ABSTRACT
Mutations in the with-no-lysine kinase 1 (WNK1), WNK4, kelch-like 3 (KLHL3), and cullin3 (CUL3) genes are known to cause the hereditary disease pseudohypoaldosteronism type II (PHAII). It was recently demonstrated that this results from the defective degradation of WNK1 and WNK4 by the KLHL3/CUL3 ubiquitin ligase complex. However, the other physiological in vivo roles of KLHL3 remain unclear. Therefore, here we generated KLHL3−/− mice that expressed β-galactosidase (β-Gal) under the control of the endogenous KLHL3 promoter. Immunoblots of β-Gal and LacZ staining revealed that KLHL3 was expressed in some organs, such as brain. However, the expression levels of WNK kinases were not increased in any of these organs other than the kidney, where WNK1 and WNK4 increased in KLHL3−/− mice but not in KLHL3+/− mice. KLHL3−/− mice also showed PHAII-like phenotypes, whereas KLHL3+/− mice did not. This clearly demonstrates that the heterozygous deletion of KLHL3 was not sufficient to cause PHAII, indicating that autosomal dominant type PHAII is caused by the dominant negative effect of mutant KLHL3. We further demonstrated that the dimerization of KLHL3 can explain this dominant negative effect. These findings could help us to further understand the physiological roles of KLHL3 and the pathophysiology of PHAII caused by mutant KLHL3.
KEYWORDS: kelch-like 3 (KLHL3), distal convoluted tubule, hypertension, kidney, NaCl cotransporter, with-no-lysine kinase (WNK)
INTRODUCTION
Pseudohypoaldosteronism type II (PHAII) is a hereditary disease that is characterized by salt-sensitive hypertension, hyperkalemia, metabolic acidosis, and thiazide sensitivity (1). Mutations in the with-no-lysine kinase 1 (WNK1) and WNK4 genes are known to cause PHAII (2). Furthermore, it has generally been considered that overactivation of the thiazide-sensitive Na-Cl cotransporter (NCC) is the main cause of PHAII (3).
Many studies have demonstrated that WNK kinases are at the top of the signaling cascade, along with oxidative stress-responsive gene 1 (OSR1), Ste20-related proline-alanine-rich kinase (SPAK), and the solute carrier family 12a (SLC12a) transporter family, which includes the NCC and Na-K-Cl cotransporter (NKCC). WNK phosphorylates and activates OSR1/SPAK, which in turn phosphorylate and activate the SLC12a transporters (4–6). The regulation of NCC by WNK-OSR1/SPAK signaling was confirmed in vivo using various genetically engineered mouse models (7–14) and overactivation of this WNK-OSR1/SPAK-NCC phosphorylation signal in the kidney causes PHAII (5, 6, 15, 16).
In 2012, mutations in KLHL3 and cullin3 (CUL3) genes were newly identified as causing PHAII (17, 18). KLHL3 is a member of the BTB-BACK-Kelch family, and combines with Cul3 to form the E3 ubiquitin ligase complex. KLHL3 binds with the acidic motif of WNK kinases and functions as a substrate adapter protein for Cul3-based E3 ligase-mediated ubiquitination (19–22). Recently, we generated KLHL3R528H/+ mice carrying autosomal-dominant type PHAII, which contained mutant KLHL3 R528H that had a reduced binding ability to WNK4. This PHAII mouse model revealed that the defective binding of WNK kinases to mutant KLHL3 results in impaired degradation of WNK kinases, leading to increased protein expression of WNK1 and WNK4 (23). These findings clearly indicated that KLHL3 is a key physiological regulator of WNK signaling via the protein degradation mechanism. However, although phosphorylation of KLHL3 is reported to regulate its binding ability to WNK4 (24, 25), the pathophysiological roles of KLHL3 aside from its involvement in PHAII remain unclear. Moreover, other substrates of the Cul3-KLHL3 E3 ligase complex are not yet known.
Previous research has largely focused on the role of WNK signaling in renal epithelial ion homeostasis. However, more recent studies have demonstrated that WNK signaling also occurs in extrarenal tissues. For example, in vascular smooth muscle cells, WNK1 and WNK3 regulate the vascular tonus through the phosphorylation of SPAK and NKCC1 (9, 26, 27), and in neuronal and brain cells, WNK kinases regulate the phosphorylation of NKCC1 and K-Cl cotransporter (KCC2) via SPAK activation, controlling intracellular Cl− levels via γ-aminobutyric acid (GABA) (28–30). In addition, several types of cross talk between WNK kinases and other signaling pathways have been reported (31–34). Thus, the WNK signaling pathway is also expected to be essential in organs other than the kidney, which raises the question whether KLHL3 also regulates the expression level of WNK kinases in these organs. However, to date, clinical features in organs other than the kidney have not been reported in patients with PHAII caused by mutant KLHL3, and the tissue distribution and intratissue localization of KLHL3 have not been well examined.
In this study, we aimed to answer these questions by generating and analyzing KLHL3−/− mice that expressed β-Gal under the control of the endogenous KLHL3 promoter. We took advantage of the expression of β-galactosidase (β-Gal) by the Klhl3 knockout allele in this mouse model to investigate the tissue distribution and intratissue localization of KLHL3, and then examined how WNK kinases are regulated in KLHL3-expressing organs. After this, we further investigated the dominant negative effect of mutant KLHL3 R528H, which causes autosomal dominant type PHAII.
RESULTS
Generation of KLHL3 knockout/lacZ knock-in mice.
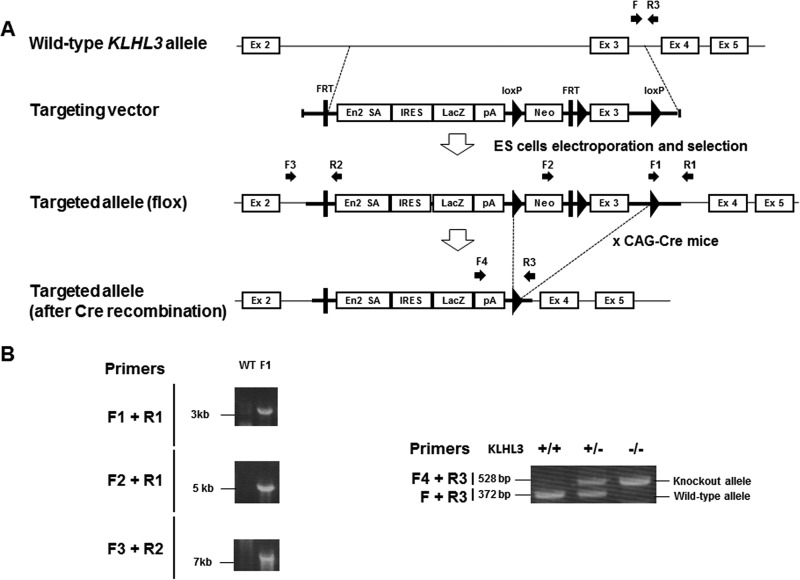
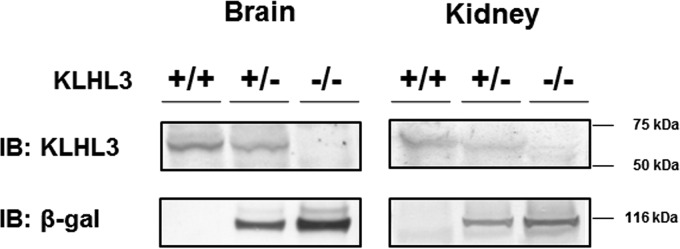
In this study, we generated KLHL3−/− mice whose knockout allele expressed β-Gal via the lacZ gene under the control of the endogenous KLHL3 promoter (i.e., KLHL3 knockout/lacZ knock-in mice). To generate these mice, we used a targeting vector that had been designed to delete exon 3 of KLHL3 but which also led to the expression of the lacZ gene under the control of the endogenous KLHL3 promoter. As shown in Fig. 1, these mice were crossed with CAG promoter-Cre transgenic mice to successfully generate KLHL3−/− mice. KLHL3 heterozygous (+/−) and homozygous (−/−) mice exhibited no gross anatomical abnormalities and no significant differences in body weight or height and also presented a normal birth rate. The absence of the KLHL3 protein in KLHL3−/− mice was confirmed by the immunoblotting of protein samples from the brain and kidney. In addition, we confirmed β-Gal protein expression by the Klhl3 knockout allele in KLHL3 knockout/lacZ knock-in mice (Fig. 2).
FIG 1.
Generation of KLHL3 knockout/lacZ knock-in mice. (A) Targeting strategy for generating KLHL3 knockout/lacZ knock-in mice. The diagram shows the wild-type KLHL3 locus, the targeting vector, and the targeted locus before and after Cre recombination. The targeting vector was designed to delete exon 3 of KLHL3. In addition, this vector led to the expression of the lacZ gene under the control of the endogenous KLHL3 promoter. EN2-SA, Engrailed-2 splice acceptor. (B) Verification of homologous recombination and genotyping PCR of the genomic DNA of the mice.
FIG 2.
Confirmation of the absence of KLHL3 expression and the presence of β-Gal expression by the knockout allele. Representative immunoblots of KLHL3 and β-Gal in the kidneys and brains of KLHL3+/+, KLHL3+/−, and KLHL3−/− mice are shown, confirming the absence of the KLHL3 protein and expression of the β-Gal protein by the Klhl3 knockout allele.
Tissue and cellular distribution of KLHL3.
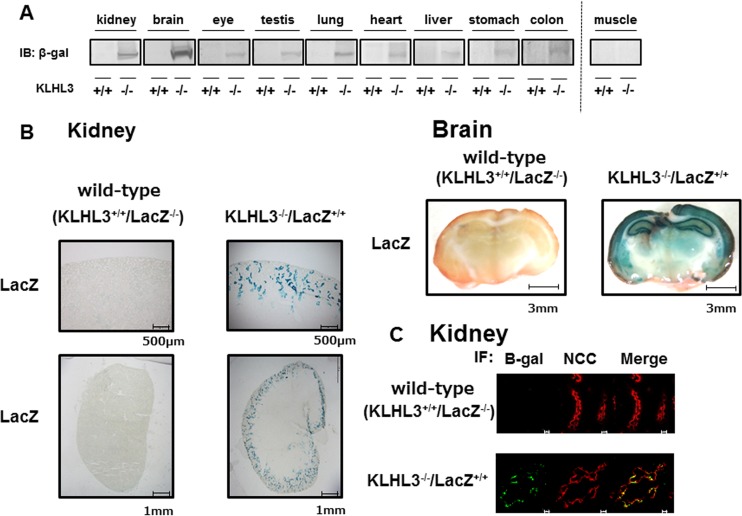
Next, we used the KLHL3 knockout/lacZ knock-in mice to investigate the tissue and cellular distribution of KLHL3. Since the KLHL3 antibody we used could only detect the KLHL3 protein in the kidney and brain, we performed immunoblots of β-Gal to investigate the distribution of KLHL3 in other tissues. This demonstrated that KLHL3 was strongly expressed in the brain and kidney, weakly expressed in the eye, testis, lung, heart, liver, stomach, and colon, and not expressed in the muscle (Fig. 3A). We then took advantage of the β-Gal expression in the knockout allele of the mouse to perform LacZ staining of the KLHL3-expressing tissue from KLHL3 knockout/lacZ knock-in mice (Fig. 3B). Although the cellular distribution of KLHL3 could not be determined in the tissues with lower levels of expression due to the poor LacZ signal, strong LacZ staining signals were observed in the hippocampus and cortex of the brain, and in the distal convoluted tubules (DCT) of the kidney, which was also confirmed by immunofluorescence with NCC. KLHL3−/− mice showed hyperplasia of the DCT, as previously reported in WNK4 transgenic PHAII model mice (35) (Fig. 3C).
FIG 3.
Tissue and intratissue distribution of KLHL3 was confirmed by an immunoblot of β-Gal and LacZ staining. (A) Immunoblotting to detect the β-Gal protein level in each organ from KLHL3 knockout/lacZ knock-in mice. Representative immunoblots of β-Gal in each of the organs of wild-type and KLHL3 homozygous knockout/lacZ knock-in mice are shown. β-Gal was strongly detected in the kidney and brain and weakly detected in the eye, testis, lung, heart, liver, stomach, and colon. (B) LacZ staining of the kidney and brain in KLHL3 knockout/lacZ knock-in mice. Strong LacZ staining was observed in the hippocampus and cortex of the brain and in the distal convoluted tubules of the kidney. (C) Double immunofluorescence of β-Gal and the Na-Cl cotransporter (NCC) in the kidneys of wild-type and KLHL3 knockout/lacZ knock-in mice. The β-Gal signal colocalized with NCC, indicating that KLHL3 was present in the distal convoluted tubules. Scale bars, 10 μm.
Protein levels of WNK kinases in the tissues of KLHL3−/− mice.
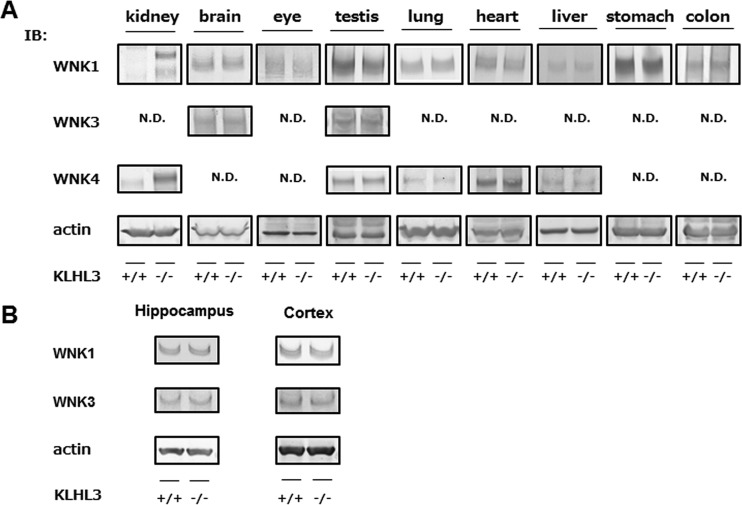
Since KLHL3 is known to play a role in the ubiquitination and degradation of WNK kinases, we expected that an absence of KLHL3 might result in increased expression levels of WNK kinases in various organs. Therefore, we investigated the protein levels of WNK1, WNK3, and WNK4 in the KLHL3-expressing organs from KLHL3−/− mice (Fig. 4A). We found that the expression levels of WNK1, WNK3, and WNK4 were not increased in the brain and other tissues that exhibited lower expression levels of KLHL3 even in the KLHL3−/− mice, but the expression of WNK1 and WNK4 was dramatically increased in the kidneys of KLHL3−/− mice. Furthermore, we separated and prepared samples of the hippocampus and cortex of the brain, where KLHL3 was expected to be present, but no significant differences in WNK1 and WNK4 protein levels were detected between wild-type and KLHL3−/− mice, as shown in Fig. 4B.
FIG 4.
WNK kinases only increased in the kidneys of KLHL3−/− mice. (A) Representative immunoblots of β-Gal and WNK1, WNK3, and WNK4 in KLHL3-expressing tissues in KLHL3+/+ and KLHL3−/− mice. In KLHL3−/− mice, the expression levels of WNK1, WNK3, and WNK4 did not increase in any organ other than the kidney, where WNK1 and WNK4 increased. N.D., not detected. (B) Immunoblotting of WNK1 and WNK3 protein levels in hippocampus and cortex from KLHL3−/− mice. No significant differences in WNK protein levels were detected between wild-type and KLHL3−/− mice. We repeated the same experiments three times with consistent results.
KLHL3−/− mice exhibited a PHAII-like phenotype.
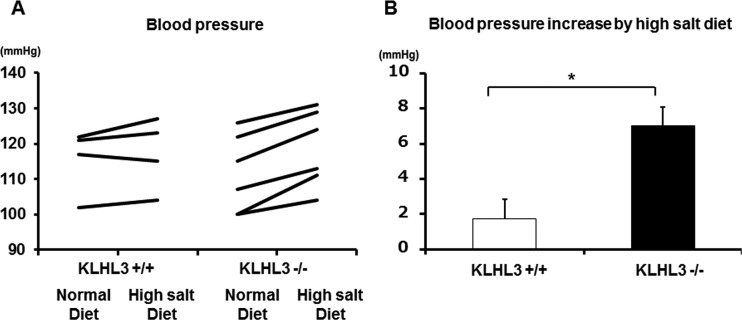
To confirm that KLHL3−/− mice are a good model of PHAII, we first checked their blood chemistry. As shown in Table 1, KLHL3−/− mice exhibited lower plasma pH and higher plasma K+ and Cl− levels than wild-type mice. However, there was no significant difference in the blood chemistry data between KLHL3+/− heterozygous mice and KLHL3+/+ mice, different from KLHL3−/− mice. We also measured the BP of KLHL3−/− mice and KLHL3+/+ mice that had been fed a normal diet and then repeated these measurements in the same mice after feeding them a high-salt diet for more than 3 days. As shown in Fig. 5, no significant difference in BP was detected under the normal diet, but the delta systolic blood pressure was significantly higher in KLHL3−/− mice than in KLHL3+/+ mice after consuming the high-salt diet, demonstrating an increased salt sensitivity of KLHL3−/− mice. Thus, KLHL3−/− mice clearly exhibited a PHAII-like phenotype.
TABLE 1.
Blood chemistry bioparameters
| Blood biochemistry parametera | Mean ± SEMb |
||
|---|---|---|---|
| KLHL3+/+ mice | KLHL3+/− mice | KLHL3−/− mice | |
| Na+ (mmol/liter) | 149.5 ± 0.6 | 148.8 ± 0.4 | 150.4 ± 1.9 |
| K+ (mmol/liter) | 4.5 ± 0.1 | 4.6 ± 0.1 | 5.2 ± 0.2* |
| Cl− (mmol/liter) | 112.0 ± 0.8 | 111.4 ± 0.6 | 116.1 ± 1.8* |
| Glu (mg/dl) | 228.0 ± 18.8 | 239.9 ± 9.5 | 222.5 ± 11.6 |
| pH | 7.297 ± 0.011 | 7.286 ± 0.009 | 7.235 ± 0.015* |
| pCO2 (mmHg) | 43.7 ± 1.1 | 43.7 ± 1.4 | 45.1 ± 2.4 |
| HCO3− (mmol/liter) | 21.6 ± 0.6 | 20.6 ± 0.4 | 19.0 ± 0.8* |
| Hb (g/dl) | 14.9 ± 0.3 | 15.0 ± 0.1 | 14.5 ± 0.2 |
Glu, glucose; Hb, hemoglobin.
*, P < 0.05 compared to KLHL3+/+ mice.
FIG 5.
Increased salt sensitivity in KLHL3−/− mice. (A) The blood pressure of KLHL3+/+ and KLHL3−/− mice after being fed a normal or a high-salt diet. (B) The observed blood pressure increased as a result of the high-salt diet. No significant difference in blood pressure was detected under a normal diet, whereas the increase of systolic blood pressure was significantly higher in KLHL3−/− mice than in KLHL3+/+ mice under a high-salt diet.
The WNK-OSR1/SPAK-NCC phosphorylation cascade in the kidneys of KLHL3 knockout mice.
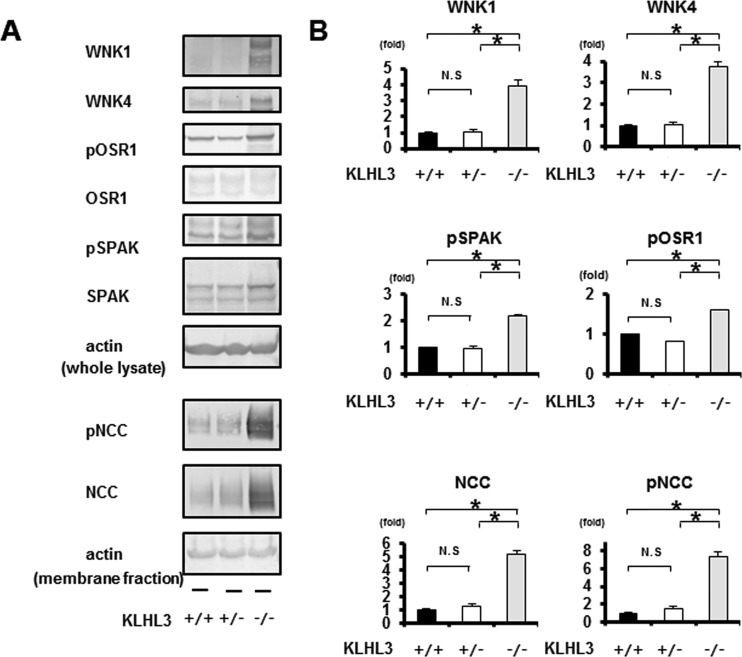
Since KLHL3−/− mice exhibited increased WNK1 and WNK4 expression levels in their kidneys, we next examined the protein expression and phosphorylation of molecules constituting the WNK signaling pathway. As shown in Fig. 6, the protein expression levels of WNK1 and WNK4 were significantly higher in the kidneys of KLHL3−/− mice compared to KLHL3+/+and KLHL3+/− mice. Accordingly, the phosphorylation of OSR1, SPAK, and NCC also increased in KLHL3−/− mice. However, although the protein expression of KLHL3 appeared to be lower in the kidneys of KLHL3+/− heterozygous mice than in KLHL3+/+ mice (Fig. 2), there was no significant difference in the protein levels of WNK1 and WNK4 in the kidneys between KLHL3+/− and KLHL3+/+ mice.
FIG 6.
Increased WNK1 and WNK4 protein levels, and the activation of the WNK-OSR1/SPAK-NCC phosphorylation signaling cascade in the kidneys of KLHL3−/− mice. (A) Representative immunoblots of WNK1 and WNK4 in the kidneys of KLHL3+/+, KLHL3+/−, and KLHL3−/− mice. The expression levels of WNK1 and WNK4 and the phosphorylation of SPAK, OSR1, and NCC were significantly higher in the kidneys of KLHL3−/− mice but not in KLHL3+/− mice. (B) Densitometric analysis. Values are expressed as a ratio of the average signal in wild-type mice. n = 3 to 6; *, P < 0.05.
Dimer formation of KLHL3 could explain the dominant negative effect of mutant KLHL3 R528H.
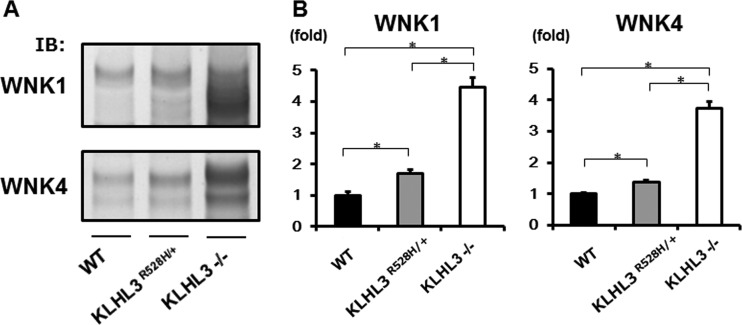
Interestingly, we found that the WNK-OSR1/SPAK-NCC phosphorylation cascade was not activated in the kidneys of KLHL3+/− mice. This finding clearly demonstrates that the heterozygous deletion of KLHL3 was not sufficient to cause PHAII in the kidney, indicating that the PHAII phenotypes previously observed in KLHL3R528H/+ heterozygous mice were caused by the dominant negative effect of the R528H KLHL3 mutant. To further investigate the mechanisms of this dominant negative effect of mutant KLHL3 R528H, we compared the protein expression levels of WNK1 and WNK4 in the kidneys of KLHL3R528H/+ and KLHL3−/− mice. As shown in Fig. 7, the protein levels of WNK1 and WNK4 were higher in the kidneys of KLHL3−/− mice than in KLHL3R528H/+ mice, indicating that KLHL3R528H/+ mice still retained some capacity for the degradation of WNK kinases.
FIG 7.
The protein levels of WNK1 and WNK4 were higher in KLHL3−/− mice than in KLHL3R528H/+ mice. (A) Protein expression levels of WNK1 and WNK4 in kidneys from wild-type, KLHL3−/−, and KLHL3R528H/+ mice. The ability to degrade WNK kinases. This was conserved in KLHL3R528H/+ mice but lost in KLHL3−/− mice. (B) Densitometric analysis. Values are expressed as a ratio of the average signal in KLHL3R528H/+ mice. n = 3 to 9; *, P < 0.05.
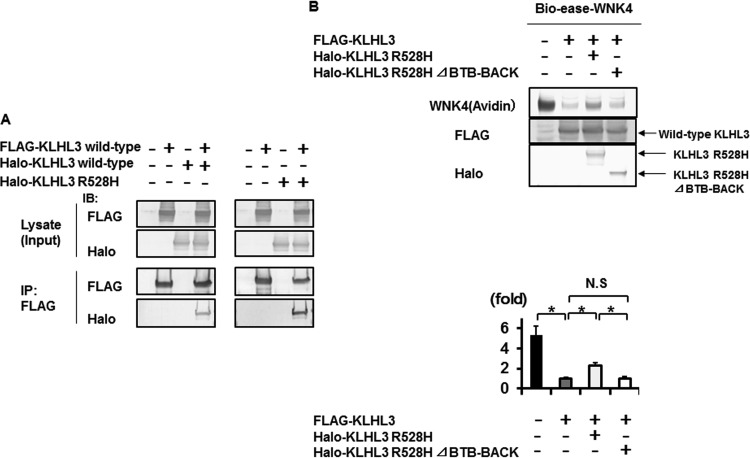
It has previously been reported that the polymerization of a disease-causing protein could explain the dominant negative effect in some cases (36). In this case, small amount of normal polymers that consist only of wild-type proteins will occur, preserving their normal function. Actually, some of the KLHL family proteins, such as Keap1, have been shown to form dimers (37). Therefore, we hypothesized that one of the mechanisms behind the dominant negative effect of mutant KLHL3 could be the formation of KLHL3 dimers. To test this hypothesis, we performed an immunoprecipitation experiment to see whether wild-type KLHL3 and mutant KLHL3 R528H could form dimers. As shown in Fig. 8A, wild-type KLHL3 could form a homodimer. In addition, we demonstrated that wild-type KLHL3 could interact with mutant KLHL3 R528H, indicating that a heterodimer of the wild-type and mutant R528H KLHL3 could be formed. To examine the dominant negative effect of KLHL3 R528H, we further performed coexpression experiments of KLHL3 R528H and wild-type KLHL3 as shown in Fig. 8B. Compared to the cells transfected with wild-type KLHL3 alone, the degradation of WNK4 protein was significantly decreased by the addition of KLHL3 R528H. In addition, this effect was cancelled when the BTB-BACK domain, containing the binding site for dimer formation, was deleted from the mutant KLHL3 R528H, suggesting that the dominant negative effect of mutant KLHL3 R528H required dimer formation of KLHL3. These results clearly indicate that mutant KLHL3 R528H has a dominant negative effect due to dimer formation.
FIG 8.
Dominant negative effect of KLHL3 R528H requires dimer formation of KLHL3. (A) FLAG-tagged KLHL3 was coimmunoprecipitated with Halo-tagged KLHL3 and Halo-tagged R528H KLHL3 in HEK293T cells. Wild-type KLHL3 could form a homodimer and heterodimer with wild-type and mutant KLHL3 R528H, respectively. IP, immunoprecipitation. (B, upper panel) Coexpression experiments of KLHL3 R528H and wild-type KLHL3. Compared to the cells transfected with wild-type KLHL3 alone, the degradation of WNK4 protein was significantly decreased by the addition of KLHL3 R528H. In addition, this effect was cancelled when the BTB-BACK domain, containing the binding site for dimer formation, was deleted from the mutant KLHL3 R528H, suggesting that the dominant negative effect of mutant KLHL3 R528H required dimer formation of KLHL3. (B, lower panel) Densitometry analysis of avidin binding to Bio-ease-WNK4.
DISCUSSION
In this study, we generated KLHL3−/− mice and confirmed that the absence of KLHL3 resulted in increased WNK1 and WNK4 levels in the kidney due to their defective degradation, which led to the activation of the downstream OSR1/SPAK-NCC phosphorylation cascade and a PHAII-like phenotype.
Recent studies have focused on WNK signaling in extrarenal tissue and have found that WNK signaling also plays a role in other tissues, including vascular smooth muscle cells (9, 26, 38), the brain (39–41), neurons (42), the intestine (43), and the heart (44). However, the tissue distribution and intratissue localization of KLHL3 remained unclear. Therefore, to investigate the physiological roles of KLHL3 in organs other than the kidney, we first investigated the tissue distribution of KLHL3. Unfortunately, the KLHL3 antibody we used could only detect KLHL3 by immunoblotting in samples from the kidney and brain. Therefore, instead, we used our KLHL3 knockout mouse model to answer these questions, in which the knockout allele expressed β-Gal as a result of the lacZ gene under the control of the endogenous KLHL3 promoter. Taking advantage of this system, we confirmed that there is strong β-Gal expression by the KLHL3 promoter in the brain and kidney and a lower expression in the eye, testis, lung, heart, liver, stomach, and colon. As expected, the expression of WNK1 and WNK4 was significantly higher in the kidneys of KLHL3−/− mice than in KLHL3+/+ mice. However, in the brain and other tissues that had lower expression levels of KLHL3, there was no difference in the expression of WNK1, WNK3, and WNK4 between KLHL3+/− and KLHL3+/+ mice.
These findings suggest that WNK protein levels in KLHL3-expressing tissues may not be solely governed by KLHL3. It is possible that the expression levels of WNK kinases are regulated only in specific KLHL3-expressing cells rather than across the entire organ, although we detected no significant differences in the WNKs of the hippocampus and cortex in the brain between wild-type and KLHL3−/− mice. Therefore, to exclude this possibility, it may be necessary to assess WNK protein levels in specific KLHL3-expressing cells in the future. Since the KLHL3 antibody we used was unable to detect a good KLHL3 signal by immunohistochemistry in extrarenal organs, the use of LacZ staining of tissue samples from β-Gal-expressing KLHL3−/− mice may be an appropriate method for detecting the expression of KLHL3 in specific types of cells in each organ. However, unfortunately, the LacZ staining of organs other than the kidney and brain has not been successful to date, likely due to the lower expression level of β-Gal. Therefore, further investigation will be required to determine the role of KLHL3 on WNK degradation in specific types of cells in each tissue.
Alternatively, it is possible that other KLHL proteins, such as KLHL2, are involved in controlling WNK protein levels. Similar to KLHL3, KLHL2 directly interacts with WNK kinases, leading to their ubiquitination and degradation (45, 46). Zeniya et al. recently reported that ubiquitination by KLHL2 is the major regulator of WNK3 protein levels in response to angiotensin II stimulation, although other in vivo roles of KLHL2 were not determined (38).
In the present study, dominant LacZ staining was observed in the hippocampus and cortex of the brain and in the distal tubules of the kidney. It was reasonable that KLHL3 was expressed in the distal convoluted tubules where NCC was localized, and this finding was consistent with previous reports (17, 23). In the brain, the WNK signal is believed to regulate the Cl− current via GABA (29, 30). The fact that GABA in the hippocampus regulates memory function (47) suggests that KLHL3 may also be important for memory through the regulation of WNK signaling. We also found that KLHL3 localized in the cortex. A recent study showed that WNK3 knockout mice were protected from focal ischemia in a middle cerebral artery occlusion-reperfusion model (48). Furthermore, a few case reports have shown that intellectual impairment occurred in sporadic cases of PHAII (1), probably due to CUL3 mutation, indicating that WNK signaling may play a role in the brain, although we cannot exclude the possibility that severe acidosis simply causes some development disorders of the brain. However, there are no reports of intellectual impairment in patients with PHAII caused by mutant KLHL3, and we did not detect an increase in the expression levels of WNK kinases in the whole lysate of the brain. Therefore, the role of KLHL3 in the brain of PHAII patients remains unclear and requires further careful investigation in the future.
We recently found that p62 interacts with KLHL3 and that WNK4 is degraded not only by proteasomes but also by p62-KLHL3-mediated selective autophagy (49). However, other proteins that interact with the Cul3-KLHL3 E3 ligase complex remain unknown. We expected that the phenotypes of KLHL3−/− mice (aside from PHAII) would give us an indication of novel substrates of KLHL3. However, unfortunately, we have not yet found any other obvious phenotypes of KLHL3−/− mice. Therefore, to investigate the novel roles of KLHL3 and new substrates of the Cul3-KLHL3 E3 ligase complex, further analysis of the KLHL3−/− mice is required, with a particular focus on the KLHL3-expressing organs that were found in the present study.
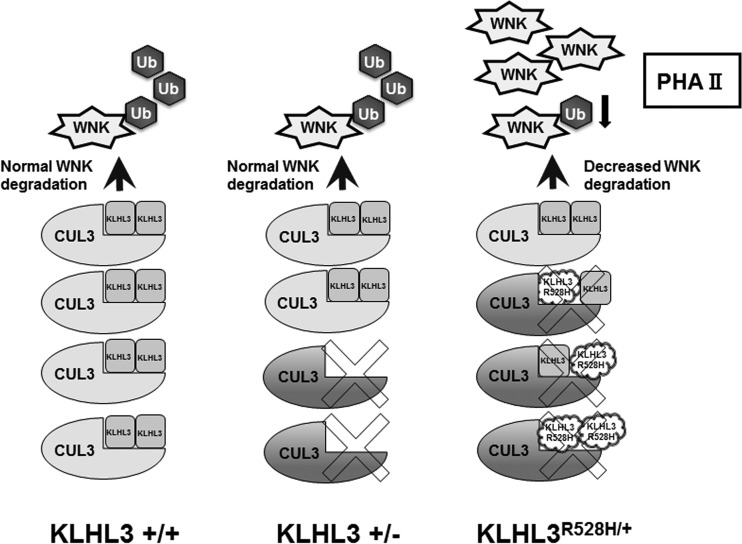
Interestingly, KLHL3+/− heterozygous mice did not show increased protein levels of WNK1 and WNK4 and did not exhibit PHAII-like phenotypes (i.e., hyperkalemia and metabolic acidosis). The lack of PHAII phenotypes in these mice clearly shows that the heterozygous deletion of KLHL3 was not sufficient to cause PHAII in the kidney, indicating that the PHAII-like phenotypes that we previously observed in KLHL3R528H/+ model mice were caused by the dominant negative effect of the mutant R528H KLHL3. It is known that KLHL family proteins, such as Keap1 and KLHL7, form a dimer by themselves (37, 50). It was previously reported that Keap1 mutations found in lung tumors suppress wild-type Keap1 activity, leading to Nrf2 activation, due to heterodimerization of mutant Keap1 with wild-type Keap1, demonstrating the mechanism of dominant negative effect of mutant Keap1 (51). However, to date, there has been no molecular evidence of the direct interaction of KLHL3, although structural analysis of its BTB-BACK domain suggests that it could form a dimer (52). In this study, we confirmed that wild-type KLHL3 forms a dimer and that wild-type and mutant R528H KLHL3 bind with each other, which constitutes dimer formation. We also demonstrated that the dominant negative effect of KLHL3 R528H required dimer formation. As shown in Fig. 9, assuming that dimers that contain at least one mutant KLHL3 exhibit a defective binding ability to WNK1 and WNK4, one-quarter of all dimers will consist only of wild-type KLHL3 and will have a normal ability to catch WNK kinases. Although mutant KLHL3 dimerization cannot be quantitatively measured in KLHL3R528H/+ heterozygous mice, the speculation presented above fits with the finding that the protein levels of WNK1 and WNK4 were higher in KLHL3−/− mice than in KLHL3R528H/+ mice (Fig. 7), since a small amount of normal dimers that consist of two wild-type KLHL3 will still occur in KLHL3R528H/+ mice. Therefore, we believe that the dimer formation of wild-type and R528H KLHL3 could explain the dominant negative effect of this mutant.
FIG 9.
Schematic representation of the mechanism of the dominant negative effect caused by KLHL3 R528H.
The majority of KLHL3 mutations in the Kelch domain show an autosomal dominant inheritance pattern, as well as mutant KLHL3 R528H. Therefore, these mutations are suggested to have dominant negative effect by the dimer formation in BTB-BACK domain in the same way as we showed in this study. On the other hand, several kinds of recessive mutations in the Kelch domain have been also reported. As we previously reported, some of the Kelch domain mutation found in recessive type resulted in instability of the KLHL3 protein (53). Therefore, these mutants might be unable to maintain enough protein expression levels to show a dominant negative effect.
KLHL3 mutations in BTB domain also shows autosomal dominant inheritance pattern; however, the precise mechanism of dominant negative effect of BTB domain mutation is not clear. One explanation might be defect of interaction of KLHL3 with Cul3. We previously showed that BTB mutation (E85A) cannot interact with Cul3 (53). Even if KLHL3 with BTB mutation could form a dimer with wild-type KLHL3, this dimer might fail to form the fully functional Cul3-KLHL3 E3 ligase complex.
KLHL3 mutations in BACK domain mainly exhibit recessive inheritance. We speculate that this could be simply explained by defective dimer formation of KLHL3 mutant at the BACK domain. Heterozygous mutation of KLHL3 BACK domain does not show any obvious phenotype in the majority of cases, similar to that of KLHL3+/− heterozygous knockout. However, a rare case of KLHL3 mutation in BACK domain exhibiting dominant inheritance has been reported, and the reason for this is still unclear.
Thus, multiple speculations can be made considering the molecular pathogenesis of KLHL3 mutations with many questions left to be clarified. Our report is the first to successfully elucidate the mechanism of dominant negative effect of KLHL3 R528H by in vivo data. There could be still unknown mechanisms in the other type of KLHL3 mutation. Further investigations are required.
In conclusion, the KLHL3 knockout mice exhibited PHAII-like phenotypes and increased WNK protein levels in the kidney, leading to activation of the WNK signaling cascade. In contrast, KLHL3+/− heterozygous mice did not show an increase in WNK kinases and activation of WNK signaling, even in the kidney. This lack of a PHAII phenotype in these mice clearly indicates that the dominant type of mutant KLHL3 has a dominant negative effect, likely due to dimer formation. These findings could be valuable for further understanding the physiological roles of KLHL3 and the pathophysiological mechanisms of PHAII caused by mutant KLHL3.
MATERIALS AND METHODS
Generation of KLHL3 knockout mice.
In this study, we generated KLHL3−/− mice that expressed β-Gal under the control of the endogenous KLHL3 promoter. To generate KLHL3−/− mice, we purchased a targeting vector containing the mouse genomic KLHL3 locus from the Knockout Mouse Project (KOMP) Repository (Davis, CA). The targeting vector was then transfected into Baltha1 embryonic stem (ES) cells (23) derived from C57BL/6 mice by electroporation, as previously reported (54). After selection with G418 at 150 μg/ml and 2 μM ganciclovir, the targeted ES cell clones were identified by PCR. Verification of homologous recombination by PCR of genomic DNA of the selected ES cell clones was performed; primers F1 (5′-GAGATGGCGCAACGCAATTAATG-3′) and R1 (5′-GGTTTTAAGCAAAGCCAAGAGGTC-3′) show an ∼3-kb band, primers F2 (5′-GGGATCTCATGCTGGAGTTCTTCG-3′) and R1 show an ∼5-kb band, and primers F3 (5′-CTGATTGAAGAATGGGCCTTCTCACG-3′) and R2 (5′-GGTGGTGTGGGAAAGGGTTCGAAG-3′) show an ∼7-kb band.
To generate KLHL3−/− mice that expressed β-Gal under the control of the endogenous KLHL3 promoter, chimeric male mice were bred with C57BL/6J females to produce heterozygous floxed (KLHL3flox/+) mice. The neo cassette was then deleted by crossing the KLHL3flox/+ mice with Cre recombinase-expressing transgenic (TG) mice (55). Genotyping of the mice was performed by PCR using the sense primer F (5′-AACTTGAGGCGCTGGGAGAAC-3′) and the antisense primer R3 (5′-CTGACACAGGTTTCAGGGATTGAGC-3′) to detect the wild-type allele. Further PCR analysis was then performed using the sense primer F4 (5′-GCTACCATTACCAGTTGGTCTGGTGTC-3′) and the antisense primer R3 to detect the upper LoxP site.
Animals.
The experiments were performed on 8- to 16-week-old mice that had been raised under a 12-h day/night cycle and had been fed a normal rodent diet and plain drinking water. This experiment was approved by the Animal Care and Use Committee of the Tokyo Medical and Dental University, Tokyo, Japan.
Plasmids.
The expression plasmids for 3×FLAG-tagged human KLHL3, Halo-tagged human KLHL3, and Halo-tagged R528H human KLHL3 have been described previously (19). The expression plasmid encoding Halo-tagged human KLHL3 R528H with deletion of BTB-BACK domain was obtained by PCR site-directed mutagenesis with the primers 5′-TGGGGAAAGCGTTGAAGAAGAAGCTTTG-3′ (forward) and 5′-CTTCTTCAACAACCTTGAATGCTTTCCCC-3′ (reverse).
Cell culture and transfections.
HEK293T cells were cultured in Dulbecco's modified Eagle's medium supplemented with 10% (vol/vol) fetal bovine serum, 2 mM l-glutamine, penicillin at 100 U/ml, and streptomycin at 0.1 mg/ml at 37°C in a humidified 5% CO2 incubator. In the coimmunoprecipitation experiments, the cells were placed in six-well plate (2.0 ml of medium/well, 60% confluence) and transiently transfected with the indicated amounts of plasmid DNA using Lipofectamine 2000 (Invitrogen). For each transfection, wild-type KLHL3 and/or mutant KLHL3 were used. In the coexpression experiments to investigate the dominant negative effect of KLHL3 R528H, the cells were placed in a 6-cm dish (4 ml of medium/dish, 60% confluence) and transfected with combinations of FLAG-tagged wild-type KLHL3 with Halo-tagged KLHL3 R528H or Halo-tagged KLHL3 R528H ΔBTB-BACK plasmid. FLAG-tagged CMV and Halo-tagged GAPDH (glyceraldehyde-3-phosphate dehydrogenase) were used as control. Bio-ease-FLAG-tagged human WNK4 and FLAG-tagged human Cul3 were also transfected to all the experiments of the dominant negative effect of KLHL3 R528H. The cells were then incubated for 24 h after the transfection.
Coimmunoprecipitation experiments.
The HEK293T cells transfected in combination with FLAG-tagged wild-type KLHL3 and Halo-tagged wild-type KLHL3 or Halo-tagged mutant KLHL3 were harvested in lysis buffer composed of 50 mM Tris-HCl (pH 7.5), 150 mM NaCl, 1% NP-40, 1 mM sodium orthovanadate, 50 mM sodium fluoride, and protease inhibitor cocktail for 30 min at 4°C. After centrifugation at 15,000 × g for 10 min, the supernatant was immunoprecipitated with anti-FLAG M2 beads (Sigma-Aldrich) for 2 h at 4°C. The beads were then washed with lysis buffer and the immunoprecipitates were eluted by boiling for 10 min in sodium dodecyl sulfate (SDS) sample buffer.
Blood measurements.
Blood for electrolyte analysis was obtained as previously described (8) and electrolyte levels were determined using an i-STAT portable clinical analyzer (Fuso Pharmaceutical Industries, Ltd.). Samples were obtained from the venous plexus near the mandible under anesthesia with light ether.
Immunoblotting.
Protein samples were extracted from the kidney and other organs, and semiquantitative immunoblotting was performed as previously described (56). For semiquantitative immunoblotting, we used entire kidney samples without the nuclear fraction (600 × g) or the crude membrane fraction (17,000 × g). The relative intensities of the immunoblot bands were analyzed and quantified using ImageJ software (National Institutes of Health). The following primary antibodies were used: rabbit anti-KLHL3 (Proteintech), rabbit anti-WNK1 (A301-515A-2; Bethyl Laboratories), sheep anti-WNK3 (57), rabbit anti-WNK4 (14); rabbit anti-phosphorylated SPAK (58), rabbit anti-SPAK (Cell Signaling Technology, Inc.), rabbit anti-phosphorylated OSR1 (23), mouse anti-OSR1 (M10; Abnova Corporation), rabbit anti-phosphorylated NCC (S71) (59), rabbit anti-NCC (60), rabbit antiactin (Cytoskeleton, Inc.), mouse anti-β-Gal (Promega Corporation), anti-FLAG (Sigma-Aldrich), anti-Halo (Promega Corporation), and streptavidin-alkaline phosphatase conjugate (Invitrogen). Western Blue (Promega Corporation) was used to detect the signals.
BP measurement.
We measured the systolic blood pressure (BP) using implantable radiotelemetric devices, as previously reported (61). Equipment for conscious, freely moving laboratory animals was purchased from Data Sciences International, which included an implantable transmitter (model TA11PA-C10), a receiver (model RPC-1), a data-processing device (Data Exchange Matrix), and an ambient pressure reference monitor (APR-1). All data were analyzed using Dataquest ART4.31.
LacZ staining.
LacZ staining was performed using the LacZ Tissue staining kit (InvivoGen, San Diego, CA). We dissected the kidneys and brains from 12-week-old KLHL3 knockout/lacZ knock-in and wild-type (control) mice into phosphate-buffered saline (PBS)–MgCl2 solution on ice. The samples were fixed in 0.5% ice-cold glutaraldehyde for 6 h on a shaker, washed with PBS solution for 1 h, and then rinsed twice more in PBS. The tissues were then cut in half and inserted into X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) solution (InvivoGen) for 2 h at 37°C in the dark. Next, serial frozen sections (10-μm sections of the brain and 5-μm sections of the kidney) were prepared using a cryostat microtome. The frozen sections were incubated in X-Gal solution for 12 h and observed under a microscope.
Immunofluorescence.
The kidneys were fixed by perfusion with periodate lysine (0.2 M) and paraformaldehyde (2%) in PBS. Immunofluorescence was then performed as previously described (54) using the primary mouse anti-β-Gal (Promega Corporation) and guinea pig-anti-NCC (62) antibodies and Alexa Fluor 488 or 546 dye-labeled (Molecular Probes; Invitrogen) secondary antibodies. Immunofluorescence images were obtained using an LSM510 Meta Confocal Microscope (Carl Zeiss, Oberkochen, Germany).
Statistical analysis.
The two groups of mice were compared by using unpaired t tests. In addition, one-way analysis of variance, followed by Tukey's post hoc test, were used to evaluate the statistical significance of comparisons between multiple groups. P values of <0.05 were considered statistically significant. Data are presented as means ± the standard errors of the means.
ACKNOWLEDGMENTS
We thank C. Iijima for help in the experiments.
This study was supported in part by Grants-in-Aid for Scientific Research (S) (25221306-00) to S.U. and Scientific Research (B) (16H05314) to E.S. from the Japanese Society for the Promotion of Science, by a Health Labor Science Research Grant from the Ministry of Health Labor and Welfare, by Challenging Exploratory Research assistance from the Ministry of Education, Culture, Sports, Science and Technology of Japan to S.U. and E.S., by grants from the Salt Science Research Foundation (1422 and 1629) to S.U. and the Takeda Science Foundation to E.S., and by a Banyu Foundation Research Grant to E.S.
REFERENCES
- 1.Gordon RD. 1986. Syndrome of hypertension and hyperkalemia with normal glomerular filtration rate. Hypertension 8:93–102. doi: 10.1161/01.HYP.8.2.93. [DOI] [PubMed] [Google Scholar]
- 2.Wilson FH, Disse-Nicodeme S, Choate KA, Ishikawa K, Nelson-Williams C, Desitter I, Gunel M, Milford DV, Lipkin GW, Achard JM, Feely MP, Dussol B, Berland Y, Unwin RJ, Mayan H, Simon DB, Farfel Z, Jeunemaitre X, Lifton RP. 2001. Human hypertension caused by mutations in WNK kinases. Science 293:1107–1112. doi: 10.1126/science.1062844. [DOI] [PubMed] [Google Scholar]
- 3.Sohara E, Uchida S. 2016. Kelch-like 3/Cullin 3 ubiquitin ligase complex and WNK signaling in salt-sensitive hypertension and electrolyte disorder. Nephrol Dial Transplant 31:1417–1424. doi: 10.1093/ndt/gfv259. [DOI] [PubMed] [Google Scholar]
- 4.Hossain Khan MZ, Sohara E, Ohta A, Chiga M, Inoue Y, Isobe K, Wakabayashi M, Oi K, Rai T, Sasaki S, Uchida S. 2012. Phosphorylation of Na-Cl cotransporter by OSR1 and SPAK kinases regulates its ubiquitination. Biochem Biophys Res Commun 425:456–461. doi: 10.1016/j.bbrc.2012.07.124. [DOI] [PubMed] [Google Scholar]
- 5.Moriguchi T, Urushiyama S, Hisamoto N, Iemura S, Uchida S, Natsume T, Matsumoto K, Shibuya H. 2005. WNK1 regulates phosphorylation of cation-chloride-coupled cotransporters via the STE20-related kinases, SPAK and OSR1. J Biol Chem 280:42685–42693. doi: 10.1074/jbc.M510042200. [DOI] [PubMed] [Google Scholar]
- 6.Vitari AC, Deak M, Morrice NA, Alessi DR. 2005. The WNK1 and WNK4 protein kinases that are mutated in Gordon's hypertension syndrome phosphorylate and activate SPAK and OSR1 protein kinases. Biochem J 391:17–24. doi: 10.1042/BJ20051180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chiga M, Rafiqi FH, Alessi DR, Sohara E, Ohta A, Rai T, Sasaki S, Uchida S. 2011. Phenotypes of pseudohypoaldosteronism type II caused by the WNK4 D561A missense mutation are dependent on the WNK-OSR1/SPAK kinase cascade. J Cell Sci 124:1391–1395. doi: 10.1242/jcs.084111. [DOI] [PubMed] [Google Scholar]
- 8.Yang SS, Morimoto T, Rai T, Chiga M, Sohara E, Ohno M, Uchida K, Lin SH, Moriguchi T, Shibuya H, Kondo Y, Sasaki S, Uchida S. 2007. Molecular pathogenesis of pseudohypoaldosteronism type II: generation and analysis of a Wnk4(D561A/+) knockin mouse model. Cell Metab 5:331–344. doi: 10.1016/j.cmet.2007.03.009. [DOI] [PubMed] [Google Scholar]
- 9.Bergaya S, Faure S, Baudrie V, Rio M, Escoubet B, Bonnin P, Henrion D, Loirand G, Achard JM, Jeunemaitre X, Hadchouel J. 2011. WNK1 regulates vasoconstriction and blood pressure response to α1-adrenergic stimulation in mice. Hypertension 58:439–445. doi: 10.1161/HYPERTENSIONAHA.111.172429. [DOI] [PubMed] [Google Scholar]
- 10.Vidal-Petiot E, Elvira-Matelot E, Mutig K, Soukaseum C, Baudrie V, Wu S, Cheval L, Huc E, Cambillau M, Bachmann S, Doucet A, Jeunemaitre X, Hadchouel J. 2013. WNK1-related familial hyperkalemic hypertension results from an increased expression of L-WNK1 specifically in the distal nephron. Proc Natl Acad Sci U S A 110:14366–14371. doi: 10.1073/pnas.1304230110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Castaneda-Bueno M, Cervantes-Perez LG, Vazquez N, Uribe N, Kantesaria S, Morla L, Bobadilla NA, Doucet A, Alessi DR, Gamba G. 2012. Activation of the renal Na+:Cl− cotransporter by angiotensin II is a WNK4-dependent process. Proc Natl Acad Sci U S A 109:7929–7934. doi: 10.1073/pnas.1200947109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rafiqi FH, Zuber AM, Glover M, Richardson C, Fleming S, Jovanovic S, Jovanovic A, O'Shaughnessy KM, Alessi DR. 2010. Role of the WNK-activated SPAK kinase in regulating blood pressure. EMBO Mol Med 2:63–75. doi: 10.1002/emmm.200900058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yang SS, Lo YF, Wu CC, Lin SW, Yeh CJ, Chu P, Sytwu HK, Uchida S, Sasaki S, Lin SH. 2010. SPAK-knockout mice manifest Gitelman syndrome and impaired vasoconstriction. J Am Soc Nephrol 21:1868–1877. doi: 10.1681/ASN.2009121295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Takahashi D, Mori T, Nomura N, Khan MZ, Araki Y, Zeniya M, Sohara E, Rai T, Sasaki S, Uchida S. 2014. WNK4 is the major WNK positively regulating NCC in the mouse kidney. Biosci Rep 34:e00107. doi: 10.1042/BSR20140047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Richardson C, Alessi DR. 2008. The regulation of salt transport and blood pressure by the WNK-SPAK/OSR1 signaling pathway. J Cell Sci 121:3293–3304. doi: 10.1242/jcs.029223. [DOI] [PubMed] [Google Scholar]
- 16.Uchida S, Sohara E, Rai T, Sasaki S. 2014. Regulation of with-no-lysine kinase signaling by Kelch-like proteins. Biol Cell 106:45–56. doi: 10.1111/boc.201300069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Boyden LM, Choi M, Choate KA, Nelson-Williams CJ, Farhi A, Toka HR, Tikhonova IR, Bjornson R, Mane SM, Colussi G, Lebel M, Gordon RD, Semmekrot BA, Poujol A, Valimaki MJ, De Ferrari ME, Sanjad SA, Gutkin M, Karet FE, Tucci JR, Stockigt JR, Keppler-Noreuil KM, Porter CC, Anand SK, Whiteford ML, Davis ID, Dewar SB, Bettinelli A, Fadrowski JJ, Belsha CW, Hunley TE, Nelson RD, Trachtman H, Cole TR, Pinsk M, Bockenhauer D, Shenoy M, Vaidyanathan P, Foreman JW, Rasoulpour M, Thameem F, Al-Shahrouri HZ, Radhakrishnan J, Gharavi AG, Goilav B, Lifton RP. 2012. Mutations in kelch-like 3 and cullin 3 cause hypertension and electrolyte abnormalities. Nature 482:98–102. doi: 10.1038/nature10814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Louis-Dit-Picard H, Barc J, Trujillano D, Miserey-Lenkei S, Bouatia-Naji N, Pylypenko O, Beaurain G, Bonnefond A, Sand O, Simian C, Vidal-Petiot E, Soukaseum C, Mandet C, Broux F, Chabre O, Delahousse M, Esnault V, Fiquet B, Houillier P, Bagnis CI, Koenig J, Konrad M, Landais P, Mourani C, Niaudet P, Probst V, Thauvin C, Unwin RJ, Soroka SD, Ehret G, Ossowski S, Caulfield M, Bruneval P, Estivill X, Froguel P, Hadchouel J, Schott JJ, Jeunemaitre X. 2012. KLHL3 mutations cause familial hyperkalemic hypertension by impairing ion transport in the distal nephron. Nat Genet 44:456–460. doi: 10.1038/ng.2218. [DOI] [PubMed] [Google Scholar]
- 19.Wakabayashi M, Mori T, Isobe K, Sohara E, Susa K, Araki Y, Chiga M, Kikuchi E, Nomura N, Mori Y, Matsuo H, Murata T, Nomura S, Asano T, Kawaguchi H, Nonoyama S, Rai T, Sasaki S, Uchida S. 2013. Impaired KLHL3-mediated ubiquitination of WNK4 causes human hypertension. Cell Rep 3:858–868. doi: 10.1016/j.celrep.2013.02.024. [DOI] [PubMed] [Google Scholar]
- 20.Wu G, Peng JB. 2013. Disease-causing mutations in KLHL3 impair its effect on WNK4 degradation. FEBS Lett 587:1717–1722. doi: 10.1016/j.febslet.2013.04.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shibata S, Zhang J, Puthumana J, Stone KL, Lifton RP. 2013. Kelch-like 3 and Cullin 3 regulate electrolyte homeostasis via ubiquitination and degradation of WNK4. Proc Natl Acad Sci U S A 110:7838–7843. doi: 10.1073/pnas.1304592110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ohta A, Schumacher FR, Mehellou Y, Johnson C, Knebel A, Macartney TJ, Wood NT, Alessi DR, Kurz T. 2013. The CUL3-KLHL3 E3 ligase complex mutated in Gordon's hypertension syndrome interacts with and ubiquitylates WNK isoforms: disease-causing mutations in KLHL3 and WNK4 disrupt interaction. Biochem J 451:111–122. doi: 10.1042/BJ20121903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Susa K, Sohara E, Rai T, Zeniya M, Mori Y, Mori T, Chiga M, Nomura N, Nishida H, Takahashi D, Isobe K, Inoue Y, Takeishi K, Takeda N, Sasaki S, Uchida S. 2014. Impaired degradation of WNK1 and WNK4 kinases causes PHAII in mutant KLHL3 knock-in mice. Hum Mol Genet 23:5052–5060. doi: 10.1093/hmg/ddu217. [DOI] [PubMed] [Google Scholar]
- 24.Yoshizaki Y, Mori Y, Tsuzaki Y, Mori T, Nomura N, Wakabayashi M, Takahashi D, Zeniya M, Kikuchi E, Araki Y, Ando F, Isobe K, Nishida H, Ohta A, Susa K, Inoue Y, Chiga M, Rai T, Sasaki S, Uchida S, Sohara E. 2015. Impaired degradation of WNK by Akt and PKA phosphorylation of KLHL3. Biochem Biophys Res Commun 467:229–234. doi: 10.1016/j.bbrc.2015.09.184. [DOI] [PubMed] [Google Scholar]
- 25.Shibata S, Arroyo JP, Castaneda-Bueno M, Puthumana J, Zhang J, Uchida S, Stone KL, Lam TT, Lifton RP. 2014. Angiotensin II signaling via protein kinase C phosphorylates Kelch-like 3, preventing WNK4 degradation. Proc Natl Acad Sci U S A 111:15556–15561. doi: 10.1073/pnas.1418342111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zeniya M, Sohara E, Kita S, Iwamoto T, Susa K, Mori T, Oi K, Chiga M, Takahashi D, Yang SS, Lin SH, Rai T, Sasaki S, Uchida S. 2013. Dietary salt intake regulates WNK3-SPAK-NKCC1 phosphorylation cascade in mouse aorta through angiotensin II. Hypertension 62:872–878. doi: 10.1161/HYPERTENSIONAHA.113.01543. [DOI] [PubMed] [Google Scholar]
- 27.Susa K, Kita S, Iwamoto T, Yang SS, Lin SH, Ohta A, Sohara E, Rai T, Sasaki S, Alessi DR, Uchida S. 2012. Effect of heterozygous deletion of WNK1 on the WNK-OSR1/ SPAK-NCC/NKCC1/NKCC2 signal cascade in the kidney and blood vessels. Clin Exp Nephrol 16:530–538. doi: 10.1007/s10157-012-0590-x. [DOI] [PubMed] [Google Scholar]
- 28.Yang L, Cai X, Zhou J, Chen S, Chen Y, Chen Z, Wang Q, Fang Z, Zhou L. 2013. STE20/SPS1-related proline/alanine-rich kinase is involved in plasticity of GABA signaling function in a mouse model of acquired epilepsy. PLoS One 8:e74614. doi: 10.1371/journal.pone.0074614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Alessi DR, Zhang J, Khanna A, Hochdorfer T, Shang Y, Kahle KT. 2014. The WNK-SPAK/OSR1 pathway: master regulator of cation-chloride cotransporters. Sci Signal 7:re3. doi: 10.1126/scisignal.2005365. [DOI] [PubMed] [Google Scholar]
- 30.Kahle KT, Khanna AR, Alper SL, Adragna NC, Lauf PK, Sun D, Delpire E. 2015. K-Cl cotransporters, cell volume homeostasis, and neurological disease. Trends Mol Med 21:513–523. doi: 10.1016/j.molmed.2015.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Moniz S, Matos P, Jordan P. 2008. WNK2 modulates MEK1 activity through the Rho GTPase pathway. Cell Signal 20:1762–1768. doi: 10.1016/j.cellsig.2008.06.002. [DOI] [PubMed] [Google Scholar]
- 32.Xu BE, Stippec S, Lenertz L, Lee BH, Zhang W, Lee YK, Cobb MH. 2004. WNK1 activates ERK5 by an MEKK2/3-dependent mechanism. J Biol Chem 279:7826–7831. doi: 10.1074/jbc.M313465200. [DOI] [PubMed] [Google Scholar]
- 33.Lee BH, Chen W, Stippec S, Cobb MH. 2007. Biological cross-talk between WNK1 and the transforming growth factor beta-Smad signaling pathway. J Biol Chem 282:17985–17996. doi: 10.1074/jbc.M702664200. [DOI] [PubMed] [Google Scholar]
- 34.DasGupta R, Kaykas A, Moon RT, Perrimon N. 2005. Functional genomic analysis of the Wnt-wingless signaling pathway. Science 308:826–833. doi: 10.1126/science.1109374. [DOI] [PubMed] [Google Scholar]
- 35.Lalioti MD, Zhang J, Volkman HM, Kahle KT, Hoffmann KE, Toka HR, Nelson-Williams C, Ellison DH, Flavell R, Booth CJ, Lu Y, Geller DS, Lifton RP. 2006. Wnk4 controls blood pressure and potassium homeostasis via regulation of mass and activity of the distal convoluted tubule. Nat Genet 38:1124–1132. doi: 10.1038/ng1877. [DOI] [PubMed] [Google Scholar]
- 36.Veitia RA. 2007. Exploring the molecular etiology of dominant-negative mutations. Plant Cell 19:3843–3851. doi: 10.1105/tpc.107.055053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ogura T, Tong KI, Mio K, Maruyama Y, Kurokawa H, Sato C, Yamamoto M. 2010. Keap1 is a forked-stem dimer structure with two large spheres enclosing the intervening, double glycine repeat, and C-terminal domains. Proc Natl Acad Sci U S A 107:2842–2847. doi: 10.1073/pnas.0914036107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zeniya M, Morimoto N, Takahashi D, Mori Y, Mori T, Ando F, Araki Y, Yoshizaki Y, Inoue Y, Isobe K, Nomura N, Oi K, Nishida H, Sasaki S, Sohara E, Rai T, Uchida S. 2015. Kelch-like protein 2 mediates angiotensin II-with no lysine 3 signaling in the regulation of vascular tonus. J Am Soc Nephrol 26:2129–2138. doi: 10.1681/ASN.2014070639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Khanna A, Walcott BP, Kahle KT. 2013. Limitations of current GABA agonists in neonatal seizures: toward GABA modulation via the targeting of neuronal Cl− transport. Front Neurol 4:78. doi: 10.3389/fneur.2013.00078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rinehart J, Vazquez N, Kahle KT, Hodson CA, Ring AM, Gulcicek EE, Louvi A, Bobadilla NA, Gamba G, Lifton RP. 2011. WNK2 kinase is a novel regulator of essential neuronal cation-chloride cotransporters. J Biol Chem 286:30171–30180. doi: 10.1074/jbc.M111.222893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhao H, Nepomuceno R, Gao X, Foley LM, Wang S, Begum G, Zhu W, Pigott VM, Falgoust LM, Kahle KT, Yang SS, Lin SH, Alper SL, Hitchens TK, Hu S, Zhang Z, Sun D. 9 February 2016. Deletion of the WNK3-SPAK kinase complex in mice improves radiographic and clinical outcomes in malignant cerebral edema after ischemic stroke. J Cereb Blood Flow Metab. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shekarabi M, Girard N, Riviere JB, Dion P, Houle M, Toulouse A, Lafreniere RG, Vercauteren F, Hince P, Laganiere J, Rochefort D, Faivre L, Samuels M, Rouleau GA. 2008. Mutations in the nervous system: specific HSN2 exon of WNK1 cause hereditary sensory neuropathy type II. J Clin Invest 118:2496–2505. doi: 10.1172/JCI34088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang Y, Viennois E, Xiao B, Baker MT, Yang S, Okoro I, Yan Y. 2013. Knockout of Ste20-like proline/alanine-rich kinase (SPAK) attenuates intestinal inflammation in mice. Am J Pathol 182:1617–1628. doi: 10.1016/j.ajpath.2013.01.028. [DOI] [PubMed] [Google Scholar]
- 44.Xie J, Yoon J, Yang SS, Lin SH, Huang CL. 2013. WNK1 protein kinase regulates embryonic cardiovascular development through the OSR1 signaling cascade. J Biol Chem 288:8566–8574. doi: 10.1074/jbc.M113.451575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang C, Meermeier NP, Terker AS, Blankenstein KI, Singer JD, Hadchouel J, Ellison DH, Yang CL. 2016. Degradation by Cullin 3 and effect on WNK kinases suggest a role of KLHL2 in the pathogenesis of familial hyperkalemic hypertension. Biochem Biophys Res Commun 469:44–48. doi: 10.1016/j.bbrc.2015.11.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Takahashi D, Mori T, Wakabayashi M, Mori Y, Susa K, Zeniya M, Sohara E, Rai T, Sasaki S, Uchida S. 2013. KLHL2 interacts with and ubiquitinates WNK kinases. Biochem Biophys Res Commun 437:457–462. doi: 10.1016/j.bbrc.2013.06.104. [DOI] [PubMed] [Google Scholar]
- 47.Sibbe M, Kulik A. 6 September 2016. GABAergic regulation of adult hippocampal neurogenesis. Mol Neurobiol. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 48.Begum G, Yuan H, Kahle KT, Li L, Wang S, Shi Y, Shmukler BE, Yang SS, Lin SH, Alper SL, Sun D. 2015. Inhibition of WNK3 kinase signaling reduces brain damage and accelerates neurological recovery after stroke. Stroke 46:1956–1965. doi: 10.1161/STROKEAHA.115.008939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mori Y, Mori T, Wakabayashi M, Yoshizaki Y, Zeniya M, Sohara E, Rai T, Uchida S. 2015. Involvement of selective autophagy mediated by p62/SQSTM1 in KLHL3-dependent WNK4 degradation. Biochem J 472:33–41. doi: 10.1042/BJ20150500. [DOI] [PubMed] [Google Scholar]
- 50.Kigoshi Y, Tsuruta F, Chiba T. 2011. Ubiquitin ligase activity of Cul3-KLHL7 protein is attenuated by autosomal dominant retinitis pigmentosa causative mutation. J Biol Chem 286:33613–33621. doi: 10.1074/jbc.M111.245126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Suzuki T, Maher J, Yamamoto M. 2011. Select heterozygous Keap1 mutations have a dominant-negative effect on wild-type Keap1 in vivo. Cancer Res 71:1700–1709. doi: 10.1158/0008-5472.CAN-10-2939. [DOI] [PubMed] [Google Scholar]
- 52.Ji AX, Prive GG. 2013. Crystal structure of KLHL3 in complex with Cullin3. PLoS One 8:e60445. doi: 10.1371/journal.pone.0060445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mori Y, Wakabayashi M, Mori T, Araki Y, Sohara E, Rai T, Sasaki S, Uchida S. 2013. Decrease of WNK4 ubiquitination by disease-causing mutations of KLHL3 through different molecular mechanisms. Biochem Biophys Res Commun 439:30–34. doi: 10.1016/j.bbrc.2013.08.035. [DOI] [PubMed] [Google Scholar]
- 54.Sohara E, Rai T, Yang SS, Uchida K, Nitta K, Horita S, Ohno M, Harada A, Sasaki S, Uchida S. 2006. Pathogenesis and treatment of autosomal-dominant nephrogenic diabetes insipidus caused by an aquaporin 2 mutation. Proc Natl Acad Sci U S A 103:14217–14222. doi: 10.1073/pnas.0602331103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sakai K, Miyazaki J. 1997. A transgenic mouse line that retains Cre recombinase activity in mature oocytes irrespective of the cre transgene transmission. Biochem Biophys Res Commun 237:318–324. doi: 10.1006/bbrc.1997.7111. [DOI] [PubMed] [Google Scholar]
- 56.Araki Y, Rai T, Sohara E, Mori T, Inoue Y, Isobe K, Kikuchi E, Ohta A, Sasaki S, Uchida S. 2015. Generation and analysis of knock-in mice carrying pseudohypoaldosteronism type II-causing mutations in the cullin 3 gene. Biol Open 4:1509–1517. doi: 10.1242/bio.013276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Oi K, Sohara E, Rai T, Misawa M, Chiga M, Alessi DR, Sasaki S, Uchida S. 2012. A minor role of WNK3 in regulating phosphorylation of renal NKCC2 and NCC cotransporters in vivo. Biol Open 1:120–127. doi: 10.1242/bio.2011048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sohara E, Rai T, Yang SS, Ohta A, Naito S, Chiga M, Nomura N, Lin SH, Vandewalle A, Ohta E, Sasaki S, Uchida S. 2011. Acute insulin stimulation induces phosphorylation of the Na-Cl cotransporter in cultured distal mpkDCT cells and mouse kidney. PLoS One 6:e24277. doi: 10.1371/journal.pone.0024277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Nishida H, Sohara E, Nomura N, Chiga M, Alessi DR, Rai T, Sasaki S, Uchida S. 2012. Phosphatidylinositol 3-kinase/Akt signaling pathway activates the WNK-OSR1/SPAK-NCC phosphorylation cascade in hyperinsulinemic db/db mice. Hypertension 60:981–990. doi: 10.1161/HYPERTENSIONAHA.112.201509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Isobe K, Mori T, Asano T, Kawaguchi H, Nonoyama S, Kumagai N, Kamada F, Morimoto T, Hayashi M, Sohara E, Rai T, Sasaki S, Uchida S. 2013. Development of enzyme-linked immunosorbent assays for urinary thiazide-sensitive Na-Cl cotransporter measurement. Am J Physiol Renal Physiol 305:F1374–F1381. doi: 10.1152/ajprenal.00208.2013. [DOI] [PubMed] [Google Scholar]
- 61.Kikuchi E, Mori T, Zeniya M, Isobe K, Ishigami-Yuasa M, Fujii S, Kagechika H, Ishihara T, Mizushima T, Sasaki S, Sohara E, Rai T, Uchida S. 2015. Discovery of novel SPAK inhibitors that block WNK kinase signaling to cation chloride transporters. J Am Soc Nephrol 26:1525–1536. doi: 10.1681/ASN.2014060560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ohno M, Uchida K, Ohashi T, Nitta K, Ohta A, Chiga M, Sasaki S, Uchida S. 2011. Immunolocalization of WNK4 in mouse kidney. Histochem Cell Biol 136:25–35. doi: 10.1007/s00418-011-0827-x. [DOI] [PubMed] [Google Scholar]