ABSTRACT
Campylobacter is among the most common worldwide causes of bacterial gastroenteritis. This organism is part of the commensal microbiota of numerous host species, including livestock, and these animals constitute potential sources of human infection. Molecular typing approaches, especially multilocus sequence typing (MLST), have been used to attribute the source of human campylobacteriosis by quantifying the relative abundance of alleles at seven MLST loci among isolates from animal reservoirs and human infection, implicating chicken as a major infection source. The increasing availability of bacterial genomes provides data on allelic variation at loci across the genome, providing the potential to improve the discriminatory power of data for source attribution. Here we present a source attribution approach based on the identification of novel epidemiological markers among a reference pan-genome list of 1,810 genes identified by gene-by-gene comparison of 884 genomes of Campylobacter jejuni isolates from animal reservoirs, the environment, and clinical cases. Fifteen loci involved in metabolic activities, protein modification, signal transduction, and stress response or coding for hypothetical proteins were selected as host-segregating markers and used to attribute the source of 42 French and 281 United Kingdom clinical C. jejuni isolates. Consistent with previous studies of British campylobacteriosis, analyses performed using STRUCTURE software attributed 56.8% of British clinical cases to chicken, emphasizing the importance of this host reservoir as an infection source in the United Kingdom. However, among French clinical isolates, approximately equal proportions of isolates were attributed to chicken and ruminant reservoirs, suggesting possible differences in the relative importance of animal host reservoirs and indicating a benefit for further national-scale attribution modeling to account for differences in production, behavior, and food consumption.
IMPORTANCE Accurately quantifying the relative contribution of different host reservoirs to human Campylobacter infection is an ongoing challenge. This study, based on the development of a novel source attribution approach, provides the first results of source attribution in Campylobacter jejuni in France. A systematic analysis using gene-by-gene comparison of 884 genomes of C. jejuni isolates, with a pan-genome list of genes, identified 15 novel epidemiological markers for source attribution. The different proportions of French and United Kingdom clinical isolates attributed to each host reservoir illustrate a potential role for local/national variations in C. jejuni transmission dynamics.
KEYWORDS: source attribution, Campylobacter, gene-by-gene approach, genomics, food-borne diseases
INTRODUCTION
Campylobacter spp. are among the main causes of foodborne bacterial gastroenteritis worldwide with nearly 236,000 reported cases in Europe in 2014 (1). Campylobacteriosis, mostly caused by Campylobacter jejuni and Campylobacter coli (2), is characterized by acute diarrhea, abdominal pain, headache, and nausea (3) and can lead to Guillain-Barré syndrome (4) and inflammatory bowel syndromes, including Crohn's disease (5, 6). Despite the improvement of surveillance services, the incidence of campylobacteriosis is underestimated. In France, the European Food Safety Authority (EFSA) reported nearly 5,000 cases annually between 2011 and 2014 (1), while the actual burden of disease is thought to be much higher at around 500,000 cases each year (7).
The ubiquity of Campylobacter, as part of the commensal microbiota of various animals, contributes to the threat this organism poses to humans. C. jejuni is commonly isolated from the digestive tracts of many mammals and wild and domestic birds (8). However, factors including the ability to form biofilms (2, 9) and colonize protozoa (10) mean that Campylobacter can be isolated from sources outside the host gut, such as food and water sources (11–14). Humans are usually infected by handling, preparation, or consumption of meat contaminated during slaughter, including pork, beef, and especially poultry (15–17). Consumption of raw milk or untreated water and contact with animals are also potential infection sources (18–20), and quantifying the relative contribution of different infection sources remains an important aim in public health.
Molecular typing methods, such as multilocus sequence typing (MLST) (21), have shown Campylobacter populations to be highly structured, providing a better understanding of how lineage clusters relate to ecology (22, 23). In particular, this has revealed the existence of host-associated genotypes that are more commonly isolated from, for example, chickens or cattle, as well as generalist genotypes that are commonly isolated from multiple hosts. This means that genotyped clinical isolates can be assigned to the reservoir host population from which they most likely originated, based on genotype and allele frequencies, allowing the investigation of the source of human infection. This source attribution approach has been applied to Campylobacter, mainly using MLST data, emphasizing a significant role for the chicken reservoir in human infection (12, 24–29).
The increasing use of whole-genome sequencing (WGS) data sets is enhancing understanding of the genetic basis of Campylobacter host ecology and transmission (9, 30–32). In terms of source attribution, data on allelic variation at loci across the genome have considerable potential to improve sensitivity, particularly for assigning the origin of isolates from generalist clonal complexes, such as sequence type 21 (ST-21) and ST-45 complexes, where seven-locus MLST has been of limited use in identifying the source population (23, 33). The evolution of genomic signatures of host association requires that strains are genetically isolated in a specific host niche for long enough for adaptation and genetic drift to lead to host-associated sequence variation in the genome. Therefore, while targeting genetic variation across the genome increases the chances of identifying variation that segregates by host, rapid zoonotic transmission may erode host association signatures. However, even with estimates of host transitions occurring as frequently as every 2 years for isolates from the ST-21 and ST-45 complexes (34), in rapidly recombining species, such as Campylobacter spp., adaptation may occur fast enough to generate variation that provides a basis for source attribution.
In this study, we take a systematic gene-by-gene approach (35, 36) to mine the entire pan-genome of a defined genomic data set of C. jejuni isolates from several animal reservoirs to identify novel epidemiological markers for source attribution. To achieve this, alleles at loci across the genomes of isolates from known hosts were probabilistically assigned to several host populations. Loci where this “self-attribution” gave a high probability of assigning isolates to the correct host population were considered good candidates for source attribution. These host-segregating marker loci were then used to attribute the origin of human clinical cases from France and the United Kingdom, as well as isolates from French pets, to host reservoir populations based on the polymorphism at these 15 loci.
RESULTS
Core, soft-core, and accessory genomes.
The principle of MLST was extended to the whole genomes of 884 C. jejuni isolates using the 1,810 loci of the pan-genome allowing definition of the core, soft-core, and accessory genomes of the C. jejuni isolates in this study. A core set of 472 genes were universally present within the 884 C. jejuni genomes. A set of 953 genes were found in at least 95% of the 884 isolates and constituted the soft-core genome of our population. Finally, 385 genes were present within less than 95% of the isolates and formed the accessory genome of the population. In these 1,810 loci, the number of alleles per locus ranged from 1 to 430 (see Table S3 in the supplemental material).
Similar highly structured genetic organization in French and worldwide agricultural C. jejuni isolate populations.
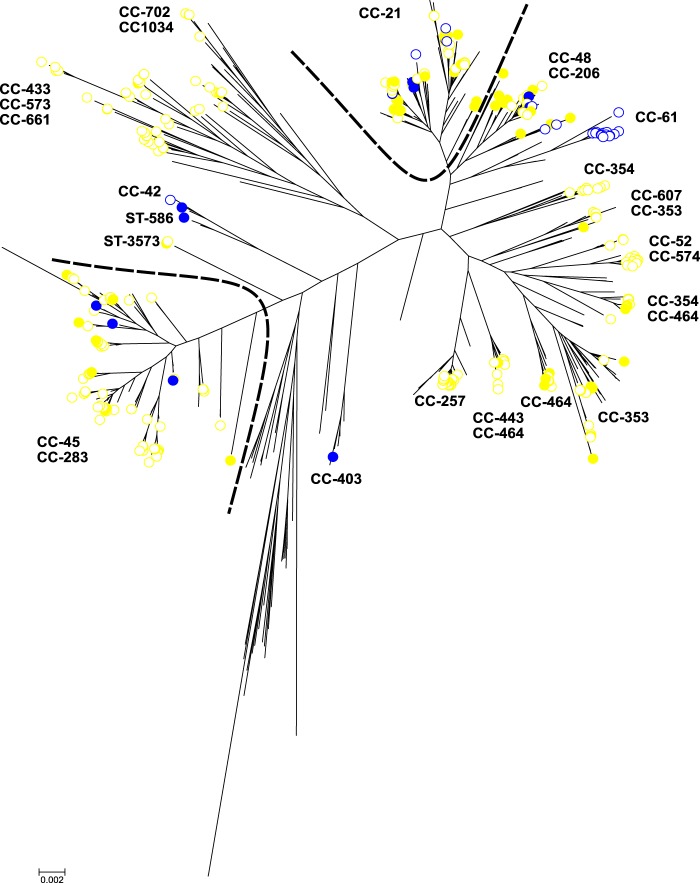
A core genome genealogical tree using an approximation of the maximum likelihood algorithm was implemented in FastTree2 to compare the population structure of chicken and ruminant isolates in different countries (Fig. 1). French agricultural C. jejuni isolates did not form distinct isolated clusters that were separate from agricultural isolates originating outside France. As in other studies (37, 38), this shows that host-associated genetic variation in C. jejuni is distributed across national boundaries and allows the use of existing reference training data sets to assign isolates from France. French isolates from chicken belonged to the ST-21 complex (n = 32), ST-45 complex (n = 9), ST-48 complex (n = 7), ST-206 complex (n = 7), ST-353 complex (n = 5), and ST-464 complex (n = 5). The French isolates from ruminants belonged to the ST-21 complex (n = 4), ST-42 complex (n = 1), ST-45 complex (n = 3), ST-48 complex (n = 3), ST-403 complex (n = 1), and ST-586 (n = 1). All the C. jejuni isolates from the different agricultural sources in France clustered with agricultural isolates from other countries except for one French isolate from cattle belonging to the ST-403 complex. Isolate structuring mirrored the clonal complex structure based on MLST designations. The cattle-associated ST-42 complex (39) was present among French isolates as were isolates belonging to the chicken specialist ST-353 complex (39).
FIG 1.
Genetic structure of 411 C. jejuni isolates from chicken and ruminants in different countries. A phylogenetic tree was constructed from 1,810 genes found in four reference strains of C. jejuni (NCTC11168, 81-176, 81116, and M1) through an approximation of the maximum likelihood algorithm using FastTree2, and visualized using MEGA6. The bar represents the number of substitutions per site. The color of the circle indicates the original host of the isolates, and the kind of circle indicates the country of origin. Isolates from chicken are shown in yellow and those from ruminants in blue. Worldwide isolates are represented by empty (white) circles, and French isolates are represented by filled-in circles. Numeric labels correspond to clonal complexes (CC) or sequence type (ST) of chicken and cattle isolates.
Selection of loci as potential host-segregating markers for source attribution.
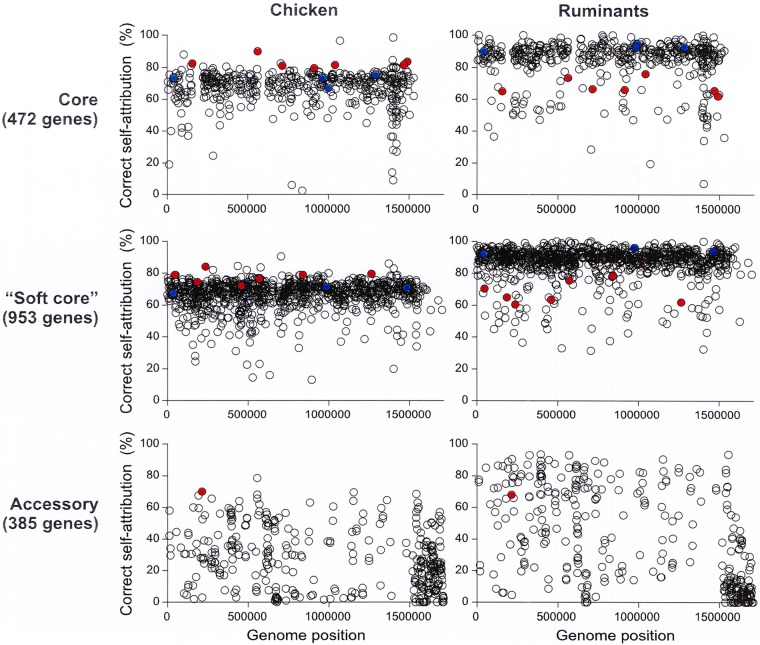
To identify potentially suitable epidemiological markers for source attribution, we assessed the host-segregating power of the 1,810 loci by quantifying their accuracy for each source in self-attribution tests. The correct self-attribution rate of chicken isolates was generally lower than that for ruminant isolates. This difference was significantly lower (P < 0.001 by t test) using the 472 genes belonging to the core genome or the 953 genes belonging to the soft-core genome (Fig. 2). These two data sets of core and soft-core genes allowed a correct self-attribution to the host of 92% and 92.5% of ruminant isolates, respectively, and 77% and 73% of chicken isolates, respectively. Thus, according to the set of genes used to perform the source attribution, 23% to 27% of chicken isolates were wrongly attributed to the ruminant reservoir, while less than 10% of ruminant isolates were misattributed. When the genes belonging to the accessory genome were used for source attribution analyses, 87% of ruminant isolates were correctly associated with their host compared to 76.5% in the chicken population. This difference of locus segregating power according to the source can lead to a bias in the source assignment and an overestimation of the ruminant involvement in campylobacteriosis. Moreover, it has also been observed that chicken isolates may less often be assigned to the right host than ruminant isolates in self-attribution tests performed using the STRUCTURE software program (26). Thus, to account for this, we focused on epidemiological marker loci allowing equivalent or higher segregation of chicken isolates compared to ruminant isolates with a minimum threshold fixed at 60% (Fig. S1).
FIG 2.
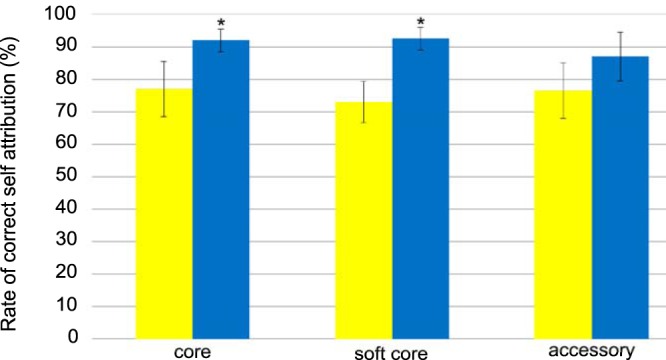
Correct host assignment accuracy in self-attribution tests of 1,810 core, soft-core, or accessory genes in C. jejuni isolates from chicken (yellow) and ruminants (blue). Twenty self-attribution tests were performed on random subsets of isolates using STRUCTURE software. Alleles at all loci in isolates of known host origin were assigned to the host training data set, and the probability of correct host population attribution was recorded. The average ± standard deviation (error bar) values of the rates of correct host assignment in self-attribution were calculated for each population. A Student t test was performed to assess statistical significance. Values were considered significantly different when P < 0.01 and are indicated by an asterisk.
In total, 17 core genes, 20 soft-core genes, and one accessory gene demonstrated ≥60% correct host self-attribution for both chicken and cattle populations, constituting candidates for host-segregating markers. Finally, the 15 loci that showed the most accurate self-attribution were selected as potential good candidates for host-segregating markers. These genes are putatively involved in metabolic activities such as amino acid or vitamin biosynthesis, energy metabolism, modification of protein, or signal transduction, and stress response to heat shock (Table 1). The correct host attribution rate for these loci ranged from 70% to 90% in chicken and 60.5% to 78.5% in ruminants (Table 1 and Fig. 3). These 15 loci thus allowed an average correct host attribution of 80.7% in chicken populations and 68.2% in ruminant populations, which corresponded to a difference of 12.5% between chicken and ruminant populations. The correct host attribution rate for MLST was good for ruminant isolates (90% to 96%), but for isolates from chicken, it ranged from 66.5% to 75% (Fig. 3). The greater difference in correct host attribution with MLST, observed between chicken and ruminant (22.3%), could lead to a bias in host assignment with the overestimation of attribution to a particular reservoir.
TABLE 1.
Function of genes identified as potential host-segregating epidemiological markers for C. jejuni
| Locus tag | Gene | Genome | Descriptiona | No. of alleles at each locus | Prevalence in chicken isolates (n = 392) | Prevalence in ruminant isolates (n = 59) | C. jejuni reference genomic position | Correct self-attribution in chicken (%) | Correct self-attribution in ruminants (%) |
|---|---|---|---|---|---|---|---|---|---|
| Cj0133 | Core | Glycoprotease family protein | 44 | 392 | 59 | 156131 | 82.5 | 65 | |
| Cj0260c | Accessory | Hypothetical protein | 55 | 313 | 59 | 213615 | 70 | 68 | |
| Cj0348 | trpB | Core | Tryptophan synthase subunit beta (EC 4.2.1.20); K01696 tryptophan synthase beta chain | 124 | 392 | 59 | 1489335 | 83.5 | 62 |
| Cj0350 | Soft core | Hypothetical protein | 47 | 392 | 59 | 236997 | 84 | 60.5 | |
| Cj0663c | hslV | Core | ATP-dependent protease peptidase subunit (EC 3.4.25.1); K01419 ATP-dependent HslUV protease | 69 | 392 | 59 | 1041635 | 81.5 | 76 |
| Cj0986c | Soft core | Pseudogene | 28 | 392 | 59 | 491470 | 82 | 73 | |
| Cj1047c | thiS | Core | Thiamine biosynthesis protein; K03154 sulfur carrier protein | 49 | 392 | 59 | 1467213 | 81.5 | 65.5 |
| Cj1169c | Core | Hypothetical protein | 43 | 392 | 59 | 561701 | 90 | 73.5 | |
| Cj1186c | petA | Soft core | Ubiquinol-cytochrome c reductase iron-sulfur subunit (EC 1.10.2.2); K00411 ubiquinol-cytochrome | 61 | 392 | 59 | 1264263 | 79.5 | 62 |
| Cj1192 | dctA | Soft core | C4-dicarboxylate transport protein; K11103 aerobic C4-dicarboxylate transport protein | 139 | 392 | 59 | 840278 | 79 | 78.5 |
| Cj1204c | atpB | Soft core | FoF1 ATP synthase subunit A (EC 3.6.3.14); K02108 F-type H+-transporting ATPase subunit a | 75 | 392 | 59 | 46678 | 79 | 70.5 |
| Cj1220 | groES | Soft core | Cochaperonin GroES; K04078 chaperonin GroES | 30 | 392 | 58 | 998123 | 80 | 61 |
| Cj1463 | flgJ | Core | Hypothetical protein | 70 | 392 | 59 | 912509 | 79.5 | 66 |
| Cj1513c | Core | Hypothetical protein | 24 | 392 | 59 | 714099 | 81 | 66.5 | |
| Cj1621 | Soft core | Hypothetical protein | 91 | 391 | 59 | 753195 | 78 | 75.5 |
Annotations were obtained from the KEGG database.
FIG 3.
Host-segregating power of each locus within the core, soft-core, and accessory genomes of C. jejuni. Each circle represents the rate of correct host assignment of one locus in self-attribution tests on chicken and ruminant isolates. Self-attribution tests were performed using the allelic diversity of chicken or ruminant isolates within their core, soft-core, or accessory genomes separately and STRUCTURE software. Alleles at all loci in isolates of known host origin were assigned to the host training data set, and the probability of correct host population attribution was recorded to determine the host-segregating power of each locus. Solid red circles represent the 15 candidates for host-segregating markers, and MLST loci are colored in blue.
Source attribution of British and French clinical cases and pet C. jejuni isolates.
A total of 506 agricultural or environmental C. jejuni isolates were used as the reference data set to assign the source of French (n = 42) and British (n = 281) clinical isolates and those from French pets (n = 55). These isolates were attributed probabilistically using the STRUCTURE software program to each potential host source population using allele information at 15 host-segregating loci (Fig. 4). A total of 56.8% of British human cases were attributed to chicken, while 37.1% of cases were attributed to ruminants and 6.1% to the environment. The same analysis applied to French clinical cases attributed an approximately equivalent proportion of cases to chicken (45.8%) and ruminants (46.9%), with 7.3% of cases attributed to environmental/wild bird sources. Analysis of the French pet isolate population revealed an equivalent attribution between the three host populations: 30.7% of the pet isolate population was attributed to chicken, 35.5% to ruminants, and 33.8% to the environment. Consistent with previous work, there was relatively low attribution to environment/wild bird sources in both countries (26). There was some evidence for differences in the contribution of chicken and cattle reservoirs of infection, although there were relatively few French samples.
FIG 4.
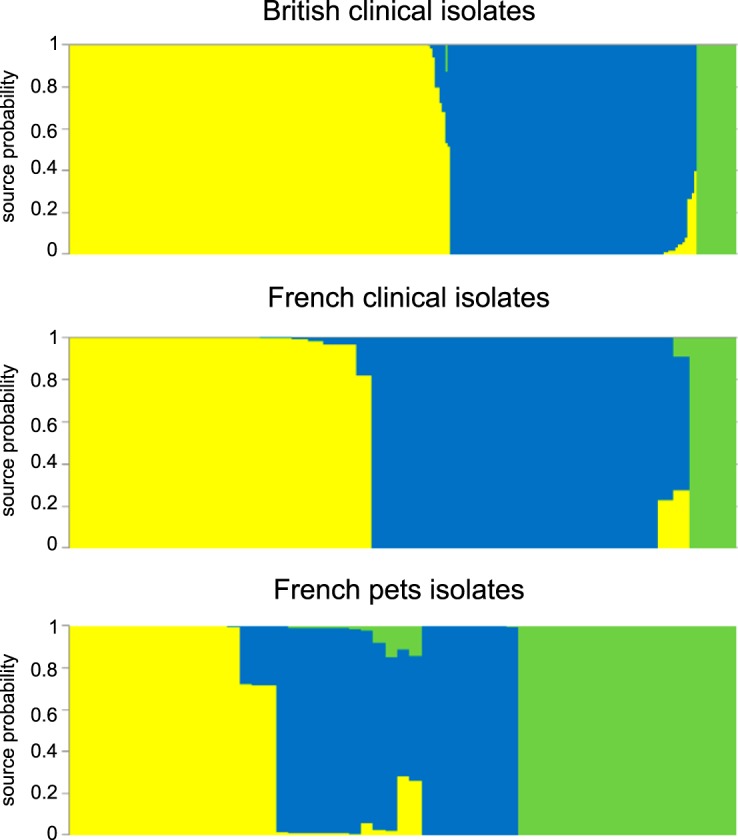
Assignment to source of British and French human clinical C. jejuni isolates and those from French pets using STRUCTURE software. Each vertical bar represents one isolate, and the color of the bar shows the estimated probability that this isolate originates from each of the potential sources. Attribution source populations are chickens (yellow), ruminants (blue), and environment/wild bird isolates (green). Isolates were ordered by assigned host.
DISCUSSION
Accurately quantifying the relative contribution of different host reservoirs to human Campylobacter infection is an ongoing challenge. Probabilistic attribution based on seven-locus MLST has provided valuable information and implicated poultry as an important source (12, 24, 27, 40). However, these techniques can act only on host association signals in seven genes. This is particularly limiting when assigning the origin of lineages that have switched hosts relatively recently and therefore have had limited time for host-associated signatures to evolve in these genes. One potential way to improve power is to target signatures at other loci across the genome. While there is host-associated genetic variation, even in the genomes of host generalist C. jejuni lineages (30), using whole-genome MLST (35) data in an existing attribution model provided little additional power over seven-locus MLST (34). One explanation for this is the relative scarcity of host-segregating markers. For example, here we found that nearly 31% of core gene alleles present in more than one ST-21 and ST-45 complex isolate genome were present in isolates from both cattle and chickens. In part because of this, signals of host association may be masked in conventional attribution models by signals of numerous non-host-segregating loci present in the population. In this study, 1,772 loci (constituting 98% of pan-genome loci) did not meet our criteria for host segregation. To account for this, we took the alternative approach of conducting gene-by-gene analysis of the genome and defining a panel of host-segregating loci.
Self-attribution tests, using STRUCTURE software, quantified the probability of correct host assignment for each locus across the genome. Consistent with analysis based on seven-locus MLST (26), fewer chicken isolates were correctly self-attributed compared to those from ruminants, with 73% to 77% and 87% to 92.5% correct self-attribution, respectively, using the different sets of genes (core genes, soft-core genes, and accessory genes). While alleles at some loci gave up to 100% correct self-attribution in one host, it was essential that selection of host-segregating marker loci was based upon the proportion of correct host segregation in both chicken and ruminant reservoirs to reduce attribution bias. Gene-by-gene assessment of the probability of correct self-attribution identified seven core genes, seven soft-core genes, and one accessory gene as candidate host-segregating epidemiological markers. The 15 chosen marker loci had various putative functions, with six loci encoding hypothetical proteins and the remaining loci involved in metabolic activities such as amino acid biosynthesis and energy metabolism, protein modification, signal transduction, and stress response. Multiple factors associated with differences in animal husbandry and host physiology make it difficult to assign a biological basis for host segregation of alleles among the marker loci. However, some of the genes are involved in acid stress response (groES) (41) or are organized in the same operon as flagellar proteins (flgJ) or as proteins involved in oxidative stress response (Cj1169c) (42).
While it is not necessary to define epidemiological markers based on functional differences, it is interesting to speculate how host colonization factors may have influenced genomic signatures of host association. There are numerous differences between the chicken and ruminant digestive tracts, including body temperature, 41°C in chicken and 38.6°C in cattle, and pH (43, 44). This can be used as a context for considering the sequence variation at host-segregating loci in this study. For example, groES, which is involved in the heat shock response (45), has been shown to contribute to the protection of C. jejuni against pH acidity during transit in the stomach (41). In addition, differences between chicken and cattle husbandry may apply different selection pressures to genes associated with survival of oxygen-intolerant Campylobacter outside the host. For example, depletion of the hypothetical protein Cj1169c and the protein Cj1170c (OMP50) in C. jejuni mutants has been shown to result in reduced colonization of chicken and higher sensitivity to oxygen (42, 46). Considering the cause of sequence variation at host-segregating loci is purely speculative in this study, but it is interesting to note that many of these loci are within the core (soft-core) genome. This is consistent with homologous sequence variation having a role in host adaptation, as previously described for C. coli (47), and not just acquisition of new genes conferring a specific functional advantage.
Consistent with previous studies (26, 48), source attribution of British and French clinical isolates using the 15 host-segregating markers in this study indicated a relatively small contribution of environmental and wild bird reservoirs as human infection sources. As in seven-locus MLST studies in United Kingdom, New Zealand, and Swiss human cases (26, 27, 49), the majority (56.8%) of British clinical cases in this study were attributed to the chicken reservoir, with 37.1% attributed to cattle. Among the most interesting findings was the higher attribution to the ruminant reservoir (46.9%) among French clinical cases, which was approximately equivalent to the contribution from chicken (45.8%). While the number of clinical isolates from France was relatively small, increasing the possibility of sampling bias, the elevated attribution to the ruminant reservoir was consistent with the role of cattle as an infection source among rural children in northeastern Scotland (40). Cultural and dietary differences could influence the relative contribution of sources of foodborne disease in France and the United Kingdom. For example, chicken consumption is higher in the United Kingdom (30 kg/person/year) compared to France (25 kg/person/year) (50), where other known infection sources, including ruminant offal and veal (51–54), form a greater proportion of the diet. Factors associated with food preparation may also be significant, but analysis of a larger data set of French clinical isolates would be necessary to achieve a more representative description of human C. jejuni contamination routes in France.
MATERIALS AND METHODS
Isolates and genome sequencing.
A collection of 212 French C. jejuni isolates was sequenced, including isolates from clinical cases, chicken, cattle, pets, and the environment. The human isolates (n = 40) were from campylobacteriosis cases occurring in 2009 in France in regions with a significant broiler chicken meat consumption pattern. Chicken isolates were collected in 2008 and 2009 during two monitoring surveys designed to be representative of the broiler chicken production in France. The first sampling survey allowed the collection of isolates from ceca (n = 11) and carcasses (n = 21) collected from 425 batches of broiler chickens slaughtered in 58 French slaughterhouses over a 12-month period in 2008 (55). During the second monitoring survey, retail meat isolates (n = 33) were collected from broiler meat sampled in retail outlets over a 6-month period in 2009 in geographic areas representing the most significant broiler meat consumption patterns in France (14, 56). Cattle isolates were collected from dairy cow feces sampled during a local survey in 10 farms located in France in 2013 (n = 13) (57). The pet isolates (n = 55) were collected from 304 pet feces sampled in four veterinary clinics, two kennels, and individuals owning pets in Brittany in France in December 2014 and during a 6-month period from April to October 2015. Finally, the environmental C. jejuni isolates were collected from seawater (n = 3), freshwater (n = 30), sediments (n = 3), or mussels (n = 3) in France between 2013 and 2015 (see Table S1 in the supplemental material).
Isolates were subcultured onto Campylobacter selective blood-free agar (Karmali; Oxoid) in microaerophilic conditions (85% N2, 10% CO2, and 5% O2) at 42°C for 48 h. The genomic DNA was extracted from 1-day single-colony cultures incubated at 37°C using the QiaAMP DNA minikit (Qiagen) and quantified using the Qubit 2.0 fluorometer and the Qubit dsDNA (double-stranded DNA) HS (high-sensitivity) assay kit (Invitrogen). Genomes were sequenced using the Ion Torrent technology (Life Technologies). Libraries were prepared using the Ion Xpress Plus fragment library kit fragmentation (Life Technologies), cleaned using Agencourt AMPure XP (Beckman Coulter), and enriched after the size selection performed on a 2% E-Gel SizeSelect (Invitrogen). Emulsion PCR on Ion OneTouch 2 system and subsequent enrichment of template particles on Ion OneTouch ES system were both performed using the Ion PI template OT2 200 kit v3 (Life Technologies). The samples were loaded on a P1 chip and sequenced with an Ion Torrent Proton machine (Life Technologies). When needed for SPAdes assembly and torrent mapping alignment program (TMAP) alignment, the read number was down sampled to fit a maximal coverage depth of 80 (coverage depth evaluated on TMAP alignment, Torrent Suite v.4.0.2) before cleaning with Trimmomatic 0.32 software (58). Assemblies were produced by either MIRA version 4.0rc1 (59) or SPAdes 3.1.1 (60). The k-mer size used by MIRA was deduced by using kmergenie version 1.5658 (61). An average of 138 contigs were obtained for the 212 C. jejuni sequenced genomes with a median value of 71 contigs. The average of the total assembled sequence length is 1,708,807 bp (Table S2).
Isolates sequenced in this study were augmented with 672 genomes of C. jejuni isolated from clinical cases, chickens, ruminants, environmental water, and wild birds in different countries and published in previous studies (30, 32, 39, 62). This gave a total of 884 C. jejuni genomes in our study data set (Table S1).
Defining core, soft-core, and accessory genomes.
The genomes sequenced in this study were stored on a web-based archive based on the BIGSdb software (36). The BLAST algorithm was used to perform gene-by-gene alignment and whole-genome MLST on the 884 C. jejuni genomes using a reference pan-genome approach (35, 63, 64). The reference pan-genome included 1,810 unique loci and was obtained from four available genomes, C. jejuni strains NCTC11168 (65), 81-176 (66), 81116 (67), and M1 (68) using a previously described method (64). Orthologous genes at these loci were defined as present in the 884 genomes if the sequence was present with >70% nucleotide identity over ≥50% of the sequence length. Individual gene sequences were aligned using MAFFT (69) and concatenated into contiguous sequence for each isolate including gaps for missing nucleotides or entire genes. An approximation of the maximum likelihood algorithm implemented in FastTree2 software (70) was used to reconstruct a phylogeny of core genome alignments, and the tree was visualized and annotated using MEGA6 software (71). Genetic variation at pan-genome loci was investigated in a presence/absence matrix with allelic diversity (64). The number of missing or incomplete genes for each locus was calculated to define the core, soft-core, and accessory genome of the C. jejuni population in this study. The core genome was defined as genes shared by all the isolates, including incomplete genes, which are a technical artifact due to the use of draft genomes. Genes shared in at least 95% of the isolates constituted the soft-core genome (72, 73), and the remaining genes constituted the accessory genome.
Identification of epidemiological markers segregating isolates by host source.
Loci where alleles segregate by host represent useful epidemiological markers for source attribution. To identify these loci, we assigned alleles at all loci in isolates of known host origin to host source training data sets and recorded the probability of correct host population in self-attribution, as in previous studies using seven MLST genes (26). Self-attribution tests focused on chicken isolates (n = 352) and ruminant isolates (n = 59) as the source of the majority of isolates in this study and major reservoirs of human infection by C. jejuni. Furthermore, generalist ST-21 and ST-45 clonal complexes are common in these hosts but have been difficult to attribute to source using seven-locus MLST (23, 26, 29, 34, 49). Host attribution was performed using STRUCTURE software, a Bayesian model-based clustering method designed to infer population structure and attribute individuals to populations using multilocus genotype data (74). Probabilistic assignment was carried out using 1,810 pan-genome loci using the “No Admixture” model with uncorrelated allele frequency model, assuming that each isolate originated in one of the putative source populations each with its own characteristic set of allelic frequencies (75). Analyses were performed with 10,000 burn-in cycles followed by 10,000 iterations with the parameters using source population information (USEPOPINFO) with test isolates differentiated from the training data set using POPFLAG. Random subsets of 20 isolates from each species were assigned to the training data, and self-attribution was performed 10 times for core, soft-core, and accessory genome loci separately.
Based on the self-attribution tests, the segregating power of each locus was calculated as the average probability of allele assignment to the correct host. Loci strongly contributing to correct assignments to chicken and ruminant populations constitute potential candidates for host-segregating epidemiological markers.
Assignment of human and pet isolates using the host-segregating markers.
Source attribution of the human isolates from France and the United Kingdom was performed using allelic profiles of host-segregating loci. The source of pet contamination was also investigated because of the potential role as vectors in Campylobacter transmission to humans (19, 76, 77). Assignment analyses were carried out separately for 42 French and 281 British clinical isolates (32) and 55 isolates from French pets. The data set used as a reference to probabilistically attribute the sources of clinical and pet isolates comprised 352 chicken isolates, 59 ruminant isolates, and 95 wild bird and environmental isolates. The same settings were used as for self-attribution tests except the burn-in period and the iterations, which were set at 100,000, consistent with published work (49).
Accession number(s).
Genome sequences generated as part of this study were deposited in SRA (SRR5123296 to SRR5123507; see Table S1), and assemblies are available in Dryad (http://datadryad.org/) at https://doi.org/10.5061/dryad.m86k3. The assemblies of genomes sequenced in earlier studies can be found in Dryad (https://doi.org/10.5061/dryad.8t80s and https://doi.org/10.5061/dryad.28n35) and NCBI (BioProject PRJNA312235).
Supplementary Material
ACKNOWLEDGMENTS
A.T. was funded by a Ph.D. studentship from the French Ministry of Defence (Direction Générale de l'Armement) and the Conseil General des Côtes d'Armor (CG22) and by a short-term mission grant from the MedVetNet European program. G.M. was supported by a NISCHR Health Research Fellowship (HF-14-13). S.K.S. was funded by Biotechnology and Biological Sciences Research Council (BBSRC) grant BB/I02464X/1, Medical Research Council (MRC) grant MR/L015080/1, and Wellcome Trust grant 088786/C/09/Z. We thank Francis Mégraud, head of the National Reference Center for Campylobacter and Helicobacter in France, and Michèle Gourmelon from the French Research Institute for Exploitation of the Sea (IFREMER) for kindly providing the French clinical and environmental isolates of C. jejuni.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.03085-16.
REFERENCES
- 1.European Food Safety Authority. 2015. The European Union summary report on trends and sources of zoonoses, zoonotic agents and food-borne outbreaks in 2014. EFSA J 13:4329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nguyen VT, Fegan N, Turner MS, Dykes GA. 2012. Role of attachment to surfaces on the prevalence and survival of Campylobacter through food systems. J Food Prot 75:195–206. doi: 10.4315/0362-028X.JFP-11-012. [DOI] [PubMed] [Google Scholar]
- 3.Zilbauer M, Dorrell N, Elmi A, Lindley KJ, Schuller S, Jones HE, Klein NJ, Nunez G, Wren BW, Bajaj-Elliott M. 2007. A major role for intestinal epithelial nucleotide oligomerization domain 1 (NOD1) in eliciting host bactericidal immune responses to Campylobacter jejuni. Cell Microbiol 9:2404–2416. doi: 10.1111/j.1462-5822.2007.00969.x. [DOI] [PubMed] [Google Scholar]
- 4.Koga M, Gilbert M, Takahashi M, Li J, Koike S, Hirata K, Yuki N. 2006. Comprehensive analysis of bacterial risk factors for the development of Guillain-Barre syndrome after Campylobacter jejuni enteritis. J Infect Dis 193:547–555. doi: 10.1086/499969. [DOI] [PubMed] [Google Scholar]
- 5.Lamhonwah AM, Ackerley C, Onizuka R, Tilups A, Lamhonwah D, Chung C, Tao KS, Tellier R, Tein I. 2005. Epitope shared by functional variant of organic cation/carnitine transporter, OCTN1, Campylobacter jejuni and Mycobacterium paratuberculosis may underlie susceptibility to Crohn's disease at 5q31. Biochem Biophys Res Commun 337:1165–1175. doi: 10.1016/j.bbrc.2005.09.170. [DOI] [PubMed] [Google Scholar]
- 6.Kalischuk LD, Inglis GD, Buret AG. 2009. Campylobacter jejuni induces transcellular translocation of commensal bacteria via lipid rafts. Gut Pathog 1:2. doi: 10.1186/1757-4749-1-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Van Cauteren D, De Valk H, Sommen C, King LA, Jourdan-Da Silva N, Weill FX, Le Hello S, Megraud F, Vaillant V, Desenclos JC. 2015. Community incidence of campylobacteriosis and nontyphoidal salmonellosis, France, 2008-2013. Foodborne Pathog Dis 12:664–669. doi: 10.1089/fpd.2015.1964. [DOI] [PubMed] [Google Scholar]
- 8.Humphrey T, O'Brien S, Madsen M. 2007. Campylobacters as zoonotic pathogens: a food production perspective. Int J Food Microbiol 117:237–257. doi: 10.1016/j.ijfoodmicro.2007.01.006. [DOI] [PubMed] [Google Scholar]
- 9.Pascoe B, Méric G, Murray S, Yahara K, Mageiros L, Bowen R, Jones NH, Jeeves RE, Lappin-Scott HM, Asakura H, Sheppard SK. 2015. Enhanced biofilm formation and multi-host transmission evolve from divergent genetic backgrounds in Campylobacter jejuni. Environ Microbiol 17:4779–4789. doi: 10.1111/1462-2920.13051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Snelling WJ, Moore JE, McKenna JP, Lecky DM, Dooley JS. 2006. Bacterial-protozoa interactions; an update on the role these phenomena play towards human illness. Microbes Infect 8:578–587. doi: 10.1016/j.micinf.2005.09.001. [DOI] [PubMed] [Google Scholar]
- 11.Engberg J, Gerner-Smidt P, Scheutz F, Moller Nielsen E, On SL, Molbak K. 1998. Water-borne Campylobacter jejuni infection in a Danish town—a 6-week continuous source outbreak. Clin Microbiol Infect 4:648–656. doi: 10.1111/j.1469-0691.1998.tb00348.x. [DOI] [PubMed] [Google Scholar]
- 12.Sheppard SK, Dallas JF, MacRae M, McCarthy ND, Sproston EL, Gormley FJ, Strachan NJ, Ogden ID, Maiden MC, Forbes KJ. 2009. Campylobacter genotypes from food animals, environmental sources and clinical disease in Scotland 2005/6. Int J Food Microbiol 134:96–103. doi: 10.1016/j.ijfoodmicro.2009.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wimalarathna HM, Richardson JF, Lawson AJ, Elson R, Meldrum R, Little CL, Maiden MC, McCarthy ND, Sheppard SK. 2013. Widespread acquisition of antimicrobial resistance among Campylobacter isolates from UK retail poultry and evidence for clonal expansion of resistant lineages. BMC Microbiol 13:160. doi: 10.1186/1471-2180-13-160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guyard-Nicodème M, Rivoal K, Houard E, Rose V, Quesne S, Mourand G, Rouxel S, Kempf I, Guillier L, Gauchard F, Chemaly M. 2015. Prevalence and characterization of Campylobacter jejuni from chicken meat sold in French retail outlets. Int J Food Microbiol 203:8–14. doi: 10.1016/j.ijfoodmicro.2015.02.013. [DOI] [PubMed] [Google Scholar]
- 15.Fravalo P, Laisney MJ, Gillard MO, Salvat G, Chemaly M. 2009. Campylobacter transfer from naturally contaminated chicken thighs to cutting boards is inversely related to initial load. J Food Prot 72:1836–1840. doi: 10.4315/0362-028X-72.9.1836. [DOI] [PubMed] [Google Scholar]
- 16.Hermans D, Pasmans F, Messens W, Martel A, Van Immerseel F, Rasschaert G, Heyndrickx M, Van Deun K, Haesebrouck F. 2012. Poultry as a host for the zoonotic pathogen Campylobacter jejuni. Vector Borne Zoonotic Dis 12:89–98. doi: 10.1089/vbz.2011.0676. [DOI] [PubMed] [Google Scholar]
- 17.Guyard-Nicodème M, Tresse O, Houard E, Jugiau F, Courtillon C, El Manaa K, Laisney MJ, Chemaly M. 2013. Characterization of Campylobacter spp. transferred from naturally contaminated chicken legs to cooked chicken slices via a cutting board. Int J Food Microbiol 164:7–14. doi: 10.1016/j.ijfoodmicro.2013.03.009. [DOI] [PubMed] [Google Scholar]
- 18.Friedman CR, Hoekstra RM, Samuel M, Marcus R, Bender J, Shiferaw B, Reddy S, Ahuja SD, Helfrick DL, Hardnett F, Carter M, Anderson B, Tauxe RV, Emerging Infections Program FoodNet Working Group . 2004. Risk factors for sporadic Campylobacter infection in the United States: a case-control study in FoodNet sites. Clin Infect Dis 38(Suppl 3):S285–S296. doi: 10.1086/381598. [DOI] [PubMed] [Google Scholar]
- 19.Olson CK, Ethelberg S, van Pelt W, Tauxe RV. 2008. Epidemiology of Campylobacter jejuni infections in industrialized nations, p 163–189. In Nachamkin I, Szymanski C, Blaser M (ed), Campylobacter, 3rd ed American Society for Microbiology, Washington, DC. doi: 10.1128/9781555815554.ch9. [DOI] [Google Scholar]
- 20.Pitkanen T. 2013. Review of Campylobacter spp. in drinking and environmental waters. J Microbiol Methods 95:39–47. doi: 10.1016/j.mimet.2013.06.008. [DOI] [PubMed] [Google Scholar]
- 21.Maiden MC, Bygraves JA, Feil E, Morelli G, Russell JE, Urwin R, Zhang Q, Zhou J, Zurth K, Caugant DA, Feavers IM, Achtman M, Spratt BG. 1998. Multilocus sequence typing: a portable approach to the identification of clones within populations of pathogenic microorganisms. Proc Natl Acad Sci U S A 95:3140–3145. doi: 10.1073/pnas.95.6.3140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.van Belkum A, Tassios PT, Dijkshoorn L, Haeggman S, Cookson B, Fry NK, Fussing V, Green J, Feil E, Gerner-Smidt P, Brisse S, Struelens M, European Society of Clinical Microbiology and Infectious Diseases (ESCMID) Study Group on Epidemiological Markers (ESGEM) . 2007. Guidelines for the validation and application of typing methods for use in bacterial epidemiology. Clin Microbiol Infect 13(Suppl 3):S1–S46. doi: 10.1111/j.1469-0691.2007.01732.x. [DOI] [PubMed] [Google Scholar]
- 23.Sheppard SK, Colles FM, McCarthy ND, Strachan NJ, Ogden ID, Forbes KJ, Dallas JF, Maiden MC. 2011. Niche segregation and genetic structure of Campylobacter jejuni populations from wild and agricultural host species. Mol Ecol 20:3484–3490. doi: 10.1111/j.1365-294X.2011.05179.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McCarthy ND, Colles FM, Dingle KE, Bagnall MC, Manning G, Maiden MC, Falush D. 2007. Host-associated genetic import in Campylobacter jejuni. Emerg Infect Dis 13:267–272. doi: 10.3201/eid1302.060620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wilson DJ, Gabriel E, Leatherbarrow AJ, Cheesbrough J, Gee S, Bolton E, Fox A, Fearnhead P, Hart CA, Diggle PJ. 2008. Tracing the source of campylobacteriosis. PLoS Genet 4:e1000203. doi: 10.1371/journal.pgen.1000203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sheppard SK, Dallas JF, Strachan NJ, MacRae M, McCarthy ND, Wilson DJ, Gormley FJ, Falush D, Ogden ID, Maiden MC, Forbes KJ. 2009. Campylobacter genotyping to determine the source of human infection. Clin Infect Dis 48:1072–1078. doi: 10.1086/597402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mullner P, Spencer SE, Wilson DJ, Jones G, Noble AD, Midwinter AC, Collins-Emerson JM, Carter P, Hathaway S, French NP. 2009. Assigning the source of human campylobacteriosis in New Zealand: a comparative genetic and epidemiological approach. Infect Genet Evol 9:1311–1319. doi: 10.1016/j.meegid.2009.09.003. [DOI] [PubMed] [Google Scholar]
- 28.Boysen L, Rosenquist H, Larsson JT, Nielsen EM, Sorensen G, Nordentoft S, Hald T. 2014. Source attribution of human campylobacteriosis in Denmark. Epidemiol Infect 142:1599–1608. doi: 10.1017/S0950268813002719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mossong J, Mughini-Gras L, Penny C, Devaux A, Olinger C, Losch S, Cauchie HM, van Pelt W, Ragimbeau C. 2016. Human campylobacteriosis in Luxembourg, 2010-2013: a case-control study combined with multilocus sequence typing for source attribution and risk factor analysis. Sci Rep 6:20939. doi: 10.1038/srep20939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sheppard SK, Didelot X, Méric G, Torralbo A, Jolley KA, Kelly DJ, Bentley SD, Maiden MC, Parkhill J, Falush D. 2013. Genome-wide association study identifies vitamin B5 biosynthesis as a host specificity factor in Campylobacter. Proc Natl Acad Sci U S A 110:11923–11927. doi: 10.1073/pnas.1305559110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gilbert MJ, Miller WG, Yee E, Zomer AL, van der Graaf-van Bloois L, Fitzgerald C, Forbes KJ, Méric G, Sheppard SK, Wagenaar JA, Duim B. 2016. Comparative genomics of Campylobacter fetus from reptiles and mammals reveals divergent evolution in host-associated lineages. Genome Biol Evol 8:2006–2019. doi: 10.1093/gbe/evw146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yahara K, Méric G, Taylor AJ, de Vries SP, Murray S, Pascoe B, Mageiros L, Torralbo A, Vidal A, Ridley AM, Komukai S, Wimalarathna H, Cody AJ, Colles FM, McCarthy N, Harris D, Bray JE, Jolley KA, Maiden MC, Bentley SD, Parkhill J, Bayliss CD, Grant A, Maskell D, Didelot X, Kelly DJ, Sheppard SK. 24 November 2016. Genome-wide association of functional traits linked with Campylobacter jejuni survival from farm to fork. Environ Microbiol doi: 10.1111/1462-2920.13628. [DOI] [PubMed] [Google Scholar]
- 33.Gripp E, Hlahla D, Didelot X, Kops F, Maurischat S, Tedin K, Alter T, Ellerbroek L, Schreiber K, Schomburg D, Janssen T, Bartholomaus P, Hofreuter D, Woltemate S, Uhr M, Brenneke B, Gruning P, Gerlach G, Wieler L, Suerbaum S, Josenhans C. 2011. Closely related Campylobacter jejuni strains from different sources reveal a generalist rather than a specialist lifestyle. BMC Genomics 12:584. doi: 10.1186/1471-2164-12-584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dearlove BL, Cody AJ, Pascoe B, Méric G, Wilson DJ, Sheppard SK. 2016. Rapid host switching in generalist Campylobacter strains erodes the signal for tracing human infections. ISME J 10:721–729. doi: 10.1038/ismej.2015.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sheppard SK, Jolley KA, Maiden MC. 2012. A gene-by-gene approach to bacterial population genomics: whole genome MLST of Campylobacter. Genes (Basel) 3:261–277. doi: 10.3390/genes3020261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jolley KA, Maiden MC. 2010. BIGSdb: scalable analysis of bacterial genome variation at the population level. BMC Bioinformatics 11:595. doi: 10.1186/1471-2105-11-595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sheppard SK, Colles F, Richardson J, Cody AJ, Elson R, Lawson A, Brick G, Meldrum R, Little CL, Owen RJ, Maiden MC, McCarthy ND. 2010. Host association of Campylobacter genotypes transcends geographic variation. Appl Environ Microbiol 76:5269–5277. doi: 10.1128/AEM.00124-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Griekspoor P, Colles FM, McCarthy ND, Hansbro PM, Ashhurst-Smith C, Olsen B, Hasselquist D, Maiden MC, Waldenstrom J. 2013. Marked host specificity and lack of phylogeographic population structure of Campylobacter jejuni in wild birds. Mol Ecol 22:1463–1472. doi: 10.1111/mec.12144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sheppard SK, Cheng L, Meric G, de Haan CP, Llarena AK, Marttinen P, Vidal A, Ridley A, Clifton-Hadley F, Connor TR, Strachan NJ, Forbes K, Colles FM, Jolley KA, Bentley SD, Maiden MC, Hanninen ML, Parkhill J, Hanage WP, Corander J. 2014. Cryptic ecology among host generalist Campylobacter jejuni in domestic animals. Mol Ecol 23:2442–2451. doi: 10.1111/mec.12742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Strachan NJ, Gormley FJ, Rotariu O, Ogden ID, Miller G, Dunn GM, Sheppard SK, Dallas JF, Reid TM, Howie H, Maiden MC, Forbes KJ. 2009. Attribution of Campylobacter infections in northeast Scotland to specific sources by use of multilocus sequence typing. J Infect Dis 199:1205–1208. doi: 10.1086/597417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Reid AN, Pandey R, Palyada K, Naikare H, Stintzi A. 2008. Identification of Campylobacter jejuni genes involved in the response to acidic pH and stomach transit. Appl Environ Microbiol 74:1583–1597. doi: 10.1128/AEM.01507-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Corcionivoschi N, Alvarez LA, Sharp TH, Strengert M, Alemka A, Mantell J, Verkade P, Knaus UG, Bourke B. 2012. Mucosal reactive oxygen species decrease virulence by disrupting Campylobacter jejuni phosphotyrosine signaling. Cell Host Microbe 12:47–59. doi: 10.1016/j.chom.2012.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zimmermann J, Saalbach I, Jahn D, Giersberg M, Haehnel S, Wedel J, Macek J, Zoufal K, Glunder G, Falkenburg D, Kipriyanov SM. 2009. Antibody expressing pea seeds as fodder for prevention of gastrointestinal parasitic infections in chickens. BMC Biotechnol 9:79. doi: 10.1186/1472-6750-9-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Umphrey JE, Staples CR. 1992. General anatomy of the ruminant digestive system. University of Florida Cooperative Extension Service, Institute of Food and Agriculture Sciences, EDIS. [Google Scholar]
- 45.Wu YL, Lee LH, Rollins DM, Ching WM. 1994. Heat shock- and alkaline pH-induced proteins of Campylobacter jejuni: characterization and immunological properties. Infect Immun 62:4256–4260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Xia Q, Muraoka WT, Shen Z, Sahin O, Wang H, Wu Z, Liu P, Zhang Q. 2013. Adaptive mechanisms of Campylobacter jejuni to erythromycin treatment. BMC Microbiol 13:133. doi: 10.1186/1471-2180-13-133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Miller WG, Englen MD, Kathariou S, Wesley IV, Wang G, Pittenger-Alley L, Siletz RM, Muraoka W, Fedorka-Cray PJ, Mandrell RE. 2006. Identification of host-associated alleles by multilocus sequence typing of Campylobacter coli strains from food animals. Microbiology 152:245–255. doi: 10.1099/mic.0.28348-0. [DOI] [PubMed] [Google Scholar]
- 48.Cody AJ, McCarthy ND, Bray JE, Wimalarathna HM, Colles FM, Jansen van Rensburg MJ, Dingle KE, Waldenstrom J, Maiden MC. 2015. Wild bird-associated Campylobacter jejuni isolates are a consistent source of human disease, in Oxfordshire, United Kingdom. Environ Microbiol Rep 7:782–788. doi: 10.1111/1758-2229.12314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kittl S, Heckel G, Korczak BM, Kuhnert P. 2013. Source attribution of human Campylobacter isolates by MLST and fla-typing and association of genotypes with quinolone resistance. PLoS One 8:e81796. doi: 10.1371/journal.pone.0081796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Magdelaine P, Spiess M, Valceschini E. 2008. Poultry meat consumption trends in Europe. World's Poultry Sci J 64:53–64. [Google Scholar]
- 51.Ghafir Y, China B, Dierick K, De Zutter L, Daube G. 2007. A seven-year survey of Campylobacter contamination in meat at different production stages in Belgium. Int J Food Microbiol 116:111–120. [DOI] [PubMed] [Google Scholar]
- 52.Noormohamed A, Fakhr MK. 2013. A higher prevalence rate of Campylobacter in retail beef livers compared to other beef and pork meat cuts. Int J Environ Res Public Health 10:2058–2068. doi: 10.3390/ijerph10052058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Korsak D, Mackiw E, Rozynek E, Zylowska M. 2015. Prevalence of Campylobacter spp. in retail chicken, turkey, pork, and beef meat in Poland between 2009 and 2013. J Food Prot 78:1024–1028. doi: 10.4315/0362-028X.JFP-14-353. [DOI] [PubMed] [Google Scholar]
- 54.Huang H, Brooks BW, Lowman R, Carrillo CD. 2015. Campylobacter species in animal, food, and environmental sources, and relevant testing programs in Canada. Can J Microbiol 61:701–721. doi: 10.1139/cjm-2014-0770. [DOI] [PubMed] [Google Scholar]
- 55.Hue O, Le Bouquin S, Laisney MJ, Allain V, Lalande F, Petetin I, Rouxel S, Quesne S, Gloaguen PY, Picherot M, Santolini J, Salvat G, Bougeard S, Chemaly M. 2010. Prevalence of and risk factors for Campylobacter spp. contamination of broiler chicken carcasses at the slaughterhouse. Food Microbiol 27:992–999. doi: 10.1016/j.fm.2010.06.004. [DOI] [PubMed] [Google Scholar]
- 56.Ministère de l'Agriculture et de la Pêche. 2010. Plan de surveillance 2009 de la contamination par Salmonella et Campylobacter des viandes fraîches de poulet au stade de la distribution. DGAL/SDSSA/N2009-8090. Ministère de l'Agriculture et de la Pêche, Paris, France. [Google Scholar]
- 57.Souillard R, Le Marechal C, Hollebecque F, Rouxel S, Barbe A, Houard E, Leon D, Poezevara T, Fach P, Woudstra C, Mahe F, Chemaly M, Le Bouquin S. 2015. Occurrence of C. botulinum in healthy cattle and their environment following poultry botulism outbreaks in mixed farms. Vet Microbiol 180:142–145. doi: 10.1016/j.vetmic.2015.07.032. [DOI] [PubMed] [Google Scholar]
- 58.Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chevreux B, Wetter T, Suhai S. Genome sequence assembly using trace signals and additional sequence information, p 45–56. In GCB '99 German Conference on Bioinformatics. http://www.bioinfo.de/isb/gcb99/talks/chevreux/. [Google Scholar]
- 60.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, Sirotkin AV, Vyahhi N, Tesler G, Alekseyev MA, Pevzner PA. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chikhi R, Medvedev P. 2014. Informed and automated k-mer size selection for genome assembly. Bioinformatics 30:31–37. doi: 10.1093/bioinformatics/btt310. [DOI] [PubMed] [Google Scholar]
- 62.Pascoe B, Meric G, Yahara K, Wimaralathna H, Murray S, Hitchings MD, Sproston EL, Carrillo CD, Taboada EN, Cooper KK, Huynh S, Cody AJ, Jolley KA, Maiden MC, McCarthy ND, Didelot X, Parker C, Sheppard SK. 2016. Local genes for local bacteria: evidence of allopatry in the genomes of transatlantic Campylobacter populations. PeerJ Preprints 4:e2638v2631. doi: 10.7287/peerj.preprints.2638v1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Maiden MC, Jansen van Rensburg MJ, Bray JE, Earle SG, Ford SA, Jolley KA, McCarthy ND. 2013. MLST revisited: the gene-by-gene approach to bacterial genomics. Nat Rev Microbiol 11:728–736. doi: 10.1038/nrmicro3093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Méric G, Yahara K, Mageiros L, Pascoe B, Maiden MC, Jolley KA, Sheppard SK. 2014. A reference pan-genome approach to comparative bacterial genomics: identification of novel epidemiological markers in pathogenic Campylobacter. PLoS One 9:e92798. doi: 10.1371/journal.pone.0092798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gundogdu O, Bentley SD, Holden MT, Parkhill J, Dorrell N, Wren BW. 2007. Re-annotation and re-analysis of the Campylobacter jejuni NCTC11168 genome sequence. BMC Genomics 8:162. doi: 10.1186/1471-2164-8-162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hofreuter D, Tsai J, Watson RO, Novik V, Altman B, Benitez M, Clark C, Perbost C, Jarvie T, Du L, Galan JE. 2006. Unique features of a highly pathogenic Campylobacter jejuni strain. Infect Immun 74:4694–4707. doi: 10.1128/IAI.00210-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Pearson BM, Gaskin DJ, Segers RP, Wells JM, Nuijten PJ, van Vliet AH. 2007. The complete genome sequence of Campylobacter jejuni strain 81116 (NCTC11828). J Bacteriol 189:8402–8403. doi: 10.1128/JB.01404-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Friis C, Wassenaar TM, Javed MA, Snipen L, Lagesen K, Hallin PF, Newell DG, Toszeghy M, Ridley A, Manning G, Ussery DW. 2010. Genomic characterization of Campylobacter jejuni strain M1. PLoS One 5:e12253. doi: 10.1371/journal.pone.0012253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Katoh K, Misawa K, Kuma K, Miyata T. 2002. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res 30:3059–3066. doi: 10.1093/nar/gkf436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Price MN, Dehal PS, Arkin AP. 2010. FastTree 2–approximately maximum-likelihood trees for large alignments. PLoS One 5:e9490. doi: 10.1371/journal.pone.0009490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol 30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kaas RS, Friis C, Ussery DW, Aarestrup FM. 2012. Estimating variation within the genes and inferring the phylogeny of 186 sequenced diverse Escherichia coli genomes. BMC Genomics 13:577. doi: 10.1186/1471-2164-13-577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Periwal V, Patowary A, Vellarikkal SK, Gupta A, Singh M, Mittal A, Jeyapaul S, Chauhan RK, Singh AV, Singh PK, Garg P, Katoch VM, Katoch K, Chauhan DS, Sivasubbu S, Scaria V. 2015. Comparative whole-genome analysis of clinical isolates reveals characteristic architecture of Mycobacterium tuberculosis pangenome. PLoS One 10:e0122979. doi: 10.1371/journal.pone.0122979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Pritchard JK, Stephens M, Donnelly P. 2000. Inference of population structure using multilocus genotype data. Genetics 155:945–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Pritchard JK, Wen X, Falush D. 2010. Documentation for structure software: version 2.3. University of Chicago, Chicago, IL. [Google Scholar]
- 76.Workman SN, Mathison GE, Lavoie MC. 2005. Pet dogs and chicken meat as reservoirs of Campylobacter spp. in Barbados. J Clin Microbiol 43:2642–2650. doi: 10.1128/JCM.43.6.2642-2650.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Mughini Gras L, Smid JH, Wagenaar JA, Koene MG, Havelaar AH, Friesema IH, French NP, Flemming C, Galson JD, Graziani C, Busani L, Van Pelt W. 2013. Increased risk for Campylobacter jejuni and C. coli infection of pet origin in dog owners and evidence for genetic association between strains causing infection in humans and their pets. Epidemiol Infect 141:2526–2535. doi: 10.1017/S0950268813000356. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.