ABSTRACT
On the basis of bioinformatic evidence, we suspected that proteins with a CYTH (CyaB thiamine triphosphatase) domain and/or a CHAD (conserved histidine α-helical domain) motif might represent polyphosphate (polyP) granule-associated proteins. We found no evidence of polyP targeting by proteins with CYTH domains. In contrast, two CHAD motif-containing proteins from Ralstonia eutropha H16 (A0104 and B1017) that were expressed as fusions with enhanced yellow fluorescent protein (eYFP) colocalized with polyP granules. While the expression of B1017 was not detectable, the A0104 protein was specifically identified in an isolated polyP granule fraction by proteome analysis. Moreover, eYFP fusions with the CHAD motif-containing proteins MGMSRV2-1987 from Magnetospirillum gryphiswaldense and PP2307 from Pseudomonas putida also colocalized with polyP granules in a transspecies-specific manner. These data indicated that CHAD-containing proteins are generally attached to polyP granules. Together with the findings from four previously polyP-attached proteins (polyP kinases), the results of this study raised the number of polyP-associated proteins in R. eutropha to six. We suggest designating polyP granule-bound proteins with CHAD motifs as phosins (phosphate), analogous to phasins and oleosins that are specifically bound to the surface of polyhydroxyalkanoate (PHA) granules in PHA-accumulating bacteria and to oil droplets in oil seed plants, respectively.
IMPORTANCE The importance of polyphosphate (polyP) for life is evident from the ubiquitous presence of polyP in all species on earth. In unicellular eukaryotic microorganisms, polyP is located in specific membrane-enclosed organelles, called acidocalcisomes. However, in most prokaryotes, polyP is present as insoluble granules that have been designated previously as volutin granules. Almost nothing is known regarding the macromolecular composition of polyP granules. Particularly, the absence or presence of cellular compounds on the surface of polyP granules has not yet been investigated. In this study, we identified a novel class of proteins that are attached to the surface of polyP granules in three model species of Alphaproteobacteria, Betaproteobacteria, and Gammaproteobacteria. These proteins are characterized by the presence of a CHAD (conserved histidine α-helical domain) motif that functions as a polyP granule-targeting signal. We suggest designating CHAD motif-containing proteins as phosins [analogous to phasins for poly(3-hydroxybutyrate)-associated proteins and to oleosins for oil droplet-associated proteins in oil seed plants]. The expression of phosins in different species confirmed their polyP-targeting function in a transspecies-specific manner. We postulate that polyP granules in prokaryotic species generally have a complex surface structure that consists of one to several polyP kinases and phosin proteins. We suggest differentiating polyP granules from acidocalcisomes by designating them as polyphosphatosomes.
KEYWORDS: polyphosphate, volutin granules, biopolymers, polyhydroxyalkanoates (PHA), Pseudomonas putida, Ralstonia eutropha, Magnetospirillum gryphiswaldense
INTRODUCTION
Many bacteria produce insoluble inclusions (granules), such as polyhydroxyalkanoates (PHAs), cyanophycins (CPs), and polyphosphates (polyPs), in their cytoplasm. It is generally accepted that these inclusions function as storage materials for carbon and energy, for nitrogen and carbon, and for phosphorous and cations, such as calcium/magnesium, respectively. Phosphorous is a rare element on earth, and phosphate is often the limiting factor for the growth of plants and microbes in natural ecosystems. Therefore, it is obvious that single cell organisms developed tools to take up more phosphate than growth requires and to store the surplus in the form of polyP granules. Phosphate can be efficiently taken up in large amounts even under unfavorable conditions, and this “behavior” is used in the enhanced biological phosphate removal (EBPR) process for wastewater treatment (for reviews on EBPR see references 1–4). Many functions in addition to that of a storage compound have been proposed for the inorganic biopolymer, polyP. (i) It can replace ATP in some energy-dependent reactions (5), (ii) it is important for motility and virulence in some pathogenic bacteria (6, 7), (iii) it can be involved in resistances against starvation, heavy metals, reactive oxygen species, and other stresses (8–12), (iv) it participates in the germination of spores in some endospore-forming species (13), and (v) it can even act as a molecular chaperone (14); for reviews on polyP, see references 5 and 15–18. It is difficult to imagine how a simple inorganic molecule such as polyP can participate in so many different functions. One reason for this might be that we do not yet know the exact molecular structures and compositions of polyP granules in different species. Only for Rhodospirillum rubrum and Agrobacterium tumefaciens was it shown that polyP is present in the form of membrane-surrounded acidic compartments called acidocalcisomes (19–22). However, little is known of the architecture of polyP granules in non-acidocalcisome-forming bacteria. Experiments using cryo-electron tomography or expression of a phospholipid-binding protein fused to fluorescent proteins gave no indication for the presence of a lipid membrane around polyP granules in Ralstonia eutropha (23, 24). On the other hand, it is likely that polyP kinases (PPKs), the enzymes that catalyze the formation of polyP by the polycondensation of phosphate residues of ATP or GTP, are bound to polyP granules in vivo. In Caulobacter crescentus, the in vivo colocalization of Ppk with formed polyP granules is well documented (25). The same was found in R. eutropha for Ppk1a and for at least three other Ppks (Ppk2c, Ppk2d, and Ppk2e) (26). Another example for the presence of specific compounds or structures on polyP granules came from cryo-electron tomography of Halothiobacillus neapolitanus cells, which revealed the presence of regularly ordered structures (lattices) between the surfaces of polyP inclusions and adjacent carboxysomes (27). These indications from the literature suggest that there could be other unknown molecules, presumably proteins, that are surface-displayed on polyP granules. A similar case is well-documented for another type of inclusion structure. Many prokaryotes synthesize and accumulate polyhydroxyalkanoates (PHAs) in the form of granular inclusions, such as poly(3-hydroxybutyrate) (PHB) granules. The hydrophobic polymer core is covered by a surface layer of several proteins, including PHA-synthesizing or -degrading enzymes and so-called phasins (24, 28–31). Phasins are small amphiphilic proteins that bind to the hydrophobic PHA surface and expose a hydrophilic phase to the cytoplasm (32–34). Since it is unlikely that a polar and charged polyanion such as polyP is present in an unshielded form, proteins at the surface of polyP granules might have a function similar to that of phasins for PHAs. Therefore, we assume that polyP granules are covered by a surface structure that might be composed of proteins. Consequently, this study aimed to identify novel polyP-bound proteins and elucidate the function they might have.
RESULTS AND DISCUSSION
Identification of two novel polyP-associated proteins in R. eutropha.
A bioinformatic approach recently showed that a subgroup of prokaryotic adenylate cyclases (Cya proteins) was structurally related to the family of mammalian thiamine triphosphatases (35). The authors identified a common protein domain that they designated as a CYTH (CyaB thiamine triphosphatase) domain. Proteins with a CYTH domain are frequently present in translated prokaryotic genome sequences and often harbor a second novel domain, a so-called CHAD (conserved histidine α-helical domain) motif (35). Remarkably, genes with sequences coding for a CYTH and/or a CHAD motif are often contextually located with genes encoding proteins for polyP and nucleotide metabolism. The assumption that proteins with a CYTH and/or a CHAD motif may be involved in polyP metabolism is further supported by the presence of a CYTH domain in the VTC4 protein in Saccharomyces cerevisiae. The VTC4 proteins of unicellular eukaryotic microorganisms are functionally homologous to polyP kinase proteins in prokaryotes (36). These correlations enable us to hypothesize that proteins with CYTH and/or CHAD motifs may function in polyP metabolism in bacteria.
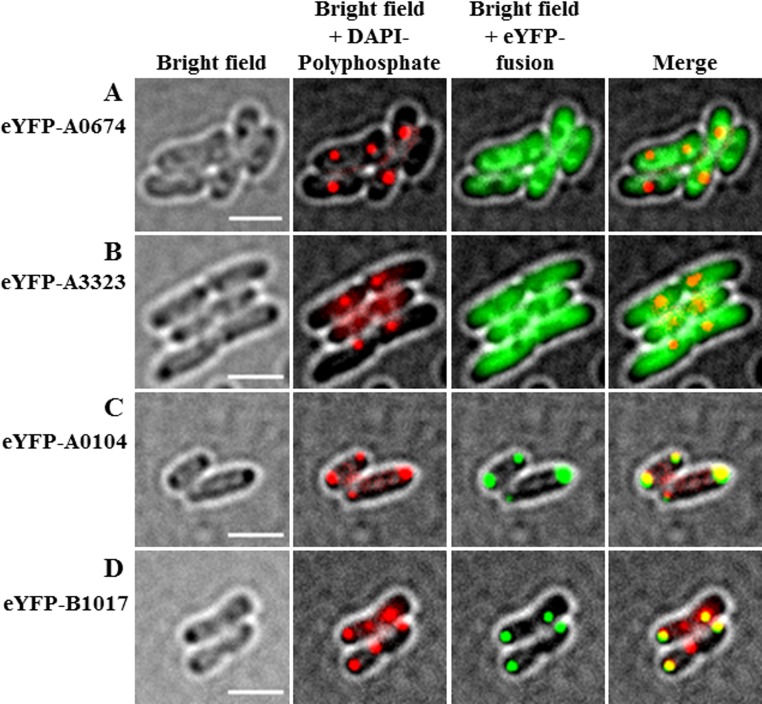
We inspected the R. eutropha H16 genome sequence (37) for genes coding for proteins with CYTH and/or CHAD motifs and identified four candidate genes (A0104, A0674, B1017, and A3323). A0674 and A3323 each code for proteins with a single CYTH domain (19.4 kDa and 22.2 kDa, respectively), A0104 codes for a 35.5-kDa protein with a single CHAD motif, and the B1017 protein (56.7 kDa) has an N-terminal CYTH domain and a C-terminal CHAD motif. With the exception of A0674 (putative adenylate cyclase class 2), no putative functions have been annotated for these CYTH/CHAD motif-containing proteins. Interestingly, the polyP kinase Ppk1b gene of R. eutropha H16 (B1019) is located near the B1017 gene (transcribed in an opposite direction). Downstream of B1017 there are genes for biopolymer hydrolases (PHB depolymerase PhaZa5 [B1014] and cyanophycinase [B1013]). A relationship between PHB and polyP metabolism is well known from the EBPR process and was recently demonstrated in R. eutropha (26). To find experimental evidence of whether any of the four proteins with CYTH and/or CHAD motifs have a function in polyP metabolism or polyP granule formation, we constructed gene fusions of the enhanced yellow fluorescent protein gene (eyfp) with each of the four candidate genes and expressed them in R. eutropha transconjugants. N-terminal fusions of the two proteins with only a CYTH domain (eYFP-A0674 and eYFP-A3323) were homogeneously distributed in recombinant R. eutropha and never colocalized with polyP granules (Fig. 1A and B). In contrast, N-terminal fusions of eYFP with A0104 (only CHAD) and with B1017 (CYTH and CHAD) formed fluorescent foci. In most cells, one or two foci per cell were formed that were located predominantly at the cell poles (Fig. 1C and D). Occasionally, a fluorescent focus was located mid-cell in the area of future cell division. When the cells were stained with 4′,6-diamidino-2-phenylindole (DAPI) and were imaged using a DAPI-polyP-specific emission wavelength, it became evident that the fusion proteins colocalized with polyP granules (Fig. 1C and D). We concluded that A0104 and B1017 are polyP-associated proteins in R. eutropha and that the CHAD motif might constitute a polyP granule-binding domain. The high isoelectric points (IEPs) of the amino acid sequences of the A0104 and B1017 proteins (A0104, 11.5; B1017, 8.9) are in agreement with in vivo binding of the basic proteins to an acidic polyP anion. Contrarily, the theoretical IEPs of A0674 and A3323 were acidic (5.1 and 4.9, respectively); therefore, it is no surprise that neither of the proteins bound to polyP granules in vivo. We conclude that CHAD motifs have a polyP granule-targeting function. The assumed in vivo attachment of A0104 to polyP granules was experimentally supported by the identification of the A0104 protein in a proteome analysis of an isolated polyP granule-enriched cell fraction (see Table S1 in the supplemental material). Remarkably, A0104 was not detected in any other cell fraction (soluble, membrane, or membrane-associated). The B1017 protein was not detected in any fraction of nutrient broth (NB)-grown cells and apparently is not expressed under the growth conditions we used. We found no evidence for a polyP-targeting or otherwise polyP-related function of the CYTH domain in our experiments.
FIG 1.
Localization of eYFP fusion proteins. N-terminal fusions of A0674 (CYTH) (A), A3323 (CYTH) (B), A0104 (CHAD) (C), and B1017 (CYTH and CHAD) (D) with eYFP were expressed in R. eutropha during growth on NB medium at 30°C. Cells were stained with DAPI for at least 10 min, immobilized with agarose, and imaged under bright field (left), at a polyP-DAPI-specific wavelength (middle left), and in the green channel (middle right). Merges of all channels are shown in the images on the right. Note the “soluble” eYFP-fusion fluorescence for YFP-A0674 and eYFP-A3323 and the colocalization of eYFP-A0104 and eYFP-B1017 foci with polyP granules. Bars, 2 μm.
Effect of overproduction of A0104 on polyP granule localization.
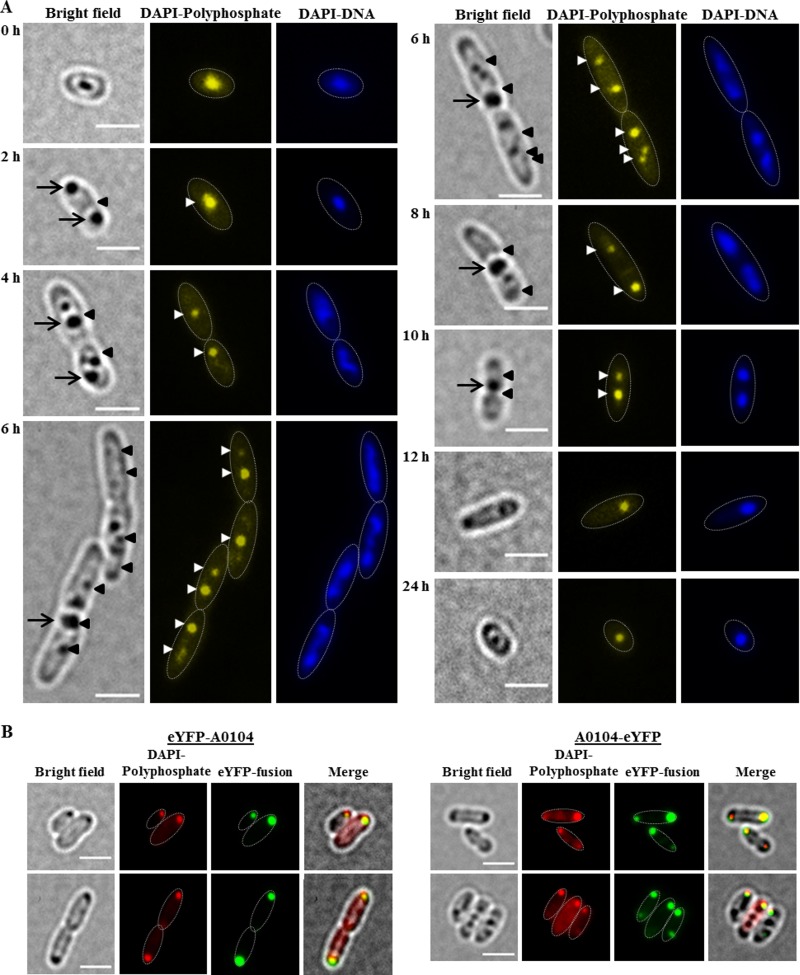
Most of the R. eutropha H16 wild-type cells had one or two polyP granules depending on the growth phase. While more than 90% of all cells from a liquid culture in the late stationary phase or cells of the lag phase had only one polyP granule, many cells from an actively growing culture harbored two polyP granules. Elongated cells that had initiated but not completed cell division had up to four polyP granules, with two granules in each cell half (Fig. 2A). For a statistical analysis of the number of polyP granules, see Table 1. Most of the polyP granules in wild-type cells were located at mid-cell or in the region between one- and three-quarters of the length axes of the cells. Staining of the cells with DAPI and imaging polyP- and DNA-specific wavelengths indicated that the polyP granules were generally located in the nucleoid region. The finding of an apparent colocalization of polyP granules with the nucleoid region is in agreement with similar findings in Caulobacter crescentus (25) and indicated that polyP granules are presumably not randomly located in prokaryotes.
FIG 2.
(A) Time course of polyP granule formation in R. eutropha H16. Cells were grown in NB medium at 30°C. Samples were taken at time points as indicated and stained with DAPI. Note the dark globular structures visible under bright field represent either PHB granules or polyP granules. PHB and polyP granules are detected under bright field only if they are in focus. In contrast, stained granules can be detected in the fluorescence channel even if they are not in perfect focus. Selected PHB and polyP granules are indicated by arrows (PHB) or arrowheads (polyP). The approximate cell shapes are highlighted in the fluorescence images by white dotted lines. (B) polyP formation and localization of eYFP-A0104 and A0104-eYFP in R. eutropha transconjugants. R. eutropha pBBR1MCS2-PphaC-eyfp-A0104 (bottom left) and R. eutropha pBBR1MCS2-PphaC-A0104-eyfp (bottom right) cells grown in NB. For better visualization, in merged figures, the DAPI-polyP-specific signals (normally yellowish) are red in color. The approximate cell shapes are highlighted in the fluorescence images by white dotted lines. Bars, 2 μm.
TABLE 1.
Numbers and subcellular localization of polyP granules in R. eutropha wild type and in recombinant strainsa
| Time (h) | No. of polyP granules per cell (%) |
|||
|---|---|---|---|---|
| WT | ΔA0104 | eYFP-A0104 (plasmid) | A0104-eYFP (chromosome integrated) | |
| 0 | 1 (>99) | 1 (>98) | 1 (>99) | 1 (>97) |
| 2 | 1 (90) | 1 (90) | 1 (70) | 1 (80) |
| 2 (10) | 2 (10) | 2 (30) | 2 (20) | |
| 4 | 1 (80) | 1 (80) | 1 (50) | 1 (25) |
| 2 (20) | 2 (20) | 2 (50) | 2 (60) | |
| 3 (5) | ||||
| 4 (10) dividing cells | ||||
| 6 | 2 (80) | 2 (80) | 1 (30) | 1 (15) |
| 3 (15) | 3 (15) | 2 (70) | 2 (80) | |
| 4 (5) dividing cells | 4 (5) dividing cells | 3 (occasionally) | 3 (5) dividing cells | |
| 8 | 1 (50) | 1 (50) | 1 (80) | 1 (45) |
| 2 (50) | 2 (50) | 2 (20) | 2 (50) | |
| 3 (5) | ||||
| 10 | 1 (80) | 1 (80) | 1 (80) | 1 (80) |
| 2 (20) | 2 (20) | 2 (20) | 2 (20) | |
| 12 | 1 (90) | 1 (90) | 1 (>99) | 1 (90) |
| 2 (10) | 2 (10) | 2 (10) | ||
| 18 | 1 (>99) | 1 (>98) | 1 (>99) | 1 (>97) |
| 24 | 1 (>99) | 1 (>98) | 1 (>99) | 1 (>97) |
| Localization of polyP at cell poles | Never (<1) (polyP granules between ¼ and ¾ of cell length axis) | Never (<2) (polyP granules between ¼ and ¾ of cell length axis) | Always (>99) (polyP granules never between ¼ and ¾ of cell length axis) | Rarely (≤3) (polyP granules between ¼ and ¾ of cell length axis) |
Cells were grown on NB medium at 30°C. The number of examined of cells varied between 35 (A0104-eYFP chromosome-integrated), 60 (ΔA0104), and 80 (all others). Cells with four polyP granules were always elongated and were undergoing or about to undergo cell division. WT, wild type.
To analyze the effect of overproduction of A0104 on the numbers and localization of polyP granules, we followed the formation of polyP granules in a strain expressing the eYFP-A0104 fusion under the constitutive phaC promoter during growth (Fig. 2B; see also Fig. S2). The numbers of polyP granules per cell were not different from those in the wild type at all stages of growth. However, the polyP granules and the granule-attached eYFP-A0104 fusion proteins were generally displaced from the mid-cell and the one- to three-quarter region (nucleoid region) to the cell pole region. Occasionally, polyP granules were located in the middle region of elongated cells in the area of future cell division. The same result was obtained when we expressed eYFP fused to the C terminus of A0104 (A0104-eYFP) (Fig. 2B). Time lapse experiments, in which we followed the fates of individual cells during cell-division, gave no indication for an active movement of polyP granules from the middle of the cell/nucleoid to the cell pole region. Rather, the overproduction of eYFP-A0104 seemed to fix polyP granules to the cell pole region.
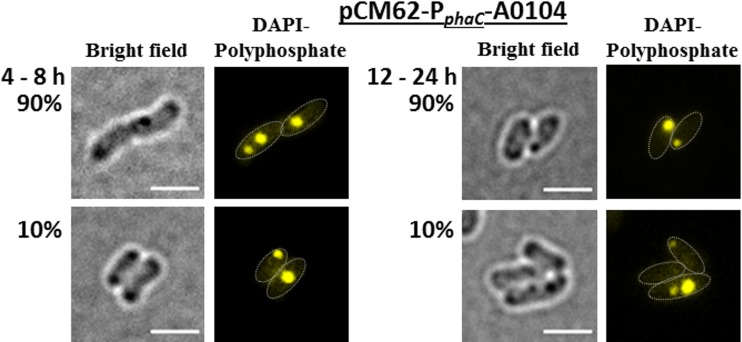
These findings indicated that the A0104 protein might have a function in the subcellular positioning of polyP granules. To investigate whether the altered localization of polyP granules was an artifact caused by the eYFP moiety of the fusion protein, we determined polyP granule formation in an R. eutropha strain in which a wild-type copy of the A0104 gene without a fusion was overexpressed from a plasmid under the constitutive phaC (PHB synthase of R. eutropha) promoter (Fig. 3). The numbers of polyP granules were not significantly changed during growth compared with those of the wild type (1 to 2 granules in most cells, occasionally up to 4 polyP granules per [elongated] cell). Only a minority of exponentially growing cells (≈10%) had cell pole-located polyP granules, while polyP granules in the other cells were located in the nucleoid region. Interestingly, the number of cells with polyP granules at the cell poles increased to 90% in the stationary growth phase. This high percentage of cells with cell pole-localized polyP granules was not observed in the wild type (Fig. 2A).
FIG 3.
polyP formation and localization of A0104 without fusion to eYFP in R. eutropha harboring pCM62-phaC-A0104. Cells were grown in NB medium at 30°C. The approximate cell shapes are highlighted in the fluorescence images by white dotted lines. Note that all cells form polyP granules. In the exponential growth phase (4 to 8 h; left), only 10% of the cells had cell pole-located polyP granules. In the stationary growth phase (12 to 24 h; right), 90% of the cells had cell pole-located polyP granules. Bars, 2 μm.
Effect of deletion of A0104 and B1017.
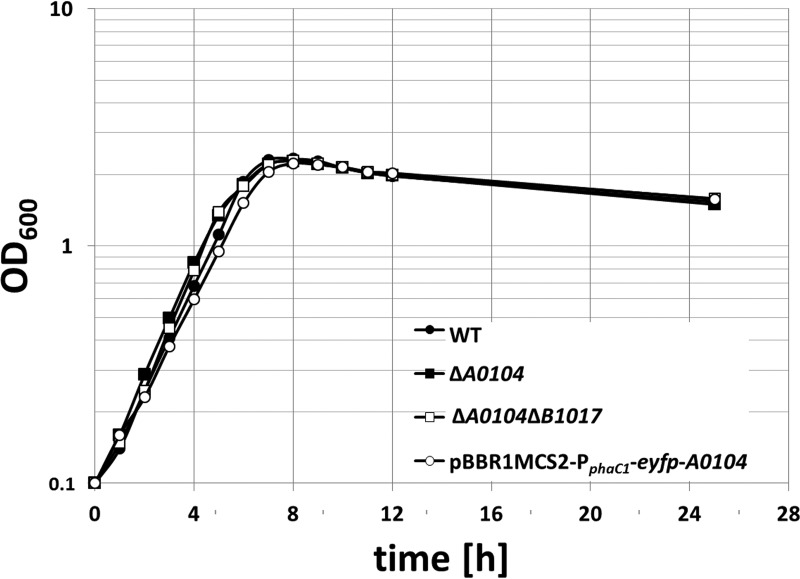
To analyze the potential function of CHAD-containing proteins, we constructed recombinant strains of R. eutropha in which the A0104 gene or both genes (A0104 and B1017) were deleted from the chromosome. The growth and formation of polyP granules were then followed for both mutant strains and compared with those of the wild type. No detectable phenotype was determined with respect to the growth of the mutant stains on NB medium (Fig. 4). Doubling times of 1.9 h ± 14 min were determined for all of the strains. The overproduction of the A0104 gene from a plasmid in a wild-type background also did not detectably affect growth. Deleting the A0104 gene, the B1017 gene, or of both genes did not detectably alter the formation and localization of polyP granules in comparison with the wild type (see Fig. S3). In summary, the A0104 and B1017 gene products are not essential for the formation and localization of polyP granules in the wild type, and their physiological functions remain obscure.
FIG 4.
Growth curves of the wild type and recombinant strains of R. eutropha H16 on NB medium. Cells were inoculated from two subsequent seed cultures. Seed cultures of the plasmid-containing strain contained 150 μg/ml kanamycin. All main cultures were grown without antibiotics. The values represent the means from results of 2 technical replicates. One of two biological replicates is shown. OD600, optical density at 600 nm.
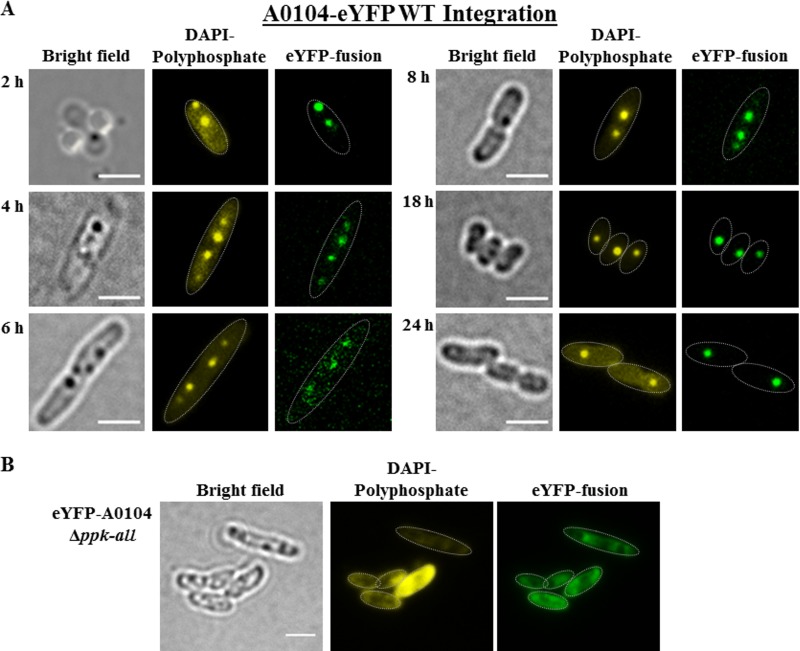
Expression of eYFP-A0104 from a chromosome-anchored gene fusion.
The results given above showed that increasing the amount of A0104 protein by plasmid-derived overexpression impacted the localization of polyP granules, at least in the stationary growth phase. To investigate whether the degree of A0104 expression determines polyP granule formation and polyP granule localization, we constructed a recombinant R. eutropha strain in which the A0104 gene was replaced by A0104-eyfp in the chromosome under the control of the native promoter. This enabled us to study the time course of A0104-eYFP expression and localization of the gene product at wild-type levels. The expression of the chromosomally anchored A0104-eyfp gene was much lower than plasmid-phaC promoter-driven expression, and it was necessary to increase the exposure time in fluorescence imaging. As shown in Fig. 5A, A0104-eYFP always colocalized with formed polyP granules. The number and subcellular localization of polyP granules were the same as for the wild type (1 to 2 polyP granules per cell in the nucleoid region, and up to 4 polyP granules in elongated cells; Table 1), and no specific cell pole localization was detected for the majority of the cells at any time during the 24-h growth experiment. In only in a minor fraction of the cells (see cell at 2-h time point of Fig. 5A) were cell pole-localized polyP granules with attached eYFP-A0104 fusion protein detectable. In conclusion, the localization of overproduced eYFP-A0104 at the cell poles is not an artifact of the fusion to eYFP, but seems to be a result of the increased expression in the plasmid expression system of A0104, and is not detected in the wild type.
FIG 5.
(A) Expression of a genome-integrated A0104-eyfp fusion in R. eutropha. Cells were grown in NB medium at 30°C. Samples were taken at the time points indicated, were stained with DAPI for at least 10 min, and were imaged. The cell shapes are highlighted by thin white lines in the fluorescence images. The distribution of the polyP granules within the cell corresponded to that of wild-type cells. Note that the dark globular structures visible in bright field represent either PHB granules or polyP granules. (B) Localization of eYFP-A0104 in a polyP granule-free background of R. eutropha. Cells were grown in NB medium at 30°C. Expression of eYFP-A0104 in R. eutropha Δppk-all. The shapes of the cells are highlighted in fluorescence images by white dotted lines. The strong (yellow) fluorescence in one cell is not typical for the culture and is seen only in a minor fraction of cells. Bars, 2 μm.
eYFP-A0104 is a soluble protein in a polyP granule-free background.
To determine whether A0104 has an affinity for a specific cellular position independent from polyP granules, we expressed eYFP-A0104 in a background lacking polyP granule formation. To this end, all seven ppk genes of R. eutropha (ppk1a, ppk1b, ppk2a, ppk2b, ppk2c, ppk2d, and ppk2e) were deleted from the genome by successive rounds of individual gene deletion experiments. The resulting 7-fold ppk-deletion strain was designated R. eutropha Δppk-all. The Δppk-all mutant did not form any polyP granules under any of the conditions. When we expressed the eYFP-A0104 fusion in the Δppk-all strain, a homogeneous distribution of the eYFP-A0104 fluorescence was determined at all stages of growth (Fig. 5B). We conclude that A0104 in the absence of polyP granules is a soluble protein. This result is in agreement with the function of A0104 as a polyP granule-binding protein. However, evidence for a specific function in the positioning of polyP granules was not obtained.
CHAD proteins in Alphaproteobacteria and Gammaproteobacteria are also polyP-associated proteins.
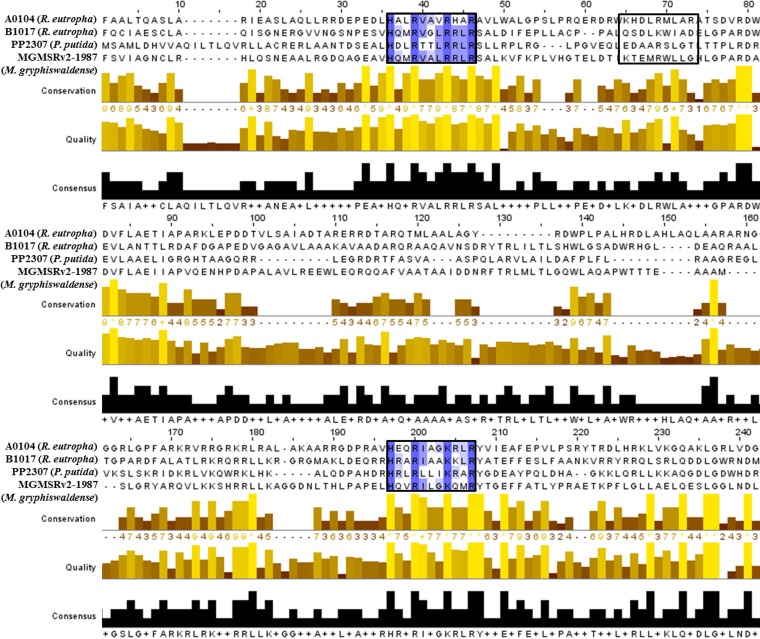
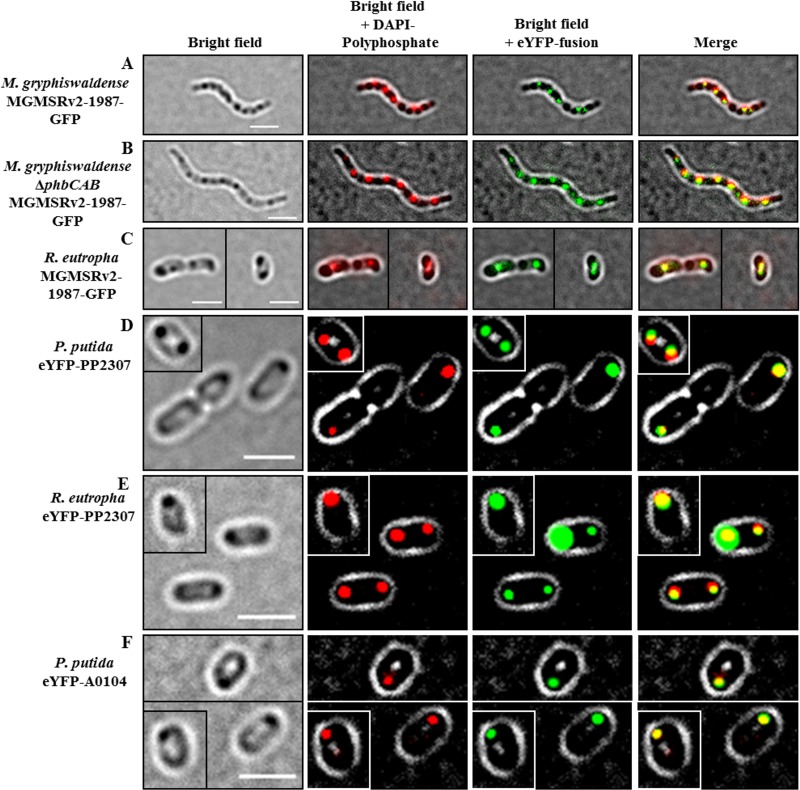
Our investigations with the betaproteobacterium Ralstonia eutropha clearly showed that the two CHAD motif-containing proteins of R. eutropha (A0104 and B1017) always colocalized with polyP granules. To investigate whether the presence of a CHAD motif also directs the protein to polyP granules in other bacteria, we chose Magnetospirillum gryphiswaldense and Pseudomonas putida as model organisms for Alphaproteobacteria and Gammaproteobacteria, respectively. An inspection of the genome sequences of both species (38, 39) revealed the presence of one protein with a CHAD motif for each species (MGMSRV2-1987, putative adenylate cyclase of M. gryphiswaldense, and PP2307, CHAD-containing protein of P. putida KT2440). MGMSRV2-1987 (56.2 kDa) has a CYTH domain in addition to the CHAD motif similar to B1017 of R. eutropha, and 35% of the amino acid sequence is identical to B1017. The PP2307 gene codes for a CHAD motif-containing protein of 29.5 kDa and is only 21% identical to the A0104 amino acid sequence. Figure 6 shows an alignment of the CHAD domains of the proteins investigated in this study. Only a moderate overall similarity was detected. However, we identified two repetitions of a short sequence motif with conserved histidine and arginine residues (HXXRXXXR/KXXR) in all four sequences. A third incomplete version of this motif was present only in the A0104 amino acid sequence. Gene fusions coding for MGMSRV2-1987-green fluorescent protein (GFP) and eYFP-PP2307 were constructed and expressed in M. gryphiswaldense and P. putida, respectively. As shown in Fig. 7A, the expressed MGMSRV2-1987-GFP fusion formed several fluorescent foci/cell that clearly colocalized with DAPI-stained polyP granules. The same result was obtained when the MGMSRV2-1987-GFP fusion was expressed in PHB-free mutants of M. gryphiswaldense in which the PHB biosynthetic genes had been deleted. A comparison of the images of MGMSRV2-1987-GFP expression in the wild type with those of a PHB-negative mutant (ΔphaCAB strain, Fig. 7B) enabled us to differentiate between polyP and PHB granules. Interestingly, polyP and PHB granules were aligned in an alternating order along the cell length axes of M. gryphiswaldense. The molecular basis for this order might be the attachment of polyP granules and PHB granules to specific molecules or molecule parts of the bacterial nucleoid. An attachment of PHB granules and polyP granules to the nucleoid has been postulated for R. eutropha and C. crescentus, respectively (25, 40, 41). Next, the MGMSRV2-1987-GFP fusion was expressed in R. eutropha; the recombinant cells showed one or two fluorescent foci that colocalized with polyP granules (Fig. 7C). Notably, the polyP granules with attached MGMSRV2-1987-GFP fusions in R. eutropha were not located at the cell poles as was determined for the expression of the R. eutropha CHAD proteins A0104 and B1017 (Fig. 1, Fig. 2B). This indicated that the localizations of the overexpressed eYFP-A0104 and eYFP-B1017 proteins at the cell poles are intrinsic properties of the respective CHAD motif-containing proteins and that the MGMSRV2-1987 protein does not have this polyP-to-cell pole-targeting property.
FIG 6.
Alignment of CHAD proteins and consensus sequence. For the alignment, the software “Multiple Sequence Alignment Clustal Omega” was applied. Refinements were conducted with the software “Jalview.” For better comparability, all four of the sequences were shortened to emphasize the conserved CHAD domain. The highlighted boxes represent a conserved motif (HXXRXXXR/KXXR), and the blue color intensity correlates with the degree of conservation. A third (incomplete) repetition of the HXXRXXXR/KXXR motif is present in the A0104 protein (black box). Furthermore, conservation, quality (yellow to brown bars), and consensus sequences (black bars) are shown.
FIG 7.
Expression of CHAD domain-containing proteins MGMSRV2-1987-GFP, eYFP-PP2307, and eYFP-A0104 in different species and backgrounds. (A to C) Expression of MGMSRV2-1987-GFP in M. gryphiswaldense wild type (A), in a PHB-negative mutant M. gryphiswaldense ΔphaCAB background (B), and in R. eutropha (C). (D and E) Expression of eYFP-PP2307 in P. putida wild type (D) and in R. eutropha (E). (F) Expression of eYFP-A0104 in P. putida. Note the colocalization of the fusion protein with DAPI-stained polyP granules in all cases and no colocalization with the PHB granules that are visible in A and C as dark inclusions under bright field. Single cells in the frames of the images in C, D, E, and F correspond to cells of the very same culture and the same micrograph. To save space, cells were packed together in one image. Bars, 2 μm.
The expression of the eYFP-PP2307 fusion in P. putida is shown in Fig. 7D. Again, the fusion protein clearly colocalized with formed polyP granules. However, as in the case of R. eutropha transconjugants overproducing eYFP-A0104, the polyP granules were located near the cell poles in eYFP-PP2307-expressing P. putida cells. The same results were obtained when the eYFP-PP2307 fusion was expressed in R. eutropha (Fig. 7E), or vice versa, when eYFP-A0104 was expressed in P. putida (Fig. 7F). In both combinations, the fusion proteins colocalized with formed polyP granules at the cell poles. In conclusion, the CHAD proteins of three species representing model species of the Alphaproteobacteria, Betaproteobacteria, and Gammaproteobacteria were exclusively bound to polyP granules. We suggest designating polyP granule-bound CHAD motif-containing proteins generally as phosins, analogous to phasins (polyhydroxyalkanoate [PHA]-associated proteins) in PHA-accumulating species and to oleosins (oil droplet-associated proteins) in oil seed plants. We assume that phosins are produced by the cells to provide a surface suitable for the highly polar and charged polyP granules. Similar to the phasins of accumulated PHA granules, phosins are not essential for polyP granule formation and presumably have no or only a minor, nonessential specific enzymatic function. No evidence was obtained in our study that phosins determine the number of formed polyP granules or the surface-to-volume ratio of polyP granules. We predict that CHAD proteins of other species are also polyP-associated proteins. We designate the A0104 protein (only CHAD) and the B1017 protein (CYTH and CHAD) of R. eutropha as polyP-targeting phosins PptAReu and PptBReu, and PP2307 (only CHAD) of P. putida and MGMSRV2-1987 of M. gryphiswaldense (CYTH and CHAD) as PptAPpu and PptBMgr, respectively.
Our findings show that polyP granules in non-acidocalcisome-forming bacteria are composed of at least two types of polymers. In addition to the polyP molecules, several polypeptides are present at the surface of polyP granules. While the presence of PPK proteins on the polyP granule surface was previously described (25, 26), the specific presence of proteins with CHAD domains with the polyP granules came as a surprise. Meanwhile, six proteins (4 PPKs [Ppk1a, Ppk2c, Ppk2d, and Ppk2e] and 2 phosins [PptA and PptB]) have been identified at the surfaces of polyP granules of R. eutropha. We assume that we have not likely identified all polyP-associated proteins. The cell pole-specific localization of polyP granules after the overproduction of eYFP-A0104 or wild-type A0104 in the stationary growth phase suggests that polyP granules or polyP granule-associated proteins might interact with other target structures of the cells. Future experiments will concentrate on identifying such potential targets of phosin proteins and not-yet-identified polyP-associated proteins. Since the R. eutropha genome has no copy of an acidocalcisome-specific hpptA gene (vacuolar H+-pyrophosphatase) (22) and polyP granules of R. eutropha were not enclosed by a membrane layer, they clearly differ from membrane-enclosed acidocalcisomes. In summary, our findings suggest that polyP granules in R. eutropha—and probably in other bacteria as well—are complexly organized organelle-like structures which we suggest designating as polyphosphatosomes.
MATERIALS AND METHODS
Bacterial strains, plasmids, and culture conditions.
The bacterial strains and plasmids used in this study are shown in Table 2. M. gryphiswaldense was grown in modified flask standard medium (FSM) at 28°C in 15-ml polypropylene tubes with sealed screw caps in a culture volume of 10 ml at microoxic conditions with moderate shaking (120 rpm) (42). R. eutropha H16 strains were grown aerobically on NB (0.8%, wt/vol) with or without sodium-gluconate (0.2%, wt/vol) at 30°C. Escherichia coli and Pseudomonas putida strains were grown aerobically in lysogeny broth (LB) medium at 37°C and 30°C, respectively.
TABLE 2.
List and characteristics of strains and plasmids used in this study
| Strain/plasmid | Relevant characteristica | Source/reference |
|---|---|---|
| Strains | ||
| Escherichia coli JM109 | Cloning strain | DSMZ 3423 |
| E. coli WM3064 | Conjugation strain for M. gryphiswaldense | William Metcalf |
| E. coli S17-1 | Conjugation strain | 44 |
| Ralstonia eutropha H16 | R. eutropha wild-type strain | DSMZ 428 |
| R. eutropha ΔA0104 (SK5693) | Chromosomal deletion of A0104 | This study |
| R. eutropha ΔB1017 (SK5971) | Chromosomal deletion of B1017 | This study |
| R. eutropha ΔA0104+ΔB1017 (SK5972) | Chromosomal deletion of A0104 and B1017 | This study |
| R. eutropha Δppk-all (SK5908) | Chromosomal deletion of seven ppk genes: Δppk1a, Δppk1b, Δppk2a, Δppk2b, Δppk2c, Δppk2d, and Δppk2e | This study |
| R. eutropha A0104-eyfp (SK6364) | Chromosomal integration of eyfp downstream of A0104 | This study |
| R. eutropha Δppk-all A0104-eyfp (SK6365) | Chromosomal integration of eyfp downstream of A0104 in Δppk-all background | This study |
| Magnetospirillum gryphiswaldense MSR-1 | M. gryphiswaldense wild-type strain | DSMZ 6361 |
| M. gryphiswaldense MSR-1 ΔphbCAB | M. gryphiswaldense with deletion of phbCAB | 45 |
| Pseudomonas putida KT2440 | P. putida wild type | DSMZ 6125 |
| Plasmids | ||
| pLO3 | Deletion vector, Tetr, sacB | 46 |
| pLO3-A0104-eyfp | Integration of A0104-eyfp | This study |
| pBBR1MCS2 | Broad host range vector, KANr | 47 |
| pBBR1MCS2-PphaC-eyfp-c1 | Universal vectors for construction of fusions C-terminal to eYFP under the PphaC promoter | 40 |
| pBBR1MCS2-PphaC-eyfp-n1 | Universal vectors for construction of fusions N-terminal to eYFP under the PphaC promoter | 40 |
| pBBR1MCS2-PphaC-eyfp-A0104 | N-terminal fusion of A0104 to eYFP | This study |
| pBBR1MCS2-PphaC-A0104-eyfp | C-terminal fusion of A0104 to eYFP | This study |
| pBBR1MCS2-PphaC-eyfp-B1017 | N-terminal fusion of B1017 to eYFP | This study |
| pBBR1MCS2-PphaC-eyfp-A0674 | N-terminal fusion of A0674 to eYFP | This study |
| pBBR1MCS2-PphaC-eyfp-A3323 | N-terminal fusion of A3323 to eYFP | This study |
| pBBR1MCS2-PphaC-eyfp-PP2307 | N-terminal fusion of PP2307 to eYFP | This study |
| pBAM160-Ptet-MGMSRV2-1987-gfp-c1 | C-terminal fusion of MGMSRV2-1987 to GFP | This study |
Resistance against kanamycin (KANr) and tetracycline (Tetr) as indicated.
Construction of gene fusions encoding fluorescent fusion proteins.
Gene fusions coding for fusion proteins with enhanced yellow fluorescent protein (eYFP) as the fluorophore were constructed as described previously using pBBR1MCS2::PphaC-eyfp-c1 or pBBR1MCS2::PphaC-eyfp-n1 as the vector (40). These plasmids enable constitutive expression of the respective fusion proteins from (in R. eutropha) the constitutive phaC1 promoter. The designations of fusion proteins (N terminal or C terminal) in this study always refer to the N or C terminus of the protein of interest. E. coli was transformed with each of constructs by standard heat shock transformation, and the plasmids were subsequently transferred via conjugation from recombinant E. coli S17-1 to R. eutropha H16. Selection was achieved by plating on mineral salts medium supplemented with 0.2% fructose and 350 μg ml−1 kanamycin. Fusion proteins with green fluorescent protein (GFP) as the fluorophore were constructed using pBAM160-Ptet-c1-gfp as the vector. This plasmid enables expression of the respective fusion protein expressed from an anhydro-tetracycline-inducible promoter after induction with anhydro-tetracycline (50 ng/ml). E. coli WM3064 (auxotroph for diaminopimelate) was transformed with this construct by standard (heat shock) transformation, and the plasmids were subsequently transferred via conjugation to R. eutropha H16. Selection was achieved by plating on mineral salts medium supplemented with 0.2% fructose and 350 μg ml−1 kanamycin. For M. gryphiswaldense, pBAM plasmids were transferred from E. coli WM3064 to M. gryphiswaldense MSR-1, and recombinant clones were selected in the presence of 5 μg/ml kanamycin. P. putida was transformed with the plasmids used by standard electroporation.
Construction of deletions and integrations into the genome.
Precise genomic deletions of R. eutropha genes were constructed using the sacB-sucrose selection method (15% sucrose used for selection) and pLO3 as the deletion vector as described previously (26, 43). The same procedure was applied to construct chromosomal integrations of eyfp in the genome. The genotypes of all generated constructs and mutants were verified by PCR of the respective DNA regions and by DNA sequencing.
Microscopy methods.
polyP granules were stained with 4′,6-diamidin-2-phenylindole (DAPI, 60 μg/ml, for at least 10 min at room temperature) and detected with the aid of a DAPI-polyP-specific filter set (excitation, 415/20 nm; emission, 520/60 nm). The nucleoid regions of DAPI-stained cells were visualized using a DAPI-DNA-specific filter set (excitation, 387/11 nm; emission, 447/60 nm). Enhanced yellow fluorescent protein (eYFP), green fluorescent protein (GFP), and dsRedEC2 were imaged using standard filters (excitation at 500/24 nm and emission at 542/27 nm for eYFP and GFP; excitation at 562/40 nm and emission at 594 nm [longpass filter] for dsRedEC2). The longpass filter was also used for visualizing Nile-red-stained PHB granules. Insoluble inclusions (polyP granules and PHB granules) were also visualized by bright field microscopy and appeared as dark globule-shaped inclusions in the gray background cytoplasm.
Supplementary Material
ACKNOWLEDGMENTS
The authors greatly acknowledge D. Pfeiffer for his input, for his help with constructing the recombinant M. gryphiswaldense strains, and for providing the replicon for constructing pBAM160-Ptet-MGMSRV2-1987-gfp-c1.
This work was supported by a grant from the Deutsche Forschungsgemeinschaft to D.J.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.03399-16.
REFERENCES
- 1.Blackall LL, Crocetti G, Saunders AM, Bond PL. 2002. A review and update of the microbiology of enhanced biological phosphorus removal in wastewater treatment plants. Antonie Van Leeuwenhoek 81:681–691. doi: 10.1023/A:1020538429009. [DOI] [PubMed] [Google Scholar]
- 2.Forbes CM, O'Leary ND, Dobson AD, Marchesi JR. 2009. The contribution of “omic”-based approaches to the study of enhanced biological phosphorus removal microbiology. FEMS Microbiol Ecol 69:1–15. doi: 10.1111/j.1574-6941.2009.00698.x. [DOI] [PubMed] [Google Scholar]
- 3.Nielsen PH, Saunders AM, Hansen AA, Larsen P, Nielsen JL. 2012. Microbial communities involved in enhanced biological phosphorus removal from wastewater–a model system in environmental biotechnology. Curr Opin Biotechnol 23:452–459. doi: 10.1016/j.copbio.2011.11.027. [DOI] [PubMed] [Google Scholar]
- 4.Shen N, Zhou Y. 2016. Enhanced biological phosphorus removal with different carbon sources. Appl Microbiol Biotechnol 100:4735–4745. doi: 10.1007/s00253-016-7518-4. [DOI] [PubMed] [Google Scholar]
- 5.Rao NN, Gómez-García MR, Kornberg A. 2009. Inorganic polyphosphate: essential for growth and survival. Annu Rev Biochem 78:605–647. doi: 10.1146/annurev.biochem.77.083007.093039. [DOI] [PubMed] [Google Scholar]
- 6.Rashid MH, Rao NN, Kornberg A. 2000. Inorganic polyphosphate is required for motility of bacterial pathogens. J Bacteriol 182:225–227. doi: 10.1128/JB.182.1.225-227.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rashid MH, Rumbaugh K, Passador L, Davies DG, Hamood AN, Iglewski BH, Kornberg A. 2000. Polyphosphate kinase is essential for biofilm development, quorum sensing, and virulence of Pseudomonas aeruginosa. Proc Natl Acad Sci U S A 97:9636–9641. doi: 10.1073/pnas.170283397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Casey WT, Nikodinovic-Runic J, Fonseca Garcia P, Guzik MW, McGrath JW, Quinn JP, Cagney G, Auxiliadora Prieto M, O'Connor KE. 2013. The effect of polyphosphate kinase gene deletion on polyhydroxyalkanoate accumulation and carbon metabolism in Pseudomonas putida KT2440. Environ Microbiol Rep 5:740–746. [DOI] [PubMed] [Google Scholar]
- 9.Keasling JD, Van Dien SJ, Trelstad P, Renninger N, McMahon K. 2000. Application of polyphosphate metabolism to environmental and biotechnological problems. Biochemistry (Mosc) 65:324–331. [PubMed] [Google Scholar]
- 10.Renninger N, Knopp R, Nitsche H, Clark DS, Keasling JD. 2004. Uranyl precipitation by Pseudomonas aeruginosa via controlled polyphosphate metabolism. Appl Environ Microbiol 70:7404–7412. doi: 10.1128/AEM.70.12.7404-7412.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nikel PI, Chavarria M, Martinez-Garcia E, Taylor AC, de Lorenzo V. 2013. Accumulation of inorganic polyphosphate enables stress endurance and catalytic vigour in Pseudomonas putida KT2440. Microb Cell Fact 12:50 doi: 10.1186/1475-2859-12-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gray MJ, Jakob U. 2015. Oxidative stress protection by polyphosphate–new roles for an old player. Curr Opin Microbiol 24:1–6. doi: 10.1016/j.mib.2014.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tocheva EI, Dekas AE, McGlynn SE, Morris D, Orphan VJ, Jensen GJ. 2013. Polyphosphate storage during sporulation in the gram-negative bacterium Acetonema longum. J Bacteriol 195:3940–3946. doi: 10.1128/JB.00712-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gray MJ, Wholey W-Y, Wagner NO, Cremers CM, Mueller-Schickert A, Hock NT, Krieger AG, Smith EM, Bender RA, Bardwell JCA, Jakob U. 2014. Polyphosphate is a primordial chaperone. Mol Cell 53:689–699. doi: 10.1016/j.molcel.2014.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kulaev IS, Vagabov VM. 1983. Polyphosphate metabolism in micro-organisms. Adv Microb Physiol 24:83–171. doi: 10.1016/S0065-2911(08)60385-9. [DOI] [PubMed] [Google Scholar]
- 16.Kulaev I, Kulakovskaya T. 2000. Polyphosphate and phosphate pump. Annu Rev Microbiol 54:709–734. doi: 10.1146/annurev.micro.54.1.709. [DOI] [PubMed] [Google Scholar]
- 17.Kornberg A, Rao NN, Ault-Riche D. 1999. Inorganic polyphosphate: a molecule of many functions. Annu Rev Biochem 68:89–125. doi: 10.1146/annurev.biochem.68.1.89. [DOI] [PubMed] [Google Scholar]
- 18.Kulakovskaya TV, Lichko LP, Ryazanova LP. 2014. Diversity of phosphorus reserves in microorganisms. Biochemistry Mosc 79:1602–1614. doi: 10.1134/S0006297914130100. [DOI] [PubMed] [Google Scholar]
- 19.Seufferheld M, Vieira M, Ruiz FA, Rodrigues CO, Moreno S, Docampo R. 2003. Identification of organelles in bacteria similar to acidocalcisomes of unicellular eukaryotes. J Biol Chem 278:29971–29978. doi: 10.1074/jbc.M304548200. [DOI] [PubMed] [Google Scholar]
- 20.Seufferheld M, Lea CR, Vieira M, Oldfield E, Docampo R. 2004. The H+-pyrophosphatase of Rhodospirillum rubrum is predominantly located in polyphosphate-rich acidocalcisomes. J Biol Chem 279:51193–51202. doi: 10.1074/jbc.M406099200. [DOI] [PubMed] [Google Scholar]
- 21.Docampo R, Moreno SNJ. 2011. Acidocalcisomes. Cell Calcium 50:113–119. doi: 10.1016/j.ceca.2011.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lander N, Cordeiro C, Huang G, Docampo R. 2016. Polyphosphate and acidocalcisomes. Biochem Soc Trans 44:1–6. doi: 10.1042/BST20150193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Beeby M, Cho M, Stubbe J, Jensen GJ. 2012. Growth and localization of polyhydroxybutyrate granules in Ralstonia eutropha. J Bacteriol 194:1092–1099. doi: 10.1128/JB.06125-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bresan S, Sznajder A, Hauf W, Forchhammer K, Pfeiffer D, Jendrossek D. 2016. Polyhydroxyalkanoate (PHA) granules have no phospholipids. Sci Rep 6:26612. doi: 10.1038/srep26612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Henry JT, Crosson S. 2013. Chromosome replication and segregation govern the biogenesis and inheritance of inorganic polyphosphate granules. Mol Biol Cell 24:3177–3186. doi: 10.1091/mbc.E13-04-0182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tumlirsch T, Sznajder A, Jendrossek D. 2015. Formation of polyphosphate by polyphosphate kinases and its relationship to poly(3-hydroxybutyrate) accumulation in Ralstonia eutropha strain H16. Appl Environ Microbiol 81:8277–8293. doi: 10.1128/AEM.02279-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Iancu CV, Morris DM, Dou Z, Heinhorst S, Cannon GC, Jensen GJ. 2010. Organization, structure, and assembly of alpha-carboxysomes determined by electron cryotomography of intact cells. J Mol Biol 396:105–117. doi: 10.1016/j.jmb.2009.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Galan B, Dinjaski N, Maestro B, de Eugenio LI, Escapa IF, Sanz JM, Garcia JL, Prieto MA. 2011. Nucleoid-associated PhaF phasin drives intracellular location and segregation of polyhydroxyalkanoate granules in Pseudomonas putida KT2442. Mol Microbiol 79:402–418. doi: 10.1111/j.1365-2958.2010.07450.x. [DOI] [PubMed] [Google Scholar]
- 29.Pötter M, Steinbüchel A. 2006. Biogenesis and structure of polyhydroxyalkanoate granules, p 109–136. In Shively JM. (ed), Inclusions in prokaryotes. Springer-Verlag, Berlin, Germany. [Google Scholar]
- 30.Jendrossek D, Pfeiffer D. 2014. New insights in the formation of polyhydroxyalkanoate granules (carbonosomes) and novel functions of poly(3-hydroxybutyrate). Environ Microbiol 16:2357–2373. doi: 10.1111/1462-2920.12356. [DOI] [PubMed] [Google Scholar]
- 31.Sznajder A, Pfeiffer D, Jendrossek D. 2015. Comparative proteome analysis reveals four novel polyhydroxybutyrate (PHB) granule-associated proteins in Ralstonia eutropha H16. Appl Environ Microbiol 81:1847–1858. doi: 10.1128/AEM.03791-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Maestro B, Galan B, Alfonso C, Rivas G, Prieto MA, Sanz JM. 2013. A new family of intrinsically disordered proteins: structural characterization of the major phasin PhaF from Pseudomonas putida KT2440. PLoS One 8:e56904. doi: 10.1371/journal.pone.0056904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kuchta K, Chi L, Fuchs H, Pötter M, Steinbüchel A. 2007. Studies on the influence of phasins on accumulation and degradation of PHB and nanostructure of PHB granules in Ralstonia eutropha H16. Biomacromolecules 8:657–662. doi: 10.1021/bm060912e. [DOI] [PubMed] [Google Scholar]
- 34.Mezzina MP, Pettinari MJ. 2016. Phasins, multifaceted polyhydroxyalkanoate granule-associated proteins. Appl Environ Microbiol 82:5060–5067. doi: 10.1128/AEM.01161-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Iyer LM, Aravind L. 2002. The catalytic domains of thiamine triphosphatase and CyaB-like adenylyl cyclase define a novel superfamily of domains that bind organic phosphates. BMC Genomics 3:33. doi: 10.1186/1471-2164-3-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hothorn M, Neumann H, Lenherr ED, Wehner M, Rybin V, Hassa PO, Uttenweiler A, Reinhardt M, Schmidt A, Seiler J, Ladurner AG, Herrmann C, Scheffzek K, Mayer A. 2009. Catalytic core of a membrane-associated eukaryotic polyphosphate polymerase. Science 324:513–516. doi: 10.1126/science.1168120. [DOI] [PubMed] [Google Scholar]
- 37.Pohlmann A, Fricke WF, Reinecke F, Kusian B, Liesegang H, Cramm R, Eitinger T, Ewering C, Pötter M, Schwartz E, Strittmatter A, Voss I, Gottschalk G, Steinbüchel A, Friedrich B, Bowien B. 2006. Genome sequence of the bioplastic-producing “Knallgas” bacterium Ralstonia eutropha H16. Nat Biotechnol 24:1257–1262. doi: 10.1038/nbt1244. [DOI] [PubMed] [Google Scholar]
- 38.Nelson KE, Weinel C, Paulsen IT, Dodson RJ, Hilbert H, Martins dos Santos VA, Fouts DE, Gill SR, Pop M, Holmes M, Brinkac L, Beanan M, DeBoy RT, Daugherty S, Kolonay J, Madupu R, Nelson W, White O, Peterson J, Khouri H, Hance I, Chris Lee P, Holtzapple E, Scanlan D, Tran K, Moazzez A, Utterback T, Rizzo M, Lee K, Kosack D, Moestl D, Wedler H, Lauber J, Stjepandic D, Hoheisel J, Straetz M, Heim S, Kiewitz C, Eisen J, Timmis KN, Dusterhoft A, Tummler B, Fraser CM. 2002. Complete genome sequence and comparative analysis of the metabolically versatile Pseudomonas putida KT2440. Environ Microbiol 4:799–808. doi: 10.1046/j.1462-2920.2002.00366.x. [DOI] [PubMed] [Google Scholar]
- 39.Wang X, Wang Q, Zhang W, Wang Y, Li L, Wen T, Zhang T, Zhang Y, Xu J, Hu J, Li S, Liu L, Liu J, Jiang W, Tian J, Li Y, Schüler D, Wang L, Li J. 2014. Complete genome sequence of Magnetospirillum gryphiswaldense MSR-1. Genome Announc 2:e00171-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pfeiffer D, Wahl A, Jendrossek D. 2011. Identification of a multifunctional protein, PhaM, that determines number, surface to volume ratio, subcellular localization and distribution to daughter cells of poly(3-hydroxybutyrate), PHB, granules in Ralstonia eutropha H16. Mol Microbiol 82:936–951. doi: 10.1111/j.1365-2958.2011.07869.x. [DOI] [PubMed] [Google Scholar]
- 41.Wahl A, Schuth N, Pfeiffer D, Nussberger S, Jendrossek D. 2012. PHB granules are attached to the nucleoid via PhaM in Ralstonia eutropha. BMC Microbiol 12:262. doi: 10.1186/1471-2180-12-262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Heyen U, Schüler D. 2003. Growth and magnetosome formation by microaerophilic Magnetospirillum strains in an oxygen-controlled fermentor. Appl Microbiol Biotechnol 61:536–544. doi: 10.1007/s00253-002-1219-x. [DOI] [PubMed] [Google Scholar]
- 43.Sznajder A, Jendrossek D. 2014. To be or not to be a poly(3-hydroxybutyrate) (PHB) depolymerase: PhaZd1 (PhaZ6) and PhaZd2 (PhaZ7) of Ralstonia eutropha, highly active PHB depolymerases with no detectable role in mobilization of accumulated PHB. Appl Environ Microbiol 80:4936–4946. doi: 10.1128/AEM.01056-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Simon R, Priefer R, Pühler A. 1983. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in Gram negative bacteria. Nat Biotechnol 1:784–791. doi: 10.1038/nbt1183-784. [DOI] [Google Scholar]
- 45.Raschdorf O, Plitzko JM, Schüler D, Müller FD. 2014. A tailored galK counterselection system for efficient markerless gene deletion and chromosomal tagging in Magnetospirillum gryphiswaldense. Appl Environ Microbiol 80:4323–4330. doi: 10.1128/AEM.00588-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lenz O, Friedrich B. 1998. A novel multicomponent regulatory system mediates H-2 sensing in Alcaligenes eutrophus. Proc Natl Acad Sci U S A 95:12474–12479. doi: 10.1073/pnas.95.21.12474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kovach ME, Elzer PH, Hill DS, Robertson GT, Farris MA, Roop RM, Peterson KM. 1995. Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene 166:175–176. doi: 10.1016/0378-1119(95)00584-1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.