Abstract
Background
Genome wide association studies (GWAS) already have identified tens of susceptible loci for nonsyndromic cleft lip with or without cleft palate (NSCL/P). However, whether these loci associated with nonsyndromic cleft palate only (NSCPO) remains unknown.
Material and Methods
In this study, we replicated 38 SNPs (Single nucleotide polymorphisms) which has the most significant p values in published GWASs, genotyping by using SNPscan among 144 NSCPO trios from Western Han Chinese. We performed the transmission disequilibrium test (TDT) on individual SNPs and gene-gene (GxG) interaction analyses on the family data; Parent-of-Origin effects were assessed by separately considering transmissions from heterozygous fathers versus heterozygous mothers to affected offspring.
Results
Allelic TDT results showed that T allele at rs742071 (PAX7) (p=0.025, ORtransmission=3.00, 95%CI: 1.09-8.25) and G allele at rs2485893 (10kb 3’ of SYT14) were associated with NSCPO (p=0.0036, ORtransmission= 0.60, 95%CI: 0.42-0.85). Genotypic TDT based on 3 pseudo controls further confirmed that rs742071 (p-value=0.03, ORtransmission=3.00, 95%CI: 1.09-8.25) and rs2485893 were associated with NSCPO under additive model (p-value= 0.02, ORtransmission= 0.66, 95%CI: 0.47-0.92). Genotypic TDT for epistatic interactions showed that rs4844913 (37kb 3’ of DIEXF) interacted with rs11119388 (SYT14) (p-value=1.80E-08) and rs6072081 (53kb 3’ of MAFB) interacted with rs6102085 (33kb 3’ of MAFB) (p-value=3.60E-04) for NSCPO, suggesting they may act in the same pathway in the etiology of NSCPO.
Conclusions
In this study, we found that rs742071 and rs2485893 were associated NSCPO from Han Chinese population; also, interactions of rs4844913:rs11119388 and rs6072081:rs6102085 for NSCPO were identified, gene-gene interactions have been proposed as a potential source of the remaining heritability, these findings provided new insights of the previous GWAS.
Key words:GWAS, NSCPO, TDT, parent-of-origin effects, epistatic interactions.
Introduction
Cleft palate (CP) is a common birth defect, which has a lower birth prevalence compared to cleft lip with/without cleft palate (CL/P): 1/2500 live births vs. 1/700; but CP shows less variability in birth prevalence across populations compared to CL/P (1).
In nonsyndromic cleft palate only (NSCPO), affected individuals have no other physical or developmental anomalies. Most studies suggest that about 50% of CP is nonsyndromic (2). Both population studies and family studies suggested that genetic factors played a critical role in the etiology of NSCPO (3,4). Among first degree relatives, the relative risks of recurrence risks were 56 for cleft palate only and 32 for any cleft lip when compared to the general population in Norway (3); several genes have been identified for syndromic forms of CP, few have been identified as influencing risk to NSCPO. The etiology of this complex trait has been widely studied in order to search for the risk factors and to design strategies for prevention.
Genome wide association studies entail the matching of a given human genome sequence with an annotated, high-resolution map of common genetic variation; They are contributing a lot to our understanding of diseases to which there is a genetic predisposition (5). Genome wide association studies (GWAS) already have identified tens of susceptible loci for cleft lip with or without cleft palate (CL/P).
However, whether these loci associated with nonsyndromic cleft palate only for Han Chinese remains unknown. In this study, we replicated 38 SNPs from 19 genes/ regions of 11 chromosomes from previous GWAS (6-10) and other studies (11-13) with prior compelling evidence contributing to NSCL/P to investigate their roles in Han Chinese population.
Material and Methods
- Samples
Our samples consisted of 144 complete case-parent trios with nonsydromic cleft palate only (NSCPO), 59 males, 82 females and 3 unknown gender of the probands. All subjects were self-identified as Western Han Chinese, they were recruited between 2008 and 2013 from the Cleft Surgery Department of West China Hospital of Stomatology, Sichuan University. Written informed consent was obtained from parents on behalf of the children and all affected individuals old enough to give their own consent in this study. The consent procedure and this study approved by the Hospital Ethics Committee (HEC) of West China Hospital of Stomatology, Sichuan University.
- Genotyping
Venous blood samples were drawn from participants and DNA was extracted by phenol chloroform extraction protocol. The SNP genotyping work was performed using a custom-by-design 2x48-Plex SNPscanTM Kit (Cat#:G0104, Genesky Biotechnologies Inc., Shanghai, China).
- Statistical analysis
The unaffected parents were underwent Hardy-Weinberg Equilibrium (HWE) analysis and minor allele frequency (MAF) determination. HWE, MAF, allelic TDT and parent-of-origin effects, were calculated using PLINK (14). Pairwise LD as both their D’ and r2 were computed for all the SNPs using the haploview program (http://www.broad.mit.edu/haploview/haploview). Genotypic TDT and Likelihood ratio test for epistatic interactions based on genotypic TDTs were determined with R Package trio (v1.4.23) (15), all two-way interactions comprised a matrix in genotype format were tested using the function colGxG, without specifying the genes, and the interactions between all 38 (38-1)/2 pairs of the SNPs in a matrix were tested. We used a Bonferonni correction for 38 tests to determine a threshold for formal significance of p=0.0013.
Results
- Transmission Disequilibrium Test
All SNPs were passed the HWE test (p >0.05) ( Table 1). Allelic TDT results showed that T allele at rs742071 (PAX7) (p=0.025, ORtransmission=3.0, 95%CI: 1.09-8.25) and G allele at rs2485893 (10kb 3’ of SYT14) were associated with NSCPO (p= 0.0036, ORtransmission= 0.60, 95%CI: 0.42-0.85) ( Table 2). Genotypic TDT based on 3 pseudo controls further confirmed that rs742071 (p-value=0.03, ORtransmission=3.0, 95%CI: 1.09-8.25) and rs2485893 are associated with NSCPO under additive model (p-value=0.02, ORtransmission=0.66, 95%CI: 0.47-0.92) ( Table 3).
Table 1. Minor allele frequency and Hardy-Weinberg Equilibrium Test of the SNPs for NSCPO.
Table 2. Allelic TDT results of the SNPs for NSCPO.
Table 3. Genotypic TDT Based on 3 Pseudo Controls for NSCPO under different genetic models.
- Parent-of-Origin Effects
There was no significant difference of minor allele transmissions between the maternal and paternal for all SNPs (data not shown). However, we found an excess of maternal transmission of the allele G at rs2485893 (p=0.026), allele A at rs8001641 (p=0.018) and allele G at rs17563 (p=0.045) compared with the paternal ( Table 4), which might warrant future investigations.
Table 4. Parent-of-origin effect of effect of the SNPs.
- Gene by Gene Interactions
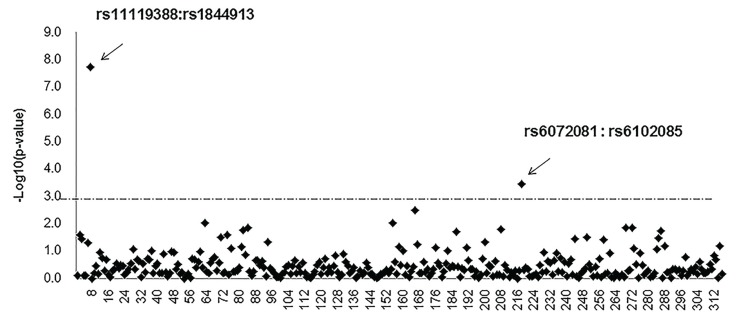
The Genotypic TDT for epistatic interactions showed that rs4844913 (43kb 3’ of DIEXF) interacted with rs11119388 (SYT14) (p=1.80E-08) and rs6072081 (53kb 3’ of MAFB) interacted with rs6102085 (33kb 3’ of MAFB) (p= 3.60E-04) for NSCPO (Fig. 1).
Figure 1.
Genotypic TDT for Epistatic Interactions of Pair-wise SNPs.
- Pair-wise Linkage Disequilibrium and Haplotype Analysis
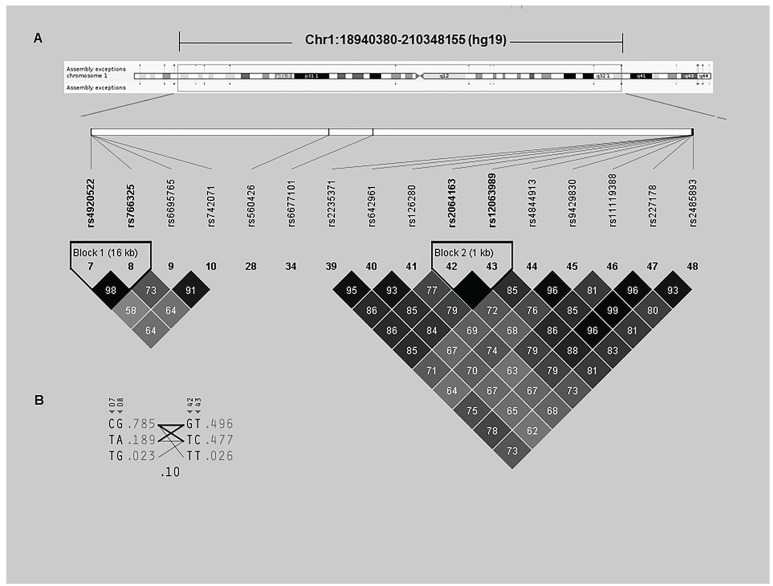
We calculated the pair-wise linkage disequilibrium (LD) of SNPs on chromosome 1 based the association results above. There were strong LD between six pairs of SNPs (rs4920552-rs766325, rs126280-rs642961, rs2064163-12063989, rs4844913-rs9429830, rs4844913-rs227178 and rs9429830-227178) with D’>0.93 and r2>0.80, which distributed on two haplotypes (Fig. 2). Based on this, we tried to test if these pairs of SNPs segregate together among NSCPO by carrying out the haplotype analysis. The results did not show any significance (data not shown).
Figure 2.
Pair-wise Linkage Disequilibrium (A) and Haplotype (B) of SNPs at Chromosome 1.
Discussion
Rs742071 is located in the intron of PAX7 which is involved in neural crest induction and is expressed in cranial neural crest cells, and mice lacking Pax7 have malformations of the nasal and maxillary structures (16). The PAX7 was a second tier GWAS hit (8), later it was confirmed by replication among European and Southeast Asian (17) and GWAS meta-analysis (10). Recently, Leslie et al. 2015 performed targeted sequencing of 13 regions from GWASs and other studies in 1,409 Asian and European trios, and carried out a series of statistical and functional analyses, the results indicated that de novo mutation p.Ala259Val disrupted PAX7 function and might contribute to CLP pathogenesis in this individual (18). Although intronic SNPs do not typically alter protein structure, associations with intronic variants have been reported for a number of complex diseases. In this study, we found rs742071 (PAX7) was associated with NSCPO and had a larger genetic effects compared with the associations with NSCL/P from previous GWASs (8,10); Motif analyses by HaploReg indicated that the T allele of rs742071 could greatly alter the affinity of Sin3Ak-20_disc4 (score: 2.1-14.1).
Rs742071 had the minor allele frequency as 0.04 among Han Chinese population, indicating it may need larger sample size to validate its significance; and with the limited sample size, rs742071 did not pass the threshold of the Bonferonni correction p value in the current study. We will add more samples to study it and other variants at PAX7 gene among NSCPO among Han Chinese population.
Rs2485893 was associated with CL/P among Asian ancestry with p value 7.86E-07 by GWAS (8). In this study, we found rs2485893 (10kb 3’ of SYT14) was also found to be associated with NSCPO. Marked parent of origin effects were seen for rs2485893 alleles, over-transmission was seen preferentially from mothers compared with fathers ( Table 4). Motif analyses by HaploReg indicated that the G allele of rs2485893 could greatly alter the affinity of AFP1 (score: 2.7-12.3).
Numerous studies have shown that highly conserved non-coding elements act as developmental enhancers in vivo (19-21). Non-coding conserved elements around rs742071 and rs2485893 therefore might represent putative regulatory elements for PAX7 and SYT14, we will perform functional studies to elucidate their roles in human craniofacial development.
Gene-gene interactions have been proposed as a potential source of the remaining heritability. Genotypic TDT for epistatic interactions showed that rs4844913 interacts with rs11119388 (SYT14) and rs6072081 interacts with rs6102085 for NSCPO, which provided new insights for the previous GWASs.
In summary, we replicated 38 SNPs contributing to NSCL/P to investigate their roles in NSCPO among Han Chinese population. In this study, we found that rs742071 and rs2485893 were associated NSCPO from Han Chinese population; also, interactions of rs4844913:rs11119388 and rs6072081:rs6102085 for NSCPO were identified, which may provide new insights for the previous GWASs.
Acknowledgments
We are particularly thankful to the patients and their families who participated in this study. This project was supported by the National Natural Science Foundation of China (No. 81600849 and 30530730) and Scientific Research Funds for Young Teachers of Sichuan University (No. 2015SCU11999). Shi-Jun Duan, Ning Huang, Zhong-Lin Jia, these authors contributed equally to this work.
References
- 1.Beaty TH, Ruczinski I, Murray JC, Marazita ML, Munger RG, Hetmanski JB. Evidence for gene-environment interaction in a genome wide study of nonsyndromic cleft palate. Genet Epidemiol. 2011;35:469–78. doi: 10.1002/gepi.20595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shi M, Wehby GL, Murray JC. Review on genetic variants and maternal smoking in the etiology of oral clefts and other birth defects. Birth Defects Res C Embryo Today. 2008;84:16–29. doi: 10.1002/bdrc.20117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sivertsen A, Wilcox AJ, Skjaerven R, Vindenes HA, Abyholm F, Harvilles E. Familial risk of oral clefts by morphological type and severity: population based cohort study of first degree relatives. BMJ. 2008;336:432–4. doi: 10.1136/bmj.39458.563611.AE. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Grosen D, Chevrier C, Skytthe A, Bille C, Mølsted K, Sivertsen A. A cohort study of recurrence patterns among more than 54,000 relatives of oral cleft cases in Denmark: support for the multifactorial threshold model of inheritance. J Med Genet. 2010;47:162–8. doi: 10.1136/jmg.2009.069385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Christensen K, Murray JC. What genome-wide association studies can do for medicine. N Engl J Med. 2007;356:1094–7. doi: 10.1056/NEJMp068126. [DOI] [PubMed] [Google Scholar]
- 6.Birnbaum S, Ludwig KU, Reutter H, Herms S, Steffens M, Rubini M. Key susceptibility locus for nonsyndromic cleft lip with or without cleft palate on chromosome 8q24. Nat Genet. 2009;41:473–7. doi: 10.1038/ng.333. [DOI] [PubMed] [Google Scholar]
- 7.Grant SF, Wang K, Zhang H, Glaberson W, Annaiah K, Kim CE. A genome-wide association study identifies a locus for nonsyndromic cleft lip with or without cleft palate on 8q24. J Pediatr. 2009;155:909–13. doi: 10.1016/j.jpeds.2009.06.020. [DOI] [PubMed] [Google Scholar]
- 8.Beaty TH, Murray JC, Marazita ML, Munger RG, Ruczinski I, Hetmanski JB. A genome-wide association study of cleft lip with and without cleft palate identifies risk variants near MAFB and ABCA4. Nat Genet. 2010;42:525–9. doi: 10.1038/ng.580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mangold E, Ludwig KU, Birnbaum S, Baluardo C, Ferrian M, Herms S. Genome-wide association study identifies two susceptibility loci for nonsyndromic cleft lip with or without cleft palate. Nat Genet. 2010;42:24–6. doi: 10.1038/ng.506. [DOI] [PubMed] [Google Scholar]
- 10.Ludwig KU, Mangold E, Herms S, Nowak S, Reutter H, Paul A. Genome-wide meta-analyses of nonsyndromic cleft lip with or without cleft palate identify six new risk loci. Nat Genet. 2012;44:968–71. doi: 10.1038/ng.2360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kondo S, Schutte BC, Richardson RJ, Bjork BC, Knight AS, Watanabe Y. Mutations in IRF6 cause Van der Woude and popliteal pterygium syndromes. Nat Genet. 2002;32:285–9. doi: 10.1038/ng985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rahimov F, Marazita ML, Visel A, Cooper ME, Hitchler MJ, Rubini M. Disruption of an AP-2alpha binding site in an IRF6 enhancer is associated with cleft lip. Nat Genet. 2008;40:1341–7. doi: 10.1038/ng.242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Suzuki S, Marazita ML, Cooper ME, Miwa N, Hing A, Jugessur A. Mutations in BMP4 are associated with subepithelial, microform, and overt cleft lip. Am J Hum Genet. 2009;84:406–11. doi: 10.1016/j.ajhg.2009.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–75. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schwender H, Taub MA, Beaty TH, Marazita ML, Ruczinski I. Rapid testing of SNPs and gene-environment interactions in case-parent trio data based on exact analytic parameter estimation. Biometrics. 2012;68:766–73. doi: 10.1111/j.1541-0420.2011.01713.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mansouri A, Stoykova A, Torres M, Gruss P. Dysgenesis of cephalic neural crest derivatives in Pax7-/- mutant mice. Development. 1996;122:831–8. doi: 10.1242/dev.122.3.831. [DOI] [PubMed] [Google Scholar]
- 17.Beaty TH, Taub MA, Scott AF, Murray JC, Marazita ML, Schwender H. Confirming genes influencing risk to cleft lip with/without cleft palate in a case-parent trio study. Hum Genet. 2013;132:771–81. doi: 10.1007/s00439-013-1283-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Leslie EJ, Taub MA, Liu H, Steinberg KM, Koboldt DC, Zhang Q. Identification of functional variants for cleft lip with or without cleft palate in or near PAX7, FGFR2, and NOG by targeted sequencing of GWAS loci. Am J Hum Genet. 2015;96:397–411. doi: 10.1016/j.ajhg.2015.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pennacchio LA, Ahituv N, Moses AM, Prabhakar S, Nobrega MA, Shoukry M. In vivo enhancer analysis of human conserved non-coding sequences. Nature. 2006;444:499–502. doi: 10.1038/nature05295. [DOI] [PubMed] [Google Scholar]
- 20.Visel A, Minovitsky S, Dubchak I, Pennacchio LA. VISTA Enhancer Browser--a database of tissue-specific human enhancers. Nucleic acids research. 2007;35:D88–92. doi: 10.1093/nar/gkl822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Visel A, Rubin EM, Pennacchio LA. Genomic views of distant-acting enhancers. Nature. 2009;461:199–205. doi: 10.1038/nature08451. [DOI] [PMC free article] [PubMed] [Google Scholar]