Abstract
Medical image can provide valuable information for preclinical research, clinical diagnosis, and treatment. As the widespread use of digital medical imaging, many researchers are currently developing medical image processing algorithms and systems in order to accommodate a better result to clinical community, including accurate clinical parameters or processed images from the original images. In this paper, we propose a web-based platform to present and process medical images. By using Internet and novel database technologies, authorized users can easily access to medical images and facilitate their workflows of processing with server-side powerful computing performance without any installation. We implement a series of algorithms of image processing and visualization in the initial version of Rayplus. Integration of our system allows much flexibility and convenience for both research and clinical communities.
Keywords: Web technology, Digital image processing, Software design, Medical displays
Introduction
Today, the medical image plays an indispensable role in preclinical research, clinical diagnosis, and treatment. Typical examples are computed tomography (CT), magnetic resonance (MR) or positron emission tomography (PET) imaging. Digital imaging and communication in medicine (DICOM) is the widely accepted standard for medical images and is supported by medical imaging equipment manufacturers from all over the world [1]. The widespread use of digital medical imaging solves the conventional problems such as losing films, spending considerable amounts of time, and expense to copy and archive the films. Moreover, the digital input promotes the development of medical image processing, which has grown into a vast field of study [2].
There are many commercial and non-commercial software systems or toolkits for medical image processing to assist diagnosis and therapy [3–5]. Common functionalities of these software tools include DICOM image presenting, processing (e.g., segmentation, de-noising, registration, fusion) and visualization, all of which require high-capacity data storage and computing. Furthermore, several systems such as virtual endoscopy and surgery planning are homing in on the particular organs or diseases [6, 7]. Currently, most of these systems are local applications that run on high-performance workstations, and the images have to be processed offline.
The objective and challenge for the medical imaging community is to develop software that accommodates a better result to research and clinical community. As healthcare systems continue to increase their use of internet and cloud technologies, software of medical imaging centers are trying to evolve and benefit from these services [8]. By using a web-based platform, with centralized storage of data and availability of the software, there are two advantages for clinicians. The first one is ubiquitous access and processing of medical images from any device, regardless of relative processing power or operating system. The second advantage is easier collaborative analysis.
Various web-based medical software tools have been published because of a strong demand for accessing DICOM images through the web in recent years [9–13]. However, the web applications are usually picture archiving and communication systems (PACS) or Medical Informatics Systems (MIS). Medical Image Access and Presentation System (MIAPS) [13] is one such instance. These systems focus on distributing and archiving medical images via wide area network (WAN) for teleradiology and data sharing but integrate few processing algorithms. Since the processed medical image can offer more accurate and helpful information of tissues and organs, researchers provide web-based software to process and visualize medical images [14–19]. Besides basic tools such as annotation, windowing, and zooming, more complicated algorithms are considered. Jacinto H. et al. [17] proposed a web-accessible visualization and processing framework for medical imaging. In their work, three forms of processing included volume slicing, segmentation and surface meshing, which were completed on server-side. On client-side, they use HTMLCanvasElement object and THREE.js to present 2D/3D images. Mahmoudi SE. et al. [18] suggested an application with algorithms to encompass noise and distortion reduction, edge detection, filtering, segmentation and registration. Most processing algorithms were coded on the server-side and then computed on the client-side. Ortega M. et al. [19] developed a web-based system for retinal image analysis, including special processing modules for retinal diseases. These systems implemented a web environment to process medical images, even having the capability to support some algorithms of computer-aided diagnosis. Nonetheless, there are two drawbacks. Primarily, computing is on client-side, where algorithms coded by ASP.NET, Java, or Javascript actually use the hardware on client-side to get the processed images. Secondly, it is difficult to extend the whole system when novel approaches are necessary.
In this work, we propose a pure web-based platform for medical image processing. The architecture of the system makes the process run on server side with C++ program and makes it possible to turn a local application into webpages rapidly.
Methods
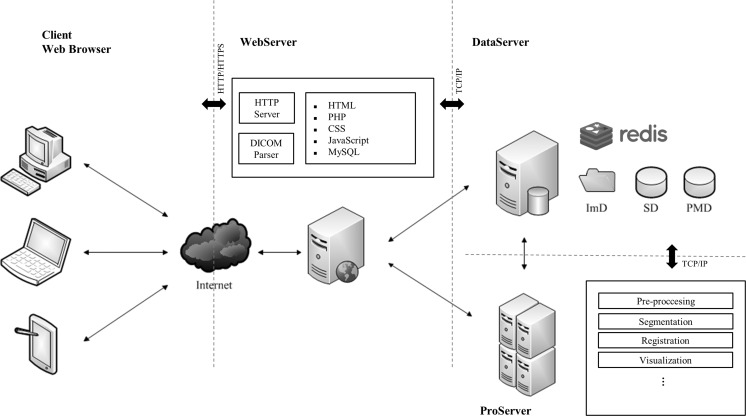
As a browser-server application, RayPlus follows a Model-View-Controller (MVC) architecture to separate internal representations of the data from the user’s interaction. Physically, the architecture essentially encapsulates three servers, which we named WebServer, DataServer, and ProServer (Fig. 1).
Fig. 1.
The architecture of RayPlus
WebServer is the only server that interacts with users via the Internet. The server generates the web pages for browsers to offer a desktop-like user interface and connects with the DataServer and the ProServer. DataServer, as its name shows, is designed for storing and retrieving desired amounts of data including user accounts, DICOM images, and processing pipelines. To perform algorithmic processing on the server side, we use a professional server with as high-performance as the ProServer to satisfy the demand of concurrently graphical calculations and memory occupations. DataServer and ProServer are implemented to be scalable and extensible for future possible upgrades for more storage and more powerful processing capabilities.
Our ideal system should look and feel like a local application and should be instantly available on different devices. For this reason, we use HTML5 and CSS3 to layout the user interaction facilities. Javascript is applied to accomplish the client-side browser controlling and alter the content that is displayed.
On the client side, the web technology covers the base functionalities as menus, toolbars and widgets. The images received from the server are directly update bitmap in HTML5 canvas element. By using the file API (Application Programming Interface), which was added to the DOM (Document Object Model) in HTML5, DICOM data is uploaded via the browser. Javascript implements the basic medical image actions include crop, window/level adjustment, invert, zoom, rotation, annotation, measurement and pseudo-color. Regarding the server side, apache HTTP (The Hypertext Transfer Protocol) server is used on WebServer to respond web requests. By various PHP scripts on WebServer, it generates the dynamic webpages to present the images or processed results from DataServer and ProServer.
When it comes to medical images, DICOM is an indispensable component. NanoDICOM [20], a lightweight PHP DICOM toolkit that follows a MIT license, is used to identify the DICOM images. To ensure that all the uploaded images are from the same scanning, three DICOM tags: patient ID, study instance UID (Unique IDentifier) and series instance UID are used as check codes. The uploaded DICOM images are divided into header information and raw data and stored separately in the database on DataServer. There are three kinds of databases on DataServer: study database (SD), processing manage database (PMD), and image database (ImD). All of them are built on DataServer. Thereinto, SD and PMD are MySQL tables while ImD is a set of files.
Obviously, the presenting and basic processing are far from meeting the need for preclinical research and clinical diagnosis. To make the system flexible, the most important part of RayPlus is offering an open platform to various applications. Redis [21], a BSD licensed key-value cache and store, is a special database in our system. It works with WebSocket to keep an efficient connection and exchange data between different servers. As an in-memory data store used for caches, Redis is built on DataServer. We set diverse key names based on a unified rule to ensure that values, usually images and user interactions, can be sent and received correctly and frequently. For each application, the communication part and the algorithm part are combined into an executable program on the server side. The website on browsers works for presenting the results and interacting with users. In the initial version of RayPlus, we implement the medical image processing modules including Anisotropic filtering, Graph-Cut segmentation, Region-Growing segmentation, IFFD (Incremental Free Form Deformation) registration, Marching-cubes surface modeling, and Ray-casting volume rendering.
Take the volume rendering (VR) as an example. Generally, the web-accessible visualization of volume data is a challenging task. There are several standards and proprietary solutions available in web applications such as VRML (Virtual Reality Modeling Language), X3D, THREE.js. These technologies are all client side graphical methods. However, to medical image 3D visualization, the traditional methods face critical issues: it is memory-consuming and rendering of large data sets takes much time as a typical series of medical image is usually larger than 100 MB.
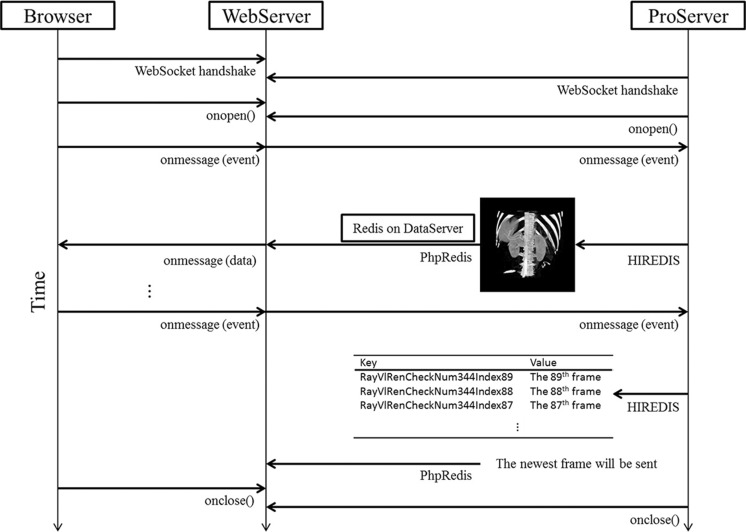
In RayPlus, we first implement a CUDA accelerated volumetric ray-casting algorithm on ProServer. Different with other web-based 3D-visual solutions, we do not transmit the whole 3D model or scene to a browser directly. Temporarily and continuously, 2D projections of 3D scenes are stored into memory as results in PNG format. When a user begins the VR module on his browser, two WebSocket handshakes will start: the browser with the WebServer and the WebServer with the ProServer. Interactions on the browser such as rotating, zooming, transfer function controlling are sent as “event” to WebServer, and then to the ProServer via socket. With the help of HIREDIS [22], the PNG format data are transmitted to Redis database on DataServer from ProServer. In VR application, each frame has a unique key in Redis to distinguish from individual keys from others, and its corresponding value is the 2D frame. The newest frame in Redis will be sent to WebServer through PhpRedis [23] with the key-value mechanism (Fig. 2). Lastly, the 2D projections are rendered on browser within Javascript.
Fig. 2.
The communication between browser and servers
On the webpage, we monitor two kinds of interactions: mouse events and widget events. Mouse events are the mouse actions like left/right button down, left/right button up and mouse move. Widget events are the changes on the web widget such as palette widget, transfer function widget, and so on. These events are packaged into a WebSocket message and unpacked on the server side. A specialized class is developed to unpack various events such as mouse left, mouse right, mouse wheel, button, and slider. By overloading callback functions, the events can be edited by the executable program.
Different applications can be combined for a particular need. Through the management of PMD, the information of each processing to a series of images is recorded, including username, time, the name of algorithm, the setting of parameters, and so on. The result of an application can be the input for another application if it is needed. The applications in RayPlus form a complex pipeline of medical image processing. For instance, a liver surgery planning system is implemented on RayPlus consists of segmentation, visualization and vessel analyzing.
Results
The link of RayPlus is available at http://rayplus.life.hust.edu.cn. The user can access the website through a username/password. An open account for testing is offered. Considering the safety of the whole system, uploading and downloading for test account is disabled. “Contact Us” tab located at the bottom of the page is for any query or assistance. The current version is implemented on the servers which hardware and software specifications are:
WebServer: Intel Xeon E3-1231 v3 CPU, two 8GB RAM; DataServer: two Intel Xeon E5-2620 v2 CPU, two 16GB RAM, two Samsung 512GB SSD and eight WD 2 TB hard drive; ProServer: two Intel Xeon E5-2680 v3 CPU, four 16GB RAM, a NVIDIA Titan X graphics card and a NVIDIA GTX750 graphics card. Three servers are connected with Intel 82599 10Gb Ethernet controller. WebServer and DataServer run Ubuntu 14 for security and robustness while ProServer runs Microsoft Windows 7 64bit for executable programs.
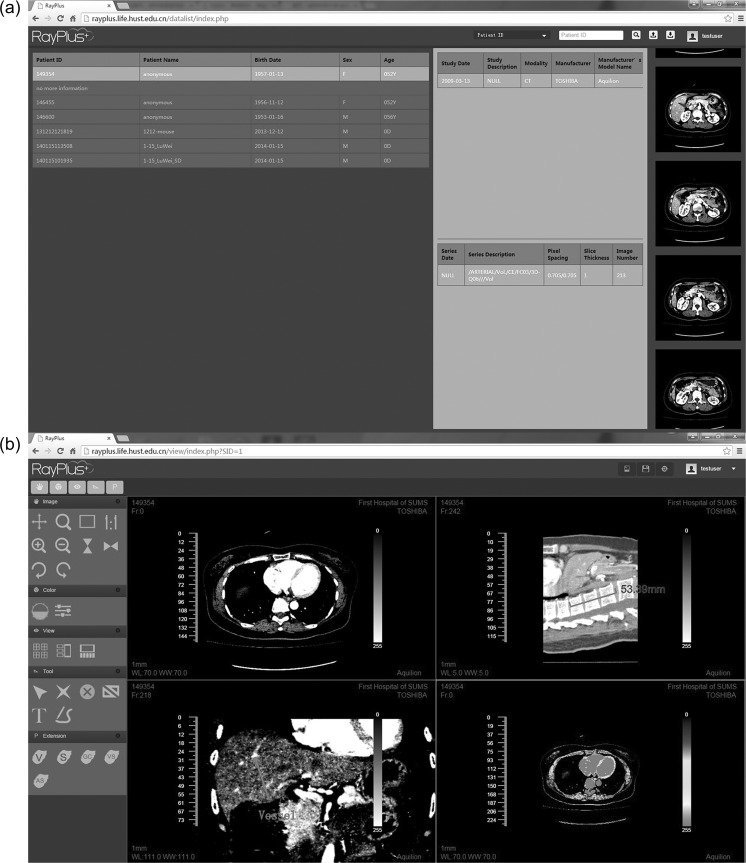
The screen captures of RayPlus on Google Chrome (v43.0.2357.134 m) are shown in Fig. 3. On the webpage of DataList (Fig. 3a), user selects the DICOM series and a preview of the images is listed. After an image series is selected, the image is presented on the webpage of Viewer (Fig. 3b). Tools are offered for crop, window level adjustment, invert, zoom, rotate, annotation and so on. The user interface is designed to be simple and easy-to-use.
Fig. 3.
The web interface of DataList and Viewer. a DataList contains basic DICOM information and a preview of the selected series. Users can search the series based on patient ID, patient name, study date, and series date. b Viewer is designed like classic medical image viewer, offering several basic tools to images and perspectives from coronal and sagittal plane
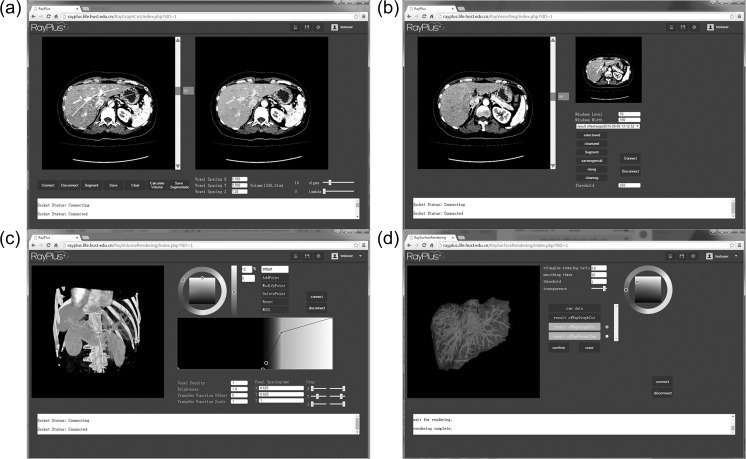
Figure 4 displays typical webpages for the applications of the developed image processing algorithms including semi/full-automatic liver segmentation (Fig. 4a), intrahepatic vessel segmentation (Fig. 4b), surface modeling (Fig. 4c), and volume rendering (Fig. 4d). The applications could combine as a system for hepatic surgical planning.
Fig. 4.
Example of processing for hepatic surgical planning. a Semi-automatic liver segmentation. b Intrahepatic vessel segmentation. c Volume rendering. d Surface modeling
After 2-week online evaluation, we asked 200 questionnaires about this system from 100 radiologists and 100 surgeons. Figure 5 presents questionnaire statistic results: 75% of the radiologists and 80% of the surgeons believe that RayPlus is valuable for clinical use. Fifty-eight percent of the radiologists and 71 % of surgeons think it is easy to operate. Furthermore, except 2 radiologists, all the respondents believe that it has bright prospect.
Fig. 5.
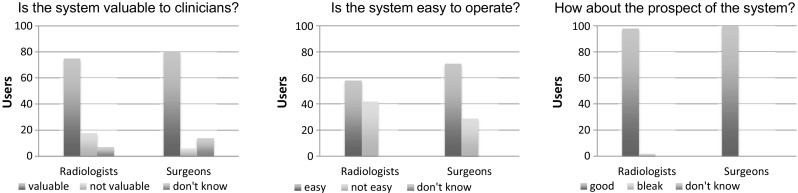
Questionnaire statistic results. a Results of system value to clinicians. b Results of whether the system is easy to operate or not. c Results of prospect of the system
Discussion
As a web-based application, RayPlus is accessible for a range of devices and achieves a far greater level of interoperability than isolated desktop systems. Moreover, maintenance and cooperation become less complicated. On one hand, RayPlus helps clinicians to access and process DICOM images at anytime and anywhere with zero installation via Internet. Once a new version or upgrade is installed on the host server, all users can access it straight away. There is no need to upgrade each client PC. On the other hand, for medical imaging researchers, rolling out new algorithms can be accomplished more easily. And as the upgrades are only performed to servers, the results are more predictable and reliable. Meanwhile, the potential disadvantages of a web-based system are obvious as well. The interface latency is limited by the network bandwidth and the high concurrency will lead to degradation in performance.
Although the initial version of RayPlus realize medical image processing on web service, two issues should be noticed and involved in the future work. An important issue is the mechanism for security and privacy protection. Currently, we use role-based access control to protect the system from vicious and illicit read/write of medical image data. In the future, we plan to employ HTTPS and SSL to enhance security protection of RayPlus. Another issue is considering the REST (REpresentational State Transfer) -based APIs on the application design. The developing system is expected to help in rapidly building new processing methods as web applications and allow for interoperability with a variety of commercial and open source medical image managers. RayPlus may facilitate easier process to medical image of DICOM and open up new avenues for medical image processing.
Conclusions
In this paper, we propose a web-based platform for medical image processing. The main goal of RayPlus is to develop a generic framework to process medical images and provide an open platform to both clinical and imaging communities. In the design of RayPlus, four main requirements are implemented:
It is usable by different users on mainstream browsers from different locations and institutions without any installation (even as ActiveX or Java Applets).
It efficiently manages DICOM images to present and process in an easy way.
It carries out all the high performance computing procedures on server side.
It has scalability and extensibility to potential algorithms.
Acknowledgements
We wish to express our gratitude to Dr. Daoyu Hu, Dr. Zhen Li, and Dr. Yaqi Shen from the Department of Radiology, Tongji Hospital, Wuhan, China, for all the inspiring discussions. This work was supported by the National Natural Science Foundation of China (Grant No. 61540041) and Ministry of Science and Technology of the People’s Republic of China (Grant No. 2013YQ030923).
References
- 1.Mildenberger P, Eichelberg M, Martin E. Introduction to the DICOM standard. Eur Radiol. 2002;12(4):920–7. doi: 10.1007/s003300101100. [DOI] [PubMed] [Google Scholar]
- 2.Setarehdan SK, Singh S: Advanced algorithmic approaches to medical image segmentation: state-of-the-art applications in cardiology, neurology, mammography and pathology. Springer Science & Business Media, 2012
- 3.Pieper S, Halle M, Kikinis R. 3D slicer. In: biomedical imaging: Nano to macro. IEEE Int. Symp. 2004;2004:632–5. [Google Scholar]
- 4.Keeve E, Jansen T, Krol Z, Ritter L, von Rymon-Lipinski B, Sader R, Zeilhofer HF, Zerfass P: JULIUS—an extendable software framework for surgical planning and image-guided navigation. In: Medical Image Computing and Computer-Assisted Intervention–MICCAI 2001. Springer; 2001. p. 1336–7
- 5.Rosset A, Spadola L, Ratib O. OsiriX: an open-source software for navigating in multidimensional DICOM images. J Digit Imaging. 2004;17(3):205–216. doi: 10.1007/s10278-004-1014-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bourquain H, Schenk A, Link F, Preim B, Prause G, Peitgen HO: HepaVision2—a software assistant for preoperative planning in living-related liver transplantation and oncologic liver surgery. In: CARS 2002 Computer Assisted Radiology and Surgery. Springer; 2002. p. 341–6
- 7.Shevchenko N, Seidl B, Schwaiger J, Markert M, Lueth T. MiMed liver: a planning system for liver surgery. In: engineering in medicine and biology society (EMBC) Annual Int Conf IEEE. 2010;2010:1882–5. doi: 10.1109/IEMBS.2010.5627120. [DOI] [PubMed] [Google Scholar]
- 8.Kagadis GC, Kloukinas C, Moore K, Philbin J, Papadimitroulas P, Alexakos C, Nagy PG, Visvikis D, Hendee WR. Cloud computing in medical imaging. Med Phys. 2013;40(7):070901. doi: 10.1118/1.4811272. [DOI] [PubMed] [Google Scholar]
- 9.Poh CL, Kitney R, Shrestha RBK. Addressing the future of clinical information systems—web-based multilayer visualization. IEEE Trans Inf Technol Biomed. 2007;11(2):127–140. doi: 10.1109/TITB.2006.875680. [DOI] [PubMed] [Google Scholar]
- 10.Hernandez JA, Acuna CJ, de Castro M, Marcos E, Lopez M, Malpica N. Web-PACS for multicenter clinical trials. IEEE Trans Inf Technol Biomed. 2007;11(1):87–93. doi: 10.1109/TITB.2006.879601. [DOI] [PubMed] [Google Scholar]
- 11.Klimes D, Kubasek M, Smid R, Dusek L. Internet-based system for anti-tumor chemotherapy evaluation. Comput. Methods Prog. Biomed. 2009;93(3):292–296. doi: 10.1016/j.cmpb.2008.10.013. [DOI] [PubMed] [Google Scholar]
- 12.Kennedy DN, Haselgrove C, Makris N, Goldin DM, Lev MH, Caplan D, Caviness VS. WebParc: a tool for analysis of the topography and volume of stroke from MRI. Med Biol Eng Comput. 2010;48(3):215–228. doi: 10.1007/s11517-009-0571-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shen H, Ma D, Zhao Y, Sun H, Sun S, Ye R, Huang L, Lang B, Sun Y. MIAPS: a web-based system for remotely accessing and presenting medical images. Comput. Methods Prog. Biomed. 2014;113(1):266–283. doi: 10.1016/j.cmpb.2013.09.008. [DOI] [PubMed] [Google Scholar]
- 14.Matsopoulos GK, Kouloulias V, Asvestas P, Mouravliansky N, Delibasis K, Demetriades D. MITIS: a WWW-based medical system for managing and processing gynecological–obstetrical–radiological data. Comput. Methods Prog. Biomed. 2004;76(1):53–71. doi: 10.1016/j.cmpb.2004.03.001. [DOI] [PubMed] [Google Scholar]
- 15.Bigler DC, Aksu Y, Miller DJ, Yang QX. STAMPS: software tool for automated MRI post-processing on a supercomputer. Comput. Methods Prog. Biomed. 2009;95(2):146–157. doi: 10.1016/j.cmpb.2009.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Papamichail D, Pantelis E, Papagiannis P, Karaiskos P, Georgiou E. A web simulation of medical image reconstruction and processing as an educational tool. J Digit Imaging. 2015;28(1):24–31. doi: 10.1007/s10278-014-9689-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jacinto H, Kechichian R, Desvignes M, Prost R, Valette S: A web interface for 3D visualization and interactive segmentation of medical images. In: Proceedings of the 17th International Conference on 3D Web Technology. ACM; 2012. p. 51–58
- 18.Mahmoudi SE, Akhondi-Asl A, Rahmani R, Faghih-Roohi S, Taimouri V, Sabouri A, et al. Web-based interactive 2D/3D medical image processing and visualization software. Comput. Methods Prog. Biomed. 2010;98(2):172–182. doi: 10.1016/j.cmpb.2009.11.012. [DOI] [PubMed] [Google Scholar]
- 19.Ortega M, Barreira N, Novo J, Penedo MG, Pose-Reino A, Gomez-Ulla F. Sirius: a web-based system for retinal image analysis. Int J Med Inform. 2010;79(10):722–732. doi: 10.1016/j.ijmedinf.2010.07.005. [DOI] [PubMed] [Google Scholar]
- 20.Nanodicom, Available at http://www.nanodicom.org. Accessed 18 November 2015.
- 21.Redis, Available at http://redis.io. Accessed 18 November 2015.
- 22.GitHub, Available at https://github.com/redis/hiredis. Accessed 18 November 2015.
- 23.GitHub, Available at https://github.com/phpredis/phpredis. Accessed 18 November 2015.