Abstract
Inflammation plays a prominent role in the events following traumatic injury to the central nervous system (CNS). The initial inflammatory response is driven by mediators such as tumor necrosis factor α and interleukin 1β, which are produced by activated astrocytes and microglia at the site of injury. These factors are regulated post-transcriptionally by RNA binding proteins (RBP) that interact with adenylate and uridylate-rich elements (ARE) in the 3′-untranslated region of the messenger RNA (mRNA). Human antigen R (HuR) is one of these RBPs and generally functions as a positive regulator of ARE-containing mRNAs. Here, we hypothesized that HuR plays an important role in the induction of cytokine and chemokines in astrocytes following traumatic injury. Using a mouse model of spinal cord injury, we found HuR to be extensively translocated to the cytoplasm in astrocytes at the level of injury, consistent with its activation. In an in vitro stretch injury model of CNS trauma, we observed a similar cytoplasmic shift of HuR in astrocytes and an attenuation of cytokine induction with HuR knockdown. RNA kinetics and luciferase assays suggested that the effect was more related to transcription than RNA destabilization. A small molecule inhibitor of HuR suppressed cytokine induction of injured astrocytes and reduced chemoattraction for neutrophils and microglia. In summary, HuR is activated in astrocytes in the early stages of CNS trauma and positively regulates the molecular response of key inflammatory mediators in astrocytes. Our findings suggest that HuR may be a therapeutic target in acute CNS trauma for blunting secondary tissue injury triggered by the inflammatory response.
Keywords: : glial cell response to injury, inflammation, molecular biological approaches, spinal cord injury
Introduction
In traumatic injuries to the brain or spinal cord, glial cells (astrocytes and microglia) in the vicinity of injury are first responders and become activated within minutes, undergoing morphological and molecular changes in response to signals in the injured tissue.1–4 Part of the molecular transformation is a rapid induction and release of an array of inflammatory mediators such as tumor necrosis factor (TNF)-α, interleukin (IL)-1β, IL-6, and matrix metalloproteases (e.g., 2, 3, 9, and 12) into the milieu of the injured tissue. These factors activate additional glial cells, recruit circulating immune cells such as neutrophils and monocytes into the injured area, and facilitate glial cell mobility. The result is an amplification of inflammatory factor production, accumulation of cytotoxic substances, disruption of the blood–brain barrier, and additional cell death, all of which substantially extend the boundary of the initial injury.1,3,5–8
A key regulatory pathway of inflammatory cytokines is at the post-transcriptional level and often mediated by adenylate and uridylate-rich elements (ARE) in the 3′- and 5′-untranslated regions (UTRs).9,10 The ARE functions as a binding target for RNA binding proteins (RBP) that regulate RNA stability and translational efficiency. Most of the cytokines that drive the early inflammatory response in central nervous system (CNS) trauma, such as interleukin 1β (IL-1β) and tumor necrosis factor α (TNF-α), are regulated at this level.9,11,12
Human antigen R (HuR) is a well characterized RBP that generally enhances gene expression by binding to the ARE of the mRNA.10,13 A hallmark feature of HuR activity is its translocation to the cytoplasm, where it associates with polysomes and enhances the translational efficiency and stability of the bound mRNA.11–13 Many HuR targets are highly relevant to CNS trauma, such as TNF-α, IL-1β, and IL-6. This observation, coupled with the known role of HuR as a positive regulator of these mRNAs in cancer and other cell systems, led us to hypothesize that HuR may play an active role in the early molecular responses to CNS injury.10
We focused on astrocytes since they, along with microglia, play a pivotal role in the inflammatory response in injured CNS tissue. To test this hypothesis, we assessed the location of HuR in astrocytes in mice subjected to spinal cord injury and found prominent cytoplasmic translocation of HuR consistent with its activation. In an in vitro stretch model for CNS injury, we observed significant attenuation of cytokine induction in astrocytes when HuR was knocked down or chemically inhibited. Conditioned media from these astrocytes showed reduced chemoattraction for neutrophils and microglial cells. In summary, HuR is a key modulator of the inflammatory response and may be a therapeutic target for reducing secondary tissue injury in early CNS trauma.
Methods
Contusion model of spinal cord injury and stretch assay
All animal procedures were reviewed and approved by the University of Alabama, Birmingham (UAB) Institutional Animal Care and Use Committee in compliance with the National Research Council Guide for the Care and Use of Laboratory Animals. Female C57/Bl6 mice at 8–12 weeks of age were anesthetized under isoflurane and the lamina was removed at thoracic level 10. Using a 0.8-mm diameter tip, a centrally located contusion injury was induced by application of 75 kilodynes of force to the exposed spinal cord using the Infinite Horizon spinal cord injury device (Precision Systems & Instrumentation, Lexington, KY). Mice used as sham controls were subjected to laminectomy only. For stretch injury, astrocytes were first isolated from pups as previously described.14 Cells were plated on Bioflex plates (Flexcell, Burlington, NC) with elastic bottoms that are deformable with air pressure. Using the 94A Cell Injury Controller (Bioengineering Facility, Virginia Commonwealth University, Richmond, VA), a 50 msec pulse of air was applied to cause a deformation of 6.3 mm (moderate injury) to the wells of the plate, causing a stretch injury to the astrocytes as previously described.15
Immunohistochemistry and immunocytochemistry
At 24 h post-injury, mice were euthanized by isoflurane and pentobarbital, then perfused with ice cold phosphate-buffered saline (PBS) for 2 min, followed by 4% paraformaldehyde (PFA) for 5 min. The spinal column was then extracted and post-fixed in PFA for 24 h before overnight decalcification in 8% hydrochloric acid and 10% ethanol in PBS. The spinal column was cryoprotected in a 10% sucrose in PBS solution for 2 h, followed by 30% sucrose in PBS for 48 h. Three millimeter samples at the epicenter of injury and adjacent rostral and caudal tissue were cryomolded in Tissue-Tek OCT (Sakura Finetek, Torrance, CA) and cut transversely into 30-μm serial sections using a cryostat (Leica, Buffalo Grove, IL). Spinal cord sections were washed with PBS and blocked with 10% goat serum for 1 h before overnight incubation using the following antibodies: HuR (3A2, mouse monoclonal; Santa-Cruz, Santa Cruz, CA) at 1:1000 and glial fibrillary acidic protein (GFAP; Z0334, rabbit polyclonal; Sigma, St. Louis, MO) at 1:2000. Nuclear labeling was performed using 4',6-diamidino-2-phenylindole.
Qualitative assessment of HuR localization was performed in a group of four injured and four sham-injured mice. For quantitative assessment of HuR localization, photomicrographs of eight random high-powered fields of epicenter, rostral, and caudal sections (3 mm relative to the epicenter) were taken from one injured mouse at 24 h using an Olympus BX41 microscope. A sham-injured mouse was similarly assessed. HuR/GFAP colocalization was then performed on each photomicrograph using ImageJ software (National Institutes of Health, Bethesda, MD) and the Green and Red Puncta colocalization Macro for ImageJ (Daniel J. Shiwarski, Ruben K. Dagda, and Charleen T. Chu). Immunocytochemistry of HuR and GFAP also was performed in stretch-injured astrocytes. Twenty-four hours after injury, astrocytes were washed with PBS and fixed with PFA for 10 min. The fixed astrocytes were washed, blocked in 10% goat serum, and incubated with anti-HuR and anti-GFAP antibodies at the same concentrations and time intervals used for immunohistochemistry. After staining, silastic membranes from Flexcell plates were cut out and inverted onto six-well plates for imaging using an Olympus IX73 inverted microscope.
HuR knockdown and MS-444 treatment
HuR knockdown in primary astrocytes were performed by transfection of a small interfering RNA (siRNA) specific to HuR, siHuR (Dharmacon, Lafayette, CO), 72 h prior to stretch injury. Astrocytes were electroporated using the Neon transfection kit (Invitrogen, Carlsbad, CA) according to the manufacturer's protocol. siHuR or control siRNA targeting green fluorescent protein was transfected into astrocytes at a concentration of 300 pmol per 1 × 106 cells. MS-444 was generously provided by Dr. Meisner-Kober (Novartis, Basel, Switzerland). MS-444 was reconstituted in pure dimethyl sulfoxide (DMSO) and was added to the cell-culture media at concentrations of 25 and 50 μM, with a final DMSO concentration of 0.5% (% v/v). The control consisted of DMSO in cell culture media at the same concentration. Astrocytes were treated with MS-444 or vehicle for 8 h prior to injury or stimulation to ensure HuR inhibition.
RNA isolation, kinetics, and protein analysis
RNA was extracted and isolated using the illustra RNAspin kit (GE Healthcare, Little Chalfont, UK) and concentrations were measured by Nanodrop (Thermo Scientific, Waltham, MA). Complementary DNA (cDNA) was reverse-transcribed from RNA using the High Capacity cDNA Reverse Transcription Kit (Applied Biosystems, Foster City, CA). Quantitative real-time polymerase chain reaction (qRT-PCR) was performed using the ViiA7 Real-Time PCR System and commercially available primer and probes (Applied Biosystems). RNA half-lives were determined as previously described.16 For protein analysis, extracts were collected in M-Per lysis buffer and quantitated using the BCA Protein Assay (Thermo Scientific, Waltham, MA). Forty micrograms of protein were analyzed by Western blot and probed with antibodies for the following: HuR (Santa Cruz, Santa Cruz, CA) at 1:1000 and actin at 1:2,000 (Cell Signaling, Danvers, MA). Astrocyte conditioned media from stretch injured astrocytes were pooled from three wells and concentrated 6-fold using Amicon Ultra 3NMWL centrifugal filters (Millipore, Billerica, MA). Cytokine expression was analyzed using enzyme-linked immunosorbent assay (ELISA) for the following cytokines: TNF-α, IL-6, C-X-C motif chemokine ligand 1 (CXCL-1), leukemia inhibitory factor (LIF), and C-C motif ligand 2 (CCL2; R&D Biosystems, Minneapolis, MN), and IL-1β (Abcam, Cambridge, UK).
Luciferase promoter assay
Luciferase reporter plasmids were generously provided by Dr. Dong, University of Pittsburgh (IL-1β promoter) and Dr. Nagaoka, Juntendo University, Japan (TREM-1 promoter). The TNF-α promoter was provided by Dr. Economou (University of California, Los Angeles) and modified as previously described.16 The Hel-N1 promoter is described elsewhere.17 The PGL2 control plasmid (E1611) was purchased from Promega (Madison, WI). Astrocytes were transfected by electroporation with siRNA, 2 μg of luciferase reporter plasmids, and 1 μg of pCMV β-galactosidase plasmid as an internal control. Twenty-four hours after stretch-injury, astrocytes were lysed and luciferase/β-galactosidase activities were assayed as previously described.17,18 Luciferase values were adjusted to β-galactosidase activity.
Transwell migration assays
Conditioned medium from stretch-injured astrocytes was used as a chemoattractant for primary neutrophils and microglia-like BV2 cells (generously provided by Dr. Tika Benveniste). Primary murine neutrophils were isolated from tibia and femur bone marrow by differential centrifugation through a Percoll gradient (52%, 65%, 78%). Neutrophils (1 × 105) were plated onto a transwell insert with a pore size of 3 μm, then placed into conditioned medium from stretch-injured astrocytes for 1 h. Neutrophils that migrated into the astrocyte conditioned media (ACM) below were labeled with a fluorescein isothyocyanate–conjugated Ly6G antibody (RB6-8C5, rat monoclonal; Abcam, Cambridge, MA) and counted by fluorescence-activated cell sorting (FACS). To investigate chemoattraction of microglia/macrophages, 2.5 × 104 BV2 cells or primary mouse microglia were seeded into the upper well of an 8 μm pore transwell insert and placed into ACM for 24 h.
Primary microglial cells (PMG) were isolated from 1- to 3-day-old pups as described previously.19 The purity of PMG was assessed by IBA1 staining. Migrated cells on the lower side of the transwell membrane were stained using the Hema 3 Stat Pack staining kit (Fisher, Kalamazoo, MI). Ten random high-powered fields were taken for each membrane and cells were counted using an Olympus BX41 microscope.
Statistical analysis
All RNA, protein, and luciferase measurements were assessed using a Student's t-test comparing control versus treated samples. For in vivo HuR localization, a one-way analysis of variance with Bonferroni multiple comparisons test was used for comparisons of merged signal between different spinal cord locations and the sham-injured control.
Results
HuR translocates from nucleus to cytoplasm in the acute phase of spinal cord injury
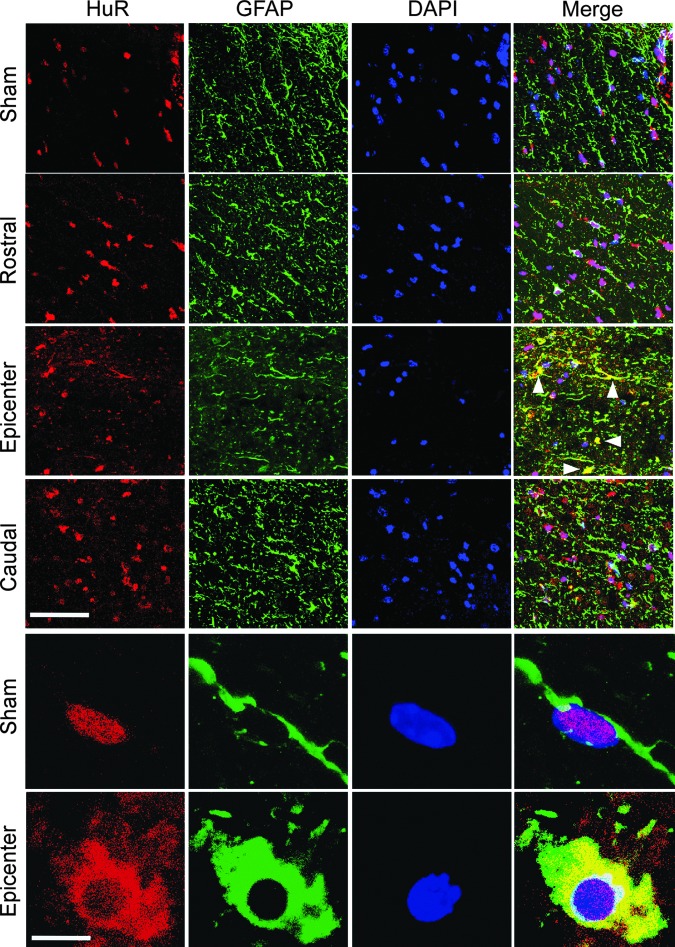
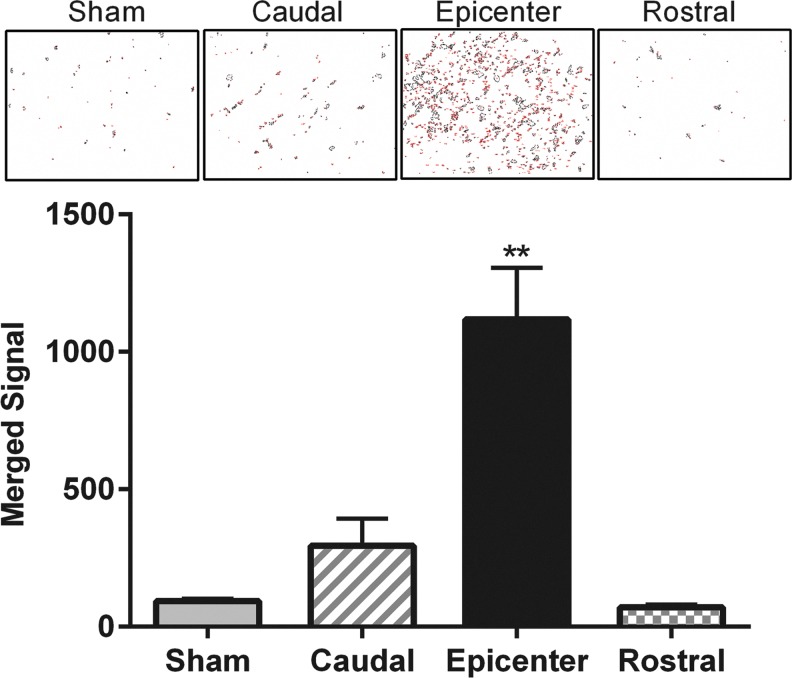
HuR is predominantly nuclear in localization, and its translocation to the cytoplasm is a hallmark of increased activity, where it promotes stabilization and enhanced translational efficiency of target mRNAs.20,21 To determine if translocation of HuR to the cytoplasm occurs in astrocytes after spinal cord injury (SCI), we subjected mice to a thoracic (T10) contusion injury. At 24 h post-injury, spinal cord sections at the epicenter (T10) and 3 mm rostral and caudal to the level of injury were assessed by confocal microscopy for HuR localization in astrocytes. Immunoreactivity with GFAP was used both to identify astrocytes and the cytoplasmic compartment. At the epicenter, we observed merged HuR and GFAP signal indicating colocalization and HuR translocation, whereas in levels rostral or caudal to the injury or in sham-injured mice, HuR remained predominantly in the nucleus (Fig. 1). Merged signal at the level of injury was detected extensively in astrocytic processes and cell bodies throughout the epicenter. A high-power view of single astrocytes demonstrates the latter (Fig. 1). It is possible that some of the colocalization might have resulted from nuclear leakage of HuR in astrocytes undergoing injury-induced necrosis. To estimate the extent of HuR and GFAP colocalization, quantitation of merged signal was done on additional sections using a colocalization macro with ImageJ (see Methods). We observed an approximately 12-fold increase in merged signal in the epicenter, compared with the sham-injured control and rostral levels (Fig. 2). There was a slight but not significant increase in merged signal in the caudal section, which may be related to extension of the injury beyond the epicenter. These findings are consistent with HuR translocation to the cytoplasm in astrocytes at the level of injury in the acute phase.
FIG. 1.
Human antigen R (HuR) translocates to the cytoplasm in astrocytes at the level of acutely injured spinal cord. Mice were subjected to a T10 contusion injury and immunohistochemistry was performed on sections obtained at the epicenter, rostral and caudal levels and at the epicenter of sham-injured mice 24 h after injury. Upper panels show lower powered views at the different indicated levels. Antibodies are shown at the top. Arrows indicate areas of merged glial fibrillary acidic protein (GFAP) and HuR signals. Scale bar, 100 μm. The lower panels show high-powered views of single astrocytes at the epicenter of injury or in a sham control. Scale bar, 10 μm. Images are from one injured and one sham-injured mouse, but similar results were observed in three additional mice. DAPI, 4′,6-diamidino-2-phenylindole. Color image is available online at www.liebertpub.com/neu
FIG. 2.
Quantitation of human antigen R (HuR) cytoplasmic translocation in injured spinal cord. Immunohistochemistry sections were analyzed for merged (yellow) signal of HuR and glial fibrillary acidic protein immunoreactivity using ImageJ software with a colocalization macro (see Methods). Representative fields where loci meeting the threshold for merged signal are shown in the upper panel. The graph shows the mean ± standard error of the mean of four independent sections from a mouse undergoing T10 contusion or sham injury, with two random fields counted per section. **p < 0.0001. Color image is available online at www.liebertpub.com/neu
HuR translocates to the cytoplasm in astrocytes subjected to stretch injury and positively modulates production of key inflammatory mediators
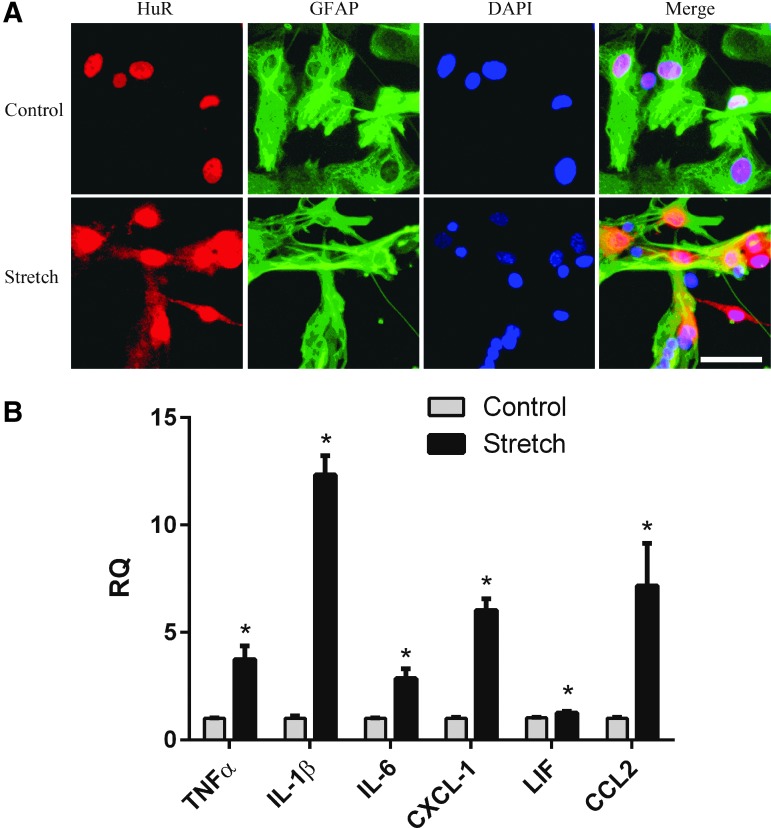
To characterize the impact of HuR on the induction of trauma-relevant inflammatory cytokines in astrocytes, we used an in vitro model of stretch injury that recapitulates viscoelastic forces sustained in CNS trauma.15 We first sought to determine if this model induces changes in HuR localization as observed in vivo. We performed immunocytochemistry 24 h after stretch injury and observed cytoplasmic translocation of HuR (Fig. 3A). In non-stretch controls, HuR remained nuclear in location. These observations are consistent with the pattern of translocation that we observed in vivo in spinal cord injury.
FIG. 3.
An in vitro stretch model of astrocytes induces cytoplasmic translocation of human antigen R (HuR) and induction of inflammatory cytokines. (A) Primary astrocytes were subjected to stretch injury and stained with HuR and glial fibrillary acidic protein (GFAP) antibodies 24 h later. Control astrocytes were similarly plated but not stretched. Scale bar, 40 μm. (B) Quantitative real-time polymerase chain reaction analysis of stretch-injured astrocytes shows induction of pro-inflammatory cytokines observed in early central nervous system trauma. The data represent the mean ± standard error of the mean of six independent samples. *p < 0.05. DAPI, 4',6-diamidino-2-phenylindole; RQ, relative quantity; TNF, tumor necrosis factor; IL, interleukin. Color image is available online at www.liebertpub.com/neu
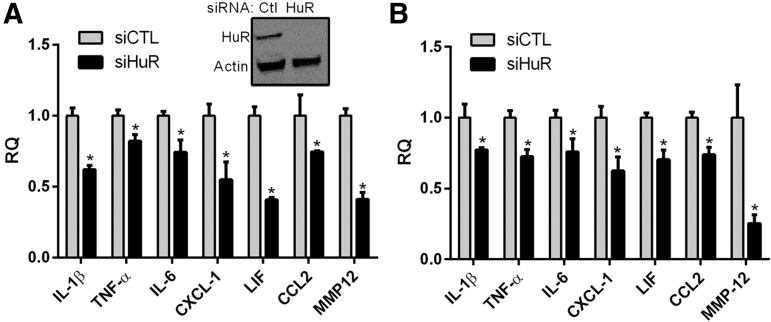
In several recent RNA sequencing studies in SCI and traumatic brain injury (TBI), the most differentially expressed mRNA targets in the acute phase were linked to inflammation.22,23 We analyzed some of the top ranking genes that were increased (several by more than a 100-fold) and found multiple HuR consensus binding sites in the 3′-UTR (Table 1).24,25 To assess the induction of these cytokines, astrocytes were subjected to stretch injury and RNA was collected 24 h post-injury. There was a significant induction of each mRNA in a pattern similar to that reported in early CNS trauma (Fig. 3B).3,26 Taken together, these findings indicate that the in vitro stretch injury model recapitulates some features of injury in vivo with regard to HuR translocation and cytokine induction. To determine the impact of HuR on cytokine induction, astrocytes were transfected with siHuR or control siRNA and subjected to stretch injury. HuR was knocked down by ∼90% (Fig. 4A). Analysis by qRT-PCR showed a significant attenuation of each mRNA (20–60%) including the matrix metalloproteinase, MMP-12, at 24 h post-injury. Protein products for these mRNAs were then measured in the conditioned media by ELISA. Astrocytes were further activated with TNF-α stimulation to increase protein production above the threshold for ELISA detection. Treatment with TNF-α results in a marked increase in cytokine production in astrocytes and this cytokine is present, along with other inflammatory cytokines, in the microenvironment of the injured spinal cord.2,4,16,27,28 HuR knockdown resulted in an approximately 20–30% reduction of all targets (Fig. 4B). Interestingly, for MMP-12, there was a 75% drop in protein expression that was disproportionately greater than the other targets, suggesting HuR may be modulating translational efficiency of this target as well as mRNA expression. This dual function of HuR has been observed with other targets in other cell systems.10
Table 1.
Top Ranking Response Genes Up-Regulated in CNS Injury Have HuR Binding Sites in the mRNA
| Gene | Name | Fold increase in SCI1 | Fold increase in TBI2 | Function in CNS injury3 | HuR Binding Sites4 |
|---|---|---|---|---|---|
| IL-1β | Interleukin 1β | 160 | 34 | I, C | 4 |
| LIF | Leukemia inhibitory factor | 32 | 16 | I, C | 20 |
| IL-6 | Interleukin 6 | 168 | 48 | I | 4 |
| TNF-α | Tumor necrosis factor alpha | 23 | 610 | I, C, E | 12 |
| CXCL1 | C-X-C motif chemokine ligand 1 | n.a. | 161 | C | 9 |
| CCL2 | C-C motif ligand 2 | 230 | 144 | C | 4 |
| MMP12 | Matrix metalloproteinase-12 | 189 | 154 | I, E | 9 |
Based on data published elsewhere ranking the relevance of gene responses in spinal cord injury.22,51
Based on data published elsewhere investigating mRNA expression in response to traumatic brain injury.23
I, inflammation; C, chemotaxis; E, edema.
Binding sites located in 3′ untranslated region of mRNA based on consensus sequences identified previously.24,25
CNS, central nervous system; HuR, human antigen R; mRNA, messenger RNA; SCI, spinal cord injury; TBI, traumatic brain injury.
FIG. 4.
Knockdown of human antigen R (HuR) in astrocytes attenuates the induction of pro-inflammatory cytokines 24 h after stretch injury. (A) Upper panel shows Western blot of astrocytes following transfection of HuR or control (Ctl) small interfering RNA (siRNA). Below is a quantitative real-time polymerase chain reaction analysis of cytokine messenger RNA levels in siRNA-treated astrocytes. (B) Enzyme-linked immunosorbent assay of cytokines in conditioned media from the injured astrocytes. All data points are the mean ± standard error of the mean of six independent samples, and values were expressed as a relative quantity (RQ) to the control siRNA. *p < 0.05. IL, interleukin; TNF, tumor necrosis factor; MMP, matrix metalloproteinase.
HuR knockdown attenuates promoter activity of cytokine targets rather than RNA stability after stretch injury of astrocytes
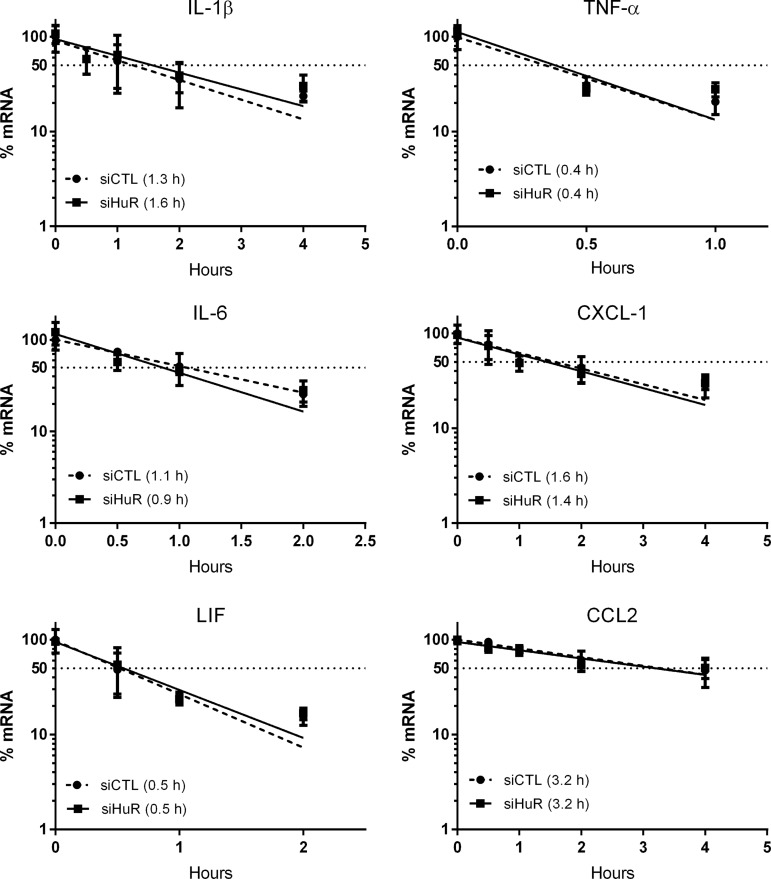
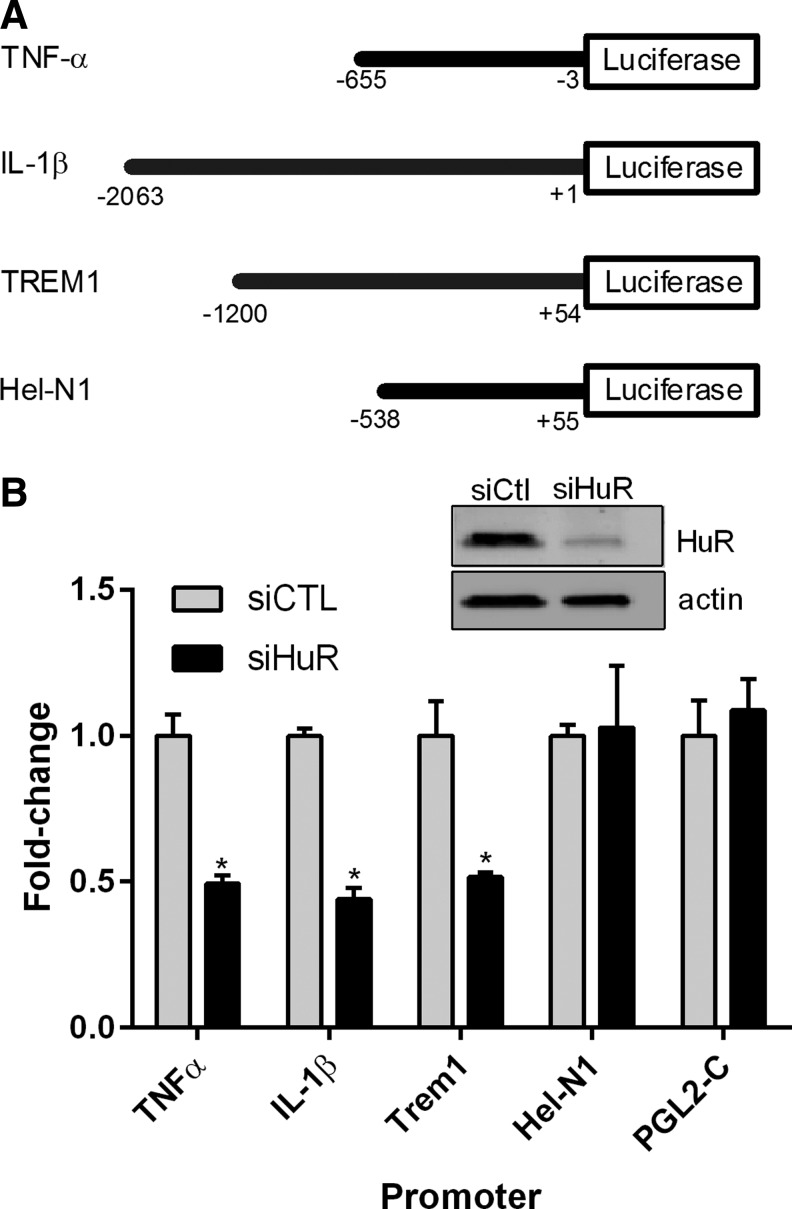
Because of the presence of HuR binding sites in the 3′-UTRs of these cytokine targets (Table 1), we next sought to determine the impact of HuR on transcript stability. We assessed the mRNA half-life of each cytokine using actinomycin D (ActD), a transcriptional inhibitor. After HuR knockdown, stretch-injured astrocytes were pulsed with ActD and mRNA levels at subsequent time intervals were quantified by qRT-PCR. Surprisingly, no change in half-life was observed (Fig. 5). The short half-life of TNF-α mRNA (0.4 h) is in keeping with prior reports and indicates transcriptional inhibition was effective.16,18 The findings suggest that the attenuated cytokine mRNA levels following HuR knockdown may be related to a transcriptional effect. To address this possibility, we assessed the promoter activity of TNF-α and IL-1β using luciferase reporters (Fig. 6).16,29 Hel-N1 and TREM-1 promoters also were assessed.17,30 The latter is a cell surface receptor that amplifies IL-1β and TNF-α production, whereas Hel-N1 is expressed in neurons.31,32 Astrocytes were cotransfected with HuR or control siRNA, the luciferase plasmids, and a transfection control (β-galactosidase). After 72 h, cells were subjected to stretch injury. Luciferase and β-galactosidase activities were then measured and expressed as a fold-change, compared with the siRNA control (Fig. 6). There was a significant reduction of luciferase activity for IL-1β, TNF-α, and TREM-1 promoters to approximately 50% of control, indicating that HuR knockdown led to suppression of promoter activity. The effect was specific as neither the Hel-N1 promoter nor the luciferase vector control (PGL2-C) was affected. In summary, the data indicate that HuR knockdown attenuated cytokine gene induction post-stretch injury, possibly through a transcriptional mechanism.
FIG. 5.
Human antigen R (HuR) knockdown in injured astrocytes does not affect the RNA stability of cytokine targets. Following small interfering RNA (siRNA) transfection and stretch injury, astrocytes were treated with actinomycin D to inhibit transcription. RNA levels were then quantitated at different time-points to generate a degradation curve. All data points are the mean ± standard error of the mean of six independent samples. The estimated half-life is shown in parentheses.
FIG. 6.
Human antigen R (HuR) knockdown attenuates promoter activity of several pro-inflammatory targets. (A) Schematic diagram of the different promoter constructs analyzed. (B) Astrocytes were transfected with small interfering RNA (siRNA), luciferase reporter, and a transfection control (β-galactosidase) followed by stretch injury. At 24 h, cells were harvested and assayed for luciferase activity. All values were adjusted to the transfection control and expressed as a fold-change over the control siRNA. A Western blot shows adequate knockdown of HuR. All data points are the mean ± standard error of the mean of three independent samples. *p < 0.05.
MS-444, a small molecule inhibitor of HuR, attenuates inflammatory factor induction after stretch injury of astrocytes
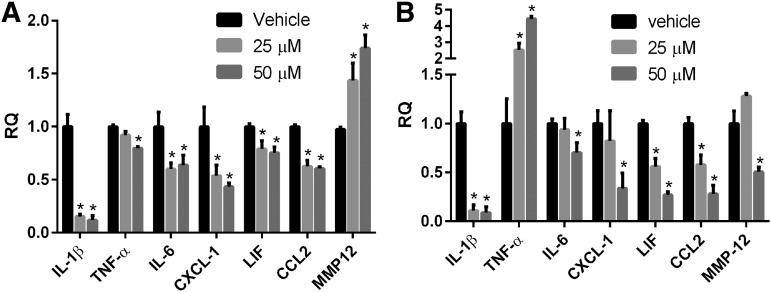
MS-444 is a small molecule that prevents HuR from dimerizing and translocating to the cytoplasm.33 We have shown previously that this inhibitor can block molecular phenotypes linked to HuR including TDP-43 and FUS expression.34 Here, we investigated the effect of MS-444 on cytokine induction in astrocytes following stretch injury. We observed significant and dose–dependent decreases in IL-1β, TNF-α, CXCL-1, and CCL2 mRNA expression, compared with vehicle treated cells 24 h after stretch injury (Fig. 7A). IL-1β showed nearly a 6-fold decline. MMP-12 mRNA, on the other hand, increased significantly. Conditioned media from these astrocytes showed a significant reduction in all cytokine levels, with a remarkable ∼6-fold decrease in IL-1β production (Fig. 7B). Despite a reduction at the mRNA level, TNF-α protein increased with MS-444 treatment in a dose–dependent fashion. This result is in contrast to the HuR knockdown finding (Fig. 4) and may represent an off-target effect of the inhibitor.
FIG. 7.
MS-444, a small molecule inhibitor of human antigen R (HuR), attenuates cytokine messenger RNA expression in stretch-injured astrocytes. (A) Twenty-four hours after injury, RNA levels were measured by quantitative real-time polymerase chain reaction analysis. (B) Enzyme-linked immunosorbent assay analysis of conditioned media from astrocytes shown in (A). All data points are the mean ± standard error of the mean of six independent samples, and values were expressed as a relative quantity (RQ) to the vehicle control. *p < 0.05. IL, interleukin; TNF, tumor necrosis factor; MMP, matrix metalloproteinase.
HuR inhibition impairs astrocyte chemoattraction of other immune cells
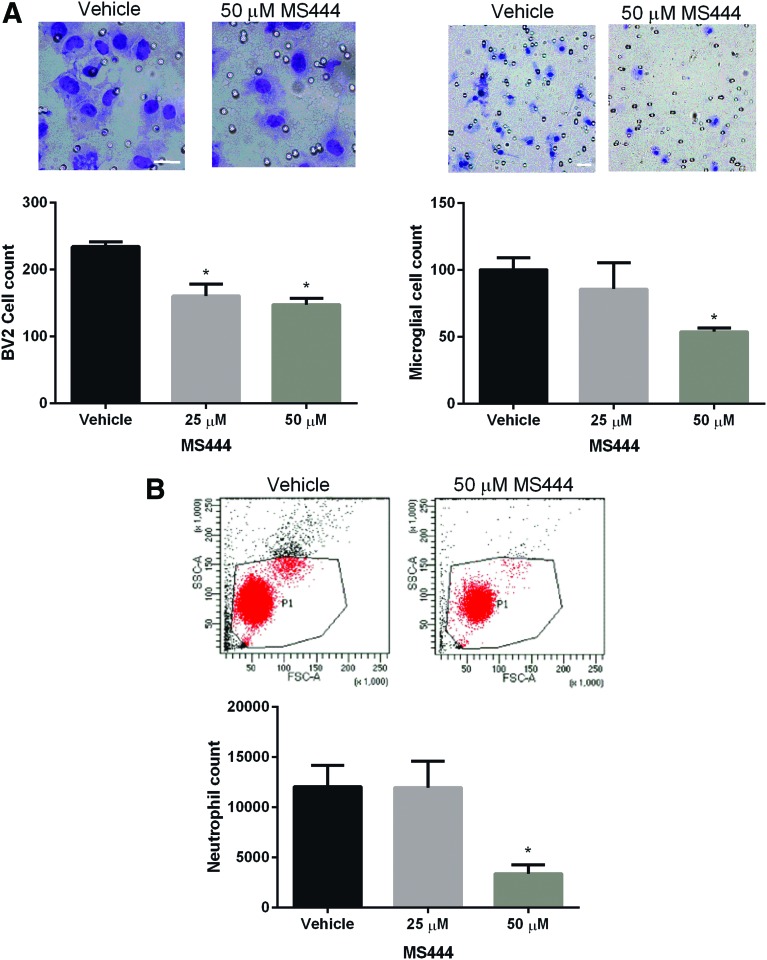
One important role of the inflammatory mediators regulated by HuR is the recruitment of immune cells such as monocytes, microglia and neutrophils. For example, CXCL1 and CCL2 are chemokines linked to recruitment of neutrophils and monocytes/macrophages, respectively.35,36 To assess the impact of HuR inhibition on chemotaxis of other immune cells, we used a transwell migration assay with conditioned media from injured astrocytes (ACM) treated with MS-444 (Fig. 8). Neutrophils, primary microglia (PMG), or microglia-derived BV2 cells were plated into the upper chamber and ACM in the lower chamber. Migrated cells were quantified by FACS (neutrophils) or direct staining (microglial cells). For microglial cells, we saw a significant dose–dependent reduction of chemotaxis to ∼45–55% of control at 50 μM of MS-444. ACM from stretch-injured astrocytes treated with MS-444 showed an approximately 3.5-fold reduction in migrated neutrophils. In summary, HuR inhibition attenuated the chemotaxis of other immune cells by injured astrocytes.
FIG. 8.
Human antigen R (HuR) inhibition in astrocytes inhibits chemoattraction of immune cells. (A) Conditioned media from injured astrocytes treated with MS-444 or vehicle was placed in the bottom well of a transwell plate and BV2 (left panel) or primary microglial cells (right panel) were seeded in the top well. At 24 h, the filters were removed and cells were stained with a Diff-Quik kit and counted under a light microscope. Representative micrographs are shown above. Scale bars: 30 μm. All data points are the mean ± standard error of the mean (SEM) of six independent samples. *p < 0.05. (B) After seeding primary neutrophils in the upper well, migrated cells were fluorescence-activated cell sorted and counted. All data points are the mean ± SEM of three independent samples. *p < 0.05. Color image is available online at www.liebertpub.com/neu
Discussion
Upregulation of inflammatory cytokines in the acute phase of SCI is an important molecular event that drives secondary immune responses and additional tissue injury. In this study, we show that the RNA binding protein HuR, which positively modulates many of these inflammatory mediators, is translocated in astrocytes at an early time-point after trauma to the spinal cord consistent with its activation. Knockdown of HuR in stretch-injured astrocytes led to significant attenuation of cytokine and chemotactic factor production in vitro. The reduction in mRNA expression appeared more related to changes in promoter activity than mRNA half-life. A small molecule inhibitor of HuR, MS-444, suppressed the mRNA production of these factors, and with the exception of TNF-α, protein production as well, suggesting that HuR can be a druggable target. Treatment of astrocytes with MS-444 led to significant attenuation of chemotaxis of other immune cells, including neutrophils and microglia. These findings highlight an important role of HuR and post-transcriptional regulation in the molecular and cellular response of astrocytes in the injured environment, and opens up new avenues of potential therapy for the early phases of CNS trauma.
In a recent report, RNA sequencing analysis of acutely injured spinal cord revealed that the 40 most relevant upregulated genes were linked to inflammation, including the ones shown in Table 1.22 Most of these genes contain AREs in the 3′-UTR that in conjunction with ARE-binding proteins, rapidly modulate their expression in response to changes in the microenvironment. ARE-binding proteins such as HuR, KH-type splicing regulatory protein (KSRP) and tristetraprolin (TTP) represent a structurally diverse group of proteins that overlap in their binding affinity to these RNA targets, but differ functionally in regard to promoting or attenuating RNA stability and translational efficiency.11–13 With few exceptions, HuR is a positive regulator of ARE-bearing mRNAs, whereas TTP and KSRP are RNA destabilizers. Since these RBPs are expressed in most cell types, the ultimate fate of the ARE-bearing mRNA is likely influenced by several factors including modulation of RNA binding affinity (e.g., TTP phosphorylation reduces binding affinity) or intracellular localization of the RBP.11,12,37
The positive regulatory effect of HuR, for example, is linked with cytoplasmic localization, where it associates with polysomes and promotes translation of the bound mRNA target. In our study, there was prominent translocation of HuR to the cytoplasm in astrocytes, specifically at the level of spinal cord injury, consistent with its activation (Fig. 1). Supportive of this positive regulatory role, we observed a significant suppression of pro-inflammatory cytokine (mRNA and protein) production in stretch-injured astrocytes with HuR knockdown (Fig. 3). While the in vitro assay does not fully recapitulate the microenvironment of an injured spinal cord, we observed that stretch-injury alone can significantly increase production of all inflammatory cytokines and induce HuR translocation. This molecular response could be secondary to cellular stress or reactivity of surviving cells to those that underwent cell death. Both mechanisms may be at play in vivo.1,38
Surprisingly, we did not see any changes in RNA half-life of these cytokine mRNAs (Fig. 5), suggesting that the attenuated RNA levels following HuR knockdown were secondary to changes in transcription. HuR, for example, may be positively modulating a transcriptional factor or cofactor necessary for optimal promoter activity. In one report, HuR suppressed levels of the transcriptional factor, c-Myc, through an interaction with the microRNA let-7.39 HuR also regulates TDP-43, an RBP that positively modulates NF-κB activity in astrocytes.34,40 We previously observed an effect on transcription with the RNA destabilizer KSRP. Knockdown of this RBP in astrocytes led to increased RNA levels of IL-1β, TNF-α, and other inflammatory mediators, without an effect on RNA stability.16 Overexpression of KSRP suppressed TNF-α promoter activity, and the furthest upstream NF-κB binding site was implicated in this negative regulation. NF-κB is a major transcriptional activator of the astroglial inflammatory response in CNS trauma. It drives the transcription of many factors regulated by HuR, including Trem1 whose promoter also was suppressed by HuR knockdown (Fig. 6).41–44 We speculate that a component of the NF-κB transcriptional unit may be regulated by HuR, KSRP and/or other ARE binding proteins.
Our findings with MMP-12 suggest an effect on translational efficiency as we observed a disproportionally large decrease in MMP-12 protein production, compared with the other targets (Fig. 4). RNA stability and translational efficiency represent two overlapping but distinct levels of post-transcriptional regulation that are governed by ARE-binding proteins. In other cell systems, HuR can modulate translational efficiency without affecting RNA stability/expression. Some examples include TDP-43, cytochrome C, and XIAP.10,34,45,46 Additional studies, such as polysome profiling, would be necessary to confirm that HuR affects translational efficiency of MMP-12 in astrocytes.
Suppression of inflammatory cytokine production and chemotaxis with the HuR inhibitor MS-444 supports the concept of HuR as a druggable target. IL-1β, a cytokine associated with neuronal toxicity, leukocyte infiltration, increased astrogliosis, and worse functional outcome in CNS trauma, was attenuated nearly 10-fold with MS-444 (Fig. 7).47–50 MMP-12 production was reduced by 50%. This protein is increased by more than 180-fold in spinal cord injury and has been linked to blood–brain barrier disruption, infiltration of immune cells, and worse outcome.51 Likewise, chemotactic factors CXCL-1 and CCL2 were suppressed by 3-fold. Consistent with these molecular effects, the conditioned media produced significantly less chemotaxis of neutrophils and microglial cells (Fig. 8). A major role of activated astrocytes in the vicinity of injured tissue injury is to recruit and activate other immune cells, such as neutrophils, neighboring microglial cells, and monocytes.1,3 This recruitment escalates the production of cytokines and reactive oxygen species, leading to further toxicity. Thus, targeting HuR in the early stages of CNS trauma might disrupt this cascade. Microglial cells are another major producer of cytokines and chemotactic factors, but the role of HuR in those cells has not yet been characterized.
The unexpected increase in TNF-α protein production following MS-444 treatment may reflect an off-target effect. The molecule was first characterized as a myosin light chain kinase inhibitor before its link to HuR inhibition, and thus may have other mechanisms of action.33,52 On the other hand, this finding also underscores the potential complexity and context-dependent nature of post-transcriptional regulation. Knockdown or overexpression of HuR in macrophages, for example, have both been shown to attenuate TNF-α translation, independent of the effect on mRNA levels.53–55 In glioma cells, on the other hand, overexpression of HuR leads to a nearly 4-fold increase in TNF-α production.18 The opposing effects of MS-444 on MMP-12 mRNA and protein provide another example of this complexity and the dissociation that can occur between post-transcriptional effects on RNA and protein expression.
Our findings bring to light the importance of post-transcriptional regulation in CNS trauma and more importantly, identify HuR as a potential therapeutic target in early injury. CNS trauma is a dynamic process, however, where the role of astrocytes and the factors they produce change over the course of injury. LIF for example, which was attenuated by HuR inhibition, promotes myelination by oligodendrocytes.56 Other mRNA targets important for tissue repair and recovery, such as brain-derived neurotrophic factor and glial cell–derived neurotrophic factor, harbor HuR binding sites in their 3′-UTRs and may be modulated by HuR.57,58 Thus, the role of HuR will likely change during the course of CNS trauma, and the therapeutic effect of its inhibition may be limited to the early phases of injury.
Acknowledgments
We wish to thank Terry Lewis, PhD, and the UAB Neuroscience Molecular Detection Core Facility (P30 NS47466) for assistance with immunohistochemistry, and Marion Spell and the UAB Center for AIDS Research flow cytometry core for their assistance with FACS analysis. This work was supported by a pilot grant from the UAB Comprehensive Neuroscience Center.
Author Disclosure Statement
No competing financial interests exist.
References
- 1.Gyoneva S. and Ransohoff R.M. (2015). Inflammatory reaction after traumatic brain injury: therapeutic potential of targeting cell-cell communication by chemokines. Trends Pharmacol. Sci. 36, 471–480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.David S. and Kroner A. (2011). Repertoire of microglial and macrophage responses after spinal cord injury. Nat. Rev. Neurosci. 12, 388–399 [DOI] [PubMed] [Google Scholar]
- 3.Donnelly D.J. and Popovich P.G. (2008). Inflammation and its role in neuroprotection, axonal regeneration and functional recovery after spinal cord injury. Exp. Neurol. 209, 378–388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sofroniew M.V. and Vinters H.V. (2010). Astrocytes: biology and pathology. Acta Neuropathol. 119, 7–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hausmann O.N. (2003). Post-traumatic inflammation following spinal cord injury. Spinal Cord 41, 369–378 [DOI] [PubMed] [Google Scholar]
- 6.Hagg T. and Oudega M. (2006). Degenerative and spontaneous regenerative processes after spinal cord injury. J. Neurotrauma 23, 264–280 [DOI] [PubMed] [Google Scholar]
- 7.Profyris C., Cheema S.S., Zang D., Azari M.F., Boyle K., and Petratos S. (2004). Degenerative and regenerative mechanisms governing spinal cord injury. Neurobiol. Dis. 15, 415–436 [DOI] [PubMed] [Google Scholar]
- 8.Pan J.Z., Ni L., Sodhi A., Aguanno A., Young W., and Hart R.P. (2002). Cytokine activity contributes to induction of inflammatory cytokine mRNAs in spinal cord following contusion. J. Neurosci. Res. 68, 315–322 [DOI] [PubMed] [Google Scholar]
- 9.Anderson P. (2010). Post-transcriptional regulons coordinate the initiation and resolution of inflammation. Nat. Rev. Immunol. 10, 24–35 [DOI] [PubMed] [Google Scholar]
- 10.Srikantan S. and Gorospe M. (2012). HuR function in disease. Front. Biosci. (Landmark Ed) 17, 189–205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Abdelmohsen K., Yuki K., Kim H.H., and Gorospe M. (2008). Posttranscriptional gene regulation by RNA-binding proteins during oxidative stress: implications for cellular senescence. Biol. Chem. 389, 243–255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Barreau C., Paillard L., and Osborne H.B. (2006). AU-rich elements and associated factors: are there unifying principles? Nucleic Acids Res. 33, 7138–7150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brennan C.M. and Steitz J.A. (2001). HuR and mRNA stability. Cell. Mol. Life Sci. 58, 266–277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Qin H., Niyongere S.A., Lee S.J., Baker B.J., and Benveniste E.N. (2008). Expression and Functional Significance of SOCS-1 and SOCS-3 in Astrocytes. J. Immunol. 181, 3167–3176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ellis E.F., McKinney J.S., Willoughby K.A., Liang S., and Povlishock J.T. (1995). A new model for rapid stretch-induced injury of cells in culture: characterization of the model using astrocytes. J. Neurotrauma 12, 325–339 [DOI] [PubMed] [Google Scholar]
- 16.Li X., Lin W.J., Chen C.Y., Si Y., Zhang X., Lu L., Suswam E., Zheng L., and King P.H. (2012). KSRP: a checkpoint for inflammatory cytokine production in astrocytes. Glia 60, 1773–1784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.King P.H. (1996). Cloning the 5' flanking sequence of Hel-N1: evidence for positive regulatory elements governing cell-specific transcription. Brain Res. 723, 141–147 [DOI] [PubMed] [Google Scholar]
- 18.Nabors L.B., Suswam E., Huang Y., Yang X., Johnson M.J., and King P.H. (2003). Tumor necrosis factor-a induces angiogenic factor up-regulation in malignant glioma cells: a role for RNA stabilization and HuR. Cancer Res. 63, 4181–4187 [PubMed] [Google Scholar]
- 19.Giulian D. and Baker T.J. (1986). Characterization of ameboid microglia isolated from developing mammalian brain. J. Neurosci. 6, 2163–2178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fan X.C. and Steitz J.A. (1998). HNS, a nuclear-cytoplasmic shuttling sequence in HuR. Proc. Natl. Acad. Sci. U.S.A. 95, 15293–15298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim H.H. and Gorospe M. (2008). Phosphorylated HuR shuttles in cycles. Cell Cycle 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen K., Deng S., Lu H., Zheng Y., Yang G., Kim D., Cao Q., and Wu J.Q. (2013). RNA-seq characterization of spinal cord injury transcriptome in acute/subacute phases: a resource for understanding the pathology at the systems level. PLoS One 8, e72567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhong J., Jiang L., Cheng C., Huang Z., Zhang H., Liu H., He J., Cao F., Peng J., Jiang Y., and Sun X. (2016). Altered expression of long non-coding RNA and mRNA in mouse cortex after traumatic brain injury. Brain Res. 1646, 589–600 [DOI] [PubMed] [Google Scholar]
- 24.Ray D., Kazan H., Chan E.T., Pena Castillo L., Chaudhry S., Talukder S., Blencowe B.J., Morris Q., and Hughes T.R. (2009). Rapid and systematic analysis of the RNA recognition specificities of RNA-binding proteins. Nat. Biotechnol. 27, 667–670 [DOI] [PubMed] [Google Scholar]
- 25.Lebedeva S., Jens M., Theil K., Schwanhäusser B., Selbach M., Landthaler M., and Rajewsky N. (2011). Transcriptome-wide analysis of regulatory interactions of the RNA-binding protein HuR. Mol. Cell 43, 340–352 [DOI] [PubMed] [Google Scholar]
- 26.McTigue D.M., Tani M., Krivacic K., Chernosky A., Kelner G.S., Maciejewski D., Maki R., Ransohoff R.M., and Stokes B.T. (1998). Selective chemokine mRNA accumulation in the rat spinal cord after contusion injury. J. Neurosci. Res. 53, 368–376 [DOI] [PubMed] [Google Scholar]
- 27.Dong Y. and Benveniste E.N. (2001). Immune function of astrocytes. Glia 36, 180–190 [DOI] [PubMed] [Google Scholar]
- 28.Esposito E., and Cuzzocrea S. Anti-TNF therapy in the injured spinal cord. Trends Pharmacol. Sci. 32, 107–115 [DOI] [PubMed] [Google Scholar]
- 29.Su D., Coudriet G.M., Hyun Kim D., Lu Y., Perdomo G., Qu S., Slusher S., Tse H.M., Piganelli J., Giannoukakis N., Zhang J., and Henry Dong H. (2009). FoxO1 links insulin resistance to proinflammatory cytokine IL-1β production in macrophages. Diabetes 58, 2624–2633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hosoda H., Tamura H., Kida S., and Nagaoka I. (2011). Transcriptional regulation of mouse TREM-1 gene in RAW264.7 macrophage-like cells. Life Sci. 89, 115–122 [DOI] [PubMed] [Google Scholar]
- 31.Bouchon A., Facchetti F., Weigand M.A., and Colonna M. (2001). TREM-1 amplifies inflammation and is a crucial mediator of septic shock. Nature 410, 1103–1107 [DOI] [PubMed] [Google Scholar]
- 32.King P.H., Levine T.D., Fremeau R.T., and Keene J.D. (1994). Mammalian homologs of Drosphila ELAV localized to a neuronal subset can bind in vitro to the 3′UTR of mRNA encoding the Id transcriptional repressor. J. Neurosci. 14, 1943–1952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Meisner N.C., Hintersteiner M., Mueller K., Bauer R., Seifert J.M., Naegeli H.U., Ottl J., Oberer L., Guenat C., Moss S., Harrer N., Woisetschlaeger M., Buehler C., Uhl V., and Auer M. (2007). Identification and mechanistic characterization of low-molecular-weight inhibitors for HuR. Nat. Chem. Biol. 3, 508–515 [DOI] [PubMed] [Google Scholar]
- 34.Lu L., Zheng L., Si Y., Luo W., Dujardin G., Kwan T., Potochick N.R., Thompson S.R., Schneider D.A., and King P.H. (2014). Hu antigen R (HuR) is a positive regulator of the RNA-binding proteins TDP-43 and FUS/TLS: implications for amyotrophic lateral sclerosis. J. Biol. Chem. 289, 31792–31804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fuentes M.E., Durham S.K., Swerdel M.R., Lewin A.C., Barton D.S., Megill J.R., Bravo R., and Lira S.A. (1995). Controlled recruitment of monocytes and macrophages to specific organs through transgenic expression of monocyte chemoattractant protein-1. J. Immunol. 155, 5769–5776 [PubMed] [Google Scholar]
- 36.Moser B., Clark-Lewis I., Zwahlen R., and Baggiolini M. (1990). Neutrophil-activating properties of the melanoma growth-stimulatory activity. J. Exp. Med. 171, 1797–1802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Meisner N.C. and Filipowicz W. (2011). Properties of the regulatory RNA-binding protein HuR and its role in controlling miRNA repression. Adv. Exp. Med. Biol. 700, 106–123 [DOI] [PubMed] [Google Scholar]
- 38.Sofroniew M.V. (2009). Molecular dissection of reactive astrogliosis and glial scar formation. Trends Neurosci. 32, 638–647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kim H.H., Kuwano Y., Srikantan S., Lee E.K., Martindale J.L., and Gorospe M. (2009). HuR recruits let-7/RISC to repress c-Myc expression. Genes Dev. 23, 1743–1748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Swarup V., Phaneuf D., Dupre N., Petri S., Strong M., Kriz J., and Julien J.P. (2011). Deregulation of TDP-43 in amyotrophic lateral sclerosis triggers nuclear factor kappaB-mediated pathogenic pathways. J. Exp. Med. 208, 2429–2447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bethea J.R., Castro M., Keane R.W., Lee T.T., Dietrich W.D., and Yezierski R.P. (1998). Traumatic spinal cord injury induces nuclear factor-kB activation. J. Neurosci. 18, 3251–3260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Engelmann C., Weih F., and Haenold R. (2014). Role of nuclear factor kappa B in central nervous system regeneration. Neural Regen. Res. 9, 707–711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shih R.-H., Wang C.Y., and Yang C.M. (2015). NF-kappaB signaling pathways in neurological inflammation: a mini review. Front. Mol. Neurosci. 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zeng H., Ornatowska M., Joo M.S., and Sadikot R.T. (2007). TREM-1 expression in macrophages is regulated at transcriptional level by NF-kappaB and PU.1. Eur. J. Immunol. 37, 2300–2308 [DOI] [PubMed] [Google Scholar]
- 45.Durie D., Lewis S.M., Liwak U., Kisilewicz M., Gorospe M., and Holcik M. (2011). RNA-binding protein HuR mediates cytoprotection through stimulation of XIAP translation. Oncogene 30, 1460–1469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kawai T., Lal A., Yang X., Galban S., Mazan-Mamczarz K., and Gorospe M. (2006). Translational control of cytochrome c by RNA-binding proteins TIA-1 and HuR. Mol. Cell. Biol. 26, 3295–3307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lin W.P., Lin J.H., Cai B., Shi J.X., Li W.J., Choudhury G.R., Wu S.Q., Wu J.Z., Wu H.P., and Ke Q.F. (2016). Effect of adenovirus-mediated RNA interference of IL-1beta expression on spinal cord injury in rats. Spinal Cord. 54, 778–784 [DOI] [PubMed] [Google Scholar]
- 48.Nesic O., Xu G.Y., McAdoo D., High K.W., Hulsebosch C., and Perez-Pol R. (2001). IL-1 receptor antagonist prevents apoptosis and caspase-3 activation after spinal cord injury. J. Neurotrauma 18, 947–956 [DOI] [PubMed] [Google Scholar]
- 49.Boato F., Rosenberger K., Nelissen S., Geboes L., Peters E.M., Nitsch R., and Hendrix S. (2013). Absence of IL-1beta positively affects neurological outcome, lesion development and axonal plasticity after spinal cord injury. J. Neuroinflammation 10, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Allan S.M., Tyrrell P.J., and Rothwell N.J. (2005). Interleukin-1 and neuronal injury. Nat. Rev. Immunol. 5, 629–640 [DOI] [PubMed] [Google Scholar]
- 51.Wells J.E., Rice T.K., Nuttall R.K., Edwards D.R., Zekki H., Rivest S., and Yong V.W. (2003). An adverse role for matrix metalloproteinase 12 after spinal cord injury in mice. J. Neurosci. 23, 10107–10115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nakanishi S., Chiba S., Yano H., Kawamoto I., and Matsuda Y. (1995). MS-444, a new inhibitor of myosin light chain kinase from Micromonospora sp. KY7123. J. Antibiot. (Tokyo). 48, 948–951 [DOI] [PubMed] [Google Scholar]
- 53.Katsanou V., Papadaki O., Milatos S., Blackshear P.J., Anderson P., Kollias G., and Kontoyiannis D.L. (2005). HuR as a negative posttranscriptional modulator in inflammation. Mol. Cell 19, 777–789 [DOI] [PubMed] [Google Scholar]
- 54.Tiedje C., Ronkina N., Tehrani M., Dhamija S., Laass K., Holtmann H., Kotlyarov A., and Gaestel M. (2012). The p38/MK2-driven exchange between tristetraprolin and HuR regulates AU-rich element-dependent translation. PLoS Genet. 8, e1002977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yiakouvaki A., Dimitriou M., Karakasiliotis I., Eftychi C., Theocharis S., and Kontoyiannis D.L. (2012). Myeloid cell expression of the RNA-binding protein HuR protects mice from pathologic inflammation and colorectal carcinogenesis. J Clin Invest 122, 48–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ishibashi T., Dakin K.A., Stevens B., Lee P.R., Kozlov S.V., Stewart C.L., and Fields R.D. (2006). Astrocytes promote myelination in response to electrical impulses. Neuron 49, 823–832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ibanez C.F. and Andressoo J.O. (2016). Biology of GDNF and its receptors—relevance for disorders of the central nervous system. Neurobiol Dis. 97, 80–89 [DOI] [PubMed] [Google Scholar]
- 58.Weishaupt N., Blesch A., and Fouad K. (2012). BDNF: the career of a multifaceted neurotrophin in spinal cord injury. Exp Neurol 238, 254–264 [DOI] [PubMed] [Google Scholar]