Abstract
There is widespread transcriptional dysregulation in Huntington’s disease (HD) brain, but analysis is inevitably limited by advanced disease and postmortem changes. However, mutant HTT is ubiquitously expressed and acts systemically, meaning blood, which is readily available and contains cells that are dysfunctional in HD, could act as a surrogate for brain tissue. We conducted an RNA-Seq transcriptomic analysis using whole blood from two HD cohorts, and performed gene set enrichment analysis using public databases and weighted correlation network analysis modules from HD and control brain datasets. We identified dysregulated gene sets in blood that replicated in the independent cohorts, correlated with disease severity, corresponded to the most significantly dysregulated modules in the HD caudate, the most prominently affected brain region, and significantly overlapped with the transcriptional signature of HD myeloid cells. High-throughput sequencing technologies and use of gene sets likely surmounted the limitations of previously inconsistent HD blood expression studies. Our results suggest transcription is disrupted in peripheral cells in HD through mechanisms that parallel those in brain. Immune upregulation in HD overlapped with Alzheimer’s disease, suggesting a common pathogenic mechanism involving macrophage phagocytosis and microglial synaptic pruning, and raises the potential for shared therapeutic approaches.
Huntington’s disease (HD), the most common monogenic neurodegenerative disorder in the developed world1, is caused by a CAG repeat expansion in the HTT gene and is characterised by motor, cognitive and psychiatric features. Onset occurs around 45 years on average and inversely correlates with CAG repeat length2. The disease progresses inexorably and, with the exception of late-onset cases, is uniformly fatal a median of 18 years from motor onset3. HD is currently incurable and no treatments slow progression.
HD research has traditionally focused on the brain due to the presence of characteristic mutant huntingtin protein aggregates4 and because the prominent symptoms and signs can be linked to neurodegeneration in the basal ganglia and cerebral cortex5. However, mutant HTT is ubiquitously expressed6 and mounting evidence suggests it has direct effects in peripheral tissues5,7, though whether these effects are distinct, or parallel those in the brain remains unclear. HD patients demonstrate peripheral immune dysfunction presymptomatically8,9,10,11, as well as weight loss that leads to cachexia with advancing disease7. There is progressive muscle wasting12, endocrine dysfunction13 liver impairment7, and cardiac dysfunction14,15,16. Mutant HTT protein aggregates can be found in the peripheral tissues of HD mice17, as well as advanced patients18. These peripheral features may contribute to CNS pathology, disease progression and mortality5,7, and strongly suggest that HD is a systemic disorder. This peripheral phenotype provides an opportunity to study mutant huntingtin’s pathogenic mechanisms. In contrast to brain tissue, availability of which is limited and from post-mortem subjects with end-stage disease19,20, peripheral tissues can be sampled minimally invasively and inexpensively from living patients, enabling longitudinal study throughout disease course.
Transcriptional dysregulation is a central feature of HD pathogenesis21. However, studies of gene expression changes in HD blood have been inconsistent. Using microarray technology, Borovecki, et al.22 identified 12 upregulated transcripts, seven of which were also upregulated in brain. However, subsequent studies did not replicate these results23,24,25. Using tag-based serial analysis of gene expression (SAGE), Mastrokolias, et al.25 found 167 genes differentially expressed by motor score, 40 of which had previously been reported in at least one microarray study.
In the current study we present a transcriptomic analysis of whole blood in human HD using RNA sequencing (RNA-Seq). We studied differential expression of individual gene transcripts and enrichment of differential expression in gene sets in two independent cohorts from Track-HD26 and Leiden. We then investigated whether transcriptional changes seen in blood parallel those from previous studies in HD brain. There was significant dysregulation of brain Weighted Gene Correlation Network Analysis (WGCNA) modules in the same direction in blood, as well as significant dysregulation of pathways. Immune gene sets were notably upregulated in both analyses and this signal overlapped with the transcriptional signature of Alzheimer’s disease (AD) brain.
Results
No differential expression of individual transcripts in HD whole blood between disease stages or states
Attempting to identify both HD specific and stage-specific changes in gene expression (mRNA) level, we compared premanifest, manifest and control subjects, whilst controlling for age and gender. Premanifest gene carriers had a mean total motor score (TMS) of 2 and total functional capacity (TFC) of 13 (Table 1), indicating no substantial motor signs. Manifest subjects demonstrated motor abnormalities that were unequivocal signs of HD. No transcripts were significantly differentially expressed (FDR < 0.05) between premanifest and manifest HD in either the Track-HD26 or the independently collected Leiden cohort, or when these cohorts were combined (results not shown). As expression changes did not differ significantly between disease stages, all mutant HTT gene carriers were combined to increase the analytical power in a comparison of HD and controls. Once again there were no individually significant transcripts in independent or combined cohorts, but the differential expression analysis in the combined cohort is given in Table S1.
Table 1. Track-HD and Leiden cohorts for RNA-Seq analysis.
| Cohort | Group | n | Mean age, y ± SD (range) | Gender (male/female) | Mean (CAG)n length ± SD (range) | Mean TMS ± SD (range) | Mean TFC ± SD (range) |
|---|---|---|---|---|---|---|---|
| Track-HD | Premanifest | 50 | 42 ± 9 (22–64) | 24/26 | 43 ± 3 (39–52) | 2 ± 2 (0–8) | 13 ± 0 (12–13) |
| Manifest | 62 | 48 ± 10 (23–64) | 26/36 | 44 ± 3 (39–59) | 23 ± 11 (5–45) | 11 ± 2 (7–13) | |
| HD | 112 | 46 ± 10 (22–64) | 50/62 | 44 ± 3 (39–59) | 14 ± 13 (0–45) | 12 ± 2 (7–13) | |
| Control | 22 | 45 ± 5 (34–53) | 9/13 | — | — | — | |
| Leiden | Premanifest | 18 | 46 ± 10 (29–63) | 5/13 | 42 ± 2 (39–47) | 3 ± 2 (0–5) | 12 ± 1 (10–13) |
| Manifest | 56 | 55 ± 11 (35–79) | 29/27 | 44 ± 3 (39–53) | 42 ± 30 (6–102) | 7 ± 5 (0–13) | |
| HD | 74 | 53 ± 11 (29–79) | 34/40 | 44 ± 3 (39–53) | 32 ± 31 (0–102) | 8 ± 5 (0–13) | |
| Control | 27 | 43 ± 11 (26–65) | 13/14 | — | — | — | |
| Combined | HD | 186 | 48 ± 11 (22–79) | 84/102 | 44 ± 3 (39–59) | 21 ± 24 (0–102) | 10 ± 4 (0–13) |
| Control | 49 | 44 ± 9 (26–65) | 22/27 | — | — | — |
Manifest subjects demonstrated motor abnormalities that were unequivocal signs of HD. Premanifest gene carriers had a total motor score of 5 or lower and a diagnostic confidence score (DCS) less than 4 on the UHDRS, indicating no substantial motor signs. The HD group consists of the combined premanifest and manifest subjects. Controls were matched for age and gender. Age and clinical scores considered for the analysis were at time of blood collection. SD – standard deviation; TFC – Total Functional Capacity; TMS – Total Motor Score.
Pathways are dysregulated in HD blood compared with controls
We next asked whether networks of genes with similar functional annotation were dysregulated in HD relative to controls. Pathway annotations were collated from publicly available gene ontology databases to form a set of generic pathways using the same method as the recent HD genome-wide association study (GWAS) of modifiers of age at onset27 (see Materials and Methods). The number of pathways significantly dysregulated in both Track-HD and Leiden blood datasets was significantly higher than would be expected by chance (Table 2). Our findings indicate shared biology between the two independent cohorts despite differences in demographic and disease stage; Leiden subjects were on average 7 years older and had correspondingly higher TMS (mean 32 versus 14 in Track-HD) and lower TFC (mean 8 versus 12 in Track-HD). The significance of the overlap was greatly increased in analyses specifying the direction of dysregulation (increased or decreased expression) (Table 2). Therefore, directional analyses were used in the combined dataset as the primary analysis.
Table 2. Overlap analysis of Track-HD and Leiden cohorts shows that a significant excess of pathways are associated with HD (p < 0.05) in both datasets.
| Reference dataset | Comparison dataset | Direction of dysregulation in HD | Number of pathways significant in both datasets (p value) |
||
|---|---|---|---|---|---|
| Generic pathways | HD brain modules | Control brain modules | |||
| Leiden | Track-HD | Nondirectional | 69 (4.6E-02) | — | — |
| Downregulated | 130 (<1.0E-03) | 4 (1.1E-01) | 24 (<1.0E-03) | ||
| Upregulated | 219 (<1.0E-03) | 9 (<1.0E-03) | 23 (<1.0E-03) | ||
| Track-HD | Leiden | Nondirectional | 69 (1.4E-01) | — | — |
| Downregulated | 130 (1.7E-02) | 4 (3.5E-02) | 24 (<1.0E-03) | ||
| Upregulated | 217 (<1.0E-03) | 10 (<1.0E-03) | 21 (<1.0E-03) | ||
Significance of overlap is greatest when directionality is taken into account. There is an excess of significantly enriched pathways and modules in the reference dataset conditional on the pathway being enriched (p < 0.05) in the comparison dataset. The generic pathways gene set is collated from publicly-available databases including GO and KEGG. HD brain modules are derived from Neueder and Bates30. Control brain modules are from the Braineac32 and Gibbs, et al.33 expression datasets.
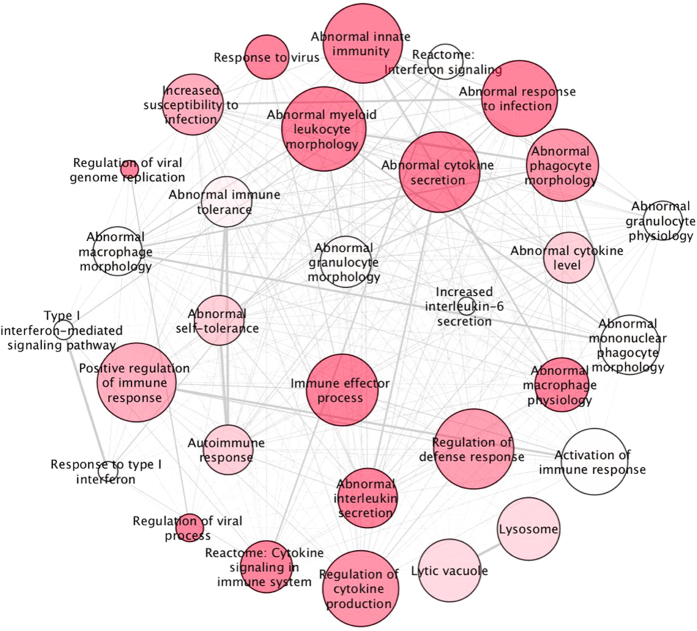
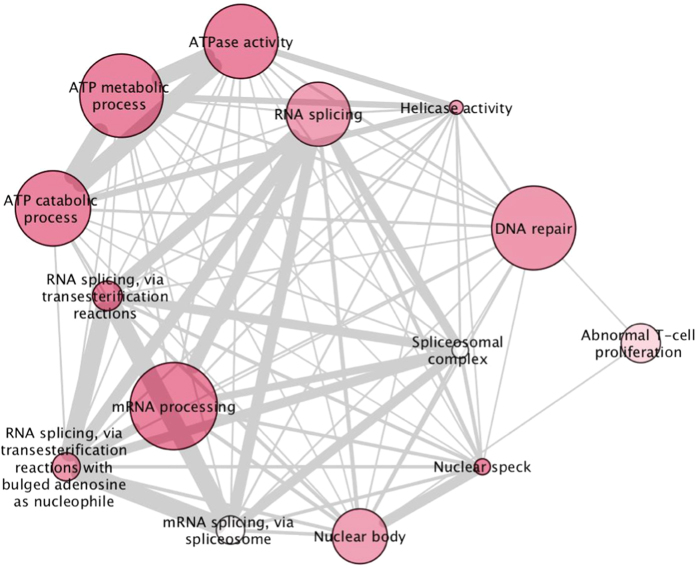
Gene set enrichment analysis (GSEA), with a false discovery rate (q-value) threshold of q < 0.05 to correct for multiple testing, identified 53 upregulated (Fig. 1 and Table S2) and 14 downregulated pathways (Fig. 2 and Table S3) that are at least nominally significant in both cohorts. Multiple immune-related pathways were upregulated, and RNA processing, ATP metabolism and DNA repair were notably downregulated. The 10 most significant pathways for each direction of dysregulation are given in Table 3 and the full list of significant pathways in Tables S2 and S3. The 10 most dysregulated genes (p < 0.01) from the significantly up or downregulated generic pathways (q < 0.05) are listed in Table S4, and a complete list of genes (p < 0.05) in all nominally significant pathways (p < 0.05) is given in Table S5. Notably, the significantly upregulated pathways contain some of the most differentially expressed transcripts (Table S1), with several more contained in pathways reaching nominal significance (p < 0.05) for dysregulation (Table S5). Genes highlighted by MGI pathways appear distinct from other pathway databases, likely because they are based on knockout studies in mice.
Figure 1. Upregulated pathways in HD versus control blood.
Schematic representation of pathways collated from publicly available databases that are significantly upregulated in HD versus controls after correction for multiple testing (q < 0.05). Modules with similar gene content and functional annotation have been consolidated. Nodal shading is inversely proportional to false discovery rate threshold (q value); deep shades have low q values and pale shading is close to the 5% threshold. The weight of connecting lines is proportional to the number of genes shared between pathways.
Figure 2. Downregulated pathways in HD versus control blood.
Schematic representation of pathways collated from publicly available databases that are significantly downregulated in HD versus controls after correction for multiple testing (q < 0.05). Modules with similar gene content and functional annotation have been consolidated. Nodal shading is inversely proportional to false discovery rate threshold (q value); deep shades have low q values and pale shading is close to the 5% threshold. The weight of connecting lines is proportional to the number of genes shared between pathways.
Table 3. The 10 most significantly up and downregulated ‘generic’ pathways in HD versus control blood GSEA.
| Direction of dysregulation in HD | Pathway | Number of dysregulated genes | p (combined) | q (combined) | p (Track-HD) | p (Leiden) | Description |
|---|---|---|---|---|---|---|---|
| Upregulated | MGI: 2419 | 434 | 3.03E-10 | 4.32E-06 | 5.10E-05 | 3.01E-05 | Abnormal Innate Immunity |
| MGI: 3009 | 432 | 5.78E-09 | 4.13E-05 | 5.96E-06 | 1.65E-04 | Abnormal Cytokine Secretion | |
| GO: 50792 | 117 | 2.59E-08 | 1.23E-04 | 1.12E-02 | 7.24E-05 | Regulation Of Viral Process | |
| GO: 9615 | 208 | 1.22E-07 | 4.36E-04 | 3.06E-02 | 5.34E-06 | Response To Virus | |
| MGI: 2451 | 278 | 1.68E-07 | 4.80E-04 | 1.26E-02 | 9.51E-06 | Abnormal Macrophage Physiology | |
| GO: 19221 | 308 | 2.38E-07 | 5.45E-04 | 4.60E-05 | 1.71E-04 | Cytokine-Mediated Signaling Pathway | |
| GO: 2252 | 365 | 3.10E-07 | 5.45E-04 | 7.01E-03 | 1.14E-04 | Immune Effector Process | |
| MGI: 5025 | 406 | 3.44E-07 | 5.45E-04 | 5.91E-05 | 2.02E-04 | Abnormal Response To Infection | |
| MGI: 1793 | 372 | 4.33E-07 | 5.82E-04 | 5.93E-05 | 2.42E-04 | Altered Susceptibility To Infection | |
| MGI: 8568 | 305 | 4.49E-07 | 5.82E-04 | 4.79E-05 | 6.25E-05 | Abnormal Interleukin Secretion | |
| Downregulated | GO: 8380 | 282 | 5.22E-08 | 7.45E-04 | 4.25E-05 | 7.24E-05 | RNA splicing |
| GO: 6397 | 359 | 2.38E-07 | 1.70E-03 | 1.48E-04 | 4.14E-04 | mRNA processing | |
| GO: 16887 | 329 | 1.37E-06 | 5.48E-03 | 1.96E-04 | 3.34E-02 | ATPase activity | |
| GO: 6200 | 333 | 1.54E-06 | 5.48E-03 | 2.42E-04 | 3.36E-02 | ATP catabolic process | |
| GO: 46034 | 361 | 5.36E-06 | 1.53E-02 | 1.74E-04 | 4.45E-02 | ATP metabolic process | |
| GO: 16607 | 144 | 9.06E-06 | 2.15E-02 | 4.68E-04 | 4.61E-03 | Nuclear speck | |
| GO: 6281 | 356 | 1.66E-05 | 2.75E-02 | 2.00E-03 | 1.18E-04 | DNA repair | |
| GO: 16604 | 271 | 2.08E-05 | 2.75E-02 | 5.59E-03 | 2.46E-03 | Nuclear Body | |
| GO: 4386 | 135 | 2.12E-05 | 2.75E-02 | 2.83E-02 | 4.81E-02 | Helicase Activity | |
| GO: 375 | 184 | 2.40E-05 | 2.86E-02 | 1.14E-03 | 2.05E-03 | RNA splicing, via transesterification reactions |
A total of 14,706 Generic pathways, each containing between 3 and 500 genes, were collated from publicly-available databases including GO and KEGG. Pathways are significantly dysregulated after multiple testing correction (q < 0.05). Enrichment p values in the current study for the Track-HD, Leiden and combined datasets are shown.
Pathway dysregulation in HD whole blood overlaps with HD myeloid cells
Through RNA-Seq, Miller, et al.28 identified transcriptional dysregulation in unstimulated monocytes from HD cases relative to controls. Their GSEA used the same set of generic pathways used here. We found a significant excess of pathways to be significantly (p < 0.05) enriched for dysregulation in both Miller, et al.28 and the combined TRACK-HD and Leiden whole blood data (Table S6). This overlap was attributable to a significant excess of pathways enriched for upregulation in both datasets. Overlap in downregulated pathways was not significantly larger than expected by chance. Pathways significantly (p < 0.05) enriched for up and downregulation in both myeloid and whole blood are listed in Tables S7 and S8. Pathways that are significantly enriched for upregulation relate mainly to immunity.
Gene co-expression modules from HD striatum are significantly enriched for dysregulation in HD blood
A limitation of using curated pathways from databases is the incomplete or incorrect annotation. One way to overcome this is to use gene co-expression, because genes that are co-expressed often have related functions. WGCNA identifies clusters (modules) of genes with highly correlated expression, constructing original, unbiased gene co-expression networks based on observed data29. HD brain expression modules were generated by Neueder and Bates30, who applied WGCNA to Hodges, et al.31 data and annotated each module that was associated with HD disease status. To further fill the annotation gap and better define functional biological pathways, we generated co-expression modules for control brain from the Braineac32 and Gibbs, et al.33 datasets.
GSEA for brain co-expression modules was applied to our combined Track-HD and Leiden blood expression dataset. Immune- and inflammatory-related brain modules were upregulated in HD blood, and notable downregulated modules included synaptic function, proteasomal degradation, mitochondrial function and transcription. The 10 most significantly up and downregulated modules in the combined dataset that were also nominally significant (p < 0.05) in both independent cohorts are given in Table 4, and the full list of modules nominally significant in both datasets in Table S9. A list of genes from the modules in Table S9 that are themselves nominally significantly dysregulated (p < 0.05) in the combined dataset is given in Table S10. In addition to reinforcing the biological conclusions from our pathway analysis, the significantly dysregulated modules from Table 4 also share genes with the top pathways, as shown in Supplementary Figures S1 and S2. We then investigated whether gene sets that are dysregulated in HD brain30 are also disrupted in peripheral blood. Table 5 lists the modules that were significantly dysregulated (after correcting for multiple testing of modules) in both HD brain30 and in our combined Track-HD and Leiden blood expression dataset. The direction of dysregulation in brain is shown by the correlation between the module eigengene and HD status (with a positive correlation corresponding to upregulation in the HD brain). Notably, two of the most significantly dysregulated modules in HD caudate30 were also significantly dysregulated in the same direction in blood (Table 5), not only in the combined dataset, but in each of the Track-HD and Leiden datasets independently; these being module 48 (CNpos2), which is upregulated in HD, and module 66 (CNneg1), which is downregulated.
Table 4. The 10 most significantly up and downregulated WGCNA brain expression modules in HD versus control blood.
| Direction of dysregulation in HD | Brain expression gene set | Module | Brain region | Annotation | Number of genes | p (Combined) | p (Track-HD) | p (Leiden) |
|---|---|---|---|---|---|---|---|---|
| Upregulated | HD | 111 | FC BA9 | Immune response | 514 | 7.8E-12 | 1.3E-04 | 7.5E-05 |
| HD | 69 (FC4pos1) | FC BA4 | Inflammatory response | 712 | 3.8E-08 | 3.1E-05 | 1.3E-03 | |
| Control (Braineac) | 712 | TCTX | Inflammatory response | 213 | 1.4E-07 | 3.4E-05 | 8.1E-04 | |
| HD | 48 (CNpos2)* | CN | Lipid metabolism/regulation of transcription | 1785 | 2.0E-07 | 3.9E-03 | 6.3E-03 | |
| Control (Braineac) | 110 | FCTX | Inflammatory response | 173 | 8.9E-07 | 1.0E-03 | 2.5E-03 | |
| Control (Braineac) | 909 | White Matter | Activation of immune response | 265 | 2.1E-06 | 1.2E-03 | 2.5E-02 | |
| Control (Braineac) | 610 | Substantia Nigra | Inflammatory response | 178 | 1.2E-05 | 8.6E-04 | 5.6E-04 | |
| Control (Braineac) | 811 | Thalamus | Inflammatory response | 142 | 1.6E-05 | 3.9E-03 | 2.9E-03 | |
| Control (Gibbs) | 56 | Pons | Lipoprotein/ immune response /GTPase regulator activity | 207 | 2.0E-05 | 2.4E-04 | 4.2E-02 | |
| Control (Braineac) | 911 | White Matter | Inflammatory response | 159 | 3.0E-05 | 8.4E-04 | 1.4E-02 | |
| Downregulated | Control (Gibbs) | 22 | CB | Pro-rich region | 831 | 1.8E-08 | 2.5E-03 | 2.1E-02 |
| Control (Gibbs) | 28 | FC | Intra-cellular transport/mitochondrion | 3178 | 2.1E-08 | 6.3E-04 | 7.7E-05 | |
| Control (Braineac) | 304 | Medulla | mRNA metabolic process | 1811 | 2.9E-08 | 5.0E-15 | 4.0E-02 | |
| HD | 66 (CNneg1)* | CN | Synapse/ion channels | 2645 | 2.7E-07 | 1.5E-04 | 2.1E-02 | |
| Control (Braineac) | 804 | Thalamus | Regulation of cell morphogenesis | 857 | 1.3E-06 | 4.0E-02 | 4.1E-04 | |
| Control (Braineac) | 522 | Putamen | Regulation of RNA splicing | 64 | 4.4E-06 | 6.3E-03 | 2.7E-04 | |
| Control (Gibbs) | 74 | Pons | Transcription/acetylation/protein transport | 1183 | 9.2E-06 | 3.9E-08 | 7.4E-04 | |
| Control (Braineac) | 702 | TCTX | Antigen processing: ubiquitination and proteasome degradation | 4602 | 3.9E-04 | 1.2E-03 | 2.5E-02 | |
| Control (Gibbs) | 48 | FC | Transcription corepressor/cell morphogenesis | 648 | 4.7E-04 | 7.8E-03 | 2.1E-02 | |
| Control (Braineac) | 202 | Hippocampus | Mitochondrial membrane | 2737 | 4.8E-04 | 1.2E-07 | 1.5E-02 |
All modules in this table are significantly dysregulated after correction for multiple testing (q < 0.05) in the combined blood sample. HD brain modules were defined by Neueder and Bates30, and Control brain modules were generated from Braineac32 and Gibbs, et al.33. Neueder and Bates30 module identifiers are given in brackets where available. *Denotes the caudate modules that were highly positively and negatively Neueder and Bates30. CN – caudate nucleus; FC – frontal cortex; FC BA4 – BA4 region of the frontal cortex; FC BA9 – BA9 region of the frontal cortex; CB – cerebellum; TCTX – temporal cortex.
Table 5. Brain expression modules significantly dysregulated both in HD brain and HD blood.
| Module number | Brain Region | Module name | Number of genes | p (combined) | p (TRACK) | p (Leiden) | cor (HD brain) | p (HD brain) | Description |
|---|---|---|---|---|---|---|---|---|---|
| 69 | FC_BA4 | FC4pos1 | 712 | 3.77E-08 | 3.05E-05 | 1.32E-03 | 0.610 | 3.77E-03 | Inflammatory response |
| 48 | CN | CNpos2 | 1785 | 2.03E-07 | 3.85E-03 | 6.33E-03 | 0.724 | 2.21E-11 | Lipid metabolism/regulation of transcription |
| 64 | CN | CNpos6 | 114 | 3.13E-04 | 1.18E-02 | 3.80E-02 | 0.463 | 2.28E-04 | Inflammatory response |
| 66 | CN | CNneg1 | 2644 | 2.71E-07 | 1.51E-04 | 2.13E-02 | −0.800 | 6.03E-15 | Synapse |
All modules in this table are significantly dysregulated after correction for multiple testing (q < 0.05) in the combined blood sample, and are nominally significantly dysregulated (p < 0.05) in both Track-HD and Leiden datasets separately. Cor(HD brain) – the correlation between module eigengene and HD status observed by Neueder and Bates30 in brain expression data, with a positive correlation corresponding to upregulation in HD. p(HD brain) is the p-value for that correlation (corrected for multiple testing of modules).
The module membership (kME) of a gene is measured by the correlation of its expression with the eigengene, which is representative of all gene expression profiles in the module 34; highly connected ‘hub’ genes have high kME values. Interestingly, among genes in module 48 (CNpos2), the Neueder and Bates30 HD caudate module that was also significantly upregulated in blood, there was a significant (p = 7.6 × 10–4) correlation between dysregulation p-value in the direction of interest (positive) in HD blood and degree of module membership (kME)30. This suggests that highly connected “hub” genes in this module may play a role in transcriptional dysregulation in HD. A similar, although much stronger, effect was noted in caudate30. There was no significant correlation in module 66 (CNneg1). Genes in module 48 (CNpos2) that are dysregulated (p < 0.05) in both blood and caudate are shown in Table S11, ranked by their kME value.
Expression changes in HD blood replicate those in HD prefrontal cortex
Labadorf, et al.35 identified dysregulated expression of immune and developmental genes in human HD postmortem prefrontal cortex (BA9). Fold changes in expression of individual genes in the combined Track-HD and Leiden data were compared to those observed in Labadorf, et al.35, and were found to be in the same direction for 8,425 out of the 15,834 genes present in both datasets. This is a highly significant (p < 2.2 × 10−16) excess (see Materials and Methods), suggesting some concordance in signal at the individual gene level. Furthermore, a significant excess of generic pathways was found to be significantly (p < 0.05) dysregulated in both datasets, most markedly in the positive (p < 0.001) direction, but also negative (p = 0.028), thus showing an overlap in biological signal. Pathways significantly upregulated in both datasets are mainly related to immune response (Table S12), a pattern also observed in the upregulated brain co-expression modules (Table S13). Pathways downregulated in both datasets are shown in Table S14, with modules in Table S15. Notably, several modules related to the synapse and neuron projection are downregulated in both datasets. The two HD-related caudate modules from Neueder and Bates30 that were significantly dysregulated in blood were also significantly dysregulated in the same direction in Labadorf, et al.35. Module 48 (CNpos2) was significantly upregulated (p < 1 × 10−16, Table S13) and module 66 (CNneg1) significantly downregulated (p < 1 × 10−16, Table S15), as are several other significant modules from Neueder and Bates30.
Pathways dysregulated in the blood of HD subjects are associated with motor score
We investigated the effect of disease severity by testing for correlation between gene expression and UHDRS total motor score (TMS) in the 112 gene positive Track-HD subjects (Table S16). After correcting for multiple testing, expression of phosphatidylcholine transfer protein (PCTP) was significantly positively correlated with TMS. However, this gene was not found to be significantly correlated with TMS by Mastrokolias et al.25.
We then tested whether generic pathways that were significantly enriched for upregulated (Table S2) or downregulated (Table S3) genes, also enriched for genes correlated with TMS in the expected direction (Tables S17 and S18) using a similar method to that previously used to test for enrichment of differentially expressed genes. Several immune related pathways were positively correlated with TMS, including MGI:2419, the most significantly dysregulated pathway in HD blood (Table S2). Downregulated pathways that correlated with TMS were related to ATP metabolism and DNA repair.
Similarly, we tested whether modules dysregulated in HD blood relative to controls (Table S9) also correlated with TMS in the expected direction (Table S19). Many modules significantly correlated with TMS, including 68 (CNpos5; p = 5.52 × 10−7) and 66 (CNneg1; p = 1.05 × 10−7), which were also dysregulated in the HD caudate30.
Mastrokolias et al.25 listed 170 genes significantly associated with TMS, of which 142 passed quality control in our RNA-Seq data. We tested for correlation between these genes and TMS in gene positive subjects from the Track-HD cohort (Table S20). 14 genes were nominally significant (p < 0.05), which is significantly higher than expected by chance (p = 7.89 × 10−3). Using the same method as for concordance with Labadorf, et al.35 (see Materials and Methods), we compared fold changes in expression of individual genes between Track-HD and Mastrokolias et al.25 Strikingly, 101 genes showed consistent direction of effect, as measured by log(FC), significantly greater than expected by chance (p = 4.78 × 10−7). Thus, we conclude that analysis of TMS in the Track-HD cohort broadly supported the associations reported in Mastrokolias et al.25.
The Alzheimer’s disease brain transcriptional signature is significantly dysregulated in HD blood
In Alzheimer’s disease, an early inflammatory response involving microglia contributes to pathogenesis36,37,38. Given the upregulation of immune-related gene sets in HD, we next asked whether co-expression modules dysregulated in Alzheimer’s disease (AD) brain were also disrupted in HD blood. Recently the International Genomics of Alzheimer’s Disease Consortium (IGAP) identified four modules from the Gibbs, et al.33 brain co-expression network that showed enrichment of signal in the GWAS of >70,000 late-onset Alzheimer’s disease (LOAD) and control subjects39. These four modules, each derived from a different brain region, are all involved in the immune response and were all significantly upregulated in our combined HD blood dataset (Table S21). Module 56, derived from pontine data, was also significantly enriched in both Track-HD and Leiden datasets independently. IGAP identified 151 genes that were present in two or more of these modules and showed the most significant enrichment with LOAD GWAS signal39. These 151 genes were also significantly enriched for upregulation in the combined HD blood dataset (p = 2.50 × 10−4).
Zhang, et al.40 identified co-expression modules that were differentially connected between LOAD and controls. Ten of these were also significantly enriched for upregulation in our HD blood expression dataset (Table S22) after correction for multiple testing (q < 0.05), with their most significant module, yellow, being particularly highly enriched (combined Track-HD and Leiden p < 1 × 10−16). Notably, this module has immune and microglia-specific functions40. This enrichment for modules from the IGAP GWAS39 and Zhang, et al.40 in the HD blood transcriptome suggests a shared immune-related mechanism between different neurodegenerative diseases, at least including HD and Alzheimer’s disease.
Discussion
HD research has focused on the brain as the most conspicuous clinical features can be clearly linked to progressive degeneration of specific brain regions4,5. However, HD is a systemic condition with peripheral expression of mutant huntingtin directly driving abnormalities such as immune dysfunction, metabolic derangement and transcriptional dysregulation that contribute to onset, progression, quality of life and mortality5,7.
We conducted RNA-Seq of whole blood in two independent cohorts of HD patients. Using gene set enrichment analysis (GSEA) with publicly-available pathway databases and WGCNA modules from HD and control brain datasets, we identified dysregulated genes and gene sets in blood that replicated in both independent cohorts and correlated with clinical motor signs (TMS). These correspond to the most significantly dysregulated modules in caudate nucleus, the most prominently affected region in HD brain. This suggests mutant huntingtin drives a common pathogenic signature in both blood and brain.
RNA-Seq more comprehensively and accurately quantifies mRNA than hybridisation-based microarrays or tag-based methods41. Expression of phosphatidylcholine transfer protein (PCTP) significantly correlated with TMS (Table S16). This protein transports phospholipids across intracellular membranes, which is of interest given the upregulation of lipid metabolic modules identified above (Tables 4 and 5) and increasing evidence for a pathological interaction between mutant huntingtin and membrane phospholipids42. However, PCTP was not significantly correlated with TMS in Mastrokolias et al.25. It is perhaps unsurprising that there was limited differential expression of individual transcripts by disease state (Table S1) or severity in either the independent or combined cohorts; the major cell types known to contribute to symptoms are not present in blood and the haematogenous cells known to be dysfunctional in HD, such as monocytes and macrophages9,43, constitute only a small proportion of circulating cells44. The variation of gene expression in blood with age, gender, cell type and time of day is also likely to contribute44,45. Our results are consistent with previous studies that have shown weak correlation at the transcript level between blood and brain46.
Despite these limitations, gene set enrichment analysis identified significantly overlapping dysregulated pathways in the Track-HD and Leiden HD blood datasets, even though they differed in age and disease severity. Thus, through grouping transcripts into biologically relevant pathways and co-expressed transcripts, we could highlight areas of dysfunctional biology in HD. The observed upregulation of immune-related pathways is consistent with that previously identified in transcriptional and functional studies5,7,25. HD patients are known to have immune dysfunction, both in the central nervous system (CNS) with microglial activation8, and peripherally with elevated proinflammatory cytokines in premanifest carriers up to 16 years before predicted onset9,43. The migration of phagocytic cells is impaired in HD10,11 and patient-derived monocytes are hyperactive on stimulation, an effect reduced by HTT lowering9. Modulation of the peripheral immune system with a type 2 cannabinoid receptor (CB2) agonist47 or bone marrow transplantation48 can increase lifespan and reduce motor deficits and synaptic loss in HD mouse models.
RNA processing pathways were downregulated, which is congruent with known decreases in miRNAs and altered expression of key miRNA processing enzymes in HD49. Consistent with the downregulation of pathways involved in energy metabolism that we observe, mitochondrial ATP is known to be reduced in HD brain50 and blood51, and PGC-1α, a member of the dysregulated ATP metabolic process pathway (Tables 3, S14 and S18), is a key protective regulator of mitochondrial genes that is repressed HD mouse models52,53. Downregulation of genes involved in DNA repair is likely to be relevant to somatic expansion that may influence disease onset and progression54. The signature of pathway dysregulation we identified in HD whole blood correlates with TMS in HD subjects from Track-HD. It also significantly overlaps with that recently found in unstimulated HD monocytes28. This enrichment was driven primarily by upregulation of immune pathways, as might be expected given that Miller, et al.28 isolated myeloid cells.
To overcome the annotation gap commonly observed with publicly-derived pathway databases and to investigate whether gene expression changes from HD brain are also present in blood, we performed GSEA using brain co-expression networks derived from HD30 and control32,33 subjects. Several HD brain modules were significantly dysregulated in HD blood, suggesting a common signature of transcriptional dysregulation between blood and brain.
Brain modules upregulated in blood were enriched for immune-related genes, confirming the results of our pathway analysis. Strikingly, two of the modules most significantly dysregulated in HD caudate, 48 (CNpos2) and 66 (CNneg1), were also significantly dysregulated in the same direction in both independent blood datasets. Compared with other brain regions, the caudate has the largest number of expression changes and the highest correlation with HD30. Module 48 (CNpos2), the second most significantly upregulated module in caudate, is enriched for transcriptional regulators, chromatin modifiers and genes involved in mRNA processing30. We also find this module to be significantly enriched for immune response genes, giving further support to the pathway results. Module 66 (CNneg1), the most significantly downregulated module in caudate, contains genes involved in neuronal function, particularly synaptic function and plasticity, and ion channels. Around half of its hub genes are implicated in synaptic function and all were significantly downregulated in Hodges, et al.31. Though synapses are not present in blood, synaptic genes may be dysregulated in circulating cells without significant pathogenic impact, or alternatively they may serve distinct functions in blood cells. Indeed, Cai, et al.46 found that the synaptic module was well preserved between brain and blood. We also found that gene expression and pathway dysregulation from HD prefrontal cortex35 was replicated in HD blood. The high degree of replication increases confidence in the shared signal between blood and brain. A significant proportion of the modules dysregulated in HD blood correlated with TMS.
Our demonstration of a transcriptional signature common to both HD blood and brain supports the use of blood cells to study aspects of HD biology. HD model systems, such as mice, only recapitulate aspects of disease and must be compared to the relevant data in human tissue55,56. Access to brain tissue is very limited and tends to be from post-mortem subjects with advanced disease, which affects RNA integrity19,20. Blood, by contrast, is readily available and can be obtained longitudinally from HD subjects. Recently, Mina, et al.57 performed WGCNA on the Leiden blood sample, finding modules related to immune response that were associated with TFC and TMS. Furthermore, by comparing biological annotations of their HD blood modules with those they derived from Hodges, et al.31 brain expression data, they showed a common signature between blood and caudate related to immune response. These analyses, using different methodology to ours, lend further support to our conclusions.
In AD, amyloid plaques are surrounded by chronically activated microglia36,37 and GWA studies have identified immune-related genes as risk factors for LOAD58. Recently Hong, et al.38 showed that early in the disease process, before plaque formation, microglia and complement activation drive synaptic loss, a process that may reflect reactivation of developmental synaptic pruning59. In HD blood we found significant upregulation of immune modules associated with AD in the IGAP GWAS39, a subset of genes with shared membership of several of these modules, and the most significant immune and microglia-related modules from Zhang, et al.40. In a co-expression network generated from prefrontal cortex of 194 HD patients, Zhang, et al.40 found that their most significant immune and microglia module was well conserved, though was not significantly dysregulated in HD and did not correlate with CAG repeat length. This may be because cortex shows less severe pathology and transcriptional dysregulation than caudate21. Overlapping immune upregulation in HD and AD suggests these two distinct neurodegenerative diseases share some common pathogenic mechanisms, including macrophage function38. Improved understanding of these mechanisms may open the way to therapeutic targets in these currently incurable diseases.
Materials and methods
All experiments we performed in accordance with the Declaration of Helsinki and approved by the University College London (UCL)/UCL Hospitals Joint Research Ethics Committee and the LUMC IRB. Peripheral blood samples were donated by genetically-diagnosed HD patients and controls, and all subjects provided informed written consent.
Cohorts
The Track-HD cohort consisted of 54 premanifest gene carriers, 63 manifest HD subjects and 23 controls. These were a representative sample from the Track-HD study (Table 1), preselected to assure a wide range of disease risk and severity. Control subjects were age and gender matched to individuals in the premanifest and manifest groups, and selected from spouses or partners to ensure consistency of environments. Track-HD enrolled participants at four study sites in London (UK), Paris (France), Leiden (Netherlands), and Vancouver (BC, Canada)26. Manifest subjects demonstrated motor abnormalities that were unequivocal signs of HD, as evidenced by total motor scores (TMS) over 5 on the Unified Huntington’s Disease Rating Scale (UHDRS). Premanifest gene carriers had a burden of pathology score (age x [CAG – 36.5))60 greater than 250, and a TMS of 5 or lower and a diagnostic confidence score (DCS) less than 4 on the UHDRS61, indicating no substantial motor signs26. Age and clinical scores considered for the analysis were at time of blood collection.
The Leiden cohort25 consisted of 18 premanifest gene carriers, 56 manifest HD subjects and 27 age and gender-matched controls. Motor onset was determined by an experienced neurologist using the same UHDRS standard as in TRACK-HD. All premanifest carriers showed no substantial motor signs, with a TMS of 5 or less and a UHDRS diagnostic confidence level less than 4. All controls were free of known medical conditions. Blood sample collection and analysis methods, described below, were identical for the two cohorts.
Sample collection
Whole blood was collected in two PAXGene Blood RNA tubes (PreAnalytix, Qiagen/BD Company) per subject, and immediately placed upright at room temperature. They were checked at 5 hours for incomplete mixing or separation, and any showing separation were remixed with a further 10 inversions. Tubes were stored overnight at −20 °C and transferred to −80 °C the following morning. They were sent on dry ice to Biorep within 30 days.
RNA preparation
Total RNA extraction was performed using the PAXGene Blood RNA kit (catalog N. 762174; PreAnalytix, Qiagen/BD Company), following the supplier’s instructions. Each solution in the kit was divided into aliquots to process batches of 12 samples. Replicate tubes for each subject were processed on different days. RNA was stored at −80 °C before proceeding with the quality measurements and further use. RNA was collected by centrifugation, washing with 70% ethanol, and resuspended in buffer. Quality measurements of total RNA were made using spectrophotometric analysis (Nanodrop), 260/280 ratio denaturing agarose gel, and the RNA 6000 Nano kit for the Agilent Bioanalyzer (catalog N. 5067-1511, Agilent Technologies). Samples were globin reduced using the GLOBINclearTM method (catalog N. AM1980, ThermoFisher Scientific). Quality control measures were made on globin-reduced samples on the Bioanalyzer RNA 6000 Nano kit (Catalog N. 5067-1511, Agilent Technologies).
Sequencing
Indexed cDNA sequencing libraries were prepared using the TruSeqTM Poly-A mRNA method (Illumina). In short, poly-A mRNA transcripts were captured from total RNA using poly-T beads and cDNA generated using random hexamer priming62. Paired-end sequencing of indexed cDNA libraries on a HiSeq 2500 generated at least 50 M reads per sample. Sequencing was performed using SBS and cluster kits from Illumina. Indexed samples were demultiplexed and FASTQ files were generated.
Quality control
Sequencing failed for six Track-HD samples, including four premanifest, one manifest and one control subject. Quality control analysis was performed using the RNA-SeQC package63, ensuring measures including rRNA rate, mapping rate, concordance mapping rate and uniqueness rate were within acceptable ranges. Globin depletion was checked by inspecting read counts mapped to HBB, HBA1 and HBA2, confirming they made up less than 2% of reads for all samples. Four Track-HD and six Leiden samples failed quality control for duplication rate over 75%, GC bias or 5’ bias, and were removed, leaving 48 premanifest, 61 manifest and 21 control subjects in the Track-HD cohort and 15 premanifest, 54 manifest and 26 control subjects in the Leiden cohort.
Gene expression analysis
RNA-Seq data were aligned to the human reference genome hg19 using TopHat264. Read counts were summarised using HTSeq, keeping any duplicates and using the Ensembl transcript/gene database (http://www.ensembl.org/info/data/ftp/index.html, obtained in gtf format, genome build GRCh38.3, gene build updated in June 2015). To remove residual batch effects the R package svaseq was used65. Using the cleaned count data, differential expression analysis was conducted using the R package DESeq266. Outlier counts were removed using a Cooks distance cutoff of 5 in DESeq2. After filtering by the mean of normalised counts, 18,257 transcripts were detected. Age and gender were used as covariates in the analysis.
Pathway analysis
Enrichment of differential expression among gene sets corresponding to biological hypotheses (pathways) was tested using the Gene Set Enrichment Analysis (GSEA) method67. Rather than defining a list of significant genes, GSEA ranks all genes in order of their differential expression statistic, and tests whether the genes in a particular gene set have a higher rank overall than would be expected by chance. The analysis is weighted by the differential expression statistic, thus giving more weight to more significant genes. Significance of enrichment was obtained by randomly permuting gene-wide association statistics among genes. One-sided p-values were calculated separately for differential upregulation and downregulation of expression in HD, and these were then converted into the corresponding chi-square statistic for use in the GSEA analysis. To avoid making a priori assumptions, we collated a large pathway set from publicly available pathway databases, including Gene Ontology (GO)68, Kyoto Encyclopedia of Genes and Genomes (KEGG)69, Mouse Genome Informatics (MGI)70, PANTHER71, BioCarta72, REACTOME73 and NCI74. This resulted in a total of 14,706 functional pathways, many with overlapping members, containing between 3 and 500 genes. To correct for multiple testing of pathways we converted the GSEA p-values into q-values75, which can be interpreted as the minimum false discovery rate at which that q-value would be counted as significant.
Gene co-expression networks
Weaknesses of relying on public databases to provide pathways for analysis include their restriction to prior biological knowledge and the poor annotation of many genes. To overcome this annotation gap, we also tested the following sets of gene co-expression modules for enrichment of dysregulation:
The set of 124 HD brain expression modules derived by Neueder and Bates30, who applied weighted gene correlation network analysis (WGCNA)34 to the Hodges, et al.31 microarray brain expression data set of 44 human HD and 36 matched control brains. They generated networks for four brain regions; the caudate nucleus (CN), BA4 region of the frontal cortex, which has motor function (FC-BA4), BA9 region of the frontal cortex, involved in association and cognitive functions (FC-BA9), and cerebellum (CB).
A set of 117 co-expression modules derived from the Gibbs, et al.33 dataset, comprising microarray expression data from 150 control individuals measured in four brain regions: cerebellum (CB), frontal cortex (FC), caudal pons (Pons) and temporal cortex (TCTX). Modules were generated using WGCNA as described in ref. 39.
We generated a set of 213 co-expression modules from Braineac32, which consists of microarray expression data for 12 brain regions from 134 control brains; occipital cortex, frontal cortex, temporal cortex, hippocampus, intralobular white matter, cerebellar cortex, thalamus, putamen, substantia nigra, and medulla (inferior olivary nucleus). For each brain region, the array data was normalised in the R statistical-programming environment using the RMA algorithm76. Principal Component Analysis (PCA) and hierarchical clustering were used to identify single outlier arrays for removal. In addition, small outlier clusters (<6 arrays) that were distinct from most of the other arrays were removed (i.e. small clusters appearing at the top of the dendrogram). Once outlier arrays were removed, the arrays were re-normalized and inspected again and re-processed if necessary until a homogenous dataset was produced. WGCNA was performed using the R package to derive modules34. Multiple probesets of the same gene were collapsed to a single value using the collapseRows() function, using default settings and based on gene annotation provided by Affymetrix77. Scale independence and mean connectivity were plotted to derive a soft threshold power of 6. Networks were unsigned.
The set of 111 co-expression modules from Zhang, et al.40, generated using microarray expression data on 1,647 postmortem samples from three brain regions of late-onset Alzheimer’s disease (LOAD) and control subjects; prefrontal cortex (BA9), primary visual cortex (BA17), and cerebellum.
Concordance of fold change in gene expression between datasets
Labadorf, et al.35 analysed the transcriptome of human postmortem prefrontal cortex Brodmann area 9 (BA9) from 20 HD subjects and 49 controls using next-generation high throughput sequencing, identifying dysregulation of immune and developmental genes. Of the 15,834 genes common to both the combined Track-HD and Leiden blood dataset and the Labadorf, et al.35 prefrontal cortex dataset, 8447 had a fold change >1 (i.e. upregulated) in blood and 7860 in cortex. Thus, if fold changes in the two datasets were assumed to be unrelated, the expected probability that a gene would show concordant fold change is equal to ((8447/15834)x(7860/15834)) + ((7387/15834)x(7974/15834)) = 0.4997. The number of genes with concordant fold change in the absence of a relationship between the datasets is thus distributed as a binomial (15834, 0.4997) distribution. In the actual data, 8425 genes were observed to have concordant direction of fold change, significantly higher than the number expected by chance (7912).
We used a similar method to test for concordance of fold change in genes between the Track-HD and Mastrokolias et al. datasets.
Data availability
All data is deposited at the European Genome-phenome Archive (EGA) and accessible through the authors or the NeurOmics consortium.
Additional Information
How to cite this article: Hensman Moss, D. J. et al. Huntington’s disease blood and brain show a common gene expression pattern and share an immune signature with Alzheimer’s disease. Sci. Rep. 7, 44849; doi: 10.1038/srep44849 (2017).
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Material
Acknowledgments
We would like to thank the patients and control subjects who donated samples and took part in these studies, as well as the Track-HD investigators, particularly Professors Blair Leavitt (UBC, Vancouver), Alexandra Durr (Pitié-Salpêtrière Hospital, Paris) and Raymund Roos (LUMC, Leiden). Some of this work was undertaken at UCLH/UCL who acknowledge support from the Department of Health’s NIHR Biomedical Research Centre. We thank R.C. van der Mast and E. van Duijn at the Psychiatry department of the LUMC for collecting blood samples from the Leiden cohort. This work was supported by funding from the European Community’s Seventh Framework Programme (FP7/2007-2013) under grant agreement n° 2012-305121 “Integrated European –omics research project for diagnosis and therapy in rare neuromuscular and neurodegenerative diseases (NEUROMICS)” [261358], the Medical Research Council UK [MR/J003832/1, 515109 MR/L02053X/1], CHDI Foundation, the BBSRC, the Dementia and Neurodegenerative Disease Network UK, the European Huntington’s Disease Network, the Huntington’s Disease Association of the UK, the Guarantors of Brain [523410], the Rosetrees Trust [JS16/M220], the Centre for Medical Systems Biology within the framework of the Netherlands Genomics Initiative/Netherlands Organization for Scientific Research and the Dutch Centre for Biomedical Genetics.
Footnotes
The authors declare no competing financial interests.
Author Contributions D.H.M. collected samples and clinical information and coordinated the analysis, M.F. wrote the M.S., K.L. performed the RNA-Seq analysis, J.M. assisted collecting samples and wrote the M.S., L.J. designed the study and co-wrote the M.S., D.L. devised the statistical measures, V.P. supervised the RNA-Seq analysis, WMCvRM, GJvO and PAC’tH provided samples and discussed results, T.S. and A.G. derived co-expression modules, T.S. produced Cytoscape figures, P.H. designed and supervised the study, performed the pathway analyses and co-wrote the M.S., S.T. designed and supervised the study and co-wrote the M.S.
References
- Evans S. J. et al. Prevalence of adult Huntington’s disease in the UK based on diagnoses recorded in general practice records. Journal of neurology, neurosurgery, and psychiatry 84, 1156–1160, doi: 10.1136/jnnp-2012-304636 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langbehn D. R., Hayden M. R. & Paulsen J. S. CAG-repeat length and the age of onset in Huntington disease (HD): a review and validation study of statistical approaches. American journal of medical genetics. Part B, Neuropsychiatric genetics : the official publication of the International Society of Psychiatric Genetics 153b, 397–408, doi: 10.1002/ajmg.b.30992 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross C. A. et al. Huntington disease: natural history, biomarkers and prospects for therapeutics. Nature reviews. Neurology 10, 204–216, doi: 10.1038/nrneurol.2014.24 (2014). [DOI] [PubMed] [Google Scholar]
- Bates G. P. et al. Huntington disease. Nature Reviews Disease Primers, 15005, doi: 10.1038/nrdp.2015.5 (2015). [DOI] [PubMed] [Google Scholar]
- van der Burg J. M., Bjorkqvist M. & Brundin P. Beyond the brain: widespread pathology in Huntington’s disease. The Lancet. Neurology 8, 765–774, doi: 10.1016/S1474-4422(09)70178-4 (2009). [DOI] [PubMed] [Google Scholar]
- Trottier Y. et al. Cellular localization of the Huntington’s disease protein and discrimination of the normal and mutated form. Nature Genetics 10, 104–110, doi: 10.1038/ng0595-104 (1995). [DOI] [PubMed] [Google Scholar]
- Carroll J. B., Bates G. P., Steffan J., Saft C. & Tabrizi S. J. Treating the whole body in Huntington’s disease. The Lancet. Neurology 14, 1135–1142, doi: 10.1016/s1474-4422(15)00177-5 (2015). [DOI] [PubMed] [Google Scholar]
- Tai Y. F. et al. Microglial activation in presymptomatic Huntington’s disease gene carriers. Brain 130, 1759–1766, doi: 10.1093/brain/awm044 (2007). [DOI] [PubMed] [Google Scholar]
- Bjorkqvist M. et al. A novel pathogenic pathway of immune activation detectable before clinical onset in Huntington’s disease. The Journal of experimental medicine 205, 1869–1877, doi: 10.1084/jem.20080178 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwan W. et al. Mutant huntingtin impairs immune cell migration in Huntington disease. Journal of Clinical Investigation 122, 4737–4747, doi: 10.1172/jci64484 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Träger U. et al. Characterisation of immune cell function in fragment and full-length Huntington’s disease mouse models. Neurobiology of disease 73, 388–398, doi: 10.1016/j.nbd.2014.10.012 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busse M. E., Hughes G., Wiles C. M. & Rosser A. E. Use of hand-held dynamometry in the evaluation of lower limb muscle strength in people with Huntington’s disease. Journal of neurology 255, 1534–1540, doi: 10.1007/s00415-008-0964-x (2008). [DOI] [PubMed] [Google Scholar]
- Saleh N. et al. Neuroendocrine disturbances in Huntington’s disease. PLoS One 4, e4962, doi: 10.1371/journal.pone.0004962 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanska D. J., Lavine L., Lanska M. J. & Schoenberg B. S. Huntington’s disease mortality in the United States. Neurology 38, 769–772 (1988). [DOI] [PubMed] [Google Scholar]
- Mihm M. J. et al. Cardiac dysfunction in the R6/2 mouse model of Huntington’s disease. Neurobiology of disease 25, 297–308, doi: 10.1016/j.nbd.2006.09.016 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pattison J. S. et al. Cardiomyocyte Expression of a Polyglutamine Preamyloid Oligomer Causes Heart Failure. Circulation 117, 2743–2751, doi: 10.1161/circulationaha.107.750232 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orth M., Cooper J. M., Bates G. P. & Schapira A. H. V. Inclusion formation in Huntington’s disease R6/2 mouse muscle cultures. Journal of Neurochemistry 87, 1–6, doi: 10.1046/j.1471-4159.2003.02009.x (2003). [DOI] [PubMed] [Google Scholar]
- Turner C., Cooper J. M. & Schapira A. H. V. Clinical correlates of mitochondrial function in Huntington’s disease muscle. Movement Disorders 22, 1715–1721, doi: 10.1002/mds.21540 (2007). [DOI] [PubMed] [Google Scholar]
- Montanini L. et al. Human RNA integrity after postmortem retinal tissue recovery. Ophthalmic Genet 34, 27–31, doi: 10.3109/13816810.2012.720342 (2013). [DOI] [PubMed] [Google Scholar]
- Tomita H. et al. Effect of agonal and postmortem factors on gene expression profile: quality control in microarray analyses of postmortem human brain. Biol Psychiatry 55, 346–352, doi: 10.1016/j.biopsych.2003.10.013 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodges A. Regional and cellular gene expression changes in human Huntington’s disease brain. Human molecular genetics 15, 965–977, doi: 10.1093/hmg/ddl013 (2006). [DOI] [PubMed] [Google Scholar]
- Borovecki F. et al. Genome-wide expression profiling of human blood reveals biomarkers for Huntington’s disease. Proceedings of the National Academy of Sciences of the United States of America 102, 11023–11028, doi: 10.1073/pnas.0504921102 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Runne H. et al. Analysis of potential transcriptomic biomarkers for Huntington’s disease in peripheral blood. Proceedings of the National Academy of Sciences of the United States of America 104, 14424–14429, doi: 10.1073/pnas.0703652104 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovrecic L. et al. Gene expression changes in blood as a putative biomarker for Huntington’s disease. Movement disorders : official journal of the Movement Disorder Society 24, 2277–2281, doi: 10.1002/mds.22477 (2009). [DOI] [PubMed] [Google Scholar]
- Mastrokolias A. et al. Huntington’s disease biomarker progression profile identified by transcriptome sequencing in peripheral blood. Eur J Hum Genet, doi: 10.1038/ejhg.2014.281 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabrizi S. J. et al. Biological and clinical manifestations of Huntington’s disease in the longitudinal TRACK-HD study: cross-sectional analysis of baseline data. The Lancet. Neurology 8, 791–801, doi: 10.1016/S1474-4422(09)70170-X (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Consortium G. M. o. H. s. D. G.-H. Identification of Genetic Factors that Modify Clinical Onset of Huntington’s Disease. Cell 162, 516–526, doi: 10.1016/j.cell.2015.07.003 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller J. R. et al. RNA-Seq of Huntington’s disease patient myeloid cells reveals innate transcriptional dysregulation associated with proinflammatory pathway activation. Human molecular genetics, doi: 10.1093/hmg/ddw142 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbs D. L. et al. Protein co-expression network analysis (ProCoNA). Journal of clinical bioinformatics 3, 11, doi: 10.1186/2043-9113-3-11 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neueder A. & Bates G. P. A common gene expression signature in Huntington’s disease patient brain regions. BMC medical genomics 7, 60, doi: 10.1186/s12920-014-0060-2 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodges A. et al. Regional and cellular gene expression changes in human Huntington’s disease brain. Human molecular genetics 15, 965–977, doi: 10.1093/hmg/ddl013 (2006). [DOI] [PubMed] [Google Scholar]
- Braineac. Braineac - The Brain eQTL Almanac, http://www.braineac.org/ (2016). [Google Scholar]
- Gibbs J. R. et al. Abundant quantitative trait loci exist for DNA methylation and gene expression in human brain. PLoS genetics 6, e1000952, doi: 10.1371/journal.pgen.1000952 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langfelder P. & Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC bioinformatics 9, 559, doi: 10.1186/1471-2105-9-559 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labadorf A. et al. RNA Sequence Analysis of Human Huntington Disease Brain Reveals an Extensive Increase in Inflammatory and Developmental Gene Expression. PLOS ONE 10, e0143563, doi: 10.1371/journal.pone.0143563 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Nicola D., Fransen N. L., Suzzi S. & Perry V. H. Regulation of microglial proliferation during chronic neurodegeneration. The Journal of neuroscience : the official journal of the Society for Neuroscience 33, 2481–2493, doi: 10.1523/JNEUROSCI.4440-12.2013 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olmos-Alonso A. et al. Pharmacological targeting of CSF1R inhibits microglial proliferation and prevents the progression of Alzheimer’s-like pathology. Brain 139, 891–907, doi: 10.1093/brain/awv379 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong S. et al. Complement and microglia mediate early synapse loss in Alzheimer mouse models. Science (New York, N.Y.), doi: 10.1126/science.aad8373 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Genomics of Alzheimer’s Disease, C. Convergent genetic and expression data implicate immunity in Alzheimer’s disease. Alzheimers Dement 11, 658–671, doi: 10.1016/j.jalz.2014.05.1757 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B. et al. Integrated systems approach identifies genetic nodes and networks in late-onset Alzheimer’s disease. Cell 153, 707–720, doi: 10.1016/j.cell.2013.03.030 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa V., Angelini C., De Feis I. & Ciccodicola A. Uncovering the complexity of transcriptomes with RNA-Seq. Journal of biomedicine & biotechnology 2010, 853916, doi: 10.1155/2010/853916 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kegel-Gleason K. B. Huntingtin interactions with membrane phospholipids: strategic targets for therapeutic intervention? Journal of Huntington’s disease 2, 239–250, doi: 10.3233/JHD-130068 (2013). [DOI] [PubMed] [Google Scholar]
- Wild E. et al. Abnormal peripheral chemokine profile in Huntington’s disease. PLoS currents 3, RRN1231, doi: 10.1371/currents.rrn1231 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitney A. R. et al. Individuality and variation in gene expression patterns in human blood. Proceedings of the National Academy of Sciences of the United States of America 100, 1896–1901, doi: 10.1073/pnas.252784499 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horvath S. et al. Aging effects on DNA methylation modules in human brain and blood tissue. Genome Biol 13, R97, doi: 10.1186/gb-2012-13-10-r97 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai C. et al. Is human blood a good surrogate for brain tissue in transcriptional studies? BMC genomics 11, 589, doi: 10.1186/1471-2164-11-589 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouchard J. et al. Cannabinoid Receptor 2 Signaling in Peripheral Immune Cells Modulates Disease Onset and Severity in Mouse Models of Huntington’s Disease. Journal of Neuroscience 32, 18259–18268, doi: 10.1523/jneurosci.4008-12.2012 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwan W. et al. Bone Marrow Transplantation Confers Modest Benefits in Mouse Models of Huntington’s Disease. Journal of Neuroscience 32, 133–142, doi: 10.1523/jneurosci.4846-11.2012 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seredenina T. & Luthi-Carter R. What have we learned from gene expression profiles in Huntington’s disease? Neurobiology of disease 45, 83–98, doi: 10.1016/j.nbd.2011.07.001 (2012). [DOI] [PubMed] [Google Scholar]
- Mochel F. et al. Early alterations of brain cellular energy homeostasis in Huntington disease models. The Journal of biological chemistry 287, 1361–1370, doi: 10.1074/jbc.M111.309849 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seong I. S. HD CAG repeat implicates a dominant property of huntingtin in mitochondrial energy metabolism. Human molecular genetics 14, 2871–2880, doi: 10.1093/hmg/ddi319 (2005). [DOI] [PubMed] [Google Scholar]
- Cui L. et al. Transcriptional repression of PGC-1alpha by mutant huntingtin leads to mitochondrial dysfunction and neurodegeneration. Cell 127, 59–69, doi: 10.1016/j.cell.2006.09.015 (2006). [DOI] [PubMed] [Google Scholar]
- Chaturvedi R. K. et al. Impairment of PGC-1alpha expression, neuropathology and hepatic steatosis in a transgenic mouse model of Huntington’s disease following chronic energy deprivation. Human molecular genetics 19, 3190–3205, doi: 10.1093/hmg/ddq229 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonson I., Ougland R. & Larsen E. DNA repair mechanisms in Huntington’s disease. Molecular neurobiology 47, 1093–1102, doi: 10.1007/s12035-013-8409-7 (2013). [DOI] [PubMed] [Google Scholar]
- Morton A. J. & Howland D. S. Large genetic animal models of Huntington’s Disease. Journal of Huntington’s disease 2, 3–19, doi: 10.3233/jhd-130050 (2013). [DOI] [PubMed] [Google Scholar]
- Ehrnhoefer D. E., Butland S. L., Pouladi M. A. & Hayden M. R. Mouse models of Huntington disease: variations on a theme. Disease Models & Mechanisms 2, 123–129, doi: 10.1242/dmm.002451 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mina E. et al. Common disease signatures between blood and brain in Huntington’s Disease. Orphanet Journal of Rare Diseases(2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyss-Coray T. & Rogers J. Inflammation in Alzheimer disease-a brief review of the basic science and clinical literature. Cold Spring Harb Perspect Med 2, a006346, doi: 10.1101/cshperspect.a006346 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong S., Dissing-Olesen L. & Stevens B. New insights on the role of microglia in synaptic pruning in health and disease. Curr Opin Neurobiol 36, 128–134, doi: 10.1016/j.conb.2015.12.004 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penney J. B. Jr., Vonsattel J. P., MacDonald M. E., Gusella J. F. & Myers R. H. CAG repeat number governs the development rate of pathology in Huntington’s disease. Ann Neurol 41, 689–692, doi: 10.1002/ana.410410521 (1997). [DOI] [PubMed] [Google Scholar]
- Group H. S. Unified Huntington’s Disease Rating Scale: reliability and consistency. Huntington Study Group. Movement disorders : official journal of the Movement Disorder Society 11, 136–142, doi: 10.1002/mds.870110204 (1996). [DOI] [PubMed] [Google Scholar]
- Illumina. TruSeq(R) RNA Sample Preparation v2 Guide, http://support.illumina.com/content/dam/illumina-support/documents/documentation/chemistry_documentation/samplepreps_truseq/truseqrna/truseq-rna-sample-prep-v2-guide-15026495-f.pdf (2014). [Google Scholar]
- DeLuca D. S. et al. RNA-SeQC: RNA-seq metrics for quality control and process optimization. Bioinformatics 28, 1530–1532, doi: 10.1093/bioinformatics/bts196 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D. et al. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol 14, R36, doi: 10.1186/gb-2013-14-4-r36 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leek J. T. svaseq: removing batch effects and other unwanted noise from sequencing data. Nucleic Acids Res 42, doi: 10.1093/nar/gku864 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love M. I., Huber W. & Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15, 550, doi: 10.1186/s13059-014-0550-8 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A. et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proceedings of the National Academy of Sciences of the United States of America 102, 15545–15550, doi: 10.1073/pnas.0506580102 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Consortium G. O. Gene Ontology Consortium, http://geneontology.org/ (2016). [Google Scholar]
- KEGG. KEGG: Kyoto Encyclopedia of Genes and Genomes, http://www.kegg.jp/ (2016). [Google Scholar]
- MGI. MGI-Mouse Genome Informatics-The international database resource for the laboratory mouse, http://www.informatics.jax.org/ (2016). [Google Scholar]
- PANTHER. PANTHER - Gene List Analysis, http://pantherdb.org/ (2016). [Google Scholar]
- BioCarta. http://www.biocarta.com/. (2016).
- REACTOME. Reactome Pathway Database, http://www.reactome.org/ (2016). [Google Scholar]
- Institute N. C. Home : Pathway Interaction Database, http://www.ncbi.nlm.nih.gov/pubmed/ (2012). [Google Scholar]
- Storey J. D. & Tibshirani R. Statistical significance for genomewide studies. Proceedings of the National Academy of Sciences of the United States of America 100, 9440–9445, doi: 10.1073/pnas.1530509100 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho B. S. & Irizarry R. A. A framework for oligonucleotide microarray preprocessing. Bioinformatics 26, 2363–2367, doi: 10.1093/bioinformatics/btq431 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Affymetrix. Affymetrix, http://www.affymetrix.com/estore/index.jsp (2016). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data is deposited at the European Genome-phenome Archive (EGA) and accessible through the authors or the NeurOmics consortium.