Abstract
Significance: In the last years, metabolic reprogramming, fluctuations in bioenergetic fuels, and modulation of oxidative stress became new key hallmarks of tumor development. In cancer, elevated glucose uptake and high glycolytic rate, as a source of adenosine triphosphate, constitute a growth advantage for tumors. This represents the universally known Warburg effect, which gave rise to one major clinical application for detecting cancer cells using glucose analogs: the positron emission tomography scan imaging.
Recent Advances: Glucose utilization and carbon sources in tumors are much more heterogeneous than initially thought. Indeed, new studies emerged and revealed a dual capacity of tumor cells for glycolytic and oxidative phosphorylation (OXPHOS) metabolism. OXPHOS metabolism, which relies predominantly on mitochondrial respiration, exhibits fine-tuned regulation of respiratory chain complexes and enhanced antioxidant response or detoxification capacity.
Critical Issues: OXPHOS-dependent cancer cells use alternative oxidizable substrates, such as glutamine and fatty acids. The diversity of carbon substrates fueling neoplastic cells is indicative of metabolic heterogeneity, even within tumors sharing the same clinical diagnosis. Metabolic switch supports cancer cell stemness and their bioenergy-consuming functions, such as proliferation, survival, migration, and invasion. Moreover, reactive oxygen species-induced mitochondrial metabolism and nutrient availability are important for interaction with tumor microenvironment components. Carcinoma-associated fibroblasts and immune cells participate in the metabolic interplay with neoplastic cells. They collectively adapt in a dynamic manner to the metabolic needs of cancer cells, thus participating in tumorigenesis and resistance to treatments.
Future Directions: Characterizing the reciprocal metabolic interplay between stromal, immune, and neoplastic cells will provide a better understanding of treatment resistance. Antioxid. Redox Signal. 26, 462–485.
Keywords: : mitochondria, cancer, ROS, immunology, metabolism, oxidative stress
Introduction
The energy metabolism field has emerged in the mid-19th century with the pioneering works from O. Warburg, who deciphered the nature and mode of action of enzymes involved in respiration (Nobel Prize in Physiology, 1931) and fermentation (prevented from receiving a 2nd Nobel Prize in 1944 by the Nazi regime), A.L. Lenhinger, defining mitochondria as the main site of oxidative phosphorylation (OXPHOS), H.A. Krebs, uncovering the tricarboxylic acid (TCA) cycle (Nobel Prize in Physiology and Medicine, 1953), and S. Weinhouse, revisiting respiratory deficiencies in cancer cells (106, 144, 228, 229, 234, 235). Warburg's observations revealed that adenosine triphosphate (ATP), the main source of energy, is mostly produced by aerobic glycolysis in cancer cells. Indeed, to make ATP, neoplastic cells rather produce lactate than oxidize pyruvate through the TCA cycle (212, 228, 229). This metabolic switch occurs under aerobic conditions, even if glycolysis is less efficient to produce ATP (2 ATP molecules instead of 36 by TCA cycle) and if it generates an acidic environment due to lactic acid production. This is the basis of the universally known Warburg effect, the gold standard process in cancer metabolism, during the last century.
Aerobic glycolysis is considered as a key feature in cancer and has recently taken place in the famous “Hallmarks of cancer” described by D. Hanahan and B. Weinberg (77). In many cancers, increased glucose uptake and enhanced glycolytic rates, as a source of ATP, represent a growth advantage for tumor cells (63). Indeed, aerobic glycolysis represents also an effective shunting by providing carbon sources to other key metabolic pathways required for nucleotide, lipid, and amino acid synthesis, building blocks that are essential for highly proliferative cells (Fig. 1). For instance, increased levels of the glycolysis intermediate, glucose-6-phosphate (G6P), favor the pentose phosphate pathway (PPP) (Fig. 1). The PPP generates reducing equivalents, in the form of nicotinamide adenine dinucleotide phosphate (NADPH), used for redox homeostasis maintenance and fatty acid (FA) biosynthesis. Moreover, it produces ribose-5-phosphate used for nucleic acid synthesis and erythrose-4-phosphate required for aromatic amino acid production (84). Moreover, glycolysis intermediates in cancer cells are also redirected toward the serine pathway, which starts with the conversion of 3-phosphoglycerate into phosphohydroxypyruvate via the phosphoglycerate dehydrogenase (123, 162) (Fig. 1). This pathway is essential for amino acid (serine and glycine) synthesis and is also involved in the folate cycle, a major source of methyl groups for one-carbon pools and purine synthesis (122). Subsequently, this pathway provides essential precursors of proteins, nucleic acids, and glutathione-dependent antioxidant capacities. Although glycolytic switch is now established as a key process in tumorigenesis, the cause and the mechanisms leading to this metabolic reprogramming are still under debate (24, 26, 115, 231). In brief, it was initially thought that mitochondria were bearing mutations and functionally defective, thus forcing tumor cells to adapt to this respiratory deficiency. However, mitochondria alterations are quite rare and electron microscopy revealed that mitochondria are active. Moreover, several studies showed that cancers cells retain OXPHOS capacity and do not suffer from respiratory defects (58, 95, 170, 214, 235, 236, 239, 253). Furthermore, it has been shown that MCF7 breast cancer cells generate 80% of their ATP through mitochondrial respiration (74). Finally, inhibiting glycolysis in neoplastic cells restores mitochondrial OXPHOS (18, 48, 135, 138), demonstrating that oxidative metabolism remains functional in most glycolytic cancer cells.
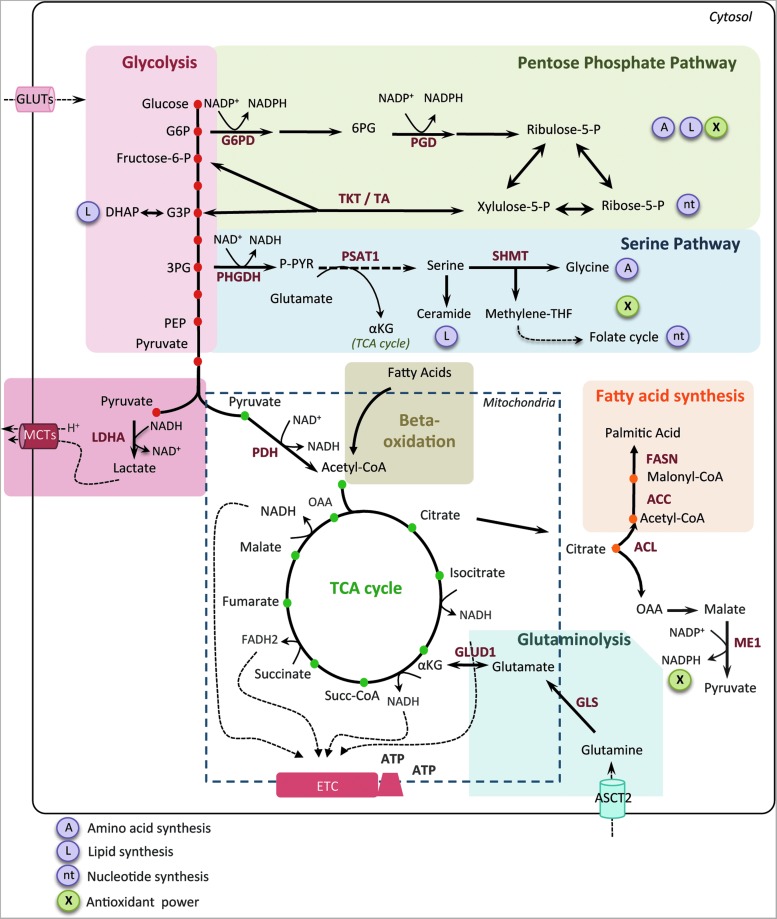
FIG. 1.
Core metabolic pathways and enzymes in cancer cells. Here are schematically represented the main metabolic pathways altered in cancers, including the glycolysis, the PPP, the serine pathway, the fatty acid synthesis, and the TCA cycle. In cancer cells, the canonical energy metabolism pathways are often truncated (glycolysis, TCA cycle) or redirected (glutaminolysis or serine and lipid biosynthesis). Briefly, glucose enters into cancer cells through glucose transporters and is phosphorylated to G6P by an irreversible reaction catalyzed by the hexokinase. G6P either proceeds through glycolysis to produce pyruvate or through the PPP to generate ribose-5-phosphate and NADPH. The PPP is connected at the first step of glycolysis starting with G6P dehydrogenase (G6PD) and has both an oxidative and nonoxidative arm. G6P oxidation produces the reducing equivalents, in the form of NADPH, important cellular antioxidant, and cofactor for fatty acid biosynthesis. Moreover, the PPP provides cancer cells with pentose sugars for the biosynthesis of nucleic acids. The first enzymes involved in the nonoxidative arm of the PPP are TKT and TA. Ribose-5-phosphate and xylulose-5-phosphate, generated by the oxidative PPP, can be further metabolized into F6P and G3P to reenter into glycolysis for ATP production, depending on the cell requirement. Thus, the PPP plays a key role in cancer cells to supply their anabolic demands and to counteract oxidative stress. The serine pathway is branched to glycolysis via 3-phosphoglycerate (3PG), which is converted by PHGDH into phosphohydroxypyruvate (P-PYR). This pathway produces serine and glycine, essential precursors for synthesis of proteins and nucleic acids through the folate cycle. Following glycolysis, pyruvate is either converted into lactate by LDHA and released through monocarboxylate transporters, MCT4 and MCT1, further causing extra cellular acidification, or converted into acetyl-CoA, through the PDH complex. Acetyl-CoA enters into TCA cycle and produces ATP, NADH, and FADH2 molecules. Reduced cofactors are then oxidized by the ETC complexes for ATP production. Glutamine and other amino acids can also replenish the TCA cycle. Indeed, the first step of glutaminolysis is the conversion of glutamine into glutamate by the GLS. Glutamate is subsequently converted into alpha-ketoglutarate (αKG) that fuels back the TCA cycle. Fatty acid degradation can also supply the TCA cycle through beta-oxidation, which produces acetyl-CoA. Reciprocally, citrate, a TCA cycle intermediate, can be used as a precursor for fatty acid synthesis and for NADPH production through the ACL. Citrate is subsequently converted to acetyl-CoA and OAA into the cytoplasm. Acetyl-CoA is used for fatty acid synthesis through its conversion to malonyl-CoA by ACC and to palmitic acid by the FASN. OAA is converted to malate, which is then decarboxylated into pyruvate, by the ME1 and produces NADPH. Mitochondria are represented by dotted line. ACC, acetyl-CoA carboxylase; ACL, ATP citrate lyase; αKG, alpha-ketoglutarate; ASCT2, amino acid transporter; ATP, adenosine triphosphate; DHAP, dihydroxyacetone phosphate; ETC, electron transport chain; FAD, flavin adenine dinucleotide; FASN, fatty acid synthase; F6P, fructose-6-phosphate; GADP, glyceraldehyde-3-phosphate; G3P, glyceraldehyde-3-phosphate; GLS, glutaminase; G6P, glucose-6-phosphate; GLUT, glucose transporter; LDHA, lactate dehydrogenase A; MCT, monocarboxylate transporter; ME1, malic enzyme; NAD, nicotinamide adenine dinucleotide; NADPH, nicotinamide adenine dinucleotide phosphate; OAA, oxaloacetate; PDH, pyruvate dehydrogenase; PEP, phospho-enol-pyruvate; 6PG, 6-phospho-gluconolactone; 3PG, 3-phopho-glycerate; PGD, phosphogluconate dehydrogenase; PHGDH, phosphoglycerate dehydrogenase; PPP, pentose phosphate pathway; P-PYR, phosphohydroxypyruvate; PSAT1, phosphoserine aminotransferase; SHMT1, serine hydroxymethyl transferase; Succ-CoA, succinyl-CoA; TCA, tricarboxylic acid; TA, transaldolase; TKT, transketolase. To see this illustration in color, the reader is referred to the web version of this article at www.liebertpub.com/ars
Physiological stresses, such as the lack of oxygen (O2), are considered as one of the main drivers of the metabolic switch in tumor cells. As early tumors expand, they become hypoxic and require blood and nutrient supplies to keep growing. Decreased dependence on aerobic respiration becomes advantageous and tumor metabolism is thus shifted toward glycolysis, therefore balancing O2 consumption with O2 supply. Under hypoxia, the hypoxia-inducible factor-1 (HIF-1) transcription factor accumulates and provides O2 to tumor cells by increasing angiogenesis, erythropoiesis, and glycolysis (21, 29, 163). HIF-1 enhances glycolysis by increasing expression of glucose transporters (GLUT1-3) and glycolytic enzymes, including hexokinase 2 (HK2) and lactate dehydrogenase A (LDHA). HIF-1 also inhibits the TCA cycle by upregulating the transcription of pyruvate dehydrogenase kinase (PDK), which inactivates the pyruvate dehydrogenase (PDH), preventing the conversion of pyruvate into acetyl coenzyme A (acetyl-CoA) (20, 103, 155, 163). HIF-1 is not only activated upon low O2 concentration but also under normoxic conditions by oncogenic and oxidative stress (7, 36, 65, 111). Aerobic glycolysis and concomitant increase in glucose uptake give rise to one major clinical application for detecting cancer cells: the positron emission tomography (PET) scan, an imaging technology, which uses glucose analog tracer. In oncology, PET scan consists in measuring disease extent, lymph node localization, prediction of relapse, and diagnostic accuracy (4, 248). Despite the major advances provided by this technique in clinical practice, some tumors do not absorb glucose and false-negative results have been diagnosed for some cancer patients. Taken together, these observations strongly suggest that glucose utilization and carbon sources in tumors could be more heterogeneous than initially thought (33, 176).
Our review will describe tumor metabolism heterogeneity, considering both cancer cells and cells of the tumor microenvironment (TME). We will focus on recent findings highlighting OXPHOS metabolism in neoplastic cancer cells, thereby arguing that glucose is not the only energy fuel and can be substituted, in part, by glutamine, serine/glycine, or FA for replenishing TCA cycle intermediates. Moreover, we will discuss the mechanisms regulating synthesis of mitochondria respiratory complexes, which are absolutely required to support OXPHOS metabolism. We will then describe the impact of metabolic variations on cancer cell functions, such as proliferation, survival, migration, invasion, and stemness. Finally, we will provide clues on the role of metabolism in the cross-talk between tumor cells and some key players of the TME, such as macrophages, lymphocytes, and fibroblasts, and we will show how metabolism can determine their differentiation.
Cancer Metabolism Heterogeneity
Over the past decade, advances in cancer research have enhanced our understanding of how metabolic reprogramming supports the anabolic requirements of cancer cell proliferation. It has become clear enough that a single metabolic program cannot be used to globally define altered tumor metabolism.
OXPHOS metabolism: modulation of reactive oxygen species levels and antioxidant response
At the beginning of the 21st century, our view of cancer metabolism gained in complexity and became more attractive, with the birth of new concepts (Fig. 2). The break with the Warburg dogma started when scientists studied cancer metabolism without comparing tumors with their normal counterparts. New studies emerged and proposed a dual capacity for glycolytic and oxidative metabolism in tumor cells (127, 139, 147). First, gene expression profiling of 176 diffuse large B-cell lymphomas revealed the existence of a subset of tumors characterized by an OXPHOS gene pattern (137). In agreement with this observation, N. Danial's laboratory highlighted two subgroups of lymphoma cells with either an OXPHOS or a non-OXPHOS metabolism based on the protein expression level of electron transport chain (ETC) subunits and TCA cycle enzymes (27). Moreover, metabolic functional analysis revealed that the OXPHOS lymphoma cell lines rely more on mitochondrial respiration to grow and survive than the non-OXPHOS cells (27). R. DeBerardinis confirmed that glucose could be both metabolized to lactate and oxidized by mitochondria, using an orthotopic mouse model of human glioblastomas (127). Shortly after, two other studies reported that similar to large B-cell lymphoma, a subset of melanoma is also critically dependent on OXPHOS rather than on glycolysis (78, 215). Indeed, the melanocyte-specific transcription factor, MITF, upregulates the peroxisome proliferator-activated receptor-γ coactivator-1α (PGC-1α), therefore resulting in increased expression of ETC proteins and enhanced OXPHOS (PGC-1α-dependent mitochondrial functions will be addressed below in the Transcriptional regulation of ETC complexes section). PGC-1α-dependent mitochondrial oxidative metabolism is essential for maintaining growth and survival of this subset of human melanomas. Loss of viability caused by suppression of PGC-1α in these melanomas is rescued by induction of glycolysis (118). These original metabolic discoveries opened up the mind of scientists in the field and pinpointed for the first time that within a group of tumors sharing the same clinical features, the metabolic program can be either glycolytic or oxidative.
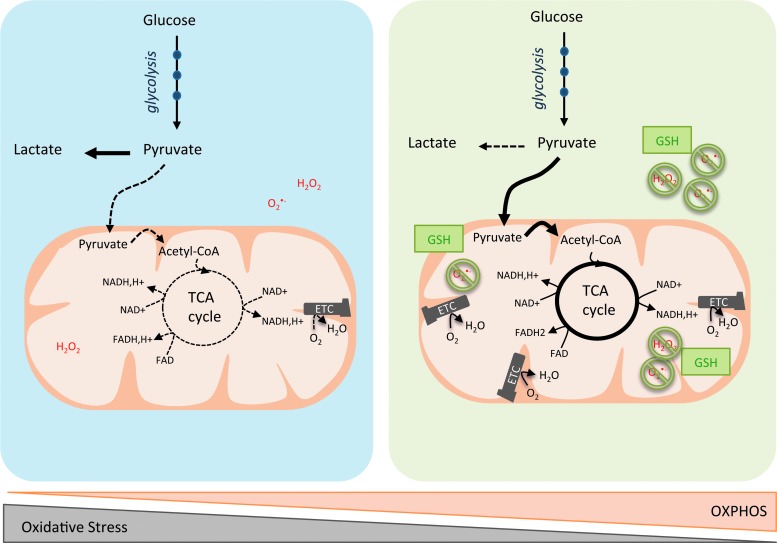
FIG. 2.
OXPHOS/glycolytic metabolism and oxidative stress heterogeneity in cancer cells. Blue left panel is a schematic representation of cancer cells relying predominantly on aerobic glycolysis. Pyruvate is preferentially oxidized into lactate (dark line). Consequently, acetyl-CoA is less incorporated into the TCA cycle (dashed line), which leads to decreased production of reducing equivalents. Some cancer cells exhibit a reciprocal phenotype, with enhancement of the OXPHOS metabolism (green right panel). Here, pyruvate is oxidized into acetyl-CoA and subsequently metabolized into the TCA cycle (dark lines), but less converted into lactate (dashed line). Mitochondrial respiration produces ATP and oxidizes electrons from reduced cofactors and reduces O2 into H2O through the ETC complexes. The various single-electron intermediates can escape and react with O2 forming ROS. OXPHOS cancer cells show elevated antioxidant programs, which help them to detoxify ROS produced by the ETC and regenerate reduced GSH. GSH, glutathione; H2O2, hydrogen peroxide; H2O, water; OXPHOS, oxidative phosphorylation; O2, oxygen; O2(−, superoxide anion radical; ROS, reactive oxygen species. To see this illustration in color, the reader is referred to the web version of this article at www.liebertpub.com/ars
Cancer cell oxidative metabolism can be associated with increased reactive oxygen species (ROS) production, thereby requiring maintenance of redox balance. By oxidizing carbohydrates, lipids, and amino acids, mitochondria produce ATP and electrons—from NADH and succinate—and reduce O2 into water via the ETC. ETCs are major sites of premature electron leakage, generating superoxide and potentially resulting in increased oxidative stress. Excessive generation of ROS or failure in antioxidant scavenging systems can disrupt cellular functions by oxidizing lipids, proteins, and DNA (141). P. Puigserver and N. Danial have both identified an enhanced antioxidant response and increased detoxification capacity in subgroups of melanoma and lymphoma characterized by high OXPHOS activity (27, 118, 215). In lymphoma, the authors reported higher levels of reduced glutathione in the OXPHOS subgroup compared with the non-OXPHOS subgroup (27). Similarly, in melanoma, ROS levels are reduced in the OXPHOS subgroup due to enhanced ROS detoxification capacities mediated by PGC-1α (215). Taken as a whole, these data suggest that OXPHOS metabolism, which relies predominantly on mitochondrial respiration, can lead to oxidative stress resistance through enhanced antioxidant response and increased detoxification capacity.
Sources of carbon: beyond glucose
In addition to the importance of mitochondria for tumor cell survival and proliferation, the use of oxidizable substrates other than glucose, such as glutamine and FA, starts to be well appreciated (Fig. 3). The diversity of carbon substrates fueling cancer cells could be indicative of metabolic heterogeneity within tumors that share the same clinical features. Glucose-independent metabolism was first hypothesized for tumors requiring amino acids for survival.
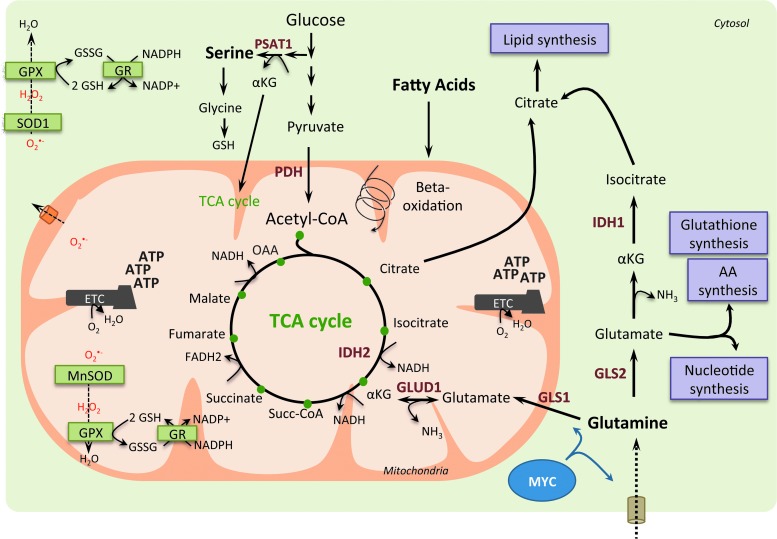
FIG. 3.
Glutamine, serine, and fatty acids as anaplerotic sources for TCA cycle intermediates in cancer cells. In cancer cells, glutamine can fuel the TCA cycle through αKG produced by glutaminolysis. Indeed, glutamine is converted to glutamate by the mitochondrial glutaminase (GLS1) or by the cytosolique isoform (GLS2), and glutamate can be converted to αKG in the mitochondria by the glutamine dehydrogenase 1 (GLUD1). MYC oncogene drives glutamine metabolism by promoting glutamine entry into mitochondria and its conversion into glutamate. Moreover, in cancer cells, fatty acids can be degraded through beta-oxidation, which generates acetyl-CoA subsequently fueling the TCA cycle. Indeed, glutaminolysis and fatty acid beta-oxidation provide intermediates to fuel the TCA cycle, resulting in the generation of reducing equivalents, such as NADH and FADH2. This provides electrons to the ETC and leads to ATP production. TCA cycle intermediates can also be directed into biosynthetic pathways (purple boxes) enabling production of macromolecules, such as lipids, amino acids, and nucleotides. Finally, OXPHOS metabolism through ETC not only produces high levels of ROS but generates also high levels of intermediates with antioxidant capacities, such as reduced GSH and NADPH. NADPH is used as a cofactor for antioxidant enzymes, such as glutathione reductase (GR), which reduces the oxidized glutathione (GSSG) into its reduced form (GSH). Thus, high production of reducing equivalents favors ROS scavenging and prevents deleterious accumulation of ROS in the mitochondrial matrix and cytoplasm. GLUD1, glutamate dehydrogenase 1; GPX, glutathione peroxidase; IDH, isocitrate dehydrogenase; MnSOD, manganese superoxide dismutase; nicotinamide adenine dinucleotide; NH3, ammonia; SOD, superoxide dismutase. To see this illustration in color, the reader is referred to the web version of this article at www.liebertpub.com/ars
Glutamine
Since the 1950s, glutamine is recognized as an important nutrient for tumor metabolism, in addition to its function in nitrogen storage in muscle (181). Among the other energy fuels, glutamine is the most abundant amino acid in the blood and the main donor of nitrogen. In some cell types, glutamine can be generated from intracellular glutamate by the glutamine synthetase (GS), the reverse reaction being catalyzed by glutaminases. This process is important for removal of ammonia or glutamate, whose accumulation is toxic (145). Recently, there is growing evidence regarding the essential role of glutamine in cancer cells to supply cellular ATP by replenishing the TCA cycle (process called anaplerosis) (41). Glutamine was also considered as a building block for protein and nucleotide synthesis, as well as for its antioxidative capacity (40). Neoplastic cells undergo growth arrest and death when they are subjected to glutamine deprivation, further demonstrating their addiction to this substrate. Interestingly, such glutamine-dependent metabolism can be driven by increased c-MYC activity (121, 249). Moreover, stimulating the expression of amino acid transporters, such as SLC1A5 and SLC38A5, in glial tumor cells (241) or repressing miR-23a and miR-23b targeting mitochondrial glutaminase in lymphoma or prostate cancer cells (61) favors glutamine-dependent metabolism. Furthermore, in tumors with PTEN mutations or expressing low levels of PTEN, the mTORC1/S6K pathway also regulates glutamine metabolism through enhanced translation of c-MYC by modulating the phosphorylation of the eukaryotic translation initiation factor, eIF4B (37). Finally, breast cancer subtypes show different exogenous glutamine addiction: basal-like breast tumors are more dependent on exogenous glutamine, compared with luminal tumors, due to reduced expression of GS (107). Another study using functional assays in 47 independent breast cancer cell lines confirmed that triple-negative cancer cells consume more glutamine than luminal samples (210). These studies suggest that basal-like breast cancers might be susceptible to glutamine-targeting therapeutics. Recently, metabolomic profiling of human breast tumors showed that the oncometabolite 2-hydroxyglutarate (2HG) accumulates in triple-negative breast cancers (209). Interestingly, this is not linked to isocitrate dehydrogenase 1 or 2 (IDH1 or IDH2) mutations, as previously shown in gliomas and leukemia (39, 158, 230), but associated with MYC pathway activation and glutaminase overexpression (209), thereby corroborating previous data on glutamine addiction in triple-negative breast cancers. In KRAS-mutated human pancreatic ductal adenocarcinoma, glutamine supports pancreatic cancer cell growth by a noncanonical pathway: while most cancer cells convert glutamine into alpha-ketoglutarate in the mitochondria to fuel the TCA cycle, pancreatic cancer cells rely on a distinct pathway in which glutamine-derived aspartate is transported into the cytoplasm, where it is converted into oxaloacetate (by the glutamic oxaloacetic transaminase 1, GOT1) and subsequently into malate and pyruvate (197). This increases the NADPH/NADP+ ratio and maintains the cellular redox state. Consequently, glutamine deprivation suppresses pancreatic cancer cell growth by increasing ROS levels (197). Finally, glutamine metabolism is also regulated by hypoxia through HIF-1 activation. HIF-1 promotes SIAH2-mediated ubiquitination and degradation of the E1 subunit of the mitochondrial complex alpha-ketoglutarate dehydrogenase that supports glutamine-dependent lipid synthesis (195, 202).
Serine/glycine
Recent advances in understanding the role of metabolism in tumorigenesis have demonstrated the relevance of serine/glycine biosynthesis (2). Indeed, although glucose and glutamine are main energy sources used to maintain glycolysis and TCA cycle, the serine anabolic pathway represents also an important feature in glucose conversion and cancer development (97). Serine can be either imported into cells or de novo synthetized from a derived branching of glycolysis (Fig. 1). The glycolytic intermediate 3-phosphoglycerate can be converted into serine following a three-step enzymatic reaction catalyzed by phosphoglycerate dehydrogenase (PHGDH), phosphoserine aminotransferase 1 (PSAT1), and phosphoserine phosphatase (PSPH) (Fig. 1). Serine can next be converted into glycine, which is a major source of methyl groups for the one-carbon pools required for the biosynthesis of protein, purines, and DNA/histone methylation, as well as glutathione. The serine/glycine pathway thus modulates cellular antioxidant capacity and is involved in redox homeostasis. Moreover, PSAT1 uses the PHGDH by-product, namely 3-phosphohydroxypyruvate, to convert glutamate into alpha-ketoglutarate, anaplerotic intermediate that fuels back the TCA cycle and sustains cancer metabolism. In cells with reduced PHGDH or PSAT1 expression, the conversion of glutamine to glutamate and to alpha-ketoglutarate is significantly reduced, while the serine biosynthesis pathway remains the major contributor of anaplerotic supplies of TCA intermediates (123). Genetic activation of serine biosynthesis drives cancer cell proliferation and predisposes normal mammary epithelial cells to cancer transformation (162). Moreover, cells lacking p53 fail to respond to serine starvation due to oxidative stress, which leads to reduced viability and impaired proliferation (125). Thus, p53 helps cancer cells to overcome serine starvation, preserving by this way their cellular antioxidant capacity. Finally, serine acts as an allosteric activator of PKM2, the less active isoform of the pyruvate kinase. Upon serine deprivation, PKM2 activity is reduced, and pyruvate is diverted to a fuel-efficient mode in the mitochondria (245). Moreover, the serine/glycine pathway is also regulated through the NRF2-ATF4 signaling pathway in nonsmall cell lung carcinoma (NSCLC). Indeed, NRF2 constitutive activation through KEAP1 mutation promotes tumorigenesis via ROS detoxification and confers a poor prognosis in human NSCLC (44).
Fatty acids
In addition to glucose and glutamine, FAs represent a particularly relevant source of energy. They are either directly incorporated into the cells from the extracellular media or obtained from hydrolyzed triglyceride droplets. Until recently, most studies on cancer metabolism focused on glycolysis, glutaminolysis, and FA synthesis rather than on FA beta-oxidation, while this pathway is one the most energetic and provides six times more ATP than glycogen oxidation (28). Inhibition of FA oxidation sensitizes most cancer cells to apoptosis, supporting the key role of FAs in cancer cell survival and suggesting that FA oxidation inhibitors could provide substantial therapeutic interest (38). FA beta-oxidation takes place in the mitochondria and generates acetyl-CoA, NADH, and FADH, cofactors used by the ETC. Some cancer cells, such as the high-OXPHOS large B-cell lymphoma described above, preferentially use FA as a substrate and metabolize palmitate, rather than glucose or glutamine, for sustaining their growth (27). FA oxidation is also essential for survival of lymphoma cells by supplying ATP pools and recycling both glutathione and NADH, thereby counteracting oxidative stress (161). FA oxidation has thus a dual role: it is essential for redox balance, but, in the meantime, it can increase ROS levels in case of nonalcoholic fatty liver disease (64). Finally, P. Carmeliet's laboratory has recently reported a new role for FA beta-oxidation in de novo nucleotide synthesis and DNA replication in endothelial cells during angiogenesis (185).
Fine-tuned regulation of mitochondrial respiratory chain complex synthesis in neoplastic cells
As previously mentioned, cancer cells with high OXPHOS activity are characterized by accumulation of ETC proteins, which are yet finely regulated. This chapter will thus describe the mechanisms involved in regulating expression of ETC-encoding genes and proteins (Fig. 4). Mitochondria biogenesis is a complex process that requires the synthesis, import, and assembly of proteins and lipids within the existing mitochondrial reticulum. It also implies replication of the mitochondrial DNA (mtDNA). ETC proteins are encoded by both nuclear and mitochondrial DNA. Nucleus-encoded factors control the transcription as well as replication of mtDNA and achieve coordination of the two genomes (183). Remarkably, human mtDNA is of small size (16,569 bp), but it is present in high number of copies (around 1000). ETC complexes are synthetized by less than 100 genes, 13 of them being encoded by mtDNA. These 13 proteins participate in the composition of 4 different ETC complexes (Complex I, III, IV, V). The rest of the subunits (> 35 subunits for Complex I, 10 subunits for Complex III and IV, and 14 subunits for Complex V) and the entire complex II (4 subunits) are encoded by nuclear DNA (3, 9, 184, 191, 224). mtDNA also encodes 22 transfer RNA (tRNA) and two ribosomal RNAs (rRNA) involved in the synthesis of ETC subunits. mtDNA is not protected by histones and is thus extremely vulnerable to oxidative damages generated in the matrix (1). Metabolic processes are regulated by a number of different mechanisms, including allosteric regulation, enzyme degradation, and reversible post-translational modifications (PTMs). In this part, we will focus on the different levels of regulation that could be responsible for enhancing OXPHOS metabolism in cancer cells.
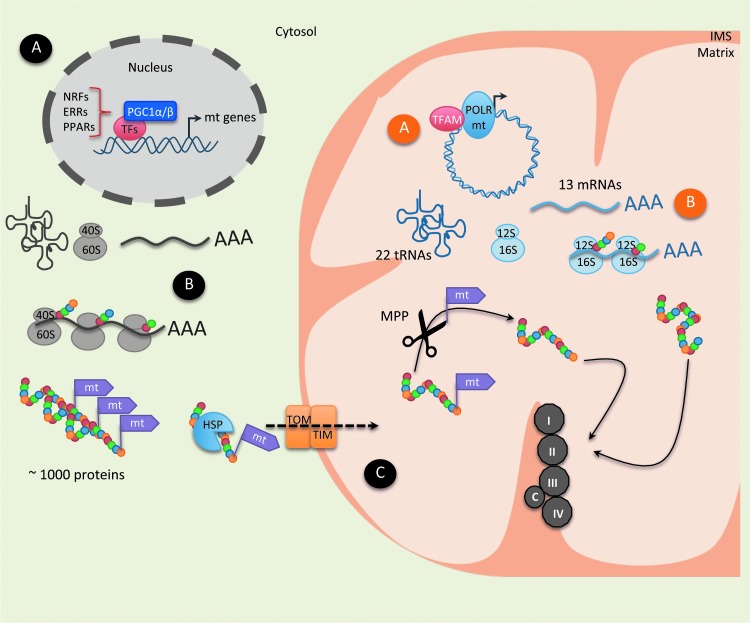
FIG. 4.
Schematic representation of the different levels of regulation of ETC complexes. The mtDNA encodes 13 ETC proteins, 22 tRNAs, as well as 12S and 16S rRNAs, whereas the nuclear DNA encodes approximately 1000 proteins belonging to ETC complexes. There are three levels of regulation: transcriptional, translational, and post-translational. (A) Transcriptional regulation: In the nucleus, PGC-1α and various transcription factors, such as NRF, ERR, and PPAR, bind to the regulatory regions of mitochondrial (mt) target genes and stimulate their expression. Inside mitochondria, TFAM, a mitochondrial DNA transcription factor, cooperates with the mtPOLR to induce the expression of mtDNA-encoded proteins. (B) Translational regulation: Mitochondrial-addressed proteins, which are transcribed in the nucleus, are exported into the cytoplasm, where their translation takes place via the cytoplasmic ribosomal (40S and 60S) machinery. While nuclear DNA encodes most of the mitochondrial proteins, few of them are encoded by mtDNA and synthesized by the mitochondrial translation system. mRNAs transcribed into the mitochondrial matrix are translated by the mitochondrial ribosomal 12S and 16S complexes. (C) Post-translational regulation: Upon translation, many mitochondrial proteins are synthesized as precursor proteins, with cleavable N-terminal presequences. The TOM complex allows the translocation of tagged proteins from the outer membrane barrier to the IMS. The tag signal and adjacent parts of the protein are recognized by the TOM complex, which works together with the TIM complex to translocate proteins into the mitochondria. Once imported, the N-terminal signal of the precursor protein is processed through MPP and mature isoforms are assembled into the IMS. Mitochondrial proteins, encoded by the mitochondrial or nuclear genomes, are finally assembled to form ETC complexes. ERR, estrogen-related receptor; HSP, heat shock protein; IMS, inner membrane space; MPPs, mitochondrial processing peptidases; mtDNA, mitochondrial DNA; mt, mitochondrial; NRF, nuclear respiratory factor; mtPOLR, mitochondrial RNA polymerase; PPAR, peroxisome proliferator-activated receptor; PGC-1α, peroxisome proliferator-activated receptor-γ coactivator-1α; TF, transcription factor; TIM, transporter inner membrane; TOM, translocase of the outer membrane; tRNA, transfer RNA; rRNA, ribosomal RNA. To see this illustration in color, the reader is referred to the web version of this article at www.liebertpub.com/ars
Transcriptional regulation of ETC complexes
Regulation of mitochondrial biogenesis represents a transcriptional challenge finely orchestrated by transcription factors and coregulators (183). The nuclear respiratory factors 1 and 2 (NRF-1 and NRF-2) were the first identified nuclear transcription factors involved in regulating the transcription of ETC-encoding genes (47, 73, 142, 182). NRF-1 and NRF-2 binding sites are evolutionary conserved in the proximal promoters of many mitochondrial genes, encoding ETC components, mitochondrial transporters, and mitochondrial ribosomal proteins (MRPs) (183). Moreover, NRF-1 activates the transcription of genes encoding the mitochondrial transcription factors A (TFAM), B1 (TFB1M), and B2 (TFB2M) that mediate replication and transcription of the mitochondrial genome (102, 219). Therefore, NRF-1 coordinates nuclear mitochondrial gene expression with mtDNA replication (68). NRF-2 was identified as a transcriptional activator of many mitochondrial genes encoding ETC components, mitochondrial import, TFAM, TFB1M, and TFB2M (220). Estrogen-related receptors (ERRs) also regulate the expression of genes involved in OXPHOS, mitochondrial dynamics, and oxidative stress defenses (42, 186, 218). Furthermore, transcriptional coregulators ensure the coordinated expression of all ETC components by interacting with and coactivate several nuclear receptors through a specific LXXLL motif (130). The PGC-1 family, originally identified in the mitochondrial-rich brown adipose tissue (164), comprises 3 members, PGC-1α, PGC-1β, and PRC (PGC-1-related coactivator), which regulate mitochondrial biogenesis in a wide variety of tissues. These 3 coactivators are able to interact with NRF-1, NRF-2, PPARs, and ERRs. While PGC-1β participates in the maintenance of basal mitochondrial function, PGC-1α promotes mitochondrial mass increase as an immediate tissue adaptation to high energetic demands (119, 182, 199). The function of PRC, the less characterized member of the family, appears to be restricted to the regulation of mitochondrial biogenesis in proliferating cells (216). PGC-1α expression level is either decreased in some cancer, including colon (50), breast (232), and ovary (251), or increased in other cancer types (112, 215), suggesting a differential regulation of ETC complexes and OXPHOS. In agreement with these observations, high and low OXPHOS subgroups of melanoma exhibit different PGC-1α expression rates (215). Tumor cells harboring high PGC-1α mRNA levels show elevated ETC protein levels and enhanced mitochondrial respiration (215). PGC-1α transcription factor is also regulated by PTMs, as discussed below (Post-translational regulation of ETC complexes section). Silencing PGC-1α is sufficient to decrease mitochondrial respiration and ETC protein expression in melanoma cells, demonstrating that PGC-1α is a key actor in OXPHOS metabolism in cancer (215). Similarly, a recent study using both mouse models and silencing strategies demonstrates that PGC-1α enhances OXPHOS metabolism, mitochondrial biogenesis, and oxygen consumption rate in invasive breast cancers (112). In addition, increased expression of c-MYC in many aggressive tumors upregulates mitochondrial biogenesis by regulating expression of the TFAM gene (116). Thus, transcriptional regulation of mitochondrial ETC complexes varies among tumor types and plays an important role in driving OXPHOS metabolism.
Translational regulation of ETC complexes
While nuclear DNA encodes most of the mitochondrial proteins, few of them are encoded by mtDNA and synthesized by the mitochondrial translation system. Although the mechanisms of translation are well characterized in the cytoplasm, little is known about mitochondrial protein translation. Impaired mitochondrial translation usually results in severe respiratory dysfunction through the lack of all mtDNA-encoded proteins (193, 244). Mitochondrial mRNAs are uncapped (72), but contain a poly(A) tail (150). Unlike cytoplasmic translation, the small subunit of mitochondrial ribosomes (mitoribosomes) binds mRNAs in a sequence-independent manner (117). The mitochondrial translation machinery consists of 2 initiation factors and 3 elongation factors. The termination process has not been entirely elucidated, although 2 release factors (mtRF1 and mtRF1a) and 1 recycling factor (mtRRF) have been partly characterized (174). The human mitoribosome consists of two rRNAs (12S and 16S) and about 81 MRPs (194). Levels of mitochondrial proteins are regulated by their own translational activators, which bind to 5′-untranslated regions (82). Translational activator imbalance alters mitochondrial protein levels and ultimately impairs respiration (55, 240), demonstrating the key role of mitochondrial-driven translation in OXPHOS activity.
Mutations or alterations of the mitochondrial translational process have not been deeply investigated in cancer cells (62), despite the recent identification of high OXPHOS tumors (27, 78, 215). In contrast, translational defects of nuclear-encoded mitochondrial proteins have been investigated for years as one of the major pathways involved in regulating protein translation, the so-called mammalian target of rapamycin (mTOR) pathway, is often deregulated in human cancers. mTOR interacts with several proteins to form two distinct complexes, namely mTORC1 and mTORC2, which have different upstream regulators, downstream effectors, and exhibit distinct functions (110). mTORC1 senses nutrient availability and is a major regulator of protein translation. N. Sonenberg has recently reported that mTORC1 controls both mitochondrial activity and biogenesis through the eukaryotic translation initiation factor, eIF4E, in vitro and in vivo (140). mTOR inhibitors impair mitochondrial respiration by inhibiting translation of TFAM, subsequently reducing expression of ETC genes (140). In addition to their transcriptional stimulatory activity, TFB1M and TFB2M have rRNA methyltransferase activity (129). Thus, these factors are indirectly involved in mitochondrial protein synthesis via their ability to methylate the mitochondrial 12S rRNA, which is important for mitoribosome activity.
Post-translational regulation of ETC complexes
Mitochondrial addressing
Most mitochondrial proteins, including proteins involved in mitochondrial translation, are synthetized in the cytoplasm as nuclear DNA encodes them. Therefore, they need to be imported into mitochondria. Cytosolic chaperones, such as heat shock proteins, HSP70 and HSP90, escort precursor proteins toward translocation channel receptors on the mitochondrial membrane. It has been suggested that cytoplasmic and mitochondrial translation machineries are in close proximity to allow efficient assembly of the ETC system (87). Nuclear DNA-encoded proteins, either tagged by an N-terminal mitochondrial targeting signal (N-MTS) (85) or containing an internal signal (23), are recognized by receptors of the outer mitochondrial membrane and imported into the mitochondria (9). The translocase of the outer mitochondrial membrane (TOM) complex allows the translocation of mitochondrial precursor proteins from the outer membrane barrier to the intermembrane space (IMS) (196). The N-MTS and its adjacent parts are recognized by the TOM complex, which works together with the translocase of the inner mitochondrial membrane (TIM) complex to transfer proteins into the mitochondria (9, 168). After translocation, the N-MTS is removed by the mitochondrial processing peptidases, and ETC mature proteins are then released into the IMS. As shown by this short description, mitochondrial protein targeting, import, processing, and assembly require a complex series of processes, which can be mutated or altered during tumorigenesis. In that sense, N-MTS mutation in one of the PDH subunits was identified as the cause of PDH import defect and PDH deficiency in humans (206). Similarly, genetic dimorphism in the N-MTS of the manganese superoxide dismutase (MnSOD) affects MnSOD import into mitochondria and is considered as a risk factor for severe alcoholic liver disease and hepatocellular carcinoma (143, 203). As translocation across the inner membrane requires an electrochemical hydrogen ion gradient (transmembrane potential ΔΨ), redox imbalance alters this process and impairs mitochondrial respiration, as observed in cancer cells (242).
Lysine acetylation
PTMs, including phosphorylation, acetylation, methylation, N-linked, and O-linked glycosylation and ubiquitination, are involved in cell signaling networks. Lysine acetylation is conserved among species and links acetyl-CoA to cellular metabolism. Acetylation was long considered as an epigenetic modification mainly occurring on chromatin-associated proteins, but recent proteomic analyses reveal that most acetylation events occur also on non-nuclear proteins, particularly on mitochondrial proteins (35). Lysine acetylation is regulated both by lysine acetyltransferases and lysine deacetylases, which add or remove acetyl groups from proteins, respectively. Lysine acetylation is one of the most prevalent mitochondrial PTMs (233). Reciprocally, metabolic intermediates modulate lysine acetylation (81, 238). Indeed, acetyl-CoA synthesis occurs in two distinct compartments defined by the permeability of the mitochondrial membrane (238). Nuclear histone acetylation levels are directly correlated with the activity of the cytosolic ATP citrate lyase, an enzyme that converts glucose-derived citrate into acetyl-CoA in human cells and is considered as a new link between nutrient metabolism and histone acetylation (237).
Lysine acetylation regulates enzyme activity as lysine residues are often present in the active sites of enzymes. For instance, acetyl-CoA synthetase 2 (ACSS2) is a mitochondrial matrix protein that produces acetyl-CoA from acetate. ACSS2 is reversibly acetylated at K642 residue (localized into its enzymatic active site) and thereby inactivated (59). In human breast cancer, the ACSS2 gene is amplified, highly expressed, and positively correlated with tumor invasiveness (187). Comparative metabolomic and lipidomic analyses demonstrated that ACSS2 promotes a switch in nutrient utilization from glucose to acetate to support FA and phospholipid biosynthesis in cancer cells. ACSS2 silencing reduces tumor growth in xenograft mouse models. Thus, acetate consumption exhibits a critical role in lipid biomass production and membrane biogenesis in cancers. This constitutes the rational basis for using acetate-based PET tracers in breast cancer diagnosis (187). In contrast to ACSS2, acetylation of the glycolytic enzyme, phosphoglycerate mutase 1 (PGAM1), enhances its activity and stimulates glycolysis (76). Moreover, glucose deprivation triggers increased levels of sirtuin 1 (SIRT1), an evolutionary conserved NAD-dependent histone deacetylase (19, 91), leading to PGAM1 deacetylation. These results suggest that SIRT1 might have a role in the regulation of glycolysis flux by preventing depletion of glycolytic intermediates used in other biosynthetic pathways, such as the serine pathway. Additionally, SIRT1 has been previously reported to affect metabolic pathways at the transcriptional level, promoting beta-oxidation and inhibition of lipogenesis through PGC-1α regulation (75). Indeed, PGC-1α activity is regulated at post-translational levels. Mitochondrial biogenesis is induced by PGC-1α phosphorylation at threonine-177 and serine-538 residues by AMPK (89). In addition, PGC-1α is activated by deacetylation by SIRT1 (171). The cross-talk between SIRT1 and PGC-1α thus represents a key regulatory process in cancer metabolism (19, 91). Thus, examining the contribution of key actors involved in acetylation regulation will likely provide novel insights into cancer metabolism.
Role of Metabolism on Cancer Cell Properties and Functions
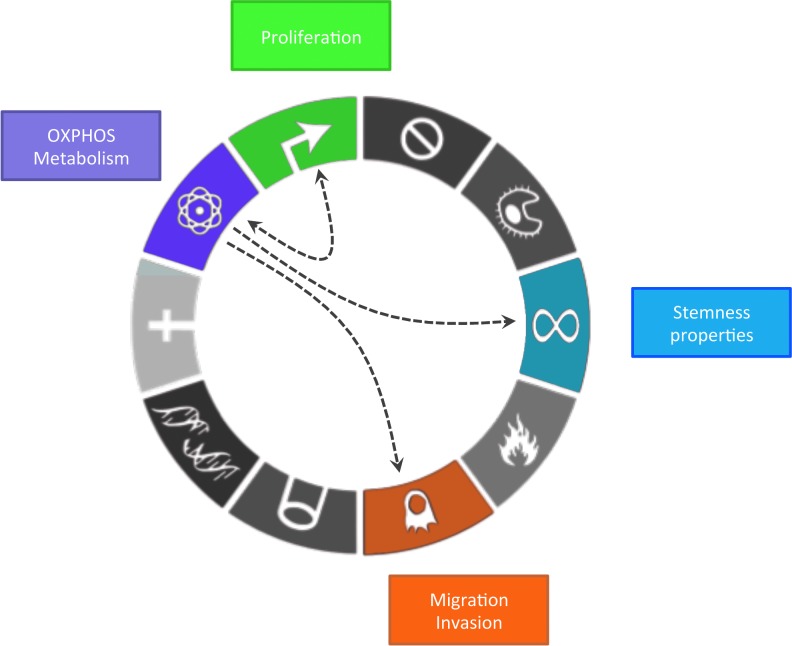
As previously shown, metabolism in cancer is much more heterogeneous than initially thought, but the impact of these metabolic variations (in particular OXPHOS regulation) in cancer development and progression remains unclear. In this chapter, we address this question. Indeed, oncogenic transformation requires metabolic adaptation to support stemness properties of cancer cells and their highly energy-consuming functions, such as proliferation, growth, migration, and invasion (Fig. 5).
FIG. 5.
OXPHOS metabolism on cancer cell functions integrated into the 10 hallmarks of cancer from Hanahan and Weinberg, 2011. OXPHOS metabolism (purple box) supports high proliferative capabilities of cancer cells (green box). Reciprocally, cell cycle regulators are able to promote OXPHOS metabolism. Moreover, OXPHOS metabolism favors migration and invasion of cancer cells (orange box). Finally, OXPHOS metabolism plays an important role as it increases cancer cell stemness (blue box). To see this illustration in color, the reader is referred to the web version of this article at www.liebertpub.com/ars
Proliferation
Cancer metabolism is often considered as an adaptation to sustain cancer cell proliferation. Since O. Warburg's studies, it has been assumed that cancer cell growth and proliferation require increased glycolytic and decreased oxidative metabolism. This metabolic switch is not only required for energy production but also for the synthesis of nucleotides, proteins, and lipids to generate the building blocks supporting uncontrolled tumor proliferation and growth. Indeed, there is clear evidence showing that both mitochondrial morphology and respiratory function are linked to cell cycle regulators such as cyclins. Cyclin D1, well known to promote nuclear DNA replication through phosphorylation and inactivation of the retinoblastoma protein (pRB), is involved in mitochondrial bioenergetic coordination during G1 progression (178). The overexpression of cyclin D1 during G1 leads to a twofold decrease in mitochondrial activity mediated by CDK4 kinase activity, independently of pRB (225). Conversely, hepatocytes lacking cyclin D1 exhibit increased mitochondrial size and activity, both associated with an increased expression of Nrf1 (225). Cyclin E controls the formation of high-energy charged mitochondria during G1/S transition (136). During G2/M transition, the cyclin B1/CDK1 complex is a key regulator of mitochondrial fission (205), respiration, and ATP production (227). Both cyclin B1/CDK1 mitochondrial abundance and kinase activity are dramatically increased during G2/M, leading to ETC complex I phosphorylation and enhanced activity (227). Finally, cyclin-dependent regulation of mitochondrial activity is consistent with enhanced OXPHOS activity during G2/M transition, while glycolysis rather supplies the energy needs during G1 (6). Notably, in colon cancer cells, ATP production during G2/M transition is predominantly dependent on mitochondrial respiration (6).
Recently, the team of L. Fajas investigated the regulation of cell cycle by mitochondria (15, 124). They observed that mice lacking E2 transcription factor 1 (E2F1), an important regulator of S-phase entry and a downstream target of pRB, often inactivated in human cancers, consume more O2 than control mice. This phenotype is associated with increased expression of genes involved in ETC composition, mitochondrial biogenesis, beta-oxidation, and uncoupling activity in muscle and brown adipose tissue (15). Computational analysis revealed the identification of E2F binding sites in the promoter of these OXPHOS genes, suggesting direct transcriptional regulation by E2F. Moreover, RB deletion affects the mitochondrial biogenesis transcription program in erythroid progenitor cells due to a decrease in PGC-1β expression (180). Exploiting a novel link between cell cycle regulators and mitochondria may help to sensitize tumors to treatment and may offer new combinatory therapeutic strategies. Given the recent success of CDK4/6 inhibitor (PD0332991) in clinical trials for HER2-positive advanced breast cancer patients (54, 152, 247), it would be interesting to investigate the impact of such compound on mitochondria morphology and function.
Despite an increasing number of studies showing the importance of mitochondrial respiration for cancer cell proliferative capacities, the reasons for this requirement remained unclear until recently. Similarly, it was unknown why pyruvate supplementation allows ETC-defective cells to proliferate (104). Two recent studies (11, 201) have highlighted these questions. They showed that respiration-deficient cells are auxotrophic for pyruvate, which acts as an electron acceptor and plays a key role in redox balance. Moreover, they demonstrated that an essential function of mitochondrial ETC complexes in cell proliferation is to enable aspartate biosynthesis (11, 201). Pyruvate stimulates aspartate synthesis in a GOT1-dependent manner, GOT1 being the cytosolic aspartate aminotransferase, and is sufficient to rescue proliferation in ETC-deficient cells (11, 201). Altogether, these studies provide a new perspective on the different roles of the ETC complexes in metabolism and proliferation capacity.
Migration and metastases
Whether and how metabolic reprogramming may play a role in tumor progression and metastatic process remain elusive. Recently, the identification of specific metabolites affecting cancer cell migration and invasion, as well as the characterization of bioenergetic profiling of invasive and metastatic cancer cells, brought new insights. Metabolic stress, such as hypoxia or oxidative stress, enhances stemness, angiogenesis, invasiveness, and metastatic potential of tumor cells and is therefore of poor prognosis. Indeed, HIF-1α activation causes E-cadherin loss and epithelium-to-mesenchymal transition (EMT), a process by which epithelial cancer cells acquire invasive properties as they loose their polarity and their cell-to-cell adhesion (36, 163). Aerobic glycolysis causes extracellular matrix (ECM) acidification, which leads to matrix metalloproteinase and cathepsin activation. As a consequence, there is increased ECM degradation (204), a process that takes part in most basic cell behaviors, including cell migration. Specific metabolites, such as glutamine, play a critical role in cancer cell invasion in vitro and in vivo. A recent study, using The Cancer Genome Atlas database, reported that the poor survival of ovarian cancer patients is significantly correlated with elevated expression of genes involved in glutaminolysis or TCA cycle and low expression of glycolytic genes (243). Highly invasive ovarian cancer cells preferentially use glutamine rather than glucose to replenish TCA cycle, which dramatically increases the oxygen consumption rate. Affecting oxygen consumption by using rotenone, a mitochondrial respiratory chain inhibitor, reduces both TCA cycle activity and invasiveness of ovarian cancer cells (243). Similarly, a study aiming at identifying the bioenergetic profiles of invasive and metastatic cancer cells uncovered that human and mouse metastatic cells are characterized by increased mitochondrial biogenesis and ATP production, as opposed to primary tumor cells (112). This was associated with elevated levels of PGC-1α. Moreover, PGC-1α silencing dramatically reduced primary tumor cell invasiveness and metastasis without affecting tumor growth. Interestingly, patients with invasive breast cancer exhibiting high PGC-1α expression have increased metastasis and decreased survival rates. Altogether, this suggests that PGC-1α-induced OXPHOS metabolism is essential for the metastatic capabilities of breast cancer cells (112). Importantly, distinct metabolic signatures of breast cancer cells were found to predict the organ site of metastasis (45). While liver-metastatic breast cancer cells exhibit enrichment in glycolytic genes, bone- and lung-metastatic cells display an increased OXPHOS metabolism and glutaminolysis associated with accumulation of TCA cycle intermediates, such as citrate and succinate. Liver-metastatic breast cancer cells maintain their glycolytic functions via HIF-1α and pyruvate dehydrogenase kinase 1 (PDK1). PDK1 expression is always high in liver metastases, while it is either low or high at the primary tumor site. This suggests that the metabolic switch influencing tropism of cancer cells may occur when they acquire metastatic properties (45). It would be interesting to investigate whether this results from intrinsic heterogeneity in cancer cell metabolic program or a clonal selection, mediated by TME, such as oxygen concentration that varies from one metastatic organ to the other.
Increased stem cell properties
Quiescent adult stem cells generally exhibit low metabolism associated with a slow proliferation rate and reduced risk of ROS-mediated cellular damage and long-lived renewal capacity (88, 175, 200). This low metabolism mainly depends on glycolysis (41, 154). Similar to cancer cells showing high glycolytic flux with some OXPHOS heterogeneity (as described above in the OXPHOS metabolism: modulation of reactive oxygen species levels and antioxidant response section), the new concept of bioenergetic heterogeneity of cancer stem cells (CSCs) is now commonly recognized. While some CSCs rely on glycolytic metabolism, there is also an enrichment of CSCs exhibiting OXPHOS metabolism that would confer resistance to chemotherapy (32, 70, 156). Recently, the mitochondrial features and metabolic properties of CSCs and noncancer stem cells (non-CSCs) have been investigated in many cancers. Lung CSCs show a lower mtDNA content, O2 and glucose consumption, ATP, and ROS intracellular concentration compared with non-CSCs (246). Similarly, leukemia CSCs exhibit lower ROS levels and OXPHOS metabolism compared with non-CSCs (108). However, ovarian CSCs show an increased expression of genes involved in glucose uptake, OXPHOS metabolism, and FA beta-oxidation (159). Moreover, CSCs from glioma or glioblastoma also depend on OXPHOS for their energy production and survival compared with their differentiated progeny, which mainly rely on aerobic glycolysis (90, 221). However, the metabolic state (OXPHOS vs. aerobic glycolysis) of CSCs is less clear in breast cancer as it is still debated due to discrepant results (51, 222). Taken as a whole, these studies highlight the heterogeneity of CSC bioenergetic properties within tumors, before any treatment.
Interestingly, there is also increasing evidence that chemotherapy influences CSC metabolic status. Metastatic melanoma is a heterogeneous tumor of neuroectodermal origin with less than 1-year median survival. Despite the encouraging initial tumor response to BRAF inhibitor vemurafenib, relapse occurs only after few months of treatment due to multiple resistance mechanisms. Among the highly proliferative melanocytes, a subpopulation of slow-cycling cells has been identified after chemotherapy. These slow proliferative cells exhibit high levels of the histone H3K4-demethylase JARID1B, high levels of mitochondrial ETC proteins, and increased OXPHOS activity (173). Similar metabolic heterogeneity and plasticity were identified in CSCs isolated from human primary pancreatic tumors and patient-derived xenografts (179). As previously reported in other cancers, pancreatic CSCs rely mainly on OXPHOS metabolism, while the non-CSCs are dependent on glycolysis (179, 217). Interestingly, the use of the OXPHOS inhibitor, metformin, to selectively kill the CSCs, leads to the positive selection of a resistant CSC subpopulation, while CSC treatment with the mitochondrial ROS inducer, menadione, does not (179). Importantly, this resistant CSC subpopulation activates a glycolytic program through an MYC/PGC-1α-dependent regulatory process (179). In conclusion, deciphering the mechanisms involved in CSC metabolic heterogeneity and adaptability would be crucial to identify key actors enabling CSCs to escape from chemotherapy.
Reciprocal Impact of Cancer Cell Metabolism on TME
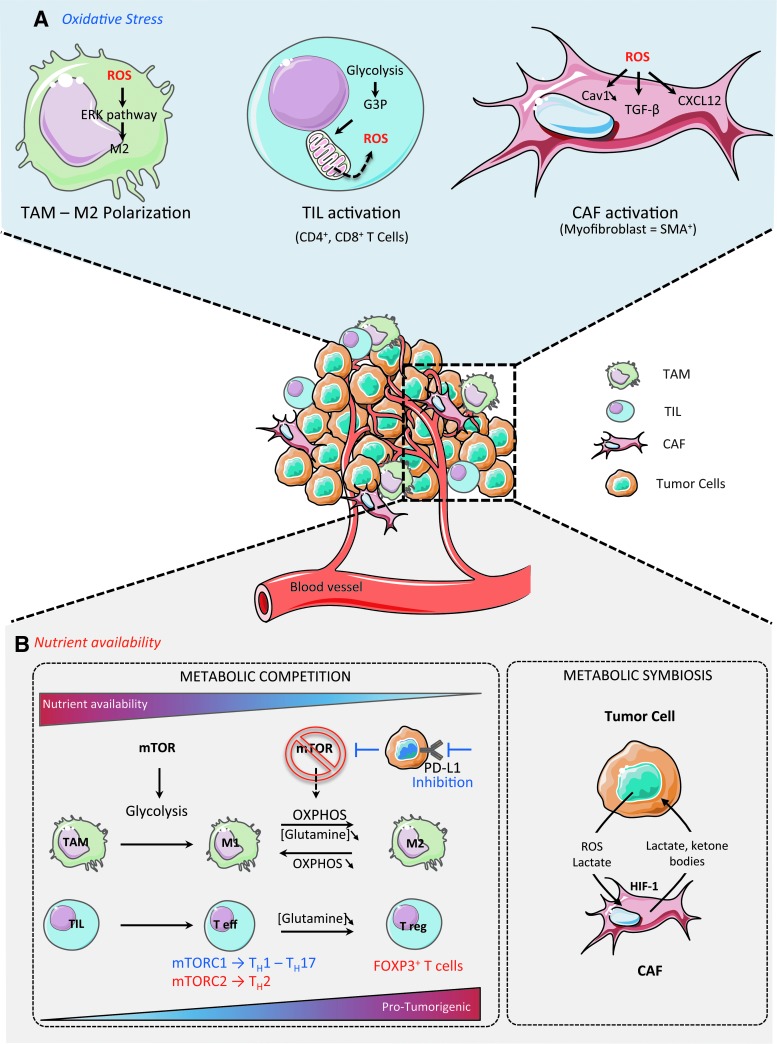
TME plays a crucial role in tumor development, growth, and spreading. The view of cancer as a heterogeneous disease was initially restricted to genetic mutations in tumor cells. However, it is now well accepted that tumor heterogeneity also results from stromal diversity (165, 189), including tumor-associated macrophages (TAMs), tumor-infiltrating lymphocytes (TILs), and carcinoma-associated fibroblasts (CAFs). The proportion and the activation state of each cell population also increase TME heterogeneity. Some recent studies pinpoint both the importance of ROS-induced mitochondrial metabolism and nutrient availability in TME regulation (Fig. 6).
FIG. 6.
Reciprocal impact of cancer cell metabolism on TME. (A) Oxidative Stress and TME. TAM: ROS signaling promotes M2 polarization through the ERK signaling pathway; TIL: TIL activation (differentiation in CD4+, CD8+ T cells) and expansion increase rates of glycolysis and generate excessive amount of glycerol-3-phosphate. G3P is oxidized by the mitochondrial G3P dehydrogenase 2, which in turn increases ROS production; CAF: ROS promote CAF activation and conversion of fibroblasts into myofibroblasts through TGF-β and CXCL12 (SDF-1)-dependent pathways. (B) Nutrient availability and TME. Both immune and stromal cells participate in a complex metabolic interplay with neoplastic cells. They can collectively adapt in a dynamic manner to the metabolic needs of cancer cells and thus participate in tumorigenesis. Metabolic competition between immune and tumor cells: Tumor cell metabolism modulates nutrient availability in TME, impacting macrophage polarization and immune response. High nutrient availability in the TME favors glycolysis through mTOR signaling and promotes M1 polarization and Teff differentiation. mTORC1 is involved in naïve CD4+ T-cell differentiation into T helper 1 (TH1) and T helper 17 (TH17) cells, supporting an antitumor effect. In contrast, mTORC2 promotes the differentiation of naïve CD4+ T cells into the protumorigenic T helper 2 (TH2) cells. Moreover, M2 macrophages exhibit an OXPHOS metabolism. Interestingly, blocking OXPHOS metabolism induces M1 polarization, while forcing OXPHOS metabolism in M1 macrophages potentiates M2 polarization. Glutamine deprivation promotes the differentiation of naïve CD4+ T cells into FOXP3+ Treg cells and thus induces a shift in the immune response balance, which becomes immunosuppressive. Glutamine deprivation also impacts M2 phenotype by promoting a protumorigenic response. Moreover, glucose availability is also another layer of TIL regulation. As progressing tumors have higher glucose consumption than the regressing ones, TILs from progressing tumors are glucose restricted and exhibit impaired effector functions. Blocking PD-L1 in tumor cells reduces their glycolysis rates by inhibiting mTOR activity, which consequently increases extracellular glucose availability for TILs. Thus, by modulating tumor cell metabolism, one can control nutrient availability for T cells, thus promoting either their antitumor or immunosuppressive functions. Metabolic symbiosis between CAFs and tumor cells: Increased ROS production by cancer cells, in particular the highly diffusible H2O2, stimulates HIF-1 signaling in CAFs. As a consequence, CAFs switch their metabolism toward aerobic glycolysis, through an HIF-1- and oxidative stress-dependent mechanism. This highly glycolytic rate in CAFs provides nutrient and energetic fuels, such as lactate and ketone bodies, to cancer cells. This symbiotic relationship between CAFs and tumor cells is reversible, thus representing a metabolic optimization in cancer treatments. CAF, carcinoma-associated fibroblast; Cav 1, caveolin 1; HIF-1, hypoxia-inducible factor-1; mTOR, mammalian target of rapamycin; SMA, smooth muscle cells; TAM, tumor-associated macrophage; Teff, effector T cell; TIL, tumor-infiltrated lymphocyte; TGF-β, transforming growth factor-β; Treg, regulatory T cell. To see this illustration in color, the reader is referred to the web version of this article at www.liebertpub.com/ars
Oxidative stress and TME activation
Tumor-associated macrophages
Macrophages are sentinel cells that maintain homeostasis by regulating innate immunity in response to several stimuli, such as pathogen-associated molecular patterns or cytokines found in surrounding tissue (14). In cancer, TAMs influence several aspects of tumor progression, such as angiogenesis, metastasis, and survival of cancer cells (13, 126). All these activities are delivered by different subpopulations of TAMs, TAM subset complexity having considerably increased in the past few years (146). To keep it simple, activated macrophages either polarize as the proinflammatory M1 phenotype or as anti-inflammatory M2 phenotype (71). The M1 phenotype is characterized by the production of inflammatory cytokines, reactive nitrogen species, and ROS (14). In contrast, the M2 phenotype is characterized by increased production of immunosuppressive cytokines and factors (148). Recently, it has been shown that ROS production is important for M2, but not for M1 differentiation (252). Indeed, inhibiting ROS production blocks the differentiation of M2 macrophages through the ERK pathway. This is also true in vivo as antioxidant treatment blocks the occurrence of M2 macrophages in lung tumor mouse models (252). Similarly, in malignant melanomas, TAMs exhibiting high levels of ROS have enhanced invasive properties, mediated by increased TNF-α secretion (120). Recently, an HIF-1-dependent metabolic switch toward glycolysis has been detected following activation of macrophages (101). Citrate withdrawal from TCA cycle is critical for lipid biosynthesis in TAMs, and succinate, another TCA cycle intermediate, is also crucial to promote inflammatory signaling. Because TAMs play major roles in suppressing antitumor responses, blocking M2 macrophages by impacting the redox balance may represent an interesting strategy for cancer treatment.
Tumor-infiltrating lymphocytes
T cells orchestrate the adaptive immune response to specific antigens. Classically, when an antigen activates a naïve CD4+ T cell, cytokines determine whether it differentiates into an effector T cell (Teff) to induce an immune response or a regulatory T cell (Treg) with immunosuppressive functions (96). The main metabolic change, occurring upon T-cell activation, corresponds to an increased requirement for energy and for biosynthesis of reducing equivalents. Activated T cells rely preferentially on glycolysis and lactate production, mediated by the PI3K/AKT pathway (57). However, mitochondria-dependent metabolism also plays an important role in T-cell responses, through ROS signaling rather than ATP production (94, 99, 188). During T-cell activation, the high glycolytic rate induces excessive amount of glycerol-3-phosphate that is oxidized in an FAD-dependent reaction, mediated by glycerol-3-phosphate dehydrogenase 2, which increases ROS production. Silencing this enzyme decreases ROS and IL-2 production and reduces T-cell activation (98). Similar defects in T-cell activation have been observed in mice engineered for specific inactivation in T cells of a component of the respiratory chain complex III (Uqcrfs1) as it leads to reduced ROS levels (188). Moreover, mitochondrial ROS are required for antigen-specific CD4+ and CD8+ T-cell expansion in vivo (188). Using mice lacking the lymphocyte expansion molecule, an orphan protein that promotes CD8+ T-cell proliferation, another study confirmed that mitochondrial ROS affect respiratory chain assembly into the inner mitochondrial membrane and CD8+ T-cell expansion, thus demonstrating that long-term protective immunity can be OXPHOS driven (151). Antineoplastic chemotherapies stimulate autophagy, which appears as an adaptive metabolic mechanism of resistance to treatment (113). Thus, inhibiting autophagy would be a new strategy for improving chemotherapy efficiency in some tumors, such as triple-negative breast cancers, lacking efficient anticancer immune response (113). Using an immunocompetent mouse model, another study demonstrated that autophagy is required for immunogenic release of ATP from dying cells to recruit immune cells following chemotherapy (134).
CAF
Although the role of fibroblasts and the concept of seed and soil have been considered in the 19th century (153), the key role of CAFs in ECM remodeling and tumor progression has been highlighted only recently (17, 36, 56, 100, 149). While normal fibroblasts inhibit tissue progression to cancer, changes in stromal components shift the antitumor balance toward a procancerous state (12, 36, 211). Fibroblasts are activated through specific communication with cancer cells and can promote tumor initiation, progression, and metastasis. A large proportion of activated CAFs detected in invasive adenocarcinomas express smooth muscle α-actin (α-SMA) and are defined as myofibroblasts (211), but other markers such as platelet-derived growth factor receptor (PDGF-R) or fibroblast activation protein (FAP) also characterize stromal components (22, 46). The cellular origins of myofibroblasts can be multiple. Myofibroblasts were thought to derive from EMT, bone marrow, or various mesenchymal cell types, including endothelial cells, pericytes, or preadipocytes (16, 66, 109, 131, 166, 177). Resident fibroblasts are also considered as one of the major source of CAFs. Indeed, ROS release from tumor cells promotes the conversion of fibroblasts into myofibroblasts in various types of solid tumors (8, 36, 92, 157, 198, 211). Various signaling pathways, including transforming growth factor-β (TGF-β)- and CXCL12 (SDF-1)/CXCR4-dependent pathways, are also involved in CAF activation (5, 25, 211, 226). Interestingly, targeting both CXCL12/CXCR4 axis and CAFs has a synergistic effect when used in combination with anti-PD-L1 immunotherapy (49). Although stromal remodeling by caveolin 1 favors tumor invasion (69), ROS accumulation in CAFs promotes loss of caveolin 1 by oxidative stress-induced autophagy (128, 132, 190). Moreover, chronic oxidative stress in TME leads to HIF-1 accumulation by inhibiting prolyl hydroxylases (65). Stimulation of HIF-1 signaling is associated with increased glycolysis in CAFs, which in turn provide nutrients such as lactate to adjacent cancer cells (see below in the Nutrient availability and TME differentiation section). ROS have been implicated in the metabolic reprogramming of both cancer cells and CAFs, allowing an adaptation to oxidative stress that ultimately promotes tumorigenesis and chemoresistance. As metabolic reprogramming may impact the redox balance, it is crucial to better understand the metabolic cross-talk between neoplastic and surrounding cells.
Nutrient availability and TME differentiation
Tumor-associated macrophages
A pioneering work on metabolism in TAMs showed that activation of murine peritoneal macrophages leads to a metabolic switch as they rely on glycolysis rather than OXPHOS (79). Oxidation of glucose, glutamine, and FA are not just sources of energy but are also involved in macrophage polarization. Indeed, M1 macrophages rely preferentially on mTOR-HIF1α-mediated glycolysis, whereas M2 macrophages exhibit an OXPHOS metabolism, involving the PGC-1β-STAT6 signaling pathway (34, 172, 208, 213). Glycolysis induced in M1 macrophages allows the synthesis of reducing equivalents and energy through the PPP (60). Decreasing aerobic glycolysis by silencing PDK1 promotes M2 polarization as it attenuates the expression of proinflammatory cytokines upon LPS or TLR2 stimulation (207). Interestingly, blocking OXPHOS metabolism induces M1 polarization, while forcing OXPHOS metabolism in M1 macrophages potentiates M2 polarization (172, 213). Macrophages also exhibit high rates of glutaminolysis, which promote phagocytosis, cytokine production, and antigen presentation (223) and replenish the TCA cycle (93). Glutamine deprivation only impacts on M2 phenotype, while it has no effect on M1 polarization. Uptake and oxidation of FA, favored by PGC-1β signaling, increase OXPHOS and M2 polarization (213). Reciprocally, inhibiting lipolysis affects M2 polarization and survival (86). Altogether, these data suggest that tumor cell metabolism should modulate nutrient availability in TME, therefore affecting macrophage polarization and immune response. Highly glycolytic tumor cells may compromise M1 polarization by inducing glucose deprivation, while FA abundance may influence M2 differentiation.
Tumor-infiltrating lymphocytes
Several studies based on metabolic profiling revealed that Teff lymphocytes are highly glycolytic and lipogenic (133), whereas Treg cells display increased lipolysis and lipid oxidation (10). The mTOR pathway, a major regulator of cell growth that senses nutrient availability, plays a crucial role in T-cell differentiation (43, 53, 80, 167). Indeed, mTORC1 is involved in naïve CD4+ T-cell differentiation into T helper 1 (TH1) and T helper 17 (TH17) cells, supporting an antitumor effect. In contrast, mTORC2 promotes the differentiation of naïve CD4+ T cells into the protumorigenic T helper 2 (TH2) cells (43). Among the nutrients involved in T-cell differentiation, glutamine and glucose play critical functions. Glutamine deprivation promotes the differentiation of naïve CD4+ T cells into FOXP3+ Treg cells, even in the presence of cytokines that promote TH1 cell expansion (105). Adding alpha-ketoglutarate to a glutamine-deprived medium inhibits the generation of Treg cells. Thus, glutamine deprivation in TME induces a shift in the immune response balance, which becomes immunosuppressive (105). T-cell receptor stimulation can activate several signaling pathways that coordinate T-cell proliferation and differentiation, together with aerobic glycolysis (192). Two recent studies support the hypothesis that tumor cells can suppress antitumor T-cell responses by modulating metabolism through glucose deprivation (30, 83). Indeed, these studies provide some clues indicating that glycolysis is essential not only for biomass and ATP production but also to sustain T-cell effector functions through transcriptional and translational regulation. Using a well-established model of regressing and progressing sarcomas in immunocompetent mice, E.L. Pearce showed that progressing tumors have higher glucose consumption than the regressing ones (30). As a consequence, TILs from progressing tumors are glucose restricted and exhibit impaired effector functions compared with TILs in regressing tumors. Interestingly, treatments of tumor-bearing mice with immune checkpoint inhibitors (such as anti-CTLA-4, PD-1, and PD-L1 antibodies) restore glucose in TME, allowing T-cell glycolysis and IFN-γ production. Moreover, blocking PD-L1 in tumor cells reduces their glycolysis rates by inhibiting mTOR activity, which consequently increases extracellular glucose availability for TILs. These observations were confirmed in a melanoma mouse model as there is an anticorrelation between glycolytic genes and IFNγ or CD40 ligand mRNA levels. Finally, hexokinase 2 overexpression in melanoma cells confers on them high glycolytic capacities (30). In conclusion, these different studies clearly demonstrated how tumor cell metabolism deeply affects TME differentiation and functions. One major pathway involved in this process is the nutrient-sensitive mTOR pathway. By modulating tumor cell metabolism, one can control nutrient availability for T cells, thus promoting either their antitumor or immunosuppressive functions.
CAF
Cancer cells produce ROS that directly impact surrounding CAFs (36, 211). Reciprocally, CAFs act as a metabolic driving force for providing energetic fuels to cancer cells and facilitate their growth and survival (128). Metabolic reprogramming is not only restricted to tumor or immune cells, but CAFs also show changes in their metabolism. Primary fibroblasts are metabolically active, even in their quiescent state induced by contact inhibition. Indeed, contact-inhibited fibroblasts use glucose to similar levels as those in proliferating cells through the PPP (114). In contrast to normal fibroblasts, CAFs use aerobic glycolysis as a source of energy. Indeed, human CAFs isolated from invasive breast and prostate cancers are able to switch their metabolism toward aerobic glycolysis, through an HIF-1- and oxidative stress-dependent mechanism (36, 211). Human CAFs from colon cancer and melanoma also exhibit increased glucose uptake and lactate production, while their O2 consumption is decreased (250). In tumors, CAFs highly express glycolytic enzymes, such as PGK1, PKM2, or LDH (Lactate dehydrogenase), to release lactate to fuel cancer cells (52, 67, 160, 226). Indeed, lactate and ketones secreted from CAFs act as oncometabolites that can fuel OXPHOS metabolism in cancer cells, a process referred to as the reverse Warburg effect (128). Mass spectrometry-based metabolic profiling comparing human CAFs from lung cancers with normal adjacent fibroblasts shows increased abundance in dipeptides arising from autophagy-induced protein breakdown in CAFs when cancer cells are highly glycolytic (31). Gene expression analysis of CAFs induced by TGF-β shows an anticorrelation between IDH3α protein levels and fibroblast-specific protein 1 expression, a specific marker of CAFs (250). Indeed, IDH3α silencing inhibits OXPHOS, but its overexpression reduces lactate production. IDH3α downregulation decreases alpha-ketoglutarate levels, which promotes HIF-1α stabilization ultimately leading to increased glycolysis (250). Regarding the function of this metabolic reprogramming, it is unlikely that aerobic glycolysis in CAFs would be induced to support proliferation as the proliferation rate of CAFs is lower than normal fibroblasts. However, it can provide enough nutrients for tumor cells. This symbiotic relationship between CAFs and tumor cells is reversible, thus representing a metabolic optimization. Indeed, nuclear magnetic resonance spectroscopic measurements reveal that lactate produced by cancer cells can be taken up by CAFs through increased monocarboxylate transporter 1 expression (169). Invasive tumors can thus be classified according to their metabolic heterogeneity detected in both cancer and stromal cells: they can correspond to the Warburg type (glycolytic neoplastic cells, but nonglycolytic stroma), the reverse Warburg type (nonglycolytic neoplastic cells, but glycolytic stroma), the glycolytic type (glycolysis in both stroma and neoplastic cells), or the OXPHOS type (nonglycolysis in both stroma and neoplastic cells) (36, 149). Taken as a whole, these observations suggest that in some tumors, PET scan imaging may detect glucose consumption in stroma rather than strictly in cancer cells.
Conclusion
Since the initial description of the glycolytic metabolism in tumors by O. Warburg, we have now reached a better understanding of the metabolic heterogeneity in cancer cells. Starting with glucose as the only essential source of carbons for neoplastic cells, there is increasing evidence showing the huge diversity of nutrients that support neoplastic cell functions and affect deeply cells of the TME. Although the metabolic field is booming, we are only at the tip of the iceberg. Molecular mechanisms underlying metabolic heterogeneity and reprogramming are still debated and remain to be fully deciphered. The emergence of functional studies revealed how metabolic reprogramming allows cancer cells to adapt themselves and to promote tumor progression and metastatic spread. Moreover, we need to consider the important role of TME, especially CAFs and immune cells, which deeply participate in the metabolic interplay with neoplastic cells. CAFs and immune cells can collectively adapt to the metabolic needs of cancer cells and thus participate in tumorigenesis. Metabolic balance between all these players also promotes resistance to treatments, including not only the conventional chemo- or radiotherapy but also the most recent and innovative immunotherapies. Characterizing the reciprocal metabolic interplay between stromal, immune, and neoplastic cells will thus provide a better understanding of tumor progression and treatment resistance.
Abbreviations Used
- ACSS
acetyl-CoA synthetase
- ATP
adenosine triphosphate
- CAF
carcinoma-associated fibroblast
- CoA
coenzyme A
- CSCs
cancer stem cells
- E2F1
E2 transcription factor 1
- ECM
extracellular matrix
- EMT
epithelium–mesenchymal transition
- ERRs
estrogen-related receptors
- ETC
electron transport chain
- FA
fatty acid
- FAD
flavin adenine dinucleotide
- G6P
glucose-6-phosphate
- GLUT
glucose transporter
- GOT
glutamic oxaloacetic transaminase
- GS
glutamine synthetase
- HIF-1
hypoxia-inducible factor-1
- HK
hexokinase
- IMS
intermembrane space
- LDH
lactate dehydrogenase
- MnSOD
manganese superoxide dismutase
- mtDNA
mitochondrial DNA
- mTOR
mammalian target of rapamycin
- NAD
nicotinamide adenine dinucleotide
- NADPH
nicotinamide adenine dinucleotide phosphate
- N-MTS
N-terminal mitochondrial targeting signal
- NRF
nuclear respiratory factor
- NSCLC
nonsmall cell lung carcinoma
- O2
oxygen
- OXPHOS
oxidative phosphorylation
- PDH
pyruvate dehydrogenase
- PDK
pyruvate dehydrogenase kinase
- PET
positron emission tomography
- PGAM
phosphoglycerate mutase
- PGC-1α
peroxisome proliferator-activated receptor-γ coactivator-1α
- PHGDH
phosphoglycerate dehydrogenase
- PPAR
peroxisome proliferator-activated receptors
- PPP
pentose phosphate pathway
- pRB
retinoblastoma protein
- PSAT1
phosphoserine aminotransferase 1
- PSPH
phosphoserine phosphatase
- PTM
post-translational modification
- ROS
reactive oxygen species
- SIRT
sirtuin
- SMA
smooth muscle cells
- TAMs
tumor-associated macrophages
- TCA
tricarboxylic acid
- TGF-β
transforming growth factor-β
- TILs
tumor-infiltrating lymphocytes
- TIM
translocase of the inner mitochondrial membrane
- TME
tumor microenvironment
- TOM
translocase of the outer mitochondrial membrane
Acknowledgments
G.G. and V.M. are funded by the Fondation ARC and the Institut National du Cancer (INCa), respectively. The authors would like to thank these funders. F.M.G's laboratory is labelized by the Ligue Nationale Contre le Cancer (LNCC), the Institut National de la Santé et de la Recherche Médicale (Inserm), and the Institut Curie. The authors are very grateful to the LNCC, Inserm, and Institut Curie for providing their support.
References
- 1.Alexeyev M, Shokolenko I, Wilson G, and LeDoux S. The maintenance of mitochondrial DNA integrity—critical analysis and update. Cold Spring Harbor Perspect Biol 5: a012641, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amelio I, Cutruzzola F, Antonov A, Agostini M, and Melino G. Serine and glycine metabolism in cancer. Trends Biochem Sci 39: 191–198, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Anderson S, Bankier AT, Barrell BG, de Bruijn MH, Coulson AR, Drouin J, Eperon IC, Nierlich DP, Roe BA, Sanger F, Schreier PH, Smith AJ, Staden R, and Young IG. Sequence and organization of the human mitochondrial genome. Nature 290: 457–465, 1981 [DOI] [PubMed] [Google Scholar]
- 4.Antoch G, Saoudi N, Kuehl H, Dahmen G, Mueller SP, Beyer T, Bockisch A, Debatin JF, and Freudenberg LS. Accuracy of whole-body dual-modality fluorine-18–2-fluoro-2-deoxy-D-glucose positron emission tomography and computed tomography (FDG-PET/CT) for tumor staging in solid tumors: comparison with CT and PET. J Clin Oncol 22: 4357–4368, 2004 [DOI] [PubMed] [Google Scholar]
- 5.Augsten M, Sjoberg E, Frings O, Vorrink SU, Frijhoff J, Olsson E, Borg A, and Ostman A. Cancer-associated fibroblasts expressing CXCL14 rely upon NOS1-derived nitric oxide signaling for their tumor-supporting properties. Cancer Res 74: 2999–3010, 2014 [DOI] [PubMed] [Google Scholar]
- 6.Bao Y, Mukai K, Hishiki T, Kubo A, Ohmura M, Sugiura Y, Matsuura T, Nagahata Y, Hayakawa N, Yamamoto T, Fukuda R, Saya H, Suematsu M, and Minamishima YA. Energy management by enhanced glycolysis in G1-phase in human colon cancer cells in vitro and in vivo. Mol Cancer Res 11: 973–985, 2013 [DOI] [PubMed] [Google Scholar]
- 7.Bardos JI. and Ashcroft M. Hypoxia-inducible factor-1 and oncogenic signalling. BioEssays 26: 262–269, 2004 [DOI] [PubMed] [Google Scholar]
- 8.Batista L, Gruosso T, and Mechta-Grigoriou F. Ovarian cancer emerging subtypes: role of oxidative stress and fibrosis in tumour development and response to treatment. Int J Biochem Cell Biol 45: 1092–1098, 2013 [DOI] [PubMed] [Google Scholar]
- 9.Becker T, Bottinger L, and Pfanner N. Mitochondrial protein import: from transport pathways to an integrated network. Trends Biochem Sci 37: 85–91, 2012 [DOI] [PubMed] [Google Scholar]
- 10.Berod L, Friedrich C, Nandan A, Freitag J, Hagemann S, Harmrolfs K, Sandouk A, Hesse C, Castro CN, Bahre H, Tschirner SK, Gorinski N, Gohmert M, Mayer CT, Huehn J, Ponimaskin E, Abraham WR, Muller R, Lochner M, and Sparwasser T. De novo fatty acid synthesis controls the fate between regulatory T and T helper 17 cells. Nat Med 20: 1327–1333, 2014 [DOI] [PubMed] [Google Scholar]
- 11.Birsoy K, Wang T, Chen WW, Freinkman E, Abu-Remaileh M, and Sabatini DM. An essential role of the mitochondrial electron transport chain in cell proliferation is to enable aspartate synthesis. Cell 162: 540–551, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bissell MJ. and Hines WC. Why don't we get more cancer? A proposed role of the microenvironment in restraining cancer progression. Nat Med 17: 320–329, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Biswas SK. Metabolic reprogramming of immune cells in cancer progression. Immunity 43: 435–449, 2015 [DOI] [PubMed] [Google Scholar]
- 14.Biswas SK. and Mantovani A. Macrophage plasticity and interaction with lymphocyte subsets: cancer as a paradigm. Nat Immunol 11: 889–896, 2010 [DOI] [PubMed] [Google Scholar]
- 15.Blanchet E, Annicotte JS, Lagarrigue S, Aguilar V, Clape C, Chavey C, Fritz V, Casas F, Apparailly F, Auwerx J, and Fajas L. E2F transcription factor-1 regulates oxidative metabolism. Nat Cell Biol 13: 1146–1152, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bochet L, Lehuede C, Dauvillier S, Wang YY, Dirat B, Laurent V, Dray C, Guiet R, Maridonneau-Parini I, Le Gonidec S, Couderc B, Escourrou G, Valet P, and Muller C. Adipocyte-derived fibroblasts promote tumor progression and contribute to the desmoplastic reaction in breast cancer. Cancer Res 73: 5657–5668, 2013 [DOI] [PubMed] [Google Scholar]
- 17.Bonnans C, Chou J, and Werb Z. Remodelling the extracellular matrix in development and disease. Nat Rev Mol Cell Biol 15: 786–801, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bonnet S, Archer SL, Allalunis-Turner J, Haromy A, Beaulieu C, Thompson R, Lee CT, Lopaschuk GD, Puttagunta L, Bonnet S, Harry G, Hashimoto K, Porter CJ, Andrade MA, Thebaud B, and Michelakis ED. A mitochondria-K+ channel axis is suppressed in cancer and its normalization promotes apoptosis and inhibits cancer growth. Cancer Cell 11: 37–51, 2007 [DOI] [PubMed] [Google Scholar]
- 19.Bordone L. and Guarente L. Calorie restriction, SIRT1 and metabolism: understanding longevity. Nat Rev Mol Cell Biol 6: 298–305, 2005 [DOI] [PubMed] [Google Scholar]
- 20.Brahimi-Horn MC, Bellot G, and Pouyssegur J. Hypoxia and energetic tumour metabolism. Curr Opin Genet Dev 21: 67–72, 2011 [DOI] [PubMed] [Google Scholar]
- 21.Brahimi-Horn MC, Chiche J, and Pouyssegur J. Hypoxia signalling controls metabolic demand. Curr Opin Cell Biol 19: 223–229, 2007 [DOI] [PubMed] [Google Scholar]
- 22.Brennen WN, Rosen DM, Wang H, Isaacs JT, and Denmeade SR. Targeting carcinoma-associated fibroblasts within the tumor stroma with a fibroblast activation protein-activated prodrug. J Natl Cancer Inst 104: 1320–1334, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brix J, Rudiger S, Bukau B, Schneider-Mergener J, and Pfanner N. Distribution of binding sequences for the mitochondrial import receptors Tom20, Tom22, and Tom70 in a presequence-carrying preprotein and a non-cleavable preprotein. J Biol Chem 274: 16522–16530, 1999 [DOI] [PubMed] [Google Scholar]
- 24.Cairns RA, Harris IS, and Mak TW. Regulation of cancer cell metabolism. Nat Rev Cancer 11: 85–95, 2011 [DOI] [PubMed] [Google Scholar]
- 25.Calon A, Tauriello DV, and Batlle E. TGF-beta in CAF-mediated tumor growth and metastasis. Semin Cancer Biol 25: 15–22, 2014 [DOI] [PubMed] [Google Scholar]
- 26.Cantor JR. and Sabatini DM. Cancer cell metabolism: one hallmark, many faces. Cancer Discov 2: 881–898, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Caro P, Kishan AU, Norberg E, Stanley IA, Chapuy B, Ficarro SB, Polak K, Tondera D, Gounarides J, Yin H, Zhou F, Green MR, Chen L, Monti S, Marto JA, Shipp MA, and Danial NN. Metabolic signatures uncover distinct targets in molecular subsets of diffuse large B cell lymphoma. Cancer Cell 22: 547–560, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Carracedo A, Cantley LC, and Pandolfi PP. Cancer metabolism: fatty acid oxidation in the limelight. Nat Rev Cancer 13: 227–232, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Casazza A, Di Conza G, Wenes M, Finisguerra V, Deschoemaeker S, and Mazzone M. Tumor stroma: a complexity dictated by the hypoxic tumor microenvironment. Oncogene 33: 1743–1754, 2014 [DOI] [PubMed] [Google Scholar]
- 30.Chang CH, Qiu J, O'Sullivan D, Buck MD, Noguchi T, Curtis JD, Chen Q, Gindin M, Gubin MM, van der Windt GJ, Tonc E, Schreiber RD, Pearce EJ, and Pearce EL. Metabolic competition in the tumor microenvironment is a driver of cancer progression. Cell 162: 1229–1241, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chaudhri VK, Salzler GG, Dick SA, Buckman MS, Sordella R, Karoly ED, Mohney R, Stiles BM, Elemento O, Altorki NK, and McGraw TE. Metabolic alterations in lung cancer-associated fibroblasts correlated with increased glycolytic metabolism of the tumor. Mol Cancer Res 11: 579–592, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen J, Li Y, Yu TS, McKay RM, Burns DK, Kernie SG, and Parada LF. A restricted cell population propagates glioblastoma growth after chemotherapy. Nature 488: 522–526, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen X, Qian Y, and Wu S. The Warburg effect: evolving interpretations of an established concept. Free Radic Biol Med 79: 253–263, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cheng SC, Quintin J, Cramer RA, Shepardson KM, Saeed S, Kumar V, Giamarellos-Bourboulis EJ, Martens JH, Rao NA, Aghajanirefah A, Manjeri GR, Li Y, Ifrim DC, Arts RJ, van der Veer BM, Deen PM, Logie C, O'Neill LA, Willems P, van de Veerdonk FL, van der Meer JW, Ng A, Joosten LA, Wijmenga C, Stunnenberg HG, Xavier RJ, and Netea MG. mTOR- and HIF-1alpha-mediated aerobic glycolysis as metabolic basis for trained immunity. Science 345: 1250684, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Choudhary C, Kumar C, Gnad F, Nielsen ML, Rehman M, Walther TC, Olsen JV, and Mann M. Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science 325: 834–840, 2009 [DOI] [PubMed] [Google Scholar]
- 36.Costa A, Scholer-Dahirel A, and Mechta-Grigoriou F. The role of reactive oxygen species and metabolism on cancer cells and their microenvironment. Semin Cancer Biol 25: 23–32, 2014 [DOI] [PubMed] [Google Scholar]
- 37.Csibi A, Lee G, Yoon SO, Tong H, Ilter D, Elia I, Fendt SM, Roberts TM, and Blenis J. The mTORC1/S6K1 pathway regulates glutamine metabolism through the eIF4B-dependent control of c-Myc translation. Curr Biol 24: 2274–2280, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 38.Currie E, Schulze A, Zechner R, Walther TC, and Farese RV., Jr. Cellular fatty acid metabolism and cancer. Cell Metab 18: 153–161, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dang L, White DW, Gross S, Bennett BD, Bittinger MA, Driggers EM, Fantin VR, Jang HG, Jin S, Keenan MC, Marks KM, Prins RM, Ward PS, Yen KE, Liau LM, Rabinowitz JD, Cantley LC, Thompson CB, Vander Heiden MG, and Su SM. Cancer-associated IDH1 mutations produce 2-hydroxyglutarate. Nature 462: 739–744, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.DeBerardinis RJ. and Cheng T. Q's next: the diverse functions of glutamine in metabolism, cell biology and cancer. Oncogene 29: 313–324, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.DeBerardinis RJ, Lum JJ, Hatzivassiliou G, and Thompson CB. The biology of cancer: metabolic reprogramming fuels cell growth and proliferation. Cell Metab 7: 11–20, 2008 [DOI] [PubMed] [Google Scholar]
- 42.Deblois G, St-Pierre J, and Giguere V. The PGC-1/ERR signaling axis in cancer. Oncogene 32: 3483–3490, 2013 [DOI] [PubMed] [Google Scholar]
- 43.Delgoffe GM, Pollizzi KN, Waickman AT, Heikamp E, Meyers DJ, Horton MR, Xiao B, Worley PF, and Powell JD. The kinase mTOR regulates the differentiation of helper T cells through the selective activation of signaling by mTORC1 and mTORC2. Nat Immunol 12: 295–303, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.DeNicola GM, Chen PH, Mullarky E, Sudderth JA, Hu Z, Wu D, Tang H, Xie Y, Asara JM, Huffman KE, Wistuba II, Minna JD, DeBerardinis RJ, and Cantley LC. NRF2 regulates serine biosynthesis in non-small cell lung cancer. Nat Genet 47: 1475–1481, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dupuy F, Tabaries S, Andrzejewski S, Dong Z, Blagih J, Annis MG, Omeroglu A, Gao D, Leung S, Amir E, Clemons M, Aguilar-Mahecha A, Basik M, Vincent EE, St-Pierre J, Jones RG, and Siegel PM. PDK1-dependent metabolic reprogramming dictates metastatic potential in breast cancer. Cell Metab 22: 577–589, 2015 [DOI] [PubMed] [Google Scholar]
- 46.Erez N, Truitt M, Olson P, Arron ST, and Hanahan D. Cancer-associated fibroblasts are activated in incipient neoplasia to orchestrate tumor-promoting inflammation in an NF-kappaB-dependent manner. Cancer Cell 17: 135–147, 2010 [DOI] [PubMed] [Google Scholar]
- 47.Evans MJ. and Scarpulla RC. Interaction of nuclear factors with multiple sites in the somatic cytochrome c promoter. Characterization of upstream NRF-1, ATF, and intron Sp1 recognition sequences. J Biol Chem 264: 14361–14368, 1989 [PubMed] [Google Scholar]
- 48.Fantin VR, St-Pierre J, and Leder P. Attenuation of LDH-A expression uncovers a link between glycolysis, mitochondrial physiology, and tumor maintenance. Cancer Cell 9: 425–434, 2006 [DOI] [PubMed] [Google Scholar]
- 49.Feig C, Jones JO, Kraman M, Wells RJ, Deonarine A, Chan DS, Connell CM, Roberts EW, Zhao Q, Caballero OL, Teichmann SA, Janowitz T, Jodrell DI, Tuveson DA, and Fearon DT. Targeting CXCL12 from FAP-expressing carcinoma-associated fibroblasts synergizes with anti-PD-L1 immunotherapy in pancreatic cancer. Proc Natl Acad Sci U S A 110: 20212–20217, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Feilchenfeldt J, Brundler MA, Soravia C, Totsch M, and Meier CA. Peroxisome proliferator-activated receptors (PPARs) and associated transcription factors in colon cancer: reduced expression of PPARgamma-coactivator 1 (PGC-1). Cancer Lett 203: 25–33, 2004 [DOI] [PubMed] [Google Scholar]
- 51.Feng W, Gentles A, Nair RV, Huang M, Lin Y, Lee CY, Cai S, Scheeren FA, Kuo AH, and Diehn M. Targeting unique metabolic properties of breast tumor initiating cells. Stem Cells 32: 1734–1745, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fiaschi T, Marini A, Giannoni E, Taddei ML, Gandellini P, De Donatis A, Lanciotti M, Serni S, Cirri P, and Chiarugi P. Reciprocal metabolic reprogramming through lactate shuttle coordinately influences tumor-stroma interplay. Cancer Res 72: 5130–5140, 2012 [DOI] [PubMed] [Google Scholar]
- 53.Finlay DK, Rosenzweig E, Sinclair LV, Feijoo-Carnero C, Hukelmann JL, Rolf J, Panteleyev AA, Okkenhaug K, and Cantrell DA. PDK1 regulation of mTOR and hypoxia-inducible factor 1 integrate metabolism and migration of CD8+ T cells. J Exp Med 209: 2441–2453, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Finn RS, Dering J, Conklin D, Kalous O, Cohen DJ, Desai AJ, Ginther C, Atefi M, Chen I, Fowst C, Los G, and Slamon DJ. PD 0332991, a selective cyclin D kinase 4/6 inhibitor, preferentially inhibits proliferation of luminal estrogen receptor-positive human breast cancer cell lines in vitro. Breast Cancer Res 11: R77, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fiori A, Perez-Martinez X, and Fox TD. Overexpression of the COX2 translational activator, Pet111p, prevents translation of COX1 mRNA and cytochrome c oxidase assembly in mitochondria of Saccharomyces cerevisiae. Mol Microbiol 56: 1689–1704, 2005 [DOI] [PubMed] [Google Scholar]
- 56.Frantz C, Stewart KM, and Weaver VM. The extracellular matrix at a glance. J Cell Sci 123: 4195–4200, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Frauwirth KA, Riley JL, Harris MH, Parry RV, Rathmell JC, Plas DR, Elstrom RL, June CH, and Thompson CB. The CD28 signaling pathway regulates glucose metabolism. Immunity 16: 769–777, 2002 [DOI] [PubMed] [Google Scholar]
- 58.Frezza C. and Gottlieb E. Mitochondria in cancer: not just innocent bystanders. Semin Cancer Biol 19: 4–11, 2009 [DOI] [PubMed] [Google Scholar]
- 59.Fujino T, Kondo J, Ishikawa M, Morikawa K, and Yamamoto TT. Acetyl-CoA synthetase 2, a mitochondrial matrix enzyme involved in the oxidation of acetate. J Biol Chem 276: 11420–11426, 2001 [DOI] [PubMed] [Google Scholar]
- 60.Galvan-Pena S. and O'Neill LA. Metabolic reprograming in macrophage polarization. Front Immunol 5: 420, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gao P, Tchernyshyov I, Chang TC, Lee YS, Kita K, Ochi T, Zeller KI, De Marzo AM, Van Eyk JE, Mendell JT, and Dang CV. c-Myc suppression of miR-23a/b enhances mitochondrial glutaminase expression and glutamine metabolism. Nature 458: 762–765, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gasparre G, Porcelli AM, Lenaz G, and Romeo G. Relevance of mitochondrial genetics and metabolism in cancer development. Cold Spring Harbor Perspect Biol 5, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gatenby RA. and Gillies RJ. Why do cancers have high aerobic glycolysis? Nat Rev Cancer 4: 891–899, 2004 [DOI] [PubMed] [Google Scholar]
- 64.Gentric G, Maillet V, Paradis V, Couton D, L'Hermitte A, Panasyuk G, Fromenty B, Celton-Morizur S, and Desdouets C. Oxidative stress promotes pathologic polyploidization in nonalcoholic fatty liver disease. J Clin Invest 125: 981–992, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gerald D, Berra E, Frapart YM, Chan DA, Giaccia AJ, Mansuy D, Pouyssegur J, Yaniv M, and Mechta-Grigoriou F. JunD reduces tumor angiogenesis by protecting cells from oxidative stress. Cell 118: 781–794, 2004 [DOI] [PubMed] [Google Scholar]
- 66.Giannoni E, Bianchini F, Calorini L, and Chiarugi P. Cancer associated fibroblasts exploit reactive oxygen species through a proinflammatory signature leading to epithelial mesenchymal transition and stemness. Antioxid Redox Signal 14: 2361–2371, 2011 [DOI] [PubMed] [Google Scholar]
- 67.Giannoni E, Taddei ML, Morandi A, Comito G, Calvani M, Bianchini F, Richichi B, Raugei G, Wong N, Tang D, and Chiarugi P. Targeting stromal-induced pyruvate kinase M2 nuclear translocation impairs oxphos and prostate cancer metastatic spread. Oncotarget 6: 24061–24074, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Gleyzer N, Vercauteren K, and Scarpulla RC. Control of mitochondrial transcription specificity factors (TFB1M and TFB2M) by nuclear respiratory factors (NRF-1 and NRF-2) and PGC-1 family coactivators. Mol Cell Biol 25: 1354–1366, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Goetz JG, Minguet S, Navarro-Lerida I, Lazcano JJ, Samaniego R, Calvo E, Tello M, Osteso-Ibanez T, Pellinen T, Echarri A, Cerezo A, Klein-Szanto AJ, Garcia R, Keely PJ, Sanchez-Mateos P, Cukierman E, and Del Pozo MA. Biomechanical remodeling of the microenvironment by stromal caveolin-1 favors tumor invasion and metastasis. Cell 146: 148–163, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Gogvadze V, Orrenius S, and Zhivotovsky B. Mitochondria in cancer cells: what is so special about them? Trends Cell Biol 18: 165–173, 2008 [DOI] [PubMed] [Google Scholar]
- 71.Gordon S. and Taylor PR. Monocyte and macrophage heterogeneity. Nat Rev Immunol 5: 953–964, 2005 [DOI] [PubMed] [Google Scholar]
- 72.Grohmann K, Amairic F, Crews S, and Attardi G. Failure to detect “cap” structures in mitochondrial DNA-coded poly(A)-containing RNA from HeLa cells. Nucleic Acids Res 5: 637–651, 1978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Gugneja S, Virbasius CM, and Scarpulla RC. Nuclear respiratory factors 1 and 2 utilize similar glutamine-containing clusters of hydrophobic residues to activate transcription. Mol Cell Biol 16: 5708–5716, 1996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Guppy M, Leedman P, Zu X, and Russell V. Contribution by different fuels and metabolic pathways to the total ATP turnover of proliferating MCF-7 breast cancer cells. Biochem J 364: 309–315, 2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Haigis MC. and Sinclair DA. Mammalian sirtuins: biological insights and disease relevance. Ann Rev Pathol 5: 253–295, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hallows WC, Yu W, and Denu JM. Regulation of glycolytic enzyme phosphoglycerate mutase-1 by Sirt1 protein-mediated deacetylation. J Biol Chem 287: 3850–3858, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Hanahan D. and Weinberg RA. Hallmarks of cancer: the next generation. Cell 144: 646–674, 2011 [DOI] [PubMed] [Google Scholar]
- 78.Haq R, Yokoyama S, Hawryluk EB, Jonsson GB, Frederick DT, McHenry K, Porter D, Tran TN, Love KT, Langer R, Anderson DG, Garraway LA, Duncan LM, Morton DL, Hoon DS, Wargo JA, Song JS, and Fisher DE. BCL2A1 is a lineage-specific antiapoptotic melanoma oncogene that confers resistance to BRAF inhibition. Proc Natl Acad Sci USA 110: 4321–4326, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Hard GC. Some biochemical aspects of the immune macrophage. Br J Exp Pathol 51: 97–105, 1970 [PMC free article] [PubMed] [Google Scholar]
- 80.Haxhinasto S, Mathis D, and Benoist C. The AKT-mTOR axis regulates de novo differentiation of CD4+Foxp3+ cells. J Exp Med 205: 565–574, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.He W, Newman JC, Wang MZ, Ho L, and Verdin E. Mitochondrial sirtuins: regulators of protein acylation and metabolism. Trends Endocrinol Metab 23: 467–476, 2012 [DOI] [PubMed] [Google Scholar]
- 82.Herrmann JM, Woellhaf MW, and Bonnefoy N. Control of protein synthesis in yeast mitochondria: the concept of translational activators. Biochim Biophys Acta 1833: 286–294, 2013 [DOI] [PubMed] [Google Scholar]
- 83.Ho PC, Bihuniak JD, Macintyre AN, Staron M, Liu X, Amezquita R, Tsui YC, Cui G, Micevic G, Perales JC, Kleinstein SH, Abel ED, Insogna KL, Feske S, Locasale JW, Bosenberg MW, Rathmell JC, and Kaech SM. Phosphoenolpyruvate is a metabolic checkpoint of anti-tumor T cell responses. Cell 162: 1217–1228, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Horecker BL. The pentose phosphate pathway. J Biol Chem 277: 47965–47971, 2002 [DOI] [PubMed] [Google Scholar]
- 85.Horwich AL, Kalousek F, Mellman I, and Rosenberg LE. A leader peptide is sufficient to direct mitochondrial import of a chimeric protein. EMBO J 4: 1129–1135, 1985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Huang SC, Everts B, Ivanova Y, O'Sullivan D, Nascimento M, Smith AM, Beatty W, Love-Gregory L, Lam WY, O'Neill CM, Yan C, Du H, Abumrad NA, Urban JF, Jr., Artyomov MN, Pearce EL, and Pearce EJ. Cell-intrinsic lysosomal lipolysis is essential for alternative activation of macrophages. Nat Immunol 15: 846–855, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Iborra FJ, Kimura H, and Cook PR. The functional organization of mitochondrial genomes in human cells. BMC Biol 2: 9, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Ito K. and Suda T. Metabolic requirements for the maintenance of self-renewing stem cells. Nat Rev Mol Cell Biol 15: 243–256, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Jager S, Handschin C, St-Pierre J, and Spiegelman BM. AMP-activated protein kinase (AMPK) action in skeletal muscle via direct phosphorylation of PGC-1alpha. Proc Natl Acad Sci USA 104: 12017–12022, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Janiszewska M, Suva ML, Riggi N, Houtkooper RH, Auwerx J, Clement-Schatlo V, Radovanovic I, Rheinbay E, Provero P, and Stamenkovic I. Imp2 controls oxidative phosphorylation and is crucial for preserving glioblastoma cancer stem cells. Genes Dev 26: 1926–1944, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Jeninga EH, Schoonjans K, and Auwerx J. Reversible acetylation of PGC-1: connecting energy sensors and effectors to guarantee metabolic flexibility. Oncogene 29: 4617–4624, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Jezierska-Drutel A, Rosenzweig SA, and Neumann CA. Role of oxidative stress and the microenvironment in breast cancer development and progression. Adv Cancer Res 119: 107–125, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Jha AK, Huang SC, Sergushichev A, Lampropoulou V, Ivanova Y, Loginicheva E, Chmielewski K, Stewart KM, Ashall J, Everts B, Pearce EJ, Driggers EM, and Artyomov MN. Network integration of parallel metabolic and transcriptional data reveals metabolic modules that regulate macrophage polarization. Immunity 42: 419–430, 2015 [DOI] [PubMed] [Google Scholar]
- 94.Jones RG. and Thompson CB. Revving the engine: signal transduction fuels T cell activation. Immunity 27: 173–178, 2007 [DOI] [PubMed] [Google Scholar]
- 95.Jose C, Bellance N, and Rossignol R. Choosing between glycolysis and oxidative phosphorylation: a tumor's dilemma? Biochim Biophys Acta 1807: 552–561, 2011 [DOI] [PubMed] [Google Scholar]
- 96.Joyce JA. and Fearon DT. T cell exclusion, immune privilege, and the tumor microenvironment. Science 348: 74–80, 2015 [DOI] [PubMed] [Google Scholar]
- 97.Kalhan SC. and Hanson RW. Resurgence of serine: an often neglected but indispensable amino Acid. J Biol Chem 287: 19786–19791, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Kaminski MM, Sauer SW, Kaminski M, Opp S, Ruppert T, Grigaravicius P, Grudnik P, Grone HJ, Krammer PH, and Gulow K. T cell activation is driven by an ADP-dependent glucokinase linking enhanced glycolysis with mitochondrial reactive oxygen species generation. Cell Rep 2: 1300–1315, 2012 [DOI] [PubMed] [Google Scholar]
- 99.Kaminski MM, Sauer SW, Klemke CD, Suss D, Okun JG, Krammer PH, and Gulow K. Mitochondrial reactive oxygen species control T cell activation by regulating IL-2 and IL-4 expression: mechanism of ciprofloxacin-mediated immunosuppression. J Immunol 184: 4827–4841, 2010 [DOI] [PubMed] [Google Scholar]
- 100.Karagiannis GS, Poutahidis T, Erdman SE, Kirsch R, Riddell RH, and Diamandis EP. Cancer-associated fibroblasts drive the progression of metastasis through both paracrine and mechanical pressure on cancer tissue. Mol Cancer Res 10: 1403–1418, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Kelly B. and O'Neill LA. Metabolic reprogramming in macrophages and dendritic cells in innate immunity. Cell Res 25: 771–784, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Kelly DP. and Scarpulla RC. Transcriptional regulatory circuits controlling mitochondrial biogenesis and function. Genes Dev 18: 357–368, 2004 [DOI] [PubMed] [Google Scholar]
- 103.Kim JW, Tchernyshyov I, Semenza GL, and Dang CV. HIF-1-mediated expression of pyruvate dehydrogenase kinase: a metabolic switch required for cellular adaptation to hypoxia. Cell Metab 3: 177–185, 2006 [DOI] [PubMed] [Google Scholar]
- 104.King MP. and Attardi G. Human cells lacking mtDNA: repopulation with exogenous mitochondria by complementation. Science 246: 500–503, 1989 [DOI] [PubMed] [Google Scholar]
- 105.Klysz D, Tai X, Robert PA, Craveiro M, Cretenet G, Oburoglu L, Mongellaz C, Floess S, Fritz V, Matias MI, Yong C, Surh N, Marie JC, Huehn J, Zimmermann V, Kinet S, Dardalhon V, and Taylor N. Glutamine-dependent alpha-ketoglutarate production regulates the balance between T helper 1 cell and regulatory T cell generation. Sci Signal 8: ra97, 2015 [DOI] [PubMed] [Google Scholar]
- 106.Krebs HA, Ruffo A, Johnson M, Eggleston LV, and Hems R. Oxidative phosphorylation. Biochem J 54: 107–116, 1953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Kung HN, Marks JR, and Chi JT. Glutamine synthetase is a genetic determinant of cell type-specific glutamine independence in breast epithelia. PLoS Genet 7: e1002229, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Lagadinou ED, Sach A, Callahan K, Rossi RM, Neering SJ, Minhajuddin M, Ashton JM, Pei S, Grose V, O'Dwyer KM, Liesveld JL, Brookes PS, Becker MW, and Jordan CT. BCL-2 inhibition targets oxidative phosphorylation and selectively eradicates quiescent human leukemia stem cells. Cell Stem Cell 12: 329–341, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Lapeire L, Hendrix A, Lambein K, Van Bockstal M, Braems G, Van Den Broecke R, Limame R, Mestdagh P, Vandesompele J, Vanhove C, Maynard D, Lehuede C, Muller C, Valet P, Gespach CP, Bracke M, Cocquyt V, Denys H, and De Wever O. Cancer-associated adipose tissue promotes breast cancer progression by paracrine oncostatin M and Jak/STAT3 signaling. Cancer Res 74: 6806–6819, 2014 [DOI] [PubMed] [Google Scholar]
- 110.Laplante M. and Sabatini DM. mTOR signaling in growth control and disease. Cell 149: 274–293, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Laurent G, Solari F, Mateescu B, Karaca M, Castel J, Bourachot B, Magnan C, Billaud M, and Mechta-Grigoriou F. Oxidative stress contributes to aging by enhancing pancreatic angiogenesis and insulin signaling. Cell Metab 7: 113–124, 2008 [DOI] [PubMed] [Google Scholar]
- 112.LeBleu VS, O'Connell JT, Gonzalez Herrera KN, Wikman H, Pantel K, Haigis MC, de Carvalho FM, Damascena A, Domingos Chinen LT, Rocha RM, Asara JM, and Kalluri R. PGC-1alpha mediates mitochondrial biogenesis and oxidative phosphorylation in cancer cells to promote metastasis. Nat Cell Biol 16: 992–1003, 100 1–1015, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Lefort S, Joffre C, Kieffer Y, Givel AM, Bourachot B, Zago G, Bieche I, Dubois T, Meseure D, Vincent-Salomon A, Camonis J, and Mechta-Grigoriou F. Inhibition of autophagy as a new means of improving chemotherapy efficiency in high-LC3B triple-negative breast cancers. Autophagy 10: 2122–2142, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Lemons JM, Feng XJ, Bennett BD, Legesse-Miller A, Johnson EL, Raitman I, Pollina EA, Rabitz HA, Rabinowitz JD, and Coller HA. Quiescent fibroblasts exhibit high metabolic activity. PLoS Biol 8: e1000514, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Levine AJ. and Puzio-Kuter AM. The control of the metabolic switch in cancers by oncogenes and tumor suppressor genes. Science 330: 1340–1344, 2010 [DOI] [PubMed] [Google Scholar]
- 116.Li F, Wang Y, Zeller KI, Potter JJ, Wonsey DR, O'Donnell KA, Kim JW, Yustein JT, Lee LA, and Dang CV. Myc stimulates nuclearly encoded mitochondrial genes and mitochondrial biogenesis. Mol Cell Biol 25: 6225–6234, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Liao HX. and Spremulli LL. Interaction of bovine mitochondrial ribosomes with messenger RNA. J Biol Chem 264: 7518–7522, 1989 [PubMed] [Google Scholar]
- 118.Lim JH, Luo C, Vazquez F, and Puigserver P. Targeting mitochondrial oxidative metabolism in melanoma causes metabolic compensation through glucose and glutamine utilization. Cancer Res 74: 3535–3545, 2014 [DOI] [PubMed] [Google Scholar]
- 119.Lin J, Puigserver P, Donovan J, Tarr P, and Spiegelman BM. Peroxisome proliferator-activated receptor gamma coactivator 1beta (PGC-1beta), a novel PGC-1-related transcription coactivator associated with host cell factor. J Biol Chem 277: 1645–1648, 2002 [DOI] [PubMed] [Google Scholar]
- 120.Lin X, Zheng W, Liu J, Zhang Y, Qin H, Wu H, Xue B, Lu Y, and Shen P. Oxidative stress in malignant melanoma enhances tumor necrosis factor-alpha secretion of tumor-associated macrophages that promote cancer cell invasion. Antioxid Redox Signal 19: 1337–1355, 2013 [DOI] [PubMed] [Google Scholar]
- 121.Liu W, Le A, Hancock C, Lane AN, Dang CV, Fan TW, and Phang JM. Reprogramming of proline and glutamine metabolism contributes to the proliferative and metabolic responses regulated by oncogenic transcription factor c-MYC. Proc Natl Acad Sci USA 109: 8983–8988, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Locasale JW. Serine, glycine and one-carbon units: cancer metabolism in full circle. Nat Rev Cancer 13: 572–583, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Locasale JW, Grassian AR, Melman T, Lyssiotis CA, Mattaini KR, Bass AJ, Heffron G, Metallo CM, Muranen T, Sharfi H, Sasaki AT, Anastasiou D, Mullarky E, Vokes NI, Sasaki M, Beroukhim R, Stephanopoulos G, Ligon AH, Meyerson M, Richardson AL, Chin L, Wagner G, Asara JM, Brugge JS, Cantley LC, and Vander Heiden MG. Phosphoglycerate dehydrogenase diverts glycolytic flux and contributes to oncogenesis. Nat Genet 43: 869–874, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Lopez-Mejia IC. and Fajas L. Cell cycle regulation of mitochondrial function. Curr Opin Cell Biol 33: 19–25, 2015 [DOI] [PubMed] [Google Scholar]
- 125.Maddocks OD, Berkers CR, Mason SM, Zheng L, Blyth K, Gottlieb E, and Vousden KH. Serine starvation induces stress and p53-dependent metabolic remodelling in cancer cells. Nature 493: 542–546, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Mantovani A, Sozzani S, Locati M, Allavena P, and Sica A. Macrophage polarization: tumor-associated macrophages as a paradigm for polarized M2 mononuclear phagocytes. Trends Immunol 23: 549–555, 2002 [DOI] [PubMed] [Google Scholar]
- 127.Marin-Valencia I, Yang C, Mashimo T, Cho S, Baek H, Yang XL, Rajagopalan KN, Maddie M, Vemireddy V, Zhao Z, Cai L, Good L, Tu BP, Hatanpaa KJ, Mickey BE, Mates JM, Pascual JM, Maher EA, Malloy CR, Deberardinis RJ, and Bachoo RM. Analysis of tumor metabolism reveals mitochondrial glucose oxidation in genetically diverse human glioblastomas in the mouse brain in vivo. Cell Metab 15: 827–837, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Martinez-Outschoorn UE, Sotgia F, and Lisanti MP. Caveolae and signalling in cancer. Nat Rev Cancer 15: 225–237, 2015 [DOI] [PubMed] [Google Scholar]
- 129.McCulloch V, Seidel-Rogol BL, and Shadel GS. A human mitochondrial transcription factor is related to RNA adenine methyltransferases and binds S-adenosylmethionine. Mol Cell Biol 22: 1116–1125, 2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.McInerney EM, Rose DW, Flynn SE, Westin S, Mullen TM, Krones A, Inostroza J, Torchia J, Nolte RT, Assa-Munt N, Milburn MV, Glass CK, and Rosenfeld MG. Determinants of coactivator LXXLL motif specificity in nuclear receptor transcriptional activation. Genes Dev 12: 3357–3368, 1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Medici D. and Kalluri R. Endothelial-mesenchymal transition and its contribution to the emergence of stem cell phenotype. Semin Cancer Biol 22: 379–384, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Mercier I, Camacho J, Titchen K, Gonzales DM, Quann K, Bryant KG, Molchansky A, Milliman JN, Whitaker-Menezes D, Sotgia F, Jasmin JF, Schwarting R, Pestell RG, Blagosklonny MV, and Lisanti MP. Caveolin-1 and accelerated host aging in the breast tumor microenvironment: chemoprevention with rapamycin, an mTOR inhibitor and anti-aging drug. Am J Pathol 181: 278–293, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Michalek RD, Gerriets VA, Jacobs SR, Macintyre AN, MacIver NJ, Mason EF, Sullivan SA, Nichols AG, and Rathmell JC. Cutting edge: distinct glycolytic and lipid oxidative metabolic programs are essential for effector and regulatory CD4+ T cell subsets. J Immunol 186: 3299–3303, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Michaud M, Martins I, Sukkurwala AQ, Adjemian S, Ma Y, Pellegatti P, Shen S, Kepp O, Scoazec M, Mignot G, Rello-Varona S, Tailler M, Menger L, Vacchelli E, Galluzzi L, Ghiringhelli F, di Virgilio F, Zitvogel L, and Kroemer G. Autophagy-dependent anticancer immune responses induced by chemotherapeutic agents in mice. Science 334: 1573–1577, 2011 [DOI] [PubMed] [Google Scholar]
- 135.Michelakis ED, Sutendra G, Dromparis P, Webster L, Haromy A, Niven E, Maguire C, Gammer TL, Mackey JR, Fulton D, Abdulkarim B, McMurtry MS, and Petruk KC. Metabolic modulation of glioblastoma with dichloroacetate. Sci Transl Med 2: 31ra34, 2010 [DOI] [PubMed] [Google Scholar]
- 136.Mitra K, Wunder C, Roysam B, Lin G, and Lippincott-Schwartz J. A hyperfused mitochondrial state achieved at G1-S regulates cyclin E buildup and entry into S phase. Proc Natl Acad Sci USA 106: 11960–11965, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Monti S, Savage KJ, Kutok JL, Feuerhake F, Kurtin P, Mihm M, Wu B, Pasqualucci L, Neuberg D, Aguiar RC, Dal Cin P, Ladd C, Pinkus GS, Salles G, Harris NL, Dalla-Favera R, Habermann TM, Aster JC, Golub TR, and Shipp MA. Molecular profiling of diffuse large B-cell lymphoma identifies robust subtypes including one characterized by host inflammatory response. Blood 105: 1851–1861, 2005 [DOI] [PubMed] [Google Scholar]
- 138.Moreno-Sanchez R, Rodriguez-Enriquez S, Marin-Hernandez A, and Saavedra E. Energy metabolism in tumor cells. FEBS J 274: 1393–1418, 2007 [DOI] [PubMed] [Google Scholar]
- 139.Moreno-Sanchez R, Rodriguez-Enriquez S, Saavedra E, Marin-Hernandez A, and Gallardo-Perez JC. The bioenergetics of cancer: is glycolysis the main ATP supplier in all tumor cells? Biofactors 35: 209–225, 2009 [DOI] [PubMed] [Google Scholar]
- 140.Morita M, Gravel SP, Chenard V, Sikstrom K, Zheng L, Alain T, Gandin V, Avizonis D, Arguello M, Zakaria C, McLaughlan S, Nouet Y, Pause A, Pollak M, Gottlieb E, Larsson O, St-Pierre J, Topisirovic I, and Sonenberg N. mTORC1 controls mitochondrial activity and biogenesis through 4E-BP-dependent translational regulation. Cell Metab 18: 698–711, 2013 [DOI] [PubMed] [Google Scholar]
- 141.Murphy MP, Holmgren A, Larsson NG, Halliwell B, Chang CJ, Kalyanaraman B, Rhee SG, Thornalley PJ, Partridge L, Gems D, Nystrom T, Belousov V, Schumacker PT, and Winterbourn CC. Unraveling the biological roles of reactive oxygen species. Cell Metab 13: 361–366, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Nagley P. Coordination of gene expression in the formation of mammalian mitochondria. Trends Genet 7: 1–4, 1991 [DOI] [PubMed] [Google Scholar]
- 143.Nahon P, Sutton A, Pessayre D, Rufat P, Degoul F, Ganne-Carrie N, Ziol M, Charnaux N, N'Kontchou G, Trinchet JC, Gattegno L, and Beaugrand M. Genetic dimorphism in superoxide dismutase and susceptibility to alcoholic cirrhosis, hepatocellular carcinoma, and death. Clin Gastroenterol Hepatol 3: 292–298, 2005 [DOI] [PubMed] [Google Scholar]
- 144.Nelson DL. and Cox MM. Lehninger, Principles of Biochemistry. 6th edition. New York, NY: W.H. Freeman, 2013 [Google Scholar]
- 145.Newsholme P, Procopio J, Lima MM, Pithon-Curi TC, and Curi R. Glutamine and glutamate—their central role in cell metabolism and function. Cell Biochem Funct 21: 1–9, 2003 [DOI] [PubMed] [Google Scholar]
- 146.Noy R. and Pollard JW. Tumor-associated macrophages: from mechanisms to therapy. Immunity 41: 49–61, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Obre E. and Rossignol R. Emerging concepts in bioenergetics and cancer research: metabolic flexibility, coupling, symbiosis, switch, oxidative tumors, metabolic remodeling, signaling and bioenergetic therapy. Int J Biochem Cell Biol 59: 167–181, 2015 [DOI] [PubMed] [Google Scholar]
- 148.Odegaard JI. and Chawla A. Alternative macrophage activation and metabolism. Ann Rev Pathol 6: 275–297, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Ohlund D, Elyada E, and Tuveson D. Fibroblast heterogeneity in the cancer wound. J Exp Med 211: 1503–1523, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Ojala D, Montoya J, and Attardi G. tRNA punctuation model of RNA processing in human mitochondria. Nature 290: 470–474, 1981 [DOI] [PubMed] [Google Scholar]
- 151.Okoye I, Wang L, Pallmer K, Richter K, Ichimura T, Haas R, Crouse J, Choi O, Heathcote D, Lovo E, Mauro C, Abdi R, Oxenius A, Rutschmann S, and Ashton-Rickardt PG. T cell metabolism. The protein LEM promotes CD8(+) T cell immunity through effects on mitochondrial respiration. Science 348: 995–1001, 2015 [DOI] [PubMed] [Google Scholar]
- 152.Ozaki A, Tanimoto T, and Saji S. Palbociclib in Hormone-Receptor-Positive Advanced Breast Cancer. N Engl J Med 373: 1672–1673, 2015 [DOI] [PubMed] [Google Scholar]
- 153.Paget S. The distribution of secondary growths in cancer of the breast. Lancet 133: 571–573, 1889 [PubMed] [Google Scholar]
- 154.Panopoulos AD, Yanes O, Ruiz S, Kida YS, Diep D, Tautenhahn R, Herrerias A, Batchelder EM, Plongthongkum N, Lutz M, Berggren WT, Zhang K, Evans RM, Siuzdak G, and Izpisua Belmonte JC. The metabolome of induced pluripotent stem cells reveals metabolic changes occurring in somatic cell reprogramming. Cell Res 22: 168–177, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Papandreou I, Cairns RA, Fontana L, Lim AL, and Denko NC. HIF-1 mediates adaptation to hypoxia by actively downregulating mitochondrial oxygen consumption. Cell Metab 3: 187–197, 2006 [DOI] [PubMed] [Google Scholar]
- 156.Parker GC, Acsadi G, and Brenner CA. Mitochondria: determinants of stem cell fate? Stem Cells Dev 18: 803–806, 2009 [DOI] [PubMed] [Google Scholar]
- 157.Parri M. and Chiarugi P. Redox molecular machines involved in tumor progression. Antioxid Redox Signal 19: 1828–1845, 2013 [DOI] [PubMed] [Google Scholar]
- 158.Parsons DW, Jones S, Zhang X, Lin JC, Leary RJ, Angenendt P, Mankoo P, Carter H, Siu IM, Gallia GL, Olivi A, McLendon R, Rasheed BA, Keir S, Nikolskaya T, Nikolsky Y, Busam DA, Tekleab H, Diaz LA, Jr., Hartigan J, Smith DR, Strausberg RL, Marie SK, Shinjo SM, Yan H, Riggins GJ, Bigner DD, Karchin R, Papadopoulos N, Parmigiani G, Vogelstein B, Velculescu VE, and Kinzler KW. An integrated genomic analysis of human glioblastoma multiforme. Science 321: 1807–1812, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Pasto A, Bellio C, Pilotto G, Ciminale V, Silic-Benussi M, Guzzo G, Rasola A, Frasson C, Nardo G, Zulato E, Nicoletto MO, Manicone M, Indraccolo S, and Amadori A. Cancer stem cells from epithelial ovarian cancer patients privilege oxidative phosphorylation, and resist glucose deprivation. Oncotarget 5: 4305–4319, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Pavlides S, Whitaker-Menezes D, Castello-Cros R, Flomenberg N, Witkiewicz AK, Frank PG, Casimiro MC, Wang C, Fortina P, Addya S, Pestell RG, Martinez-Outschoorn UE, Sotgia F, and Lisanti MP. The reverse Warburg effect: aerobic glycolysis in cancer associated fibroblasts and the tumor stroma. Cell Cycle 8: 3984–4001, 2009 [DOI] [PubMed] [Google Scholar]
- 161.Pike LS, Smift AL, Croteau NJ, Ferrick DA, and Wu M. Inhibition of fatty acid oxidation by etomoxir impairs NADPH production and increases reactive oxygen species resulting in ATP depletion and cell death in human glioblastoma cells. Biochim Biophys Acta 1807: 726–734, 2011 [DOI] [PubMed] [Google Scholar]
- 162.Possemato R, Marks KM, Shaul YD, Pacold ME, Kim D, Birsoy K, Sethumadhavan S, Woo HK, Jang HG, Jha AK, Chen WW, Barrett FG, Stransky N, Tsun ZY, Cowley GS, Barretina J, Kalaany NY, Hsu PP, Ottina K, Chan AM, Yuan B, Garraway LA, Root DE, Mino-Kenudson M, Brachtel EF, Driggers EM, and Sabatini DM. Functional genomics reveal that the serine synthesis pathway is essential in breast cancer. Nature 476: 346–350, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Pouyssegur J. and Mechta-Grigoriou F. Redox regulation of the hypoxia-inducible factor. Biol Chem 387: 1337–1346, 2006 [DOI] [PubMed] [Google Scholar]
- 164.Puigserver P, Wu Z, Park CW, Graves R, Wright M, and Spiegelman BM. A cold-inducible coactivator of nuclear receptors linked to adaptive thermogenesis. Cell 92: 829–839, 1998 [DOI] [PubMed] [Google Scholar]
- 165.Quail DF. and Joyce JA. Microenvironmental regulation of tumor progression and metastasis. Nat Med 19: 1423–1437, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Radisky DC, Levy DD, Littlepage LE, Liu H, Nelson CM, Fata JE, Leake D, Godden EL, Albertson DG, Nieto MA, Werb Z, and Bissell MJ. Rac1b and reactive oxygen species mediate MMP-3-induced EMT and genomic instability. Nature 436: 123–127, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Rao RR, Li Q, Odunsi K, and Shrikant PA. The mTOR kinase determines effector versus memory CD8+ T cell fate by regulating the expression of transcription factors T-bet and Eomesodermin. Immunity 32: 67–78, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Rapaport D. Finding the right organelle. Targeting signals in mitochondrial outer-membrane proteins. EMBO Rep 4: 948–952, 2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Rattigan YI, Patel BB, Ackerstaff E, Sukenick G, Koutcher JA, Glod JW, and Banerjee D. Lactate is a mediator of metabolic cooperation between stromal carcinoma associated fibroblasts and glycolytic tumor cells in the tumor microenvironment. Exp Cell Res 318: 326–335, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Reitzer LJ, Wice BM, and Kennell D. Evidence that glutamine, not sugar, is the major energy source for cultured HeLa cells. J Biol Chem 254: 2669–2676, 1979 [PubMed] [Google Scholar]
- 171.Rodgers JT, Lerin C, Haas W, Gygi SP, Spiegelman BM, and Puigserver P. Nutrient control of glucose homeostasis through a complex of PGC-1alpha and SIRT1. Nature 434: 113–118, 2005 [DOI] [PubMed] [Google Scholar]
- 172.Rodriguez-Prados JC, Traves PG, Cuenca J, Rico D, Aragones J, Martin-Sanz P, Cascante M, and Bosca L. Substrate fate in activated macrophages: a comparison between innate, classic, and alternative activation. J Immunol 185: 605–614, 2010 [DOI] [PubMed] [Google Scholar]
- 173.Roesch A, Vultur A, Bogeski I, Wang H, Zimmermann KM, Speicher D, Korbel C, Laschke MW, Gimotty PA, Philipp SE, Krause E, Patzold S, Villanueva J, Krepler C, Fukunaga-Kalabis M, Hoth M, Bastian BC, Vogt T, and Herlyn M. Overcoming intrinsic multidrug resistance in melanoma by blocking the mitochondrial respiratory chain of slow-cycling JARID1B(high) cells. Cancer Cell 23: 811–825, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Rorbach J, Richter R, Wessels HJ, Wydro M, Pekalski M, Farhoud M, Kuhl I, Gaisne M, Bonnefoy N, Smeitink JA, Lightowlers RN, and Chrzanowska-Lightowlers ZM. The human mitochondrial ribosome recycling factor is essential for cell viability. Nucleic Acids Res 36: 5787–5799, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Rossi DJ, Jamieson CH, and Weissman IL. Stems cells and the pathways to aging and cancer. Cell 132: 681–696, 2008 [DOI] [PubMed] [Google Scholar]
- 176.Rossignol R, Gilkerson R, Aggeler R, Yamagata K, Remington SJ, and Capaldi RA. Energy substrate modulates mitochondrial structure and oxidative capacity in cancer cells. Cancer Res 64: 985–993, 2004 [DOI] [PubMed] [Google Scholar]
- 177.Rynne-Vidal A, Jimenez-Heffernan JA, Fernandez-Chacon C, Lopez-Cabrera M, and Sandoval P. The mesothelial origin of carcinoma associated-fibroblasts in peritoneal metastasis. Cancers 7: 1994–2011, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Sakamaki T, Casimiro MC, Ju X, Quong AA, Katiyar S, Liu M, Jiao X, Li A, Zhang X, Lu Y, Wang C, Byers S, Nicholson R, Link T, Shemluck M, Yang J, Fricke ST, Novikoff PM, Papanikolaou A, Arnold A, Albanese C, and Pestell R. Cyclin D1 determines mitochondrial function in vivo. Mol Cell Biol 26: 5449–5469, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Sancho P, Burgos-Ramos E, Tavera A, Bou Kheir T, Jagust P, Schoenhals M, Barneda D, Sellers K, Campos-Olivas R, Grana O, Viera CR, Yuneva M, Sainz B, Jr., and Heeschen C. MYC/PGC-1alpha Balance Determines the Metabolic Phenotype and Plasticity of Pancreatic Cancer Stem Cells. Cell Metab 22: 590–605, 2015 [DOI] [PubMed] [Google Scholar]
- 180.Sankaran VG, Orkin SH, and Walkley CR. Rb intrinsically promotes erythropoiesis by coupling cell cycle exit with mitochondrial biogenesis. Genes Dev 22: 463–475, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Sauer LA, Stayman JW, 3rd, and Dauchy RT. Amino acid, glucose, and lactic acid utilization in vivo by rat tumors. Cancer Res 42: 4090–4097, 1982 [PubMed] [Google Scholar]
- 182.Scarpulla RC. Metabolic control of mitochondrial biogenesis through the PGC-1 family regulatory network. Biochim Biophys Acta 1813: 1269–1278, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Scarpulla RC. Nuclear control of respiratory chain expression by nuclear respiratory factors and PGC-1-related coactivator. Ann N Y Acad Sci 1147: 321–334, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Schatz G. Mitochondria: beyond oxidative phosphorylation. Biochim Biophys Acta 1271: 123–126, 1995 [DOI] [PubMed] [Google Scholar]
- 185.Schoors S, Bruning U, Missiaen R, Queiroz KC, Borgers G, Elia I, Zecchin A, Cantelmo AR, Christen S, Goveia J, Heggermont W, Godde L, Vinckier S, Van Veldhoven PP, Eelen G, Schoonjans L, Gerhardt H, Dewerchin M, Baes M, De Bock K, Ghesquiere B, Lunt SY, Fendt SM, and Carmeliet P. Fatty acid carbon is essential for dNTP synthesis in endothelial cells. Nature 520: 192–197, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Schreiber SN, Emter R, Hock MB, Knutti D, Cardenas J, Podvinec M, Oakeley EJ, and Kralli A. The estrogen-related receptor alpha (ERRalpha) functions in PPARgamma coactivator 1alpha (PGC-1alpha)-induced mitochondrial biogenesis. Proc Natl Acad Sci USA 101: 6472–6477, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Schug ZT, Peck B, Jones DT, Zhang Q, Grosskurth S, Alam IS, Goodwin LM, Smethurst E, Mason S, Blyth K, McGarry L, James D, Shanks E, Kalna G, Saunders RE, Jiang M, Howell M, Lassailly F, Thin MZ, Spencer-Dene B, Stamp G, van den Broek NJ, Mackay G, Bulusu V, Kamphorst JJ, Tardito S, Strachan D, Harris AL, Aboagye EO, Critchlow SE, Wakelam MJ, Schulze A, and Gottlieb E. Acetyl-CoA synthetase 2 promotes acetate utilization and maintains cancer cell growth under metabolic stress. Cancer Cell 27: 57–71, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Sena LA, Li S, Jairaman A, Prakriya M, Ezponda T, Hildeman DA, Wang CR, Schumacker PT, Licht JD, Perlman H, Bryce PJ, and Chandel NS. Mitochondria are required for antigen-specific T cell activation through reactive oxygen species signaling. Immunity 38: 225–236, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Shalapour S. and Karin M. Immunity, inflammation, and cancer: an eternal fight between good and evil. J Clin Invest 125: 3347–3355, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Shen XJ, Zhang H, Tang GS, Wang XD, Zheng R, Wang Y, Zhu Y, Xue XC, and Bi JW. Caveolin-1 is a modulator of fibroblast activation and a potential biomarker for gastric cancer. Int J Biol Sci 11: 370–379, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Smeitink JA, Loeffen JL, Triepels RH, Smeets RJ, Trijbels JM, and van den Heuvel LP. Nuclear genes of human complex I of the mitochondrial electron transport chain: state of the art. Hum Mol Genet 7: 1573–1579, 1998 [DOI] [PubMed] [Google Scholar]
- 192.Smith-Garvin JE, Koretzky GA, and Jordan MS. T cell activation. Ann Rev Immunol 27: 591–619, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Smits P, Smeitink J, and van den Heuvel L. Mitochondrial translation and beyond: processes implicated in combined oxidative phosphorylation deficiencies. J Biomed Biotechnol 2010: 737385, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Smits P, Smeitink JA, van den Heuvel LP, Huynen MA, and Ettema TJ. Reconstructing the evolution of the mitochondrial ribosomal proteome. Nucleic Acids Res 35: 4686–4703, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Soh H, Wasa M, and Fukuzawa M. Hypoxia upregulates amino acid transport in a human neuroblastoma cell line. J Pediatr Surg 42: 608–612, 2007 [DOI] [PubMed] [Google Scholar]
- 196.Sollner T, Rassow J, Wiedmann M, Schlossmann J, Keil P, Neupert W, and Pfanner N. Mapping of the protein import machinery in the mitochondrial outer membrane by crosslinking of translocation intermediates. Nature 355: 84–87, 1992 [DOI] [PubMed] [Google Scholar]
- 197.Son J, Lyssiotis CA, Ying H, Wang X, Hua S, Ligorio M, Perera RM, Ferrone CR, Mullarky E, Shyh-Chang N, Kang Y, Fleming JB, Bardeesy N, Asara JM, Haigis MC, DePinho RA, Cantley LC, and Kimmelman AC. Glutamine supports pancreatic cancer growth through a KRAS-regulated metabolic pathway. Nature 496: 101–105, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Sosa V, Moline T, Somoza R, Paciucci R, Kondoh H, and ME LL. Oxidative stress and cancer: an overview. Ageing Res Rev 12: 376–390, 2013 [DOI] [PubMed] [Google Scholar]
- 199.St-Pierre J, Lin J, Krauss S, Tarr PT, Yang R, Newgard CB, and Spiegelman BM. Bioenergetic analysis of peroxisome proliferator-activated receptor gamma coactivators 1alpha and 1beta (PGC-1alpha and PGC-1beta) in muscle cells. J Biol Chem 278: 26597–26603, 2003 [DOI] [PubMed] [Google Scholar]
- 200.Suda T, Takubo K, and Semenza GL. Metabolic regulation of hematopoietic stem cells in the hypoxic niche. Cell Stem Cell 9: 298–310, 2011 [DOI] [PubMed] [Google Scholar]
- 201.Sullivan LB, Gui DY, Hosios AM, Bush LN, Freinkman E, and Vander Heiden MG. Supporting aspartate biosynthesis is an essential function of respiration in proliferating cells. Cell 162: 552–563, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Sun RC. and Denko NC. Hypoxic regulation of glutamine metabolism through HIF1 and SIAH2 supports lipid synthesis that is necessary for tumor growth. Cell Metab 19: 285–292, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Sutton A, Imbert A, Igoudjil A, Descatoire V, Cazanave S, Pessayre D, and Degoul F. The manganese superoxide dismutase Ala16Val dimorphism modulates both mitochondrial import and mRNA stability. Pharmacogenetics Genomics 15: 311–319, 2005 [DOI] [PubMed] [Google Scholar]
- 204.Swietach P, Vaughan-Jones RD, and Harris AL. Regulation of tumor pH and the role of carbonic anhydrase 9. Cancer Metastasis Rev 26: 299–310, 2007 [DOI] [PubMed] [Google Scholar]
- 205.Taguchi N, Ishihara N, Jofuku A, Oka T, and Mihara K. Mitotic phosphorylation of dynamin-related GTPase Drp1 participates in mitochondrial fission. J Biol Chem 282: 11521–11529, 2007 [DOI] [PubMed] [Google Scholar]
- 206.Takakubo F, Cartwright P, Hoogenraad N, Thorburn DR, Collins F, Lithgow T, and Dahl HH. An amino acid substitution in the pyruvate dehydrogenase E1 alpha gene, affecting mitochondrial import of the precursor protein. Am J Hum Genet 57: 772–780, 1995 [PMC free article] [PubMed] [Google Scholar]
- 207.Tan Z, Xie N, Cui H, Moellering DR, Abraham E, Thannickal VJ, and Liu G. Pyruvate dehydrogenase kinase 1 participates in macrophage polarization via regulating glucose metabolism. J Immunol 194: 6082–6089, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Tannahill GM, Curtis AM, Adamik J, Palsson-McDermott EM, McGettrick AF, Goel G, Frezza C, Bernard NJ, Kelly B, Foley NH, Zheng L, Gardet A, Tong Z, Jany SS, Corr SC, Haneklaus M, Caffrey BE, Pierce K, Walmsley S, Beasley FC, Cummins E, Nizet V, Whyte M, Taylor CT, Lin H, Masters SL, Gottlieb E, Kelly VP, Clish C, Auron PE, Xavier RJ, and O'Neill LA. Succinate is an inflammatory signal that induces IL-1beta through HIF-1alpha. Nature 496: 238–242, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Terunuma A, Putluri N, Mishra P, Mathe EA, Dorsey TH, Yi M, Wallace TA, Issaq HJ, Zhou M, Killian JK, Stevenson HS, Karoly ED, Chan K, Samanta S, Prieto D, Hsu TY, Kurley SJ, Putluri V, Sonavane R, Edelman DC, Wulff J, Starks AM, Yang Y, Kittles RA, Yfantis HG, Lee DH, Ioffe OB, Schiff R, Stephens RM, Meltzer PS, Veenstra TD, Westbrook TF, Sreekumar A, and Ambs S. MYC-driven accumulation of 2-hydroxyglutarate is associated with breast cancer prognosis. J Clin Invest 124: 398–412, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Timmerman LA, Holton T, Yuneva M, Louie RJ, Padro M, Daemen A, Hu M, Chan DA, Ethier SP, van 't Veer LJ, Polyak K, McCormick F, and Gray JW. Glutamine sensitivity analysis identifies the xCT antiporter as a common triple-negative breast tumor therapeutic target. Cancer Cell 24: 450–465, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Toullec A, Gerald D, Despouy G, Bourachot B, Cardon M, Lefort S, Richardson M, Rigaill G, Parrini MC, Lucchesi C, Bellanger D, Stern MH, Dubois T, Sastre-Garau X, Delattre O, Vincent-Salomon A, and Mechta-Grigoriou F. Oxidative stress promotes myofibroblast differentiation and tumour spreading. EMBO Mol Med 2: 211–230, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Vander Heiden MG, Cantley LC, and Thompson CB. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science 324: 1029–1033, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Vats D, Mukundan L, Odegaard JI, Zhang L, Smith KL, Morel CR, Wagner RA, Greaves DR, Murray PJ, and Chawla A. Oxidative metabolism and PGC-1beta attenuate macrophage-mediated inflammation. Cell Metab 4: 13–24, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Vaupel P, Kallinowski F, and Okunieff P. Blood flow, oxygen and nutrient supply, and metabolic microenvironment of human tumors: a review. Cancer Res 49: 6449–6465, 1989 [PubMed] [Google Scholar]
- 215.Vazquez F, Lim JH, Chim H, Bhalla K, Girnun G, Pierce K, Clish CB, Granter SR, Widlund HR, Spiegelman BM, and Puigserver P. PGC1alpha expression defines a subset of human melanoma tumors with increased mitochondrial capacity and resistance to oxidative stress. Cancer Cell 23: 287–301, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Vercauteren K, Gleyzer N, and Scarpulla RC. Short hairpin RNA-mediated silencing of PRC (PGC-1-related coactivator) results in a severe respiratory chain deficiency associated with the proliferation of aberrant mitochondria. J Biol Chem 284: 2307–2319, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Viale A, Pettazzoni P, Lyssiotis CA, Ying H, Sanchez N, Marchesini M, Carugo A, Green T, Seth S, Giuliani V, Kost-Alimova M, Muller F, Colla S, Nezi L, Genovese G, Deem AK, Kapoor A, Yao W, Brunetto E, Kang Y, Yuan M, Asara JM, Wang YA, Heffernan TP, Kimmelman AC, Wang H, Fleming JB, Cantley LC, DePinho RA, and Draetta GF. Oncogene ablation-resistant pancreatic cancer cells depend on mitochondrial function. Nature 514: 628–632, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Villena JA. and Kralli A. ERRalpha: a metabolic function for the oldest orphan. Trends Endocrinol Metab 19: 269–276, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Virbasius JV. and Scarpulla RC. Activation of the human mitochondrial transcription factor A gene by nuclear respiratory factors: a potential regulatory link between nuclear and mitochondrial gene expression in organelle biogenesis. Proc Natl Acad Sci USA 91: 1309–1313, 1994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Virbasius JV, Virbasius CA, and Scarpulla RC. Identity of GABP with NRF-2, a multisubunit activator of cytochrome oxidase expression, reveals a cellular role for an ETS domain activator of viral promoters. Genes Dev 7: 380–392, 1993 [DOI] [PubMed] [Google Scholar]
- 221.Vlashi E, Lagadec C, Vergnes L, Matsutani T, Masui K, Poulou M, Popescu R, Della Donna L, Evers P, Dekmezian C, Reue K, Christofk H, Mischel PS, and Pajonk F. Metabolic state of glioma stem cells and nontumorigenic cells. Proc Natl Acad Sci USA 108: 16062–16067, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Vlashi E, Lagadec C, Vergnes L, Reue K, Frohnen P, Chan M, Alhiyari Y, Dratver MB, and Pajonk F. Metabolic differences in breast cancer stem cells and differentiated progeny. Breast Cancer Res Treat 146: 525–534, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Wallace C. and Keast D. Glutamine and macrophage function. Metabolism 41: 1016–1020, 1992 [DOI] [PubMed] [Google Scholar]
- 224.Wallace DC. Mitochondrial diseases in man and mouse. Science 283: 1482–1488, 1999 [DOI] [PubMed] [Google Scholar]
- 225.Wang C, Li Z, Lu Y, Du R, Katiyar S, Yang J, Fu M, Leader JE, Quong A, Novikoff PM, and Pestell RG. Cyclin D1 repression of nuclear respiratory factor 1 integrates nuclear DNA synthesis and mitochondrial function. Proc Natl Acad Sci USA 103: 11567–11572, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Wang J, Ying G, Wang J, Jung Y, Lu J, Zhu J, Pienta KJ, and Taichman RS. Characterization of phosphoglycerate kinase-1 expression of stromal cells derived from tumor microenvironment in prostate cancer progression. Cancer Res 70: 471–480, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 227.Wang Z, Fan M, Candas D, Zhang TQ, Qin L, Eldridge A, Wachsmann-Hogiu S, Ahmed KM, Chromy BA, Nantajit D, Duru N, He F, Chen M, Finkel T, Weinstein LS, and Li JJ. Cyclin B1/Cdk1 coordinates mitochondrial respiration for cell-cycle G2/M progression. Dev Cell 29: 217–232, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Warburg O. On respiratory impairment in cancer cells. Science 124: 269–270, 1956 [PubMed] [Google Scholar]
- 229.Warburg O, Wind F. and Negelein, E. The metabolism of tumors in the body. J Gener Physiol 8: 519–530, 1927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Ward PS, Patel J, Wise DR, Abdel-Wahab O, Bennett BD, Coller HA, Cross JR, Fantin VR, Hedvat CV, Perl AE, Rabinowitz JD, Carroll M, Su SM, Sharp KA, Levine RL, and Thompson CB. The common feature of leukemia-associated IDH1 and IDH2 mutations is a neomorphic enzyme activity converting alpha-ketoglutarate to 2-hydroxyglutarate. Cancer Cell 17: 225–234, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Ward PS. and Thompson CB. Metabolic reprogramming: a cancer hallmark even warburg did not anticipate. Cancer Cell 21: 297–308, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Watkins G, Douglas-Jones A, Mansel RE, and Jiang WG. The localisation and reduction of nuclear staining of PPARgamma and PGC-1 in human breast cancer. Oncol Rep 12: 483–488, 2004 [PubMed] [Google Scholar]
- 233.Weinert BT, Wagner SA, Horn H, Henriksen P, Liu WR, Olsen JV, Jensen LJ, and Choudhary C. Proteome-wide mapping of the Drosophila acetylome demonstrates a high degree of conservation of lysine acetylation. Sci Signal 4: ra48, 2011 [DOI] [PubMed] [Google Scholar]
- 234.Weinhouse S. On respiratory impairment in cancer cells. Science 124: 267–269, 1956 [DOI] [PubMed] [Google Scholar]
- 235.Weinhouse S. Oxidative metabolism of neoplastic tissues. Adv Cancer Res 3: 269–325, 1955 [DOI] [PubMed] [Google Scholar]
- 236.Weinhouse S. The Warburg hypothesis fifty years later. Z Krebsforsch klin Onkol Cancer Res Clin Oncol 87: 115–126, 1976 [DOI] [PubMed] [Google Scholar]
- 237.Wellen KE, Hatzivassiliou G, Sachdeva UM, Bui TV, Cross JR, and Thompson CB. ATP-citrate lyase links cellular metabolism to histone acetylation. Science 324: 1076–1080, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Wellen KE. and Thompson CB. A two-way street: reciprocal regulation of metabolism and signalling. Nat Rev Mol Cell Biol 13: 270–276, 2012 [DOI] [PubMed] [Google Scholar]
- 239.Wenner CE, Spirtes MA, and Weinhouse S. Metabolism of neoplastic tissue. II. A survey of enzymes of the citric acid cycle in transplanted tumors. Cancer Res 12: 44–49, 1952 [PubMed] [Google Scholar]
- 240.Weraarpachai W, Antonicka H, Sasarman F, Seeger J, Schrank B, Kolesar JE, Lochmuller H, Chevrette M, Kaufman BA, Horvath R, and Shoubridge EA. Mutation in TACO1, encoding a translational activator of COX I, results in cytochrome c oxidase deficiency and late-onset Leigh syndrome. Nat Genet 41: 833–837, 2009 [DOI] [PubMed] [Google Scholar]
- 241.Wise DR, DeBerardinis RJ, Mancuso A, Sayed N, Zhang XY, Pfeiffer HK, Nissim I, Daikhin E, Yudkoff M, McMahon SB, and Thompson CB. Myc regulates a transcriptional program that stimulates mitochondrial glutaminolysis and leads to glutamine addiction. Proc Natl Acad Sci USA 105: 18782–18787, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Wright G, Terada K, Yano M, Sergeev I, and Mori M. Oxidative stress inhibits the mitochondrial import of preproteins and leads to their degradation. Exp Cell Res 263: 107–117, 2001 [DOI] [PubMed] [Google Scholar]
- 243.Yang L, Moss T, Mangala LS, Marini J, Zhao H, Wahlig S, Armaiz-Pena G, Jiang D, Achreja A, Win J, Roopaimoole R, Rodriguez-Aguayo C, Mercado-Uribe I, Lopez-Berestein G, Liu J, Tsukamoto T, Sood AK, Ram PT, and Nagrath D. Metabolic shifts toward glutamine regulate tumor growth, invasion and bioenergetics in ovarian cancer. Mol Syst Biol 10: 728, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Yao P. and Fox PL. Aminoacyl-tRNA synthetases in medicine and disease. EMBO Mol Med 5: 332–343, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Ye J, Mancuso A, Tong X, Ward PS, Fan J, Rabinowitz JD, and Thompson CB. Pyruvate kinase M2 promotes de novo serine synthesis to sustain mTORC1 activity and cell proliferation. Proc Natl Acad Sci USA 109: 6904–6909, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Ye XQ, Li Q, Wang GH, Sun FF, Huang GJ, Bian XW, Yu SC, and Qian GS. Mitochondrial and energy metabolism-related properties as novel indicators of lung cancer stem cells. Int J Cancer 129: 820–831, 2011 [DOI] [PubMed] [Google Scholar]
- 247.Yu Q, Geng Y, and Sicinski P. Specific protection against breast cancers by cyclin D1 ablation. Nature 411: 1017–1021, 2001 [DOI] [PubMed] [Google Scholar]
- 248.Yuan Y, Gu ZX, Tao XF, and Liu SY. Computer tomography, magnetic resonance imaging, and positron emission tomography or positron emission tomography/computer tomography for detection of metastatic lymph nodes in patients with ovarian cancer: a meta-analysis. Eur J Radiol 81: 1002–1006, 2012 [DOI] [PubMed] [Google Scholar]
- 249.Yuneva M, Zamboni N, Oefner P, Sachidanandam R, and Lazebnik Y. Deficiency in glutamine but not glucose induces MYC-dependent apoptosis in human cells. J Cell Biol 178: 93–105, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Zhang D, Wang Y, Shi Z, Liu J, Sun P, Hou X, Zhang J, Zhao S, Zhou BP, and Mi J. Metabolic reprogramming of cancer-associated fibroblasts by IDH3alpha downregulation. Cell Rep 10: 1335–1348, 2015 [DOI] [PubMed] [Google Scholar]
- 251.Zhang Y, Ba Y, Liu C, Sun G, Ding L, Gao S, Hao J, Yu Z, Zhang J, Zen K, Tong Z, Xiang Y, and Zhang CY. PGC-1alpha induces apoptosis in human epithelial ovarian cancer cells through a PPARgamma-dependent pathway. Cell Res 17: 363–373, 2007 [DOI] [PubMed] [Google Scholar]
- 252.Zhang Y, Choksi S, Chen K, Pobezinskaya Y, Linnoila I, and Liu ZG. ROS play a critical role in the differentiation of alternatively activated macrophages and the occurrence of tumor-associated macrophages. Cell Res 23: 898–914, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Zu XL. and Guppy M. Cancer metabolism: facts, fantasy, and fiction. Biochem Biophys Res Commun 313: 459–465, 2004 [DOI] [PubMed] [Google Scholar]