ABSTRACT
Lysobacter enzymogenes is a ubiquitous soil gammaproteobacterium that produces a broad-spectrum antifungal antibiotic, known as heat-stable antifungal factor (HSAF). To increase HSAF production for use against fungal crop diseases, it is important to understand how HSAF synthesis is regulated. To gain insights into transcriptional regulation of the HSAF synthesis gene cluster, we generated a library with deletion mutations in the genes predicted to encode response regulators of the two-component signaling systems in L. enzymogenes strain OH11. By quantifying HSAF production levels in the 45 constructed mutants, we identified two strains that produced significantly smaller amounts of HSAF. One of the mutations affected a gene encoding a conserved bacterial response regulator, PilR, which is commonly associated with type IV pilus synthesis. We determined that L. enzymogenes PilR regulates pilus synthesis and twitching motility via a traditional pathway, by binding to the pilA promoter and upregulating pilA expression. Regulation of HSAF production by PilR was found to be independent of pilus formation. We discovered that the pilR mutant contained significantly higher intracellular levels of the second messenger cyclic di-GMP (c-di-GMP) and that this was the inhibitory signal for HSAF production. Therefore, the type IV pilus regulator PilR in L. enzymogenes activates twitching motility while downregulating antibiotic HSAF production by increasing intracellular c-di-GMP levels. This study identifies a new role of a common pilus regulator in proteobacteria and provides guidance for increasing antifungal antibiotic production in L. enzymogenes.
IMPORTANCE PilR is a widespread response regulator of the two-component system known for regulating type IV pilus synthesis in proteobacteria. Here we report that, in the soil bacterium Lysobacter enzymogenes, PilR regulates pilus synthesis and twitching motility, as expected. Unexpectedly, PilR was also found to control intracellular levels of the second messenger c-di-GMP, which in turn inhibits production of the antifungal antibiotic HSAF. The coordinated production of type IV pili and antifungal antibiotics has not been observed previously.
KEYWORDS: antibiotics, HSAF, Lysobacter, PilR, type IV pili
INTRODUCTION
The genus Lysobacter, belonging to the family Xanthomonadaceae, is ubiquitous in the environment (1). Among more than 30 described Lysobacter species, Lysobacter enzymogenes is the best studied (2, 3). Two L. enzymogenes strains, C3 and OH11, produce antifungal antibiotics, which are applied to control crop fungal diseases (4–6). One antibiotic, i.e., heat-stable antifungal factor (HSAF), a polycyclic tetramate macrolactam with a distinct chemical structure, has broad-spectrum antifungal activity (7, 8). It is synthesized via a unique biosynthetic pathway, in which a hybrid polyketide synthase and a nonribosomal peptide synthetase, encoded by the lafB gene (originally described as hsaf pks/nrps), within the HSAF biosynthesis cluster catalyze the linkage of ornithine to two polyketides (9, 10). HSAF inhibits fungal pathogens by targeting sphingolipid biosynthesis, which is a distinct target, compared to the targets of other antifungal agents (11), thus making HSAF particularly attractive for antifungal control.
Understanding the mechanisms regulating HSAF biosynthesis in L. enzymogenes is important for the purpose of increasing antibiotic production. Some initial insights into HSAF regulation have been obtained; however, the regulatory picture is far from complete. We and our collaborators have shown that HSAF levels are increased when L. enzymogenes is grown in poorer medium, e.g., 0.1× tryptic soy broth (TSB), compared to regular 1× TSB (8, 11, 12). This observation suggests that HSAF synthesis depends on extracellular stimuli. In support of this hypothesis, two two-component systems (TCSs) that affect HSAF biosynthesis in L. enzymogenes have been identified (12–14). One of these TCSs, i.e., RpfC-RpfG, activates HSAF production in response to extracellular levels of the fatty acid signaling molecule diffusible signaling factor 3 (DSF3) (12, 13). Another member of a TCS family, PilG, which is an orphan response regulator (RR) protein, was found to negatively regulate HSAF biosynthesis in response to as yet unknown stimuli (14).
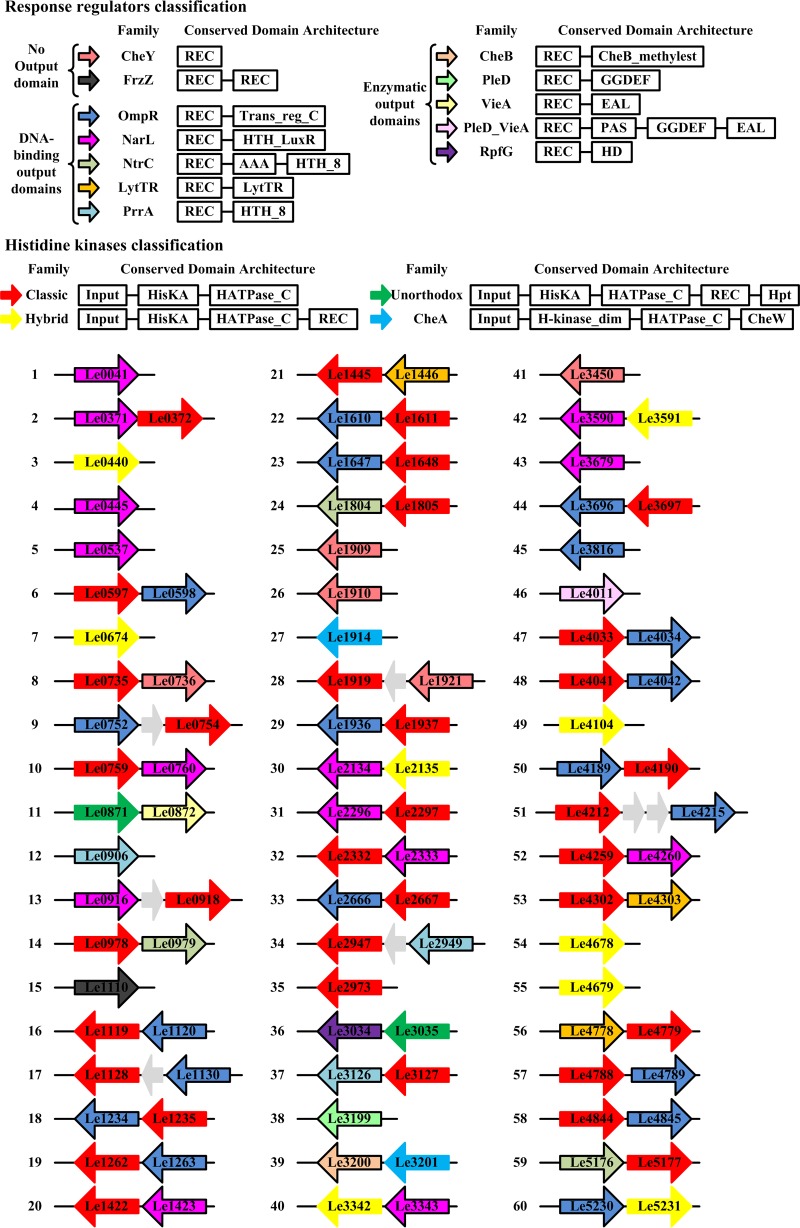
According to our genomic survey, strain L. enzymogenes OH11 encodes 48 putative histidine kinases (HKs) and 53 RRs (Fig. 1). We hypothesized that some of the remaining TCSs in L. enzymogenes might also be involved in regulating HSAF biosynthesis. To analyze the roles of these remaining TCSs, we decided to knock out each RR gene. As a result, we generated a genome-wide library of the in-frame RR deletion mutants in L. enzymogenes. By screening this deletion library, we unexpectedly found that PilR, the RR associated with regulation of type IV pilus (T4P) genes, is involved in regulating HSAF production. Here we show that PilR is a bona fide regulator of T4P synthesis and twitching motility in L. enzymogenes and that it regulates HSAF independently of T4P. Our findings suggest that the PilS-PilR TCS affects HSAF production via the cyclic di-GMP (c-di-GMP) signaling pathway, with c-di-GMP being a ubiquitous bacterial second messenger (15). In addition to the discovery of a new TCS involved in HSAF regulation and its unexpected role in controlling c-di-GMP signaling, our study has uncovered an antagonistic relationship between twitching motility and antibiotic production in L. enzymogenes.
FIG 1.
Identification of two-component systems (TCSs) in L. enzymogenes OH11. The histidine kinases (HKs) and response regulators (RRs) were classified according to the P2CS database (40). HKs and RRs belonging to various families are depicted in different colors.
RESULTS
Generation and analysis of the RR deletion library in L. enzymogenes.
To investigate the potential role of L. enzymogenes TCSs in HSAF production, we analyzed the genome of strain OH11 for the presence of TCSs. Using the Pfam database, we identified 48 putative HKs and 53 putative RRs, which represent 41 paired HK-RR TCSs and 19 orphan TCSs (7 HKs and 12 RRs) (Fig. 1; also see Table S1 in the supplemental material). As expected, the RRs fell into three categories, based on their output domains. Group I, which harbors RRs with only receiver domains and no identifiable output domains, has 6 representatives in L. enzymogenes. Group II contains 42 representatives, each of which has an N-terminal receiver domain linked to a C-terminal DNA-binding domain. Group III contains 5 RRs that possess N-terminal receiver domains attached to C-terminal domains with various enzymatic activities, most of which contain GGDEF, EAL, or HD-GYP domains involved in c-di-GMP synthesis or hydrolysis (16).
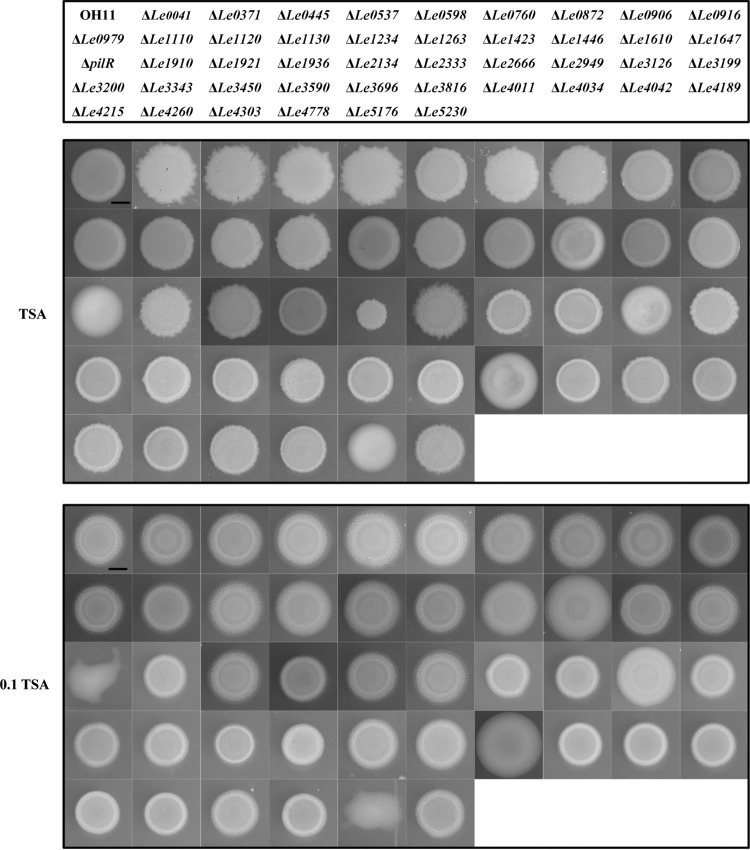
We generated a deletion mutant library with each of the remaining RR-encoding genes. Forty-five genes were individually deleted. Genes encoding six RRs (Le0736, Le0752, Le2296, Le3679, Le4789, and Le4845) could not to be deleted despite several attempts, which suggests that these RRs are potentially essential for bacterial survival under our experimental conditions. We compared the growth rates of the generated RR mutants in the medium for maximal HSAF production (0.1× TSB) and found that none of the mutants showed significant growth defects, compared to the wild-type strain, although several mutants had different colony morphologies, compared to the wild-type strain (Fig. 2; Fig. S1).
FIG 2.
L. enzymogenes RR deletion mutants displaying no significant growth defects in HSAF-inducing medium. TSA is the nutrient-rich medium used as the control, and 0.1 TSA is the HSAF-inducing medium. Scale bar, 2 mm. The growth curves of each mutant in liquid 0.1× TSB are shown in Fig. S1 in the supplemental material.
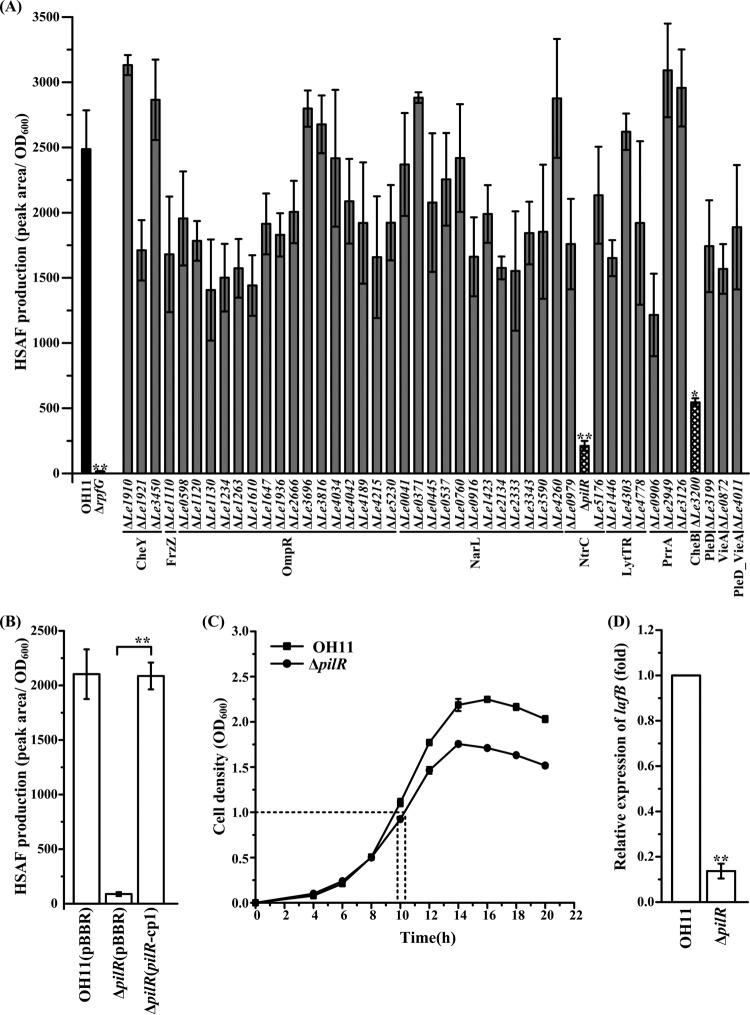
We quantified HSAF production in each RR mutant by high-performance liquid chromatography (HPLC). Two RR proteins (RpfG and PilG) were known to control HSAF levels, based on our earlier work (13, 14). In the present work, we used the rpfG deletion mutant (ΔrpfG) as a control and confirmed that HSAF levels were significantly decreased in the mutant. In addition, we found two new mutants, with mutations in the pilR and Le3200 genes, that exhibited significant reductions in HSAF levels, compared to the wild-type strain (Fig. 3A; Table S2).
FIG 3.
Quantification of HSAF produced by the L. enzymogenes RR mutants. (A) HSAF production, measured by HPLC and normalized to OD600 values. Data from triplicate experiments are shown. *, P < 0.05; **, P < 0.01. (B) Complementation of the ΔpilR mutant with the plasmid-borne pilR gene, rescuing HSAF production. Error bars represent standard deviations. ΔpilR(pBBR), the pilR mutant carrying an empty vector (pBBR1-MCS5); ΔpilR(pilR-cp1), the pilR mutant with plasmid pilR-pBBR, carrying the intact pilR gene. **, P < 0.01. (C) Representative growth curves of wild-type and ΔpilR strains in the HSAF-inducing medium (0.1× TSB). The dashed lines indicate the time points at which cells reached an OD600 of 1.0, when they were collected for qRT-PCR analysis, as shown in panel D. (D) qRT-PCR analysis of lafB mRNA levels. The lafB mRNA level in the wild-type strain OH11 was set as 1. **, P < 0.01.
Indirect activation of HSAF biosynthesis by PilR.
In this study, we focused on one of the newly found RRs involved in HSAF synthesis regulation, namely, PilR; Le3200 will be subject to a separate study. PilR belongs to the PilS-PilR TCS, which is conserved in proteobacteria and is involved in the regulation of T4P synthesis and twitching motility (17–19). This TCS also plays a role in bacterial attachment to surfaces and biofilm formation (20–23).
To ascertain the role of PilR in the regulation of HSAF biosynthesis, we complemented the pilR mutant with plasmid-borne pilR. The complemented strain produced similar amounts of HSAF, compared to the wild-type strain (Fig. 3B). To investigate the level at which PilR affects HSAF production, we measured the levels of the transcript of lafB (originally described as hsaf pks/nrps), the key HSAF biosynthetic gene (9). Results of the quantitative reverse transcription (qRT)-PCR analysis showed that lafB mRNA levels were significantly lower in the pilR mutant, compared to the wild-type strain (Fig. 3C and D), which suggests that PilR regulates HSAF biosynthesis at the level of gene expression.
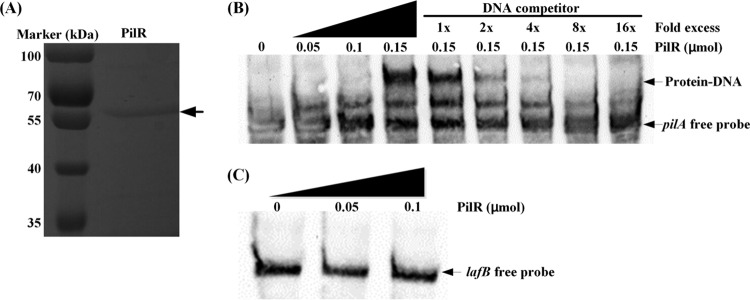
Next, we tested the ability of PilR to bind to the lafB promoter, using an electrophoretic mobility shift assay (EMSA). To this end, we overexpressed and purified PilR as a His6-fusion (Fig. 4A). As a positive control, we used the 541-bp promoter region upstream of L. enzymogenes pilA, which was chosen on the basis of the previously characterized PilR-regulated pilA promoter from Pseudomonas aeruginosa (24). The EMSA revealed the PilR-DNA complex with the L. enzymogenes pilA probe (Fig. 4B). This complex could be competitively inhibited by excess unlabeled pilA probe, which suggests that the interactions are specific (Fig. 4B). Under similar conditions, however, no protein-DNA complex was observed between PilR and the lafB promoter (Fig. 4C), which suggests that PilR affects HSAF biosynthesis gene expression indirectly.
FIG 4.
EMSA showing binding of PilR to the pilA promoter but not the lafB promoter. (A) SDS-PAGE analysis of the purified His-tagged PilR. (B) PilR binding to the pilA promoter in vitro. The unlabeled pilA probe provided in excess with respect to the labeled probe competitively inhibits formation of the PilR-DNA complex. Arrows indicate free DNA and PilR-DNA complexes. (C) No detection of PilR binding to the lafB promoter.
PilS-PilR TCS in L. enzymogenes regulation of T4P-driven twitching motility.
Since PilR was found to bind to the L. enzymogenes pilA promoter, we expected it to be involved in the formation of T4P-driven twitching motility (14, 25). Consistent with this expectation, the pilR mutant produced no motile cells that could migrate away from the margin of the colony, which is in contrast to the wild-type strain (Fig. 5A). The impairment of the pilR mutant in twitching motility could be rescued by the pilR-expressing plasmid but not the empty vector (Fig. 5A). Furthermore, the pilA mRNA levels were greatly downregulated in the ΔpilR strain, compared to the wild-type strain (Fig. 5B and C). These results demonstrate that L. enzymogenes PilR acts as a bona fide regulator of T4P synthesis and twitching motility.
FIG 5.
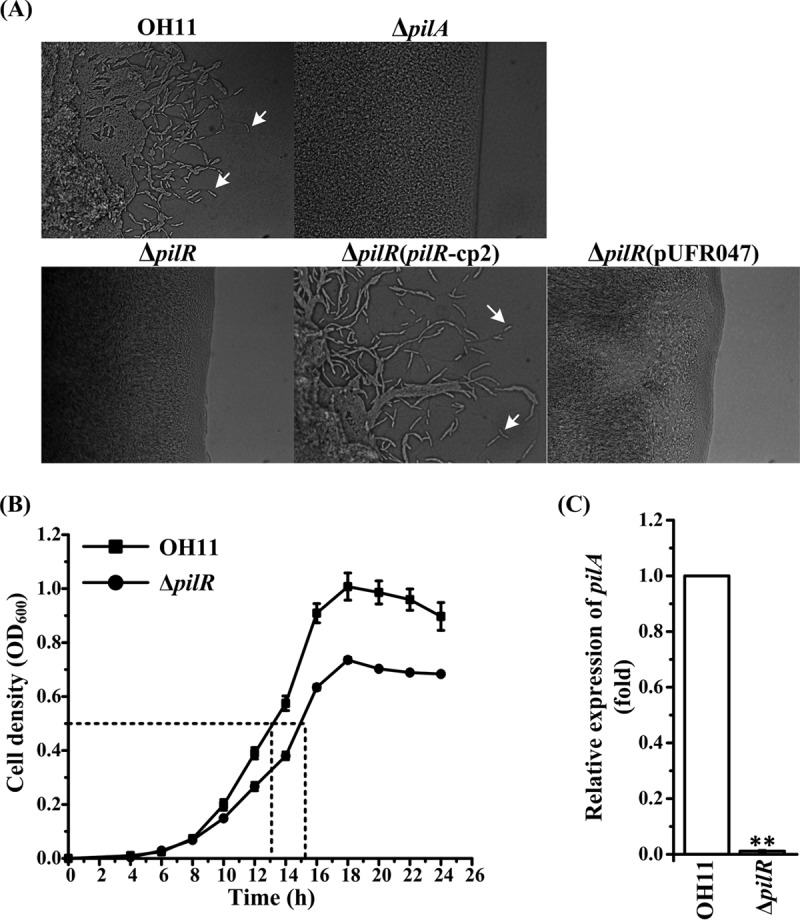
PilR involvement in regulating twitching motility in L. enzymogenes. (A) Indicated by arrows are motile cells at the margin of a colony, which is characteristic of twitching motility in L. enzymogenes (25). ΔpilR(pURF047), the ΔpilR mutant containing an empty vector; ΔpilR(pilR-cp2), the ΔpilR mutant with plasmid pilR-pUFR047, containing the intact pilR gene. ΔpilA, the strain lacking T4P and deficient in twitching motility (25), was used as a control. (B) Growth curves of the wild-type strain and the ΔpilR mutant in 0.05× TSB (the medium optimal for twitching motility) (25). The dashed lines indicate cells at an OD600 of 0.5, which were collected for qRT-PCR analysis, as shown in panel C. (C) qRT-PCR analysis of pilA mRNA in the wild-type and ΔpilR strains. The pilA mRNA level in the wild-type OH11 strain was set as 1. **, P < 0.01. Three replicates were used for each treatment, and the experiment was performed three times.
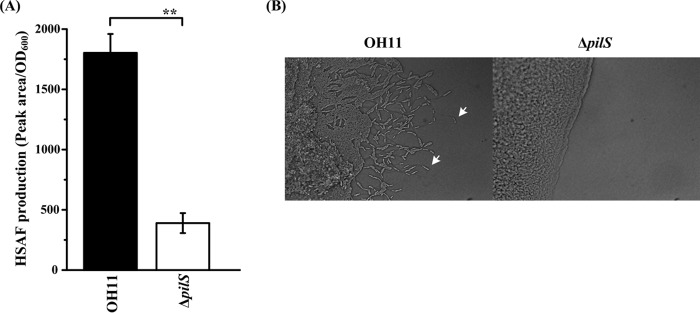
To test whether the L. enzymogenes PilS, the predicted HK of PilR, acts upstream of PilR in the same signal transduction cascade, we created an in-frame deletion in the pilS gene (Table 1). HSAF quantification and twitching motility tests showed that the pilS deletion caused a significant drop in HSAF production (Fig. 6A) and complete loss of twitching motility (Fig. 6B). These results are consistent with the key role of the PilS-PilR TCS in coordinate regulation of twitching motility and HSAF biosynthesis in L. enzymogenes.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Characteristicsa | Source |
|---|---|---|
| Strains | ||
| Lysobacter enzymogenes | ||
| Wild-type | ||
| OH11 | Wild-type; Kmr | 6 |
| In-frame deletion mutants | ||
| ΔLe0041 | In-frame deletion of Le0041; Kmr | This study |
| ΔLe0371 | In-frame deletion of Le0371; Kmr | This study |
| ΔLe0445 | In-frame deletion of Le0445; Kmr | This study |
| ΔLe0537 | In-frame deletion of Le0537; Kmr | This study |
| ΔLe0598 | In-frame deletion of Le0598; Kmr | This study |
| ΔLe0760 | In-frame deletion of Le0760; Kmr | This study |
| ΔLe0872 | In-frame deletion of Le0872; Kmr | This study |
| ΔLe0906 | In-frame deletion of Le0906; Kmr | This study |
| ΔLe0916 | In-frame deletion of Le0916; Kmr | This study |
| ΔLe0979 | In-frame deletion of Le0979; Kmr | This study |
| ΔLe1110 | In-frame deletion of Le1110; Kmr | This study |
| ΔLe1120 | In-frame deletion of Le1120; Kmr | This study |
| ΔLe1130 | In-frame deletion of Le1130; Kmr | This study |
| ΔLe1234 | In-frame deletion of Le1234; Kmr | This study |
| ΔLe1263 | In-frame deletion of Le1263; Kmr | This study |
| ΔLe1423 | In-frame deletion of Le1423; Kmr | This study |
| ΔLe1446 | In-frame deletion of Le1446; Kmr | This study |
| ΔLe1610 | In-frame deletion of Le1610; Kmr | This study |
| ΔLe1647 | In-frame deletion of Le1647; Kmr | This study |
| ΔpilR | In-frame deletion of pilR; Kmr | This study |
| ΔLe1910 | In-frame deletion of Le1910; Kmr | This study |
| ΔLe1921 | In-frame deletion of Le1921; Kmr | This study |
| ΔLe1936 | In-frame deletion of Le1936; Kmr | This study |
| ΔLe2134 | In-frame deletion of Le2134; Kmr | This study |
| ΔLe2333 | In-frame deletion of Le2333; Kmr | This study |
| ΔLe2666 | In-frame deletion of Le2666; Kmr | This study |
| ΔLe2949 | In-frame deletion of Le2949; Kmr | This study |
| ΔLe3126 | In-frame deletion of Le3126; Kmr | This study |
| ΔLe3199 | In-frame deletion of Le3199; Kmr | This study |
| ΔLe3200 | In-frame deletion of Le3200; Kmr | This study |
| ΔLe3343 | In-frame deletion of Le3343; Kmr | This study |
| ΔLe3450 | In-frame deletion of Le3450; Kmr | This study |
| ΔLe3590 | In-frame deletion of Le3590; Kmr | This study |
| ΔLe3696 | In-frame deletion of Le3696; Kmr | This study |
| ΔLe3816 | In-frame deletion of Le3816; Kmr | This study |
| ΔLe4011 | In-frame deletion of Le4011; Kmr | This study |
| ΔLe4034 | In-frame deletion of Le4034; Kmr | This study |
| ΔLe4042 | In-frame deletion of Le4042; Kmr | This study |
| ΔLe4189 | In-frame deletion of Le4189; Kmr | This study |
| ΔLe4215 | In-frame deletion of Le4215; Kmr | This study |
| ΔLe4260 | In-frame deletion of Le4260; Kmr | This study |
| ΔLe4303 | In-frame deletion of Le4303; Kmr | This study |
| ΔLe4778 | In-frame deletion of Le4778; Kmr | This study |
| ΔLe5176 | In-frame deletion of Le5176; Kmr | This study |
| ΔLe5230 | In-frame deletion of Le5230; Kmr | This study |
| ΔpilS | In-frame deletion of pilS; Kmr | This study |
| ΔrpfG | In-frame deletion of rpfG; Kmr | 13 |
| ΔpilR ΔpilA | In-frame deletion of pilR and pilA; Kmr | This study |
| ΔpilR ΔlafB | In-frame deletion of pilR and lafB; Kmr | This study |
| Complementary strains | ||
| OH11(pBBR) | OH11 harboring plasmid pBBR1-MCS5; Gmr, Kmr | This study |
| ΔpilR(pBBR) | ΔpilR harboring plasmid pBBR1-MCS5; Gmr, Kmr | This study |
| ΔpilR ΔpilA(pBBR) | ΔpilR ΔpilA harboring plasmid pBBR1-MCS5; Gmr, Kmr | This study |
| ΔpilR(pUFR047) | ΔpilR harboring plasmid pUFR047; Apr, Gmr, Kmr | This study |
| ΔpilR ΔlafB(pUFR047) | ΔpilR ΔlafB harboring plasmid pUFR047; Apr, Gmr, Kmr | This study |
| ΔpilR(pilR-cp1) | ΔpilR harboring plasmid pilR-pBBR; Gmr, Kmr | This study |
| ΔpilR(pilR-cp2) | ΔpilR harboring plasmid pilR-pUFR047; Apr, Gmr, Kmr | This study |
| ΔpilR ΔpilA(pilR-cp1) | ΔpilR ΔpilA harboring plasmid pilR-pBBR; Gmr, Kmr | This study |
| ΔpilR ΔlafB(pilR-cp2) | ΔpilR ΔlafB harboring plasmid pilR-pUFR047; Apr, Gmr, Kmr | This study |
| ΔpilR(slr-pBBR) | ΔpilR harboring plasmid slr-pBBR; Gmr, Kmr | This study |
| ΔpilR(yhjH-pBBR) | ΔpilR harboring plasmid yhjH-pBBR; Gmr, Kmr | This study |
| Escherichia coli | ||
| DH5α | Host strain for molecular cloning | Laboratory collection |
| BL21(DE3) | Host strain for protein expression | Laboratory collection |
| Plasmids | ||
| pEX18GM | Suicide vector with sacB gene; Gmr | 41 |
| pBBR1-MCS5 | Broad-host-range vector with Plac promoter | 42 |
| pUFR047 | Low-copy-number plasmid; Gmr, Apr | 43 |
| pET30a | Protein expression vector; Kmr | Novagen |
| slr-pBBR | pBBR1-MCS5 cloned with Gm promoter and 1,032-bp fragment containing intact slr1143; Gmr | This study |
| yhjH-pBBR | pBBR1-MCS5 cloned with Gm promoter and 768-bp fragment containing intact yhjH; Gmr | This study |
| 0041-pEX18 | pEX18GM with two flanking fragments of Le0041; Gmr | This study |
| 0371-pEX18 | pEX18GM with two flanking fragments of Le0041; Gmr | This study |
| 0445-pEX18 | pEX18GM with two flanking fragments of Le0445; Gmr | This study |
| 0537-pEX18 | pEX18GM with two flanking fragments of Le0537; Gmr | This study |
| 0598-pEX18 | pEX18GM with two flanking fragments of Le0598; Gmr | This study |
| 0760-pEX18 | pEX18GM with two flanking fragments of Le0760; Gmr | This study |
| 0872-pEX18 | pEX18GM with two flanking fragments of Le0872; Gmr | This study |
| 0906-pEX18 | pEX18GM with two flanking fragments of Le0906; Gmr | This study |
| 0916-pEX18 | pEX18GM with two flanking fragments of Le0916; Gmr | This study |
| 0979-pEX18 | pEX18GM with two flanking fragments of Le0979; Gmr | This study |
| 1110-pEX18 | pEX18GM with two flanking fragments of Le1110; Gmr | This study |
| 1120-pEX18 | pEX18GM with two flanking fragments of Le1120; Gmr | This study |
| 1130-pEX18 | pEX18GM with two flanking fragments of Le1130; Gmr | This study |
| 1234-pEX18 | pEX18GM with two flanking fragments of Le1234; Gmr | This study |
| 1263-pEX18 | pEX18GM with two flanking fragments of Le1263; Gmr | This study |
| 1423-pEX18 | pEX18GM with two flanking fragments of Le1423; Gmr | This study |
| 1446-pEX18 | pEX18GM with two flanking fragments of Le1446; Gmr | This study |
| 1610-pEX18 | pEX18GM with two flanking fragments of Le1610; Gmr | This study |
| 1647-pEX18 | pEX18GM with two flanking fragments of Le1647; Gmr | This study |
| pilR-pEX18 | pEX18GM with two flanking fragments of pilR; Gmr | This study |
| 1910-pEX18 | pEX18GM with two flanking fragments of Le1910; Gmr | This study |
| 1921-pEX18 | pEX18GM with two flanking fragments of Le1921; Gmr | This study |
| 1936-pEX18 | pEX18GM with two flanking fragments of Le1936; Gmr | This study |
| 2134-pEX18 | pEX18GM with two flanking fragments of Le2134; Gmr | This study |
| 2333-pEX18 | pEX18GM with two flanking fragments of Le2333; Gmr | This study |
| 2666-pEX18 | pEX18GM with two flanking fragments of Le2666; Gmr | This study |
| 2949-pEX18 | pEX18GM with two flanking fragments of Le2949; Gmr | This study |
| 3126-pEX18 | pEX18GM with two flanking fragments of Le3126; Gmr | This study |
| 3199-pEX18 | pEX18GM with two flanking fragments of Le3199; Gmr | This study |
| 3200-pEX18 | pEX18GM with two flanking fragments of Le3200; Gmr | This study |
| 3343-pEX18 | pEX18GM with two flanking fragments of Le3343; Gmr | This study |
| 3450-pEX18 | pEX18GM with two flanking fragments of Le3450; Gmr | This study |
| 3590-pEX18 | pEX18GM with two flanking fragments of Le3590; Gmr | This study |
| 3696-pEX18 | pEX18GM with two flanking fragments of Le3696; Gmr | This study |
| 3816-pEX18 | pEX18GM with two flanking fragments of Le3816; Gmr | This study |
| 4011-pEX18 | pEX18GM with two flanking fragments of Le4011; Gmr | This study |
| 4034-pEX18 | pEX18GM with two flanking fragments of Le4034; Gmr | This study |
| 4042-pEX18 | pEX18GM with two flanking fragments of Le4042; Gmr | This study |
| 4189-pEX18 | pEX18GM with two flanking fragments of Le4189; Gmr | This study |
| 4215-pEX18 | pEX18GM with two flanking fragments of Le4215; Gmr | This study |
| 4260-pEX18 | pEX18GM with two flanking fragments of Le4260; Gmr | This study |
| 4303-pEX18 | pEX18GM with two flanking fragments of Le4303; Gmr | This study |
| 4778-pEX18 | pEX18GM with two flanking fragments of Le4778; Gmr | This study |
| 5176-pEX18 | pEX18GM with two flanking fragments of Le5176; Gmr | This study |
| 5230-pEX18 | pEX18GM with two flanking fragments of Le5230; Gmr | This study |
| pilS-pEX18 | pEX18GM with two flanking fragments of pilS; Gmr | This study |
| pilR-pBBR | pBBR1-MCS5 cloned with 1,540-bp fragment containing intact pilR; Gmr | This study |
| pilR-pUFR047 | pUFR047 cloned with 1,540-bp fragment containing intact pilR; Apr, Gmr | This study |
| pilR-pET30(a) | pET30a cloned with fragment containing full-length pilR; Kmr | This study |
Kmr, kanamycin resistant; Gmr, gentamicin resistant; Apr, ampicillin resistant.
FIG 6.
L. enzymogenes PilS involvement in regulating HSAF biosynthesis (A) and twitching motility (B). Three technical replicates were used for each treatment, and the biological experiment was performed three times. Vertical bars represent standard errors. **, P < 0.01, relative to the wild-type OH11 strain. ΔpilS, the pilS deletion mutant. Arrows indicate motile cells at the margins of a colony.
PilR regulation of HSAF biosynthesis via a c-di-GMP signaling pathway.
Since PilR regulates HSAF gene transcription indirectly, we turned to the transcription factor Clp, which was identified by us earlier as a major contributor to HSAF gene expression (25). We hypothesized that PilR may act upstream of Clp. The Clp proteins from Xanthomonas species, which are closely related to Lysobacter, bind c-di-GMP and sense intracellular c-di-GMP levels (26). Therefore, it is possible that L. enzymogenes PilR affects either clp gene expression or c-di-GMP levels. Our proteomics data suggest that the levels of the Clp protein in the pilR mutant and the wild-type strain do not significantly differ (Fig. S2); therefore, we looked at the potential role of PilR in changing c-di-GMP levels.
Prior to exploring the PilR-c-di-GMP link, we wanted to test whether intracellular c-di-GMP levels play any role in HSAF production. To this end, we introduced into L. enzymogenes a potent diguanylate cyclase (c-di-GMP synthase), i.e., Slr1143 from Synechocystis sp., and a potent c-di-GMP phosphodiesterase, i.e., YhjH (PdeH) from Escherichia coli (27, 28). The slr1143 and yhjH genes were constitutively expressed from the broad-host-range vectors (Table 1). As shown in Fig. 7A, introduction of the phosphodiesterase gene yhjH into the pilR mutant caused a significant increase in the HSAF yield, while introduction of the diguanylate cyclase gene slr1143 slightly decreased the HSAF yield (Fig. 7A). These findings suggest that lower HSAF production in the pilR mutant may have been caused by elevated c-di-GMP levels. To test this prediction, we measured, by liquid chromatography-mass spectrometry (LC-MS), the intracellular c-di-GMP levels in the wild-type strain and the ΔpilR strain. In accord with our expectations, the intracellular c-di-GMP levels in the pilR mutant were significantly elevated, compared to the levels in the wild-type strain (Fig. 7B). To gain an additional piece of evidence indicating that elevated c-di-GMP levels are inhibitory to HSAF production, we introduced the plasmid-borne slr1143 gene into the wild-type strain and found that HSAF production was significantly decreased (Fig. S3). These results strongly suggest that elevated c-di-GMP levels are inhibitory for HSAF production and that PilR regulates HSAF biosynthesis via a c-di-GMP signaling pathway.
FIG 7.
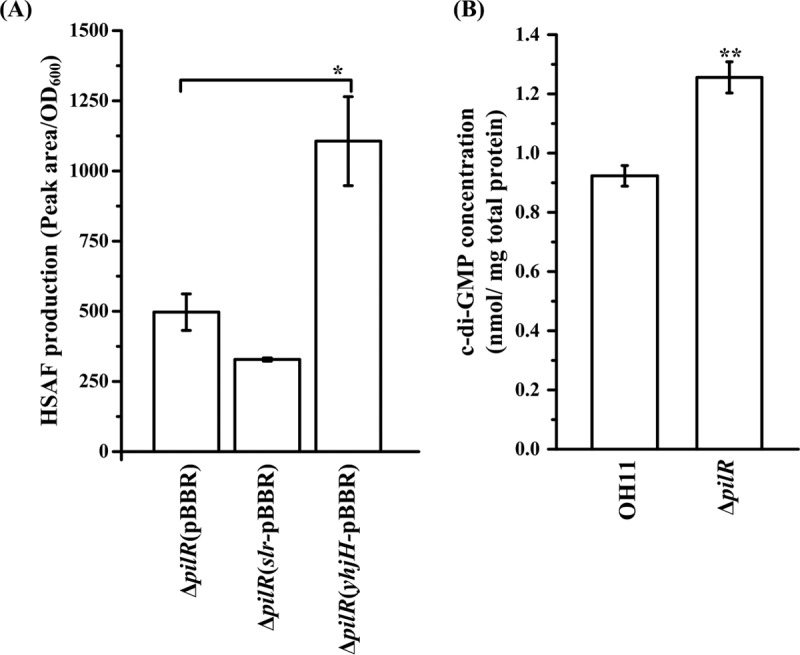
Intracellular c-di-GMP levels affecting HSAF production in the pilR mutant. (A) The c-di-GMP phosphodiesterase YhjH increased, while the diguanylate cyclase Slr1143 decreased, HSAF production in the ΔpilR mutant. ΔpilR(pBBR), ΔpilR(slr-pBBR), and ΔpilR(yhjH-pBBR) are pilR mutant strains containing an empty vector, the plasmid-borne slr1143, and yjhH, respectively. *, P < 0.05. (B) The pilR mutant had significantly elevated intracellular c-di-GMP levels. Three technical replicates were used for each treatment, and the biological experiment was performed three times. **, P < 0.01.
Because several PilR-type transcription regulators, e.g., FleQ from P. aeruginosa (29, 30) and XbmR from Xanthomonas citri (31), have been shown to bind c-di-GMP directly, we also tested, using microscale thermophoresis, the ability of L. enzymogenes PilR to bind c-di-GMP. However, we found no evidence of c-di-GMP binding (Fig. S4).
Independence of regulation of HSAF biosynthesis and twitching motility by PilR.
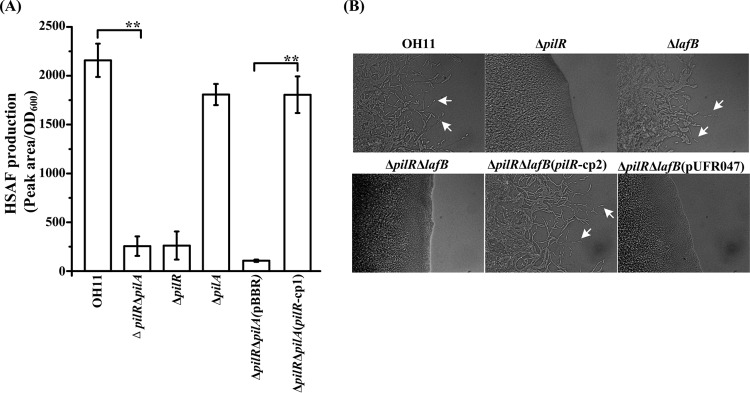
The results of the experiments described above suggest that L. enzymogenes PilR controls HSAF biosynthesis and twitching motility via two independent pathways. To verify this conclusion, we generated and tested two double mutants, i.e., ΔpilR ΔpilA and ΔpilR ΔlafB, which were impaired in motility and HSAF synthesis, respectively (Table 1). Then we introduced the plasmid-borne pilR gene into these double mutants and quantified HSAF production and motility. As shown in Fig. 8A, the ΔpilR ΔpilA double mutant lacking T4P was rescued with respect to HSAF production by the pilR-expressing plasmid, which shows that T4P are not involved in the PilR-dependent regulation of HSAF production. Similarly, twitching motility of the ΔpilR ΔlafB double mutant was fully restored by the pilR-expressing plasmid (Fig. 8B), which indicates that HSAF production does not affect motility. Furthermore, the ΔpilA mutant made with the wild-type genetic background produced HSAF levels similar to the levels of the wild-type strain (Fig. 8A), while the HSAF-deficient ΔlafB mutant was unaffected with respect to twitching motility, compared to the wild-type strain (Fig. 8B). Taken together, our results show that PilR coordinates T4P-driven twitching motility and HSAF production in L. enzymogenes via independent pathways (Fig. 9).
FIG 8.
Independent PilR regulation of HSAF production and twitching motility. (A) HSAF quantification in the wild-type strain and mutants. ΔpilRΔpilA(pBBR) and ΔpilRΔpilA(pilR-cp1) indicate the ΔpilR ΔpilA double mutant containing an empty vector (pBBR1-MCS5) and the plasmid pilR-pBBR, carrying the intact pilR gene, respectively. **, P < 0.01. (B) Twitching motility of the ΔpilR ΔlafB double mutant containing an empty vector (pURF047) or plasmid pilR-pUFR047, carrying the intact pilR gene. Arrows indicate motile cells at the margins of a colony.
FIG 9.
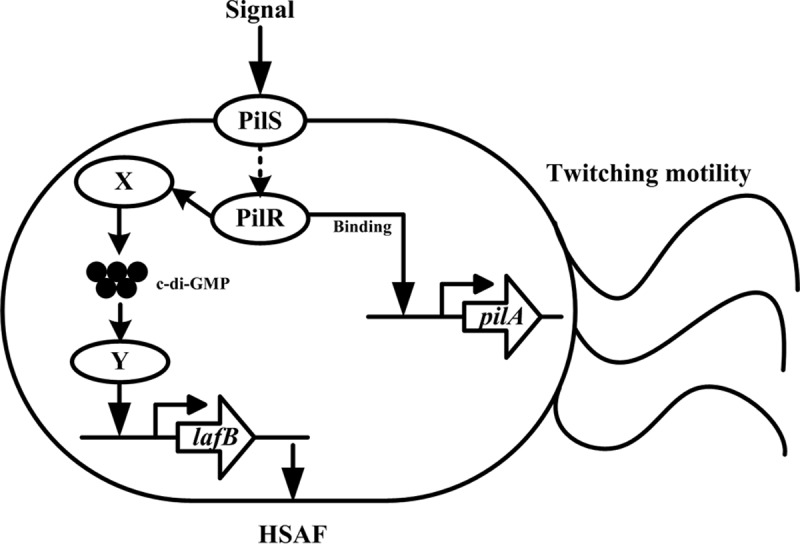
Proposed model of dual regulation by L. enzymogenes PilR. PilR forms a TCS with its cognate histidine kinase, PilS. Upon activation, PilR directly activates pilA transcription, which is required for T4P twitching motility. PilR affects the synthesis (or degradation) of c-di-GMP (system X) (indicated by an arrow), which in turn affects the activity of a transcription factor (Y) that regulates lafB gene expression and HSAF biosynthesis.
DISCUSSION
L. enzymogenes is a biocontrol bacterium that produces HSAF, a promising antifungal agent (7, 8). Because of its agricultural applications, L. enzymogenes is emerging as an important model for studying the regulation of HSAF biosynthesis. Previous studies identified several regulators of HSAF production (12, 14, 25, 32). To expand the range of potential factors affecting HSAF synthesis and to gain insights into the mechanisms of such regulation, we systematically deleted genes encoding RRs of TCSs in L. enzymogenes. We found two new regulators, i.e., PilR and Le3200, the latter of which will be characterized in a separate study.
Finding PilR as a regulator of HSAF production was unexpected, because PilR is a highly conserved RR of T4P synthesis and twitching motility in proteobacteria but it has not been known to affect secondary metabolite synthesis. In this study, we confirmed that, according to expectations, L. enzymogenes PilR functions as an activator of pilA expression and is required for twitching motility. We also showed that PilS, a cognate HK of PilR, acts in the expected manner. The PilR-mediated regulation of HSAF production turned out to be indirect and independent of the regulation of T4P genes. Because several NtrC-type RRs to which PilR belongs, including P. aeruginosa FleQ (29, 30) and X. citri XbmR (31), bind c-di-GMP directly, in this work we tested L. enzymogenes PilR for c-di-GMP binding; however, no binding was detected. Intriguingly, we found that the pilR deletion resulted in elevated intracellular c-di-GMP levels, which proved to be inhibitory for HSAF production. The latter conclusion was confirmed by our manipulation of c-di-GMP levels via heterologous diguanylate cyclase (Slr1143) and c-di-GMP phosphodiesterase (YhjH/PdeH). Our finding of c-di-GMP as an inhibitory stimulus for HSAF production suggests that a strain with a constitutive or induced system for decreasing c-di-GMP levels could show improved HSAF yields in industrial applications.
The mechanisms underlying the inhibitory role of c-di-GMP in HSAF gene expression remain to be explored. One candidate for mediating such regulation is Clp, whose requirement for HSAF gene regulation was noted by us earlier (25). Clp is a c-di-GMP-responsive transcription factor that has been characterized in Xanthomonas species (33) but not yet in Lysobacter. It remains to be determined whether L. enzymogenes Clp activates HSAF gene expression directly and whether it responds to intracellular c-di-GMP levels.
Which c-di-GMP signaling systems are controlled by PilR and in turn affect HSAF gene expression remain unknown. Like many environmental proteobacteria, L. enzymogenes contains a large set of enzymes (26 enzymes) that are potentially involved in c-di-GMP synthesis and hydrolysis (14). Why elevated c-di-GMP levels are inhibitory for HSAF production also remains unknown. Consistent with the notion that c-di-GMP signaling plays an important role in HSAF production is our earlier observation that HSAF production was lower in the rpfG mutant (13). The RpfG protein is an RR containing an HD-GYP domain, which is predicted, based on its similarity to RpfG from Xanthomonas (34), to have c-di-GMP phosphodiesterase activity. Our work also contributes to the growing realization of the importance of c-di-GMP signaling pathways for the production of secondary metabolites in diverse bacteria. Earlier studies with Streptomyces coelicolor and P. aeruginosa identified the engagement of c-di-GMP pathways in the regulation of pigment and antibiotic synthesis (35).
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
The complete list of bacterial strains and plasmids used in this study is presented in Table 1. L. enzymogenes OH11 (6) was used as the wild-type strain. The deletion mutants in the RR genes were made in the OH11 background and designated ΔLe# (the number sign indicates the gene number). Escherichia coli strains DH5α and BL21(DE3), which were used for plasmid maintenance and protein overexpression, respectively, were routinely grown at 37°C in Luria broth (LB) supplemented with appropriate antibiotics (25 μg/ml gentamicin [Gm] or 100 μg/ml ampicillin [Ap]) and 100 μg/ml 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal). L. enzymogenes was grown at 28°C in LB or TSB. When required, antibiotics were added at the final concentrations of 25 μg/ml kanamycin (Km) or 150 μg/ml Gm.
Bioinformatics analysis.
The putative HKs and RRs in L. enzymogenes strain OH11 (9) were identified by using the Pfam 28.0 database (36).
Genetic methods.
In-frame deletions in L. enzymogenes OH11 were generated via double-crossover homologous recombination, as described previously (37). The primers used are listed in Table 2. In brief, the flanking regions of each gene were amplified by PCR and cloned into the suicide vector pEX18Gm (Table 1). The deletion constructs were transformed into the wild-type strain OH11 or its derivatives by electroporation. The single-crossover recombinants were selected on LB plates supplemented with Km and Gm. The recombinants were then cultured for 6 h in liquid LB without antibiotics and subsequently were grown on LB plates containing 10% (wt/vol) sucrose and Km, for double-crossover enrichment. The sucrose-resistant, Km-resistant, Gm-sensitive colonies representing double-crossover recombinants were picked. In-frame gene deletions were verified by PCR using appropriate primers (Table 2).
TABLE 2.
Primers used in this study
| Primer | Sequencea | Purpose |
|---|---|---|
| In-frame deletion | ||
| 0041-F1 | GGGGTACCGGCTTCCCGTTTCACCCTG (KpnI) | To amplify 920-bp upstream homologue arm of Le0041 |
| 0041-R1 | CCCAAGCTTGATCCAGCGCAGTCCGTGA (HindIII) | |
| 0041-F2 | CCCAAGCTTCGGCGAAGGGGCGTTGAT (HindIII) | To amplify 646-bp downstream homologue arm of Le0041 |
| 0041-R2 | GCTCTAGATGGAGCGTGTCGGGCTGGTC (XbaI) | |
| 0371-F1 | GGGGTACCTGCTGATGCTCGCCCACG (KpnI) | To amplify 500-bp upstream homologue arm of Le0371 |
| 0371-R1 | CCCAAGCTTATGCCCGGCATCATCAGGT (HindIII) | |
| 0371-F2 | CCCAAGCTTGGGTTGATGCGCCGGAAGGA (HindIII) | To amplify 336-bp downstream homologue arm of Le0371 |
| 0371-R2 | GCTCTAGAGGTGCTGAGCCTGTCCAACGAG (XbaI) | |
| 0445-F1 | GGGGTACCCGGCATTTCGTGCGTAGCG (KpnI) | To amplify 997-bp upstream homologue arm of Le0445 |
| 0445-R1 | CCCAAGCTTCTCGGAAGCCACGGTCAAGG (HindIII) | |
| 0445-F2 | CCCAAGCTTTCAGGTCGAGCAGGAGCAGG (HindIII) | To amplify 1,132-bp downstream homologue arm of Le0445 |
| 0445-R2 | GCTCTAGACGGCGAGCAAGCCGTTCTT (XbaI) | |
| 0537-F1 | CCCGGTACCAGCAGCAACAGCCAGCCGAT (KpnI) | To amplify 787-bp upstream homologue arm of Le0537 |
| 0537-R1 | CCCTCTAGAACGGTCAGGGCGAAGGTCAT (XbaI) | |
| 0537-F2 | CCCTCTAGAATCACGAACACTCGAATCAT (XbaI) | To amplify 766-bp downstream homologue arm of Le0537 |
| 0537-R2 | CCCAAGCTTTTTTGAAGAATGGAGACGCG (HindIII) | |
| 0598-F1 | CCCGGTACCCTGCCGGTCCTGCACGGTCT (KpnI) | To amplify 592-bp upstream homologue arm of Le0598 |
| 0598-R1 | CCCTCTAGAGGCCTGGGTTACGTGGTCG (XbaI) | |
| 0598-F2 | CCCTCTAGAAGGTCGTAGGTGCCGTCGTG (XbaI) | To amplify 647-bp downstream homologue arm of Le0598 |
| 0598-R2 | CCCAAGCTTTTGCCATCGTGTTGTCCCCT (HindIII) | |
| 0752-F1 | CGGAATTCGGCATAGCCGATCACCTCG (EcoRI) | To amplify 502-bp upstream homologue arm of Le0752 |
| 0752-R1 | CCCAAGCTTTCGCGTCGTCTTCCACCAG (HindIII) | |
| 0752-F2 | CCCAAGCTTCAACACCATCGCGGTGTACG (HindIII) | To amplify 354-bp downstream homologue arm of Le0752 |
| 0752-R2 | GCTCTAGACCGCCTTGAACAGCACCCA (XbaI) | |
| 0760-F1 | CGGGGTACCAGAGTCGGTCGTCGTGGGCG (KpnI) | To amplify 346-bp upstream homologue arm of Le0760 |
| 0760-R1 | CCCAAGCTTCGAGGGCACGGTCAAGAATT (HindIII) | |
| 0760-F2 | CCCAAGCTTGGCATGCGGATGTCGCTGAGCACCA (HindIII) | To amplify 738-bp downstream homologue arm of Le0760 |
| 0760-R2 | GCTCTAGATCACCGCGATGATGCTGAACC (XbaI) | |
| 0872-F1 | GGGGTACCCGTAGGCGTCGGAGATGGTC (KpnI) | To amplify 546-bp upstream homologue arm of Le0872 |
| 0872-R1 | CCCAAGCTTGCGGCTGCGGCTGAAAGGCT (HindIII) | |
| 0872-F2 | CCCAAGCTTGGTTGGGGTTCTTCGTCGGC (HindIII) | To amplify 528-bp downstream homologue arm of Le0872 |
| 0872-R2 | GCTCTAGACTTCCCGTTCGCTCCCGTAC (XbaI) | |
| 0906-F1 | GGGGTACCTGCGGACAAGGTGGTGGACT (KpnI) | To amplify 378-bp upstream homologue arm of Le0906 |
| 0906-R1 | CCCAAGCTTCGTAGGTCTCGTCGTCGTCG (HindIII) | |
| 0906-F2 | CCCAAGCTTGGCGGCAACATCTCGGCGAC (HindIII) | To amplify 683-bp downstream homologue arm of Le0906 |
| 0906-R2 | GCTCTAGAATTCGCTCCTGTTCGCCGCC (XbaI) | |
| 0916-F1 | CGGGGTACCGGTGCGTGGAAAGGGTCAGG (KpnI) | To amplify 932-bp upstream homologue arm of Le0916 |
| 0916-R1 | CCCAAGCTTGTGCTCGGCATCAGCGTCAA (HindIII) | |
| 0916-F2 | CCCAAGCTTAGGTAGATGCGGGCTTCGC (HindIII) | To amplify 788-bp downstream homologue arm of Le0916 |
| 0916-R2 | GCTCTAGACGTGTTCGGGTTCACCTTGC (XbaI) | |
| 0979-F1 | GGGGTACCTTGTTCCTGCCGCTGGTGTC (KpnI) | To amplify 386-bp upstream homologue arm of Le0979 |
| 0979-R1 | CCCAAGCTTTTTCTCCAGCAACGCCAGCC (HindIII) | |
| 0979-F2 | CCCAAGCTTTGGGACGCAACACGCTCACG (HindIII) | To amplify 286-bp downstream homologue arm of Le0979 |
| 0979-R2 | GCTCTAGAATTATGGCGGCGATGCGGGC (XbaI) | |
| 1110-F1 | CGGAATTCGAGAACAACCCGCTGCCGAG (EcoRI) | To amplify 873-bp upstream homologue arm of Le1110 |
| 1110-R1 | CCCAAGCTTGTCCACCACCATCACCCGCG (HindIII) | |
| 1110-F2 | CCCAAGCTTCGAGGAGCGATTGCTGGTGA (HindIII) | To amplify 589-bp downstream homologue arm of Le1110 |
| 1110-R2 | GCTCTAGACCCACAGCAGGAACACCAATC (XbaI) | |
| 1120-F1 | GGGGTACCGCAGGAGCAGGAATCGCCGC (KpnI) | To amplify 725-bp upstream homologue arm of Le1120 |
| 1120-R1 | CCCAAGCTTTGTCGGGCAGTTCGTCGCGC (HindIII) | |
| 1120-F2 | CCCAAGCTTGCTACCGCCTCGCCGTGCCG (HindIII) | To amplify 856-bp downstream homologue arm of Le1120 |
| 1120-R2 | GCTCTAGAGCGAGGTGCGGCGGATGCGG (XbaI) | |
| 1130-F1 | GGGGTACCCCAGCACCACGCACGGCACC (KpnI) | To amplify 728-bp upstream homologue arm of Le1130 |
| 1130-R1 | CCCAAGCTTCGCAGCACCACCGAGGTCTG (HindIII) | |
| 1130-F2 | CCCAAGCTTAGACCGTGTGGGGACGAGGC (HindIII) | To amplify 406-bp downstream homologue arm of Le1130 |
| 1130-R2 | GCTCTAGAGGCAGGCGCAGGAACAGGTG (XbaI) | |
| 1234-F1 | CCCGGTACCTGTAGCCCCAGCCGTAGAAC (KpnI) | To amplify 677-bp upstream homologue arm of Le1234 |
| 1234-R1 | CCCTCTAGACGAAGCCCTGGTAACGCAGC (XbaI) | |
| 1234-F2 | CCCTCTAGACGGGCTACATGATCGAGGC (XbaI) | To amplify 749-bp downstream homologue arm of Le1234 |
| 1234-R2 | CCCAAGCTTCGGTTCAACGATTCCACGG (HindIII) | |
| 1263-F1 | CCCGGTACCGACGAGCACGCCGAACAAGG (KpnI) | To amplify 675-bp upstream homologue arm of Le1263 |
| 1263-R1 | CCCTCTAGACGAACGCCGAGGAAGTGCTG (XbaI) | |
| 1263-F2 | CCCTCTAGAGCAGGTCGGCTTGGTCTTCG (XbaI) | To amplify 594-bp downstream homologue arm of Le1263 |
| 1263-R2 | CCCAAGCTTATCAGCGAGCAGCCCAGGCG (HindIII) | |
| 1423-F1 | GGGGTACCGTGTCGGAGGAACGCAACCG (KpnI) | To amplify 828-bp upstream homologue arm of Le1423 |
| 1423-R1 | CCCAAGCTTCGTCGGGCATCATCAGGTCG (HindIII) | |
| 1423-F2 | CCCAAGCTTCAAGTCGCACCTGGGCAACG (HindIII) | To amplify 516-bp downstream homologue arm of Le1423 |
| 1423-R2 | GCTCTAGACACAGCAGGGAGCGGGAAAG (XbaI) | |
| 1446-F1 | GGGGTACCCAAGCCGCATTTCCTGTTCAA (KpnI) | To amplify 612-bp upstream homologue arm of Le1446 |
| 1446-R1 | CCCAAGCTTCGCCAACGGTTCGTCGTCA (HindIII) | |
| 1446-F2 | CCCAAGCTTCGAGGAATCGCTCAAGTCGC (HindIII) | To amplify 1,130-bp downstream homologue arm of Le1446 |
| 1446-R2 | GCTCTAGACTCATCGGCACCAGGGTCA (XbaI) | |
| 1610-F1 | CCCGGTACCTCGTTCCTGCGCTCGTGGTC (KpnI) | To amplify 676-bp upstream homologue arm of Le1610 |
| 1610-R1 | CCCTCTAGACAGCCAGTTCGGGTTCGTCTTC (XbaI) | |
| 1610-F2 | CCCTCTAGACGGGGTCGGCTATCTGCTGC (XbaI) | To amplify 487-bp downstream homologue arm of Le1610 |
| 1610-R2 | CCCAAGCTTCTTTCTCGGTGGTCAGGATC (HindIII) | |
| 1647-F1 | CCCGGTACCTAAAAAAAGTTCATCCGCCG (KpnI) | To amplify 535-bp upstream homologue arm of Le1647 |
| 1647-R1 | CCCTCTAGACGCATACCGCCTCCGAAAGC (XbaI) | |
| 1647-F2 | CCCTCTAGAACTACCACTTCGACCCGCAG (XbaI) | To amplify 681-bp downstream homologue arm of Le1647 |
| 1647-R2 | CCCAAGCTTCAGCATCAAGCCGAGGAAGC (HindIII) | |
| pilR-F1 | CCCGGTACCTAGGAGTGATTGGTTGCTTC (KpnI) | To amplify 688-bp upstream homologue arm of pilR |
| pilR-R1 | CCCTCTAGAGAGAACCGCTACAACAAGAC (XbaI) | |
| pilR-F2 | CCCTCTAGAGTTTCAGCCATTACGCCCTC (XbaI) | To amplify 756-bp downstream homologue arm of pilR |
| pilR-R2 | CCCAAGCTTGCCCACGAGATCCGCAATCC (HindIII) | |
| 1910-F1 | GGGGTACCATGGCAACGGAATCCTCAA (KpnI) | To amplify 901-bp upstream homologue arm of Le1910 |
| 1910-R1 | CCCAAGCTTCATCACCAGATCGGGCAACT (HindIII) | |
| 1910-F2 | CCCAAGCTTTGCTGGATAAGTGAACCGATGA (HindIII) | To amplify 471-bp downstream homologue arm of Le1910 |
| 1910-R2 | GCTCTAGAGACGAAATGGGCGTAGCG (XbaI) | |
| 1921-F1 | GGGGTACCCGGAACAACTGGAATCGCTC (KpnI) | To amplify 335-bp upstream homologue arm of Le1921 |
| 1921-R1 | CCCAAGCTTGCGATGCGTTGGCGGATCAC (HindIII) | |
| 1921-F2 | CCCAAGCTTCCCTGCTGGAGATGCTGCCTAC (HindIII) | To amplify 1,039-bp downstream homologue arm of Le1921 |
| 1921-R2 | GCTCTAGACGGGAAACGCCTGCAACA (XbaI) | |
| 1936-F1 | CGGAATTCAGGGGTGGTGTGTGATGGCC (EcoRI) | To amplify 249-bp upstream homologue arm of Le1936 |
| 1936-R1 | GCTCTAGAGCTGTTCTCCGTCGCCAACC (XbaI) | |
| 1936-F2 | GCTCTAGAGCCACCTCTACAACCTGCGC (XbaI) | To amplify 226-bp downstream homologue arm of Le1936 |
| 1936-R2 | CGAGCTCAGGAGGATGCCCAGCGTGAT (SacI) | |
| 2134-F1 | GGGGTACCCGCCAACACCTTCATCCCGC (KpnI) | To amplify 781-bp upstream homologue arm of Le2134 |
| 2134-R1 | CCCAAGCTTACACCAGCGAAGAGAAGCCC (HindIII) | |
| 2134-F2 | CCCAAGCTTACGGGGTCGGCGGGTTGAGT (HindIII) | To amplify 470-bp downstream homologue arm of Le2134 |
| 2134-R2 | GCTCTAGACTGCTCGATTACCGCCTGGG (XbaI) | |
| 2333-F1 | CGGAATTCCTTTGTCGGTGGTGGTGCTGAA (EcoRI) | To amplify 683-bp upstream homologue arm of Le2333 |
| 2333-R1 | CCCAAGCTTCGATGTCGGGTTCCAGGTTCA (HindIII) | |
| 2333-F2 | CCCAAGCTTGCAACCGGATCGAGGCGTAT (HindIII) | To amplify 533-bp downstream homologue arm of Le2333 |
| 2333-R2 | GCTCTAGAGGCGGAAGGTCGTAATGGAAGT (XbaI) | |
| 2666-F1 | CCCGGTACCCGCCCGCATCGCTTGATA (KpnI) | To amplify 899-bp upstream homologue arm of Le2666 |
| 2666-R1 | CCCTCTAGATCCTGGTCGGCAAACTGC (XbaI) | |
| 2666-F2 | CCCTCTAGACTTGCGGATCTGCGGTTCGT (XbaI) | To amplify 704-bp downstream homologue arm of Le2666 |
| 2666-R2 | CCCAAGCTTGAAACCATCCGCATCGAAGG (HindIII) | |
| 2949-F1 | GGGGTACCGGCGACGATGGGCTTGCT (KpnI) | To amplify 651-bp upstream homologue arm of Le2949 |
| 2949-R1 | CCCAAGCTTCGCCAGCACCTGGCAGAAC (HindIII) | |
| 2949-F2 | CCCAAGCTTGTGGACCGGCGCACCTTG (HindIII) | To amplify 940-bp downstream homologue arm of Le2949 |
| 2949-R2 | GCTCTAGAGAACGGGCGGACTTGATG (XbaI) | |
| 3126-F1 | GGGGTACCGGCTGGGTCGGGCTGGAATC (KpnI) | To amplify 329-bp upstream homologue arm of Le3126 |
| 3126-R1 | CCCAAGCTTGTTCGTCGTCCTCGTCGTCG (HindIII) | |
| 3126-F2 | CCCAAGCTTTCAAGACCTTGGAGTGGGAACG (HindIII) | To amplify 416-bp downstream homologue arm of Le3126 |
| 3126-R2 | GCTCTAGACTCGGTCGGTGTTCGTCCAT (XbaI) | |
| 3199-F1 | GGGGTACCACCACACCATCGCCCAGGAC (KpnI) | To amplify 178-bp upstream homologue arm of Le3199 |
| 3199-R1 | CCCAAGCTTGGGAGTGGGGAAGGAGGGTT (HindIII) | |
| 3199-F2 | CCCAAGCTTGCTGCTGATGCTGGATGTCG (HindIII) | To amplify 465-bp downstream homologue arm of Le3199 |
| 3199-R2 | GCTCTAGATGCGTTTCCTGGGTGTCTGT (XbaI) | |
| 3200-F1 | GGGGTACCGGAATGAACCACGCCACAGC (KpnI) | To amplify 557-bp upstream homologue arm of Le3200 |
| 3200-R1 | CCCAAGCTTCAGTCCTTCCAGCAACCGCG (HindIII) | |
| 3200-F2 | CCCAAGCTTTATTCGCCCAGACCCAGACC (HindIII) | To amplify 299-bp downstream homologue arm of Le3200 |
| 3200-R2 | GCTCTAGAGGTGGATGCGGTAGTGGTGC (XbaI) | |
| 3343-F1 | GGGGTACCGCCTGGACCGGATCGGGATT (KpnI) | To amplify 315-bp upstream homologue arm of Le3343 |
| 3343-R1 | CCCAAGCTTAGGTGCTGCGGCTGTTCGTC (HindIII) | |
| 3343-F2 | CCCAAGCTTTGCTCAGGCGGACAAAAGG (HindIII) | To amplify 966-bp downstream homologue arm of Le3343 |
| 3343-R2 | GCTCTAGAAGCAGACGAGGACAGCCCAT (XbaI) | |
| 3450-F1 | GGGGTACCGGCGTGTCCCTGCTCGGCAT (KpnI) | To amplify 360-bp upstream homologue arm of Le3450 |
| 3450-R1 | CCCAAGCTTCGTCTTCTGCGGTGAGGGCC (HindIII) | |
| 3450-F2 | CCCAAGCTTCCGTTCAGCGAGACCGACCT (HindIII) | To amplify 269-bp downstream homologue arm of Le3450 |
| 3450-R2 | GCTCTAGACAAAACGCTCCGCCGCCACT (XbaI) | |
| 3590-F1 | GGGGTACCGGAATCCTGTGCGGTCGTCTTG (KpnI) | To amplify 280-bp upstream homologue arm of Le3590 |
| 3590-R1 | GGGGTACCGGAATCCTGTGCGGTCGTCTTG (HindIII) | |
| 3590-F2 | CCCAAGCTTTCTGTCGCCGCAGCAGTTCC (HindIII) | To amplify 392-bp downstream homologue arm of Le3590 |
| 3590-R2 | GCTCTAGACCGCTGTCCGCAGGTTTGTC (XbaI) | |
| 3696-F1 | CCCGGTACCATCCCTGCCCCCATCGCTAC (KpnI) | To amplify 678-bp upstream homologue arm of Le3696 |
| 3696-R1 | CCCTCTAGAAGGATGTGGTCGCTGGGTTT (XbaI) | |
| 3696-F2 | CCCTCTAGAGCTACATCAAGACCGTGCGC (XbaI) | To amplify 605-bp downstream homologue arm of Le3696 |
| 3696-R2 | CCCAAGCTTCGCACAGCAGCAGCAACGCC (HindIII) | |
| 3816-F1 | GGGGTACCTCTGGTCGGAAGTGCTCG (KpnI) | To amplify 596-bp upstream homologue arm of Le3816 |
| 3816-R1 | CCCAAGCTTGGTGGACTGCTGAAATGGC (HindIII) | |
| 3816-F2 | CCCAAGCTTTGATCGGCTCGCTGACCAAC (HindIII) | To amplify 656-bp downstream homologue arm of Le3816 |
| 3816-R2 | GCTCTAGACAACGCACTCATGCTGCTTCAC (XbaI) | |
| 4011-F1 | GGGGTACCCGGTGAACTGCCGCTACTGC (KpnI) | To amplify 1,098-bp upstream homologue arm of Le4011 |
| 4011-R1 | CCCAAGCTTGCCTCCATCATCACCAGCACC (HindIII) | |
| 4011-F2 | CCCAAGCTTGCGGCTACATGGAAGACCTGA (HindIII) | To amplify 724-bp downstream homologue arm of Le4011 |
| 4011-R2 | GCTCTAGAGCGATGATGAGCGGCAACC (XbaI) | |
| 4034-F1 | CCCGGTACCAGACCAGGAAATACAGCGGC (KpnI) | To amplify 755-bp upstream homologue arm of Le4034 |
| 4034-R1 | CCCTCTAGAGCCAAATCCTCAGCCGCGAC (XbaI) | |
| 4034-F2 | CCCTCTAGAGCCGTGCTTGCCCAGGTAAT (XbaI) | To amplify 566-bp downstream homologue arm of Le4034 |
| 4034-R2 | CCCAAGCTTGCGGCGGGCTGCAAAAAAAT (HindIII) | |
| 4042-F1 | CCCGGTACCGACAGGTTGCTCGGGCTCAG (KpnI) | To amplify 685-bp upstream homologue arm of Le4042 |
| 4042-R1 | CCCTCTAGAATGGGCTATGTGCTGGAGAC (XbaI) | |
| 4042-F2 | CCCTCTAGAAGGTAGTCGGCGGTCTTGGC (XbaI) | To amplify 718-bp downstream homologue arm of Le4042 |
| 4042-R2 | CCCAAGCTTGGATGCCGAAACCGAAGCCG (HindIII) | |
| 4104-F1 | GGGGTACCCAGGGCGATGTAGGCGTTGC (KpnI) | To amplify 851-bp upstream homologue arm of Le4104 |
| 4104-R1 | CCCAAGCTTAAGGCTCGGCTGGTGGGGTC (HindIII) | |
| 4104-F2 | CCCAAGCTTAGGTGGCGGGCGAGACGATC (HindIII) | To amplify 269-bp downstream homologue arm of Le4104 |
| 4104-R2 | GCTCTAGAGGGAAACCGCCGAGCCAATC (XbaI) | |
| 4189-F1 | CCCGGTACCCCAAGAACAGCCTCACAGCG (KpnI) | To amplify 1,511-bp upstream homologue arm of Le4189 |
| 4189-R1 | CCCTCTAGACGCAGGGCAAAGGACACCAT (XbaI) | |
| 4189-F2 | CCCTCTAGATACCGCTTCTCCGCCTCGCT (XbaI) | To amplify 621-bp downstream homologue arm of Le4189 |
| 4189-R2 | CCCAAGCTTCAGCAGCCACAGGTTTTCCG (HindIII) | |
| 4215-F1 | CCCGGTACCATCAGCACGAAGCCCCAGCG (KpnI) | To amplify 611-bp upstream homologue arm of Le4215 |
| 4215-R1 | CCCTCTAGATGGGCTATTCGCTGGACAAC (XbaI) | |
| 4215-F2 | CCCTCTAGAATGGCGGATTCGTCTTCTAC (XbaI) | To amplify 1,065-bp downstream homologue arm of Le4215 |
| 4215-R2 | CCCAAGCTTTCTATTCGGGCTGCTTCAAC (HindIII) | |
| 4260-F1 | GGGGTACCATGCCGACGACCAGGAACA (KpnI) | To amplify 662-bp upstream homologue arm of Le4260 |
| 4260-R1 | CCCAAGCTTGAGGGGACGATCAAGAACCAC (HindIII) | |
| 4260-F2 | CCCAAGCTTCCTCCAGGCCGGACATCA (HindIII) | To amplify 857-bp downstream homologue arm of Le4260 |
| 4260-R2 | GCTCTAGAGGCTCAACGCCGAACTGC (XbaI) | |
| 4303-F1 | GGGGTACCGCCGCACTTCCTCTACAACACC (KpnI) | To amplify 659-bp upstream homologue arm of Le4303 |
| 4303-R1 | CCCAAGCTTGCGCAATGCCTCGACCAA (HindIII) | |
| 4303-F2 | CCCAAGCTTGCTGAGCGTGAGCCAGACCTT (HindIII) | To amplify 477-bp downstream homologue arm of Le4303 |
| 4303-R2 | GCTCTAGAGCGATGCGTTCGGTGATGC (XbaI) | |
| 4778-F1 | GGGGTACCTCAACGAGGACACCGAGCGC (KpnI) | To amplify 540-bp upstream homologue arm of Le4778 |
| 4778-R1 | CCCAAGCTTATCAGCACGCTCAACGGGCG (HindIII) | |
| 4778-F2 | CCCAAGCTTTCGGTGGAACTGGCGGTGGG (HindIII) | To amplify 619-bp downstream homologue arm of Le4778 |
| 4778-R2 | GCTCTAGACACCCATCCCGACGCCTACG (XbaI) | |
| 5176-F1 | GGGGTACCCTCGGAAGAACTGGGCAAGG (KpnI) | To amplify 859-bp upstream homologue arm of Le5176 |
| 5176-R1 | CCCAAGCTTGTGGTGTCGGCGGTGAAGTT (HindIII) | |
| 5176-F2 | CCCAAGCTTACCGCCTCAACACCATCCAG (HindIII) | To amplify 598-bp downstream homologue arm of Le5176 |
| 5176-R2 | GCTCTAGAGTGATAGAACAGCAGCGGCG (XbaI) | |
| 5230-F1 | GGGGTACCGGGCGATGGAAGCAAGGGTG (KpnI) | To amplify 1,257-bp upstream homologue arm of Le5230 |
| 5230-R1 | CCCAAGCTTGCCAGCAGGGCGAGAATGT (HindIII) | |
| 5230-F2 | CCCAAGCTTCCGGGGCTGATCAAGGCG (HindIII) | To amplify 1,070-bp downstream homologue arm of Le5230 |
| 5230-R2 | GCTCTAGAGCCTGCCTCGGGCTTGTC (XbaI) | |
| pilS-F1 | CCCGGTACCTAGGAGTGATTGGTTGCTTC (EcoRI) | To amplify 999-bp upstream homologue arm of pilS |
| pilS-R1 | CCCTCTAGAGAGAACCGCTACAACAAGAC (HindIII) | |
| pilS-F2 | CCCTCTAGAGTTTCAGCCATTACGCCCTC (HindIII) | To amplify 970-bp downstream homologue arm of pilS |
| pilS-R2 | CCCAAGCTTGCCCACGAGATCCGCAATCC (XbaI) | |
| 0445-F | TCCCACAACAGCCGACAGCC | To confirm mutant construction of ΔLe0445 |
| 0445-R | CCACCTTCACCCATCGTCCAAT | |
| 0537-F | ATCGCCGGATTCCGTTATG | To confirm mutant construction of ΔLe0537 |
| 0537-R | GGTATCGGTGATCGTGAGCC | |
| 0598-F | GCACCAGCAGGAACAGCAGC | To confirm mutant construction of ΔLe0598 |
| 0598-R | GGCTTTGTAACCGTGCGTATCG | |
| 0760-F | CGATGCGAAAGCGGAGATGG | To confirm mutant construction of ΔLe0760 |
| 0760-R | CGAACTGCTCGGCGACATCC | |
| 0906-F | GCAACCACAGGCATGGACACTT | To confirm mutant construction of ΔLe0906 |
| 0906-R | CACCTGATGCTGATCGGATTGC | |
| 0916-F | CGATGTCCGCTTGCGTATCAG | To confirm mutant construction of ΔLe0916 |
| 0916-R | CAACCAACAGTTCCCGCCCTAT | |
| 1120-F | TGCGGGAATGATCGAAACGG | To confirm mutant construction of ΔLe1120 |
| 1120-R | CCGAACAGGCCGAGCAGGAT | |
| 1130-F | TGAAGCGATTCGGGTCCAGC | To confirm mutant construction of ΔLe1130 |
| 1130-R | TGAGGTACAACCGCACCAGCA | |
| 1234-F | AGCCGTAGAACTTGCCCGACAC | To confirm mutant construction of ΔLe1234 |
| 1234-R | TGGACACGCGGTAGAACACCC | |
| 1446-F | CGTCCAAGACCGACTCCAGC | To confirm mutant construction of ΔLe1446 |
| 1446-R | TCACAGGTGCTTCAAGGTCTCG | |
| 1610-F | AGATGCTGGGCGAGCGTTTCC | To confirm mutant construction of ΔLe1610 |
| 1610-R | CGTCGCGGATCACGTACCACA | |
| 1647-F | CGTCGCACAAGCACAAGAAGC | To confirm mutant construction of ΔLe1647 |
| 1647-R | CGATGCCGAGCAGCACGAA | |
| pilR-F | CGGAGGCGATACTGGGAATG | To confirm mutant construction of ΔpilR |
| pilR-R | GGAGGGCGTAATGGCTGAAAC | |
| 1921-F | CCGGGACCATTTCCATGTCG | To confirm mutant construction of ΔLe1921 |
| 1921-R | AAGTGCTTGCGGGCGTTGC | |
| 2333-F | ACGCCTGAGCCTGCTGGTCT | To confirm mutant construction of ΔLe2333 |
| 2333-R | GGTTCGGATCGGGAAGGAGAA | |
| 3200-F | GGACCCCGCAGTGAGGATAGG | To confirm mutant construction of ΔLe3200 |
| 3200-R | CGCTGGGAGTGGGGAAGGAG | |
| 4104-F | GGTCCGCAGCATGGAAGCA | To confirm mutant construction of ΔLe4104 |
| 4104-R | CGAGCCAATCGGCGCTGTAC | |
| 4215-F | ATCACCGTGTCGTCGGGATTG | To confirm mutant construction of ΔLe4215 |
| 4215-R | GTTTCCCTTCATTTCCCTGCTCC | |
| 4778-F | CAGAACCCACCCTCGGAAAGC | To confirm mutant construction of ΔLe4778 |
| 4778-R | CGACGTGTTGAGCCAGGAAGG | |
| Construction of complementary plasmids | ||
| pilR-cpF | CCCAAGCTTCGCACGGCAAGCAGAAAA (HindIII) | To amplify 1,540-bp fragment containing intact sequence of pilR |
| pilR-cpR | GCTCTAGAAGGGCGGGAACGACCCTGT (XbaI) | |
| Protein expression | ||
| pilR-pET-F | GGAATTCCATATGGCTGAAACCCGTAGTGCATT (NdeI) | To amplify fragment of intact pilR sequence |
| pilR-pET-R | CCCAAGCTTGTCGATCCCGAGCTTCTTCA (HindIII) | |
| Biotin-labeled probe for EMSA analysis | ||
| pilA-biotin-F | 5′-Biotin-CGCCACGTAGCCGCCCGCCG-3′ | To amplify 541-bp biotin probe of promoter region of pilA |
| pilA-biotin-R | 5′-GGTGTATCCCCTAGGAGTGA-3′ | |
| pilA-cold-F | 5′-CGCCACGTAGCCGCCCGCCG-3′ | To amplify 541-bp cold probe of promoter region of pilA |
| pilA-cold-R | 5′-GGTGTATCCCCTAGGAGTGA-3′ | |
| lafB-biotin-F | 5′-Biotin-AATGATCCGCGTCGCAGGAT-3′ | To amplify 491-bp biotin probe of promoter region of lafB |
| lafB-biotin-R | 5′-CAGCAGCGGGTGGGCGCAGT-3′ | |
| lafB-cold-F | 5′-AATGATCCGCGTCGCAGGAT-3′ | To amplify 491-bp cold probe of promoter region of lafB |
| lafB-cold-R | 5′-CAGCAGCGGGTGGGCGCAGT-3′ | |
| qRT-PCR analysisb | ||
| pilA-qRT-F | TACAACTTCACCGCCAACAG | |
| pilA-qRT-R | TCAGGGTGAACTTGATGTCG | |
| lafB-qRT-F | ACTATTTGTTGGGCGACGAC | |
| lafB-qRT-F | GTAACCGAACAGGGTGCAA | |
| 16S-qRT-F | ACGGTCGCAAGACTGAAACT | |
| 16S-qRT-F | AAGGCACCAATCCATCTCTG |
Underlined nucleotide sequences are restriction sites, and the restriction enzymes are indicated at the end of primers.
From reference 25.
Complementation constructs for each mutant were generated as described previously (12). In brief, the DNA fragments containing full-length genes along with their upstream promoter regions were amplified by PCR and cloned into the broad-host-range vectors pBBR1-MCS5 and pUFR047 (Table 1).
Twitching motility assays.
L. enzymogenes twitching motility was assayed as described previously (14, 25). Briefly, bacteria were inoculated at the edge of a sterilized coverslip containing a thin layer of 0.05× tryptic soy agar (TSA). After 24 h of incubation, the margin of the bacterial culture on the microscope slide was observed. Cell clusters growing away from the main colony represented motile cells (14). Three slides for each treatment were used, and each experiment was performed three times.
HSAF extraction and quantification.
HSAF was extracted from 25-ml L. enzymogenes cultures grown for 48 h at 28°C in 0.1× TSB, with shaking at 200 rpm. HSAF was detected by HPLC, as described previously (12), and quantified per unit of optical density at 600 nm (OD600), as described previously (25). Three biological replicates were used, and each was assayed in three technical replicates.
RNA extraction and qRT-PCR.
Cells were grown in 0.1× TSB or 0.05× TSB and collected at an OD600 of 1.0. RNA was extracted using a bacterial RNA kit (Omega, China), according to the manufacturer's protocol. Real-time qRT-PCR was performed using the 16S rRNA gene as an internal control, as described previously (12, 32). Primers for qRT-PCR are listed in Table 2. The primers used for measuring pilA and lafB mRNA were reported previously (12, 25).
Protein purification and EMSA.
The full-length pilR coding sequence was amplified and cloned into the expression vector pET30a(+) to generate a pilR-His6 fusion [plasmid PilR-pET30(a)]. E. coli BL21(DE3) [PilR-pET30(a)] was grown at 37°C, with shaking at 200 rpm, until the OD600 was 0.6. pilR expression was induced with isopropyl β-d-1-thiogalactopyranoside (0.5 mM final concentration), followed by incubation at 37°C for 6 h. The cells were collected by centrifugation, resuspended in 25 ml of lysis buffer, i.e., phosphate-buffered saline (PBS) containing 10 mM phenylmethylsulfonyl fluoride (PMSF) (a protease inhibitor), and lysed by sonication (Branson 250 digital sonifier). Following centrifugation at 13,000 rpm for 30 min at 4°C, the soluble extract was collected and mixed with preequilibrated Ni2+ resin (GE Health) for 1 h at 4°C. A column containing resin with bound PilR-His6 was washed extensively with resuspension buffer, i.e., 50 mM PBS containing 30 mM imidazole and 300 mM NaCl. The PilR-His6 protein was eluted with 250 mM imidazole. Finally, the protein eluent was transferred into an ultrafiltration device and concentrated by centrifugation at 3,000 × g.
An EMSA was performed as follows. The fragments containing promoter regions of pilA or lafB were amplified by PCR using biotin-5′-end-labeled primers (Table 2). The biotin-end-labeled target DNA and protein extract were incubated in binding reactions for the test system for 20 min at room temperature, according to the protocols of the LightShift chemiluminescent EMSA kit (Thermo). The binding reaction mixtures were then loaded onto a polyacrylamide (8%) gel, electrophoresed in 0.5× Tris-borate-EDTA (TBE) buffer, transferred to a nylon membrane, and cross-linked. Finally, the biotinylated DNA fragments were detected by chemiluminescence with a VersaDoc imaging system (Bio-Rad).
c-di-GMP extraction and quantification.
Cultures were grown in 0.1× TSB at 28°C until the cell density reached an OD600 of 1.5. Cells from 2-ml cultures were harvested for protein quantification by the bicinchoninic acid (BCA) assay (TransGen). Cells from 8 ml of culture were used for c-di-GMP extraction with 0.6 M HClO4 and 2.5 M K2CO3, as described previously (29, 38). The samples were analyzed by LC-MS, as described previously (29, 38, 39).
Supplementary Material
ACKNOWLEDGMENTS
We appreciate critical review of the manuscript by Shan-Ho Chou (National Chung Hsing University) and Liangcheng Du (University of Nebraska-Lincoln). We thank Gary Y. Yuen (University of Nebraska-Lincoln) for useful suggestions on manuscript organization. We also thank Lvyan Ma and Shiwei Wang (Chinese Academy of Sciences) for technical support in c-di-GMP measurements.
This study was supported by the National Basic Research 973 Program of China (grant 2015CB150600 to G.Q.), Fundamental Research Funds for the Central Universities (grants Y0201600126 and KYTZ201403 to G.Q.), the Special Fund for Agro-Scientific Research in the Public Interest (grant 201303015 to G.Q. and F.L.), the National Natural Science Foundation of China (grant 31572046 to G.Q.), the Jiangsu Provincial Key Technology Support Program (grants BE2014386 and BE2015354 to F.L.), the Basal Research Funds from JAAS [grant ZX(15)1006 to F.L.], Jiangsu Agricultural Science and Technology Innovation Funds [grant CX(16)1049 to F.L.], the 948 Project of the Ministry of Agriculture (grant 2014-Z24 to F.L.), and the National Pear Industry Technology System (grant CARS-29-09 to G.Q. and F.L.). G.X. and G.Q. were visiting scholars in M.G.'s laboratory, supported by the China Scholarship Council and the Nanjing Agricultural University, respectively.
G.Q. and F.L. conceived the project, M.G., G.Q, and F.L. designed the experiments, Y.C., J.X., Z.S., and G.X. carried out the experiments, Y.C., M.G., G.Q., and F.L. analyzed the data, G.Q. wrote the manuscript draft, and M.G. and F.L. revised the manuscript.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.03397-16.
REFERENCES
- 1.Christensen P, Cook FD. 1978. Lysobacter, a new genus of nonhiting, gliding bacteria with a high base ratio. Int J Syst Bacteriol 28:367–393. doi: 10.1099/00207713-28-3-367. [DOI] [Google Scholar]
- 2.Hayward AC, Fegan N, Fegan M, Stirling GR. 2010. Stenotrophomonas and Lysobacter: ubiquitous plant-associated gamma-proteobacteria of developing significance in applied microbiology. J Appl Microbiol 108:756–770. doi: 10.1111/j.1365-2672.2009.04471.x. [DOI] [PubMed] [Google Scholar]
- 3.de Bruijn I, Cheng X, de Jager V, Exposito RG, Watrous J, Patel N, Postma J, Dorrestein PC, Kobayashi D, Raaijmakers JM. 2015. Comparative genomics and metabolic profiling of the genus Lysobacter. BMC Genomics 16:991. doi: 10.1186/s12864-015-2191-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang Z, Yuen GY. 1999. Biological control of Bipolaris sorokiniana on tall fescue by Stenotrophomonas maltophilia strain C3. Phytopathology 89:817–822. doi: 10.1094/PHYTO.1999.89.9.817. [DOI] [PubMed] [Google Scholar]
- 5.Jochum CC, Osborne LE, Yuen GY. 2006. Fusarium head blight biological control with Lysobacter enzymogenes. Bio Control 39:336–344. doi: 10.1016/j.biocontrol.2006.05.004. [DOI] [Google Scholar]
- 6.Qian GL, Hu BS, Jiang YH, Liu FQ. 2009. Identification and characterization of Lysobacter enzymogenes as a biological control agent against some fungal pathogens. Agric Sci China 8:68–75. doi: 10.1016/S1671-2927(09)60010-9. [DOI] [Google Scholar]
- 7.Li S, Jochum CC, Yu F, Zaleta-Rivera K, Du L, Harris SD, Yuen GY. 2008. An antibiotic complex from Lysobacter enzymogenes strain C3: antimicrobial activity and role in plant disease control. Phytopathology 98:695–701. doi: 10.1094/PHYTO-98-6-0695. [DOI] [PubMed] [Google Scholar]
- 8.Yu F, Zaleta-Rivera K, Zhu X, Huffman J, Millet JC, Harris SD, Yuen G, Li XC, Du L. 2007. Structure and biosynthesis of heat-stable antifungal factor (HSAF), a broad-spectrum antimycotic with a novel mode of action. Antimicrob Agents Chemother 51:64–72. doi: 10.1128/AAC.00931-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lou L, Qian G, Xie Y, Hang J, Chen H, Zaleta-Rivera K, Li Y, Shen Y, Dussault PH, Liu F, Du L. 2011. Biosynthesis of HSAF, a tetramic acid-containing macrolactam from Lysobacter enzymogenes. J Am Chem Soc 133:643–645. doi: 10.1021/ja105732c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li Y, Chen H, Ding Y, Xie Y, Wang H, Cerny RL, Shen Y, Du L. 2014. Iterative assembly of two separate polyketide chains by the same single-module bacterial polyketide synthase in the biosynthesis of HSAF. Angew Chem Int Ed Engl 53:7524–7530. doi: 10.1002/anie.201403500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li S, Du L, Yuen G, Harris SD. 2006. Distinct ceramide synthases regulate polarized growth in the filamentous fungus Aspergillus nidulans. Mol Biol Cell 17:1218–1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Qian G, Wang Y, Liu Y, Xu F, He YW, Du L, Venturi V, Fan J, Hu B, Liu F. 2013. Lysobacter enzymogenes uses two distinct cell-cell signaling systems for differential regulation of secondary-metabolite biosynthesis and colony morphology. Appl Environ Microbiol 79:6604–6616. doi: 10.1128/AEM.01841-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Han Y, Wang Y, Tombosa S, Wright S, Huffman J, Yuen G, Qian G, Liu F, Shen Y, Du L. 2015. Identification of a small molecule signaling factor that regulates the biosynthesis of the antifungal polycyclic tetramate macrolactam HSAF in Lysobacter enzymogenes. Appl Microbiol Biotechnol 99:801–811. doi: 10.1007/s00253-014-6120-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhou X, Qian G, Chen Y, Du L, Liu F, Yuen GY. 2015. PilG is involved in the regulation of twitching motility and antifungal antibiotic biosynthesis in the biological control agent Lysobacter enzymogenes. Phytopathology 105:1318–1324. doi: 10.1094/PHYTO-12-14-0361-R. [DOI] [PubMed] [Google Scholar]
- 15.Romling U, Galperin MY, Gomelsky M. 2013. Cyclic di-GMP: the first 25 years of a universal bacterial second messenger. Microbiol Mol Biol Rev 77:1–52. doi: 10.1128/MMBR.00043-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hengge R. 2009. Principles of c-di-GMP signalling in bacteria. Nat Rev Microbiol 7:263–273. doi: 10.1038/nrmicro2109. [DOI] [PubMed] [Google Scholar]
- 17.Ishimoto KS, Lory S. 1992. Identification of pilR, which encodes a transcriptional activator of the Pseudomonas aeruginosa pilin gene. J Bacteriol 174:3514–3521. doi: 10.1128/jb.174.11.3514-3521.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hobbs M, Collie ES, Free PD, Livingston SP, Mattick JS. 1993. PilS and PilR, a two-component transcriptional regulatory system controlling expression of type 4 fimbriae in Pseudomonas aeruginosa. Mol Microbiol 7:669–682. doi: 10.1111/j.1365-2958.1993.tb01158.x. [DOI] [PubMed] [Google Scholar]
- 19.Boyd JM, Koga T, Lory S. 1994. Identification and characterization of PilS, an essential regulator of pilin expression in Pseudomonas aeruginosa. Mol Gen Genet 243:565–574. doi: 10.1007/BF00284205. [DOI] [PubMed] [Google Scholar]
- 20.Wu SS, Kaiser D. 1997. Regulation of expression of the pilA gene in Myxococcus xanthus. J Bacteriol 179:7748–7758. doi: 10.1128/jb.179.24.7748-7758.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kehl-Fie TE, Porsch EA, Miller SE, St Geme JW III. 2009. Expression of Kingella kingae type IV pili is regulated by σ54, PilS, and PilR. J Bacteriol 191:4976–4986. doi: 10.1128/JB.00123-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dunger G, Guzzo CR, Andrade MO, Jones JB, Farah CS. 2014. Xanthomonas citri subsp. citri type IV pilus is required for twitching motility, biofilm development, and adherence. Mol Plant Microbe Interact 27:1132–1147. doi: 10.1094/MPMI-06-14-0184-R. [DOI] [PubMed] [Google Scholar]
- 23.Dunger G, Llontop E, Guzzo CR, Farah CS. 2016. The Xanthomonas type IV pilus. Curr Opin Microbiol 30:88–97. doi: 10.1016/j.mib.2016.01.007. [DOI] [PubMed] [Google Scholar]
- 24.Jin S, Ishimoto KS, Lory S. 1994. PilR, a transcriptional regulator of piliation in Pseudomonas aeruginosa, binds to a cis-acting sequence upstream of the pilin gene promoter. Mol Microbiol 14:1049–1057. doi: 10.1111/j.1365-2958.1994.tb01338.x. [DOI] [PubMed] [Google Scholar]
- 25.Wang Y, Zhao Y, Zhang J, Zhao Y, Shen Y, Su Z, Xu G, Du L, Huffman JM, Venturi V, Qian G, Liu F. 2014. Transcriptomic analysis reveals new regulatory roles of Clp signaling in secondary metabolite biosynthesis and surface motility in Lysobacter enzymogenes OH11. Appl Microbiol Biotechnol 98:9009–9020. doi: 10.1007/s00253-014-6072-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chin KH, Lee YC, Tu ZL, Chen CH, Tseng YH, Yang JM, Ryan RP, McCarthy Y, Dow JM, Wang AH, Chou SH. 2010. The cAMP receptor-like protein CLP is a novel c-di-GMP receptor linking cell-cell signaling to virulence gene expression in Xanthomonas campestris. J Mol Biol 396:646–662. doi: 10.1016/j.jmb.2009.11.076. [DOI] [PubMed] [Google Scholar]
- 27.Ryjenkov DA, Tarutina M, Moskvin OV, Gomelsky M. 2005. Cyclic diguanylate is a ubiquitous signaling molecule in bacteria: insights into biochemistry of the GGDEF protein domain. J Bacteriol 187:1792–1798. doi: 10.1128/JB.187.5.1792-1798.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schmidt AJ, Ryjenkov DA, Gomelsky M. 2005. The ubiquitous protein domain EAL is a c-di-GMP-specific phosphodiesterase: enzymatically active and inactive EAL domains. J Bacteriol 187:4774–4781. doi: 10.1128/JB.187.14.4774-4781.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hickman JW, Harwood CS. 2008. Identification of FleQ from Pseudomonas aeruginosa as a c-di-GMP-responsive transcription factor. Mol Microbiol 69:376–389. doi: 10.1111/j.1365-2958.2008.06281.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Matsuyama BY, Krasteva PV, Baraquet C, Harwood CS, Sondermann H, Navarro MV. 2016. Mechanistic insights into c-di-GMP-dependent control of the biofilm regulator FleQ from Pseudomonas aeruginosa. Proc Natl Acad Sci U S A 113:E209–E218. doi: 10.1073/pnas.1523148113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yaryura PM, Conforte VP, Malamud F, Roeschlin R, de Pino V, Castagnaro AP, McCarthy Y, Dow JM, Marano MR, Vojnov AA. 2015. XbmR, a new transcription factor involved in the regulation of chemotaxis, biofilm formation and virulence in Xanthomonas citri subsp. citri. Environ Microbiol 17:4164–4176. doi: 10.1111/1462-2920.12684. [DOI] [PubMed] [Google Scholar]
- 32.Qian G, Xu F, Venturi V, Du L, Liu F. 2014. Roles of a solo LuxR in the biological control agent Lysobacter enzymogenes strain OH11. Phytopathology 104:224–231. doi: 10.1094/PHYTO-07-13-0188-R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tao F, He YW, Wu DH, Swarup S, Zhang LH. 2010. The cyclic nucleotide monophosphate domain of Xanthomonas campestris global regulator Clp defines a new class of cyclic di-GMP effectors. J Bacteriol 192:1020–1029. doi: 10.1128/JB.01253-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Slater H, Alvarez-Morales A, Barber CE, Daniels MJ, Dow JM. 2000. A two-component system involving an HD-GYP domain protein links cell-cell signalling to pathogenicity gene expression in Xanthomonas campestris. Mol Microbiol 38:986–1003. [DOI] [PubMed] [Google Scholar]
- 35.Liang ZX. 2015. The expanding roles of c-di-GMP in the biosynthesis of exopolysaccharides and secondary metabolites. Nat Prod Rep 32:663–683. doi: 10.1039/C4NP00086B. [DOI] [PubMed] [Google Scholar]
- 36.Finn RD, Coggill P, Eberhardt RY, Eddy SR, Mistry J, Mitchell AL, Potter SC, Punta M, Qureshi M, Sangrador-Vegas A, Salazar GA, Tate J, Bateman A. 2016. The Pfam protein families database: towards a more sustainable future. Nucleic Acids Res 44:D279–D285. doi: 10.1093/nar/gkv1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Qian G, Wang Y, Qian D, Fan J, Hu B, Liu F. 2012. Selection of available suicide vectors for gene mutagenesis using chiA (a chitinase encoding gene) as a new reporter and primary functional analysis of chiA in Lysobacter enzymogenes strain OH11. World J Microbiol Biotechnol 28:549–557. doi: 10.1007/s11274-011-0846-8. [DOI] [PubMed] [Google Scholar]
- 38.Zhu B, Liu C, Liu S, Cong H, Chen Y, Gu L, Ma LZ. 2016. Membrane association of SadC enhances its diguanylate cyclase activity to control exopolysaccharides synthesis and biofilm formation in Pseudomonas aeruginosa. Environ Microbiol 18:3440–3452. doi: 10.1111/1462-2920.13263. [DOI] [PubMed] [Google Scholar]
- 39.Thormann KM, Duttler S, Saville RM, Hyodo M, Shukla S, Hayakawa Y, Spormann AM. 2006. Control of formation and cellular detachment from Shewanella oneidensis MR-1 biofilms by cyclic di-GMP. J Bacteriol 188:2681–2691. doi: 10.1128/JB.188.7.2681-2691.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ortet P, Whitworth DE, Santaella C, Achouak W, Barakat M. 2015. P2CS: updates of the prokaryotic two-component systems database. Nucleic Acids Res 43:D536–D541. doi: 10.1093/nar/gku968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hoang TT, Karkhoff-Schweizer RR, Kutchma AJ, Schweizer HP. 1998. A broad-host-range Flp-FRT recombination system for site-specific excision of chromosomally-located DNA sequences: application for isolation of unmarked Pseudomonas aeruginosa mutants. Gene 212:77–86. doi: 10.1016/S0378-1119(98)00130-9. [DOI] [PubMed] [Google Scholar]
- 42.Kovach ME, Elzer PH, Hill DS, Robertson GT, Farris MA, Roop RM II, Peterson KM. 1995. Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene 166:175–176. doi: 10.1016/0378-1119(95)00584-1. [DOI] [PubMed] [Google Scholar]
- 43.De Feyter R, Yang Y, Gabriel DW. 1993. Gene-for-genes interactions between cotton R genes and Xanthomonas campestris pv. malvacearum avr genes. Mol Plant Microbe Interact 6:225–237. doi: 10.1094/MPMI-6-225. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.