promote the process of tumor metastasis. MMP9 and MMP2, two of the most important proteases related with tumor metastasis, are capable of degrading the primary structure of the extracellular matrix and basement membrane to promote tumor metastasis.22 Induced overexpression of KiSS-1 in HT-1080 cells resulted in reduced transcription and activity of MMP9 and thereby a marked reduction in invasion in vitro. Consequently, the reduced synthesis of MMP9 induces certain inhibitory effects on the mobility and invasion of cancer cells.23 In addition, KiSS-1 treatment has been shown to elicit a strong and sustained phosphorylation of ERK1/2 and pAKT.24 Recently, Arab et al25 have found that TCF21 binds the promoter of the melanoma metastasis-suppressing gene, KiSS1, and enhances its expression through interaction with E12 and with TCF12. Loss of TCF21 expression results in loss of KiSS1 expression through loss of direct interaction of TCF21 at the KiSS1 promoter. Additionally, microRNA (miR)-21 can downregulate the expression of TCF21, and the expression of KiSS-1 is downregulated in Caki-1 cells with TCF21 gene silencing.3 In CRC, KiSS-1 overexpression significantly decreases the cell proliferation and invasiveness of HCT-119 cells as well as enhancing cell apoptosis. Overexpression of KiSS-1 also results in blockade of synthesis of MMP9 and PI3K, as well as the inhibition of phosphorylation of Akt. Furthermore, treatment with PI3K and Akt agonists attenuates the effect of KiSS-1 on the biological activity of CRC cells.24 Taken together, our results suggest that TCF21 induces Kiss-1 expression and subsequently results in downregulation of MMPs and inactivation of PI3K/AKT signaling to exert its inhibitory function on CRC.
Conclusion
Collectively, our data reveal that decreased levels of TCF21 predict a poor prognosis in CRC patients and that overexpression of TCF21 can suppress the proliferation and invasiveness of CRC cells; moreover, the TCF21 gene exerts its function probably by inducing Kiss-1 and reducing the expression of MMPs by blocking the PI3K/Akt pathway. Restoration of TCF21 levels may become a potential strategy to modulate CRC growth and metastasis in the clinic.
Acknowledgments
This work was supported by grants: National Natural Science Foundation of China (81372792 and 81602683, Major Projects of Science and Technology in Hunan Province (2015SK2044, Science and Technology Planning Project of Hunan Provincial Health Department (B2014-090).
Footnotes
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Deen KI, Silva H, Deen R, Chandrasinghe PC. Colorectal cancer in the young, many questions, few answers. World J Gastrointest Oncol. 2016;8(6):481–488. doi: 10.4251/wjgo.v8.i6.481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang J, Gao X, Wang M, Zhang J. Clinicopathological significance and biological role of TCF21 mRNA in breast cancer. Tumour Biol. 2015;36(11):8679–8683. doi: 10.1007/s13277-015-3476-1. [DOI] [PubMed] [Google Scholar]
- 3.Zhang H, Guo Y, Shang C, Song Y, Wu B. miR-21 downregulated TCF21 to inhibit KISS1 in renal cancer. Urology. 2012;80(6):1298–1302. doi: 10.1016/j.urology.2012.08.013. [DOI] [PubMed] [Google Scholar]
- 4.Ye YW, Jiang ZM, Li WH, et al. Down-regulation of TCF21 is associated with poor survival in clear cell renal cell carcinoma. Neoplasma. 2012;59(6):599–605. doi: 10.4149/neo_2012_076. [DOI] [PubMed] [Google Scholar]
- 5.Yang Z, Li DM, Xie Q, Dai DQ. Protein expression and promoter methylation of the candidate biomarker TCF21 in gastric cancer. J Cancer Res Clin Oncol. 2015;141(2):211–220. doi: 10.1007/s00432-014-1809-x. [DOI] [PubMed] [Google Scholar]
- 6.Gao X, Yang J, Wang M, Zhang J. TCF21 genetic polymorphisms and breast cancer risk in Chinese women. Oncotarget. 2016 doi: 10.18632/oncotarget.9825. Epub 2016 Jun 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dai Y, Duan H, Duan C, et al. Down-regulation of TCF21 by hypermethylation induces cell proliferation, migration and invasion in colorectal cancer. Biochem Biophys Res Commun. 2016;469(3):430–436. doi: 10.1016/j.bbrc.2015.09.109. [DOI] [PubMed] [Google Scholar]
- 8.Seal BS, Sullivan SD, Ramsey SD, et al. Systemic therapy for colorectal cancer: patterns of chemotherapy and biologic therapy use in nationally representative US claims database. BioDrugs. 2014;28(2):229–236. doi: 10.1007/s40259-013-0073-y. [DOI] [PubMed] [Google Scholar]
- 9.Luo Y, Wang X, Wang H, et al. High Bak expression is associated with a favorable prognosis in breast cancer and sensitizes breast cancer cells to paclitaxel. PLoS One. 2015;10(9):e138955. doi: 10.1371/journal.pone.0138955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tessema M, Willink R, Do K, et al. Promoter methylation of genes in and around the candidate lung cancer susceptibility locus 6q23-25. Cancer Res. 2008;68(6):1707–1714. doi: 10.1158/0008-5472.CAN-07-6325. [DOI] [PubMed] [Google Scholar]
- 11.Smith LT, Lin M, Brena RM, et al. Epigenetic regulation of the tumor suppressor gene TCF21 on 6q23-q24 in lung and head and neck cancer. Proc Natl Acad Sci U S A. 2006;103(4):982–987. doi: 10.1073/pnas.0510171102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shivapurkar N, Stastny V, Xie Y, et al. Differential methylation of a short CpG-rich sequence within exon 1 of TCF21 gene: a promising cancer biomarker assay. Cancer Epidemiol Biomarkers Prev. 2008;17(4):995–1000. doi: 10.1158/1055-9965.EPI-07-2808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Richards KL, Zhang B, Sun M, et al. Methylation of the candidate biomarker TCF21 is very frequent across a spectrum of early-stage nonsmall cell lung cancers. Cancer. 2011;117(3):606–617. doi: 10.1002/cncr.25472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Costa VL, Henrique R, Danielsen SA, et al. TCF21 and PCDH17 methylation: an innovative panel of biomarkers for a simultaneous detection of urological cancers. Epigenetics. 2011;6(9):1120–1130. doi: 10.4161/epi.6.9.16376. [DOI] [PubMed] [Google Scholar]
- 15.Valletti A, Gigante M, Palumbo O, et al. Genome-wide analysis of differentially expressed genes and splicing isoforms in clear cell renal cell carcinoma. PLoS One. 2013;8(10):e78452. doi: 10.1371/journal.pone.0078452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Weiss D, Stockmann C, Schrodter K, Rudack C. Protein expression and promoter methylation of the candidate biomarker TCF21 in head and neck squamous cell carcinoma. Cell Oncol (Dordr) 2013;36(3):213–224. doi: 10.1007/s13402-013-0129-5. [DOI] [PubMed] [Google Scholar]
- 17.Gooskens SL, Gadd S, Guidry AJ, et al. TCF21 hypermethylation in genetically quiescent clear cell sarcoma of the kidney. Oncotarget. 2015;6(18):15828–15841. doi: 10.18632/oncotarget.4682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dhar DK, Naora H, Kubota H, et al. Downregulation of KiSS-1 expression is responsible for tumor invasion and worse prognosis in gastric carcinoma. Int J Cancer. 2004;111(6):868–872. doi: 10.1002/ijc.20357. [DOI] [PubMed] [Google Scholar]
- 19.Schmid K, Wang X, Haitel A, et al. KiSS-1 overexpression as an independent prognostic marker in hepatocellular carcinoma: an immunohistochemical study. Virchows Arch. 2007;450(2):143–149. doi: 10.1007/s00428-006-0352-9. [DOI] [PubMed] [Google Scholar]
- 20.Nagai K, Doi R, Katagiri F, et al. Prognostic value of metastin expression in human pancreatic cancer. J Exp Clin Cancer Res. 2009;28:9. doi: 10.1186/1756-9966-28-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ji K, Ye L, Mason MD, Jiang WG. The Kiss-1/Kiss-1R complex as a negative regulator of cell motility and cancer metastasis (Review) Int J Mol Med. 2013;32(4):747–754. doi: 10.3892/ijmm.2013.1472. [DOI] [PubMed] [Google Scholar]
- 22.Araujo RJ, Lira GA, Vilaca JA, et al. Prognostic and diagnostic implications of MMP-2, MMP-9, and VEGF-alpha expressions in colorectal cancer. Pathol Res Pract. 2015;211(1):71–77. doi: 10.1016/j.prp.2014.09.007. [DOI] [PubMed] [Google Scholar]
- 23.Pandurangan AK, Dharmalingam P, Sadagopan SK, Ganapasam S. Luteolin inhibits matrix metalloproteinase 9 and 2 in azoxymethane-induced colon carcinogenesis. Hum Exp Toxicol. 2014;33(11):1176–1185. doi: 10.1177/0960327114522502. [DOI] [PubMed] [Google Scholar]
- 24.Chen S, Chen W, Zhang X, Lin S, Chen Z. Overexpression of KiSS-1 reduces colorectal cancer cell invasion by downregulating MMP-9 via blocking PI3K/Akt/NF-kappaB signal pathway. Int J Oncol. 2016;48(4):1391–1398. doi: 10.3892/ijo.2016.3368. [DOI] [PubMed] [Google Scholar]
- 25.Arab K, Smith LT, Gast A, et al. Epigenetic deregulation of TCF21 inhibits metastasis suppressor KISS1 in metastatic melanoma. Carcinogenesis. 2011;32(10):1467–1473. doi: 10.1093/carcin/bgr138. [DOI] [PMC free article] [PubMed] [Google Scholar]