Abstract
The 40S subunit in 48S complexes formed at the initiation codon of mRNA is bound to eukaryotic initiation factor (eIF) 3, eIF1, eIF1A, and an eIF2/GTP/Met-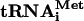 ternary complex and can therefore not join a 60S subunit directly to form an 80S ribosome. We report that eIF5-induced hydrolysis of eIF2-bound GTP in 48S complexes led to release of eIF2-GDP but not eIF3 or eIF1. eIF5B did not influence factor release in the absence of 60S subunits. Therefore eIF3 and eIF1 dissociate from 40S subunits during, rather than before, the eIF5B-mediated subunit joining event. In the absence of eIF1, eIF5-stimulated hydrolysis of eIF2-bound GTP occurred at the same rate in 43S pre-initiation and 48S initiation complexes. GTP hydrolysis in 43S complexes assembled with eIF1 was much slower than in 43S or 48S complexes assembled without eIF1. Establishment of codon-anticodon base-pairing in 48S complexes relieved eIF1's inhibition. Thus, in addition to its role in initiation codon selection during 48S complex formation, eIF1 also participates in maintaining the fidelity of the initiation process at a later stage, hydrolysis of eIF2-bound GTP, by inhibiting premature GTP hydrolysis and by linking establishment of codon-anticodon base-pairing with GTP hydrolysis.
ternary complex and can therefore not join a 60S subunit directly to form an 80S ribosome. We report that eIF5-induced hydrolysis of eIF2-bound GTP in 48S complexes led to release of eIF2-GDP but not eIF3 or eIF1. eIF5B did not influence factor release in the absence of 60S subunits. Therefore eIF3 and eIF1 dissociate from 40S subunits during, rather than before, the eIF5B-mediated subunit joining event. In the absence of eIF1, eIF5-stimulated hydrolysis of eIF2-bound GTP occurred at the same rate in 43S pre-initiation and 48S initiation complexes. GTP hydrolysis in 43S complexes assembled with eIF1 was much slower than in 43S or 48S complexes assembled without eIF1. Establishment of codon-anticodon base-pairing in 48S complexes relieved eIF1's inhibition. Thus, in addition to its role in initiation codon selection during 48S complex formation, eIF1 also participates in maintaining the fidelity of the initiation process at a later stage, hydrolysis of eIF2-bound GTP, by inhibiting premature GTP hydrolysis and by linking establishment of codon-anticodon base-pairing with GTP hydrolysis.
Keywords: Eukaryotic initiation factor 2, eukaryotic initiation factor 3, eukaryotic initiation factor 5, eukaryotic initiation factor 5B, GTP hydrolysis, translation
Translation initiation in eukaryotes leads to the assembly of an 80S ribosome containing aminoacylated initiator tRNA (Met-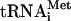 ) in the P (peptidyl) site with its anticodon base-paired to the initiation codon of mRNA. It involves at least eleven eukaryotic initiation factors (eIFs). eIF2, GTP, and Met-
) in the P (peptidyl) site with its anticodon base-paired to the initiation codon of mRNA. It involves at least eleven eukaryotic initiation factors (eIFs). eIF2, GTP, and Met-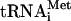 form a ternary complex, which together with eIF3, eIF1, and eIF1A binds to the 40S subunit to form a 43S complex (Benne and Hershey 1978). After binding to the 5′-end of mRNA, mediated by eIF4A, eIF4B, and eIF4F, the 43S complex scans downstream until it encounters an AUG triplet in a favorable context, where it stops and forms a stable 48S complex with established codon-anticodon base-pairing. eIF1 plays a key role in initiation codon selection (Yoon and Donahue 1992; Pestova et al. 1998; Pestova and Kolupaeva 2002). Finally, a 60S subunit joins the 48S complex but cannot do so directly, presumably because the interface surface of the 40S subunit is occupied by eIF1, eIF1A, eIF2, and eIF3, which must be displaced.
form a ternary complex, which together with eIF3, eIF1, and eIF1A binds to the 40S subunit to form a 43S complex (Benne and Hershey 1978). After binding to the 5′-end of mRNA, mediated by eIF4A, eIF4B, and eIF4F, the 43S complex scans downstream until it encounters an AUG triplet in a favorable context, where it stops and forms a stable 48S complex with established codon-anticodon base-pairing. eIF1 plays a key role in initiation codon selection (Yoon and Donahue 1992; Pestova et al. 1998; Pestova and Kolupaeva 2002). Finally, a 60S subunit joins the 48S complex but cannot do so directly, presumably because the interface surface of the 40S subunit is occupied by eIF1, eIF1A, eIF2, and eIF3, which must be displaced.
The first step in ribosomal subunit joining is hydrolysis of eIF2-bound GTP and release of eIF2-GDP from 48S complexes (Das and Maitra 2002). eIF2 consists of α, β, and γ subunits. The structure of eIF2γ is characteristic of GTP-binding proteins (Schmitt et al. 2002), but eIF2 does not have intrinsic GTPase activity. Hydrolysis of eIF2-bound GTP is induced by eIF5, a GTPase-activating protein (GAP) specific for eIF2, which provides an “arginine finger” for the catalytic center of eIF2γ (Das and Maitra 2002). eIF5 binds directly to eIF2β but induces the GTPase activity of eIF2 only in those eIF2 ternary complexes that are bound to 40S subunits (Raychaudhuri et al. 1985). A few biochemical experiments indicate that eIF5-induced hydrolysis of eIF2-bound GTP may be weakly AUG-dependent (e.g., Raychaudhuri et al. 1985) but they were done in a minimal system containing only 40S subunits, eIF2 ternary complex, and eIF5, in which AUG triplets also stimulated the prior stage, binding of ternary complexes to 40S subunits. Although in vitro data concerning a link between initiation codon recognition and hydrolysis of eIF2-bound GTP are limited, strong genetic data support the importance of a stringent connection between them: Mutations in eIF5 (which increase its GAP activity), in eIF2γ (which increase eIF2 dissociation from Met-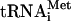 even without GTP hydrolysis) and in eIF2β (which increase eIF2's intrinsic GTPase activity) all enhance initiation at non-AUG codons in yeast (Huang et al. 1997). Thus premature hydrolysis of eIF2-bound GTP and release of eIF2 from initiation complexes reduce the fidelity of initiation and should be avoided.
even without GTP hydrolysis) and in eIF2β (which increase eIF2's intrinsic GTPase activity) all enhance initiation at non-AUG codons in yeast (Huang et al. 1997). Thus premature hydrolysis of eIF2-bound GTP and release of eIF2 from initiation complexes reduce the fidelity of initiation and should be avoided.
It has been widely assumed that eIF5-induced hydrolysis of eIF2-bound GTP and release of eIF2-GDP from 48S complexes lead to release of all other factors (e.g., Das and Maitra 2002). If this were so, hydrolysis of eIF2-bound GTP should logically suffice for subunit joining. However, it is sufficient only for 60S subunits to join minimal 48S complexes formed on AUG triplets, but not to 48S complexes formed on native mRNA with eIF2, eIF3, eIF1, and eIF1A. In this case, eIF5B is also required (Pestova et al. 2000).
It has been suggested that eIF5B's role could be to adjust the position of Met-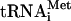 on the 40S subunit as a prerequisite for subunit joining (Roll-Mecak et al. 2001). However, eIF5B's role in subunit joining could also be because, contrary to the common assumption, one or more of eIF1, eIF1A, and eIF3 are not displaced from the 40S subunit after eIF5-induced eIF2-GDP release. Although eIF1, eIF1A, and eIF3 can all bind to 40S subunits independent of the eIF2 ternary complex (Fraser et al. 2004; Maag and Lorsch 2003), contrary to other reports (e.g., Trachsel and Staehelin 1979), in the absence of eIF2 ternary complex, these factors do not protect 40S subunits from joining with 60S subunits (Chaudhuri et al. 1999; Kolupaeva et al. 2005). Therefore, even if eIF1, eIF1A, and eIF3 were not released from 48S complexes after GTP hydrolysis, their presence should not prevent subunit joining. However, we recently found that eIF3 stably binds to 40S subunits in the presence of single-stranded RNA, forming ternary complexes in which RNA is most likely bound in the mRNA-binding cleft of the 40S subunit, and this prevents 40S subunits from joining with 60S subunits even in the absence of eIF2 ternary complex (Kolupaeva et al. 2005). These data suggest that association of eIF3 with 40S subunits in mRNA-containing 48S complexes after release of eIF2-GDP would potentially block subunit joining, and the role of eIF5B could be to displace eIF3 and possibly eIF1 and eIF1A from the 40S subunit after release of eIF2. This could occur in at least two ways. eIF5B could either actively displace these factors, yielding a free 40S subunit/Met-
on the 40S subunit as a prerequisite for subunit joining (Roll-Mecak et al. 2001). However, eIF5B's role in subunit joining could also be because, contrary to the common assumption, one or more of eIF1, eIF1A, and eIF3 are not displaced from the 40S subunit after eIF5-induced eIF2-GDP release. Although eIF1, eIF1A, and eIF3 can all bind to 40S subunits independent of the eIF2 ternary complex (Fraser et al. 2004; Maag and Lorsch 2003), contrary to other reports (e.g., Trachsel and Staehelin 1979), in the absence of eIF2 ternary complex, these factors do not protect 40S subunits from joining with 60S subunits (Chaudhuri et al. 1999; Kolupaeva et al. 2005). Therefore, even if eIF1, eIF1A, and eIF3 were not released from 48S complexes after GTP hydrolysis, their presence should not prevent subunit joining. However, we recently found that eIF3 stably binds to 40S subunits in the presence of single-stranded RNA, forming ternary complexes in which RNA is most likely bound in the mRNA-binding cleft of the 40S subunit, and this prevents 40S subunits from joining with 60S subunits even in the absence of eIF2 ternary complex (Kolupaeva et al. 2005). These data suggest that association of eIF3 with 40S subunits in mRNA-containing 48S complexes after release of eIF2-GDP would potentially block subunit joining, and the role of eIF5B could be to displace eIF3 and possibly eIF1 and eIF1A from the 40S subunit after release of eIF2. This could occur in at least two ways. eIF5B could either actively displace these factors, yielding a free 40S subunit/Met-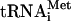 complex before subunit joining, or it could mediate subunit joining on 48S complexes containing eIF3, eIF1, and eIF1A so that these factors dissociate during the actual subunit joining event.
complex before subunit joining, or it could mediate subunit joining on 48S complexes containing eIF3, eIF1, and eIF1A so that these factors dissociate during the actual subunit joining event.
We have investigated these questions and report here that eIF5-induced hydrolysis of eIF2-bound GTP in 48S complexes assembled on mRNA led to release of only eIF2, whereas eIF1 and eIF3 remained bound to 40S subunits. eIF5B does not influence factor displacement in the absence of 60S subunits, and eIF3 and eIF1 are released only during the actual subunit joining event mediated by eIF5B. In the absence of eIF1, eIF5-stimulated hydrolysis of eIF2-bound GTP did not depend on establishment of codon-anticodon interaction and occurred at the same rate in 43S and 48S complexes. In the presence of eIF1, GTP hydrolysis in 43S complexes was much slower than in 43S or 48S complexes assembled in the absence of eIF1. Establishment of codon-anticodon interaction in 48S complexes assembled in the presence of eIF1 relieved eIF1's inhibition. eIF1 therefore plays the role of a negative regulator, which inhibits premature GTP hydrolysis and links codon-anticodon base-pairing with hydrolysis of eIF2-bound GTP. Thus, in addition to its role in initiation codon selection at the stage of 48S complex formation, eIF1 also participates in ensuring the fidelity of initiation at the stage of hydrolysis of eIF2-bound GTP.
Results
eIF1 couples codon-anticodon base-pairing with hydrolysis of eIF2-bound GTP
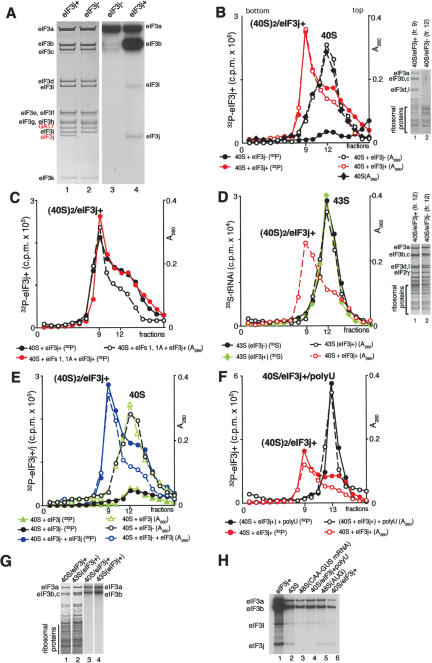
Composition of eIF3 Prior to hydrolysis of eIF2-bound GTP, the 40S subunit in 48S complexes is bound by eIF2, eIF3, eIF1, and eIF1A. The largest factor, eIF3, is thought to contain 12 different polypeptides, but can be purified without the eIF3j subunit (Fig. 2A, below; Fraser et al. 2004), which dissociates from the rest of eIF3 during sucrose density gradient centrifugation in buffer containing 0.4 M KCl, but remains stoichiometrically bound if this stage is omitted. In addition to the known subunits, eIF3 from HeLa cells (Fig. 2A, below) and rabbit reticulocytes (data not shown) contained an additional polypeptide, which yielded tryptic peptides WISDWNLTTEK, QQWQQLYDTLNAWK, and NSLLSLSDT, identical to residues 150-160, 345-360 and 366-374 of GA-17 (GenBank NP_006351), respectively. It is a protein of unknown function that contains a PCI domain like those in eIF3a, eIF3c, and eIF3e. We will refer to eIF3 that contains eIF3j as eIF3j+ and to eIF3 that does not as eIF3j-. Although eIF3j+ and eIF3j- differed in their ability to bind 40S subunits in the absence of other factors (Fraser et al. 2004; this study), they were equally active in 43S and 48S complex formation (Kolupaeva et al. 2005; this study).
Figure 2.
Subunit composition of eIF3, phosphorylation of eIF3 by cAMP-dependent kinase, and eIF3's binding to 40S subunits. (A) Coomassie staining of HeLa eIF3j+ and eIF3j- and autoradiography of eIF3j- and eIF3j+ 32P-phosphorylated by cAMP-dependent kinase. eIF3 subunits are indicated. (B,C) Binding of 32P-eIF3j+ and 32P-eIF3j- to 40S subunits in the absence (B) and presence (C) of eIF1 and eIF1A, assayed by sucrose density gradient centrifugation and analyzed by optical density, Cerenkov counting (B,C) and gel electrophoresis (B, right). (D) 43S complex formation in the presence of 40S subunits, eIF2, [35S]Met-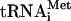 , eIF1, eIF1A, and eIF3j+ or eIF3j- assayed by sucrose density gradient centrifugation and analyzed by optical density, scintillation counting, and gel electrophoresis (right). The optical density of eIF3j+/40S subunit complexes subjected to sucrose density gradient centrifugation is included for comparison of their mobility and that of 43S complexes. (E) Influence of rec-eIF3j subunit on binding of 32P-eIF3j- to 40S subunits and binding of 32P-eIF3j itself to 40S subunits assayed by sucrose density gradient centrifugation and analyzed by optical density and Cerenkov counting. (F) Influence of poly(U) RNA on binding of 32P-eIF3j+ to 40S subunits assayed by sucrose density gradient centrifugation and analyzed by optical density and Cerenkov counting. Sedimentation was from right to left in all cases. Ribosomal complexes are indicated above appropriate peaks. Upper fractions from sucrose gradients have been omitted for clarity. (G) Comparison of the relative amounts of 40S subunits and 32P-eIF3j+ in binary eIF3j+/40S subunit complexes and in 43S complexes separated by sucrose density gradient centrifugation and analyzed by fluorescent SYPRO staining (lanes 1,2) and autoradiography (lanes 3,4) after gel electrophoresis. (H) Presence of the eIF3j subunit from 32P-eIF3j+ in different ribosomal complexes isolated by sucrose density gradient centrifugation.
, eIF1, eIF1A, and eIF3j+ or eIF3j- assayed by sucrose density gradient centrifugation and analyzed by optical density, scintillation counting, and gel electrophoresis (right). The optical density of eIF3j+/40S subunit complexes subjected to sucrose density gradient centrifugation is included for comparison of their mobility and that of 43S complexes. (E) Influence of rec-eIF3j subunit on binding of 32P-eIF3j- to 40S subunits and binding of 32P-eIF3j itself to 40S subunits assayed by sucrose density gradient centrifugation and analyzed by optical density and Cerenkov counting. (F) Influence of poly(U) RNA on binding of 32P-eIF3j+ to 40S subunits assayed by sucrose density gradient centrifugation and analyzed by optical density and Cerenkov counting. Sedimentation was from right to left in all cases. Ribosomal complexes are indicated above appropriate peaks. Upper fractions from sucrose gradients have been omitted for clarity. (G) Comparison of the relative amounts of 40S subunits and 32P-eIF3j+ in binary eIF3j+/40S subunit complexes and in 43S complexes separated by sucrose density gradient centrifugation and analyzed by fluorescent SYPRO staining (lanes 1,2) and autoradiography (lanes 3,4) after gel electrophoresis. (H) Presence of the eIF3j subunit from 32P-eIF3j+ in different ribosomal complexes isolated by sucrose density gradient centrifugation.
Hydrolysis of eIF2-bound GTP in 43S and in 48S complexes formed on AUG triplets To investigate the factor-dependence of the influence of codon-anticodon basepairing on eIF5-induced hydrolysis of eIF2-bound GTP, we compared GTP hydrolysis in 43S complexes and in 48S complexes assembled on AUG triplets (i.e., with the codon-anticodon interaction in its simplest form) and on model mRNAs with various combinations of factors. Results for eIF3j- and eIF3j+ were identical, so they are both described as eIF3 in this section. eIF1 maintains the fidelity of initiation codon selection and binds on the interface surface of 40S subunit (Pestova and Kolupaeva 2002; Lomakin et al. 2003). This position suggests that eIF1 likely plays this role by influencing the conformation of the 40S subunit platform and the positions of mRNA and Met-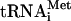 in initiation complexes. Regarding eIF1's role in initiation codon selection and its potential mode of action, we were interested in the influence of this particular factor on hydrolysis of eIF2-bound GTP. To do so, ternary complexes were assembled using purified individual native Met-
in initiation complexes. Regarding eIF1's role in initiation codon selection and its potential mode of action, we were interested in the influence of this particular factor on hydrolysis of eIF2-bound GTP. To do so, ternary complexes were assembled using purified individual native Met-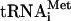 to avoid the influence of RNA contaminants in unfractionated tRNA that might bind 43S complexes and mimic the codon-anticodon interaction. Although AUG triplets do not influence 43S complex formation in the presence of eIF3, eIF1, and eIF1A and have only a minor stimulatory effect in the presence of eIF3 and eIF1A (Kolupaeva et al. 2005), to avoid any indirect stimulation by AUG triplets of GTP hydrolysis due to enhanced binding of ternary complexes to 40S subunits, 43S complexes were separated from unbound ternary complexes by sucrose density gradient centrifugation after assembly. This allowed equal amounts of 43S complexes to be used in reaction mixtures with different combinations of other translation components. For 48S complex formation, purified 43S complexes were incubated with AUG triplets or CAA-GUS mRNA and various combinations of eIF4A, eIF4B, eIF4F, eIF1A, and eIF1. Because its binding to 43S complexes does not withstand centrifugation, eIF1A was added to purified 43S complexes in 48S complex assembly reactions and control reactions containing 43S complexes. Although eIF5 is thought to stimulate GTP hydrolysis catalytically (Das and Maitra 2002), we used it at a concentration that exceeded that of 43S/48S complexes to avoid any possibility that potential differences in recycling of eIF5 in the presence of different translation components could affect GTP hydrolysis.
to avoid the influence of RNA contaminants in unfractionated tRNA that might bind 43S complexes and mimic the codon-anticodon interaction. Although AUG triplets do not influence 43S complex formation in the presence of eIF3, eIF1, and eIF1A and have only a minor stimulatory effect in the presence of eIF3 and eIF1A (Kolupaeva et al. 2005), to avoid any indirect stimulation by AUG triplets of GTP hydrolysis due to enhanced binding of ternary complexes to 40S subunits, 43S complexes were separated from unbound ternary complexes by sucrose density gradient centrifugation after assembly. This allowed equal amounts of 43S complexes to be used in reaction mixtures with different combinations of other translation components. For 48S complex formation, purified 43S complexes were incubated with AUG triplets or CAA-GUS mRNA and various combinations of eIF4A, eIF4B, eIF4F, eIF1A, and eIF1. Because its binding to 43S complexes does not withstand centrifugation, eIF1A was added to purified 43S complexes in 48S complex assembly reactions and control reactions containing 43S complexes. Although eIF5 is thought to stimulate GTP hydrolysis catalytically (Das and Maitra 2002), we used it at a concentration that exceeded that of 43S/48S complexes to avoid any possibility that potential differences in recycling of eIF5 in the presence of different translation components could affect GTP hydrolysis.
In the absence of eIF1, neither the rate nor the extent of GTP hydrolysis was stimulated by AUG triplets, and hydrolysis was nearly complete within the first 5 min (Fig. 1A). When eIF1 was present, AUG triplets stimulated the rate but not the extent of eIF5-induced hydrolysis of eIF2-bound GTP so that equal amounts of GTP were hydrolyzed after 30 min in both instances (Fig. 1B). The use of equal amounts of preformed purified 43S complexes allowed data obtained with and without eIF1 to be compared (Fig. 1A,B). The rate of GTP hydrolysis in 43S complexes in the presence of eIF1 was lower than in 43S complexes in its absence, which in turn was similar to the rate in 48S complexes assembled with or without eIF1, and the biggest difference in the extent of GTP hydrolyzed in the presence and in the absence of AUG triplets was observed during the first 2 min of incubation. Stimulation by AUG triplets was specific: Triplets corresponding to near-cognate (UUG) or noncognate (CAA) initiation codons did not enhance GTP hydrolysis in otherwise identical reactions (Fig. 1C). These observations indicate that in the absence of codon-anticodon base-pairing, eIF1 inhibits premature GTP hydrolysis and that establishment of codon-anticodon base-pairing relieves this inhibition. The absence of eIF1A or the presence of eIF4A, eIF4B, and eIF4F did not influence eIF1's ability to inhibit hydrolysis of eIF2-bound GTP in the absence of AUG triplets, and in all cases AUG triplets enhanced the rate of eIF5-induced hydrolysis of GTP when eIF1 was present (Fig. 1D,E). Although eIF1A is lost from 43S complexes during centrifugation, for reactions that did not contain eIF1A (Fig. 1D), initial 43S complexes were nevertheless assembled without eIF1A. Very little GTP hydrolysis occurred in reactions lacking eIF5 (Fig. 1A-E) or 40S subunits (data not shown).
Figure 1.
Influence of initiation factors and codon-anticodon base-pairing on hydrolysis of eIF2-bound GTP. 43S complexes were assembled from 40S subunits, eIF1A, eIF3, and eIF2/[32P]GTP/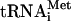 ternary complex; purified by sucrose density gradient centrifugation; and incubated with AUG, UUG, CAA triplets, mRNAs, and eIF1, eIF1A, eIF4A, eIF4B, eIF4F, eIF5, and ΔeIF5B as indicated. GTP hydrolysis was assayed by release of 32Pi. Data represent the average of five or more experiments.
ternary complex; purified by sucrose density gradient centrifugation; and incubated with AUG, UUG, CAA triplets, mRNAs, and eIF1, eIF1A, eIF4A, eIF4B, eIF4F, eIF5, and ΔeIF5B as indicated. GTP hydrolysis was assayed by release of 32Pi. Data represent the average of five or more experiments.
eIF5-induced hydrolysis of eIF2-bound GTP in 43S complexes and in 48S complexes assembled on CAA-GUS mRNA The eIF1-dependent stimulatory effect of codon-anticodon base-pairing on GTP hydrolysis was further investigated using a model mRNA. CAA-GUS mRNA comprises a GUS-reporter gene and a 5′-UTR with one GAA and twenty CAA triplets (Pestova and Kolupaeva 2002). This 5′-UTR is unstructured and does not contain potential near-cognate initiation codons, which allows 48S complexes to form efficiently on the initiation codon in the absence of eIF1 (Pestova and Kolupaeva 2002). In a reaction mixture containing eIF2, eIF3, eIF4A, eIF4B, eIF4F, and eIF1A but no eIF1, eIF5-induced hydrolysis of eIF2-bound GTP was not stimulated by this mRNA (Fig. 1F). As with AUG triplets, inclusion of this mRNA relieved the inhibitory effect of eIF1 on eIF5-induced hydrolysis of eIF2-bound GTP (Fig. 1G). The rates of GTP hydrolysis in reaction mixtures that contained eIF1 and this mRNA and in those lacking eIF1 were similar (Fig. 1F,G).
To confirm that the stimulatory effect of CAA-GUS mRNA was specific and that it was due to codon-anticodon base-pairing, this mRNA was replaced by CAA-stem-GUS mRNA, which contains a stable stem (-13.1 kCal) in the 5′-UTR [with the sequence 5′-G(CAA)14GGGGCUGCGCCUGCAGCCCC(CAA)4CCAUG, in which base-paired regions are underlined] that prevents assembly of 48S complexes in the reconstituted system (Pestova and Kolupaeva 2002). Consistent with its inability to support 48S complex assembly, this mRNA also did not stimulate eIF5-induced hydrolysis of eIF2-bound GTP in the presence of eIF1 (Fig. 1G). It even slightly inhibited GTP hydrolysis in the absence of eIF1 and more so in its presence (Fig. 1F,G). Taken together, these results indicate that it is the presence of eIF1 that couples codon-anticodon base-pairing with hydrolysis of eIF2-bound GTP.
Inclusion in reaction mixtures of eIF5B587-1220 (ΔeIF5B, which is fully active in subunit joining; Pestova et al. 2000) did not influence eIF5-stimulated hydrolysis of eIF2-bound GTP in 43S complexes or in 48S complexes assembled on CAA-GUS mRNA, with or without eIF1 (Fig. 1H,I). GTP hydrolysis in reactions lacking eIF5 was low (Fig. 1F-I).
Release of eIF1, eIF2, and eIF3 from 48S complexes after eIF5-induced hydrolysis of eIF2-bound GTP
Phosphorylation of eIF3 by cAMP-dependent protein kinase eIF3 is the principal factor involved in ribosomal subunit anti-association (Trachsel and Staehelin 1979; Goumans et al. 1980). To be able to quantify eIF3's association with 40S subunits before and after hydrolysis of eIF2-bound GTP, we phosphorylated it using cAMP-dependent protein kinase (Fig. 2A). The phosphorylation patterns of eIF3j+ and eIF3j- differed. Although eIF3a was labeled equally strongly in eIF3j- and eIF3j+, phosphorylation of eIF3b and eIF3c in eIF3j- was very weak, whereas in eIF3j+, phosphorylation of eIF3b was even stronger than that of eIF3a, and eIF3j and eIF3l were also weakly phosphorylated (Fig. 2A, lanes 3,4). As a result, the specific activity of 32P-phosphorylated eIF3j+ was ∼2.5 times that of eIF3j-. In yeast, eIF3j bridges eIF3a and eIF3b (Valasek et al. 2001) and could induce conformational changes that make some residues in eIF3b accessible to phosphorylation. eIF3 phosphorylation did not affect its function in 40S subunit binding and 43S, 48S, or 80S complex formation, so this issue will not be addressed further.
Binding of eIF3 to 40S subunits eIF3j+ and eIF3j- differ in their ability to bind to 40S subunits: Unlike eIF3j+, eIF3j- alone does not bind to them (Fraser et al. 2004). In our experiments, eIF3j+ alone also bound to 40S subunits, but contrary to the previous report, binary eIF3j+/40S subunit complexes migrated faster than 40S subunits (Fig. 2B), 43S complexes (Fig. 2D), and even 48S complexes (data not shown), and had an estimated sedimentation coefficient of ∼60S. Inclusion of eIF1 and eIF1A with eIF3j+ did not prevent formation of this fast migrating complex (Fig. 2C). Both forms of eIF3 were equally active in formation of 43S complexes of similar mobility in the presence of eIF1, eIF1A, eIF2, and [35S]-Met-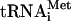 (Fig. 2D). The eIF3:eIF2:40S subunit ratio in 43S complexes was calculated by comparison with known amounts of 40S subunits, eIF2, and eIF3 using gel electrophoresis (data not shown). Amounts of protein were determined by PhosphorImager analysis after staining with fluorescent SYPRO stain. As expected, 43S complexes contained equimolar amounts of 40S subunits, eIF2, and eIF3 in both instances.
(Fig. 2D). The eIF3:eIF2:40S subunit ratio in 43S complexes was calculated by comparison with known amounts of 40S subunits, eIF2, and eIF3 using gel electrophoresis (data not shown). Amounts of protein were determined by PhosphorImager analysis after staining with fluorescent SYPRO stain. As expected, 43S complexes contained equimolar amounts of 40S subunits, eIF2, and eIF3 in both instances.
The composition of eIF3j+/40S complexes was estimated by comparing the amounts of 40S subunits and eIF3 in them and in 43S complexes (Fig. 2G). eIF3j+/40S subunit complexes contained about half as much eIF3 as 43S complexes, i.e., approximately two 40S subunits per molecule of eIF3j+. Incubation of 40S subunits with eIF3j+ reconstituted from eIF3j- and recombinant eIF3j (rec-eIF3j) but not with rec-eIF3j alone also led to formation of the fast migrating peak, just as with eIF3j+ (Fig. 2B,E) even though rec-eIF3j alone bound to 40S subunits, as previously reported (Fraser et al. 2004). Substoichiometric amounts of eIF3j+ in eIF3j+/40S complexes suggest that their mobility is likely due to their containing two 40S molecules rather than to extensive conformational change caused by the eIF3j subunit. Formation of 40S dimers (e.g., Peterson et al. 1979; Goumans et al. 1980) and association of eIF3 with them (Trachsel and Staehelin 1979) has previously been reported. Our data do not allow the possibilities that eIF3j+ actively promotes dimer formation or that it stabilizes pre-existing dimers to be distinguished. The peak of free 40S subunits had a small faster migrating shoulder (Fig. 2B), which likely represents dimers. It is possible that they occur more often in solution before centrifugation and that binding of eIF3j+ to one 40S subunit stabilizes pre-existing dimers and enhances formation of the ∼60S peak. It is reasonable to assume that dimer formation could occlude the eIF3-binding site on one subunit. Various HeLa and rabbit reticulocyte 40S subunit preparations all formed ∼60S complexes when incubated with eIF3j+. The failure to detect formation of a fast migrating peak with eIF3j+ previously (Fraser et al. 2004) may be due to differences in buffer composition and/or centrifugation conditions. We emphasize that the 40S subunits used here were fully active in all stages of initiation, including subunit joining.
The eIF3j subunit remained associated with eIF3j+/40S subunit complexes and 43S complexes that had assembled with 32P-eIF3j+, but not with 48S complexes formed on CAA-GUS mRNA with the same factors (eIF2, eIF3, eIF1, and eIF1A) that were present in 43S complexes (Fig. 2H). eIF3 has been reported to lose a 35kD component upon binding 40S subunits (Benne and Hershey 1976), which in retrospect may have been eIF3j. To investigate whether it is the presence of mRNA in ribosomal complexes or establishment of codon-anticodon base-pairing that weakens eIF3j's association with ribosomal complexes, we investigated if it is present in other ribosomal complexes. As we recently found (Kolupaeva et al. 2005), eIF3 stably binds to 40S subunits in the presence of U-rich single-stranded RNA, forming ternary complexes in which RNA is most likely bound in the mRNA-binding cleft of the 40S subunit. Addition of poly(U) to eIF3j+/40S complexes yielded a slower migrating ternary complex (Fig. 2F) containing stoichiometric amounts of 40S subunits and eIF3 (data not shown) that no longer contained eIF3j (Fig. 2H). On the other hand, assembly of 48S complexes on AUG triplets did not lead to dissociation of eIF3j (Fig. 2H). We conclude that it is the presence of RNA in the mRNA-binding cleft of the 40S subunit rather than codon-anticodon base-pairing that weakens eIF3j's association with ribosomal complexes.
Factor release from 48S complexes assembled on AUG triplets Because the composition of eIF3 and the presence of RNA on the 40S subunit strongly influence the eIF3/40S interaction, we investigated the eIF5-induced release of factors from 48S complexes assembled with both eIF3j- and eIF3j+ on AUG triplets and on mRNAs with 5′-UTRs and coding sequences of different lengths. 48S complexes were initially assembled on AUG triplets with eIF2, [35S]Met-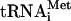 (to permit monitoring of its association), unlabeled GTP or 32P-GTP (to permit monitoring of GTP hydrolysis), eIF1, eIF1A, and either of eIF3j+ and eIF3j- forms of 32P-labeled or unlabeled eIF3. 48S complexes were incubated with eIF5 alone or with ΔeIF5B to induce hydrolysis of eIF2-bound GTP and separated by sucrose density gradient centrifugation. Because of the quantities required, we used
(to permit monitoring of its association), unlabeled GTP or 32P-GTP (to permit monitoring of GTP hydrolysis), eIF1, eIF1A, and either of eIF3j+ and eIF3j- forms of 32P-labeled or unlabeled eIF3. 48S complexes were incubated with eIF5 alone or with ΔeIF5B to induce hydrolysis of eIF2-bound GTP and separated by sucrose density gradient centrifugation. Because of the quantities required, we used 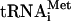 transcripts instead of native tRNA, which is expensive and requires extensive purification to remove RNA contaminants. Although we previously reported that
transcripts instead of native tRNA, which is expensive and requires extensive purification to remove RNA contaminants. Although we previously reported that 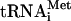 transcripts can perform all functions of native
transcripts can perform all functions of native 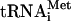 (Pestova and Hellen 2001), the most crucial experiments were repeated with individual native Met-
(Pestova and Hellen 2001), the most crucial experiments were repeated with individual native Met-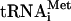 . Factor release from 48S complexes assembled with transcript and native Met-
. Factor release from 48S complexes assembled with transcript and native Met-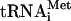 did not differ, so this issue will not be discussed further. Release of eIF1A was not investigated because its binding to 40S subunits is labile and does not withstand centrifugation even prior to hydrolysis of eIF2-bound GTP (Pestova et al. 1998).
did not differ, so this issue will not be discussed further. Release of eIF1A was not investigated because its binding to 40S subunits is labile and does not withstand centrifugation even prior to hydrolysis of eIF2-bound GTP (Pestova et al. 1998).
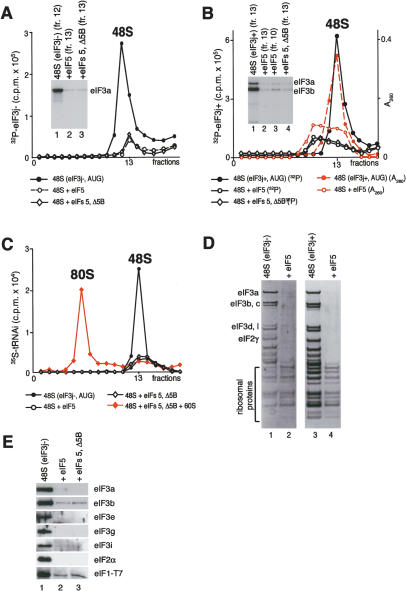
Incubation with eIF5 of 48S complexes assembled on AUG triplets in the presence of eIF3j- led to near-complete hydrolysis of eIF2-bound GTP (data not shown) and consequent near-total dissociation of eIF2 from 40S subunits (Fig. 3D [lane 2], E). It also led to 90%-95% release of 32P-labeled (Fig. 3A) or unlabeled eIF3 and eIF1 (Fig. 3D [lane 2], E). Incubation of eIF5 with 48S complexes assembled on AUG triplets with eIF3j+ also led to nearly complete hydrolysis of eIF2-bound GTP (data not shown), to a reduction in the OD260 of the peak of individual 40S subunits, and to appearance of a second peak at ∼60S (Fig. 3B). Neither peak contained eIF2, and a little eIF3 was associated with individual 40S subunits (Fig. 3B,D, lane 4; data not shown). The ∼60S peak (Fig. 3B) contained sub-stoichiometric amounts of eIF3 and may comprise 40S subunit dimers, like those detected earlier (Fig. 2B). The higher counts in peaks that corresponded to 48S complexes assembled with 32P-eIF3j+ reflect its higher specific radioactivity (Fig. 2A).
Figure 3.
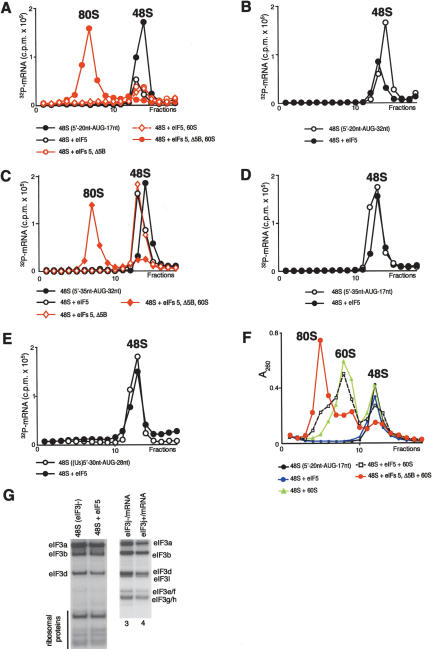
Factor release from 48S complexes assembled on AUG triplets after hydrolysis of eIF2-bound GTP. (A) Release of 32P-eIF3j- and (B) 32P-eIF3j+ from 48S complexes assembled on AUG triplets in the presence of eIF2, eIF3, eIF1, and eIF1A, incubated with eIF5 and ΔeIF5B, as indicated, and assayed after sucrose density gradient centrifugation by Cerenkov counting, optical density measurement, and gel electrophoresis/autoradiography of peak fractions (inset panels). (C) Association of [35S]Met-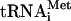 with ribosomal complexes formed after incubating 48S complexes with eIF5, ΔeIF5B, and 60S subunits, as indicated, and separated by sucrose density gradient centrifugation. Upper fractions of gradients have been omitted for clarity. Ribosomal complexes are indicated above appropriate peaks. (D,E) Association of eIF1, eIF2, eIF3j-, and eIF3j+ with 40S subunits before and after treatment with eIF5 and ΔeIF5B of 48S complexes assembled on AUG triplets, analyzed by fluorescent SYPRO staining (D) or immunoblotting (E), after gel electrophoresis of peak fractions obtained after sucrose density gradient centrifugation of ribosomal complexes. eIF2 and eIF3 subunits, eIF1, and ribosomal proteins are indicated.
with ribosomal complexes formed after incubating 48S complexes with eIF5, ΔeIF5B, and 60S subunits, as indicated, and separated by sucrose density gradient centrifugation. Upper fractions of gradients have been omitted for clarity. Ribosomal complexes are indicated above appropriate peaks. (D,E) Association of eIF1, eIF2, eIF3j-, and eIF3j+ with 40S subunits before and after treatment with eIF5 and ΔeIF5B of 48S complexes assembled on AUG triplets, analyzed by fluorescent SYPRO staining (D) or immunoblotting (E), after gel electrophoresis of peak fractions obtained after sucrose density gradient centrifugation of ribosomal complexes. eIF2 and eIF3 subunits, eIF1, and ribosomal proteins are indicated.
Inclusion of ΔeIF5B with eIF5 did not affect the release of eIF1, eIF2, or eIF3 (Fig. 3A,B,E; data not shown). Hydrolysis of eIF2-bound GTP also led to release after centrifugation of 70%-80% Met-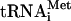 from 48S complexes assembled with eIF3j+ or eIF3j- (Fig. 3C; data not shown). This release was in part due to the stringency of sucrose density gradient centrifugation, because in filter binding assays ∼65%-80% of [35S]Met-
from 48S complexes assembled with eIF3j+ or eIF3j- (Fig. 3C; data not shown). This release was in part due to the stringency of sucrose density gradient centrifugation, because in filter binding assays ∼65%-80% of [35S]Met-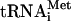 remained associated with 40S subunits after incubation of 48S complexes with eIF5 for 10 min (data not shown). In the case of 48S complexes assembled with eIF3j+, Met-
remained associated with 40S subunits after incubation of 48S complexes with eIF5 for 10 min (data not shown). In the case of 48S complexes assembled with eIF3j+, Met-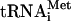 was present only in the slower-migrating peak of 40S subunits. Incubation of 48S complexes with eIF5, ΔeIF5B, and 60S subunits led to formation of 80S ribosomes (Fig. 3C).
was present only in the slower-migrating peak of 40S subunits. Incubation of 48S complexes with eIF5, ΔeIF5B, and 60S subunits led to formation of 80S ribosomes (Fig. 3C).
We conclude that eIF5-stimulated GTP hydrolysis in 48S complexes assembled on AUG triplets with eIF3j-led to release of eIF1, eIF2, and eIF3. eIF3j+ may also be released after GTP hydrolysis and later be rebound by 40S subunits into 40S dimer/eIF3j+ complexes.
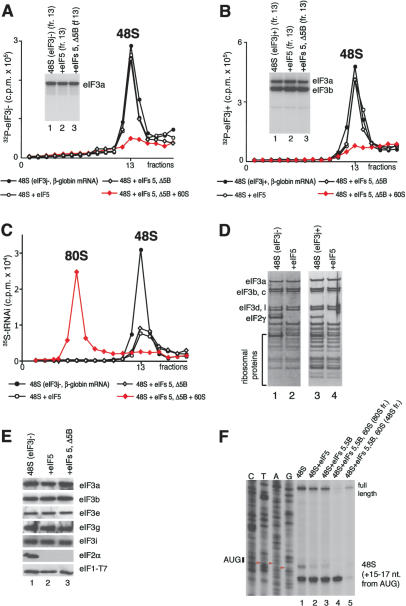
Factor release from 48S complexes assembled on globin mRNA To investigate the influence of mRNA on eIF5-induced release of factors, 48S complexes were assembled on native capped, polyadenylated globin mRNA in the presence of eIF2, [35S]Met-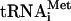 , unlabeled GTP or 32P-GTP, eIF1, eIF1A, eIF4A, eIF4B, eIF4F, and either eIF3j+ or eIF3j- forms of 32P-labeled or unlabeled eIF3. In all instances, incubation of complexes with eIF5 led to near-complete hydrolysis of eIF2-bound GTP (data not shown) and dissociation of eIF2 (Fig. 4D,E). In contrast to 48S complexes assembled on AUG triplets, neither eIF1, eIF3j- or eIF3j+ were released from 48S complexes assembled on mRNA after hydrolysis of eIF2-bound GTP (Fig. 4A,B,D,E). Inclusion of eIF5B with eIF5 did not affect the release of eIF1, eIF2, and eIF3 (Fig. 4A,B,E). However, incubation of 48S complexes with eIF5, ΔeIF5B, and 60S subunits led to their conversion into 80S ribosomes that contained Met-
, unlabeled GTP or 32P-GTP, eIF1, eIF1A, eIF4A, eIF4B, eIF4F, and either eIF3j+ or eIF3j- forms of 32P-labeled or unlabeled eIF3. In all instances, incubation of complexes with eIF5 led to near-complete hydrolysis of eIF2-bound GTP (data not shown) and dissociation of eIF2 (Fig. 4D,E). In contrast to 48S complexes assembled on AUG triplets, neither eIF1, eIF3j- or eIF3j+ were released from 48S complexes assembled on mRNA after hydrolysis of eIF2-bound GTP (Fig. 4A,B,D,E). Inclusion of eIF5B with eIF5 did not affect the release of eIF1, eIF2, and eIF3 (Fig. 4A,B,E). However, incubation of 48S complexes with eIF5, ΔeIF5B, and 60S subunits led to their conversion into 80S ribosomes that contained Met-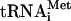 (Fig. 4C) and β-globin mRNA (Fig. 4F, lane 4) but not eIF3 (Fig. 4A,B). Toe-printing of purified 48S complexes showed that treatment with eIF5 and ΔeIF5B in the absence of 60S subunits did not alter β-globin mRNA's association with 40S subunits (Fig. 4D, lanes 1-3). In the presence of eIF5, ΔeIF5B, and 60S subunits, 48S complexes were almost all converted into mRNA-containing 80S ribosomes (Fig. 4F, lane 4,5). When 48S complexes were assembled on mRNA, eIF3 and eIF1 remain associated with 40S subunits even after hydrolysis of eIF2-bound GTP. As with 48S complexes assembled on AUG triplets, hydrolysis of eIF2-bound GTP weakened the association of Met-
(Fig. 4C) and β-globin mRNA (Fig. 4F, lane 4) but not eIF3 (Fig. 4A,B). Toe-printing of purified 48S complexes showed that treatment with eIF5 and ΔeIF5B in the absence of 60S subunits did not alter β-globin mRNA's association with 40S subunits (Fig. 4D, lanes 1-3). In the presence of eIF5, ΔeIF5B, and 60S subunits, 48S complexes were almost all converted into mRNA-containing 80S ribosomes (Fig. 4F, lane 4,5). When 48S complexes were assembled on mRNA, eIF3 and eIF1 remain associated with 40S subunits even after hydrolysis of eIF2-bound GTP. As with 48S complexes assembled on AUG triplets, hydrolysis of eIF2-bound GTP weakened the association of Met-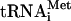 with mRNA-bound ribosomal complexes so that ∼70% of it was released during centrifugation (Fig. 4C).
with mRNA-bound ribosomal complexes so that ∼70% of it was released during centrifugation (Fig. 4C).
Figure 4.
Release of initiation factors from 48S complexes assembled on β-globin mRNA after hydrolysis of eIF2-bound GTP. (A,B) Association of 32P-eIF3j-(A) and 32P-eIF3j+ (B) with 48S complexes assembled on β-globin mRNA in the presence of eIF2, eIF3, eIF1, eIF1A, eIF4A, eIF4B, and eIF4F before and after incubation with eIF5, ΔeIF5B, and 60S subunits, as indicated, and assayed after sucrose density gradient centrifugation by Cerenkov counting and gel electrophoresis/autoradiography of peak fractions (inset panels). (C) Association of [35S]Met-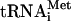 with ribosomal complexes formed after incubating 48S complexes with eIF5, ΔeIF5B, and 60S subunits, as indicated, and separated by sucrose density gradient centrifugation. Upper fractions of gradients have been omitted for clarity. Ribosomal complexes are indicated above appropriate peaks. (D,E) Association of eIF1, eIF2, eIF3j- and eIF3j+ with 40S subunits before and after treatment with eIF5 and ΔeIF5B of 48S complexes assembled on β-globin mRNA, analyzed by fluorescent SYPRO staining (D) or immunoblotting (E), after gel electrophoresis of peak fractions obtained after sucrose density gradient centrifugation of ribosomal complexes. eIF2 and eIF3 subunits, eIF1, and ribosomal proteins are indicated. (F) Association of β-globin mRNA with 48S complexes before and after incubation with eIF5, eIF5, and ΔeIF5B, or eIF5, ΔeIF5B, and 60S subunits, and association with 80S ribosomes formed by incubating 48S complexes with eIF5, ΔeIF5B, and 60S subunits, indicated by toeprints 15-17 nt from the initiation codon. The red arrowheads indicate the positions of A, U, and G nucleotides of the β-globin mRNA initiation codon.
with ribosomal complexes formed after incubating 48S complexes with eIF5, ΔeIF5B, and 60S subunits, as indicated, and separated by sucrose density gradient centrifugation. Upper fractions of gradients have been omitted for clarity. Ribosomal complexes are indicated above appropriate peaks. (D,E) Association of eIF1, eIF2, eIF3j- and eIF3j+ with 40S subunits before and after treatment with eIF5 and ΔeIF5B of 48S complexes assembled on β-globin mRNA, analyzed by fluorescent SYPRO staining (D) or immunoblotting (E), after gel electrophoresis of peak fractions obtained after sucrose density gradient centrifugation of ribosomal complexes. eIF2 and eIF3 subunits, eIF1, and ribosomal proteins are indicated. (F) Association of β-globin mRNA with 48S complexes before and after incubation with eIF5, eIF5, and ΔeIF5B, or eIF5, ΔeIF5B, and 60S subunits, and association with 80S ribosomes formed by incubating 48S complexes with eIF5, ΔeIF5B, and 60S subunits, indicated by toeprints 15-17 nt from the initiation codon. The red arrowheads indicate the positions of A, U, and G nucleotides of the β-globin mRNA initiation codon.
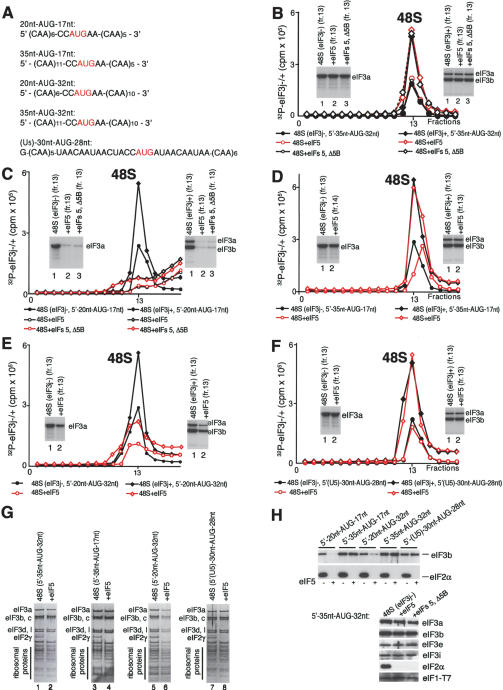
Factor release from 48S complexes assembled on mRNAs with 5′-UTRs and coding regions of different lengths The stable association of eIF1 and eIF3 with 48S complexes assembled on globin mRNA after hydrolysis of eIF2-bound GTP could be because mRNA stabilizes the ribosome/factor interaction directly (through mRNA/eIF3 interaction) or indirectly (by inducing conformational changes in the 40S subunit), or because of the presence of eIF4A, eIF4B, and eIF4F. If mRNA stabilizes the eIF3/40S interaction, then it is important to know the length that is required. We therefore studied factor release from 48S complexes assembled on (CAA)n-AUG-(CAA)m mRNAs, which have unstructured 5′-UTRs and coding regions of different lengths (Fig. 5A). 48S complexes were assembled with Met-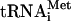 , eIF1, eIF1A, eIF2, eIF3j+, or eIF3j-, but without eIF4A, eIF4B, and eIF4F, because, in contrast to globin mRNA, 48S complexes form on such mRNAs efficiently in their absence (Pestova and Kolupaeva 2002). As with globin mRNA, eIF3 and eIF1 (Fig. 5B,G,H; data not shown) were not released from 40S subunits after eIF5-stimulated hydrolysis of eIF2-bound GTP in 48S complexes assembled on 35nt-AUG-32nt mRNA (which has a 35-nt-long 5′-UTR and a 35-nt-long coding region). Stable association of eIF-3 and eIF1 with 40S subunits after GTP hydrolysis was therefore due to the presence of mRNA and not of eIF4A, eIF4B, and eIF4F in ribosomal complexes. However, eIF3 and eIF1 (Fig. 5C,H; data not shown) were released from 48S complexes assembled on 20nt-AUG-17nt mRNA in which the 5′-UTR and coding region were both shortened from 35 to 20 nucleotides (nt) in length. eIF1 and eIF3 were not released from a 55-nt-long mRNA with a 35-nt 5′-UTR in which only the coding region had been truncated and were released partially (∼60%) from a 55-nt-long mRNA with a 35-nt coding region and a 5′-UTR of 20 nt (Fig. 5D,E,G,H; data not shown). The eIF5-induced dissociation of eIF1, eIF3j+, and eIF3j- from 48S complexes during centrifugation therefore depended particularly strongly on the length of the 5′-UTR of mRNAs, so that a 35-nt-long 5′-UTR was sufficient to promote retention of eIF3 and eIF1 on 40S subunits whereas a 5′-UTR of 20 nt was not. In all cases treatment of 48S complexes with eIF5 led to the expected release of eIF2 (Fig. 5G,H). As with 48S complexes assembled on AUG triplets and globin mRNA, inclusion of ΔeIF5B with eIF5 did not affect factor release (Fig. 5B,C,H; data not shown).
, eIF1, eIF1A, eIF2, eIF3j+, or eIF3j-, but without eIF4A, eIF4B, and eIF4F, because, in contrast to globin mRNA, 48S complexes form on such mRNAs efficiently in their absence (Pestova and Kolupaeva 2002). As with globin mRNA, eIF3 and eIF1 (Fig. 5B,G,H; data not shown) were not released from 40S subunits after eIF5-stimulated hydrolysis of eIF2-bound GTP in 48S complexes assembled on 35nt-AUG-32nt mRNA (which has a 35-nt-long 5′-UTR and a 35-nt-long coding region). Stable association of eIF-3 and eIF1 with 40S subunits after GTP hydrolysis was therefore due to the presence of mRNA and not of eIF4A, eIF4B, and eIF4F in ribosomal complexes. However, eIF3 and eIF1 (Fig. 5C,H; data not shown) were released from 48S complexes assembled on 20nt-AUG-17nt mRNA in which the 5′-UTR and coding region were both shortened from 35 to 20 nucleotides (nt) in length. eIF1 and eIF3 were not released from a 55-nt-long mRNA with a 35-nt 5′-UTR in which only the coding region had been truncated and were released partially (∼60%) from a 55-nt-long mRNA with a 35-nt coding region and a 5′-UTR of 20 nt (Fig. 5D,E,G,H; data not shown). The eIF5-induced dissociation of eIF1, eIF3j+, and eIF3j- from 48S complexes during centrifugation therefore depended particularly strongly on the length of the 5′-UTR of mRNAs, so that a 35-nt-long 5′-UTR was sufficient to promote retention of eIF3 and eIF1 on 40S subunits whereas a 5′-UTR of 20 nt was not. In all cases treatment of 48S complexes with eIF5 led to the expected release of eIF2 (Fig. 5G,H). As with 48S complexes assembled on AUG triplets and globin mRNA, inclusion of ΔeIF5B with eIF5 did not affect factor release (Fig. 5B,C,H; data not shown).
Figure 5.
Association of eIF3 with 40S subunits before and after hydrolysis of eIF2-bound GTP in 48S complexes assembled on mRNAs with different lengths. (A) mRNA sequences. (B-F) Release of 32P-eIF3j- and 32P-eIF3j+ from 48S complexes assembled with eIF1, eIF1A, eIF2, and eIF3 on 35nt-AUG-32nt (B), 20nt-AUG-17nt (C), 35nt-AUG-17nt (D), 20nt-AUG-32nt (E), and (U5)-30nt-AUG-28nt (F) mRNAs after incubation with eIF5 or eIF5 and ΔeIF5B, as indicated, and separated by sucrose density gradient centrifugation, followed by Cerenkov counting and autoradiography after gel electrophoresis of peak fractions (inset panels). (G,H) Association of eIF1, eIF2, and eIF3j- with 40S subunits before and after treatment with eIF5 and ΔeIF5B of 48S complexes assembled on different mRNAs (as indicated), analyzed by fluorescent SYPRO staining (G) or immunoblotting (H), after gel electrophoresis of peak fractions obtained after sucrose density gradient centrifugation of ribosomal complexes. eIF2 and eIF3 subunits, eIF1, and ribosomal proteins are indicated.
mRNA release from 48S complexes assembled on mRNAs with 5′-UTRs and coding regions of different lengths A significant observation is that the ability of different mRNAs to promote the continued stable binding of eIF3 and eIF1 with 40S subunits after hydrolysis of eIF2-bound GTP inversely correlated with dissociation of those mRNAs from 48S complexes. Data are presented only for 48S complexes assembled with eIF3j-. A 40-nt-long mRNA was released from 48S complexes after incubation with eIF5, whereas a 55-nt-long mRNA with a 35-nt coding region was only partially released, and 55-nt- and 70-nt-long mRNAs with extended 5′-UTRs remained bound to 40S subunits (Fig. 6A-D). eIF3 therefore stabilizes mRNA/40S subunit association after hydrolysis of eIF2-bound GTP. Inclusion of ΔeIF5B with eIF5 did not influence the loss of mRNA from initiation complexes, and their inclusion with 60S subunits led to formation of 80S ribosomes on all mRNAs (Fig. 6A,C; data not shown). Although hydrolysis of eIF2-bound GTP led to release of eIF3 from 48S complexes assembled on the 40-nt-long mRNA, eIF5 alone was not sufficient to promote subunit joining on this mRNA and eIF5B was absolutely required (Fig. 6A).
Figure 6.
Association of mRNA with 40S subunits and 80S ribosomes before and after treatment with eIF5, ΔeIF5B, and 60S subunits of 48S complexes assembled on 20nt-AUG-17nt (A), 20nt-AUG-32nt (B), 35nt-AUG-32nt (C), 35nt-AUG-17nt (D), and (U5)-30nt-AUG-28nt (E) mRNAs, as indicated. Association of 32P-mRNA with ribosomal complexes was assayed by Cerenkov counting after sucrose density gradient centrifugation. (F) 80S complex formation after treatment with eIF5, ΔeIF5B, and 60S subunits (as indicated) of 48S complexes assembled on 20nt-AUG-17nt mRNA. Ribosomal complexes were separated by sucrose density gradient centrifugation and assayed by measuring optical density. Ribosomal complexes are indicated above appropriate peaks. Upper fractions from gradients have been omitted for clarity. (G) UV cross-linking of 32P-(U5)-30nt-AUG-28nt mRNA to components of sucrose density gradient-purified 48S complexes before and after induction of hydrolysis of eIF2-bound GTP and in binary complexes with eIF3j- and eIF3j+. UV cross-linked eIF3 subunits and ribosomal proteins are indicated.
Although it is possible that hydrolysis of eIF2-bound GTP causes dissociation of eIF3 and the 40-nt-long mRNA in solution prior to centrifugation, the trailing of eIF3 in this case (Fig. 5C) and the slight trailing observed with the 55-nt mRNA with a 35-nt coding region (Fig. 5E) suggest that eIF3's dissociation could occur mainly during centrifugation. If 48S complexes formed on the 40-nt-long mRNA dissociated before centrifugation, then inclusion of 60S subunits in reaction mixtures with eIF5 would lead to formation of empty 80S ribosomes. However, only very few 80S ribosomes were formed in this case after incubation of 48S complexes with eIF5 and 60S subunits compared to reaction mixtures that also contained ΔeIF5B (Fig. 6F), which indicates that most 40S subunits were still engaged in initiation complexes, and that dissociation of 48S complexes assembled on the 40-nt mRNA after GTP hydrolysis therefore occurred mostly during centrifugation. Although this mRNA was not long enough to impart stability to the eIF3/40S interaction during centrifugation, association of eIF3 with 40S subunits was most likely stabilized enough to prevent subunit joining in solution.
eIF3/mRNA interaction in 48S complexes before and after hydrolysis of eIF2-bound GTP Data presented above indicate that the mRNA in 48S complexes is responsible for eIF3's stable association with 40S subunits after hydrolysis of eIF2-bound GTP. Before considering the implications of this observation, it is necessary to confirm that eIF3 occupies the same position on 40S subunits after GTP hydrolysis as before and did not simply attach randomly after GTP hydrolysis to exposed regions of mRNA that are not bound by the 40S subunit. We recently found (Kolupaeva et al. 2005) that eIF3 can be specifically cross-linked in 48S complexes to (U5)-30nt-AUG-28nt mRNA which contains a 30-nt-long 5′-UTR and U residues at positions -14, -8, -4, +5, and +11 relative to the A (+1) of the AUG triplet (Fig. 5A). This mRNA was long enough to ensure stable association of eIF3 and itself with 40S subunits after hydrolysis of eIF2-bound GTP (Figs. 5F, 6E). To investigate eIF3's interaction with this mRNA in 48S complexes, it was transcribed in vitro in the presence of 32P-ATP and 4-thioUTP, which can be activated by low energy radiation, yielding specific “zero-length” cross-links that represent direct contacts of mRNA with ribosomal components. In 48S complexes, all 4-thiouridines of this mRNA can be specifically cross-linked to several ribosomal proteins and to eIF3a, eIF3b, and eIF3d with distinct efficiencies (Fig. 6G, lane 1; Kolupaeva et al. 2005). The cross-linking patterns for 48S complexes assembled with eIF3j+ or eIF3j- did not differ (data not shown). Hydrolysis of eIF2-bound GTP did not alter the cross-linking patterns of mRNA with ribosomal proteins or eIF3 in 48S complexes (Fig. 6G, lane 2) and therefore did not dramatically alter the position of mRNA on 40S subunits or its interaction with eIF3. Thus, eIF3's position on the 40S subunit is not changed by GTP hydrolysis. The cross-linking pattern of eIF3 with mRNA in 48S complexes and in eIF3/RNA binary complexes differ (Fig. 6G, cf. lanes 1,2 and 3,4). Thus eIF3b and eIF3d are cross-linked to mRNA more efficiently in the latter, and cross-linking of mRNA to eIF3e/f, eIF3g/h and eIF3l occurred only in binary complexes. UV cross-linking of eIF3j+ and eIF3j- did not differ and the eIF3j subunit was not cross-linked to mRNA in binary complexes. We did not investigate the cross-linking of eIF3 to mRNA from -30 to -20 nt and from +20 to +35 nt, which are important for stabilizing the eIF3/40S subunit interaction, so a firm conclusion about the mechanism of stabilization by mRNA cannot yet be made.
Discussion
Hydrolysis of eIF2-bound GTP
Although eIF5 binds directly to eIF2 ternary complex off the 40S subunit, this interaction is not sufficient to induce eIF2's GTPase activity. It is also widely accepted that eIF5-induced hydrolysis of eIF2-bound GTP is stimulated by codon-anticodon base-pairing (Raychaudhuri et al. 1985). However, we report here that in the absence of eIF1, eIF5 induces rapid hydrolysis of eIF2-bound GTP in 43S complexes (i.e., without codon-anticodon base-pairing) and that base-pairing in 48S complexes assembled without eIF1 did not stimulate GTP hydrolysis. The presence of eIF1 in 43S complexes reduced the rate of eIF5-induced GTP hydrolysis compared to that in 43S/48S complexes assembled without eIF1. Establishment of codon-anticodon base-pairing in 48S complexes assembled with eIF1 relieved its inhibition, and hydrolysis of eIF2-bound GTP in 48S complexes assembled with eIF1 occurred at the same rate as in 43S/48S complexes assembled without it. In yeast, premature hydrolysis of eIF2-bound GTP before 43S complexes reach the initiation codon led to initiation at inappropriate sites (Huang et al. 1997). Therefore, in addition to its role in initiation codon selection at the stage of 48S complex formation (Pestova and Kolupaeva 2002), eIF1 also participates in ensuring the fidelity of initiation at the later stage of ribosomal subunit joining by inhibiting premature GTP hydrolysis and by linking establishment of codon-anticodon base-pairing with hydrolysis of eIF2-bound GTP. The fact that in the presence of eIF1, CAA-Stem-GUS mRNA even inhibited hydrolysis of eIF2-bound GTP compared to hydrolysis in free 43S complexes suggests that when eIF1 is present, GTP hydrolysis in scanning 43S complexes occurs even more slowly than when they are free. The presence or absence of eIF4A, eIF4B, eIF4F, eIF1A, and eIF5B did not affect eIF1's ability to link codon-anticodon base-pairing with hydrolysis of eIF2-bound GTP.
eIF1 increases the scanning processivity of 43S complexes and plays a key role in initiation codon selection, for example by enabling 43S complexes to reject codon-anticodon mismatches (Yoon and Donahue 1992; Pestova and Kolupaeva 2002). eIF1's binding site on the interface surface of the 40S subunit platform (Lomakin et al. 2003) suggests that it most likely plays its monitoring function indirectly, by influencing the conformation of the 40S subunit platform and the positions of mRNA and Met-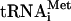 in ribosomal complexes. The conformation of 43S complexes bound by eIF1 is therefore optimal for scanning and for rejection of codon-anticodon mismatches but not for eIF5-induced hydrolysis of eIF2-bound GTP. eIF1 therefore acts as a negative regulator, in this instance of hydrolysis of eIF2-bound GTP, just as it does when monitoring initiation codon selection. Recognition of the initiation codon by a 43S complex containing eIF1 may restore the structural requirements within the complex for efficient hydrolysis to occur. When discussing the potential mechanism of action of eIF1 in initiation codon selection, we suggested that codon-anticodon base-pairing may lead to dissociation of eIF1 from ribosomal complexes or at least in weakening of its binding to the 40S subunit (Lomakin et al. 2003). Dissociation of eIF1 from ribosomal complexes upon establishment of codon-anticodon base-pairing would explain the relief of eIF1's inhibition of GTP hydrolysis after initiation codon recognition. In fact, association of eIF1 with 48S complexes was apparent even after sucrose density gradient centrifugation (Figs. 3E, 4E). This may mean that eIF1 did not dissociate from its site on the 40S subunit after codon-anticodon base-pairing, but despite being bound to eIF1, ribosomal complexes could nevertheless undergo the conformational changes that are required for fast GTP hydrolysis, or alternatively, that eIF1 was ejected from its position on the 40S subunit but remained associated with ribosomal complexes through its interaction with eIF3. The molecular basis for stimulation of eIF5-induced hydrolysis of eIF2-bound GTP by the 40S subunit and by codon-anticodon base-pairing remains to be elucidated. The 40S subunit together with factors might induce essential conformational changes in eIF2 and/or change the relative orientation of eIF2 and eIF5, so that eIF5 becomes able to activate the catalytic center of eIF2.
in ribosomal complexes. The conformation of 43S complexes bound by eIF1 is therefore optimal for scanning and for rejection of codon-anticodon mismatches but not for eIF5-induced hydrolysis of eIF2-bound GTP. eIF1 therefore acts as a negative regulator, in this instance of hydrolysis of eIF2-bound GTP, just as it does when monitoring initiation codon selection. Recognition of the initiation codon by a 43S complex containing eIF1 may restore the structural requirements within the complex for efficient hydrolysis to occur. When discussing the potential mechanism of action of eIF1 in initiation codon selection, we suggested that codon-anticodon base-pairing may lead to dissociation of eIF1 from ribosomal complexes or at least in weakening of its binding to the 40S subunit (Lomakin et al. 2003). Dissociation of eIF1 from ribosomal complexes upon establishment of codon-anticodon base-pairing would explain the relief of eIF1's inhibition of GTP hydrolysis after initiation codon recognition. In fact, association of eIF1 with 48S complexes was apparent even after sucrose density gradient centrifugation (Figs. 3E, 4E). This may mean that eIF1 did not dissociate from its site on the 40S subunit after codon-anticodon base-pairing, but despite being bound to eIF1, ribosomal complexes could nevertheless undergo the conformational changes that are required for fast GTP hydrolysis, or alternatively, that eIF1 was ejected from its position on the 40S subunit but remained associated with ribosomal complexes through its interaction with eIF3. The molecular basis for stimulation of eIF5-induced hydrolysis of eIF2-bound GTP by the 40S subunit and by codon-anticodon base-pairing remains to be elucidated. The 40S subunit together with factors might induce essential conformational changes in eIF2 and/or change the relative orientation of eIF2 and eIF5, so that eIF5 becomes able to activate the catalytic center of eIF2.
Factor release from 48S complexes upon hydrolysis of eIF2-bound GTP
The 40S subunit in naturally occurring 48S complexes is, at a minimum, associated with eIF1, eIF1A, eIF2, and eIF3, which must be displaced to allow subunit joining. eIF5-induced hydrolysis of eIF2-bound GTP is not sufficient to promote joining of 60S subunits to mRNA-containing 48S complexes, and this process also requires eIF5B.
The interactions of eIF3 and to a lesser extent of eIF1A with the 40S subunit are thought to account for these factors' ribosomal anti-association activities (Trachsel and Staehelin 1979; Goumans et al. 1980). eIF3's ability to bind 40S subunits in the absence of eIF2 ternary complex was recently reported to depend on its weakly bound eIF3j subunit (Fraser et al. 2004). We found that despite their different ribosome binding activities, eIF3j+ and eIF3j- were equally active in 43S, 48S, and 80S complex formation. Significantly, eIF3j was present in binary eIF3/40S subunit complexes and in 43S complexes assembled with eIF3j+, but was absent from 48S complexes. eIF3j's association with eIF3 was therefore weakened by the presence of mRNA on the 40S subunit. Although eIF3j is not essential for initiation in vitro, it might be involved in the initial docking of eIF3 to the 40S subunit, which may enhance initiation in vivo, as has been reported for yeast eIF3j (Valasek et al. 2001). The shuttling of eIF3j on and off ribosomal complexes reported here might be an important dynamic change in eIF3's composition during initiation.
Contrary to the previously reported ribosomal anti-association activity of eIF3, recent data (Chaudhuri et al. 1999; Kolupaeva et al. 2005) showed that in the absence of eIF2 ternary complex, neither eIF3j- nor eIF3j+ alone or in combination with eIF1 and eIF1A have such activity. However, it is very significant that, in the absence of eIF2 ternary complexes, >25-nt-long ssRNA promotes stable association of eIF3 in eIF3/RNA/40S subunit complexes with RNA most likely bound to the mRNA-binding cleft of the 40S subunit, that can no longer bind to 60S subunits (Kolupaeva et al. 2005). Hydrolysis of eIF2-bound GTP and release of eIF2 from 48S complexes assembled on mRNA may therefore not necessarily lead to release of eIF3, and its presence on the 40S subunit in this case would likely prevent subunit joining.
We found that hydrolysis of eIF2-bound GTP and release of eIF2 led to release of eIF3 from 48S complexes assembled on AUG triplets, as previously reported (Peterson et al. 1979). However, contrary to other reports (Benne and Hershey 1978; Trachsel and Staehelin 1979) but consistent with our hypothesis, eIF3 (and eIF1) remained stably bound to 40S subunits after hydrolysis of eIF2-bound GTP and release of eIF2 from 48S complexes assembled on β-globin mRNA. The presence/absence of the eIF3j subunit and inclusion of eIF5B with eIF5 did not influence factor release. The lability of eIF1A's binding to 48S complexes even before hydrolysis of eIF2-bound GTP (Thomas et al. 1980; Pestova et al. 1998) did not allow us to determine the stage at which it is released. The exact reason for the discrepancy between our data and previous reports concerning factor release from 48S complexes assembled on mRNA is not obvious, but might be due to differences in the efficiency of 48S complex formation. Earlier studies were done without eIF4F, so if 48S complex formation were consequently inefficient, then the ribosomal peak in sucrose density gradients would contain mostly 43S rather than 48S complexes, which are difficult to resolve by this method. Release of eIF3 from 43S complexes could thus be mistaken for its release from 48S complexes assembled on native mRNA.
The stabilizing effect of mRNA on binding of eIF3 and eIF1 to the 40S subunit depended on its length, and in particular, on the length of its 5′-UTR. Thus eIF3 and eIF1 were released from 48S complexes assembled on mRNA with 20-nt-long 5′-UTR and coding regions after hydrolysis of eIF2-bound GTP, during sucrose density gradient centrifugation. They were not released or were partially released from 48S complexes assembled on mRNAs with an extended (30-nt) 5′-UTR or (35-nt) coding region, respectively. It is important that the release of eIF3 and eIF1 correlated with the release of mRNA from 48S complexes. Thus, like eIF3 and eIF1, 40-nt mRNA dissociated from 48S complexes whereas mRNA with an extended 5′-UTR yielded 48S complexes from which neither eIF3, eIF1, nor mRNA dissociated after GTP hydrolysis. However, the fact that only very few 40S subunits were able to form empty 80S ribosomes after incubating eIF5 and 60S subunits with 48S complexes assembled on the 40-nt mRNA and that in this case eIF3 was trailing in sucrose density gradients indicate that even in this case, prior to centrifugation, eIF3 and this mRNA remain stably associated with 40S subunits after hydrolysis of eIF2-bound GTP. We suggest that the mutual stabilization by eIF3 and mRNA of their association with 40S subunits after GTP hydrolysis could explain the requirement for eIF5B in subunit joining. The lower complexity of eIF3 in yeast and the smaller number of RNA-binding subunits in it may account for the fact that in yeast, eIF5B is not essential (Choi et al. 1998). This mutual stabilization may also play an important role in maintaining the integrity of initiation complexes after hydrolysis of eIF2-bound GTP and release of eIF2, prior to subunit joining. The fact that in the absence of 60S subunits, eIF5B did not influence factor release suggests that eIF3 and eIF1 are released not before but during the subunit joining event itself.
UV cross-linking experiments also showed that eIF3 interacts with mRNA in a similar way and occupies the same position on the 40S subunit in 48S complexes before and after GTP hydrolysis. Investigation of interactions of mRNA from nucleotides -30 to -20 and nucleotides +20 to +35 with 40S subunit and eIF3 components of the 48S complex is required to elucidate the mechanism by which mRNA stabilizes the eIF3/40S subunit interaction. It could involve direct simultaneous binding of mRNA to the 40S subunit and eIF3, and/or induction of conformational change in the 40S subunit. However, accumulating data suggesting that eIF3 binds to the solvent side of the 40S subunit (Srivastava et al. 1992; Valasek et al. 2003) suggest that the part of mRNA outside the mRNA-binding cleft on the 40S subunit interface surface may participate directly in binding to eIF3 in 48S complexes. As suggested above, the mutual stabilization by eIF3 and mRNA of their association with the 40S subunit may be particularly important following eIF5-induced hydrolysis of eIF2-bound GTP and release of eIF2-GDP from the 48S complex, to maintain the association of mRNA and initiator tRNA with the initiation complex prior to subunit joining.
A model for factor release during subunit joining
Data presented here indicate that eIF5-induced hydrolysis of eIF2-bound GTP and eIF5B are not sufficient to dissociate eIF3 and eIF1 from 48S complexes. Although our present techniques did not allow us to investigate the release of eIF1A, recent genetic data in yeast suggest that eIF1A could be released together with eIF5B after 80S ribosome assembly (Olson et al. 2003). We therefore propose a model for factor release from 48S complexes (Fig. 7) in which eIF5-induced hydrolysis of eIF2-bound GTP releases only eIF2-GDP, whereas eIF3, eIF1, and likely eIF1A are displaced during the subsequent subunit-joining step mediated by eIF5B. Subunit joining in eukaryotes is thus in many ways similar to this process in prokaryotes (Fig. 7). Eukaryotic eIF1A and eIF5B are orthologs of prokaryotic initiation factors IF1 and IF2, respectively. It is thus probable that like IF1 (Carter et al. 2001), eIF1A also binds to the ribosomal A site. We previously reported that eIF1 is a functional homologue of the C-terminal domain (CTD) of IF3: Both ensure the fidelity of initiation codon selection by dissociating pseudo-initiation complexes assembled on non-AUG triplets (Petrelli et al. 2001; Pestova and Kolupaeva 2002). Moreover, eIF1 binds near the P site in an analogous region to the IF3-CTD-binding site on the 30S subunit (Dallas and Noller 2001; Lomakin et al. 2003). The eIF3c subunit binds to eIF1 (Asano et al. 1998; Fletcher et al. 1999) but eIF3's exact position on the 40S subunit is not known. The disposition of initiator tRNA and initiation factors on the interface surface of the small subunit before subunit joining may therefore be similar in prokaryotes and eukaryotes. In prokaryotes, IF2 first promotes binding of initiator tRNA to the 30S subunit and then promotes joining of a 50S subunit to the 30S subunit/IF1/IF2/IF3 complex; all three factors are released during or after the actual joining event (Gualerzi et al. 2001). There are therefore striking parallels between subunit joining in eukaryotes (in which hydrolysis of eIF2-bound GTP and release of eIF2-GDP are followed by eIF5B-mediated joining of a 60S subunit to a 40S subunit bound by Met-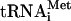 eIF1, eIF3, and probably eIF1A) and in prokaryotes (in which IF2 is thought to enhance joining of a 50S subunit to a 30S subunit bound by fMet-tRNA and IF1, IF2, and IF3).
eIF1, eIF3, and probably eIF1A) and in prokaryotes (in which IF2 is thought to enhance joining of a 50S subunit to a 30S subunit bound by fMet-tRNA and IF1, IF2, and IF3).
Figure 7.
Comparative ribosomal subunit joining model for prokaryotes (A) and eukaryotes (B).
Materials and methods
Purification of factors and ribosomal subunits
Ribosomal subunits and initiation factors were purified as described (Pestova et al. 1996; Pestova et al. 1998; Pestova et al. 2000). Recombinant His-tagged eIF3j (rec-eIF3j) was expressed in Escherichia coli and purified by chromatography on Ni2+-NTA (Qiagen) and Mono Q (Amersham Pharmacia).
eIF3j- was purified from a 0%-40% (NH4)2SO4 precipitation fraction of the 0.5 M-KCl wash of HeLa ribosomes by chromatography on DEAE and phosphocellulose (Pestova et al. 1996). The fraction containing eIF3 was centrifuged through a 10%-30% sucrose density gradient in buffer A (20 mM Tris at pH 7.5, 0.1 mM EDTA, 2 mM DTT) plus 0.4 M KCl in a SW41 rotor for 22 h at 40,000 r.p.m. Fractions containing eIF3 were purified on Mono Q from which eIF3 eluted at ∼430 mM KCl. Purified eIF3j- contained all subunits except eIF3j, which dissociated during sucrose density gradient centrifugation. eIF3j+ was obtained when this step was omitted. The subunit composition of eIF3 was analyzed by electrophoresis in 4%-12% NuPAGE Bis-Tris gel (Invitrogen). The nature of eIF3 subunits was confirmed by LC-nanospray tandem mass spectrometry of peptides derived by in-gel tryptic digestion at an in-house facility. eIF3j+ containing rec-eIF3j was reconstituted from eIF3j- by incubating with a five times molar excess of rec-eIF3j for 15 min at 37°C in buffer A plus 100 mM KCl, followed by purification from un-incorporated rec-eIF3j on Superdex 75 (Amersham Pharmacia).
eIF3j+, eIF3j- and rec-eIF3j were phosphorylated (specific activity [spec. act.] of 400,000, 180,000, and 30,000 cpm/pmol, respectively) using the catalytic subunit of cAMP-dependent protein kinase (New England BioLabs), followed by purification on Mono Q.
Synthetic and in vitro transcribed mRNAs
The plasmid for CAA-stem-GUS mRNA was derived from the CAA-GUS transcription vector (Pestova and Kolupaeva 2002) using polymerase chain reactions. The (U5)-30nt-AUG-28nt transcription vector was made by inserting annealed oligo-nucleotides into BamHI-HindIII sites of pBR322. All mRNAs were transcribed using T7 RNA polymerase. For UV cross-linking experiments (U5)-30nt-AUG-28nt mRNA (spec. act. 50,000 c.p.m./pmol) was transcribed in the presence of 4thio-UTP and α32P-ATP (111 Tbq/mmol). HPLC-purified AUG, UUG, or CAA triplets and PAGE-purified synthetic 5′-20nt-AUG-17nt, 5′-20nt-AUG-32nt, 5′-35nt-AUG-17nt, and 5′-35nt-AUG-32nt mRNAs were from Dharmacon. For sucrose density gradient experiments these and (U5)-30nt-AUG-28nt mRNAs were labeled with T4 polynucleotide kinase and γ32P-ATP (259 Tbq/mmol) (spec. act. 400,000 c.p.m./pmol mRNA).
Preparation of aminoacylated transcript and native 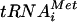
In vitro transcribed 35S-Met-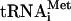 (100,000 c.p.m./pmol) was synthesized, purified, and aminoacylated as described (Pestova and Hellen 2001). Native aminoacylated [35S]-Met-
(100,000 c.p.m./pmol) was synthesized, purified, and aminoacylated as described (Pestova and Hellen 2001). Native aminoacylated [35S]-Met-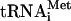 (100,000 c.p.m./pmol) was purified from total rabbit tRNA (Novagen) by a combination of gel-filtration on Superdex 75 and reverse phase chromatography on a Waters 3.9 × 300 mm Delta Pak C4 column equilibrated in buffer I (20 mM NH4 Ac at pH 5.5, 10 mM Mg Ac, 400 mM NaCl), which was then subjected to a 0%-30% gradient of buffers I and II (buffer I with 60% methanol) (Cayama et al. 2000). The peak of [35S]-Met-
(100,000 c.p.m./pmol) was purified from total rabbit tRNA (Novagen) by a combination of gel-filtration on Superdex 75 and reverse phase chromatography on a Waters 3.9 × 300 mm Delta Pak C4 column equilibrated in buffer I (20 mM NH4 Ac at pH 5.5, 10 mM Mg Ac, 400 mM NaCl), which was then subjected to a 0%-30% gradient of buffers I and II (buffer I with 60% methanol) (Cayama et al. 2000). The peak of [35S]-Met-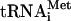 eluted at 6.5% buffer II.
eluted at 6.5% buffer II.
Activities of eIF3j+ and eIF3j-
eIF3/40S subunit complexes were assembled by incubating 30 pmol 40S subunit with 100 pmol unlabeled or 32P-phosphorylated eIF3j+, eIF3j-, or eIF3j+ containing rec-eIF3j with/without 150 pmol eIF1 and eIF1A, or 20 μg polyU RNA in 200 μL buffer B (20 mM Tris at pH 7.5, 100 mM KAc, 2 mM DTT, 2.5 mM MgCl2, 0.5 mM spermidine) for 10 min at 37°C.
Rec-eIF3j/40S complexes were formed by incubating 30 pmol 40S subunits and 200 pmol 32P-Rec-eIF3j in 200 μL buffer B. 43S complexes were assembled by incubating 30 pmol 40S subunits, 100 pmol unlabeled or 32P-eIF3j+ or eIF3j-, 50 pmol [35S]-Met-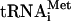 , 100 pmol eIF2, 150 pmol eIF1, and eIF1A in 200 μL buffer B plus 0.4 mM GTP. 48S complexes were assembled by incubating 30 pmol 40S subunits, 100 pmol 32P-eIF3j+, 50 pmol [35S]-Met-
, 100 pmol eIF2, 150 pmol eIF1, and eIF1A in 200 μL buffer B plus 0.4 mM GTP. 48S complexes were assembled by incubating 30 pmol 40S subunits, 100 pmol 32P-eIF3j+, 50 pmol [35S]-Met-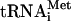 , 100 pmol eIF2, 150 pmol eIF1, and eIF1A and either 1 nM AUG triplet or 50 pmol CAA-GUS mRNA in 200 μL buffer B plus 0.4 mM GTP. Ribosomal complexes were purified by centrifugation through 10%-30% sucrose density gradients in a SW55 rotor at 50,000 rpm for 95 min. Optical density of fractionated gradients was measured at 260 nm and association of 32P-eIF3 and [35S]-Met-
, 100 pmol eIF2, 150 pmol eIF1, and eIF1A and either 1 nM AUG triplet or 50 pmol CAA-GUS mRNA in 200 μL buffer B plus 0.4 mM GTP. Ribosomal complexes were purified by centrifugation through 10%-30% sucrose density gradients in a SW55 rotor at 50,000 rpm for 95 min. Optical density of fractionated gradients was measured at 260 nm and association of 32P-eIF3 and [35S]-Met-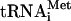 with 40S subunits was assayed by Cerenkov or scinitillation counting of an aliquot of each fraction. Peak fractions were applied to NuPAGE 4%-12% Bis-Tris-Gel and autoradiographed or stained with fluorescent SYPRO protein stain (Molecular Probes).
with 40S subunits was assayed by Cerenkov or scinitillation counting of an aliquot of each fraction. Peak fractions were applied to NuPAGE 4%-12% Bis-Tris-Gel and autoradiographed or stained with fluorescent SYPRO protein stain (Molecular Probes).
GTP hydrolysis assays
Ternary eIF2/GTP/Met-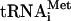 complexes (spec. act. 150,000 c.p.m./pmol) containing native or transcribed Met-
complexes (spec. act. 150,000 c.p.m./pmol) containing native or transcribed Met-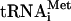 and γ32P-GTP (74 Tbq/mmol) were prepared and purified (Pestova and Hellen 2001). 43S complexes (spec. act. 150,000 c.p.m./pmol) were assembled from 150 pmol 40S subunits, 200 pmol eIF2 ternary complex, 300 pmol eIF3 with or without 500 pmol eIF1A by incubation in buffer B for 10 min at 37°C and purified by centrifugation through 10%-30% sucrose density gradients in buffer B.
and γ32P-GTP (74 Tbq/mmol) were prepared and purified (Pestova and Hellen 2001). 43S complexes (spec. act. 150,000 c.p.m./pmol) were assembled from 150 pmol 40S subunits, 200 pmol eIF2 ternary complex, 300 pmol eIF3 with or without 500 pmol eIF1A by incubation in buffer B for 10 min at 37°C and purified by centrifugation through 10%-30% sucrose density gradients in buffer B.
43S/48S complexes were assembled by incubating multiples of 30 μL reaction mixtures containing 0.8 pmol purified 43S complex with different combinations of 0.5 nmol AUG, CAA, or UUG triplet, 3 pmol CAA-GUS or CAA-stem-GUS mRNA, 10 pmol eIF1A, 10 pmol eIF4A, 5 pmol eIF4B, 2 pmol eIF4F, and 10 pmol eIF1 in buffer B plus 1 mM ATP for 5 min at 37°C. Hydrolysis of eIF2-bound GTP was initiated by adding 3 pmol eIF5 per reaction. Aliquots (30 μL) were removed at indicated time points and assayed for 32P-Pi release (Pestova et al. 2000).
Analysis of factor and mRNA release from 43S/48S complexes
48S complexes were assembled by incubating 30 pmol 40S subunits, 50 pmol [35S]-Met-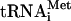 , 100 pmol eIF2, 50 pmol unlabeled or 32P-phosphorylated eIF3j+ or eIF3j-, 150 pmol eIF1, and eIF1A with different combinations of 50 pmol eIF4F, 100 pmol eIF4A, 50 pmol eIF4B, 1 nm AUG triplet, and 50 pmol unlabeled or 32P-labeled mRNA (as indicated in the text) in 200 μL buffer B plus 1 mM ATP + 0.4 mM unlabeled or γ[32P]GTP for 10 min at 37°C. Hydrolysis of eIF2-GTP was induced by adding 200 pmol eIF5 and incubating for 20 min more. For 80S complex formation, 50 pmol 60S subunits and 70 pmol ΔeIF5B were added with eIF5. Ribosomal complexes were purified by centrifugation through 10%-30% sucrose density gradients in a SW55 rotor at 50,000 rpm for 95 min. The optical density of fractionated gradients was measured at 260 nm and association of 32P-eIF3, 32P-mRNA, and 35S-Met-
, 100 pmol eIF2, 50 pmol unlabeled or 32P-phosphorylated eIF3j+ or eIF3j-, 150 pmol eIF1, and eIF1A with different combinations of 50 pmol eIF4F, 100 pmol eIF4A, 50 pmol eIF4B, 1 nm AUG triplet, and 50 pmol unlabeled or 32P-labeled mRNA (as indicated in the text) in 200 μL buffer B plus 1 mM ATP + 0.4 mM unlabeled or γ[32P]GTP for 10 min at 37°C. Hydrolysis of eIF2-GTP was induced by adding 200 pmol eIF5 and incubating for 20 min more. For 80S complex formation, 50 pmol 60S subunits and 70 pmol ΔeIF5B were added with eIF5. Ribosomal complexes were purified by centrifugation through 10%-30% sucrose density gradients in a SW55 rotor at 50,000 rpm for 95 min. The optical density of fractionated gradients was measured at 260 nm and association of 32P-eIF3, 32P-mRNA, and 35S-Met-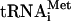 with 40S subunits was assayed by Cerenkov or scinitillation counting of an aliquot of each fraction. Peak fractions were applied to NuPAGE 4%-12% Bis-Tris-Gel and autoradiographed or stained with SYPRO protein stain. Association of eIF1, eIF2, and eIF3 with 40S subunits was confirmed by immunoblotting (see Supplemental Material).
with 40S subunits was assayed by Cerenkov or scinitillation counting of an aliquot of each fraction. Peak fractions were applied to NuPAGE 4%-12% Bis-Tris-Gel and autoradiographed or stained with SYPRO protein stain. Association of eIF1, eIF2, and eIF3 with 40S subunits was confirmed by immunoblotting (see Supplemental Material).
Analysis of 48S/80S complexes by primer extension
48S/80S complexes assembled on β-globin mRNA were purified by sucrose density gradient centrifugation and their position was determined by primer extension (Pestova et al. 1998).
UV cross-linking
48S complexes assembled on (U5)-30nt-AUG-28nt mRNA synthesized in the presence of 4-thioUTP and [32P]ATP were incubated with/without eIF5, purified by sucrose density gradient centrifugation, irradiated at 360 nm for 30 min on ice using a UV-Stratalinker (Stratagene), treated with RNase A, and analyzed by autoradiography.
Acknowledgments
We thank A. Hinnebusch, N. Sonenberg, S. Kanner, R. Callahan, T. Tang, and R. Schneider for generously providing reagents (eIF3j expression vector and antibodies against eIF3 and eIF2 subunits). This work was supported by grant R01GM63940 from the NIGMS. A.U. received a DAAD (German Academic Exchange Service) postdoctoral fellowship.
Supplemental material is available at http://www.genesdev.org.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.1255704.
References
- Asano K.S., Phan, L., Anderson, J., and Hinnebusch, A.G. 1998. Complex formation by all five homologues of mammalian translation initiation factor 3 subunits from Saccharomyces cerevisiae. J. Biol. Chem. 273: 18573-18585. [DOI] [PubMed] [Google Scholar]
- Benne R. and Hershey, J.W.B. 1976. Purification and characterization of initiation factor IF-E3 from rabbit reticulocytes. Proc. Natl. Acad. Sci. 73: 3005-3009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ____. 1978. The mechanism of action of protein synthesis initiation factors from rabbit reticulocytes. J. Biol. Chem. 253: 3078-3087. [PubMed] [Google Scholar]
- Carter A.P., Clemons, W.M., Brodersen, D.E., Morgan-Warren, R.J., Hartsch., T., Wimberly, B.T., and Ramakrishnan, V. 2001. Crystal structure of an initiation factor bound to the 30S ribosomal subunit. Science 291: 498-501. [DOI] [PubMed] [Google Scholar]
- Cayama E., Yepez, A., Rotondo, F., Bandeira, E., Ferreras, A.C., and Triana-Alonso, F.J. 2000. New chromatographic and biochemical strategies for quick preparative isolation of tRNA. Nucleic Acids Res. 28: E64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhuri J., Chowdury, D., and Maitra, U. 1999. Distinct function of eukaryotic translation initiation factors eIF1A and eIF3 in the formation of the 40S ribosomal preinitiation complex. J. Biol. Chem. 274: 17975-17980. [DOI] [PubMed] [Google Scholar]
-
Choi S.K., Lee, J.H., Zoll, W.L., Merrick, W.C., and Dever, T.E. 1998. Promotion of Met-
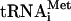 binding to ribosome by yIF2, a bacterial IF2 homolog in yeast. Science 280: 1757-1760. [DOI] [PubMed] [Google Scholar]
binding to ribosome by yIF2, a bacterial IF2 homolog in yeast. Science 280: 1757-1760. [DOI] [PubMed] [Google Scholar] - Dallas A. and Noller, H.F. 2001. Interaction of translation initiation factor 3 with the 30S ribosomal subunit. Mol. Cell 8: 855-864. [DOI] [PubMed] [Google Scholar]
- Das S. and Maitra, U. 2002. Functional significance and mechanism of eIF5-promoted GTP hydrolysis in eukaryotic translation initiation. Prog. Nucl. Acids Res. Mol. Biol. 70: 207-231. [DOI] [PubMed] [Google Scholar]
- Fletcher M., Pestova, T.V., Hellen, C.U.T., and Wagner, G. 1999. Structure and interactions of the translation initiation factor eIF1. EMBO J. 18: 2631-2637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser C.S., Lee, J.Y., Mayeur, G.L., Bushell, M., Doudna, J.A., and Hershey, J.W. 2004. The j-subunit of human translation initiation factor eIF3 is required for the stable binding of eIF3 and its subcomplexes to 40S ribosomal subunits in vitro. J. Biol. Chem. 279: 8946-8956. [DOI] [PubMed] [Google Scholar]
- Goumans H., Thomas, A., Verhoeven, A., Voorma, H.O., and Benne, R. 1980. The role of eIF-4C in protein synthesis initiation complex formation. Biochim. Biophys. Acta 608: 39-46. [DOI] [PubMed] [Google Scholar]
- Gualerzi C.O., Brandi, L., Caserta, E., Garofalo, C., Lammi, M., La Teana, A., Petrelli, D., Spurio, R., Tomsic, J., and Pon, C.L. 2001. Initiation factors in the early events of mRNA translation in bacteria. Cold Spring Harbor Symp. Quant. Biol. 66: 363-376. [DOI] [PubMed] [Google Scholar]
- Huang H.-K., Youn, H., Hannig, E.M., and Donahue, T.F. 1997. GTP hydrolysis controls stringent selection of the AUG start codon during translation initiation in Saccharomyces cerevisiae. Genes & Dev. 11: 2396-2413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolupaeva V.G., Unbehaun, A., Lomakin, I.B., Hellen, C.U.T., and Pestova, T.V. 2005. Binding of eukaryotic initiation factor 3 to ribosomal 40S subunits and its role in ribosomal dissociation and anti-association. RNA (in press). [DOI] [PMC free article] [PubMed]
- Lomakin I.B., Kolupaeva, V.G., Marintchev, A., Wagner, G., and Pestova, T.V. 2003. Position of eukaryotic initiation factor eIF1 on the 40S ribosomal subunit determined by directed hydroxyl radical probing. Genes & Dev. 17: 2786-2797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maag D. and Lorsch, J.R. 2003. Communication between eukaryotic translation initiation factors 1 and 1A on the yeast small ribosomal subunit. J. Mol. Biol. 330: 917-924. [DOI] [PubMed] [Google Scholar]
- Olsen D.S., Savner, E.M., Mathew, A., Zhang, F., Krishnamoorthy, T., Phan, L., and Hinnebusch, A.G. 2003. Domains of eIF1A that mediate binding to eIF2, eIF3 and eIF5B and promote ternary complex recruitment in vivo. EMBO J. 22: 193-204. [DOI] [PMC free article] [PubMed] [Google Scholar]
-
Pestova T.V. and Hellen, C.U.T. 2001. Preparation and activity of synthetic unmodified mammalian
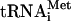 in initiation of translation in vitro. RNA 7: 1496-1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
in initiation of translation in vitro. RNA 7: 1496-1505. [DOI] [PMC free article] [PubMed] [Google Scholar] - Pestova T.V. and Kolupaeva, V.G. 2002. The roles of individual eukaryotic translation initiation factors in ribosomal scanning and initiation codon selection. Genes & Dev. 16: 2906-2922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pestova T.V., Hellen, C.U.T., and Shatsky, I.N. 1996. Canonical eukaryotic initiation factors determine initiation of translation by internal ribosomal entry. Mol. Cell. Biol. 16: 6859-6869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pestova T.V., Borukhov, S.I., and Hellen, C.U.T. 1998. Eukaryotic ribosomes require eIFs 1 and 1A to locate initiation codons. Nature 394: 854-859. [DOI] [PubMed] [Google Scholar]
- Pestova T.V., Lomakin, I.B., Lee, J.H., Choi, S.K., Dever, T.E., and Hellen, C.U.T. 2000. The joining of ribosomal subunits in eukaryotes requires eIF5B. Nature 403: 332-335. [DOI] [PubMed] [Google Scholar]
- Peterson D.T., Merrick, W.C., and Safer, B. 1979. Binding and release of radiolabeled eukaryotic initiation factors 2 and 3 during 80S initiation complex formation. J. Biol. Chem. 254: 2509-2516. [PubMed] [Google Scholar]
- Petrelli D., LaTeana, A., Garofalo, C., Spurio, R., Pon, C.L., and Gualerzi, C.O. 2001. Translation initiation factor IF3: Two domains, five functions, one mechanism? EMBO J. 20: 4560-4569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raychaudhuri P., Chaudhuri, A., and Maitra, U. 1985. Eukaryotic initiation factor 5 from calf liver is a single polypeptide chain protein of Mr = 62,000. J. Biol. Chem. 260: 2132-2139. [PubMed] [Google Scholar]
- Roll-Mecak A., Shin, B.S., Dever, T.E., and Burley, S.K. 2001. Engaging the ribosome: Universal IFs of translation. Trends Biochem. Sci. 26: 705-709. [DOI] [PubMed] [Google Scholar]
- Schmitt E., Blanquet, S., and Mechulam, Y. 2002. The large subunit of initiation factor aIF2 is a close structural homolog of elongation factors. EMBO J. 21: 1821-1832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava S., Verschoor, A., and Frank, J. 1992. Eukaryotic initiation factor 3 does not prevent association through physical blockage of the ribosomal subunit interface. J. Mol. Biol. 220: 301-304. [DOI] [PubMed] [Google Scholar]
- Thomas A., Goumans, H., Voorma, H.O., and Benne, R. 1980. The mechanism of action of eukaryotic initiation factor 4C in protein synthesis. Eur. J. Biochem. 107: 39-45. [DOI] [PubMed] [Google Scholar]
- Trachsel H. and Staehelin, T. 1979. Initiation of mammalian protein synthesis. The multiple functions of the initiation factor eIF-3. Biochim. Biophys. Acta 565: 305-314. [DOI] [PubMed] [Google Scholar]
- Valasek L., Phan, L., Schoenfeld, L.W., Veraskova, V., and Hinnebusch, A.G. 2001. Related eIF3 subunits TIF32 and HCR1 interact with an RNA recognition motif in PRT1 required for eIF3 integrity and ribosome binding. EMBO J. 20: 891-904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valasek L., Mathew, A.A., Shin, B.-S., Nielsen, K.H., Szamecz, B., and Hinnebusch, A.G. 2003. The yeast eIF3 subunits TIF32/a, NIP1/c, and eIF5 make critical connections with the 40S ribosome in vivo. Genes & Dev. 17: 786-799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon H. and Donahue, T.F. 1992. The sui1 suppressor locus in Saccharomyces cerevisiae encodes a translation factor that functions during tRNAiMet recognition of the start codon. Mol. Cell. Biol. 12: 248-260. [DOI] [PMC free article] [PubMed] [Google Scholar]