Abstract
Many bacterial pathogens are well characterized but, in some cases, little is known about the populations from which they emerged. This limits understanding of the molecular mechanisms underlying disease. The crop pathogen Pseudomonas syringae sensu lato has been widely isolated from the environment, including wild plants and components of the water cycle, and causes disease in several economically important crops. Here, we compared genome sequences of 45 P. syringae crop pathogen outbreak strains with 69 closely related environmental isolates. Phylogenetic reconstruction revealed that crop pathogens emerged many times independently from environmental populations. Unexpectedly, differences in gene content between environmental populations and outbreak strains were minimal with most virulence genes present in both. However, a genome-wide association study identified a small number of genes, including the type III effector genes hopQ1 and hopD1, to be associated with crop pathogens, but not with environmental populations, suggesting that this small group of genes may play an important role in crop disease emergence. Intriguingly, genome-wide analysis of homologous recombination revealed that the locus Psyr 0346, predicted to encode a protein that confers antibiotic resistance, has been frequently exchanged among lineages and thus may contribute to pathogen fitness. Finally, we found that isolates from diseased crops and from components of the water cycle, collected during the same crop disease epidemic, form a single population. This provides the strongest evidence yet that precipitation and irrigation water are an overlooked inoculum source for disease epidemics caused by P. syringae.
Keywords: Disease emergence, pathoadaptation, crop diseases, Pseudomonas syringae, type III secreted effectors
Data Summary
We confirm all supporting data, code and protocols have been provided within the article or through supplementary data files. Sequencing reads have been submitted to the NCBI Small Read Archive as Bioproject PRJNA320409 with biosample accession numbers SAMN04942971 to SAMN04943055 and SAMN05301579 to SAMN05301583 and can be accessed at the following link: http://www.ncbi.nlm.nih.gov/bioproject?LinkName=sra_bioproject&from_uid=2500232.
Impact Statement
Just like human diseases, new crop diseases emerge without warning and sometimes spread rapidly around the globe causing devastation. Where these pathogens originally came from is often unknown. The bacterial species Pseudomonas syringae consists of a group of genetically diverse bacteria including strains that are important crop pathogens as well as strains isolated from wild plants and components of the water cycle, such as clouds, rain and freshwater. The existence of these environmental strains, which are closely related to crop pathogens, suggests that crop pathogenic P. syringae possibly emerged from a diverse pre-existing P. syringae population that was present in the environment before the development of modern agriculture. Here, we found evidence for this hypothesis by sequencing and comparing the genomes of crop pathogenic and environmental strains, we inferred their evolutionary relationships and we identified genes with putative key roles in emergence of crop disease.
Introduction
Successful disease prevention and management rely on a detailed understanding of the ecological and evolutionary processes driving disease emergence. In the case of bacterial crop diseases, a lot has been learned about crop pathogen virulence genes and their function (Lindeberg et al., 2008; Tampakaki et al., 2011; O'Brien et al., 2011) but little is known about the genetic basis of crop disease emergence (Vinatzer et al., 2014) and the conditions that promote it (Stukenbrock & McDonald, 2008). For diseases caused by host-restricted obligate pathogens such as Puccinia striiformis f. sp. tritici and Puccinia graminis. f. sp. tritici, the causal agents of stripe rust and stem rust, respectively (Chen, 2005; Singh et al., 2011), these issues can be addressed relatively easily because dissemination patterns and ecology are restricted to one or a small number of plant hosts. Where infection can be caused by isolates in multiple environmental sources, it can be more difficult to pinpoint the source. For many human pathogens, the role of environmental reservoirs in disease epidemiology has been well described (Struve & Kogfelt, 2004; Whiley et al., 2013; Grosso-Becerra et al., 2014; Hazen et al., 2015), but for facultative saprophytic crop pathogens, with environmental reservoirs, dissemination routes and interactions within multiple habitats are mostly uncharacterized (Woolhouse et al., 2001; Johnson et al., 2015).
In the past 10 years, MLSA studies have revealed considerable genetic diversity among environmental isolates that are closely related to epidemic, clonal crop-pathogenic lineages of the plant pathogen Pseudomonas syringae (sensu lato) (Morris et al., 2008, 2010; Monteil et al., 2012, 2014b). P. syringae is one of the economically most important bacterial crop pathogens and a well-characterized model species for molecular plant–microbe interactions (Hirano & Upper, 2000; O'Brien et al., 2011). Environmental isolates have been collected from wild plants as well as non-plant reservoirs including soil, plant debris, and components of the water cycle including clouds, precipitation and surface water (Morris et al., 2013; Berge et al., 2014). Host range analysis revealed that some crop-pathogenic epidemic clones (referred to as ‘crop pathogens’ from here on) within P. syringae, such as the most common lineage of the tomato pathogen P. syringae pv. tomato (Pto), have a narrow host range limited to tomato (Cai et al., 2011a, b). Conversely, lineages such as the cantaloupe pathogen P. syringae pv. aptata (Pap) have a broad host range, infecting various plant families (Morris et al., 2000; Berge et al., 2014). In the case of Pto, MLSA revealed the existence of closely related isolates from natural freshwater sources and recombination events between these environmental isolates, Pto and other crop pathogens (Monteil et al., 2013). The environmental lineages were found to be equipped with some of the same virulence genes as the crop pathogen Pto, in particular genes coding for type III secretion (T3S) effectors, the best studied and most important class of virulence genes in P. syringae (Lindeberg et al., 2008). Moreover, the environmental isolates had a wider host range than Pto but were less virulent on tomato (Monteil et al., 2013). Taken together, these results are consistent with the evolution of highly virulent crop pathogens with a relatively narrow host range from a population of ancestors with a wider host range. This potentially occurs through the acquisition of genomic elements that promote virulence on the crop hosts but reduce virulence (or fitness) on other hosts. However, what these genomic elements might be, and whether they were acquired by horizontal gene transfer, remains unknown.
The increasing availability of large genomic datasets provides new opportunities for investigating pathotypes in multiple niches (Vinatzer et al., 2014). By comparing the genomes of P. syringae crop pathogens and isolates from environmental reservoirs, it should be possible to identify the genetic basis of disease emergence and the genomic regions that are horizontally transferred between strains, in particular between crop pathogens and their environmental relatives. Therefore, we sequenced the genomes of 107 isolates of crop-pathogenic and environmental P. syringae and analysed them together with 86 publically available P. syringae genomes. We investigated two P. syringae phylogroups (Berge et al., 2014) with contrasting host ranges (Buell et al., 2003) and disease etiology (Feil et al., 2005): phylogroup 1a, which includes Pto and other related crop pathogens and environmental isolates; and a subset of phylogroup 2d, for which we sampled exclusively closely related Pap isolates from diseased cantaloupe and the environment. Importantly, each phylogroup also includes one intensively studied model pathogen isolate: P. syringae pv. tomato (Pto) DC3000 in phylogroup 1a and P. syringae pv. syringae (Psy) B728a in phylogroup 2d. For both of these isolates, virulence traits have been investigated for decades and closed genome sequences are available (O'Brien et al., 2011). Phylogenetic reconstruction, core and accessory genome analysis, and genome-wide association approaches were then used to characterize the evolutionary relationships between crop pathogens and environmental relatives, the population structure of these phylogroups and the genetic basis of crop adaptation (Sheppard et al., 2013; Pascoe et al., 2015). The results provide new insight into crop pathogen emergence, crop adaptation and pathogen dissemination.
Methods
Isolates and sequencing.
Genomes of 92 P. syringae isolates from phylogroups 1a and 2d (Berge et al., 2014) were chosen for sequencing, whereby phylogroup 1a was sampled maximizing genetic diversity avoiding multiple crop pathogen strains with identical MLSA sequences, while for phylogroup 2d only one subset was sampled with isolates that were identical at two MLSA loci. These datasets were augmented with 12 genome sequences from the same phylogroups available in public databases, including reference genomes from Pto strain DC3000 (Buell et al., 2003) and Psy strain B728a (Feil et al., 2005). Most of the isolates sequenced in this study were described previously (Morris et al., 2008; Morris et al., 2010; Monteil et al., 2012; Monteil et al., 2014b) with 36 isolates collected from diseased crops (defined as cultivated lands) and 56 isolates collected from streams and rivers (11 isolates), precipitation (15 isolates), irrigation water (11 isolates) or epilithic biofilms (12 isolates) and leaf litter (seven isolates). Table S1 (available in the online Supplementary Material) provides a detailed list. Genomes representing other P. syringae phylogroups were included in the analyses: three of them were sequenced in this study while 86 genomes were downloaded from public databases to give a total of 193 genome sequences (Table S1).
For genomes sequenced in this study, DNA was extracted using the Gentra Puregene bacteria kit (Qiagen; cat. no.: 158567), according to the manufacturer’s instructions. The library preparation was performed with the Nextera XT DNA sample prep kit from Illumina following the manufacturer’s instructions for denaturing and normalization steps. High-throughput genome sequencing was performed using the 151×151 PE Rapid Run mode of a HiSeq 2500 sequencer (Illumina) using Illumina Truseq sequencing reagents. The quality of resulting sequencing reads was examined using FastQC (Andrews, 2010). TrimGalore (Krueger, 2015) was used to trim sequence reads and remove poor-quality data, using command-line options ‘-q 30 and -paired’. Illumina adapter sequences were removed using CutAdapt (Martin, 2011). Cleaned sequencing reads were assembled using the de novo assembly algorithm Velvet (Zerbino & Birney, 2008) (version 1.2.08). The value of k was optimized by assembling over a range of values and choosing the assembly with maximal N50. The minimum output contig size was set to 200 bp with the scaffolding option switched off, and all other program settings were left unchanged. The average number of contigs and the standard error in 95 newly sequenced P. syringae genomes was 474±32 for an average total assembled sequence size of 5 969 322±18 142 bp. Sequence reads in FastQ format deposited in the Sequence Read Archive (SRA) are available via BioProject accession PRJNA320409.
Core and accessory genome.
Analysing core and accessory genome variation, genealogies and recombination patterns, we investigated the evolutionary relationships linking outbreak strains to their relatives in the environment. A reference pan-genome approach (Méric et al., 2014) and gene-by-gene alignment (Sheppard et al., 2012) was implemented using BIGSdb open source software (Jolley & Maiden, 2010). First, a reference gene list was assembled from four publicly available genomes, Pto DC3000 (Buell et al., 2003) and Psy B728a (Feil et al., 2005), P. syringae pv. phaseolicola 1448a (Joardar et al., 2005) and the environmental strain CC1557 (Hockett et al., 2014) (Table S1). The total number of genes in these isolates was 20 955 and after removal of 13 471 allelic variants that shared >70 % nucleotide similarity across ≥50 % of the genes' length, the final reference pan-genome list contained 7484 unique loci. Each locus was searched in the 193 genomes of all isolates using the blast algorithm and setting parameters for locus match to a minimum of 70 % sequence similarity over a minimum of 50 % of the query sequence length. The average core genome nucleotide sequence similarity within P. syringae is considerably higher than the blast match criteria. Therefore, these blast parameters ensure relatively low stringency for identifying homologous genes as in existing whole-genome MLST methodology (Jolley & Maiden, 2010; Sheppard et al., 2012; Maiden et al., 2013; Méric et al., 2014). Consistent with whole-genome MLST (Berge et al., 2014), a matrix was produced summarizing the presence/absence and allelic diversity of reference pan-genome genes, based upon these blast parameters. Each gene of the reference pan-genome that was not, or only partially, detected in a genome was indicated as missing or truncated, and this number was calculated at each locus for all P. syringae genomes. However, truncated gene sequences detected at the end or beginning of a contig were considered as present but were not counted as alleles. For each pair of isolates, the number of shared genes and alleles (identical sequences at a given locus over the whole sequence length) was calculated and the core genome for each species, and for the genus, was defined as the complement of genes that were present in all isolates.
Population genetic structure.
Phylogenetic trees were reconstructed from the alignment of the core genome. The core genome was determined based on the 5619 coding sequences of the reference genome Pto DC3000 and its two plasmids. Genes in the core genome were aligned individually using mafft (Katoh & Toh, 2008) and concatenated to produce contiguous sequence alignments.
For analysis of the entire P. syringae species complex (193 genomes), core loci for which some of the sequences were truncated were kept in the analysis, which accounts for a total of 1889 genes. A recompiled version of FastTree 2.1.7 (Price et al., 2010) was used to reconstruct an approximation of a maximum-likelihood tree. With this configuration, the minimum branch length was one substitution for every 2 000 000 bp (1000 times higher than the default FastTree parameters). The software was run with the Jukes–Cantor model of nucleotide evolution and gaps from truncated sequence alignments were considered as missing data. The tree was visualized and annotated using FigTree v1.4.2 (http://tree.bio.ed.ac.uk/software/figtree).
For the analysis of phylogroups 1a and 2d, trees were reconstructed from alignments including only non-truncated sequences for all core loci (respectively 810 and 2147 loci). Genealogies were inferred using ClonalFrame, a model-based approach for inference of microevolution in bacteria that accounts for recombination events that can disrupt phylogenetic reconstruction (Didelot & Falush, 2007). This program differentiates mutation and recombination events on each branch of the tree based on the density of polymorphisms. Clusters of polymorphisms are likely to have arisen from recombination, and scattered polymorphisms are likely to have arisen from mutation. The program was run with 20 000 burn-in iterations, followed by 50 000 and 100 000 sampling iterations for phylogroups 2d and 1a, respectively, until convergence. The consensus tree represents combined data from three independent runs with 75 % consensus required for inference of relatedness. Recombination events were defined as sequences with a length of >50 bp with a probability of recombination of 75 % over the length, reaching 95 % in at least one site.
Associations of lineages with isolation sources were investigated applying the HierBAPS clustering model (Corander et al., 2004; Cheng et al., 2013). This method reveals nested genetic population structures and any association of strain metadata with genetically divergent clusters and the substructure within them. Therefore, we were able to specify the genetic boundaries between P. syringae lineages at different resolutions and test if they were associated with a unique isolation source or not. The same alignments used for each tree reconstruction (whole P. syringae diversity, 1a or 2d) were used to infer genetically divergent clusters increasing levels of resolution from 1 to 4. The mixture partition was inferred setting the prior to 10 k panmictic subpopulations in which individuals were uniformly distributed and the structure, which maximizes the posterior distributions, was obtained using a stochastic search algorithm.
Genome-wide association mapping.
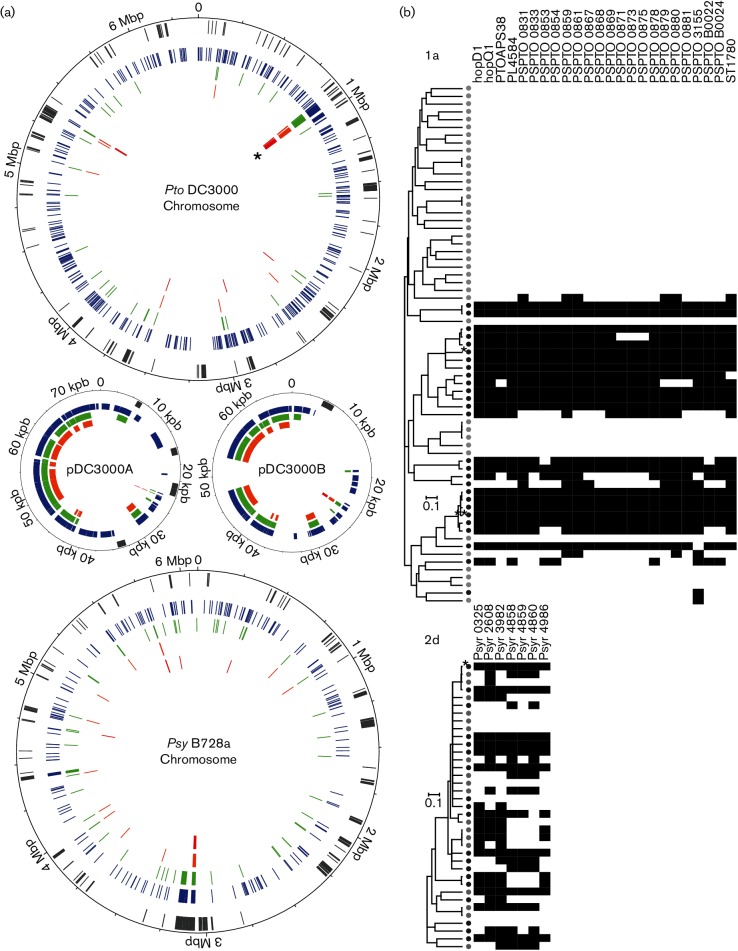
We sought to determine the genetic basis of crop pathogen emergence by comparing genomes of crop pathogens and their relatives from other sources. We used a recently developed genome-wide association study (GWAS) method that is adapted for bacterial populations (Sheppard et al., 2013; Pascoe et al., 2015; Yahara et al., 2016b) and excludes associations due to confounding population structure. The whole-genome sequence of each isolate was fragmented into unique overlapping 30 bp 'words'. For each word, the method examines the extent of association with the phenotype. To test the significance of association of each word after controlling for the effect of population structure and clonal inheritance of genetic variants, here determined using ClonalFrame (Didelot & Falush, 2007), the method computes P values by comparing the observed association score with a null distribution of the score calculated through Monte Carlo simulations (Martins & Garland, 1991; Garland et al., 2005). To account for multiple testing, only words with a probability below 5×10−4 were considered significant. The distribution of source-associated words for which homologues were found in reference genome Pto DC3000 (Buell et al., 2003) was visualized using Artemis (Rutherford et al., 2000) and DNAPlotter (Carver et al., 2009) (see Fig. 3).
Fig. 3.
Distribution of genes associated with P. syringae crop pathogen strains within reference genomes and within populations. (a) The 30 bp long words that were found to be significantly associated with crop pathogens in either phylogroup 1a or 2d were mapped on the genome of the reference crop pathogens Pto DC3000 and Psy B728a, respectively. A total of 73 299 and 5970 crop pathogen-associated words in Pto DC3000 and Psy B728a were distributed in 571 and 222 genes, respectively. The list of these genes and the distribution of mapped words is given in Table S4. For each chromosome and plasmid, the first grey circle represents virulence genes (listed in Lindeberg et al., 2008). The next four coloured circles correspond to the genes in which words were mapped from the highest to the lowest probability (blue, green, orange and red corresponding to P value cutoffs of 5×10−4, 5×10−5, 5×10−6 and 5×10−7, respectively). The 25 kb region containing hopQ1 and hopD1 described in the text is indicated with an asterisk. (b) Heatmaps showing the presence of genes for which at least one word was significantly associated with crop pathogens with a probability inferior or equal to 5×10−7. Black boxes denote that at least one word was mapped for the corresponding isolate and gene, while white boxes denote that not a single word was present with this probability. Isolates were organized following the core genome trees built with ClonalFrame (Didelot & Falush, 2007) in Fig. 1(b). The trees are drawn to scale, with branch lengths proportional to the number of substitutions per site. Dark and light grey circles symbolize crop pathogen strains and environmental isolates, respectively.
Inference of homologous recombination.
The extent of homologous recombination was inferred between crop pathogens and related environmental isolates and ancestors of phylogroups 1a and 2d, to investigate genetic fluxes at a fine scale and the evolutionary origin of genes associated with disease outbreak populations. First, chromosome painting (Lawson et al., 2012; Yahara et al., 2013) was used to build a co-ancestry matrix summarizing the number of recombination-derived ‘chunks’ of DNA from each donor to each recipient isolate. Using this matrix, fineSTRUCTURE (Lawson et al., 2012) was used to conduct model-based clustering of individuals by a Bayesian Markov chain Monte Carlo (MCMC) approach that explores the space of possible partitions. In parallel, we applied the Ordered Painting approach (Yahara et al., 2014) to identify hotspots of recombination within phylogroups. In this method, the extent of genealogical changes for a specific site due to recombination compared with the average genome genealogy is represented by the distance statistic Hi representing recombination hotness at each polymorphic site i (Yahara et al., 2016a). These atypical changes, for example, above the top percentile of the whole-genome distribution, are expected to indicate hotspots of recombination.
Results
Population structure of the P. syringae species complex
Before determining the population structure of P. syringae sensu lato, we determined whether the 193 P. syringae genome sequences (sequenced in this study or publically available) formed a clade that is distinct from genomes of other Pseudomonas species. A genealogy of 629 available Pseudomonas species genomes was reconstructed based on 52 ribosomal protein gene sequences (Tables S1 and S2). The phylogenetic tree confirmed P. syringae as a largely monophyletic species complex. The one exception was phylogroup 13 [isolate UB246 (Berge et al., 2014)], which clustered with Pseudomomas fluorescens based on population structure analysis inferred with BAPS, a Bayesian statistical clustering method (Fig. S1).
To analyse evolutionary relationships with greater resolution within the P. syringae species complex, a core genome of the 193 isolates (Table S2) was determined by aligning all genome sequences with the annotated genes of the fully sequenced and annotated reference genome Pto DC3000 (Buell et al., 2003). The P. syringae genealogy reconstructed from 1889 core gene loci (a total of 108 393 bp for which 38 484 were variable sites) confirmed genealogies previously inferred by MLSA and by previous core genome analysis (Berge et al., 2014). Importantly for the goal of this study, it also revealed recent common ancestry of all lineages within phylogroups 1a and 2d, respectively (Fig. 1a). To determine the extent to which these phylogroups reflected distinct genetically divergent clusters within the P. syringae species complex, we performed a hierarchical clustering analysis of the core genome using the HierBAPS method that estimates nested population structures (Cheng et al., 2013). Based on clustering at the lowest level of resolution, clade designations were congruent with most phylogroups, including phylogroups 2d and 1a.
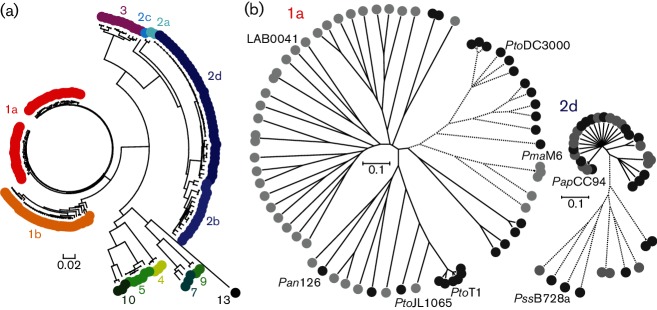
Fig. 1.
Population structure of 193 isolates from the P. syringae species complex. (a) Unrooted phylogenetic tree reconstructed from 1889 genes detected in all isolates using an approximation of the maximum-likelihood algorithm and the GTR model for nucleotide substitution. Bar, the number of substitutions per site. Isolates are coloured according to the monophyletic groups described by Berge et al. (2014). (b) Genealogies inferred by ClonalFrame (Didelot & Falush, 2007) for phylogroups 1a and 2d based only on those genes with no evidence of homologous recombination (810 and 2293 genes, respectively). The trees are drawn to scale, with branch lengths corresponding to the number of substitutions per site. Dark and light grey circles symbolize isolation sources: crop pathogens and environmental strains, respectively. Solid and dotted branches symbolize subpopulations within each phylogroup determined with the first level of HierBAPS hierarchy (Corander et al., 2008; Cheng et al., 2013).
To investigate the role of environmental reservoirs in the emergence of crop pathogens, we then focused on phylogroup 1a, for which we had sequenced representative isolates of all crop pathogens available to us and all available environmental relatives in order to include as much genetic diversity as possible. Individual genealogies were built and Bayesian analysis of population structures (Cheng et al., 2013) was performed. Since homologous recombination may have had an impact on population genetic structure, clonal relationships of crop pathogens and their relatives were first investigated using ClonalFrame (Didelot & Falush, 2007), which accounts for recombination when reconstructing the genealogy. At the lowest degree of resolution, the population structure analysis of all core genes identified three genetic clusters, each containing both crop pathogens and environmental relatives (Fig. 1b). The ClonalFrame analysis also revealed that some monophyletic groups of lineages within these clusters correspond to one isolation source only. For example, the Pto DC3000 crop pathogen clusters only with other crop pathogens, and the LAB0041 isolate from an alpine epthilic biofilm clusters only with other environmental isolates. Importantly, however, several crop pathogens, such as the tomato pathogens Pto T1 and Pto JL1065 and the snapdragon pathogen Pan 126, are interspersed with lineages from rain or surface water within the same group. This shared ancestry between some crop pathogens and some environmental isolates, together with the long external, short internal branches of the ClonalFrame tree (Fig. 1b), and a high degree of reticulation in the NeighborNet network (Fig. S2a), are all consistent with a scenario of multiple emergences of different crop pathogens and environmental lineages from recombining ancestral populations.
For phylogroup 2d, we investigated the different components of the water cycle in pathogen dissemination. Isolates from diseased crops, collected during a cantaloupe blight epidemic in France, were sequenced together with isolates from precipitation, surface water, irrigation water and ground water (Morris et al., 2000; Morris et al., 2008). Specifically, we chose isolates that were identical at two MLSA loci and we wanted to determine whether crop and environmental isolates would cluster together, or separately, based on core genome sequences. ClonalFrame and population structure analysis revealed that most of these isolates clustered together with a star-like genealogy without any separation between crop isolates and environmental isolates. The genomes of crop and environmental isolates in the first cluster were extremely similar to each other (average nucleotide identities ranging from 99.50 to 99.97 % within each clade). To confirm these results, sequencing reads were aligned against the Psy B728a genome and SNPs were identified. This approach revealed that some of the crop isolates differed from their most similar environmental relatives by as few as three SNPs per million base pairs (data not shown), suggesting very recent exchanges of P. syringae between cantaloupes and water cycle components. Moreover, ClonalFrame showed that some core genes experienced recent homologous recombination between crop and environmental isolates, which was supported by the NeighborNet network shown in Fig. S2b.
Core and accessory genome variation and pathogen emergence in phylogroups 2d and 1a
Genome-wide genetic differences between crop pathogens and environmental isolates were investigated using a reference pan-genome approach (Méric et al., 2014). In short, four fully assembled and annotated genome sequences from four different phylogroups were chosen as references: Pto DC3000 (Buell et al., 2003), Psy B728a (Feil et al., 2005), Pph 1448a (Joardar et al., 2005) and CC1557 (Hockett et al., 2014). The pan-genome of these four reference genomes, defined as the total set of gene families present in the four genomes, was then determined and found to consist of 7484 unique genes. Finally, the reference pan-genome was aligned against all the other genomes. This analysis revealed that every genome in phylogroup 1a contained a set of 3576 genes of the reference pan-genome, thus representing the 1a core genome. Every genome of phylogroup 2d contained a set of 4147 core genes representing the 2d core genome. In total, 3062 genes were present in both phylogroups, representing their combined core genome. At the remaining 4422 loci, genes were either present or absent, presenting the combined accessory genome (Fig. 2a). Phylogroup 2d had a lower average p-distance between allele pairs, and on average fewer unique alleles per gene: 0.201±0.001 compared to 0.480±0.003 for phylogroup 1a (Student’s t-test, P<0.001, Fig. S3a,b). Note that differences in the genetic diversity of phylogroups 2d and 1a reflect our sampling strategy, maximizing genetic diversity for phylogroup 1a while prioritizing a single genetic lineage for phylogroup 2d.
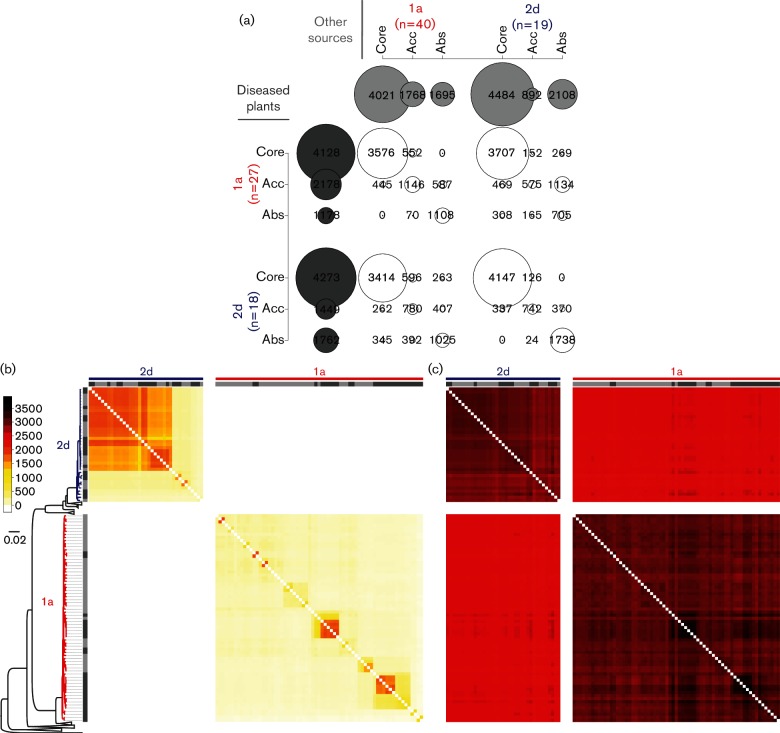
Fig. 2.
Core genome similarity and accessory genome variation within and between phylogroups 2d and 1a. (a) Overlap between the core and accessory genomes calculated for phylogroups 1a and 2d. Core genes (Core), accessory genes (Acc) and absent genes (Abs) were defined as genes detected in 100 % of the isolates, detected in less than 100 % but more than 0 % of the isolates, and absent in 100 % of the isolates, respectively. The radius of each circle is proportional to the number of detected genes. Dark grey circles represent genes from strains isolated from diseased crops while light grey circles represent genes from environmental isolates. White circles represent overlapping genes from both populations. Matrices show pairwise comparison of core genome similarity (b) and accessory genome variation (c) between 104 isolates ordered according to the phylogenetic tree presented in Fig. 1. Entire matrices with all 193 P. syringae isolates are shown in Fig. S4. Dark grey boxes symbolize crop pathogens while isolates from other sources are symbolized by light grey boxes. Heatmap colours ranging from white, through yellow, red to black represent values from the lowest to the highest number of shared alleles or genes, in the first and second heatmap, respectively. The minimum number of shared alleles in the core genome ranged from 0 to 3010 while the minimum number of shared accessory genes ranged from 1105 to 2508.
To determine the extent of genes associated with isolation sources, we compared gene content of crop pathogens and environmental isolates. It is striking that none of the genes that are core to crop pathogen populations are absent in environmental populations, and that none of the genes that are core to environmental populations are absent in crop pathogen populations (Fig. 2a). This suggests that there is weak ecological differentiation between crop pathogens and their environmental relatives. Moreover, for both phylogroups the majority of accessory genes present in environmental isolates are also present in crop pathogens and vice versa. This result reveals that there is no strong barrier to gene flow between environmental isolates and crop pathogens. Importantly, about 61 % of T3S effector genes detected in crop pathogen populations were present in environmental relatives as well (Table S3, Fig. S5).
Pairwise genome comparisons showed that patterns of core genome allelic similarity (Fig. 2b) and accessory genome similarity, measured as similarity with regard to presence and absence of accessory genes (Fig. 2c), reflected genealogies rather than isolation host or source (crop versus environment). Extending this analysis beyond phylogroups 1a and 2d to P. syringae genome sequences in phylogroups 2b and 3 confirmed the same trend (Fig. S4). Therefore, pairwise genome comparisons confirmed the absence of strong barriers to gene flow between crop pathogens and environmental relatives as well as between pathogens of different hosts.
Candidate genes associated with pathoadaption
Although the previous analyses clearly showed that crop pathogens are not genetically isolated from their environmental relatives, it is still possible that at least a small set of genes, or alleles, may be more frequently associated with either crop pathogens or environmental isolates. This would suggest that crop pathogens are adapted to a pathogenic lifestyle (pathoadaptation) that is different from the adaptation of environmental isolates to a lifecycle in non-agricultural environments. Therefore, genetic elements overrepresented either in crop pathogen strains or in environmental isolates were determined using a GWAS (Sheppard et al., 2013; Pascoe et al., 2015), which identifies 30 bp DNA sequences (words) in the core and accessory genome, taking into account the clonal frame and vertical inheritance and core genes. Based on the analysis of 67 and 37 isolates within phylogroups 1a and 2d, 73 299 and 5970 words, respectively, were identified that were over-represented in crop pathogens. These mapped to 571 genes in phylogroup 1a and 222 genes in phylogroup 2d, 74 % of which are annotated with a putative function (Table S4). Genes containing pathogen-associated elements were mapped to reference genomes of the crop pathogens Pto DC3000 and Psy B728a, for 1a and 2d isolates, respectively, using Artemis (Rutherford et al., 2000) and DNAPlotter (Carver et al., 2009) (Fig. 3a). Associated genes were dispersed across the genome with evidence of seven (phylogroup 1a) and five (phylogroup 2d) hotspots of strong pathogen association, with a P value less than 5×10−6.
For phylogroup 1a isolates, only 7 % of the mapped words were associated with known virulence genes, with 22 % of the total associated words mapping to a single 25 kb region consisting of 21 adjacent genes of the Pto DC3000 genome (Fig. 3a). This included the T3S effector genes hopD1 and hopQ1 (Lindeberg et al., 2008). These genes were present in all but two of crop pathogen isolates and absent from all environmental isolates (Fig. 3b). Importantly, after aligning raw reads of these two isolates against the Pto DC300 genome (data not shown), relic fragments of hopD1 and hopQ1 were even detected in the only two crop pathogens, P. syringae pv. apii BS252 and P. syringae pv. antirrhini 126, that did not contain the intact genes. Other crop pathogen-associated hotspots in phylogroup 1a contained genes encoding putative proteins related to: (i) replication, integration, recombination and repair of DNA; (ii) carbohydrate, lipid and amino acid transport; and (iii) energy production and conservation (Table S4).
Interestingly, no T3S effector genes were associated with crop pathogen isolates in phylogroup 2d and, compared to the 1a phylogroup, the repertoire of T3S effector genes was generally smaller in phylogroup 2d (Fig. S5). Predicted functions of genes that were significantly associated with crop pathogen isolates in 2d were: (i) DNA transcription and translation regulation; (ii) uptake of sparse substrates linked to TonB-dependent transporters; (iii) the conversion of energy into storage molecules; (iv) secondary metabolite production and export; and (v) type I secretion systems (Table S4). Note that sampling within phylogroup 2d focused on a subset of very similar isolates to address questions about pathogen dissemination. Additional sampling would be necessary for more robust inference of pathogen associations in this lineage.
Homologous recombination and pathoadaptation
In recombining bacteria, the acquisition of DNA from other lineages can confer novel functions, such as those related to pathoadaptation (Ochman et al., 2000). ClonalFrame analysis and patterns of reticulation using simple NeighborNet genealogical reconstructions (Fig. S2) suggested recent homologous recombination among various crop pathogens and environmental isolates. A more detailed analysis was thus carried out to investigate inter- and intra-phylogroup homologous recombination and to quantify recombination landscapes across the genome.
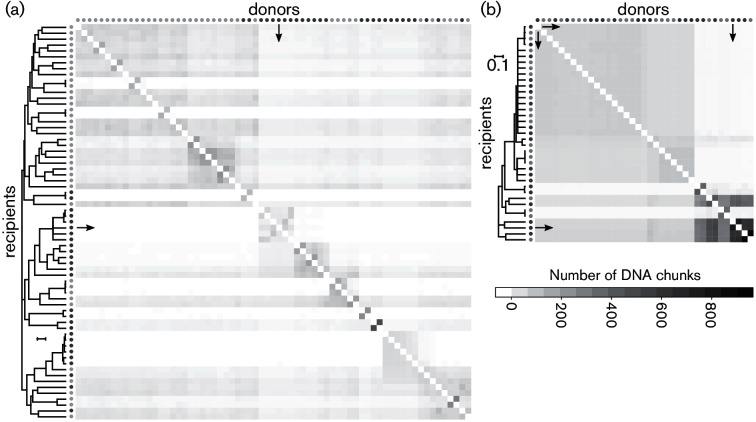
Gene flow within and between phylogroups was quantified by characterizing DNA donated and received among the strains using chromosome painting (Yahara et al., 2013). The number of recombination-derived chunks of DNA, defined as genetic material donated from a nearest ‘donor’ to a ‘recipient’ haplotype, was summarized into a co-ancestry matrix. A co-ancestry matrix with all 1a and 2d strains confirmed the barrier to gene flow between the two phylogroups for which the number of DNA chunks was under one per genome on average. However, gene flow was observed within each phylogroup (Fig. 4). Both co-ancestry matrices not only showed admixture between ancestors of all lineages but also showed that gene flow occurred between crop pathogens and environmental isolates in both directions, as for Pap CC94 (Fig. 4b). However, in phylogroup 1a isolates, gene flow was asymmetrical with some lineages being principally donors or recipients. For example, Pto DC3000 was mostly a donor and not a recipient to isolates from both isolation sources (Fig. 4a) in contrast to Psy B728a from phylogroup 2d, which received twice as many DNA chunks from ancestors of other crop and environmental isolates compared to what the strain donated to other strains (Fig. 4b).
Fig. 4.
Inference of genetic fluxes within P. syringae populations. Co-ancestry matrices were determined by Chromosome Painting and fineSTRUCTURE for phylogroups 1a (a) and 2d (b). The expected number of ‘chunks’ imported from a donor genome (column) to a recipient genome (row) is given by the colour of each cell of the matrices. The trees are drawn to scale, with branch lengths proportional to the number of substitutions per site. Arrows in (a) point to Pto DC3000, while arrows in (b) point to Pap CC94 and Psy B728a at the top and bottom of the tree, respectively. Dark and light grey circles symbolize crop pathogen strains and environmental isolates, respectively.
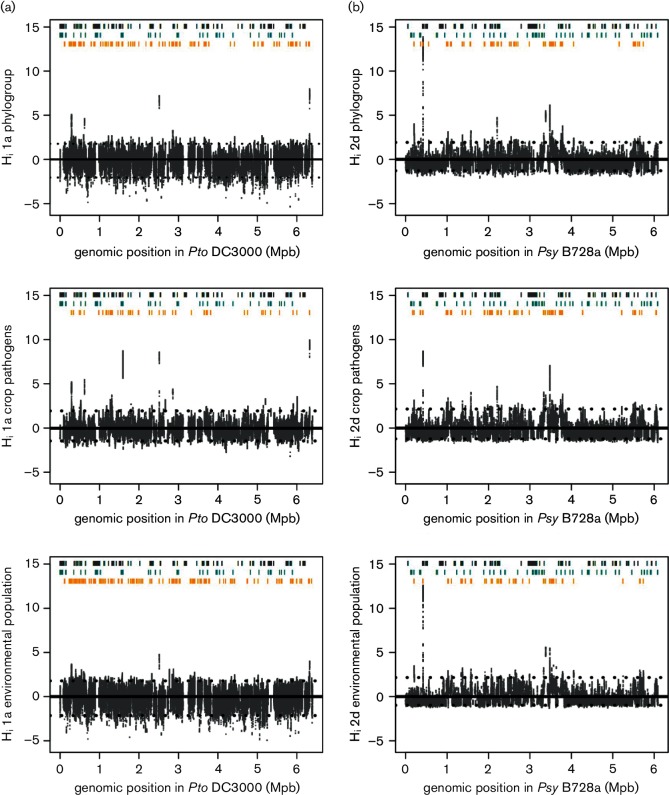
In parallel to characterizing the direction of gene flow, recombination hotspots across the genome were identified based upon a per-site estimate of intensity of recombination (Hi) (Yahara et al., 2016a), which refers to a normalized value quantifying the extent of genealogical changes due to recombination compared to the average genealogy (Fig. 5). For phylogroups 2d and 1a, respectively, 144 and 244 recombining genes had Hi values in the upper 2.5 % for at least one base position (Table S5). Evidence for a role of recombination in pathoadaptation was seen in phylogroup 1a where 72 genes recombined in crop pathogens. A total of 8 % were known virulence genes, including genes coding for T3S effectors (hopAA1-1, hopAH1 and hopB1), T4 pili, chemotaxis, pioverdine production and levansucrase (lsc-1). The highest rates of recombination in crop pathogens were found for those genes that were also identified as hotspots of recombination in environmental isolates and were mostly associated with hypothetical proteins or a peptide ABC transporter permease (PSPTO 265, 561, 2271, 2587 and 5552). In phylogroup 2d, 116 genes recombined in crop isolates and were mainly associated with metabolism, regulators (lysR, lclR, tetR), transporters and the type 1 secretion system (involving TolC and HlyD proteins), while the 23 genes recombining in environmental isolates were associated with other functions among which were several T3S effectors and structural components (e.g. hopM1, hrcN, hrpQ, hrcV and hrpK1). As observed within phylogroup 1a, hotspots of recombination in 2d isolates from crops were sometimes hotspots in environmental isolates as well. Hotspots in 2d were associated with genes coding for hypothetical proteins (PSYR 392 and 393), a tRNA-dihydroudine synthetase A (PSYR 1936) and a binding-protein dependent transport system (PSYR 2903 and 2904). Importantly, 18 orthologous genes were found to be recombining in both phylogroups, some of them coding for ABC transporters, proteins involved in antibiotic resistance (PSPTO 3132, 3302 and 5191), chemotaxis, extracellular solute binding (PSPTO 2962 and 3302), glutamate racemase (murI), porins (oprD) and a heavy metal translocating ATPase (cad A_1) (Table S5).
Fig. 5.
Inference of homologous recombination hotspots within P. syringae populations. Homologous recombination hotspots were inferred in phylogroups 1a (a) and 2d (b) as described by Yahara et al. (2014). A total of 177 790 and 141 997 SNPs were used, respectively, representing the chromosome of each reference genome. For each group, the extent of recombination was estimated from the whole phylogroup, from the crop pathogen strains only and, finally, considering only isolates from environmental reservoirs. The x-axis indicates the position in the reference genomes Pto DC3000 (Buell et al., 2003) and Psy B728a (Feil et al., 2005). The y-axis indicates the empirical distribution of the distance statistic Hi representing the intensity of normalized recombination. The solid line represents the average value of Hi in a genome The dotted lines represent the top and bottom 2.5 percentiles. Grey lines represent virulence genes (Lindeberg et al., 2008). Turquoise lines represent genes associated with being a crop pathogen using the GWAS approach (Table S4). Orange lines represent genes associated with hotspots of recombination. The genes in the regions showing more intense recombination are given in Table S5.
Finally, we sought to identify those genes associated with crop pathogens (based on GWAS) and hotspots of recombination, and determine whether they have been previously characterized as virulence genes. Comparisons flagged the PSPTO 5191/PSYR 0346 gene, a member of the AcrB/AcrD/AcrF family of membrane proteins, implicated in multiple antibiotic resistance in Salmonella typhimurium (Piddock, 2006), that is not only recombining in both phylogroups but also associated with pathoadaptation in 2d. Other genes associated with pathoadaptation in phylogroup 2d were also recombination hotspots in crop pathogens, such as PSYR 195, 336, 1992 and 3131 coding for a hypothetical protein, an outer membrane autotransporter barrel, a zinc-containing alcohol dehydrogenase superfamily protein and a secretion protein HlyD, respectively. However, only PSYR 1794 and PSYR 3151 coding for a non-ribosomal peptide and the protein E of a type II secretion system, respectively, were also known virulence genes (Lindeberg et al., 2008).
In phylogroup 1a, the T3S effector gene hopAH1 was the only known virulence gene located within a recombination hotspot and that had also been associated with pathoadaptation in GWAS. Other crop pathogen-associated genes that are not known virulence genes corresponded to hotspots of homologous recombination, including the ferric iron reductase protein fhuF, an acyltransferase PSPTO 0997 (COG accession COG1835) and a transcriptional regulator from the GntR family (SMART accession smart00895).
Discussion
The bacterial plant pathogen P. syringae is well known as a model organism to study the molecular basis of plant–microbe interactions (Alfano & Collmer, 2004; Xin & He, 2013). Moreover, there is no other bacterial plant pathogen species for which so much is known about genetic diversity outside of agricultural environments (Morris et al., 2008, Morris et al., 2010; Monteil et al., 2012; Berge et al., 2014), which has made P. syringae a model for studying crop pathogen emergence as well (Mohr et al., 2008; Yan et al., 2008; Cai et al., 2011a, b; Diallo et al., 2012; Morris et al., 2013; Bartoli et al., 2015). Here, we applied for the first time a population genomics approach to P. syringae crop pathogens and their close environmental relatives and gained new insight into crop pathogen ancestry, emergence, crop adaptation and dissemination.
Phylogenetic reconstruction based on a small number of genes had already suggested that epidemic P. syringae crop pathogens in phylogroup 1a, such as Pto, have close relatives in non-agricultural environments (Monteil et al., 2013). After sequencing the genomes of representative crop pathogens and their environmental relatives in phylogroups 1a and 2d, we have now shown that several crop pathogens, including Pto T1 and Pap CC94, are more closely related to environmental isolates than to other crop pathogens. Moreover, by investigating patterns of recombination and population structure at the whole-genome level within each phylogroup, we show that several crop pathogens emerged from an ancestral recombining population independently from each other.
A fundamental question is when these emergence events occurred. Accurate molecular clock estimates are not possible within the sample frame of this study as it does not include a longitudinal sample. However, almost identical pathogen isolates have been sampled from crops dozens of years apart (Cai et al., 2011a; Clarke et al., 2015) and this is consistent with a mutation rate as low as one mutation per million base pairs per year. Therefore, considering that some of the P. syringae crop pathogen lineages in phylogroup 1a have diverged substantially from each other, more than one mutation per 1000 bp in some MLSA loci, it is likely that their most recent ancestor existed before humans started domesticating crop plants and before the advent of agriculture 5000–10 000 years ago. Therefore, the inferred ancestral population may have existed in non-agricultural plant communities and environmental reservoirs.
For phylogroup 2d, our analysis focused on a subset of isolates selected based on their identity at two MLSA loci excluding most of the genetic diversity that is known to exist in this phylogroup. This sample frame allowed comparison of contemporaneous isolates collected from diseased cantaloupe, sampled during a cantaloupe blight epidemic in France, and their closest relatives isolated from irrigation water, precipitation and ground water. Based on whole-genome analysis, isolates with almost identical genome sequences clustered together despite their different isolation sources and with some isolates from cantaloupes collected as far apart as 350 km. This result is consistent with frequent migration events between cantaloupe production fields and components of the water cycle and suggests that rain and irrigation water are involved in the dissemination of crop-pathogenic P. syringae between geographically distant fields. Although P. syringae had been reported in rain before (Constantinidou et al., 1990; Morris et al., 2008; Morris et al., 2010; Monteil et al., 2014a), no strong genetic linkage between the presence of P. syringae in rain or irrigation water on the one hand and P. syringae isolated from diseased crops on the other was possible without genome sequences. Additionally, in the specific case of phylogroup 2d isolates, which have a wide host range (Morris et al., 2000), rain and irrigation water may also be the original inoculum source of epidemics by transporting the pathogen from colonized wild and crop plants over long distances to crop fields and starting new outbreaks. This has important implications for crop disease prevention programmes since it is commonplace to link new P. syringae disease outbreaks to contaminated seed and nearby weeds but not to components of the water cycle or irrigation water (McCarter et al., 1983; Gitaitis & Walcott, 2007).
To understand the molecular basis of crop disease emergence, it is necessary to determine what differentiates epidemic crop pathogen isolates from their close relatives that are not epidemic pathogens. Bulk shotgun sequencing and virulence analysis of a small number of environmental isolates in phylogroup 1a had already revealed that these isolates contain well-known virulence genes such as T3S effector genes and that some of the environmental isolates are almost as virulent as Pto on tomato and other plant species (Monteil et al., 2013). By extending comparison of gene content to multiple whole genomes, we show here that all environmental relatives in phylogroup 1a are equipped with T3S systems and with repertoires of T3S effectors and other virulence genes similar to those of crop pathogens. Moreover, extending our virulence assays to all environmental relatives in phylogroup 1a, we confirmed that these isolates are all pathogenic on tomato, although less virulent than Pto (data not shown). We thus conclude that although the analysed environmental isolates were originally mostly collected from water, they appear to be adapted to life in association with plants and they possibly are pathogens of wild plants. These observations raise many questions about the role of wild plants and crop plants in the emergence and diversification of virulence traits in plant pathogenic populations. In particular, considering the genetic diversity of virulence-gene-equipped environmental populations, it is not clear why there are not more frequent emergence events. For example, there is only a single Pto lineage that has spread successfully worldwide on tomato (Cai et al., 2011a). It is therefore possible that there is a genetic barrier to emergence whereby only rare combinations of virulence genes allow emergence of an epidemic clone.
To test this hypothesis, a GWAS (Sheppard et al., 2013) was performed to identify genomic regions that show a statistically significant association with crop pathogens. Intriguingly, only two of the 58 T3S effector genes, hopD1 and hopQ1, were found to be pathogen-associated in phylogroup 1a and not a single T3S effector was found to be pathogen-associated in phylogroup 2d. Like some other T3S effectors, HopD1 interferes with the immune response triggered by other effectors (Block et al., 2014) and HopQ1 interferes with immunity triggered by microbial-associated molecular patterns, specifically the immune response triggered by the bacterial flagellum (Li et al., 2013a, b; Hann et al., 2014). Moreover, just as for many other effectors, deletion of either hopQ1 or hopD1 from Pto DC3000 has been shown to reduce bacterial growth on some plant genotypes under laboratory conditions (Wei et al., 2007). Therefore, the fact that only hopQ1 and hopD1 were identified as crop pathogen-associated in the GWAS suggests that the specific contribution to virulence by these two effectors is in some way more relevant in the life cycle of epidemic crop pathogens compared to the life cycle of bacteria associated with non-agricultural environments, while the other T3S effectors that are more evenly shared by crop pathogens and their environmental relatives play a role in fitness in agricultural as well as non-agricultural environments.
In phylogroup 2d, although our sampling strategy may have inflated associations, the GWAS revealed fewer statistically significant associations with source of isolation with lower P values compared to associations in phylogroup 1a. In particular, the distribution of the top GWAS hits within the group of most closely related isolates (Fig. 3b) showed no clear association at all. This is probably a result of our sampling frame and is consistent with our earlier conclusion that the sequenced 2d isolates represent a single population of P. syringae that regularly transfers between cantaloupe, other plant hosts and components of the water cycle.
Finally, our previous analysis of phylogroup 1a isolates (Monteil et al., 2013), and analysis of the kiwifruit pathogen P. syringae pv. actinidiae, suggested that crop pathogens emerge from recombining P. syringae populations. While it has been suggested that this recombination mainly occurs within P. syringae pathogen populations specific to a host plant species, such as kiwifruit (McCann et al., 2013), we provide evidence that the recombining P. syringae populations may not be host-specific and could include environmental P. syringae residing, for example, in wild plants and in decaying plant material, where P. syringae populations reach densities as high as 106 c.f.u. g–1 (Monteil et al., 2012). Importantly, using quantitative analysis of homologous recombination, we demonstrate that recombination between crop pathogens and their environmental relatives is as frequent as recombination between isolates within either niche (Fig. 4). This result reveals that P. syringae crop pathogens can acquire new genes from environmental populations and vice versa, possibly including potential virulence genes, which might exist at low frequency in environmental populations. In fact, although hopD1 and hopQ1 were not found in the environmental isolates analysed here, they may exist at low frequencies in these populations. One possible emergence scenario is that when a strain receives these genes in a recombination event from a donor, its fitness on crops increases, and it emerges as a highly virulent crop pathogen. Surprisingly, however, hopAH1 was the only T3S effector located in one of the identified hotspots of homologous recombination while hopD1 and hopQ1 were not. Their exclusive presence in crop pathogens that are phylogenetically distinct and their absence from closely related environmental isolates is nonetheless consistent with acquisition by horizontal gene transfer. This conclusion is supported by inferred phylogenies based on hopD1 and on hopQ1 (Fig. S6) that were incongruent with the core genome tree, suggesting that these genes were in fact subject to recombination.
Among the genes found to be located within hotspots of recombination in both analysed phylogroups, and more frequently present in crop pathogen isolates than environmental isolates, was the gene with locus tag PSPTO 5191/PSYR 0346. Functional predictions using blast p of the translated sequences identified strong signatures of a conserved domain specific of the acriflavin resistance protein family (Pfam accession PF00873, E value 4.03×10–143; TIGRFAM00915, 8.04×10–81, >90 % length of sequence). Therefore, this putative membrane protein could be part of an aminoglycoside efflux system involved in either toxin production or resistance processes. This gene has been implicated in multiple antibiotic resistances in human pathogens such as Salmonella Typhimurium (Piddock, 2006) but its potential role in conferring resistance to antibiotics and/or other agropesticides in P. syringae is unknown. It remains the case that recombination was found to be most frequent between closely related isolates with little genetic exchange between 1a and 2d isolates, consistent with well-known homology dependence of recombination (Hanage et al., 2009).
In conclusion, the extensive comparison of multiple P. syringae crop pathogens and their environmental relatives using several population genomics approaches clearly showed that not only does P. syringae frequently move between crop hosts and non-agricultural environments, but also P. syringae genes move just as frequently between one crop pathogen and the other as between crop pathogens and environmental relatives. This suggests that most virulence genes, including T3S effectors, are equally important for fitness on crops and non-crop hosts and that pathogen populations in environmental reservoirs could be important sources of virulence genes for crop pathogens. Intriguingly, the frequency of a small number of virulence genes, and of some genes of yet unknown function, is significantly higher in crop pathogens compared to their frequency in environmental relatives. Therefore, these genes appear to play a particularly important role in crop disease emergence and/or for fitness in agricultural settings. These findings provide a new basis for an improved understanding of crop pathogen emergence and control.
Acknowledgements
We are grateful to Haiji Liu (Virginia Tech) for assistance in DNA extraction, Zhongmeng Bao (Cornell University) for assistance in preparation of some DNA sequencing libraries, and Caroline Guilbaud (INRA) for assistance in preparing, verifying and shipping strains. We thank Susan Murray (Swansea University) who helped to run one of the bioinformatics analyses. The computational calculations were done at the Human Genome Center at the Institute of Medical Science (University of Tokyo, Japan) and HPC Wales (UK). C.L.M. received support from INRA and the European Union, in the framework of the Marie-Curie FP7 COFUND People Programme, through the award of an AgreenSkills’ fellowship (under grant agreement no. 267196). Research in B.A.V.’s laboratory and genome sequencing was funded by the National Science Foundation of the USA (grants IOS-1354215 and DEB-1241068). Funding for work in the Vinatzer laboratory was also provided in part by the Virginia Agricultural Experiment Station and the Hatch Program of the National Institute of Food and Agriculture, U.S. Department of Agriculture. Work carried out in the Sheppard laboratory was supported by the Medical Research Council (MRC) grant MR/L015080/1, and the Wellcome Trust grant 088786/C/09/Z. G.M. was supported by a NISCHR Health Research Fellowship (HF-14-13). S.K.S., C.L.M. and B.A.V. conceived and designed the overall project. C.L.M. performed experiments. C.E.M. contributed strains. B.A.V. and B.S. contributed genome sequences. D.J.S. generated genome assemblies. C.L.M., L.M., K.Y., D.J.S. and G.M. analysed the data. S.K.S., K.Y. and D.J.S. contributed analytical tools. C.L.M., B.A.V. and S.K.S. wrote the paper.
Supplementary Data
Supplementary Data
Supplementary Data
Supplementary Data
Supplementary Data
Supplementary Data
Supplementary Data
Supplementary Data
Supplementary Data
Supplementary Data
Supplementary Data
Abbreviations:
- GWAS
genome-wide association study
- T3S
type 3 secretion
References
- Alfano J. R., Collmer A.(2004). Type III secretion system effector proteins: Double agents in bacterial disease and plant defense. Annual Review of. Phytopathology 42385–414. [DOI] [PubMed] [Google Scholar]
- Andrews S.(2010). FastQC: A quality control tool for high throughput sequence data. Bioinformatics. http://www.bioinformatics.babraham.ac.uk/projects/fastqc/. [Google Scholar]
- Bartoli C., Lamichhane J. R., Berge O., Guilbaud C., Varvaro L., Balestra G. M., Vinatzer B. A., Morris C. E.(2015). A framework to gauge the epidemic potential of plant pathogens in environmental reservoirs: the example of kiwifruit canker. Mol Plant Pathol 16137–149. 10.1111/mpp.12167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berge O., Monteil C. L., Bartoli C., Chandeysson C., Guilbaud C., Sands D. C., Morris C. E.(2014). A user's guide to a data base of the diversity of Pseudomonas syringae and its application to classifying strains in this phylogenetic complex. PLoS One 9e105547. 10.1371/journal.pone.0105547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Block A., Toruño T. Y., Elowsky C. G., Zhang C., Steinbrenner J., Beynon J., Alfano J. R.(2014). The Pseudomonas syringae type III effector HopD1 suppresses effector-triggered immunity, localizes to the endoplasmic reticulum, and targets the Arabidopsis transcription factor NTL9. New Phytol 2011358–1370. 10.1111/nph.12626 [DOI] [PubMed] [Google Scholar]
- Buell C. R., Joardar V., Lindeberg M., Selengut J., Paulsen I. T., Gwinn M. L., Dodson R. J., Deboy R. T., Durkin A. S., et al. (2003). The complete genome sequence of the Arabidopsis and tomato pathogen Pseudomonas syringae pv. tomato DC3000. Proc Natl Acad Sci U S A 10010181–10186. 10.1073/pnas.1731982100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai R., Lewis J., Yan S., Liu H., Clarke C. R., Campanile F., Almeida N. F., Studholme D. J., Lindeberg M., et al. (2011a). The plant pathogen Pseudomonas syringae pv. tomato is genetically monomorphic and under strong selection to evade tomato immunity. PLoS Pathogens 7e1002130 10.1371/journal.ppat.1002130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai R., Yan S., Liu H., Leman S., Vinatzer B. A.(2011b). Reconstructing host range evolution of bacterial plant pathogens using Pseudomonas syringae pv. tomato and its close relatives as a model. Infection Genetics and. Evolution 111738–1751. [DOI] [PubMed] [Google Scholar]
- Carver T., Thomson N., Bleasby A., Berriman M., Parkhill J.(2009). DNAPlotter: circular and linear interactive genome visualization. Bioinformatics 25119–120. 10.1093/bioinformatics/btn578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X. M.(2005). Epidemiology and control of stripe rust Puccinia striiformis f. sp tritici on wheat. Canadian Journal of Plant Pathology 27314–337. [Google Scholar]
- Cheng L., Connor T. R., Siren J., Aanensen D. M., Corander J.(2013). Hierarchical and Spatially Explicit Clustering of DNA Sequences with BAPS Software. Molecular Biology and. Evolution 301224–1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke C. R., Studholme D. J., Hayes B., Runde B., Weisberg A., Cai R., Wroblewski T., Daunay M. C., Wicker E., et al. (2015). Genome-enabled phylogeographic investigation of the quarantine pathogen Ralstonia solanacearum race 3 biovar 2 and screening for sources of resistance against its core effectors. Phytopathology 105597–607. 10.1094/PHYTO-12-14-0373-R [DOI] [PubMed] [Google Scholar]
- Constantinidou H. A., Hirano S. S., Baker L. S., Upper C. D.(1990). Atmospheric dispersal of ice nucleation-active bacteria: the role of rain. Phytopathology 80934–937. 10.1094/Phyto-80-934 [DOI] [Google Scholar]
- Corander J., Waldmann P., Marttinen P., Sillanpää M. J.(2004). BAPS 2: enhanced possibilities for the analysis of genetic population structure. Bioinformatics 202363–2369. 10.1093/bioinformatics/bth250 [DOI] [PubMed] [Google Scholar]
- Corander J., Marttinen P., Sirén J., Tang J.(2008). Enhanced Bayesian modelling in BAPS software for learning genetic structures of populations. BMC Bioinformatics 9. 10.1186/1471-2105-9-539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demba Diallo M., Monteil C. L., Vinatzer B. A., Clarke C. R., Glaux C., Guilbaud C., Desbiez C., Morris C. E.(2012). Pseudomonas syringae naturally lacking the canonical type III secretion system are ubiquitous in nonagricultural habitats, are phylogenetically diverse and can be pathogenic. ISME J 61325–1335. 10.1038/ismej.2011.202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Didelot X., Falush D.(2007). Inference of bacterial microevolution using multilocus sequence data. Genetics 1751251–1266. 10.1534/genetics.106.063305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feil H., Feil W. S., Chain P., Larimer F., DiBartolo G., Copeland A., Lykidis A., Trong S., Nolan M., et al. (2005). Comparison of the complete genome sequences of Pseudomonas syringae pv. syringae B728a and pv. tomato DC3000. Proc Natl Acad Sci U S A 10211064–11069. 10.1073/pnas.0504930102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garland T., Bennett A. F., Rezende E. L.(2005). Phylogenetic approaches in comparative physiology. J Exp Biol 2083015–3035. 10.1242/jeb.01745 [DOI] [PubMed] [Google Scholar]
- Gitaitis R., Walcott R.(2007). The epidemiology and management of seedborne bacterial diseases. Annu Rev Phytopathol 45371–397. 10.1146/annurev.phyto.45.062806.094321 [DOI] [PubMed] [Google Scholar]
- Grosso-Becerra M. V., Santos-Medellín C., González-Valdez A., Méndez J. L., Delgado G., Morales-Espinosa R., Servín-González L., Alcaraz L. D., Soberón-Chávez G.(2014). Pseudomonas aeruginosa clinical and environmental isolates constitute a single population with high phenotypic diversity. BMC Genomics 15. 10.1186/1471-2164-15-318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanage W. P., Fraser C., Tang J., Connor T. R., Corander J.(2009). Hyper-recombination, diversity, and antibiotic resistance in pneumococcus. Science 3241454–1457. 10.1126/science.1171908 [DOI] [PubMed] [Google Scholar]
- Hann D. R., Domínguez-Ferreras A., Motyka V., Dobrev P., Schornack S., Jehle A., Felix G., Chinchilla D., Rathjen J. P., Boller T.(2014). The Pseudomonas type III effector HopQ1 activates cytokinin signaling and interferes with plant innate immunity. New Phytol 201585–598. 10.1111/nph.12544 [DOI] [PubMed] [Google Scholar]
- Hazen T. H., Lafon P. C., Garrett N. M., Lowe T. M., Silberger D. J., Rowe L. A., Frace M., Parsons M. B., Bopp C. A., et al. (2015). Insights into the environmental reservoir of pathogenic Vibrio parahaemolyticus using comparative genomics. Front Microbiol 6204. 10.3389/fmicb.2015.00204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano S. S., Upper C. D.(2000). Bacteria in the leaf ecosystem with emphasis on Pseudomonas syringae-a pathogen, ice nucleus, and epiphyte. Microbiol Mol Biol Rev 64624–653. 10.1128/MMBR.64.3.624-653.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hockett K. L., Nishmura M., Karsrud E., Dougherty K. M., Baltrus D. A.(2014). P. syringae CC1557: a highly virulent strain with an unusually small type III effector repertoire that includes a novel effector. Am Phytopath Society. [DOI] [PubMed] [Google Scholar]
- Joardar V., Lindeberg M., Jackson R. W., Selengut J., Dodson R., Brinkac L. M., Daugherty S. C., Deboy R., Durkin A. S., et al. (2005). Whole-genome sequence analysis of Pseudomonas syringae pv. phaseolicola 1448A reveals divergence among pathovars in genes involved in virulence and transposition. J Bacteriol 1876488–6498. 10.1128/JB.187.18.6488-6498.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson P. T., de Roode J. C., Fenton A.(2015). Why infectious disease research needs community ecology. Science 349. 10.1126/science.1259504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jolley K. A., Maiden M. C.(2010). BIGSdb: Scalable analysis of bacterial genome variation at the population level. BMC Bioinformatics 11. 10.1186/1471-2105-11-595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K., Toh H.(2008). Recent developments in the MAFFT multiple sequence alignment program. Brief Bioinform 9286–298. 10.1093/bib/bbn013 [DOI] [PubMed] [Google Scholar]
- Krueger F.(2015). Trim Galore!: A wrapper tool around Cutadapt and FastQC to consistently apply quality and adapter trimming to FastQ files. http://www.bioinformatics.babraham.ac.uk/projects/trim_galore/.
- Lawson D. J., Hellenthal G., Myers S., Falush D.(2012). Inference of population structure using dense haplotype data. PLoS Genet 8. 10.1371/journal.pgen.1002453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Chiang Y.-H., Coaker G.(2013a). The HopQ1 effector’s nucleoside hydrolase-like domain is required for bacterial virulence in arabidopsis and tomato, but not host recognition in Tobacco. PLoS ONE 8e59684 10.1371/journal.pone.0059684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Yadeta K. A., Elmore J. M., Coaker G.(2013). The Pseudomonas syringae Effector HopQ1 Promotes Bacterial Virulence and Interacts with Tomato 14-3-3 Proteins in a Phosphorylation-Dependent Manner. PLANT PHYSIOLOGY 1612062–2074. 10.1104/pp.112.211748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindeberg M., Myers C. R., Collmer A., Schneider D. J.(2008). Roadmap to new virulence determinants in Pseudomonas syringae: Insights from comparative genomics and genome organization. Mol Plant Microbe In 21685–700. [DOI] [PubMed] [Google Scholar]
- Maiden M. C., Jansen van Rensburg M. J., Bray J. E., Earle S. G., Ford S. A., Jolley K. A., McCarthy N. D.(2013). MLST revisited: the gene-by-gene approach to bacterial genomics. Nat Rev Microbiol 11728–736. 10.1038/nrmicro3093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin M.(2011). Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.journal 1710. 10.14806/ej.17.1.200 [DOI] [Google Scholar]
- Martins E. P., Garland T.(1991). Phylogenetic analyses of the correlated evolution of continuous characters: a simulation study. Evolution 45534–557. 10.2307/2409910 [DOI] [PubMed] [Google Scholar]
- McCann H. C., Rikkerink E. H., Bertels F., Fiers M., Lu A., Rees-George J., Andersen M. T., Gleave A. P., Haubold B., et al. (2013). Genomic analysis of the Kiwifruit pathogen Pseudomonas syringae pv. actinidiae provides insight into the origins of an emergent plant disease. PLoS Pathog 9e1003503. 10.1371/journal.ppat.1003503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarter S. M., Jones J. B., Gitaitis R. D., Smitley D. R.(1983). Survival of Pseudomonas syringae pv. tomato in association with tomato seed, soil, host tissue, and epiphytic weed hosts in Georgia. Phytopathology 731393–1398. 10.1094/Phyto-73-1393 [DOI] [Google Scholar]
- Méric G., Yahara K., Mageiros L., Pascoe B., Maiden M. C., Jolley K. A., Sheppard S. K.(2014). A reference pan-genome approach to comparative bacterial genomics: identification of novel epidemiological markers in pathogenic Campylobacter. PLoS One 9e92798. 10.1371/journal.pone.0092798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohr T. J., Liu H., Yan S., Morris C. E., Castillo J. A., Jelenska J., Vinatzer B. A.(2008). Naturally occurring nonpathogenic isolates of the plant pathogen Pseudomonas syringae lack a type III secretion system and effector gene orthologues. J Bacteriol 1902858–2870. 10.1128/JB.01757-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monteil C. L., Guilbaud C., Glaux C., Lafolie F., Soubeyrand S., Morris C. E.(2012). Emigration of the plant pathogen Pseudomonas syringae from leaf litter contributes to its population dynamics in alpine snowpack. Environ Microbiol 142099–2112. 10.1111/j.1462-2920.2011.02680.x [DOI] [PubMed] [Google Scholar]
- Monteil C. L., Cai R., Liu H., Llontop M. E., Leman S., Studholme D. J., Morris C. E., Vinatzer B. A. (2013). Nonagricultural reservoirs contribute to emergence and evolution of Pseudomonas syringae crop pathogens. New Phytol 199800–811. 10.1111/nph.12316 [DOI] [PubMed] [Google Scholar]
- Monteil C. L., Bardin M., Morris C. E.(2014a). Features of air masses associated with the deposition of Pseudomonas syringae and Botrytis cinerea by rain and snowfall. ISME J 82290–2304. 10.1038/ismej.2014.55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monteil C. L., Lafolie F., Laurent J., Clement J. C., Simler R., Travi Y., Morris C. E.(2014b). Soil water flow is a source of the plant pathogen Pseudomonas syringae in subalpine headwaters. Environ Microbiol 162038–2052. 10.1111/1462-2920.12296 [DOI] [PubMed] [Google Scholar]
- Morris C. E., Glaux C., Latour X., Gardan L., Samson R., Pitrat M.(2000). The Relationship of host range, physiology, and genotype to virulence on cantaloupe in pseudomonas syringae from cantaloupe blight epidemics in France. Phytopathology 90636–646. 10.1094/PHYTO.2000.90.6.636 [DOI] [PubMed] [Google Scholar]
- Morris C. E., Sands D. C., Vinatzer B. A., Glaux C., Guilbaud C., Buffière A., Yan S., Dominguez H., Thompson B. M.(2008). The life history of the plant pathogen Pseudomonas syringae is linked to the water cycle. ISME J 2321–334. 10.1038/ismej.2007.113 [DOI] [PubMed] [Google Scholar]
- Morris C. E., Sands D. C., Vanneste J. L., Montarry J., Oakley B., Guilbaud C., Glaux C.(2010). Inferring the evolutionary history of the plant pathogen Pseudomonas syringae from its biogeography in headwaters of rivers in North America, Europe, and New Zealand. MBio 1e00107-10–e00107-20. 10.1128/mBio.00107-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris C. E., Monteil C. L., Berge O.(2013). The life history of Pseudomonas syringae: linking agriculture to earth system processes. Annu Rev Phytopathol 5185–104. 10.1146/annurev-phyto-082712-102402 [DOI] [PubMed] [Google Scholar]
- O'Brien H. E., Thakur S., Guttman D. S.(2011). Evolution of plant pathogenesis in Pseudomonas syringae: a genomics perspective. Annu Rev Phytopathol 49269–289. 10.1146/annurev-phyto-072910-095242 [DOI] [PubMed] [Google Scholar]
- Ochman H., Lawrence J. G., Groisman E. A.(2000). Lateral gene transfer and the nature of bacterial innovation. Nature 405299–304. 10.1038/35012500 [DOI] [PubMed] [Google Scholar]
- Pascoe B., Meric G., Murray S., Mageiros L., Yahara K., Bowen R., Jones N. H., Jeeves R. E., Lappin-Scott H. M., et al. (2015). Enhanced biofilm formation evolves from divergent genetic backgrounds in host generalist Campylobacter jejuni. Environ Microbiol 174779–4789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piddock L.(2006). Multidrug-resistance efflux pumps - not just for resistance. Nat Rev Microbiol 4629–636. 10.1038/nrmicro1464 [DOI] [PubMed] [Google Scholar]
- Price M. N., Dehal P. S., Arkin A. P.(2010). FastTree 2—approximately maximum-likelihood trees for large alignments. PLoS One 5e9490. 10.1371/journal.pone.0009490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutherford K., Parkhill J., Crook J., Horsnell T., Rice P., Rajandream M. A., Barrell B.(2000). Artemis: sequence visualization and annotation. Bioinformatics 16944–945. 10.1093/bioinformatics/16.10.944 [DOI] [PubMed] [Google Scholar]
- Sheppard S. K., Jolley K. A., Maiden M. C.(2012). A Gene-By-Gene Approach to Bacterial Population Genomics: Whole Genome MLST of Campylobacter. Genes 3261–277. 10.3390/genes3020261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheppard S. K., Didelot X., Meric G., Torralbo A., Jolley K. A., Kelly D. J., Bentley S. D., Maiden M. C., Parkhill J., Falush D.(2013). Genome-wide association study identifies vitamin B5 biosynthesis as a host specificity factor in Campylobacter. Proc Natl Acad Sci U S A 11011923–11927. 10.1073/pnas.1305559110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh R. P., Hodson D. P., Huerta-Espino J., Jin Y., Bhavani S., Njau P., Herrera-Foessel S., Singh P. K., Singh S., Govindan V.(2011). The emergence of Ug99 races of the stem rust fungus is a threat to world wheat production. Annu Rev Phytopathol 49465–481. 10.1146/annurev-phyto-072910-095423 [DOI] [PubMed] [Google Scholar]
- Struve C., Krogfelt K. A.(2004). Pathogenic potential of environmental Klebsiella pneumoniae isolates. Environ Microbiol 6584–590. 10.1111/j.1462-2920.2004.00590.x [DOI] [PubMed] [Google Scholar]
- Stukenbrock E. H., McDonald B. A.(2008). The origins of plant pathogens in agro-ecosystems. Annu Rev Phytopathol 4675–100. 10.1146/annurev.phyto.010708.154114 [DOI] [PubMed] [Google Scholar]
- Tampakaki A. P., Skandalis N., Gazi A. D., Bastaki M. N., Sarris P. F., Charova S. N., Kokkinidis M., Panopoulos N. J.(2011). Playing the “Harp”: evolution of our understanding of hrp/hrc genes. Annu Rev Phytopathol 48347–370. [DOI] [PubMed] [Google Scholar]
- Vinatzer B. A., Monteil C. L., Clarke C. R.(2014). Harnessing population genomics to understand how bacterial pathogens emerge, adapt to crop hosts, and disseminate. Annu Rev Phytopathol 52,19–43. 10.1146/annurev-phyto-102313-045907 [DOI] [PubMed] [Google Scholar]
- Wei C. F., Kvitko B. H., Shimizu R., Crabill E., Alfano J. R., Lin N. C., Martin G. B., Huang H. C., Collmer A.(2007). A Pseudomonas syringae pv. tomato DC3000 mutant lacking the type III effector HopQ1-1 is able to cause disease in the model plant Nicotiana benthamiana. Plant J 5132–46. 10.1111/j.1365-313X.2007.03126.x [DOI] [PubMed] [Google Scholar]
- Whiley H., van den Akker B., Giglio S., Bentham R.(2013). The role of environmental reservoirs in human campylobacteriosis. Int J Environ Res Public Health 105886–5907. 10.3390/ijerph10115886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woolhouse M. E., Taylor L. H., Haydon D. T.(2001). Population biology of multihost pathogens. Science 2921109–1112. 10.1126/science.1059026 [DOI] [PubMed] [Google Scholar]
- Xin X. F., He S. Y.(2013). Pseudomonas syringae pv. tomato DC3000: a model pathogen for probing disease susceptibility and hormone signaling in plants. Annu Rev Phytopathol 51473–498. 10.1146/annurev-phyto-082712-102321 [DOI] [PubMed] [Google Scholar]
- Yahara K., Furuta Y., Oshima K., Yoshida M., Azuma T., Hattori M., Uchiyama I., Kobayashi I.(2013). Chromosome painting in silico in a bacterial species reveals fine population structure. Molecular Biology and. Evolution 301454–1464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yahara K., Didelot X., Ansari M. A., Sheppard S. K., Falush D.(2014). Efficient inference of recombination hot regions in bacterial genomes. Mol Biol Evol 311593–1605. 10.1093/molbev/msu082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yahara K., Didelot X., Jolley K. A., Kobayashi I., Maiden M. C., Sheppard S. K., Falush D.(2016a). The landscape of realized homologous recombination in pathogenic bacteria. Mol Biol Evol 33456–471. 10.1093/molbev/msv237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yahara K., Taylor A., de Vries S., Murray S., Pascoe B., Mageiros L., Torralbo A., Vidal A., Ridley A., et al. (2016b). Genome-wide association of functional traits linked with Campylobacter jejuni survival from farm to fork. PeerJ Preprints 4e2300v1. [DOI] [PubMed] [Google Scholar]
- Yan S., Liu H., Mohr T. J., Jenrette J., Chiodini R., Zaccardelli M., Setubal J. C., Vinatzer B. A.(2008). Role of recombination in the evolution of the model plant pathogen Pseudomonas syringae pv. tomato DC3000, a very atypical tomato strain. Applied and Environmental. Microbiology 743171–3181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zerbino D. R., Birney E.(2008). Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res 18821–829. 10.1101/gr.074492.107 [DOI] [PMC free article] [PubMed] [Google Scholar]
Data Bibliography
- 1. Studholme, D., Monteil C., Swingle B. and Vinatzer, B. A. NCBI, Pseudomonas syringae BioProject (ID 320409) (2016).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.