ABSTRACT
As a prominent characteristic of cell life, the regulation of cell quiescence is important for proper development, regeneration, and stress resistance and may play a role in certain degenerative diseases. However, the mechanism underlying quiescence remains largely unknown. Encysted embryos of Artemia are useful for studying the regulation of this state because they remain quiescent for prolonged periods during diapause, a state of obligate dormancy. In the present study, SET domain-containing protein 4, a histone lysine methyltransferase from Artemia, was identified, characterized, and named Ar-SETD4. We found that Ar-SETD4 was expressed abundantly in Artemia diapause embryos, in which cells were in a quiescent state. Meanwhile, trimethylated histone H4K20 (H4K20me3) was enriched in diapause embryos. The knockdown of Ar-SETD4 reduced the level of H4K20me3 significantly and prevented the formation of diapause embryos in which neither the cell cycle nor embryogenesis ceased. The catalytic activity of Ar-SETD4 on H4K20me3 was confirmed by an in vitro histone methyltransferase (HMT) assay and overexpression in cell lines. This study provides insights into the function of SETD4 and the mechanism of cell quiescence regulation.
KEYWORDS: SETD4, H4K20me3, cell quiescence, diapause, Artemia
INTRODUCTION
Quiescence is a reversible cell cycle arrest in which cells enter the G0 phase but retain the ability to reenter the cell cycle in response to appropriate stimulation (1, 2). For a long time, cellular quiescence was considered a passive and dormant state; however, new findings revealed that it is actually a heterogeneous state that is regulated by a complex network of intrinsic and extrinsic factors (3–5). Studies show that the ability of stem cells to maintain quiescence is important for tissue homoeostasis as well as their self-renewal, differentiation, development, regeneration, and stress resistance (6–8). Dysregulation and loss of quiescence often result in stem cell depletion and can cause certain degenerative diseases (9, 10). Recently, epigenetic studies showed that the chromatin structure is involved in maintaining the reversibility of quiescence and that histone methylation contributes to the control of gene expression associated with quiescence regulation (11–13). In addition, methylation of lysine 4 of histone H3 (H3K4), H3K36, and H3K79 is frequently associated with active transcription, whereas methylation of H3K9, H3K27, and H4K20 is associated with a negative regulation of transcription (14–16). Analyses of histone modifications revealed that di- and trimethylation of H4K20 increase during quiescence, whereas other histone modifications are present at similar levels in proliferating and quiescent primary human fibroblasts, which display tight chromatin compaction (17).
The family of SET domain-containing protein (SETD) methyltransferases catalyzes the methylation of lysine residues of histones, with the exception of histone H3K79 methylation, which is performed by the disruptor of telomeric silencing (DOT1) family (18, 19). The SET domain was originally identified in three Drosophila melanogaster proteins, namely, suppressor of variegation 3-9, enhancer of zeste, and Trithorax (20), which contain a structurally homologous and globular SET domain. The C-terminal segment of the SET domain contains a series of β strands folded into several discrete sheets that surround a knot-like structure (21). This topologically unusual knot-like arrangement, known as a pseudoknot, is critical for the catalytic activities of SET domain-containing proteins and their interaction with the cofactor S-adenosyl-l-methionine (Adomet) (22, 23). Based on the sequences surrounding the SET domain, proteins containing this domain are classified into seven subfamilies, namely, Suv, Ash, Trx, E(z), PRDM, SMYD, and SETD (24). In addition to having similar sequence motifs surrounding the SET domain, proteins within a subfamily often share a high level of similarity within the SET domain itself. SETD4 belongs to the SETD subfamily, members of which contain an insertion of 100 to 300 residues in the middle of the SET domain (I-SET) and a C-terminal RubisCO LSMT substrate-binding (Rubis-subs-bind) domain. The Rubis-subs-bind domain is critical for the binding of SETD proteins to histones H3 and H4 (25).
Trimethylation of H3K4 by the MLL1 methyltransferase is dispensable for maintaining the quiescence of hematopoietic stem cells and supporting leukemogenesis (26). In addition, trimethylation of H3K9 by the Suv39h methyltransferase has emerged as a hallmark of pericentric heterochromatin in mammals (27), and trimethylation of H3K27 by the EZH1 and EZH2 methyltransferases is essential for hair follicle homeostasis and wound repair (28). SETD3-mediated methylation of H3K36 activates transcription and induces muscle cell differentiation (29). Furthermore, DOT1L-mediated methylation of H3K79 is important for maintaining normal hematopoiesis in mice (30, 31), and Suv4-20h1 and Suv4-20h2, which catalyze di- and trimethylation of H4K20, respectively, promote chromatin compaction and quiescence in primary human fibroblasts (17, 32). Taken together, those studies suggest that H3K4, H3K9, H3K27, H3K36, H3K79, and H4K20 methyltransferases play important roles in regulating quiescence and gene expression in an epigenetic manner. Although accumulated evidence has shown that SET domain proteins catalyze histone methylation and are involved in regulating the chromatin structure and gene expression, to our knowledge, the function and catalytic activity of SETD4 have not yet been reported.
Here, Artemia was used as a model system to examine the regulation of cell quiescence because this primitive crustacean undergoes quiescence for prolonged periods during diapause, a state of obligate dormancy (33, 34). Artemia is found in severely hypersaline environments, such as salt lakes, which are among the most hostile environments on earth. Under unfavorable conditions, mature females produce and release encysted embryos that enter diapause, a state of obligate dormancy, whereas they release swimming nauplius larvae under favorable conditions. Here, Artemia SETD4 (Ar-SETD4) was identified and characterized in Artemia diapause embryos. We found that the levels of Ar-SETD4 and trimethylated H4K20 (H4K20me3) were enriched in diapause embryos in which cells were quiescent. RNA interference (RNAi)-mediated knockdown of Ar-SETD4 in vivo reduced the levels of H4K20me3 significantly and resulted in the release of pseudodiapause embryos in which neither cell division nor embryogenesis had ceased. In contrast, the level of H4K20me3 was enhanced with the supplementation of Ar-SETD4 by an in vitro histone methyltransferase (HMT) assay, and the overexpression of Ar-SETD4 in cell lines upregulated the trimethylation of H4K20 and caused cell division to cease.
RESULTS
Cells of Artemia diapause embryos are kept in a quiescent state.
As a survival strategy, Artemia possesses two independent reproductive pathways that enable adaptation to widely fluctuating environments (35). Under favorable conditions, mature Artemia females release swimming nauplius larvae by the ovoviviparous pathway; alternatively, they produce and release encysted embryos that enter diapause by the oviparous pathway under unfavorable conditions (Fig. 1A). The cells, which number about 4,000 in diapause embryos, can be kept in the quiescent state without cell divisions for long periods lasting many years (36). Furthermore, the diapause phase of encysted embryos can be terminated by environmental stimuli that signal favorable conditions, upon which they activate into postdiapause embryos and proceed to hatch out as nauplii.
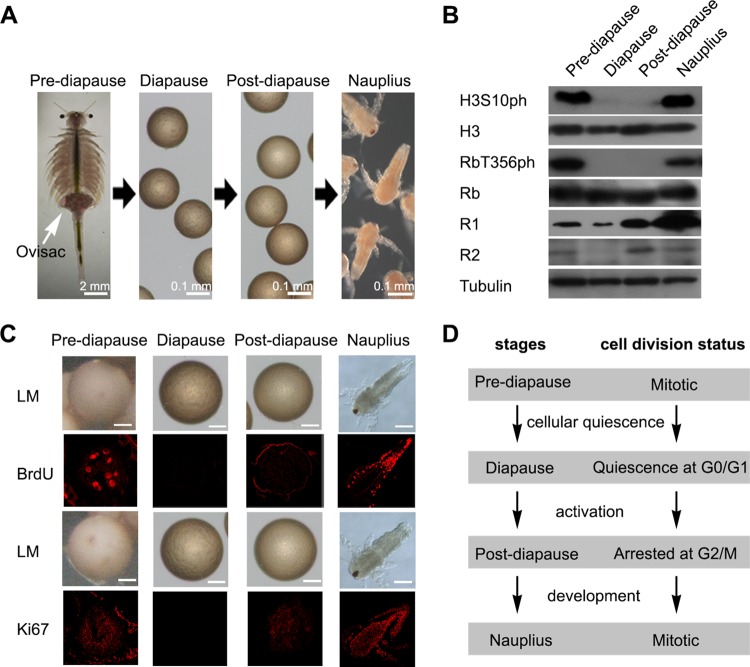
FIG 1.
Stages during diapause formation and cell quiescence in Artemia diapause embryos. (A) The stages during diapause formation are prediapause, where the embryos are in the ovisac (white arrow); diapause; postdiapause; and nauplius. (B) Western blot analysis of the phosphorylation of the cell proliferation markers H3S10ph and RbT356ph and the DNA synthesis markers R1 and R2 at each developmental stage. Tubulin was used as a loading control. (C) BrdU incorporation assays and immunofluorescence analysis of Ki67 at each developmental stage. Light microscopy (LM) images were taken to show the overall morphology. Red, incorporated BrdU and Ki67 detected by Alexa Fluor 647-conjugated secondary antibody. Bars, 50 μm. (D) Diagram of cell cycle progression in embryos during diapause formation and termination.
To determine the cell division state at each developmental stage during diapause formation, Western blot analyses were used to examine histone H3 at Ser10 (H3S10ph) and the phosphorylation of retinoblastoma (Rb) at Thr356 (RbT356ph). H3S10ph and RbT356ph are proliferation markers for mitotically dividing cells during mitosis and G1/S phase, respectively (37, 38). H3S10ph and RbT356ph were not detected in the diapause and postdiapause stages but were abundant in the prediapause and larval stages (Fig. 1B). Moreover, ribonucleotide reductase catalyzes the first unique, rate-limiting step of DNA synthesis, and both its large (R1) and small (R2) subunits are constitutively expressed in cycling cells at approximately equal levels to enable the delivery of the dinucleotide triphosphates (dNTPs) required for DNA replication (39, 40). In diapause embryos, R1 expression was inhibited and R2 was absent, indicating that DNA synthesis was turned off in diapause embryonic cells (Fig. 1B). A previous report indicated that the cell cycle was arrested at G2/M phase in the postdiapause stage (41). In this study, the result of a 5-bromo-2′-deoxyuridine (BrdU) incorporation assay showed that BrdU signals could not be detected in the diapause stage and were faint in postdiapause embryos compared to those in both prediapause embryos and hatched larvae (Fig. 1C). However, immunofluorescence analysis showed that the proliferation marker Ki67, which marks cells in all active phases of the cell cycle, was completely absent in diapause embryos but was expressed in prediapause and postdiapause embryos and hatched larvae (Fig. 1C). These results indicated that the cells in diapause embryos were in a quiescent state during the long diapause period. Based on our results, the cell cycle states during diapause formation were concluded (Fig. 1D).
Identification and characterization of Ar-SETD4 in diapause embryos of Artemia.
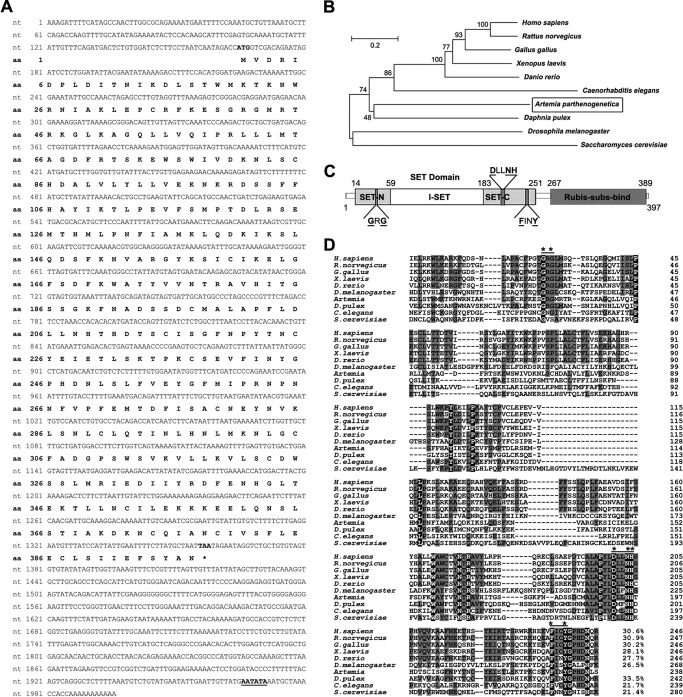
Although SETD4 was identified over a decade ago, its biological function has remained unclear. To investigate the role of SETD4 during diapause formation of Artemia, the cDNA of Ar-SETD4 was cloned from Artemia. The full length of Ar-SETD4 is 1,996 bp, with a 1,194-bp open reading frame encoding a 397-amino-acid (aa) protein (Fig. 2A). The deduced amino acid sequence of the protein is similar to those of other SETD family members, and phylogenetic analysis suggested that Ar-SETD4 could be grouped with SETD4 in the arthropod Daphnia pulex (Fig. 2B). The deduced amino acid sequence shows that Ar-SETD4 contains a catalytic SET domain (aa 14 to 251) at the N terminus and a substrate-binding Rubis-subs-bind domain (aa 267 to 393) at the C terminus, joined by a linker region (Fig. 2C). These domains occur in SETD4 proteins in many other species ranging from plants to humans (25, 42). The SET domain is comprised of SET-N (aa 14 to 58), I-SET (aa 59 to 182), and SET-C (aa 183 to 251). Although the amino acid sequences of the SET domains in SETD4 show low overall sequence similarity (<35%) across different species, the amino acids predicted to interact with Adomet and play a role in catalytic activity in Artemia are entirely conserved, including Gly40 and Gly42 in the GXG motif; Asp205, Asn208, and His209 in the DXXNH motif; and Phe241 and Tyr244 in the FXXY motif (Fig. 2D).
FIG 2.
Nucleotide sequences of Ar-SETD4 cDNA and its deduced amino acid sequence and comparison with those of SETD4 proteins from other species. (A) Nucleotide sequence of the Ar-SETD4 cDNA and its deduced amino acid sequence. Numbers on the left indicate the nucleotide and amino acid positions. The start (ATG) and stop (TAA) codons are shown in boldface type, and the putative polyadenylation signal (AATATA) is underlined. The asterisk denotes the termination of amino acids. (B) Phylogenetic analysis of SETD4 proteins from other species. The tree was constructed by using the neighbor-joining method. Percent bootstrap values for 1,000 replicates are shown at the branching points. The bar shows the branch length corresponding to the mean number of differences (0.2) per residue along each branch. (C) Schematic representation of the structure of Ar-SETD4 showing the SET and Rubis-subs-bind domains. The SET domain is comprised of SET-N, I-SET, and SET-C. The three typical motifs GXG, DXXNH, and FXXY (GRG, DLLNH, and FINY in Artemia) are indicated by dark gray segments, and the conserved amino acids are in boldface type and underlined. The numbers indicate amino acid positions. (D) Alignment of the amino acid sequence of the SET domain of Ar-SETD4 with those of SETD4 proteins from other species. Numbers on the right indicate the amino acid positions. The three typical motifs GXG, DXXNH, and FXXY (GRG, DLLNH, and FINY in Artemia) are indicated by black lines. * indicates a complete conservation of amino acids in these motifs. The black- and gray-shaded areas represent conserved and identical amino acids, respectively. The similarities are shown at the end of the alignment.
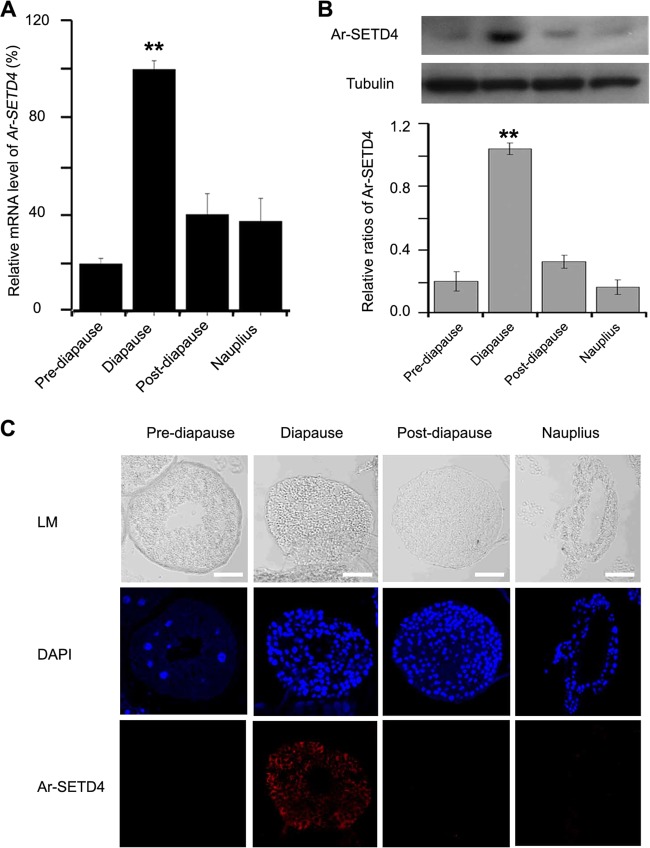
Real-time quantitative PCR was used to analyze the mRNA level of Ar-SETD4 at each developmental stage during diapause formation. We found that the Ar-SETD4 mRNA level in diapause embryos was ∼2- to 3-fold higher than those in other stages (prediapause embryos, postdiapause embryos, and hatched nauplii) (Fig. 3A). Meanwhile, the expression of the Ar-SETD4 protein was analyzed by Western blotting using polyclonal antibodies specific for Ar-SETD4. The results showed that the level of Ar-SETD4 was significantly higher in diapause embryos than in prediapause and postdiapause embryos and hatched larvae (Fig. 3B). Furthermore, this result was supported by data from immunofluorescence analysis in which Ar-SETD4 was present in diapause embryos but was completely absent in prediapause and postdiapause embryos and hatched larvae (Fig. 3C). These results suggest that Ar-SETD4 may play important functions in diapause embryo formation in Artemia.
FIG 3.
Expression of Ar-SETD4 at each developmental stage during diapause formation in Artemia. (A) Real-time quantitative PCR analysis of Ar-SETD4 mRNA expression at each developmental stage. The mRNA amounts were normalized to those of tubulin mRNA. The means of data from three independent biological replicates are shown; error bars represent the standard errors of the means. **, P < 0.01 by a one-tailed t test. (B) Western blot analysis of Ar-SETD4 expression at each developmental stage. Tubulin was used as a loading control. The relative band intensities were quantified by using ImageJ software, and the ratio of Ar-SETD4 to tubulin is presented. The values are means ± standard deviations (n = 3). Statistical significance was determined by a one-tailed t test (**, P < 0.01). (C) Immunofluorescence analysis of Ar-SETD4 at each developmental stage. Light microscopy (LM) images were taken to show the overall morphology. Red, Ar-SETD4 detected by Alexa Fluor 647-conjugated secondary antibody; blue, nuclei counterstained with DAPI. Bars, 50 μm.
Ar-SETD4 is associated with cell quiescence during diapause formation.
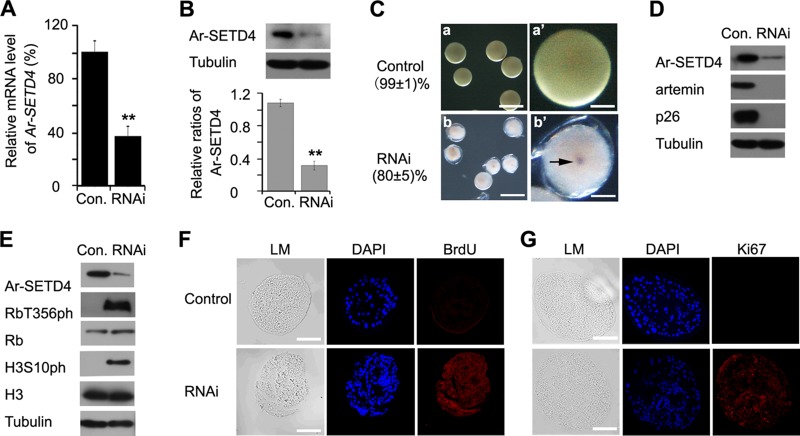
To elucidate the function of Ar-SETD4 in the regulation of diapause formation and cell quiescence in Artemia, RNAi was used to knock down Ar-SETD4 gene expression. Just before ovarian development, a double-strand RNA (dsRNA) designed from the Ar-SETD4 cDNA sequence was injected. After the injection of diapause-destined Artemia adults with 1 μg of Ar-SETD4 dsRNA, the Ar-SETD4 mRNA level decreased to <30% of that in control Artemia adults injected with a green fluorescent protein (GFP)-specific dsRNA (GFP dsRNA) (Fig. 4A), and the Ar-SETD4 protein expression level was also decreased significantly (Fig. 4B). Female Artemia adults injected with GFP dsRNA released diapause embryos (Fig. 4Ca and a′), whereas those injected with Ar-SETD4 dsRNA produced pseudodiapause embryos (Fig. 4Cb and b′), in which embryogenesis did not cease and a faint eyespot could be observed. Furthermore, the diapause-specific proteins p26 and artemin (43, 44) were expressed abundantly in control diapause embryos but were absent in knockdown embryos (Fig. 4D). These results indicated that the knockdown of Ar-SETD4 inhibited the formation of Artemia diapause embryos.
FIG 4.
Effect of knockdown of Ar-SETD4 on diapause formation in Artemia. (A) Real-time quantitative PCR analysis of the expression level of Ar-SETD4 mRNA in embryos produced by females treated with GFP-specific (control [Con.]) or Ar-SETD4-specific RNAi. The mRNA amounts were normalized to those of tubulin gene mRNA. Data are presented as the means ± standard deviations of data from 10 replicates. **, P < 0.01 by a one-tailed t test. (B) Western blot analysis of Ar-SETD4 protein expression in control and Ar-SETD4 knockdown embryos. Tubulin was used as a loading control. The relative band intensities were quantified by using ImageJ software, and the ratio of Ar-SETD4 to tubulin is presented. The values are means ± standard deviations (n = 3). Statistical significance was determined by a one-tailed t test (**, P < 0.01). (C) Phenotypes of embryos injected with 1 μg of Ar-SETD4 dsRNA (b and b′) or GFP dsRNA (a and a′) as a control. Bars, 200 μm (a and b) and 50 μm (a′ and b′). Embryos treated with GFP dsRNA remained in diapause, whereas those treated with Ar-SETD4 dsRNA entered pseudodiapause. The arrow indicates the faint eyespot. (D) Western blot analyses of artemin and p26 (diapause-specific proteins) after RNAi. Tubulin was used as a loading control. (E) Western blot analyses of H3S10ph and RbT356ph (cell proliferation markers) after RNAi. Tubulin was used as a loading control. (F and G) Detection of mitosis in control and Ar-SETD4 knockdown embryos using a BrdU incorporation assay (F) and immunofluorescence analysis of Ki67 (G). Light microscopy (LM) images were taken to show the overall morphology. Red, BrdU and Ki67 detected by Alexa Fluor 647-conjugated secondary antibody; blue, nuclei counterstained with DAPI. Bars, 50 μm.
Next, the cell cycle state was examined after knockdown of the Ar-SETD4 gene. Although RbT356ph and H3S10ph were not detected in control embryos, they were easily detected in Ar-SETD4 knockdown embryos (Fig. 4E). Furthermore, BrdU incorporation and Ki67 expression were observed in pseudodiapause embryos released from Artemia adults injected with Ar-SETD4 dsRNA but not in diapause embryos released from control Artemia adults (Fig. 4F and G). In contrast to the control embryos, neither the cell cycle nor embryogenesis was halted in the pseudodiapause embryos induced by the knockdown of Ar-SETD4. These results indicate that Ar-SETD4 has important functions in the regulation of cell quiescence and diapause formation.
Ar-SETD4 is responsible for the trimethylation of H4K20 and H3K79 during diapause formation in Artemia.
As mentioned above, Ar-SETD4 belongs to the SET domain-containing family of proteins, which mostly catalyze the transfer of methyl groups from the cofactor Adomet to specific lysine residues of histones and are involved in the regulation of chromatin and gene expression (20–29). In order to investigate the function of Ar-SETD4, methylations of H3 and H4 at each stage during diapause formation and after knockdown of the Ar-SETD4 gene were analyzed by Western blotting. The specificity of anti-H3K79me3 and anti-H4K20me3 was examined. The results indicated that the anti-H3K79me3 and anti-H4K20me3 antibodies exclusively specifically recognize H3K79me3 and H4K20me3, respectively, with no cross-reaction (see Fig. S1 in the supplemental material). We found that the levels of H3K9me3, H3K79me1, and H3K79me3 were higher in diapause and postdiapause embryos than in prediapause embryos and nauplii (Fig. 5). Notably, H4K20me3 was significantly abundant in the diapause embryos (P < 0.01 by a one-tailed t test) (Fig. 5), in which the cells were quiescent. These results indicate that a high level of H4K20me3 associated with a high level of Ar-SETD4 might be involved in the regulation of cell quiescence during diapause formation in Artemia.
FIG 5.
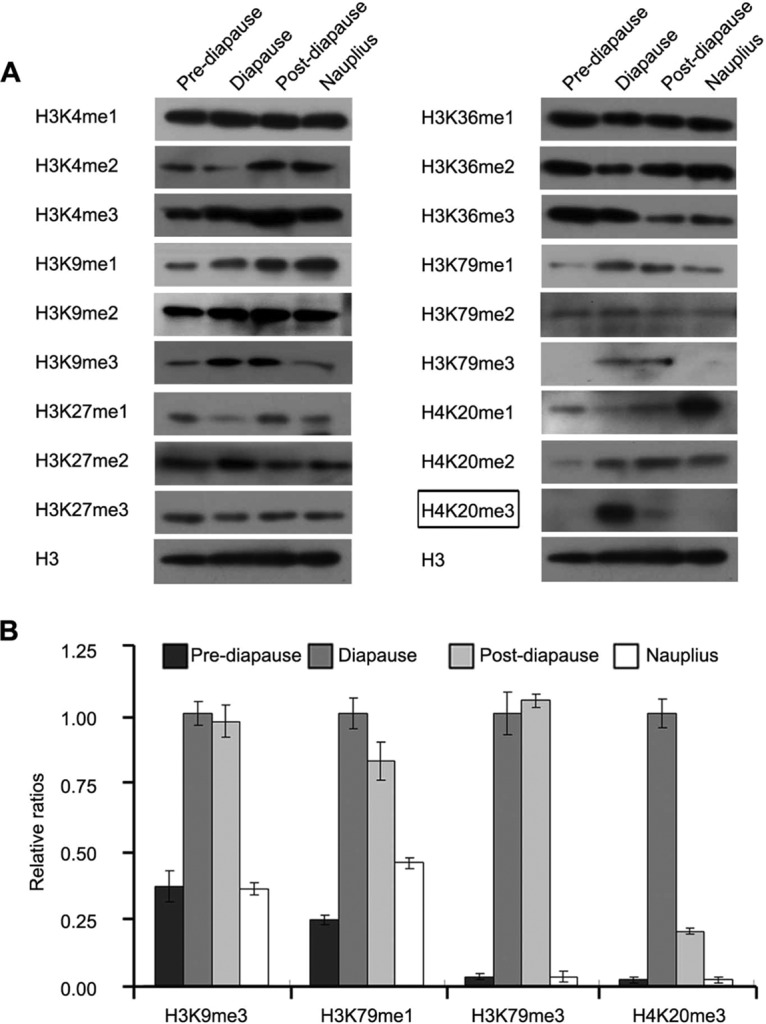
Methylation of H3 and H4 at various lysine residues during diapause formation. (A) Western blot analyses of the levels of methylation of H3 and H4 at various positions during each developmental stage. H3 was used as a loading control. At the diapause stage, high levels of H4K20me3 were specific for the diapause embryo stage (indicated by a box). (B) The relative band intensities in panel A were quantified by using ImageJ software, and the ratios of H3K9me3, H3K79me1, H3K79me3, and H4K20me3 to H3 are presented. The values are means ± standard deviations (n = 3). Statistical significance was determined by a one-tailed t test.
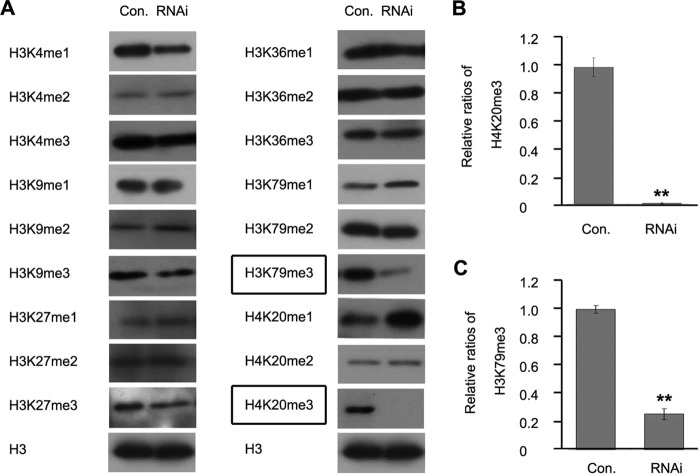
To investigate the function of Ar-SETD4 in histone methylation, the levels of methylation at various positions of H3 and H4 were analyzed after the knockdown of Ar-SETD4. As shown in Fig. 6, the knockdown of Ar-SETD4 reduced the levels of H3K79me3 and H4K20me3 significantly but had no effect on the methylation patterns of other histone residues. These results indicated that Ar-SETD4 is involved in the regulation of H4K20me3 and H3K79me3 during diapause formation in Artemia.
FIG 6.
Effect of knockdown of Ar-SETD4 on methylation of H3 and H4 in Artemia. (A) Western blot analysis of the levels of lysine methylation of H3 and H4 after RNAi-mediated knockdown of Ar-SETD4. H3 was used as a loading control. The levels of H4K20me3 and H3K79me3 were decreased after RNAi (indicated by boxes). (B and C) The relative band intensities in panel A were quantified by using ImageJ software, and the ratios of H4K20me3 and H3K79me3 to H3 are presented. The values are means ± standard deviations (n = 3). Statistical significance was determined by a one-tailed t test (**, P < 0.01).
Ar-SETD4 catalyzes the trimethylation of H4K20 directly in vitro and in vivo.
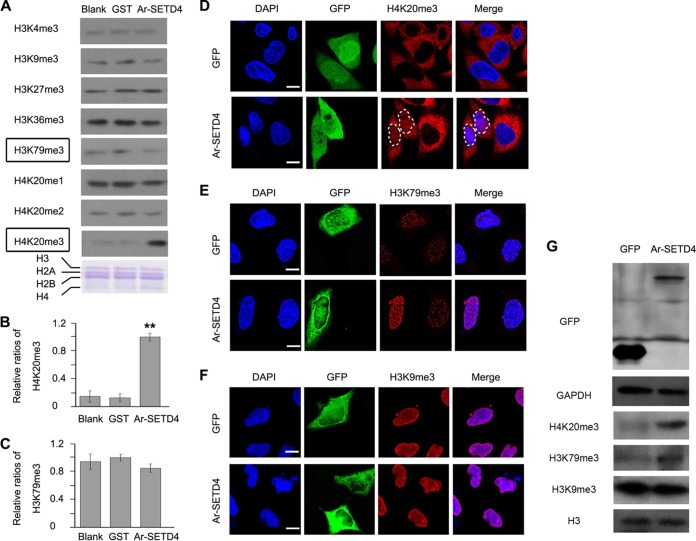
To characterize the HMT activity of Ar-SETD4 in vitro, we performed an in vitro HMT assay with a purified glutathione S-transferase (GST)–Ar-SETD4 fusion protein. Core histones and S-adenosyl-l-methionine were utilized as the substrate and methyl donor, respectively. The results showed that the level of H4K20me3 was enhanced with the supplementation of Ar-SETD4 but that this enhancement did not occur for H3K79me3, H4K20me1, H4K20me2, or any of the other histone residues examined (Fig. 7A to C). Notably, the level of H4K20me3 was 6- to 7-fold higher with the supplementation of Ar-SETD4 than in the blank and GST protein groups (Fig. 7B). Furthermore, as the substrates of Ar-SETD4, peptide fragments of H4 from Lys 12 to Tyr 28, KGGAKRHRK20VLRDNIQY, with H4K20me0, H4K20me1, and H4K20me2 modification on lysine (K; boldface, underlined) were synthesized. The results of dot blot and mass spectrometry analyses showed that Ar-SEDT4 catalyzed the modification of H4K20me2 into H3K20me3 specifically (see Fig. S2 in the supplemental material).
FIG 7.
Ar-SETD4 catalyzes the trimethylation of H4K20. (A to C) Ar-SETD4 catalyzes the trimethylation of H4K20 directly in vitro. (A) An in vitro HMT assay was done in the presence of core histones only (Blank), histones and GST (GST), and histones and GST–Ar-SETD4 (Ar-SETD4). Adomet was added as the methyl donor, and Western blotting was then performed to detect histone lysine methylations. As a loading control, histone proteins, H3, H2A, H2B, and H4 (indicated) were used and stained with Coomassie brilliant blue. The level of H4K20me3 was increased, but the level of H3K79me3 did not change with the supplementation of Ar-SETD4 (indicated by boxes). (B and C) The relative band intensities in panel A were quantified by using ImageJ software, and the ratios of H4K20me3 and H3K79me3 to H3 are presented. The values are means ± standard deviations (n = 3). Statistical significance was determined by a one-tailed t test (**, P < 0.01). (D to G) Ar-SETD4 catalyzes the trimethylation of H4K20 in vivo. MKN45 cells were transfected with the pEGFP-C1 (GFP) and pEGFP-C1-Ar-SETD4 (Ar-SETD4) plasmids and then fixed with methanol (for anti-H3K79me3) or 4% paraformaldehyde (for anti-H4K20me3 and anti-H3K9me3). Immunofluorescent signals of H4K20me3 (D), H3K79me3 (E), and H3K9me3 (F) were detected (1 of 20 fields is shown representatively). Note that the signals of anti-H4K20me3 located in the cytoplasm were nonspecific. The nucleus areas of cells with Ar-SETD4 overexpression are marked by dashed circles. Green, signals of GFP and GFP–Ar-SETD4; red, H4K20me3, H3K79me3, and H3K9me3 detected by Alexa Fluor 594-conjugated secondary antibody; blue, nuclei counterstained with DAPI. Merge indicates that blue and red signals were merged. Bars, 20 μm. (G) Western blot analysis of the levels of H420me3, H3K79me3, and H3K9me3 after Ar-SETD4 overexpression in MKN45 cells. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and H3 were used as loading controls.
Since gene overexpression has not been available for Artemia to date, we tested the trimethylation of H4K20 by Ar-SETD4 in human cell lines. GFP-fused Ar-SETD4 was overexpressed in a human gastric cancer cell line, MKN45. The results of immunofluorescence and Western blot analyses showed that the level of H4K20me3 (the specific signals are in the nucleus) was increased significantly in cells with overexpressed Ar-SETD4 compared to that in control cells (Fig. 7D and G). However, in contrast to the results of in vitro assays, an increased H3K79me3 level was also observed in Ar-SETD4-overexpressing cells (Fig. 7E and G). As a specific control for the trimethylation of H4K20 and H3K79, the level of H3K9me3 in Ar-SETD4-overexpressing cells did not change compared to the control (Fig. 7F and G). Consistent results were also observed in human fibrosarcoma HT1080 cells (see Fig. S3 in the supplemental material). These results suggest that Ar-SETD4 catalyzes the trimethylation of H4K20 directly but may also induce the trimethylation of H3K79 indirectly. Furthermore, mutants of Ar-SETD4 with a deletion of the SET domain or Sub domain were constructed, and their enzymatic activity of trimethylation on H3K79 and H4K20 was tested in MKN45 cells. These mutants abolished the enzymatic activity of trimethylation on H3K79 and H4K20 by Ar-SETD4 (Fig. S4). These results indicated that both the SET domain and the Sub domain are essential for the catalytic activity of Ar-SETD4.
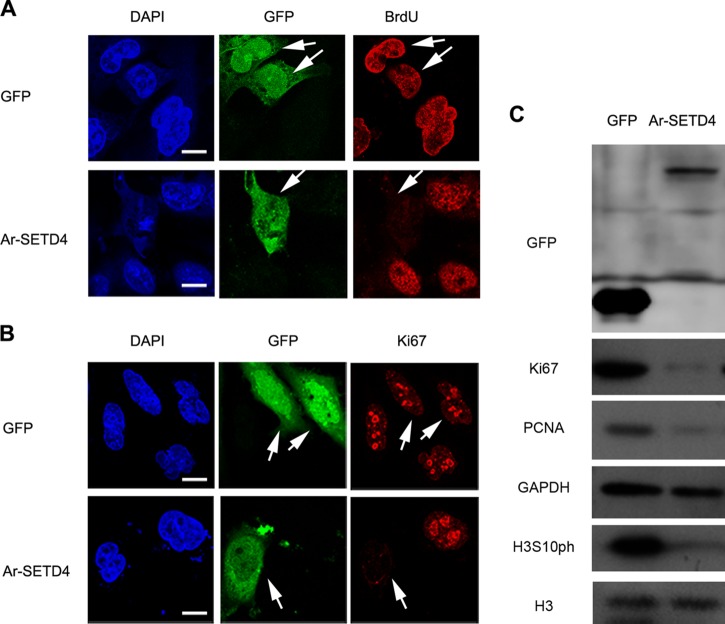
To explore the function of Ar-SETD4 in quiescence regulation in cells other than Artemia cells, Ar-SETD4 was overexpressed in MKN45 cells, and the effect on cell proliferation was determined. Analysis of BrdU incorporation and immunofluorescence of the proliferation marker Ki67 indicated that cells overexpressing Ar-SETD4 had no mitotic activity, in contrast to control cells (Fig. 8A and B). Similar to the results obtained with diapause embryos of Artemia, levels of Ki67, PCNA, and H3S10ph were decreased markedly in cells with Ar-SETD4 overexpression in contrast to the two mutants and control cells (Fig. 8C; see also Fig. S5 in the supplemental material). Based on our results, Ar-SETD4 regulates cell quiescence and mediates the trimethylation of H4K20 and H3K79 during diapause formation in Artemia.
FIG 8.
Cell division ceases after overexpression of Ar-SETD4. MKN45 cells were transfected with the pEGFP-C1 (GFP) and pEGFP-C1-Ar-SETD4 (Ar-SETD4) plasmids and then fixed with 4% paraformaldehyde. (A) For the BrdU incorporation assay, cells were incubated in medium containing 12 μM BrdU for 30 h and then fixed with 4% paraformaldehyde. (B) Detection of immunofluorescent signals of Ki67. The arrows indicate cells with GFP or Ar-SETD4 overexpression. Green, signals of GFP and GFP–Ar-SETD4; red, BrdU and Ki67 detected by Alexa Fluor 594-conjugated secondary antibody; blue, nuclei counterstained with DAPI. Bars, 20 μm. (C) Western blot analysis of the cell proliferation marker Ki67, PCNA, and H3S10ph after Ar-SETD4 overexpression. GAPDH and H3 were used as loading controls.
DISCUSSION
Artemia embryos are an excellent model for studying the regulation of cell quiescence, because they remain in this state for prolonged periods during diapause. Our previous studies attempted to elucidate the molecular mechanisms involved in the regulation of cell division during diapause embryo formation and termination by examining the roles of p90RSK, AMP-activated protein kinase, polo-like kinase 1, and H3K56ac (histone 3 acetylated on lysine 56) (41, 45–48). These experiments indicated that the mechanisms underlying cell quiescence are complex and that epigenetic regulation plays an important role in the process of diapause formation and termination (48). Here, we found that Ar-SETD4 regulates cell quiescence and mediates the trimethylation of H4K20 and H3K79, leading to the initiation of diapause formation in Artemia and the cessation of cell division in MKN45 cells.
Members of the SET domain-containing family of proteins mostly catalyze the transfer of methyl groups from the cofactor Adomet to specific lysine residues of histones. Based on structural analysis, Ar-SETD4 contains conserved GXG and H/R/DXXNH motifs, which are required for interactions with Adomet. In addition, the SET-C subdomain of Ar-SETD4 is composed of two β strands that fold into a pseudoknot, which is critical for its catalytic activity and interaction with Adomet. Histone lysine methylation is a functionally complex process that can either activate or repress transcription, depending on the sequence-specific lysine methylation site and the methylation state of the ε-amino group of the target lysine residue. Consistent with their high substrate specificity, SET domains of SETD subfamily members show low overall sequence similarity, and the residues at the active site are not conserved. In particular, the residues in the I-SET region that are involved directly in interactions with the substrate are significantly different among subfamily members (49, 50). The I-SET region of Ar-SETD4 is comprised of several α-helices, the lengths of which vary from those of other SETD family members and lack any consistent pattern of sequence conservation across SET domain proteins, suggesting that they are suited to the role of mediating the recognition of different target sequences. Furthermore, it has been suggested that the F/Y switch establishes SET domain product specificities. For example, a Y245A or Y305F mutation in SET7/9 alters its specificity from an H3K4 monomethylase to a tri- or dimethylase, respectively (51), whereas an F281Y mutation in the lysine-binding pocket of DIM-5 converts this protein from an H3K9 trimethylase to an H3K9 mono- or dimethylase (52). In support of this view, Ar-SETD4 is an H3K79 and H4K20 trimethylase (Phe in the F/Y switch). Together, variations in the insertion sequences or domains flanking the SET domain and the F/Y switch determine the target lysine or degree of methylation activity of SET domain-containing proteins.
Many SET domain family proteins play a role in the regulation of the chromatin structure and gene expression by catalyzing the methylation of histone proteins (20–29). However, the biological function of SETD4 is currently unknown. It has been difficult to elucidate the function of SETD4 proteins, in part because they are expressed only in quiescent cells with different suppressors of cell division, and very few suitable animal models are available. In the present study, we used encysted embryos of Artemia to demonstrate that Ar-SETD4 is responsible for the trimethylation of histones H4K20 and H3K79 and regulates the cell quiescence required to initiate diapause formation. In addition, the overexpression of Ar-SETD4 enhanced the levels of H4K20me3, H3K79me3, or both in cancer cells. This study indicated that Ar-SETD4 is involved in the regulation of cell quiescence and mediates the trimethylation of H4K20 and H3K79.
In contrast to H4K20me1 catalysis by SET8/PR-Set7, the mechanism of catalysis of H4K20me2 and H4K20me3 remains largely unclear (53, 54). The available evidence suggests that SMYD5 is recruited to a subset of Toll-like receptor-responsive promoters through its association with NCoR corepressor complexes, where it catalyzes the trimethylation of H4K20 (55). A previous study showed that the knockout of both Suv4-20h1 and Suv4-20h2 leads to a loss of H4K20me2 and H4K20me3 and is perinatally lethal (56). Here, the knockdown of Ar-SETD4 in Artemia led to a loss of H4K20me3, and the overexpression of Ar-SETD4 induced an increased level of H4K20me3 in cancer cells. Given the sequence similarity between Artemia and human SETD4 proteins, recombinant human SETD4 (Hs-SETD4) was also tested for activity in the trimethylation of H4K20. Hs-SETD4 was overexpressed in the human cell line MKN45. Immunofluorescence analysis showed that the overexpression of Hs-SETD4 but not GFP promotes the trimethylation of H4K20 (see Fig. S6 in the supplemental material), consistent with data from similar experiments with Ar-SETD4. Furthermore, in an in vitro HMT assay, trimethylation of H4K20, but not mono- or dimethylation of this histone residue, was catalyzed specifically and directly by Ar-SETD4. However, we observed that the expression levels of Suv4-20h2 were similar in both control and Ar-SETD4-overexpressing cells (Fig. S7). Hence, this indicates that the catalysis of H4K20me3 by Ar-SETD4 is Suv4-20h2 independent in the case of Artemia diapause embryo formation. It is possible that there are two types of catalysis of H4K20me3 by the two different enzymes Suv4-20h2 and SETD4. In the case of Artemia, Ar-SETD4 catalyzes H4K20me3 to regulate cell quiescence for diapause formation.
DOT1-catalyzed methylation of H3K79 is a marker of active gene transcription in species ranging from yeast to mammals (57, 58). A previous study showed that the inhibition of H3K79 methylation selectively inhibits the proliferation, self-renewal, and metastatic potential of breast cancer cells (59). Like H4K20, H3K79 exists in mono-, di-, and trimethylated states. A previous mass spectrometry (MS) analysis revealed that H3K79me1 is more abundant than H3K79me2 in mice and humans, whereas H3K79me3 is rarely detected (60, 61). Because only one enzyme (DOT1) is responsible for the methylation of H3K79, it is unclear whether mono-, di-, or trimethylated H3K79 regulates diverse cellular processes such as DNA replication, transcription, proliferation, differentiation, and development. A previous study suggested that a DOT1-containing complex is capable of trimethylating H3K79 and linking it to the Wnt signaling pathway (62). In addition, a more recent study showed that two homologues of DOT1 from African trypanosomes, DOT1A and DOT1B, catalyze the mono- or dimethylation and trimethylation of H3K79, respectively (63). In contrast to its mono- and dimethylation, the function and mechanism of catalysis of the trimethylation of H3K79 are more complex and are currently unclear.
In this study, we found that the knockdown of Ar-SETD4 induced decreases in the levels of H4K20me3 and H3K79me3 but not those of H3K79me1 and H3K79me2, suggesting that the catalysis of H3K79me3 is different from that of H3K79me1 and H3K79me2. Although Ar-SETD4 catalyzed the direct trimethylation of H4K20 but not H3K79 in an in vitro methylation system, the increase of the H3K79me3 level accompanied by an increase of the H4K20me3 level in diapause embryos of Artemia and Ar-SETD4-overexpressing cells as well as decreases of H4K20me3 and H3K79me3 levels were observed simultaneously in Ar-SETD4 knockdown Artemia embryos. Furthermore, we did not find that DOT1 responds to Ar-SETD4 overexpression in MKN45 cells in this study (see Fig. S8 in the supplemental material). These results suggested that SETD4 regulated the trimethylation of H3K79 indirectly, possibly mediated by unknown factors other than DOT1. Diapause and postdiapause are different states in the development of Artemia through the oviparous pathway, and high levels of H3K79me3 and H4K20me3 were observed in diapause embryos. In contrast, a high level of H3K79me3 was detected only in postdiapause embryos in which cells escape from quiescence and arrest in G2/M phase (41). These results suggested that Ar-SETD4 is involved in the regulation of cell quiescence and mediates the trimethylation of H4K20 and H3K79 during diapause formation in Artemia.
MATERIALS AND METHODS
Antibodies.
The following antibodies were used: anti-H3K4me1 (catalogue number R130313; HuaAn Biotechnology), anti-H3K4me2 (catalogue number 1347-1; Epitomics), anti-H3K4me3 (catalogue number 2207275; Millipore), anti-H3K9me1 (catalogue number ab9045; Abcam), anti-H3K9me2 (catalogue number 1349-1; Epitomics), anti-H3K9me3 (catalogue number ab1773; Abcam), anti-H3K27me1 (catalogue number 7693s; Cell Signaling Technology), anti-H3K27me2 (catalogue number 9728s; Cell Signaling Technology), anti-H3K27me3 (catalogue number ab6174; Abcam), anti-H3K36me1 (catalogue number ab9048; Abcam), anti-H3K36me2 (catalogue number ab9049; Abcam), anti-H3K36me3 (catalogue number ab9050; Abcam), anti-H3K79me1 (catalogue number ab2886; Abcam), anti-H3K79me2 (catalogue number 9757s; Cell Signaling Technology), anti-H3K79me3 (catalogue number ab2621; Abcam), anti-H4K20me1 (catalogue number sc-134221; Santa Cruz Biotechnology), anti-H4K20me2 (catalogue number GTX630545; GeneTeX), anti-H4K20me3 (catalogue number 5737s; Cell Signaling Technology), antitubulin (catalogue number T6199; Sigma), anti-Hs-SUV4-20h2 (catalogue number sc-366867; Santa Cruz Biotechnology), anti-H3 (catalogue number ab1791; Abcam), anti-H3S10ph (catalogue number ab5176; Abcam), anti-Rb (catalogue number ab6075; Abcam), anti-RbT356ph (catalogue number ab76298; Abcam), anti-Ki67 (catalogue number ab16667; Abcam), anti-BrdU (catalogue number B8434; Sigma), anti-R1 (catalogue number ab81085; Abcam), anti-R2 (catalogue number ab57653; Abcam), and anti-PCNA (catalogue number ab92552; Abcam). The anti-Ar-SETD4 antibody was raised in rabbit (HuaAn Biotechnology) against peptides containing partial amino acid sequences (aa 184 to 197 [TGSSGKFNADSSDC] and aa 352 to 366 [CILEKKKEELQNSLS]), which represented the best epitopes.
Cell culture.
Cells of the human gastric cancer cell line MKN45 and the fibrosarcoma cell line HT1080 were purchased from the Tumor Cell Bank of the Chinese Academy of Sciences (Shanghai, China). These cells were cultured in RPMI 1640 medium (Corning) supplemented with 10% fetal bovine serum (Gibco), 100 IU/ml penicillin, and 100 μg/ml streptomycin (Sigma-Aldrich) at 37°C in a humidified atmosphere with 5% CO2.
Culture of Artemia parthenogenetica.
Diapause embryos of Artemia parthenogenetica were harvested from Gahai Lake, Qinghai Province, China. The embryos were activated by soaking in saturated brine for 48 h and freezing at −20°C for 3 months. Subsequently, postdiapause embryos were hydrated at 4°C for 5 h, and the sedimentations were incubated in 2.5% artificial seawater (Blue Starfish, Zhejiang, China) at 28°C with continuous light exposure. For oviparous Artemia, swimming nauplii were reared in 8% (wt/vol) artificial seawater under a light/dark cycle of 4 h/20 h (12:00 to 16:00 light). For ovoviviparous Artemia, the swimming nauplii were reared in 4% (wt/vol) artificial seawater under a light/dark cycle of 16 h/8 h (7:00 to 23:00 light). The water was maintained at 28°C and was supplemented with Chlorella powder (Fuqing King Dnarmsa Spirulina Co. Ltd.) as brine shrimp food every 2 days.
Molecular cloning of Ar-SETD4.
Two sets of degenerate primers (ArSETD4 dF1 [GCNYTNGCHCCRTAYYTDGA], ArSETD4 dR1 [AAKCCRTAYTCYARVARVAG], ArSETD4 dF2 [TWYYTRGAYMTKYTDAAYCAC], and ArSETD4 dR2 [CKBTGRTTRTCRTGRGGVCC]) were designed based on the alignment of SETD4 sequences from different organisms. Total RNA was extracted from Artemia by using TRIzol reagent (Invitrogen), according to the manufacturer's instructions, and then quantified by using a Genova UV-visible spectrophotometer at 260 nm. First-strand cDNA was synthesized from 1 μg of total RNA by using the SuperScript/First-Strand cDNA synthesis kit (Invitrogen), according to the manufacturer's protocol, and was used as a template for PCR. A cDNA fragment of ∼140 bp was amplified and subcloned into the pUCm-T vector (TaKaRa Bio) for sequencing analysis (Sangon Biotech). Rapid amplification of cDNA ends (RACE) was performed by using primers 5′ArSETD4 R1 (TAAAGACTTCTGAGCACTTCGG), 5′ArSETD4 R2 (TTTCATAACAGTTTGTGTAAGG), 3′ArSETD4 F1 (AATCCTTACACAAACTGTTATG), and 3′ArSETD4 F2 (AACCCCGAAGTGCTCAGAAGTC) and adapter primers from the FirstChoice RLM-RACE kit (Ambion), according to the manufacturer's protocol. Fragments of 870 bp and 1,160 bp were obtained by 5′ RACE and 3′ RACE, respectively. The full-length Ar-SETD4 cDNA sequence was obtained from the overlapping RACE fragments. The sequenced cDNA and the deduced peptide were analyzed by using EditSeq v5.00 (DNAStar), and a BLAST analysis (NCBI) was performed to confirm their homologies. The online analysis packages PSIPRED (http://bioinf.cs.ucl.ac.uk/psipred) and Phyre2 (http://www.sbg.bio.ic.ac.uk/phyre2) were used to predict the structure and functional domains. The phylogenetic tree of SETD4 proteins from Homo sapiens (GenBank accession number NP_059134.1), Rattus norvegicus (accession number NP_001107219), Gallus gallus (accession number NP_001025965), Xenopus laevis (accession number NP_001079674), Danio rerio (accession number NP_001039027), Drosophila melanogaster (accession number NP_995955), Daphnia pulex (accession number EFX73381), Artemia parthenogenetica (accession number KP749757), Caenorhabditis elegans (accession number NP_490849), and Saccharomyces cerevisiae (accession number EGA74690) was constructed by using the neighbor-joining method.
RNAi in Artemia.
For dsRNA preparation, the PET-T7 plasmid containing two inverted T7 promoter sites flanking the multiple-cloning sites was used as the expression vector (41). To obtain the reconstructed plasmid expressing Ar-SETD4 dsRNA, a 557-bp fragment in the coding region of the Ar-SETD4 gene was amplified with specific primers (shSETD4 F [GCTCTAGAATGGTCGACAGAATAGATC] and shSETD4 R [CGGGATCCCTCCCAGTTATATGTACTG]; XbaI and EcoRI sites are underlined) and subcloned into PET-T7 at the XbaI and EcoRI sites. The recombinant plasmid was transformed into Escherichia coli DH5α, sequenced to confirm the inserted nucleotide sequence, and then transformed into E. coli HT115 cells to express the dsRNA. The plasmid expressing GFP dsRNA was constructed as described previously and was used as a negative control (64). The dsRNA was produced and purified as described previously by Yodmuang et al. (65). Artemia adults (diapause destined) were injected with 1 μg of Ar-SETD4 dsRNA or GFP dsRNA by using an Ultra-MicroPump II instrument equipped with a Micro4TM MicroSyringe pump controller (World Precision Instruments). After injection, the animals were cultured in 8% artificial seawater with 4 h of light exposure per day. One week later, RNA and proteins were extracted as reported previously (64), and real-time PCR (RT-PCR) and Western blot analyses were then performed to assess RNAi efficiency. Three independent groups were analyzed.
Real-time quantitative PCR.
Specimens from different developmental stages were snap-frozen in liquid nitrogen, and total RNA was then prepared from homogenized specimens by using TRIzol reagent (Invitrogen). The extracted RNA was quantified by measuring the absorbance at 260 nm with a Genova UV-visible spectrophotometer. First-strand cDNAs were prepared from the total RNA specimens as described above. After reverse transcription, real-time PCRs were performed on the Bio-Rad MiniOpticon real-time PCR system using SYBR Premix Ex Taq (TaKaRa Bio) and gene-specific primers to amplify Ar-SETD4 and tubulin as an internal control (RT-ArSETD4 F [AGGAATGAGAACAAGAAAAGG] and RT-ArSETD4 R [AAGTAAATACAACACCAAAGC] for Ar-SETD4 and Tubulin F [GCAGTGGTCTACAAGGTTTC] and Tubulin R [ATCAAAACGAAGGCTGGCGGTG] for tubulin). The relative amounts of mRNAs were analyzed by using the comparative threshold cycle (CT) method, as described previously by Schmittgen and Livak (66). All data were expressed as the means ± standard errors (SE) of data from three independent repetitions. All statistical analyses were performed by one-way analysis of variance, and a P value of <0.01 was considered significant.
Western blot analysis.
To examine the specificity of anti-H3K79me3 and anti-H4K20me3 antibodies, peptides of histone H3 (VREIAQDFK79TDLRFQSSAV; the modification site is underlined, and the associated number indicates the position on the histone) with various methylation states on K79 (H3K79me0, H3K79me1, H3K79me2, and H3K79me3) and histone H4 (KGGAKRHRK20VLRDNIQY) with various methylation states on K20 (H4K20me0, H4K20me1, H4K20me2, and H4K20me3) were synthesized (GL Biochem [Shanghai] Ltd., Shanghai, China). The corresponding peptides with serial dilutions (20 ng, 100 ng, and 500 ng) were dotted onto a polyvinylidene difluoride (PVDF) membrane and blotted separately with anti-H3K79me1, anti-H3K79me2, anti-H3K79me3, anti-H4K20me1, anti-H4K20me2, or anti-H4K20me3 antibodies.
The open reading frame of Ar-SETD4 was PCR amplified by using primers C1-ArSETD4 F (CCGCTCGAGCTATGGTCGACAGAATAGATC) and C1-ArSETD4 R (CGGGATCCTTAATTAGCATAAGAAAATTC), the SET domain was PCR amplified with primers C1-ArSETD4 F and C1-ArSET domain R (CGGGATCCTTAAGACAGATTGTCATGAGG), the Sub domain was PCR amplified with primers C1-ArSub domain F (CCGCTCGAGCTTTTGTACCTTTTGAAATG) and C1-ArSETD4 R, and these constructs were then inserted into the pEGFP-C1 plasmid. MKN45 cells were plated onto 6-well slides at a density of 1 × 105 cells per well. The following day, the cells were transfected with the pEGFP-C1 and pEGFP-C1-Ar-SETD4 plasmids by using Attractene reagent (Qiagen). At 24 h posttransfection, the cells were harvested. Proteins were extracted from each sample by using TRIzol reagent (Invitrogen), according to the manufacturer's instructions. Each protein sample (25 μg) was subjected to SDS-PAGE and then transferred to a nitrocellulose membrane at 0.15 V/cm2 for 1 h in transfer buffer (25 mM Tris [pH 8.3], 192 mM glycine, and 10% methanol). After blocking in blocking buffer (Roche), the membrane was incubated with the primary antibody (diluted in blocking buffer) at 4°C overnight. After two washes in Tris-buffered saline–Tween (TBST) (20 mM Tris [pH 7.6], 137 mM NaCl) for 10 min, the membrane was incubated with horseradish peroxidase-conjugated goat anti-rabbit IgG (diluted 1:25,000) or goat anti-mouse IgG (diluted 1:25,000) (Roche) for 40 min. After washing, detection was performed by using the BM chemiluminescence Western blot kit (Roche).
BrdU incorporation assay.
DNA synthesis was determined by the incorporation of BrdU (Sangon Biotech). Prediapause embryos and nauplii were incubated in artificial seawater containing 1 mM BrdU for 24 h. Diapause and postdiapause embryos were decapsulated by using a 3% sodium hypochlorite solution before BrdU incorporation, fixed with 4% paraformaldehyde, paraffin embedded, and then cut into 6-μm sections by using a microtome (EM UC6; Leica). For the BrdU incorporation assay with MKN45 cells, the cells were incubated in medium containing 12 μM BrdU for 30 h and then fixed with 4% paraformaldehyde. The samples were incubated with a mouse monoclonal anti-BrdU antibody (1:750) (Sigma-Aldrich) at 4°C overnight and then incubated with an anti-mouse Alexa Fluor 647-conjugated (for Artemia samples) or Alexa Fluor 594-conjugated (for MKN45 cells) secondary antibody (1:200) (Invitrogen). The signals were detected by confocal microscopy.
In vitro HMT assays.
The open reading frame of Ar-SETD4 was cloned by using primers for GST–Ar-SETD4 (forward primer CGGGATCCATGGTCGACAGAATAGATC and reverse primer CCGCTCGAGTTAATTAGCATAAGAAAATTC) and then inserted into the pGEX-4T-1 plasmid. GST and GST–Ar-SETD4 were expressed in bacteria and purified by using GST-Sepharose beads for subsequent in vitro HMT assays. The in vitro HMT assays were modified versions of protocols described previously (67) and were performed by using 50 μl of methylase activity buffer (50 mM Tris [pH 8.5], 20 mM KCl, 10 mM MgCl2, 10 mM β-mercaptoethanol, and 250 mM sucrose) containing either 10 μg of core histones (mixture of H1, H3, H2B, H2A, and H4 from calf thymus) (catalogue number LS002544; Worthington) or 1 μg of synthesized H4 peptide with H4K20me0, H4K20me1, or H4K20me2 modifications (the sequence is described above) as the substrates, 10 mM Adomet (Sangon Biotech) was used as a methyl donor, and GST or GST–Ar-SETD4 was used as the enzyme. After incubation for 60 min at 30°C, the reactions were stopped by boiling in 2× SDS loading buffer. The reaction products were then examined by Western blot and dot blot analyses with modification-specific antibodies.
Mass spectrometry analyses.
The products of the in vitro methyltransferase reaction were separately subjected to a Nanospray Flex ionization source followed by tandem mass spectrometry (MS/MS) with the Thermo LTQ-Orbitrap Velos Pro system and an FTMS (Fourier transform ion cyclotron resonance mass analyzer) combined with a Thermo LTQ-Orbitrap Elite-equipped ion trap analyzer. The parameters for the FTMS were data collection at a scan resolution of 60,000 for full-scan MS, with positive polarity and profile as the data type, and we then proceeded to isolate the top 5 ions for MS/MS by collision-induced dissociation (CID) (isolation width of 1.0 m/z, 35% collision energy, 0.25 activation Q, and 10-ms activation time) and higher-energy collision dissociation (HCD). The scan range was set as a 300 m/z first mass and a 2,000 m/z last mass. The Seguest HT search engine configured with Proteome Discoverer 1.4 workflow software (Thermo Fischer Scientific, Bremen, Germany) was used for mass spectrometry data analyses. The search parameters included 10-ppm and 0.8-Da mass tolerances for MS and MS/MS, respectively. Furthermore, the peptides were extracted with high peptide confidence.
Immunofluorescence.
The open reading frames of Ar-SETD4 (C1-ArSETD4 primers [sequences are given above]) and Hs-SETD4 (forward primer CCGCTCGAGCTATGCAGAAAGGAAAAGG and reverse primer CGGGATCCTCAGGTAAAAGCTGTTTG) were cloned and then inserted into the pEGFP-C1 plasmid. MKN45 or HT1080 cells were plated onto 6-well slides at a density of 1 × 105 cells per well. The following day, the cells were transfected with the pEGFP-C1 and pEGFP-C1-Ar-SETD4 plasmids by using Attractene reagent (Qiagen). At 24 h posttransfection, the cells were fixed with 4% paraformaldehyde or frozen in 100% methanol and then permeabilized with 0.2% Triton X-100. The samples were incubated with the appropriate antibodies, and the nuclei were counterstained with 4′,6-diamidino-2-phenylindole (DAPI) (Sangon Biotech). All confocal images were collected by using a Zeiss LSM 510 or Zeiss 5 Live confocal microscope equipped with a 63×, 1.4-numerical-aperture (NA) objective lens. Excitation at 488 nm and observation at 505 to 550 nm were used to detect Alexa Fluor 488 staining (Invitrogen), excitation at 594 nm and observation at 617 nm were used to detect Alexa Fluor 594 staining (Invitrogen), and excitation at 650 nm and observation at 665 nm were used to detect Alexa Fluor 647 staining (Invitrogen).
Accession number(s).
The nucleotide sequence of Ar-SETD4 and its peptide was submitted to GenBank under accession number KP749757.
Supplementary Material
ACKNOWLEDGMENTS
We thank Zhiyuan Shen (Department of Radiation Oncology, Rutgers Robert Wood Johnson Medical School, Rutgers, The State University of New Jersey, New Brunswick, NJ) and Binghui Shen (College of Life Sciences, Zhejiang University, Hangzhou, China) for insightful comments and suggestions. We are grateful to She-Long Zhang (College of Life Sciences, Zhejiang University, Hangzhou, China) for technical assistance with the confocal microscope. We are also grateful to the members of Core Facilities of Zhejiang University School of Medicine (Hangzhou, China) for technical assistance with flow cytometry.
This work was supported by the National Natural Science Foundation of China (grants 41476106 and 31470437) and National Major Research and Development Projects (2016YFA0101201).
We declare no conflicts of interest.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/MCB.00453-16.
REFERENCES
- 1.Coller HA, Sang L, Roberts JM. 2006. A new description of cellular quiescence. PLoS Biol 4:e83. doi: 10.1371/journal.pbio.0040083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yao G. 2014. Modelling mammalian cellular quiescence. Interface Focus 4:20130074. doi: 10.1098/rsfs.2013.0074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nakamura-Ishizu A, Takizawa H, Suda T. 2014. The analysis, roles and regulation of quiescence in hematopoietic stem cells. Development 141:4656–4666. doi: 10.1242/dev.106575. [DOI] [PubMed] [Google Scholar]
- 4.Cheung TH, Rando TA. 2013. Molecular regulation of stem cell quiescence. Nat Rev Mol Cell Biol 14:329–340. doi: 10.1038/nrm3591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Orford KW, Scadden DT. 2008. Deconstructing stem cell self-renewal: genetic insights into cell-cycle regulation. Nat Rev Genet 9:115–128. doi: 10.1038/nrg2269. [DOI] [PubMed] [Google Scholar]
- 6.Tothova Z, Kollipara R, Huntly BJ, Lee BH, Castrillon DH, Cullen DE, McDowell EP, Lazo-Kallanian S, Williams IR, Sears C, Armstrong SA, Passegué E, DePinho RA, Gilliland DG. 2007. FoxOs are critical mediators of hematopoietic stem cell resistance to physiologic oxidative stress. Cell 128:325–339. doi: 10.1016/j.cell.2007.01.003. [DOI] [PubMed] [Google Scholar]
- 7.Shan T, Zhang P, Liang X, Bi P, Yue F, Kuang S. 2014. Lkb1 is indispensable for skeletal muscle development, regeneration, and satellite cell homeostasis. Stem Cells 32:2893–2907. doi: 10.1002/stem.1788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhao M, Perry JM, Marshall H, Venkatraman A, Qian P, He XC, Ahamed J, Li L. 2014. Megakaryocytes maintain homeostatic quiescence and promote post-injury regeneration of hematopoietic stem cells. Nat Med 20:1321–1326. doi: 10.1038/nm.3706. [DOI] [PubMed] [Google Scholar]
- 9.Janzen V, Forkert R, Fleming HE, Saito Y, Waring MT, Dombkowski DM, Cheng T, DePinho RA, Sharpless NE, Scadden DT. 2006. Stem-cell ageing modified by the cyclin-dependent kinase inhibitor p16INK4a. Nature 443:421–426. [DOI] [PubMed] [Google Scholar]
- 10.Rando TA. 2006. Stem cells, ageing and the quest for immortality. Nature 441:1080–1086. doi: 10.1038/nature04958. [DOI] [PubMed] [Google Scholar]
- 11.Fitzsimons CP, van Bodegraven E, Schouten M, Lardenoije R, Kompotis K, Kenis G, van den Hurk M, Boks MP, Biojone C, Joca S, Steinbusch HW, Lunnon K, Mastroeni DF, Mill J, Lucassen PJ, Coleman PD, van den Hove DL, Rutten BP. 2014. Epigenetic regulation of adult neural stem cells: implications for Alzheimer's disease. Mol Neurodegener 9:25. doi: 10.1186/1750-1326-9-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Segalés J, Perdiguero E, Muñoz-Cánoves P. 21 October 2014. Epigenetic control of adult skeletal muscle stem cell functions. FEBS J doi: 10.1111/febs.13065. [DOI] [PubMed] [Google Scholar]
- 13.Liu L, Cheung TH, Charville GW, Hurgo BM, Leavitt T, Shih J, Brunet A, Rando TA. 2013. Chromatin modifications as determinants of muscle stem cell quiescence and chronological aging. Cell Rep 4:189–204. doi: 10.1016/j.celrep.2013.05.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lachner M, O'Sullivan RJ, Jenuwein T. 2003. An epigenetic road map for histone lysine methylation. J Cell Sci 116:2117–2124. doi: 10.1242/jcs.00493. [DOI] [PubMed] [Google Scholar]
- 15.Black JC, Van Rechem C, Whetstine JR. 2012. Histone lysine methylation dynamics: establishment, regulation, and biological impact. Mol Cell 48:491–507. doi: 10.1016/j.molcel.2012.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang Y, Reinberg D. 2001. Transcription regulation by histone methylation: interplay between different covalent modifications of the core histone tails. Genes Dev 15:2343–2360. doi: 10.1101/gad.927301. [DOI] [PubMed] [Google Scholar]
- 17.Evertts AG, Manning AL, Wang X, Dyson NJ, Garcia BA, Coller HA. 2013. H4K20 methylation regulates quiescence and chromatin compaction. Mol Biol Cell 24:3025–3037. doi: 10.1091/mbc.E12-07-0529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Feng Q, Wang H, Ng HH, Erdjument-Bromage H, Tempst P, Struhl K, Zhang Y. 2002. Methylation of H3-lysine 79 is mediated by a new family of HMTases without a SET domain. Curr Biol 12:1052–1058. doi: 10.1016/S0960-9822(02)00901-6. [DOI] [PubMed] [Google Scholar]
- 19.Nguyen AT, Zhang Y. 2011. The diverse functions of DOT1 and H3K79 methylation. Genes Dev 25:1345–1358. doi: 10.1101/gad.2057811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Alvarez-Venegas R, Avramova Z. 2002. SET-domain proteins of the Su (var) 3-9, E (z) and trithorax families. Gene 285:25–37. doi: 10.1016/S0378-1119(02)00401-8. [DOI] [PubMed] [Google Scholar]
- 21.Qian C, Zhou MM. 2006. SET domain protein lysine methyltransferases: structure, specificity and catalysis. Cell Mol Life Sci 63:2755–2763. doi: 10.1007/s00018-006-6274-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Trievel RC, Beach BM, Dirk LM, Houtz RL, Hurley JH. 2002. Structure and catalytic mechanism of a SET domain protein methyltransferase. Cell 111:91–103. doi: 10.1016/S0092-8674(02)01000-0. [DOI] [PubMed] [Google Scholar]
- 23.Xiao B, Wilson JR, Gamblin SJ. 2003. SET domains and histone methylation. Curr Opin Struct Biol 13:699–705. doi: 10.1016/j.sbi.2003.10.003. [DOI] [PubMed] [Google Scholar]
- 24.Zhang L, Ma H. 2012. Complex evolutionary history and diverse domain organization of SET proteins suggest divergent regulatory interactions. New Phytol 195:248–263. doi: 10.1111/j.1469-8137.2012.04143.x. [DOI] [PubMed] [Google Scholar]
- 25.Trievel RC, Flynn EM, Houtz RL, Hurley JH. 2003. Mechanism of multiple lysine methylation by the SET domain enzyme Rubisco LSMT. Nat Struct Biol 10:545–552. doi: 10.1038/nsb946. [DOI] [PubMed] [Google Scholar]
- 26.Mishra BP, Zaffuto KM, Artinger EL, Org T, Mikkola HK, Cheng C, Djabali M, Ernst P. 2014. The histone methyltransferase activity of MLL1 is dispensable for hematopoiesis and leukemogenesis. Cell Rep 7:1239–1247. doi: 10.1016/j.celrep.2014.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schotta G, Lachner M, Sarma K, Ebert A, Sengupta R, Reuter G, Reinberg D, Jenuwein T. 2004. A silencing pathway to induce H3-K9 and H4-K20 trimethylation at constitutive heterochromatin. Genes Dev 18:1251–1262. doi: 10.1101/gad.300704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ezhkova E, Lien WH, Stokes N, Pasolli HA, Silva JM, Fuchs E. 2011. EZH1 and EZH2 cogovern histone H3K27 trimethylation and are essential for hair follicle homeostasis and wound repair. Genes Dev 25:485–498. doi: 10.1101/gad.2019811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Eom GH, Kim KB, Kim JH, Kim JY, Kim JR, Kee HJ, Kim DW, Choe N, Park HJ, Son HJ, Choi SY, Kook H, Seo SB. 2011. Histone methyltransferase SETD3 regulates muscle differentiation. J Biol Chem 286:34733–34742. doi: 10.1074/jbc.M110.203307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nguyen AT, He J, Taranova O, Zhang Y. 2011. Essential role of DOT1L in maintaining normal adult hematopoiesis. Cell Res 21:1370–1373. doi: 10.1038/cr.2011.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Feng Y, Yang Y, Ortega MM, Copeland JN, Zhang M, Jacob JB, Fields TA, Vivian JL, Fields PE. 2010. Early mammalian erythropoiesis requires the Dot1L methyltransferase. Blood 116:4483–4491. doi: 10.1182/blood-2010-03-276501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bierhoff H, Dammert MA, Brocks D, Dambacher S, Schotta G, Grummt I. 2014. Quiescence-induced LncRNAs trigger H4K20 trimethylation and transcriptional silencing. Mol Cell 54:675–682. doi: 10.1016/j.molcel.2014.03.032. [DOI] [PubMed] [Google Scholar]
- 33.Sleger H. 1991. Enzyme activities through development: a synthesis of the activity and control of the various enzymes as the embryo matures, p 37–73. In Browne RA, Sorgeloos P, Trotman C (ed), Artemia biology. CRC Press, Boca Raton, FL. [Google Scholar]
- 34.Clegg JS, Trotman CAN. 2002. Physiological and biochemical aspects of Artemia ecology, p 129–170. In Abatzopoulos THJ, Beardmore JA, Clegg JS, Sorgeloos P (ed), Artemia, basic and applied biology. Kluwer Academic Publishers, Dordrecht, The Netherlands. [Google Scholar]
- 35.MacRae TH. 2010. Gene expression, metabolic regulation and stress tolerance during diapauses. Cell Mol Life Sci 67:2405–2424. doi: 10.1007/s00018-010-0311-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Drinkwater LE, Clegg JS. 1991. Experimental biology of cyst diapause, p 93–117. In Browne RA, Sorgeloos P, Trotman C (ed), Artemia biology. CRC Press, Boca Raton, FL. [Google Scholar]
- 37.Mateescu B, England P, Halgand F, Yaniv M, Muchardt C. 2004. Tethering of hp1 proteins to chromatin is relieved by phosphoacetylation of histone h3. EMBO Rep 5:490–496. doi: 10.1038/sj.embor.7400139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Beck TN, Kaczmar J, Handorf E, Nikonova A, Dubyk C, Peri S, Lango M, Ridge JA, Serebriiskii IG, Burtness B, Golemis EA, Mehra R. 2015. Phospho-t356rb1 predicts survival in HPV-negative squamous cell carcinoma of the head and neck. Oncotarget 6:18863–18874. doi: 10.18632/oncotarget.4321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nordlund P, Reichard P. 2006. Ribonucleotide reductases. Annu Rev Biochem 75:681–706. doi: 10.1146/annurev.biochem.75.103004.142443. [DOI] [PubMed] [Google Scholar]
- 40.Pontarin G, Ferraro P, Håkansson P, Thelander L, Reichard P, Bianchi V. 2007. p53R2-dependent ribonucleotide reduction provides deoxyribonucleotides in quiescent human fibroblasts in the absence of induced DNA damage. J Biol Chem 282:16820–16828. doi: 10.1074/jbc.M701310200. [DOI] [PubMed] [Google Scholar]
- 41.Dai JQ, Zhu XJ, Liu FQ, Xiang JH, Nagasawa H, Yang WJ. 2008. Involvement of p90 ribosomal S6 kinase in termination of cell cycle arrest during development of Artemia-encysted embryos. J Biol Chem 283:1705–1712. doi: 10.1074/jbc.M707853200. [DOI] [PubMed] [Google Scholar]
- 42.Faria JA, Corrêa NC, de Andrade C, de Angelis Campos AC, Dos Santos Samuel de Almeida R, Rodrigues TS, de Goes AM, Gomes DA, Silva FP. 2013. SET domain-containing protein 4 (SETD4) is a newly identified cytosolic and nuclear lysine methyltransferase involved in breast cancer cell proliferation. J Cancer Sci Ther 5:58–65. [PMC free article] [PubMed] [Google Scholar]
- 43.Sun Y, Mansour M, Crack JA, Gass GL, MacRae TH. 2004. Oligomerization, chaperone activity, and nuclear localization of p26, a small heat shock protein from Artemia franciscana. J Biol Chem 279:39999–40006. doi: 10.1074/jbc.M406999200. [DOI] [PubMed] [Google Scholar]
- 44.Chen T, Villeneuve TS, Garant KA, Amons R, MacRae TH. 2007. Functional characterization of artemin, a ferritin homolog synthesized in Artemia embryos during encystment and diapause. FEBS J 274:1093–1101. doi: 10.1111/j.1742-4658.2007.05659.x. [DOI] [PubMed] [Google Scholar]
- 45.Duan RB, Zhang L, Chen DF, Yang F, Yang JS, Yang WJ. 2014. Two p90 ribosomal S6 kinase isoforms are involved in the regulation of mitotic and meiotic arrest in Artemia. J Biol Chem 289:16006–16015. doi: 10.1074/jbc.M114.553370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhu XJ, Dai JQ, Tan X, Zhao Y, Yang WJ. 2009. Activation of an AMP-activated protein kinase is involved in post-diapause development of Artemia franciscana encysted embryos. BMC Dev Biol 9:21. doi: 10.1186/1471-213X-9-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li R, Chen DF, Zhou R, Jia SN, Yang JS, Clegg JS, Yang WJ. 2012. Involvement of polo-like kinase 1 (Plk1) in mitotic arrest by inhibition of mitogen-activated protein kinase-extracellular signal-regulated kinase-ribosomal S6 kinase 1 (MEK-ERK-RSK1) cascade. J Biol Chem 287:15923–15934. doi: 10.1074/jbc.M111.312413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhou R, Yang F, Chen DF, Sun YX, Yang JS, Yang WJ. 2013. Acetylation of chromatin-associated histone H3 lysine 56 inhibits the development of encysted Artemia embryos. PLoS One 8:e68374. doi: 10.1371/journal.pone.0068374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Del Rizzo PA, Trievel RC. 2011. Substrate and product specificities of SET domain methyltransferases. Epigenetics 6:1059–1067. doi: 10.4161/epi.6.9.16069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Marmorstein R. 2003. Structure of SET domain proteins: a new twist on histone methylation. Trends Biochem Sci 28:59–62. doi: 10.1016/S0968-0004(03)00007-0. [DOI] [PubMed] [Google Scholar]
- 51.Xiao B, Jing C, Wilson JR, Walker PA, Vasisht N, Kelly G, Howell S, Taylor IA, Blackburn GM, Gamblin SJ. 2003. Structure and catalytic mechanism of the human histone methyltransferase SET7/9. Nature 421:652–656. doi: 10.1038/nature01378. [DOI] [PubMed] [Google Scholar]
- 52.Zhang X, Yang Z, Khan SI, Horton JR, Tamaru H, Selker EU, Cheng X. 2003. Structural basis for the product specificity of histone lysine methyltransferases. Mol Cell 12:177–185. doi: 10.1016/S1097-2765(03)00224-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jørgensen S, Schotta G, Sørensen CS. 2013. Histone H4 lysine 20 methylation: key player in epigenetic regulation of genomic integrity. Nucleic Acids Res 41:2797–2806. doi: 10.1093/nar/gkt012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sanders SL, Portoso M, Mata J, Bähler J, Allshire RC, Kouzarides T. 2004. Methylation of histone H4 lysine 20 controls recruitment of Crb2 to sites of DNA damage. Cell 119:603–614. doi: 10.1016/j.cell.2004.11.009. [DOI] [PubMed] [Google Scholar]
- 55.Stender JD, Pascual G, Liu W, Kaikkonen MU, Do K, Spann NJ, Boutros M, Perrimon N, Rosenfeld MG, Glass CK. 2012. Control of proinflammatory gene programs by regulated trimethylation and demethylation of histone H4K20. Mol Cell 48:28–38. doi: 10.1016/j.molcel.2012.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Schotta G, Sengupta R, Kubicek S, Malin S, Kauer M, Callén E, Celeste A, Pagani M, Opravil S, De La Rosa-Velazquez IA, Espejo A, Bedford MT, Nussenzweig A, Busslinger M, Jenuwein T. 2008. A chromatin-wide transition to H4K20 monomethylation impairs genome integrity and programmed DNA rearrangements in the mouse. Genes Dev 22:2048–2061. doi: 10.1101/gad.476008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.van Leeuwen F, Gafken PR, Gottschling DE. 2002. Dot1p modulates silencing in yeast by methylation of the nucleosome core. Cell 109:745–756. doi: 10.1016/S0092-8674(02)00759-6. [DOI] [PubMed] [Google Scholar]
- 58.Steger DJ, Lefterova MI, Ying L, Stonestrom AJ, Schupp M, Zhuo D, Vakoc AL, Kim JE, Chen J, Lazar MA, Blobel GA, Vakoc CR. 2008. DOT1L/KMT4 recruitment and H3K79 methylation are ubiquitously coupled with gene transcription in mammalian cells. Mol Cell Biol 28:2825–2839. doi: 10.1128/MCB.02076-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhang L, Deng L, Chen F, Yao Y, Wu B, Wei L, Mo Q, Song Y. 2014. Inhibition of histone H3K79 methylation selectively inhibits proliferation, self-renewal and metastatic potential of breast cancer. Oncotarget 5:10665–10677. doi: 10.18632/oncotarget.2496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jones B, Su H, Bhat A, Lei H, Bajko J, Hevi S, Baltus GA, Kadam S, Zhai H, Valdez R, Gonzalo S, Zhang Y, Li E, Chen T. 2008. The histone H3K79 methyltransferase Dot1L is essential for mammalian development and heterochromatin structure. PLoS Genet 4:e1000190. doi: 10.1371/journal.pgen.1000190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sweet SM, Li M, Thomas PM, Durbin KR, Kelleher NL. 2010. Kinetics of re-establishing H3K79 methylation marks in global human chromatin. J Biol Chem 285:32778–32786. doi: 10.1074/jbc.M110.145094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mohan M, Herz HM, Takahashi YH, Lin C, Lai KC, Zhang Y, Washburn MP, Florens L, Shilatifard A. 2010. Linking H3K79 trimethylation to Wnt signaling through a novel Dot1-containing complex (DotCom). Genes Dev 24:574–589. doi: 10.1101/gad.1898410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Janzen CJ, Hake SB, Lowell JE, Cross GA. 2006. Selective di- or trimethylation of histone H3 lysine 76 by two DOT1 homologs is important for cell cycle regulation in Trypanosoma brucei. Mol Cell 23:497–507. doi: 10.1016/j.molcel.2006.06.027. [DOI] [PubMed] [Google Scholar]
- 64.Dai L, Chen DF, Liu YL, Zhao Y, Yang F, Yang JS, Yang WJ. 2011. Extracellular matrix peptides of Artemia cyst shell participate in protecting encysted embryos from extreme environments. PLoS One 6:e20187. doi: 10.1371/journal.pone.0020187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yodmuang S, Tirasophon W, Roshorm Y, Chinnirunvong W, Panyim S. 2006. YHV-protease dsRNA inhibits YHV replication in Penaeus monodon and prevents mortality. Biochem Biophys Res Commun 341:351–356. doi: 10.1016/j.bbrc.2005.12.186. [DOI] [PubMed] [Google Scholar]
- 66.Schmittgen TD, Livak KJ. 2008. Analyzing real-time PCR data by the comparative CT method. Nat Protoc 3:1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- 67.Rea S, Eisenhaber F, O'Carroll D, Strahl BD, Sun ZW, Schmid M, Opravil S, Mechtler K, Ponting CP, Allis CD, Jenuwein T. 2000. Regulation of chromatin structure by site-specific histone H3 methyltransferases. Nature 406:593–699. doi: 10.1038/35020506. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.