Abstract
RNase III proteins play key roles in microRNA (miRNA) biogenesis. The nuclear RNase III Drosha cleaves primary miRNAs (pri-miRNAs) to release hairpin-shaped pre-miRNAs that are subsequently cut by the cytoplasmic RNase III Dicer to generate mature miRNAs. While Dicer (class III) and other simple RNase III proteins (class I) have been studied intensively, the class II enzyme Drosha remains to be characterized. Here we dissected the action mechanism of human Drosha by generating mutants and by characterizing its new interacting partner, DGCR8. The basic action mechanism of Drosha was found to be similar to that of human Dicer; the RNase III domains A and B form an intramolecular dimer and cleave the 3′ and 5′ strands of the stem, respectively. Human Drosha fractionates at ∼650 kDa, indicating that Drosha functions as a large complex. In this complex, Drosha interacts with DGCR8, which contains two double-stranded RNA (dsRNA)-binding domains. By RNAi and biochemical reconstitution, we show that DGCR8 may be an essential component of the pri-miRNA processing complex, along with Drosha. Based on these results, we propose a model for the action mechanism of class II RNase III proteins.
Keywords: microRNA, Drosha, DGCR8, processing
MicroRNAs (miRNAs) are single-stranded RNAs of ∼22 nucleotides (nt) in length (ranging 19∼25 nt) generated from endogenous transcripts that can form local hairpin structures (Ambros et al. 2003). MiRNAs act as guide molecules by basepairing with the target mRNAs, and lead to translational repression and/or mRNA cleavage. MiRNAs direct diverse regulatory pathways, including developmental timing control, hematopoietic cell differentiation, apoptosis, cell proliferation, and organ development (Bartel 2004). Hundreds of miRNA genes were recently found in eukaryotes and viruses (Bartel 2004; Murchison and Hannon 2004; Pfeffer et al. 2004), making them one of the largest gene families.
Our knowledge of miRNA biogenesis has significantly advanced in recent years (Kim 2004; Murchison and Hannon 2004). In animals, miRNAs are transcribed as long primary transcripts (pri-miRNAs) by RNA polymerase II (Lee et al. 2004b), which are cropped into the hairpin-shaped pre-miRNAs by the nuclear RNase III Drosha (Lee et al. 2003). The processing intermediates, pre-miRNAs, are exported out of the nucleus by exportin-5 (Exp5) (Yi et al. 2003; Bohnsack et al. 2004; Lund et al. 2004) and they are subsequently cleaved by the cytoplasmic RNase III Dicer into ∼22-nt miRNA duplexes (Bernstein et al. 2001; Grishok et al. 2001; Hutvagner et al. 2001; Ketting et al. 2001; Knight and Bass 2001). One strand of the duplex usually remains as a mature miRNA (Khvorova et al. 2003; Schwarz et al. 2003).
Pri-miRNA processing is a critical event in miRNA biogenesis. This initial processing event predetermines mature miRNA sequences by generating one end of mature miRNA (Lee et al. 2003; Lund et al. 2004). The other end is formed by Dicer that measures ∼21 nt from the terminus of the stem. In addition, Drosha facilitates the overall production rate of miRNA by generating the 3′ protruding ends, which are recognized efficiently by Exp5 and Dicer (Basyuk et al. 2003; Lee et al. 2003; Lund et al. 2004).
RNase III proteins are grouped into three classes based on their domain organization (Supplementary Figure S1). Class I proteins include RNase III proteins found in bacteria and yeasts. Each of them contains one RNase III domain (RIIID) and one double-stranded RNA (dsRNA)-binding domain (dsRBD). Class II proteins such as Drosha possess two RIIIDs and a dsRBD. Drosha homologs are found only in animals. These large proteins (130-160 kDa) possess extended N termini whose functions remain to be determined. The N-terminal portion of Drosha contains a proline-rich region as well as a serine/arginine-rich region. Class III includes Dicer homologs that are conserved in Schizosaccharomyces pombe, plants, and animals. Dicer homologs are ∼200 kDa and contain multiple domains. Apart from two RIIIDs and a dsRBD, Dicer has a long N terminus containing a DExH RNA helicase/ATPase domain, as well as DUF283 and the PAZ domain. The PAZ domain is also found in a group of highly conserved proteins, referred to as Argonaute proteins (also known as PPD proteins). Structural and biochemical studies of the PAZ domain from Drosophila Ago1 and Ago2 suggest that the PAZ domain binds to the 3′ protruding end of small RNA (Lingel et al. 2003; Song et al. 2003; Yan et al. 2003). The roles of the other domains in Dicer are unclear. Although Dicer associates with several other proteins (Argonaute proteins in various organisms, RDE-4 in Caenorhabditis elegans, R2D2 in Drosophila, and dFMR1 in Drosophila) (Hammond et al. 2001; Ishizuka et al. 2002; Tabara et al. 2002; Liu et al. 2003), these interacting proteins do not seem to be required for the cleavage reaction itself because purified human Dicer and Drosophila Dicer-2 can catalyze the cleavage reaction (Zhang et al. 2002, 2004; Liu et al. 2003).
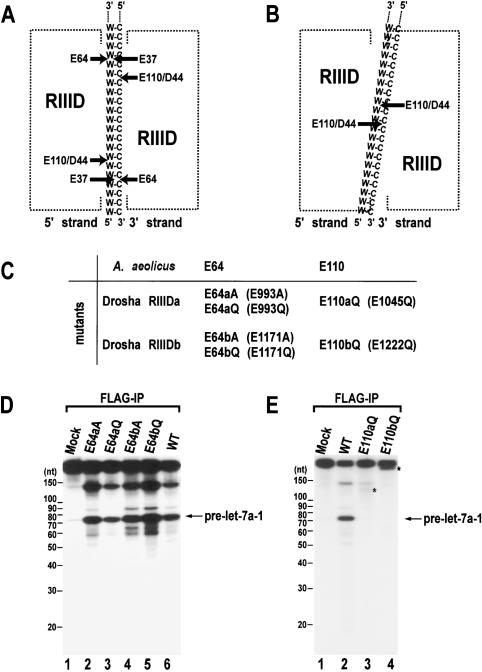
The first model for the action mechanism of RNase III proteins was constructed based on the X-ray structure of Aquifex aeolicus (Aa) and the mutagenesis of the Escherichia coli (Ec) RNase III (Blaszczyk et al. 2001). According to this model, two compound processing centers are formed between two RIIIDs (Fig. 1A). Each center contains two catalytic sites that cleave two nearby phosphodiester bonds on opposite RNA strands. One catalytic site is composed of residues E40, D44, D107, and E110, coordinating a single metal ion, Mn2+ or Mg2+. Another is made of residues E37 and E64. This model was disputed in a recent study by Filipowicz and colleagues (Zhang et al. 2004). By generating mutations in residues significant in catalysis, it was shown that both human Dicer (class III) and E. coli RNase III (class I) have single processing centers, which are comprised of residues corresponding to E40, D44, D107, and E110 of Aa RNase III (Fig. 1B). Residues corresponding to E37 and E64 of Aa RNase III do not participate in catalysis. Another important finding in that study was that Dicer is likely to function as a monomer, and that the single processing center is formed by the intramolecular dimerization of the two RIIIDs of Dicer. The N-terminal RNase III domain (RIIIDa) and the C-terminal domain (RIIIDb) of Dicer are discrete both in their sequences and in their roles (Zhang et al. 2004). Dicer RIIIDa cleaves the 3′ strand, whereas Dicer RIIIDb cuts the 5′ strand of pre-miRNA (Fig. 1B). The 5′ and 3′ strands refer to the strands that possess the 5′-phosphate and 3′-hydroxyl groups, respectively, at the terminus of dsRNA. In human Dicer, the RIIIDa along with the N-terminal region (presumably the PAZ domain) is proposed to interact with the 3′ end of the 3′ strand. Bacterial RNase III protein contains only one RIIID, so that they act as a true dimer to create a single processing center.
Figure 1.
Site-directed mutagenesis of human Drosha. (A) A model for the cleavage mechanism of RNase III (Blaszczyk et al. 2001). Two processing centers are formed between two RIIIDs. Each center contains two catalytic sites that cleave two nearby phosphodiester bonds on the opposite RNA strands of dsRNA (W-C, Watson-Crick base pairs). The 5′ strand indicates the strand that contains the 5′ phosphate group at the terminus of stem. The 3′ strand means the strand that possesses the 3′-hydroxyl group at the 3′ end. (B) The “single processing center” model where only two catalytic sites are formed in a single processing center (Zhang et al. 2004). (C) Naming of the single amino acid mutants in the RIIIDs of human Drosha. (D) In vitro processing of pri-let-7a-1. The substrate was incubated with E64aA, E64aQ, E64bA, E64bQ, or wild-type (WT) Drosha-Flag proteins that were expressed in HEK293T cells and prepared by immunoprecipitation with anti-Flag antibody. The RNA size markers (Decade Marker, Ambion) are indicated on the left side of the gel. (E) In vitro processing of pri-let-7a-1. The substrate was incubated with E110aQ, E110bQ, or wild-type (WT) Drosha-Flag proteins that were immobilized on anti-Flag beads. The asterisks indicate fragments that accumulate when the particular mutant was used in the assay.
Unlike class I and III proteins that have been intensively studied, class II proteins have not been characterized in detail. Specifically, it is still not clear (1) whether class II enzymes contain a single processing center like other RNase III proteins, (2) whether class II proteins can act alone like human Dicer or require cofactor(s) for cleavage, and (3) how the domains and the cofactors, if any, interact with each other to build up a functional enzyme. In the present study, we investigated the action mode of human Drosha by mutagenesis, and we characterize the Drosha protein and its new interacting partner, DGCR8.
Results
Pri-miRNA processing by the RNase III domain mutants
In order to understand the action mechanism of Drosha, a series of site-directed mutagenesis was carried out in the residues of the RIIIDs of human Drosha. One of the major differences between the two models is concerning the involvement of Glu64 and Glu37. These residues were hypothesized to participate in the cleavage reaction based on a structural study (Fig. 1A; Blaszczyk et al. 2001). However, the mutations of the equivalent residues to alanine in human Dicer and Ec-RNase III did not affect the enzymatic activities (Zhang et al. 2004). The residues Glu993 and Glu1171 of human Drosha correspond to Glu64 of Aa-RNase III. We mutated Glu993 and Glu1171 of Drosha to either alanine or glutamine to generate mutants E993A, E993Q, E1171A, and E1171Q, which are referred to here as E64aA, E64aQ, E64bA, and E64bQ, respectively (Fig. 1C). The proteins were expressed in HEK293T cells by transient transfection of the corresponding expression plasmids. The expression levels of the mutants are comparable to that of wild-type protein (data not shown). Mutant and wild-type proteins were immunoprecipitated using anti-Flag antibody-conjugated agarose and used for in vitro processing assay (Lee et al. 2003). All of the E64 mutants were capable of processing pri-let-7a-1 at an efficiency that is comparable to that of wild type (Fig. 1D). This result demonstrates that the residues equivalent to E64 do not participate in catalysis, not only in class I and III proteins but also in class II RNase III proteins.
E110 equivalents have been shown to be critical in Ec-RNase III, human Dicer, and Drosophila Dicer-2 (Lee et al. 2004a; Zhang et al. 2004). To test whether this residue is also important in Drosha, Glu1045 and Glu1222 of human Drosha, which are equivalent to Glu110 of Aa-RNase III, were mutated to glutamine. These mutants were named E110aQ and E110bQ, respectively (Fig. 1C). The proteins were expressed transiently, immunoprecipitated, and employed for in vitro processing assay, as described for E64 mutants. Neither of the E110 mutants, E110aQ and E110bQ, were capable of producing pre-let-7a-1 (Fig. 1E), indicating that these residues are critical in class II RNase III enzymes. Notably, these mutants demonstrate intriguing cleavage patterns, accumulating long fragments of different sizes (indicated with asterisks in Fig. 1E, lanes 3,4). The sequences of the pri-let-7a-1 used in this study are shown in Supplementary Figure S2.
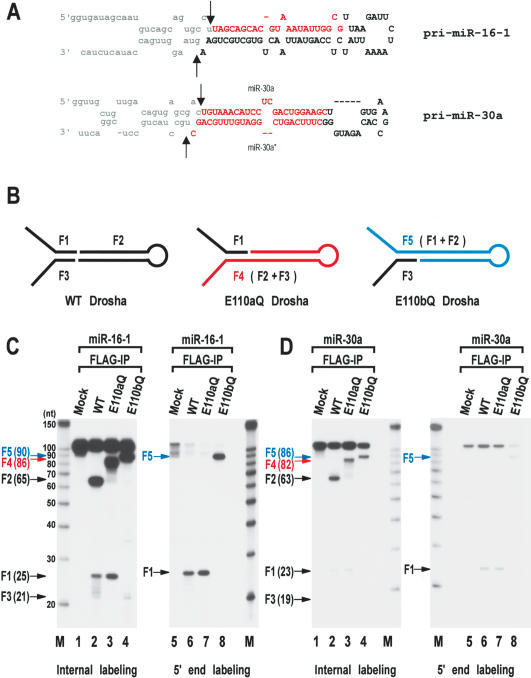
In order to determine the identity of the accumulating fragments and to simplify the analysis, we generated shorter substrates derived from pri-miR-16-1 and pri-miR-30a (Fig. 2A). The short pri-miRNAs were designed to contain ∼20-nt sequences upstream and downstream of the cleavage sites. The cleavage sites at pri-miR-30a have been previously reported (Lee et al. 2003). To determine the cleavage sites at pri-miR-16-1, pre-miR-16-1 was cloned by the directional cloning method originally described by Tuschl and others (Elbashir et al. 2001). Briefly, pri-miR-15a∼16-1 was incubated with immobilized Drosha-Flag protein to produce pre-miR-16-1. The ∼65-nt fragment was gel-purified, ligated to the 3′ and 5′ adapters, and amplified by RT-PCR using the primers that are complementary to the adapters. The PCR product was subcloned and sequenced. Our result shows that pre-miR-16-1 is a 65-nt-long stem-loop with a 2-nt 3′ overhang (Fig. 2A). The 5′ end of pre-miR-16-1 is identical to that of mature miR-16-1.
Figure 2.
Processing of the “minimal” pri-miRNA with E110 mutants. (A) Sequences and secondary structure of the short pri-miRNAs derived from pri-miR-16-1 and pri-miR-30a. These RNAs are 111 nt and 105 nt, respectively. The cleavage sites (the 5′ and 3′ ends of pre-miRNAs) are indicated with arrows. Actual cleavage sites of pri-miR-16-1 were determined by directional cloning of pre-miR-16-1. Pre-miR-16-1 was prepared by in vitro processing, ligated to the 5′ and 3′ adapters, amplified by RT-PCR, and subcloned into pGEM-T-easy vector. Ten clones were randomly chosen and sequenced. All the clones were identical at the 5′ end. Five of the 10 clones had the same 3′ end shown here. Three other clones were 1 nt shorter and two other clones were 2 nt shorter at the 3′ end, which is likely to be due to exonucleolytic trimming. The cleavage sites at miR-30a were reported (Lee et al. 2003). Pre-miR-16-1 shown here is 65 nt; pre-miR-30a is 63 nt. (B) Schematic representation of the processing products from in vitro processing by wild-type (WT), E110aQ, and E110bQ Drosha. Wild-type Drosha generates F1, F2, and F3 E110aQ produces F1 and F4 and E110bQ generates F3 and F5. (C) In vitro processing of pri-miR-16-1. The short pri-miR-16-1 was incubated with wild-type (WT) Drosha, E110aQ, or E110bQ proteins that were expressed in HEK293T cells and immobilized on anti-Flag affinity gel. The substrate was either internally labeled during transcription (lanes 1-4) or labeled at the 5′ end after transcription (lanes 5-8). The size of each fragment is indicated in the bracket next to the arrow. The RNA size markers (Ambion) are indicated on the side of the gel. (D)In vitro processing of pri-miR-30a. The short pri-miR-30a was incubated with wild-type Drosha, E110aQ, or E110bQ proteins as in C. The substrate was either internally labeled during transcription (lanes 1-4) or labeled at the 5′ end after transcription (lanes 5-8).
The short pri-miR-16-1 was designed to contain 23-nt upstream sequences and 21-nt downstream sequences flanking the cleavage sites. Similarly, the short pri-miR-30a contains 21 nt and 19 nt at the 5′ and 3′ sides of pre-miR-30a, respectively. The templates for in vitro transcription were generated by PCR amplification of pri-miRNA sequences from genomic constructs. The forward primers possess T7 promoter sequences in addition to the pri-miRNA sequences to allow in vitro transcription directly from the PCR products. Two additional Gs were included at the 5′ end of pri-miRNA because T7 RNA polymerase prefers GG at the initiation site for efficient transcription. These “minimal” pri-miR-16-1 and pri-miR-30a are 111 nt and 105 nt in length, respectively. They were cleaved efficiently by wild-type Drosha at two sites, yielding three kinds of fragments (Fig. 2B); the 5′ flanking fragment (F1), pre-miRNA (F2), and the 3′ flanking fragment (F3) (lane 2 in Fig. 2C,D). When the RNA was internally labeled using [α-32P] UTP, all three types of fragments could be observed. Fragment F3 from pri-miR-30a appears to be less stable than other fragments, for an unknown reason. If the RNA is labeled at the 5′ end using T4 polynucleotide kinase and [γ-32P] ATP (lanes 5-8 in Fig. 2C,D), only the 5′-most fragments are visualized.
Mutant E110aQ produces the 5′ flanking fragment (F1) without generating significant amounts of pre-miRNA (F2) or the 3′ flanking fragment (F3) (Fig. 2C,D, lanes 3,7). Instead, F4 is generated, which corresponds to F2 and F3, combined. Therefore the mutation at E110 in the RIIIDa affects the cleavage at the 3′ (lower) strand, indicating that the residue E110a (E1045 in the RIIIDa of human Drosha) is responsible for cutting the 3′ (lower) strand of the pri-miRNA hairpin (Fig. 2B). Mutant E110bQ, on the other hand, generates F5, which corresponds to F1 and F2, combined (Fig. 2C,D, lanes 4,8). These data show that the mutation at E110b impairs the cleavage at the 5′ (upper) strand (Fig. 2B). Thus, the residue E110b (E1222 in the RIIIDb of human Drosha) is critical for the cleavage of the 5′ (upper) strand of the pri-miRNA hairpin. The wild-type Drosha appears not to generate fragments F4 and F5 as intermediate cleavage products, suggesting coordinate processing at both sites.
Because processing was affected by E110 mutants but not by E64 mutants, our results are consistent with the recent model (Zhang et al. 2004); two RIIIDs interact with each other to build up one processing center that contains two catalytic sites where E110 plays a major role (Fig. 1B). Each catalytic site in the processing center cuts one strand each of the dsRNA at nearby sites, generating a short (∼2-nt) 3′ overhang. Thus, our data demonstrate that class II enzyme Drosha shares the common cleavage mechanism with class I and class III RNase III enzymes.
Human Drosha is part of an ∼650-kDa complex
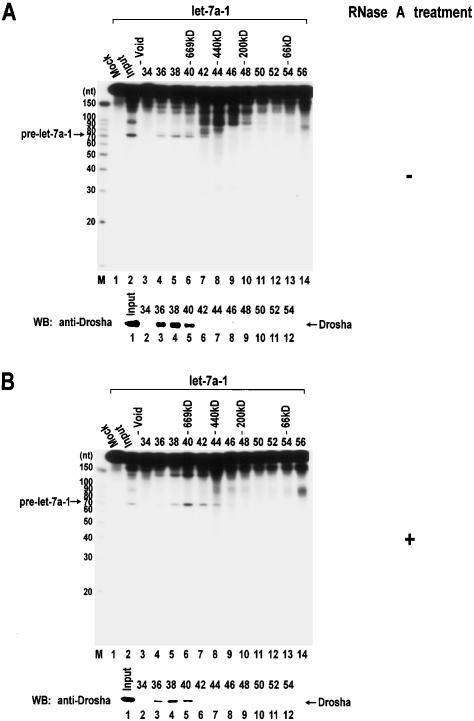
Despite the similarities, unambiguous differences do exist among different classes of RNase III proteins. For instance, class I Ec-RNase III produces ∼11-nt fragments, whereas class III Dicer typically generates ∼22-nt cleavage products. Class II Drosha cleaves at the stem of a local hairpin structure embedded in long pri-miRNAs, releasing ∼70-nt stem-loop pre-miRNAs. It is currently unknown how Drosha recognizes the substrate specifically. So as to further understand the action mode of Drosha, we first determined the size of Drosha by gel filtration chromatography. Nuclear extract from HEK293T cells was fractionated through Sephacryl-S300 HR, and the enzyme activity was assayed by incubating pri-let-7a-1 in each fraction (Fig. 3). The pri-miRNA processing activity was found in high-molecular-weight fractions, corresponding to a molecular mass of >700 kDa (Fig. 3A). However, following treatment of the extract with 50 μg/mL of RNase A for 30 min at 4°C, the activity peak shifted to ∼650 kDa. These data indicate that Drosha may function in a large complex that weighs ∼650 kDa.
Figure 3.
Gel exclusion chromatography of Drosha. (A) Nuclear extract was prepared from twenty-five 10-cm plates of HEK293T cells and fractionated through a Sephacryl-S300 HR column. (Top panel) Each fraction was 1.75 mL in volume, from which 20 μL was taken for in vitro processing of pri-let-7a-1. The fraction number and the protein molecular mass standards (Sigma) are indicated at the top of the gel. “Mock” means that lysis buffer was used instead of protein fractions for the in vitro processing assay. (Bottom panel) The fractions were also analyzed by SDS-PAGE, and Drosha proteins were detected by Western blotting using anti-Drosha antibody. (B) Nuclear extract was treated with 50 μg/mL of RNase A for 30 min at 4°C before loading to the column.
Proteins in each fraction were analyzed by Western blot analysis using anti-Drosha antibody (lower panels in Fig. 3A,B). Drosha cofractionates with pri-miRNA processing activity as expected. Following RNase A treatment, the Drosha protein peak also shifted slightly towards lower-molecular-weight fractions. However, the shift in protein level is not as dramatic as that in enzymatic activity level. This suggests that a considerable portion of Drosha proteins are in large complex(es) of >700 kDa that are catalytically inactive and that are resistant to RNase A-treatment. The Drosha proteins in these high-molecular-weight fractions may be involved in distinct function(s) in other RNA processing pathway(s).
Human DGCR8, a dsRBD-containing protein, is an essential component of the Drosha complex
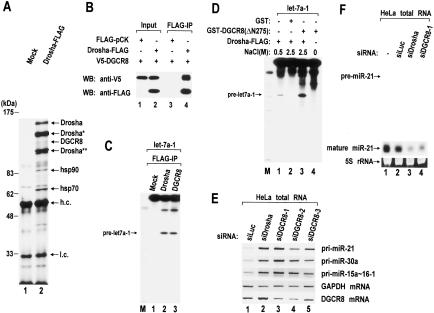
The size of pri-miRNA processing complex (∼650 kDa) suggests that Drosha may interact with other proteins to form a functional complex. To identify the components of the human Drosha complex, we carried out immunoprecipitation from total extract of HEK293T cells. The cells were transfected transiently with a plasmid expressing human Drosha protein fused to a Flag tag at the C terminus (Lee et al. 2003). The Drosha protein was then pulled down using anti-Flag antibody conjugated to agarose beads (Fig. 4A). Shorter polypeptides derived from Drosha cDNA (Fig. 4A, Drosha★ and Drosha★★) were expressed, apart from the full-length protein (Drosha). It is not clear yet which portion of the full-length protein is missing in these shorter proteins and what caused the shortening. Two other prominent bands correspond to heat shock proteins (Hsp90 and Hsp70). These proteins may associate with Drosha only in the cytoplasm, because the interaction was not detected when nuclear extract was used instead of total cell extract (data not shown). Therefore it is unlikely that Hsp90 and Hsp70 are part of the nuclear processing complex.
Figure 4.
Role of human DGCR8 in pri-miRNA processing. (A) Immunoprecipitation of the Drosha complex with antibody against Flag. Drosha-Flag protein was expressed in HEK293T cells by transfection of pCK-Drosha-Flag. Total cell extract was prepared from four 10-cm dishes and used for immunoprecipitation with anti-Flag antibody-conjugated agarose beads. (Lane 1) Extract prepared from the same amount of “mock” transfected cells was used as a control. Proteins were separated on 7.5% denaturing polyacrylamide gels and visualized by silver staining. The specific bands were analyzed by MALDI-TOF mass spectrometry. Two forms of truncated Drosha proteins were marked with one or two asterisks (Drosha★ and Drosha★★). Heavy and light chains of anti-Flag antibody are abbreviated as h.c. and l.c. The position of the molecular mass markers is indicated on the left in kilodaltons. (B) Coimmunoprecipitation of DGCR8 and Drosha. V5-tagged DGCR8 and Flag-tagged Drosha proteins were coexpressed in HEK293T cells. Flag-pCK is the backbone for Drosha expression and was used here to match the total amount of plasmids in each transfection. Immunoprecipitation was carried out by incubating total cell extract with anti-Flag antibody-conjugated agarose beads in buffer D-K′250. For Western blot analysis, anti-V5 antibody was used to visualize the V5-DGCR8 protein. (C) In vitro processing of pri-let-7a-1 using Drosha-Flag or Flag-DGCR8 immunoprecipitates prepared in buffer D-K′100. (D) Reconstitution of pri-miRNA processing activity using Drosha-Flag immunoprecipitate and recombinant GST-DGCR8 protein. Pri-miRNA processing activity was measured by in vitro processing of pri-let-7a-1. Drosha-Flag immunoprecipitate was prepared by washing with either buffer A (0.5 M NaCl) or buffer B (2.5 M NaCl); 1.5 μg of recombinant proteins (GST or GST-DGCR8) was added to 1.5 μg of the Drosha-Flag immunoprecipitate. GST-DGCR8 (ΔN275) protein contains 276-773 amino acids of human DGCR8. (E) RT-PCR of pri-miRNA. The siRNA duplex against luciferase, Drosha, or DGCR8 was transfected to HeLa cells. After 72 h, total RNA was prepared and used for RT-PCR. (F) Northern blot analysis of miR-21. RNAi was carried out by transfection of siRNA duplexes to HeLa cells as in E. 5S rRNA bands stained with ethidium bromide are presented as a loading control.
In addition to the proteins described above, human Drosha coprecipitated with human DGCR8 (Fig. 4A). DGCR8 is a protein of unknown function which contains two dsRBDs and a putative WW domain. In order to confirm its interaction with Drosha, the DGCR8 protein fused to a V5 tag was expressed along with Drosha-Flag protein in HEK293T cells and immunoprecipitated using anti-Flag antibody (Fig. 4B). Western blot analysis using anti-V5 antibody shows that V5-DGCR8 is associated specifically with Drosha-Flag (Fig. 4B). DGCR8 was then tested for pri-miRNA processing activity (Fig. 4C). To do this, the Flag-DGCR8 protein or the Drosha-Flag protein was expressed in HEK293T cells and immunoprecipitated from total cell extract. An in vitro processing assay showed that Flag-DGCR8 immunoprecipitate can process pri-let-7a-1 efficiently. This result indicates that Flag-DGCR8 protein forms a functional complex with Drosha. We then tested whether DGCR8 is the only essential cofactor for Drosha in pri-miRNA processing and whether other factors are also required (Fig. 4D). Drosha-Flag protein was immobilized on anti-Flag antibody beads and washed intensively with high-salt buffer containing 2.5M NaCl and 1% Triton X-100 to remove other proteins associated with Drosha (Fig. 4D, lane 2). The purified Drosha was unable to execute pri-miRNA processing (Fig. 4D, lane 2). However, when Drosha was supplemented with GST-fused human DGCR8 protein prepared in E. coli (Supplementary Fig. S3), which itself does not possess catalytic activity (Fig. 4D, lane 4), Drosha processed pri-let-7a-1 efficiently (Fig. 4D, lane 3). The GST-DGCR8 protein used in this experiment contains 276-773 amino acids of human DGCR8, which is fully active in a processing assay (data not shown). Although we cannot formally exclude the possibility that small amounts of other cofactor(s) remained in the preparation of purified Drosha, this reconstitution experiment indicates that DGCR8 and Drosha may be necessary and sufficient to constitute a functional complex for pri-miRNA processing.
To confirm the function of human DGCR8 in vivo, DGCR8 was depleted in HeLa cells by transfecting three different siRNA duplexes against DGCR8 mRNA. The pri-miRNA level was determined by RT-PCR (Fig. 4E), and the pre-miRNA and mature miRNA level was measured by Northern blot analysis (Fig. 4F). When DGCR8 was depleted, pri-miRNA accumulated (Fig. 4E) whereas pre-miRNA and mature miRNA diminished (Fig. 4F). Thus, we conclude that human DGCR8 is critical for the processing of pri-miRNA into pre-miRNA.
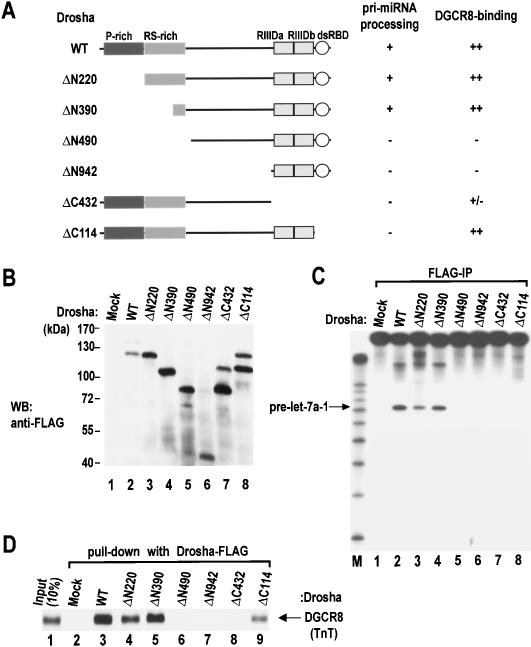
Domains of human Drosha essential for processing and complex formation
Drosha is a large protein with multiple domains (Fig. 5A). To investigate the biochemical role of each domain, we generated a series of deletion mutants. These mutants were expressed transiently in HEK293T cells (Fig. 5B) and immunoprecipitated using anti-Flag antibody to test for the enzymatic activity (Fig. 5C). Deletions of the N-terminal region that remove the P-rich region (ΔN220) and most of the RS-rich region (ΔN390) did not affect the enzymatic activity (Fig. 5C, lanes 3,4). Therefore these regions are dispensable for pri-miRNA processing itself. These regions, however, may possess other functional domains such as the nuclear localization signal. The remainder of the mutants (ΔN490, ΔC432, and ΔC114) turned out to be inactive in our assay, indicating that the middle region as well as the RIIIDs and the dsRBD are required for pri-miRNA processing (Fig. 5C, lanes 5-8). The expression level of mutant ΔN942 was significantly lower than those of the other mutants, making it difficult to assess the importance of the residues deleted in this mutant.
Figure 5.
Domain mapping of human Drosha. (A) Schematic representation of Drosha mutants. Results from the pri-miRNA processing assay and the DGCR8 binding assay are summarized at the right. (B) Western blot analysis of the mutant proteins. Expression plasmids were transfected into HEK293T cells. Forty-eight hours post-transfection, total cell extract was analyzed by Western blot analysis using anti-Flag antibody. (C) In vitro processing of pri-let-7a-1. The substrate was incubated with wild-type (WT) Drosha or mutant proteins that were prepared by immunoprecipitation in Buffer D-K′100 from transiently transfected HEK293T cells. (D) In vitro binding assay. Drosha protein (wild type [WT] or mutants) were immobilized on anti-Flag beads and incubated with radiolabeled DGCR8 that had been prepared in TnT-coupled reticulocyte lysate system (Promega). Drosha and DGCR8 were incubated with rotation in buffer D-K′250 for 90 min at 4°C.
The mutants were also tested for binding to DGCR8 (Fig. 5D). The Flag-tagged mutants were immobilized on anti-Flag beads and incubated with the full-length DGCR8 protein prepared in a coupled transcription-and-translation system (TnT). Wild-type Drosha as well as the mutants, ΔN220, ΔN390, and ΔC114, interact efficiently with DGCR8. However, deletion of the middle region including a part of the RS-rich region and the RIIIDs severely affected the binding activity (ΔN490 and ΔC432) (Fig. 5D, lanes 6-8), indicating that DGCR8 interacts with Drosha through the middle region and the RIIIDs of the Drosha protein. The WW domain is generally known as an interaction module to specific proline-rich sequences. The WW domain of DGCR8, however, is unlikely to serve as the binding site to the P-rich region of Drosha, because the proline-rich region of Drosha is dispensable for the interaction.
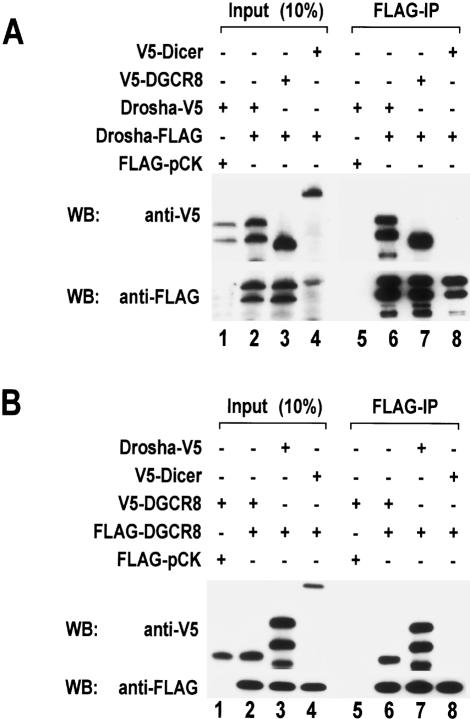
Drosha molecules interact with one another
Because the Drosha complex is ∼650 kDa, it is expected to be composed of multiple subunits. The reconstitution experiment described above (Fig. 4D), however, suggests that Drosha and DGCR8 may be the only two components of the complex. This raises the possibility that multiple copies of Drosha and/or DGCR8 proteins may exist in the complex. Thus we carried out coimmunoprecipitation experiments by transfecting two kinds of Drosha-expression plasmids containing different tags (either Flag or V5) into HEK293T cells (Fig. 6A). When Drosha-Flag was pulled down with anti-Flag antibody, Drosha-V5 was coimmunoprecipitated with Drosha-Flag (Fig. 6A, lane 6), suggesting that Drosha molecules interact with one another and that the Drosha complex may contain more than one copy of Drosha protein. This interaction appears to be specific, because Drosha does not coimmunoprecipitate with human Dicer (Fig. 6A, lane 8), the Argonaute protein eIF2C2 (data not shown), or an mRNA-binding protein Upf3 (data not shown).
Figure 6.
Homodimerization of Drosha and DGCR8. (A) Flag-tagged and V5-tagged Drosha proteins were coexpressed in HEK293T cells by transient transfection of the indicated plasmids. Forty-eight hours post-transfection, total cell extract was prepared and used for immunoprecipitation with anti-Flag affinity gel. Proteins were analyzed by Western blotting with anti-V5 antibody (upper panel) or anti-Flag antibody (lower panel). V5-Dicer was used as a control. (B) Flag-tagged and V5-tagged DGCR8 proteins were coprecipitated. Immunoprecipitation and Western blot analysis were done as in A.
We also investigated whether DGCR8 molecules interact with one another by a similar coimmunoprecipitation experiment (Fig. 6B). Flag-DGCR8 coprecipitated with V5-DGCR8 (Fig. 6B, lane 6), almost as efficiently as with Drosha-V5 (Fig. 6B, lane 7). These interactions are specific, as DGCR8 does not bind to Dicer (Fig. 6B, lane 8). Thus, the Drosha-DGCR8 complex may contain more than one copy of each protein.
To rule out the possibility that homodimerization was bridged by a single-stranded RNA (ssRNA) molecule, the extract was treated with RNase A (50 μg/mL) prior to immunoprecipitation. The efficiency of coprecipitation was not reduced by this treatment (Supplementary Fig. S4). We also examined whether the interactions are not due to artificial aggregation of overexpressed proteins. Firstly, less proteins were expressed by transfecting reduced amounts of plasmids. At lower concentrations, V5-tagged proteins were still coprecipitated with Flag-tagged proteins (Supplementary Fig. S5). We also carried out gel-filtration analysis of coexpressed Flag- and V5-tagged Drosha, followed by immunoprecipitation. The enzyme activity and protein profiles were analyzed by in vitro processing assay and Western blot analysis, respectively. Both the activity and the protein profile of overexpressed Drosha (Supplementary Fig. S6) are similar to those of endogenous Drosha (Fig. 3), indicating that the Drosha-Drosha interaction is unlikely to be an artifact of overexpression.
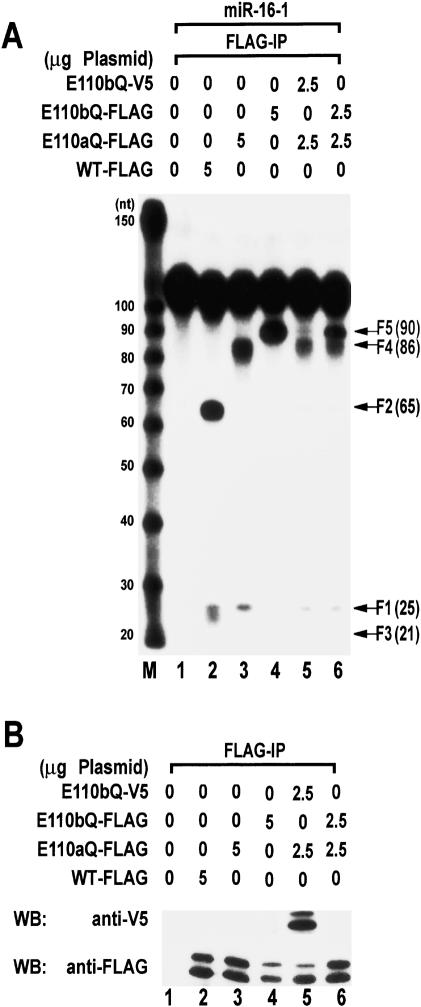
Intramolecular dimerization of the two RNase III domains in Drosha
If the pri-miRNA processing complex indeed contains two molecules of Drosha, the processing center could be assembled either intramolecularly or intermolecularly. To distinguish between the two possibilities, the mutants E110aQ and E110bQ were coexpressed, immunoprecipitated, and employed in processing assays (Fig. 7A). If both mutant proteins are expressed at much higher levels than endogenous Drosha and if most mutant molecules are engaged in dimerization, they would constitute E110aQ/E110aQ homodimer, E110aQ/E110bQ or E110bQ/E110aQ heterodimer, and E110bQ/E110bQ homodimer. Expression and interaction of the mutant proteins were analyzed by immunoprecipitation and Western blot analysis (Fig. 7B). When E110aQ-Flag and E110bQ-V5 were coexpressed and precipitated, the intramolecular dimerization model predicts the production of F4 and F5 but no F2 (pre-miRNA). But if intermolecular dimerization is the true assembly mode, equal amounts of F4 and F2 are predicted to be made without producing F5. In our processing assay, E110aQ-Flag and E110bQ-V5 produced F4 and F5 without generating significant amounts of F2 (Fig. 7A, lane 5). Therefore, it is likely that the RIIIDs of Drosha are engaged in intramolecular dimerization. It should be noted, however, that the amount of F5 produced in this assay is reproducibly much lower than that of F4 (Fig. 7A, lane 5). Because F5 can be produced only by E110bQ-V5 that coprecipitates with E110aQ-Flag, this result suggests that only a minority of active Drosha proteins may be engaged in Drosha-Drosha homodimerization in this experiment.
Figure 7.
Intramolecular dimerization between the RIIIDa and RIIIDb of human Drosha. (A) Flag-tagged or V5-tagged Drosha proteins were expressed in HEK293T cells by transient transfection of the indicated plasmids. Forty-eight hours post-transfection, immunoprecipitation was carried out using anti-Flag antibody from total cell extract. Processing activity was determined by incubating the immunoprecipitates with internally labeled pri-miR-16-1. Wild-type (WT) enzyme produces F1 (the 5′ flanking fragment), F2 (pre-miRNA), and F3 (the 3′ flanking fragment). Fragment F4 corresponds to F2 and F3 combined, whereas fragment F5 corresponds to F1 and F2 combined. Refer to Figure 2B for graphical explanation. (B) Proteins were analyzed by immunoprecipitation with anti-Flag antibody and Western blotting with anti-V5 antibody (upper panel) or anti-Flag antibody (lower panel).
Discussion
By demonstrating the fundamental similarities in the basic mechanism of the RNase III-mediated reaction, our study supports and expands the “single processing center” model recently proposed by Filipowicz and colleagues (Zhang et al. 2004; Supplementary Fig. S7A,B). Residues equivalent to E64 of Aa-RNase III do not participate in the catalytic reaction, whereas residues corresponding to E110 of Aa-RNase III are critical for cleavage. Thus, in this model, two nearby catalytic sites constitute a “single processing center” where the E110 equivalent residues play a key role. The dsRNA substrate is positioned along the shallow valley aslant the dimer of the two RIIIDs, which is rotated ∼30° compared to the position in the previous model of Blaszczyk and colleagues (2001).
As in class III enzyme human Dicer, the two RIIIDs of human Drosha form an intramolecular dimer where the two domains are distinct in their roles. The RIIIDa cuts the 3′ strand while the RIIIDb cleaves the 5′ strand, independently of each other (Fig. 2; Supplementary Fig. S7A,B). This result suggests that the Drosha protein is capable of orienting itself on pri-miRNA in a way that each RIIID is positioned on the correct strand. How does this specific orientation take place? As for human Dicer, it was proposed that the RIIIDa together with the PAZ domain recognizes the 3′ protruding end of dsRNA and places the processing center at ∼20 base pairs from the terminus (Zhang et al. 2004). It can be envisioned that the Drosha RIIIDs and the middle region may recognize the terminal loop or the bottom ssRNA region of pri-miRNA. Alternatively, DGCR8 may help Drosha to be correctly positioned on pri-miRNA. It would be interesting to know which domain (of which protein) is responsible for the specific orientation of the two RIIIDs on pri-miRNA and which part(s) of pri-miRNA provide the basis for such specificity.
Despite the similarities among RNase III proteins, Drosha differs from other RNase III proteins in many aspects, particularly in its substrate specificity and cleavage site selection (Bernstein et al. 2001; Lee et al. 2003). Dicer can act on any dsRNAs with simple preference toward the terminus of dsRNA (Elbashir et al. 2001; Bernstein et al. 2001; Provost et al. 2002; Zhang et al. 2002). As a result, Dicer produces ∼22-nt fragments progressively from the terminus. Drosha is apparently less promiscuous than Dicer. Pri-miRNAs contain local hairpin structures as well as long flanking sequences at both sides. Mutations disturbing the stem structure of pri-miRNA affect processing efficiency (Lee et al. 2003; Zeng and Cullen 2003). It appears that all the cis-acting elements required for processing are located close to the stem-loop, because Drosha can process efficiently the “minimal” pri-miRNAs that contain only ∼20 nt outside the cleavage sites (Fig. 2). However, it is still unclear how Drosha recognizes its target and makes a cut at the specific sites. Since there is no common sequence element found in pri-miRNAs, Drosha and/or its cofactor(s) is thought to recognize the common structural feature(s) among diverse pri-miRNAs. It would be interesting to decipher the structural elements that define a “pri-miRNA”. The “minimal” pri-miR-16-1 and pri-miR-30a described in this study will provide a useful tool for further biochemical analyses.
At present, it is unclear which domain of the Drosha protein is involved in substrate recognition. Part of the interaction between the substrate and the enzyme should occur around the catalytic sites. This interaction is likely to be fortified by the dsRBD of Drosha (Supplementary Fig. S7). In fact, the deletion of dsRBD abolished enzymatic activity (Fig. 5). In the case of class I RNase III proteins, two dsRBDs are involved in RNA-enzyme interaction. Human Dicer contains only one dsRBD, but the PAZ domain is thought to interact with the 3′ overhang of the dsRNA terminus. Thus, a single dsRBD of Drosha protein may not be sufficient for strong interaction with the substrate. Additional RNA-binding module(s) may be required for the recognition of the substrate.
DGCR8 may provide such an RNA-binding module and thereby may serve as an essential component of the Drosha complex. The DGCR8 gene was originally identified in the “DiGeorge syndrome chromosomal region (DGCR)” at human chromosome 22q11 (Shiohama et al. 2003). Monoallelic deletion of this region is associated with a complicated clinical phenotype, including DiGeorge syndrome/conotruncal anomaly face syndrome/velocardiofacial syndrome, although it remains unknown whether DGCR8 is involved in this genetic disorder.
A model of the pri-miRNA cleavage by Drosha, integrating our results and other previous studies on RNase III proteins, is presented in Supplementary Figure S7. Two RIIIDs of Drosha form an intramolecular dimer to create one processing center containing two catalytic sites. Because of the helical (twisted) structure of dsRNA, the RIIIDa makes a cut at the 3′ strand while the RIIIDb cleaves the 5′ strand, resulting in an ∼2-nt 3′ overhang. The dsRBD of Drosha holds the stem, and the middle region of Drosha interacts with DGCR8. The RIIIDs may also contribute to the binding to DGCR8. DGCR8 may interact with the stem to stabilize the substrate binding. Alternatively but not exclusively, DGCR8 may also interact with the single-stranded region of RNA so that the complex is correctly oriented on pri-miRNA.
The reconstitution experiment (Fig. 4D) suggests that Drosha and DGCR8 may be the only essential components of the pri-miRNA processing complex. However, the active complex is ∼650 kDa, which may accommodate multiple subunits. One possibility is that additional auxiliary factors are present in the Drosha-DGCR8 complex (Supplementary Fig. S7C, proteins X and Y). These proteins may regulate and/or assist the function of the main components. Another nonmutually exclusive possibility is that multiple copies of Drosha and DGCR8 may be included in the Drosha-DGCR8 complex. This complex may accommodate two molecules of Drosha (160 kDa) and two or more copies of DGCR8 (120 kDa on SDS-PAGE). Thus it is possible that the entire complex may be a dimer of heterodimers, resulting in a heterotetrameric complex of Drosha and DGCR8 (Supplementary Fig. S7D). However, our current data cannot distinguish clearly between these two possibilities (“heterodimer model” and “heterotetramer model”). In fact, these two forms (heterodimer and heterotetramer) may convert to each other dynamically. It remains to be determined which form plays a major role in pri-miRNA processing reaction. The functional significance of homodimerization should be investigated by further mutagenesis and biochemical analyses. For a full understanding of the Drosha-DGCR8 complex, it would also be critical to solve the structure of the individual components and the whole complex.
Materials and methods
Cell culture and transfection
HeLa cells and HEK293T cells were cultured in DMEM (WelGENE) supplemented with 10% FBS (WelGENE). Transfection was carried out by the calcium-phosphate method.
Immunoprecipitation
HEK293T cells grown in a 10-cm dish were collected in 500 μL of ice-cold buffer D-K′100 (20 mM Tris at pH 8.0, 100 mM KCl, 0.2 mM EDTA, 0.2 mM PMSF) 48 h post-transfection. The cells were sonicated on ice and centrifuged at 13,200 rpm for 15 min at 4°C. The supernatant was incubated with 10 μL of anti-Flag antibody conjugated to agarose beads (anti-Flag M2 affinity gel, Sigma) with constant rotation for 90 min at 4°C. The beads were washed six times in buffer D-K′100, drained, and used for in vitro processing. When the immunoprecipitate is to be used for Western blot analysis, buffer D-K′250 (20 mM Tris at pH 8.0, 250 mM KCl, 0.2 mM EDTA, 0.2 mM PMSF) was used for immunoprecipitation instead of buffer D-K′100.
In vitro processing of pri-miRNAs
In vitro processing of pri-miRNAs using whole-cell extract or Flag-immunoprecipitates was carried out as described (Lee et al. 2003). Briefly, 30 μL of processing reaction contained 15 μL of whole-cell extract or the beads from immunoprecipitation, 6.4 mM MgCl2, 1 U/μL of Ribonuclease Inhibitor (TAKARA), and the labeled transcripts of 1 × 104 to 1 × 105 cpm. The reaction mixture was incubated at 37°C for 90 min. RNA was extracted from the reaction mixture by phenol extraction and analyzed on 10%-12.5% denaturing polyacrylamide gel.
Western blot analysis
Protein samples were typically analyzed on 7.5% SDS-polyacrylamide gels and transferred to Hybond-C Extra membrane (Amersham). Primary antibodies used in this study are rabbit anti-Drosha antibody raised against recombinant Drosha protein prepared in E. coli, mouse anti-Flag M2 antibody (Sigma), and mouse anti-V5 antibody (Invitrogen).
Cloning and sequencing of pre-miR-16-1
Pre-miR-16-1 was prepared by in vitro processing as described (Lee et al. 2003). Briefly, pri-miR-15a∼16-1 was transcribed in vitro and processed using the immunoprecipitated Drosha-Flag at 37°C for 90 min. The ∼65-nt band presumably containing pre-miR-16-1 was gel-purified and ligated to the 3′ adapter. The ligated product was gel-purified and ligated to the 5′ adapter. The 3′ and 5′ adapters used for the cloning are 5′-pUUUaaccgcgaattccagidT-3′ (uppercase, RNA; lowercase, DNA; p, phosphate; idT, inverted deoxythymidine) and 5′-acggaattcctcac tAAA-3′ (uppercase, RNA; lowercase, DNA), respectively. Reverse transcription was performed using reverse transcription primer (5′-ACTGGAATTCGCGGTTAAA-3′). Forward primer (5′-CAGCCAACGGAATTCCTCCTCACTAAA-3′) and reverse primer (reverse transcription primer) were used for RCR amplification. The PCR product was subcloned into pGEM-T-easy (Promega), and 10 clones were sequenced.
Generation of minimal pri-miRNAs
To produce “minimal pri-miR-16-1”, the region including 23 nt upstream of the 5′ cleavage site and 21 nt downstream of the 3′ cleavage site was amplified by PCR. To generate “minimal pri-miR-30a”, the region covering 21 nt upstream of the 5′ cleavage site and 19 nt downstream of the 3′ cleavage site was amplified by PCR. The forward primer was designed to contain the T7 promoter sequences in addition to miRNA sequences so that the PCR product will have the promoter at the 5′ end of pri-miRNA sequences. For efficient transcription, two additional Gs were included between the promoter and the pri-miRNA sequence. The PCR product was used directly as the template for in vitro transcription. For pri-miR-16-1, 5′-TAATACGACTCACTATAGGTGATAGCAATGTCAGCAGTG-3′ (forward) and 5′-GTAGAGTATGGTCAACCTTA-3′ (reverse) were used for PCR amplification. For pri-miR-30a, 5′-TAATACGACTCACTATAGGTTGCTGTTGACAGTGAGCGA-3′ (forward) and 5′-AAGTCCGAGGCAGTAGGCAG-3′ (reverse) were used for PCR amplification.
Labeling of pri-miRNAs
For internal labeling, [α-32P] UTP was included in transcription as described (Lee et al. 2002). For 5′ end labeling, nonradioactive pri-miRNA was synthesized and dephosphorylated by calf intestine phosphatase (TAKARA). The transcript was labeled at the 5′ end using T4 polynucleotide kinase (TAKARA) and [γ-32P] ATP.
Gel exclusion chromatography
HEK293T cells grown in twenty-five 10-cm plates were resuspended in RSB-100-digitonin buffer (10 mM Tris at pH 7.4, 100 mM NaCl, 35 μg/mL digitonin) (800 μL for each plate) and incubated at 4°C for 15 min to disrupt the plasma membrane. The cells were transferred to 1.75-mL tubes and centrifuged at 3000g for 5 sec. Supernatant was removed, and the pellets containing the nuclei were resuspended in 8 mL of lysis buffer (20 mM Tris at pH 8.0, 100 mM KCl, 0.2 mM EDTA, 0.2 mM PMSF). The nuclei were disrupted by sonication and centrifuged at 13,200 rpm at 4°C for 60 min. The supernatant was filtered through a 0.22-μm filter membrane and concentrated using Centricon YM-30 (Millipore) to the final volume of 2 mL. This nuclear extract was treated with 10 μL of 10 mg/mL RNase A (to the final concentration of 50 μg/mL) and incubated at 4°C for 30 min. Finally, 1.75 mL of the nuclear extract was loaded on a Sephacryl S-300 HR (Sigma) column and fractionated. Each fraction was 1.75 mL in volume. In vitro processing of pri-let-7a-1 was carried out using 20 μL from each fraction. Fifty microliters from each fraction was subject to Western blot analysis using anti-Drosha antibody to detect endogenous Drosha protein. To analyze the complex formation of the overexpressed Drosha proteins, the nuclear extract was prepared from HEK293T cells transfected with of Drosha-Flag and Drosha-V5 expression plasmids (1.5 μg each plasmid per 10-cm plate) 48 h post-transfection. Following gel filtration chromatography, 250 μL of each fraction was taken for immunoprecipitation with anti-Flag antibody, and the immunoprecipitate was used for in vitro processing of pri-let-7a-1. To analyze the expression and dimerization of the Drosha-Flag and Drosha-V5 proteins, 1.3 mL from each fraction was used for immunoprecipitation with anti-Flag antibody. The immunoprecipitate from each fraction was analyzed by Western blot analysis using anti-Flag antibody and anti-V5 antibody.
Identification of Drosha interacting proteins
To express Flag-tagged Drosha proteins, 10 μg of pCK-Drosha-Flag was transfected to HEK293T cells grown on 10-cm dishes. For “mock” transfection, the same amount of pCK plasmid was transfected. To purify the Drosha complex, total cell extract was prepared from four 10-cm dishes in 2 mL of lysis buffer (20 mM HEPES-KOH at pH 7.9, 350 mM KCl, 0.2 mM EDTA, 0.5 mM DTT, 0.2 mM PMSF, 5% glycerol) by sonication followed by centrifugation. This extract was incubated with anti-Flag M2 affinity gel (Sigma) at 4°C for 2 h. After washing six times with lysis buffer, the beads were drained and boiled with 40 μL of protein sample buffer (two times). The protein samples were loaded on 7.5% SDS-PAGE, followed by silver staining. Specific bands were sliced, digested by trypsin, and identified by MALDI-TOF mass spectrometry.
RNA interference
siRNA duplexes targeting Drosha and DGCR8 mRNAs were transfected to HeLa cells using RNAiFect transfection reagent (QIAGEN) as described in the manufacturer's instructions. Target sequences of siDrosha, siDGCR8-1, siDGCR8-2, and siDGCR8-3 are 5′-AACGAGUAGGCUUCGUGACUU-3′, 5′-AACAUCGGACAAGAGUGUGAU-3′, 5′-AACGAUGAUGACCAAGAUUAA-3′, and 5′-AACAAUUUGGAGCUAGAUGAA-3′, respectively. SiDrosha, siDGCR8-2, and siDGCR8-3 were manufactured by Samchully Pham, and siDGCR8-1 was purchased from Dharmacon. Total RNA was prepared 3 d after transfection and used for RT-PCR or Northern blot analysis.
RT-PCR
Five micrograms of HeLa total RNA was used for the first-strand cDNA synthesis with SUPERSCRIPT II and oligo-dT primers (Invitrogen). To detect the expression level of pri-miRNAs and GAPDH mRNA, the following primers were used. For pri-miR-30a, 5′-ATTGCTGTTTGAATGAGGCTTCAGTACTTT-3′ (forward) and 5′-TTCAGCTTTGTAAAAATGTATCAAAGAGAT-3′ (reverse) were used for PCR amplification. For pri-miR-15a∼16-1, 5′-CCTTGGAGTAAAGTAGCAGCACATAATG-3′ (forward) and 5′-ATATACATTAAAACACAACTGTAGAGTATG-3′ (reverse) were used for PCR amplification. For pri-miR-21, 5′-ACCAGACAGAAGGACCAGAGTTTCTGATTA-3′ was used for the first-strand cDNA synthesis, and this primer (reverse) and 5′-GTTCGATCTTAACAGGCCAGAAATGCCTGG-3′ (forward) were used for PCR amplification. For GAPDH mRNA, 5′-CCCATCACCATCTTCCAGGAGTGAGTGGAAGAC-3′ (forward) and 5′-CGCCCCACTTGATTTTGGAGGGATCTCGCCTACCG-3′ were used for PCR amplification. For DGCR8 mRNA, 5′-ATGGAGACAGATGAGAGCCCC-3′ (forward) and 5′-CAAGATGGGCCTCTCGGACTC-3′ were used for PCR amplification.
Northern blot analysis
Fifty micrograms of HeLa total RNA were resolved on 12.5% urea-polyacrylamide gels and transferred electronically to Zeta-probe membrane (Bio-Rad). An oligonucleotide complementary to miR-21 was end-labeled and used as the probe for Northern. The sequence of the oligonucleotide is 5′-TCAACATCAGTCTGATAAGCTA-3′.
Recombinant DGCR8
To prepare recombinant DGCR8 (ΔN275), the partial length (276-773 amino acids) of human DGCR8 cDNA was amplified from HEK293T cDNA by PCR (forward primer: 5′-GAATTCGATGGAGAGACAAGTGTGCAGC-3′, reverse primer: 5′-GCGGCCGCTCACACGTCCACGGTGCACAG-3′), subcloned into pGEM-T easy vector (Promega) and sequenced. The human DGCR8 partial-length cDNA was subcloned into pGEX-4T3 vector (Amersham), which has GST-tag at the N terminus, using EcoRI and NotI sites. The expression clone of DGCR8 was transformed to E. coli BL21-RIL strain. The expression and purification of recombinant DGCR8 were conducted according to the manufacturer's protocol.
Reconstitution of pri-miRNA processing activity
HEK293T cells transfected with Drosha-Flag expression plasmid (pCK-Drosha-Flag) were harvested 2 d after transfection. Total cell extract was prepared in buffer A (20 mM Tri-HCl at pH 8.0, 500 mM NaCl, 0.2 mM EDTA, 0.2 mM PMSF) by sonication followed by centrifugation. The whole-cell extract was incubated with anti-Flag M2 affinity gel (Sigma) at 4°C for 2 h. After washing twice with buffer A, the Flag immunoprecipitates were drained and incubated with 1 mL of buffer B (20 mM Tris-HCl at pH 8.0, 2.5 M NaCl, 0.2 mM EDTA, 0.2 mM PMSF, 1% Triton X-100) at 4°C for 1 h in order to detach the Drosha-interacting proteins from Drosha-Flag complex. The beads were washed five times with buffer B and twice with buffer A. After washing, the immunoprecipitates were drained and used for in vitro processing with 1.5 μg of recombinant proteins (GST or GST-DGCR8).
In vitro protein binding assay
Immunopurified Drosha-Flag proteins (∼2 μg) bound to 15 μL of anti-Flag affinity gels were incubated with in vitro translated DGCR8 protein in 1 mL of buffer D-′K250. After incubation for 90 min at 4°C, the resin was washed six times with 1 mL of buffer D-K′250. The bound fraction was eluted by boiling the beads in 40 μL of SDS-PAGE sample buffer and run on SDS-PAGE.
Acknowledgments
We thank Drs. Patrick Provost, Gideon Dreyfuss, and Zissimos Mourelatos for Dicer cDNA, hUpf3 cDNA, and eIF2C2 cDNA, respectively. This work was supported by grants from the Basic Research Program of the Korea Science & Engineering Foundation (R02-2004-000-10173-0), the BK21 Research Studentship, and a grant (KRF-2002-041-C00204) from the Korea Research Foundation.
Supplemental material is available at http://www.genesdev.org.
Article published online ahead of print. Article and publication date are at http://www.genesdev.org/cgi/doi/10.1101/gad.1262504.
References
- Ambros V., Bartel, B., Bartel, D.P., Burge, C.B., Carrington, J.C., Chen, X., Dreyfuss, G., Eddy, S.R., Griffiths-Jones, S., Marshall, M., et al. 2003. A uniform system for microRNA annotation. RNA 9: 277-279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel D.P. 2004. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 116: 281-297. [DOI] [PubMed] [Google Scholar]
- Basyuk E., Suavet, F., Doglio, A., Bordonne, R., and Bertrand, E. 2003. Human let-7 stem-loop precursors harbor features of RNase III cleavage products. Nucleic Acids Res. 31: 6593-6597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein E., Caudy, A.A., Hammond, S.M., and Hannon, G.J. 2001. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature 409: 363-366. [DOI] [PubMed] [Google Scholar]
- Blaszczyk J., Tropea, J.E., Bubunenko, M., Routzahn, K.M., Waugh, D.S., Court, D.L., and Ji, X. 2001. Crystallographic and modeling studies of RNase III suggest a mechanism for double-stranded RNA cleavage. Structure (Camb) 9: 1225-1236. [DOI] [PubMed] [Google Scholar]
- Bohnsack M.T., Czaplinski, K., and Gorlich, D. 2004. Exportin 5 is a RanGTP-dependent dsRNA-binding protein that mediates nuclear export of pre-miRNAs. RNA 10: 185-191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbashir S.M., Lendeckel, W., and Tuschl, T. 2001. RNA interference is mediated by 21- and 22-nucleotide RNAs. Genes & Dev. 15: 188-200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grishok A., Pasquinelli, A.E., Conte, D., Li, N., Parrish, S., Ha, I., Baillie, D.L., Fire, A., Ruvkun, G., et al. 2001. Genes and mechanisms related to RNA interference regulate expression of the small temporal RNAs that control C. elegans developmental timing. Cell 106: 23-34. [DOI] [PubMed] [Google Scholar]
- Hammond S.M., Boettcher, S., Caudy, A.A., Kobayashi, R., and Hannon, G.J. 2001. Argonaute2, a link between genetic and biochemical analyses of RNAi. Science 293: 1146-1150. [DOI] [PubMed] [Google Scholar]
- Hutvagner G., McLachlan, J., Pasquinelli, A.E., Balint, E., Tuschl, T., and Zamore, P.D. 2001. A cellular function for the RNA-interference enzyme Dicer in the maturation of the let-7 small temporal RNA. Science 293: 834-838. [DOI] [PubMed] [Google Scholar]
- Ishizuka A., Siomi, M.C., and Siomi, H. 2002. A Drosophila fragile X protein interacts with components of RNAi and ribosomal proteins. Genes & Dev. 16: 2497-2508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ketting R.F., Fischer, S.E., Bernstein, E., Sijen, T., Hannon, G.J., and Plasterk, R.H. 2001. Dicer functions in RNA interference and in synthesis of small RNA involved in developmental timing in C. elegans. Genes & Dev. 15: 2654-2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khvorova A., Reynolds, A., and Jayasena, S.D. 2003. Functional siRNAs and miRNAs exhibit strand bias. Cell 115: 209-216. [DOI] [PubMed] [Google Scholar]
- Kim V.N. 2004. MicroRNA precursors in motion: Exportin-5 mediates their nuclear export. Trends Cell Biol. 14: 156-159. [DOI] [PubMed] [Google Scholar]
- Knight S.W. and Bass, B.L. 2001. A role for the RNase III enzyme DCR-1 in RNA interference and germ line development in Caenorhabditis elegans. Science 293: 2269-2271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y., Jeon, K., Lee, J.T., Kim, S., and Kim, V.N. 2002. MicroRNA maturation: Stepwise processing and subcellular localization. EMBO J. 21: 4663-4670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y., Ahn, C., Han, J., Choi, H., Kim, J., Yim, J., Lee, J., Provost, P., Radmark, O., Kim, S., et al. 2003. The nuclear RNase III Drosha initiates microRNA processing. Nature 425: 415-419. [DOI] [PubMed] [Google Scholar]
- Lee Y.S., Nakahara, K., Pham, J.W., Kim, K., He, Z., Sontheimer, E.J., and Carthew, R.W. 2004a. Distinct roles for Drosophila Dicer-1 and Dicer-2 in the siRNA/miRNA silencing pathways. Cell 117: 69-81. [DOI] [PubMed] [Google Scholar]
- Lee Y., Kim, M., Han, J., Yeom, K.H., Lee, S., Baek, S.H., and Kim, V.N. 2004b. MicroRNA genes are transcribed by RNA polymerase II. EMBO J. 23: 4051-4060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lingel A., Simon, B., Izaurralde, E., and Sattler, M. 2003. Structure and nucleic-acid binding of the Drosophila Argonaute 2 PAZ domain. Nature 426: 465-469. [DOI] [PubMed] [Google Scholar]
- Liu Q., Rand, T.A., Kalidas, S., Du, F., Kim, H.E., Smith, D.P., and Wang, X. 2003. R2D2, a bridge between the initiation and effector steps of the Drosophila RNAi pathway. Science 301: 1921-1925. [DOI] [PubMed] [Google Scholar]
- Lund E., Guttinger, S., Calado, A., Dahlberg, J.E., and Kutay, U. 2004. Nuclear export of microRNA precursors. Science 303: 95-98. [DOI] [PubMed] [Google Scholar]
- Murchison E.P. and Hannon, G.J. 2004. miRNAs on the move: miRNA biogenesis and the RNAi machinery. Curr. Opin. Cell Biol. 16: 223-229. [DOI] [PubMed] [Google Scholar]
- Pfeffer S., Zavolan, M., Grasser, F.A., Chien, M., Russo, J.J., Ju, J., John, B., Enright, A.J., Marks, D., Sander, C., et al. 2004. Identification of virus-encoded microRNAs. Science 304: 734-736. [DOI] [PubMed] [Google Scholar]
- Provost P., Dishart, D., Doucet, J., Frendewey, D., Samuelsson, B., and Radmark, O. 2002. Ribonuclease activity and RNA binding of recombinant human Dicer. EMBO J. 21: 5864-5874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz D.S., Hutvagner, G., Du, T., Xu, Z., Aronin, N., and Zamore, P.D. 2003. Asymmetry in the assembly of the RNAi enzyme complex. Cell 115: 199-208. [DOI] [PubMed] [Google Scholar]
- Shiohama A., Sasaki, T., Noda, S., Minoshima, S., and Shimizu, N. 2003. Molecular cloning and expression analysis of a novel gene DGCR8 located in the DiGeorge syndrome chromosomal region. Biochem. Biophys. Res. Commun. 304: 184-190. [DOI] [PubMed] [Google Scholar]
- Song J.J., Liu, J., Tolia, N.H., Schneiderman, J., Smith, S.K., Martienssen, R.A., Hannon, G.J., and Joshua-Tor, L. 2003. The crystal structure of the Argonaute2 PAZ domain reveals an RNA binding motif in RNAi effector complexes. Nat. Struct. Biol. 10: 1026-1032. [DOI] [PubMed] [Google Scholar]
- Tabara H., Yigit, E., Siomi, H., and Mello, C.C. 2002. The dsRNA binding protein RDE-4 interacts with RDE-1, DCR-1, and a DExH-box helicase to direct RNAi in C. elegans. Cell 109: 861-871. [DOI] [PubMed] [Google Scholar]
- Yan K.S., Yan, S., Farooq, A., Han, A., Zeng, L., and Zhou, M.M. 2003. Structure and conserved RNA binding of the PAZ domain. Nature 426: 468-474. [DOI] [PubMed] [Google Scholar]
- Yi R., Qin, Y., Macara, I.G., and Cullen, B.R. 2003. Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes & Dev. 17: 3011-3016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng Y. and Cullen, B.R. 2003. Sequence requirements for micro RNA processing and function in human cells. RNA 9: 112-123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., Kolb, F.A., Brondani, V., Billy, E., and Filipowicz, W. 2002. Human Dicer preferentially cleaves dsRNAs at their termini without a requirement for ATP. EMBO J. 21: 5875-5885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., Kolb, F.A., Jaskiewicz, L., Westhof, E., and Filipowicz, W. 2004. Single processing center models for human Dicer and bacterial RNase III. Cell 118: 57-68. [DOI] [PubMed] [Google Scholar]