Abstract
Various proteoforms may be generated from a single gene due to primary structure alterations (PSAs) such as genetic variations, alternative splicing, and post-translational modifications (PTMs). Top-down mass spectrometry is capable of analyzing intact proteins and identifying patterns of multiple PSAs, making it the method of choice for studying complex proteoforms. In top-down proteomics, proteoform identification is often performed by searching tandem mass spectra against a protein sequence database that contains only one reference protein sequence for each gene or transcript variant in a proteome. Because of the incompleteness of the protein database, an identified proteoform may contain unknown PSAs compared with the reference sequence. Proteoform characterization is to identify and localize PSAs in a proteoform. Although many software tools have been proposed for proteoform identification by top-down mass spectrometry, the characterization of proteoforms in identified proteoform-spectrum-matches still relies mainly on manual annotation. We propose to use the Modification Identification Score (MIScore), which is based on Bayesian models, to automatically identify and localize PTMs in proteoforms. Experiments showed that the MIScore is accurate in identifying and localizing one or two modifications.
Introduction
The expression of a gene may result in many proteoforms,1 which often contain some primary structure alterations (PSAs), such as amino acid substitutions, insertions/deletions of amino acids or exons, and post-translational modifications (PTMs), compared with the reference protein sequence in the Swiss-Prot2 or RefSeq3 database. Because many PSAs alter protein structure, function, and protein-protein interactions, they play a vital role in biological processes and are closely related to many diseases such as heart failure4 and age-dependent memory impairment.5 Researchers have been actively developing experimental and computational methods for identifying proteoforms with PSAs.6
Bottom-up mass spectrometry (MS) has dominated proteomics studies for more than two decades. However, protein digestion in bottom-up MS cleaves long proteins into short peptides, limiting its ability to identify the combinatorial pattern of multiple PSAs in a complex proteoform.7 In addition, only a fraction of peptides can be confidently identified, and the PSAs on those unidentified peptides cannot be observed. By contrast, top-down MS analyzes intact proteins and provides whole protein sequence coverage, making it the method of choice for studying complex proteoforms with PSAs. Over the past five years, high accuracy and high resolution mass spectrometers (e.g., Orbitrap), which are required for top-down MS, have become available to many laboratories. Developments in protein separation and MS instrumentation have boosted the applications of top-down MS, which open a window into the poorly explored world of proteoforms.1
There are three main approaches to identifying proteoforms by top-down MS: extended databases, blind PSA search, and the combination of the first two. In the first approach, an extended proteoform database is constructed that includes all known proteoforms, against which tandem mass (MS/MS) spectra are searched. The second approach is similar to blind PTM search in bottom-up MS, in which MS/MS spectra are searched against an ordinary protein database, such as a Swiss-Prot protein database, to identify proteoforms with unknown PSAs. These two approaches can be combined, that is, MS/MS spectra are searched against an extended proteoform database to identify proteoforms with known and/or unknown PSAs. ProteinGoggle8 and the absolute mass search mode of ProSightPC9 exemplify the first approach. Various methods have been proposed using the second approach, such as spectral alignment,10,11 precursor ion independent search (PIITA),12 and tag-based methods. 13 ProSightPC provides the Δm and biomarker search modes that are based on the third approach. MS-Align-E14 is another example of the third approach, which is capable of identifying proteoforms with both variable and unknown PTMs. Although the first approach is fast, it often misses many identifications because of the existence of unknown PSAs. As a result, the second and third approaches are more efficient in exploring the world of unknown complex proteoforms.
In the third approach, the objective is to map a top-down MS/MS spectrum to a proteoform of the target gene in the database that shares the maximum number of PSAs with the target proteoform. PSAs shared by the database and target proteoforms are known PSAs; those in the target proteoform only are unknown or novel PSAs. Although proteoform characterization, which identifies and localizes PSAs, is an indispensable step in top-down MS data analysis, existing proteoform identification tools often report only the database proteoform, but fail to characterize the target proteoform.
In bottom-up MS, many methods have been proposed for the automated identification and localization of PTMs, particularly for the localization of phosphorylation, such as A-score, 15 PTM score,16 Phosphorylation Localization Score,17 SLoMo,18 PhosphoRS19 and Mascot Delta Score.20 After a mass shift in a peptide-spectrum match is identified, these methods identify the PTM based on the mass shift and compute a confidence score for each possible site of the PTM.21 In addition, there are methods that refine predicted PTMs and their locations, such as PTMFinder22 and iPTMClust.23 However, the methods have some limitations: PTMFinder uses a peptide-level approach, which favours modified peptides with high-abundance; iPTMClust cannot handle peptides with multiple PTMs.
In top-down MS, software tools such as ProSightPC9 provide graphical user interfaces for manually characterizing complex proteoforms, but they are inefficient in analyzing high throughput data. Software tools for automated characterization of proteoforms are still lacking.
Dang et al. described three types of confidence scores in proteoform identification and characterization by top-down MS:24 protein identification scores, PTM localization scores, and proteoform characterization scores. The last two are used in proteoform characterization. The methods for PTM localization on peptides, such as A-score, can be extended to compute PTM localization scores in proteoform characterization. However, most of the methods were designed for single PTM localization, not for the characterization of complex proteoforms with multiple PSAs. LeDuc et al.25 proposed a Bayesian approach for proteoform identification, in which C-scores are computed for candidate proteoforms in an extended proteoform database. C-scores are proteoform characterization scores when the target proteoform does not contain unknown PSAs and the candidate proteoforms are limited to those in the extended database.
We limit this study to the identification and localization of PTMs in proteoforms and use Bayesian models to compute the Modification Identification Score (MIScore), which is a PTM level, not proteoform level, score. While a PTM localization score is the confidence score of a potential site of a given PTM; an MIScore is the probability that the reported modification and site are correct. The computation of posterior probabilities in the proposed models is simpler and faster than that in the C-score method. We give efficient algorithms for computing MIScores as well as a divide and conquer method for the localization of two modifications. One limitation of the MIScore method is that it can identify at most 2 modifications from an unknown mass shift. Experiments showed that the MIScore method was accurate in identifying and localizing modifications in proteoforms.
Methods
Data sets
The MIScore method was tested on two top-down MS/MS data sets: one from Escherichia coli K-12 MG1655 (EC) and the other from Salmonella typhimurium 14028s (ST). In addition, a Salmonella typhimurium 14028s bottom-up MS/MS data set was used for the validation of identified modification sites.
EC data set
Escherichia coli K-12 MG1655 was grown in M9 minimal medium at 37°C with shaking. Cells were harvested at OD600 of 0.6 by centrifugation (2 400 g, 15 min) at 4°C, and washed with ammonium bicarbonate buffer (100 mM, pH 8). Cell pellets (1.5 g, wet weight) were reconstituted in the ammonium bicarbonate buffer plus 1 mM PMSF. The suspension was lysed with bead beating (0.1 mm Zirconia beads) at the maximum speed for 3 min. The cell debris and beads were removed by centrifugation (10 000 g, 5 min). The supernatant represented the soluble protein extract. No reduction and alkylation of cysteine residues were performed in the sample preparation. The protein extract was separated by a Waters NanoAquity LC system with a custom packed column (80 cm × 75 μm i.d., C5, 5 μm particle diameter, 300 Å pore size). Mobile phase A was composed of 0.5% acetic acid, 0.01% TFA, 5% isopropanol, 10% ACN, and 84.5% water. Mobile phase B consisted of 0.5% acetic acid, 0.01% TFA, 9.9% water, 45% isopropanol, and 45% ACN. The operating flow rate was 0.3 μl/min. The LC system was equilibrated with 100% mobile phase A for 5 minutes, and then increased to 20% mobile phase B in 1 minute. A 250 minute linear gradient was set from 20% mobile phase B to 55% mobile phase B. All the related MS analysis was performed using an LTQ Orbitrap Velos mass spectrometer (Thermo Scientific, San Jose, CA). FTMS MS and MSn AGC target values were 106 and 2 × 105, respectively. For the LC-MS/MS analysis with higher-energy C-trap dissociation (HCD) fragmentation, a parent spectrum was collected at a 60K resolution at m/z of 400 and was followed by high resolution (60K at m/z of 400) HCD MS/MS spectra of the 6 most intense ions, isolated with a 3 m/z window, from the parent mass spectrum. FT MS/MS employed 45% normalized collision energy for HCD. Mass calibration was performed prior to analysis according to the method recommended by the instrument manufacturer. A total of 3 704 HCD MS/MS spectra were collected.
ST data set
Cultures of Salmonella typhimurium 14028s were grown in low-phosphate, low-magnesium, low-pH minimal medium (LPM) for infection-like condition. Protein samples of the cultures were collected and divided into two portions: one for top-down MS/MS analysis and the other for bottom-up MS/MS analysis. No reduction and alkylation of cysteine residues were performed in the preparation of the samples. In the top-down MS/MS experiment, the protein samples were separated by a reversed phase liquid chromatography (RPLC) system and then analyzed by an LTQ Orbitrap Velos mass spectrometer. The most 8 intense ions in each MS spectrum were selected to generate high resolution (60K) collision-induced dissociation (CID) MS/MS spectra. In the bottom-up MS/MS experiment, the protein samples were digested using trypsin and analyzed by an high-performance liquid chromatography (HPLC) system coupled with an LTQ Orbitrap Velos mass spectrometer. The 6 most intensity ions in each MS spectrum were selected for CID MS/MS analysis. Finally, a total of 7 400 top-down and 106 350 bottom-up MS/MS were collected. (See Ref 26 for details of the experiments.)
Binary representation of peptides and spectra
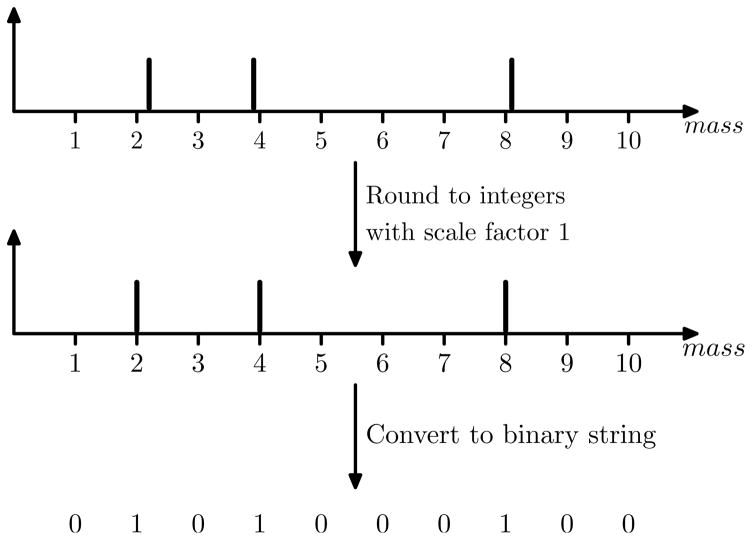
An MS/MS spectrum is represented by a precursor mass and a list of peaks. The precursor mass corresponds to the molecular mass of the proteoform, and each peak (m/z, intensity) corresponds to a fragment ion of the proteoform. The m/z value and intensity are the mass-to-charge ratio and abundance of the fragment ion, respectively. In preprocessing of top-down spectra, m/z values are converted into neutral masses of fragment ions by a deconvolution algorithm.27–29 The neutral masses and the precursor mass are discretized by multiplying the masses by a scale factor and rounding the resulting values to integers. A scale factor 274.335215 was used in the experiments.14 In practice, the scale factor is determined by the accuracy of m/z values in top-down MS/MS spectra. For simplicity, peak intensities are ignored in the following description of the method.
Let M be the discretized precursor mass of an MS/MS spectrum S. We represent spectrum S as a binary string s1s2 … sM, where sj = 1 if j is a discretized neutral fragment mass in S; and sj = 0, otherwise (Figure 1). Let F be a proteoform matched to spectrum S. The molecular mass of F equals M (within an error tolerance), and the proteoform F is represented as a binary string f1f2 … fM, where fj = 1 if j is the discretized neutral mass of a theoretical fragment ion of F; and fj = 0, otherwise. For example, when only b- and y-ions are used in the generation of theoretical spectra, a proteoform AGR (without modifications) has four theoretical neutral fragment ions (b1, b2, y1 and y2) whose masses are 71.04, 128.06, 174.11, 231.13 Dalton (Da). In addition, the molecular mass of the proteoform is 302.17 Da. After discretization with a scale factor 1, the integer molecular mass is 302 and the four integer neutral fragment masses are 71, 128, 174, 231. The protein AGR is represented by the following binary string:
Figure 1.
Illustration of the conversion from a deconvoluted spectrum of neutral masses to a binary string. A spectrum (top) has three neutral fragment masses 2.2, 3.9, and 8.1 Da (peak intensities are ignored), and its precursor mass is 10.1 Da. The precursor and fragment masses are discretized by multiplying by a scale factor 1 and rounding to integers, resulting in a spectrum with a precursor mass 10 and three fragment masses 2, 4 and 8. The discretized spectrum is converted to a binary string 0101000100. The length of the string is the same to the integer precursor mass; the three 1s correspond to the three fragment masses.
The shared mass count of S and F is the number of matched 1s in the binary strings of S and F, denoted as Score(S, F). When the precursor masses of S and F do not match, Score(S, F) = −∞. Notations in this paper are summarized in Table 1.
Table 1.
Symbol definitions
| Symbol | Definition |
|---|---|
| S | A top-down tandem mass spectrum |
| Score(S, F) | The shared mass count between spectrum S and a proteoform F. |
| P | The unmodified protein sequence of the target proteoform with length m |
| Fi | The proteoform of P in which the ith amino acid is modified. The molecular mass of Fi matches the precursor mass of S. |
| Pr(Fi) | The prior probability of proteoform Fi |
| Pr(S) | The probability of the data (spectrum). In Bayesian models, it is usually computed as the sum of the prior probabilities of all hypotheses multiplied by their likelihoods. |
| Pr(S|Fi) | The likelihood, the conditional probability of observing S given Fi |
| Pr(Fi|S) | The posterior probability, the probability for Fi after taking into account S |
| Ti,j | The proteoform of P in which the first i amino acids are truncated and the jth amino acid is modified. The molecular mass of Ti,j matches the precursor mass of S. |
| 𝒬i | The set of proteoforms of P with a pair of ordered modifications (xi, yi) |
| 𝒬ij | The set of proteoforms satisfying that the first modification xi occurs on the first j amino acids and the second modification yi on the last m − j amino acids |
| ℱ1 | The set of all proteoforms of P with one common modification whose molecular masses match the precursor mass of S |
| ℱ2 | The set of all proteoforms of P with two common modifications whose precursor masses match the precursor mass of S |
Single modifications
When a top-down MS/MS spectrum is matched to a proteoform in the database and the target proteoform contains unknown modifications compared with the database proteoform, the resulting proteoform-spectrum match (PrSM) (between the database proteoform and the spectrum) contains some mass shifts identified based on matched theoretical and experimental fragment masses.11 When the target proteoform contains one unknown modification (and no other types of PSAs) and one mass shift is reported in the PrSM, the mass of the modification equals (within an error tolerance) the mass shift. Because the type of the modification can be generally determined by the mass shift, the remaining task is to find the location of the modification. Following the approach proposed in Ref 25, we use a Bayesian model to compute the confidence score for each candidate site of the modification, that is, the probability that the modification is on the site. For simplicity, we use the following assumptions: (a) the database proteoform is an unmodified protein, (b) the target proteoform is not truncated, and (c) the modification can occur on any amino acid of the protein.
Suppose a top-down MS/MS spectrum S is generated from a proteoform containing m amino acids and a modification. Let P be the unmodified protein sequence of the target proteoform, and F1, F2, … , Fm all possible modified proteoforms of P with the modification. The modification in Fi is on the ith amino acid. By Bayes’ theorem,
where Pr(Fi|S) is the posterior probability for proteoform Fi given spectrum S, Pr(S|Fi) is the conditional probability of observing spectrum S given proteoform Fi, and Pr(S) is the probability of the data S (Table 1). The probability Pr(S) is computed as the sum of the prior probabilities Pr(Fj) multiplied by their likelihoods Pr(S|Fj). In practice, the uniform distribution is used for the prior probability of each proteoform, that is, Pr(Fj) = 1/m for j = 1, 2, … , m.
Below we describe how to obtain the values of Pr(S|Fi) for i = 1, 2, … , m, which are needed for computing the confidence scores of candidate sites. Let X0 be a random variable that represents if a mass that does not match any theoretical fragment masses of a protein in a given proteome database is observed in a top-down MS/MS spectrum of the protein. Let X1 be a random variable that represents if a theoretical fragment mass of a protein in a given proteome database is observed in a top-down MS/MS spectrum of the protein. The random variable (X0 or X1) equals 1 if the mass is observed; otherwise, 0. A matched pair (sj, fj) in the binary strings of S = s1s2 … sM and Fi = f1f2 … fM has four possible values (0, 0), (0, 1), (1, 0), and (1, 1). Let z00, z01, z10, z11 be the numbers of (0, 0), (0, 1), (1, 0), and (1, 1) pairs in the binary strings, respectively. For example, the numbers of (0, 0), (0, 1), (1, 0) and (1, 1) pairs in S = 0101000100 and Fi = 0101001000 are 6, 1, 1, and 2, respectively. That is, z00 = 6, z01 = 1, z10 = 1 and z11 = 2. The number z11 is the same as Score(S, Fi), the shared mass count between S and Fi. By assuming the values in s1s2 … sM and those in f1f2 … fM are independent, the likelihood is computed as follows:
| (1) |
where z00, z01, z10 and z11 are exponents.
To simplify the analysis, we assume that only two types of fragment ions (one is N-terminal and the other is C-terminal) are used for generating the binary string of Fi, and that all neutral fragment masses are distinct. As a result, the number of 1s in the binary string of Fi is 2m−2, where m is the number of amino acids in Fi. Suppose that z01 and z11 are known, the values of z00 and z10 are computed as follows: z00 = M−z01−z10−z11 = M−z01−(2m−2); z10 = 2m − 2 − z11.
The four probabilities for X0 = 0, X0 = 1, X1 = 0, and X1 = 1 are estimated from training data sets of identified PrSMs without modifications. (See Section “Estimation of parameters.”) The probability Pr(S|Fi) is determined by the values of m, M, z01, and z11. Because m and M are known, the probability Pr(S|Fi) can be computed if z01 and z11 are obtained. In practice, an error tolerance is allowed to match a theoretical fragment mass to an experimental one. In this case, the value z01 is replaced by the number of fj in the binary string of Fi such that fj = 0 and the corresponding mass of fj matches an experimental fragment mass within the error tolerance, denoted by RandMatch(S, Fi). Similarly, z11 is replaced by the number of matched theoretical fragment masses within an error tolerance, denoted by TheoMatch(S, Fi). For a given error tolerance, the number of fj satisfying that the corresponding mass of fj matches an experimental fragment mass within the error tolerance is fixed, that is, RandMatch(S, Fi)+TheoMatch(S, Fi) are the same for 1 ≤ i ≤ m. As a result, the value RandMatch(S, Fi) can be obtained from TheoMatch(S, Fi). Similarly, z01 + z11 equals the number of neutral masses in S, and the value z01 can be obtained from z11 = Score(S, Fi). Based on the observation, we discuss the computation of Score(S, Fi) and TheoMatch(S, Fi) only in the following analysis.
When Score(S, Fi−1) (or TheoMatch(S, Fi−1)) is given, it takes only several operations to compute Score(S, Fi) (or TheoMatch(S, Fi)) because the theoretical spectra of Fi−1 and Fi are almost the same. The number of operations for computing all probabilities Pr(S|Fi), for i = 1, 2, … , m, is proportional to n + m, where n is the number of masses in S and m is the number amino acids in P. The probability Pr(Fi|S) is a modification localization score. Because the type of the modification is known, we also report it as the MIScore. After obtaining the MIScores for all potential sites, the best scoring site is reported.
Modifications near N or C termini
A proteoform may have an unknown N-terminal (or C-terminal) truncation and an unknown modification near the N-terminus (or C-terminus). The truncation and the type of the modification need to be determined simultaneously. Below we use a proteoform with an N-terminal truncation and a modification near the N-terminus as an example to illustrate how to use a Bayesian model to solve the problem. To simplify the description, we assume that all modifications can occur on any amino acid of the protein.
Let P be an unmodified protein sequence in the database with m amino acids and S an MS/MS spectrum generated from a modified proteoform of P with an N-terminal truncation and a modification near the N-terminus. Let Ti,j, 0 ≤ i < j ≤ m, be the proteoform of P in which the first i amino acids at the N-terminus are truncated and the modification is on the jth amino acid. The proteoform Ti,j is valid if the mass difference between the precursor mass of S and the molecular mass of the truncated unmodified protein sequence (the last n − i amino acids) matches the mass of a common modification (within an error tolerance). The list of common modifications is specified by the user. The prior probability of an invalid proteoform is 0; the uniform distribution is assumed for the prior probabilities of valid proteoforms. By Bayes’ theorem
Two modifications
When a mass shift in an identified PrSM results from a combination of two modifications, the sum of the masses of the two modifications equals (within an error tolerance) the mass shift. However, the mass shift may be explained by many combinations of two modifications. For example, a mass shift 56.0626 Da can be explained by a methylation site (14.01565 Da) and a trimethylation site (42.04695 Da) or by two dimethylation sites (28.0313 Da each). We propose to solve the problem with two steps: (1) determine the types and order of the two modifications and (2) localize the two modifications. Finally, we report an ordered pair of modifications, localized sites of the modifications, and their MIScores.
At the first step, we consider only common modifications specified by the user. If a mass shift in a PrSM cannot be explained by two common modifications, it will be annotated by a unknown mass shift and the proteoform will not be fully characterized. To simplify the analysis, we assume that the PrSM contains only one unknown mass shift. Given a mass shift d, we find all possible ordered pairs of two common modifications: (x1, y1), (x2, y2),… , (xk, yk), such that the sum of the masses of xi and yi is similar to d (within an error tolerance). Because the pairs are ordered, we treat (methylation, trimethylation) and (trimethylation, methylation) as two different pairs. For i = 1, 2, … , k, let 𝒬i be the set of all candidate proteoforms of protein P with two modifications (xi, yi) satisfying that xi is closer to the N-terminus of the protein than yi. We compute the probability that the spectrum is generated from a proteoform in 𝒬i, that is, the probability that the types and order of the modifications is (xi, yi), as follows:
| (2) |
In practice, we assume that all the candidate proteoforms with two common modifications follow a uniform distribution.
We describe a dynamic programming algorithm that efficiently computes the distribution of the shared mass counts between S and all Q ∈ 𝒬i, which are required for the computation of the probability Pr(Q ∈ 𝒬i|S). In the algorithm, the shared mass count between a prefix of a proteoform and an MS/MS spectrum needs to be computed. A length l prefix R of a proteoform is represented by a binary string by treating it as a special proteoform with l+1 amino acids: the first l amino acids are the same as those in the prefix and the l+1th amino acid is a special one representing the remaining amino acids. The residue mass of the special amino acid is the sum of the residue masses of the remaining amino acids. The shared mass count Score(R, S) is the number of matched 1s in the binary strings of R and S.
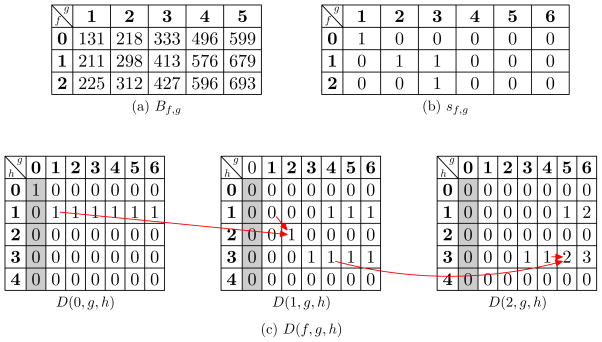
We fill out a three-dimensional table D(f, g, h) for f = 0, 1, and 2. The value D(f, g, h) represents the number of different prefixes R of proteoforms in 𝒬i satisfying that (1) R contains the first f modifications of the pair (xi, yi) (when f = 1, R contains the modification xi; when f = 2, the ordered modifications xi and yi), (2) the length of R is g, and (3) Score(R, S) = h (Figure 2).
Figure 2.
The three-dimensional table D(f, g, h) for a discretized spectrum with a precursor mass 848 and four neutral fragment masses 131, 413, 421, 550, a protein sequence MSDYCH, and an ordered pair of modifications (phosphorylation, methylation). A scale factor 1 is used in the computation. (a) B0,g is the sum of the masses of the first g residues of the protein. B1,g is the sum of B0,g and the mass of phosphorylation (80 Da). B2,g is the sum of B0,g and the masses of phosphorylation (80 Da) and methylation (14 Da). (b) Table sf,g is generated based on Bf,g using Equation (3). (c) D(f, g, h) is filled out by the dynamic programming algorithm in Figure S1 in the supplementary material. The shaded areas are initialized using Equation (4). The second residue S is a modification site of phosphorylation, and the value D(1, 2, 2) is computed as D(0, 1, 2−s1,2+D(1, 1, 2−s1,2) = D(0, 1, 1)+D(1, 1, 1). Similarly, the fifth residue C is modification site of methylation, and the value D(2, 5, 3) is computed as D(1, 4, 3 − s2,5) + D(2, 4, 3 − s2,5) = D(1, 4, 3) + D(2, 4, 3).
To simplify the analysis, we assume that all theoretical N- and C-terminal fragment masses of a proteoform are distinct. When an N-terminal theoretical neutral fragment mass and a C-terminal one of a proteoform Q ∈ 𝒬i are the same and matched to a neutral fragment mass in S, the proposed algorithm treats them as two matched theoretical fragment masses and reports an approximation of Score(S,Q). In addition, we assume that CID spectra are studied and that only b- or y-ions are used for the generation of theoretical spectra.
The masses of xi and yi are denoted as MX and MY, respectively. Let Bg denote the neutral mass of the bg ion (the b-ion containing g amino acids) of Q. Let Bf,g be the neutral mass of the bg ion with f modifications in (xi, yi) for f = 0, 1, 2, that is, B0,g = Bg, B1,g = Bg +Mx, B2,g = Bg +Mx +My (Figure 2a). When the bg ion with f modifications is a product ion of a proteoform Q, the neutral mass of the complementary y ion is M − Bf,g, where M is the molecular mass of Q. We define
| (3) |
In addition, we set s0,m = s1,m = s2,m = 0, where m is the length of Q. Figure 2b shows an example of table sf,g.
Let t = min{n, 2m−2} be the largest shared mass count score between S and a proteoform in 𝒬i. In the initialization, we set
| (4) |
The values initialized in Figure 2c are shown in shaded areas. We use the following recurrence functions to compute the values D(f, g, h) that are not initialized.
Finally, the values in D(2,m, h) for h = 0, 1, … , t are reported as the distribution of the shared mass counts between S and all Q ∈ 𝒬i. The dynamic programming algorithm is given in Figure S1 in the supplementary material. When error tolerances of fragment masses are allowed, the algorithm can be modified to compute distributions of TheoMatch(S,Q) for Q ∈ 𝒬i by introducing error tolerances in Formula (3). Based on the distribution of TheoMatch(S,Q), the confidence score for each modification type pair is obtained, and the modification type pair with the highest confidence score is reported. The number of operations of the algorithm is proportional to m2.
After the types and order of the two modifications (xi, yi) are determined, a divide and conquer method is employed to localize the two modifications. We assume that all proteoforms in 𝒬i follow a uniform distribution. Let 𝒬ij be the set of proteoforms satisfying that xi occurs on the first j amino acids and yi on the last n−j amino acids. When S is generated from a proteoform Q with a pair of modifications (xi, yi),
The denominator and numerator of the right-hand side of the equation are determined by the shared mass count distribution of PrSMs between S and proteoforms in 𝒬i and that between S and proteoforms in 𝒬ij. The first is computed using the algorithm for determining the ordered modification pair; the second can be efficiently calculated using a similar dynamic programming algorithm (Figure S2 in the supplementary material). Suppose the highest probability among Pr(Q ∈ 𝒬ij |S,Q ∈ 𝒬i) for j = 1, 2, … , m is obtained from the set 𝒬ij*. In this case, the proteoform is broken into two sub-proteins: the first contains the first j* amino acids and the second the last m−j* amino acids. The two modifications are treated as single ones in their corresponding sub-proteins for localization, resulting in two probabilities Pr(xi on k1|S,Q ∈ 𝒬ij*) and Pr(yi on k2|S,Q ∈ 𝒬ij*) for the best localization sites k1 and k2 of the two modifications xi and yi. Finally, we report two probabilities as the MIScores:
| (5) |
| (6) |
Determination of the number of modifications
A mass shift in a PrSM may be explained by one or two modifications. For example, a mass shift 28.0313 Da can be explained by a dimethylation site (28.0313 Da) or two methylation sites (14.01565 Da each). We use a Bayesian model to determine the number of modifications that best explain a mass shift. To simplify the analysis, we assume that the PrSM (P, S) has only one unknown mass shift. Let ℱ1 (ℱ2) be the set of all proteoforms of P with one (two) common modifications whose molecular masses match the precursor mass of S. The probability that the target proteoform F contains one modification is estimated as
In the computation, all proteoforms in ℱ1 have the same prior probability, and all proteoforms in ℱ2 have the same prior probability. The ratio r between the prior probabilities of the proteoforms with one modification and those with two modifications (r = Pr(F ∈ ℱ1)/Pr(F ∈ ℱ2)) is a user-specified parameter.
Multiple modifications
The methods for identifying two modifications from a mass shift can be extended to multiple modifications. When a mass shift results from K modifications, the number of ordered K modification types that can explain the mass shift is an exponential function with respect to K, making the proposed method inefficient. This dynamic programming algorithm in Figure S1 in the supplementary material is modified to fill out a table D(f, g, h) for f = 0, 1, … , K. We extend the definitions of Bf,g and sf,g for f = 3, 4, … , K and fill out the table using the following recurrence function:
The number of operations of the algorithm is proportional to ntK.
This divide and conquer method is employed to localize K ordered modifications. Let ℘i be the set of proteoforms of P with ordered modifications xi,1, xi,2, … , xi,K. Let ℘ij be the set of proteoforms satisfying that the first modification occurs on the first j amino acids and all other modifications on the last m − j amino acids. Using this method, we find a position j* with the highest probability Pr(F ∈ ℘ij*|S, F ∈ ℘i) to divide the protein into two parts. The first modification is localized as a single modification on the first j* amino acids, and the other K −1 modifications are localized using the divide and conquer method progressively.
Results
The MIScore method was implemented in C++ and tested on a desktop with a 3.4 GHz CPU (Intel Core i7-3770) and 16 GB memory.
Training and test PrSMs
The proteome database of Escherichia coli K-12 MG1655 was downloaded from UniProt (Jun 18, 2015 version, 4 305 entries). All EC top-down MS/MS spectra were deconvoluted by MS-Deconv28 and searched against a target-decoy concatenated database by TopPIC.30 In database searches, the error tolerances for precursor and fragment masses were set as 15 ppm, and at most 2 unknown mass shifts were allowed in a PrSM. (The parameters used in TopPIC are summarized in Table S1 in the supplementary material.)
A total of 1 533 PrSMs were identified with a 1% spectrum level false discovery rate (FDR), including 767 PrSMs without modifications. We further removed PrSMs that contain less than 15 matched fragment ions, resulting in 1 277 PrSMs including 610 PrSMs without modifications (Table S2 in the supplementary material). Because of the stringent filtering, the 610 PrSMs without modifications were treated as correct ones. They are randomly divided into two groups with the same size: one for training parameters and the other for generating test PrSMs.
Test PrSMs with modifications were generated from the identified PrSMs without modifications. Given an identified PrSM without modifications, we change the protein sequence to introduce a modification with two steps: (a) randomly select a modification and an amino acid on which the modification can occur in the protein sequence, and (b) replace the amino acid with a special amino acid “X”, whose residue mass equals the difference between the masses of the amino acid residue and the modification. For instance, if the selected amino acid is an alanine (71.0371 Da) and the selected modification is methylation (+14.0156 Da), the residue mass value of “X” (“X” is a glycine) that replaces the alanine residue is 71.0371 − 14.0156 = 57.0215 Da, resulting in a PrSM with a methylation on the amino acid “X”. To generate PrSMs with a truncation at the N (or C) terminus and a modification near the N (or C) terminus, we limit the replacement to the 15 amino acids at the N (or C) terminus and add a random peptide (no longer than 20 amino acids) to the N (or C) terminus. PrSMs with two modifications can be generated in a similar way.
Using four common modifications (acetylation, methylation, oxidation, and phosphorylation), we generated 6 100 test PrSMs with one modification, 3 050 test PrSMs with one modification near the N-terminus and an N-terminal truncation, 3 050 test PrSMs with one modification near the C-terminus and a C-terminal truncation, and 6 100 test PrSMs with two modifications from the 305 PrSMs without modifications. These PrSMs were used as a gold standard in the experiments.
Estimation of parameters
The 305 training PrSMs without modifications were used to estimate the four probabilities: Pr(X0 = 0), Pr(X0 = 1), Pr(X1 = 0) and Pr(X1 = 1). (See Section Methods.) For a protein P and its matched spectrum S, we compute TheoMatch(S, P) and RandMatch(S, P) with an error tolerance of 15 ppm. In addition, we converted the protein into its binary representation with a scale factor 274.335215. Let N01 be the sum of RandMatch(S, P) of the training PrSMs and N0 the total number of 0s in the binary strings of the proteins in the PrSMs. The probability Pr(X0 = 1) is estimated as and Pr(X0 = 0) = 1 − Pr(X0 = 1). Let N11 be the sum of TheoMatch(S, P) of the PrSMs and N1 the total number of 1s in the binary strings of the proteins in the PrSMs. The probability Pr(X1 = 1) is estimated as and Pr(X1 = 0) = 1 − Pr(X1 = 1). The estimated probabilities are listed in Table S3 in the supplementary material.
The 305 training PrSMs were used to compare the performance of the Bayesian model for determining the number of modifications with different settings of the ratio r. For each PrSM, we generated two pairs of proteoforms with modifications. In the first pair, one proteoform has a dimethylation site and the other has two methylation sites. In the second pair, one proteoform has two oxidation sites and the other has a dioxidation site. By setting the ratio r to 0.5, 0, 6, … , 1, 1.1, … , 2, we used the proposed method to report the number of modifications for each modified proteoform and calculate the accuracy of reported modification numbers. The ratio r = 0.8 achieved the best accuracy 83.9% (Figure S3 in the supplementary material) and was used in the experiments.
Identification of single modifications
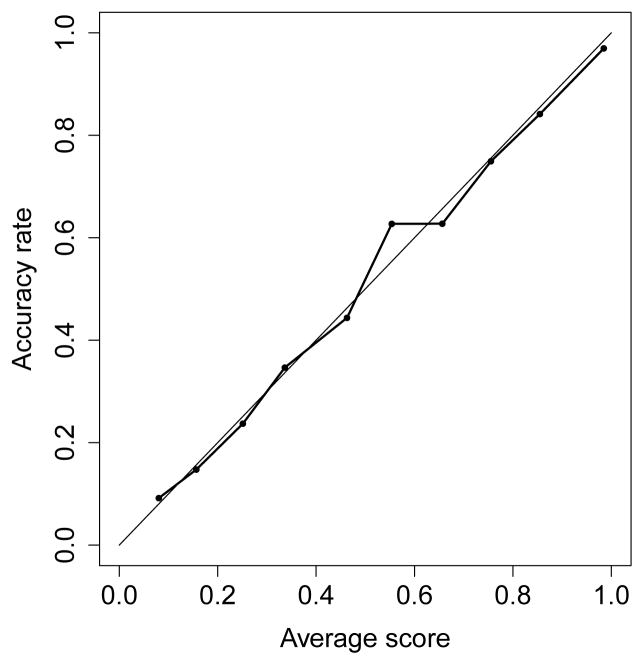
The MIScore method was employed to analyze the 6 100 test PrSMs with one modification, in which the correct location of each modification is known. The proposed model reported for each PrSM a site with the highest MIScore. A total of 3 038 (49.8%) modification sites were localized to a site with an MIScore ≥ 0.45, of which 2 381 (39.0%) were correct (Figure S4 in the supplementary material). Many modification sites were not identified with a high MIScore because some top-down MS/MS spectra had low sequence coverage and failed to provide enough fragment masses for confident localization of modifications. We divided the reported sites into 10 groups with scores in [0, 0.1], (0.1, 0.2], … , (0, 9, 1.0]. If the reported scores are accurate, the accuracy rate of the sites in each group should be similar to their average scores because the scores are the accuracy rates estimated by the model. Figure 3 shows that the accuracy rates are similar to the average scores for these 10 groups, demonstrating that the MIScores reported by the model were accurate.
Figure 3.
The modification sites reported by the MIScore method from the 6 100 PrSMs with one modification are grouped into bins with width 0.1 based on their MIScores. The average identification score and accuracy rate of the modification sites in each bin are compared.
Identification of modifications near N or C termini
The MIScore method was used to analyze the 6 100 test PrSMs with a truncation at the N or C terminus and one modification near the N or C terminus. If the correct truncation and modification site are reported, we say the result is correct; otherwise, incorrect. A total of 2 874 (47.1%) modification sites were localized to a site with an MIScore ≥ 0.45, of which 2 107 (34.5%) were correct (Figure S5 in the supplementary material). Similar to the previous experiment, the reported modification sites were divided into 10 groups based on their MIScores, and the average of the scores in each group was compared with the accuracy rate of the corresponding sites (Figure S6 in supplementary information). The results showed that the model reported accurate MIScores for modifications near N or C termini.
Identification of two modifications
For each of the 6 100 test PrSMs with two modifications, the proposed method reported an ordered modification pair, their best locations, and three scores: the first one is the confidence score that the modification pair is correct (Equation (2)); the other two are the MIScores of the two modifications (Equations (5) and (6)).
First, we evaluated the accuracy of the confidence scores of reported modification pairs. We divided the reported modification pairs into 10 groups with scores in [0, 0.1], (0.1, 0.2], … , (0, 9, 1.0], and computed the accuracy rate for the modification pairs in each group (Figure S7 in supplementary information). The average confidence score is approximately the same to the accuracy rate in each group, demonstrating that the reported confidence scores were accurate.
Second, we evaluated the accuracy of reported MIScores. A total of 6 154 (50.4%) modification sites were localized to a site with an MIScore ≥ 0.45, of which 4 798 (39.3%) were correct (Figure S8 in the supplementary material). Similar to single modifications, we divided the reported modification sites into 10 groups based on their scores and compared the average scores and accuracy rates of the groups. The results showed that the accuracy rates were similar to the average scores (Figure S9 in supplementary information), and that the reported MIScores were accurate.
Modifications in the EC data set
Among the 1 277 PrSMs identified by TopPIC30 from the EC data set, 667 PrSMs contain mass shifts. A total of 318 PrSMs contain a mass shift about ±1 Da, which may be caused by ±1 Da errors introduced in the deconvolution of precursor masses. The MIScore method was employed to characterize the proteoforms in the remaining 349 PrSMs from 74 proteins. Four PTMs (acetylation, methylation, oxidation, and phosphorylation) were chosen as common PTMs (Table 2). If a mass shift in the PrSMs can be explained by one common PTM, the MIScore method reports the type of the PTM and the site with the best score. If several sites have the same best MIScore, all the sites are reported. For a PrSM with one mass shift, the mass shift equals the difference between the precursor mass of the spectrum and the molecular mass of the protein or a truncated form of the protein. The error tolerance (15 ppm) of the precursor mass is used for mapping a mass shift to a modification. Because ±1 Da errors are often observed in deconvoluted precursor masses, ±1 Da errors are also allowed in the mapping. If the error tolerance of the precursor mass is δ, a modification with mass m1 is mapped to a mass shift m2 if min{|m1 − m2|, |m1 − m2 + 1.00235|, |m1 − m2 − 1.00235|} ≤ δ, where 1.00235 Da is the average mass difference between two isotopomers whose neutron numbers differ by 1. Similarly, a mass shift is mapped to two modifications if the difference between the mass shift and the sum of the masses of the two modifications satisfies the condition described above. In this case, the types and best scoring sites of the two modifications are reported. Because ±1 Da errors are observed more frequently in large fragment masses than small ones, they are allowed for fragment masses that are larger than an empirical threshold 5 000 Da, and not allowed for those less than the threshold. The running time of the analysis was about 204 seconds.
Table 2.
Four PTMs are treated as common PTMs in proteoform characterization
| PTM | Modified amino acids at the N-terminus | Modified amino acids | Monoisotopic mass mass (Da) |
|---|---|---|---|
| Acetylation | All 20 amino acids | K | 42.01 |
| Methylation | All 20 amino acids | HKNQRILDEST | 14.01 |
| Oxidation | DKNPYRC* | DKNPYRC* | 15.99 |
| Phosphorylation | STY | STY | 79.96 |
Artifacts introduced in sample preparations and mass spectrometry experiments are not included.
A total of 116 and 13 mass shifts in the 349 PrSMs match the mass of one common PTM and a combination of two common PTMs, respectively. Of the 116 mass shifts explained by single PTMs, 28 were localized to a site with an MIScore no less than 0.9 and 10 were localized to two candidate sites with the same MIScore no less than 0.45. For the 13 mass shifts explained by PTM pairs (26 PTMs in total), 10 PTMs were localized to a site with an MIScore no less than 0.9 (Table S4 in the supplementary material). The reason that only a small number of PTMs were confidently identified and localized is that most identified PrSMs had many missing fragment peaks, lacking enough information to localize PTMs to one or two sites.
The 28 mass shifts that are explained by single PTMs and localized to single sites correspond to 15 PTM sites (methylation: 6, oxidation: 8, phosphorylation: 1) in 11 proteins. The 10 mass shifts explained by single PTMs and each localized to two sites correspond to 4 PTMs (methylation: 3, acetylation: 1) in 3 proteins. We compared the reported modification sites with the annotations of the proteins in the Swiss-Prot database. The N-terminal methylation site K82 in the protein RL7 ECOLI (UniProt ID: P0A7K2) and the N-terminal methylation site A2 in the protein RL33 ECOLI (UniProt ID: P0A7N9) were supported by the annotations. One main reason for the lack of support by the annotations is that the annotation of the EC proteome is incomplete in the Swiss-Prot database. In addition, two N-terminal methylation sites were reported (M1 in the protein PTHP ECOLI, UniProt ID: P0AA04; M1 in the protein RL23 ECOLI, UniProt ID: P0ADZ0). Because N-terminal methylation has been found in many proteins,31 these sites may be new identified N-terminal methylation sites.
The 5 mass shifts explained by two PTMs correspond to 8 PTM sites (1 oxidation pair and 3 methylation pairs). Manual inspection showed that the oxidation pair may be explained by a dioxidation and that the two methylation pairs may be explained by dimethylation sites.
Comparison with the Mascot Delta Score
The Mascot Delta Score (MD-score)20 is computed based on the difference between the scores reported by Mascot32 for the best and second best modified peptides with different modification sites and the identical peptide sequence for a bottom-up MS/MS spectrum. We tested the MD-score method using the 38 PrSMs identified from the EC data set each of which contains an unknown mass shift that is explained by a PTM and localized to either one site with an MIScore ≥ 0.9 or two sites each with an MIScore ≥ 0.45 (Table S4 in the supplementary material). The 38 spectra were converted to MGF files containing charge +1 fragment m/z values and divided into four groups based on the PTMs reported by the MIScore method: 28 spectra with methylation, 8 with oxidation, 1 with acetylation, and 1 with phosphorylation. The four groups of spectra were searched separately against the Swiss-Prot EC proteome database using the Mascot server at http://www.matrixscience.com. For each group, the corresponding PTM was set as the variable PTM. Other parameters of Mascot are shown in Table S5 in the supplementary material.
Mascot identified 13 PrSMs with an E-value ≤ 0.05, of which 4 contained a localized N-terminal modification (all were methylation) and 9 contained a localized modification not at the N-terminus. Mascot reported MD-scores for only the latter 9 PrSMs, not for the N-terminal ones (Table S6 in the supplementary material). Because Mascot treated the top-down MS/MS spectra as bottom-up ones and these top-down spectra contained many fragment peaks, it automatically removed many low abundance peaks from the spectra before database search. It may be the main reason that Mascot identified only about 34.2% of the test spectra. The 4 N-terminal methylation sites reported by Mascot are consistent with those reported by the MIScore method. The two methods reported the same localization site for only one of the 9 modification sites with MD-scores and different localization sites for the other 8. By manual inspection of the results reported by the two methods, we found that the main reason for the different localized sites is that the MD-score method uses only a set of high abundance peaks, not all peaks, for localizing PTM sites.
Modifications in the ST data set
The proteome database of Salmonella typhimurium 14028s were downloaded from UniProt (Jul 30, 2015 version, 5369 entries). All top-down MS/MS spectra of the ST data set were deconvoluted by MS-Deconv28 and searched against the proteome database concatenated with a decoy database by TopPIC30 using the parameters in Table S1 in the supplementary material.
After filtering with a 1% spectrum-level FDR and a threshold 15 for the number of matched fragment ions, TopPIC30 identified 1 413 PrSMs without mass shifts and 1 278 PrSMs with mass shifts (Table S7 in the supplementary material). Those with mass shifts were analyzed by the MIScore method using the same parameters in the analysis of the EC data set except for the PTM cysteinylation. Ansong et al. showed that cysteinylation is often observed in ST in response to infection-like conditions,26 so cysteinylation was also treated as a common PTM (mass: 119.00 Da; modified residue: cysteine). The running time of the analysis was about 994 seconds.
A total of 132 mass shifts match the mass of one common PTM, of which 58 were localized to a site with an MIScore no less than 0.9. These mass shifts correspond to 41 PTM sites (acetylation: 10, methylation: 2, oxidation: 2, cysteinylation: 27) in 33 proteins. And 11 mass shifts explained by single PTMs were localized to two sites each with an MIScore no less than 0.45. These mass shifts correspond to 8 PTM sites (acetylation: 4, oxidation: 3, cysteinylation: 1) in 6 proteins (Table S8 in the supplementary material). In addition, 14 mass shift matches the mass of a combination of two common PTMs, but no localized sites with high MIScores were reported.
To further validate the localized PTMs, the bottom-up data set generated from the same sample was searched against the Salmonella typhimurium 14028s proteome database concatenated with a decoy database using MS-GF+.33 A total of five rounds of database searches were performed to identify peptides with PTMs. In MS-GF+, the high-resolution mode was used (the error tolerances for precursor and fragment masses were 20 ppm and 0.1 Da, respectively); no fixed PTMs were used; non-tryptic termini were allowed, and the default settings were used for the other parameters (Table S9 in the supplementary material). In the first round, cysteinylation was set as a variable PTM. With a 5% Q-value cutoff, 52 825 peptide-spectrum matches were identified. Of the 29 cysteinylation sites identified by the proposed method, 8 (all of them are from mass shifts localized to single sites) were supported by identified peptide-spectrum matches (Table S10 in the supplementary material). The site C239 in the protein Transaldolase (UniProt ID: A0A0F6AWC3) was covered by identified peptides without modifications. Proteoforms without modification on the sites were also identified by the top-down MS analysis, showing that there exist two proteoforms (one modified and the other unmodified) of the protein in the sample. The site C36 in the protein Triosephosphate isomerase (UniProt ID: A0A0F6B9R1) was also covered by identified peptides without modifications, and proteoforms without the modification were not identified by top-down MS. The remaining 19 cysteinylation sites were not supported by identified peptides because the bottom-up MS/MS spectra covered only about 18.2% of the sequences of identified proteins. When cysteinylation sites were covered by both identified proteoforms and peptides, the peptides supported most of the PTM sites (8 out of 10) identified by the proposed method. In the other four rounds, similar analyses of the bottom-up MS/MS spectra were performed to find peptides supporting identified acetylation, methylation, oxidation, and phosphorylation sites. However, because of the low protein sequence coverage of identified peptides, only two acetylation sites (K314 in Elongation factor Tu, UniProt ID: A0A0F6B9X6; K226 in Cysteine synthase, UniProt ID: A0A0F6B4H6) were covered by identified peptides, which were not modified and did not support the reported PTM sites.
Discussion and conclusions
In this paper, we proposed several Bayesian models that determine the types of modifications, localize modifications, and identify truncations for proteoforms with unknown mass shifts. The experiments on the test PrSMs generated from the EC data set showed that MIScores reported by the models were accurate for proteoforms with one or two modifications. In addition, the MIScore method identified and localized many modifications from mass shifts in PrSMs reported from the EC and ST data sets, of which some were supported Swiss-Prot annotations and some by bottom-up MS/MS spectra.
Several parameters, such as the probability Pr(X0 = 1) in Equation (1) and the ratio r, are used in the MIScore method. When a new data set is analyzed, we can train these parameters using PrSMs without modifications identified from the data set to improve the accuracy of reported MIScores with two steps: (a) A proteoform identification tool is used to report PrSMs without modifications from the data set, and (b) the methods described in Section “Estimation of parameters” are employed to estimate the parameters.
The MIScore method is faster than the C-score method because the proposed dynamic programming algorithms significantly speed up the computation of probabilities. For example, when a mass shift identified in a PrSM is explained by two modifications whose types are known and each of which has n candidate sites, a total of n2 proteoforms need to be considered in the localization of two modifications. In the C-score method, each of the n2 proteoforms needs to be explicitly generated to compute the conditional probability that the spectrum is observed given the proteoform (the likelihood in Table 1) because of the lack of efficient algorithms. By contrast, the dynamic programming algorithm in the MIScore method can efficiently compute the probabilities of the n2 proteoforms in one run without explicitly generating them. Let q be the ratio between the running time for computing all probabilities in the MIScore method and that for computing one probability in the C-score method. In practice, q is much smaller than n2 and the speed of the MIScore method is about n2/q faster than the C-score method.
Top-down spectral deconvolution algorithms may introduce ±1 Da errors in reported precursor masses. Since precursor masses are used to compute the mass shifts of unknown modifications, the errors in precursor masses may result in incorrect identifications of modifications. Increasing the accuracy of deconvoluted precursor masses is essential to improving the accuracy of proteoform characterization.
A simple shared mass count score is used for computing MIScores. Peak intensities and errors in matched theoretical and experimental masses also provide valuable information for proteoform characterization. Incorporating these information into the proposed models will further improve the accuracy of MIScores, but the incorporation also makes it complex to compute posterior probabilities in the models. Designing efficient algorithms for computing posterior probabilities using these complex probabilistic models is a future research direction.
Many possible modifications need to be considered in proteoform characterization in proteome-level analyses of complex species. Including all these modifications may increase the possibility of reporting incorrectly characterized modifications. One possible solution to the problem is to divide identified PrSMs in a proteome-level analysis into groups, each of which has one or several common modifications that are expected to be observed based on domain knowledge. Using protein-specific modifications can improve the accuracy of proteoform characterization.
The MIScore method still has many limitations in analyzing complex proteins and complex species such as humans. First, the number of modifications the MIScore method can identify from an unknown mass shift is limited to 1 or 2. Second, protein samples of complex eukaryotic species may contain many proteoforms generated from alternative splicing, which the MIScore method cannot characterize. Third, the accuracy of the MIScore method heavily replies on the accuracy of reported precursor masses. When the molecular mass of the target proteoform is very large and a large error, e.g. 0.5 Da, is introduced into the measured precursor mass, the MIScore method may fail to find the correct modifications. Fourth, when a protein has heterogeneous modifications and many possible modification sites, liquid chromatography or other separation techniques may fail to separate multiple proteoforms with similar molecular masses of the same protein, resulting in multiplexed MS/MS spectra. The MIScore method cannot accurately characterize unknown mass shifts identified by these multiplexed spectra. Fifth, a mass shift identified in a ultramodified protein may result from a combination of three or more modifications because of missing peaks. The mass shift can be explained by many combinations of modification types and sites and there are no enough matched peaks to distinguish the target proteoforms from other candidates. As a result, the MIScore method may fail to characterize and localize these modifications.
Supplementary Material
Figure S1. An algorithm for computing the distribution of shared mass counts between a spectrum and the proteoforms of a protein with a pair of ordered modifications.
Figure S2. An algorithm for computing the distribution of shared mass count scores between spectrum S and 𝒬i,j.
Figure S3. Comparison of the accuracy of the model for determining the number of modifications with different settings 0.5, 0.6, … , 1, 1.1, … , 2 of the ratio r.
Figure S4. Percentages of identified modification sites and correctly identified ones by the MIScore method from the 6 100 PrSMs with one modification with various MIScore cutoff values.
Figure S5. Percentages of identified modification sites and correctly identified ones by the MIScore method from the 6 100 PrSMs with a truncation at the N (or C) terminus and one PTM near the N (or C) terminus with various MIScore cutoff values.
Figure S6. The modification sites reported by the MIScore method from the 6 100 PrSMs with a truncation at the N (or C) terminus and one PTM near the N (or C) terminus are grouped into bins with width 0.1 based on their modification identification scores. The average identification score and accuracy rate of the modification sites in each bin are compared.
Figure S7. The ordered modification pairs reported by the MIScore method from the 6 100 PrSMs with two modifications are grouped into bins with width 0.1 based on their confidence scores. The average confidence score and accuracy rate of the ordered modification pairs in each bin are compared.
Figure S8. Percentages of identified modification sites and correctly identified ones by the MIScore method from the 6 100 PrSMs with two modifications with various MIScore cutoff values.
Figure S9. The modification sites reported by the MIScore method from the 6 100 PrSMs with two modifications are grouped into bins with width 0.1 based on their modification identification scores. The average identification score and accuracy rate of the modification sites in each bin are compared.
Table S1. Parameters used in TopPIC.
Table S3. Probabilities estimated from the 305 training PrSMs of the EC data set.
Table S5. Parameters used in Mascot for searching the 38 top-down MS/MS spectra against the Swiss-Prot EC proteome database.
Table S9. Parameters used in MS-GF+.
Table S10. A total of 8 cysteinylation sites reported from ST data set that are supported peptides identified from the bottom-up data set by MS-GF+.
Table S2. A total of 1 277 PrSMs with at least 15 matched fragment ions are reported from EC data set by TopPIC with a 1% spectrum level FDR.
Table S4. PTM sites localized by the MIScore method in the EC data set: 28 mass shifts are explained by one PTM and localized to one site with an MIScore ≥ 0.9; 10 mass shifts are explained by one PTM and localized to two candidate sites with the same MIScore ≥ 0.45; 5 mass shifts are explained by two PTMs and each PTM is localized to a site with an MIScore ≥ 0.9.
Table S6. A total of 13 of the 38 top-down MS/MS spectra are identified by Mascot with an E-value cutoff 0.05.
Table S7. A total of 2 691 PrSMs with at least 15 matched fragment ions are reported from the ST data set by TopPIC with a 1% spectrum level FDR.
Table S8. PTM sites localized by the MIScore method in the ST data set: 58 mass shifts are explained by one PTM and localized to one site with an MIScore ≥ 0.9; 11 mass shifts are explained by one PTM and localized to two candidate sites with the same MIScore ≥ 0.45.
Acknowledgments
The research was supported by the National Institute of General Medical Sciences, National Institutes of Health (NIH) through Grant R01GM118470. The ST data set was generated by EMSL, a DOE Office of Science User Facility sponsored by the Office of Biological and Environmental Research and located at Pacific Northwest National Laboratory. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
The MIScore method has been added as a component of the software TopPIC, which is freely available at http://proteomics.informatics.iupui.edu/software/toppic/.
References
- 1.Smith LM, Kelleher NL Consortium for Top Down Proteomics. Proteoform: a single term describing protein complexity. Nature Methods. 2013;10:186–187. doi: 10.1038/nmeth.2369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bairoch A, Boeckmann B, Ferro S, Gasteiger E. Swiss-Prot: juggling between evolution and stability. Briefings in Bioinformatics. 2004;5:39–55. doi: 10.1093/bib/5.1.39. [DOI] [PubMed] [Google Scholar]
- 3.Pruitt KD, Tatusova T, Maglott DR. NCBI reference sequences (RefSeq): a curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Research. 2007;35:D61–D65. doi: 10.1093/nar/gkl842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dong X, Sumandea CA, Chen YC, Garcia-Cazarin ML, Zhang J, Balke CW, Sumandea MP, Ge Y. Augmented phosphorylation of cardiac troponin I in hypertensive heart failure. Journal of Biological Chemistry. 2012;287:848–857. doi: 10.1074/jbc.M111.293258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Peleg S, et al. Altered histone acetylation is associated with age-dependent memory impairment in mice. Science. 2010;328:753–756. doi: 10.1126/science.1186088. [DOI] [PubMed] [Google Scholar]
- 6.Tran JC, et al. Mapping intact protein isoforms in discovery mode using top-down proteomics. Nature. 2011;480:254–258. doi: 10.1038/nature10575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wu S, Lourette NM, Tolić N, Zhao R, Robinson EW, Tolmachev AV, Smith RD, Paša-Tolić L. An integrated top-down and bottom-up strategy for broadly characterizing protein isoforms and modifications. Journal of Proteome Research. 2009;8:1347–1357. doi: 10.1021/pr800720d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li L, Tian Z. Interpreting raw biological mass spectra using isotopic mass-to-charge ratio and envelope fingerprinting. Rapid Communications in Mass Spectrometry. 2013;27:1267–1277. doi: 10.1002/rcm.6565. [DOI] [PubMed] [Google Scholar]
- 9.Zamdborg L, LeDuc RD, Glowacz KJ, Kim YB, Viswanathan V, Spaulding IT, Early BP, Bluhm EJ, Babai S, Kelleher NL. ProSight PTM 2. 0: improved protein identification and characterization for top down mass spectrometry. Nucleic Acids Research. 2007;35:W701–W706. doi: 10.1093/nar/gkm371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Frank AM, Pesavento JJ, Mizzen CA, Kelleher NL, Pevzner PA. Interpreting top-down mass spectra using spectral alignment. Analytical Chemistry. 2008;80:2499–2505. doi: 10.1021/ac702324u. [DOI] [PubMed] [Google Scholar]
- 11.Liu X, Sirotkin Y, Shen Y, Anderson G, Tsai YS, Ting YS, Goodlett DR, Smith RD, Bafna V, Pevzner PA. Protein identification using top-down spectra. Molecular & Cellular Proteomics. 2012;11:M111.008524. doi: 10.1074/mcp.M111.008524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tsai YS, Scherl A, Shaw JL, MacKay CL, Shaffer SA, Langridge-Smith PRR, Goodlett DR. Precursor ion independent algorithm for top-down shotgun proteomics. Journal of the American Society for Mass Spectrometry. 2009;20:2154–2166. doi: 10.1016/j.jasms.2009.07.024. [DOI] [PubMed] [Google Scholar]
- 13.Shen Y, Tolić N, Hixson KK, Purvine SO, Anderson GA, Smith RD. De novo sequencing of unique sequence tags for discovery of post-translational modifications of proteins. Analytical Chemistry. 2008;80:7742–7754. doi: 10.1021/ac801123p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu X, Hengel S, Wu S, Tolić N, Paša-Tolić L, Pevzner PA. Identification of ultramodified proteins using top-down tandem mass spectra. Journal of Proteome Research. 2013;12:5830–5838. doi: 10.1021/pr400849y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Beausoleil SA, Villn J, Gerber SA, Rush J, Gygi SP. A probability-based approach for high-throughput protein phosphorylation analysis and site localization. Nature Biotechnology. 2006;24:1285–1292. doi: 10.1038/nbt1240. [DOI] [PubMed] [Google Scholar]
- 16.Olsen JV, Blagoev B, Gnad F, Macek B, Kumar C, Mortensen P, Mann M. Global, in vivo, and site-specific phosphorylation dynamics in signaling networks. Cell. 2006;127:635–648. doi: 10.1016/j.cell.2006.09.026. [DOI] [PubMed] [Google Scholar]
- 17.Albuquerque CP, Smolka MB, Payne SH, Bafna V, Eng J, Zhou H. A multidimensional chromatography technology for in-depth phosphoproteome analysis. Molecular & Cellular Proteomics. 2008;7:1389–1396. doi: 10.1074/mcp.M700468-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bailey CM, Sweet SMM, Cunningham DL, Zeller M, Heath JK, Cooper HJ. SLoMo: automated site localization of modifications from ETD/ECD mass spectra. Journal of Proteome Research. 2009;8:1965–1971. doi: 10.1021/pr800917p. [DOI] [PubMed] [Google Scholar]
- 19.Taus T, Kcher T, Pichler P, Paschke C, Schmidt A, Henrich C, Mechtler K. Universal and confident phosphorylation site localization using phosphoRS. Journal of Proteome Research. 2011;10:5354–5362. doi: 10.1021/pr200611n. [DOI] [PubMed] [Google Scholar]
- 20.Savitski MM, Lemeer S, Boesche M, Lang M, Mathieson T, Bantscheff M, Kuster B. Confident phosphorylation site localization using the Mascot Delta Score. Molecular & Cellular Proteomics. 2011;10:M110.003830. doi: 10.1074/mcp.M110.003830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chalkley RJ, Clauser KR. Modification site localization scoring: strategies and performance. Molecular & Cellular Proteomics. 2012;11:3–14. doi: 10.1074/mcp.R111.015305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tanner S, Payne SH, Dasari S, Shen Z, Wilmarth PA, David LL, Loomis WF, Briggs SP, Bafna V. Accurate annotation of peptide modifications through unrestrictive database search. Journal of Proteome Research. 2008;7:170–181. doi: 10.1021/pr070444v. [DOI] [PubMed] [Google Scholar]
- 23.Chung C, Emili A, Frey BJ. Non-parametric Bayesian approach to post-translational modification refinement of predictions from tandem mass spectrometry. Bioinformatics. 2013;29:821–829. doi: 10.1093/bioinformatics/btt056. [DOI] [PubMed] [Google Scholar]
- 24.Dang X, Scotcher J, Wu S, Chu RK, Toli N, Ntai I, Thomas PM, Fellers RT, Early BP, Zheng Y, et al. The first pilot project of the consortium for top-down proteomics: A status report. PROTEOMICS. 2014;14:11301140. doi: 10.1002/pmic.201300438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.LeDuc RD, Fellers RT, Early BP, Greer JB, Thomas PM, Kelleher NL. The C-score: a Bayesian framework to sharply improve proteoform scoring in high-throughput top down proteomics. Journal of Proteome Research. 2014;13:3231–3240. doi: 10.1021/pr401277r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ansong C, Wu S, Meng D, Liu X, Brewer HM, Kaiser BLD, Nakayasu ES, Cort JR, Pevzner P, Smith RD, Heffrong F, N AJ, Paša-Tolićb L. Top-down proteomics reveals a unique protein S-thiolation switch in Salmonella Typhimurium in response to infection-like conditions. Proceedings of the National Academy of Sciences. 2013;110:10153–10158. doi: 10.1073/pnas.1221210110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Horn DM, Zubarev RA, McLafferty FW. Automated reduction and interpretation of high resolution electrospray mass spectra of large molecules. Journal of the American Society for Mass Spectrometry. 2000;11:320–332. doi: 10.1016/s1044-0305(99)00157-9. [DOI] [PubMed] [Google Scholar]
- 28.Liu X, Inbar Y, Dorrestein PC, Wynne C, Edwards N, Souda P, Whitelegge JP, Bafna V, Pevzner PA. Deconvolution and database search of complex tandem mass spectra of intact proteins: a combinatorial approach. Molecular & Cellular Proteomics. 2010;9:2772–2782. doi: 10.1074/mcp.M110.002766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kou Q, Wu S, Liu X. A new scoring function for top-down spectral deconvolution. BMC Genomics. 2014;15:1140. doi: 10.1186/1471-2164-15-1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.TopPIC: Top-Down Mass Spectrometry Based Proteoform Identification and Characterization. doi: 10.1093/bioinformatics/btw398. http://proteomics.informatics.iupui.edu/software/toppic/ [DOI] [PMC free article] [PubMed]
- 31.Stock A, Clarke S, Clarke C, Stock J. N-terminal methylation of proteins: structure, function and specificity. FEBS letters. 1987;220:8–14. doi: 10.1016/0014-5793(87)80866-9. [DOI] [PubMed] [Google Scholar]
- 32.Perkins DN, Pappin DJ, Creasy DM, Cottrell JS. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis. 1999;20:3551–67. doi: 10.1002/(SICI)1522-2683(19991201)20:18<3551::AID-ELPS3551>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 33.Kim S, Pevzner PA. MS-GF+ makes progress towards a universal database search tool for proteomics. Nature Communications. 2014;5:5277. doi: 10.1038/ncomms6277. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. An algorithm for computing the distribution of shared mass counts between a spectrum and the proteoforms of a protein with a pair of ordered modifications.
Figure S2. An algorithm for computing the distribution of shared mass count scores between spectrum S and 𝒬i,j.
Figure S3. Comparison of the accuracy of the model for determining the number of modifications with different settings 0.5, 0.6, … , 1, 1.1, … , 2 of the ratio r.
Figure S4. Percentages of identified modification sites and correctly identified ones by the MIScore method from the 6 100 PrSMs with one modification with various MIScore cutoff values.
Figure S5. Percentages of identified modification sites and correctly identified ones by the MIScore method from the 6 100 PrSMs with a truncation at the N (or C) terminus and one PTM near the N (or C) terminus with various MIScore cutoff values.
Figure S6. The modification sites reported by the MIScore method from the 6 100 PrSMs with a truncation at the N (or C) terminus and one PTM near the N (or C) terminus are grouped into bins with width 0.1 based on their modification identification scores. The average identification score and accuracy rate of the modification sites in each bin are compared.
Figure S7. The ordered modification pairs reported by the MIScore method from the 6 100 PrSMs with two modifications are grouped into bins with width 0.1 based on their confidence scores. The average confidence score and accuracy rate of the ordered modification pairs in each bin are compared.
Figure S8. Percentages of identified modification sites and correctly identified ones by the MIScore method from the 6 100 PrSMs with two modifications with various MIScore cutoff values.
Figure S9. The modification sites reported by the MIScore method from the 6 100 PrSMs with two modifications are grouped into bins with width 0.1 based on their modification identification scores. The average identification score and accuracy rate of the modification sites in each bin are compared.
Table S1. Parameters used in TopPIC.
Table S3. Probabilities estimated from the 305 training PrSMs of the EC data set.
Table S5. Parameters used in Mascot for searching the 38 top-down MS/MS spectra against the Swiss-Prot EC proteome database.
Table S9. Parameters used in MS-GF+.
Table S10. A total of 8 cysteinylation sites reported from ST data set that are supported peptides identified from the bottom-up data set by MS-GF+.
Table S2. A total of 1 277 PrSMs with at least 15 matched fragment ions are reported from EC data set by TopPIC with a 1% spectrum level FDR.
Table S4. PTM sites localized by the MIScore method in the EC data set: 28 mass shifts are explained by one PTM and localized to one site with an MIScore ≥ 0.9; 10 mass shifts are explained by one PTM and localized to two candidate sites with the same MIScore ≥ 0.45; 5 mass shifts are explained by two PTMs and each PTM is localized to a site with an MIScore ≥ 0.9.
Table S6. A total of 13 of the 38 top-down MS/MS spectra are identified by Mascot with an E-value cutoff 0.05.
Table S7. A total of 2 691 PrSMs with at least 15 matched fragment ions are reported from the ST data set by TopPIC with a 1% spectrum level FDR.
Table S8. PTM sites localized by the MIScore method in the ST data set: 58 mass shifts are explained by one PTM and localized to one site with an MIScore ≥ 0.9; 11 mass shifts are explained by one PTM and localized to two candidate sites with the same MIScore ≥ 0.45.