Abstract
The checkpoint kinases Chk1 and Chk2 are central to the induction of cell cycle arrest, DNA repair, and apoptosis as elements in the DNA-damage checkpoint. We report here that in several human tumor cell lines, Chk1 and Chk2 control the induction of the p53 related transcription factor p73 in response to DNA damage. Multiple experimental systems were used to show that interference with or augmentation of Chk1 or Chk2 signaling strongly impacts p73 accumulation. Furthermore, Chk1 and Chk2 control p73 mRNA accumulation after DNA damage. We demonstrate as well that E2F1 directs p73 expression in the presence and absence of DNA damage. Chk1 and Chk2, in turn, are vital to E2F1 stabilization and activity after genotoxic stress. Thus, Chk1, Chk2, E2F1, and p73 function in a pathway mediating p53-independent cell death produced by cytotoxic drugs. Since p53 is often obviated through mutation as a cellular port for anticancer intervention, this pathway controlling p53 autonomous pro-apoptotic signaling is of potential therapeutic importance.
Keywords: Apoptosis, Chk1, Chk2, DNA damage, E2F1, p73
The discovery of the p53 gene family member p73 has added complexity to the current view of p53 signaling. The p73 isoforms share a similar configuration of domains and significant homology with some regions of p53, especially within their DNA-binding domains (Kaghad et al. 1997; Kartasheva et al. 2002). However, in vivo p53 and p73 seem to play very different roles and the intersection of their various functions is as yet not understood. While p53 mice are famously tumor prone, p73 null mice show no defect in tumor suppression. Rather p73 loss confers defects in neuronal development and immune function (Donehower et al. 1992; Yang et al. 2000). Further, while p53 sustains mutations in over 50% of human cancers and these tend to cluster within the DNA-binding domain, p73 is rarely found to be mutated (Hollstein et al. 1999; Melino et al. 2002).
In contrast to p53, p73 can be expressed as a variety of isoforms due to alternate promoter usage and alternative splicing. There are two N-terminally distinct isoforms, TA which possesses a transactivation domain with ∼25% homology to the p53 transactivation domain, and a second isoform lacking this transactivation domain (called ΔN) that is directed from a downstream promoter between exons 3 and 4. ΔN isoforms are thought to act in a dominant negative manner against full-length transcriptionally active (TA) p73 as well as p53 (Stiewe et al. 2002; Zaika et al. 2002), although in some experimental settings ΔN isoforms of p73 themselves display transcriptional activation ability (Liu et al. 2004). At the C terminus, alternative splicing generates multiple isoforms designated p73α, p73β, and p73γ with increasing truncation, but the in vivo functions of such C-terminal variants are unknown.
Among the most important functions of p53 is the induction of apoptosis in response to cellular stress (Johnstone et al. 2002). Nevertheless, the role of p73 in this process is not clear. In mouse embryos fibroblasts expressing E1A, DNA-damage-facilitated apoptosis mediated by p53 requires p73 and the other family member p63, suggesting that at least in some conditions these p53 homologs play a role in apoptosis in vivo (Flores et al. 2002). Given that p53 is commonly inactivated in human tumors while p73 for the most part remains unmutated, it is of great interest to determine whether and when p73 can mediate p53-independent apoptosis and cell cycle arrest. RNA interference studies have demonstrated that p73 is able is induce apoptosis, as its down-regulation protected a colorectal cancer cell line from apoptosis induced by a panel of chemotherapeutic agents (Irwin et al. 2003). Similarly, expression of dominant-negative p73 protects p53-null cells from drug-induced apoptosis (Bergamaschi et al. 2003). The precise downstream targets involved in p73-mediated apoptosis are not clear. p73 can bind to the p53AIP1 promoter (Costanzo et al. 2002) and p53-independent induction of this gene has been observed (Bergamaschi et al. 2003). Moreover, overexpression of two p53 coactivators, ASPP1 and ASPP2, can induce several p53 target genes via p73 activation in p53 null cell lines (Bergamaschi et al. 2004).
Given p73's demonstrated role in p53 independent apoptosis, delineation of the signaling pathway leading to p73 activation in response to stress is a central question. Transcriptional and posttranscriptional regulators of p73 have been identified. The E2F1 transcription factor can induce p73 mRNA and p73 is required for E2F1-induced apoptosis (Irwin et al. 2000; Lissy et al. 2000; Stiewe and Putzer 2000). Furthermore, E2F1 binds to the p73 promoter in response to DNA damage and there recruits the histone acetyltransferase PCAF (Pediconi et al. 2003). p73 protein is stabilized in response to cisplatin and gamma irradiation through c-Abl-mediated phosphorylation, and co-expression of c-Abl and p73 enhances apoptosis (Agami et al. 1999; Gong et al. 1999; Yuan et al. 1999; Tsai and Yuan 2003). The p38 MAPK cascade is also a part of c-Abl-mediated p73 stabilization (Sanchez-Prieto et al. 2002). The mismatch repair factors MLH1 and PMS2 also play a role in p73 induction by DNA damage (Gong et al. 1999; Shimodaira et al. 2003). p73 is phosphorylated at Thr 86 by cyclin/cdk complexes which can repress p73 function (Fulco et al. 2003; Gaiddon et al. 2003). A novel HECT type E3 ubiquitin liagase, NEDL2, has been shown to stabilize p73 (Miyazaki et al. 2003). Other factors which can regulate p73 function independent of protein level include Chk1 (Gonzalez et al. 2003) and Akt (Basu et al. 2003), which regulate p73 transcriptional activity positively and negatively, respectively.
In contrast to that of p73, the mechanism of p53 stabilization by DNA damage has been intensively studied. Both p53 and Mdm2 are phosphorylated in response to DNA damage and such phosphorylation can prevent their interacting with each other as well as inhibiting the ability of Mdm2 to target p53 for proteasome-mediated degradation (for reviews, see Prives 1998; Appella and Anderson 2001). In particular, the ATM kinase has been shown to phosphorylate both p53 and Mdm2 (Banin et al. 1998; Canman et al. 1998; Khosravi et al. 1999; de Toledo et al. 2000; Maya et al. 2001). The checkpoint kinases Chk1 and Chk2 have also been shown to phosphorylate p53 at N-terminal sites including Ser 20 located within the Mdm2 interacting region on p53 (Chehab et al. 2000; Shieh et al. 2000).
Chk1 and Chk2 are protein kinases that function as effectors in a DNA-damage-response pathway whose general features are conserved between yeast and humans (Bartek and Lukas 2003). In addition to p53, they have been reported to phosphorylate a number of substrates, including Cdc25A, Cdc25B, and Cdc25C (Matsuoka et al. 1998), BRCA1 (Lee et al. 2000), and others. Each of these substrates possesses one or more Chk1/2 phosphorylation sites that conform to consensus sequences (similar for both kinases) that were determined using peptide libraries (O'Neill et al. 2002). Interestingly, while p53 does not possess such a Chk consensus site, Chk2 null mice have been shown to display defects in p53 stabilization (Hirao et al. 2000), although different studies report somewhat varying results in this regard (Takai et al. 2002). In contrast, siRNA-mediated down-regulation of Chk1 and Chk2 in some human tumor cell lines (Ahn et al. 2003) or deletion of both Chk2 alleles in HCT116 cells (Jallepalli et al. 2003) does not impact p53 stabilization or activation. Despite the complexity and perhaps controversy in evaluation of the role of Chk kinases in regulating p53, it is important to note that Chk2 null mice display profound defects in apoptosis in several tissues (Hirao et al. 2000, 2002; Takai et al. 2002).
Since Chk1 and Chk2 are not obligate or universal regulators of p53 and yet at least Chk2 is known to play a critical role in apoptosis in mice (Chk1 null mice die early in embryogenesis), we therefore sought other targets for Chk regulation. Here we show that Chk1 and Chk2 control p73 accumulation after DNA damage. Their ability to regulate expression of E2F1, a factor involved in regulation of TAp73 transcription, is critical for induction of p73 and most likely for their roles in apoptosis as well.
Results
Generation and characterization of a TAp73-specific antisera
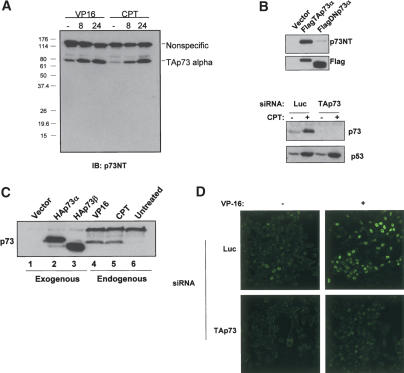
We raised a polyclonal antibody (p73NT) against a peptide derived from the transactivation domain of TAp73 but absent from transcripts derived from the ΔN promoter element. Crude serum was affinity purified using baculovirus-derived purified TAp73β protein. Immunoblotting of HCT116 cells treated with VP16 or camptothecin for 8 or 24 h revealed a single dominant polypeptide that was induced by DNA damage migrating just below an 80-kDa polypeptide marker (Fig. 1A). To determine the identity of this isoform, we first confirmed that our antibody only recognizes TAp73 isoforms by expressing Flag-tagged TAp73α and ΔNp73α in H1299 cells. As expected, p73NT only reacted with TAp73α while the anti-Flag antibody reacted with both isoforms (Fig. 1B, top). Next, we transfected HCT116 cells with a control siRNA or an siRNA specific for TAp73 and induced p73 with camptothecin for 24 h. The TAp73 siRNA greatly diminished reactivity with p73NT but had no effect on p53, further confirming that the dominant form present in HCT116 is TAp73 (Fig. 1B, bottom). To investigate the C-terminal identity of this TAp73 species, H1299 cells were transfected with HA-TAp73α and HA-TAp73β and their migration was compared to that of endogenous p73 from HCT116 cells treated with VP16 or CPT (Fig. 1C). The induced TAp73 species migrated just below the tagged p73α but well above the β isoform (Fig. 1C, cf. lanes 2,3 and 4,5), leading to the conclusion that the major isoform induced by DNA damage in HCT116 is TAp73α. This is consistent with previous work detailing the migration characteristics of various p73 isoforms working with antibodies targeting C-terminal epitopes (Marin et al. 1998). Further substantiating the specificity of our antiserum, HCT116 cells were transfected with control or TAp73 siRNA and were treated with VP16. In cells transfected with control siRNA, treatment with VP16 caused an increase in signal by immunofluorescent staining with p73NT, while those cells transfected with TAp73 siRNA showed no change in p73NT staining (Fig. 1D). Having thus characterized the specificity of the antibody, p73NT was used for subsequent experiments in this study.
Figure 1.
Validation of p73NT polyclonal antibody and endogenous p73 isoform characterization. (A) Extracts of HCT116 cells that were treated with VP16 (10 μM) or camptothecin (CPT; 300 nM) for 8 or 24 h were subjected to SDS-PAGE and immunoblotting with p73NT. (B, top panels) Extracts of H1299 cells transfected with expression vectors for Flag-tagged TAp73α and ΔNp73α were immunoblotted with p73NT (top) or with Flag (bottom) antibodies. (Bottom panels) Immunoblot of extracts of HCT116 cells that were transfected with TA-p73 specific (TAp73) or control (Luc) siRNA and then treated with camptothecin (300 nM) for 24 h. (C, lanes 1-3) H1299 cells were transfected with empty vector or vectors expressing HA-TAp73α or HA-TAp73β as indicated. (Lanes 4-6) HCT116 cells were exposed to VP16 (10 μM), camptothecin (300 nM), or vehicle (DMSO) for 24 h. Lysates were prepared and immunoblotted with p73NT. (D) p73NT immunoflourescent staining of HCT116 cells transfected with p73 or control siRNA and then treated with VP16 (10 μM) for 24 h.
Chk1 and Chk2 regulate p73 accumulation after DNA damage
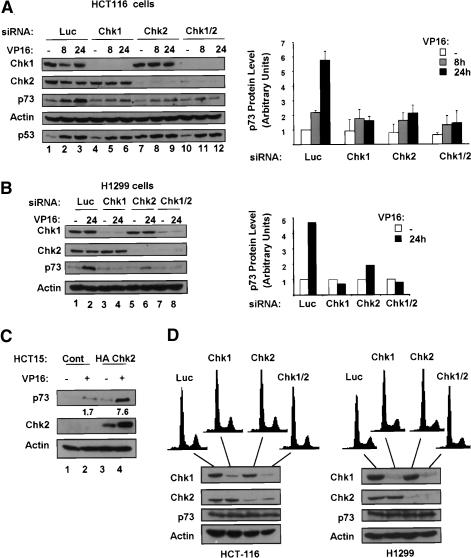
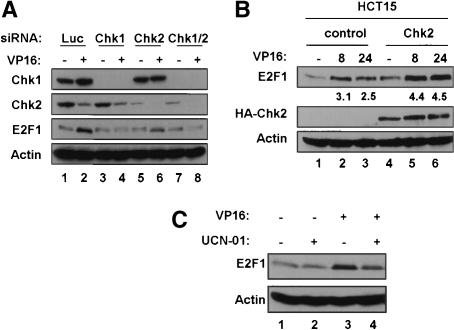
To determine whether p73 accumulation is affected by Chk1 or Chk2, HCT116 cells were transfected with siRNAs targeting luciferase, Chk1, Chk2, or Chk1 and Chk2 together and then were treated with VP16. The relevant siRNAs resulted in efficient and specific reduction in the levels of their respective Chk kinases (Fig. 2A, cf. lanes 1-3 and 4-6 for Chk1, and cf. lanes 1-3 and 7-9 for Chk2). Treatment with VP16 caused a time dependent increase in p73 protein in cells transfected with control siRNA; however, reduction of Chk1, Chk2, or both kinases together, while having little effect on basal p73 levels, caused a striking reduction in DNA-damage-induced p73 protein induction. By contrast, as we had previously demonstrated (Ahn et al. 2003) treatment with these siRNAs had little effect on p53 accumulation (Fig. 2A). In a second cell line, H1299, down-regulation of the Chk kinases also affected p73 accumulation, although the impact of Chk1 down-regulation was relatively more pronounced (Fig. 2B). These experiments indicate strongly that Chk1 and Chk2 mediate p73 protein induction following DNA damage.
Figure 2.
Chk1 and Chk2 control p73 induction in response to DNA damage. Immunoblotting was performed as in Figure 1A with extracts of HCT116 (A) and H1299 (B) cells that were transfected with control luciferase (Luc), Chk1, Chk2, or Chk1 and Chk2 (Chk1/2) siRNAs and treated with 10 μM VP16 for 8 or 24 h. Graphs on right show results of densitometric analysis of two experiments in A and the experiment shown in B. (C) Extracts of HCT15-control or HCT15-HAChk2 cells treated with 10 μM VP16 were immunoblotted with anti-HA and p73NT antibodies. Numbers indicate fold induction of p73 after DNA damage as determined by densitometry. (D) HCT1116 (left) or H1299 (right) cells were transfected with control (Luc), Chk1, Chk2, or Chk1 and Chk2 siRNAs as indicated. At 24 h after the second transfection cells were collected and processed for FACS analysis or immunoblotting.
siRNA experiments may be confounded by off-target effects (Jackson et al. 2003; Persengiev et al. 2004). Therefore a second system for manipulation of Chk2 function was examined. HCT15 cells carry the R145W Chk2 allele that encodes a greatly destabilized protein such that these cells express only very low levels of endogenous Chk2 (Lee et al. 2001). This cell line has been used in reconstitution experiments to study various functions of Chk2 (Falck et al. 2001; Lou et al. 2003). p73 induction was compared in HCT15 stable cell lines expressing either control empty vector or a vector expressing HA-Chk2 following treatment with VP16 (Fig. 2C). Consistent with the siRNA results, reexpression of Chk2 enhanced p73 accumulation in response to VP16. This observation further strengthens the observation that Chk2 function is linked to p73 induction by DNA damage. Also, expression of Chk2 augments p73 levels in the absence of DNA damage. To ensure that the increased levels of p73 induced by DNA damage are not simply due to higher resting levels, we confirmed that Chk2 affects the fold increase in p73 after VP16 as well. In the absence of Chk2, p73 levels were induced 1.7-fold by VP16 treatment while in its presence they were induced 7.6-fold.
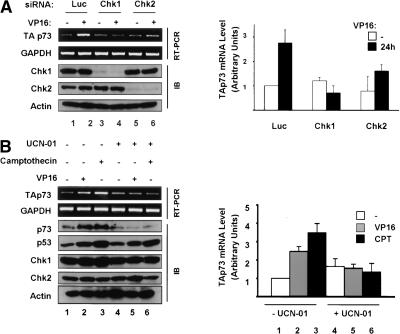
These results are further supported by experiments performed with the G2 checkpoint inhibitor UCN-01 that is thought to target Chk1 in vivo although other kinases including Chk2 may also be affected (Busby et al. 2000; Graves et al. 2000). Pretreatment of HCT116 cells with UCN-01 for 1.5 h prior to the addition of VP16 or CPT completely prevented the induction of p73 (see Fig. 4B, below). Taken in the context of the experiments with siRNA and HCT15 cells described above, results with this inhibitor support the likelihood that the Chk kinases play an important role in p73 accumulation following DNA damage.
Figure 4.
Chk1 and Chk2 control TAp73 RNA levels after DNA damage. (A) H1299 cells were transfected with control (Luc), Chk1, or Chk2 siRNAs and treated with 10 μM VP16 for 24 h. Parallel cultures were collected for either immunoblotting (IB panels) or RNA extraction using primers specific for TAp73 mRNA or control for GAPDH (RT-PCR panels). (B) HCT116 cells were pretreated for 1.5 h with 150 nM UCN-01 before addition of 10 μM VP16 or 300 nM Camptothecin for 15 h. Cells were then processed for RT-PCR analysis of TAp73 and GAPDH mRNAs or for immunoblotting (IB) against the indicated proteins as in A. Graphs on right represent analyses of three experiments.
Since Chk1 and Chk2 regulate multiple cell cycle transitions within the stress response, it was important to exclude the possibility that Chk1 and Chk2 knockdown might result in altered cell cycle profiles which would confound our observations with respect to p73. FACS analysis of cells exposed to control or Chk kinase siRNAs showed that Chk silencing in both HCT116 and H1299 cells did not change cell cycle distribution (Fig. 2D). This indicates that the dependence of p73 induction after DNA damage upon Chk1 and Chk2 is not secondary to changes in cell cycle distribution induced by reduced levels of Chk kinases.
TAp73 mRNA accumulates after DNA damage
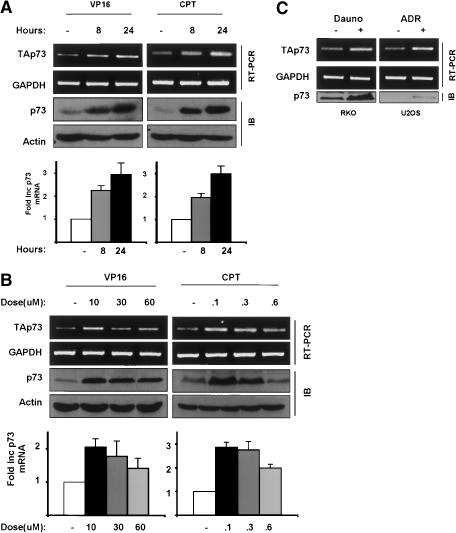
The manner by which p73 is induced by DNA damage is yet an unsettled question since several groups have published conflicting results on the existence of p73 mRNA regulation by genotoxicity (Gong et al. 1999; Kartasheva et al. 2002; Pediconi et al. 2003). Thus, before examining the mechanistic basis for p73 regulation by Chk1 and Chk2, the level of control at which the DNA-damage checkpoint impinges upon p73 was examined. Asynchronously growing HCT116 cells were treated with VP16 or CPT at the same doses employed in Figure 1 for 8 or 24 h and parallel cultures were prepared for protein or RNA extraction. Using a previously described strategy, semiquantitative RT-PCR was performed on cDNA produced via a p73 exon 4 specific primer to ensure efficient reverse transcription of extreme 5′ templates which may not be consistently well represented in oligo-dT derived cDNA (Kartasheva et al. 2002). VP16 and CPT induced a highly reproducible two- to threefold enrichment of TAp73 mRNA in a time dependent manner over 24 h (Fig. 3A). In fact a reverse dose dependency was observed for VP16 and CPT whereby lower doses of drug induced greater levels of TAp73 mRNA and protein (Fig. 3B). Similar data were obtained with H1299 cells (data not shown). This dose dependence of p73 protein accumulation was observed previously with a number of chemotherapeutic agents (Irwin et al. 2003). Examination of other cell lines and DNA damaging agents revealed a similar two- to threefold mRNA induction in RKO cells treated with daunorubicin (Fig. 3C, left) or U2OS cells exposed to adriamycin (Fig. 3C, right). This extent of TAp73 mRNA up-regulation is consistent with other published measurements of p73's transcriptional activation by chemotherapeutic agents both by quantitative RT-PCR and Northern blotting (Marabese et al. 2003; Pediconi et al. 2003). Thus multiple agents that cause DNA damage result in increased levels of TAp73 mRNA.
Figure 3.
p73 mRNA levels increase after DNA damage. (A) HCT116 cells were treated with 10 μM VP16 or 300 nM camptothecin for 8 or 24 h and processed for semiquantitative RT-PCR to measure TAp73 and GAPDH RNA levels (top two panels) and immunoblotting (IB) to detect p73 and actin (bottom two panels). (B) HCT116 cells were treated with the indicated doses of VP16 and camptothecin (CPT) and processed as in A for p73 RNA and protein with indicated controls. (C) RKO and U2OS cells were treated with 0.22 μM daunorubicin (dauno) for 10 h or 2 μM adriamycin (Adr) for 24 h, respectively, and semiquantative RT-PCR and Western blotting for p73 was performed as in A. Bar graphs represent an analysis of two experiments.
Chk1 and Chk2 control activation of TAp73 transcription after stress
Having demonstrated that Chk1 and Chk2 are vital to p73 protein induction after genotoxic stress and that TAp73 mRNA is induced after DNA damage, we then determined whether TAp73 mRNA accumulation is also dependent on Chk1 and Chk2. H1299 cells were transfected with Chk1 and Chk2 siRNAs as before and treated for 24 h with VP16 (Fig. 4A). A strikingly similar pattern to that for p73 protein data was observed. In the response to VP16, Chk1 or Chk2 down-regulation was sufficient to markedly reduce accumulation of TAp73 mRNA (Fig. 4A). To extend this observation, the G2 checkpoint inhibitor UCN-01 described above was employed. HCT116 cells were pretreated with UCN-01 for 1.5 h prior to overnight exposure to VP16 or CPT (Fig. 4B). As expected, in cells not pretreated with UCN-01 TAp73 was up-regulated at both the protein and mRNA levels (Fig. 4B, lanes 1-3). However, in the UCN-01 administered cells p73 protein and mRNA was not induced by VP16 or CPT (Fig. 4B, lanes 4-6). UCN-01 treatment was not associated with change in Chk1 or Chk2 protein levels. Interestingly as well, UCN-01 had virtually no effect on p53 induction under these conditions. Thus, Chk1 and Chk2 play roles in p73 mRNA accumulation after DNA damage.
E2F1 is stabilized by DNA damage in a Chk1- and Chk2-dependent manner
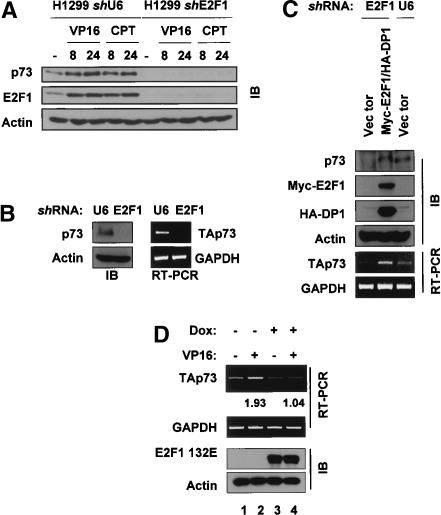
Recent data showing that E2F1 is responsible for TAp73 promoter activation after DNA damage (Pediconi et al. 2003) and that Chk2 phosphorylates E2F1 (Stevens et al. 2003) prompted us to investigate a signaling pathway whereby E2F1 mediates Chk1 and Chk2 modulation of p73 gene transcription. To examine the role of E2F1 in p73 induction by DNA damage, H1299 cell lines stably transfected with an empty control vector (shU6, as the vector uses the U6 RNA promoter) or an anti-E2F1 small hairpin RNA (shE2F1) were obtained (Ma et al. 2003). The extent of p73 protein induction after VP16 or camptothecin was determined in each cell line. Strikingly, while p73 was induced after either treatment in control cells, the E2F1 shRNA-expressing cells, which have very low levels of E2F1, did not induce p73 after stress and had undetectable basal levels of p73 as well (Fig. 5A). To confirm that this was due to down-regulation of p73 mRNA, TAp73 levels in control U6 and E2F1 shRNA cells were compared, and we determined that the E2F1 hairpin expressing cells had significantly reduced TAp73 mRNA (Fig. 5B). To verify that this was not due to an irrelevant effect on cells (for example, fortuitous plasmid integration within the p73 locus in the shE2F1 stable line), E2F1 and DP1 were cotransfected into the shE2F1 cells (Fig. 5C). Indeed, restoration of E2F1 returned p73 protein and RNA levels to those in control cells, indicating that E2F1 controls p73 transcription even in the absence of DNA damage. In addition, we determined that shU6 and shE2F1 H1299 cells have indistinguishable cell cycle profiles (data not shown).
Figure 5.
E2F1 regulates basal and DNA-damage-induced levels of p73 mRNA and protein. (A) H1299 cells stably transfected with either empty vector (U6) or a vector expressing an anti-E2F1 shRNA (E2F1) were treated with VP16 or camptothecin (CPT) and immunoblotting was performed for p73, E2F1, and actin. (B) Asynchronously growing H1299-shU6 and H1299-shE2F1 cells were collected for Western blotting (left) or RT-PCR for p73 (right). (C) Empty vector or constructs expressing 6myc-E2F1 and HA-DP1 were cotransfected into H1299-shE2F1 cells (E2F1 lanes) and 24 h later extracts were prepared for immunoblotting with the indicated antibodies or RT-PCR for p73 and GADPH. Protein lysates or RNA from H1299-shU6 cells (lane labeled U6) were included for comparison of p73 levels in H1299 cells. (D) Saos2 cells expressing E2F1 132E under the control of a doxycyline (dox)-inducible promoter were exposed to 2 μg/uL dox for 24 h before the addition of 10 μM VP-16 for 24 h. Parallel cultures were collected for RT-PCR and Western blotting as above. Numbers indicate fold inducion of p73 after DNA damage as determined by densitometry.
To determine whether TAp73 mRNA induction after DNA damage is also regulated by E2F1, we employed Saos2 cells expressing an inducible mutant form of E2F1 (E2F1 132E) that is defective in binding to DNA (Johnson et al. 1993). When E2F1 132E was expressed in these cells TAp73 mRNA levels were not increased by VP16 treatment (Fig. 5D) whereas in the absence of mutant E2F1 TAp73 levels were increased approximately twofold, consistent with our previous observations shown in Figure 3. Here too, expression of E2F1 32E caused no significant changes in cell cycle distribution (data not shown).
In a manner analogous to p53, E2F1 undergoes protein stabilization in response to DNA damage mediated by the ATM kinase (Blattner et al. 1999; Lin et al. 2001). In addition a consensus Chk kinase phosphorylation sequence in E2F1 occurs at Ser 364, and mutation of this site was reported to prevent E2F1 stabilization by VP16 (Stevens et al. 2003). In view of these data and the critical role of E2F1 in control of p73 transcription, the impact of Chk1 and Chk2 inhibition on E2F1 induction by DNA damage was investigated. We first determined the effect on E2F1 stabilization by expression of Chk1 and Chk2 siRNA in H1299 cells (Fig. 6A). Indeed, Chk kinase dependence was observed for E2F1 induction as we had demonstrated for p73 protein and mRNA accumulation. While reduction of Chk1 completely prevented E2F1 induction, Chk2 down-regulation had a somewhat less drastic effect, with E2F1 being only slightly increased after VP16 treatment and down-regulation of Chk1 and Chk2 together prevented any increase in E2F1. Note that in this experiment, as observed previously, there was a reduction in detectable Chk2 protein after cells were treated with VP16, which may reflect a loss of reactivity with the anti-Chk2 antibody (Matsuoka et al. 1998; Ahn et al. 2003).
Figure 6.
Chk1 and Chk2 mediate DNA-damage-induced E2F1 stabilization. (A) H1299 cells were transfected with control (Luc), Chk1, or Chk2 siRNAs and treated (+) or not (-) with 10 μM VP16 for 24 h. Cell extracts were prepared for immunoblotting with the indicated antibodies. (B) HCT15-control or HCT15-HAChk2 cells were treated with 10 μM VP16 for 8 or 24 h and immunoblotting of cell extracts was performed for E2F1 and HA-Chk2. Numbers indicate fold induction of p73 after DNA damage as determined by densitometry. (C) HCT116 cells were pretreated with 150 nM UCN-01 for 1 h before the addition of 10 μM VP16 or 300 nM CPT for 15 h and immunoblotting of extracts was performed for E2F1 and actin.
This dependence was also tested in two other systems. First, reconstitution of Chk2 in HCT15 cells markedly enhanced E2F1 stabilization by VP16 (Fig. 6B). As seen with p73 in these cells, basal levels of E2F1 were also somewhat enhanced by Chk2 in the absence of DNA damage. Fold induction was therefore calculated, and we confirmed that Chk2 not only affects basal E2F1 levels but also the extent of induction after DNA damage. Second, incubation of HCT116 cells with UCN-01 prior to the addition of VP16 also prevented E2F1 stabilization (Fig. 6C).
Chk1 and Chk2 regulate TAp73 promoter activation by E2F1
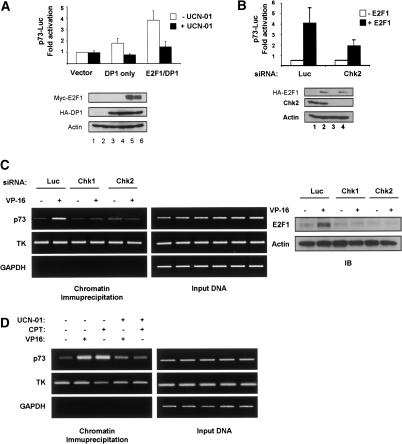
Given that induction of E2F1 by DNA damage is dependent on Chk1 and Chk2, we next examined the ability of the Chk kinases to regulate E2F1 transcriptional activity specifically toward the p73 promoter. First, reporter assays were performed using H1299 cells transfected with E2F1 along with a construct containing the luciferase gene under the control of the TAp73 promoter in the presence or absence of UCN-01 (Fig. 7A, upper panel). Cotransfection of E2F1 and the promoter construct induced luciferase activity approximately fourfold. However, when cotransfected in the presence of UCN-01, the ability of E2F1 to induce luciferase activity via the p73 promoter was markedly reduced, suggesting that Chk1 may regulate E2F1 transcriptional activation of the p73 promoter. Although luciferase assays were normalized to an internal control, immunoblotting of a representative experiment was performed to confirm comparable expression of E2F1 and DP1 in the treated and untreated samples (Fig. 7A, lower panel).
Figure 7.
Chk1 and Chk2 regulate TAp73 promoter activation by E2F1. (A, upper panel) H1299 cells were transfected in the presence or absence of 150 nM UCN-01 with constructs expressing Myc-E2F1 (100 ng), HA-DP1 (100 ng), p73-Luc (500 ng), and a pCMV-RL construct (10 ng) for 24 h. (Lower panels) Luciferase assays were performed by standard methods and data were normalized to control Renilla luciferase signal. A representative immunoblot of MycE2F1, HA-DP1, and actin. (B, upper panel) shE2F1 H1299 cells were transfected with the indicated siRNA as in Figure 1, then transfected with 5 ng HA-E2F1, 1 μg p73-Luc, and 10 ng pCMV-RL constructs for 24 h. Luciferase assays were performed by standard methods and data were normalized to control Renilla luciferase signal. (Lower panels) A representative immunoblot of HA-E2F1, Chk2, and actin. (C, left panels) H1299 cells were transfected as in Figure 1, and treated with 10 μM VP16 for 24 h. Chromatin immunoprecipitation was performed using an anti-E2F1 antibody (C20). Precipitated DNA was amplified with primer pairs for each promoter as indicated for p73 and Thymidine Kinase (TK). Nonspecific amplification was controlled for by performing PCR of a region within the GAPDH promoter. (Middle panels) Input DNA PCR products (1% of total). (Right panels) Immunoblotting of cross-linked protein extracts for E2F1 and Actin. (D, left panels) H1299 cells were exposed to 150 nM UCN-01 for 1 h prior to the addition of 10 μM VP16 or 300 nM CPT for 24 h. Chromatin immunoprecipitation was performed as in C. (Right panels). Input DNA samples as in C.
UCN-01 is thought to be a more potent inhibitor of Chk1 than Chk2. Therefore to determine whether Chk2 can similarly regulate E2F1 a second reporter assay was carried out in which H1299 shE2F1 cells were transfected with control luciferase or Chk2 specific siRNA as in Figure 1 along with transfection of E2F1 and the p73-promoter luciferase constructs. Activation of the p73 reporter construct by E2F1 was significantly reduced upon Chk2 knockdown (Fig. 7B, upper panel). Immunoblotting of proteins in extracts of similarly transfected cells is shown (Fig. 7B, lower panel). These data suggest that Chk1 and Chk2 can regulate TAp73 promoter activation by E2F1.
To gain more physiologically relevant information by examining the transcriptional activity of endogenous E2F1 after DNA damage in vivo we performed E2F1 chromatin immunoprecipitation (ChIP) assays. H1299 cells were transfected as above with control, Chk1, or Chk2 siRNAs. In the control siRNA transfected cells, treatment with VP16 markedly enhanced TAp73 promoter association by E2F1 in ChIP assay. In contrast, Chk1 or Chk2 knockdown essentially ablated the increase in TAp73 promoter signal following VP16 treatment (Fig. 7C, left panels). Consistent with a previous report the promoter of another E2F1 target gene, thymidine kinase (TK), did not show any DNA-damage-associated changes (Pediconi et al. 2003). As a negative control the GAPDH promoter was not detected. Confirming our previous results, Chk1 and Chk2 siRNAs prevented the induction of E2F1 protein levels by DNA damage (Fig. 7C, right panel). These data strongly suggest that Chk kinase activity is required for increased TAp73 promoter association following genotoxic stress. Moreover, the amount of TAp73 promoter DNA recovered by ChIP assay closely tracks E2F1 protein levels. Taken together, our results are consistent with a crucial role for Chk1 and Chk2 in mediating E2F1 activation after DNA damage.
We next examined the effect of UCN-01 on E2F1 association with the p73 promoter in H1299 cells exposed to VP16 or CPT. In the cells not exposed to UCN-01, these genotoxins induced E2F1 accumulation at the TAp73 promoter (Fig. 7D), and UCN-01 reduced this increase in E2F1 ChIP signal at the p73 promoter. Again, no change was seen with another E2F1 target gene, TK, and E2F1 association with the GAPDH promoter region could not be detected. As we had seen before (Fig. 4B) UCN01 treatment did not affect Chk protein levels (data not shown). While we cannot deny the possibility that the rabbit polyclonal antibody against E2F1 employed in the ChIP assay may weakly cross-react with other E2F family members, E2F1 is the only E2F family member induced by DNA damage (Lin et al. 2001) and E2F1 activates p73 transcription much more strongly than the other activator E2Fs (Irwin et al. 2000). Taken together, our data strongly implicate E2F1 as the critical Chk kinase-regulated transcription factor that activates the TAp73 promoter in the DNA-damage response.
Chk1, Chk2, E2F1, and p73 impact p53-independent apoptosis
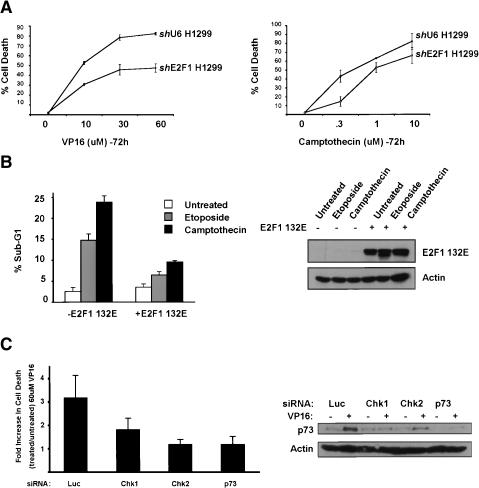
Returning to the fact that Chk2 ablation profoundly affects apoptosis in mice, we considered the possibility that our results may tie p73 through E2F to this observation. Therefore we examined whether down-regulation of E2F1, the Chk kinases, or p73 impede DNA-damage-induced apoptosis. For E2F1, induction of cell death by various doses of VP16 or camptothecin in the shU6 and shE2F1 H1299 cells was compared (Fig. 8A). Consistent with a role for E2F1 (and by extension p73) in p53-independent apoptosis, shE2F1 cells showed decreased VP16-induced cell death across a range of drug concentrations (Fig. 8A, left panel). Interestingly, shE2F1 cells were markedly more resistant to camptothecin-generated death at lower doses (0.3 μM) than at higher doses (1 μM and 10 μM; Fig. 8A, right panel). This closely parallels the results we obtained comparing induction of p73 at these doses of camptothecin (see Fig. 3B, right panels).
Figure 8.
Chk1, Chk2, E2F1, and p73 play a role in p53-independent apoptosis of H1299 cells. (A) Asynchronously growing H1299-shU6 and H1299-shE2F1 cells were treated with the indicated doses of VP16 or camptothecin for 72 h and scored for viability by trypan blue exclusion assay. Data represent three independent experiments. (B, left panel) Saos2-132E cells were exposed or not to 2 ng/mL doxycyline for 24 h before the addition of 10 μM VP16 or 300 nM CPT for 72 h. Cells were then collected and processed for FACS analysis. (Right panels) Extracts were blotted for E2F1 and actin to assure mutant E2F1 induction. (C, left panel) H1299 cells were transfected with Luc, Chk1, Chk2, or p73 siRNAs as in Figure 2 and then treated with 60 μM VP16 for 48 h followed by trypan blue exclusion assay. Data represent five independent experiments. (Right panels) Western blotting of extracts from H1299 cells treated as on the left.
The role of E2F1 in p53-independent apoptosis was also investigated using the dominant negative E2F1 32E mutant. Consistent with the ability of this mutant to block the DNA-damage-associated transcriptional activation of TAp73, E2F1132E significantly reduced apoptosis induced by either VP16 or CPT (Fig. 8B, left panel) again suggesting that E2F1 plays an important role in p53-independent apoptosis.
To study the role of the other factors in p53-independent apoptosis, control, Chk1, Chk2, and p73 siRNAs were transfected into H1299 cells which were subsequently untreated or treated with VP16 for 48 h (Fig. 8C). In control transfected cells, VP16 engendered a threefold increase in cell death. Importantly, in cells transfected with either Chk2 or p73 siRNAs VP16 failed to induce an increase in cell death compared with transfected, untreated cells. Chk1 down-regulation also decreased the fold increase in apoptosis but did not fully abolish it. This may be due to the fact that Chk1 as compared to Chk2 has crucial functions in maintaining cellular viability, so that p73-specific functions are partially offset. Our observations are consistent with an apoptotic defect induced by loss of physiological levels of Chk1, Chk2, and p73, and, taken together, are also consistent with each acting in a common pathway.
Discussion
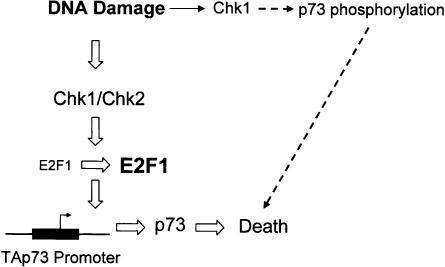
p53 is mutated in one half of human cancers and inactivated by indirect mechanisms in a large percentage of the remainder. Logically then, pathways mediating cell death in the absence of p53 would be of interest for therapeutic intervention. Here, the outline of such a pathway begins to take shape (Fig. 9). First, Chk1 and Chk2 are required for induction of p73 following DNA damage. This was demonstrated by siRNA knockdown of both kinases, reexpression of Chk2 in a mutant background, and inhibition of checkpoint activity via a chemical inhibitor, UCN-01 (Fig. 2). Conclusive evidence on the mode(s) of p73 induction by DNA damage has been lacking. We showed that both transcriptional induction (Fig. 3) as well as protein stabilization (data not shown) of p73 occur coincidently and that stress activation of the TAp73 promoter is dependent upon Chk1 and Chk2 (Fig. 4). We also identified E2F1 as a critical interdicting transcription factor that mediates Chk kinase-dependent transcriptional regulation of p73. To explore the E2F1-p73 connection, we found that an E2F1-depleted cell line lacks p73 induction in response to DNA damage and has undetectable basal TAp73 expression at both the protein and mRNA levels. We further showed that both Chk1 and Chk2 regulate E2F1 induction following stress in a number of systems. UCN-01 as well as Chk2 siRNA can hinder the ability of E2F1 to activate the p73 promoter by luciferase assay. Importantly, Chk1 or Chk2 knockdown significantly impacts association with the TAp73 promoter in E2F1 ChIP assays after DNA damage. Finally, we showed that all the members of this presumptive signaling pathway are shown to function in cell death induced by VP16 (Fig. 7). Our data therefore are consistent with an E2F1-mediated pathway linking Chk kinases to TAp73 and apoptosis (Fig. 8).
Figure 9.
A schematic diagram for Chk kinase activation of TAp73 after DNA damage.
Previously, E2F1 null thymocytes were reported to be resistant to VP16 (Lin et al. 2001) and the H1299-shE2F1 cell lines are also refractory to death by flavopiridol (Ma et al. 2003). Moreover, E2F1 induction has been correlated with sensitivity to p53 independent apoptosis (Meng et al. 1999). Integrating all of these observations produces strong evidence that Chk1 and Chk2 control a circuit activating the pro-apoptotic transcription factor p73 through stabilization and activation of E2F1, an activator of the TAp73 promoter.
Many interesting questions surface from these experiments. Concerning Chk1 and Chk2 it will be of interest to determine which are the specific functions of Chk1 versus Chk2. The published data and those presented here show that they very likely share many roles. Our data suggest that under certain circumstances, a full complement of both Chk1 and Chk2 is essential, implying not only functional commonality but also synergism. Note, however, that we observed that when cells are treated with camptothecin, which has a mechanism of action distinct from VP-16, Chk1 depletion much more strongly impacts p73 accumulation than Chk2 (data not shown). This is in contrast to VP16, where we saw a requirement for both kinases. These differences might be explained in a number of ways. Certain DNA-damaging agents may activate Chk1 or Chk2 individually. Alternatively, different modes of checkpoint activation could engage different spectra of Chk1 and Chk2 functions so that their ability to compensate the other's loss is dependent on the genotoxin. A more banal explanation is that the strength of Chk1 or Chk2 activation varies by DNA damage so that very strong activation cannot be suppressed by RNAi. Regardless of the explanation our studies point to intriguing similarities as well as differences in how the DNA-damage checkpoint utilizes Chk1 and Chk2.
For E2F1, it will be of interest of determine how phosphorylation at Ser 364 regulates both protein stability and transcriptional activity. As E2F1's F-box adaptor Skp2 binds within the first 41 amino acids of E2F1 it is not immediately clear why a distal C-terminal phosphorylation event should regulate stability (Marti et al. 1999). Interestingly, Hofmann et al. mapped a destabilizing element in E2F1 encompassing amino acids 363-378; perhaps phosphorylation within this domain mitigates its destabilization function (Hofmann et al. 1996). Consistent with our data the region surrounding Ser 364 seems to be vital for E2F1-induced apoptosis and the activation of p73, indicating that this region also affects transcriptional activity (Hallstrom and Nevins 2003; Stevens et al. 2003). Identification of other DNA-damage-induced E2F target genes will be important in understanding the nature of p53-independent apoptosis.
With respect to p73, our data indicate that p73 is regulated at both the transcriptional and posttranscriptional (data not shown) level following DNA damage and that the transcriptional component is controlled through Chk1- and Chk2-mediated E2F1 activation and stabilization. Intriguingly, Chk1 but not Chk2 was recently shown to phosphorylate p73 at Ser 47 (Gonzalez et al. 2003). A role for this modification in DNA-damage-mediated induction of p73 expression was not investigated in that report. Future studies will determine if direct regulation of p73 protein stability by the Chk kinases plays a role in the DNA-damage response (Fig. 9). Since these interesting questions await future investigation, it is also not possible at this time to determine the relative contribution of transcriptional versus posttranscriptional regulation to the ultimate level of p73 protein achieved following checkpoint activation. What is clear from our data however is that Chk1 and Chk2 play a critical role in p73 protein induction and that modulation of E2F1 makes an important contribution.
Of additional interest is how p73 mediates p53-independent apoptosis and what are the target genes involved. Moreover, are these targets shared with p53 and how does p73-dependent apoptosis differ from p53-dependent cell death? More simply, are there both qualitative and quantitative differences in how these highly related transcription factors induce cell death?
Previoulsy published evidence suggests that both p73 and p63 are required for p53-dependent apoptosis (Flores et al. 2002). Therefore, it will be of interest to determine whether Chk1 and Chk2 also mediate p63 induction following DNA damage. Intriguingly, induction of pro-apoptotic p53 target genes is compromised in p63-/-;p73-/- MEFs—a phenotype shared with cells lacking Chk2 (Hirao et al. 2000, 2002; Flores et al. 2002; Takai et al. 2002). Perhaps the p53 functional defects observed in Chk2 null mice are a consequence of faulty p63-p73 regulation.
Initially, evidence connected Chk1 and Chk2 with Ser 20-phosphorylation-mediated stabilization of p53 thereby leading to activation of cell cycle arrest and apoptosis. Subsequent papers have supported or challenged this hypothesis. The data presented here continue to dispute the generality of this pathway but also interestingly shows that E2F1 closely adheres to such a scheme. Chk1 and Chk2 are crucial for E2F1 stabilization as well as E2F1 target gene induction. Our data implicate the Chk-E2F1-p73 pathway as central to p53-independent apoptosis, bringing together new modulators of chemosensitivity in human cells.
Materials and methods
Cell lines, drugs, and plasmids
HCT15 cell derivatives expressing either empty pcDNA3 (HCT15-control) or pcDNA3-HAChk2 (HCT15-HAChk2) (Lou et al. 2003) were kindly provided by J. Chen (Mayo Clinic, Rochester, MN). These cells were cultured in RPMI 1640 medium with 10% fetal bovine serum (FBS) and 400 μg/mL G418. H1299 shU6 and H1299 shE2F1 cells (Ma et al. 2003) were generously provided by W.D. Cress (H. Lee Moffitt Cancer Center and Research Institute, Tampa, FL) and grown in Dulbecco's modified Eagle's medium (DMEM) with 10% FBS and 400 μg/mL G418. Saos2-132E cells (Phillips et al. 1999) were a gift from K. Vousden (Beatson Institute for Cancer Research, Glasgow, UK) and were grown in DMEM with 10% FBS containing 400 μg/mL G418 and 50 μg/mL Hygromycin. HCT116 and H1299 cells were obtained from ATCC and maintained in DMEM with 10% FBS. UCN-01 was provided by Kyowa Pharmaceuticals. VP16 and camptothecin (Oncogene Research Products) were dissolved in DMSO. The HA-DP1 plasmid was provided by M. Classon (Massachusetts General Hospital Cancer Center, Charlestown, MA). Plasmids expressing Flag-TAp73, Flag-ΔNp73, and E2F1 were the generous gift of T. Tanaka (Columbia University, New York, NY). The p73 promoter reporter construct (Seelan et al. 2002) was kindly provided by W. Liu (Mayo Clinic, Rochester, MN). HA-E2F1 plasmid was provided by J. Nevins (Duke University Medical Center, Durham, NC).
Western blot analysis
For protein analysis, cells were washed once in cold PBS and then scraped in TEGN buffer (10 mM Tris at pH 8, 1 mM EDTA, 10% glycerol, 0.5% NP40, 400 mM NaCl, 1 mM DTT, 0.5 mM phenylmethylsulfonylfluoride, and protease inhibitor mixture containing 1 M Benzamidine, 3 mg/mL Leupeptin, 100 mg/mL Bacitracin, and 1 mg/mL α2 macroglobulin) and incubated on ice for 15 min. Lysates were then cleared by centrifugation at 13,000 rpm for 10 min. Total protein concentration was determined via the Bio-Rad protein assay (Bio-Rad Laboratories) and equal amounts of protein were separated on 10% SDS polyacrylamide gels and transferred to nitrocellulose membranes (Protran, Schleicher and Schuell). Membranes were then blocked for 1 h in a PBS solution containing 5% nonfat milk powder and 0.1% Tween-20 and then probed with primary antibody overnight in 1% milk, 0.1% Tween-20 PBS. After washing, membranes were incubated for 1 h with horseradish peroxidase-linked secondary antibody (Sigma) in 1% milk, 0.1% Tween-20 PBS. Finally, after three 5-min washes in 0.1% PBS-Tween 20, proteins were visualized by enhanced chemiluminesence (Amerhsam Biosciences).
RNA interference, transfection, and antibodies
siRNA duplexes were synthesized by Qiagen Inc. Sequences for Luc (Elbashir et al. 2001), Chk1 and Chk2 (Ahn et al. 2003), and p73 (Irwin et al. 2003) have been described previously. For RNAi experiments, HCT116 or H1299 cells were plated at 30% confluence and transfected twice 24 h apart with 2 μg of siRNA duplex using Lipofectamine 2000 (Invitrogen). The cells were then left untreated or exposed to 10 μM VP16 or 300 nM camptothecin. At 72 h after the first transfection, cells were lysed in TEGN buffer or pelleted and frozen in liquid N2 for RNA extraction. For plasmid transfection, H1299 cells were plated at greater than 90% confluencey and transfected with up to 5 μg of DNA with Lipofectamine 2000 at a DNA:lipid ratio of 1:1.2. For the experiment with H1299 shE2F1 cells, cultures were transfected with 1.75 μg of 6Myc-E2F1 and 1.75 μg HA-DP1 for 24 h. Cells were then washed and incubated in fresh medium for an additional 24 h.
Anti-Chk1 (G4), anti-E2F1 (C20 or KH-95), anti-Myc (9E10) (Santa Cruz Biotechnology), anti-Chk2 (ProSci Inc.), anti-Actin (rabbit polyclonal) and anti-Flag M2 antibodies (Sigma) and anti-HA (HA.11 Covance) antisera were used where indicated. p53 was detected with a mixture of 1801 and DO-1 monoclonal antibodies.
The p73 polyclonal antibody was raised commercially (Covance) against a peptide DSTYFDLPQSSRGNNE (SynPep) and crude rabbit serum was purified by p73β affinity chromatography as follows: Sf9 cells were infected with a baculovirus expressing HA-p73β as described (Gaiddon et al. 2001). Cell extracts were prepared and HA-tagged p73 protein immunopurified as previously described (Jayaraman et al. 1997). CnBr activated Sepharose 4B beads (Pharmacia) were incubated in 1 mM HCl for 15 min followed by three washes in 1 mM HCl. Beads and p73β protein were then mixed in coupling buffer (4 mM NaHCO3, 20 mM NaCl at pH 8.3) and rocked overnight at 4°C. Remaining active groups were blocked with 0.2 M glycine in coupling buffer (pH 8) for 2 h at room temperature. Beads were then washed with two cycles of 1 volume of coupling buffer and 1 volume acetate buffer (0.1 M Na acetate, 0.5 M NaCl at pH 4.0) followed by a final coupling buffer wash. They were then transferred to a column and washed with 10 column volumes each of 10 mM Tris (pH 7.5), 100 mM glycine (pH 2.5), 10 mM Tris (pH 8.8) followed by a final single volume wash with 10 mM Tris (pH 7.5). Crude serum (1 mL) was then diluted 1:10 in 10 mM Tris (pH 7.5), centrifuged for 2 min (4000 rpm) and the supernatant was then passed over the HAp73β protein column three times. The beads were then washed in 20 volumes of 10 mM Tris (pH 7.5) followed by 20 volumes of 10 mM Tris (pH 7.5) with 500 mM NaCl. Antibody was then eluted with 10 volumes of 100 mM glycine (pH 2.5) into 1 volume of 1 M Tris (pH 9.0). Fractions were then dialyzed overnight at 4°C in PBS (pH 7.5).
Densitometric analyses of immunoblots and ethidium bromide gel images were quantified using Image J (NIH). Calculated intensities normalized to actin (immunoblot) or GAPDH (RT-PCR) were used to generate induction in arbitrary units.
RT-PCR
RNA was extracted from cell pellets using RNeasy Mini Kit (Qiagen) and quantitated by ultraviolet spectrophometry. Total RNA (4 μg) was reverse transcribed using the Superscript 3 First Strand Synthesis System for RT-PCR (Invitrogen) using specific primers for p73 and GAPDH (Kartasheva et al. 2002). PCR was then performed using the Expand High Fidelity PCR system (Roche Biochemicals). Conditions for linear amplification were established through template and cycle curves. The cycling conditions for p73 and GAPDH were as follows: a denaturation step at 96°C for 3 min followed by 27 cycles (for TAp73) or 20 cycles (for GAPDH) at 96°C for 30 sec, 55°C for 30 sec and 70°C for 50 sec, as well as a final extension of 72°C for 7 min. PCR products (40% of reaction mixture) were then separated on 2.5% agarose gels and bands were visualized with ethidium bromide.
Immunoflouresence staining
Cells were fixed with 4% paraformaldehyde in PBS for 20 min at room temperature. After fixation cells were permeabilized in PBS containing 0.2% Triton X-100 for 5 min. Cells were then blocked in PBS containing 5% bovine serum albumin for 30 min at room temperature before incubation with anti-p73 antibody (1:100) for 2 h, followed by 1 h incubation with indicated secondary antibody. Nuclei were visualized by DAPI (4′-6′-diamidino-2-phenylindole; Sigma) staining.
Luciferase assay
H1299 cells were seeded in 12-well plates at >90% confluency. Each well was then transfected with constructs expressing E2F1 or DP1 (100 ng each), and a p73-Luc reporter plasmid (500 ng) along with a Renilla construct pRL-CMV (10 ng). Immediately after transfection UCN-01 was added to the culture media at a final concentration of 150 nM. At 24 h after transfection a luciferase assay was performed according to the manufacturer's instructions using the Dual Luciferase Assay kit (Promega). Data were normalized to a Renilla reporter signal.
Cell death assay and FACS analysis
Equivalent numbers of H1299-shU6 and H1299-shE2F1 cells were plated in 6-well plates. At 24 h after plating VP16 or camptothecin was added at the indicated concentrations. Seventy-two hours later cells (floating and attached) were collected and centrifuged (1500 rpm for 10 min). Cells were then diluted in DMEM without FBS and mixed 1:5 with 0.4% Trypan Blue Stain (Sigma). Cells were incubated for 5 min at room temperature and then at least 300 cells from each well were counted using a standard hemocytometer. For RNAi experiments, cells were transfected as above except that 24 h after the second transfection cells were treated with VP16 (60 μM) and collected and stained 48 h after the second transfection. For calculations, percentage of cell death in each transfection not exposed to drug was set to one and fold increase was then calculated as the ratio of death in untreated to treated for each siRNA used.
For FACS analysis, cell were collected and pelleted at 1500 rpm for 10 min and the resuspended in 300 μL of cold PBS followed by fixation with ice-cold methanol. At least 24 hours after fixation, cell were rehydrated in PBS for at least 30 min and then repelleted as above. Cell were then reconstituted in 1 mL of PBS containing 60 μg/mL propidium iodide and 50 μg/mL RNAse A. Cell cycle profiles were obtained using a Becton Dickinson FACScalibur flow cytometer and CellQuest software.
Chromatin immunoprecipitation (ChIP) assay
For ChIP experiments, H1299 cells were seeded into 6-well plates at 30% confluence and transfected twice prior to VP16 treatment as described in RNA Interference, or were treated with UCN-01 (150 nM) prior to VP16 or CPT. At 24 hours after DNA damage treatment cells were washed once in 1× phosphate-buffered saline (PBS) and then treated with 1% formaldehyde in PBS at 37°C for 10 min followed by the addition of glycine to a final concentration of 0.125 M for 5 min. Cells were then scraped on ice and centrifuged at 1500 rpm for 5 min at 4°C. After washing with PBS cell pellets were resuspended in 1 mL of cold lysis buffer (10 mM HEPES at pH 7.5, 1 mM EDTA at pH 8.0, 400 mM NaCl, 10% glycerol, 0.5% NP-40, 0.5 mM phenylmethylsulfonyl fluoride, and protease inhibitors [1 mM benzamidine, 3 mg/mL leupeptin, 0.1 mg/mL bacitracin, 1 mg/mL macroglobulin]) followed by centrifugation at 11,500 rpm in a microfuge for 5 min at 4°C to remove protein not cross-linked to chromatin. The resulting pellets were then resuspended again in 1 mL of cold lysis buffer and sonicated with a Heat Systems-Ultrasonics, Inc. W-220 sonicator at 20% output for a total of 2 min (pulsed: 30 sec on/30 sec off). The resulting solution was cleared by centrifugation at maximum speed in a microfuge for 10 min at 4°C. At this point 5% of the solution was taken and stored at -20°C for the input chromatin sample. The chromatin solution was precleared for 2-4 h with 80 μL of a 50% slurry of Protein A/G Sepharose beads (Pharmacia). The beads used for immunoprecipitation were preblocked overnight at 4°C with sheared salmon sperm DNA (0.3 mg/mL) and BSA (1 mg/mL) and then incubated with 20 μg of anti-E2F1 polyclonal Ab (C-20, Santa Cruz) for 4-6 h. Immunoprecipitations were performed overnight at 4°C. Immune complexes were eluted from the beads by adding 75 μL elution buffer (50 mM Tris at pH 8.0, 1% SDS, 10 mM EDTA at pH 8.0) and heating at 65°C for 10 min. Elutions were performed twice and the eluates pooled. Cross-links were then reversed by placing the eluates at 65°C overnight along with 5% of the cross-linked whole-cell extract sample diluted in the elution buffer (1:4) to generate the input chromatin samples. The DNA was then purified using the Qiagen QIAquick PCR, and PCR was performed using the Expand High Fidelity PCR system (Roche Biochemicals). Final primer concentration was 0.5 μM. Approximately 1% of the input chromatin sample and 10% of the ChIP sample were used as template in each reaction. Reaction mixtures were initially melted at 94°C for 5 min followed by 27 cycles of 94°C/30 sec, 56°C/1 min, 72°C/1 min, and a final extension of 72°C for 7 min. Amplicons are all in the 150-300-bp range. Samples were resolved on 3% agarose gels contraining ethidium bromide. Primer sequences: TAp73 promoter: forward, 5′-TGAGCCATGAAGATGTGCGAG-3′; reverse, 5′-GCTGCTTATGGTCTGATGCTTATG-3′. Thymidine Kinase promoter: forward, 5′-TCCCGGATTCCTCCCACGAG-3′; reverse, 5′-TGCGCCTCCGGGAAGTTCAC-3′. GAPDH promoter: forward, 5′-AAAAGCGGGGAGAAAGTAGG-3′; reverse, 5′-CTAGCCTCCCGGGTTTCTCT-3′.
Acknowledgments
We thank Ella Freulich for expert technical assistance. We are indebted to J. Chen (Mayo Clinic, MN) and D. Cress (Moffit Cancer Center, FL) for generously providing cell lines. This work was supported by NIH grant CA87497.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.1221004.
References
- Agami R., Blandino, G., Oren, M., and Shaul, Y. 1999. Interaction of c-Abl and p73α and their collaboration to induce apoptosis. Nature 399: 809-813. [DOI] [PubMed] [Google Scholar]
- Ahn J., Urist, M., and Prives, C. 2003. Questioning the role of checkpoint kinase 2 in the p53 DNA damage response. J. Biol. Chem. 278: 20480-20489. [DOI] [PubMed] [Google Scholar]
- Appella E. and Anderson, C.W. 2001. Post-translational modifications and activation of p53 by genotoxic stresses. Eur. J. Biochem. 268: 2764-2772. [DOI] [PubMed] [Google Scholar]
- Banin S., Moyal, L., Shieh, S., Taya, Y., Anderson, C.W., Chessa, L., Smorodinsky, N.I., Prives, C., Reiss, Y., Shiloh, Y., et al. 1998. Enhanced phosphorylation of p53 by ATM in response to DNA damage. Science 281: 1674-1677. [DOI] [PubMed] [Google Scholar]
- Bartek J. and Lukas, J. 2003. Chk1 and Chk2 kinases in checkpoint control and cancer. Cancer Cell 3: 421-429. [DOI] [PubMed] [Google Scholar]
- Basu S., Totty, N.F., Irwin, M.S., Sudol, M., and Downward, J. 2003. Akt phosphorylates the Yes-associated protein, YAP, to induce interaction with 14-3-3 and attenuation of p73-mediated apoptosis. Mol. Cell 11: 11-23. [DOI] [PubMed] [Google Scholar]
- Bergamaschi D., Gasco, M., Hiller, L., Sullivan, A., Syed, N., Trigiante, G., Yulug, I., Merlano, M., Numico, G., Comino, A., et al. 2003. p53 polymorphism influences response in cancer chemotherapy via modulation of p73-dependent apoptosis. Cancer Cell 3: 387-402. [DOI] [PubMed] [Google Scholar]
- Bergamaschi D., Samuels, Y., Jin, B., Duraisingham, S., Crook, T., and Lu, X. 2004. ASPP1 and ASPP2: Common activators of p53 family members. Mol. Cell. Biol. 24: 1341-1350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blattner C., Sparks, A., and Lane, D. 1999. Transcription factor E2F-1 is upregulated in response to DNA damage in a manner analogous to that of p53. Mol. Cell. Biol. 19: 3704-3713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busby E.C., Leistritz, D.F., Abraham, R.T., Karnitz, L.M., and Sarkaria, J.N. 2000. The radiosensitizing agent 7-hydroxystaurosporine (UCN-01) inhibits the DNA damage checkpoint kinase hChk1. Cancer Res. 60: 2108-2112. [PubMed] [Google Scholar]
- Canman C.E., Lim, D.S., Cimprich, K.A., Taya, Y., Tamai, K., Sakaguchi, K., Appella, E., Kastan, M.B., and Siliciano, J.D. 1998. Activation of the ATM kinase by ionizing radiation and phosphorylation of p53. Science 281: 1677-1679. [DOI] [PubMed] [Google Scholar]
- Chehab N.H., Malikzay, A., Appel, M., and Halazonetis, T.D. 2000. Chk2/hCds1 functions as a DNA damage checkpoint in G(1) by stabilizing p53. Genes & Dev. 14: 278-288. [PMC free article] [PubMed] [Google Scholar]
- Costanzo A., Merlo, P., Pediconi, N., Fulco, M., Sartorelli, V., Cole, P.A., Fontemaggi, G., Fanciulli, M., Schiltz, L., Blandino, G., et al. 2002. DNA damage-dependent acetylation of p73 dictates the selective activation of apoptotic target genes. Mol. Cell 9: 175-186. [DOI] [PubMed] [Google Scholar]
- de Toledo S.M., Azzam, E.I., Dahlberg, W.K., Gooding, T.B., and Little, J.B. 2000. ATM complexes with HDM2 and promotes its rapid phosphorylation in a p53-independent manner in normal and tumor human cells exposed to ionizing radiation. Oncogene 19: 6185-6193. [DOI] [PubMed] [Google Scholar]
- Donehower L.A., Harvey, M., Slagle, B.L., McArthur, M.J., Montgomery Jr., C.A., Butel, J.S., and Bradley, A. 1992. Mice deficient for p53 are developmentally normal but susceptible to spontaneous tumours. Nature 356: 215-221. [DOI] [PubMed] [Google Scholar]
- Elbashir S.M., Harborth, J., Lendeckel, W., Yalcin, A., Weber, K., and Tuschl, T. 2001. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature 411: 494-498. [DOI] [PubMed] [Google Scholar]
- Falck J., Mailand, N., Syljuasen, R.G., Bartek, J., and Lukas, J. 2001. The ATM-Chk2-Cdc25A checkpoint pathway guards against radioresistant DNA synthesis. Nature 410: 842-847. [DOI] [PubMed] [Google Scholar]
- Flores E.R., Tsai, K.Y., Crowley, D., Sengupta, S., Yang, A., McKeon, F., and Jacks, T. 2002. p63 and p73 are required for p53-dependent apoptosis in response to DNA damage. Nature 416: 560-564. [DOI] [PubMed] [Google Scholar]
- Fulco M., Costanzo, A., Merlo, P., Mangiacasale, R., Strano, S., Blandino, G., Balsano, C., Lavia, P., and Levrero, M. 2003. p73 is regulated by phosphorylation at the G2/M transition. J. Biol. Chem. 278: 49196-49202. [DOI] [PubMed] [Google Scholar]
- Gaiddon C., Lokshin, M., Ahn, J., Zhang, T., and Prives, C. 2001. A subset of tumor-derived mutant forms of p53 down-regulate p63 and p73 through a direct interaction with the p53 core domain. Mol. Cell. Biol. 21: 1874-1887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaiddon C., Lokshin, M., Gross, I., Levasseur, D., Taya, Y., Loeffler, J.P., and Prives, C. 2003. Cyclin-dependent kinases phosphorylate p73 at threonine 86 in a cell cycle-dependent manner and negatively regulate p73. J. Biol. Chem. 278: 27421-27431. [DOI] [PubMed] [Google Scholar]
- Gong J.G., Costanzo, A., Yang, H.Q., Melino, G., Kaelin Jr., W.G., Levrero, M., and Wang, J.Y. 1999. The tyrosine kinase c-Abl regulates p73 in apoptotic response to cisplatin-induced DNA damage. Nature 399: 806-809. [DOI] [PubMed] [Google Scholar]
- Gonzalez S., Prives, C., and Cordon-Cardo, C. 2003. p73α regulation by Chk1 in response to DNA damage. Mol. Cell. Biol. 23: 8161-8171. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Graves P.R., Yu, L., Schwarz, J.K., Gales, J., Sausville, E.A., O'Connor, P.M., and Piwnica-Worms, H. 2000. The Chk1 protein kinase and the Cdc25C regulatory pathways are targets of the anticancer agent UCN-01. J. Biol. Chem. 275: 5600-5605. [DOI] [PubMed] [Google Scholar]
- Hallstrom T.C. and Nevins, J.R. 2003. Specificity in the activation and control of transcription factor E2F-dependent apoptosis. Proc. Natl. Acad. Sci. 100: 10848-10853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirao A., Kong, Y.Y., Matsuoka, S., Wakeham, A., Ruland, J., Yoshida, H., Liu, D., Elledge, S.J., and Mak, T.W. 2000. DNA damage-induced activation of p53 by the checkpoint kinase Chk2. Science 287: 1824-1827. [DOI] [PubMed] [Google Scholar]
- Hirao A., Cheung, A., Duncan, G., Girard, P.M., Elia, A.J., Wakeham, A., Okada, H., Sarkissian, T., Wong, J.A., Sakai, T., et al. 2002. Chk2 is a tumor suppressor that regulates apoptosis in both an ataxia telangiectasia mutated (ATM)-dependent and an ATM-independent manner. Mol. Cell. Biol. 22: 6521-6532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann F., Martelli, F., Livingston, D.M., and Wang, Z. 1996. The retinoblastoma gene product protects E2F-1 from degradation by the ubiquitin-proteasome pathway. Genes & Dev. 10: 2949-2959. [DOI] [PubMed] [Google Scholar]
- Hollstein M., Hergenhahn, M., Yang, Q., Bartsch, H., Wang, Z.Q., and Hainaut, P. 1999. New approaches to understanding p53 gene tumor mutation spectra. Mutat. Res. 431: 199-209. [DOI] [PubMed] [Google Scholar]
- Irwin M., Marin, M.C., Phillips, A.C., Seelan, R.S., Smith, D.I., Liu, W., Flores, E.R., Tsai, K.Y., Jacks, T., Vousden, K.H., et al. 2000. Role for the p53 homologue p73 in E2F-1-induced apoptosis. Nature 407: 645-648. [DOI] [PubMed] [Google Scholar]
- Irwin M.S., Kondo, K., Marin, M.C., Cheng, L.S., Hahn, W.C., and Kaelin Jr., W.G., 2003. Chemosensitivity linked to p73 function. Cancer Cell 3: 403-410. [DOI] [PubMed] [Google Scholar]
- Jackson A.L., Bartz, S.R., Schelter, J., Kobayashi, S.V., Burchard, J., Mao, M., Li, B., Cavet, G., and Linsley, P.S. 2003. Expression profiling reveals off-target gene regulation by RNAi. Nat. Biotechnol. 21: 635-637. [DOI] [PubMed] [Google Scholar]
- Jallepalli P.V., Lengauer, C., Vogelstein, B., and Bunz, F. 2003. The Chk2 tumor suppressor is not required for p53 responses in human cancer cells. J. Biol. Chem. 278: 20475-20479. [DOI] [PubMed] [Google Scholar]
- Jayaraman L., Freulich, E., and Prives, C. 1997. Functional dissection of p53 tumor suppressor protein. Methods Enzymol. 283: 245-256. [DOI] [PubMed] [Google Scholar]
- Johnson D.G., Schwarz, J.K., Cress, W.D., and Nevins, J.R. 1993. Expression of transcription factor E2F1 induces quiescent cells to enter S phase. Nature 365: 349-352. [DOI] [PubMed] [Google Scholar]
- Johnstone R.W., Ruefli, A.A., and Lowe, S.W. 2002. Apoptosis: A link between cancer genetics and chemotherapy. Cell 108: 153-164. [DOI] [PubMed] [Google Scholar]
- Kaghad M., Bonnet, H., Yang, A., Creancier, L., Biscan, J.C., Valent, A., Minty, A., Chalon, P., Lelias, J.M., Dumont, X., et al. 1997. Monoallelically expressed gene related to p53 at 1p36, a region frequently deleted in neuroblastoma and other human cancers. Cell 90: 809-819. [DOI] [PubMed] [Google Scholar]
- Kartasheva N.N., Contente, A., Lenz-Stoppler, C., Roth, J., and Dobbelstein, M. 2002. p53 induces the expression of its antagonist p73 Δ N, establishing an autoregulatory feedback loop. Oncogene 21: 4715-4727. [DOI] [PubMed] [Google Scholar]
- Khosravi R., Maya, R., Gottlieb, T., Oren, M., Shiloh, Y., and Shkedy, D. 1999. Rapid ATM-dependent phosphorylation of MDM2 precedes p53 accumulation in response to DNA damage. Proc. Natl. Acad. Sci. 96: 14973-14977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J.S., Collins, K.M., Brown, A.L., Lee, C.H., and Chung, J.H. 2000. hCds1-mediated phosphorylation of BRCA1 regulates the DNA damage response. Nature 404: 201-204. [DOI] [PubMed] [Google Scholar]
- Lee S.B., Kim, S.H., Bell, D.W., Wahrer, D.C., Schiripo, T.A., Jorczak, M.M., Sgroi, D.C., Garber, J.E., Li, F.P., Nichols, K.E., et al. 2001. Destabilization of CHK2 by a missense mutation associated with Li-Fraumeni Syndrome. Cancer Res. 61: 8062-8067. [PubMed] [Google Scholar]
- Lin W.C., Lin, F.T., and Nevins, J.R. 2001. Selective induction of E2F1 in response to DNA damage, mediated by ATM-dependent phosphorylation. Genes & Dev. 15: 1833-1844. [PMC free article] [PubMed] [Google Scholar]
- Lissy N.A., Davis, P.K., Irwin, M., Kaelin, W.G., and Dowdy, S.F. 2000. A common E2F-1 and p73 pathway mediates cell death induced by TCR activation. Nature 407: 642-645. [DOI] [PubMed] [Google Scholar]
- Liu G., Nozell, S., Xiao, H., and Chen, X. 2004. ΔNp73β is active in transactivation and growth suppression. Mol. Cell. Biol. 24: 487-501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lou Z., Minter-Dykhouse, K., Wu, X., and Chen, J. 2003. MDC1 is coupled to activated CHK2 in mammalian DNA damage response pathways. Nature 421: 957-961. [DOI] [PubMed] [Google Scholar]
- Ma Y., Cress, W.D., and Haura, E.B. 2003. Flavopiridol-induced apoptosis is mediated through up-regulation of E2F1 and repression of Mcl-1. Mol. Cancer Ther. 2: 73-81. [PubMed] [Google Scholar]
- Marabese M., Vikhanskaya, F., Rainelli, C., Sakai, T., and Broggini, M. 2003. DNA damage induces transcriptional activation of p73 by removing C-EBPα repression on E2F1. Nucleic Acids Res. 31: 6624-6632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marin M.C., Jost, C.A., Irwin, M.S., DeCaprio, J.A., Caput, D., and Kaelin Jr., W.G. 1998. Viral oncoproteins discriminate between p53 and the p53 homolog p73. Mol. Cell. Biol. 18: 6316-6324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marti A., Wirbelauer, C., Scheffner, M., and Krek, W. 1999. Interaction between ubiquitin-protein ligase SCFSKP2 and E2F-1 underlies the regulation of E2F-1 degradation. Nat. Cell Biol. 1: 14-19. [DOI] [PubMed] [Google Scholar]
- Matsuoka S., Huang, M., and Elledge, S.J. 1998. Linkage of ATM to cell cycle regulation by the Chk2 protein kinase. Science 282: 1893-1897. [DOI] [PubMed] [Google Scholar]
- Maya R., Balass, M., Kim, S.T., Shkedy, D., Leal, J.F., Shifman, O., Moas, M., Buschmann, T., Ronai, Z., Shiloh, Y., et al. 2001. ATM-dependent phosphorylation of Mdm2 on serine 395: Role in p53 activation by DNA damage. Genes & Dev. 15: 1067-1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melino G., De Laurenzi, V., and Vousden, K.H. 2002. p73: Friend or foe in tumorigenesis. Nat. Rev. Cancer 2: 605-615. [DOI] [PubMed] [Google Scholar]
- Meng R.D., Phillips, P., and El-Deiry, W.S. 1999. p53-independent increase in E2F-1 expression enhances the cytotoxic effects of etoposide and of adriamycin. Int. J. Oncol. 14: 5-14. [PubMed] [Google Scholar]
- Miyazaki K., Ozaki, T., Kato, C., Hanamoto, T., Fujita, T., Irino, S., Watanabe, K., Nakagawa, T., and Nakagawara, A. 2003. A novel HECT-type E3 ubiquitin ligase, NEDL2, stabilizes p73 and enhances its transcriptional activity. Biochem. Biophys. Res. Commun. 308: 106-113. [DOI] [PubMed] [Google Scholar]
- O'Neill T., Giarratani, L., Chen, P., Iyer, L., Lee, C.H., Bobiak, M., Kanai, F., Zhou, B.B., Chung, J.H., and Rathbun, G.A. 2002. Determination of substrate motifs for human Chk1 and hCds1/Chk2 by the oriented peptide library approach. J. Biol. Chem. 277: 16102-16115. [DOI] [PubMed] [Google Scholar]
- Pediconi N., Ianari, A., Costanzo, A., Belloni, L., Gallo, R., Cimino, L., Porcellini, A., Screpanti, I., Balsano, C., Alesse, E., et al. 2003. Differential regulation of E2F1 apoptotic target genes in response to DNA damage. Nat. Cell Biol. 5: 552-558. [DOI] [PubMed] [Google Scholar]
- Persengiev S.P., Zhu, X., and Green, M.R. 2004. Nonspecific, concentration-dependent stimulation and repression of mammalian gene expression by small interfering RNAs (siRNAs). RNA 10: 12-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips A.C., Ernst, M.K., Bates, S., Rice, N.R., and Vousden, K.H. 1999. E2F-1 potentiates cell death by blocking anti-apoptotic signaling pathways. Mol. Cell 4: 771-781. [DOI] [PubMed] [Google Scholar]
- Prives C. 1998. Signaling to p53: Breaking the MDM2-p53 circuit. Cell 95: 5-8. [DOI] [PubMed] [Google Scholar]
- Sanchez-Prieto R., Sanchez-Arevalo, V.J., Servitja, J.M., and Gutkind, J.S. 2002. Regulation of p73 by c-Abl through the p38 MAP kinase pathway. Oncogene 21: 974-979. [DOI] [PubMed] [Google Scholar]
- Seelan R.S., Irwin, M., van der Stoop, P., Qian, C., Kaelin Jr., W.G., and Liu, W. 2002. The human p73 promoter: Characterization and identification of functional E2F binding sites. Neoplasia 4: 195-203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shieh S.Y., Ahn, J., Tamai, K., Taya, Y., and Prives, C. 2000. The human homologs of checkpoint kinases Chk1 and Cds1 (Chk2) phosphorylate p53 at multiple DNA damage-inducible sites. Genes & Dev. 14: 289-300. [PMC free article] [PubMed] [Google Scholar]
- Shimodaira H., Yoshioka-Yamashita, A., Kolodner, R.D., and Wang, J.Y. 2003. Interaction of mismatch repair protein PMS2 and the p53-related transcription factor p73 in apoptosis response to cisplatin. Proc. Natl. Acad. Sci. 100: 2420-2425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens C., Smith, L., and La Thangue, N.B. 2003. Chk2 activates E2F-1 in response to DNA damage. Nat. Cell Biol. 5: 401-409. [DOI] [PubMed] [Google Scholar]
- Stiewe T. and Putzer, B.M. 2000. Role of the p53-homologue p73 in E2F1-induced apoptosis. Nat. Genet. 26: 464-469. [DOI] [PubMed] [Google Scholar]
- Stiewe T., Theseling, C.C., and Putzer, B.M. 2002. Transactivation-deficient Δ TA-p73 inhibits p53 by direct competition for DNA binding: Implications for tumorigenesis. J. Biol. Chem. 277: 14177-14185. [DOI] [PubMed] [Google Scholar]
- Takai H., Naka, K., Okada, Y., Watanabe, M., Harada, N., Saito, S., Anderson, C.W., Appella, E., Nakanishi, M., Suzuki, H., et al. 2002. Chk2-deficient mice exhibit radioresistance and defective p53-mediated transcription. EMBO J. 21: 5195-5205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai K.K. and Yuan, Z.M. 2003. c-Abl stabilizes p73 by a phosphorylation-augmented interaction. Cancer Res. 63: 3418-3424. [PubMed] [Google Scholar]
- Yang A., Walker, N., Bronson, R., Kaghad, M., Oosterwegel, M., Bonnin, J., Vagner, C., Bonnet, H., Dikkes, P., Sharpe, A., et al. 2000. p73-deficient mice have neurological, pheromonal and inflammatory defects but lack spontaneous tumours. Nature 404: 99-103. [DOI] [PubMed] [Google Scholar]
- Yuan Z.M., Shioya, H., Ishiko, T., Sun, X., Gu, J., Huang, Y.Y., Lu, H., Kharbanda, S., Weichselbaum, R., and Kufe, D. 1999. p73 is regulated by tyrosine kinase c-Abl in the apoptotic response to DNA damage. Nature 399: 814-817. [DOI] [PubMed] [Google Scholar]
- Zaika A.I., Slade, N., Erster, S.H., Sansome, C., Joseph, T.W., Pearl, M., Chalas, E., and Moll, U.M. 2002. ΔNp73, a dominant-negative inhibitor of wild-type p53 and TAp73, is up-regulated in human tumors. J. Exp. Med. 196: 765-780. [DOI] [PMC free article] [PubMed] [Google Scholar]