Abstract
In an genomics course sponsored by the Howard Hughes Medical Institute (HHMI), undergraduate students have isolated and sequenced the genomes of more than 1,150 mycobacteriophages, creating the largest database of sequenced bacteriophages able to infect a single host, Mycobacterium smegmatis, a soil bacterium. Genomic analysis indicates that these mycobacteriophages can be grouped into 26 clusters based on genetic similarity. These clusters span a continuum of genetic diversity, with extensive genomic mosaicism among phages in different clusters. However, little is known regarding the primary hosts of these mycobacteriophages in their natural habitats, nor of their broader host ranges. As such, it is possible that the primary host of many newly isolated mycobacteriophages is not M. smegmatis, but instead a range of closely related bacterial species. However, determining mycobacteriophage host range presents difficulties associated with mycobacterial cultivability, pathogenicity and growth. Another way to gain insight into mycobacteriophage host range and ecology is through bioinformatic analysis of their genomic sequences. To this end, we examined the correlations between the codon usage biases of 199 different mycobacteriophages and those of several fully sequenced mycobacterial species in order to gain insight into the natural host range of these mycobacteriophages. We find that UPGMA clustering tends to match, but not consistently, clustering by shared nucleotide sequence identify. In addition, analysis of GC content, tRNA usage and correlations between mycobacteriophage and mycobacterial codon usage bias suggests that the preferred host of many clustered mycobacteriophages is not M. smegmatis but other, as yet unknown, members of the mycobacteria complex or closely allied bacterial species.
Keywords: bacteriophages, codon bias, host range, mycobacteria, UPGMA clustering, viral tRNAs
Data Summary
All genomic sequence data analyzed in this study were downloaded from NCBI GenBank via links provided at phagesdb.org. The GenBank sequence accession numbers are provided in Tables S1 and S2 (available in the online Supplementary Material) for bacteriophage and Mycobacterial genome sequences respectively.
Impact Statement
Through a course in bacteriophage discovery and genomics, thousands of undergraduate students isolated and sequenced the genomes of bacterial viruses (bacteriophages) able to infect the bacterial host, Mycobacterium smegmatis, thus creating the largest database of bacteriophages able to infect a single host type. However, little is known about the genetic organization of these phages or of their natural hosts in the wild. Here we use bioinformatic analyses to identify relationships among these phages and sequenced mycobacterial species. Based on our bioinformatic analyses, we report that M. smegmatis is unlikely to be the preferred host for many of these newly isolated bacteriophages. Instead we suggest that many isolated mycobacteriophages infect similar, but as yet unknown, mycobacterial species or have recently gained the ability to infect Mycobacteria.
Introduction
Bacteriophages are the most populous organisms in the biosphere, but surprisingly little is known about their natural diversity and host ranges (Dennehy, 2014). One of the best-studied groups of phages are the mycobacteriophages, which infect mycobacterial hosts such as Mycobacterium tuberculosis and Mycobacterium smegmatis. To date, students participating in the Howard Hughes Medical Institute (HHMI)-sponsored Science Education Alliance–Phage Hunters Advancing Genomics and Evolutionary Science (SEA-PHAGES) initiative (seaphages.org) have isolated almost 7000 mycobacteriophages from soil samples using the host M. smegmatis (phagesdb.org). Of these, more than 1150 mycobacteriophage genomes have been fully sequenced and annotated for open reading frames (ORFs), tRNA genes and other features (Pope et al., 2015). One surprising finding is that despite having the ability to infect the same host many mycobacteriophages share little or no genetic similarity (Pope et al., 2015; Brüssow & Hendrix, 2002). Moreover, extensive genomic mosaicism makes it impossible to determine the phylogeny of mycobacteriophages (Pedulla et al., 2003; Cresawn et al., 2011). Instead mycobacteriophages exist in constellations of closely related phages, termed clusters, constituting a continuous spectrum of genetic diversity (Pope et al., 2015; Grose & Casjens, 2014).
Despite the expanding knowledge of mycobacteriophage diversity and genetic content, little is known about their life history and ecology. Some of these newly isolated phages infect and form plaques on M. tuberculosis and Mycobacterium bovis (Rybniker et al., 2006; Jacobs-Sera et al., 2012), but we have little insight into the broader host ranges of these phages or of their preferred hosts in the wild. The SEA-PHAGES ecological data is mainly limited to the geographic coordinates of isolation, the date of isolation, and occasionally the uncurated discovery notes. As such, analysis of mycobacteriophage genomic sequences may be one of the best ways of acquiring ecological and evolutionary insight.
In this study, we analyzed the codon usage patterns and DNA GC content of 199 different mycobacteriophage genomes to determine if codon usage and DNA GC content patterns suggest evolutionary relationships or possible preferred hosts. Codon usage bias refers to the differences in the frequency of use of synonymous codons during protein synthesis. Despite having multiple synonymous codons for a given amino acid, organisms do not use these codons randomly or at equal frequencies (Sharp & Li, 1987; Hilterbrand et al., 2012), suggesting that codon usage bias may affect organismal fitness and/or function (Kudla et al., 2009; Parmley & Hurst, 2007). Since phages do not encode ribosomes, they are entirely dependent on their host’s translational machinery for replication. Efficient translation of a phage's proteins within a host is optimized by the phage's ability to match the codon usage patterns of their hosts (Carbone, 2008; Lucks et al., 2008). Hence we expect a correlation between the codon usage patterns of the phage and its host. An exception to this pattern may occur when phages encode their own tRNAs (Bailly-Bechet et al., 2007; Chithambaram et al., 2014). Consequently, the mycobacteriophage codon usage patterns will most closely resemble that of the preferred host, except in cases where the phage encodes its own tRNA for a particular amino acid.
Methods
Mycobacteriophage genomic analysis.
Genome sequence data was obtained from the SEA-PHAGES initiative (phagesdb.com) for phages that were previously clustered into groups according to their nucleotide similarity (Pope et al., 2015). Two genomes are placed in the same cluster if: (1) dot plot sequence similarity is >50 % of the smaller of the two genomes; (2) average nucleotide identity is >70 %; (3) the bioinformatics program Splitstree (Huson & Bryant, 2006) assigns the two genomes to a clearly defined group; and (4) the two genomes show a high degree of genome module similarity based on pairwise sequence alignments (Hatfull et al., 2010). Phages that do not meet all of these criteria are not assigned to a cluster and are termed ‘singletons’ because they have no close relatives. The SEA-PHAGES initiative identified 26 different mycobacteriophage clusters and numerous subclusters and singletons (phagesdb.com) (Pope et al., 2015). Members of a cluster tend to share genome architectures in addition to sequence similarities, and have similar genome lengths and numbers of genes per genome (Pope et al., 2015).
Another layer of genomic analysis is the assignment of genes into ‘phams’, or groups of closely related sequences, using the program Phamerator (Cresawn et al., 2011). Two genes share a pham if the amino acid sequence identity given by ClustalW alignments is >32.5 % and the if BlastP E-value is <10–50 (Cresawn et al., 2011). As of he time of writing, the total number of mycobacteriophage phams assigned is more than 21 000, which suggests a tremendous wealth of biologically novel genes (phagesdb.com).
Selection of mycobacteriophages and mycobacterial species.
Mycobacteriophages were selected for this study based on the availability of a fully annotated genome in the NCBI GenBank as of June 2013. In most clusters, all fully sequenced mycobacteriophages available were selected for further study. However in the A cluster, some subclusters contained more phages than many of the other clusters. Therefore, in order to avoid redundancy and biased results, 10 phages from each of the A1 and A4 subclusters of the A cluster were selected at random. A total of 199 complete mycobacteriophage genomes were downloaded from NCBI GenBank (Table S1). Seven species of the genus Mycobacterium, M. smegmatis mc2155, M. bovis BCG, M. tuberculosis H3R7v, Mycobacterium avium K-10, Mycobacterium leprae TN, Mycobacterium ulcerans AGY99 and Mycobacterium abscessus bolletii 50594, were selected based on previously known infectivity patterns with the selected mycobacteriophages (Rybniker et al., 2006; Jacobs-Sera et al., 2012) and their full genomes were downloaded from NCBI GenBank (Table S2).
Analysis of codon ssage bias.
The relative synonymous codon usage order (RSCU) is the ratio of the observed frequency of codons to the frequency expected if all synonymous codons were used equally (Sharp & Li, 1987). RSCU values were calculated for all mycobacteriophage and mycobacterial genomes using the program mega 6.1 (megasoftware.net) (Tamura et al., 2013). The RSCU values for all bacterial genomes were obtained from a previously reported analysis on the codon bias database (CBDB; cbdb.info) (Hilterbrand et al., 2012). Synonymous codon usage order (SCUO) is a measure determining the synonymous codon usage bias within and across genomes (Wan et al., 2004, 2006). SCUO is a newer method of analyzing codon usage bias, and is based on Shannon's information theory. We selected this method for several reasons. Since we have no a priori knowledge of bacteriophage gene expression levels, and do not have validated reference genomes, codon analysis methods based on reference genomes, such as the Codon Adaption Index (CAI), may not provide a robust analysis. Second, SCUO takes genome GC composition into account, which may be more appropriate given that mycobacteriophage and mycobacterial genomes are highly GC-biased. Furthermore, since our comparisons are mainly between genomes, including comparisons between bacteriophage and bacteria genomes, rather than among genes, we chose SCUO since it is considered to be a more robust method for between-genome comparisons. The SCUO for each genome was calculated using the program INCA 2.1 (bioinfo.hr/research/inca/).
tRNA abundance and synonymous codon usage order.
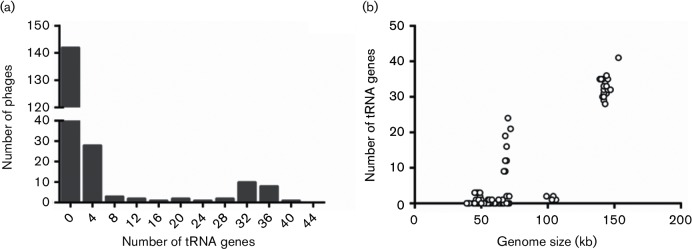
Genes encoding tRNAs are often found in mycobacteriophage genomes (Bailly-Bechet et al., 2007) (Figs 1, 2). Bacteriophages use their host’s translational machinery to reproduce, which limits successful propagation to the tRNA pool found in the host (Bailly-Bechet et al., 2007; Plotkin & Kudla, 2011). Because of this, bacteriophages encoding their own tRNAs are predicted to have higher codon usage biases than bacteriophages that do not encode their own tRNAs. We used the software tRNAscan-SE 1.21 to identify tRNA genes in mycobacteriophage genomes downloaded from NCBI GenBank (Lowe et al., 1997; Schattner et al. 2005) (lowelab.ucsc.edu/tRNAscan-SE). SCUOs were determined for genomes that encode and do not encode their own tRNAs in order to determine if tRNA prevalence is correlated with codon usage bias.
Fig. 1.
Histogram of tRNA-encoding genes found in the 199 mycobacteriophages analyzed in this study.
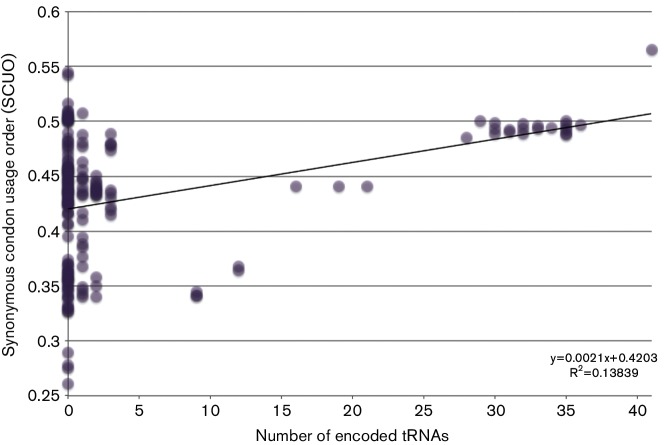
Fig. 2.
The synonymous codon usage order (SCUO) was calculated for each genome. SCUO values for each mycobacteriophage were compared with the quantity of encoded tRNA genes. tRNA abundance does not predict SCUO. However, if a phage does have tRNA genes, it tends to have a more biased genome.
Cluster analysis based on phage and host RSCU values.
Cluster analysis has been used to study the patterns of codon usage bias of genes within a genome, as well as across organisms (Sharp & Li, 1987). The RSCU for each bacteriophage was compared with those of all other bacteriophages using Pearson's correlation coefficient. A distance matrix was constructed, where the distance value (d)=(1−r)×100 where r is the Pearson coefficient. In this study, we used the software dendroUPGMA to construct unweighted pair group method with arithmetic mean (UPGMA) dendrograms to cluster all genomes and structural proteins according to their RSCU values (Garcia-Vallvé et al., 1999) (genomes.urv.cat/UPGMA). From this point forward, we will refer to the above-mentioned clusters as UPGMA clusters or nodes in order to distinguish these from the SEA-PHAGES clusters and subclusters created on the basis of genetic similarities.
Results and Discussion
Mycobacteriophage GC content and comparisons with mycobacterial GC content
Despite a universal bias towards GC→AT mutations (Hildebrand et al., 2010; Hershberg & Petrov, 2010; Ran et al., 2014), genome GC content ranges widely among in prokaryotes (Foerstner et al., 2005; Bohlin et al., 2010, Bentley & Parkhill, 2004), and some organisms such as the Mycobacteria, have high DNA GC content. Studies have shown that DNA GC content is correlated with genome length (Mitchell, 2007; Musto et al., 2006; Pedulla et al., 2003), phylogeny (Hershberg & Petrov, 2010) and ecological and environmental factors (Foerstner et al., 2005). The Actinobacteria, the phylum in which the Mycobacteria are classified, are known for their high DNA GC content. This high DNA GC content may reflect the complexities of the soil habitat characteristic of many actinobacterial species (Foerstner et al., 2005; Tringe et al., 2005).
While the high DNA GC content of the mycobacteriophages with small genomes seems at odds with the correlation between GC content and genome size, this trait is likely to be a result of correlations between the mycobacteriophages and their high-DNA-GC-content mycobacterial hosts (e.g. M. smegmatis DNA GC content: 68 mol%) (Bahir et al., 2009; Andersson & Sharp, 1996; Xia & Yuen, 2005). Since DNA GC content constrains codon usage and, therefore, may affect translational efficiency, it is expected that virus DNA GC content will match that of their hosts (Carbone 2008; Bahir et al. 2009). If this is true, the mismatch between the 68 mol% DNA GC content of M. smegmatis and many of the mycobacteriophages is inconsistent. Only phages from the B and K clusters have DNA GC contents this high (Table S3). If the other Mycobacteria (other than M. leprae) are considered, they have DNA GC contents ranging from 65 mol% to 69 mol% (Table S2). Mycobacteriophage clusters that fall within this range include B, C, G, K, I, N, O and P (Table S3). All other clusters fall below this threshold, including sub-60 mol% DNA GC content clusters D, L and H.
The differences between the DNA GC content of isolation host M. smegmatis and the DNA GC contents of many phage clusters may indicate that M. smegmatis is not the preferred host of many of these phages. Given the tremendous diversity of microbes in the soil (Fierer & Jackson, 2006), it is likely that soils contain numerous permissive hosts for a given phage type, and that phages are able to shift from one host to another as host populations wax and wane. It may be that many mycobacteriophages isolated on M. smegmatis actually prefer other close relatives of M. smegmatis which also contain high (but not quite as high) DNA GC contents, such as Corynebacteria (53.5 mol%), Rhodococcus and Gordonia.
Expanding upon the previous work of Hatfull and colleagues, it is evident that across the mycobacteriophages discovered to date, there is considerable variation in the DNA GC percentage from cluster to cluster and between subclusters, but little variation between phages belonging to the same subcluster (Jacobs-Sera et al., 2012). Using the A cluster as an example, an analysis of variance of DNA GC content with subcluster as a factor revealed significant differences among subclusters [degrees of freedom (DF)=11 409; P<0.0001, F=555.4]. These findings tend to support the idea that subcluster-level differentiation represents rapid diversification and host or niche specialization.
Encoding tRNAs and codon bias
Of our selected 199 mycobacteriophages, 88 encode genes for tRNAs (Fig. 1, Table S1). The frequency of tRNA genes encoded by the mycobacteriophages varies considerably. Most mycobacteriophage encode zero tRNA genes, but C cluster phage Myrna encodes 41 tRNA genes (Fig. 1, Table S1). In general, the C cluster phages encode the most tRNA genes, which is expected given that they also possess the largest genomes among the mycobacteriophages. A one-way ANOVA of the number of tRNA genes against genome size was highly significant (F=774.9, P<0.0001, DF=1198; Fig. 1). A similar result has been reported by Bailly-Bechet and colleagues in their analysis of bacteriophages infecting a wide variety of host types (Bailly-Bechet et al., 2007).
It may be that phages with larger capsids are able to incorporate greater numbers of tRNA genes because space constraints are less stringent. Perhaps larger genomes experience reduced deletional bias that is often characteristic of bacteriophage genomes (Mira et al., 2001; Lawrence et al., 2001). An observation that may have some bearing on this issue is the fact that despite having similar sized genomes, the C cluster phages vary tremendously in the number of tRNA genes they encode.
SCUOs for each genome and gene within the genome were determined using INCA v2.1. Phages that encode a large number of genes for tRNAs were found to also have high SCUO values, indicating a high codon usage bias within that phage genome, as predicted (Fig. 2). Similar correlations between genome codon bias and the presence of tRNA genes have been found for prasinoviruses (Michely et al., 2013), coliphages (Chithambaram et al., 2014) and mimiviruses (Colson et al., 2013). Remarkably, phages that do not possess any genes for tRNAs can exhibit SCUO values just as high as phages that do possess tRNAs (Fig. 2). This finding indicates that these phages have preferred hosts with similar biases for efficient protein translation.
UPGMA clustering of the mycobacteriophages and their potential hosts
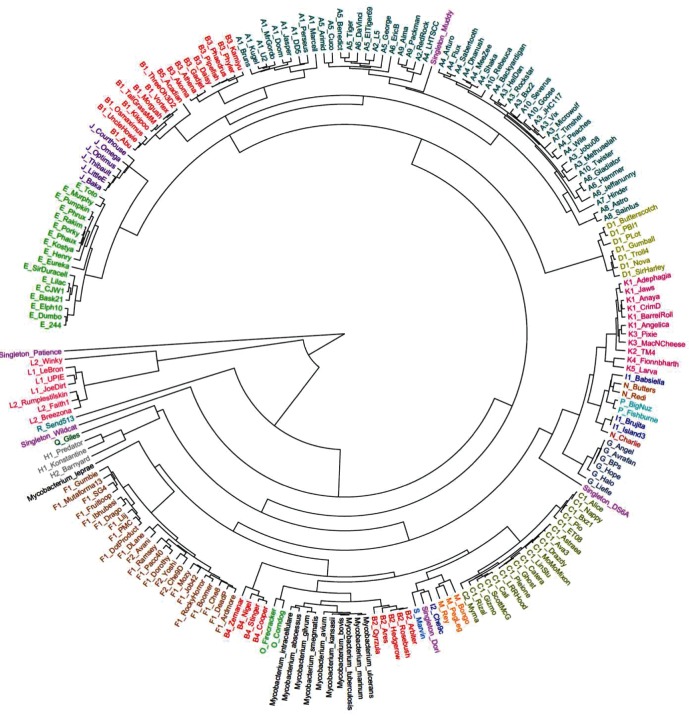
We used UPGMA cluster analysis to characterize the frequency of codon usage in mycobacteriophages and to group mycobacteriophage and mycobacterial genomes and proteins based on their RSCU values. A 59-dimensional comparison was performed using the RSCU values for each codon (excluding stop codons, methionine and tryptophan) for a given mycobacteriophage or Mycobacterium (Fig. 3). Dendrograms were reconstructed using the UPGMA method to reconstruct UPGMA clusters based on the Pearson correlation between the RSCU values of each codon for each genome (Garcia-Vallvé et al., 1999). As predicted, mycobacteriophages belonging to the same genomic cluster generally shared a UPGMA cluster, but this was not the case for all genomes analyzed. The B and I clusters stand out for being split between multiple, widely separated, branches of the UPGMA dendrogram (Fig. 3). However, mycobacteriophages belonging to the same subcluster possessed strong similarities in codon bias, and this is most notable throughout the B cluster phages, despite their being split among different UPGMA nodes.
Fig. 3.
Relative synonymous codon usage (RSCU) was calculated for each mycobacteriophage and mycobacterial species and used to reconstruct this UPGMA-dendrogram. Colors correspond to the cluster designations assigned by phagesdb.org.
Genomic subclusters B1, B3 and B5 are split between branches stemming from a shared UPGMA node with the A and J clusters, highlighting their distinct codon usage bias between the B cluster. The B2 subcluster is found on a branch that shares a UPGMA node with the C, M and O clusters and all but one of the Mycobacteria. This is significant because the B2 phages have been known to infect M. tuberculosis. The B4 subcluster is split off from one step above this node. The B3 subcluster is of particular interest because, based upon BlastN alignments, we see regions of high and low similarity with the A1, J and O clusters, which may indicate recombination with phages of these clusters. Nonetheless, recombination does not explain the divergent UPGMA clustering. Instead, it may be the case that adaptation to different host types resulted in similar nucleotide sequences, but distinct codon usage patterns, among the B cluster phages.
On the other hand, the smaller cluster I is split between: (1) the I1 subcluster, which shares a UPGMA node with N and P clusters; and (2) the sole I2 phage, which shares a UPGMA node with the Singleton Dori on a widely separate branch. Analysis of the BlastN alignments suggests that the I2 phage does not share a recent evolutionary history with the I1 subcluster, and the main reason for their sharing the same cluster is a recombination event between the progenitors of the two groups.
The clustering based on RSCU values reveals another dimension of diversity within the mycobacteriophage population (Fig. 3). Across all of the genomes analyzed, three distinct groups emerge after UPGMA analysis: a lone group containing Patience; a second group containing all of the mycobacteriophages belonging to the L genomic cluster; and the rest of the mycobacteriophages, which share a third group. Within the largest UPGMA-cluster, the Mycobacteria cluster, with the exception of M. leprae. Mycobacteria that are closely related phylogenetically, such as M. tuberculosis and M. bovis, share a UPGMA cluster on close connecting branches (Gao & Gupta, 2012; Tortoli, 2012). One compelling observation is that M. leprae, the most distant relation of the Mycobacteria, shares a node with the F cluster phages. Based on this finding, we speculate that there is an evolutionary history of infection among the F cluster phages and M. leprae. However, generally speaking, the lack of correlations between mycobacteriophage and mycobacterial host DNA GC content and codon usage patterns suggests that M. smegmatis is not the preferred host for these mycobacteriophages.
The A cluster is the largest mycobacteriophage cluster, containing approximately 249 phages (Pope et al., 2011; Jacobs-Sera et al., 2012). In our UPGMA analysis, the A cluster phages appear to be divided among three distinct branches. The first branch harbors only A1-phages and shares a node with some of the B cluster mycobacteriophages. The second and third branches of the A cluster mycobacteriophages share a common node, but are separated throughout the branches in a way that would not be predicted when looking at average nucleotide identities alone. Specifically, the A6 subcluster is found on the two UPGMA branches of the A cluster. It would appear that the A cluster phages share a high degree of nucleotide similarity, but do not subcluster together as distinctly as other mycobacteriophage clusters, reflecting the fact that genomic clusterization of the mycobacteriophages is mainly a useful organizational scheme rather than an attribute with a strong biological basis. In the long run, phylogenetic analysis of the mycobacteriophages, and other phages, may be most effective at the level of the gene.
Mycobacterium leprae and the mycobacteriophages of the F cluster
Mycobacterium leprae is the causative agent of leprosy or Hansen’s disease, and has a long history of infecting humans (Bhat & Prakash, 2012). As such, it is significant that the F cluster mycobacteriophages share a similar codon bias pattern with M. leprae (Fig. 3). We conducted an analysis of variance (ANOVA) on the hierarchical UPGMA clustering of RSCU values of each codon among the 23 F cluster phages and M. leprae, and found no significant differences among groups. Only the utilization of codons encoding leucine, isoleucine, valine and asparagine showed significantly different RSCU values between the F cluster phages and M. leprae. Also differences in the RSCU values for the codons of these amino acids is not uniform across all of the phages – most different are Boomer, Che8, Che9D, SG4 and RockyHorror mycobacteriophages. Moreover, there have been no reported encoded tRNA-genes in the F cluster phages. Although carrying out an infection-assay with the F cluster phages and M. leprae TN would be difficult, it is plausible that these phages would be able to infect M. leprae in the wild. These could be linked to the ability of the F cluster phages to infect M. smegmatis, and thus an adaptation for the host-range of these phages. It would be interesting to see if this is found for other members of the Mycobacteria genera.
The singleton mycobacteriophages and host range expansion
The mycobacteriophage Patience, which is the most genetically distinct mycobacteriophage (Table S1) (Pope et al., 2014), is found at the base of the dendrogram as the sole member of a branch separate from all other mycobacteriophages and Mycobacteria (Fig. 3). Compared with the other singleton phages, the singleton Patience is essentially the ‘singleton of the singletons’, although since the time of analysis another closely related phage (Madruga) has been discovered. Given that Patience has the lowest DNA GC content (50.4 mol%) of any phage in this study, it is tempting to speculate that Patience formerly infected a host with a lower DNA GC content, and has recently emerged in Mycobacteria (Pope et al., 2014; Dennehy, 2009). Despite the mismatch in DNA GC content between Patience and M. smegmatis (68 mol%), Patience does not seem to suffer impaired growth on M. smegmatis (Pope et al., 2014; Hatfull, 2015). While Patience's robust growth on M. smegmatis seems to imply that differences in codon utilization do not hinder Patience's growth, we note that this growth is achieved under relatively benign laboratory conditions in high-nutrient media. It may be that under more challenging conditions, Patience would be unable to reproduce to high levels because of inefficient translation.
Moreover, Pope et al. (2014) point out that Patience does experience codon selection, which is shown by the robust positive correlation between codon selection (adaptive codon enrichment) and the level of gene expression. Finally, a considerable fraction (29 out of 109) of the predicted ORFs were not observed to express peptide products. It is possible that these observations stem from translational failures, although there is no direct evidence to support this.
Another noteworthy singleton, Muddy, shares a similar codon usage pattern with mycobacteriophages belonging to a distinct subset of genomic subclusters within the A cluster (Fig. 3). Interestingly, Muddy is 93.2 % identical at the nucleotide level (E value=0.0) to the phage vB_MapS_FF47, which was isolated from bovine feces using the bacterium Mycobacterium avium subspecies paratuberculosis ATCC 19698 (Basra et al., 2014). Although it was isolated on M. avium ATCC 19698, FF47 was not able to infect six out of eight M. avium strains tested, but was able to infect M. smegmatis mc2155 (Basra et al., 2014). Despite their high degree of similarity, phages FF47 and Muddy were isolated from locations that are approximately 13 890 kilometers apart, Durban, South Africa and Guelph, Canada, respectively (Basra et al., 2014). While Muddy shares an UPGMA node with the A cluster phages, it shares little nucleotide sequence similarity with these phages. We interpret this result as indicating that while Muddy and the A cluster phages do not share an evolutionary history, and hence have little shared sequence identity, they do share similar codon usage patterns due to similar selective pressure, most likely a shared host.
Singleton mycobacteriophage DS6A is the only known mycobacteriophage to infect only mycobacteria of the TB complex (M. tuberculosis, M. bovis, M. africanum, etc.) (Jacobs-Sera et al., 2012; Hatfull et al., 2010). On analysis of the RSCU values, we find that this phage branches off a larger branch containing phages belonging to the G, N, I, P and K clusters. Intriguingly, only mycobacteriophages of the A1, A2, A3, A9, B1, B2, G, K and M subclusters are able to infect M. tuberculosis at relatively high efficiencies of plating (i.e. >10−4) (Jacobs-Sera et al., 2012; Sampson et al., 2009). The observation that the G and K cluster mycobacteriophages share a UPGMA node with the Singleton phage DS6A is suggestive that there is an underlying genetic similarity between these mycobacteriophages, which enables them to infect M. tuberculosis. We speculate that these mycobacteriophages with high-efficiency of M. tuberculosis plating contain the requisite gene(s) permitting infection of M. tuberculosis, or can easily acquire mutations allowing infection of M. tuberculosis. Curiously, the I cluster mycobacteriophages, which shares a subnode of the G cluster with the K, N and P cluster phages, are unable to infect TB Complex bacteria. Presumably, in the past, the I cluster phages possessed the ability to infect M. tuberculosis, but have since lost that ability due to one or more mutations (Jacobs-Sera et al., 2012). To our knowledge, P and N cluster phages have not been tested on M. tuberculosis, but it would be interesting to see if these phages have the ability to infect this specie
Based on the UPGMA clustering we would not expect the A1, A2, A3, A9, B1, B2 and M cluster phages to be able to infect M. tuberculosis, but they can. We speculate that either the common ancestor of these mycobacteriophage clusters possessed the ability to infect M. tuberculosis but it was subsequently lost in several diversifying lineages or that these clusters acquired the ability to infect M. tuberculosis through mutation or horizontal gene transfer.
Conclusions
The SEA-PHAGES program has made is possible to conduct large-scale comparative genomic studies of mycobacteriophages. Such a vast collection of sequences allows for large-scale comparative genomic studies that aim to account for the high genetic diversity, dynamic nature and mosaicism of these phages. Codon usage bias is one way of understanding this. Despite cluster organization not representing phylogenetic groupings, we were able to combine the knowledge of cluster assignment with codon usage in order to make inferences about mycobacteriophage host range.
The similarities of codon bias profiles in the mycobacteriophages sheds light on their ability to infect Mycobacteria. Our analysis of the mycobacteriophage genomes suggests that, due to the lack of similarities in the DNA GC contents and codon utilization patterns among many mycobacteriophages, the preferred host of many mycobacteriophages is not M. smegmatis, despite their having been isolated on M. smegmatis.
Further investigation into the structural similarities between DS6A, the only mycobacteriophage cultivated from M. tuberculosis, with the mycobacteriophage of the G, I, K, L, N and P clusters may allow identification of mechanisms of M. tuberculosis infection, such as host attachment proteins, host receptors and host-specific adaptations. Understanding the mechanisms of bacteriophage infectivity is a necessary step in using these phages therapeutically against M. tuberculosis.
Acknowledgements
This manuscript was significantly improved based on useful comments provided by three anonymous reviewers. The authors are also grateful to Catherine Putonti (Loyola University Chicago) for excellent feedback on an earlier version of the manuscript. We appreciate advice and assistance from Jose Anadón (Queens College CUNY), Lucia Barker (American Society of Human Genetics), Graham Hatfull (University of Pittsburgh), Deborah Jacobs-Sera (University of Pittsburgh), Allison Johnson (Virginia Commonwealth University), Tuajuanda Jordan (St. Mary’s College of Maryland), Razi Khaja (National Center for Biotechnology Information), Welkin Pope (University of Pittsburgh), Daniel Russell (University of Pittsburgh), and Louise Temple (James Madison University). Thanks also to the past and present members of the Dennehy Bacteriophage Evolutionary Ecology Research Laboratory. The Howard Hughes Medical Institute Science Education Alliance provided support for the SEA PHAGES program as administered at Queens College CUNY. J. J. D. was supported by grants from the Professional Staff Congress of the City University of New York and the National Science Foundation (Division of Environmental Biology Awards #0804039 and #1148879).
Supplementary Data
Abbreviations:
- ORF
open reading frame
- RSCU
relative synonymous codon usage order
- SCUO
synonymous codon usage order
- CAI
codon adaption index
References
- Andersson G. E., Sharp P. M.(1996). Codon usage in the Mycobacterium tuberculosis complex. Microbiol 142915–925. 10.1099/00221287-142-4-915 [DOI] [PubMed] [Google Scholar]
- Bahir I., Fromer M., Prat Y., Linial M.(2009). Viral adaptation to host: a proteome-based analysis of codon usage and amino acid preferences. Mol Syst Biol 5311. 10.1038/msb.2009.71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailly-Bechet M., Vergassola M., Rocha E.(2007). Causes for the intriguing presence of tRNAs in phages. Genome Res 171486–1495. 10.1101/gr.6649807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basra S., Anany H., Brovko L., Kropinski A. M., Griffiths M. W.(2014). Isolation and characterization of a novel bacteriophage against Mycobacterium avium subspecies paratuberculosis. Arch Virol 1592659–2674. 10.1007/s00705-014-2122-3 [DOI] [PubMed] [Google Scholar]
- Bentley S. D., Parkhill J.(2004). Comparative genomic structure of prokaryotes. Annu Rev Genet 38771–792. 10.1146/annurev.genet.38.072902.094318 [DOI] [PubMed] [Google Scholar]
- Bhat R. M., Prakash C.(2012). Leprosy: an overview of pathophysiology. Interdiscip Perspect Infect Dis 2012181089. 10.1155/2012/181089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohlin J., Snipen L., Hardy S. P., Kristoffersen A. B., Lagesen K., Dønsvik T., Skjerve E., Ussery D. W.(2010). Analysis of intra-genomic GC content homogeneity within prokaryotes. BMC Genomics 11464. 10.1186/1471-2164-11-464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brosch R., Gordon S. V., Garnier T., Eiglmeier K., Frigui W., Valenti P., Dos Santos S., Duthoy S., Lacroix C., et al. (2007). Genome plasticity of BCG and impact on vaccine efficacy. Proc Natl Acad Sci USA 1045596–5601. 10.1073/pnas.0700869104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brüssow H., Hendrix R. W.(2002). Phage genomics: small is beautiful. Cell 10813–16. [DOI] [PubMed] [Google Scholar]
- Carbone A.(2008). Codon bias is a major factor explaining phage evolution in translationally biased hosts. J Mol Evol 66210–223. 10.1007/s00239-008-9068-6 [DOI] [PubMed] [Google Scholar]
- Chithambaram S., Prabhakaran R., Xia X.(2014). Differential codon adaptation between dsDNA and ssDNA phages in Escherichia coli. Mol Biol Evol 311606–1617. 10.1093/molbev/msu087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole S. T., Eiglmeier K., Parkhill J., James K. D., Thomson N. R., Wheeler P. R., Honoré N., Garnier T., Churcher C., et al. (2001). Massive gene decay in the leprosy bacillus. Nature 4091007–1011. 10.1038/35059006 [DOI] [PubMed] [Google Scholar]
- Cole S. T., Brosch R., Parkhill J., Garnier T., Churcher C., Harris D., Gordon S. V., Eiglmeier K., Gas S., et al. (1998). Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 393537–544. 10.1038/31159 [DOI] [PubMed] [Google Scholar]
- Colson P., Fournous G., Diene S. M., Raoult D.(2013). Codon usage, amino acid usage, transfer RNA and amino-acyl-tRNA synthetases in Mimiviruses. Intervirology 56364–375. 10.1159/000354557 [DOI] [PubMed] [Google Scholar]
- Cresawn S. G., Bogel M., Day N., Jacobs-Sera D., Hendrix R. W., Hatfull G. F.(2011). Phamerator: a bioinformatic tool for comparative bacteriophage genomics. BMC Bioinformatics 12395. 10.1186/1471-2105-12-395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennehy J. J.(2009). Bacteriophages as model organisms for virus emergence research. Trends Microbiol 17450–457. 10.1016/j.tim.2009.07.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennehy J. J.(2014). What ecologists can tell virologists. Annu Rev Microbiol 68117–135. 10.1146/annurev-micro-091313-103436 [DOI] [PubMed] [Google Scholar]
- Fierer N., Jackson R. B.(2006). The diversity and biogeography of soil bacterial communities. Proc Natl Acad Sci U S A 103626–631. 10.1073/pnas.0507535103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foerstner K. U., von Mering C., Hooper S. D., Bork P.(2005). Environments shape the nucleotide composition of genomes. EMBO Rep 61208–1213. 10.1038/sj.embor.7400538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallien S., Perrodou E., Carapito C., Deshayes C., Reyrat J.-M., Van Dorsselaer A., Poch O., Schaeffer C., Lecompte O., et al. (2010). Ortho-proteogenomics: Multiple proteomes investigation through orthology and a new MS-based protocol. Genome Research 19128–135. 10.1101/gr.081901.108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao B., Gupta R. S.(2012). Phylogenetic framework and molecular signatures for the main clades of the phylum Actinobacteria. Microbiol Mol Biol Rev 7666–112. 10.1128/MMBR.05011-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Vallvé S., Palau J., Romeu A.(1999). Horizontal gene transfer in glycosyl hydrolases inferred from codon usage in Escherichia coli and Bacillus subtilis. Mol Biol Evol 161125–1134. 10.1093/oxfordjournals.molbev.a026203 [DOI] [PubMed] [Google Scholar]
- Grose J. H., Casjens S. R.(2014). Understanding the enormous diversity of bacteriophages: the tailed phages that infect the bacterial family Enterobacteriaceae. Virology 468-470421–443. 10.1016/j.virol.2014.08.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatfull G. F., Science Education Alliance Phage Hunters Advancing Genomics and Evolutionary Science (SEA-PHAGES) Program, KwaZulu-Natal Research Institute for Tuberculosis and HIV (K-RITH) Mycobacterial Genetics Course, University of California—Los Angeles Research Immersion Laboratory in Virology, Phage Hunters Integrating Research and Education (PHIRE) Program (2013). Complete genome sequences of 63 mycobacteriophages. Genome Announc 1,e00847-13. 10.1128/genomeA.00847-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatfull G. F.(2015). Dark matter of the biosphere: the amazing world of bacteriophage diversity. J Virol 898107–8110. 10.1128/JVI.01340-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatfull G. F., Science Education Alliance Phage Hunters Advancing Genomics and Evolutionary Science Program, KwaZulu-Natal Research Institute for Tuberculosis and HIV Mycobacterial Genetics Course Students, Phage Hunters Integrating Research and Education Program (2012). Complete genome sequences of 138 mycobacteriophages. J Virol 862382–2384. 10.1128/JVI.06870-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatfull G. F., Jacobs-Sera D., Lawrence J. G., Pope W. H., Russell D. A., Ko C. C., Weber R. J., Patel M. C., Germane K. L., et al. (2010). Comparative genomic analysis of 60 Mycobacteriophage genomes: genome clustering, gene acquisition, and gene size. J Mol Biol 397119–143. 10.1016/j.jmb.2010.01.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry M., O'Sullivan O., Sleator R. D., Coffey A., Ross R. P., McAuliffe O., O'Mahony J. M.(2010). In silico analysis of Ardmore, a novel mycobacteriophage isolated from soil. Gene 4539–23. 10.1016/j.gene.2009.12.007 [DOI] [PubMed] [Google Scholar]
- Hershberg R., Petrov D. A.(2010). Evidence that mutation is universally biased towards AT in bacteria. PLoS Genet 6,e1001115. 10.1371/journal.pgen.1001115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hildebrand F., Meyer A., Eyre-Walker A.(2010). Evidence of selection upon genomic GC-content in bacteria. PLoS Genet 6,e1001107. 10.1371/journal.pgen.1001107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilterbrand A., Saelens J., Putonti C.(2012). CBDB: the codon bias database. BMC Bioinformatics 1362. 10.1186/1471-2105-13-62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huson D. H., Bryant D.(2006). Application of phylogenetic networks in evolutionary studies. Mol Biol Evol 23254–267. 10.1093/molbev/msj030 [DOI] [PubMed] [Google Scholar]
- Jacobs-Sera D., Marinelli L. J., Bowman C., Broussard G. W., Guerrero Bustamante C., Boyle M. M., Petrova Z. O., Dedrick R. M., Pope W. H., et al. (2012). On the nature of mycobacteriophage diversity and host preference. Virology 434187–201. 10.1016/j.virol.2012.09.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kallimanis A., Karabika E., Mavromatis K., Lapidus A., Labutti K. M., Liolios K., Ivanova N., Goodwin L., Woyke T., et al. (2011). Complete genome sequence of Mycobacterium sp. strain (Spyr1) and reclassification to Mycobacterium gilvum Spyr1. Stand Genomic Sci 5144–153. 10.4056/sigs.2265047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim B. J., Kim B. R., Hong S. H., Seok S. H., Kook Y. H., Kim B. J.(2013). Complete genome sequence of Mycobacterium massiliense clinical strain Asan 50594, belonging to the type II genotype. Genome Announc 1,e00429–13. 10.1128/genomeA.00429-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim B. J., Choi B. S., Lim J. S., Choi I. Y., Lee J. H., Chun J., Kook Y. H., Kim B. J.(2012). Complete genome sequence of Mycobacterium intracellulare strain ATCC 13950T. J Bacteriol 1942750. 10.1128/JB.00295-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kudla G., Murray A. W., Tollervey D., Plotkin J. B.(2009). Coding-sequence determinants of gene expression in Escherichia coli. Science 324255–258. 10.1126/science.1170160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence J. G., Hendrix R. W., Casjens S.(2001). Where are the pseudogenes in bacterial genomes? Trends Microbiol 9535–540. 10.1016/S0966-842X(01)02198-9 [DOI] [PubMed] [Google Scholar]
- Li L., Bannantine J. P., Zhang Q., Amonsin A., May B. J., Alt D., Banerji N., Kanjilal S., Kapur V.(2005). The complete genome sequence of Mycobacterium avium subspecies paratuberculosis. Proc Natl Acad Sci U S A 10212344–12349. 10.1073/pnas.0505662102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe T. M., Eddy S. R.(1997). tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res 25955–964. 10.1093/nar/25.5.0955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucks J. B., Nelson D. R., Kudla G. R., Plotkin J. B.(2008). Genome landscapes and bacteriophage codon usage. PLoS Comput Biol 4,e1000001. 10.1371/journal.pcbi.1000001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michely S., Toulza E., Subirana L., John U., Cognat V., Maréchal-Drouard L., Grimsley N., Moreau H., Piganeau G.(2013). Evolution of codon usage in the smallest photosynthetic eukaryotes and their giant viruses. Genome Biol Evol 5848–859. 10.1093/gbe/evt053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mira A., Ochman H., Moran N. A.(2001). Deletional bias and the evolution of bacterial genomes. Trends Genet 17589–596. 10.1016/S0168-9525(01)02447-7 [DOI] [PubMed] [Google Scholar]
- Mitchell D.(2007). GC content and genome length in Chargaff compliant genomes. Biochem Biophys Res Commun 353207–210. 10.1016/j.bbrc.2006.12.008 [DOI] [PubMed] [Google Scholar]
- Musto H., Naya H., Zavala A., Romero H., Alvarez-Valín F., Bernardi G.(2006). Genomic GC level, optimal growth temperature, and genome size in prokaryotes. Biochem Biophys Res Commun 3471–3. 10.1016/j.bbrc.2006.06.054 [DOI] [PubMed] [Google Scholar]
- Parmley J. L., Hurst L. D.(2007). How do synonymous mutations affect fitness? Bioessays 29515–519. 10.1002/bies.20592 [DOI] [PubMed] [Google Scholar]
- Pedulla M. L., Ford M. E., Houtz J. M., Karthikeyan T., Wadsworth C., Lewis J. A., Jacobs-Sera D., Falbo J., Gross J., et al. (2003). Origins of highly mosaic mycobacteriophage genomes. Cell 113171–182. 10.1016/S0092-8674(03)00233-2 [DOI] [PubMed] [Google Scholar]
- Plotkin J. B., Kudla G.(2011). Synonymous but not the same: the causes and consequences of codon bias. Nat Rev Genet 1232–42. 10.1038/nrg2899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pope W. H., Jacobs-Sera D., Russell D. A., Peebles C. L., Al-Atrache Z., Alcoser T. A., Alexander L. M., Alfano M. B., Alford S. T., et al. (2011). Expanding the diversity of mycobacteriophages: insights into genome architecture and evolution. PLoS One 6,E16329. 10.1371/journal.pone.0016329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pope W. H., Jacobs-Sera D., Russell D. A., Rubin D. H., Kajee A., Msibi Z. N., Larsen M. H., Jacobs W. R., Lawrence J. G., et al. (2014). Genomics and proteomics of mycobacteriophage Patience, an accidental tourist in the Mycobacterium neighborhood. MBio 5,e02145. 10.1128/mBio.02145-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pope W. H., Bowman C. A., Russell D. A., Jacobs-Sera D., Asai D. J., Cresawn S. G., Jacobs W. R., Hendrix R. W., Lawrence J. G., et al. (2015). Whole genome comparison of a large collection of mycobacteriophages reveals a continuum of phage genetic diversity. Elife 4,e06416–e06416. 10.7554/eLife.06416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ran W., Kristensen D. M., Koonin E. V.(2014). Coupling between protein level selection and codon usage optimization in the evolution of bacteria and archaea. MBio 5,e00956-14. 10.1128/mBio.00956-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rybniker J., Kramme S., Small P. L.(2006). Host range of 14 mycobacteriophages in Mycobacterium ulcerans and seven other mycobacteria including Mycobacterium tuberculosis – application for identification and susceptibility testing. J Med Microbiol 5537–42. 10.1099/jmm.0.46238-0 [DOI] [PubMed] [Google Scholar]
- Sampson T., Broussard G. W., Marinelli L. J., Jacobs-Sera D., Ray M., Ko C. C., Russell D., Hendrix R. W., Hatfull G. F.(2009). Mycobacteriophages BPs, Angel and Halo: comparative genomics reveals a novel class of ultra-small mobile genetic elements. Microbiology 1552962–2977. 10.1099/mic.0.030486-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schattner P., Brooks A. N., Lowe T. M.(2005). The tRNAscan-SE, snoscan and snoGPS web servers for the detection of tRNAs and snoRNAs. Nucleic Acids Res 33W686–689. 10.1093/nar/gki366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp P. M., Li W. H.(1987). The codon adaptation Index–a measure of directional synonymous codon usage bias, and its potential applications. Nucleic Acids Res 151281–1295. 10.1093/nar/15.3.1281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stinear T. P., Mve-Obiang A., Small P. L., Frigui W., Pryor M. J., Brosch R., Jenkin G. A., Johnson P. D., Davies J. K., et al. (2004). Giant plasmid-encoded polyketide synthases produce the macrolide toxin of Mycobacterium ulcerans. Proc Natl Acad Sci U S A 1011345–1349. 10.1073/pnas.0305877101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stinear T. P., Seemann T., Harrison P. F., Jenkin G. A., Davies J. K., Johnson P. D., Abdellah Z., Arrowsmith C., Chillingworth T., et al. (2008). Insights from the complete genome sequence of Mycobacterium marinum on the evolution of Mycobacterium tuberculosis. Genome Res 18729–741. 10.1101/gr.075069.107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K., Stecher G., Peterson D., Filipski A., Kumar S.(2013). mega6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol 302725–2729. 10.1093/molbev/mst197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tortoli E.(2012). Phylogeny of the genus Mycobacterium: many doubts, few certainties. Infect Genet Evol 12827–831. 10.1016/j.meegid.2011.05.025 [DOI] [PubMed] [Google Scholar]
- Tringe S. G., von Mering C., Kobayashi A., Salamov A. A., Chen K., Chang H. W., Podar M., Short J. M., Mathur E. J., et al. (2005). Comparative metagenomics of microbial communities. Science 308554–557. 10.1126/science.1107851 [DOI] [PubMed] [Google Scholar]
- Wan X. F., Xu D., Kleinhofs A., Zhou J.(2004). Quantitative relationship between synonymous codon usage bias and GC composition across unicellular genomes. BMC Evol Biol 419. 10.1186/1471-2148-4-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan X.-F., Zhou J., Xu D.(2006). CodonO: a new informatics method for measuring synonymous codon usage bias within and across genomes. Int J Gen Syst 35109–125. 10.1080/03081070500502967 [DOI] [Google Scholar]
- Xia X., Yuen K. Y.(2005). Differential selection and mutation between dsDNA and ssDNA phages shape the evolution of their genomic AT percentage. BMC Genet 620. 10.1186/1471-2156-6-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
Data Bibliography
- 1.Alferez, G. I., Bryan, W. J., Byington, E. L., & other authors . NCBI GenBank, http://www.ncbi.nlm.nih.gov/nuccore/JF704105 - GenBank Accession #: JF704105 (2012).
- 2.Bambawale, V., Bieberich, J. C., Borowski, A. L., & other authors . NCBI GenBank, http://www.ncbi.nlm.nih.gov/nuccore/JF704116 - GenBank Accession #: JF704116 (2012).
- 3.Brosch, R., Gordon, S. V., Garnier, T., & other authors. Genome plasticity of BCG and impact on vaccine efficacy. Proc Natl Acad Sci USA 104, 5596-5601. GenBank Accession #: NC_008769 (2007). [DOI] [PMC free article] [PubMed]
- 4.Cole, S. T., Brosch, R., Parkhill, J., & other authors. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 393, 537-544. GenBank Accession #: NC_000962 (2001). [DOI] [PubMed]
- 5.Cole, S. T., Eiglmeier, K., Parkhill, J., & other authors. Massive gene decay in the leprosy bacillus. Nature 409, 1007-1011. GenBank Accession #: NC_002677 (2001). [DOI] [PubMed]
- 6.Copeland, A., Lucas S., Lapidus, A., & other authors. NCBI GenBank, http://www.ncbi.nlm.nih.gov/nuccore/FJ641182 - GenBank Accession #: FJ641182 (2009).
- 7.Gallien, S., Perrodou, E., Carapito, C., & other authors. Ortho-proteogenomics: multiple proteomes investigation through orthology and a new MS-based protocol. Genome Res 19, 128-135. GenBank Accession #: NC_018289 (2010). [DOI] [PMC free article] [PubMed]
- 8.Hatfull, G.F., Jacobs-Sera, D., Lawrence, J.G. & other authors. Comparative genomic analysis of 60 mycobacteriophage genomes: genome clustering, gene acquisition, and gene size. J Mol Biol 397, 119-143. - GenBank Accession #s: EU744252, EU744251, AY500152, AY129332, DQ398048, AY129334, EU816589, DQ398049, DQ398044, EU770221, AY129337, EU826471, DQ398053, EU826467, EU826469, EU826466, FJ168660, FJ168661, DQ398047, DQ398051, FJ168662, DQ398041, AY129331, EU816591, EU816588, EU816590, AY129330, FJ174690, DQ398045, FJ174692, DQ398050, FJ174693, AY129336, EU568876, DQ398042, FJ174691, EU770222, AY129339, FJ168659, AY129333, AY129338, AF068845, AY129335, EU203571, DQ398052 (2010). [DOI] [PMC free article] [PubMed]
- 9.Hatfull, SEA-PHAGES, & other authors. Complete genome sequences of 138 mycobacteriophages. J Virol 86, 2382–2384. - GenBank Accession #s: JF704093, JN699015, JN243856, JF792674, JN243857, JN083852, JN408459, JF704107, JN698999, JF937092, JN049605, JF704097, JF937094, JN699019, JF957060, JN831654, JN699005, JF704110, JF704091, JN699017, JN638753, JN006064, JN699010, JN699009, JF704103, JN618996, JN699004, JN698991, JN699006, JN699003, JF704095, JN698992, JN699018, JN699011, JF704104, JN699007, JF704092, JN412588, JF704096, JN412592, JN699626, JN699627, JN699013, JN624850, JN699014, JF937107, JF937091, JN391441, JN412590, JF937096, JN382248, JN006062, JF937106, JN006061, JN698996, JF937093, JN859129, JN542517, JN398368, JF937098, JF937102, JN020142, JF704117, JN699012, JF704115, JN699002, JN412593, JN699001, JF937090, JN698997, JF937101, JF957059, JN201525, JF704106, JN643714, JN185608, JN831653, JN243855, JF704108, JF704113, JF744988, JN680858, JN699628, JF937105, KC576783, JN256079, JN624851, JN698993, JN412591, JF704112, JF704100, JN698995, JN698994,JN412589 (2012). [DOI] [PMC free article] [PubMed]
- 10.Hatfull, SEA-PHAGES, & other authors. The complete genome sequences of 63 mycobacteriophages. Genome Announc 1, e00847-13. - GenBank Accession #s: JX042578, JQ684677, KC661275, JX015524, JX307704, JX411619, KC661279, JQ512844, KC748970, KC691257, JQ911768, KC748968, KC691255, KC748971, KC748969, KC661277, JX411620, KC661280, JQ809702, JX042579, KC691254, KC661276, KC900379, KC691256, KF024728 (2013).
- 11.Henry, M., O'Sullivan, O., Sleator, R. D., & other authors. In silico analysis of Ardmore, a novel mycobacteriophage isolated from soil. Gene 453, 9-23. GenBank Accession #: GU060500 (2010). [DOI] [PubMed]
- 12.Jacobs-Sera, D., Zellars, M., Wells, D. & other authors. NCBI GenBank, http://www.ncbi.nlm.nih.gov/nuccore/GU339467.1 GenBank Accession #: GU339467 (2010).
- 13.Kallimanis, A., Karabika, E., Mavromatis, K., & other authors. Complete genome sequence of Mycobacterium sp. strain (Spyr1) and reclassification to Mycobacterium gilvum Spyr1. Stand Genomic Sci 5, 144-153. GenBank Accession #: NC_014814 (2011) [DOI] [PMC free article] [PubMed]
- 14.Kim, B. J., Choi, B. S., Lim, J. S., & other authors. Complete genome sequence of Mycobacterium intracellulare strain ATCC 13950. J Bacteriol 194, 2750. GenBank Accession #: NC_016946 (2012). [DOI] [PMC free article] [PubMed]
- 15.Kim, B. J., Kim, B. R., Hong, S.H., & other authors. Complete genome sequence of Mycobacterium massiliense clinical strain Asan 50594, belonging to the Type II genotype. Genome Announc, 1, e00429-13. GenBank Accession #: CP004374 (2013). [DOI] [PMC free article] [PubMed]
- 16.Li, L., Bannantine, J., Zhang, Q., & other authors. The complete genome sequence of Mycobacterium avium subspecies paratuberculosis. Proc Nat Acad Sc USA 102, 12344-12349. GenBank Accession #: NC_002944 (2005). [DOI] [PMC free article] [PubMed]
- 17.Pope, W. H., Jacobs-Sera, D., Russell, D. A. & other authors . Expanding the diversity of mycobacteriophages: insights into genome architecture and evolution. PLoS One 6, E16329. GenBank Accession #s: HM152765, GQ303263, GQ303260, GQ303262, GQ303265, GQ303261, HM152764, HM152767, HM152763, GQ303266 (2011). [DOI] [PMC free article] [PubMed]
- 18.Pope, W.H., Bowman, C.A., Russell, D.A. & other authors. Whole genome comparison of a large collection of mycobacteriophages reveals a continuum of phage genetic diversity. eLife 4, e06416-e06416. - GenBank Accession #s: JN698998, JN153085, JF937099, JN699016, JX307705, JN020140, JN572689, JN408461, JF957058, JF704098, KC661281, KC661272, JF704101, JF704111, JF704114, JX307702, KC661271, JQ809701, JX307703, JF937104 (2015). [DOI] [PMC free article] [PubMed]
- 19.Sampson, T., Broussard, G. W., Marinelli, L. J., & other authors. Mycobacteriophages BPs, Angel and Halo: comparative genomics reveals a novel class of ultra-small mobile genetic elements. Microbiology-SGM 155, 2962-2977. GenBank Accession #: FJ973624 (2009). [DOI] [PMC free article] [PubMed]
- 20.Stinear, T. P., Mve-Obiang, A., Small, P. L., & other authors. Giant plasmid-encoded polyketide synthases produce the macrolide toxin of Mycobacterium ulcerans. Proc Natl Acad Sci USA 101, 1345-1349. GenBank Accession #:NC_005916 (2004). [DOI] [PMC free article] [PubMed]
- 21.Stinear, T. P., Seemann, T., Harrison, P. F., & other authors. Insights from the complete genome sequence of Mycobacterium marinum on the evolution of Mycobacterium tuberculosis Genome Res 18, 729-741. GenBank Accession #: NC_010612 (2008). [DOI] [PMC free article] [PubMed]
- 22.Veyrier, F. J. and Behr, M. A. NCBI GenBank, http://www.ncbi.nlm.nih.gov/nuccore/NC_022663 - GenBank Accession #: NC_022663 (2013).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.