ABSTRACT
Lactobacillus plantarum is a lactic acid bacterium that can degrade food tannins by the successive action of tannase and gallate decarboxylase enzymes. In the L. plantarum genome, the gene encoding the catalytic subunit of gallate decarboxylase (lpdC, or lp_2945) is only 6.5 kb distant from the gene encoding inducible tannase (L. plantarum tanB [tanBLp], or lp_2956). This genomic context suggests concomitant activity and regulation of both enzymatic activities. Reverse transcription analysis revealed that subunits B (lpdB, or lp_0271) and D (lpdD, or lp_0272) of the gallate decarboxylase are cotranscribed, whereas subunit C (lpdC, or lp_2945) is cotranscribed with a gene encoding a transport protein (gacP, or lp_2943). In contrast, the tannase gene is transcribed as a monocistronic mRNA. Investigation of knockout mutations of genes located in this chromosomal region indicated that only mutants of the gallate decarboxylase (subunits B and C), tannase, GacP transport protein, and TanR transcriptional regulator (lp_2942) genes exhibited altered tannin metabolism. The expression profile of genes involved in tannin metabolism was also analyzed in these mutants in the presence of methyl gallate and gallic acid. It is noteworthy that inactivation of tanR suppresses the induction of all genes overexpressed in the presence of methyl gallate and gallic acid. This transcriptional regulator was also induced in the presence of other phenolic compounds, such as kaempferol and myricetin. This study complements the catalog of L. plantarum expression profiles responsive to phenolic compounds, which enable this bacterium to adapt to a plant food environment.
IMPORTANCE Lactobacillus plantarum is a bacterial species frequently found in the fermentation of vegetables when tannins are present. L. plantarum strains degrade tannins to the less-toxic pyrogallol by the successive action of tannase and gallate decarboxylase enzymes. The genes encoding these enzymes are located close to each other in the chromosome, suggesting concomitant regulation. Proteins involved in tannin metabolism and regulation, such GacP (gallic acid permease) and TanR (tannin transcriptional regulator), were identified by differential gene expression in knockout mutants with mutations in genes from this region. This study provides insights into the highly coordinated mechanisms that enable L. plantarum to adapt to plant food fermentations.
KEYWORDS: fermentation, food phenolics, lactic acid bacteria, tannins
INTRODUCTION
Vegetable tannins are present in a variety of plants utilized as food and feed. Tannase (tannin acyl hydrolase, EC 3.1.1.20) catalyzes the hydrolysis of ester linkages in hydrolyzable tannins. The products of hydrolysis are glucose and gallic acid. Although gallic acid is widely distributed in nature, it is not a preferred substrate for bacterial growth. In fact, only bacteria of the genus Pseudomonas have been reported to utilize free gallic acid as a sole carbon and energy source under aerobic conditions (1). In addition, there are microorganisms that decarboxylate gallic acid nonoxidatively but do not possess appropriate mechanisms by which to further degrade the pyrogallol produced by this dead-end pathway (2).
Lactobacillus plantarum is the species most frequently encountered in the fermentation of plant materials where tannins are present, e.g., olives, must, and a variety of vegetable fermentation products. Apart from Streptococcus gallolyticus (3, 4), species of the L. plantarum group are the only lactic acid bacteria that possess tannase activity (5, 6). The biochemical pathway used by L. plantarum to hydrolyze tannic acid produces gallic acid and glucose, and the gallic acid formed is subsequently decarboxylated to pyrogallol (7). This metabolic transformation involves the successive action of tannase and gallate decarboxylase enzymes.
Iwamoto et al. (2008) (8) reported the identification of a gene (named tanBLp, tanLp1, or lp_2956 in L. plantarum WCFS1) encoding an intracellular tannase that was present in all the L. plantarum strains analyzed (9). Gallate decarboxylase was identified as a nonoxidative aromatic acid decarboxylase composed of three different subunits (subunits B, C, and D) (10). L. plantarum is the only bacterium in which the gene encoding subunit C (lpdC, or lp_2945) is separated in the chromosome from the genes encoding subunits B (lpdB, or lp_0271) and D (lpdD, or lp_0272) (2). A recent report demonstrated that LpdC is the only protein required to yield gallate decarboxylase activity, although LpdB is also essential for activity (2). Like tannase, which showed activity only on esters derived from gallic and protocatechuic acids, the purified LpdC protein showed decarboxylase activity only against both hydroxybenzoic acids (2).
The identification of the L. plantarum decarboxylase enzyme involved in tannin degradation completes the description of the first route of degradation of a phenolic compound in lactic acid bacteria. In this pathway, tannase and subunit C of gallate decarboxylase are encoded by genes only 6.5 kb distant from each other on the L. plantarum chromosome, suggesting common regulation. The present study reports the regulation of the tannase and decarboxylase gene clusters in L. plantarum WCFS1 in the presence of gallic acid and one of its esters, as determined by using a gene deletion strategy and differential gene expression.
RESULTS
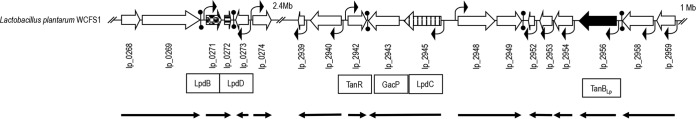
Genetic organization of the region containing the genes involved in tannin metabolism in L. plantarum WCFS1.
Two enzymatic activities had been reported to be involved in the metabolism of tannins in L. plantarum WCFS1: tannase and gallate decarboxylase. The tannase enzyme is encoded by the lp_2956 (tanBLp) gene, and gallate decarboxylase is encoded by lp_0271 (lpdB), lp_0272 (lpdD), and lp_2945 (lpdC). Remarkably, the genes encoding tannase and the catalytic subunit of gallate decarboxylase (lpdC) are only 6.5 kb distant from each other in the L. plantarum WCFS1 genome (Fig. 1). In view of this genomic context, the transcriptional profile of this chromosomal region was determined. Table S2 in the supplemental material shows the characteristics of the open reading frames (ORFs) annotated in this region.
FIG 1.
Genetic organization of the L. plantarum WCFS1 chromosomal region containing tannase- and gallate decarboxylase-encoding genes (GenBank accession no. NC_004567, positions 243093 to 252815 and 2617779 to 2635121). Arrows represent genes. Shaded arrows represent genes encoding gallate decarboxylase subunits. The gene coding for tannase protein is represented by a filled arrow. The locations of putative promoters and transcription terminators are indicated. The sizes and directions of the transcripts revealed by reverse transcription are also shown.
In the LpdB-LpdD chromosomal region, the lp_0269–lpdB and lpdD–lp_0273 intergenic regions rendered no reverse transcription-PCR (RT-PCR) product (see Fig. S1 in the supplemental material, lanes 4 and 8, respectively), indicating that these intergenic regions were not transcribed. In fact, potential terminators were found within each intergenic region (ΔG, −7.3 and −10.3 kcal/mol, respectively). In contrast, RT-PCR amplifications of the lp_0268–lp_0269 and lpdB–lpdD intergenic regions rendered DNA fragments of the expected sizes (Fig. S1, lanes 2 and 6, respectively), showing that lp_0268 and lp_2069, as well as the lpdB and lpdD genes, are cotranscribed (Fig. 1). Since quantitative PCR (qPCR) experiments were not performed, the amount of intergenic transcript detected in relation to the individual transcripts is unknown; therefore, monocistronic transcripts might also exist.
The RT-PCR approach, performed on the LpdC and tannase regions, yielded no RT-PCR products for the regions encompassing lp_2940–tanR, tanR–gacP, lpdC–lp_2948, lp_2949–lp_2952, lp_2953–lp_2954, lp_2954–tanBLp, and tanBLp–lp_2958 (Fig. S1 in the supplemental material, lanes 14, 16, 20, 24, 28, 30, and 32). Potential terminators were found between tanR and gacP, between lp_2949 and lp2952, and between tanBLp and lp_2958 (ΔG, −6.0, −8.2, and −11.4 kcal/mol, respectively). The catalytic LpdC subunit of gallate decarboxylase was cotranscribed with GacP (Fig. S1, lane 18), whereas the TanBLp tannase was transcribed alone (Fig. 1). RT-PCRs for the negative controls failed to yield any amplification product, while DNA template controls uniformly yielded the PCR product of the expected size. These results indicate that subunits B and D of gallate decarboxylase are cotranscribed (lpdB-lpdD), whereas subunit C (lpdC) is cotranscribed with gacP as a bicistronic mRNA from the PlpdC promoter. In contrast, tannase (tanBLp) is transcribed from its own promoter (PtanBLp) as a monocistronic mRNA (Fig. 1).
Tannin metabolism displayed by knockout mutants with mutations in the tannase-gallate decarboxylase region.
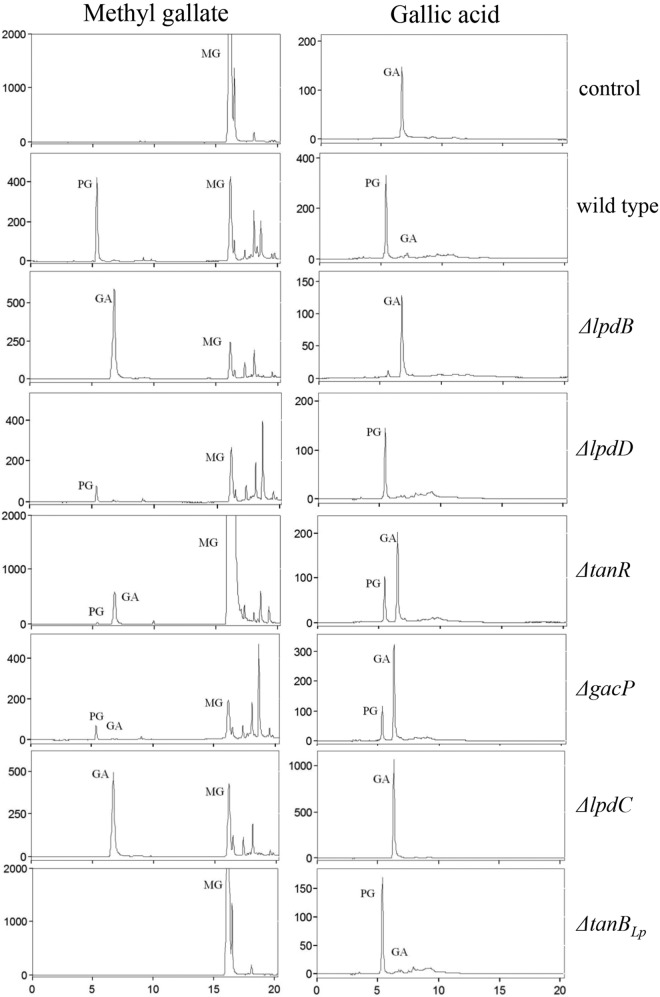
To investigate the involvement of these L. plantarum genes in tannin metabolism, L. plantarum WCFS1 knockout mutants were constructed by insertion-duplication mutagenesis. These knockout mutants were obtained for 10 genes (lpdB, lpdD, lp_0273, lp_0274, lp_2940, tanR, gacP, lpdC, lp_2953, and tanBLp). The wild-type and mutant strains were grown in MRS medium supplemented with 3 mM methyl gallate or gallic acid. After 10 days of incubation, samples were collected, and the phenolic compounds present in the supernatants were extracted and were analyzed by high-performance liquid chromatography (HPLC) (Fig. 2). With regard to the metabolism of methyl gallate and gallic acid, mutants with mutations in the lpdD, lp_0273, lp_0274, lp_2940, or lp_2953 gene exhibited the same behavior as the L. plantarum WCFS1 wild-type strain. However, this metabolic profile was not observed in the other mutants analyzed. As described previously, the lpdB and lpdC knockout mutants growing in a medium containing gallic acid were unable to transform this phenolic compound (2), whereas they accumulated gallic acid in the presence of methyl gallate due to inactivation of the gallate decarboxylase activity (Fig. 2). However, knockout of lpdD, encoding subunit D of gallate decarboxylase, did not affect decarboxylase activity (Fig. 2). As expected, methyl gallate metabolism was inhibited only in tannase knockout mutants (Fig. 2).
FIG 2.
Effects of disruption of several L. plantarum WCFS1 genes on methyl gallate and gallic acid metabolism. HPLC chromatograms of L. plantarum cultures incubated in 3 mM methyl gallate (MG) or gallic acid (GA) for 10 days at 30°C are shown for L. plantarum WCFS1 (wild type) and its lpdB, lpdD, tanR, gacP, lpdC, and tanBLp mutants. Results for an uninoculated medium are also shown (control). The MG, GA, and pyrogallol (PG) detected are indicated. Chromatograms were recorded at 280 nm.
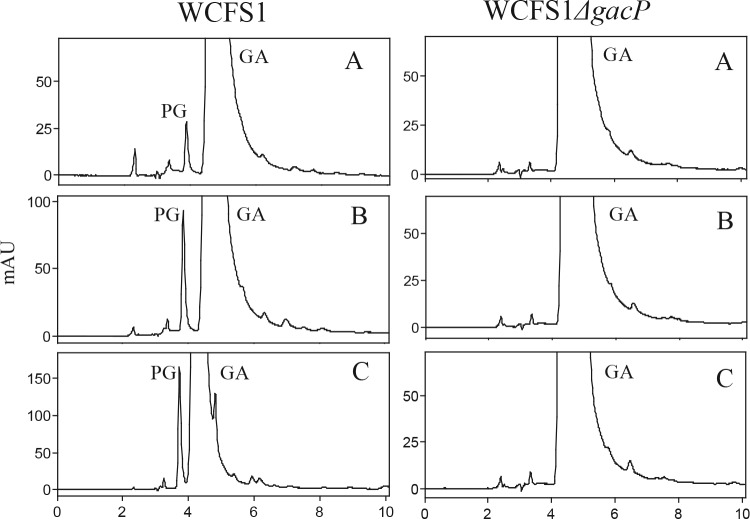
In addition to these mutants, the metabolism of methyl gallate and gallic acid displayed by the tanR and gacP knockout mutants was investigated. TanR is annotated as a LysR family transcriptional regulator, and its inactivation led to significant reductions in the transformation of methyl gallate (>80%) and gallic acid (nearly 50%) (Fig. 2). Inactivation of gacP (which codes for a protein annotated as a cation transporter) produced a significant reduction (nearly 33%) in the decarboxylation of gallic acid only (Fig. 2). The reduced, but clear, pyrogallol production in growing cells of the gacP mutant during a prolonged incubation (Fig. 2) was rather surprising, since we identified gacP as the sole gene coding for an ion transporter induced by gallic acid in a recent transcriptomic analysis (11). To confirm that the product of gacP was the sole gallate transporter, gallic acid metabolism was tracked in resting cells obtained from cultures of the gacP knockout mutant induced by gallic acid. It was observed that the mutant strain did not produce pyrogallol after a 60-min incubation. The failure of the gacP mutant to produce pyrogallol in these assays (Fig. 3) rules out the involvement of another transporter in gallic acid transport inside the cell.
FIG 3.
Gallic acid (GA) metabolism in resting cells of L. plantarum. Gallic acid (2 mM) was added to washed cells of L. plantarum WCFS1 or its gacP knockout mutant (WCFS1ΔgacP) grown in MRS medium supplemented with 3 mM gallic acid. The gallic acid and pyrogallol (PG) present in the supernatants after 15 (A), 30 (B), or 60 (C) min of incubation were detected by HPLC analysis. Chromatograms were recorded at 280 nm.
Transcriptional regulation of the genes involved in tannin metabolism by methyl gallate and gallic acid.
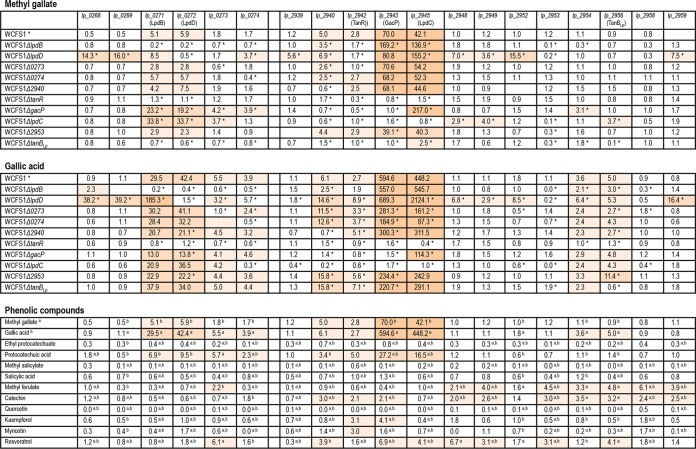
The effect of methyl gallate or gallic acid on the expression of genes located in the tannase and gallate decarboxylase regions of L. plantarum WCFS1 was analyzed by qPCR. Figure 4 shows the relative quantification (RQ) values of these genes normalized to the expression of the reference gene, ldh, and relative to the expression of the calibrator sample (in the absence of phenolic compounds). The RQ value of the calibrator sample is 1. Both methyl gallate and gallic acid greatly induced (>40-fold) the expression of the lpdC gene, as well as that of gacP, which is cotranscribed with it. Similarly, both compounds induced the expression of lpdB, lpdD, and tanR. In addition, gallic acid induced the expression of tanBLp and lp_2954 (coding for a hypothetical membrane protein).
FIG 4.
Relative transcriptional expression of genes putatively related to tannin metabolism in L. plantarum WCFS1 inoculated onto a medium containing methyl gallate, gallic acid, or one of several phenolic compounds. These compounds were added at 3 mM (for methyl gallate, gallic acid, ethyl protocatechuate, protocatechuic acid, methyl salicylate, salicylic acid, methyl ferulate, and catechin), 0.3 mM (quercetin), 10 μM (kaempferol and myricetin), and 1 mM (resveratrol) concentrations. Relative expression on methyl gallate and gallic acid was assayed for the L. plantarum WCFS1 wild-type strain and for 10 mutants with knockouts of genes putatively involved in tannin metabolism. RQ values were calculated by the comparative CT method using the 7500 Fast System relative quantification software with the L. plantarum ldh gene as the endogenous gene and growth in the absence of added phenolic compounds as the growth condition calibrator. The experiments were carried out in triplicate, and the mean values are shown. Uncolored boxes show RQ values lower than 2 (an RQ value of 2 indicates a 2-fold increase in gene expression over an RQ value of 1, which is assigned to expression in the absence of phenolic compounds). Light-, medium- and dark-pink boxes show 2- to 10-fold, 10- to 50-fold, and >50-fold increases in RQ values, respectively, over those in the absence of phenolic compounds. Asterisks indicate significant differences between the wild-type strain and the mutant strain in response to the presence of methyl gallate or gallic acid (P, ≤0.05 by Duncan's test). Superscript letters indicate significant differences between the presence of methyl gallate (a) or gallic acid (b) and the presence of other phenolic compounds (P, ≤0.05 by Duncan's test).
The expression pattern shown by the wild-type L. plantarum WCFS1 strain in response to both phenolic compounds was altered in the knockout mutants. Thus, the expression of lpdB and lpdD was not induced by methyl gallate or gallic acid in the lpdB knockout mutant. In contrast, inactivation of the catalytic C subunit gene (lpdC) increased the induction of subunits B and D. Although no definite function has been assigned to gallate decarboxylase subunit D (LpdD), its inactivation significantly induced the expression of adjacent genes not currently related to tannin metabolism.
Knockout mutations of the lp_0273, lp_0274, and lp_2940 genes did not give rise to relevant expression changes. In contrast, it should be noted that inactivation of tanR (lp_2942), encoding a LysR family transcriptional regulator, prevents the induction of all genes overexpressed in the presence of methyl gallate and gallic acid. In the gacP (lp_2943) knockout mutant, the lp_2940 and tanR genes were not induced; however, the presence of methyl gallate increased the expression of gallate decarboxylase. Finally, the induction of all genes triggered by methyl gallate in the wild-type strain was prevented in the tannase knockout mutant.
Expression profile of the L. plantarum WCFS1 region required for tannin metabolism in response to other phenolic compounds.
It has been demonstrated previously that tannase from L. plantarum is an esterase acting on gallate and protocatechuate esters (20). Similarly, L. plantarum gallate decarboxylase is also able to decarboxylate gallic and protocatechuic acids. In view of this, it is reasonable to hypothesize that the presence of protocatechuic acid or its derivative esters could induce a transcriptional response of tannase and decarboxylase enzymes. Figure 4 shows that protocatechuic acid induced the expression of genes encoding decarboxylase subunits, mainly lpdC (16-fold), as well as the gacP, lp_2940, and tanR genes. However, ethyl protocatechuate, which is a protocatechuate ester, failed to induce any of the genes tested.
In order to determine if other hydroxybenzoic acids or their derivative esters, which are not substrates for these enzymes, are able to induce the expression of these genes, salicylic acid and methyl salicylate were assayed. At a 3 mM concentration, neither of the two compounds produced a change in the relative expression of the genes analyzed.
Phenolic compounds with different structures were also assayed for their potential influence on the gene expression of this chromosomal region. Interestingly, methyl ferulate, a hydroxycinnamic ester, elicited low induction of several genes, including tanBLp. The presence of methyl ferulate was able to induce genes that were not induced previously in the L. plantarum WCFS1 wild-type strain (i.e., lp_2948, lp_2949, lp_2953, lp_2958, and lp_2959). Interestingly, Lp_2953 is 53.3% identical to a feruloyl esterase from Lactobacillus johnsonii (12) (Table S2 in the supplemental material). The expression of methyl ferulate-induced genes was also increased in the presence of the flavonol catechin. The different flavonols assayed exhibited different expression patterns. None of the genes analyzed changed its expression level in the presence of quercetin. However, the transcriptional regulator TanR was induced by catechin, kaempferol, and myricetin. Kaempferol also induced, though slightly, the GacP transport protein. Resveratrol, a stilbene, is the only compound assayed that was able to increase the expression of the three genes gacP, lpdC, and tanBLp.
DISCUSSION
L. plantarum is the only bacterium in which the genes encoding the different gallate decarboxylase subunits are separated in the chromosome. Transcriptional studies of these decarboxylase genes detected RNA spanning the intergenic regions between lpdB and lpdD, as well as between lpdC and gacP, confirming that gallate decarboxylase-encoding genes are transcribed, at least, in two independent polycistronic mRNAs. This transcriptional pattern differs from those of other bacterial nonoxidative aromatic acid decarboxylases, which are encoded by three contiguous genes cotranscribed as an operon (10, 13, 14).
Although there are genes in the tannase-gallate decarboxylase region that are transcribed independently of the genes dedicated to tannin metabolism, the possibility that these genes play a role in tannin degradation could not be discarded. Therefore, 10 genes were knocked out by an insertion-duplication strategy, and the tannin metabolism of the corresponding mutants was analyzed. Aside from the tannase, LpdC, and LpdB mutants, only the TanR and GacP mutants showed a metabolism different from that of the wild-type strain. Inactivation of TanR led to a general change in the expression pattern of the genes studied, preventing the induction of all genes overexpressed in the presence of methyl gallate and gallic acid. In contrast, the reduced decarboxylation of gallic acid observed in the ΔgacP mutant can be ascribed to inactivation of gallic acid transport via the GacP transporter rather than to general expression changes. These gene expression and metabolic profiles, together with the failure of resting cells of the ΔgacP mutant to metabolize gallic acid into pyrogallol (Fig. 3), show that GacP is the antiporter protein involved in the uptake of gallic acid by the cell and the excretion of pyrogallol to the culture medium, as proposed recently (11). It follows that the reduced gallic acid metabolism observed in growing ΔgacP cells could be due to the diffusion of a fraction of the gallic acid in its uncharged form through the membrane and its subsequent conversion to pyrogallol by the active gallate decarboxylase.
The most striking expression changes were observed when the tanR regulator was inactivated. Inactivation of TanR, annotated as a transcriptional regulator (LysR family), resulted in the inhibition of all the genes induced in response to methyl gallate or gallic acid. The mechanisms through which LysR-type transcriptional regulators modulate gene expression are diverse. KaeR, a LysR-type transcriptional regulator with the ability to mediate transcriptional activation in response to flavonoid exposure, has been identified in Lactobacillus brevis (15). KaeR and the three genes under its control were upregulated by the addition of kaempferol to the growth medium. Interestingly, the L. plantarum WCFS1 gallate decarboxylase subunits (LpdB, LpdC, and LpdD) showed high identity to the proteins encoded by the three genes controlled by KaeR, which are clustered in L. brevis. Although the corresponding L. plantarum genes are atypically organized (separated) in the genome, a mechanism similar to that for L. brevis KaeR could be inferred for L. plantarum TanR, considering that they are 41% identical, and both are LysR transcriptional activators. The transcription activation mediated by KaeR is dependent on the presence of KaeR and the inducer molecule (the flavonoid kaempferol) (15). In the case of TanR, gallic acid, as well as methyl gallate, seems to act as an inducer.
Different aromatic molecules present in the environment show structural resemblance to the substrates or ligands of transcriptional regulators. In L. brevis, the flavonols kaempferol and myricetin have the potential to induce the expression of aromatic decarboxylase genes (15). In Bacillus subtilis, salicylic acid induced the bsdBCD genes, encoding a nonoxidative aromatic acid decarboxylase (16). However, in this study, only protocatechuic acid, a substrate of gallate decarboxylase, was able to induce decarboxylase genes. The flavonols tested (kaempferol, quercetin, and myricetin) did not induce decarboxylase or tannase genes at the concentration assayed. Kaempferol, the flavonol catechin, and resveratrol induce gacP, which codes for the gallic acid transporter. However, the induction observed was at much lower levels than the upregulation induced by gallic acid (11). Overall, these results suggest that while the main function of this transporter is to act as a gallic acid/pyrogallol exchanger, it could also be involved in the transport of other phenolic compounds or their metabolites, a possibility that deserves further investigation.
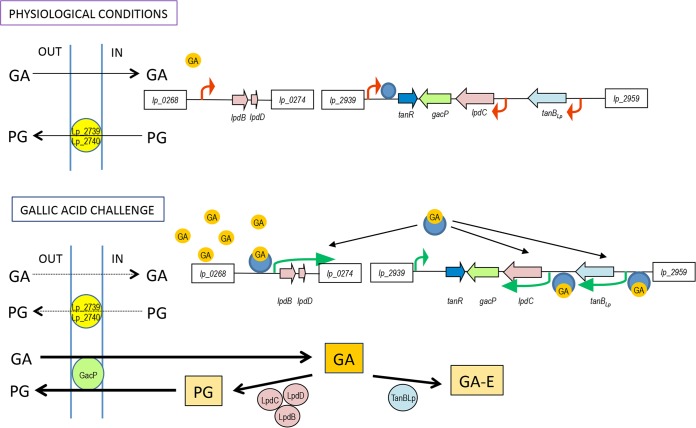
Taking all the data together, we propose a model for tannase and gallate decarboxylase regulation in L. plantarum WCFS1 (Fig. 5) similar to the model proposed previously for the hydroxyarylic acid decarboxylase operon in Enterobacteriaceae (14). In this model, TanR is constantly present in the cell in a definite small amount under physiological conditions. It could be assumed that the minimal amount of TanR is sufficient to repress its own expression by binding, possibly downstream of its transcription start site. The presence of gallic acid at very low concentrations under physiological conditions inside the cell is insufficient to induce the gallate decarboxylase and tannase operons. This low concentration of gallic acid could be due to the diffusion of its uncharged form from the external environment to the cytoplasm. The small amount of pyrogallol formed could be extruded by the action of unspecific transporters, such as Lp_2739 and Lp_2740; these ABC transporters were downregulated in the presence of high gallic acid concentrations and could be involved in the traffic of pyrogallol across the membrane (11). On external exposure to high gallic acid concentrations, the level of gallic acid increases inside the cell by the diffusion of its uncharged form. TanR is positively autoregulated upon interaction with the inducer gallic acid. The TanR gene is transcribed when the TanR protein is dissociated from its promoter. TanR protein could bind with DNA upstream of the promoter of target genes, probably by inducing a conformational change, and this binding results in the progression of the transcription of the gallate decarboxylase and tannase operons.
FIG 5.
Schematic model proposed for the regulation of the tannase and gallate decarboxylase operons in L. plantarum WCFS1. Genes encoding tannase (tanBLp), gallate decarboxylase (lpdBCD), gallate transport protein (gacP), and the tannin transcriptional regulator (tanR) are represented by arrows. The unspecific ABC transporters Lp_2739 and Lp_2740 are shown in the membrane. Dark blue circles represent TanR not bound to gallic acid. Gallic acid is represented by orange circles. The thin green and red arrows represent the progression of transcription and the blockage of the tannase and gallate decarboxylase operons, respectively. GA-E, gallic acid esters; GA, gallic acid; PG, pyrogallol.
Environmental cues are sensed by bacteria directly or indirectly through proteins, mainly by regulators, enabling them to adapt and survive under particular conditions. Different regulators act in highly coordinated ways that enable the bacteria to better fit into a plant food environment. Ligand-responsive transcriptional regulators, such as TanR, could be modulated by small-molecule ligands, usually of an aromatic nature, such as gallic acid or methyl gallate, for better adaptation. The results described here complement the catalog of L. plantarum expression profiles in the presence of gallic acid and can be used for deepening our knowledge of the mechanisms used by L. plantarum to overcome the toxicity of phenolic compounds in food.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
L. plantarum WCFS1, used throughout this study, was kindly provided by Michiel Kleerebezem (NIZO Food Research, The Netherlands). This strain is a colony isolate of L. plantarum NCIMB 8826, which was isolated from human saliva. Lactic acid bacteria were routinely grown on MRS broth. Where appropriate, erythromycin was added to the culture medium at 10 μg/ml and lincomycin at 100 μg/ml.
Tannin degradation assays.
Tannase and gallate decarboxylase activities on L. plantarum cultures were assayed by growing the strains in RPM medium, described previously (17), supplemented with 3 mM methyl gallate or gallic acid, in darkness for 10 days. The phenolic compounds present in the supernatant were extracted by ethyl acetate and were analyzed by HPLC with diode array detection (DAD) as described previously (2). Where indicated, these activities were also assayed in a system using resting cells according to a protocol described previously (11).
Construction of L. plantarum WCFS1 knockout mutant strains.
To investigate the participation in tannin degradation of genes located in the vicinity of the genes encoding tannase and gallate decarboxylase, insertion-duplication mutagenesis was employed. Internal fragments from genes located in the gallate decarboxylase (subunits B and D) chromosomal region (lp_0268 to lp_0274) or in the tannase and gallate decarboxylase (subunit C) region (lp_2939 to lp_2960) were cloned into the suicide vector pUCE191 (18). When pUCE191 and its derivatives were used as donor DNA, L. plantarum transformants were selected by plating with 100 μg/ml lincomycin and 10 μg/ml erythromycin, and Escherichia coli transformants by plating with ampicillin at 100 μg/ml. pUCE191 derivative plasmids, constructed in E. coli, were used to transform L. plantarum WCFS1 competent cells by electroporation according to the method of Aukrust and Blom (1992) (19). Knockout mutants were selected by plating on MRS plates containing erythromycin and lincomycin. The correct insertion of the donor pUCE191 derivative plasmid into the L. plantarum WCFS1 chromosome was checked by PCR analysis using primers flanking the target region combined with vector-specific primers (see Table S1 in the supplemental material).
RT-PCR.
L. plantarum WCFS1 cells were grown in MRS medium in the presence of 3 mM methyl gallate. Fifty milliliters of culture was collected in triplicate at the exponential phase of growth. Total RNA was extracted from cultures as described previously (9). The RNA extracted from the three different cultures was treated with DNase and was retrotranscribed using the High Capacity cDNA reverse transcription kit (Applied Biosystems) according to the manufacturer's instructions. The cDNAs from biological replicates were mixed, and the mixture was used for the RT-PCR experiments. The lp_0268–lp_0269, lp_0269–lpdB, lpdB–lpdD, lpdD–lp_0273, lp_0273–lp_0274, lp_2939–lp_2940, lp_2940–tanR, lp_2942–gacP, gacP–lpdC, lpdC–lp_2948, lp_2948–lp_2949, lp_2949–lp_2952, lp_2952–lp_2953, lp_2953–lp_2954, lp_2954–tanBLp, tanBLp–lp_2958, and lp_2958–lp_2959 intergenic regions were analyzed by PCR amplification using cDNA as a template with specific pairs of primers (Table S1 in the supplemental material). PCR amplifications were performed using 1 μl of the cDNA template and 0.5 μM (each) corresponding primers. Amplifications were performed for 35 cycles (94°C for 30 s, 55°C for 45 s, and 72°C for 1 min); the resulting amplicons were separated on 2% agarose gels. As a control, PCR of DNase-treated RNA was performed with the same primers to check for DNA contamination.
qPCR assays.
Quantitative gene expression was analyzed in an ABI Prism 7500 Fast Real-Time PCR system by using cDNAs obtained from three biological replicates of each sample. Specific primer pairs were designed with the Primer Express program, version 3.0, to amplify gene internal regions and the endogenous control gene (ldh) (Table S1 in the supplemental material). The SYBR green method was used, and each assay was performed in triplicate using the SYBR green real-time PCR master mix (Applied Biosystems). Amplification was initiated at 95°C for 10 min, followed by 40 cycles of 95°C for 15 s and annealing-extension at 60°C for 1 min. Control PCRs were included to confirm the absence of primer dimer formation and to verify that there was no DNA contamination. All quantitative PCR (qPCR) assays amplified a single product, as determined by melting-curve analysis and by electrophoresis. A standard curve was plotted with cycle threshold (CT) values obtained from the amplification of known quantities of cDNA and was used to determine the efficiency (E) as follows: E = 10−1/slope. In order to measure L. plantarum gene expression, amplification of the endogenous control gene was performed simultaneously, and its relative expression was compared with that of the target gene.
Relative quantification (RQ) expression levels were calculated with the Applied Biosystems 7500 Fast System relative quantification software using the L. plantarum ldh gene as the endogenous gene and growth in the absence of phenolic compounds as the growth condition calibrator. Results were analyzed using the comparative CT method [also called the double delta CT (2−ΔΔCT) method]. Amplifications were performed in triplicate from biological triplicates, and the main RQ value (eliminating the lowest and highest values) and standard deviation were calculated.
Statistical analyses.
RQ values were subjected to one-way analysis of variance (ANOVA) by Statgraphics Centurion XVI software, version 16.1.17 (Statistical Graphics Corporation, Rockville, MD). Differences between samples were compared by using Duncan's multiple-range tests at a probability level (P) of ≤0.05.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to M. V. Santamaría and J. M. Barcenilla for their assistance.
N. Jiménez is a recipient of an FPI Fellowship from MINECO. This work was supported by grants AGL2014-52911-R, AGL2011-22745 (MINECO), and RM2008-00002 (Instituto Nacional de Investigación Agraria y Alimentaria).
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.03387-16.
REFERENCES
- 1.Nogales J, Canales A, Jiménez-Barbero J, Serra B, Pingarrón JM, García JL, Díaz E. 2011. Unravelling the gallic acid degradation pathway in bacteria: the gal cluster from Pseudomonas putida. Mol Microbiol 79:359–374. doi: 10.1111/j.1365-2958.2010.07448.x. [DOI] [PubMed] [Google Scholar]
- 2.Jiménez N, Curiel JA, Reverón I, de las Rivas B, Muñoz R. 2013. Uncovering the Lactobacillus plantarum WCFS1 gallate decarboxylase involved in tannin degradation. Appl Environ Microbiol 79:4253–4263. doi: 10.1128/AEM.00840-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jiménez N, Reverón I, Esteban-Torres M, López de Felipe F, de las Rivas B, Muñoz R. 2014. Genetic and biochemical approaches towards unravelling the degradation of gallotannins by Streptococcus gallolyticus. Microb Cell Fact 13:154. doi: 10.1186/s12934-014-0154-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sly LI, Cahill M, Osawa R, Fujisawa T. 1997. The tannin-degrading species Streptococcus gallolyticus and Streptococcus caprinus are subjective synonyms. Int J Syst Bacteriol 47:893–894. doi: 10.1099/00207713-47-3-893. [DOI] [PubMed] [Google Scholar]
- 5.Nishitani Y, Sasaki E, Fujisawa T, Osawa R. 2004. Genotypic analysis of lactobacilli with a range of tannase activities isolated from human feces and fermented foods. Syst Appl Microbiol 27:109–117. doi: 10.1078/0723-2020-00262. [DOI] [PubMed] [Google Scholar]
- 6.Vaquero I, Marcobal A, Muñoz R. 2004. Tannase activity by lactic acid bacteria isolated from grape must and wine. Int J Food Microbiol 96:199–204. doi: 10.1016/j.ijfoodmicro.2004.04.004. [DOI] [PubMed] [Google Scholar]
- 7.Rodríguez H, de las Rivas B, Gómez-Cordovés MC, Muñoz R. 2008. Degradation of tannic acid by cell-free extracts of Lactobacillus plantarum. Food Chem 107:664–670. doi: 10.1016/j.foodchem.2007.08.063. [DOI] [PubMed] [Google Scholar]
- 8.Iwamoto K, Tsuruta H, Nishitaini Y, Osawa R. 2008. Identification and cloning of a gene encoding tannase (tannin acylhydrolase) from Lactobacillus plantarum ATCC 14917T. Syst Appl Microbiol 31:269–277. doi: 10.1016/j.syapm.2008.05.004. [DOI] [PubMed] [Google Scholar]
- 9.Jiménez N, Esteban-Torres M, Mancheño JM, de las Rivas B, Muñoz R. 2014. Tannin degradation by a novel tannase enzyme present in some Lactobacillus plantarum strains. Appl Environ Microbiol 80:2991–2997. doi: 10.1128/AEM.00324-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lupa B, Lyon D, Gibbs MD, Reeves RA, Wiegel J. 2005. Distribution of genes encoding the microbial non-oxidative reversible hydroxyarylic acid decarboxylases/phenol carboxylases. Genomics 86:342–351. doi: 10.1016/j.ygeno.2005.05.002. [DOI] [PubMed] [Google Scholar]
- 11.Reverón I, de las Rivas B, Matesanz R, Muñoz R, López de Felipe F. 2015. Molecular adaptation of Lactobacillus plantarum WCFS1 to gallic acid revealed by genome-scale transcriptomic signature and physiological analysis. Microb Cell Fact 14:160. doi: 10.1186/s12934-015-0345-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lai KK, Lorca GL, González CF. 2009. Biochemical properties of two cinnamoyl esterases purified from a Lactobacillus johnsonii strain isolated from stool samples of diabetes-resistant rats. Appl Environ Microbiol 75:5018–5029. doi: 10.1128/AEM.02837-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lupa B, Lyon D, Shaw LN, Sieprawska-Lupa M, Wiegel J. 2008. Properties of the reversible nonoxidative vanillate/4-hydroxybenzoate decarboxylase from Bacillus subtilis. Can J Microbiol 54:75–81. doi: 10.1139/W07-113. [DOI] [PubMed] [Google Scholar]
- 14.Roy A, Ranjan A. 2016. HosA, a MarR family transcriptional regulator, represses nonoxidative hydroxyarylic acid decarboxylase operon and is modulated by 4-hydroxybenzoic acid. Biochemistry 55:1120–1134. doi: 10.1021/acs.biochem.5b01163. [DOI] [PubMed] [Google Scholar]
- 15.Pande SG, Pagliani FA, Gardner CL, Wrench A, Narvel R, González CF, Lorca GL. 2011. Lactobacillus brevis responds to flavonoids through KaeR, a LysR-type of transcriptional regulator. Mol Microbiol 81:1623–1639. doi: 10.1111/j.1365-2958.2011.07796.x. [DOI] [PubMed] [Google Scholar]
- 16.Duy NV, Mäder U, Tran NP, Cavin J-F, Tam LT, Alberecht D, Hecker M, Anterlmann H. 2007. The proteome and transcriptome analysis of Bacillus subtilis in response to salicylic acid. Proteomics 7:698–710. doi: 10.1002/pmic.200600706. [DOI] [PubMed] [Google Scholar]
- 17.Rozès N, Peres C. 1998. Effects of phenolic compounds on the growth and the fatty acid composition of Lactobacillus plantarum. Appl Microbiol Biotechnol 49:108–111. doi: 10.1007/s002530051145. [DOI] [Google Scholar]
- 18.Arrecubieta C, García E, López R. 1995. Sequence and transcriptional analysis of a DNA region involved in the production of capsular polysaccharide in Streptococcus pneumoniae type 3. Gene 167:1–7. doi: 10.1016/0378-1119(95)00657-5. [DOI] [PubMed] [Google Scholar]
- 19.Aukrust T, Blom H. 1992. Transformation of Lactobacillus strains used in meat and vegetable fermentations. Food Res Int 25:253–261. doi: 10.1016/0963-9969(92)90121-K. [DOI] [Google Scholar]
- 20.Curiel JA, Rodríguez H, Acebrón I, Mancheño JM, de las Rivas B, Muñoz R. 2009. Production and physicochemical properties of recombinant Lactobacillus plantarum tannase. J Agric Food Chem 57:6224–6230. doi: 10.1021/jf901045s. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.