Abstract
Background
Rapid diagnostic tests (RDTs) for histidine rich protein 2 (HRP2) are often used to determine whether persons with fever should be treated with anti-malarials. However, Plasmodium falciparum parasites with a deletion of the hrp2 gene yield false-negative RDTs and there are concerns the sensitivity of HRP2-based RDTs may fall when the intensity of transmission decreases.
Methods
This observational study enrolled 9226 patients at three health centres in Rwanda from April 2014 to April 2015. It then compared the sensitivity of RDTs based on HRP2 and the Plasmodium lactate dehydrogenase (pLDH) to microscopy (thick smears) for the diagnosis of malaria. PCR was used to determine whether deletions of the histidine-rich central repeat region of the hrp2 gene (exon 2) were associated with false-negative HRP2-based RDTs.
Results
In comparison to microscopy, the sensitivity and specificity of HRP2- and pLDH-based RDTs were 89.5 and 86.2% and 80.2 and 94.3%, respectively. When the results for both RDTs were combined, sensitivity rose to 91.8% and specificity was 85.7%. Additionally, when smear positivity fell from 46 to 3%, the sensitivity of the HRP2-based RDT fell from 88 to 67%. Of 370 samples with false-negative HRP2 RDT results for which PCR was performed, 140 (38%) were identified as P. falciparum by PCR. Of the isolates identified as P. falciparum by PCR, 32 (23%) were negative for the hrp2 gene based on PCR. Of the 32 P. falciparum isolates negative for hrp2 by PCR, 17 (53%) were positive based on the pLDH RDT.
Conclusion
This prospective study of RDT performance coincided with a decline in the intensity of malaria transmission in Kibirizi (fall in slide positivity from 46 to 3%). This decline was associated with a decrease in HRP2 RDT sensitivity (from 88 to 67%). While P. falciparum isolates without the hrp2 gene were an important cause of false-negative HRP2-based RDTs, most were identified by the pLDH-based RDT. Although WHO does not recommend the use of combined HRP2/pLDH testing in sub-Saharan Africa, these results suggest that combination HRP2/pLDH-based RDTs could reduce the impact of false-negative HRP2-based RDTs for detection of symptomatic P. falciparum malaria.
Electronic supplementary material
The online version of this article (doi:10.1186/s12936-017-1768-1) contains supplementary material, which is available to authorized users.
Keywords: Malaria, Plasmodium falciparum, Rapid diagnostic test (RDT), Histidine rich protein 2 (HRP2), Plasmodium lactate dehydrogenase (pLDH), Sensitivity, Polymerase chain reaction (PCR), Rwanda
Background
Rapid diagnostic tests (RDTs) have become the focal point of malaria control. The central role of RDTs is the result of a recent paradigm shift in malaria case management, based on the World Health Organization (WHO) 2010 recommendation that all persons thought to have malaria should have their diagnosis confirmed by microscopy or an RDT before treatment [1]. However, the value of a “test before you treat” policy depends on accurate diagnosis. False-negative tests may delay the provision of life-saving treatment for individual patients and may simultaneously increase the number of persons capable of infecting mosquitoes in the community.
Although microscopy has been used most commonly to detect malaria parasites, it requires equipment, reagents and skilled microscopists [2]. Thus, in parts of sub-Saharan Africa where microscopy is inaccessible or of low quality, RDTs have become the primary tool for the parasitologic diagnosis or confirmation of malaria [3, 4]. Since 2005, the proportion of suspected malaria cases examined using a diagnostic test (microscopy or RDT) in sub-Saharan Africa rose from 36% in 2005 to 41% in 2010 and 65% in 2014. In 2014, RDTs accounted for 71% of the diagnostic tests performed in sub-Saharan Africa [3]. Given the central role RDTs now play in determining whether persons with symptoms have malaria and should be treated, it is increasingly important to understand the factors that influence their performance.
RDTs are immunochromatographic tests which detect proteins released from parasitized red blood cells. Most of the RDTs used currently to diagnose P. falciparum infections target HRP2 [5]. HRP2-based RDTs are specific for P. falciparum because P. falciparum is the only human parasite that produces HRP2 [3]. In contrast, RDTs targeting pan-lactate dehydrogenase (pLDH) or aldolase can detect all the Plasmodium species that infect humans, although they are reported to be less sensitive than HRP2-based tests, especially with low parasite densities [6, 7]. In regions where P. falciparum predominates and non-falciparum infections occur as mixed infections with P. falciparum, including most of sub-Saharan Africa, WHO generally recommends HRP2-based RDTs. In contrast, pLDH and aldolase-based RDTs are recommended in areas where non-falciparum infections predominate. Currently, WHO suggests restricting combined HRP2/pLDH RDTs to regions where P. falciparum and non-falciparum infections occur as single-species infections [5].
Although the sensitivity of HRP2-based RDTs has been reported as >90% for P. falciparum, several groups have reported decreases in the sensitivity of HRP2-based RDTs after decreases in the intensity of transmission [7–9]. For example, in Zanzibar, the sensitivity of HRP2-based RDTs in relation to thick smears fell from 93 to 79% as the percent of malaria-attributable fever episodes in the population decreased from 30 to <3% [9].
Plasmodium falciparum parasites without the central repeat region of the hrp2 gene may cause false-negative RDTs [10, 11] because they fail to produce the HRP2 target molecule for HRP2-based RDTs. Isolates with hrp2-negative P. falciparum have now been identified in the blood of infected human subjects in South America, Asia and Africa [10–13]. However, despite the diagnostic threat and malaria control concerns posed by parasites without hrp2, there is a paucity of data on the frequency of those parasites and the factors driving (responsible for) their selection.
Preliminary studies from Mali have found seasonal fluctuations in the prevalence of false-negative RDTs and suggest the peak prevalence of hrp2-negative isolates is at the end of the dry season [8]. However, it is not clear whether the seasonal variation in RDT sensitivity for P. falciparum infection observed in Mali occurs in East Africa or elsewhere. There is also a need to determine whether hrp2-negative parasites can be identified using pLDH-based RDTs.
To address these knowledge gaps, this study compared the sensitivities of HRP2- and pLDH-based RDTs at sites with varying intensities of malaria transmission in Rwanda to determine whether deletions of hrp2 were responsible for false-negative HRP2-based RDTs.
Methods
Study design and sites
This cross-sectional study was conducted at three primary health centres in Rwanda: Busogo Health Centre (HC) in the Musanze district of the Northern Province, Rukara HC in the Kayonza district of the Eastern Province and Kibirizi HC in the Gisagara district of the Southern Province (Fig. 1). These health centres were selected to provide sites with varying prevalences of infection consistent with different intensities of transmission. For example, in 2013, slide positivity rates for symptomatic patients were 32.0, 10.8 and 3.5% for the Kibirizi, Rukara and Busogo HCs, respectively [14]. Historically, malaria transmission has occurred year-round in Rwanda’s endemic regions with two peaks (May–June and November–December) after seasonal rains in March–April and September–October. Plasmodium falciparum is the predominant parasite and mosquitoes of the Anopheles gambiae complex are the primary vectors.
Fig. 1.
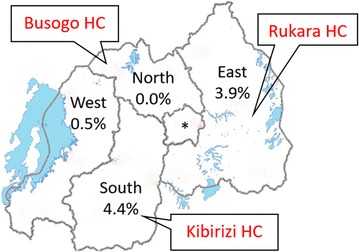
Study sites and household-level parasite prevalence in children under 5 years of age, by province. Parasite prevalence data from 2014 to 2015 DHS [16]. Asterisk notes the city of Kigali with a parasite prevalence of 0.0%. HC health center
The major malaria control interventions in Rwanda are long-lasting insecticidal nets (LLINs) impregnated with pyrethroids, indoor residual spraying (IRS) with carbamates and prompt treatment of confirmed cases with artemisinin-based combination therapy (artemether–lumefantrine) [15]. Between 2009 and 2011, over 6.1 million LLINs were distributed in Rwanda (total population 10.5 million) with the goal of achieving universal coverage. Subsequently, the 2014–2015 Rwandan Demographic and Health Survey (DHS) found 83% of households owned at least one treated bed net and 43% of households had one net for every two people sleeping in the house [16]. IRS has also been implemented in districts selected by the Malaria and Other Parasitic Diseases Division (MOPDD) of the Ministry of Health including Gisagara, where the Kibirizi HC is located. During this study, two rounds of IRS (September/ October 2014 and February/March 2015) were performed in Kibirizi and coverage was reported to be greater than 98% [15]. For patients with uncomplicated malaria, artemether–lumefantrine has been the first-line treatment in Rwanda since 2006.
Patient enrollment and study procedures
Convenience sampling was used to enroll patients who presented to these three health centres with symptoms consistent with uncomplicated malaria and were referred to the outpatient laboratory for blood smears in accordance with Rwanda malaria treatment guidelines. Study personnel were not involved in the decision to refer patients to the laboratory. Patients were excluded from this study if they were diagnosed as having severe malaria or other conditions requiring urgent diagnosis or treatment. Eligible patients were recruited Monday to Friday from 8 a.m. to 4 p.m.
At the time of enrollment, demographic and clinical information was collected and a finger-prick blood sample was obtained for thick and thin blood smears, an RDT and filter paper blots which were stored for molecular testing. Study personnel were experienced nurses and laboratory technicians trained in the performance and interpretation of blood smears and RDTs for malaria.
Laboratory procedures
Rapid diagnostic tests
The RDT used in this study was the First Response® Malaria pLDH/HRP2 Combo test (Premier Medical Corporation Limited, India; catalogue number I16FRC) which was provided by the Rwandan Ministry of Health. This RDT uses a buffer solution containing dye-labelled monoclonal antibodies to HRP2 and pLDH. The antigen–antibody complexes are then captured by monoclonal antibodies to the target antigens, which are immobilized on the test strip. The combo test has three lines of immobilized antibodies on the test strip: a species-specific antibody for P. falciparum HRP2, a genus (Plasmodium)-specific antibody for pLDH at a different position on the test strip and a control antibody at a third position. Testing was performed and interpreted according to the manufacturer’s instructions in the package insert. Faint bands at the test line positions were interpreted as positive [9, 17].
Microscopy
Thick and thin blood smears were collected on the same slide, air-dried and stained with 10% Giemsa for 10 min after the thin smears had been fixed with methanol. Thick smears were examined under oil immersion (1000× magnification) by two independent laboratory technicians. Asexual parasitaemia at any parasite density was reported as a positive smear. Blood smears were considered negative if no parasites were observed after the examination of 100 oil immersion fields. Disparities in slide readings (positive vs negative) and differences >30% in parasite densities were resolved by a third reader. Note that study microscopists did not have access to the subjects’ RDT results or previous microscopy results. All laboratory technicians were qualified at the diploma level or above in the medical laboratory sciences.
Filter paper blots
Blood samples collected on filter paper (Whatman 3 MM) were air-dried thoroughly, placed in individual sealed plastic bags with desiccant and stored at room temperature prior to molecular analysis by PCR.
Amplification of the central repeat region of the hrp2 gene (exon 2) and 18S rRNA parasite DNA sequences
PCR was performed to test for Plasmodium DNA in samples that were negative using the HRP2 RDT but positive by microcopy or the pLDH RDT. Within 3 months of sample collection, DNA was isolated from dried blood spots on filter papers using six 3 mm punches and the QIAamp DNA mini kit (Qiagen, Germantown, MD). Positive and negative controls were used with each round of PCR. After PCR, amplicons were visualized by electrophoresis using 2% agarose gels stained with ethidium bromide.
PCR protocol
A sequential, 3-step approach to PCR was used to assign each sample to one of four categories: (1) DNA from P. falciparum with the hrp2 gene, (2) DNA from P. falciparum without the central histidine-rich repeat region of the hrp2 gene, (3) DNA from Plasmodium other than P. falciparum and (4) samples without Plasmodium DNA. This stepwise protocol is depicted in Fig. 2 and described below.
Fig. 2.
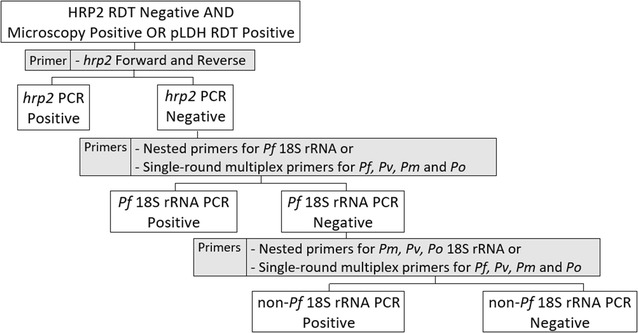
PCR amplification of samples negative with the HRP2 RDT which were positive by microscopy or by pLDH RDT. PCR was performed to test for Plasmodium DNA in samples that were negative by HRP2 RDT but positive by either microcopy or by pLDH RDT. PCR was performed using a tiered, 3-stage approach. Rows 2–4 of the flowchart reflect PCR performed in stages 1–3, respectively. The primers used at each stage are listed in the gray boxes to the right of the flowchart. If a sample was identified as positive by PCR at any stage, no further PCR testing was pursued. Samples positive by PCR for the hrp2 gene were considered P. falciparum isolates containing the hrp2 gene. Samples negative by PCR for the hrp2 gene and positive by PCR for Pf 18S rRNA were considered P. falciparum isolates lacking the hrp2 gene. Samples negative by PCR for the hrp2 gene and Pf 18S rRNA but positive by PCR for Pv, Pm or Po 18S rRNA were considered non-Pf malaria. Samples negative by PCR for the hrp2 gene, Pf 18S rRNA and Pv, Pm and Po 18S rRNA were considered negative for Plasmodium DNA. Pf Plasmodium falciparum, Pv P. vivax, Pm P. malariae, Po P. ovale
For samples negative with the HRP2 RDT but positive by microcopy or the pLDH RDT, primers for the conserved 5′ and 3′ regions of the hrp2 gene (Additional file 1: Table S1) were used to amplify the histidine-rich central repeat region of the hrp2 gene [10]. Samples positive for the hrp2 gene with this PCR were designated as P. falciparum with the hrp2 gene and no further PCR testing was performed.
Samples PCR-negative for the hrp2 gene were examined by PCR using primers for conserved regions of P. falciparum 18S rRNA [18, 19]. Samples positive in this PCR for Pf 18S rRNA were designated as P. falciparum without the hrp2 gene and no further testing was performed.
In contrast, samples negative for P. falciparum 18S rRNA were re-examined by PCR using primers for conserved regions of 18S rRNA in Plasmodium vivax, Plasmodium malariae and Plasmodium ovale [18, 19]. Samples positive using primers for 18S rRNA from P. vivax, P. malariae or P. ovale were designated non-P. falciparum malaria parasites and no further testing was performed. Samples negative in this PCR were considered negative for Plasmodium DNA and no further testing was performed.
The primers used are listed in Additional file 1 and a sample gel is presented in Additional file 2. Because the prevalence of non-falciparum Plasmodium parasites was greater than anticipated, amplification of 18S rRNA was changed from nested PCR described by Singh et al. [19] to multiplex PCR capable of detecting P. falciparum, P. vivax, P. malariae and P. ovale in a single-round of amplification to minimize the number of rounds of amplification required for species identification [18]. Primers targeting 18S rRNA have been reported to detect parasitemia at parasite densities as low as 1 parasite per microlitre [18, 20].
Statistical methods
Microsoft Excel was used for data entry and Stata (version 13) for analysis. The sensitivity, specificity and positive and negative predictive values of the HRP2 and pLDH RDTs were compared to microscopy (thick smears) which was used as the reference standard. RDT results were considered true positives or true negatives if they were concordant with microscopy. Negative RDT results were considered false-negatives if microscopy was positive. Positive RDTs were considered false-positives if microscopy was negative. Differences with a probability of less than 0.05 (P < 0.05) were accepted as significant.
Results
The flow of patients through the study is outlined in Fig. 3 and Table 1 provides a summary of baseline characteristics for the patients enrolled at each site. Of the 9226 patients screened, seven did not provide informed consent for their participation. Of the 9219 patients who consented to participate in the study, 462 were not included in the analysis because of incomplete test results (invalid or unrecorded RDT or microscopy results, other missing data or unreadable blood smears). Thus, 8757 sets of paired RDT and microscopy results for 8757 patients were included in the analysis (Fig. 3).
Fig. 3.
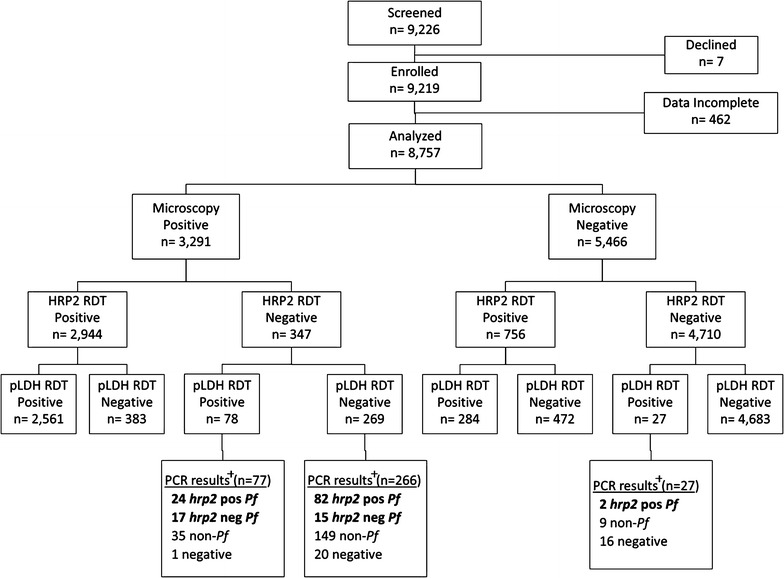
Laboratory results for patients enrolled at all three health centers. Samples positive by PCR targeting the hrp2 gene were labelled hrp2 pos Pf, samples negative by PCR targeting the hrp2 gene and positive by PCR targeting Pf 18S rRNA were labelled hrp2 neg Pf, samples negative by PCR targeting the hrp2 gene and Pf 18S rRNA but positive by PCR targeting Pv, Pm or Po 18S rRNA were labelled non-Pf and samples negative by PCR targeting the hrp2 gene, Pf 18S rRNA and Pv, Pm and Po 18S rRNA were labelled negative. Pf Plasmodium falciparum, Pv P. vivax, Pm P. malariae, Po P. ovale, RDT rapid diagnostic test, HRP2 histidine rich protein 2, pLDH Plasmodium lactate dehydrogenase, PCR polymerase chain reaction, Pf Plasmodium falciparum, non-Pf non-Plasmodium falciparum
Table 1.
Baseline characteristics
| Characteristic | Sites | ||
|---|---|---|---|
| Kibirizi n = 5148 |
Rukara n = 2696 |
Busogo n = 913 |
|
| Altitude (m) | 1718 | 1591 | 2247 |
| Average annual rainfall (mm) [36] | 81 | 75 | 129 |
| EIRa [15] | – | 20.9 | <1b |
| IRS with carbamates | Sep ‘14; Feb ‘15 | ||
| Dates of study | Apr ‘14–Apr ‘15 | Oct ‘14–Mar ‘15 | Nov ‘14–Apr ‘15 |
| Mean age (95% CI) | 22.3 (21.7–22.8) | 20.8 (20.1–21.5) | 20.5 (19.5–21.6) |
| Age <5 years, % (95% CI) | 19.4 (18.4–20.6) | 17.5 (16.2–19.0) | 22.3 (19.8–25.2) |
| SPR if age <5 years, % (95% CI) | 32.1 (29.1–35.1) | 41.6 (37.2–46.1) | 2.4 (0.3–4.6) |
| SPR if age ≥5 years, % (95% CI) | 36.0 (34.5–37.5) | 55.3 (53.2–57.4) | 11.6 (9.2–13.9) |
EIR entomological inoculation rate, IRS indoor residual spraying, CI confidence interval, SPR slide positivity rate
aEIR defined as the number of infective Anopheles mosquito bites per person per year
bData provided from closest sentinel site, Bungwe (no sentinel site in Busogo HC’s district)
Study sites
Patients were enrolled from three sites across Rwanda which vary in malaria endemicity. When the study sites were selected, malaria transmission was believed to be lowest in Busogo and highest in Kibirizi. The rationale for enrolling subjects in Kibirizi for 12 months was to determine whether RDT sensitivity varied seasonally. However, Kibirizi HC is within a district targeted for IRS by Rwanda’s MOPDD and two rounds of IRS were performed in Kibirizi during this study: in September 2014 and February 2015. Overall, the fractions of referred patients with positive thick smears for Plasmodium species were 10% (87/913) at Busogo, 35% (1778/5148) at Kibirizi and 53% (1426/2696) at Rukara. At the Kibirizi HC, the monthly slide positivity rate declined from 46% in April 2014 to 3% in April 2015 after two rounds of IRS (Fig. 4).
Fig. 4.
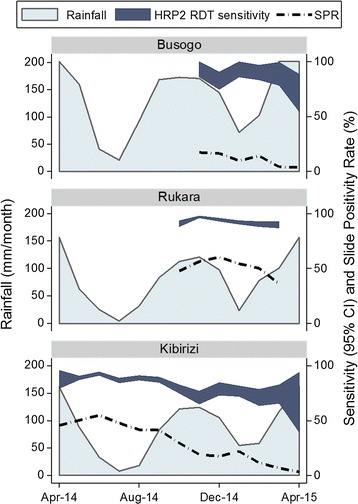
HRP2 RDT sensitivity, slide positivity rate and rainfall by study site and month. Rainfall is plotted in millimeters per month on the left y axis. Sensitivity and slide positivity rate are plotted on the right y axis. The navy band represents the 95% confidence interval for sensitivity. The dashed line represents the slide positivity rate. There was significant variation in HRP2 RDT sensitivity across the study sites and a decline in RDT sensitivity was noted following a decline in malaria transmission. In Rukara, monthly estimates of HRP2 RDT sensitivity ranged from 90 to 97%. Conversely, in Kibirizi, a site subject to two rounds of IRS during the study period, the sensitivity of the HRP2 RDT declined from 88 to 67% as the slide positivity rate fell from 46 to 3%. SPR slide positivity rate; CI confidence interval
Sensitivity and specificity: HRP2 RDT vs combined HRP2 + pLDH RDT
Among the 8757 patients for whom microscopy and RDT test results were available, 3291 (38%) were positive for Plasmodium species by thick smear. Among those specimens, the HRP2 RDT identified 2944 positives and the pLDH-based RDT identified 2639 positives. The overall sensitivities and specificities of the RDTs in comparison to microscopy were 89.5% (95% confidence interval [CI] 88.4–90.5) and 86.2% (95% CI 85.2–87.1) for the HRP2 RDT and 80.2% (95% CI 78.8–81.5) and 94.3% (95% CI 93.7–94.9) for the pLDH RDT, respectively. The pLDH RDT detected 78 microscopy-positive samples that were missed by the HRP2 RDT. Thus, when the pLDH RDT results were considered with the HRP2 RDT results, RDT sensitivity increased from 89.5 to 91.8% (95% CI 90.8–92.7) and there was no significant difference (P = 0.29) in RDT specificity. When the HRP2 and pLDH RDTs were considered together, RDT specificity was 85.7% (95% CI 84.7–86.6) (Table 2).
Table 2.
RDT sensitivity and specificity for the diagnosis of malaria compared to microscopy as the gold standard
| RDT target | Sensitivity, % (95% CI) | All sites | ||
|---|---|---|---|---|
| Kibirizi | Rukara | Busogo | ||
| HRP2 RDT | 86.2 (84.5–87.8) | 93.6 (92.2–94.8) | 87.4 (78.5–93.5) | 89.5 (88.9–90.0) |
| pLDH RDT | 81.4 (79.5–83.2) | 79.9 (77.8–82) | 59.8 (48.7–70.1) | 80.2 (79.5–80.9) |
| HRP2 + pLDH RDT | 90.5 (89.0–91.8) | 93.8 (92.4–95.0) | 87.4 (78.5–93.5) | 91.8 (91.3–92.3) |
| RDT target | Specificity, % (95% CI) | All sites | ||
|---|---|---|---|---|
| Kibirizi | Rukara | Busogo | ||
| HRP2 RDT | 84.7 (83.5–85.9) | 83.4 (81.2–85.4) | 96.2 (94.7–97.4) | 86.2 (85.7–86.6) |
| pLDH RDT | 92.7 (91.8–93.6) | 95.4 (94.1–96.5) | 99.0 (98.1–99.6) | 94.3 (94.0–94.6) |
| HRP2 + pLDH RDT | 84.0 (82.7–85.2) | 83.3 (81.1–85.3) | 96.2 (94.7–97.4) | 85.7 (85.2–86.1) |
CI confidence interval, RDT rapid diagnostic test, HRP2 histidine rich protein 2, pLDH Plasmodium lactate dehydrogenase
Variation in sensitivity of the HRP2 RDT and the slide positivity rate
Variations in the sensitivity of the HRP2 RDT were noted by site and month (Fig. 4). The sensitivity of the HRP2 RDT was greatest at Rukara (90–97%), the site with the highest fraction of positive blood smears (slide positivity rate). In contrast, at Kibirizi, when the smear positivity rate decreased from 55% in June 2014 to 3% in April 2015, the sensitivity of the HRP2 RDT fell from 93 to 67% (Chi square for trend = 37.2, P < 0.001). At Busogo, the monthly sensitivity of the HRP2 RDT ranged from 71 to 93% (Chi square for trend = 0.4, P = 0.522).
False-negative HRP2 RDTs and PCR for the hrp2 gene
PCR studies were performed for 343 of the 347 samples with false-negative HRP2 RDTs (Fig. 3). Of the 343 samples examined, Plasmodium DNA was detected using primers for the histidine-rich central repeat region of the hrp2 gene or conserved loci in Plasmodium 18S rRNA [10, 18, 19] in 322 (94%) of samples. Of these 322 samples, 138 (43%) were positive by PCR for P. falciparum DNA using primers for the histidine-rich central repeat region of the hrp2 gene or P. falciparum 18S rRNA and 184 (57%) were positive only by PCR for 18S rRNA from non-falciparum species. Of the 138 P. falciparum samples with false-negative RDTs, 106 (77%) were positive for the P. falciparum hrp2 gene and 32 (23%) were negative on PCR for the central repeat region of the hrp2 gene but positive for P. falciparum 18S rRNA (consistent with hrp2 deletion).
In Kibirizi, PCR for the hrp2 gene was negative for 26 of the 110 (24%) of the P. falciparum infections with false-negative RDTs and the proportion of hrp2-negative isolates did not increase as the slide positivity rate decreased (Fig. 5).
Fig. 5.
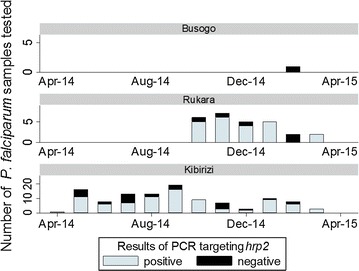
hrp2 PCR for P. falciparum isolates with false-negative HRP2 RDTs. Of the 138 P. falciparum samples with false-negative HRP2 RDTs, 106 were positive by PCR for the P. falciparum hrp2 gene (light blue bars) and 32 were negative by PCR for the central repeat region of the hrp2 gene (black bars). In Kibirizi, improved malaria control was not associated with an increased frequency of false-negative RDTs due to hrp2-negative P. falciparum isolates
pLDH RDT detection of samples with false-negative HRP2 RDTs
The pLDH RDT was positive for 41 of the 138 (30%) P. falciparum infections with false-negative HRP2 RDTs. Notably, the pLDH RDT was positive for the majority (53%) of the P. falciparum samples with false-negative HRP2 RDTs that were negative by PCR for the hrp2 gene.
pLDH RDT results for samples positive by microscopy
In Kibirizi, the proportion of microscopy positive samples that were negative by pLDH RDT increased as slide positivity decreased (Fig. 6). The proportion of microscopy positive and pLDH negative samples rose from 13.9% (95% CI 12.7–16.0) during April to August 2014 to 38.6% (95% CI 32.1–45.6) during December 2014 to April 2015.
Fig. 6.
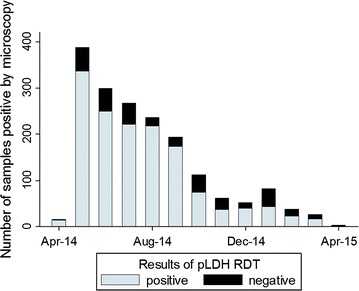
pLDH RDT results for microcopy positive samples in Kibirizi. Of the 1778 microscopy positive samples from Kibirizi, 1447 samples were positive by the pLDH RDT (light blue bars) and 331 samples were negative by the pLDH RDT (black bars). Improved malaria control was associated with an increased in the proportion of the microscopy positive samples that were negative by the pLDH RDT
pLDH RDT positive samples negative by microscopy and HRP2 RDT
PCR studies were also performed on 27 samples positive the pLDH RDT but negative by microscopy and the HRP2 RDT. Of these 27 samples, 2 were positive by PCR for hrp2 and 9 were positive only by PCR for 18S rRNA from non-falciparum species.
Discussion
In sub-Saharan Africa, HRP2 RDTs are the test used most commonly for parasitologic confirmation of malaria before treatment [5]. However, several reports have noted significant declines in the sensitivity of HRP2 RDTs after declines in the intensity of transmission [7–9]. In Mali, preliminary studies found seasonal declines in RDT sensitivity were associated with peaks in the prevalence of hrp2-negative P. falciparum isolates at the end of the dry season [8]. It is not clear if the association between declining RDT sensitivity and increasing prevalence of hrp2-negative isolates observed in Mali occurs elsewhere.
Thus, this study compared the sensitivity of HRP2 RDTs at 3 sites with varying intensities of transmission in Rwanda to determine whether deletions of hrp2 were responsible for false-negative HRP2-based RDTs. RDT performance was examined in relation to microscopy which was considered the gold standard. Samples with false-negative HRP2 RDTs (positive smear, negative RDT) were re-examined using PCR to test for the hrp2 gene.
Consistent with previous reports, this study found that HRP2-based RDTs were more sensitive than pLDH-based RDTs, although less specific [6, 7, 17]. However, when the HRP2 and pLDH RDTs were considered together, sensitivity increased slightly, without a decline in specificity. Using both RDTs, sensitivity was 91.8% and specificity was 85.7%.
Notably, the sensitivity of HRP2-based RDTs varied across the study sites and there was a decrease in the sensitivity of the HRP2 RDT after a fall in malaria transmission. In Rukara, monthly estimates of HRP2 RDT sensitivity ranged from 90 to 97%. Conversely, in Kibirizi, the sensitivity of the HRP2 RDT declined from 88 to 67% after two rounds of IRS as slide positivity rate for symptomatic patients fell from 46 to 3%.
Although IRS appeared to reduce the incidence of malaria in Kibirizi, a recent study in an area of The Gambia with high LLIN coverage found no additional benefit from adding IRS. Of note, in The Gambia, ≥93% LLIN coverage was achieved for all sleeping spaces and IRS coverage was 83–86%. [21]. In contrast, the 2014–2015 Rwandan DHS found that IRS achieved >98% coverage of targeted areas while only 81% of households had at least one LLIN and 43% of households had an LLIN for every two persons [16].
Potential explanations for false-negative HRP2 RDTs and the decline of RDT sensitivity in Kibirizi include: loss (deletion) of the hrp2 gene and low parasite densities [6, 10, 11].
Parasites lacking the hrp2 gene are a potential source of false-negative HRP2 RDTs. The hrp2 gene is absent in P. falciparum isolates with hrp2 gene deletions and in non-falciparum Plasmodium parasites. PCR analysis was used to identify isolates without the hrp2 gene and to confirm the presence P. falciparum DNA. Of 138 P. falciparum infections with false-negative HRP2 RDTs, 32 were negative by PCR for hrp2 (consistent with deletion of the hrp2 gene).
Plasmodium falciparum isolates lacking the hrp2 gene appear to be a significant source of false-negative RDTs in Rwanda. However, in this study, most P. falciparum isolates lacking the hrp2 gene were detected by the pLDH-based RDT and improved malaria control was not associated with an increased frequency of false-negative RDTs due to hrp2-negative P. falciparum isolates (Fig. 5).
For the majority (106/138 = 77%) of P. falciparum samples with false-negative HRP2 RDT results, PCR for hrp2 was positive. P. falciparum isolates containing the hrp2 gene may produce a false-negative RDT if the parasite density is below the threshold for RDT detection. Although this study lacks quantitative data on parasite density, a positive pLDH RDT may provide a crude estimate of the parasite density (≥200–1000) parasites per microlitre [22]. Because HRP2-based RDTs are more sensitive than pLDH-based RDTs at low parasite densities, a positive pLDH RDT suggests the parasite density was at or above the threshold for HRP2 RDT detection [6, 7, 23, 24]. Conversely, microscopy positive/pLDH negative P. falciparum samples may reflect low density infections.
In this study, P. falciparum infections with parasite densities below the threshold for detection may be responsible for many of the false-negative RDTs. Of the 106 P. falciparum isolates with false-negative HRP2 RDTs and the hrp2 gene (confirmed by PCR), most (77%) were negative by pLDH RDT. Additionally, in Kibirizi, the proportion of microscopy positive/pLDH RDT negative samples increased as the slide positivity rate fell (Fig. 6). The proportion of microscopy positive/pLDH negative samples rose from 13.9% (95% CI 12.7–16.0) during April to August 2014 to 38.6% (95% CI 32.1–45.6) during December 2014 to April 2015. This increase in the proportion of microscopy positive/pLDH negative samples may reflect an increase in the proportion of low density infections. Thus, the pLDH RDT data suggest that a decline in parasite density may have contributed to the decrease in HRP2 RDT sensitivity as malaria control improved in Kibirizi.
Other potential causes for false-negative RDTs which were not examined in this study include partial deletions of the hrp2 gene, prozone effects due to excess antigen, sequence variability of P. falciparum hrp2 and circulating antibodies to HRP2 which have been reported to interfere with RDT detection of HRP2 [11, 25–28]. While the primers used in this study amplified only exon 2 of the hrp2 gene, the hrp2 gene is also known to have chromosomal breaking points outside exon 2 [28].
There have been several reports of prozone-like effects with HRP2-based RDTs in patients with hyperparasitaemia. Although the mechanism of prozone-like effects for antigen detection tests is not well defined, one plausible explanation is that the amount of antigen may exceed the binding capacity of the dye-labelled antibodies used for antigen detection. In this situation, unlabelled target antigen reaches the test strip and saturates the binding capacity of the capture antibodies affixed to the test strip. As a result, antigen captured by dye-labelled antibodies may be unable to bind to the test strip to form a visible band [29]. However, false-negative HRP2 test lines attributed to the prozone effect have been described only in samples with ≥288,000 parasites/µL [30, 31]. While this study lacks data on parasite density, results of previous studies suggest that hyperparasitaemia is unlikely to have been a significant cause of false-negative RDTs in this study [31].
Of the 343 samples with false-negative HRP2 RDTs that were tested by PCR, 21 were negative by PCR for both hrp2 and 18S rRNA of the four Plasmodium parasites known to cause human infection. Sub-optimal PCR sensitivity may have occurred as a result of inadequate DNA sample, degradation of DNA sample, presence of PCR inhibitors and deletion or mutation of the targeted DNA [20]. Other possible explanations for these discrepancies include false-positive microscopy results and pLDH RDT cross-reactivity with other infectious agents, such as African trypanosomes [32].
Importantly, there is potential for confusion about the RDT used in this study because Premier Medical Corporation Ltd. submitted two different products with the name “First Response® Malaria Ag. pLDH/HRP2 Combo Card Test” to WHO for testing. The RDT used in this study, catalogue number I16FRC, was tested in rounds 1, 2 and 5. However, a different product was tested in round 6 (catalogue number PI16FRC). Product I16FRC did not meet WHO recommended procurement criteria during round 5 of WHO RDT lot testing because the panel detection score (PDS) for P. vivax at 200 parasites per microlitre was 74.5 (below the WHO criterion of ≥75). In contrast, product I16FRC had a satisfactory PDS score of 85.0 for P. falciparum (please note that PDS is not equivalent to sensitivity) [6]. These data are available at: http://www.rdt-interactive-guide.org/ [33]. Because P. falciparum is the predominant species in Rwanda, the marginally low sensitivity of this RDT for P. vivax would not be expected to have a significant impact of the finding of this study [5].
Finally, the authors recognize the lack of testing for additional single-copy genes is a theoretical limitation because the reported sensitivity of PCR is greater for multi-copy genes (18S rRNA) than single copy genes (hrp2) (1 vs 10–100 parasite per microlitre) [34]. However, several factors suggest the parasite densities of the P. falciparum samples that were negative by PCR for hrp2 were above the threshold for detection by PCR for single-copy genes: all patients had symptoms consistent with malaria infection, DNA was extracted within 3 months of sample collection and most P. falciparum isolates without the hrp2 gene were detected by the pLDH RDT. Most symptomatic individuals in malaria-endemic areas have parasite densities ≥1000 parasites per microlitre [35] and prompt extraction of DNA limits the time for DNA degradation which may disproportionately reduce the sensitivity of PCR for single-copy genes. Additionally, based on the results from round 5 of WHO RDT quality testing, a positive pLDH RDT suggests the parasite density was above the threshold for detection by PCR for single-copy genes (≥200 parasites per microlitre). The pLDH RDT of the First Response® Malaria pLDH/HRP2 Combo test (catalogue number I16FRC) had a sensitivity of 31% for wild-type (clinical) P. falciparum smear-positive samples with 200 parasites per microlitre. In contrast, with parasite densities of 2000 per microlitre, the sensitivity of the pLDH RDT was 100% [22].
Ultimately, the factors driving the decline in RDT sensitivity as malaria control improves are not clear. If parasite density declines as malaria control improves, the decrease in RDT sensitivity could be driven in part by an increase in the number of infections with parasite densities below the RDT threshold for detection. In addition, there are concerns that hrp2-negative parasites may have an increased impact on RDT performance as malaria control improves [10, 11]. Conversely, in high transmission settings, hrp2 negative parasites may have less impact on RDT sensitivity because individuals are commonly infected with more than one P. falciparum parasite strain (genotype) and the RDT will yield a true-positive result if any one parasite (genotype) is hrp2-positive [10]. However, the number of parasite genotypes infecting individual subjects, the multiplicity of infection (MOI), declines as malaria control improves. Conversely, individuals infected by only a single parasite genotype negative for the hrp2 gene will produce false-negative results when tested with an HRP2-based RDT [10]. In the present study, the decrease in HRP2 RDT sensitivity as malaria control improved was not associated with an increased frequency of hrp2-negative P. falciparum isolates. However, it may have been associated with an increase in the frequency of low density infections.
Conclusions
This study provides new information on the performance of RDTs in Rwanda and supports previously raised concerns that the sensitivity of HRP2 RDTs may decline as malaria control improves. In addition, the results of this study suggest that the use of three-band RDTs (HRP2, pLDH and control bands) may improve sensitivity without decreasing specificity. This study also found that P. falciparum isolates lacking the hrp2 gene are an important source of false-negative HRP2 RDTs in Rwanda. Further investigations are warranted to better define the prevalence of these isolates and the factors responsible for the decline in RDT sensitivity as malaria control improves.
Authors’ contributions
CTK, NU, SR, CK and DK contributed substantially to the conception and design of this study. CTK and NU coordinated and supervised field work. CTK, EM and JPH supervised and performed the laboratory analysis. CTK and DK performed the data analysis and prepared the first draft of the manuscript. All authors read and approved the final manuscript.
Acknowledgements
The authors thank all the patients, caretakers and health centre staff for contributing their time to participate in the study. The authors are also grateful to Mr. Kamana Gatera Emmanuel, Mr. Prosper Gahamanyi and Ms. Vestine Uwimana for their assistance with microscopy, Mr. Tharcisse Munyaneza and Mr. Jean Bosco Mucaca for their assistance with laboratory training, microscopy quality assurance and PCR, Mr. Guillaume Karangwa for his assistance with data entry and management and Dr. Eric Tongren for his guidance on study design and implementation.
Competing interests
The authors declare that they have no competing interests.
Availability of data and materials
Data are available from the corresponding author upon request.
Ethics, consent and permissions
Ethical approvals for this study were obtained from the Rwanda National Ethics Committee (FWA 00001973, IRB 00001497) and the Tulane University Institutional Review Board (FWA 00002055, IRB 00000324). An informed consent discussion was held with all potential subjects or their guardians and individual verbal consent was obtained prior to enrollment. For children less than 18 years of age, consent was provided by a parent or guardian. For children older than 7 years of age, individual verbal assent was also obtained. A written statement about this study was provided to all subjects in the local language, Kinyarwanda and was also available in English. Based on Rwandan Ministry of Health guidelines, all patients infected with malaria parasites were offered treatment with artemether–lumefantrine. Study documents and blood samples are stored at the National Reference Laboratory. Consistent with Health and Human Services regulation 45 CFR 46.117(c), the requirement for written consent was waived by the Rwanda and Tulane IRBs for this study because the research presented no more than minimal risk to the subjects. Written consent was not sought because the only record linking the subject and the research would be the consent document and the principal risk would be the potential harm resulting from a breach of confidentiality.
Funding
This work was supported by the Rwanda Ministry of Health through the Malaria & Other Parasitic Diseases Division and by a Global Health Fellowship Award to CK supported by the Fogarty International Center, Office of AIDS Research, National Cancer Center, National Heart, Blood, and Lung Institute, and the NIH Office of Research for Women’s Health through the Fogarty Global Health Fellows Program Consortium comprised of the University of North Carolina, John Hopkins, Morehouse and Tulane (1R25TW009340-01).
Abbreviations
- RDT
rapid diagnostic test
- WHO
World Health Organization
- HRP2
histidine-rich protein 2
- pLDH
Plasmodium lactate dehydrogenase
- HC
health centre
- LLINs
long-lasting insecticidal nets
- IRS
indoor residual spraying
- DHS
Demographic and Health Survey
- MOPDD
Malaria and Other Parasitic Diseases Division
- PCR
polymerase chain reaction
- CI
confidence interval
Additional files
Additional file 1. Primer sequences used to amplify hrp2 and P. falciparum, P. vivax, P. malariae and P. ovale 18S rRNA, expected product sizes and PCR conditions.
Additional file 2. PCR products visualized on an agarose gel. DNA is from two thick-smear positive subjects with negative HRP2-based RDTs. Lane 1: 100 bp marker. Lane 2: PCR targeting hrp2 with DNA from subject #1 (shows amplicon of expected size of ~900 bp). Lanes 3–5: PCR with DNA from subject #2. Lane 3 shows the absence of hrp2 amplicons. Lane 4 shows results of multiplex PCR for 18S rRNA with an amplicon of 276 bp (expected size for P. falciparum). Lane 5 shows results of nested PCR with species-specific primers for P. falciparum 18S rRNA with an amplicon of the expected size of 205 bp.
Contributor Information
Christina T. Kozycki, Email: christina.kozycki@gmail.com
Noella Umulisa, Email: numulisa@gmail.com.
Stephen Rulisa, Email: s.rulisa@gmail.com.
Emil I. Mwikarago, Email: emil.ivank@gmail.com
Jean Pierre Musabyimana, Email: musajampiereb@gmail.com.
Jean Pierre Habimana, Email: habimanajp@gmail.com.
Corine Karema, Email: ckarema@gmail.com.
Donald J. Krogstad, Email: krogstad@tulane.edu
References
- 1.WHO. Guidelines for the treatment of malaria. Geneva: World Health Organization; 2010. http://apps.who.int/iris/bitstream/10665/162441/1/9789241549127_eng.pdf. Accessed 21 Feb 2017.
- 2.Ochola LB, Vounatsou P, Smith T, Mabaso MLH, Newton C. The reliability of diagnostic techniques in the diagnosis and management of malaria in the absence of a gold standard. Lancet Infect Dis. 2006;6:582–588. doi: 10.1016/S1473-3099(06)70579-5. [DOI] [PubMed] [Google Scholar]
- 3.WHO. World malaria report 2015. Geneva: World Health Organization; 2015. http://www.who.int/malaria/publications/world-malaria-report-2015/report/en/. Accessed 21 Feb 2017.
- 4.Kahama-Maro J, D’Acremont V, Mtasiwa D, Genton B, Lengeler C. Low quality of routine microscopy for malaria at different levels of the health system in Dar es Salaam. Malar J. 2011;10:332. doi: 10.1186/1475-2875-10-332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.WHO. Universal access to malaria diagnostic testing—an operational manual. Geneva: World Health Organization; 2013. http://www.who.int/malaria/publications/atoz/9789241502092/en/. Accessed 21 Feb 2017.
- 6.WHO. Summary results of WHO product testing of malaria RDTs: rounds 1–6 (2008–2015). Geneva: World Health Organization; 2015. http://apps.who.int/iris/bitstream/10665/204119/1/9789241510042_eng.pdf. Accessed 21 Feb 2017.
- 7.Abba K, Deeks JJ, Olliaro P, Naing CM, Jackson SM, Takwoingi Y, et al. Rapid diagnostic tests for diagnosing uncomplicated P. falciparum malaria in endemic countries. Cochrane Database Syst Rev. 2011;7:CD008122. doi: 10.1002/14651858.CD008122.pub2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Koita OA, Ndiaye J, Nwakanma D, Sangare L, Ndiaye D, Joof F, et al. Seasonal changes in the frequency of false negative rapid diagnostic tests based on histidine rich protein 2 (HRP2) Am J Trop Med Hyg. 2013;80(Suppl 5):1. [Google Scholar]
- 9.Shakely D, Elfving K, Aydin-Schmidt B, Msellem MI, Morris U, Omar R, et al. The usefulness of rapid diagnostic tests in the new context of low malaria transmission in Zanzibar. PLoS ONE. 2013;8:e72912. doi: 10.1371/journal.pone.0072912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Koita OA, Doumbo OK, Ouattara A, Tall LK, Konare A, Diakite M, et al. False-negative rapid diagnostic tests for malaria and deletion of the histidine-rich repeat region of the hrp2 gene. Am J Trop Med Hyg. 2012;86:194–198. doi: 10.4269/ajtmh.2012.10-0665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gamboa D, Ho MF, Bendezu J, Torres K, Chiodini PL, Barnwell JW, et al. A large proportion of P. falciparum isolates in the Amazon region of Peru lack pfhrp2 and pfhrp3: implications for malaria rapid diagnostic tests. PLoS ONE. 2010;5:e8091. doi: 10.1371/journal.pone.0008091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kumar N, Pande V, Bhatt RM, Shah NK, Mishra N, Srivastava B, et al. Genetic deletion of HRP2 and HRP3 in Indian Plasmodium falciparum population and false negative malaria rapid diagnostic test. Acta Trop. 2013;125:119–121. doi: 10.1016/j.actatropica.2012.09.015. [DOI] [PubMed] [Google Scholar]
- 13.Parr JB, Verity R, Doctor SM, Janko M, Carey-Ewend K, Turman BJ, et al. Pfhrp2-deleted Plasmodium falciparum parasites in the Democratic Republic of Congo: a national cross-sectional survey. J Infect Dis. 2016 doi: 10.1093/infdis/jiw538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Government of Rwanda Ministry of Health: In: Health Management Information Systems. 2013 (Unpublished raw data).
- 15.Malaria & Other Parasitic Diseases Division. Annual vector control report: July 2013–June 2014. Rwanda Biomedical Center; 2014. http://www.moh.gov.rw/fileadmin/templates/MOH-Reports/RwandaMalariaVector_control_annual_report_2013_2014__1_.pdf. Accessed 21 Feb 2017.
- 16.National Institute of Statistics of Rwanda MoHR, and ICF International. Rwanda Demographic and Health Survey 2014–15. 2016. http://dhsprogram.com/publications/publication-FR316-DHS-Final-Reports.cfm. Accessed 21 Feb 2017.
- 17.Hopkins H, Bebell L, Kambale W, Dokomajilar C, Rosenthal PJ, Dorsey G. Rapid diagnostic tests for malaria at sites of varying transmission intensity in Uganda. J Infect Dis. 2008;197:510–518. doi: 10.1086/526502. [DOI] [PubMed] [Google Scholar]
- 18.Padley D, Moody AH, Chiodini PL, Saldanha J. Use of a rapid, single-round, multiplex PCR to detect malarial parasites and identify the species present. Ann Trop Med Parasitol. 2003;97:131–137. doi: 10.1179/000349803125002977. [DOI] [PubMed] [Google Scholar]
- 19.Singh B, Bobogare A, Cox-Singh J, Snounou G, Abdullah MS, Rahman HA. A genus- and species-specific nested polymerase chain reaction malaria detection assay for epidemiologic studies. Am J Trop Med Hyg. 1999;60:687–692. doi: 10.4269/ajtmh.1999.60.687. [DOI] [PubMed] [Google Scholar]
- 20.Mixson-Hayden T, Lucchi NW, Udhayakumar V. Evaluation of three PCR-based diagnostic assays for detecting mixed Plasmodium infection. BMC Res Notes. 2010;3:88. doi: 10.1186/1756-0500-3-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pinder M, Jawara M, Jarju LB, Salami K, Jeffries D, Adiamoh M, et al. Efficacy of indoor residual spraying with dichlorodiphenyltrichloroethane against malaria in Gambian communities with high usage of long-lasting insecticidal mosquito nets: a cluster-randomised controlled trial. Lancet. 2015;385:1436–1446. doi: 10.1016/S0140-6736(14)61007-2. [DOI] [PubMed] [Google Scholar]
- 22.WHO. Malaria rapid diagnostic test performance: results of WHO product testing of malaria RDTs: round 5 (2013). Geneva: World Health Organization; 2014. http://apps.who.int/iris/bitstream/10665/128678/1/9789241507554_eng.pdf. Accessed 21 Feb 2017.
- 23.Gatton ML, Rees-Channer RR, Glenn J, Barnwell JW, Cheng Q, Chiodini PL, et al. Pan-Plasmodium band sensitivity for Plasmodium falciparum detection in combination malaria rapid diagnostic tests and implications for clinical management. Malar J. 2015;14:115. doi: 10.1186/s12936-015-0629-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Heutmekers M, Gillet P, Cnops L, Bottieau E, Van Esbroeck M, Maltha J, et al. Evaluation of the malaria rapid diagnostic test SDFK90: detection of both PfHRP2 and Pf-pLDH. Malar J. 2012;11:359. doi: 10.1186/1475-2875-11-359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee N, Baker J, Andrews KT, Gatton ML, Bell D, Cheng Q, et al. Effect of sequence variation in Plasmodium falciparum histidine-rich protein 2 on binding of specific monoclonal antibodies: implications for rapid diagnostic tests for malaria. J Clin Microbiol. 2006;44:2773–2778. doi: 10.1128/JCM.02557-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ho MF, Baker J, Lee N, Luchavez J, Ariey F, Nhem S, et al. Circulating antibodies against Plasmodium falciparum histidine-rich proteins 2 interfere with antigen detection by rapid diagnostic tests. Malar J. 2014;13:480. doi: 10.1186/1475-2875-13-480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gillet P, Mori M, Van Esbroeck M, Van den Ende J, Jacobs J. Assessment of the prozone effect in malaria rapid diagnostic tests. Malar J. 2009;8:271. doi: 10.1186/1475-2875-8-271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cheng Q, Gatton ML, Barnwell J, Chiodini P, McCarthy J, Bell D, et al. Plasmodium falciparum parasites lacking histidine-rich protein 2 and 3: a review and recommendations for accurate reporting. Malar J. 2014;13:283. doi: 10.1186/1475-2875-13-283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Luchavez J, Baker J, Alcantara S, Belizario V, Jr, Cheng Q, McCarthy JS, et al. Laboratory demonstration of a prozone-like effect in HRP2-detecting malaria rapid diagnostic tests: implications for clinical management. Malar J. 2011;10:286. doi: 10.1186/1475-2875-10-286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gerstl S, Dunkley S, Mukhtar A, De Smet M, Baker S, Maikere J. Assessment of two malaria rapid diagnostic tests in children under five years of age, with follow-up of false-positive pLDH test results, in a hyperendemic falciparum malaria area, Sierra Leone. Malar J. 2010;9:28. doi: 10.1186/1475-2875-9-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gillet P, Scheirlinck A, Stokx J, De Weggheleire A, Chauque HS, Canhanga OD, et al. Prozone in malaria rapid diagnostics tests: how many cases are missed? Malar J. 2011;10:166. doi: 10.1186/1475-2875-10-166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gillet P, Mumba Ngoyi D, Lukuka A, Kande V, Atua B, van Griensven J, et al. False positivity of non-targeted infections in malaria rapid diagnostic tests: the case of human African Trypanosomiasis. PLOS Negl Trop Dis. 2013;7:e2180. doi: 10.1371/journal.pntd.0002180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.FIND. Malaria RDT product testing: interactive guide. 2016. http://www.rdt-interactive-guide.org/. Accessed 21 Feb 2017.
- 34.Bell DR, Wilson DW, Martin LB. False-positive results of a Plasmodium falciparum histidine-rich protein 2—detecting malaria rapid diagnostic test due to high sensitivity in a community with fluctuating low parasite density. Am J Trop Med Hyg. 2005;73:199–203. [PubMed] [Google Scholar]
- 35.Nwakanma DC, Gomez-Escobar N, Walther M, Crozier S, Dubovsky F, Malkin E, et al. Quantitative detection of Plasmodium falciparum DNA in saliva, blood, and urine. J Infect Dis. 2009;199:1567–1574. doi: 10.1086/598856. [DOI] [PubMed] [Google Scholar]
- 36.Rwanda Meteorology Agency . Rainfall by monitoring station, 2010–2014. Kigali: Rwanda Meteorology Agency; 2015. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data are available from the corresponding author upon request.


