Abstract
5D images of engrailed (en) and eye gone (eyg) gene expressions during the course of the eye-antenna disc primordium (EADP) formation of Drosophila embryos from embryonic stages 13 through 16 were recorded via light sheet microscopy and analyzed to reveal the cell dynamics involved in the development of the EADP. Detailed analysis of the time-lapsed images revealed the process of EADP formation and its invagination trajectory, which involved an inversion of the EADP anterior-posterior axis relative to the body. Furthermore, analysis of the en-expression pattern in the EADP provided strong evidence that the EADP is derived from one of the en-expressing head segments.
The larval eye-antenna disc (EAD) in Drosophila has served as a popular experimental system for the study of many biological processes1,2,3,4,5,6. It develops into the adult compound eyes, ocelli, antenna, maxillary palp, and the head capsule that surrounds these organs7. When the larva is first hatched, the EAD can be morphologically recognized to contain about 70 cells8. Several studies have mapped and followed the development of the precursors for the EAD (eye-antenna disc primordium, EADP) during embryo development9,10,11,12,13,14.
The development of the EADP is related to the morphogenesis of the Drosophila visual system, which is composed of the larva eye (the Bolwig’s organ), the adult eye (developed from the EADP) and the optic lobe of the brain. The Drosophila visual system is thought to be originated from the dorsal head region in the blastoderm embryo. Analysis using gynandromorphs and somatic clones15,16 estimated the number of founder cells for each EAD in blastoderm stage to be 23 and 6, respectively. Fate mapping experiments, by gynandromorph mapping, photoactivation tracing, and cell ablation, suggest that the EADP can be tracked back to the dorsal lateral head region in blastoderm embryo7,10,13,17. In stage 4, sine oculis (so) is expressed in a broad unpaired head area, known as the visual primordium, that is believed to give rise to the Drosophila visual system11,14,18,19. In stage 5, it partitions to form bilaterally symmetrical procephalic lobes14. In stage 11, the optic lobe placode is formed from the lateral part of the visual primordium11. In stages 12 and 13, the optic lobe placode invaginates by apical constriction and then splits into the optic lobe (defined by so expression) and Bolwig’s organ (defined by mAb22C10 and FasII expression)11. The EADP can be identified by the expression of the Pax6 gene eyeless (ey) in stage 12 as two symmetrical regions on the dorsolateral surface in the head14,19. Ey expression in the EADP continues throughout the embryonic stage19. The Pax gene eye gone (eyg) and its sister gene twin of eyegone (toe) are expressed in the EADP beginning in stage 1520,21. Lineage mapping showed that ey and eyg expressing cells contributed to the entire EAD22. Hence, the EADP can be identified by ey, eyg and toe expressions.
Although the EADP is thought to be derived from the visual primordium, which is defined by so expression, ey and so expressions do not overlap23. Consistently, loss of so does not affect ey and toy expressions and the formation of the EADP24. Other genes in the retinal determination network, namely, dachshund (dac) and eyes absent (eya), are not expressed in the EADP23. These observations strongly suggest that the EADP formation is independent of the retinal determination network genes so, dac and eya. Its origin and the course of formation need further clarification.
Earlier studies of the morphological change of head primordium in late embryonic period suggested that the EADP invaginates in stage 15 to form a V-shape structure11,12,19,23,25, later fused with other head segments (e.g. the antennal segment)12,13, and subsequently evaginated from the posterior dorsal pouch as a sac-like structure before hatching11,12. However, these earlier studies of the EADP were hindered by the lack of specific marker and relied on histology or the general imaginal cell marker escargot (esg)13,26,27. Furthermore, the organization and movement of the EADP were deduced from snapshots of individual embryos at discrete time points, rather than from continuous observation of a single embryo. The movement of EADP precursors is particularly difficult to follow because the embryonic head region undergoes rapid and complicated morphogenetic movements (known as head involution) in stage 15.
In this study, we examined fluorescence reporters to mark and track EADP cells via light sheet microscopy28,29, which provided the desired 5D (three dimensional spatial, in conjunction with temporal and multi-fluorescence channel) images. The extraordinary capability to achieve large field of view, fast acquisition speed and great penetration depth of the dual-illumination light sheet microscopy enabled us to capture and analyze the cellular dynamics of the EADP formation and movement.
Results
Markers for the EADP
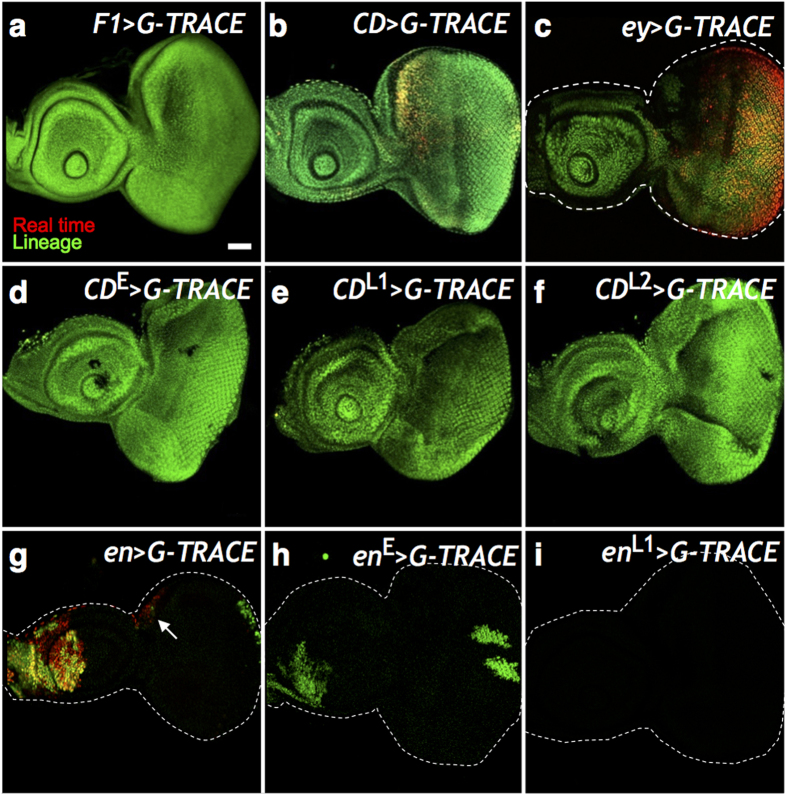
The enhancers in the eyg-toe locus have been dissected22, of which the CD enhancer mimics the eyg expression from embryonic stage to the larval EAD, the F1 enhancer is expressed in the EADP, but not in later stages, and the EX enhancer is expressed in the EADP and first instar (L1) eye disc. Lineage tracing30 via CD enhancer to drive the induction of clones has shown that the CD-expressing (CD+) cells contributed to the entire EAD22. We used the G-TRACE method31 to confirm the lineage distribution. G-TRACE showed that F1, CD, and ey lineage cells contributed to the entire EAD (Fig. 1a–c). Our results showed that eyg expression (represented by the F1 and CD enhancers) and ey expression in the embryonic stage can be used to identify the EADP. We further combined GAL80ts 32 and used temperature shifts to induce clones in different developmental stages. The results showed that the lineage of CD+ cells in the embryonic, L1, and second instar (L2) stages contributed to the entire EAD (Fig. 1d–f), indicating that CD is expressed in all cells in the EADP and continued to be expressed throughout the EAD development. The CD-GAL4 induced lineage clones in embryonic stage contributed to the entire L3 EAD, including the disc proper and the peripodial membrane (Supplementary Fig. S1). The result indicated that the CD-expression in the EADP in the embryonic stage included all the EAD progenitor cells. Therefore, we used the CD enhancer as a marker for the EADP.
Figure 1. Stage-specific lineage analysis.
The G-TRACE reporters RFP and GFP were driven by (a) F1-GAL4 (F1 > G-TRACE), (b) CD-GAL4 (CD > G-TRACE), and (c) ey-GAL4 (ey > G-TRACE). Late third instar (L3) EAD were examined. The RFP (red) represents real time expression, while GFP (green) represents lineage expression. (a) F1 was expressed only in embryo, therefore showed no real time expression in the L3 EAD. (b) CD real time expression mimics eyg expression in the equator and the anterior dorsal region in the L3 eye disc. (c) ey real time expression in the L3 eye disc. (a–c) The lineage expression (green) of F1, CD, and ey comprised entire EAD. (d–f) Stage-specific CD > G-TRACE in embryonic, L1 and L2 stages (designated as CDE, CDL1 and CDL2, respectively). CD+ cells in the embryonic, L1, and L2 stages contributed to the entire EAD. Since the GAL80ts blocked the GAL4 activity in L3, there was no real time expression in the L3 EAD. (g) The G-TRACE reporters driven by en (en > G-TRACE). en+ cells contributed to the posterior compartment of the antenna and eye disc. Real-time en expression in L3 was in the posterior compartment of the antenna disc and in the ocellar region (white arrow) in the eye disc. (h–i) Stage-specific en > G-TRACE in embryonic and L1 stages (designated as enE and enL1, respectively). en+ cells in the embryonic stage contributed to the posterior compartment of the antenna and eye discs. Note the inverted anterior-posterior axis of antenna disc relative to that in the eye disc. en+ cells in L1 did not contribute to the L3 EAD, suggesting that en was not expressed in L1 EAD. Scale bar is 50 μm and the anterior is to the left in all frames.
However, the CD enhancer is not activated until late stage 14; hence, it cannot be used to trace the earlier stages of the EADP development. To trace the earlier stages of the EADP development, we used en-GAL4 driven UAS-H2B-RFP (en > RFP). In contrast to previous report12, en is expressed in the EADP. G-TRACE showed that en-expressing (en+) cells contributed to the posterior compartment of the antenna disc, ocellar region, and the posterior part of the eye disc (Fig. 1g). The temperature shift experiments of en-GAl4 driven G-TRACE (en > G-TRACE) showed that cells expressing en in the embryonic stage (enE) contributed to regions same as the en > G-TRACE result (Fig. 1h). Interestingly, en+ cells in L1 (enL1) did not contribute to the EAD at all (Fig. 1h), suggesting that the en expression in the EAD is interrupted during L1 and resumed in later larval stages (Fig. 1h,i).
Since en expression is not limited to the EADP12,33,34,35,36,37, we used live-imaging of Drosophila embryos to simultaneously follow the movement of both CD+ and en+ cells. When the CD-GFP expression appeared in the EADP, we traced backward in time to define the en+ cells that were associated with the EADP. These cells are termed the EADP-engaged en-expressing cells (EECs). Since en expression (from late stage 8)33 in the procephalic region precedes ey (from stage 12), EECs potentially can allow us to trace the earliest stage of the EADP formation. However, the en > RFP expression in the head region was so weak in the early stage, that we were able to trace only as early as stage 13.
The origin and movement of EECs through a 5-phase process
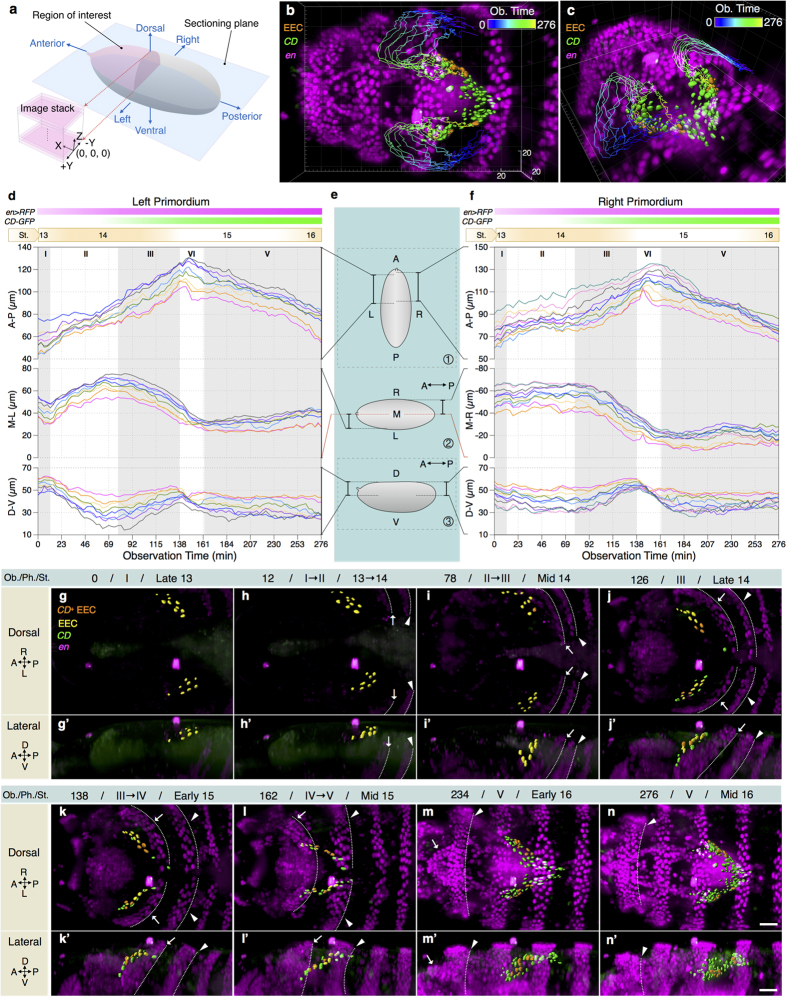
We followed simultaneously the CD+ and en+ cells via dual-illumination light sheet microscopy (Supplementary Fig. S2) and quantitatively analyzed the 5D image data to study the cell dynamics of EADP cells. Our region of interest was the dorsal half of the head region, as shown in Fig. 2a. Each dual fluorescence channel 3D image was acquired within 40 seconds, every 3 minutes, for a total period of 4 to 7 hours, depending on the time point when the embryo started to wriggle within the vitelline membrane to disable clear imaging (Supplementary Movie 1 and Supplementary Fig. S3). EECs were tracked back to their initial locations. The 3D trajectory and the surface rendering of each EEC and CD+ cell were reconstructed from their first appearance through the last trackable observation time point (Fig. 2b and c). Although the numbers of EECs in the left and the right EADP in a single embryo were not identical (4 to14 cells in each primordium in the 6 embryos that we have analyzed), their migration patterns in different embryos were consistent (Supplementary Fig. S4).
Figure 2. The origin and movement of EECs.
(a) A schematic illustration of the region of interest, embryo orientation and the 3D coordinates of an image stack. The imaging plane was parallel to the sectioning plane. (b) The trajectories of EECs, over the observation time 0 to 276 min, displayed with the 3D fluorescence patterns of en > RFP (magenta) and CD-GFP (green) in dorsal view. EECs are labeled in orange. (c) A tilted view of the figure in (b). (d) A composite chart of the trajectories of EECs in the left primordium, decomposed along 3 mutually orthogonal axes, namely, the anterior-posterior (A-P), the medial-lateral (M-L), and the dorsal-ventral (D-V) axes. The timing of en > RFP and CD-GFP expression and the corresponding developmental stages are marked on top. (e) The schematic diagrams showing the locations, the orientations, and the ranges associated with the corresponding diagrams in (d) and (f). Diagrams ① and ② represent an embryo in the dorsal view while diagram ③ shows the corresponding lateral view. (f) A similar chart as (d) for the right primordium. (g–n) The dorsal views of EADP cells movement from embryonic stage 13 to stage 16. The observation time points (Ob. Time, listed on the top of the corresponding figures) of the images from (g–n) are 0, 12, 78, 126, 138, 162, 234, and 276 min, respectively. (g’–n’) The lateral view of the left primordium corresponding to figures (g–n). The corresponding phase (Ph.) and the equivalent embryonic stage (St.) are listed next to the observation time. The dorsal ridge and the first thoracic segment are indicated by “ → ” and “▸”, respectively. Their posterior boundaries are indicated by dotted lines. In frames (m,m’), (n,n’), the dorsal ridge has involuted inside the embryo so that only the posterior boundaries of the first thoracic segment are visible. Grid size in the background is 20 μm in (b,c). Scale bars are 10 μm in frames (n) and (n’). Frames (g–n) and (g’–n’) are in the same scale.
Based on our observations, we propose that the course of the EADP formation can be phenomenologically divided into the following 5 phases (Fig. 2d,f; Supplementary Movie 2).
Phase I (from observation time 0 to 12 min.): EECs emerged as two en+ stripes, one on each side near the periphery of the procephalic regions (Fig. 2g) and showed slight random movements. The dorsal ridge and the epidermal segments were barely discernible from the dorsal view due to their weak fluorescence signals and their relatively ventral position.
Phase II (from observation time 13 to 78 min.): In this phase, dorsal closure and head involution began, and a few EECs started to express CD-GFP. The dorsal ridge and en+ epidermal segments appeared (Fig. 2h). EECs started to migrate anteriorly and slightly ventrally, as well as toward the midline (Fig. 2g,g’ and also d and f).
Phase III (observation time 79 to 135 min.): The dorsal ridge and the epidermal segments began to fuse at the dorsal midline and moved anteriorly. EECs continued to move toward the dorsal midline (Fig. 2i,i’). More EECs expressed CD-GFP.
Phase IV (observation time 136 to 162 min.): The dorsal ridge and the epidermal segments moved anteriorly and over the cephalic region (Fig. 2k,k’). EECs rearranged into slender stripes along the A-P axis, and reversed their anterior trajectories to move posteriorly. EECs also moved ventrally, as if they were pressed downward by the sliding of the dorsal ridge above them. At the end of this phase, EECs were completely internalized (Fig. 1l,l’). A few non-EEC CD+ s appeared around EECs (Fig. 2l,l’) and migrated collectively with EECs (Supplementary Movie 3).
Phase V (observation time 163 to 276 min.): EECs and the CD+ cells kept moving posteriorly after being overwhelmed by the epidermal segments. The epidermal sheets fused completely at dorsal midline as the dorsal closure terminated at the end of stage 15. All the EECs expressed CD-GFP at the observation time 234 (Fig. 2m,m’). The number of CD+ cells increased significantly (Supplementary Fig. S5a), and the sac-like structure of the EADP was readily distinguishable (Fig. 2n,n’).
During the observation period, the fluorescence intensity of en > RFP in EECs increased with time before internalization, but slightly decreased thereafter (Supplementary Fig. S5b). In contrast, the CD-GFP intensity in EECs continuously increased with time. Severe fluctuations of fluorescence signals were observed during the head involution because of the rapid morphogenetic movement. It is noteworthy that we did not see any cell division in the EADP throughout the period of observation, indicating that the increase in CD+ cells is not due to cell division but due to the expression of CD (representing eyg transcription). No mitotic cells (stained by anti-phospho-histone H3; pHH3) were detected to co-localize with CD+ cells (Supplementary Fig. S6). No spatial preference of the position of new non-EEC CD+ cells relative to EECs was discerned. The spatial distribution of the subsequently-formed non-EEC CD+ cells do not seem to bear any correlation relative to that of EECs.
Spatiotemporal mapping of CD expression during the course of the EADP formation
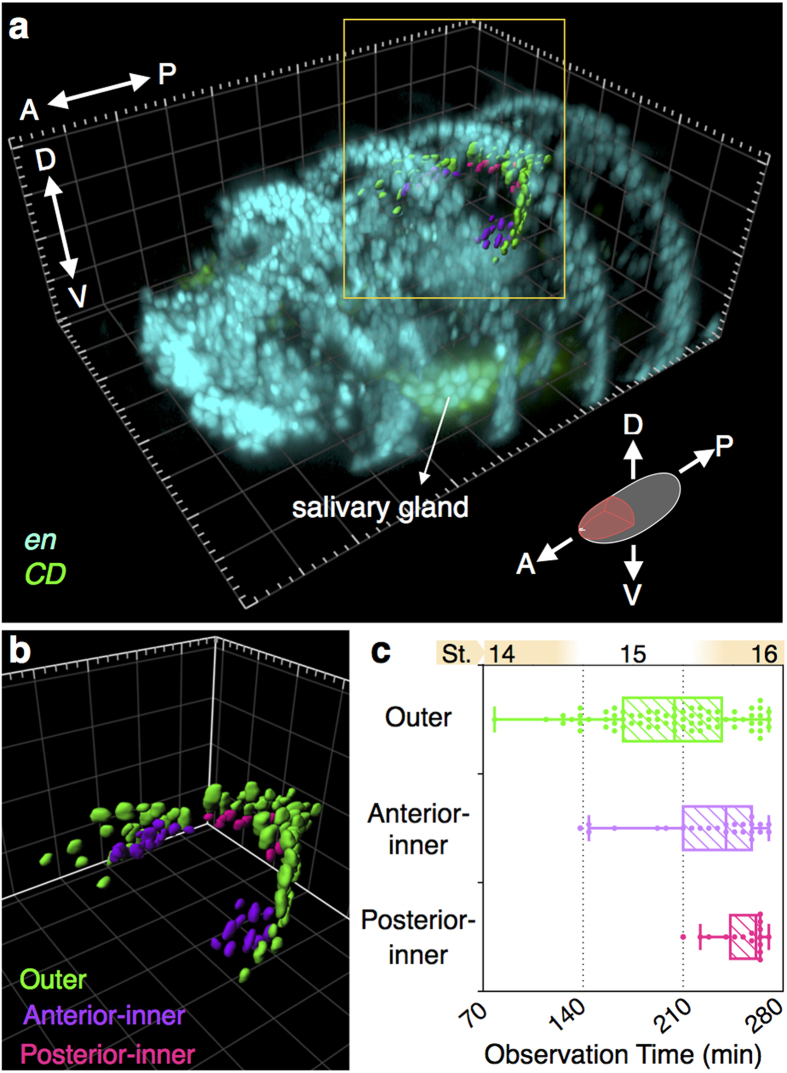
The number of CD+ cells increased progressively without any cell division, suggesting that the cells progressively expressed CD. We examined the spatiotemporal pattern of CD expression during the course of the EADP formation, and analyzed the correlation between the expression timing and spatial distribution within the EADP. In stage 16, the EADP cells can be characterized as belonging to either an outer, an anterior-inner, or a posterior-inner group based on their locations within the the EADP structure (Fig. 3a). From the EADP in stage 16, we back-traced the origin of each CD+ cells in each group and analyzed their onset of CD expression time (Fig. 3b and Supplementary Movie 4). The result shows that most of the cells in the outer layer expressed CD-GFP earlier than those in the inner layer, and cells in the posterior-inner group expressed CD-GFP much later than those in other groups. The relationship between the onset of CD-GFP expression and the spatial distribution of CD+ cells in the EADP was compatible in the three embryos we analyzed (Supplementary Fig. S7). Most, but not all, of the EECs were confined in the outer layer (Fig. 4). Whether the temporal expression of CD determined its final spatial destination, or its location determined the timing of expression, awaits further study.
Figure 3. The spatiotemporal relationship of CD expression in the EADP.
(a) A 3D volume rendering of an en > RFP (cyan) and CD-GFP (green) co-expressing embryo from a tilted view at stage 16. The orientation and the imaging area of the data is displayed as the pink region in the schematic embryo. CD+ EADP cells are shown in 3D surface rendering and categorized into outer, anterior-inner, and posterior-inner groups, which are color-coded in green, purple, and pink, respectively. Some ventral en+ CD+ cells belonged to the salivary gland. (b) An enlarged version of the EADP without showing the en- and CD-expression patterns. (c) The distribution of the onset of CD expression of EADP cells. The data points outside the whiskers are regarded as outliers. Grid size in the background in (a) and (b) is 20 μm. Abbreviation: A, anterior; D, dorsal; St., embryonic stage.
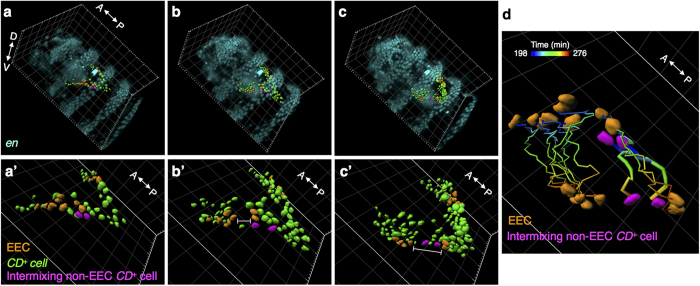
Figure 4. The cell intermixing in EECs during the EADP formation.
(a–c) The 3D volume rendering of fluorescence signals from an en > RFP CD-GFP co-expressing embryo at observation time 198, 240, and 276 min in tilted view. The corresponding embryonic stage was about stage 16. To highlight the EADP, CD+ EADP cells are shown in surface rendering. The dataset is the same as in Fig. 2. (a’–c’) Enlargements of CD+ EADP cells in (a–c), respectively; CD+ EECs are shown in orange, and CD+ cells are shown in green. Two intermixing non-EEC CD+ cells are highlighted in magenta in the left primordium. Gaps between EECs in the left primordium are indicated by the white I-bars in (b’) and (c’). (d) The trajectories of EECs and two intermixing cells from time points 198 to 276 min. The temporal sequence of each trajectory is color-coded. The surface renderings of EECs and the intermixing non-EEC CD+ cells, are labeled in orange and magenta, respectively, at both the starting point and the end point. The trajectories of intermixing non-EEC CD+ cells are highlighted in thicker lines. Grid size in the background of all frames is 20 μm.
EECs allow cell intermixing during the EADP formation
Before the internalization, EECs generally moved collectively as a cluster. However, we observed that in some cases a group of EECs could be trespassed by non-EEC CD+ cells during the EADP morphogenesis (Fig. 4a–c). The trajectory of each cell, examined from a tilted angle, revealed how the non-EEC CD+ cells trespassed the continuity of EECs (Fig. 4d and Supplementary Movie 5). Unfortunately, the severe image blurring, due to the embryonic wriggling at later times, disabled further analysis. Surface rendering, at those time points, of EADP images which were not affected seriously by sample wriggling were selected and reconstructed (Supplementary Fig. S8). The morphological change of the EADP indicates that a folding process had taken place to condense the structure of the EADP.
Discussion
We monitored the course of the EADP formation in Drosophila embryos from stage 13 to stage 16 through long-term 5D live imaging, which provided unprecedented clear observation of the cell dynamics during the EADP formation. Our lineage analysis showed that the CD enhancer from the eyg gene can be used as a faithful marker for the EADP. Although CD expression begins only in stage 15, the use of en > RFP allowed us to trace EECs, hence the formation of the EADP, to as early as stage 13. The position and the migration path of EADP cells were consistent with the expression of ey14,19. Previous studies23,38,39 indicate that various genes, including the eyeless (ey), twin of eyeless (toy), sine oculis (so), eye gone (eyg), homothorax (hth) and teashirt (tsh) are expressed uniformly in the EADP in the first instar larval stage (L1). Gene expressions are not restricted in the eye and the antenna until the second instar larval stage (L2). The EADP has been suggested to be a homogeneous structure in L1, which later segregate to form the eye and the antennal primordium from L2. Thus, specific markers for the eye and the antenna are not available in the embryonic stage.
The origin of EECs in stage 13 is spatially separated from the en antenna stripe and anteriorly adjacent to the optic lobes (Fig. 5a, Supplementary Fig. S9, and Supplementary Movie 6), which is consistent with the area of the presumptive eye field described in previous studies19,40,41. Although the EEC and the en antennal stripe arise at different locations at the first sign of en > RFP that we can detect, we cannot fully rule out the possibility that the two groups of cells share a common origin before en > RFP becomes detectable.
Figure 5. A schematic summary of the location and migration pattern of EECs.
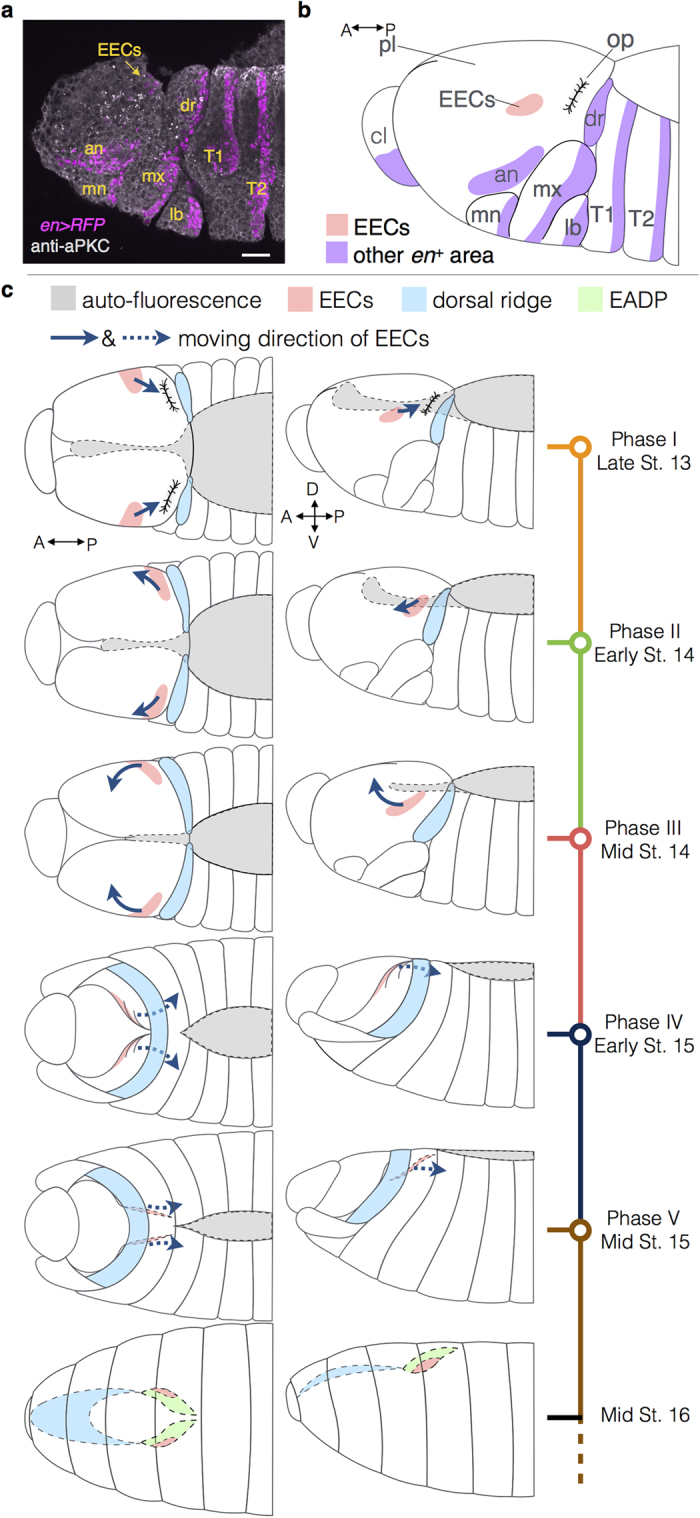
(a) The en expression pattern in lateral view in late stage 13. Scale bar: 20 μm. (b) A schematic diagram of the en expression pattern in (a). EECs and several landmarks, including other en+ sections in procephalic ectoderm (highlighted in purple) and the invagination of optic lobe, are indicated. EECs localize anteriorly adjacent to the optic lobe without connections to other en+ areas. (c) Migration patterns of EECs and the dorsal ridge in different phases in dorsal (left column) and lateral (right column) views. The corresponding embryonic stages are listed below the phase in timeline labels. Solid arrowed lines indicate the direction of EECs’ movement along the surface of the procephalic lobe, whereas the dashed arrowed lines indicate the EECs’ movement in the interior of the embryo, as well as their subsequent interior movement to form part of the EADP (highlighted in green and marked with dashed borders). The areas shaded in gray represent the auto-fluorescence in the amnioserosa and serve as a reference of the embryo morphogenesis in different stages. Anterior is to the left in all figures. Abbreviation: St., embryonic stage, pl, procephalic lobe; op, optic lobe; dr, dorsal ridge; an, antennal; cl, clypeolabral; mn, mandibular; mx, maxillary; lb, labial; T1, the first thoracic segment; T2, the second thoracic segment.
The migration pattern of EECs shows that the invagination of the EADP does not follow the invagination of optic lobes in stage 14 as described previously40, and neither does it through evagination from the dorsal pouch via involution of the dorsal ridge as previously proposed12. Rather, EADP cells move into the interior of the embryo with the aid of the relative motion between the procephalic lobe and the dorsal ridge during the head involution, such that the EADP is pushed under the dorsal ridge. (Fig. 5b).
EECs form a part of the EADP. They were located in a posterior lateral position in the EADP in stage 14 and gradually changed to an anterior lateral position in the EADP. The spatial arrangement of EECs changed from a transverse orientation (approximately parallel to the L-R axis) to a longitudinal orientation (approximately parallel to the A-P axis) during phase I through mid-phase IV before the internalization. We propose a transposition model (Supplementary Fig. S10) to explain why the anterior-posterior orientation of the antenna disc is reversed relative to the body.
After the EADP internalization, we observed in some (but not all) cases that EECs got split into several cell groups due to insertion of non-EEC CD+ cells as they moved posteriorly in stage 15 and 16. The allowance of separation in EECs conflicts with the absence of cell intermixing between compartments reported earlier42,43. The phenomenon suggests that the boundary property of the en-defined posterior compartment may not be strictly retained during the EADP morphogenesis.
Whether the EAD has a single-segmental or multi-segmental origin has been a long unsettled issue7,12,34,35,44. Since en is expressed in the posterior compartment of all embryonic segments, it has been used in the analysis of the segmental origin of the embryonic head region12,35,44. Our G-TRACE results showed that the en+ cells in the embryonic stage contributed to the posterior compartment of the antenna disc and the posterior part of the eye disc (Fig. 1). Relevant to the eye and antenna, en has expression in a pair of antennal stripes and a pair of antennal spots12,34,35; the former was regarded as contributing to the antennae, and the latter was regarded as derived from the en antennal stripes in stage 13. We have not observed the en antennal stripes fuse with the EADP, as defined by CD-GFP expression. EECs are the only en+ cell group in the EADP. In newly hatched first instar larva, the eye and antennal portion of the EAD has 42 and 35 cells, respectively8. In our observation, the number of CD+ cells reached a plateau of 85–90 per primordium in stage 16, while the number of EECs within the primordium remained at about 10 per primordium since stage 15. These observations show that there is no further contribution from non-EEC en+ cells to the formation of the EADP after stage 16. However, non-EEC CD+ cells before late stage 14 were not trackable, so the possibility that non-EEC CD+ cells originated from multi-head segments in an early time cannot be fully excluded. Overall, these results strongly suggest that the EADP is derived from one of the en+ head segments, which is defined by EECs.
Methods
Fly Strains
en2.4-GAL445, UAS-G-TRACE31, and tubP-GAL80ts 32 were from the Bloomington stock center. CD-GFP, CD-GAL4, and F1-GAL4 have been reported22. ey-GAL446 was a gift from Walter Gehring (University of Basel, Switzerland). ubi-GFP-CAAX47 was from the KYOTO Stock Center (DGRC). All fly grown and crosses were performed according to the standard procedure at 25 °C unless otherwise noted.
Experimental procedure for stage-specific G-TRACE analysis via temperature shift
For stage-specific lineage tracing, CD-GAL4; tub-GAL80ts or en-GAL4; tub-GAL80ts virgins were crossed with UAS-G-TRACE males. Flies were allowed to lay eggs for 4 hours in 17 °C or 29 °C in all stage-specific experiments. The lineage distributions in EAD were examined in L3. A diagram to illustrate experimental principles and timelines is shown in Supplementary Fig. S11. Fixation and staining of larval EAD were performed as described previously48. Fixed EAD were imaged by Zeiss LSM780. The morphology of each EAD was determined by differential interference contrast (DIC) image.
Experimental procedure for 5D imaging via dual-illumination light sheet microscopy
To track the en > RFP CD-GFP co-expressing cells, en-Gal4 and CD-GFP; UAS-H2B-RFP were crossed to generate the embryos co-expressing en > RFP and CD-GFP. The embryos were dechorinated at 7 h after egg deposition (AED), which was about embryonic stage 12. The dechorinated embryos were gently mixed with 0.5% agarose with low gelling point agarose (Sigma Aldrich, A9045) at 30˚C. The sample mixture was molded by a capillary with ~1 mm diameter and pushed out after congealing. The mounted embryo was immersed in a PBS-filled chamber and imaged by dual-illumination light sheet microscope at 9 h AED from dorsal view. A 3D image stack was acquired (with an image acquisition time of 40 sec.) every 3 min. The observation and image recording were stopped when the embryos started to wriggle. All imaging experiments were performed at 25 °C.
Image processing and quantification
5D images were processed and presented via Imaris (Bitplane Inc.). EADP cells were identified by the CD-expressing V-shaped structure in the dorsal embryo in stage 16. en > RFP CD-GFP co-expressing cells were distinguished by fluorescence co-localization. Some en > RFP CD-GFP co-expressing cells belonging to the salivary glands and the foregut were not analyzed. EADP cells were manually tracked back to the initial observation time from the image stacks to quantify the CD-expression time and the CD+ cell number in each trackable time point. The inner and the outer layers of EADP cells were classified by their positions in stage 16 when the two-layer structure became obvious. The anterior and posterior sections of the inner layer were demarcated by the waist of the EADP (Supplementary Fig. S7).
Additional Information
How to cite this article: Huang, Y. S. et al. 5D imaging via light sheet microscopy reveals cell dynamics during the eye-antenna disc primordium formation in Drosophila. Sci. Rep. 7, 44945; doi: 10.1038/srep44945 (2017).
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Material
Acknowledgments
This work has been supported by grants from the Ministry of Science and Technology, Taiwan (Project No. MOST 104-2221-E-010-010- to A.C. and MOST 103-2311-B-001-035 to Y.H.S.) and the Top University Project from Taiwan Ministry of Education to A.C. The authors would also like to specially thank the software support from Sixclear, LLC, Austin, TX, USA, the help on imaging facility from Su Ping Lee, Institute of Molecular Biology, Academia Sinica, Taipei, Taiwan, and the consultation on mechanical design from Chih-Hsien Lin, Force Precision Instrument Co., Ltd, Taipei, Taiwan.
Footnotes
The authors declare no competing financial interests.
Author Contributions Y.S.H. designed and established the dual-illumination light sheet microscope. Y.S.H, Y.C.T., C.H.C. and S.H.P. performed in vivo imaging experiments. Y.S.H. and H.Y.K. performed G-TRACE experiments. Y.S.H., Y.C.T., C.H.C. S.H.P and H.Y.K. prepared the samples and maintained the fly strains. Y.S.H. and Y.C.T. processed and analyzed the data. Y.S.H., Y.H.S. and A.C. wrote the manuscript. All authors reviewed the manuscript.
References
- Morata G. How Drosophila appendages develop. Nature Reviews Molecular Cell Biology 2, 89–97 (2001). [DOI] [PubMed] [Google Scholar]
- Hariharan I. K. & Bilder D. Regulation of imaginal disc growth by tumor-suppressor genes in Drosophila. Annu. Rev. Genet. 40, 335–361, doi: 10.1146/annurev.genet.39.073003.100738 (2006). [DOI] [PubMed] [Google Scholar]
- Roignant J.-Y. & Treisman J. E. Pattern formation in the Drosophila eye disc. The International journal of developmental biology 53, 795 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silies M., Yuva-Aydemir Y., Rut Franzdottir S. & Klämbt C. The eye imaginal disc as a model to study the coordination of neuronal and glial development. Fly 4, 71–79, doi: 10.4161/fly.4.1.11312 (2010). [DOI] [PubMed] [Google Scholar]
- Worley M. I., Setiawan L. & Hariharan I. K. Regeneration and transdetermination in Drosophila imaginal discs. Annu. Rev. Genet. 46, 289–310, doi: 10.1146/annurev-genet-110711-155637 (2012). [DOI] [PubMed] [Google Scholar]
- Beira J. V. & Paro R. The legacy of Drosophila imaginal discs. Chromosoma 1–20, doi: 10.1007/s00412-016-0595-4 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haynie J. L. & Bryant P. J. Development of the eye-antenna imaginal disc and morphogenesis of the adult head in Drosophila melanogaster. J. Exp. Zool. 237, 293–308, doi: 10.1002/jez.1402370302 (1986). [DOI] [PubMed] [Google Scholar]
- Mandaravally Madhavan M. & Schneiderman H. Histological analysis of the dynamics of growth of imaginal discs and histoblast nests during the larval development of Drosophila melanogaster. Wilhelm Roux’s archives of developmental biology 183, 269–305, doi: 10.1007/BF00848459 (1977). [DOI] [PubMed] [Google Scholar]
- Struhl G. A homoeotic mutation transforming leg to antenna in Drosophila. Nature 292, 635–638 (1981). [DOI] [PubMed] [Google Scholar]
- Jürgens G., Lehmann R., Schardin M. & Nüsslein-Volhard C. Segmental organisation of the head in the embryo of Drosophila melanogaster. Rouxs Arch. Dev. Biol. 195, 359–377, doi: 10.1007/bf00402870 (1986). [DOI] [PubMed] [Google Scholar]
- Green P., Hartenstein A. Y. & Hartenstein V. The embryonic development of the Drosophila visual system. Cell Tissue Res. 273, 583–598 (1993). [DOI] [PubMed] [Google Scholar]
- Younossi-Hartenstein A., Tepass U. & Hartenstein V. Embryonic origin of the imaginal discs of the head of Drosophila melanogaster. Rouxs Arch. Dev. Biol. 203, 60–73 (1993). [DOI] [PubMed] [Google Scholar]
- Namba R. & Minden J. S. Fate mapping of Drosophila embryonic mitotic domain 20 reveals that the larval visual system is derived from a subdomain of a few cells. Dev. Biol. 212, 465–476, doi: 10.1006/dbio.1999.9349 (1999). [DOI] [PubMed] [Google Scholar]
- Chang T., Mazotta J., Dumstrei K., Dumitrescu A. & Hartenstein V. Dpp and Hh signaling in the Drosophila embryonic eye field. Development 128, 4691–4704 (2001). [DOI] [PubMed] [Google Scholar]
- Garcia‐Bellido A. & Merriam J. R. Cell lineage of the imaginal discs in Drosophila gynandromorphs. J. Exp. Zool. 170, 61–75 (1969). [DOI] [PubMed] [Google Scholar]
- Wieschaus E. & Gehring W. Gynandromorph analysis of the thoracic disc primordia in Drosophila melanogaster. Wilhelm Roux’s archives of developmental biology 180, 31–46, doi: 10.1007/bf00848883 (1976). [DOI] [PubMed] [Google Scholar]
- Struhl G. A blastoderm fate map of compartments and segments of the Drosophila head. Dev. Biol. 84, 386–396, doi: 10.1016/0012-1606(81)90407-3 (1981). [DOI] [PubMed] [Google Scholar]
- Cheyette B. N. R. et al. The Drosophila sine oculis locus encodes a homeodomain-containing protein required for the development of the entire visual system. Neuron 12, 977–996, doi: 10.1016/0896-6273(94)90308-5 (1994). [DOI] [PubMed] [Google Scholar]
- Daniel A., Dumstrei K., Lengyel J. A. & Hartenstein V. The control of cell fate in the embryonic visual system by atonal, tailless and EGFR signaling. Development 126, 2945–2954 (1999). [DOI] [PubMed] [Google Scholar]
- Jang C.-C. et al. Two Pax genes, eye gone and eyeless, act cooperatively in promoting Drosophila eye development. Development 130, 2939–2951 (2003). [DOI] [PubMed] [Google Scholar]
- Yao J.-G. et al. Differential requirements for the Pax6(5a) genes eyegone and twin of eyegone during eye development in Drosophila. Dev. Biol. 315, 535–551, doi: 10.1016/j.ydbio.2007.12.037 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L.-H., Chiu S.-J. & Sun Y. H. Temporal switching of regulation and function of eye gone (eyg) in Drosophila eye development. Dev. Biol. 321, 515–527, doi: 10.1016/j.ydbio.2008.06.038 (2008). [DOI] [PubMed] [Google Scholar]
- Kumar J. P. & Moses K. Expression of evolutionarily conserved eye specification genes during Drosophila embryogenesis. Dev. Genes Evol. 211, 406–414 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halder G. et al. Eyeless initiates the expression of both sine oculis and eyes absent duringDrosophilacompound eye development. Development 125, 2181–2191 (1998). [DOI] [PubMed] [Google Scholar]
- Quiring R., Walldorf U., Kloter U. & Gehring W. Homology of the eyeless gene of Drosophila to the small eye gene in mice and Aniridia in humans. Science 265, 785–789, doi: 10.1126/science.7914031 (1994). [DOI] [PubMed] [Google Scholar]
- Fuse N., Hirose S. & Hayashi S. Diploidy of Drosophila imaginal cells is maintained by a transcriptional repressor encoded by escargot. Genes Dev. 8, 2270–2281, doi: 10.1101/gad.8.19.2270 (1994). [DOI] [PubMed] [Google Scholar]
- Fuse N., Hirose S. & Hayashi S. Determination of wing cell fate by the escargot and snail genes in Drosophila. Development 122, 1059–1067 (1996). [DOI] [PubMed] [Google Scholar]
- Huisken J., Swoger J., Bene F. D., Wittbrodt J. & Stelzer E. H. K. optical sectioning deep inside live embryo by selective plane illumination microscopy. Science 305, 3, doi: 10.1126/Science.1100035 (2004). [DOI] [PubMed] [Google Scholar]
- Weber M. & Huisken J. Light sheet microscopy for real-time developmental biology. Curr. Opin. Genet. Dev. 21, 1–7, doi: 10.1016/j.gde.2011.09.009 (2011). [DOI] [PubMed] [Google Scholar]
- Ito K., Awano W., Suzuki K., Hiromi Y. & Yamamoto D. The Drosophila mushroom body is a quadruple structure of clonal units each of which contains a virtually identical set of neurones and glial cells. Development 124, 761–771 (1997). [DOI] [PubMed] [Google Scholar]
- Evans C. J. et al. G-TRACE: rapid Gal4-based cell lineage analysisIn Drosophila. Nat. Methods 6, 603–605, doi: 10.1038/nmeth.1356 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGuire S. E., Le P. T., Osborn A. J., Matsumoto K. & Davis R. L. Spatiotemporal rescue of memory dysfunction in Drosophila. Science 302, 1765–1768, doi: 10.1126/science.1089035 (2003). [DOI] [PubMed] [Google Scholar]
- Urbach R., Schnabel R. & Technau G. M. The pattern of neuroblast formation, mitotic domains and proneural gene expression during early brain development in Drosophila. Development 130, 3589–3606, doi: 10.1242/dev.00528 (2003). [DOI] [PubMed] [Google Scholar]
- Schmidt-Ott U. & Technau G. M. Expression of en and wg in the embryonic head and brain of Drosophila indicates a refolded band of seven segment remnants. Development 116, 111–125 (1992). [DOI] [PubMed] [Google Scholar]
- Rogers B. T. & Kaufman T. C. Structure of the insect head as revealed by the EN protein pattern in developing embryos. Development 122, 3419–3432 (1996). [DOI] [PubMed] [Google Scholar]
- Jones N. A., Kuo Y. M., Sun Y. H. & Beckendorf S. K. The Drosophila Pax gene eye gone is required for embryonic salivary duct development. Development 125, 4163–4174 (1998). [DOI] [PubMed] [Google Scholar]
- Ntini E. & Wimmer E. A. Unique establishment of procephalic head segments is supported by the identification of cis-regulatory elements driving segment-specific segment polarity gene expression in Drosophila. Dev. Genes Evol. 221, 1–16, doi: 10.1007/s00427-011-0354-8 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C.-W. & Sun Y. H. Segregation of eye and antenna fates maintained by mutual antagonism in Drosophila. Development 139, 3413–3421, doi: 10.1242/dev.078857 (2012). [DOI] [PubMed] [Google Scholar]
- Singh A., Kango-Singh M. & Sun Y. H. Eye suppression, a novel function of teashirt, requires Wingless signaling. Development 129, 4271–4280 (2002). [DOI] [PubMed] [Google Scholar]
- Chang T., Shy D. & Hartenstein V. Antagonistic relationship between Dpp and EGFR signaling in Drosophila head patterning. Dev. Biol. 263, 103–113, doi: 10.1016/S0012-1606(03)00448-2 (2003). [DOI] [PubMed] [Google Scholar]
- Friedrich M. Ancient mechanisms of visual sense organ development based on comparison of the gene networks controlling larval eye, ocellus, and compound eye specification in Drosophila. Arthropod Structure & Development 35, 357–378, doi: 10.1016/j.asd.2006.08.010 (2006). [DOI] [PubMed] [Google Scholar]
- Martin A. C. & Wieschaus E. F. Tensions divide. Nat. Cell Biol. 12, 5–7, doi: 10.1038/ncb0110-5 (2010). [DOI] [PubMed] [Google Scholar]
- Monier B., Pelissier-Monier A., Brand A. H. & Sanson B. An actomyosin-based barrier inhibits cell mixing at compartmental boundaries in Drosophila embryos. Nat. Cell Biol. 12, 60–65, doi: 10.1038/ncb2005 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt-Ott U., González-Gaitán M. & Technau G. M. Analysis of neural elements in head-mutant Drosophila embryos suggests segmental origin of the optic lobes. Rouxs Arch. Dev. Biol. 205, 31–44, doi: 10.1007/bf00188841 (1995). [DOI] [PubMed] [Google Scholar]
- Tabata T., Schwartz C., Gustavson E., Ali Z. & Kornberg T. B. Creating a Drosophila wing de novo, the role of engrailed, and the compartment border hypothesis. Development 121, 3359–3369 (1995). [DOI] [PubMed] [Google Scholar]
- Hauck B., Gehring W. J. & Walldorf U. Functional analysis of an eye specific enhancer of the eyeless gene in Drosophila. Proc. Natl. Acad. Sci. USA 96, 564–569 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo T. & Hayashi S. Mitotic cell rounding accelerates epithelial invagination. Nature 494, 125–129, doi: 10.1038/nature11792 (2013). [DOI] [PubMed] [Google Scholar]
- Pai C.-Y. et al. The Homothorax homeoprotein activates the nuclear localization of another homeoprotein, Extradenticle, and suppresses eye development in Drosophila. Genes Dev. 12, 435–446 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.