ABSTRACT
Peptidoglycan hydrolases (PGHs) have been suggested as novel therapeutics for the treatment of bovine mastitis. However, activity in the presence of cow's milk is an important requirement for drugs administered into the bovine udder. We have screened a library of >170 recombinant PGHs, including engineered bacteriophage endolysins, for enzymes with activity against Staphylococcus aureus in milk, using a microtiter plate-based protocol. Nine suitable PGH constructs were identified by this approach and further compared in time-kill assays for their efficacy against S. aureus in heat-treated milk. The three most active enzymes (lysostaphin, Ami2638A, and CHAPK_CWT-LST) reduced S. aureus in milk to undetectable numbers within minutes at nanomolar concentrations. Due to their different peptidoglycan cleavage sites, these PGH constructs revealed synergistic activity in most combinations, as demonstrated by checkerboard assays, spot assays, and time-kill experiments. Furthermore, they proved active against a selection of staphylococcal mastitis isolates from different geographical regions when applied individually or in synergistic combination. The most effective PGH combination completely eradicated S. aureus from milk, with no more bacteria being detected within 24 h after addition of the enzymes, corresponding to a reduction of >9 log units compared to the control. Efficacy was also retained at different inoculum levels (3 versus 6 log CFU/ml) and when S. aureus was grown in milk as opposed to broth prior to the experiments. In raw cow's milk, CHAPK_CWT-LST showed reduced efficacy, whereas both Ami2638A and lysostaphin retained their activity, reducing bacterial numbers by >3.5 log units within 3 h.
IMPORTANCE Staphylococci and S. aureus in particular are a major cause of bovine mastitis, an inflammation of the mammary gland in cows associated with high costs and risks for consumers of milk products. S. aureus-induced mastitis, commonly treated by intramammary infusion of antibiotics, is characterized by low cure rates and increasing antibiotic resistance in bacteria. Therefore, alternative treatment options are highly desirable. PGHs, including bacteriophage endolysins, rapidly and specifically kill selected pathogens by degrading their cell wall and are refractory to resistance development, therefore holding promise as novel antibacterial agents. This study employed a screening approach to identify PGH constructs with high staphylolytic activity in cow's milk within a large collection of enzymes. Our results suggest that the most promising enzymes identified by this strategy hold potential as novel mastitis therapeutics and support their further characterization in animal models.
KEYWORDS: antibiotic resistance, antimicrobial agents, bovine mastitis, peptidoglycan hydrolases
INTRODUCTION
Staphylococcus is a genus of Gram-positive cocci that includes both human and animal pathogens. Besides its important role as a human pathogen (1), Staphylococcus aureus (in addition to various coagulase-negative staphylococcal species [CoNS]) is a major causative agent of bovine mastitis, an infection of the mammary gland in cows, which represents the most widespread and costly disease in animal agriculture (2). Loss in milk production and quality, veterinary treatment, and premature culling of animals due to intramammary infections amount to annual costs of approximately $200 per cow (3). As a contagious mastitis pathogen, S. aureus can be transmitted from cow to cow, often during the milking process via contaminated equipment, and can cause both acute and, most frequently, chronic subclinical mastitis (4). Especially in the latter case, infections are difficult to cure, since the pathogens frequently persist intracellularly and in the form of dormant small-colony variants (SCVs), avoiding the host's immune response and treatment by antimicrobial agents (5, 6). In the absence of pathogen-specific antimicrobials, treatment of mastitis has historically been limited to the use of antibiotics such as cephalosporins, penicillin, and pirlimycin (administered via intramammary infusion), which are often less than 50% successful (7). Another important factor contributing to this low cure rate is the increasing prevalence of antibiotic-resistant strains such as methicillin-resistant S. aureus (MRSA) in both human clinics and agricultural settings (8), warranting the need for alternative antimicrobial agents for effective treatment of Staphylococcus-induced bovine mastitis.
Peptidoglycan hydrolases (PGHs) and bacteriophage endolysins in particular represent such novel types of antimicrobials and have received increasing attention due to their high killing efficacy, high specificity for the target pathogen, low chance of resistance development, and activity against dormant bacteria (9–12). Endolysins are enzymes encoded by bacteriophage and produced inside the bacterial host cell at the end of the phage's lytic cycle, from which they get access to the cell wall and degrade their peptidoglycan (PG) substrate, ultimately resulting in cell lysis and release of progeny virions. Due to the absence of an outer membrane in Gram-positive organisms, PGHs can also act from the outside against these bacteria, and their antimicrobial potential has been demonstrated both in vitro and in multiple animal infection models (9, 13). PGHs from a Gram-positive background feature a modular design, consisting of enzymatically active domains (EADs), which define the enzyme's cleavage sites within the PG, and cell wall binding domains (CBDs), which confer specificity on the genus, species, or serovar level. Staphylococcal phage endolysins typically comprise one C-terminal genus-specific SH3b-type CBD (14) and two EADs: a cysteine-histidine-dependent amidohydrolase/peptidase (CHAP) or M23 endopeptidase domain and an amidase domain (15). This modular architecture makes these enzymes highly amenable to protein engineering, enabling the creation of chimeric fusion proteins with novel enzymatic and antimicrobial properties (9, 16, 17). While phage endolysins have evolved to cleave highly conserved bonds within the PG, making resistance development unlikely, resistance against nonendolysin PGHs such as lysostaphin (18) has been described. This bacteriocin targets the variable pentaglycine bridge of staphylococcal PG, and consequently, lysostaphin-resistant mutant strains featuring variations in this PG region have been reported (19, 20). Various studies using animal models suggest that both lysostaphin and engineered staphylococcal phage endolysins hold promise as potential therapeutics for treatment of bovine mastitis (3, 17, 21, 22). Recently, Becker et al. were able to demonstrate in cultured bovine mammary gland epithelial cells and in a mouse model of mastitis that fusion of PGHs to short, positively charged or amphipathic peptides termed protein transduction domains (PTDs) rendered the enzymes active against intracellular S. aureus, an important achievement in the fight against chronic intramammary infections (17). Besides efficacy against drug-resistant, dormant, and intracellular pathogens, activity in the presence of milk is an important requirement for mastitis therapeutics administered through the intramammary route and often the limiting factor when selecting PGHs for this purpose. When 9 unique staphylococcal PGHs were compared for their activity in cow's milk against S. aureus, only one enzyme (lysostaphin) was able to reduce bacterial numbers by more than 1 log unit (M. Schmelcher, unpublished data). In a mastitis mouse model, two chimeric endolysins effectively killed intramammary S. aureus in synergistic combination with lysostaphin. However, when applied individually, they showed only low efficacy, presumably due to their merely moderate activity in milk (22).
In this study, we developed a microtiter plate-based screening method allowing rapid identification of PGHs with activity in cow's milk within a large enzyme library. The three most suitable candidate PGHs identified by this method were characterized with regard to their antimicrobial efficacy against staphylococci in milk under various conditions and their synergistic effects when applied in combination.
RESULTS
Microtiter plate-based screening method identifies PGH constructs with high staphylolytic activity in cow's milk.
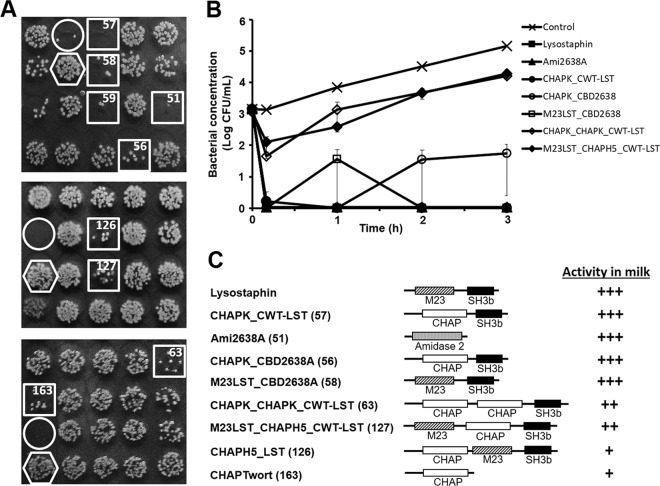
In an effort to identify PGH constructs featuring high staphylolytic activity in cow's milk as potential therapeutics against bovine mastitis, we established a protocol which allows for rapid screening of large enzyme libraries in a 96-well plate format. In this approach, promising enzymes are detected on the basis of reduced colony density on agar plates within spots of S. aureus suspensions that had been exposed to different freshly produced PGHs in milk (Fig. 1). Applying this protocol to a collection of >170 staphylococcal PGH constructs in Escherichia coli (comprising parental and engineered bacteriophage endolysins and other PGHs), we were able to select 9 promising candidate enzymes (including the positive control, lysostaphin) for further analysis. In 3 independent rounds of screening with each construct being included 3 times in different positions on each screening plate, these 9 constructs consistently yielded visibly reduced S. aureus colony densities compared to the negative control (an E. coli strain not expressing any PGH) (Fig. 1A). The 9 candidates are listed in Table 1 and include (besides lysostaphin) the constructs CHAPK_CWT-LST, a fusion protein consisting of the CHAP endopeptidase domain of the phage endolysin LysK (23, 24) and the SH3b-type cell wall targeting domain (CWT) of lysostaphin; Ami2638A, a truncation construct harboring the amidase domain of the 2638A endolysin (25); CHAPK_CBD2638A, a fusion of the LysK CHAP domain with the SH3b CBD of the 2638A endolysin; M23LST_CBD2638A, a fusion of the M23 endopeptidase domain of lysostaphin and the CBD of 2638A; CHAPK_CHAPK_CWT-LST, a construct containing the duplicated CHAP domain of LysK fused to the CWT of lysostaphin; M23LST_CHAPH5_CWT-LST, a fusion protein featuring an N-terminal M23 domain of lysostaphin, a centrally located CHAP domain from LysH5 (an endolysin reportedly active in cow's milk) (26), and a C-terminal CWT of lysostaphin; CHAPH5_LST, which includes the CHAP domain of LysH5 fused to lysostaphin; and CHAPTwort, a truncation construct consisting of the CHAP domain of the phage Twort endolysin (27). Of note, this selection of 9 PGH constructs comprises enzymes with diverse domain architectures, ranging from single EADs without a cell wall binding module to chimeric multidomain fusion proteins, and includes EADs targeting multiple PG cleavage sites (amidase, CHAP endopeptidases, and lysostaphin M23 endopeptidase). Furthermore, it includes constructs encoded by vectors derived from different plasmid backbones and expressed by different E. coli strains (Table 1). In order to comparatively analyze the 9 candidate enzymes, they were produced in E. coli on a large scale and purified by immobilized metal ion affinity chromatography (IMAC) via their 6×His affinity tags. As demonstrated by SDS-PAGE, all preparations contained target proteins of the expected molecular weight and of >90% purity, with the exception of CHAPK_CHAPK_CWT-LST (see Fig. S1 in the supplemental material). For the latter, several contaminating bands at lower molecular weights (presumably degradation products) were detected in addition to the expected band at 54.3 kDa. Time-kill assays were performed with the 9 purified candidates against mastitis-inducing S. aureus in ultra-heat-treated (UHT) cow's milk. When the PGH constructs were added to milk spiked with 103 CFU/ml (an inoculum mimicking bacterial concentrations observed at early stages of infection or for subclinical S. aureus-induced bovine mastitis) at a concentration of 4 μM, 7 out of 9 enzymes immediately reduced bacterial numbers to undetectable levels, and no bacteria were detected until the end of the experiment at 3 h postinoculation (corresponding to a reduction by >4.5 log units compared to the buffer control) (Fig. S2A). CHAPH5_LST and CHAPTwort were less effective, resulting in bacterial concentrations 1.3 and 1.8 log units below the control after 3 h, respectively. To further differentiate between the 7 most active constructs, time-kill assays were performed with lower enzyme concentrations (360 nM, corresponding to 10 μg/ml of lysostaphin). Under these conditions, complete eradication of S. aureus was achieved with lysostaphin, Ami2638A, and CHAPK_CWT-LST, and bacterial concentrations remained below the detection limit until the end of the experiment (Fig. 1B). Also, CHAPK-CBD2638A and M23LST_CBD2638A caused CFU numbers to drop below the detection limit immediately after addition of the enzymes; however, S. aureus was occasionally detected at low concentrations (∼1.5 log CFU/ml) at later time points during the course of the experiment. CHAPK_CHAPK_CWT-LST and M23LST_CHAPH5_CWT-LST were clearly less effective, keeping bacterial concentrations approximately 1 log unit below those of the control. When tested at 10-fold-lower concentrations (36 nM), none of the 5 most effective enzymes was able to clear the milk from S. aureus within 3 h (Fig. S2B). Based on their performance in the time-kill assays at different concentrations, the 9 candidates were ranked (Fig. 1C), and the three most effective enzymes (lysostaphin, CHAPK_CWT-LST, and Ami2638A) were selected for further characterization.
FIG 1.
Identification of PGH constructs with high activity against S. aureus in milk. (A) Three sections from screening plates used for identification of promising PGH constructs (see the text for details). E. coli strains harboring different PGH constructs were grown in 96-well plates. Following protein production, cells were harvested and enzymes were liberated by exposure of pellets to chloroform vapor. UHT milk spiked with S. aureus Newbould 305 was added to the wells and incubated for 2 h before aliquots of each well were spotted on agar plates. Spots with largely reduced S. aureus colony density after overnight incubation (squares) suggest the presence of promising PGH constructs in the corresponding wells. A lysostaphin-expressing E. coli strain (circles) and a strain harboring no PGH construct (hexagons) served as positive and negative controls, respectively. Numbers in the squares correspond to PGH constructs shown in panel C. (B) Comparative time-kill assay with S. aureus Newbould 305 cells in UHT milk using purified PGH constructs (360 nM) that had been identified as promising candidates by the screening as exemplified in panel A or buffer as control. Bacterial concentrations at different time points after enzyme/buffer addition were determined by serial dilution plating. It should be noted that symbols for lysostaphin, Ami2638A, and CHAPK_CWT-LST appear on top of each other since these constructs caused instantaneous reduction of S. aureus to undetectable levels. Error bars represent standard errors of the means from 3 experiments. (C) Schematics of candidate PGH constructs rated for their activity in milk based on log reduction of S. aureus cells compared to the control after 3 h in time-kill assays as exemplified in panel B. Construct numbers are given in parentheses. Note that construct 59 (A) is an N-terminally 6×His-tagged version of the positive control lysostaphin and was therefore not included in the ranking. +, <3 log reduction at 4 μM; ++, ≥3 log reduction at 4 μM but <3 log reduction at 360 nM; +++, ≥3 log reduction at 360 nM. M23, M23 endopeptidase domain; CHAP, cysteine, histidine-dependent amidohydrolase/peptidase domain; Amidase 2, amidase 2 domain; SH3b, bacterial SH3 cell wall binding domain.
TABLE 1.
PGH constructs characterized in this study
| PGH | Backbone plasmid | E. coli strain | Mol mass (kDa) |
|---|---|---|---|
| Lysostaphina | pET-21a | BL21-Gold(DE3) | 28.2 |
| CHAPK_CWT-LSTb | pQE-30 | XL1-Blue MRF′ | 32.2 |
| Ami2638Ab | pQE-30 | XL1-Blue MRF′ | 29.1 |
| CHAPK_CBD2638Ab | pQE-30 | XL1-Blue MRF′ | 35.7 |
| M23LST_CBD2638Ab | pQE-30 | XL1-Blue MRF′ | 30.7 |
| CHAPK_CHAPK_CWT-LSTb | pQE-30 | SURE | 54.3 |
| M23LST_CHAPH5_CWT-LSTa | pET-21a | BL21-Gold(DE3) | 47.8 |
| CHAPH5_LSTa | pET-21a | BL21-Gold(DE3) | 47.6 |
| CHAPTworta | pET-21a | BL21-Gold(DE3) | 17.6 |
Construct contains a C-terminal 6×His tag.
Construct contains an N-terminal 6×His tag followed by a tobacco etch virus protease cleavage site.
Mixtures of PGH constructs featuring different PG cleavage sites exhibit synergistic antibacterial activity against multiple S. aureus mastitis isolates.
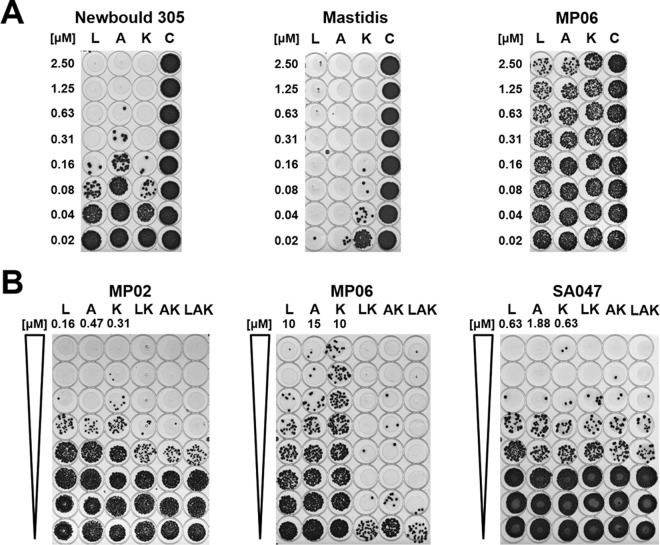
Lysostaphin and the 2 PGH constructs CHAPK_CWT-LST and Ami2638A together feature 3 different cleavage sites in the S. aureus PG, as has been determined previously by mass spectrometry analysis of PG digestion products (15, 23). The M23 endopeptidase domain of lysostaphin cleaves the second and third glycyl-glycine bond within the pentaglycine bridge; CHAPK, the catalytic domain of CHAPK_CWT-LST, cuts the d-Ala-Gly bond between the stem peptide and the pentaglycine bridge; and Ami2638A targets the MurNAc-l-Ala bond between the sugar moiety and the stem peptide (Fig. S3A). This finding led us to explore possible synergistic effects when using these enzymes in combination. In preparation for the synergy studies, the MICs of the three enzymes against S. aureus Newbould 305 were determined using a classical broth microdilution method (Fig. S4). The obtained MICs in tryptic soy broth (TSB) medium were 0.063 ± 0.000 μM (standard deviation [SD]) for lysostaphin, 0.125 ± 0.000 μM for Ami2638A, and 0.667 ± 0.289 μM for CHAPK_CWT-LST (n = 3). Based on these values, checkerboard assays with pairwise combinations of the three PGHs in a broth microdilution format were conducted (Fig. S3B to D). The sums of fractional inhibitory concentrations (ΣFICs), which are a measure for synergistic, additive, or inhibitory effects exhibited by combinations of antimicrobial agents, were calculated from these experiments. The ΣFICs obtained here were 0.460 ± 0.036 (SD, n = 3) for lysostaphin and CHAPK_CWT-LST (Lyso:CHAP, indicating strong synergy), 0.590 ± 0.043 (n = 3) for Ami2638A and CHAPK_CWT-LST (Ami:CHAP, synergy), and 0.714 ± 0.119 (n = 5) for lysostaphin and Ami2638A (Lyso:Ami, weak or no synergy). It should be noted that, for Lyso:Ami, reproducibility was lower than for the other combinations, and turbid wells sometimes occurred randomly at higher PGH concentrations (Fig. S3D). Relating to a potential use of the 3 PGHs as mastitis therapeutics, we aimed at reproducing these effects in cow's milk. To this end, we employed a modified version of the classical checkerboard assay, using a 2-dimensional spot assay format (Fig. S5). The sums of fractional bactericidal concentrations (ΣFBCs; corresponding to the ΣFIC in the classical assay) obtained in these experiments were 0.376 ± 0.029 (SD, n = 3) for Lyso:CHAP and 0.397 ± 0.096 (n = 3) for Ami:CHAP, both suggesting strong synergy in milk, and 0.949 ± 0.094 (n = 3) for Lyso:Ami. As a next step, we investigated the susceptibility of a collection of staphylococcal mastitis isolates from different geographic regions to our 3 PGH constructs in milk. For this purpose, 1-dimensional spot assays were performed to determine minimum bactericidal concentrations (MBCs; exemplified in Fig. 2A) and synergistic effects (ΣFBCs; Fig. 2B). The results of these experiments are summarized in Table 2. The majority of tested strains were highly susceptible to all 3 individually applied enzymes, with MBCs below 1 μM. Staphylococcus chromogenes MP02 and Staphylococcus warneri MP06 exhibited greater tolerance against the individual constructs (MBC, >2.5 μM); however, both strains were highly susceptible to synergistic PGH mixtures (as exemplified for MP06 in Fig. 2B). For most strains, the combinations Lyso:CHAP, Ami:CHAP, and Lyso:Ami:CHAP revealed strong synergy (ΣFBC, ≤0.5), whereas Lyso:Ami was not or only weakly synergistic, which is in agreement with the results of the checkerboard assays. The mean ΣFBCs for the Lyso:CHAP, Ami:CHAP, Lyso:Ami, and Lyso:Ami:CHAP combinations were 0.441 ± 0.196, 0.475 ± 0.209, 0.963 ± 0.153, and 0.428 ± 0.195, respectively, with Lyso:Ami being significantly (P < 0.01) different from the other combinations. While the triple combination Lyso:Ami:CHAP yielded the lowest mean ΣFBC value, the differences from Lyso:CHAP and Ami:CHAP were not statistically significant.
FIG 2.
Spot assays in milk with PGH constructs and different staphylococcal mastitis isolates. (A) Determination of minimum bactericidal concentrations (MBCs). Twofold serial dilutions of lysostaphin (L), Ami2638A (A), and CHAPK_CWT-LST (K) or buffer as control (C) were incubated with staphylococci (5 × 105 CFU/ml) in milk in 96-well plates for 2 h at 37°C. Aliquots of 5 μl from each well were spotted on TSB agar cast in the sterile lids of 96-well plates and incubated overnight. In each case, the PGH concentration yielding the first cleared spot was defined as the MBC. Examples for medium (S. aureus Newbould 305), high (S. aureus Mastidis), and low (S. warneri MP06) susceptibility are shown. (B) Determination of synergistic effects in milk when using mixtures of PGH constructs. Twofold serial dilutions of single enzymes and mixtures of lysostaphin and CHAPK_CWT-LST (LK), Ami2638A and CHAPK_CWT-LST (AK), and all three enzymes (LAK) were prepared in milk and tested in spot assays as described for panel A. For each individual PGH construct, the highest concentration used (i.e., the concentration in the first well) is indicated on top of the respective lane. The concentration of each enzyme in a mixture was half (for LK and AK) or one-third (for LAK) of the concentration of the respective single PGH construct in the same row. Results for S. chromogenes MP02 (synergy), S. warneri MP06 (strong synergy), and S. aureus SA047 (no synergy) are shown.
TABLE 2.
Susceptibilities of different staphylococcal mastitis isolates in milk to individual PGH constructs and synergistic effects of PGH combinations
| Staphylococcus strain | Origina | Susceptibility to single PGHb: |
Synergistic effect of PGH combination (ΣFBC)c: |
|||||
|---|---|---|---|---|---|---|---|---|
| Lyso | Ami | CHAP | Lyso:CHAP | Ami:CHAP | Lyso:Ami | Lyso:Ami:CHAP | ||
| S. aureus Newbould 305 | 1 | ++++ | +++ | ++++ | 0.250 ± 0.000 | 0.239 ± 0.162 | 1.207 ± 0.293 | 0.162 ± 0.052 |
| S. aureus SA001 | 2 | ++++ | ++++ | +++ | 0.250 ± 0.000 | 0.302 ± 0.073 | 0.902 ± 0.169 | 0.282 ± 0.046 |
| S. aureus SA002 | 2 | ++++ | ++++ | +++ | 0.302 ± 0.073 | 0.479 ± 0.323 | 1.000 ± 0.000 | 0.448 ± 0.073 |
| S. aureus SA003 | 2 | ++++ | ++++ | ++++ | 0.302 ± 0.073 | 0.302 ± 0.073 | 1.138 ± 0.239 | 0.282 ± 0.046 |
| S. aureus SA009 | 2 | ++++ | ++++ | ++++ | 0.500 ± 0.000 | 0.604 ± 0.146 | 0.805 ± 0.169 | 0.448 ± 0.073 |
| S. aureus SA019 | 2 | ++++ | ++++ | ++++ | 0.427 ± 0.104 | 0.375 ± 0.177 | 1.061 ± 0.500 | 0.407 ± 0.131 |
| S. aureus SA020 | 2 | +++++ | ++++ | ++++ | 0.354 ± 0.000 | 0.854 ± 0.207 | 0.854 ± 0.207 | 0.356 ± 0.058 |
| S. aureus SA021 | 2 | ++++ | ++++ | ++ | 0.177 ± 0.000 | 0.177 ± 0.000 | 0.854 ± 0.207 | 0.178 ± 0.029 |
| S. aureus SA026 | 2 | ++++ | ++++ | + | 0.265 ± 0.125 | 0.302 ± 0.073 | 0.707 ± 0.000 | 0.257 ± 0.082 |
| S. aureus SA028 | 2 | +++++ | ++++ | ++++ | 0.707 ± 0.000 | 0.707 ± 0.000 | 0.804 ± 0.207 | 0.472 ± 0.223 |
| S. aureus SA029 | 2 | +++++ | ++++ | ++++ | 0.500 ± 0.000 | 0.750 ± 0.354 | 1.000 ± 0.000 | 0.750 ± 0.354 |
| S. aureus SA031 | 2 | ++++ | ++++ | ++++ | 0.354 ± 0.000 | 0.375 ± 0.177 | 1.000 ± 0.000 | 0.472 ± 0.223 |
| S. aureus SA033 | 2 | ++++ | ++++ | ++ | 0.213 ± 0.052 | 0.188 ± 0.088 | 1.061 ± 0.500 | 0.204 ± 0.065 |
| S. aureus SA047 | 2 | ++++ | ++++ | ++++ | 0.750 ± 0.357 | 0.750 ± 0.357 | 1.061 ± 0.500 | 0.750 ± 0.357 |
| S. aureus SA048 | 2 | ++++ | ++++ | ++++ | 0.375 ± 0.177 | 0.375 ± 0.177 | 1.000 ± 0.000 | 0.375 ± 0.177 |
| S. aureus SA049 | 2 | ++++ | ++++ | ++++ | 0.375 ± 0.177 | 0.375 ± 0.177 | 0.707 ± 0.000 | 0.250 ± 0.000 |
| S. aureus Mastidis | 3 | +++++ | +++++ | ++++ | 0.677 ± 0.457 | 0.604 ± 0.146 | 1.000 ± 0.000 | 0.448 ± 0.073 |
| S. aureus M5512VL | 4 | +++++ | +++++ | ++++ | 0.667 ± 0.289 | 0.500 ± 0.000 | 1.000 ± 0.000 | 0.466 ± 0.060 |
| S. aureus M5702 | 4 | +++++ | +++++ | +++++ | 0.500 ± 0.354 | 0.333 ± 0.000 | 0.667 ± 0.000 | 0.333 ± 0.000 |
| S. aureus M6020VR | 4 | +++++ | +++++ | ++++ | 0.750 ± 0.354 | 1.000 ± 0.000 | 1.000 ± 0.000 | 1.000 ± 0.000 |
| S. aureus M3783 | 4 | +++++ | +++++ | +++++ | 0.604 ± 0.120 | 0.604 ± 0.120 | 1.000 ± 0.000 | 0.565 ± 0.075 |
| S. aureus G68 | 4 | ++++ | ++++ | ++++ | 0.845 ± 0.207 | 0.500 ± 0.000 | 0.854 ± 0.207 | 0.647 ± 0.208 |
| S. aureus M2071 | 4 | +++++ | ++++ | ++++ | 0.368 ± 0.126 | 0.638 ± 0.120 | 1.138 ± 0.239 | 0.587 ± 0.075 |
| S. chromogenes MP02 | 5 | +++++ | ++++ | +++++ | 0.500 ± 0.000 | 0.500 ± 0.000 | 1.276 ± 0.239 | 0.500 ± 0.000 |
| S. epidermidis MP04 | 5 | − | − | − | 0.156 ± 0.133 | 0.500 ± 0.000 | 1.000 ± 0.000 | 0.500 ± 0.000 |
| S. hyicus MP01 | 5 | +++++ | ++++ | +++ | 0.375 ± 0.177 | 0.500 ± 0.000 | 1.179 ± 0.408 | 0.375 ± 0.177 |
| S. simulans MP03 | 5 | +++++ | +++++ | ++++ | 0.604 ± 0.146 | 0.302 ± 0.073 | 0.805 ± 0.169 | 0.257 ± 0.082 |
| S. warneri MP06 | 5 | − | − | − | 0.141 ± 0.155 | 0.133 ± 0.166 | 1.000 ± 0.000 | 0.141 ± 0.155 |
| S. xylosus MP05 | 5 | +++++ | ++++ | ++++ | 0.500 ± 0.000 | 0.500 ± 0.000 | 0.854 ± 0.207 | 0.500 ± 0.000 |
Strains were obtained from sources as follows: 1, ATCC 29740; 2, Yasunori Tanji, Tokyo Institute of Technology, Yokohama, Japan; 3, Roger Stephan, University of Zurich, Zurich, Switzerland; 4, Hans Ulrich Graber, Agroscope, Liebefeld, Switzerland; 5, Max Paape, ARS, USDA, Beltsville, MD, USA.
Susceptibility was rated based on minimal bactericidal concentrations (MBCs) in milk. −, MBC > 2.5 μM; +, 1.9 μM < MBC ≤ 2.5 μM; ++, 1.3 μM < MBC ≤ 1.9 μM; +++, 0.7 μM < MBC ≤ 1.3 μM; ++++, 0.1 μM < MBC ≤ 0.7 μM; +++++, MBC < 0.1 μM.
Sums of fractional bactericidal concentrations (ΣFBC) of enzyme combinations in milk, as determined by spot assays. Average values and SDs from at least 2 independent experiments are shown.
Synergistic enzyme mixtures effectively kill S. aureus in UHT milk.
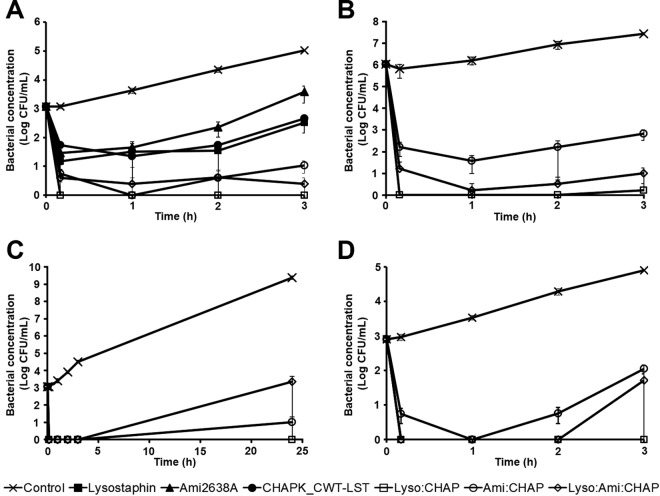
Having demonstrated synergy for various enzyme combinations in checkerboard and spot assays, we further explored if these synergistic effects also hold true in a different experimental setup, i.e., time-kill assays in UHT milk. To this end, enzyme concentrations were adjusted so that the 3 individually applied PGHs caused similar reductions in bacterial numbers over a period of 3 h and did not clear S. aureus from the milk. Efficacy of the individual PGH constructs at these predetermined concentrations was then compared to that of PGH mixtures at half (for mixtures of 2 enzymes) or one-third (for mixtures of all 3 enzymes) of these concentrations (Fig. 3A). After 3 h, lysostaphin (106.5 nM), Ami2638A (159.8 nM), and CHAPK_CWT-LST (106.5 nM) reduced bacterial concentrations by 2.64, 1.73, and 2.37 log units, respectively, compared to the control. These reductions were statistically significant (P < 0.01), whereas there were no significant differences between treatments with the 3 individual enzymes. The PGH combinations Lyso:CHAP (53.3 + 53.3 nM), Ami:CHAP (79.9 + 53.3 nM), and Lyso:Ami:CHAP (35.5 + 53.3 + 35.5 nM) reduced CFU by >5 (numbers below the detection limit), 4.24, and 4.80 log units, respectively. For all combination treatments, S. aureus concentrations at 3 h were significantly (P < 0.01) lower than those of the control and any of the individual enzyme treatments, whereas there was no significant difference between the three combination treatments. To investigate the therapeutic potential of these synergistic PGH mixtures also under conditions mimicking more acute stages of bovine mammary gland infection, time-kill assays were performed in milk inoculated with 1,000-fold-higher numbers (106 CFU/ml) of S. aureus. When applied at the same concentrations as described above (defined as 1× concentrations), the combinations Lyso:CHAP, Ami:CHAP, and Lyso:Ami:CHAP reduced bacterial numbers by 4.74, 3.55, and 3.98 log units compared to the control at 3 h, respectively (data not shown). At 3-times-higher concentrations (3×), reductions were even higher, with 7.22, 4.61, and 6.44 log units, respectively (Fig. 3B). We further explored whether the enzyme combinations were able to keep bacterial numbers in the milk at low levels also during extended periods of incubation. To this end, time-kill assays as described above were performed over a period of 24 h, using 3× PGH concentrations. At the experimental endpoint, S. aureus cells in the control sample had reached a concentration of 9.39 log CFU/ml. The combination Lyso:Ami:CHAP caused a reduction by 6.03 log units compared to the control, whereas Lyso:CHAP kept bacteria below the detection limit even after 24 h, suggesting complete eradication of the pathogens from the milk (Fig. 3C). In the case of Ami:CHAP, S. aureus was detected only sporadically, with an average of 1 log CFU/ml. Since S. aureus has been reported to feature altered surface properties following growth in milk versus growth medium (28), we further investigated whether this would affect its susceptibility to our enzyme mixtures. For this purpose, S. aureus Newbould 305 was precultured in UHT milk instead of TSB, and time-kill assays were performed as described above. Under these conditions, all PGH combinations were less effective when used at 1× concentrations than under standard conditions employing S. aureus grown in TSB. The combinations Lyso:CHAP, Ami:CHAP, and Lyso:Ami:CHAP reduced bacterial numbers by 2.26, 1.45, and 1.59 log units, respectively (data not shown). However, when doubling PGH concentrations (2×), Lyso:CHAP caused reduction of S. aureus cells to undetectable numbers, whereas for Ami:CHAP and Lyso:Ami:CHAP, bacteria were detectable after 3 h at 2.05 and 1.72 log CFU/ml, respectively (Fig. 3D).
FIG 3.
Activity of synergistic PGH mixtures in time-kill assays against S. aureus in UHT milk. (A) Assay in milk spiked with 103 CFU/ml of S. aureus Newbould 305 (low inoculum) using 106.5 nM (3 μg/ml) lysostaphin (Lyso), 106.5 nM CHAPK_CWT-LST (CHAP), 159.8 nM Ami2638A (Ami), and mixtures of the PGHs at the ratios 50:50 (for Lyso:CHAP and Ami:CHAP) or 33:33:33 (for Lyso:Ami:CHAP). (B) Assay in milk spiked with 106 CFU/ml (high inoculum) using PGH mixtures at 3 times the concentrations used in panel A (159.8 nM Lyso and CHAP and 239.7 nM Ami in mixtures of 2 enzymes; 106.5 nM Lyso and CHAP and 159.8 nM Ami in the mixture of all 3 enzymes). (C) Assays conducted over 24 h in milk spiked with 103 CFU/ml, using PGH mixtures at the same concentrations as in panel B. (D) Assays with S. aureus grown in milk to a concentration of 103 CFU/ml, using PGH mixtures at double the concentrations used in panel A (106.5 nM Lyso and CHAP and 159.8 nM Ami in mixtures of 2 enzymes and 71 nM Lyso and CHAP and 106.5 nM Ami in the mixture of all 3 enzymes). Buffer served as a control in all experiments. Error bars represent standard errors of the means from 3 experiments.
Ami2638A and lysostaphin retain their staphylolytic activity in raw bovine milk.
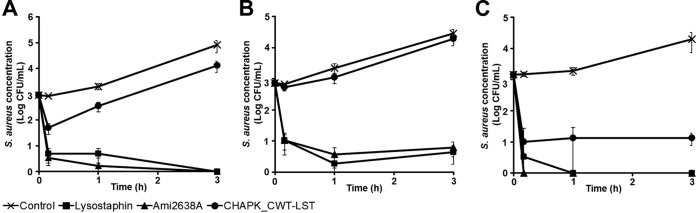
In order to avoid possible effects of the complex microflora present in raw bovine milk and reduce the possibly high variability between different batches of raw milk, the experiments described above were conducted in commercial heat-treated sterile milk. However, assays in UHT milk may inadequately mimic the situation in the bovine udder after intramammary infusion of PGHs. Even though the treatment is usually performed on evacuated udders (i.e., after milking or even at dry-off), and the therapeutics are infused in a relatively large volume of carrier substance (such as buffer), residual raw milk components may still impair the efficacy of the enzymes inside the mammary glands. In an effort to simulate this scenario and determine the effect of raw milk components on each of the 3 enzymes lysostaphin, Ami2638A, and CHAPK_CWT-LST, time-kill assays were performed in fresh raw cow's milk diluted with phosphate-buffered saline (PBS) at ratios of 1/3 and 1/10. In 1/3-dilution raw milk inoculated with 103 CFU/ml of S. aureus, both lysostaphin and Ami2638A reduced bacteria to undetectable numbers within 3 h at 3× concentrations (corresponding to a reduction of approximately 5 log units compared to the control), whereas the activity of CHAPK_CWT-LST was clearly diminished (yielding only a 0.79 log reduction) (Fig. 4A). Even in the worst-case scenario of 100% raw milk, lysostaphin and Ami2638A retained high activity and reduced S. aureus numbers to <1 log CFU/ml (corresponding to 3.82 and 3.66 log reductions, respectively). In contrast, CHAPK_CWT-LST retained only marginal activity under these conditions (Fig. 4B). In 10-fold-diluted raw milk, also CHAPK_CWT-LST retained high activity, causing a 3.17 log reduction within 3 h, whereas Ami2638A and lysostaphin completely eradicated S. aureus from the milk within minutes and 1 h, respectively (Fig. 4C). Staphylococcal background concentration in the raw milk used for these experiments was found to be negligible (<20 CFU/ml) compared to the inoculum (103 CFU/ml).
FIG 4.
Activity of lysostaphin, Ami2638A, and CHAPK_CWT-LST against S. aureus in raw milk. (A) Time-kill assay in 33% raw milk (diluted in PBS) spiked with 103 CFU/ml of S. aureus Newbould 305 using 319.5 nM (9 μg/ml) of lysostaphin, 319.5 nM CHAPK_CWT-LST, and 479.5 nM Ami2638A. (B) Time-kill assay as described in panel A but using 100% raw milk instead of diluted milk. (C) Time-kill assay as described for panel A but using 10% raw milk (diluted in PBS).
DISCUSSION
In the past, research on PGHs and their applications focused primarily on individual enzymes that had often been arbitrarily selected and may not have been the best possible choices for the specific purposes for which they were being used. This particularly applies to situations where activity against pathogens in specific niches, such as the bovine udder, is desired. From our experience, high antimicrobial activity in milk appears to be a rare trait among staphylococcal PGHs. In a series of experiments prior to this study, a collection of 9 unique staphylococcal PGHs featuring diverse enzymatic and antibacterial properties (15) had been analyzed for their activity against S. aureus in UHT milk. Even at protein concentrations as high as 200 μg/ml (∼7 μM for lysostaphin), 7 of these 9 PGH constructs showed only marginal or no staphylolytic activity in time-kill assays, the phi11 endolysin caused a moderate (∼1 log) reduction in CFU, and only lysostaphin completely eradicated S. aureus from the milk (Schmelcher, unpublished). The screening protocol that we have established was used for the first time in this study to identify enzyme constructs optimally suited for applications where high activity in milk is desired. The two most effective constructs, Ami2638A and CHAPK_CWT-LST, displayed similar staphylolytic activity as the reportedly strong antimicrobial lysostaphin, eradicating S. aureus from whole cow's milk within minutes at nanomolar concentrations (Fig. 1). Only a few other endolysin-derived PGH constructs with staphylolytic activity in cow's milk had been reported previously (22, 26, 29, 30). Even though quantitative comparison of results from different labs is difficult due to the use of different protocols and reagents, our 2 selected enzymes (out of >170 screened constructs) are the strongest PGH constructs reported to date with regard to staphylolytic activity in milk, besides lysostaphin. These enzymes are expected to be useful for the treatment of bovine mastitis and may also find application as antimicrobial food additives in milk processing, where contamination with staphylococci poses a significant health risk for consumers (29, 31).
It is important to note that the screening approach described here is not suitable as a method for quantitative comparison between multiple PGH constructs. Particularly when heterogeneous enzyme libraries containing different E. coli strains and plasmid backbones are used, as in this study, there might be inherent biases due to various factors such as different growth and expression rates and protein solubility. Interestingly, the selection of the 9 strongest enzymes (Table 1) represents all combinations of backbone plasmids and E. coli strains present in the entire library, suggesting that none of these plasmid-strain combinations is prohibitive in the identification of strong PGH constructs by our screening method. It also seems unlikely that PGHs with high activity, such as lysostaphin, Ami2638A, and CHAPK_CWT-LST, would remain undetected after multiple rounds of screening. Exceptions may be proteins with very low solubility or poor production in E. coli, properties which might also reduce their application as commercially produced antimicrobial agents. In conclusion, our screening method can serve as a useful tool to identify promising PGH candidates demonstrating high activity in milk from large libraries but needs to be followed up with more quantitative methods, e.g., time-kill assays, to further characterize the candidates identified. Although not demonstrated, our method would likely be suitable for identifying PGH constructs with high activity under various conditions, as defined by the medium used for coincubation of liberated enzymes and pathogens, and would likely be applicable also against other Gram-positive pathogens in a genus- or species-specific manner.
Interestingly, most of the 9 PGH candidates identified by our screening method (Fig. 1C) are derivatives of one or more of the aforementioned parental enzymes (15) that had previously demonstrated weak staphylolytic activity in milk. This finding suggests that activity in milk relies on additional factors other than just the origin of individual lytic domains/modules, such as protein size, tertiary structure, and/or domain arrangement, and supports the added value of protein engineering for creating enzymes with novel and desirable properties. Despite the fact that there are known mechanisms of resistance to lysostaphin, it is still a strong antimicrobial with one important advantage over phage endolysins: as lysostaphin is a bacteriocin naturally exhibiting lytic activity against staphylococci when exposed externally, it can be reasoned that lysostaphin has been optimized by evolution to effectively diffuse through various environments, potentially unaffected by components such as food matrices. In contrast, phage endolysins naturally cause lysis from within the bacterial cell and do not rely on diffusion through the environment to obtain their target peptidoglycan. In the case of endolysins acting against Gram-positive bacteria, it has been shown that they even remain bound to cellular debris after the lysis event via their high-affinity CBDs (32). Therefore, it is not surprising that many of the 9 candidate PGHs contain lysostaphin components, as is the case for CHAPK_CWT-LST, a construct similar in domain architecture to the previously described PRF-119 and HY-133 proteins (33, 34).
Synergistic effects are often observed when PGHs with different PG cleavage specificities are used in combination. This can be explained by an increased deleterious effect when several bonds within the 3-dimensional PG network are attacked simultaneously. It is also possible that one enzyme facilitates access to its substrate bond for the second enzyme, allowing for faster cell wall degradation (9). The benefits of synergy include lower enzyme concentrations required for effective treatment and likely a reduced chance of resistance development, since two simultaneous mutations would be required to evoke resistance. Also, resensitization of strains to agents against which they show high tolerance when exposed individually has been demonstrated (35). For the latter reasons, the use of lysostaphin as a therapeutic may be advisable in synergistic combination with other antimicrobials, while it is not favored as an isolated agent. The phage endolysin LysK, whose lytic activity reportedly relies primarily on its CHAP domain (23), and lysostaphin have previously been demonstrated to exhibit strong synergy when used in combination (36). This is in agreement with our results with the Lyso:CHAP combination, which showed ΣFICs and ΣFBCs below 0.5 in both checkerboard and 1-dimensional spot assays and was also highly effective in time-kill assays in milk, similar to the Ami:CHAP and Lyso:Ami:CHAP combinations. These combined PGHs also showed high activity against the Staphylococcus epidermidis and S. warneri strains tested in this study, which had shown high tolerance against all individual PGHs (Table 2). Given these and previous findings, it was unexpected that the Lyso:Ami combination exhibited only weak or no synergy. While there is no definitive explanation for this effect, protein-protein interactions between these two enzymes leading to partial inactivation or reduction in effective concentrations (e.g., through precipitation or unspecific binding to polystyrene surfaces) may play a role. The triple combination Lyso:Ami:CHAP, which would be most advisable to use therapeutically from an “avoiding resistance development” perspective, appeared slightly more effective in the spot assays than the synergistic dual combinations (even though the difference was not statistically significant), whereas Lyso:CHAP showed the greatest effect in time-kill assays (Fig. 3). Such inconsistencies on a quantitative level between different methods used for analyzing synergy have been reported previously (37, 38) and underline the necessity to characterize synergistic interactions in more than one assay format (39). Similarly, it is important to test PGHs and combinations thereof against multiple bacterial strains. Peptidoglycan structure slightly varies between different species of the genus Staphylococcus (40), and even within each species, strain-specific variations of other surface structures such as capsules and teichoic acids may affect the efficacy of the PGHs. This is consistent with our results shown in Table 2.
Decreased staphylolytic activity of phage-derived PGHs in raw cow's milk as opposed to heat-treated milk may be expected since S. aureus is known to aggregate and associate with fat globules in the milk (an effect that can be eliminated by heat treatment) (41, 42). In fact, there is only one report to date describing staphylolytic activity of a chimeric PGH (CHAPSH3b) in raw milk, which instantaneously reduced S. aureus at a concentration of 1.65 μM and kept numbers approximately 1 log unit below those of the control after 2 h. While these results were encouraging, they represented a clear decrease in activity compared to heat-treated milk (29). In this study, both lysostaphin and Ami2638A caused a reduction in bacterial numbers in 100% raw milk to <10 CFU/ml within 3 h (>3.5 log reduction), at enzyme concentrations (319.5 nM and 479.5 nM, respectively) similar to those sufficient to eradicate S. aureus in UHT milk (360 nM). In contrast, the efficacy of CHAPK_CWT-LST was clearly compromised in raw milk, although this effect was alleviated with increasing dilution of the milk, suggesting that the enzyme may retain considerable activity in a mastitis treatment scenario, where residual milk inside the depleted udder is diluted by the infusion of the therapeutic.
In conclusion, we have identified staphylococcal PGH constructs with high activity against staphylococci in cow's milk by employing a 96-well plate-based screening approach. The 3 strongest candidates exhibited activity against multiple staphylococcal mastitis isolates, acted synergistically in most enzyme combinations, proved effective in milk under various conditions, and retained activity in (diluted) raw milk. These results warrant further characterization of these enzymes with regard to their potential as mastitis therapeutics. This may include fusion to protein transduction domains in an effort to render them active against bacteria persisting inside mammary gland epithelial cells and macrophages (17), testing them for potential activity against nonstaphylococcal mastitis-causing pathogens, determining their ability to degrade staphylococcal biofilms (15), and investigating their efficacy in a mouse model of bovine mastitis (22).
MATERIALS AND METHODS
Strains, culture conditions, plasmids, and constructs.
One hundred seventy-four strains from a comprehensive E. coli library of PGH constructs available in our laboratory collection were selected for the screening approach described below. These strains carry PGH constructs with presumptive lytic activity against S. aureus, including parental staphylococcal bacteriophage endolysins, parental staphylococcal PGHs of nonphage origin, truncated PGHs, chimeric fusion proteins comprising functional domains from different origin, and otherwise engineered PGHs. Plasmid backbones of the various constructs include pET-21a (EMD Biosciences, San Diego, CA) and pQE-30 (Qiagen, Hilden, Germany), and E. coli strains containing these plasmids include BL21-Gold(DE3) (Agilent Technologies, Santa Clara, CA), XL1-Blue MRF′ (Stratagene, San Diego, CA), and SURE (Stratagene, San Diego, CA). The majority of the constructs bear N- or C-terminal 6×His tags, and production of all proteins is regulated by isopropyl-β-d-1-thiogalactopyranoside (IPTG)-inducible promoters. All E. coli strains were grown in Luria-Bertani (LB) medium or on LB agar at 37°C supplemented with suitable antibiotics for plasmid selection. Staphylococcal strains (bovine mastitis isolates) used in this study (listed in Table 2) were grown in tryptic soy broth (TSB) at 37°C.
Milk, growth media, and chemicals.
Commercial ultra-heat-treated (UHT) whole cow's milk used for screening of PGH constructs and time-kill assays was purchased from a local Swiss retailer. Bulk tank raw milk used for time-kill assays was freshly obtained from the Strickhof facility (Lindau, Switzerland) and stored on ice until used for the experiments. Components of growth medium were purchased from Merck (Zug, Switzerland) or Biolife (Milan, Italy), and chemicals were purchased from Fluka (Buchs, Switzerland) or Sigma-Aldrich (Buchs, Switzerland).
Microtiter plate-based screening for identification of PGH constructs with high staphylolytic activity in cow's milk.
A newly developed 96-well plate-based screening protocol was applied to identify enzymes from within our PGH library with activity against S. aureus in cow's milk. For this purpose, 90 μl of modified LB medium optimized for protein expression (LB-PE) (43) supplemented with ampicillin (100 mg/liter) for plasmid selection was added to each well of a 96-well plate (SPL Life Science, Pocheon-si, South Korea) and inoculated with E. coli strains harboring different PGH constructs using sterile toothpicks. Each strain was inoculated in triplicate at different positions to account for possible variability across the plate. Strains harboring empty plasmids without PGH constructs were included as negative controls, and a strain carrying C-terminally 6×His-tagged lysostaphin in pET-21a (36) served as a positive control. Lysostaphin is a nonendolysin PGH with reportedly strong activity against S. aureus in bovine milk (21). Cultures were grown under agitation at 37°C for 7.5 h, followed by addition of 100 μM (final concentration) isopropyl β-d-1-thiogalactopyranoside (IPTG) in a volume of 10 μl for induction of protein production and further incubation at 19°C for 18 h. E. coli cells were harvested by centrifugation (30 min, 2,000 × g, 4°C), and supernatants were removed. After one freeze-thaw cycle at −80°C, cell pellets were exposed to saturated chloroform vapor for disruption of cytoplasmic membranes and liberation of PGHs. This was achieved by placing the 96-well plate upside-down on a metal rack approximately 0.5 cm above a layer of tissue papers soaked in chloroform within a closed container for 10 min. We found that this treatment also completely inactivates E. coli cells (data not shown). Following evaporation of residual chloroform, 200 μl UHT whole cow's milk at 37°C spiked with 105 CFU/ml of S. aureus Newbould 305 (ATCC 29740), a bovine mastitis isolate (44), was added to each well and mixed. The plate was incubated at 37°C for 2 h under agitation. Aliquots from each well were subsequently diluted 10-fold with phosphate-buffered saline (PBS), 5 μl of each sample was spotted on tryptic soy agar (TSA), and agar plates were incubated overnight at 37°C. Staphylolytic activity of PGH constructs was visually assessed based on the density of S. aureus colonies within the respective spots compared to the controls. The entire assay was performed three times. PGHs consistently yielding largely reduced colony density compared to the negative control were selected for comparative functional analysis.
Protein production and purification.
Production in E. coli and purification of selected 6×His-tagged PGH constructs were performed essentially as previously described (15). Briefly, E. coli strains harboring constructs of interest were grown in LB-PE supplemented with suitable antibiotics at 37°C to an optical density at 600 nm (OD600) of 0.5 and then stored on ice for 30 min. Protein production was induced by addition of 0.5 mM IPTG, cultures were incubated for 18 h at 19°C, cells were harvested by centrifugation, and pellets were stored at −80°C until further use. Thawed cell pellets were resuspended in 10 ml of lysis buffer (50 mM NaH2PO4, 300 mM NaCl, 10 mM imidazole, 30% glycerol, pH 8.0), and cells were disrupted using a Stansted pressure cell homogenizer (SPCH-10-230V; Stansted Fluid Power, Harlow, United Kingdom) at a pressure of 100 MPa. Cell debris was removed from crude extracts by centrifugation at 10,000 × g for 30 min at 4°C, and cleared extracts were incubated with 2 ml of low-density nickel resin (ABT, Madrid, Spain) per liter of expression culture for 1 h at 4°C under agitation to allow binding of His-tagged target proteins to the nickel resin. The resin was transferred into Econo-Pac gravity flow columns (Bio-Rad, Cressier, Switzerland) and washed with 25 column volumes (CV) of lysis buffer, and target proteins were eluted with elution buffer (50 mM NaH2PO4, 300 mM NaCl, 250 mM imidazole, 30% glycerol, pH 8.0) in 500-μl fractions. Protein concentrations were measured with a NanoDrop ND-100 spectrophotometer (NanoDrop Technologies, Wilmington, DE), and fractions with high concentrations (>2 mg/ml) were combined and dialyzed against dialysis buffer (50 mM NaH2PO4, 300 mM NaCl, 30% glycerol, pH 7.5). Protein preparations were filtered (0.2 μm), final concentrations were measured, and purity was assessed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) using 10 to 20% Criterion TGX stain-free gels (Bio-Rad, Cressier, Switzerland).
Time-kill assays in UHT and raw bovine milk.
Time-kill assays in UHT milk performed essentially as previously described (26) were used to quantitatively compare the staphylolytic activities of selected PGH constructs and PGH mixtures. For standard assays, S. aureus Newbould 305 was grown to an OD600 of 0.5 in TSB at 37°C under agitation. UHT milk at 37°C was inoculated with S. aureus from this culture at a concentration of 103 CFU/ml or 106 CFU/ml. PGH constructs at defined concentrations in the same volume of dialysis buffer or buffer alone as a control were then added to the inoculated milk, and the milk was incubated at 37°C. Samples were taken immediately before and after addition of enzymes/buffer and at various time points up to 24 h postinoculation, and bacterial concentrations of all samples were determined by serial dilution plating on TSA. The sterility of noninoculated milk was verified by direct plating on TSA. TSA plates were incubated at 37°C for 18 h under aerobic conditions. In order to determine the effect of S. aureus growth in milk as opposed to TSB on the susceptibility of the bacteria to the enzymes, UHT milk at 37°C was inoculated with approximately 10 CFU/ml of S. aureus, and a series of preliminary experiments was performed in order to determine the time required for the bacterial concentration in the milk to reach 103 CFU/ml. Milk at this predetermined time point was then used for time-kill assays as described above. In addition to experiments with UHT milk, staphylolytic activity of selected PGHs was also determined in raw bovine milk. Staphylococcal background concentration in the raw milk was determined by direct plating on Baird Parker agar supplemented with rabbit plasma fibrinogen (Biolife, Milan, Italy). Baird Parker plates were incubated at 37°C for 18 h under aerobic conditions. Total aerobic, mesophilic viable counts were determined by serial dilution plating of raw milk samples on plate count (PC) agar. Time-kill assays in raw milk were performed as described above for UHT milk. Either undiluted raw milk or raw milk diluted 1/3 or 1/10 with PBS was used. Samples taken at different time points were plated on Baird Parker agar with egg yolk tellurite supplement (Biolife, Milan, Italy) in appropriate dilutions for determination of staphylococcal concentrations. All experiments were performed at least 3 times.
MIC and checkerboard assays.
MICs of PGH constructs against S. aureus Newbould 305 were determined using a classical microdilution broth method (45). Twofold serial dilutions in TSB of proteins to be tested were prepared in 96-well plates. Exponentially growing S. aureus in TSB was added to each well at a final concentration of 105 CFU/well (final volume of 200 μl), and the plate was incubated without shaking at 37°C for 20 h. The MIC is defined as the lowest concentration at which no visible bacterial growth occurs. Checkerboard assays for determination of synergistic effects of PGH mixtures in a pairwise manner were performed as previously described (36). Linear dilution series of both enzymes were prepared in TSB, starting at the predetermined MICs as highest final concentrations, and mixed in two dimensions on a 96-well plate. S. aureus Newbould 305 was added to each well (105 CFU/well in a final volume of 200 μl), and the plate was then incubated without shaking at 37°C for 20 h. The sum of fractional inhibitory concentrations of the two enzymes (ΣFIC = FICA + FICB) was calculated for each clear well along the inhibitory line. A ΣFIC value below 0.5 indicates a strong synergistic effect (46). All assays were performed at least in triplicate.
Spot assays for determination of MBCs and synergy in milk.
Spot assays were used to determine minimum bactericidal concentrations (MBCs) of individual PGH constructs and synergistic effects of PGH combinations in milk against multiple staphylococcal mastitis isolates (Table 2). Similarly to the MIC assay described above, 2-fold serial dilutions of enzymes were mixed with exponentially growing S. aureus cells (105 CFU/well) in UHT milk at 37°C. Following incubation for 2 h under agitation, 5-μl aliquots from each well were spotted on TSB agar and incubated overnight at 37°C. Pictures of agar plates were taken using a Molecular Imager Gel Doc XR+ imaging system (Bio-Rad) equipped with a white light transilluminator. The MBC is defined as the lowest concentration yielding a cleared spot. A 2-dimensional spot assay representing a variation of the classical microdilution broth checkerboard assay was applied to assess synergistic activity of PGH constructs in milk. To this end, enzyme dilution series and S. aureus cells were mixed in 2 dimensions on a 96-well plate as described above, using milk instead of TSB. The plates were incubated for 2 h at 37°C under agitation, followed by spotting of aliquots on TSB agar. Following overnight incubation, synergistic effects were determined as described above for the classical checkerboard assay, by calculating the sum of fractional bactericidal concentrations (ΣFBC). In addition, 1-dimensional spot assays (as used for determination of MBCs) were employed for assessing synergistic effects of PGH combinations against multiple staphylococcal strains. To this end, starting concentrations (x, y, and z) of dilution series of single PGH constructs to be tested were adjusted in such a way that the spots corresponding to the MBCs (i.e., the first cleared spots) of all enzymes appeared in the same row of the agar plate. These single enzymes were then compared to a dilution series of a mixture of two PGH constructs starting at 1/2x + 1/2y concentrations (or 1/3x + 1/3y + 1/3z concentrations when mixtures of all 3 enzymes were tested). The ΣFBC was calculated from the difference in rows containing the first cleared spot for the PGH mixtures compared to the single PGH constructs. If, due to the inherent variability of the assay, the first cleared spots of the single enzymes did not appear in the same row, the average of rows was calculated and used for comparison with the PGH mixture. If sporadic colonies emerged at higher PGH concentrations (e.g., in the third spot from the left of the first row of the SA047 plate in Fig. 2B), a cutoff value was defined and applied to the entire plate, so that spots with a number of colonies below the cutoff were considered cleared spots. All assays were done at least in duplicate.
Statistical analysis.
One-way analysis of variance (ANOVA) with a post hoc Tukey honestly significant difference (HSD) test was applied for comparisons of means in time-kill assays and synergy tests.
Supplementary Material
ACKNOWLEDGMENTS
We are thankful to Hans Graber, Roger Stephan, and Yasunori Tanji for the gift of staphylococcal mastitis isolates and to Hans-Peter Renfer for providing raw milk samples.
M.J.L. is an advisor for Micreos B.V., a company producing phage-based antimicrobials. The other authors declare no conflicts of interest.
This research received no specific grant from any funding agency in the public, commercial, or not-for-profit sectors.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.03445-16.
REFERENCES
- 1.Götz F, Bannerman T, Schleifer KH. 2006. The genera Staphylococcus and Macrococcus, p 5–75. In Falkow S, Rosenberg E, Schleifer KH, Stackebrandt E (ed), The prokaryotes, vol 4 Springer, New York, NY. [Google Scholar]
- 2.Sordillo LM, Streicher KL. 2002. Mammary gland immunity and mastitis susceptibility. J Mammary Gland Biol Neoplasia 7:135–146. doi: 10.1023/A:1020347818725. [DOI] [PubMed] [Google Scholar]
- 3.Kerr DE, Plaut K, Bramley AJ, Williamson CM, Lax AJ, Moore K, Wells KD, Wall RJ. 2001. Lysostaphin expression in mammary glands confers protection against staphylococcal infection in transgenic mice. Nat Biotechnol 19:66–70. doi: 10.1038/83540. [DOI] [PubMed] [Google Scholar]
- 4.Taponen S, Pyörälä S. 2009. Coagulase-negative staphylococci as cause of bovine mastitis—not so different from Staphylococcus aureus? Vet Microbiol 134:29–36. doi: 10.1016/j.vetmic.2008.09.011. [DOI] [PubMed] [Google Scholar]
- 5.Proctor RA, von Eiff C, Kahl BC, Becker K, McNamara P, Herrmann M, Peters G. 2006. Small colony variants: a pathogenic form of bacteria that facilitates persistent and recurrent infections. Nat Rev Microbiol 4:295–305. doi: 10.1038/nrmicro1384. [DOI] [PubMed] [Google Scholar]
- 6.Hebert A, Sayasith K, Senechal S, Dubreuil P, Lagace J. 2000. Demonstration of intracellular Staphylococcus aureus in bovine mastitis alveolar cells and macrophages isolated from naturally infected cow milk. FEMS Microbiol Lett 193:57–62. [DOI] [PubMed] [Google Scholar]
- 7.Deluyker HA, Van Oye SN, Boucher JF. 2005. Factors affecting cure and somatic cell count after pirlimycin treatment of subclinical mastitis in lactating cows. J Dairy Sci 88:604–614. doi: 10.3168/jds.S0022-0302(05)72724-7. [DOI] [PubMed] [Google Scholar]
- 8.Vanderhaeghen W, Hermans K, Haesebrouck F, Butaye P. 2010. Methicillin-resistant Staphylococcus aureus (MRSA) in food production animals. Epidemiol Infect 138:606–625. doi: 10.1017/S0950268809991567. [DOI] [PubMed] [Google Scholar]
- 9.Schmelcher M, Donovan DM, Loessner MJ. 2012. Bacteriophage endolysins as novel antimicrobials. Future Microbiol 7:1147–1171. doi: 10.2217/fmb.12.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pastagia M, Schuch R, Fischetti VA, Huang DB. 2013. Lysins: the arrival of pathogen-directed anti-infectives. J Med Microbiol 62:1506–1516. doi: 10.1099/jmm.0.061028-0. [DOI] [PubMed] [Google Scholar]
- 11.Roach DR, Donovan DM. 2015. Antimicrobial bacteriophage-derived proteins and therapeutic applications. Bacteriophage 5:e1062590. doi: 10.1080/21597081.2015.1062590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gutierrez D, Ruas-Madiedo P, Martinez B, Rodriguez A, Garcia P. 2014. Effective removal of staphylococcal biofilms by the endolysin LysH5. PLoS One 9:e107307. doi: 10.1371/journal.pone.0107307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fischetti VA. 2010. Bacteriophage endolysins: a novel anti-infective to control Gram-positive pathogens. Int J Med Microbiol 300:357–362. doi: 10.1016/j.ijmm.2010.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Whisstock JC, Lesk AM. 1999. SH3 domains in prokaryotes. Trends Biochem Sci 24:132–133. doi: 10.1016/S0968-0004(99)01366-3. [DOI] [PubMed] [Google Scholar]
- 15.Schmelcher M, Shen Y, Nelson DC, Eugster MR, Eichenseher F, Hanke DC, Loessner MJ, Dong S, Pritchard DG, Lee JC, Becker SC, Foster-Frey J, Donovan DM. 2015. Evolutionarily distinct bacteriophage endolysins featuring conserved peptidoglycan cleavage sites protect mice from MRSA infection. J Antimicrob Chemother 70:1453–1465. doi: 10.1093/jac/dku552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Daniel A, Euler C, Collin M, Chahales P, Gorelick KJ, Fischetti VA. 2010. Synergism between a novel chimeric lysin and oxacillin protects against infection by methicillin-resistant Staphylococcus aureus. Antimicrob Agents Chemother 54:1603–1612. doi: 10.1128/AAC.01625-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Becker SC, Roach DR, Chauhan VS, Shen Y, Foster-Frey J, Powell AM, Bauchan G, Lease RA, Mohammadi H, Harty WJ, Simmons C, Schmelcher M, Camp M, Dong S, Baker JR, Sheen TR, Doran KS, Pritchard DG, Almeida RA, Nelson DC, Marriott I, Lee JC, Donovan DM. 2016. Triple-acting lytic enzyme treatment of drug-resistant and intracellular Staphylococcus aureus. Sci Rep 6:25063. doi: 10.1038/srep25063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schindler CA, Schuhardt VT. 1964. Lysostaphin: a new bacteriolytic agent for the Staphylococcus. Proc Natl Acad Sci U S A 51:414–421. doi: 10.1073/pnas.51.3.414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.DeHart HP, Heath HE, Heath LS, LeBlanc PA, Sloan GL. 1995. The lysostaphin endopeptidase resistance gene (epr) specifies modification of peptidoglycan cross bridges in Staphylococcus simulans and Staphylococcus aureus. Appl Environ Microbiol 61:1475–1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kusuma C, Jadanova A, Chanturiya T, Kokai-Kun JF. 2007. Lysostaphin-resistant variants of Staphylococcus aureus demonstrate reduced fitness in vitro and in vivo. Antimicrob Agents Chemother 51:475–482. doi: 10.1128/AAC.00786-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wall RJ, Powell A, Paape MJ, Kerr DE, Bannerman DD, Pursel VG, Wells KD, Talbot N, Hawk HW. 2005. Genetically enhanced cows resist intramammary Staphylococcus aureus infection. Nat Biotechnol 23:445–451. doi: 10.1038/nbt1078. [DOI] [PubMed] [Google Scholar]
- 22.Schmelcher M, Powell AM, Becker SC, Camp MJ, Donovan DM. 2012. Chimeric phage lysins act synergistically with lysostaphin to kill mastitis-causing Staphylococcus aureus in murine mammary glands. Appl Environ Microbiol 78:2297–2305. doi: 10.1128/AEM.07050-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Becker SC, Dong S, Baker JR, Foster-Frey J, Pritchard DG, Donovan DM. 2009. LysK CHAP endopeptidase domain is required for lysis of live staphylococcal cells. FEMS Microbiol Lett 294:52–60. doi: 10.1111/j.1574-6968.2009.01541.x. [DOI] [PubMed] [Google Scholar]
- 24.O'Flaherty S, Coffey A, Meaney W, Fitzgerald GF, Ross RP. 2005. The recombinant phage lysin LysK has a broad spectrum of lytic activity against clinically relevant staphylococci, including methicillin-resistant Staphylococcus aureus. J Bacteriol 187:7161–7164. doi: 10.1128/JB.187.20.7161-7164.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Abaev I, Foster-Frey J, Korobova O, Shishkova N, Kiseleva N, Kopylov P, Pryamchuk S, Schmelcher M, Becker SC, Donovan DM. 2013. Staphylococcal phage 2638A endolysin is lytic for Staphylococcus aureus and harbors an inter-lytic-domain secondary translational start site. Appl Microbiol Biotechnol 97:3449–3456. doi: 10.1007/s00253-012-4252-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Obeso JM, Martinez B, Rodriguez A, Garcia P. 2008. Lytic activity of the recombinant staphylococcal bacteriophage PhiH5 endolysin active against Staphylococcus aureus in milk. Int J Food Microbiol 128:212–218. doi: 10.1016/j.ijfoodmicro.2008.08.010. [DOI] [PubMed] [Google Scholar]
- 27.Becker SC, Swift S, Korobova O, Schischkova N, Kopylov P, Donovan DM, Abaev I. 2015. Lytic activity of the staphylolytic Twort phage endolysin CHAP domain is enhanced by the SH3b cell wall binding domain. FEMS Microbiol Lett 362:1–8. doi: 10.1093/femsle/fnu019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mamo W, Rozgonyi F, Hjerten S, Wadström T. 1987. Effect of milk on surface properties of Staphylococcus aureus from bovine mastitis. FEMS Microbiol Lett 48:195–200. doi: 10.1111/j.1574-6968.1987.tb02541.x. [DOI] [Google Scholar]
- 29.Rodriguez-Rubio L, Martinez B, Donovan DM, Garcia P, Rodriguez A. 2013. Potential of the virion-associated peptidoglycan hydrolase HydH5 and its derivative fusion proteins in milk biopreservation. PLoS One 8:e54828. doi: 10.1371/journal.pone.0054828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mao J, Schmelcher M, Harty WJ, Foster-Frey J, Donovan DM. 2013. Chimeric Ply187 endolysin kills Staphylococcus aureus more effectively than the parental enzyme. FEMS Microbiol Lett 342:30–36. doi: 10.1111/1574-6968.12104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rodriguez-Rubio L, Gutierrez D, Donovan DM, Martinez B, Rodriguez A, Garcia P. 2016. Phage lytic proteins: biotechnological applications beyond clinical antimicrobials. Crit Rev Biotechnol 36:542–552. doi: 10.3109/07388551.2014.993587. [DOI] [PubMed] [Google Scholar]
- 32.Loessner MJ, Kramer K, Ebel F, Scherer S. 2002. C-terminal domains of Listeria monocytogenes bacteriophage murein hydrolases determine specific recognition and high-affinity binding to bacterial cell wall carbohydrates. Mol Microbiol 44:335–349. doi: 10.1046/j.1365-2958.2002.02889.x. [DOI] [PubMed] [Google Scholar]
- 33.Idelevich EA, von Eiff C, Friedrich AW, Iannelli D, Xia G, Peters G, Peschel A, Wanninger I, Becker K. 2011. In vitro activity against Staphylococcus aureus of a novel antimicrobial agent, PRF-119, a recombinant chimeric bacteriophage endolysin. Antimicrob Agents Chemother 55:4416–4419. doi: 10.1128/AAC.00217-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Idelevich EA, Schaumburg F, Knaack D, Scherzinger AS, Mutter W, Peters G, Peschel A, Becker K. 2016. The recombinant bacteriophage endolysin HY-133 exhibits in vitro activity against different African clonal lineages of the Staphylococcus aureus complex, Including Staphylococcus schweitzeri. Antimicrob Agents Chemother 60:2551–2553. doi: 10.1128/AAC.02859-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Djurkovic S, Loeffler JM, Fischetti VA. 2005. Synergistic killing of Streptococcus pneumoniae with the bacteriophage lytic enzyme Cpl-1 and penicillin or gentamicin depends on the level of penicillin resistance. Antimicrob Agents Chemother 49:1225–1228. doi: 10.1128/AAC.49.3.1225-1228.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Becker SC, Foster-Frey J, Donovan DM. 2008. The phage K lytic enzyme LysK and lysostaphin act synergistically to kill MRSA. FEMS Microbiol Lett 287:185–191. doi: 10.1111/j.1574-6968.2008.01308.x. [DOI] [PubMed] [Google Scholar]
- 37.Lewis RE, Diekema DJ, Messer SA, Pfaller MA, Klepser ME. 2002. Comparison of Etest, chequerboard dilution and time-kill studies for the detection of synergy or antagonism between antifungal agents tested against Candida species. J Antimicrob Chemother 49:345–351. doi: 10.1093/jac/49.2.345. [DOI] [PubMed] [Google Scholar]
- 38.Odds FC. 2003. Synergy, antagonism, and what the chequerboard puts between them. J Antimicrob Chemother 52:1. doi: 10.1093/jac/dkg301. [DOI] [PubMed] [Google Scholar]
- 39.Schmelcher M, Powell AM, Camp MJ, Pohl CS, Donovan DM. 2015. Synergistic streptococcal phage lambdaSA2 and B30 endolysins kill streptococci in cow milk and in a mouse model of mastitis. Appl Microbiol Biotechnol 99:8475–8486. doi: 10.1007/s00253-015-6579-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schleifer KH, Kandler O. 1972. Peptidoglycan types of bacterial cell walls and their taxonomic implications. Bacteriol Rev 36:407–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kuang Y, Jia H, Miyanaga K, Tanji Y. 2009. Effect of milk on antibacterial activity of tetracycline against Escherichia coli and Staphylococcus aureus isolated from bovine mastitis. Appl Microbiol Biotechnol 84:135–142. doi: 10.1007/s00253-009-2008-6. [DOI] [PubMed] [Google Scholar]
- 42.O'Flaherty S, Coffey A, Meaney WJ, Fitzgerald GF, Ross RP. 2005. Inhibition of bacteriophage K proliferation on Staphylococcus aureus in raw bovine milk. Lett Appl Microbiol 41:274–279. doi: 10.1111/j.1472-765X.2005.01762.x. [DOI] [PubMed] [Google Scholar]
- 43.Schmelcher M, Loessner MJ. 2014. Use of bacteriophage cell wall-binding proteins for rapid diagnostics of Listeria. Methods Mol Biol 1157:141–156. doi: 10.1007/978-1-4939-0703-8_12. [DOI] [PubMed] [Google Scholar]
- 44.Newbould FH. 1974. Antibiotic treatment of experimental Staphylococcus aureus infections of the bovine mammary gland. Can J Comp Med 38:411–416. [PMC free article] [PubMed] [Google Scholar]
- 45.Jones RN, Barry AL, Gavan TL, Washington JA II. 1985. Susceptibility tests: microdilution and macrodilution broth procedures, p 972–977. In Lennette EH, Balows A, Hausler WJ Jr, Shadomy HJ (ed), Manual of clinical microbiology. American Society for Microbiology, Washington, DC. [Google Scholar]
- 46.Hall MJ, Middleton RF, Westmacott D. 1983. The fractional inhibitory concentration (FIC) index as a measure of synergy. J Antimicrob Chemother 11:427–433. doi: 10.1093/jac/11.5.427. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.