Abstract
The C. elegans nervous system mediates protective physiological and behavioral responses amid infection. However, it remains largely unknown how the nervous system responds to reactive oxygen species (ROS) activated by pathogenic microbes during infection. Here, we show superoxide dismutase-1 (SOD-1), an enzyme that converts superoxide into less toxic hydrogen peroxide and oxygen, functions in the gustatory neuron ASER to mediate C. elegans pathogen avoidance response. When C. elegans first encounters pathogenic bacteria P. aeruginosa, SOD-1 is induced in the ASER neuron. After prolonged P. aeruginosa exposure, ASER-specific SOD-1 expression is diminished. In turn, C. elegans starts to vacate the pathogenic bacteria lawn. Genetic knockdown experiments reveal that pathogen-induced ROS activate sod-1 dependent behavioral response non cell-autonomously. We postulate that the delayed aversive response to detrimental microbes may provide survival benefits by allowing C. elegans to temporarily utilize food that is tainted with pathogens as an additional energy source. Our data offer a mechanistic insight into how the nervous system mediates food-seeking behavior amid oxidative stress and suggest that the internal state of redox homeostasis could underlie the behavioral response to harmful microbial species.
Food is often tainted with pathogens1,2. Ingestion of pathogenic microbes triggers reactive oxygen species (ROS) production in the intestine3,4,5,6. Although ROS can serve as signaling molecules to activate protective signaling pathways6,7,8,9,10,11, elevation of ROS (e.g., •O2−, •OH) causes oxidative stress. The targets of free-radical mediated damage are proteins, lipids and DNA10,11. Therefore, the failure to maintain cellular and systemic ROS concentration results in physiological decline, as oxidized proteins threaten cellular physiology.
Maintaining physiological homeostasis is essential to survival. In C. elegans, the nervous system plays a major role in modulating stress response. For example, the thermal sensory neuron AFD senses heat stress and activates heat shock response at distal tissue via serotonin signaling12. The octopamine receptor OCTR-1 acts in the ASH and ASI sensory neurons to downregulate ER unfolded protein response. In turn, it further suppresses the innate immune signaling activation in the intestine13. TRX-1, a thioredoxin, promotes nuclear localization of intestinal SKN-1 in a cell non-autonomous fashion from distal ASJ A neurons14. These mechanisms demonstrate the importance of the nervous system in detecting adverse environmental cues and maintaining physiological homeostasis.
Microbes in the digestive tract also modulate host metabolic activity and physiology15. For example, mammalian gut indigenous bacteria increase the secretion of peripheral serotonin and subsequently enhance gastrointestinal movement16. Although C. elegans can grow axenically on defined culturing ingredients without bacteria as its food source, recent studies show that bacterial diet plays an important role in aging and intestine health17,18,19,20. Microbiota (the community of microbial species) in the intestine can be commensal or pathogenic21. Prior to entering the intestine of C. elegans, bacteria are normally broken down in the pharynx. However, several human opportunistic pathogens including Pseudomonas aeruginosa, Serratia marcescens and Salmonella typhimurium have been found to colonize the C. elegans intestine17,22,23,24. Therefore, the C. elegans host and pathogen model offers a unique tool to study pathogenic microbiota and their role in affecting host physiology25,26,27.
When C. elegans is transferred to a petri dish that contains P. aeruginosa PA14, C. elegans will initially remain inside the bacterial lawn and feed on P. aeruginosa. Ingestion allows the accumulation of P. aeruginosa in the intestine17, which in turn triggers ROS production4,5. After prolonged exposure to P. aeruginosa, C. elegans will vacate the P. aeruginosa lawn24,28. Several sensory modalities including olfactory, chemosensory, and mechanosensory have been shown to mediate C. elegans behavioral response to pathogens28,29,30. In addition, it has been demonstrated that neuropeptide receptor NPR-1, which encodes C. elegans homolog of mammalian neuropeptide Y receptor, is one of the behavioral determinants of pathogen susceptibility24,28,31. However, how ROS mediate C. elegans physiological and behavioral responses to pathogens remains elusive.
Superoxide dismutase-1 (SOD-1) is an antioxidant enzyme. SOD-1 catalyzes the dismutation of superoxide into less toxic hydrogen peroxide and oxygen. Mutations in sod-1 are linked to a familial form of amyotrophic lateral sclerosis (ALS), a progressive neurodegenerative disease that affects nerve cells32,33,34. Although the pathological mechanism of ALS is still under debate, one of the prevailing hypotheses suggests that the degeneration of nerve cells is caused by chronic oxidative stress in the nervous system35,36,37.
A recent study in budding yeast highlights a dual role of SOD-1 in maintaining respiratory homeostasis and glucose metabolism38. In growth media with high glucose concentration, SOD-1 suppresses the degradation of casein kinase 1, a signaling molecule that represses respiratory metabolism. In C. elegans, SOD-1 is elevated by environmental ROS challenge. Lack of SOD-1 activity causes life span reduction39. Here, we postulate that SOD-1 may act as a sensor of redox homeostasis and mediate behavioral response to food. As a result, SOD-1 may alleviate oxidative stress triggered by foodborne agents such as pathogenic microbes.
In this study, we examined how SOD-1 mediates pathogen avoidance behavior in C. elegans. When C. elegans first encounters P. aeruginosa, SOD-1 is elevated in the gustatory neuron ASER. The elevation of SOD-1 eventually returns to baseline level after prolonged exposure to P. aeruginosa. In turn, C. elegans starts to avoid P. aeruginosa. We also found that SOD-1 is localized at the sensory cilium of ASER. By introducing a mutation that disrupts the structure of sensory cilium and by performing RNA interference to a gene that enhances ROS production, we were able to block SOD-1 induction in the ASER neuron. The lack of SOD-1 and the ablation of ASER neuron exacerbate P. aeruginosa aversion. This suggests that SOD-1 functions in the ASER neuron to inhibit the aversive response to P. aeruginosa. Together, our findings demonstrate that SOD-1 modulates pathogen avoidance behavior by integrating the external sensory stimuli and the internal state of redox reaction. In the arms race between host and pathogen, the delayed aversive response to food that contains small quantities of pathogens may allow the host to activate protective responses and prepare for possible infection.
Results
sod-1 mutant animals elicit a heightened P. aeruginosa lawn avoidance phenotype
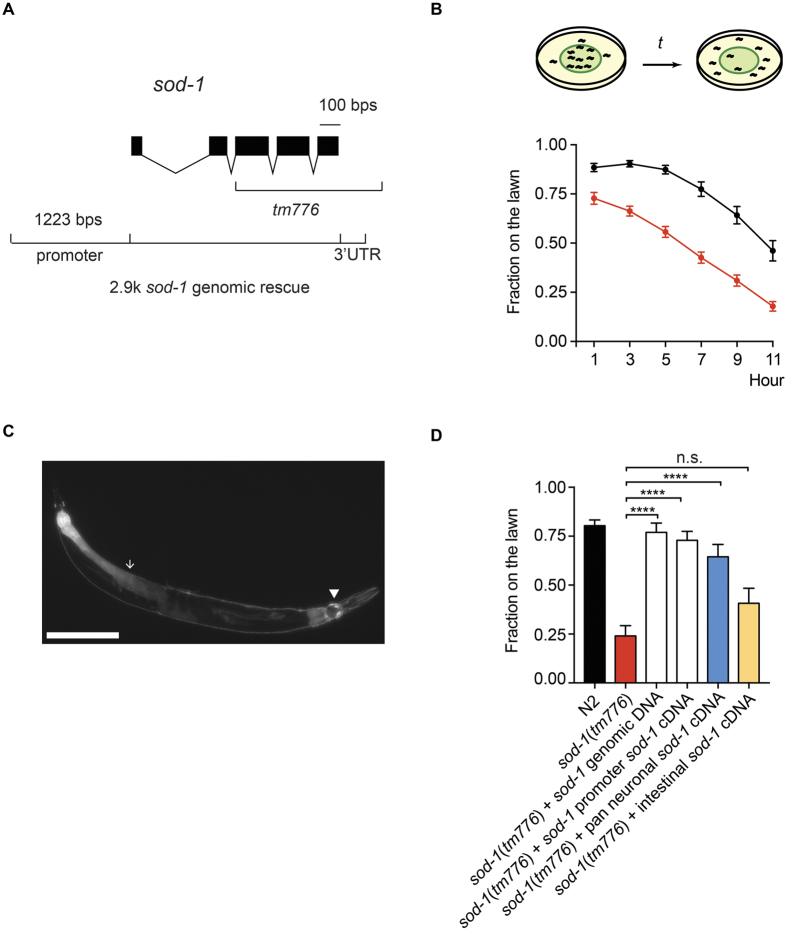
To explore if SOD-1 mediates behavioral response to pathogens, we first exposed sod-1(tm776) mutant to a lawn of P. aeruginosa and observed its behavior over time (Fig. 1A and B). Based on the sequencing results, sod-1(tm776) contains a lesion that deletes the majority of the sod-1 coding region and the 3′UTR (Fig. 1A). Therefore, we hypothesized that sod-1(tm776) may act as a null mutation. We found that sod-1(tm776) mutant animals leave the P. aeruginosa lawn earlier than wild type N2 control. For example, after 7 hour exposure to P. aeruginosa, only 42% of the sod-1(tm776) animals remained inside of P. aeruginosa lawn compared to 77% lawn occupancy of the wild type animals (Fig. 1B). We then generated sod-1 rescue constructs by introducing a 2.9 kb sod-1 genomic fragment and by expressing sod-1 cDNA under control of the sod-1 promoter (Fig. 1A). We found that both sod-1 genomic and sod-1 cDNA constructs rescued the sod-1(tm776) pathogen avoidance phenotype to the wild type level. At 7 hour, 76% of sod-1 genomic rescue and 72% of sod-1 cDNA rescue animals remained on the P. aeruginosa lawn (Fig. 1D). Thus, we concluded that SOD-1 represses the avoidance response to P. aeruginosa.
Figure 1. SOD-1 modulates P. aeruginosa avoidance behavior.
(A) The 2.9 kb region of sod-1 genomic fragment. The deleted region of the sod-1(tm776) mutant is indicated. (B) Top: schematic of P. aeruginosa avoidance assay. C. elegans was transferred to plates containing a lawn of P. aeruginosa. Lawn occupancy was scored over time. Bottom: time course of the P. aeruginosa lawn occupancy. Black indicates wild-type N2 strain; red indicates sod-1(tm776) mutant strain. N = 20. Error bars represent standard error of the mean. (C) Fluorescence micrograph of bosEx1[sod-1p::GFP]. GFP fluorescence is found in the nervous system (triangle) and in the intestine (arrow). Scale bar indicates 50 μm. (D) P. aeruginosa lawn occupancy of C. elegans strains assayed at t = 7 h. sod-1 genomic rescue contains the 2.9 kb sod-1 genomic fragment indicated in (A). sod-1 cDNA rescue contains sod-1 cDNA driven by the sod-1 promoter. Pan-neuronal and intestinal rescue constructs are sod-1 cDNA under control of the unc-119 promoter and the ges-1 promoter. ****Represents p < 0.0001, n.s. not significant, as determined by one-way ANOVA, followed by Bonferroni’s multiple comparison test. N = 12. Error bars represent standard error of the mean.
SOD-1 functions in the nervous system to modulate the behavioral response to pathogen
We investigated the expression pattern of SOD-1 by generating a strain that carries the sod-1p::GFP transgene. We observed GFP signals in the intestine. We also observed GFP signals in the head and tail neurons. GFP fluorescence signals were also located along the ventral and dorsal nerve cords (Fig. 1C). Therefore, SOD-1 is present in the nervous system and in the intestine.
To investigate the tissue-specific requirement of sod-1 in C. elegans, we performed tissue-specific sod-1 rescue experiments using pan-neuronal (unc-119) and intestinal (ges-1) promoters40,41. We found that pan-neuronal expression of sod-1 strongly rescues the pathogen avoidance phenotype of sod-1(tm776) mutant. By contrast, intestinal expression of sod-1 did not rescue the sod-1(tm776) phenotype (Fig. 1D). These results demonstrate that SOD-1 functions in the nervous system to regulate the behavioral response to P. aeruginosa.
SOD-1 functions in the gustatory neuron ASER to delay the avoidance response to P. aeruginosa
Sensory neurons play a major role in modulating C. elegans behavioral response to P. aeruginosa28,29,30. We noticed that some of the head neurons where SOD-1 is expressed elicit the morphology that resembles amphid sensory neurons (Fig. 1C). To identify these neurons, we generated a sod-1p::RFP reporter strain and used amphid sensory neuron GFP reporters to perform colocalization experiments. Consistent with results from a previous investigation39, we confirmed that SOD-1 is present in the ADL neuron pair (Fig. 2A) and is occasionally present in the ASI and ASK neuron pairs (data not shown). In addition, we discovered consistent RFP signals in an additional amphid sensory neuron. This additional sensory neuron is localized more ventrally and posteriorly to the ASK-ADL-ASI cluster. By performing a colocalization experiment using flp-6p::GFP (an ASE neuron pair marker), we confirmed the additional sensory neuron is the gustatory neuron ASER (Fig. 2C).
Figure 2. SOD-1 functions in the ASER neuron to delay P. aeruginosa avoidance response.
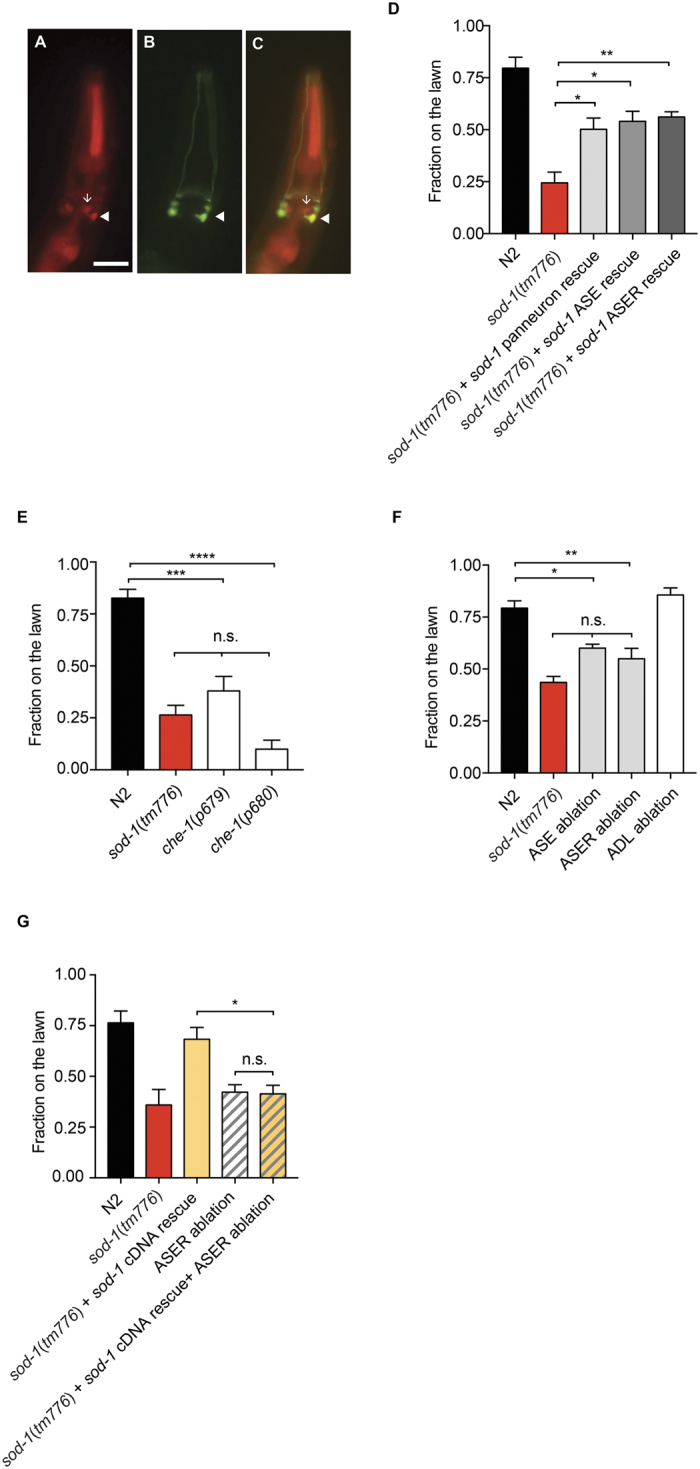
(A–C) Colocalization of sod-1p::RFP and flp-6p::GFP reporters. (A) Fluorescence micrograph of sod-1p::RFP. (B) Fluorescence micrograph of the ASE neuron pair marker flp-6p::GFP. (C) Merge of (A) and (B). Triangle represents ASER. Arrow represents ADLR. Scale bar indicates 10 μm. (D–G) P. aeruginosa lawn occupancy of C. elegans strains assayed at t = 7 h. N = 9–18. Error bars represent standard error of the mean. (D) sod-1 ASE rescue construct is flp-6p::sod-1 cDNA. sod-1 ASER rescue construct is gcy-5p::sod-1 cDNA. **Represents p < 0.01, *represents p < 0.05, as determined by one-way ANOVA, followed by Tukey’s multiple comparison test. (E) che-1 mutants elicit heightened P. aeruginosa avoidance. ****Represents p < 0.0001, ***represents p < 0.001, n.s. not significant, as determined by one-way ANOVA, followed by Tukey’s multiple comparison test. (F) ASE, ASER and ADL ablation are transgenic animals that carry an extra-chromosomal array of flp-6p::csp-1b, gcy-5p::csp-1b or sre-1p::csp-1b, respectively. **Represents p < 0.01, *represents p < 0.05, n.s. not significant, as determined by one-way ANOVA, followed by Tukey’s multiple comparison test. (G) ASER ablation abolishes the delayed P. aeruginosa avoidance phenotype contributed by SOD-1 cDNA rescue in sod-1(tm776) background. ASER ablation contains integrated gcy-5p::csp-1b. sod-1 cDNA rescue contains integrated bosIs2 [sod-1p::sod-1 cDNA::GFP]. *Represents p < 0.05, n.s. represents not significant, as determined by one-way ANOVA, followed by Tukey’s multiple comparison test.
The ASE neuron pair is a pair of bilaterally positioned gustatory neurons that detect water-soluble chemicals42. ASE neuron pair contains ciliated dendritic endings, which are exposed to the environment via an opening of the cuticle at the anterior tip, called buccal cavity43. The left/right asymmetrical expression of guanylate cyclases in ASEL (left) and ASER (right) neurons allows the ASEL and ASER neurons to discern different tastes44,45. To determine if SOD-1 is required in the ASE neurons to regulate C. elegans pathogen avoidance behavior, we expressed sod-1 cDNA in the ASE neuron pair and in the ASER neuron. Expressing SOD-1 in the ASER neuron rescued the sod-1(tm776) pathogen avoidance phenotype. ASER-specific sod-1 rescue elicited a similar lawn occupancy phenotype compared to the pan-neuronal rescue of sod-1 (Fig. 2D). Our results confirm that SOD-1 functions in the ASER neuron to modulate behavioral response to P. aeruginosa.
In order to determine if the ASE neuron pair plays a role in behavioral response to P. aeruginosa, we acquired che-1 mutants and examined their pathogen avoidance phenotype. CHE-1 encodes a C2H2-type zinc-finger transcriptional factor and is required for ASE cell fate determination. che-1 mutant animals elicit no morphological defects in chemosensory neurons. Instead, the ASE neuron pair fails to express ASEL- and ASER-specific guanylate cyclases in the che-1 mutant background43,45. We observed an enhanced pathogen avoidance phenotype in both che-1(p679) and che-1(p680) loss-of-function mutants (Fig. 2E). These results suggest that the ASE neuron pair inhibits behavioral avoidance to P. aeruginosa.
To further test if the ASER neuron itself modulates C. elegans pathogen avoidance behavior, we performed ablation experiments using CSP-1b caspase to induce apoptosis in the neuron of our interest28,46. We generated transgenic animals that carry csp-1b under control of the flp-6 (ASE pair), gcy-5 (ASER), and sre-1 (ADL pair) promoters. We observed an enhanced pathogen avoidance phenotype in animals without the ASER neuron. However, animals without the ADL neuron pair did not elicit enhanced pathogen avoidance behavior (Fig. 2F). We also obtained a transgenic strain that ablates the ASEL neuron45. The ablation of ASEL neuron did not enhance the pathogen avoidance phenotype (Supplementary Figure 1). Together, these data suggest that the ASER neuron represses behavioral response to P. aeruginosa.
Finally, we introduced integrated sod-1 rescue and ASER ablation transgenes into the sod-1(tm776) background and observed its P. aeruginosa lawn occupancy phenotype. By ablating the ASER neuron, we abolished the delayed pathogen avoidance phenotype contributed by the sod-1 rescue in sod-1(tm776) background (Fig. 2G). We concluded that SOD-1 functions in the ASER neuron to inhibit the aversive response to P. aeruginosa.
SOD-1 diminishes oxidative stress triggered by P. aeruginosa
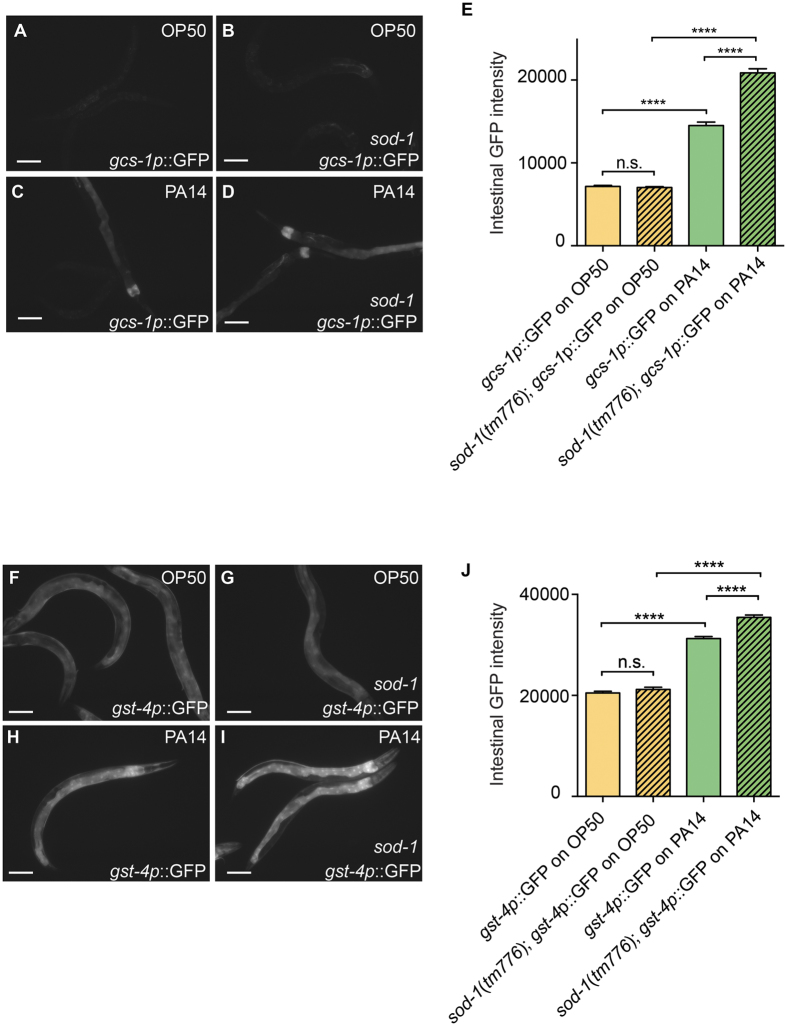
Ingestion of P. aeruginosa triggers ROS production and activates gcs-1p::GFP and gst-4p::GFP reporters in the intestine4,47. gcs-1 encodes γ-glutamine cysteine synthetase heavy chain and gst-4 encodes glutathione S-transferase. Both GCS-1 and GST-4 are antioxidant enzymes that alleviate oxidative stress48,49. To examine if SOD-1 alleviates pathogen-induced oxidative stress, we exposed gcs-1p::GFP, gst-4p::GFP with or without the sod-1(tm776) deletion to a lawn of E. coli OP50 or to a lawn of P. aeruginosa PA14. When animals were fed on E. coli, gcs-1 and gst-4 fluorescence signals were not elevated. In contrast, we observed increased gcs-1 and gst-4 fluorescence signals in the intestine at 2 hour when animals were fed on P. aeruginosa. We found that sod-1 mutation further enhances the fluorescence intensity of gcs-1 and gst-4 reporters when exposed to P. aeruginosa (Fig. 3). Our results demonstrate that SOD-1 alleviates oxidative stress triggered by P. aeruginosa.
Figure 3. SOD-1 alleviates pathogen-induced oxidative stress in the intestine.
(A–D) Fluorescence micrographs of oxidative stress reporter gcs-1p::GFP after 2 hour E. coli. (OP50) exposure or 2 hour P. aeruginosa (PA14) exposure. (A,C) gcs-1p::GFP in wild type. (B,D) gcs-1p::GFP in sod-1 (tm776) mutant background. Scale bar indicates 50 μm. (E) Average fluorescence intensity of gcs-1p::GFP. ****Represents p < 0.0001, n.s. not significant, as determined by student’s t test. N = 20–25. Error bars represent standard error of the mean. (F–I) Fluorescence micrographs of oxidative stress reporter gst-4p::GFP after 2 hour E. coli. (OP50) exposure or 2 hour P. aeruginosa (PA14) exposure. (F,H) gst-4p::GFP in wild type. (G,I) gst-4p::GFP in sod-1 (tm776) mutant background. Scale bar indicates 50 μm. (J) Average fluorescence intensity of gst-4p::GFP. ****Represents p < 0.0001, n.s. not significant, as determined by student’s t test. N = 20–25. Error bars represent standard error of the mean.
SOD-1 is localized at the sensory cilium and is induced by P. aeruginosa in the ASER neuron
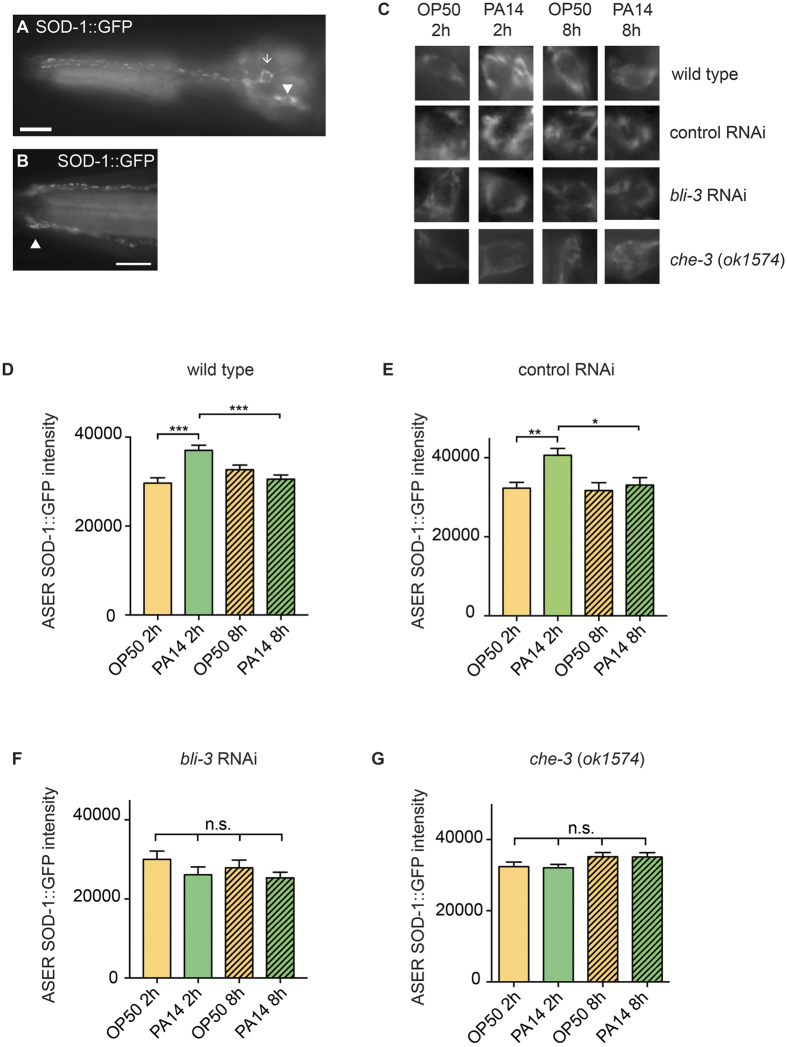
SOD-1 alleviates pathogen-induced oxidative stress. Thus, we sought to determine if SOD-1 is induced by P. aeruginosa. We first generated an integrated sod-1p::sod-1 cDNA::GFP line. We then examined the SOD-1::GFP expression pattern under normal growth conditions. We found that SOD-1::GFP fusion protein is localized as distinct puncta along the dendrite, at the sensory cilium and in the cell body of the ASER neuron (Fig. 4A). We also observed weak SOD-1::GFP signals in the intestine (Supplementary Figure 2).
Figure 4. SOD-1 is temporarily induced by P. aeruginosa in the ASER.
(A) Fluorescence micrographs of bosIs2 [sod-1p::sod-1 cDNA::GFP] strain. The transgenic worms were grown on E. coli OP50 prior to analysis. Triangle represents ASER. Arrow represents ADL. SOD-1::GFP is localized as distinct puncta along the dendrite of the ASER neuron. Scale bar indicates 10 μm. (B) Triangle indicates the ASER cilium. The transgenic worms were grown on E. coli OP50 prior to analysis. Scale bar indicates 5 μm. (C) Fluorescence micrographs of the ASER neuron cell body. Wild type: sod-1(tm776); bosIs2. Control RNAi: sod-1(tm776); bosIs2 fed with bacteria that contain RNAi control vector. bli-3 RNAi: sod-1(tm776); bosIs2 fed with bacteria that contain bli-3 RNAi. che-3(ok1574) represents che-3(ok1574); sod-1(tm776); bosIs2. (D–G) Average fluorescence intensity of SOD-1::GFP in the ASER neuron. ***Represents p < 0.001, **represents p < 0.01, *represents p < 0.05, n.s. not significant, as determined by one-way ANOVA, followed by Tukey’s multiple comparison test. N = 20–25. Error bars represent standard error of the mean.
Further, we transferred the SOD-1::GFP strain to a lawn of E. coli OP50 or to a lawn of P. aeruginosa PA14 and examined if SOD-1::GFP is induced. The activation of gcs-1 and gst-4 reporters indicates a marked increase of ROS in the intestine after 2 hour P. aeruginosa exposure (Fig. 3). However, we found no elevation of SOD-1::GFP signals in the intestine after 2 hour. Instead, intestinal SOD-1 expression was elevated by P. aeruginosa at a later time point (Supplementary Figure 2).
Because SOD-1 functions in the gustatory neuron ASER to modulate behavioral response to pathogens, we sought to examine if SOD-1 is induced in the ASER by P. aeruginosa. After 2 hour P. aeruginosa exposure, we found a 23% increase of SOD-1::GFP intensity in the ASER. However, the increase of SOD-1::GFP was short-lived. The elevated ASER SOD-1::GFP signals diminished and returned to the baseline level by 8 hour (Fig. 4C and D). In addition, we found that the reduction of SOD-1 in the ASER neuron correlates to the initiation of pathogen avoidance behavior (Fig. 1B). Wild type C. elegans remained on the P. aeruginosa lawn and started to vacate the lawn after 7 hour. In contrast, sod-1(tm776) mutant started to vacate away the P. aeruginosa lawn shortly after the mutant reached the source of P. aeruginosa (Fig. 1B and Supplementary Figure 3). Together, our data suggest that SOD-1 acts as a reactive sensor of P. aeruginosa. The elevation of SOD-1 in gustatory neuron ASER suppresses the aversive response to P. aeruginosa.
Reactive oxygen species and sensory modality contribute to SOD-1 induction in the ASER neuron
When responding to microbial challenges, NADPH oxidase promotes the generation of ROS6,11. BLI-3 is the C. elegans NADPH oxidase homolog and is present predominantly in the hypodermis and intestine50. BLI-3 initiates pathogen-induced oxidative stress response in the intestine4,8. To investigate if pathogen-induced ROS activate SOD-1 expression, we raised SOD-1::GFP animals with bacteria that either contain RNAi control vector or bli-3 RNAi. We then transferred the RNAi treated animals to a lawn of E. coli OP50 or to a lawn of P. aeruginosa PA14 and measured the fluorescence signals of SOD1::GFP at 2 hour and 8 hour. In control RNAi, we found P. aeruginosa elevates SOD-1::GFP expression in the ASER neuron at 2 hour. The ASER-specific SOD-1::GFP signals returned to the baseline level by 8 hour (Fig. 4C and E). These results are similar to what we observed in wild type (Fig. 4C and D). In contrast, bli-3 RNAi blocked the induction of SOD-1::GFP by P. aeruginosa (Fig. 4C and F). Because BLI-3 activates ROS in the intestine4,8 and neuron cells are refractory to feeding RNAi treatment51, we conclude that ROS activate ASER-specific SOD-1 expression via a non cell-autonomous mechanism.
SOD-1::GFP is localized as distinct puncta along the dendrite and at the sensory cilium of the ASER neuron (Fig. 4A and B). We reasoned that sensory cues from external P. aeruginosa may induce SOD-1 expression. To test this hypothesis, we introduced SOD-1::GFP into the che-3(ok1574) deletion background. che-3 encodes a dynein heavy chain motor protein. Mutation in che-3 elicits chemosensory defects due to the disruption of ciliary structures in amphid sensory neurons52,53. As indicated by SOD-1::GFP, we found that che-3 mutant elicits abnormal (e.g. looping and additional branching) ciliary structure of ASER neuron (Supplementary Figure 4). We further exposed che-3 (ok1574); SOD-1::GFP animals to P. aeruginosa. In contrast to wild type (Fig. 4C and D), we did not observe elevation of SOD-1::GFP in ASER of the che-3 mutant (Fig. 4C and G). This suggests that external P. aeruginosa elevates SOD-1 expression in the ASER neuron. A recent report reveals that metabolites of P. aeruginosa modulate C. elegans pathogen avoidance response via activation of daf-7 in ASJ sensory neuron29. It remains to be determined if bacterial metabolites or bacterial toxins serve as the external cues to elevate SOD-1 expression in the ASER gustatory neuron.
Antagonistic sensory modality functions in parallel to activate aversive response to pathogens
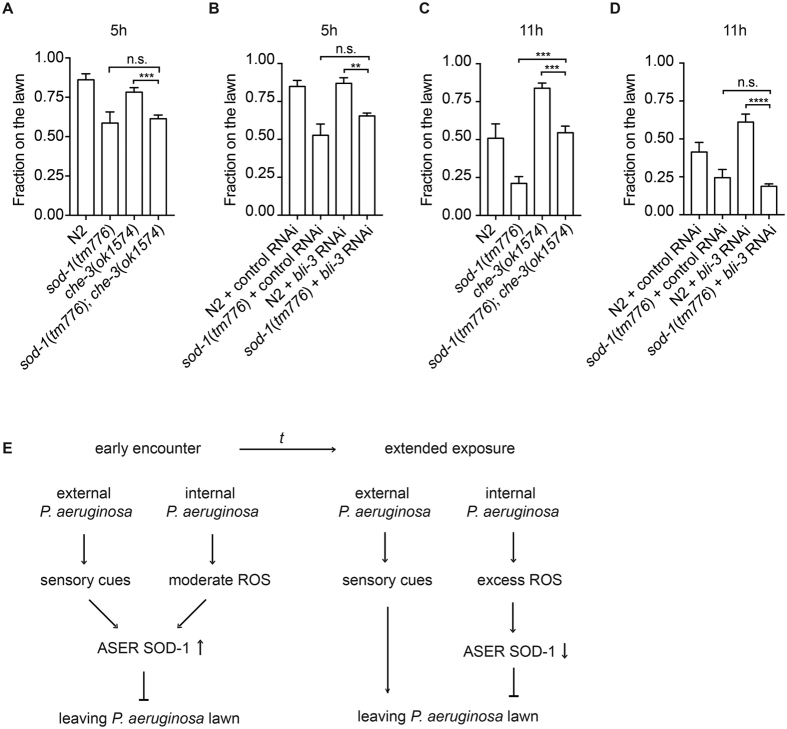
To examine if sensory modalities and ROS play a role in modulating C elegans aversive behavior to pathogens, we investigated the pathogen avoidance phenotypes of che-3 mutant animals and bli-3 RNAi animals. We found that che-3 mutant animals and bli-3 RNAi animals elicit delayed aversive responses to P. aeruginosa (Fig. 5). We further tested if the sod-1 mutation can reverse the delayed behavioral phenotype of che-3(ok1574) and bli-3 RNAi. At 5 hour, we found that sod-1(tm776); che-3(ok1574) and sod-1(tm776); bli-3 RNAi elicit heightened pathogen avoidance phenotype similar to sod-1(tm776) (Fig. 5A and B). These results are consistent with our observations that SOD-1 is induced in the ASER via CHE-3 and BLI-3 dependent signaling (Fig. 4C,E–G). Intriguingly, at 11 hour, as sod-1 mutant continued to block the behavioral response caused by bli-3 RNAi, sod-1(tm776); che-3(ok1574) double mutant elicited intermediate behavioral phenotype (Fig. 5C and D). Since mutation of che-3 disrupts many sensory neurons, the intermediate phenotype of sod-1; che-3 double mutant may be contributed by the inactivation of additional sensory modality28,29,30. Our results suggest that ROS mediate P. aeruginosa avoidance in a sod-1 dependent manner. On the other hand, external sensory stimuli initially elevate SOD-1 expression in the gustatory neuron ASER. After prolonged P. aeruginosa exposure, antagonistic sensory modality promotes pathogen avoidance behavior and allows C. elegans to exit the lawn of P. aeruginosa (Fig. 5E).
Figure 5. SOD-1 integrates signals from internal ROS and external stimuli to repress P. aeruginosa avoidance behavior.
(A–D) P. aeruginosa lawn occupancy of C. elegans strains assayed at 5 h and 11 h. ****Represents p < 0.0001, ***represents p < 0.001, **represents p < 0.01, n.s. not significant, as determined by one-way ANOVA, followed by Tukey’s multiple comparison test. N = 9–15. Error bars represent standard error of the mean. (E) When C. elegans first encounters P. aeruginosa, internal ROS and external sensory stimuli elevate SOD-1 expression in ASER. In turn, the induction of SOD-1 in ASER blocks behavioral avoidance of P. aeruginosa. After prolonged exposure to P. aeruginosa, SOD-1 expression in ASER returns to basal level. The inhibition of P. aeruginosa avoidance behavior is possibly lifted via a mechanism that is in parallel of the sod-1 dependent response.
Together, our results demonstrate that SOD-1 functions in the gustatory neuron ASER to mediate C. elegans pathogen avoidance behavior. SOD-1 is localized along the dendrite, at the cilium and in the cell body of the ASER neuron. SOD-1 is induced by P. aeruginosa. Elevation of SOD-1 inhibits the avoidance response towards P. aeruginosa. After prolonged P. aeruginosa exposure, reduction of SOD-1 in the ASER allows C. elegans to vacate the P. aeruginosa lawn. We also found that ROS modulate P. aeruginosa avoidance in a sod-1 dependent and non cell-autonomous manner. The activation of antagonistic signaling modalities in the sensory system may contribute to behavior plasticity and play a modulatory role in regulating the timing of pathogen avoidance.
Discussion
Interactions between the brain, the gut and enteric microbes are vital for maintaining physiological homeostasis15,54. Microbiota in the intestine affect obesity, type 2 diabetes and inflammatory bowel disease manifestation55,56,57. Alterations in brain-gut interaction are also associated with stress response and behavior58,59,60,61. Recent advancement in human microbiome research suggests that the composition of enteric microbes and the signaling triggered by enteric microbes from gut to the central nervous system is important to our health16,57. Therefore, it has been coined that the gut and enteric microbes are our “second brain”62. Currently, the lack of whole-animal models has impeded the progress to understand how gut microbes modulate brain physiology.
C. elegans is a well-established organism to study neurobiology63,64,65,66,67 and host-microbe interactions28,68,69,70,71,72. Compared to the mammalian brain, which contains billions of neurons, there are only 302 neurons in C. elegans. Most importantly, the neural circuitry of C. elegans has been extensively characterized to the resolution of a single neuron73,74. C. elegans can grow axenically or can feed on bacteria. Prior to entering the intestine of C. elegans, bacteria are normally broken down in the pharynx. However, many human opportunistic pathogens, including P. aeruginosa, colonize the worm intestine17,22,75,76. When C. elegans first encounters P. aeruginosa, C. elegans will remain inside the bacterial lawn and feed on P. aeruginosa28. Ingestion allows the accumulation of P. aeruginosa in the intestine17 and triggers ROS production4,5.
We found after 2 hour exposure to P. aeruginosa, the fluorescence signals of oxidative stress reporters gcs-1p::GFP and gst-4p::GFP are elevated in the intestine (Fig. 3). These results support that P. aeruginosa triggers ROS generation at the mucosal membrane in the intestine. In addition, we abolished the elevation of SOD-1 in the ASER neuron by knocking down bli-3 outside of the nervous system (Fig. 4C and F). This suggests that ROS induce SOD-1 expression in the ASER via a non cell-autonomous mechanism. ROS may act directly or may activate long-range signaling molecules at the brain-gut axis to modulate the pathogen avoidance response. Given the small size of C. elegans, many chemicals from the environment activate stress response in distal tissue via diffusion39,77,78. Excess intestinal ROS may cause elevation of systemic ROS and subsequently induce SOD-1 in the nervous system. In addition, the intestine-derived neuropeptides are potential long-range signaling molecules to mediate SOD-1 expression in the ASER. Further study is needed to confirm if neuroendocrine signaling plays a role in modulating SOD-1 levels in the nervous system.
Our results reveal that external P. aeruginosa mediates SOD-1 induction in the ASER neuron (Fig. 4C and G). the dendritic ending of ASER neuron is directly exposed to the environment. Therefore, ASER neuron may serve to detect bacterial toxins or metabolites secreted by P. aeruginosa. It has been postulated that the C. elegans behavioral response to pathogens is controlled by external sensory modalities and intracellular stress signaling79. Consistent with this hypothesis, our findings suggest that external sensory stimuli modulate ROS-induced sod-1 dependent pathogen avoidance behavior in C. elegans.
A recent study in budding yeast highlights a role of SOD-1 in maintaining respiratory homeostasis and glucose metabolism38. SOD-1 suppresses the degradation of casein kinase 1, a signaling molecule that represses respiratory metabolism. Our results suggest that in a multicellular organism, SOD-1 is induced in a gustatory neuron by ROS (Fig. 4). SOD-1 represses C. elegans behavioral response to P. aeruginosa, which in turn triggers generation of more ROS in the intestine (Fig. 1 and Supplementary Figure 2). After prolonged P. aeruginosa exposure, the reduction of neuronal SOD-1 allows C. elegans to vacate the P. aeruginosa lawn (Figs 1 and 4). By reducing ROS, we were unable to ameliorate the heightened pathogen avoidance phenotype of sod-1 deletion (Fig. 5). Our data indicate that SOD-1 plays a key role in the nervous system to mediate pathogen avoidance behavior in response to the internal state of redox homeostasis.
When food sources are scarce, sod-1 dependent behavior may provide survival benefits by delaying the aversive response thus allowing C. elegans to take advantage of food that is contaminated with harmful microbes. Our previous findings reveal that naturally occurring polymorphisms in npr-1 (a neuropeptide Y receptor homolog) and hecw-1 (a neuron specific E3 ligase) can lead to changes in behavior that may facilitate the adaptation of C. elegans to microbes in nature28. In a recent survey of microbiome in C. elegans natural habitats, it reveals that 22% of the bacterial species are detrimental to C. elegans growth. Wild P. aeruginosa strains are among the identified detrimental species, and these wild P. aeruginosa strains activate C. elegans pathogen response2. Given that 78% of the natural bacteria species are possible food sources, the ability to continue feeding when small quantities of toxic bacteria are present is beneficial to survival. We hypothesized that the activation of SOD-1 in the gustatory neuron may provide an evolutionarily conserved strategy by allowing animals to take advantage of non-ideal food.
SOD-1 is an enzyme that converts superoxide into less toxic hydrogen peroxide and oxygen. Investigation of mammalian SOD-1 reveals that SOD-1 is a stable enzyme with a slow turnover rate in the central nervous system. The slow turnover rate of wild type SOD-1 may explain the susceptibility of the nervous system in ALS neurodegenerative proteinopathies80. Although SOD-1 expression is not elevated in the intestine during the early stage of pathogen exposure, we found SOD-1 is elevated in the intestine at 8 hour (Supplementary Figure 2). This suggests that during prolonged exposure to pathogens, ROS are generated continuously. We speculate that excess systemic ROS may activate the turnover of SOD-1 in the nervous system. Our data demonstrate a link between SOD-1 protein level and animal behaviors. Given the high degree of genomic conservation, our results may shed new light on the function of SOD-1 in humans and lead to a more thorough understanding of ALS pathology.
Materials and Methods
Strains
C. elegans strains were maintained at 20 °C using standard methods81. Strains were maintained at 20 °C then shifted to 22.5 °C for P. aeruginosa PA14 lawn avoidance assays. The sod-1(tm776) mutant strain was derived from GA187 strain (obtained from the Caenorhabditis Genetics Center) by backcrossing it six times to N2. Transgenic strains were isolated by microinjecting various plasmids, (typically at 50–100 μg/ml) together with one of the following coinjection markers, rol-6dm, myo-2p::mstrawberry, unc-122p::GFP, and unc-122p::mcherry in wild type or mutant animals. UV integration of extrachromosomal array was performed following the protocol originated from S. Mitani. The integrated lines were then backcrossed six times to N2 prior to the analysis.
P. aeruginosa PA14 avoidance assay
A 100 mL solution of LB was inoculated with a single colony of P. aeruginosa PA14 and grown overnight without shaking at 37 °C until O.D. reaches 0.2–0.3. 30 μl of this culture was used to seed the center of the 100-mm NGM plate. Seeded plates were incubated for 24 h at room temperature (22.5 °C) prior to the experiment. Approximately 30 larval stage 4 (L4) animals were transferred onto plates containing the P. aeruginosa PA14 lawn at 22.5 °C, and lawn occupancy was measured at the indicated times. Three plates of each genotype were performed in each experiment and all experiments were performed at least three times. Upon being transferred to the P. aeruginosa PA14-containing plates, animals explored the plate for about 10–15 minutes until they found the bacterial lawn and then remained in the lawn. Subsequently, lawn occupancy was measured over time as the lawn avoidance behavior is observed28.
Molecular cloning
The genomic region of sod-1 was amplified by PCR using primers 5′-GAACACCAAACCGGACTGACCAAGT-3′ and 5′-GTTTATGACGCAAAGCGTACGGACAATCTC-3′. The 2.9 kb genomic fragment was cloned into a Topo® (Invitrogen) vector. The sod-1 promoter region was amplified by using a 5′ primer containing 5′-GAACACCAAACCGGACTGACCAAGT-3′ and 3′ primer containing 5′-CAAAGTTGTAGATTCAGTATTTTAGATCGGTG-3′. The 1223 bp fragment was subsequently cloned into the pPD95.75 vector (Fire Lab Vector Kit, Addgene) to generate the sod-1 promoter GFP reporter. sod-1 cDNA clones are gifts from Y. Kohara (yk524g1, yk593d7, yk1381e03 and yk1715f05). The unc-119, ges-1, flp-6, sre-1, gcy-5 promoters were generated using primers as described previously40,41,82,83,84. csp-1b cDNA is a gift from D. Denning and H.R. Horvitz46. Detailed primer sets and methods used for cloning are available upon request.
Microscopy
Animals were mounted in M9 with levamisole (10 mM) onto slides with a 3% agarose pad. The slides were viewed using an AxioImager Z1 fluorescence microscope (Zeiss) with 10x/0.25, 40x/0.75 and 63x/1.4 (oil) objectives. The fluorescence signals were recorded by a CCD camera in a 16 bit format without saturation. The images were captured and analyzed by ProgRes imaging software. Fluorescence intensity was measured and calculated using Image-Pro software.
RNAi treatments
RNAi treatments were performed by feeding C. elegans RNAi constructs and reagents described previously85,86. HT115 E. coli carrying RNAi clones in the pL4440 vector were cultured overnight in LB liquid media with antibiotics. NGM plates containing 1 mM IPTG and antibiotics were seeded with the feeding RNAi bacterial culture. The plates were incubated at 20 °C for bacteria growth. Five L2 larvae were placed onto each seeded NGM plate, and the progeny of these animals were scored for blister phenotype prior to P. aeruginosa PA14 avoidance assay to confirm the effectiveness of the RNAi treatment.
Statistical analysis
Statistical analysis was performed using GraphPad Prism software.
Additional Information
How to cite this article: Horspool, A. M. and Chang, H. C. Superoxide dismutase SOD-1 modulates C. elegans pathogen avoidance behavior. Sci. Rep. 7, 45128; doi: 10.1038/srep45128 (2017).
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Material
Acknowledgments
We thank the Caenorhabditis Genetics Center (CGC), which is supported by the NIH (P40 OD010440), to provide us many strains. We thank D. Denning and H. R. Horvitz for the csp-1b cDNA. We thank Y. Kohara for the sod-1 cDNA. We thank S. Yoon for backcrossing mutant alleles and integrated transgenic lines. We thank T. Middelkoop and S. Luo for reading the manuscript and discussion.
Footnotes
The authors declare no competing financial interests.
Author Contributions A.M.H. performed the experiments, analyzed the results and commented on the manuscript. H.C.C. designed and performed the experiments, analyzed the results and wrote the paper.
References
- Frezal L. & Felix M. A. C. elegans outside the Petri dish. Elife 4, 1–19 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samuel B. S., Rowedder H., Braendle C., Felix M. A. & Ruvkun G. Caenorhabditis elegans responses to bacteria from its natural habitats. Proc Natl Acad Sci USA 113, E3941–3949 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corcionivoschi N. et al. Mucosal reactive oxygen species decrease virulence by disrupting Campylobacter jejuni phosphotyrosine signaling. Cell Host Microbe 12, 47–59 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoeven R., McCallum K. C., Cruz M. R. & Garsin D. A. Ce-Duox1/BLI-3 generated reactive oxygen species trigger protective SKN-1 activity via p38 MAPK signaling during infection in C. elegans. PLoS Pathog 7, e1002453 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipinski S. et al. DUOX2-derived reactive oxygen species are effectors of NOD2-mediated antibacterial responses. J Cell Sci 122, 3522–3530 (2009). [DOI] [PubMed] [Google Scholar]
- Ha E. M., Oh C. T., Bae Y. S. & Lee W. J. A direct role for dual oxidase in Drosophila gut immunity. Science 310, 847–850 (2005). [DOI] [PubMed] [Google Scholar]
- Bae Y. S., Choi M. K. & Lee W. J. Dual oxidase in mucosal immunity and host-microbe homeostasis. Trends Immunol 31, 278–287 (2010). [DOI] [PubMed] [Google Scholar]
- Chavez V., Mohri-Shiomi A. & Garsin D. A. Ce-Duox1/BLI-3 generates reactive oxygen species as a protective innate immune mechanism in Caenorhabditis elegans. Infect Immun 77, 4983–4989 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chavez V., Mohri-Shiomi A., Maadani A., Vega L. A. & Garsin D. A. Oxidative stress enzymes are required for DAF-16-mediated immunity due to generation of reactive oxygen species by Caenorhabditis elegans. Genetics 176, 1567–1577 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Autreaux B. & Toledano M. B. ROS as signalling molecules: mechanisms that generate specificity in ROS homeostasis. Nat Rev Mol Cell Biol 8, 813–824 (2007). [DOI] [PubMed] [Google Scholar]
- Schieber M. & Chandel N. S. ROS function in redox signaling and oxidative stress. Curr Biol 24, R453–462 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prahlad V., Cornelius T. & Morimoto R. I. Regulation of the cellular heat shock response in Caenorhabditis elegans by thermosensory neurons. Science 320, 811–814 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J., Singh V., Kajino-Sakamoto R. & Aballay A. Neuronal GPCR controls innate immunity by regulating noncanonical unfolded protein response genes. Science 332, 729–732 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCallum K. C. et al. TRX-1 Regulates SKN-1 Nuclear Localization Cell Non-autonomously in Caenorhabditis elegans. Genetics 203, 387–402 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Backhed F., Ley R. E., Sonnenburg J. L., Peterson D. A. & Gordon J. I. Host-bacterial mutualism in the human intestine. Science 307, 1915–1920 (2005). [DOI] [PubMed] [Google Scholar]
- Yano J. M. et al. Indigenous bacteria from the gut microbiota regulate host serotonin biosynthesis. Cell 161, 264–276 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irazoqui J. E. et al. Distinct pathogenesis and host responses during infection of C. elegans by P. aeruginosa and S. aureus. PLoS Pathog 6, e1000982 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onken B. & Driscoll M. Metformin induces a dietary restriction-like state and the oxidative stress response to extend C. elegans Healthspan via AMPK, LKB1, and SKN-1. PLoS One 5, e8758 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabreiro F. et al. Metformin retards aging in C. elegans by altering microbial folate and methionine metabolism. Cell 153, 228–239 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang S. & Curran S. P. Adaptive capacity to bacterial diet modulates aging in C. elegans. Cell Metab 19, 221–231 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macdonald T. T. & Monteleone G. Immunity, inflammation, and allergy in the gut. Science 307, 1920–1925 (2005). [DOI] [PubMed] [Google Scholar]
- Labrousse A., Chauvet S., Couillault C., Kurz C. L. & Ewbank J. J. Caenorhabditis elegans is a model host for Salmonella typhimurium. Curr Biol 10, 1543–1545 (2000). [DOI] [PubMed] [Google Scholar]
- Kurz C. L. et al. Virulence factors of the human opportunistic pathogen Serratia marcescens identified by in vivo screening. EMBO J 22, 1451–1460 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy K. C., Andersen E. C., Kruglyak L. & Kim D. H. A polymorphism in npr-1 is a behavioral determinant of pathogen susceptibility in C. elegans. Science 323, 382–384 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mylonakis E. & Aballay A. Worms and flies as genetically tractable animal models to study host-pathogen interactions. Infect Immun 73, 3833–3841 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemaitre B. & Ausubel F. M. Animal models for host-pathogen interactions. Curr Opin Microbiol 11, 249–250 (2008). [DOI] [PubMed] [Google Scholar]
- Irazoqui J. E., Urbach J. M. & Ausubel F. M. Evolution of host innate defence: insights from Caenorhabditis elegans and primitive invertebrates. Nat Rev Immunol 10, 47–58 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang H. C., Paek J. & Kim D. H. Natural polymorphisms in C. elegans HECW-1 E3 ligase affect pathogen avoidance behaviour. Nature 480, 525–529 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meisel J. D., Panda O., Mahanti P., Schroeder F. C. & Kim D. H. Chemosensation of bacterial secondary metabolites modulates neuroendocrine signaling and behavior of C. elegans. Cell 159, 267–280 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Lu H. & Bargmann C. I. Pathogenic bacteria induce aversive olfactory learning in Caenorhabditis elegans. Nature 438, 179–184 (2005). [DOI] [PubMed] [Google Scholar]
- Styer K. L. et al. Innate immunity in Caenorhabditis elegans is regulated by neurons expressing NPR-1/GPCR. Science 322, 460–464 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saccon R. A., Bunton-Stasyshyn R. K., Fisher E. M. & Fratta P. Is SOD1 loss of function involved in amyotrophic lateral sclerosis? Brain 136, 2342–2358 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurney M. E. et al. Motor neuron degeneration in mice that express a human Cu, Zn superoxide dismutase mutation. Science 264, 1772–1775 (1994). [DOI] [PubMed] [Google Scholar]
- Rosen D. R. et al. Mutations in Cu/Zn superoxide dismutase gene are associated with familial amyotrophic lateral sclerosis. Nature 362, 59–62 (1993). [DOI] [PubMed] [Google Scholar]
- Fischer L. R., Li Y., Asress S. A., Jones D. P. & Glass J. D. Absence of SOD1 leads to oxidative stress in peripheral nerve and causes a progressive distal motor axonopathy. Exp Neurol 233, 163–171 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller F. L. et al. Absence of CuZn superoxide dismutase leads to elevated oxidative stress and acceleration of age-dependent skeletal muscle atrophy. Free Radic Biol Med 40, 1993–2004 (2006). [DOI] [PubMed] [Google Scholar]
- Oeda T. et al. Oxidative stress causes abnormal accumulation of familial amyotrophic lateral sclerosis-related mutant SOD1 in transgenic Caenorhabditis elegans. Hum Mol Genet 10, 2013–2023 (2001). [DOI] [PubMed] [Google Scholar]
- Reddi A. R. & Culotta V. C. SOD1 integrates signals from oxygen and glucose to repress respiration. Cell 152, 224–235 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doonan R. et al. Against the oxidative damage theory of aging: superoxide dismutases protect against oxidative stress but have little or no effect on life span in Caenorhabditis elegans. Genes Dev 22, 3236–3241 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Libina N., Berman J. R. & Kenyon C. Tissue-specific activities of C. elegans DAF-16 in the regulation of lifespan. Cell 115, 489–502 (2003). [DOI] [PubMed] [Google Scholar]
- Shivers R. P., Kooistra T., Chu S. W., Pagano D. J. & Kim D. H. Tissue-specific activities of an immune signaling module regulate physiological responses to pathogenic and nutritional bacteria in C. elegans. Cell Host Microbe 6, 321–330 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bargmann C. I. & Horvitz H. R. Chemosensory neurons with overlapping functions direct chemotaxis to multiple chemicals in C. elegans. Neuron 7, 729–742 (1991). [DOI] [PubMed] [Google Scholar]
- Uchida O., Nakano H., Koga M. & Ohshima Y. The C. elegans che-1 gene encodes a zinc finger transcription factor required for specification of the ASE chemosensory neurons. Development 130, 1215–1224 (2003). [DOI] [PubMed] [Google Scholar]
- Chang S., Johnston R. J. Jr. & Hobert O. A transcriptional regulatory cascade that controls left/right asymmetry in chemosensory neurons of C. elegans. Genes Dev 17, 2123–2137 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortiz C. O. et al. Lateralized gustatory behavior of C. elegans is controlled by specific receptor-type guanylyl cyclases. Curr Biol 19, 996–1004 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denning D. P., Hatch V. & Horvitz H. R. Both the caspase CSP-1 and a caspase-independent pathway promote programmed cell death in parallel to the canonical pathway for apoptosis in Caenorhabditis elegans. PLoS Genet 9, e1003341 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papp D., Csermely P. & Soti C. A role for SKN-1/Nrf in pathogen resistance and immunosenescence in Caenorhabditis elegans. PLoS Pathog 8, e1002673 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- An J. H. & Blackwell T. K. SKN-1 links C. elegans mesendodermal specification to a conserved oxidative stress response. Genes Dev 17, 1882–1893 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliveira R. P. et al. Condition-adapted stress and longevity gene regulation by Caenorhabditis elegans SKN-1/Nrf. Aging Cell 8, 524–541 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Hoeven R., Cruz M. R., Chavez V. & Garsin D. A. Localization of the Dual Oxidase BLI-3 and Characterization of Its NADPH Oxidase Domain during Infection of Caenorhabditis elegans. PLoS One 10, e0124091 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy S., Wang D. & Ruvkun G. A conserved siRNA-degrading RNase negatively regulates RNA interference in C. elegans. Nature 427, 645–649 (2004). [DOI] [PubMed] [Google Scholar]
- Wicks S. R., de Vries C. J., van Luenen H. G. & Plasterk R. H. CHE-3, a cytosolic dynein heavy chain, is required for sensory cilia structure and function in Caenorhabditis elegans. Dev Biol 221, 295–307 (2000). [DOI] [PubMed] [Google Scholar]
- Signor D. et al. Role of a class DHC1b dynein in retrograde transport of IFT motors and IFT raft particles along cilia, but not dendrites, in chemosensory neurons of living Caenorhabditis elegans. J Cell Biol 147, 519–530 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer E. A. Gut feelings: the emerging biology of gut-brain communication. Nat Rev Neurosci 12, 453–466 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jostins L. et al. Host-microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature 491, 119–124 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin J. et al. A metagenome-wide association study of gut microbiota in type 2 diabetes. Nature 490, 55–60 (2012). [DOI] [PubMed] [Google Scholar]
- Blanton L. V. et al. Gut bacteria that prevent growth impairments transmitted by microbiota from malnourished children. Science 351 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cryan J. F. & Dinan T. G. Mind-altering microorganisms: the impact of the gut microbiota on brain and behaviour. Nat Rev Neurosci 13, 701–712 (2012). [DOI] [PubMed] [Google Scholar]
- Diaz Heijtz R. et al. Normal gut microbiota modulates brain development and behavior. Proc Natl Acad Sci USA 108, 3047–3052 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bravo J. A. et al. Ingestion of Lactobacillus strain regulates emotional behavior and central GABA receptor expression in a mouse via the vagus nerve. Proc Natl Acad Sci USA 108, 16050–16055 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster J. A. & McVey Neufeld K. A. Gut-brain axis: how the microbiome influences anxiety and depression. Trends Neurosci 36, 305–312 (2013). [DOI] [PubMed] [Google Scholar]
- Ridaura V. & Belkaid Y. Gut microbiota: the link to your second brain. Cell 161, 193–194 (2015). [DOI] [PubMed] [Google Scholar]
- Bargmann C. I. Comparative chemosensation from receptors to ecology. Nature 444, 295–301 (2006). [DOI] [PubMed] [Google Scholar]
- Hart A. C. & Chao M. Y. In The Neurobiology of Olfaction Frontiers in Neuroscience(ed. Menini A.) (2010). [Google Scholar]
- Sasakura H. & Mori I. Behavioral plasticity, learning, and memory in C. elegans. Curr Opin Neurobiol 23, 92–99 (2013). [DOI] [PubMed] [Google Scholar]
- Hammarlund M. & Jin Y. Axon regeneration in C. elegans. Curr Opin Neurobiol 27, 199–207 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong X., Shen K. & Bulow H. E. Intrinsic and extrinsic mechanisms of dendritic morphogenesis. Annu Rev Physiol 77, 271–300 (2015). [DOI] [PubMed] [Google Scholar]
- Garsin D. A. et al. Long-lived C. elegans daf-2 mutants are resistant to bacterial pathogens. Science 300, 1921 (2003). [DOI] [PubMed] [Google Scholar]
- Kim D. H. et al. A conserved p38 MAP kinase pathway in Caenorhabditis elegans innate immunity. Science 297, 623–626 (2002). [DOI] [PubMed] [Google Scholar]
- Pradel E. et al. Detection and avoidance of a natural product from the pathogenic bacterium Serratia marcescens by Caenorhabditis elegans. Proc Natl Acad Sci USA 104, 2295–2300 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson C. E., Kooistra T. & Kim D. H. An essential role for XBP-1 in host protection against immune activation in C. elegans. Nature 463, 1092–1095 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Troemel E. R., Felix M. A., Whiteman N. K., Barriere A. & Ausubel F. M. Microsporidia are natural intracellular parasites of the nematode Caenorhabditis elegans. PLoS Biol 6, 2736–2752 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarrell T. A. et al. The connectome of a decision-making neural network. Science 337, 437–444 (2012). [DOI] [PubMed] [Google Scholar]
- White J. G., Southgate E., Thomson J. N. & Brenner S. The structure of the nervous system of the nematode Caenorhabditis elegans. Philos Trans R Soc Lond B Biol Sci 314, 1–340 (1986). [DOI] [PubMed] [Google Scholar]
- Mahajan-Miklos S., Tan M. W., Rahme L. G. & Ausubel F. M. Molecular mechanisms of bacterial virulence elucidated using a Pseudomonas aeruginosa-Caenorhabditis elegans pathogenesis model. Cell 96, 47–56 (1999). [DOI] [PubMed] [Google Scholar]
- Couillault C. & Ewbank J. J. Diverse bacteria are pathogens of Caenorhabditis elegans. Infect Immun 70, 4705–4707 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong J., Boyd W. A. & Freedman J. H. Molecular characterization of two homologs of the Caenorhabditis elegans cadmium-responsive gene cdr-1: cdr-4 and cdr-6. J Mol Biol 376, 621–633 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z., Luo J., Li Y. & Ma L. The BLI-3/TSP-15/DOXA-1 dual oxidase complex is required for iodide toxicity in Caenorhabditis elegans. G3 (Bethesda) 5, 195–203 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melo J. A. & Ruvkun G. Inactivation of conserved C. elegans genes engages pathogen- and xenobiotic-associated defenses. Cell 149, 452–466 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crisp M. J. et al. In vivo kinetic approach reveals slow SOD1 turnover in the CNS. J Clin Invest 125, 2772–2780 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner S. The genetics of Caenorhabditis elegans. Genetics 77, 71–94 (1974). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altun-Gultekin Z. et al. A regulatory cascade of three homeobox genes, ceh-10, ttx-3 and ceh-23, controls cell fate specification of a defined interneuron class in C. elegans. Development 128, 1951–1969 (2001). [DOI] [PubMed] [Google Scholar]
- Li C., Kim K. & Nelson L. S. FMRFamide-related neuropeptide gene family in Caenorhabditis elegans. Brain Res 848, 26–34 (1999). [DOI] [PubMed] [Google Scholar]
- Yu S., Avery L., Baude E. & Garbers D. L. Guanylyl cyclase expression in specific sensory neurons: a new family of chemosensory receptors. Proc Natl Acad Sci USA 94, 3384–3387 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser A. G. et al. Functional genomic analysis of C. elegans chromosome I by systematic RNA interference. Nature 408, 325–330 (2000). [DOI] [PubMed] [Google Scholar]
- Rual J. F. et al. Toward improving Caenorhabditis elegans phenome mapping with an ORFeome-based RNAi library. Genome Res 14, 2162–2168 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.