ABSTRACT
Brown rot fungi are wood-degrading fungi that employ both oxidative and hydrolytic mechanisms to degrade wood. Hydroxyl radicals that facilitate the oxidative component are powerful nonselective oxidants and are incompatible with hydrolytic enzymes unless they are spatially segregated in wood. Differential gene expression has been implicated in the segregation of these reactions in Postia placenta, but it is unclear if this two-step mechanism varies in other brown rot fungi with different traits and life history strategies that occupy different niches in nature. We employed proteomics to analyze a progression of wood decay on thin wafers, using brown rot fungi with significant taxonomic and niche distances: Serpula lacrymans (Boletales; “dry rot” lumber decay) and Gloeophyllum trabeum (order Gloeophyllales; slash, downed wood). Both fungi produced greater oxidoreductase diversity upon wood colonization and greater glycoside hydrolase activity later, consistent with a two-step mechanism. The two fungi invested very differently, however, in terms of growth (infrastructure) versus protein secretion (resource capture), with the ergosterol/extracted protein ratio being 7-fold higher with S. lacrymans than with G. trabeum. In line with the native substrate associations of these fungi, hemicellulase-specific activities were dominated by mannanase in S. lacrymans and by xylanase in G. trabeum. Consistent with previous observations, S. lacrymans did not produce glycoside hydrolase 6 (GH6) cellobiohydrolases (CBHs) in this study, despite taxonomically belonging to the order Boletales, which is distinguished among brown rot fungi by having CBH genes. This work suggests that distantly related brown rot fungi employ staggered mechanisms to degrade wood, but the underlying strategies vary among taxa.
IMPORTANCE Wood-degrading fungi are important in forest nutrient cycling and offer promise in biotechnological applications. Brown rot fungi are unique among these fungi in that they use a nonenzymatic oxidative pretreatment before enzymatic carbohydrate hydrolysis, enabling selective removal of carbohydrates from lignin. This capacity has independently evolved multiple times, but it is unclear if different mechanisms underpin similar outcomes. Here, we grew fungi directionally on wood wafers and we found similar two-step mechanisms in taxonomically divergent brown rot fungi. The results, however, revealed strikingly different growth strategies, with S. lacrymans investing more in biomass production than secretion of proteins and G. trabeum showing the opposite pattern, with a high diversity of uncharacterized proteins. The “simplified” S. lacrymans secretomic system could help narrow gene targets central to oxidative brown rot pretreatments, and a comparison of its distinctions with G. trabeum and other brown rot fungi (e.g., Postia placenta) might offer similar traction in noncatabolic genes.
KEYWORDS: Basidiomycetes, Gloeophyllum trabeum, glycoside hydrolase, proteomics, Serpula lacrymans
INTRODUCTION
Brown rot wood-degrading fungi are common pests in lumber and are a dominant agent of wood decay in forests, particularly in conifer-dominated woodlands. These fungi decompose wood via a carbohydrate-selective mechanism, using a combination of free radical oxidation and glycoside hydrolase-mediated saccharification (1). Wood oxidation during brown rot is mediated by nonselective hydroxyl radicals, requiring spatial and/or temporal segregation from hydrolytic enzymes in order to limit deactivation by hydroxyl radicals (2).
The overall two-step mechanism is thought to be shared by all brown rot fungi, despite multiple independent evolutionary origins of brown rot (3, 4). Genomic comparisons, however, suggest that differences in the decay mechanisms driving a two-step oxidative-enzymatic mechanism may exist among brown rot clades (5, 6). Differences among brown rot species are evident in some measures of decay, such as lignin selectivity indices (7). There are also examples where fungi in different clades vary in their abilities to utilize certain substrates, such as Antrodia and Gloeophyllum clades; both efficiently degrade wood, but Antrodia clade fungi fail to efficiently degrade corn stover, unlike Gloeophyllum trabeum (8). Although these genomic and phenotypic variations are known, the phylogenetic variations in the brown rot biochemical mechanisms have not been resolved.
A logical approach to help resolve variations is to resolve spatially the temporal sequence of the two-step brown rot mechanism to enable comparisons. Brown rot fungi temporally stagger oxidative and hydrolytic chemistries via differential gene expression, where wood oxidation is followed by glycoside hydrolase upregulation (9, 10). This staggered pattern was established via both transcriptomics and enzyme assays in Postia placenta (Polyporales), but this space-for-time approach has not yet been applied to study the staggered mechanisms of other brown rot fungi from other distinct clades (9).
In this study, we employed a wood wafer setup to compare the temporal progressions of decay by two taxonomically divergent brown rot fungi, Gloeophyllum trabeum and Serpula lacrymans. Specifically, we compared differences in secretome composition, enzymatic activity, and mode of colonization over the course of wood decay. Both fungi are ubiquitous lumber pests, but they evolved from distinct evolutionary lineages, with key differences in plant cell wall deconstruction genes, including the notable presence in S. lacrymans of cellobiohydrolases (CBHs) that distinguish its clade (Boletales) from all other brown rot clades (10). Serpula lacrymans is also distinct in that it is not commonly observed in nature, unlike the ubiquitous G. trabeum (11, 12). The “dry rot fungus,” as S. lacrymans is known, can initiate decay with minimal ground contact, as well as in wood with low moisture content, and perhaps with less of a need for combative ability due to its adaptations to dry environments (13). Our design and analyses therefore test the hypothesis that the agents deployed to create a two-step brown rot mechanism differ between these fungi.
RESULTS
Secretome composition.
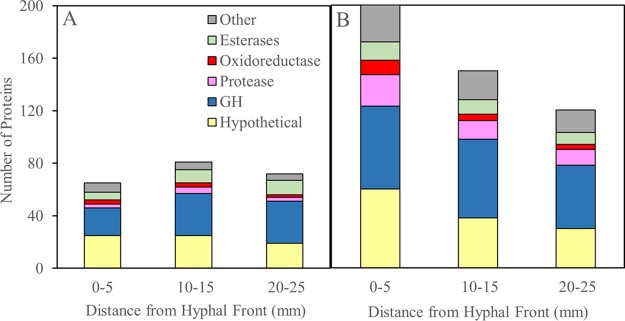
In S. lacrymans, 93 proteins (identified as peptides exclusively matched to a protein) were identified among all colonized wafer sections. G. trabeum, on the other hand, produced 209 proteins in total and about 3 times more proteins than S. lacrymans from the section nearest the hyphal front (Fig. 1). The proteins identified totaled 65, 81, and 72 in S. lacrymans cultures at distances of 0 to 5, 10 to 15, and 20 to 25 mm behind the hyphal front, respectively, and totaled 200, 150, and 120 in the equivalent wafer sections colonized by G. trabeum.
FIG 1.
Secretome composition in spruce wafer sections degraded by Serpula lacrymans (A) and Gloeophyllum trabeum (B), expressed as the total number of proteins belonging to one of 6 categories identified in each sample. Note the difference in scale for both fungi due to the larger amount of protein diversity found in G. trabeum. Data used to generate this figure are listed in Table 3. GH, glycoside hydrolase.
Hypothetical proteins and glycoside hydrolases (GHs) were the most abundant protein categories. Hypothetical protein diversity was greatest near the hyphal front for both fungal species, with 12 and 34 hypothetical proteins found only within 15 mm of the hyphal front in S. lacrymans and G. trabeum cultures, respectively (Table 1). GH diversity followed the same pattern in G. trabeum (Table 2) but not S. lacrymans (Table 3). Twenty GHs from G. trabeum were exclusive to the first 15 mm behind the hyphal front and consisted of chitinases, noncellulolytic β-glucanases, hemicellulases, and a GH28 pectinase (Table 2).
TABLE 1.
Hypothetical proteins found exclusively in the early decay stages (0 to 5 and 10 to 15 mm behind the hyphal front) in spruce wafer extracts from Serpula lacrymans and Gloeophyllum trabeum cultures
| Domaina | Unique hypothetical proteinsb |
|---|---|
| Serpula lacrymans | |
| GATase domain | 1 |
| Cytoskeletal protein | 1 |
| YjgF | 1 |
| No domain | 9 |
| Total | 12 |
| Gloeophyllum trabeum | |
| Acyl-CoA binding | 1 |
| Aldo-keto reductase | 1 |
| Cerato-platanin | 1 |
| Cupredoxin | 1 |
| Delta endotoxin | 1 |
| DnaJ | 1 |
| Lipase | 1 |
| No domain | 27 |
| Total | 34 |
Protein domains were identified by BLAST searches of the Swiss-Prot database (27).
Hypothetical proteins listed were found in extracts from either 0 to 5 or 10 to 15 mm behind the hyphal front.
TABLE 2.
Glycoside hydrolases found in extracts of spruce wafer sections degraded by Gloeophyllum trabeum and the number of unique peptides identified for each protein in each wafer section extract
| G. trabeum no.a | GH familyb | Putative functionc | SignalP resultsd | No. of unique peptides at distance (mm)e: |
||
|---|---|---|---|---|---|---|
| 0–5 | 10–15 | 20–25 | ||||
| 81012 | 1 | β-Glucosidase | No | 13 | 20 | 15 |
| 115191 | 2 | Mannanase | Yes | 12 | 13 | 11 |
| 44548 | 3 | β-Glucosidase | Yes | 2 | 0 | 0 |
| 71534 | 3 | β-Glucosidase | Yes | 0 | 6 | 0 |
| 122002 | 3 | Exo-1,4-β-xylosidase | Yes | 8 | 10 | 7 |
| 75899 | 3 | β-Glucosidase | No | 7 | 8 | 7 |
| 69843 | 3 | β-Glucosidase | Yes | 10 | 13 | 13 |
| 68070 | 5 | Exo-1,3-β-glucanase | Yes | 2 | 0 | 0 |
| 118009 | 5 | Unknown | Yes | 2 | 2 | 0 |
| 120257 | 5 | Endo-1,4-β-glucanase | Yes | 2 | 4 | 2 |
| 41779 | 5 | Exo-1,3-β-glucanase | No | 1 | 4 | 3 |
| 114574 | 5 | Endo-1,4-β-mannanase | Yes | 1 | 3 | 3 |
| 42111 | 5 | Exo-1,3-β-glucanase | No | 2 | 4 | 4 |
| 135369 | 5 | Endo-1,4-β-mannanase | Yes | 3 | 8 | 6 |
| 59826 | 5 | Exo-1,3-β-glucanase | No | 9 | 9 | 7 |
| 110405 | 5 | Endo-1,4-β-mannosidase | Yes | 3 | 7 | 10 |
| 63180 | 5 | Endo-1,4-β-glucanase | Yes | 6 | 14 | 45 |
| 149061 | 10 | Endo-1,4-β-xylanase | Yes | 3 | 0 | 0 |
| 122601 | 10 | Endo-1,4-β-xylanase | Yes | 6 | 1 | 0 |
| 138785 | 10 | Endo-1,4-β-xylanase | Yes | 3 | 7 | 5 |
| 107452 | 10 | Endo-1,4-β-xylanase | Yes | 15 | 36 | 49 |
| 138821 | 12 | Endo-1,4-β-glucanase | Yes | 8 | 18 | 49 |
| 73036 | 13 | Amylase | Yes | 9 | 0 | 0 |
| 61700 | 15 | Amylase | Yes | 11 | 7 | 7 |
| 52752 | 16 | β-Glucan synthesis | No | 4 | 0 | 0 |
| 66072 | 16 | β-Glucan synthesis | No | 2 | 0 | 0 |
| 81698 | 16 | β-Glucan synthesis | No | 7 | 7 | 2 |
| 82316 | 18 | Chitinase | No | 2 | 3 | 0 |
| 122074 | 18 | Chitinase, CBM 5 | Yes | 0 | 3 | 0 |
| 118998 | 18 | Chitinase, CBM 5 | Yes | 3 | 3 | 2 |
| 45026 | 18 | Chitinase | No | 2 | 3 | 3 |
| 81165 | 18 | Chitinase, CBM 5 | Yes | 1 | 5 | 3 |
| 91195 | 20 | β-N-Acetylglucosaminidase | Yes | 4 | 1 | 0 |
| 116582 | 20 | β-N-Acetylglucosaminidase | Yes | 2 | 1 | 0 |
| 81761 | 27 | α-Galactosidase, CBM 35 | Yes | 1 | 2 | 0 |
| 117566 | 27 | α-Galactosidase | Yes | 11 | 15 | 10 |
| 104965 | 28 | Endo-polygalacturonase | Yes | 8 | 3 | 0 |
| 110574 | 28 | Polygalacturonase | Yes | 2 | 6 | 2 |
| 54367 | 28 | Exo-polygalacturonase | Yes | 0 | 3 | 3 |
| 117232 | 28 | Exo-rhamnogalacturonase | Yes | 1 | 3 | 3 |
| 6650 | 28 | Exo-polygalacturonase | No | 2 | 6 | 4 |
| 138836 | 28 | Exo-polygalacturonase | Yes | 4 | 6 | 7 |
| 61165 | 29 | α-l-Fucosidase | No | 3 | 5 | 2 |
| 46629 | 30 | Unknown | Yes | 3 | 4 | 4 |
| 119185 | 31 | α-Glucosidase | Yes | 10 | 10 | 9 |
| 141329 | 31 | α-Xylosidase | Yes | 10 | 18 | 14 |
| 81512 | 35 | β-Galactosidase | No | 8 | 13 | 10 |
| 111095 | 35 | β-Galactosidase | Yes | 16 | 24 | 19 |
| 100356 | 37 | Trehalase | No | 0 | 2 | 3 |
| 69366 | 43 | Unknown, CBM 35 | Yes | 0 | 3 | 0 |
| 58475 | 43 | Endo-1,5-α-l-arabinase | Yes | 3 | 3 | 4 |
| 107755 | 47 | α-1,2-Mannosidase | Yes | 6 | 6 | 4 |
| 111463 | 51 | α-l-Arabinofuranosidase | Yes | 3 | 6 | 3 |
| 134804 | 51 | α-l-Arabinofuranosidase | Yes | 2 | 9 | 4 |
| 137578 | 55 | Endo-1,3-β-glucosidase | No | 5 | 2 | 1 |
| 126879 | 55 | β-1,3-Glucosidase | Yes | 8 | 12 | 15 |
| 39602 | 71 | Endo-1,3-α-glucosidase | Yes | 2 | 0 | 0 |
| 113553 | 72 | β-1,3-Glycanosyltransferase, CBM 43 | Yes | 3 | 4 | 7 |
| 68888 | 74 | Xyloglucanase | Yes | 3 | 10 | 7 |
| 121201 | 78 | Unknown | Yes | 2 | 2 | 0 |
| 136552 | 78 | Unknown | Yes | 7 | 6 | 4 |
| 44058 | 79 | β-Glucuronidase | No | 1 | 6 | 2 |
| 116837 | 79 | β-Glucuronidase | No | 3 | 11 | 10 |
| 120232 | 88 | Glucuronyl hydrolase | Yes | 2 | 2 | 0 |
| 81814 | 92 | Unknown | Yes | 7 | 11 | 10 |
| 108097 | 95 | α-Fucosidase | Yes | 6 | 9 | 7 |
| 121307 | 115 | α-Glucuronidase | Yes | 5 | 0 | 0 |
| 121308 | 115 | α-Glucuronidase | Yes | 15 | 20 | 14 |
Protein identification (ID) numbers from the Department of Energy (DOE) Joint Genome Institute (JGI) MycoCosm database (28).
Glycoside hydrolase families, as defined in the CAZy database (29).
Putative functions determined by BLAST searches of Swiss-Prot database (27). CBM, carbohydrate-binding molecule.
Secretion signals for detected protein sequences were detected using SignalP algorithm (30).
Unique peptide counts for proteins found in wafer sections 0 to 5, 10 to 15, and 20 to 25 mm from the hyphal front.
TABLE 3.
Glycoside hydrolases found in extracts of spruce wafer sections degraded by Serpula lacrymans and the number of unique peptides identified for each protein in each wafer section extract
| S. lacrymans no.a | GH familyb | Putative functionc | SignalP resultsd | No. of unique peptides at distance (mm)e: |
||
|---|---|---|---|---|---|---|
| 0–5 | 10–15 | 20–25 | ||||
| 62623 | 1 | β-Glucosidase | No | 3 | 4 | 4 |
| 107546 | 2 | β-Mannosidase | Yes | 4 | 9 | 21 |
| 158615 | 3 | β-Glucosidase | No | 0 | 2 | 1 |
| 431585 | 3 | β-Glucosidase | Yes | 0 | 3 | 4 |
| 446061 | 3 | β-Glucosidase | Yes | 0 | 5 | 4 |
| 491435 | 3 | β-Glucosidase | Yes | 0 | 6 | 10 |
| 172091 | 3 | Exo-1,3-β-xylosidase | No | 3 | 7 | 8 |
| 433208 | 5 | Endo-1,4-β-glucanase | No | 0 | 2 | 4 |
| 480589 | 5 | Endo-1,4-β-mannosidase | Yes | 1 | 1 | 3 |
| 412480 | 5 | Exo-1,3-β-glucosidase | Yes | 1 | 2 | 0 |
| 349170 | 10 | Endo-1,4-β-xylanase | Yes | 3 | 6 | 4 |
| 361860 | 20 | β-N-Acetyl-hexosamine | Yes | 0 | 2 | 2 |
| 457293 | 27 | α-Galactosidase | Yes | 1 | 2 | 3 |
| 414701 | 28 | Exo-polygalacturonase | Yes | 0 | 1 | 2 |
| 463474 | 28 | Exo-polygalacturonase | Yes | 1 | 3 | 2 |
| 463527 | 29 | α-l-Fucosidase | No | 1 | 2 | 2 |
| 453109 | 31 | α-Glucosidase | Yes | 1 | 3 | 4 |
| 451973 | 31 | Unknown | Yes | 1 | 4 | 5 |
| 154347 | 35 | β-Galactosidase | Yes | 0 | 3 | 2 |
| 453704 | 35 | β-Galactosidase | No | 2 | 9 | 7 |
| 167797 | 35 | β-Galactosidase | Yes | 3 | 8 | 10 |
| 431618 | 37 | Unknown | Yes | 0 | 2 | 3 |
| 469468 | 37 | Trehalase | Yes | 1 | 2 | 2 |
| 445922 | 43 | Unknown, CBM 35 domain | Yes | 0 | 3 | 2 |
| 102103 | 47 | α-1,2-Mannosidase | Yes | 7 | 8 | 9 |
| 412847 | 51 | α-Arabinofuranosidase | Yes | 1 | 3 | 3 |
| 94292 | 55 | Exo-1,3-β-glucanase | No | 0 | 0 | 2 |
| 446424 | 79 | β-Glucuronidase | Yes | 0 | 1 | 2 |
| 468146 | 79 | β-Glucuronidase | Yes | 1 | 1 | 2 |
| 361158 | 92 | Unknown | Yes | 2 | 4 | 5 |
| 413209 | 92 | Unknown | No | 3 | 6 | 8 |
| 442669 | 95 | α-Fucosidase | Yes | 2 | 6 | 8 |
| 77415 | 115 | Unknown | Yes | 1 | 11 | 11 |
Protein identification (ID) numbers from the Department of Energy (DOE) Joint Genome Institute (JGI) MycoCosm database (28).
Glycoside hydrolase families, as defined in the CAZy database (29).
Putative functions determined by BLAST searches of Swiss-Prot database (27). CBM, carbohydrate-binding molecule.
Secretion signals for detected protein sequences were detected using SignalP algorithm (30).
Unique peptide counts for proteins found in wafer sections 0 to 5, 10 to 15, and 20 to 25 mm from the hyphal front.
Oxidoreductases (ORs) were most abundant in the early decay stages in both fungi, and a total of 6 ORs and 1 OR were exclusive to the section 0 to 5 mm from the hyphal front in G. trabeum and S. lacrymans, respectively (Table 4). S. lacrymans produced a putative copper radical oxidase in all sections, whereas this class of enzyme was not found in G. trabeum cultures. G. trabeum, on the other hand, produced a putative glucose-methanol-choline (GMC) oxidoreductase and an auxiliary activity 9 (AA9) family protein in all sections, which were not found in S. lacrymans cultures.
TABLE 4.
Total number of proteins divided into 6 functional categories observed in spruce wafer sections degraded by Serpula lacrymans and Gloeophyllum trabeum
| Categorya | Total no. of proteins by distance (mm) from hyphal frontb |
|||||||
|---|---|---|---|---|---|---|---|---|
|
Serpula lacrymans |
Gloeophyllum trabeum |
|||||||
| 0–5 | 10–15 | 20–25 | Avg | 0–5 | 10–15 | 20–25 | Avg | |
| Other | 7 | 6 | 5 | 6.0 | 28 | 22 | 17 | 22.3 |
| Esterases | 6 | 10 | 11 | 9.0 | 14 | 11 | 9 | 11.3 |
| OR | 3 | 3 | 2 | 2.7 | 11 | 5 | 4 | 6.7 |
| Protease | 3 | 5 | 3 | 3.7 | 24 | 14 | 12 | 16.7 |
| GH | 21 | 32 | 32 | 28.3 | 63 | 60 | 48 | 57.0 |
| Hypothetical | 25 | 25 | 19 | 23.0 | 60 | 38 | 30 | 42.7 |
| Totals | 65 | 81 | 72 | 72.7 | 200 | 150 | 120 | 156.7 |
OR, oxidoreductase, GH, glycoside hydrolase.
Values shown in this table are depicted in Fig. 1.
Fungal growth, protein secretion, and ergosterol.
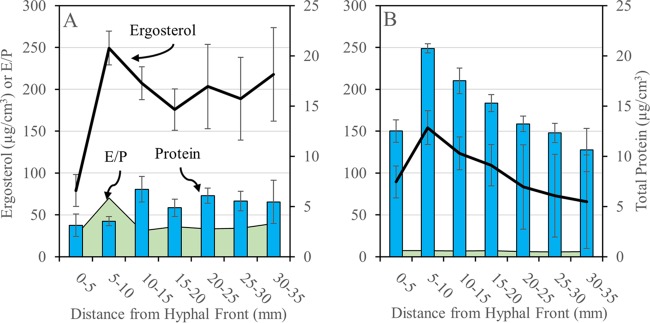
Serpula lacrymans and Gloeophyllum trabeum advanced up spruce wafers during the course of incubation at average rates of 4.6 and 2.6 mm/day, respectively. Total protein in wood extracts was generally higher in G. trabeum, rapidly increasing to a maximum of 5 to 10 mm behind the front, followed by a steady decline as wood became more degraded. S. lacrymans extracts showed the same rapid increase but at lower levels and without a decline in the later stages (Fig. 2). Because differences in total protein levels between sections of equal distance from the hyphal front for the two fungi could result from a simple distinction in growth rates (nearly 2 times higher for S. lacrymans), ergosterol profiles for each fungus were measured, showing that fungal biomass closely tracked with protein levels and resulted in nearly flat ergosterol/protein (E/P) ratios in all sections for both fungi (Fig. 2). The only exception was an increase at 5 to 10 mm in S. lacrymans. Overall, E/P ratios were 3.5 to 9.5 times higher in S. lacrymans (average, 38.6) than equivalent wafer sections from G. trabeum cultures (average, 5.7) (Fig. 2).
FIG 2.
Total protein (blue bars) and ergosterol (black lines) extracted from spruce wafer sections degraded by Serpula lacrymans (A) and Gloeophyllum trabeum (B). Ergosterol/protein ratios (E/P, green area) were calculated from total protein and ergosterol values per wafer section. Error bars for total protein values are standard deviations of the results from three assays of a single protein extract, and error bars for total ergosterol are standard deviations of the averages of three (S. lacrymans) and six (G. trabeum) wafer sections. *, ergosterol values are higher than those found in nature (31) due to dense surface growth on high surface/volume wood wafers.
Endoglucanase- and hemicellulase-specific activities.
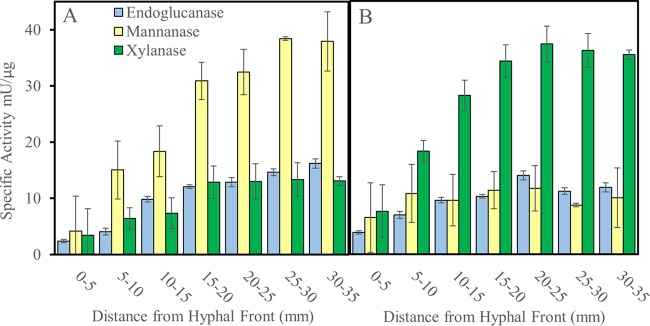
Endoglucanase- and hemicellulase-specific activities were low in the early stages of decay and increased and then plateaued in more-degraded wood, but the hemicellulase activities of the two test fungi had distinct patterns (Fig. 3). For S. lacrymans, mannanase-specific activity dominated the hemicellulase main-chain depolymerization activity and was much higher in degraded wood than the mannanase activity of G. trabeum. Conversely, xylanase activity was higher in more-decayed sections in G. trabeum than in S. lacrymans (Fig. 3). One putative xylanase from GH family 10 and one putative mannanase from GH family 5 were found in S. lacrymans wafer extracts (Table 5). Four of each (GH10 xylanases and GH5 mannanases) were found in G. trabeum extracts.
FIG 3.
Polysaccharide-degrading enzyme activities detected in protein extracts of spruce wafer sections degraded by Serpula lacrymans (A) and Gloeophyllum trabeum (B). Specific activities are the amount of enzyme required to liberate one micromole glucose-, mannose-, or xylose-reducing equivalents per minute per microgram of total protein for endoglucanase, mannanase, or xylanase values, respectively. Error bars represent standard deviations of the results from three assays of a single protein extract.
TABLE 5.
GH5 putative β-mannanases and GH10 putative xylanases found in protein extracts of spruce wafers sections degraded by Serpula lacrymans and Gloeophyllum trabeum
| Organism by GH family | Protein IDa | Putative functionb | No. of unique peptides at distance (mm)c: |
||
|---|---|---|---|---|---|
| 0–5 | 10–15 | 20–25 | |||
| GH5 | |||||
| S. lacrymans | |||||
| 433208 | Endo-β-1,4-glucanase | 0 | 2 | 4 | |
| 480589 | Endo-β-1,4-mannosidase | 1 | 1 | 3 | |
| 412480 | Exo-β-1,3-glucanase | 1 | 2 | 0 | |
| G. trabeum | |||||
| 63180 | Endo-β-1,4-glucanase | 6 | 14 | 45 | |
| 120257 | Endo-β-1,4-glucanase | 2 | 4 | 2 | |
| 110405 | Endo-β-1,4-mannanase | 3 | 7 | 10 | |
| 118009 | Endo-β-1,4-mannosidase | 2 | 2 | 0 | |
| 68070 | Exo-β-1,3-glucanase | 2 | 0 | 0 | |
| 59826 | Exo-β-1,3-glucanase | 9 | 9 | 7 | |
| 42111 | Exo-β-1,3-glucanase | 2 | 4 | 4 | |
| 41779 | Exo-β-1,3-glucanase | 1 | 4 | 3 | |
| 135369 | β-1,4-Mannanase | 3 | 8 | 6 | |
| 114574 | β-Mannanase | 1 | 3 | 3 | |
| GH10 | |||||
| S. lacrymans | |||||
| 349170 | Endo-β-1,4-xylanase | 3 | 6 | 4 | |
| G. trabeum | |||||
| 107452 | Endo-β-1,4-xylanase | 15 | 36 | 49 | |
| 138785 | Endo-β-1,4-xylanase | 3 | 7 | 5 | |
| 122601 | Endo-β-1,4-xylanase | 6 | 1 | 0 | |
| 149061 | Endo-β-1,4-xylanase | 3 | 0 | 0 | |
Protein identification (ID) numbers from the Department of Energy (DOE) Joint Genome Institute (JGI) MycoCosm database (28).
Putative functions determined by BLAST searches of Swiss-Prot database (27).
The number of unique peptides for each protein is listed for each wafer section pool (mm from the hyphal front).
DISCUSSION
S. lacrymans and G. trabeum had enzyme secretion patterns consistent with a two-step decay mechanism as observed in P. placenta (9), showing greater oxidoreductase diversity at the hyphal front and increased endoglucanase and hemicellulase activities farther behind the hyphal front. Some GHs were exclusively produced at the hyphal front by G. trabeum, including a putative GH28 pectinase. In P. placenta, a GH28 was the most highly upregulated transcript during wood colonization, suggesting that early pectin decomposition is shared by P. placenta and G. trabeum (9).
While consistent with a two-step decay mechanism, the colonization strategies of S. lacrymans and G. trabeum differed. This included a difference in the number of proteins observed, with S. lacrymans producing less than half (44%) of the number of proteins found in G. trabeum. The greatest distinction was near the hyphal front, where G. trabeum produced a more than 3 times greater variety of proteins than S. lacrymans. The discrepancy may be a result of differing growth rates, as each 5-mm section of wood represents approximately 1 and 1.9 days of growth/protein secretion for S. lacrymans and G. trabeum, respectively. These differences may be due to a difference in water translocation efficiency between the two fungi (13), despite relatively constant moisture contents (30 to 35%) along the wafers once the wood is colonized. Hyphae in the slower-growing fungus (G. trabeum) may include a wider age range in sections of equal distance from the hyphal front, resulting in a higher diversity of proteins.
Based on microscopic analysis of wafer sections degraded by Postia placenta (9), section age should coincide with increased fungal biomass. This held true for S. lacrymans, where older sections had high relative ergosterol levels, but in G. trabeum, we found the opposite. The ergosterol/protein values were also 7 times greater for S. lacrymans than for G. trabeum. Although there is a caveat when testing sequenced monokaryotic strains rather than dikaryotic strains, this distinction suggests that there may be a fundamental difference in biomass versus protein investment by these two fungi.
In addition to protein quantity, secretome composition also varied between the two fungi and was particularly notable among hypothetical proteins at the hyphal front, where G. trabeum produced over 2 times the number of proteins found at the S. lacrymans hyphal front. The number of hypothetical proteins in G. trabeum decreased by half from 5 to 25 mm behind the hyphal front; none of these have a proven biological function, but some are significantly homologous to characterized basidiomycete proteins. GT_129666, for example, is 39% identical to volvatoxin 2 (VVA2), a membrane-binding cytolytic toxin found in Volvariella volvacea (14, 15), and could be employed as a combative protein toxin in G. trabeum. Protein toxins may serve to clear territory for G. trabeum, and high hypothetical protein diversity near the hyphal front may be a result of a combative, rather than a stress tolerance, strategy that would likely characterize S. lacrymans. Pore-forming proteins are known to be produced by other wood-degrading basidiomycetes, such as Laetiporus sulphureus, and these may be common tools of combat in fungi (16); however, this conjecture requires further support.
The later stages of decay were also characterized by differing secretome compositions, particularly in the hemicellulase profiles of the two fungi. Most notably, S. lacrymans produced greater mannanase-specific activity despite the presence of only one putative GH5 mannanase (SL_480589), compared to four GH5 mannanases in the G. trabeum secretome. Greater mannanase abundance may partly explain this discrepancy, or there may be a structural difference leading to increased catalytic potential among GH5 mannanases in S. lacrymans. The cause of the discrepancy in hemicellulase profiles remains unresolved, but the reason behind observed patterns may also relate to species niches. In nature, S. lacrymans is found on glucomannan-rich conifer wood (17) and thus may have evolved more efficient enzymes for glucomannan conversion. Gloeophyllum trabeum, which produced greater xylanase activity despite growth on a xylan-poor substrate, is more of a substrate generalist, with niches including xylan-rich angiosperm wood (11). S. lacrymans also produced no cellobiohydrolase in this study, consistent with previous work failing to identify CBH in wood protein extracts and showing only low levels of expression despite its existence in the genome sequence. These results suggest that CBH plays only a minor, if any, role in decay by S. lacrymans (18).
These differences in the S. lacrymans and G. trabeum secretomes suggest that there are variations in the biochemical routes to brown rot, and this was evident in the oxidoreductase profiles of the two fungi. Serpula lacrymans produced a putative copper radical oxidase not found in G. trabeum, and G. trabeum instead produced a putative GMC oxidoreductase and a xyloglucan-specific AA9 family protein (19), a class of protein encoded in the S. lacrymans genome but not secreted on spruce wood (6). Those differences suggest that these fungi rely on different electron sources for H2O2 production for the brown rot mechanism, and they partially explain the enhanced xylanase activity in G. trabeum (20, 21).
Overall, this work supports a two-step brown rot decay mechanism common in S. lacrymans and G. trabeum, a strategy seen in other brown rot fungi (9) and white rot fungi, where the secretion of enzymatic oxidants is more prolific in the early decay stages (22, 23). Given our data, we propose that multiple biochemical manifestations of brown rot exist among the independent lineages of brown rot, driven by species-specific niches. This makes the brown rot mechanism difficult to reconstruct as a single mechanism but also indicates the presence of a more diverse pool of catabolic enzymes among brown rot fungi for various applications. It also implies that the broad outcome of brown rot must be, in part, predictable as a function of environment, given the convergence among taxonomically divergent fungal species on the same general strategy.
MATERIALS AND METHODS
Microcosm setup and harvest.
Gloeophyllum trabeum ATCC 11539 and Serpula lacrymans S7.3 were cultivated on 60-mm by 25-mm by 3-mm spruce wafers in modified ASTM standard soil block microcosms, as previously described (9). In brief, wafers were propped vertically within microcosms atop a hyphal mat and incubated in the dark at room temperature until fungal hyphae had advanced 45 to 50 mm up the wafers (2 to 3 weeks). Wafers with straight horizontal hyphal fronts were harvested, and average growth rates were determined by dividing total growth up the wafers by the total incubation time, less a 3-day lag (n = 60). Surface hyphae were removed and wafers were sectioned into seven 5-mm sections starting at the hyphal front and extending 35 mm behind it. Wafer sections were chopped into smaller pieces with sterilized razor blades prior to protein or ergosterol extractions.
Protein extraction, purification, and assay.
Spruce wafers degraded by either G. trabeum or S. lacrymans (n = 30 per species) were processed as described above, and sections were pooled with equivalents (i.e., same distance behind the hyphal front) from other wafers. Sections were suspended in 50 ml of cold 0.5 M NaCl and gently shaken for 24 h at 4°C. The extracts were filtered through Miracloth, centrifuged at 4,000 rpm for 10 min, and filtered through sterile 0.2-μm-pore polyvinylidene difluoride (PVDF) filters. Protein concentrations were determined using a Bio-Rad protein assay kit (Hercules, CA, USA).
Cellulases and hemicellulases were measured by the dinitrosalicylic acid (DNS) method using solutions of 1.5% carboxymethyl cellulose (endoglucanase), 2% birchwood xylan (xylanase), and 0.5% locust bean gum (mannanase) (24). Triplicate portions of protein extract were incubated with substrate at 50°C for 30 min in 50 mM citrate (pH 5.3). The absorbance at 540 nm was measured, and reducing sugars were determined as glucose-, xylose-, and mannose-reducing equivalents for endoglucanase, xylanase, and mannanase activities, respectively.
Mass spectrometry.
Extracts from spruce wafer sections 0 to 5, 10 to 15, and 20 to 25 mm from the visible hyphal front were generated as described above and concentrated using 3,000-kDa-cutoff polyether sulfone membranes. Extracts were tricarboxylic acid (TCA)-acetone precipitated and reconstituted in 7 M urea, 2 M thiourea, 0.4 M triethylammonium bicarbonate (TEAB) (pH 8.5), 20% methanol, and 4 mM Tris(2-carboxyethyl)phosphine (TCEP). Twenty-microgram aliquots were reduced for 1 h at 37°C and alkylated in 8 mM methyl methanethiosulfonate. Samples were diluted 4-fold with ultrapure water, mixed with trypsin at 1:50, incubated for 16 h at 37°C, dried, and cleaned with a 4-ml Extract-Clean C18 SPE cartridge from Grace-Davidson (Deerfield, IL), with eluates dried in vacuo.
Tryptic peptides were rehydrated in water, acetonitrile, and formic acid at a ratio of 98 to 1.9 to 0.1, respectively. Each sample was subjected to a Paradigm Platinum Peptide Nanotrap precolumn (0.15 by 50 mm, 400-μl volume; Michrom Bioresources, Inc., Auburn, CA, USA), followed by an analytical capillary column (100 μm by 12 cm) packed with C18 resin (5 μm, 200 Å; MagicC18AG; Michrom Bioresources, Inc.) at a flow rate of 600 nl/min. Peptides were fractionated on a 60-min (5 to 35% acetonitrile) gradient on a Paradigm MS4 high-performance liquid chromatograph (HPLC; Michrom Bioresources, Inc.). Mass spectrometry (MS) was performed on an LTQ mass spectrometer (Thermo Electron Corp., San Jose, CA, USA). Ionized peptides eluting from the capillary column were subjected to an ionizing voltage (1.8 kV) and selected for tandem MS (MS/MS) using a data-dependent procedure alternating between an MS scan and five MS/MS scans for the five most abundant precursor ions. Identified proteins were filtered using Scaffold 4.0 (Proteome Software, Portland, OR, USA). Positive protein identification was restricted to peptides with a 95.0% peptide identification probability threshold against the S. lacrymans and G. trabeum predicted proteins and a minimum number of 2 observed peptides/protein.
Ergosterol analysis.
Total ergosterol, a biomarker used to determine fungal biomass, was extracted from individual spruce wafer sections using established methods (25). Ergosterol was measured by HPLC, using previously described parameters (26), with detection of absorbance at 282 nm.
Supplementary Material
ACKNOWLEDGMENTS
This work was funded in part by the U.S. Department of Energy Office of Science (Early Career Grant DE-SC0004012 to J.S.S., from the Office of Biological and Ecological Research [BER], and BER grant DE-SC0012742 to J.S.S.). This work was also supported by the National Science Foundation Graduate Research Fellowship Program under grant 00039202 to G.N.P.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.02987-16.
REFERENCES
- 1.Baldrian P, Valaskova V. 2008. Degradation of cellulose by basidiomycetous fungi. FEMS Microbiol Rev 32:501–521. doi: 10.1111/j.1574-6976.2008.00106.x. [DOI] [PubMed] [Google Scholar]
- 2.Hyde SM, Wood PM. 1997. A mechanism for production of hydroxyl radicals by the brown-rot fungus Coniophora puteana: Fe(III) reduction by cellobiose dehydrogenase and Fe(II) oxidation at a distance from the hyphae. Microbiology 143:259–266. doi: 10.1099/00221287-143-1-259. [DOI] [PubMed] [Google Scholar]
- 3.Korripally P, Timokhin VI, Houtman CJ, Mozuch MD, Hammel KE. 2013. Evidence from Serpula lacrymans that 2,5-dimethoxyhydroquinone is a lignocellulolytic agent of divergent brown rot basidiomycetes. Appl Environ Microbiol 79:2377–2383. doi: 10.1128/AEM.03880-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hibbett DS, Donoghue MJ, Olmstead R. 2001. Analysis of character correlations among wood decay mechanisms, mating systems, and substrate ranges in homobasidiomycetes. Syst Biol 50:215–242. doi: 10.1080/10635150121079. [DOI] [PubMed] [Google Scholar]
- 5.Riley R, Salamov AA, Brown DW, Nagy LG, Floudas D, Held BW, Levasseur A, Lombard V, Morin E, Otillar R, Lindquist EA, Sun H, LaButti KM, Schmutz J, Jabbour D, Luo H, Baker SE, Pisabarro AG, Walton JD, Blanchette RA, Henrissat B, Martin F, Cullen D, Hibbett DS, Grigoriev IV. 2014. Extensive sampling of basidiomycete genomes demonstrates inadequacy of the white-rot/brown-rot paradigm for wood decay fungi. Proc Natl Acad Sci U S A 111:9923–9928. doi: 10.1073/pnas.1400592111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lundell TK, Makela MR, de Vries RP, Hilden KS. 2014. Genomics, lifestyles and future prospects of wood-decay and litter-decomposing basidiomycota, p 329–370. In Martin FM. (ed), Advances in botanical research: fungi, vol 70 Elsevier Press, Oxford, United Kingdom. [Google Scholar]
- 7.Schilling JS, Kaffenberger JT, Liew FJ, Song Z. 2015. Signature wood modifications reveal decomposer community history. PLoS One 10:e0120679. doi: 10.1371/journal.pone.0120679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kaffenberger JT, Schilling JS. 2013. Using a grass substrate to compare decay among two clades of brown rot fungi. Appl Microbiol Biotechnol 97:8831–8840. doi: 10.1007/s00253-013-5142-0. [DOI] [PubMed] [Google Scholar]
- 9.Zhang JW, Presley GN, Hammel KE, Ryu JS, Menke JR, Figueroa M, Hu DH, Orr G, Schilling JS. 2016. Localizing gene regulation reveals a staggered wood decay mechanism for the brown rot fungus Postia placenta. Proc Natl Acad Sci U S A 113:10968–10973. doi: 10.1073/pnas.1608454113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Floudas D, Binder M, Riley R, Barry K, Blanchette RA, Henrissat B, Martinez AT, Otillar R, Spatafora JW, Yadav JS, Aerts A, Benoit I, Boyd A, Carlson A, Copeland A, Coutinho PM, de Vries RP, Ferreira P, Findley K, Foster B, Gaskell J, Glotzer D, Gorecki P, Heitman J, Hesse C, Hori C, Igarashi K, Jurgens JA, Kallen N, Kersten P, Kohler A, Kues U, Kumar TKA, Kuo A, LaButti K, Larrondo LF, Lindquist E, Ling A, Lombard V, Lucas S, Lundell T, Martin R, McLaughlin DJ, Morgenstern I, Morin E, Murat C, Nagy LG, Nolan M, Ohm RA, Patyshakuliyeva A, et al. . 2012. The Paleozoic origin of enzymatic lignin decomposition reconstructed from 31 fungal genomes. Science 336:1715–1719. doi: 10.1126/science.1221748. [DOI] [PubMed] [Google Scholar]
- 11.Gilbertson RL, Ryvarden L. 1986. North American polypores, vol 1 Fungiflora, Oslo, Norway. [Google Scholar]
- 12.Palfreyman JW, Gartland JS, Sturrock CJ, Lester D, White NA, Low GA, Bech-Andersen J, Cooke DEL. 2003. The relationship between ‘wild’ and ‘building’ isolates of the dry rot fungus Serpula lacrymans. FEMS Microbiol Lett 228:281–286. doi: 10.1016/S0378-1097(03)00783-3. [DOI] [PubMed] [Google Scholar]
- 13.Jennings DH, Bravery AF. 1991. Serpula lacrymans: fundamental biology and control strategies. Wiley, New York, NY. [Google Scholar]
- 14.Lin SC, Lo YC, Lin JY, Liaw YC. 2004. Crystal structures and electron micrographs of fungal volvatoxin A2. J Mol Biol 343:477–491. doi: 10.1016/j.jmb.2004.08.045. [DOI] [PubMed] [Google Scholar]
- 15.Weng YP, Lin YP, Hsu CI, Lin JY. 2004. Functional domains of a pore-forming cardiotoxic protein, volvatoxin A2. J Biol Chem 279:6805–6814. [DOI] [PubMed] [Google Scholar]
- 16.Tateno H, Goldstein IJ. 2003. Molecular cloning, expression, and characterization of novel hemolytic lectins from the mushroom Laetiporus sulphureus, which show homology to bacterial toxins. J Biol Chem 278:40455–40463. doi: 10.1074/jbc.M306836200. [DOI] [PubMed] [Google Scholar]
- 17.White NA, Low GA, Singh J, Staines H, Palfreyman JW. 1997. Isolation and environmental study of ‘wild’ Serpula lacrymans and Serpula himantioides from the Himalayan forests. Mycol Res 101:580–584. doi: 10.1017/S0953756296003000. [DOI] [Google Scholar]
- 18.Eastwood DC, Floudas D, Binder M, Majcherczyk A, Schneider P, Aerts A, Asiegbu FO, Baker SE, Barry K, Bendiksby M, Blumentritt M, Coutinho PM, Cullen D, de Vries RP, Gathman A, Goodell B, Henrissat B, Ihrmark K, Kauserud H, Kohler A, LaButti K, Lapidus A, Lavin JL, Lee YH, Lindquist E, Lilly W, Lucas S, Morin E, Murat C, Oguiza JA, Park J, Pisabarro AG, Riley R, Rosling A, Salamov A, Schmidt O, Schmutz J, Skrede I, Stenlid J, Wiebenga A, Xie XF, Kues U, Hibbett DS, Hoffmeister D, Hogberg N, Martin F, Grigoriev IV, Watkinson SC. 2011. The plant cell wall-decomposing machinery underlies the functional diversity of forest fungi. Science 333:762–765. doi: 10.1126/science.1205411. [DOI] [PubMed] [Google Scholar]
- 19.Kojima Y, Varnai A, Ishida T, Sungawa N, Petrovic DM, Igarashi K, Jellison J, Goodell B, Alfredsen G, Westereng B, Eijsink VGH, Yoshida M. 2016. A lytic polysaccharide monooxygenase with broad xyloglucan specificity from the brown rot fungus Gloeophyllum trabeum and its action on cellulose-xyloglucan complexes. Appl Environ Microbiol 82:6557–6572. doi: 10.1128/AEM.01768-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kersten P, Cullen D. 2014. Copper radical oxidases and related extracellular oxidoreductases of wood-decay Agaricomycetes. Fungal Genet Biol 72:124–130. doi: 10.1016/j.fgb.2014.05.011. [DOI] [PubMed] [Google Scholar]
- 21.Daniel G, Volc J, Filonova L, Plihal O, Kubatova E, Halada P. 2007. Characteristics of Gloeophyllum trabeum alcohol oxidase, an extracellular source of H2O2 in brown rot decay of wood. Appl Environ Microbiol 73:6241–6253. doi: 10.1128/AEM.00977-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hori C, Gaskell J, Igarashi K, Kersten P, Mozuch M, Samejima M, Cullen D. 2014. Temporal alterations in the secretome of the selective ligninolytic fungus Ceriporiopsis subvermispora during growth on aspen wood reveal this organism's strategy for degrading lignocellulose. Appl Environ Microbiol 80:2062–2070. doi: 10.1128/AEM.03652-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kuuskeri J, Hakkinen M, Laine P, Smolander OP, Tamene F, Miettinen S, Nousiainen P, Kemell M, Auvinen P, Lundell T. 2016. Time-scale dynamics of proteome and transcriptome of the white-rot fungus Phlebia radiata: growth on spruce wood and decay effect on lignocellulose. Biotechnol Biofuels 9:192. doi: 10.1186/s13068-016-0608-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ghose TK. 1987. Measurement of cellulase activities. Pure Appl Chem 59:257–268. [Google Scholar]
- 25.Newell SY, Arsuffi TL, Fallon RD. 1988. Fundamental procedures for determining ergosterol content of decaying plant-material by liquid chromatography. Appl Environ Microbiol 54:1876–1879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schilling JS, Jellison J. 2005. Oxalate regulation by two brown rot fungi decaying oxalate-amended and non-amended wood. Holzforschung 59:681–688. [Google Scholar]
- 27.UniProt Consortium. 2015. UniProt: a hub for protein information. Nucleic Acids Res 43:D204–D212. doi: 10.1093/nar/gku989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Grigoriev IV, Nikitin R, Haridas S, Kuo A, Ohm R, Otillar R, Riley R, Salamov A, Zhao XL, Korzeniewski F, Smirnova T, Nordberg H, Dubchak I, Shabalov I. 2014. MycoCosm portal: gearing up for 1000 fungal genomes. Nucleic Acids Res 42:D699–D704. doi: 10.1093/nar/gkt1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lombard V, Ramulu HG, Drula E, Coutinho PM, Henrissat B. 2014. The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res 42:D490–D495. doi: 10.1093/nar/gkt1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Petersen TN, Brunak S, von Heijne G, Nielsen H. 2011. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat Methods 8:785–786. doi: 10.1038/nmeth.1701. [DOI] [PubMed] [Google Scholar]
- 31.Song ZW, Vail A, Sadowsky MJ, Schilling JS. 2014. Quantitative PCR for measuring biomass of decomposer fungi in planta. Fungal Ecol 7:39–46. doi: 10.1016/j.funeco.2013.12.004. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.