Abstract
Although our understanding of the genetics and pathology of congenital lung diseases such as surfactant protein deficiency, cystic fibrosis, and alpha-1 antitrypsin deficiency is extensive, treatment options are lacking. Because the lung is a barrier organ in direct communication with the external environment, targeted delivery of gene corrective technologies to the respiratory system via intratracheal or intranasal routes is an attractive option for therapy. CRISPR/Cas9 gene-editing technology is a promising approach to repairing or inactivating disease-causing mutations. Recent reports have provided proof of concept by using CRISPR/Cas9 to successfully repair or inactivate mutations in animal models of monogenic human diseases. Potential pulmonary applications of CRISPR/Cas9 gene editing include gene correction of monogenic diseases in pre- or postnatal lungs and ex vivo gene editing of patient-specific airway stem cells followed by autologous cell transplant. Strategies to enhance gene-editing efficiency and eliminate off-target effects by targeting pulmonary stem/progenitor cells and the assessment of short-term and long-term effects of gene editing are important considerations as the field advances. If methods continue to advance rapidly, CRISPR/Cas9-mediated gene editing may provide a novel opportunity to correct monogenic diseases of the respiratory system.
Keywords: genetic lung disease, CRISPR/Cas9, gene editing, lung progenitor cells
Approximately 22% of all pediatric hospital admissions are for respiratory disorders, and congenital causes of respiratory diseases are frequently lethal despite significant advances in clinical care and a better understanding of pathogenic mechanisms (1–3). Some of the most common congenital lung diseases include surfactant protein (SP) deficiency disorders, cystic fibrosis (CF) and alpha-1 antitrypsin (AAT) deficiency. Whereas CF and AAT deficiency present beyond the neonatal period and worsen into childhood or adulthood, leading to shortened life expectancy, SP deficiency presents most often immediately after birth, with severe respiratory failure (4–6). Treatment options are limited and include supportive and compassionate care or lung transplant (7, 8). Together, these genetic pulmonary diseases lead to significant lifelong morbidity and mortality, and new alternate treatment strategies are needed.
Because many of these disorders are caused by monogenic mutations, they are ideal candidates for gene-editing technologies. Furthermore, the lung is a barrier organ and is exposed to the external environment, making it an attractive target for treatment delivery systems via intratracheal or nasal routes to selectively target respiratory epithelium and other pulmonary cell lineages (9–11). Recent advances in gene-editing technology using CRISPR/Cas9 offer an exciting new approach to repairing or inactivating disease-causing gene mutations (12–14). The application of this technology is growing rapidly, and several groups have recently demonstrated successful repair of mutations in tissue and animal models of monogenic diseases such as CF, Duchenne muscular dystrophy (DMD), and ornithine transcarbamylase deficiency (OTC) and cataracts (15–20).
The purpose of this perspective is to provide clinicians and investigators with insight into the potential application of CRISPR/Cas9 in the treatment of monogenic lung diseases at a time when this technology is evolving rapidly. We will discuss the recent advances in and advantages of CRISPR/Cas9 as a gene-editing tool and the potential application of CRISPR/Cas9 gene editing in SP deficiency, CF, and AAT deficiency; elaborate on the knowledge gaps and limitations; and propose strategies to address these barriers for successful translation into human clinical trials.
Genetic Basis of Monogenic Lung Diseases
Surfactant Protein Deficiency
Inherited SP deficiency is caused by genetic mutations in several genes, including SFTPB, SFTPC, and ATP-binding cassette protein member 3 (ABCA3), important for surfactant metabolism (21). Surfactant protein B (SFTPB) and surfactant protein C (SFTPC) are hydrophobic proteins that maintain adsorption of surfactant lipids at the alveolar air–liquid interface and are necessary for lowering surface tension. ABCA3 is a transmembrane transporter protein which is located on lamellar bodies. ABCA3 is essential for lipid transport. The lamellar bodies are specialized organelles involved in surfactant packaging and recycling. SFTPB is encoded by a single SFTPB gene on chromosome 2 that is composed of 11 coding exons and is highly expressed in alveolar type 2 (AT2) cells. All known mutations of the SFTPB gene are inherited in an autosomal-recessive manner, and no known spontaneous mutations have been reported. Of the 30 loss-of-function recessive mutations reported, a frameshift mutation in codon 121 is the most common (21–23). SFTPC deficiency is caused by autosomal gain-of-function mutations in the SFTPC gene located on chromosome 8, of which the I73T missense mutation is the most common (24, 25). Sporadic diseases caused by spontaneous mutations in SFTPC have also been reported. The human ABCA3 gene, consisting of 30 coding exons, is located on chromosome 16. More than 70 recessive loss-of-function mutations in ABCA3 have been identified so far, and it is the most common genetic cause of SP deficiency (26, 27).
CF
CF is an autosomal-recessive disorder of the cystic fibrosis transmembrane conductance regulator (CFTR) gene located on chromosome 7. It results in abnormal conductance of chloride ion channel activity in airway ciliated epithelial cells and leads to the accumulation of excess thickened mucus and impaired mucociliary clearance, predisposing to recurrent infections, respiratory failure, and early death (28). Nearly 2,000 mutations of the CFTR gene have been identified thus far. The ∆F508 mutation caused by deletion of phenyl-alanine at position 508 in exon 11 is present in 90% of patients with CF. It results in the production of misfolded CFTR protein that is degraded before reaching its site of action on the cell membrane. The different CF mutations are grouped into five classes on the basis of the pathogenic mechanism. Clinical features and severity of illness are variable and depend on the genetic variant (28, 29). Through universal newborn screening programs, the majority of patients with CF are identified early in life and recent development of novel therapies has significantly extended the life of these patients (3, 30, 31).
AAT Deficiency
AAT is a serine protease inhibitor that is secreted from the liver and inhibits neutrophil elastase, proteases, and defensins in the lung. AAT deficiency results from synthesis and accumulation of abnormal AAT and leads to unopposed release of neutrophil elastase in the lung, leading to early-onset emphysema and shortened life expectancy. Accumulation of misfolded AAT also causes hepatocyte injury, which further exacerbates AAT deficiency. AAT deficiency is caused by single-base substitution mutations in the A1AT gene on chromosome 14, resulting in single amino acid modifications. The “Z” mutation caused by G342L on exon 5 and the “S” mutation caused by G264V on exon 3 are the most common AAT deficiency mutations (4, 32).
Need for Alternate Therapy
Genetic lung diseases are fatal and they represent a significant health care burden. Despite extensive understanding of the genetics, definitive treatment options are lacking. Because many genetic lung diseases are monogenic in nature, they are promising targets for treatment using gene-editing technologies. Moreover, pulmonary genetic disorders are uniquely amenable to targeted gene-editing therapy because the lung is a barrier organ and is in direct contact with the external environment. In addition, in the case of SFTPB and SFTPC deficiency syndromes, the proteins are expressed primarily in the lungs, and extrapulmonary organs are unaffected by the underlying mutations, leaving open the possibility that lung-specific therapy could lead to a clinical cure.
CRISPR/Cas9 Overview
CRISPR/Cas9 technology is an adaptation of the bacterial defense mechanism against foreign DNA. Of the different types of CRISPR systems identified so far, type 2 CRISPR is of most interest. The bacterial CRISPR locus consists of short palindromic repeats within the bacterial genome, into which the foreign DNA is incorporated in multiple small pieces. Upon reexposure to foreign DNA, the CRISPR locus is transcribed into small RNAs that guide the Cas9 endonuclease to the specific site in the foreign DNA on the basis of DNA-RNA sequence complementarity and generates a double-stranded break (DSB), helping protect the host bacterium from viral or other foreign DNA complexes (13, 33).
The CRISPR/Cas9 system generally consists of two components. The first component is the Cas9 endonuclease, which cleaves the DNA to create specific DSB. Cas9 consists of two domains, the N-terminal RuvC-like nuclease domain and the HNH-like nuclease domain; each binds to the opposite sides of the DNA at the desired location to create a DSB. The second component is the guide RNA (gRNA), which is composed of a crRNA that is complementary to the target DNA sequence in the gene of interest and an RNA sequence, called the transCRISPR RNA or transactivating crRNA, that forms the scaffold for the system. In addition to the gRNA, which targets the Cas9 enzyme to a specific DNA sequence, another important component required for the Cas9 to bind to the desired DNA site is the protospacer adjacent motif (PAM), which is a short DNA stretch of three to five nucleotides. The PAM sequence must be present immediately downstream (3′) of the target sequence for Cas9 to bind to and cleave the target sequence. The PAM sequences vary depending on the bacterial species or variants from which Cas9 is derived; in theory, this allows expansion of the spectrum of genomic loci that can be targeted. As a consequence, any gene sequence that has a PAM sequence immediately downstream can be genetically modified by CRISPR/Cas9.
After the formation of a Crispr-mediated DSB, there are two mechanisms by which the cleaved DNA can be repaired. The first mechanism is repair by nonhomologous end joining (NHEJ), which is mediated by ligation of the exposed fragmented region of the DSB by an error-prone process that results in small insertions or deletions (indels). Using this method, gene knockouts can be created by using a single gRNA targeted against the desired gene locus, followed by formation of indels by NHEJ, causing frameshift mutations. Alternatively, two gRNAs targeting different sites can be used to delete a larger section of the gene. In contrast, the second type of repair mechanism uses homologous DNA repair (HDR) through the use of homology regions between the fragmented ends of a DSB and a donor DNA template to precisely repair the gene of interest. It is this capability that may be of most use in correcting respiratory monogenic diseases such as CF or SP deficiencies. HDR can also be used to insert a different DNA sequence to encode a reporter gene of interest, such as EGFP (Figure 1).
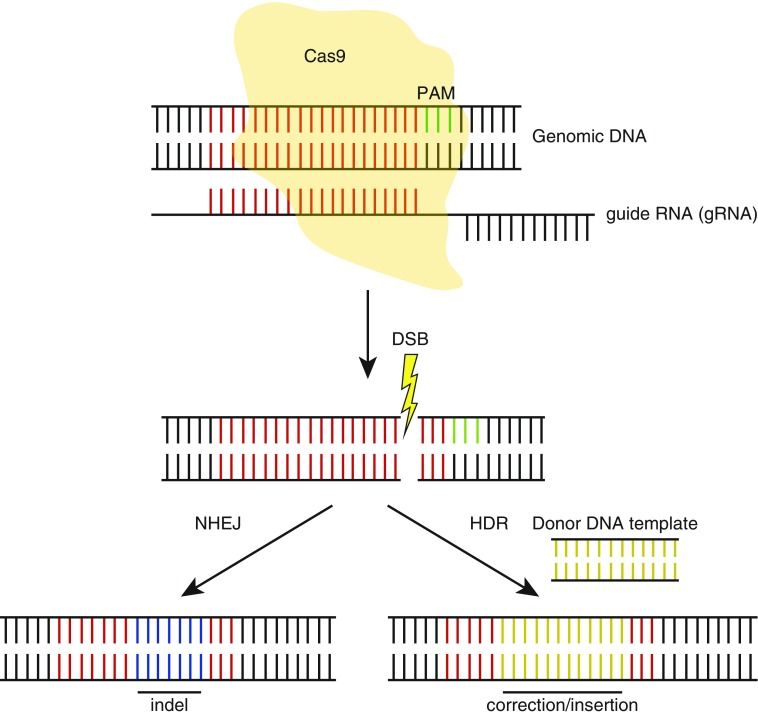
Figure 1.
Schematic representation of CRISPR/Cas9 gene editing. Guide RNA (gRNA) guides Cas9 to the target sequence (red) to create a double-stranded break (DSB) adjacent to the protospacer adjacent motif (PAM) (green). DNA repair by nonhomologous end joining (NHEJ) causes indel (blue) formation, whereas homology-directed repair (HDR) in the presence of a donor DNA template (brown) results in gene correction or insertion.
Earlier forms of gene-editing tools consisting of zinc finger nucleases and transcription activator-like effector nucleases, were based on the fusion of synthetic zinc finger or transcription activator-like effector DNA binding domains with a bacterial nonspecific restriction endonuclease to create DSB at the desired location (34, 35). A major limitation of these techniques is the laborious process involved in the synthesis of zinc fingers and transcription activator-like effectors and their relative inefficiency because of limited binding sites. In contrast, CRISPR/Cas9 is much simpler to design and has higher efficiency because it uses an RNA-DNA binding feature rather than the protein-DNA binding feature of zinc finger nucleases and transcription activator-like effector nucleases. Given these technical advantages, many groups have successfully used CRISPR/Cas9 technology in model organisms. The technique has been used to manipulate genes in vitro and in vivo in plants and animals and to create knockout or knockin models, add protein tags, and insert loxP/FRT sites to investigate disease mechanisms (36, 37). As the list of CRISPR-based disease models continues to grow, opportunities increase to use these models to explore the therapeutic capability of CRISPR/Cas9 to correct disease-causing gene mutations and to assess specific phenotypic changes and pathophysiological mechanisms.
CRISPR as a Therapeutic Strategy
Therapeutic application of CRISPR/Cas9 to correct disease-causing mutations has been investigated in vivo for DMD, OTC, hereditary tyrosinemia, and cataract. Three independent studies used CRISPR/Cas9 and NHEJ to excise the mutated exon 23 in a DMD mouse model to achieve exon skipping in the Dystrophin gene (16–18). This resulted in the synthesis of truncated, yet functional, dystrophin protein. Gene-corrected DMD mice demonstrated decreased muscle fibrosis and functional recovery, as evidenced by improved grip strength and reduced serum creatine kinase. Gene correction also occurred in cardiac muscle after systemic injection, which is critical because cardiac failure is the most common cause of death in patients with DMD. Importantly, deep sequencing for potential off-target sites showed minimal to no evidence of off-target effects in all three studies. Gene correction using CRIPSR/Cas9 and HDR was also used as a therapeutic strategy in a mouse model of urea cycle defect disorder (19). Intravenous injection of CRISPR/Cas9 and a donor DNA strand into newborn mice with partial deficiency of the OTC enzyme resulted in 10% gene correction in hepatocytes. The gene correction resulted in improved survival and reduced ammonia levels in mice challenged with a high-protein diet. Finally, a mouse model of cataract caused by a dominant mutation in the Crygc gene was corrected using CRISPR/Cas9 in the germline (20). Gene correction occurred in 30% of the live-born pups by NHEJ or HDR-mediated repair, and all genetically corrected mice lacked cataracts by gross and histological analysis. Off-target mutations were noted in 2 of 12 mice tested. Furthermore, progeny of the genetically corrected mice were free of cataracts. Thus, multiple studies have confirmed the therapeutic potential of CRISPR/Cas9 in a wide range of monogenic diseases.
Potential Application for CRISPR Gene Editing in Monogenic Lung Diseases
SP Deficiency
SPs are expressed primarily in AT2 cells of the lung. To use CRISPR/Cas9 to correct SP mutations in the lung, gene-editing reagents could be delivered intratracheally or intranasally. Because most cases of SP deficiency are caused by single base substitutions, a relatively short donor DNA template may be sufficient for HDR-mediated gene correction. Such an approach was used by Mahiny and colleagues in a mouse model to correct SFTPB deficiency by intratracheal administration of nuclease-encoding chemically modified mRNA using an adeno-associated virus (AAV) 6 vector (38). However, the limitation of this model is that it is a compound transgenic mouse model in which SFTPB is expressed conditionally and does not ideally represent the disease-causing mutations found in humans (39).
An alternative SP deficiency model includes dominant mutations in the SFTPC gene, such as the I73T mutation, which cause a misfolded proteins response, leading to AT2 cell dysfunction and death (40). In mice, SFTPC expression is not required for survival and lung function in normal physiologic conditions. Thus, a simple deletion of the mutant SFTPC gene could prevent the formation of the misfolded precursor SFTPC protein and may improve survival and lung function in individuals with this mutation. However, with only a single normal allele of SFTPC, such individuals may remain at risk of exacerbated surfactant dysfunction during episodes of infection or other injurious stimuli (41). Thus, CRISPR/Cas9-induced gene deletion may be an efficient strategy for the treatment of some dominant genetic mutations, restoring homeostasis of tissues by removing deleterious alleles without the need for HDR-mediated gene correction.
Previous reports have suggested that genetic repair by HDR works more efficiently in proliferating cells (42, 43). In line with this evidence, increased efficiency and lower off-target DSB were noted in newborn mice livers from the OTC model targeted for gene correction using CRISPR/Cas9, as compared with corrected adult mouse livers (19). For these reasons, therapeutic gene editing during the fetal period, when most cells are still proliferating, is an attractive strategy. Prenatal therapy may provide the best opportunity to treat the disease before the rapid disease progression that occurs immediately after birth in SP deficiency and thus improve both mortality and morbidity. Furthermore, widespread availability of prenatal diagnostics for early diagnosis of these conditions increases the feasibility of such an approach (44).
A major shortcoming of gene therapy is the immunogenic potential of viral vectors and immune-mediated diminution of the transgene and/or viral vectors acquired either through a previous natural infection or after gene therapy exposure. This is particularly important if repetitive administration, which induces an immune response and decreases efficiency through neutralizing antibodies, is required (45, 46). This limitation could be overcome by the delivery of gene-editing reagents early in life before exposure to natural infection and when the immune system is still immature (47). Therapeutic intervention in the fetal period may induce immune tolerance and improve efficiency. Finally, prenatal application is feasible via the intraamniotic, vitelline vein, or by fetal intratracheal injection (48–51). The presence of fetal breathing movements combined with direct contact between the developing airways and amniotic fluid suggests that CRISPR/Cas9 administered via the intraamniotic route could reach the lung epithelium. Taken together, genome editing in fetal lungs represents a promising therapeutic strategy for lethal SP deficiency disorders.
CF
The Clevers group has shown that intestinal organoids from pediatric patients that are homozygous for CFTR ∆F508 lack the ability to respond to forskolin stimulation by swelling, mimicking important aspects of CF disease in vitro (15). Importantly, on CRISPR/Cas9-mediated gene correction of the ∆F508 mutation, normal response to forskolin-induced swelling was rescued in these organoid cultures. The authors proposed colonic transplant of genetically corrected organoids as a potential strategy for gene therapy (52). Whether a similar strategy using airway basal stem cells could be used to correct airway-related lung diseases remains to be investigated. Airway organoids have been developed by harvesting basal cells from the mouse and human lung and culturing them in a three-dimensional matrix, where they can differentiate into both secretory and multiciliated epithelial lineages (53, 54). Such an organoid system could be used to treat CF, if a suitable transplant assay can be developed. Because CF is caused by a constellation of mutations, the development of individualized autologous pulmonary models using patient-specific cells would allow for a personalized approach in patients. Patients with CF are now identified early because of universal newborn screening programs, and harvesting basal airway cells early in childhood, rather than later, for gene editing could provide advantages for therapy.
AAT
AAT is a compelling candidate for gene therapy because serum AAT levels correlate directly with improvement in pulmonary function, and the efficiency of gene editing can be assessed by measuring these levels. Current gene therapy strategies consist of targeting skeletal myocytes to augment AAT synthesis and preclude augmentation in hepatocytes because of ongoing liver damage caused by accumulation of aberrant AAT (55, 56). This limitation could be overcome by CRISPR/Cas9 repairing the gene mutation within hepatocytes, to enable synthesis of normal AAT and thus treat both liver and lung disease. Moreover, portal venous injection of recombinant AAV containing human AAT in mice results in efficient transgene expression (57), indicating that portal vein injection of CRISPR/Cas9 could enable liver-directed therapy.
In summary, gene therapy for monogenic lung diseases is an attractive strategy, and the potential pulmonary therapeutic targets for CRISPR/Cas9 gene-editing approaches are varied and important (Tables 1 and 2). The fact that the lung is a barrier organ in direct contact with the external environment allows for multiple delivery options, including viral, RNA, or recombinant protein. This attribute also provides easy access to patient-specific pulmonary stem/progenitor cells for ex vivo gene-editing approaches, although engraftment of corrected cells remains a challenge. Finally, targeted gene editing in fetal lungs is also feasible because of the direct contact of the fetal airway with amniotic fluid and because of the presence of fetal breathing movements during late gestation.
Table 1.
Recent Developments in Gene and Protein Modifier Therapy for Monogenic Lung Diseases
| Disease | Type of Therapy | Method of Application | Outcome | Reference |
|---|---|---|---|---|
| SP-B deficiency | mRNA supplementation | Repeated intratracheal administration of chemically modified mRNA in knockout mice | Increased survival, lung compliance, restoration of lung histology, and decreased inflammation | 69 |
| Gene correction | Intratracheal nuclease-encoding chemically modified mRNA and AAV6 donor in knockout mice | Increased life expectancy and lung function, restoration of lung histology, and reduced inflammation | 38 | |
| CF | Gene supplementation | Intranasal/inhalation administration using viral and nonviral vectors | Significant, albeit modest, improvement in FEV1 in clinical trials. | 10, 11 |
| Protein modifiers | Oral administration of lumacaftor-ivacaftor | Clinical improvement in FEV1 | 70 | |
| Gene correction | In vitro transfection of patient-derived intestinal organoids | Restoration of forskolin-induced swelling | 15 | |
| AAT | Gene supplementation | Intramuscular administration using viral vectors | Increased serum AAT levels and improved lung function | 55, 56 |
Definition of abbreviations: AAT, alpha-1 antitrypsin; AAV, adeno-associated virus; CF, cystic fibrosis; mRNA, messenger RNA; SP-B, surfactant protein B.
Table 2.
Potential CRISPR/Cas9 Gene-Editing Approaches in the Lung
| Prenatal Gene Editing | Postnatal In Vivo Gene Editing | Postnatal Ex Vivo Gene Editing | |
|---|---|---|---|
| Target cells | Developing progenitor cells | In situ tissue-specific stem cells (i.e., basal cells for cystic fibrosis and alveolar type II cells for surfactant protein deficiency) | Harvested stem cells |
| Advantages | Higher efficiency | Relative technical ease | Patient specific |
| Immune tolerance | Avoids in vivo viral exposure | ||
| Disease prevention | Ability to select for corrected cells before engraftment | ||
| Limitations | Technical difficulties including specialized equipment | Lower efficiency | Challenges with reengraftment |
| Application | Genetic surfactant protein deficiency | Cystic fibrosis, alpha-1 antitrypsin deficiency | Depends on availability of engraftment assay but could include cystic fibrosis |
Future Directions
Although the simplicity and ease of use of CRISPR/Cas9 has provided the basis for therapeutic gene-editing approaches, there remain multiple technical and biological roadblocks to overcome. An important question that requires further assessment is at what levels of efficiency does gene editing result in physiological correction of a given monogenic disease. This is likely to be different for every disease and tissue, but some experiments suggest that in a model of CF, at least 25% of surface epithelial cells should be targeted for restoration of normal mucus transport (58). However, the percentage of AT2 cells, airway epithelial cells, and hepatocytes that need to be genetically corrected in vivo to produce adequate amounts of SFTPC, CFTR, and AAT, respectively, is unknown. Higher off-target effects could also become more prevalent when higher efficiencies are attempted. Hence, the lowest required dose of CRISPR/Cas9 to produce the required physiological change while averting off-target effects will need to be determined empirically for each disease-causing mutation. Another important consideration is that gene editing may not provide a long-lasting response because of the natural turnover of cell populations in a given organ or tissue. This possibility may necessitate the targeting of longer-lasting stem/progenitor populations or repeat administrations. Whether either of these approaches is more efficient or technically feasible requires further investigation.
The possible off-target effects of CRISPR/Cas9 gene editing also remain unclear. Although most off-targeting events are rare and in many cases could be silent mutations, it is possible that the mutation may express later in life and could potentially be deleterious. Therefore, both short-and long-term safety studies will need to be analyzed carefully. Although recent studies report rare off-target effects, an ideal strategy would be to completely eliminate the potential for any off-target effects caused by unknown long-term effects that cannot be fully assessed (12, 15). Strategies being developed to eliminate off-target effects include the use of paired Cas9 nickases to induce single-stranded breaks, and other endonucleases that do not require transactivating crRNA and cleave DNA in a staggered pattern to create sticky ends, which is particularly suited for HDR in nondividing cells (59–62).
In addition to highlighting the off-target effects of CRISPR/Cas9, previous studies using lentiviral methods for gene therapy approaches to treat severe combined immune deficiency reported the development of cancers, possibly caused by lentiviral-induced insertional mutagenesis (63). Recent advances in AAV-mediated delivery systems are likely to circumvent many of these issues (64). Moreover, the ability of the lung to be targeted easily by RNA or recombinant protein approaches would also decrease such delivery-associated problems. Innate pulmonary barriers such as the airway mucus barrier, immunological defense mechanisms, and cellular tight junctions impose challenges to effective delivery of genetic tools directly to lung epithelial cells. Airway mucus is the first physical barrier for gene-editing tools to overcome before reaching the target cells, and in diseased airways such as in CF, it is particularly dense and inhibitory to the introduction of exogenous DNA. Recent developments in lipid nanoparticles as tools for mucus penetration and gene delivery may enhance transfection efficiency (65, 66) (Figure 2).
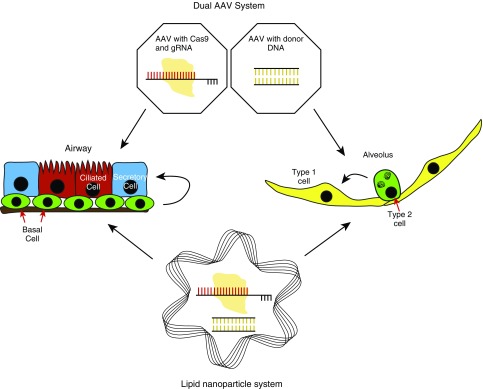
Figure 2.
CRISPR/Cas9 delivery methods. Viral vectors using adeno-associated virus (AAV) and nonviral vectors using lipid nanoparticles are potentially effective methods for delivery of gene-editing tools into both airway and alveolar epithelial cells.
Ethical Considerations
Germline editing is a concern and a source of much ethical discussion. As recommended by national and international summits on the ethical issues of germline editing, robust research is needed to evaluate changes in cells and tissues that have undergone gene editing, to assist scientific, governmental, and public communities make informed decisions (67, 68).
Conclusions
CRISPR/Cas9-mediated precise genome editing presents an exciting new potential as a therapeutic option in monogenic congenital disorders of the lung that currently lack treatment options. Potential methods of gene editing include intratracheal delivery of CRISPR/Cas9 complex or delivery of autologous stem/progenitor cells that have been genetically corrected ex vivo. Prenatal gene editing using CRISPR/Cas9 may represent a promising therapeutic strategy for SP deficiency disorders that are quickly lethal after birth. Although clinical translation of gene editing for monogenic lung diseases is appealing, several key questions related to targeted delivery of reagents to lung epithelial cells and optimizing gene-editing efficiencies to achieve adequate physiological effects remain to be investigated. Prevention of off-target mutagenesis represents the most important concern of gene editing, and recent developments such as the use of modified Cas9 or other enzymes may reduce these concerns in the future. Although a great deal of progress has been made in a short time, much is still needed, including the development of standard methods to assess physiological efficacy as well as accurate assessment of off-target effects, to adapt gene-editing technologies for therapeutic use. This will require close collaboration among basic scientists, pulmonologists, and geneticists.
Acknowledgments
Acknowledgments
The authors thank the Nemours Foundation for research support. The Morrisey laboratory is supported by funding from the National Institutes of Health.
Footnotes
Originally Published in Press as DOI: 10.1165/rcmb.2016-0301PS on October 25, 2016
Author disclosures are available with the text of this article at www.atsjournals.org.
References
- 1.Witt WP, Weiss AJ, Elixhauser A.Overview of Hospital Stays for Children in the United States, 2012Statistical Brief No. 187. Healthcare Cost and Utilization Project (H-CUP) Statistical Briefs. Agency for Healthcare Research and Quality (US); 2014 [accessed 2016 Aug 15]. Available from http://www.hcup-us.ahrq.gov/reports/statbriefs/sb187-Hospital-Stays-Children-2012.pdf
- 2.Nogee LM. Genetic basis of children’s interstitial lung disease. Pediatr Allergy Immunol Pulmonol. 2010;23:15–24. doi: 10.1089/ped.2009.0024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cystic Fibrosis FoundationCystic Fibrosis Foundation Patient Registry, 2014 Annual Data Report. Cystic Fibrosis Foundation. 2014. [accessed 2016 Aug 15]. Available from https://www.cff.org/2014_CFF_Annual_Data_Report_to_the_Center_Directors.pdf/
- 4.Tanash HA, Nilsson PM, Nilsson JA, Piitulainen E. Survival in severe alpha-1-antitrypsin deficiency (PiZZ) Respir Res. 2010;11:44–50. doi: 10.1186/1465-9921-11-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Spoonhower KA, Davis PB. Epidemiology of cystic fibrosis. Clin Chest Med. 2016;37:1–8. doi: 10.1016/j.ccm.2015.10.002. [DOI] [PubMed] [Google Scholar]
- 6.Hamvas A. Inherited surfactant protein-B deficiency and surfactant protein-C associated disease: clinical features and evaluation. Semin Perinatol. 2006;30:316–326. doi: 10.1053/j.semperi.2005.11.002. [DOI] [PubMed] [Google Scholar]
- 7.Hamvas A, Nogee LM, Mallory GB, Jr, Spray TL, Huddleston CB, August A, Dehner LP, deMello DE, Moxley M, Nelson R, et al. Lung transplantation for treatment of infants with surfactant protein B deficiency. J Pediatr. 1997;130:231–239. doi: 10.1016/s0022-3476(97)70348-2. [DOI] [PubMed] [Google Scholar]
- 8.Palomar LM, Nogee LM, Sweet SC, Huddleston CB, Cole FS, Hamvas A. Long-term outcomes after infant lung transplantation for surfactant protein B deficiency related to other causes of respiratory failure. J Pediatr. 2006;149:548–553. doi: 10.1016/j.jpeds.2006.06.004. [DOI] [PubMed] [Google Scholar]
- 9.Moss RB, Milla C, Colombo J, Accurso F, Zeitlin PL, Clancy JP, Spencer LT, Pilewski J, Waltz DA, Dorkin HL, et al. Repeated aerosolized AAV-CFTR for treatment of cystic fibrosis: a randomized placebo-controlled phase 2B trial. Hum Gene Ther. 2007;18:726–732. doi: 10.1089/hum.2007.022. [DOI] [PubMed] [Google Scholar]
- 10.Alton EW, Armstrong DK, Ashby D, Bayfield KJ, Bilton D, Bloomfield EV, Boyd AC, Brand J, Buchan R, Calcedo R, et al. UK Cystic Fibrosis Gene Therapy Consortium. Repeated nebulisation of non-viral CFTR gene therapy in patients with cystic fibrosis: a randomised, double-blind, placebo-controlled, phase 2b trial. Lancet Respir Med. 2015;3:684–691. doi: 10.1016/S2213-2600(15)00245-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee TW, Southern KW, Perry LA, Penny-Dimri JC, Aslam AA. Topical cystic fibrosis transmembrane conductance regulator gene replacement for cystic fibrosis-related lung disease. Cochrane Database Syst Rev. 2016;6:CD005599. doi: 10.1002/14651858.CD005599.pub5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mali P, Esvelt KM, Church GM. Cas9 as a versatile tool for engineering biology. Nat Methods. 2013;10:957–963. doi: 10.1038/nmeth.2649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Doudna JA, Charpentier E. Genome editing. The new frontier of genome engineering with CRISPR-Cas9. Science. 2014;346:1258096. doi: 10.1126/science.1258096. [DOI] [PubMed] [Google Scholar]
- 14.Sander JD, Joung JK. CRISPR-Cas systems for editing, regulating and targeting genomes. Nat Biotechnol. 2014;32:347–355. doi: 10.1038/nbt.2842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schwank G, Koo BK, Sasselli V, Dekkers JF, Heo I, Demircan T, Sasaki N, Boymans S, Cuppen E, van der Ent CK, et al. Functional repair of CFTR by CRISPR/Cas9 in intestinal stem cell organoids of cystic fibrosis patients. Cell Stem Cell. 2013;13:653–658. doi: 10.1016/j.stem.2013.11.002. [DOI] [PubMed] [Google Scholar]
- 16.Tabebordbar M, Zhu K, Cheng JK, Chew WL, Widrick JJ, Yan WX, Maesner C, Wu EY, Xiao R, Ran FA, et al. In vivo gene editing in dystrophic mouse muscle and muscle stem cells. Science. 2016;351:407–411. doi: 10.1126/science.aad5177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nelson CE, Hakim CH, Ousterout DG, Thakore PI, Moreb EA, Castellanos Rivera RM, Madhavan S, Pan X, Ran FA, Yan WX, et al. In vivo genome editing improves muscle function in a mouse model of Duchenne muscular dystrophy. Science. 2016;351:403–407. doi: 10.1126/science.aad5143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Long C, Amoasii L, Mireault AA, McAnally JR, Li H, Sanchez-Ortiz E, Bhattacharyya S, Shelton JM, Bassel-Duby R, Olson EN. Postnatal genome editing partially restores dystrophin expression in a mouse model of muscular dystrophy. Science. 2016;351:400–403. doi: 10.1126/science.aad5725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang Y, Wang L, Bell P, McMenamin D, He Z, White J, Yu H, Xu C, Morizono H, Musunuru K, et al. A dual AAV system enables the Cas9-mediated correction of a metabolic liver disease in newborn mice. Nat Biotechnol. 2016;34:334–338. doi: 10.1038/nbt.3469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wu Y, Liang D, Wang Y, Bai M, Tang W, Bao S, Yan Z, Li D, Li J. Correction of a genetic disease in mouse via use of CRISPR-Cas9. Cell Stem Cell. 2013;13:659–662. doi: 10.1016/j.stem.2013.10.016. [DOI] [PubMed] [Google Scholar]
- 21.Nogee LM. Genetic mechanisms of surfactant deficiency. Biol Neonate. 2004;85:314–318. doi: 10.1159/000078171. [DOI] [PubMed] [Google Scholar]
- 22.Nogee LM, de Mello DE, Dehner LP, Colten HR. Brief report: deficiency of pulmonary surfactant protein B in congenital alveolar proteinosis. N Engl J Med. 1993;328:406–410. doi: 10.1056/NEJM199302113280606. [DOI] [PubMed] [Google Scholar]
- 23.Cole FS, Hamvas A, Rubinstein P, King E, Trusgnich M, Nogee LM, deMello DE, Colten HR. Population-based estimates of surfactant protein B deficiency. Pediatrics. 2000;105:538–541. doi: 10.1542/peds.105.3.538. [DOI] [PubMed] [Google Scholar]
- 24.Nogee LM, Dunbar AE, III, Wert S, Askin F, Hamvas A, Whitsett JA. Mutations in the surfactant protein C gene associated with interstitial lung disease. Chest. 2002;121:20S–21S. doi: 10.1378/chest.121.3_suppl.20s. [DOI] [PubMed] [Google Scholar]
- 25.Litao MK, Hayes D, Jr, Chiwane S, Nogee LM, Kurland G, Guglani L. A novel surfactant protein C gene mutation associated with progressive respiratory failure in infancy. Pediatr Pulmonol. 2017;52:57–68. doi: 10.1002/ppul.23493. [DOI] [PubMed] [Google Scholar]
- 26.Shulenin S, Nogee LM, Annilo T, Wert SE, Whitsett JA, Dean M. ABCA3 gene mutations in newborns with fatal surfactant deficiency. N Engl J Med. 2004;350:1296–1303. doi: 10.1056/NEJMoa032178. [DOI] [PubMed] [Google Scholar]
- 27.Bullard JE, Wert SE, Whitsett JA, Dean M, Nogee LM. ABCA3 mutations associated with pediatric interstitial lung disease. Am J Respir Crit Care Med. 2005;172:1026–1031. doi: 10.1164/rccm.200503-504OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rowe SM, Miller S, Sorscher EJ. Cystic fibrosis. N Engl J Med. 2005;352:1992–2001. doi: 10.1056/NEJMra043184. [DOI] [PubMed] [Google Scholar]
- 29.Cutting GR. Cystic fibrosis genetics: from molecular understanding to clinical application. Nat Rev Genet. 2015;16:45–56. doi: 10.1038/nrg3849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Grosse SD, Rosenfeld M, Devine OJ, Lai HJ, Farrell PM. Potential impact of newborn screening for cystic fibrosis on child survival: a systematic review and analysis. J Pediatr. 2006;149:362–366. doi: 10.1016/j.jpeds.2006.04.059. [DOI] [PubMed] [Google Scholar]
- 31.Marshall BC, Nelson EC. Accelerating implementation of biomedical research advances: critical elements of a successful 10 year Cystic Fibrosis Foundation healthcare delivery improvement initiative. BMJ Qual Saf. 2014;23:i95–i103. doi: 10.1136/bmjqs-2013-002790. [DOI] [PubMed] [Google Scholar]
- 32.Crystal RG. Alpha 1-antitrypsin deficiency, emphysema, and liver disease. Genetic basis and strategies for therapy. J Clin Invest. 1990;85:1343–1352. doi: 10.1172/JCI114578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ran FA, Hsu PD, Wright J, Agarwala V, Scott DA, Zhang F. Genome engineering using the CRISPR-Cas9 system. Nat Protoc. 2013;8:2281–2308. doi: 10.1038/nprot.2013.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Urnov FD, Rebar EJ, Holmes MC, Zhang HS, Gregory PD. Genome editing with engineered zinc finger nucleases. Nat Rev Genet. 2010;11:636–646. doi: 10.1038/nrg2842. [DOI] [PubMed] [Google Scholar]
- 35.Christian M, Cermak T, Doyle EL, Schmidt C, Zhang F, Hummel A, Bogdanove AJ, Voytas DF. Targeting DNA double-strand breaks with TAL effector nucleases. Genetics. 2010;186:757–761. doi: 10.1534/genetics.110.120717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shen B, Zhang J, Wu H, Wang J, Ma K, Li Z, Zhang X, Zhang P, Huang X. Generation of gene-modified mice via Cas9/RNA-mediated gene targeting. Cell Res. 2013;23:720–723. doi: 10.1038/cr.2013.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sánchez-Rivera FJ, Papagiannakopoulos T, Romero R, Tammela T, Bauer MR, Bhutkar A, Joshi NS, Subbaraj L, Bronson RT, Xue W, et al. Rapid modelling of cooperating genetic events in cancer through somatic genome editing. Nature. 2014;516:428–431. doi: 10.1038/nature13906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mahiny AJ, Dewerth A, Mays LE, Alkhaled M, Mothes B, Malaeksefat E, Loretz B, Rottenberger J, Brosch DM, Reautschnig P, et al. In vivo genome editing using nuclease-encoding mRNA corrects SP-B deficiency. Nat Biotechnol. 2015;33:584–586. doi: 10.1038/nbt.3241. [DOI] [PubMed] [Google Scholar]
- 39.Melton KR, Nesslein LL, Ikegami M, Tichelaar JW, Clark JC, Whitsett JA, Weaver TE. SP-B deficiency causes respiratory failure in adult mice. Am J Physiol Lung Cell Mol Physiol. 2003;285:L543–L549. doi: 10.1152/ajplung.00011.2003. [DOI] [PubMed] [Google Scholar]
- 40.Cameron HS, Somaschini M, Carrera P, Hamvas A, Whitsett JA, Wert SE, Deutsch G, Nogee LM. A common mutation in the surfactant protein C gene associated with lung disease. J Pediatr. 2005;146:370–375. doi: 10.1016/j.jpeds.2004.10.028. [DOI] [PubMed] [Google Scholar]
- 41.Glasser SW, Burhans MS, Korfhagen TR, Na CL, Sly PD, Ross GF, Ikegami M, Whitsett JA. Altered stability of pulmonary surfactant in SP-C-deficient mice. Proc Natl Acad Sci USA. 2001;98:6366–6371. doi: 10.1073/pnas.101500298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Saleh-Gohari N, Helleday T. Conservative homologous recombination preferentially repairs DNA double-strand breaks in the S phase of the cell cycle in human cells. Nucleic Acids Res. 2004;32:3683–3688. doi: 10.1093/nar/gkh703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mao Z, Bozzella M, Seluanov A, Gorbunova V. DNA repair by nonhomologous end joining and homologous recombination during cell cycle in human cells. Cell Cycle. 2008;7:2902–2906. doi: 10.4161/cc.7.18.6679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stuhrmann M, Bohnhorst B, Peters U, Bohle RM, Poets CF, Schmidtke J. Prenatal diagnosis of congenital alveolar proteinosis (surfactant protein B deficiency) Prenat Diagn. 1998;18:953–955. doi: 10.1002/(sici)1097-0223(199809)18:9<953::aid-pd364>3.0.co;2-j. [DOI] [PubMed] [Google Scholar]
- 45.Halbert CL, Standaert TA, Wilson CB, Miller AD. Successful readministration of adeno-associated virus vectors to the mouse lung requires transient immunosuppression during the initial exposure. J Virol. 1998;72:9795–9805. doi: 10.1128/jvi.72.12.9795-9805.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Myint M, Limberis MP, Bell P, Somanathan S, Haczku A, Wilson JM, Diamond SL. In vivo evaluation of adeno-associated virus gene transfer in airways of mice with acute or chronic respiratory infection. Hum Gene Ther. 2014;25:966–976. doi: 10.1089/hum.2014.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Carlon MS, Vidović D, Dooley J, da Cunha MM, Maris M, Lampi Y, Toelen J, Van den Haute C, Baekelandt V, Deprest J, et al. Immunological ignorance allows long-term gene expression after perinatal recombinant adeno-associated virus-mediated gene transfer to murine airways. Hum Gene Ther. 2014;25:517–528. doi: 10.1089/hum.2013.196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Joyeux L, Danzer E, Limberis MP, Zoltick PW, Radu A, Flake AW, Davey MG. In utero lung gene transfer using adeno-associated viral and lentiviral vectors in mice. Hum Gene Ther Methods. 2014;25:197–205. doi: 10.1089/hgtb.2013.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Boelig MM, Kim AG, Stratigis JD, McClain LE, Li H, Flake AW, Peranteau WH. The intravenous route of injection optimizes engraftment and survival in the murine model of in utero hematopoietic cell transplantation. Biol Blood Marrow. 2016;22:991–999. doi: 10.1016/j.bbmt.2016.01.017. [DOI] [PubMed] [Google Scholar]
- 50.David AL, Peebles DM, Gregory L, Themis M, Cook T, Coutelle C, Rodeck CH. Percutaneous ultrasound-guided injection of the trachea in fetal sheep: a novel technique to target the fetal airways. Fetal Diagn Ther. 2003;18:385–390. doi: 10.1159/000071984. [DOI] [PubMed] [Google Scholar]
- 51.Carlon M, Toelen J, Van der Perren A, Vandenberghe LH, Reumers V, Sbragia L, Gijsbers R, Baekelandt V, Himmelreich U, Wilson JM, et al. Efficient gene transfer into the mouse lung by fetal intratracheal injection of raav2/6.2. Mol Ther. 2010;18:2130–2138. doi: 10.1038/mt.2010.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yui S, Nakamura T, Sato T, Nemoto Y, Mizutani T, Zheng X, Ichinose S, Nagaishi T, Okamoto R, Tsuchiya K, et al. Functional engraftment of colon epithelium expanded in vitro from a single adult Lgr5⁺ stem cell. Nat Med. 2012;18:618–623. doi: 10.1038/nm.2695. [DOI] [PubMed] [Google Scholar]
- 53.Rock JR, Onaitis MW, Rawlins EL, Lu Y, Clark CP, Xue Y, Randell SH, Hogan BL. Basal cells as stem cells of the mouse trachea and human airway epithelium. Proc Natl Acad Sci USA. 2009;106:12771–12775. doi: 10.1073/pnas.0906850106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Danahay H, Pessotti AD, Coote J, Montgomery BE, Xia D, Wilson A, Yang H, Wang Z, Bevan L, Thomas C, et al. Notch2 is required for inflammatory cytokine-driven goblet cell metaplasia in the lung. Cell Reports. 2015;10:239–252. doi: 10.1016/j.celrep.2014.12.017. [DOI] [PubMed] [Google Scholar]
- 55.Brantly ML, Spencer LT, Humphries M, Conlon TJ, Spencer CT, Poirier A, Garlington W, Baker D, Song S, Berns KI, et al. Phase I trial of intramuscular injection of a recombinant adeno-associated virus serotype 2 α-antitrypsin (AAT) vector in AAT-deficient adults. Hum Gene Ther. 2006;17:1177–1186. doi: 10.1089/hum.2006.17.1177. [DOI] [PubMed] [Google Scholar]
- 56.Flotte TR, Trapnell BC, Humphries M, Carey B, Calcedo R, Rouhani F, Campbell-Thompson M, Yachnis AT, Sandhaus RA, McElvaney NG, et al. Phase 2 clinical trial of a recombinant adeno-associated viral vector expressing α1-antitrypsin: interim results. Hum Gene Ther. 2011;22:1239–1247. doi: 10.1089/hum.2011.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Song S, Embury J, Laipis PJ, Berns KI, Crawford JM, Flotte TR. Stable therapeutic serum levels of human alpha-1 antitrypsin (AAT) after portal vein injection of recombinant adeno-associated virus (rAAV) vectors. Gene Ther. 2001;8:1299–1306. doi: 10.1038/sj.gt.3301422. [DOI] [PubMed] [Google Scholar]
- 58.Zhang L, Button B, Gabriel SE, Burkett S, Yan Y, Skiadopoulos MH, Dang YL, Vogel LN, McKay T, Mengos A, et al. CFTR delivery to 25% of surface epithelial cells restores normal rates of mucus transport to human cystic fibrosis airway epithelium. PLoS Biol. 2009;7:e1000155. doi: 10.1371/journal.pbio.1000155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Shen B, Zhang W, Zhang J, Zhou J, Wang J, Chen L, Wang L, Hodgkins A, Iyer V, Huang X, et al. Efficient genome modification by CRISPR-Cas9 nickase with minimal off-target effects. Nat Methods. 2014;11:399–402. doi: 10.1038/nmeth.2857. [DOI] [PubMed] [Google Scholar]
- 60.Qi LS, Larson MH, Gilbert LA, Doudna JA, Weissman JS, Arkin AP, Lim WA. Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell. 2013;152:1173–1183. doi: 10.1016/j.cell.2013.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chavez A, Scheiman J, Vora S, Pruitt BW, Tuttle M, P R Iyer E, Lin S, Kiani S, Guzman CD, Wiegand DJ, et al. Highly efficient Cas9-mediated transcriptional programming. Nat Methods. 2015;12:326–328. doi: 10.1038/nmeth.3312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zetsche B, Gootenberg JS, Abudayyeh OO, Slaymaker IM, Makarova KS, Essletzbichler P, Volz SE, Joung J, van der Oost J, Regev A, et al. Cpf1 is a single RNA-guided endonuclease of a class 2 CRISPR-Cas system. Cell. 2015;163:759–771. doi: 10.1016/j.cell.2015.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hacein-Bey-Abina S, Von Kalle C, Schmidt M, McCormack MP, Wulffraat N, Leboulch P, Lim A, Osborne CS, Pawliuk R, Morillon E, et al. LMO2-associated clonal T cell proliferation in two patients after gene therapy for SCID-X1. Science. 2003;302:415–419. doi: 10.1126/science.1088547. [DOI] [PubMed] [Google Scholar]
- 64.Senís E, Fatouros C, Große S, Wiedtke E, Niopek D, Mueller AK, Börner K, Grimm D. CRISPR/Cas9-mediated genome engineering: an adeno-associated viral (AAV) vector toolbox. Biotechnol J. 2014;9:1402–1412. doi: 10.1002/biot.201400046. [DOI] [PubMed] [Google Scholar]
- 65.Mastorakos P, da Silva AL, Chisholm J, Song E, Choi WK, Boyle MP, Morales MM, Hanes J, Suk JS. Highly compacted biodegradable DNA nanoparticles capable of overcoming the mucus barrier for inhaled lung gene therapy. Proc Natl Acad Sci USA. 2015;112:8720–8725. doi: 10.1073/pnas.1502281112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yin H, Song CQ, Dorkin JR, Zhu LJ, Li Y, Wu Q, Park A, Yang J, Suresh S, Bizhanova A, et al. Therapeutic genome editing by combined viral and non-viral delivery of CRISPR system components in vivo. Nat Biotechnol. 2016;34:328–333. doi: 10.1038/nbt.3471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.LaBarbera AR. Proceedings of the international summit on human gene editing: a global discussion-Washington, D.C., December 1-3, 2015. J Assist Reprod Genet. 2016;33:1123–1127. doi: 10.1007/s10815-016-0753-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Baltimore D, Berg P, Botchan M, Carroll D, Charo RA, Church G, Corn JE, Daley GQ, Doudna JA, Fenner M, et al. Biotechnology. A prudent path forward for genomic engineering and germline gene modification. Science. 2015;348:36–38. doi: 10.1126/science.aab1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kormann MS, Hasenpusch G, Aneja MK, Nica G, Flemmer AW, Herber-Jonat S, Huppmann M, Mays LE, Illenyi M, Schams A, et al. Expression of therapeutic proteins after delivery of chemically modified mRNA in mice. Nat Biotechnol. 2011;29:154–157. doi: 10.1038/nbt.1733. [DOI] [PubMed] [Google Scholar]
- 70.Wainwright CE, Elborn JS, Ramsey BW, Marigowda G, Huang X, Cipolli M, Colombo C, Davies JC, De Boeck K, Flume PA, et al. TRAFFIC Study Group; TRANSPORT Study Group. Lumacaftor-ivacaftor in patients with cystic fibrosis homozygous for phe508del cftr. N Engl J Med. 2015;373:220–231. doi: 10.1056/NEJMoa1409547. [DOI] [PMC free article] [PubMed] [Google Scholar]