Abstract
BAF and PBAF are two related mammalian chromatin remodeling complexes essential for gene expression and development. PBAF, but not BAF, is able to potentiate transcriptional activation in vitro mediated by nuclear receptors, such as RXRα, VDR, and PPARγ. Here we show that the ablation of PBAF-specific subunit BAF180 in mouse embryos results in severe hypoplastic ventricle development and trophoblast placental defects, similar to those found in mice lacking RXRα and PPARγ. Embryonic aggregation analyses reveal that in contrast to PPARγ-deficient mice, the heart defects are likely a direct result of BAF180 ablation, rather than an indirect consequence of trophoblast placental defects. We identified potential target genes for BAF180 in heart development, such as S100A13 as well as retinoic acid (RA)-induced targets RARβ2 and CRABPII. Importantly, BAF180 is recruited to the promoter of these target genes and BAF180 deficiency affects the RA response for CRABPII and RARβ2. These studies reveal unique functions of PBAF in cardiac chamber maturation.
Keywords: Polybromo protein BAF180, PBAF (SWI/SNF6b), retinoic acid (RA) signaling, chromatin remodeling, heart, placenta
Mammalian SWI/SNF chromatin remodeling complexes (Olave et al. 2002b; Wang 2003; Roberts and Orkin 2004) represent an important class of regulatory factors that make up the molecular machinery (Levine and Tjian 2003) responsible for directing spatial and temporal patterns of gene expression during growth, differentiation, and development. Evolutionarily conserved and diversified throughout eukaryotes, various chromatin remodeling complexes facilitate nucleosome mobilization and assist transcription factors to access the DNA template. In mammalian cells, there are two major forms of SWI/SNF complexes (BAF and PBAF) that are structurally related but functionally distinct. In particular, recent studies have established that PBAF, but not BAF, is able to potentiate transcriptional activation in vitro mediated by nuclear receptors, such as RXRα, VDR, and PPARγ (Lemon et al. 2001), suggesting that these two cofactors may also provide diversified and specific functions in vivo.
BAF and PBAF share eight common subunits, but are distinguished by three unique polypeptide components. BAF180 (PB1) (Xue et al. 2000), which contains six bromodomains, is only present in PBAF, while BAF250 (ARID1A) (Nie et al. 2000) and OSA2 (ARID1B) (Inoue et al. 2002; Kato et al. 2002; Nie et al. 2003), which contain ARID (AT-rich interactive domain) and multiple LXXLL motifs, are only found in BAF. Bromodomain motifs have been shown to bind selectively to acetylated histone tails (Dhalluin et al. 1999; Jacobson et al. 2000), and the LXXLL motif is thought to mediate interactions with regulatory domains of select transcription activators (Heery et al. 1997; Torchia et al. 1997). These structural features suggest that BAF180 and BAF250 could be essential for mediating distinct cofactor activities at select sites on chromatin in a gene- or cell type-specific manner. The BAF180 and BAF250 subunits could therefore represent key regulatory components that functionally distinguish PBAF from BAF.
In an effort to dissect individual roles for BAF versus PBAF in regulating gene expression, cell differentiation, and organ development, we generated knockout (KO) mice lacking either BAF180, BAF250, or OSA2. Since the chromatin remodeling and transcriptional cofactor functions of PBAF have previously been characterized in vitro (Lemon et al. 2001), here, we have focused on the in vivo functional analyses of the PBAF-specific subunit BAF180. We found that BAF180 deficiency in mouse embryos causes severe hypoplastic ventricle development and trophoblast placental defects. A battery of embryonic aggregation experiments were performed to determine whether the heart defects in BAF180 deficient embryos are a direct result of BAF180 ablation, or an indirect consequence of the trophoblast placental defects. Mircoarray gene expression profiling and quantitative RT-PCR were employed to identify potential target genes for BAF180 involved in heart development, and chromatin immunoprecipitation experiments were carried out to measure the occupancy of BAF180 at the promoters of select target genes. Together, these findings reveal unique functions of PBAF in mammalian development and provide new insights into the mechanisms of cardiac chamber maturation involving a specific chromatin remodeling complex.
Results
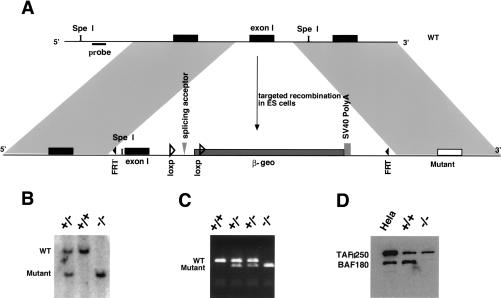
We applied a targeted knockout strategy in mice to generate null alleles of BAF180 by modifying a vector originally designed for gene trapping (Fig. 1A) (Skarnes 2000). Homologous recombination was confirmed by Southern blot (Fig. 1B) and PCR (Fig. 1C) analyses, whereas the absence of BAF180 expression was determined by Western blot analysis (Fig. 1D) and in situ hybridization (Fig. 2B; data not shown). Since β-geo (Friedrich and Soriano 1991), a fusion protein of β-galactosidase and neomycin phosphotransferase, is spliced in frame to a 5′ exon of BAF180, β-galactosidase activity should reflect the expression of endogenous BAF180. Indeed, X-Gal staining showed widespread expression of this PBAF subunit throughout embryogenesis (Fig. 2A), in accord with our in situ hybridization analysis (data not shown). Notably, BAF180 is expressed in extraembryonic tissues, including yolk sac and placenta. In placenta, its expression is most prominent in trophoblast cells within the labyrinthine layer at mid-gestation (Fig. 2B).
Figure 1.
Generation of BAF180-deficient mice. (A) KO scheme applied for generation of a BAF180 null allele in ES cells. Wild-type genomic structure (WT, top line) is replaced by a neo-resistant knockout allele (KO, neor, bottom line) through homologous recombination within the grey fragments. Cre-mediated recombination between the two loxP sites converts the KO allele to neo-sensitive and lacZ negative (neos). (B) For BAF180 KO Southern analysis, Spe I digestion produces a 7.8-kb fragment for wild type (WT) allele and a 5.9-kb fragment for a mutant allele. The probe used is indicated by a small black box located between the Spe I sites and distal to the 5′ recombination region. (C) PCR-based genotyping analyses of the progenies of BAF180 heterozygous crosses. (D) Western blot indicated that the mouse BAF180 protein is absent in cells derived from homozygous BAF180 KO mice. TAFII250 served as a positive control.
Figure 2.
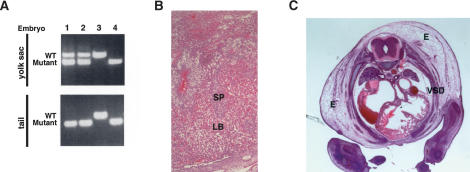
BAF180 expression in mouse tissues. (A) X-Gal staining of β-galactosidase activity revealed that BAF180 is expressed ubiquitously at E11.5 embryo. (B) In situ hybridization of E14.5 placenta with a BAF180-specific antisense RNA probe. (LB) Labyrinthine; (SP) spongiotrophoblast.
Genotype analysis indicated that BAF180 heterozygous animals (BAF180+/-, Hets) are apparently normal, and back-crosses with wild-type (BAF180+/+, WT) animals yielded about 50% Het progenies. Intercrosses of BAF180+/- mice resulted in no viable homozygous (BAF180-/-, null) pups. Further analysis indicated that most of the null embryos died between embryonic day 12.5 (E12.5) and E15.5 of gestation (Table 1). Null embryos that survived to E13.5 and E14.5 appeared pale and displayed prominent subcutaneous edema along their backs (Fig. 3A,B). Histological examination of null embryos revealed that most organs were present and appeared normal, except that the liver and lung were smaller than in wild-type embryos, a finding suggesting a delayed development of these two organs.
Table 1.
Embryonic lethality caused by BAF180 deficiency
| Age | WT | Het | Null (dead) |
|---|---|---|---|
| 10.5 dpc or earlier | 24 | 51 | 22 |
| 11.5 dpc | 31 | 54 | 18 (2) |
| 12.5 dpc | 36 | 66 | 27 (5) |
| 13.5 dpc | 37 | 75 | 14 (14) |
| 14.5 dpc | 45 | 92 | 22 (23) |
| 15.5 dpc | 12 | 26 | 0 (10) |
| Post-natal | 13 | 39 | 0 |
(WT) Wild type; (Het) heterozygous; (dpc) days post-coitum.
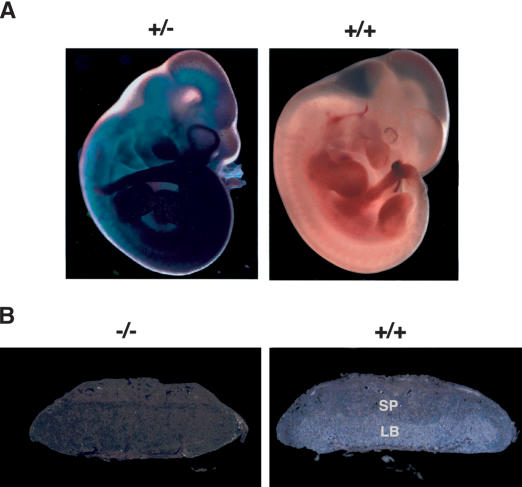
Figure 3.
Cardiac defects in BAF180-deficient embryos. (A) Wild-type (WT) and null BAF180 embryos at E14.5. Note the edema along the back of the mutant embryo. (B) Transverse sections of a wild-type (WT) and a mutant E14.5 heart. The mutant heart has severe hypoplastic myocardial wall and ventricular septal defects. (C) Tunnel assays indicated that neither BAF180 wild-type (WT) nor null E14.5 heart tissues showed any detectable cell death. A DNase I-treated heart tissue section was used as a positive control. (D) In situ hybridization of E14.5 hearts indicated that the expression pattern of MLC-2a is largely normal in a BAF180 null heart. (E) Edema; (VS) ventricular septum; (VSD) ventricular septal defect.
Generalized edema is often a sign of failure in the circulatory system due to cardiac defects. Indeed, freshly isolated hearts from null embryos pumped at a slower rate and with less blood volume compared with wild-type hearts. Histological sections of >15 BAF180 null embryos all revealed severe hypoplasia of the cardiac ventricular free walls as well as a ventricular septal defect (Fig. 3B). In contrast, the atrio-ventricular valves and the neural crest-derived aortic and pulmonary artery out-flow tracts and valves were normal and properly developed. Mouse embryonic heart ordinarily develops into its mature form at E14.5 in order to provide sufficient blood flow for the rapid growth of the fetus. Thus, it seems likely that the defective heart of BAF180 null embryos would have difficulty sustaining normal circulation, leading to the observed embryonic lethality.
To better characterize the nature of the heart defects observed in BAF180 null embryos, we first examined whether there is any evidence of inappropriate cell death events involved. Tunnel assays indicated that neither the wild-type nor the null heart tissues showed any detectable apoptotic cells (Fig. 3C), suggesting that it is a failure in cell growth or differentiation rather than cell death that causes the heart defects. We next checked the expression of several chamber-specific markers. Previous studies suggested that the thin myocardial wall observed in RXRα KO mice can be partially attributed to an overexpression of atria-specific marker myosin light chain-2, atrial isoform (MLC-2a) in ventricular tissues, thus causing an atria-like ventricle phenotype (Dyson et al. 1995). In situ hybridization indicated that, unlike the situation in RXRα KO mice, the expression pattern of MLC-2a is largely normal in a BAF180 null heart (Fig. 3D, cf. left and right panels), with slightly more expression in ventricle layers compared with its expression in wild-type heart. The expression pattern of myosin light chain 2, ventricular isoform (MLC-2v) and atrial natriuretic peptide also appear largely normal in BAF180 KO heart (data not shown). These data suggest that it is more likely these defects are caused by cell growth arrest rather than improper differentiation or cell death.
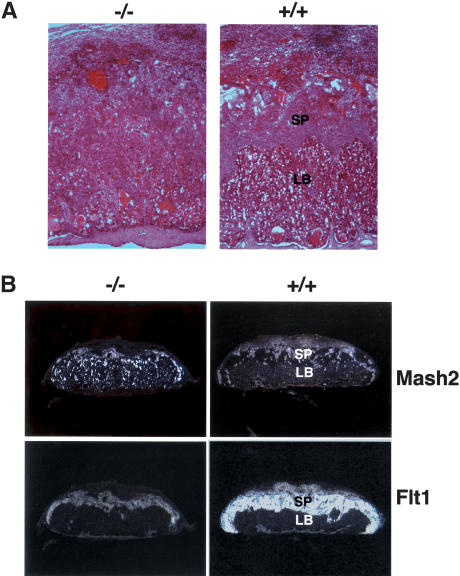
Since BAF180 is highly expressed in placenta, primarily within the labyrinthine trophoblasts (Fig. 2B), we set out to examine whether there are also defects in the placenta. Indeed, histological comparisons (Fig. 4A) indicated that placenta from BAF180 null fetuses is abnormal. The spongiotrophoblast and labyrinthine layers were indistinct. Within the labyrinthine layer, there was an accumulation of large clusters of packed trophoblasts replacing maternal and fetal blood space, likely compromising fetal-maternal exchange. Further in situ hybridization analyses (Fig. 4B) indicated that Mash2, a trophoblast-specific marker, normally expressed most prominently in the spongiotrophoblast layer in wild-type placenta, was instead expressed throughout BAF180 null placentas. At the same time, the expression of the spongiotrophoblast layer-specific marker Flt1 in null placentas indicates that the spongiotrophoblastic layer was thinner and less vascularized than in wild-type mice.
Figure 4.
Placental defects in BAF180-deficient embryos. (A) Compared with wild type (WT) at E13.5, placentas from BAF180 null fetuses have indistinct spongiotrophoblast and labyrinthine layers. (B) In situ hybridization of E14.5 placentas with placental markers Mash2 and Flt1. (LB) Labyrinthine; (SP) spongiotrophoblast.
Comparison of the heart and placental defects observed in BAF180 null embryos with those in RXRα-deficient embryos (Kastner et al. 1994; Sucov et al. 1994) reveals that the phenotypes resulting from these two gene knockouts are very similar. In addition, ablation of PPARγ also resulted in severe placental and heart defects. Interestingly, the placental defects of PPARγ knockout fetuses were found to be responsible for the thin myocardial wall phenotype, thus establishing an unknown axis between placenta and heart (Baraks et al. 1999). Since RXRα and PPARγ are two highly related nuclear receptors that can form heterodimers to perform essential transcriptional activities in the retinoic acid (RA) signaling pathway (Kliewer et al. 1992; Barak et al. 1999; Ross et al. 2000), it has been suggested that the trophoblast placental defects in RXRα null embryos might also be the cause of the observed heart defects. Given the well-established functional connection between PBAF and transcriptional activators RXRα and PPARγ from previous in vitro biochemical studies (Lemon et al. 2001), we set out to examine whether the observed heart defects were due directly to the loss of BAF180.
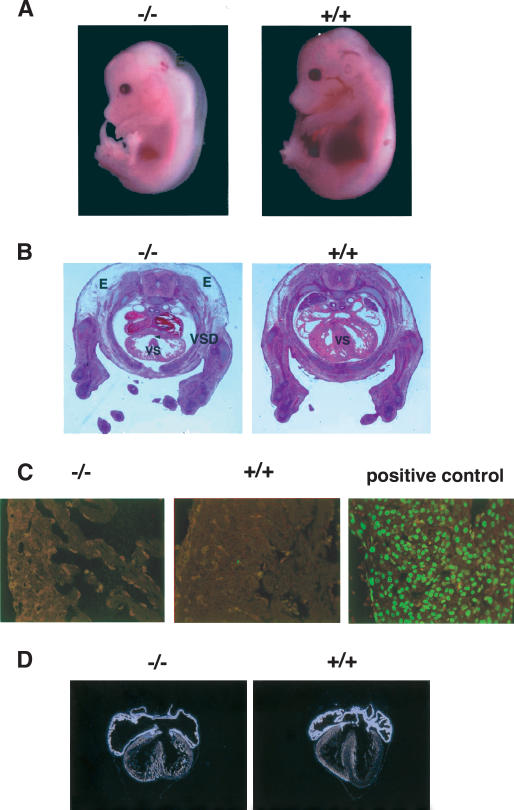
First, we tested the possibility that the heart defects observed in BAF180 null embryos were indirectly the result of placental defects by carrying out tetraploid aggregation (Rossant and Cross 2001; Nagy et al. 2003) experiments to rescue the placenta. When morula stage tetraploid embryos are aggregated with diploid embryos, tetraploid cells are thought to contribute primarily to extraembryonic lineages of the embryo including the placenta and yolk sac endoderm, but not to the embryo proper. In this study, wild-type tetraploid embryos were aggregated with diploid embryos derived from BAF180 intercrosses. The aggregated embryos were transferred into the uterus of pseudo-pregnant mice and allowed to develop to E14.5 stage. Chimeric placenta, yolk sac, and part of the embryo proper were then dissected for genotypic and histological analyses. PCR genotyping revealed substantial amounts of wild-type BAF180 DNA in the yolk sac, but no detectable wild-type BAF180 DNA in the tail from null embryo-derived chimeras (Fig. 5A, lanes 1,2), consistent with the expected preferential contribution of wild-type tetraploid cells to extraembryonic tissues of the BAF180-/- embryos. Histological analysis revealed that chimeric placenta derived from these null embryo aggregates displayed a significantly expanded labyrinthine layer and appeared largely normal (Fig. 5B). In stark contrast, of all null embryos from recovered chimeras (10 out of 42), no recognizable rescue of the heart defects was observed (Fig. 5C). Thus, our data suggest that for the BAF180 KOs, the observed heart defects are unlikely an indirect consequence of trophoblast placental defects.
Figure 5.
Wild-type tetraploid embryo rescue did not restore normal heart development in BAF180 null embryos. (A) Contribution of BAF180 wild-type (WT) alleles in chimeras derived from aggregation between wild-type tetraploid and BAF180 mutant diploid embryos is analyzed by PCR. Lanes 1 and 2 identified two embryos derived from BAF180 nulls aggregated with wild-type tetraploid embryos. Lanes 3 and 4 identified two controls from normal intercross of BAF180 Hets. (B) A chimeric placenta derived from aggregation of a null BAF180 embryo with two wild-type (WT) tetraploid embryos appears to be normal. (C) Tetraploid aggregation could not rescue the heart defects. Histological analysis of the heart from the embryo identified in A, lane 1. All 10 null embryos identified from 42 recovered chimeras showed very similar phenotypes. (E) Edema; (LB) labyrinthine; (SP) spongiotrophoblast; (VSD) ventricular septal defect.
To further confirm that the observed heart defects in BAF180-/- mice are independent of the placental defects, we performed a complementary fusion experiment in which we aggregated diploid BAF180 null embryos with wild-type embryonic stem (ES) cells. In this case, wild-type ES cells are expected to contribute primarily to the embryo proper, but not to the placental trophoblast or yolk sac endoderm (Rossant and Cross 2001; Nagy et al. 2003). Thus the ES cell derivatives should rescue the embryonic heart phenotype, but the placental defects should remain. To distinguish between heterozygous and null embryos aggregated with wild-type ES cells, two different mutant alleles (neor and neos) were used (Fig. 1A; Materials and Methods).
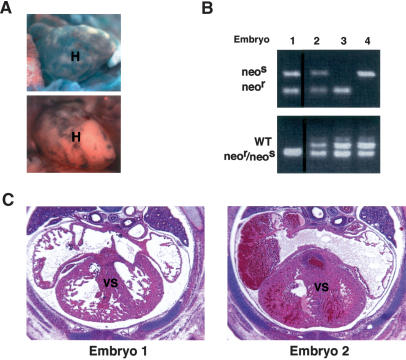
Embryos derived from intercrosses between neor and neos Hets were used in aggregations with wild-type ES cells, and aggregated embryos recovered at E14.5 were dissected for analyses. Chimeric X-Gal staining indicated the variable contribution of wild-type ES cells to these embryos (Fig. 6A, cf. upper and lower panels). The presence of only neor or neos allele identified by PCR (Fig. 6B, upper panel, lanes 3,4) indicated that these E14.5-stage chimeras were derived from aggregates of Het embryos with wild-type ES cells. Null embryos were identified by the presence of both neor and neos alleles (Fig. 6B, upper panel, lanes 1,2), whereas variable wild-type ES cell contribution to null embryos was revealed by the presence of wild-type BAF180 (Fig. 6B, lower panel, lanes 1,2). As expected, when we observed no detectable amounts of wild-type ES cell contribution (Fig. 6B, lower panel, lane 1), no significant rescue of the heart defects was obtained (Fig. 6C, left panel), whereas substantial ES cell contribution (Fig. 6B, lower panel, lane 2) largely restored normal heart development (Fig. 6C, right panel). These aggregation data suggest that, unlike the situation observed with PPARγ KO mutants, the heart defects resulting from BAF180 depletion appear to be independent of the trophoblast placental defects. Thus, the thin myocardial wall syndrome observed in the BAF180 null embryos is most likely a direct result of ablating BAF180 during heart development.
Figure 6.
Wild-type ES cell rescue restored the normal heart development of BAF180 null embryos. (A) X-Gal staining demonstrated variable chimeric contribution of wild-type (WT) ES cells in an embryonic heart derived from aggregation between wild-type ES cells and BAF180 mutant diploid embryos. Less chimeric X-Gal staining represented more ES contribution. (B) PCR analysis demonstrated contribution of BAF180 wild-type (WT) alleles in chimeras derived from aggregation between wild-type ES cells and mutant diploid embryos. PCR was performed with DNA samples obtained from embryonic limb, tail, brain, yolk sac, and placenta (only results from brain tissues were illustrated). Embryo 1 is a null embryo with little wild-type ES contribution, while embryo 2 is a null embryo with substantial ES contribution. Embryos 3 and 4 are two Het embryos aggregated with wild-type ES cells. (C) Contribution of wild-type (WT) ES cells into BAF180 null embryos rescued the heart defects. Histological analysis of the hearts from the two null embryos identified in B. (H) Heart; (VS) ventricular septum.
To better understand the molecular mechanisms underlying the hypoplastic chamber maturation caused by BAF180 ablation, we compared the gene expression profiles of wild-type and mutant hearts by microarray analysis using Murine U74Av2 chips from Affymetrix. RNA prepared from four wild-type or mutant E12.5 hearts was used as one probe set, and three independent sets of probes were used for hybridization. In total, nine sets of comparison profiles were obtained from these experiments. As listed in Table 2, a large percentage of the genes affected by BAF180 ablation comprise structural and metabolic genes. Importantly, numerous genes mediating cell growth and proliferation, particularly those involved in the insulin pathway (LeRoith and Roberts 2003), such as Grb10 (growth factor receptor-bound protein 10), insulin-like growth factor-binding protein 2 and 5, and endothelin receptor type B, were significantly down-regulated. In contrast, growth arrest-specific 1 and 2 genes were up-regulated (Table 2), consistent with the notion that the heart defects caused by BAF180 deficiency are possibly due to growth arrest instead of cell death. Disruption of the RA or erythropoietin pathway (Wu et al. 1999) in mice can cause very similar hypoplastic heart defects, and interestingly, BAF180 ablation caused an increase in the cellular RA-binding protein (CRABP), retinol-binding protein, and erythropoietin receptor in embryonic heart. These data suggest that somewhere downstream, RA and erythropoietin signaling events might be compromised in BAF180 mutant hearts, thus causing a positive feedback loop to increase the expression of these genes. Interestingly, S100A13, a member of a large family of calcium-modulated proteins with essential intracellular and extracellular functions (Donato 2001), previously identified as being up-regulated by the core subunit Brg-1 of both PBAF and BAF complexes (Liu et al. 2001), was also down-regulated by BAF180 ablation in heart. In addition, deletion of BAF180 caused little change in the expression of other members of the SWI/SNF complexes, such as BAF250 (ARIDa), BAF155 (SMARCC1), and BAF60a (SMARCD1).
Table 2.
Genes regulated in BAF180 mutant hearts
| Down-regulated genes | Up-regulated genes |
|---|---|
| Structure | Heat-shock protein, 70 kDa |
| β-1-globin | Metallothionein 1 |
| Vascular smooth muscle α-actin | Calcyclin (100A6) |
| Procollagen C-proteinase enhancer protein | Keratin complex 1, acidic |
| Potassium voltage-gated channel, Isk-related subfamily, member 1 | Coproporphyrinogen oxidase |
| Clast1, amyloid-like protein | Small inducible cytokine A2 |
| Cysteine-rich intestinal protein | Placenta-specific ATP-binding cassette transporter (Abcp) |
| Solute carrier family 8 (sodium/calcium exchanger), member 1 | β-galactoside-specific lectin |
| Elastin, tropoelastin | Uroporphyrinogen decarboxylase |
| Myosin alkali light chain (exon 1) (ventricular/slow muscle isoform) | Prostacyclin synthase |
| Junctional adhesion molecule (Jam) | Erythrocyte membrane protein Rh50 (Rhag) gene |
| Phospholemman precursor | Transforming growth factor, β 3 |
| Lumican | Lamin A |
| Solute carrier family 8 (sodium/calcium exchanger), member 1 | galK mRNA for galactokinase |
| Cardiac troponin T isoform A3b | Ferritin H subunit |
| MaxiK potassium channel β subunit gene, Ca++-activated K+channel | Chloride channel protein 3 (CLCN3) mRNA |
| Metabolism | Hexokinase II, exon 1 |
| Mouse cytochrome β-558 | Carbonic anhydrase II (CAII) |
| Aromatic-L-amino acid decarboxylase | Fibroblast inducible secreted protein |
| Lysozyme M | Erythropoietin receptor |
| Lzp-s mRNA for lysozyme P | Mutant p53 mRNA |
| Beta-1-globin | Translation initiation factor eIF2 γ Y |
| Carbonic anhydrase 3 gene 5′ UTR and exon 1 | Glutathione S-transferase |
| Lactate dehydrogenase 2, B chain | Heat-shock protein, 105 kDa |
| S100 calcium-binding protein A13 | CRABP mRNA for cellular retinoic acid-binding protein |
| osf-2 mRNA for osteoblast-specific factor 2 | Retinoblastoma 1 |
| Procollagen C-proteinase enhancer protein | Protein tyrosine phosphatase, receptor type, R |
| Muscle glycogen synthase (Gys) | Osteosarcoma oncogene |
| Purine nucleoside phosphorylase (Np-b) | Erythroid Kruppel-like factor |
| Ulip protein | Growth arrest-specific 1 |
| Annexin XI | Growth arrest-specific 2 |
| Isocitrate dehydrogenase 2 | Hemoglobin β, pseudogene bh3 |
| Sarco(endo)plasmic reticulum calcium ATPase (SERCA2) | Cyclin G |
| Hemoglobin, β adult major chain | Retinol-binding protein 1 |
| Growth | |
| Growth factor receptor-bound protein 10 (2 hits) | |
| Insulin-like growth factor-binding protein 2 | |
| Insulin-like growth factor-binding protein 5 | |
| rho7, Ras-like protein | |
| Small inducible cytokine subfamily D, 1 | |
| Endothelin receptor type B, 7-tm protein | |
| Adrenomedullin receptor | |
| Calcitonin receptor related (7-tm: G-protein-coupled receptors) | |
| Secreted frizzled related protein sFRP-2 (Sfrp2) | |
| Wingless-related MMTV integration site 5A | |
| Kras oncogene-associated gene | |
| M97216 mouse DNA-binding protein? | |
| Fibroblast growth factor receptor 2 | |
| Diazepam-binding inhibitor | |
| Ras-like GTP-binding protein Rad mRNA | |
| Gene expression | |
| Eukaryotic translation elongation factor 1 α 2 |
See Supplemental Material for more detail.
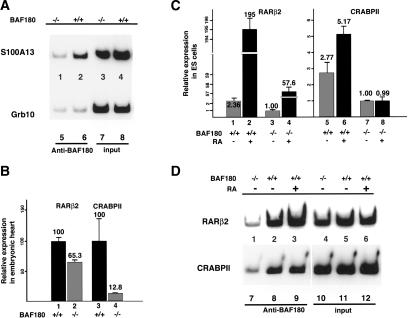
Having identified a number of putative target genes regulated by BAF180 by gene expression profiling experiments, we next set out to determine whether BAF180 is directly associated with the transcription of some of these genes in vivo. The down-regulated expression of these genes was first confirmed by either quantitative PCR or in situ hybridization or both (Supplementary Fig. S1; data not shown). We used chromatin immunoprecipitation (ChIP) assays to determine the occupancy of BAF180 at potential target genes. Due to the technical difficulties of large-scale growth of primary heart tissue cells, BAF180 wild-type and null ES cell lines were used in this study. The promoter sequences of Grb10 and S100A13 (described above) were first chosen for this ChIP analysis. As shown in Figure 7A, radioactive PCR revealed that the S100A13 promoter sequence was significantly enriched in immunoprecipitates from wild-type ES cells when using BAF180 antibodies (Fig. 7A, cf. lanes 2 and 1). These data reveal that BAF180 protein directly associates with the promoter of S100A13 in vivo and suggest that S100A13 may be a bona fide target gene for BAF180. In contrast, no enrichment of the Grb10 promoter sequence was observed in the ChIP assay (Fig. 7A, cf. lanes 6 and 5), suggesting that the down-regulation of Grb10 by BAF180 deficiency may be a secondary effect, or that BAF180 does not bind to the particular promoter region amplified.
Figure 7.
Identification of BAF180 target genes. (A) ChIP assays show binding of BAF180 to promoter regions of S100A13 but not Grb10. (B) Quantitative RT-PCR analysis show decreased expression of both RARβ2 and CRABPII in BAF180 null embryonic hearts. (C) Quantitative RT-PCR analysis of RARβ2 and CRABPII expression in ES cells. Loss of BAF180 led to decreased expression of these two genes, and abolished the RA response for CRABPII and decreased RA response for RARβ2. (D) ChIP assays show recruitment of BAF180 to promoter regions of RARβ2 and CRABPII.
Based on our previous in vitro studies (Lemon et al. 2001) and the in vivo analyses presented here, it seems likely that PBAF plays a role in RA signaling during heart development, presumably by serving as a cofactor for RXRα, PPARγ, and other RA-related nuclear receptors. However, our microarray experiments did not identify specific down-regulated gene targets involved in RA signaling. This could be partly due to the very low expression level of genes involved in this pathway, thus falling below our detection limit. For example, we failed to detect the expression of RA target genes and numerous RA related receptors present in the U74Av2 chip, including CRABPII. And other target genes like RARβ2 are not even present on this chip. Moreover, RA function can be highly tissue- and stage-specific during heart development, with RA possibly even exerting opposite effects on gene expression in different cardiac tissues (Chen et al. 2002; Stuckmann and Lassar 2002). However, because of the striking similarities between the phenotypes of BAF180, RXRα, and PPARγ null mice, and the observation that PBAF mediates the transcriptional activity of RXRα and PPARγ in vitro, we proceeded to examine whether BAF180 is required for the transcription of select RA target genes in vivo. RARβ2 (de The et al. 1990) and CRABPII (Durand et al. 1992) are two well-established direct RA targets. Quantitative RT-PCR revealed that ablation of BAF180 caused modest down-regulation of RARβ2 in heart, and five- to eightfold down-regulation of CRABPII (Fig. 7B). RARβ2 and CRABPII are also expressed in ES cells, although RARβ2 expression is extremely low. As expected, loss of BAF180 resulted in decreased expression of both RARβ2 and CRABPII in KO ES cells (Fig. 7C, cf. lanes 1 and 3, and lanes 5 and 7). In wild-type cells, RA treatment induced ∼100-fold the expression of RARβ2 (Fig. 7C, cf. lanes 1 and 2) and about twofold expression of CRABPII (Fig. 7C, cf. lanes 5 and 6). Importantly, loss of BAF180 abolished the RA response for CRABPII (Fig. 7C, cf. lanes 5,6 and 7,8) and decreased the RA response for RARβ2 (Fig. 7C, cf. lanes 1,2 and 3,4). These data, taken together, support the notion that BAF180 is required for the proper expression of these genes in vivo.
We next examined whether BAF180 is directly involved in the transcription of these two genes, and therefore, targeted directly to their promoter DNA sequences in vivo. ChIP analysis revealed that BAF180 protein is present at the RARβ2 promoter, and its occupancy increases in the presence of RA (Fig. 7D, lanes 2,3). BAF180 protein is also recruited to the promoter region of CRABPII. However, RA treatment did not promote significant additional BAF180 recruitment (Fig. 7D, lanes 8,9), presumably due to the relatively low sensitivity of the assay. Together, these data suggest that BAF180 (PBAF complex) is likely involved in transcription of RA target genes in vivo and that the heart defects in BAF180 null embryos may be at least partially due to a disruption of the RA signaling pathway.
Discussion
In this study, we have demonstrated that BAF180, a key distinguishing subunit in the chromatin remodeling complex PBAF, is required for cardiac and placental development. BAF180 deletion in mouse embryos results in severe hypoplasia of the ventricular free walls as well as a ventricular septal defect. Loss of BAF180 also causes placental defects. In particular, within the placental labyrinthine layer, large clusters of packed trophoblasts replace maternal and fetal blood space, likely compromising maternal-fetal exchange. Interestingly, embryonic aggregation experiments suggest that the observed heart defects are a direct result of ablating BAF180 in heart tissues and not caused by a secondary consequence of trophoblast placental defects. We identified potential target genes of BAF180 involved in heart development. We found that BAF180 deficiency leads to a decreased expression of select target genes, such as S100A13 and RA targets RARβ2 and CRABPII in heart tissues. Importantly, loss of BAF180 abolished RA response for CRABPII and decreased RA response for RARβ2 in ES cells. Furthermore, we found that BAF180 is directly recruited to the promoter of S100A13, RARβ2, and CRABPII. Thus, our findings revealed unique functions of a PBAF-specific subunit (BAF180) in mammalian embryonic development.
The specific functions of BAF180 in cardiac chamber maturation is in contrast to the roles of several core subunits present in both PBAF and BAF complexes, such as Brg-1, BAF155, and INI1 (DiRenzo et al. 2000; Klochendler-Yeivin et al. 2000; Roberts et al. 2000; Guidi et al. 2001; Kim et al. 2001). The null embryos of these core subunits typically die around peri-implantation stage, shortly after the embryonic cells start to differentiate, indicating a crucial role for these proteins in cell growth and differentiation. In contrast, BAF180 ablation did not lead to any observable effect on embryogenesis until mid-gestation. Most embryonic organs in BAF180 null embryos develop normally; even the general morphology and cell lineages within the hypoplastic heart appear undisturbed. On the other hand, our preliminary data obtained with BAF250 indicate that although the BAF-specific protein is not considered the core subunit of the BAF complex, its deficiency results in much more severe lethal phenotypes than those obtained for the BAF180 KO (data not shown). For example, even BAF250 Hets show obvious neural tube closure defects that were also observed in some Brg-1 and BAF155 Hets. These data suggest that the early lethal phenotypes observed with ablation of various core SWI/SNF chromatin remodeling subunits can be largely attributed to the loss of BAF, rather than PBAF function. Thus, the BAF180 studies presented here provide strong evidence to support the notion that PBAF and BAF indeed perform distinct cofactor functions in vivo, as well as in vitro (Lemon et al. 2001).
The essential extraembryonic functions of certain proteins for proper embryonic organ development have been recognized in several recent studies (Barak et al. 1999; Rossant and Cross 2001; Geng et al. 2003; Wu et al. 2003). In contrast to the PPARγ situation, our aggregation rescue experiments suggest that the BAF180 requirement for heart chamber maturation is independent of its function in trophoblast placental development. Since BAF180 KO embryos show almost identical phenotypes to those RXRα null fetuses, it will be interesting to test whether placental rescue can restore the normal heart development of RXRα null fetuses. In any case, our studies reveal the remarkable complexity and specificity of interactions between transcriptional activators and cognate cofactors during extraembryonic and embryonic development.
There are a growing number of gene KOs that cause similar hypoplastic heart chamber phenotypes (for reviews, see Sucov 1998; Olson and Schneider 2003), including RXRα and PPARγ, but the mechanisms for these genes to direct myocardial cell growth and maturation are likely to be distinct. These genes may function differently in a myocardial cell autonomous or non-cell autonomous manner. They may also direct the growth and differentiation of myocardial cells via their functions in epicardial or endocardial tissues, or even through the trophoblast placenta. For example, recent studies suggest that RA or erythropoietin signaling-related factors secreted from epicardial cells may be essential for the proper proliferation of myocardial cells (Chen et al. 2002; Stuckmann and Lassar 2002). However, the relevant downstream target genes that can account for the observed thin myocardium syndrome still remain poorly understood. The extracellular functions of S100 proteins in myocardial cell development (Donato 2001; Most et al. 2003) suggest that the S100A13 gene identified as a possible BAF180 target may indeed represent a physiologically relevant gene that contributes to the observed heart defects. Future investigation of target cardiac tissue(s) and genes regulated by BAF180 may help elucidate specific molecular mechanisms governing PBAF functions in cardiac chamber maturation.
Numerous studies have revealed possible differential functions of individual subunits within SWI/SNF complexes in mediating mammalian transcription during development (Olave et al. 2002b; Wang 2003; Roberts and Orkin 2004). However, most of the individual subunits under investigation are common to both BAF and PBAF complexes. Thus it is not clear whether the observed differential functions of these subunits can be attributed to their roles in BAF, PBAF, or both. For example, Brg-1 and BAF57 are shown to have distinct roles in CD4/CD8 expression during thymic development (Chi et al. 2002). Knock-down of the common subunit BAF47 (which presumably inhibits the activities of both BAF and PBAF complexes) can affect a subgroup of genes involved in the cellular response to viral infection (Cui et al. 2004). In addition to these individual subunits having differential functions within a cell type, there appear to be isoforms of BAF subunits that are thought to mediate the functions of the SWI/SNF complexes in a tissue-specific manner, such as the neuron-specific BAF53b (Olave et al. 2002a). Detailed structural and functional analyses of the unique and distinguishing subunits in SWI/SNF complexes will ultimately help us better understand how these complex chromatin remodeling cofactors work individually or in concert to guide gene expression, cell growth, and organ development.
Materials and methods
Generation of BAF180 KO mice
A 3-kb and a 4-kb genomic fragment that is 5′ and 3′ to the exon I (exon of interest, predicted exon 11 of mouse BAF180) were amplified by PCR and inserted into the targeting vector (Fig. 1). A 0.5-kb fragment of the exon I region was PCR amplified and inserted in front of β-geo. The vector was linearized by Not I digestion, and electroporated into ES cells as described (Skarnes 2000). More than 90% of the colonies obtained were positive homozygous recombination clones. Three independent BAF180 deficient mouse lines were obtained from blastocyst injection of these ES cell clones.
Histology, in situ hybridization, and Tunnel assay
All procedures for histology followed standard protocols. The procedure for in situ hybridization was performed as described (Shelton et al. 2000). Tunnel assay followed a standard protocol (Clontech).
Embryonic rescue by wild-type tetraploid embryo or wild-type ES cell aggregation
All procedures for aggregation experiments followed detailed published protocols (Nagy et al. 2003). A multiporator was used to generate tetraploid embryos (Eppendorf). The aggregation plates were prepared from center-well organ culture dishes (Falcon). Embryos were covered with 1 mL KSOM + 1/2 amino acids medium (Specialty medium), and 2 mL of water was added to the surrounding ring to keep a moisture balance.
Normally, two wild-type tetraploid embryos were aggregated with one embryo derived from intercrosses of BAF180 Hets. For ES cell aggregation, a cluster of eight to 16 ES cells were aggregated with one mutant embryo. In order to distinguish between heterozygous and null embryos aggregated with wild-type ES cells, two different mutant alleles were used (Fig. 1A). To obtain the second mutant allele of BAF180, our initial KO allele (neor) was crossed with Cre transgenic mice, so the DNA sequence between the two loxP sites was excised to produce the first generation of mutant allele 2 (neos identified in Fig. 4D, schematized in Fig. 1A). These neos Hets were then back-crossed with wild-type mice to remove the Cre gene. neos allele was neo-sensitive and completely lacZ negative, and neos/neos null embryos displayed phenotypes identical to those of neor/neor nulls. Embryos derived from intercrosses between neor and neos Hets were used with ES cell aggregation. Two sets of three-primer PCR analysis were used for genotyping. An initial set of three-primer PCR genotyping (Figs. 1C, 4A, 5B, lower panel) was designed to distinguish the BAF180 wild-type allele from its mutants, but not between the two mutant alleles. A second three-primer set for PCR analysis identified the presence of the two mutant alleles, but not the wild-type allele (Fig. 5B, upper panel). These primer sequences are available upon request.
DNA microarray analyses
Gene expression profiles of wild-type and mutant hearts were obtained by microarray analysis using Murine U74Av2 chips with standard procedures (Affymetrix). RNA prepared from four wild-type or mutant E12.5 hearts was used as one probe set, and three independent sets of probes were used for hybridization. In total, nine sets of comparison profiles were obtained from these experiments.
ChIP
ES cells growing at late exponential stage were cross-linked with formaldehyde for 10 min at room temperature, and the procedure for ChIP was as described (Liu et al. 2001). 0.5% or 1% formaldehyde treatment gave similar results. For RA induction, a mixture of all-trans RA (final concentration 2 × 10-6 M) and 9-cis RA (final concentration 5 × 10-8 M) was added to ES cell culture for 2.5 h before formaldehyde treatment. About 1.0 μCi of 32P-dCTP was added to the PCR reaction, and 30-33 PCR cycles were applied. PCR products were run through a PAGE gel and exposed to a PhosphorImager screen. Normally, a 2-h exposure was sufficient to obtain clear signals. PCR primers for ChIP are as follows: S100A13: GCAGTAGCAGTCCCTCTAACACAGA; GCAGTCAGGAAAAGTAACTCACCG. Grb10: GGAAGCATTGCCATCCCAAATGGAAT; CCACGGACGAGTTAATATCGTTGTTC; RARβ2: CATGATTCGGGGCTGGGAAAAAGA; AATCCACTGAGGCAGGCTTTGAGA. CRABPII: TCAGTTCCACTCCCCAACCAAAGC; GCTCTGTGCTGGAGCTACAAGTGTT.
Quantitative RT-PCR
RNA was extracted with Tri Reagent (Sigma) from E13 or E14 embryonic hearts or ES cells cultured in the same condition as for ChIP assay. cDNA was prepared using Superscript II Reverse Transcriptase with standard procedure (Invitrogen). Quantitative RT-PCR was performed using a DNA Engine Opticon 2 (MJ Research). β-Actin was used as an internal control for quantification.
Acknowledgments
We thank M. Haggart, R. Bassel-Duby, D. Pangilinan, K. Pinson, and P. Tate for technical assistance; W. Wang and B. Vick for providing antibodies against BAF180; K. Zhao, W. Liu, and K. Geles for advice on chromatin immunoprecipitation protocol; Y. Isogai for help on microarray analyses; and H. Sucov, M. Sachs, J. Reiter, O. Kelly, and M. Sohaskey for their comments on the manuscript. This work was supported in part by grants from the National Institutes of Health (R.T.). W. Zhai is partially supported by a postdoctoral fellowship from the Hereditary Disease Foundation. Z. Wang was the recipient of a postdoctoral fellowship award from the American Cancer Society, California division, and is currently supported by a postdoctoral traineeship award from the Prostate Cancer Research Program, Department of Defense.
Supplemental material is available at http://www.genesdev.org.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.1238104.
References
- Barak Y., Nelson, M.C., Ong, E.S., Jones, Y.Z., Ruiz-Lozano, P., Chien, K.R., Koder, A., and Evans, R.M. 1999. PPAR γ is required for placental, cardiac, and adipose tissue development. Mol. Cell 4: 585-595. [DOI] [PubMed] [Google Scholar]
- Chen T.H., Chang, T.C., Kang, J.O., Choudhary, B., Makita, T., Tran, C.M., Burch, J.B., Eid, H., and Sucov, H.M. 2002. Epicardial induction of fetal cardiomyocyte proliferation via a retinoic acid-inducible trophic factor. Dev. Biol. 250: 198-207. [DOI] [PubMed] [Google Scholar]
- Chi T.H., Wan, M., Zhao, K., Taniuchi, I., Chen, L., Littman, D.R., and Crabtree, G.R. 2002. Reciprocal regulation of CD4/CD8 expression by SWI/SNF-like BAF complexes. Nature 418: 195-199. [DOI] [PubMed] [Google Scholar]
- Cui K., Tailor, P., Liu, H., Chen, X., Ozato, K., and Zhao, K. 2004. The chromatin-remodeling BAF complex mediates cellular antiviral activities by promoter priming. Mol. Cell. Biol. 24: 4476-4486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de The H., Vivanco-Ruiz, M.M., Tiollais, P., Stunnenberg, H., and Dejean, A. 1990. Identification of a retinoic acid responsive element in the retinoic acid receptor β gene. Nature 343: 177-180. [DOI] [PubMed] [Google Scholar]
- Dhalluin C., Carlson, J.E., Zeng, L., He, C., Aggarwal, A.K., and Zhou, M.M. 1999. Structure and ligand of a histone acetyltransferase bromodomain. Nature 399: 491-496. [DOI] [PubMed] [Google Scholar]
- DiRenzo J., Shang, Y., Phelan, M., Sif, S., Myers, M., Kingston, R., and Brown, M. 2000. BRG-1 is recruited to estrogen-responsive promoters and cooperates with factors involved in histone acetylation. Mol. Cell. Biol. 20: 7541-7549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donato R. 2001. S100: A multigenic family of calcium-modulated proteins of the EF-hand type with intracellular and extracellular functional roles. Int. J. Biochem. Cell Biol. 33: 637-668. [DOI] [PubMed] [Google Scholar]
- Durand B., Saunders, M., Leroy, P., Leid, M., and Chambon, P. 1992. All-trans and 9-cis retinoic acid induction of CRABPII transcription is mediated by RAR-RXR heterodimers bound to DR1 and DR2 repeated motifs. Cell 71: 73-85. [DOI] [PubMed] [Google Scholar]
- Dyson E., Sucov, H.M., Kubalak, S.W., Schmid-Schonbein, G.W., DeLano, F.A., Evans, R.M., Ross Jr., J., and Chien, K.R. 1995. Atrial-like phenotype is associated with embryonic ventricular failure in retinoid X receptor α -/- mice. Proc. Natl. Acad. Sci. 92: 7386-7390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedrich G. and Soriano, P. 1991. Promoter traps in embryonic stem cells: a genetic screen to identify and mutate developmental genes in mice. Genes & Dev. 5: 1513-1523. [DOI] [PubMed] [Google Scholar]
- Geng Y., Yu, Q., Sicinska, E., Das, M., Schneider, J.E., Bhattacharya, S., Rideout, W.M., Bronson, R.T., Gardner, H., and Sicinski, P. 2003. Cyclin E ablation in the mouse. Cell 114: 431-443. [DOI] [PubMed] [Google Scholar]
- Guidi C.J., Sands, A.T., Zambrowicz, B.P., Turner, T.K., Demers, D.A., Webster, W., Smith, T.W., Imbalzano, A.N., and Jones, S.N. 2001. Disruption of Ini1 leads to peri-implantation lethality and tumorigenesis in mice. Mol. Cell. Biol. 21: 3598-3603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heery D.M., Kalkhoven, E., Hoare, S., and Parker, M.G. 1997. A signature motif in transcriptional co-activators mediates binding to nuclear receptors. Nature 387: 733-736. [DOI] [PubMed] [Google Scholar]
- Inoue H., Furukawa, T., Giannakopoulos, S., Zhou, S., King, D.S., and Tanese, N. 2002. Largest subunits of the human SWI/SNF chromatin remodeling complex promote transcriptional activation by steroid hormone receptors. J. Biol. Chem. 27: 27. [DOI] [PubMed] [Google Scholar]
- Jacobson R.H., Ladurner, A.G., King, D.S., and Tjian, R. 2000. Structure and function of a human TAFII250 double bromodomain module. Science 288: 1422-1425. [DOI] [PubMed] [Google Scholar]
- Kastner P., Grondona, J.M., Mark, M., Gansmuller, A., LeMeur, M., Decimo, D., Vonesch, J.L., Dolle, P., and Chambon, P. 1994. Genetic analysis of RXR α developmental function: Convergence of RXR and RAR signaling pathways in heart and eye morphogenesis. Cell 78: 987-1003. [DOI] [PubMed] [Google Scholar]
- Kato H., Tjernberg, A., Zhang, W., Krutchinsky, A.N., An, W., Takeuchi, T., Ohtsuki, Y., Sugano, S., de Bruijn, D.R., Chait, B.T., et al. 2002. SYT associates with human SNF/SWI complexes and the C-terminal region of its fusion partner SSX1 targets histones. J. Biol. Chem. 277: 5498-5505. [DOI] [PubMed] [Google Scholar]
- Kim J.K., Huh, S.O., Choi, H., Lee, K.S., Shin, D., Lee, C., Nam, J.S., Kim, H., Chung, H., Lee, H.W., et al. 2001. Srg3, a mouse homolog of yeast SWI3, is essential for early embryogenesis and involved in brain development. Mol. Cell. Biol. 21: 7787-7795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kliewer S.A., Umesono, K., Noonan, D.J., Heyman, R.A., and Evans, R.M. 1992. Convergence of 9-cis retinoic acid and peroxisome proliferator signalling pathways through heterodimer formation of their receptors. Nature 358: 771-774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klochendler-Yeivin A., Fiette, L., Barra, J., Muchardt, C., Babinet, C., and Yaniv, M. 2000. The murine SNF5/INI1 chromatin remodeling factor is essential for embryonic development and tumor suppression. EMBO Rep. 1: 500-506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemon B., Inouye, C., King, D.S., and Tjian, R. 2001. Selectivity of chromatin-remodelling cofactors for ligand-activated transcription. Nature 414: 924-928. [DOI] [PubMed] [Google Scholar]
- LeRoith D. and Roberts Jr., C.T. 2003. The insulin-like growth factor system and cancer. Cancer Lett. 195: 127-137. [DOI] [PubMed] [Google Scholar]
- Levine M. and Tjian, R. 2003. Transcription regulation and animal diversity. Nature 424: 147-151. [DOI] [PubMed] [Google Scholar]
- Liu R., Liu, H., Chen, X., Kirby, M., Brown, P.O., and Zhao, K. 2001. Regulation of CSF1 promoter by the SWI/SNF-like BAF complex. Cell 106: 309-318. [DOI] [PubMed] [Google Scholar]
- Most P., Boerries, M., Eicher, C., Schweda, C., Ehlermann, P., Pleger, S.T., Loeffler, E., Koch, W.J., Katus, H.A., Schoenenberger, C.A., et al. 2003. Extracellular S100A1 protein inhibits apoptosis in ventricular cardiomyocytes via activation of the extracellular signal-regulated protein kinase 1/2 (ERK1/2). J. Biol. Chem. 278: 48404-48412. [DOI] [PubMed] [Google Scholar]
- Nagy A., Gertsenstein, M., Vintersten, K., and Behringer, R. 2003. Manipulating the mouse embryo, pp. 453-506. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Nie Z., Xue, Y., Yang, D., Zhou, S., Deroo, B.J., Archer, T.K., and Wang, W. 2000. A specificity and targeting subunit of a human SWI/SNF family-related chromatin-remodeling complex. Mol. Cell. Biol. 20: 8879-8888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nie Z., Yan, Z., Chen, E.H., Sechi, S., Ling, C., Zhou, S., Xue, Y., Yang, D., Murray, D., Kanakubo, E., et al. 2003. Novel SWI/SNF chromatin-remodeling complexes contain a mixed-lineage leukemia chromosomal translocation partner. Mol. Cell. Biol. 23: 2942-2952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olave I., Wang, W., Xue, Y., Kuo, A., and Crabtree, G.R. 2002a. Identification of a polymorphic, neuron-specific chromatin remodeling complex. Genes & Dev. 16: 2509-2517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olave I.A., Reck-Peterson, S.L., and Crabtree, G.R. 2002b. Nuclear actin and actin-related proteins in chromatin remodeling. Annu. Rev. Biochem. 71: 755-781. [DOI] [PubMed] [Google Scholar]
- Olson E.N. and Schneider, M.D. 2003. Sizing up the heart: Development redux in disease. Genes & Dev. 17: 1937-1956. [DOI] [PubMed] [Google Scholar]
- Roberts C.W. and Orkin, S.H. 2004. The SWI/SNF complex—Chromatin and cancer. Nat. Rev. Cancer 4: 133-142. [DOI] [PubMed] [Google Scholar]
- Roberts C.W., Galusha, S.A., McMenamin, M.E., Fletcher, C.D., and Orkin, S.H. 2000. Haploinsufficiency of Snf5 (integrase interactor 1) predisposes to malignant rhabdoid tumors in mice. Proc. Natl. Acad. Sci. 97: 13796-13800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross S.A., McCaffery, P.J., Drager, U.C., and De Luca, L.M. 2000. Retinoids in embryonal development. Physiol. Rev. 80: 1021-1054. [DOI] [PubMed] [Google Scholar]
- Rossant J. and Cross, J.C. 2001. Placental development: Lessons from mouse mutants. Nat. Rev. Genet. 2: 538-548. [DOI] [PubMed] [Google Scholar]
- Shelton J.M., Lee, M.H., Richardson, J.A., and Patel, S.B. 2000. Microsomal triglyceride transfer protein expression during mouse development. J. Lipid Res. 41: 532-537. [PubMed] [Google Scholar]
- Skarnes W.C. 2000. Gene trapping methods for the identification and functional analysis of cell surface proteins in mice. Methods Enzymol. 328: 592-615. [DOI] [PubMed] [Google Scholar]
- Stuckmann I. and Lassar, A.B. 2002. Erythropoietin and retinoic acid signaling in the epicardium is required for cardiac myocyte proliferation. Cold Spring Harb. Symp. Quant. Biol. 67: 45-48. [DOI] [PubMed] [Google Scholar]
- Sucov H.M. 1998. Molecular insights into cardiac development. Annu. Rev. Physiol. 60: 287-308. [DOI] [PubMed] [Google Scholar]
- Sucov H.M., Dyson, E., Gumeringer, C.L., Price, J., Chien, K.R., and Evans, R.M. 1994. RXR α mutant mice establish a genetic basis for vitamin A signaling in heart morphogenesis. Genes & Dev. 8: 1007-1018. [DOI] [PubMed] [Google Scholar]
- Torchia J., Rose, D.W., Inostroza, J., Kamei, Y., Westin, S., Glass, C.K., and Rosenfeld, M.G. 1997. The transcriptional co-activator p/CIP binds CBP and mediates nuclear-receptor function. Nature 387: 677-684. [DOI] [PubMed] [Google Scholar]
- Wang W. 2003. The SWI/SNF family of ATP-dependent chromatin remodelers: Similar mechanisms for diverse functions. Curr. Top. Microbiol. Immunol. 274: 143-169. [DOI] [PubMed] [Google Scholar]
- Wu H., Lee, S.H., Gao, J., Liu, X., and Iruela-Arispe, M.L. 1999. Inactivation of erythropoietin leads to defects in cardiac morphogenesis. Development 126: 3597-3605. [DOI] [PubMed] [Google Scholar]
- Wu L., de Bruin, A., Saavedra, H.I., Starovic, M., Trimboli, A., Yang, Y., Opavska, J., Wilson, P., Thompson, J.C., Ostrowski, M.C., et al. 2003. Extra-embryonic function of Rb is essential for embryonic development and viability. Nature 421: 942-947. [DOI] [PubMed] [Google Scholar]
- Xue Y., Canman, J.C., Lee, C.S., Nie, Z., Yang, D., Moreno, G.T., Young, M.K., Salmon, E.D., and Wang, W. 2000. The human SWI/SNF-B chromatin-remodeling complex is related to yeast rsc and localizes at kinetochores of mitotic chromosomes. Proc. Natl. Acad. Sci. 97: 13015-13020. [DOI] [PMC free article] [PubMed] [Google Scholar]