Abstract
The Atacama Desert hosts diverse ecosystems including salt flats and shallow Andean lakes. Several heavy metals are found in the Atacama Desert, and microorganisms growing in this environment show varying levels of resistance/tolerance to copper, tellurium, and arsenic, among others. Herein, we report the genome sequence and comparative genomic analysis of a new Exiguobacterium strain, sp. SH31, isolated from an altiplanic shallow athalassohaline lake. Exiguobacterium sp. SH31 belongs to the phylogenetic Group II and its closest relative is Exiguobacterium sp. S17, isolated from the Argentinian Altiplano (95% average nucleotide identity). Strain SH31 encodes a wide repertoire of proteins required for cadmium, copper, mercury, tellurium, chromium, and arsenic resistance. Of the 34 Exiguobacterium genomes that were inspected, only isolates SH31 and S17 encode the arsenic efflux pump Acr3. Strain SH31 was able to grow in up to 10 mM arsenite and 100 mM arsenate, indicating that it is arsenic resistant. Further, expression of the ars operon and acr3 was strongly induced in response to both toxics, suggesting that the arsenic efflux pump Acr3 mediates arsenic resistance in Exiguobacterium sp. SH31.
Keywords: Exiguobacterium, polyextremophile, stress, comparative genomics, Chilean Altiplano
Introduction
Extremophiles are microorganisms from all three domains of life (Bacteria, Archaea and Eukarya) that grow in the most hostile environments found on Earth, where they must withstand conditions including extreme pH, temperature, salinity, pressure, UV radiation, and the presence of heavy metals or toxic compounds. In some cases, extremophiles face more than one extreme condition simultaneously, and are called polyextremophiles (Seufferheld et al., 2008; Bowers et al., 2009; Farías et al., 2013; Kurth et al., 2015). To survive under extreme conditions, microbes finely tune gene expression, modulating the levels of proteins implicated in the response to stress (Duché et al., 2002). Since their discovery, extremophiles have attracted researchers because of their unique physiology, ability to adapt to different environments, and for their potential use in biotechnology (Rothschild and Mancinelli, 2001).
The Andean Plateau region in West Central South America, the Altiplano, hosts a rich microbial diversity and remains an untapped resource for understanding the genetic diversity and distribution of polyextremophiles (Catalán et al., 2006). High-altitude Andean Lakes (HAALs) are exposed to some of the highest levels of solar radiation on Earth (Albarracín et al., 2015). Further, water bodies found in the Altiplano (i.e., shallow lakes, ponds, streams, etc.) are highly diverse, showing different chemical compositions, temperatures, evaporation rates, and depths, among other conditions. Together, these factors drive the changing community structure of rich polyextreme microbiota that populates these lakes (Márquez-García et al., 2009; Dorador et al., 2013; Cordero et al., 2014). Despite these harsh conditions, almost 500 strains of prokaryotes (archaea, cyanobacteria and eubacteria) and lower eukaryotes (fungi and yeast) have been isolated from bacterioplankton, benthos, microbial mats, and soils surrounding HAALs (Dib et al., 2008; Flores et al., 2009; Ordoñez et al., 2009; Di Capua et al., 2011; Lynch et al., 2012; Bull and Asenjo, 2013; Bull et al., 2016). Microbial communities that evolved in HAALs tolerate a wide range of chemical and physical stresses, including wide fluctuations in temperature, low nutrient levels, alkalinity, and hypersalinity (up to 30%). Further, high concentrations of heavy metals and metalloids, especially arsenic (up to 200 ppm), are found in HAALs, primarily because of the geological foundation (Fernandez-Zenoff et al., 2006; Zenoff et al., 2006; Dib et al., 2009; Farias et al., 2009; Albarracín et al., 2012).
Salar de Huasco is a saline wetland located at 3,800 m above sea level in the Chilean Altiplano (-20.303436, -68.880918) and is considered an athalassohaline system. It exhibits typical conditions of the Altiplano: low temperatures (mean annual temperature <5°C), low atmospheric pressure, high solar radiation (up to 72 Wm-2 UVA radiation: Hernández et al., 2016), negative water balance, and variable salinity ranging from freshwater to saturated salt waters. Despite these conditions, communities composed of microbial groups from all major branches of the microbial tree of life, including Proteobacteria, Cyanobacteria, Bacteroidetes, and Archaea, thrive and grow in water bodies of the Salar de Huasco (Dorador et al., 2008a,b, 2009, 2010, 2013; Aguilar et al., 2016; Molina et al., 2016). It has been proposed that this exceptional microbial diversity results from species diversification, caused by the adaptation of microorganisms to the constantly changing environment (Albarracín et al., 2016). Consequently, Salar de Huasco represents a potential reservoir of undescribed microbial diversity that could serve to study and identify novel mechanisms of stress resistance.
The Exiguobacterium genus (Collins et al., 1983) includes psychrotrophic, mesophilic, and moderate thermophilic species with variable morphologies (ovoid, rods, double rods, and chains) that grow in a wide range of habitats, including cold and hot environments with temperatures ranging from -12 to 55°C (Vishnivetskaya and Kathariou, 2005; Vishnivetskaya et al., 2007). Taxonomic and phylogenetic classification of the genus by 16S rRNA gene sequence analysis shows that Exiguobacterium forms two distinct groups: Group I includes strains isolated from cold or low temperature environments, and Group II from hot springs or slightly alkaline and marine environments (Vishnivetskaya et al., 2009). One exception is Exiguobacterium sp. GIC31, which belongs to group II, but was isolated from a glacier in Greenland (Vishnivetskaya et al., 2014).
Members of the Exiguobacterium genus display traits that could be of biotechnological interest, e.g., bioremediation and agriculture applications. For example, strains Z8 and 2Sz can neutralize highly alkaline industry wastewater and remove pesticides, respectively (Lopez et al., 2005; Kumar et al., 2006); strain WK6 reduces arsenate to arsenite (Anderson and Cook, 2004), and other strains reduce chrome and mercury in a wide range of temperatures, pH, and salt concentrations (Pattanapipitpaisal et al., 2002; Petrova et al., 2002; Okeke, 2008); Exiguobacterium oxidotolerans T-2-2T shows high catalase activity in response to H2O2 as a function of the growth phase and oxygen levels, which could be used to remove peroxides used in the bleaching industry (Takebe et al., 2007); Exiguobacterium sp. S17, isolated from Laguna Socompa in the Argentinian altiplanic desert, tolerates high arsenic concentrations (Belfiore et al., 2013; Ordoñez et al., 2013). Further, members of the Exiguobacterium genus are a source of enzymes that exhibit a broad range of thermal stability (Usuda et al., 1998; Suga and Koyama, 2000; Hwang et al., 2005; Hara et al., 2007; Kasana and Yadav, 2007).
In the present study, we aimed to understand the phylogenetic placement and to identify genetic determinants required for the response to stress in a new psychro-halotolerant strain isolated from Salar de Huasco, Exiguobacterium sp. SH31. We investigated whether Exiguobacterium sp. SH31 belongs to Group II, and to what extent species from this genus share functional features based on the group to which they belong. Additionally, we searched the genomes of Exiguobacterium sp. SH31 and other species for the presence of genes encoding proteins known to be involved in stress and metal resistance, e.g., arsenic, copper, cadmium, and oxidative and UV stress. Finally, we show that Exiguobacterium SH31 is resistant to arsenic, which might be mediated, at least in part, by the Acr3 arsenic efflux pump.
Materials and Methods
Bacterial Growth Conditions and DNA Extraction
Exiguobacterium sp. SH31 was isolated from Salar de Huasco, an altiplanic shallow saline wetland in the Atacama Desert, Chile, located between -20.303436, -68.880918 at an altitude of 3,800 m. Salinity of the sampling point was 12.3% NaCl, with 20.6 mS/cm of conductivity, pH 8.8 and 8°C at the sampling point. Strain SH31 was grown for 48 h at room temperature (∼25°C) with shaking at 120 rpm in 1 L of YP culture media (2 g/L yeast extract, 5 g/L Peptone and 25 g/L NaCl), with a 10% v/v inoculum. DNA extraction was performed with the AxyPrep Bacterial Genomic DNA miniprep kit (AXYGEN) following the manufacturer’s instructions. DNA quantity and quality was determined spectrophotometrically (OD260/280 ratio).
DNA Hybridization Assay
To assess the relatedness of Exiguobacterium SH31 to the type strain Exiguobacterium aurantiacum DSM 6208T, we performed a hybridization assay in 2 X SSC + 5% formamide medium at 69°C. We evaluated the results per the recommendations of the ad hoc committee for the definition of bacterial species (threshold value of 70% DNA–DNA similarity; Wayne et al., 1987). The assay was performed at the Leibniz Institute DSMZ – German Collection of Microorganisms and Cell Cultures1.
Sequencing and Assembly
Purified genomic DNA was used to construct two different types of sequencing libraries, namely a single-end (prepared from 500 ng of fragmented DNA (nebulization)) and a mate paired-end (prepared with 5 μg of DNA fragmented by hydroshear), using the GS FLX Titanium Rapid Library, GS FLX Titanium Rapid Library prep Paired-End Adaptors, and FastStart PCR System kits, following the manufacturer’s recommendations (average fragments size was between 500 and 900 bp). An equimolar mix was then sequenced using the Roche 454 FLX platform. SFF files were separated into two FASTQ files, one for each library type, using sff_extract utility from seq_crumbs2. Single-end reads were trimmed at <Q17 and all the resulting reads of <70 bp were discarded. Reads longer than the targeted length (500 bp) were clipped at the 3′ end so that only reads <500 bp were used. Mate paired-end reads were separated into two groups, corresponding to each sequenced end of the fragment (5′ and 3′ ends reads of the sequenced fragment) and both groups were saved as backward sense reads. Trimming was performed as described for single-end reads. Only those paired reads that conserved their mates were employed for the assembly. The final number of reads obtained after this procedure was 81.675 single-end and 61.226 mate paired-reads (approximately 11X and 4X depth coverage respectively), with a read length average of 200 bp after trimming internal adapters. Assembly was performed using both mate paired and single-end reads as in the Celera genome assembler version 7.0 (Myers et al., 2000); unitigger BOG, unitigger error ratio 0.03, with the final parameter values by default. The final assembly included 120 contigs with a length of 3,031,801 bp. The Whole Genome Shotgun project has been deposited at DDBJ/ENA/GenBank under the accession LYTG00000000. The version described in this paper is LYTG01000000 (Bioproject PRJNA319980).
Exiguobacterium Genomic Dataset
All available genome sequences used in the analyses have been deposited in GenBank as of December 2015 (Accession numbers in Supplementary Table S1), including a status of complete, scaffolds, or contigs. The resulting 33 genomes were organized into 18 described species plus 15 not classified at the species level. All the genome sequences (including the new Exiguobacterium sp. Strain SH31 from this study), were re-annotated using a combination of ab initio and similarity methods as implemented in Prokka version 1.10 (Seemann, 2014) and NCBI Prokaryotic Genome Annotation Pipeline (Tatusova et al., 2016). Some genomes were also annotated using the following databases: Gene3d 3.5.0, HAMAP 201511.02, PANTHER 10.0, PFam 28.0, PIRSF 3.01, PRINTS 42.0, ProDom 2006.1, prosite 20.113, SignalP 4.0, Smart 6.2, SuperFamily 1.75, TigerFam 15.0, and TMHMM 2.0, as implemented in InterProScan.5.16-55.0 (Jones et al., 2014; Mitchell et al., 2014).
Circular sp. SH31 Genome Map Generation
The plotMyGBK3 pipeline was used to generate a circular map of the Exiguobacterium sp. SH31genome. The plotMyGBK is written in Python and extracts contigs from a.gbk file using the SeqlO module generating a.faa file that is then used to search for genes on the conserved domains database (CDD) through RPS-blast using an e-value of 1E-2 to avoid false positives. Then, genes were classified according to the Clusters of Orthologous Groups (COG) nomenclature (Marchler-Bauer et al., 2003). Finally, COG annotations were used to plot genes, GC content, GC skew, tRNA and rRNA positions, using the OmicCircos v 1.8.1. R package4 (Hu et al., 2015).
Orthologous Gene Search
Bi-directional best hit searches using Blast was used to infer homology between reference genes from Swiss-prot and the predicted genes from our Exiguobacterium dataset (Altschul et al., 1990). A minimum e-value of 1E-05 and a query coverage filter of 85% was used to avoid partial alignments. This strategy was used to compare stress-response genes and genes involved in metal/metalloid resistance/tolerance.
Phylogenetic Analysis
Thirty phylogenetic gene markers implemented in AMPHORA2 were used (dnaG, frr, infC, nusA, pgk, pyrG, rplA, rplB, rplC, rplD, rplE, rplF, rplK, rplL, rplN, rplP, rplS, rplT, rpmA, rpoB, rpsB, rpsC, rpsE, rpsI, rpsJ, rpsK, rpsM, rpsS, smpB, and tsf) from the data set (Wu and Scott, 2012). All nucleotide sequences were translation aligned using MAFFT (Katoh and Standley, 2013) as implemented in Geneious v 7.1.9 software (Kearse et al., 2012). The alignments were then concatenated using Seqotron version 1.0.1 (Fourment and Holmes, 2016). A distribution of probable trees using Bayesian Inference as implemented in MrBayes 3.2.5 was used. Two separate runs of 20 million generations were executed (four chains each run; sampling every 1000 generations; nst = 6, nucmodel = 4by4, rates = gamma ngammacat = 4). Then, the tree distributions were summarized as the Maximum a Posteriori estimate removing the first 5 thousand trees as implemented in SumTrees from DendroPy 4.1.0 (Sukumaran and Holder, 2010). The phylogeny and stress-response gene matrix was plotted using the R package ggtree v 1.0.21 (Yu et al., 2016).
Average Nucleotide Identity Analysis
Genome-wide nucleotide identity trends were explored in the genome dataset by estimating all-against-all pairwise Average Nucleotide Identity (ANI). The ANIm approach that uses MUMmer (NUCmer) was used to align the input sequences as implemented in pyANI (Kurtz et al., 20045). The average between any given pair was used as the final value. Heat maps were generated using pheatmap V 1.0.8 R package (Kolde, 2015). Genomic clusters were defined using hierarchical clustering (method: average) based on a 75% ANI threshold, which represents a conservative boundary for genus/species level classification (Qin et al., 2014).
Pan Genome Analysis
Protein-coding gene families were defined as gene clusters with at least 70% sequence identity using three different methods (COG triangle, orthoMCL and bidirectional best-hit) as implemented in GET_HOMOLOGUES v 2.0 (Contreras-Moreira and Vinuesa, 2013). The core genome was defined as the set of genes present in at least 95% of the genomes. Likewise, the accessory genome set was defined as genes present in less than 95% of genomes. This analysis was performed at two levels: the complete 34 genome dataset, and on each of the ANI 75% clusters. To gain insights into the functional potential of the accessory genome, the core genome was subtracted from each of the 34 genomes and annotated the remaining genes using Gene Ontology (GO) terms (Ashburner et al., 2000). All plots were generated in the ggplot2 R package (Wickham, 2009).
Functional Analysis of the Core and Accessory Genomes
The core and accessory genomes were separated for each of the six 75% ANI clusters, and GO terms were assigned to both sets. Gene locus tags were used to map GO terms from the Pfam entries (supplied by InterProScan5 annotated file) for each genome. GO terms for each gene cluster was converted to their parent GO terms at the second level on a GO graph (go-basic.obo obtained from http://geneontology.org/page/download-ontology) (The Gene Ontology Consortium, 2015). Statistically significant functional terms between core and accessory genomes were determined by applying the Mann–Whitney–Wilcoxon test (p-value <0.05). Results were also plotted with the ggplot2 R package (Wickham, 2009).
Growth Curves, Minimum Inhibitory Concentration (MIC), and Expression of Arsenic Resistance Genes
Growth curve assays were performed to determine the ability of Exiguobacterium SH31 to grow at different concentrations of arsenite (AsIII) and arsenate (AsV). Briefly, a liquid culture of Exiguobacterium SH31 grown to an OD600 = 0.4 (LB medium) was diluted by adding liquid media supplemented with different concentrations of arsenic species: AsIII (NaAsO2; 4, 6, 10 mM) and AsV (Na2HAsO4; 25, 50, 100 mM) until reaching an OD600 ≈ 0.004. Growth (25°C) was monitored in a microplate reader (TECAN Infinite 200 PRO Nanoquant) at 600 nm every 60 min over a 48-h period. MIC was determined from a bacterial culture grown in LB medium (OD600 ≈ 0.004) by taking a 190 μL aliquot and diluting it with solutions of AsIII and AsV to final concentrations of 1–15 mM and 5–500 mM, respectively (200 μL final volume). Then, samples were incubated at 25°C for 72 h with constant agitation, and the OD600 was measured in a TECAN Infinite 200 PRO Nanoquant.
Finally, to determine the relative expression of genes known to be involved in arsenic resistance and present in Exiguobacterium SH31, the transcript levels of arsB, arsR, arsC and acr3, were quantified by qRT-PCR. Exiguobacterium SH31 was grown in LB medium at 25°C with constant agitation until reaching an OD600 of 0.4. The culture was divided into three fractions: control, 3 mM NaAsO2, and 20 mM Na2HAsO4, and incubated for 20 min. RNA extraction was carried out using the Phenol/Chloroform method and RNA integrity was monitored using 1% agarose electrophoresis. cDNA was synthesized using the M-MLV Reverse Transcriptase kit (Promega) and Random Primer oligonucleotides hexamers (InvitrogenTM). The PCR reaction was carried out as follows: 10 min at 95°C followed by 40 amplification cycles (95°C × 30 s, 58°C × 30 s; 72°C × 30 s), and a final step of 95°C × 15 s; 25°C × 1 s; 70°C × 15 s and 95°C × 1 s) using primers specific for each gene (Primers in Supplementary Table S2). Transcript levels were quantified using the Brilliant II SYBR Green QPCR Master mix kit (Agilent Technologies) on a Stratagene Mx3000P thermal cycler. Relative expression was normalized using 16S rRNA gene expression levels (Pfaffl, 2001).
Modeling and Alignment of ACR3
Structural modeling and visualization of Acr3 from strains SH31 and S17 strains was performed using iterative threading assembly refinement (I-TASSER software) (Yang et al., 2015).
Results
Strain SH31 showed a 99% similarity at the 16S rRNA gene sequence with Exiguobacterium aurantiacum DSM 6208 (type strain) and Exiguobacterium sp. S17 (Argentinian Altiplano), and was classified as a member of the Exiguobacterium genus. However, an in vitro DNA–DNA hybridization assay revealed that SH31 does not belong to the same species than E. aurantiacum DSM 6208 (39.5% similarity; threshold is 70% for species; Wayne et al., 1987; Goris et al., 2007).
Genome Assembly and Nucleotide Identity Analysis
Exiguobacterium genomes range from 2.8 to 3.3 Mb in length with an average GC content of 46–53%. The genome of Exiguobacterium sp. strain SH31 resulted in 120 contigs totaling 3.03 Mb in length with a GC content of 51.6%. In agreement with what was found in other isolates, 2992 coding sequences (CDS; including 902 hypothetical proteins), 23 ribosomal RNA (rRNA) genes, and 49 transference RNA (tRNA) genes were identified in strain SH31. Most of the CDS found in Exiguobacterium sp. strain SH31 were annotated using the COG system (2,079 genes annotated; Figure 1A).
FIGURE 1.
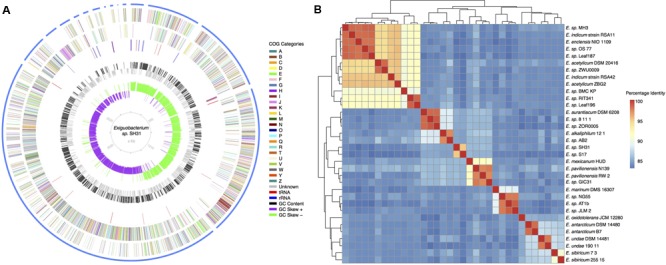
(A) Circular map of the Exiguobacterium sp. SH31 draft genome (3031801 bases, 3214 genes, 23 rRNA and 51 tRNA). Gene functions were annotated based on COG categories: (A) RNA processing and modification 0.01%; (B) Chromatin structure and dynamics 0.03%; (C) Energy production and conversion 4.5%; (D) Cell cycle control, cell division, chromosome partitioning 1.9%; (E) Amino acid transport and metabolism 10.3%; (F) Nucleotide transport and metabolism 1.6%; (G) Carbohydrate transport and metabolism 5.6%; (H) Coenzyme transport and metabolism 3.6%; (I) Lipid transport and metabolism 1.4%; (J) Translation, ribosomal structure and biogenesis 3.3%; (K) Transcription 4.8%; (L) Replication, recombination and repair 3.9%; (M) Cell wall/membrane/envelope biogenesis 4.3%; (N) Cell motility 1.3%; (O) Posttranslational modification, protein turnover, chaperones 4.4%; (P) Inorganic ion transport and metabolism 12.6%; (Q) Secondary metabolites biosynthesis, transport and catabolism 2.5%; (R) General function prediction only 19.3%; (S) Unknown function 3.4%; (T) Signal transduction mechanisms 7.1%; (U) Intracellular trafficking, secretion, and vesicular transport 0.9%; (V) Defense mechanisms 3.3%; (W) Extracellular structures 0.0%; (Y) Nuclear structure 0.0%; (Z) Cytoskeleton 0.02%. (B) Average Nucleotide Identity of 34 Exiguobacterium isolates. Exiguobacterium sp. SH31 is very distinct and is most similar to isolate Exiguobacterium S17.
To understand the overall genetic similarity of strain SH31 with members of the Exiguobacterium genus, an ANI analysis was conducted (Konstantinidis and Tiedje, 2005a,b; Goris et al., 2007; Richter and Rosselló-Mora, 2009) against other 33 publicly available Exiguobacterium genomes (Supplementary Table S1). The analysis revealed distinct genomic clusters and that strain S17 was the most similar isolate to SH31 with a 95% ANI (Figure 1B), suggesting that they belong to the same species, according to the classification defined by Goris et al. (2007). Strain S17 was isolated from a stromatolite sample of Lake Socompa in the Argentinian Altiplano (-24.525681, -68.207912), an environment with similar conditions to Salar de Huasco (Cabrol et al., 2007; Ordoñez et al., 2009; Albarracín et al., 2015). In general, as previously reported (Vishnivetskaya et al., 2009), isolates tended to cluster by habitat, i.e., strains isolated from cold environments were more similar to each other than to strains isolated from marine/alkaline/temperate environments, and vice versa. Using the proposed boundaries for species delimitation, i.e., 95% ANI (Varghese et al., 2015), 21 clusters were recognized. Because at a 95% ANI clusters were mostly composed of singletons, we used a more conservative estimate for species/genus boundary. Using a 75% ANI cut-off value, six clusters were obtained: cluster I (12 isolates) E. strain MH3, E. indicum RSA11, E. enclensis NIO-1103, E. strain OS-77, E. strain leaf 187, E. acetylicum DSM 20416, E. strain ZWU0009, E. indicum RSA42, E. acetylicum ZBG2, E. strain BCM-KP, E. strain RIT341, and E. strain leaf 196; cluster II (4 isolates) E. marinum DSM 16307, E. strain NG55, E. strain AT1b, and E. strain JLM-2; cluster III (7 isolates) E. oxidotolerans JCM 12280, E. antarcticum DSM 14480, E. antarcticum B7, E. undae DSM 14481, E. undae 190-11, E. sibiricum 7-3, and E. sibiricum 255-15; cluster IV (5 isolates) E. aurantiacum DSM 6208, E. strain 8-11-1, E. strain ZOR0005, E. alkaliphilum 12/1, and E. strain AB2; cluster V (2 isolates) E. strain SH31 and E. strain S17; cluster VI (4 isolates) E. mexicanum HUD, E. pavilionensis N139, E. pavilionensis RW-2, and E. strain GIC31.
Because Exiguobacterium sp. SH31 thrives in an environment where it is exposed to multiple stress conditions including UV radiation, high salinity, and high temperature, among others, we hypothesized that its genome encoded known genetic determinants required for stress resistance/tolerance. Using BLAST searches against SwissProt protein sets for known stress related-genes, we found that strain SH31 possesses several genes encoding proteins related to the response or adaptation to oxidative stress detoxification (katA, katE, sodA, gpo, ahpC, ahpF, ohrR); DNA repair systems (uvrABC, mutLS, mutM, ruvAB, rec systems, ssb, lexA); photolyases (phrB, cry-DASH); photoprotective pigments (car, crt systems); osmotic stress (opu systems, putP, gltA, glnA, proC); heat shock (dnaJK, groLS, htpG, hrcA); cold shock (csp); capsule biosynthesis (capAD, pgaB, pgaC); and resistance to toxic compounds such as antibiotics (marR, liaRS) and heavy metals (arsenic arsA, arsB; cadmium cadA, cadC; copper copA, csoR; mercury merA, merB, merR; tellurium tehB, terC; chromium chrR, srpC). In agreement with the ANI analysis, strain SH31 shared many of these determinants with its closest relative, strain S17, including the acr3 gene that encodes an accessory or secondary arsenic efflux pump (see Ordoñez et al., 2015).
Phylogenetic Analysis of Exiguobacterium sp. Strain SH31
We further investigated if members of the Exiguobacterium genus showed a similar pattern as in the ANI analysis, but under a phylogenetic framework. To avoid the detrimental effects of horizontally transferred genes in phylogenetic inference, 30 genes related to ribosome and replication were selected to infer the phylogenetic relationships of the group (Bayesian Inference, partitioned analysis; see Materials and Methods). Interestingly, the topology of the tree mirrors the classification proposed by Vishnivetskaya et al. (2009) and is consistent with the ANI results (Figure 2; column 1 and 2). Some species seem to be monophyletic (E. undae; E. antarcticum; E. sibiricum), while others do not form monophyletic groups (E. indicum; E. acetylicum), suggesting that more extensive sampling and clearer taxonomic classification schemes are needed.
FIGURE 2.
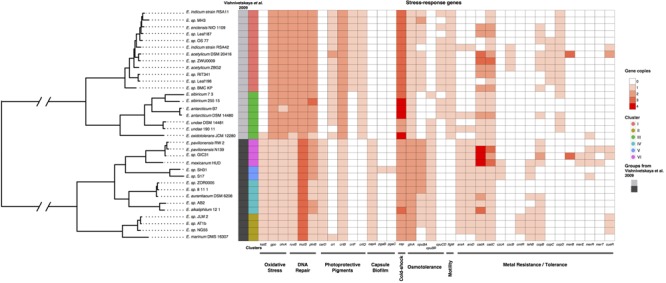
Multi-locus phylogenetic analysis of 34 Exiguobacterium isolates. Tree: Mid-point rooted phylogeny of Exiguobacterium recovers Group I and II phylogenetic groups. Note that Exiguobacterium sp. SH31 is part of Group II. Matrix: Distribution and copy number of stress-related genes among Exiguobacterium genomes. The first two columns indicate Vishnivetskaya et al., 2009 groupings and 75% ANI clustering, respectively. Stress-related genes are color-coded by copy number.
A previous work investigated the genomes of several Exiguobacterium strains to determine if there could be group-specific genes by using southernblot hybridization and Exiguobacterium sibiricum 255-15 as the reference strain. Group II does not contain the selected genes or may contain them with <70% identity (Vishnivetskaya et al., 2009). Given the current availability of genome sequences, we set out to confirm if the group pattern found by Vishnivetskaya et al. (2009) was also present in our dataset. Using a bidirectional blast hit approach, 15 of the 16 genes described by Vishnivetskaya et al. (2009) were found to be present in both groups. This finding was confirmed by annotating those genes using the InterProScan5 suite, with similar results. The only gene found exclusively in one group was flgM (Table 1).
Table 1.
Genes used as conservative markers of the two distinctive Exiguobacterium groups by Vishnivetskaya et al. (2009).
| Function | Gene ID∗ | Genes | Detectable homologous |
|||
|---|---|---|---|---|---|---|
|
Vishnivetskaya et al., 2009 |
This work |
|||||
| Group I | Group II | Group I | Group II | |||
| Replication and repair | Exig_0006 | gyrA | + | - | + | + |
| Exig_0005 | gyrB | + | + | + | + | |
| Exig_2932 | srmB/csh | + | - | + | + | |
| Exig_3033 | dnaB | + | - | + | + | |
| Transcription | Exig_2463 | flgM | + | - | + | - |
| Exig_0137 | Anti-sigma factor | + | - | + | + | |
| Exig_0136 | rpoE | + | - | + | + | |
| Translation and biogenesis | Exig_0119 | infA | + | + | + | + |
| Exig_1837 | infB | + | + | + | + | |
| Exig_0047 | RNA-binding S4 | + | - | + | + | |
| Exig_2427 | Sigma54 S30EA | + | + | + | + | |
| Posttranslational modification, | Exig_2147 | tig | + | - | + | + |
| chaperones | Exig_0781 | dnaK | + | + | + | + |
| Exig_2768 | groL | + | + | + | + | |
| Signal transduction | Exig_0364 | uspA | + | - | + | + |
| Lipid metabolism | Exig_2596 | desA | + | + | + | + |
∗Gene IDs are given as defined in http://genome.ornl.gov/microbial/exig/.
Using the same bidirectional BLAST-based approach and validation with InterProScan5, the genomes of all 34 Exiguobacterium strains were searched for the presence of stress-response genes. The analysis showed that there are differences between the two groups (Figure 2; matrix), which were observed in katE, ohrA and gpo (oxidative stress compound detoxification), carD and crtB (pigment biosynthesis), capA (capsule biosynthesis), glnA (osmoadaptation), csp (cold shock), mutS and ruvB (DNA repair) genes. The copy number of stress-response genes correlated with the environmental conditions from where each strain was isolated, e.g., E. oxydotolerans genome encodes proteins required for the response to oxidative stress and E. antarcticum and E. sibiricum for cold shock. A full list of stress-response genes analyzed and their copy numbers is found in Supplementary Table S3.
Pan Genome and Gene Ontology Analysis
As the strains used for the analyses come from disparate environments (Supplementary Table S1), we evaluated whether the core and accessory genome varied in size and biological function (by GO). Exiguobacterium genomes are highly divergent (Figure 1B), and hence, a pan genome analysis was conducted (at 70% identity; pan genome size = 7716). The analysis showed that the core genome is composed of 1,798 genes (23.3%) and the accessory genome of 5,918 genes (76.7%). The pan genome size and functional composition was also evaluated according to clusters defined at 75% ANI. Overall, pan genome size ranged from 22 to 50%, was consistent between groups, and increased according to the number of genomes within a cluster (Table 2). Cluster V, which only includes the genomes of strains S17 and SH31, had the largest accessory genome sampled so far in the genus, explaining the increased genome size of strains in this cluster. These results highlight the high degree of variability in gene content among strains from the Exiguobacterium genus, and suggest that a more thorough sampling of the Altiplano region would likely yield novel gene families.
Table 2.
Pan genome results of the six Exiguobacterium strains clusters.
| Cluster | # Strains | Pan genome | Core genome∗ | Accessory genome∗ |
|---|---|---|---|---|
| I | 12 | 4684 | 2722 (58,1%) | 1962 (41,9%) |
| II | 4 | 3557 | 2754 (77,4%) | 803 (22,6%) |
| III | 7 | 4310 | 2615 (60,7%) | 1695 (39,3%) |
| IV | 5 | 3649 | 2604 (71,4%) | 1045 (28,6%) |
| V | 2 | 3632 | 2466 (67,8%) | 1166 (32,2%) |
| VI | 4 | 3658 | 2814 (76,9%) | 844 (23,1%) |
Asterisks indicate the number of gene families (percentage of core or accessory genes on the pan genome of each cluster).
To further investigate the composition of the accessory genome, we interrogated gene distribution according to GO categories. Accessory genes related to the GO categories Binding and Catalytic Activity were more abundant within the accessory genome, while other functions such as Antioxidant Activity, Biological Regulation, and Cell Part were homogeneously distributed (Supplementary Figure S1A). Interestingly, the genomes of cluster V (strains SH31 and S17) showed a higher proportion of genes belonging to GO categories such as Catalytic activity, Cell part, Membrane, Metabolic process, Structural molecule activity, and Cellular process (Supplementary Figure S1B).
Metal/Metalloid Response Genes in Exiguobacterium Genomes
The genomes of all 34 strains were searched for the presence and patterns of metal/metalloid response genes, and whether their distribution matched the phylogenetic and ANI classifications. The analysis revealed the presence of several genes involved in metal/metalloid response, of which most were related to cobalt, zinc, cadmium, copper, mercury, tellurium, chromium, and arsenic resistance (Supplementary Table S4). Previous studies showed that strain S17 is resistant to arsenic (Belfiore et al., 2013; Ordoñez et al., 2015) and that it encodes a secondary arsenic efflux pump, named Acr3. Because SH31 and S17 are closely related, we investigated if strain SH31 and other members of the Exiguobacterium genus also encoded the genetic determinants required for arsenic resistance, including acr3. For this purpose, strain S17 was used as a reference. Based on the presence and identity of the genes and operon organization, two distinct patterns were observed. In most cases, these patterns matched the two phylogenetic groups described before, with the exception of E. marinum DSM 16307 from group II and E. indicum RSA42, E. antarcticum DSM 14480, and E. undae 190-11 from group I (Figure 3). The acr3 (arsenite efflux pump) gene is less frequent in Exiguobacterium genomes and was only found in strains S17 and SH31, showing an identity of 94%.
FIGURE 3.
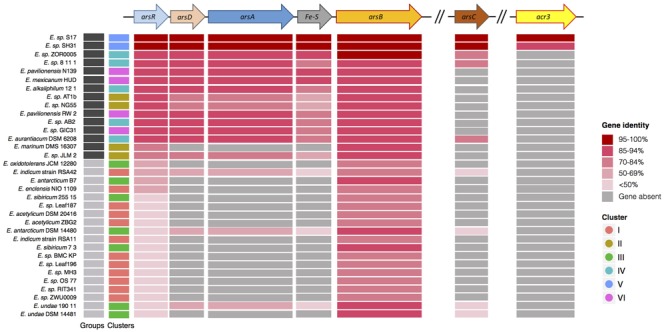
Arsenic resistance genes in sequenced Exiguobacterium strains Genetic organization of the genes colored by function. arsR arsenical resistance operon repressor; arsD arsenical resistance operon trans-acting repressor; arsA arsenical pump-driving ATPase; Fe–S putative iron sulfur protein; arsB arsenical pump membrane protein; arsC arsenate reductase; acr3 arsenite efflux pump. Heat scale shows identity percentage of all strains sequence, using as reference the Exiguobacterium S17 genes. The first two columns indicate Vishnivetskaya et al. (2009) groupings and 75% ANI clustering, respectively.
Arsenic Resistance and Expression of Genes Required for Arsenic Detoxification
To determine if the presence of genes required for arsenic detoxification in strain SH31 correlated with increased arsenic resistance, we conducted growth curves with different As (III) and As (V) concentrations (Figure 4) and determined the MIC for each compound. Strain SH31 was able to grow in media containing up to 10 mM As (III) and 100 mM As(V), indicating that it is resistant to arsenic. A similar result was observed for strain S17.
FIGURE 4.
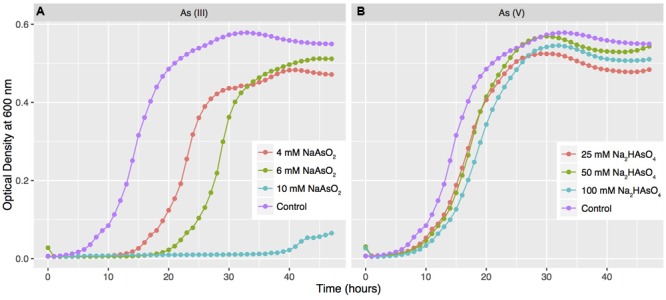
Arsenic resistance of strain SH31. Strain SH31 was grown in LB medium supplemented with different concentrations of As (III) (A) and As (V) (B). OD600 readings were recorded every hour during 48 h. The results represent the mean of three experiments. Growth in media without arsenic was used as a control.
The high arsenic resistance of strain S17 has been explained in part by the presence of the acr3 gene (Ordoñez et al., 2015). A structural analysis showed that Acr3 from strains SH31 and S17 have an amino acid sequence identity of 94% and that key amino acids are conserved between both proteins. For example, residues Arg264, Asp109, and Glu294 are part of the core transmembrane segment and together with Cys107 could be required for As (III) binding. However, some variations were observed at different sections of the protein, (locus_02597-SP17) i.e., residues H92Y, R187Q, A311D, S34V, and S164A. We speculate that S34V, and S164A could be regulated by serine kinases (Supplementary Figure S2).
To gain insights into the mechanism of arsenic resistance, strain SH31 was grown to mid-exponential growth phase, challenged with 3 mM As (III) or 20 mM As (V), and the transcript levels of genes required for arsenic resistance were quantified by qRT-PCR (Figure 5) and compared to the levels in untreated cells (control). The expression of arsB, encoding a membrane efflux transporter, increased in the presence of both As (III) and As (V), although in the later case a greater increase was observed (200 fold). Previous studies found a similar trend and showed that As (V) exposure resulted in higher transcript and protein levels of ArsB than with As (III) (Cleiss-Arnold et al., 2010; Belfiore et al., 2013; Andres and Bertin, 2016). No significant differences in the transcript levels of arsC, encoding arsenate reductase, were observed after As (V) exposure. Conversely, after As (III) exposure, arsC expression was strongly repressed. Previous studies reported that arsC is part of the ars operon in most bacteria (Páez-Espino et al., 2009; Kruger et al., 2013; Andres and Bertin, 2016); however, in the Exiguobacterium genus, arsC is not within the context of an operon (Figure 3). The expression of arsR, encoding the negative regulator of the ars operon, increased after As (III) exposure. As ArsR directly detects As (III) (Slyemi and Bonnefoy, 2012), this suggests that in strain SH31 As (III) exposure results in increased expression of the ars operon by decreasing the levels of ArsR.
FIGURE 5.
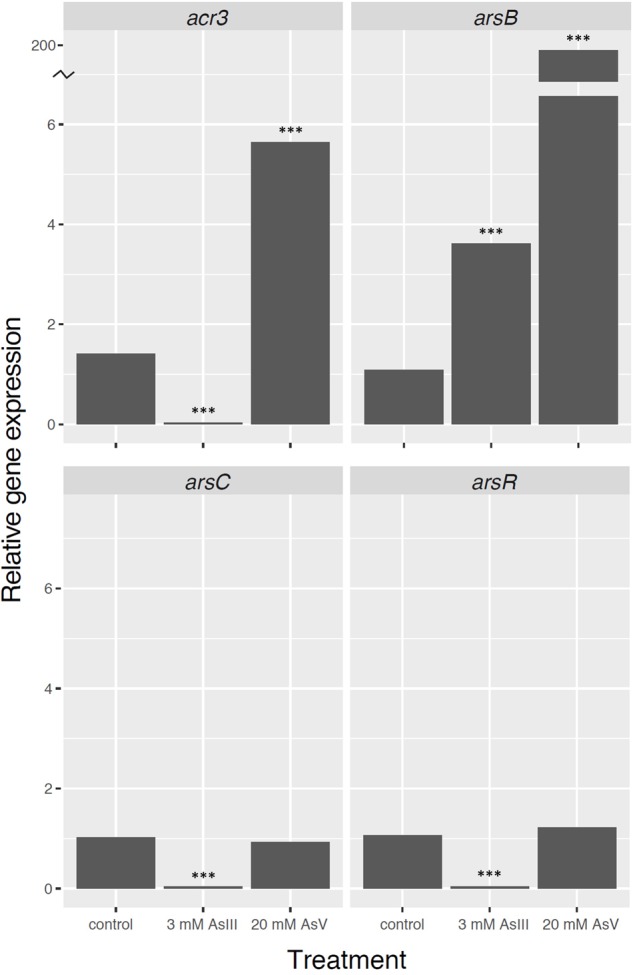
Expression of genes required for arsenic resistance in Exiguobacterium sp. SH31 in response to As(III) and As(V) exposure. Expression of the 16S rRNA gene was used as a normalizer. ∗∗∗p < 0.001.
The expression of acr3 increased after As (V), but not As (III) exposure, in strain SH31. Interestingly, it was proposed that acr3 expression is constitutive in strain S17 and does not change in response to arsenic (Ordoñez et al., 2015). This suggests that the expression of genes required for arsenic resistance could be different in both strains, although we cannot rule out that the observed result is caused by different experimental conditions between the studies.
Discussion
The Chilean Altiplano harbors a series of ‘extreme’ environmental conditions that configures a specific and unique microbiota (e.g., Dorador et al., 2013; Albarracín et al., 2016). Exiguobacterium is a frequently isolated from different environments including other saline wetlands (e.g., Cuatro Ciénagas: Rebollar et al., 2012), where it has been determined that genetic differentiation of Exiguobacterium are defined by the ecology of diverse habitats.
Members of the Exiguobacterium genus carry a number of stress-related genetic determinants, including metal/metalloid resistance genes, which largely correlate with the phylogenetic structure (Figure 2). In particular, Exiguobacterium SH31 (Group II) encodes proteins required for arsenic resistance, including Acr3, which has only been described in Exiguobacterium S17 (Ordoñez et al., 2015) (Figures 3, 5). We also found that the accessory genome of Exiguobacterium is large, and that the presence of genes involved in interactions with the environment (e.g., catalytic activity, binding, metabolic process) is variable (Supplementary Figure S1). Altogether, these results suggest that the Exiguobacterium genus is cosmopolitan, diverse, and possibly ancient, with a broad repertoire of genetic elements that allow them to efficiently adapt to local environmental conditions (Figure 2).
Exiguobacterium strains showed variability in the GO categories related to adaptation, such as catalytic activity, transporter activity, nucleic acid binding, transcription factor activity, metabolic process, etc. Conversely, less variability was observed in the GO categories related to fundamental cellular functions, e.g., structural molecule activity, membrane constituents, macromolecular complex, cell parts, and biological regulation. In agreement with our findings, Yumoto et al. (2004) reported that E. oxidotolerans JCM 12280 differed from other closely related strains in their physiological and biochemical characteristics, e.g., growth temperature range, acid production from several substrates and enzymatic activities. Our analyses support these results, showing that E. oxidotolerans JCM 12280 appears to be distinct based on its phylogenetic position, ANI similarity, and stress-response gene repertoire.
In agreement with the high levels of radiation that it must withstand in its environment (Hernández et al., 2016; Molina et al., 2016), the genome of strain SH31 comprises several genes encoding proteins required for UV radiation resistance, including the uvr, mut, rec and ruv systems; the ssb, lexA, phrB and cry-DASH genes, photoprotective pigments and those required for detoxifying oxidative stress (Kurth et al., 2015). Surprisingly, strain SH31 did not show any highly relevant distinctive features. For example, previous studies found four copies of ssb in the genome of strain S17 (Ordoñez et al., 2013, 2015). In Deinococcus radiodurans, the presence of high copy numbers of ssb was proposed to be responsible for its superior radioresistance and DNA repair efficiency (Eggington et al., 2004; Cox and Battista, 2005; Norais et al., 2009). Long read sequencing technologies such as PacBio or Oxford Nanopores could aid in determining with certainty whether SH31 has experienced the duplication of gene families involved in stress response.
Some Exiguobacterium strains are resistant to NaCl and heavy metals such as Pb, Cu, and Hg (Petrova et al., 2002; Bian et al., 2011). In particular, Exiguobacterium isolated from permafrost sediments in Russia and Canada that contained 0.001 – 2.9% of mercury carried the mer operon (Petrova et al., 2002). Our results show that all of the Exiguobacterium strains included in the analyses have the merA gene, but that only 6 strains had additional genes of the operon (Supplementary Table S4). The mer operon can be located in the chromosome, plasmids, or transposons, and has been studied as a mechanism of horizontal dissemination that confers a broad spectrum of mercury resistance among Bacillus and related species (Bogdanova et al., 2001). Other strains such as ZM-2, isolated from agricultural soils irrigated with tannery effluents in India, resist up to 12.4 mM of potassium chromate (Alam and Malik, 2008), and some isolates tolerate up to 1.5 mM and reduce 0.75 mM of Cr(VI) (Sarangi and Krishnan, 2008). Additionally, some Exiguobacterium isolates are able to reduce Cr(VI) anaerobically in a large range of temperatures, pH and salt concentrations (Pattanapipitpaisal et al., 2002; Okeke, 2008). Our comparative genomics approach confirms that all 34 Exiguobacterium strains possess the genes encoding proteins required for chromate reduction and efflux, namely chrR and srpC, respectively, which could be responsible for chromate resistance in this genus.
Exiguobacterium sp. SH31 is more closely related to the arsenic resistant strain S17 (Belfiore et al., 2013). The high arsenic resistance of strain S17 has been explained by an increased number of genes encoding proteins required to detoxify this toxic compound and by the presence of the acr3 gene, which confers increased arsenite and arsenate resistance (10 and 150 mM, respectively; Belfiore et al., 2013; Ordoñez et al., 2015). In agreement, strain SH31 is also arsenic resistant, which could be explained by the presence of several proteins required for its detoxification, including Acr3.
In the post-genomic era, high-throughput technologies have enabled researchers with the power to interrogate genomes from the dark matter of the microbial tree of life (Wu et al., 2009). Yet, the traditional use of morphology, phenotype, biochemical traits, and single-gene inferences to classify microorganisms implies that our current understanding of what constitutes a species or genus is imperfect (Hugenholtz et al., 2016). Herein, we show that using an extreme cut-off of 75% ANI in a relatively small dataset yielded as many as six clusters in an otherwise unified genus. While comparative genomics studies of polyextremohiles are needed to understand their distribution, evolutionary history, and biotechnological potential, thorough sampling designs, metagenomics-based studies, and functional assays such as those based on metabolomics, transcriptomics, and proteomics, will enable researchers to develop a systems level understanding of the patterns and processes leading to molecular adaptation.
Author Contributions
JF, PA, CD, and FR performed field work, processed samples and sequenced data; JC-S, EC-N, and CPS conceived and designed the study; JC-S, FR, and CS performed the experiments; JC-S, SV, DA, and EC-N analyzed data; RQ, FM, EC-N, CD, FR, and CPS contributed with reagents/materials/analysis tools; JC-S, CP-E, EC-N, FR, and CPS wrote the paper. All authors read and approved the final manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
EC-N would like to thank the George Washington University’s high-performance computing facility, Colonial One, for providing data storage, support, and computing power for genomic analyses (colonialone.gwu.edu/). The authors would like to thank Eduardo H. Morales for critical reading and editing of the manuscript.
Funding. This research was sponsored by CONICYT (Comisión Nacional para la Investigación Científica y Tecnológica) grants: CPS was funded by CONICYT FONDECYT 1160315 and Universidad Andrés Bello UNAB DI-786-15/R grant; FR was funded by CONICYT FONDECYT 11100414; EC-N was funded by CONICYT 2014 PAI 82140008 and FONDECYT 11160905; CD was funded by FONDECYT 1110953 and 1140179; JC-S and CP-E were founded by 2015 CONICYT National Doctoral Fellowship. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2017.00456/full#supplementary-material
References
- Aguilar P., Acosta E., Dorador C. Y., Sommaruga R. (2016). Large differences in bacterial community composition among three nearby extreme waterbodies of the high Andean plateau. Front. Microbiol. 7:976 10.3389/fmicb.2016.00976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alam M., Malik A. (2008). Chromate resistance, transport and bioreduction by Exiguobacterium sp. ZM-2 isolated from agricultural soil irrigated with tannery effluent. J. Basic Microbiol. 48 416–420. 10.1002/jobm.200800046 [DOI] [PubMed] [Google Scholar]
- Albarracín V., Gärtner W., Farias M. E. (2016). Forged under the sun: life and art of extremophiles from Andean Lakes. Photochem. Photobiol. 1 14–28. 10.1111/php.12555 [DOI] [PubMed] [Google Scholar]
- Albarracín V., Kurth D., Ordoñez O., Belfiore C., Luccini E., Salum G., et al. (2015). High-up: a remote reservoir of microbial extremophiles in central Andean wetlands. Front. Microbiol. 6:1404 10.3389/fmicb.2015.01404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albarracín V., Pathak G., Douki T., Cadet J., Borsarelli C., Gärtner W., et al. (2012). Extremophilic Acinetobacter strains from high-altitude lakes in Argentinean Puna: remarkable UV-B resistance and efficient DNA damage repair. Orig. Life Evol. Biosph. 42 201–221. 10.1007/s11084-012-9276-3 [DOI] [PubMed] [Google Scholar]
- Altschul S., Gish W., Miller W., Myers E., Lipman D. (1990). Basic local alignment search tool. J. Mol. Biol. 215 403–410. 10.1016/S0022-2836(05)80360-2 [DOI] [PubMed] [Google Scholar]
- Anderson C. R., Cook G. M. (2004). Isolation and characterization of arsenate-reducing bacteria from arsenic-contaminated sites in New Zealand. Curr. Microbiol. 48 341–347. 10.1007/s00284-003-4205-3 [DOI] [PubMed] [Google Scholar]
- Andres J., Bertin P. (2016). The microbial genomics of arsenic. FEMS Microbiol. Rev. 40 299–322. 10.1093/femsre/fuv050 [DOI] [PubMed] [Google Scholar]
- Ashburner M., Ball C., Blake J., Botstein D., Butler H., Cherry J., et al. (2000). Gene ontology: tool for the unification of biology. Nat. Genet. 25 25–29. 10.1038/7556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai Y., Müller D. B., Srinivas G., Garrido-Oter R., Potthoff E., Rott M., et al. (2015). Functional overlap of the Arabidopsis leaf and root microbiota. Nature 528 364–369. 10.1038/nature16192 [DOI] [PubMed] [Google Scholar]
- Belfiore C., Ordoñez O., Farías M. E. (2013). Proteomic approach of adaptive response to arsenic stress in Exiguobacterium sp. S17, an extremophile strain isolated from a high-altitude Andean Lake stromatolite. Extremophiles 17 421–431. 10.1007/s00792-013-0523-y [DOI] [PubMed] [Google Scholar]
- Bian G., Zhang Y., Qin S., Xing K., Xie H., Jiang J. (2011). Isolation and biodiversity of heavy metal tolerant endophytic bacteria from halotolerant plant species located in coastal shoal of Nantong. Wei Sheng Wu Xue Bao 51 1538–1547. [PubMed] [Google Scholar]
- Bogdanova E., Minakhin L., Bass I., Volodin A., Hobman J., Nikiforov V. (2001). Class II broad-spectrum mercury resistance transposons in gram-positive bacteria from natural environments. Res. Microbiol. 152 503–514. 10.1016/S0921-2508(01)01224-4 [DOI] [PubMed] [Google Scholar]
- Bowers K., Mesbah N., Wiegel J. (2009). Biodiversity of poly-extremophilic Bacteria: does combining the extremes of high salt, alkaline pH and elevated temperature approach a physico-chemical boundary for life? Saline Systems 5:9 10.1186/1746-1448-5-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bull A., Asenjo J. (2013). Microbiology of hyper-arid environments: recent insights from the Atacama Desert. Chile. Antonie Van Leeuwenhoek 103 1173–1179. 10.1007/s10482-013-9911-7 [DOI] [PubMed] [Google Scholar]
- Bull A. T., Asenjo J., Goodfellow M. Y., Gómez-Silva B. (2016). The Atacama Desert: technical resources and the growing importance of novel microbial diversity. Annu. Rev. Microbiol. 70 215–234. 10.1146/annurev-micro-102215-095236 [DOI] [PubMed] [Google Scholar]
- Cabria G., Argayosa V., Lazaro J., Argayosa A., Arcilla C. (2014). Draft genome sequence of haloalkaliphilic Exiguobacterium sp. strain AB2 from Manleluag Ophiolitic Spring. Philippines. Genome Announc. 2:e00840-14. 10.1128/genomeA.00840-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabrol N., Wettergreen D., Warren-Rhodes K., Grin E., Moersch J., Diaz G., et al. (2007). Life in the Atacama: searching for life with rovers (science overview). J. Geophys. Res. Biogeosci. 112 G04S02. 10.1029/2006JG000298 [DOI] [Google Scholar]
- Carneiro A., Ramos R., Dall’Agnol H., Pinto A., de Castro Soares S., Santos A., et al. (2012). Genome sequence of Exiguobacterium antarcticum B7, isolated from a biofilm in Ginger Lake, King George Island, Antarctica. J. Bacteriol. 194 6689–6690. 10.1128/JB.01791-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catalán J., Camarero L., Felip M., Pla S., Ventura M., Buchaca T., et al. (2006). High mountain lakes: extreme habitats and witnesses of environmental change. Limnetica 25 551–584. [Google Scholar]
- Cleiss-Arnold J., Koechler S., Proux C., Fardeau M., Dillies M., Coppee J., et al. (2010). Temporal transcriptomic response during arsenic stress in Herminiimonas arsenicoxydans. BMC Genomics 11:709 10.1186/1471-2164-11-709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins M., Lund B., Farrow J., Schleifer K. (1983). Chemotaxonomic study of an alkalophilic bacterium, Exiguobacterium aurantiacum gen. nov., sp. nov. J. Gen. Microbiol. 129 2037–2042. 10.1099/00221287-129-7-2037 [DOI] [Google Scholar]
- Contreras-Moreira B., Vinuesa P. (2013). GET_HOMOLOGUES, a versatile software package for scalable and robust microbial pangenome analysis. Appl. Environ. Microbiol. 79 7696–7701. 10.1128/AEM.02411-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordero R., Seckmeyer G., Damiani A., Riechelmann S., Rayas J., Labbe F., et al. (2014). The world’s highest levels of surface UV. Photochem. Photobiol. Sci. 13 70–81. 10.1039/c3pp50221j [DOI] [PubMed] [Google Scholar]
- Cox M., Battista J. (2005). Deinococcus radiodurans - the consummate survivor. Nat. Rev. Microbiol. 3 882–892. 10.1038/nrmicro1264 [DOI] [PubMed] [Google Scholar]
- Dastager S., Mawlankar R., Sonalkar V., Thorat M., Mual P., Verma A., et al. (2015). Exiguobacterium enclense sp. nov., isolated from sediment. Int. J. Syst. Evol. Microbiol. 65 1611–1616. 10.1099/ijs.0.000149 [DOI] [PubMed] [Google Scholar]
- Di Capua C., Bortolotti A., Farías M. E., Cortez N. (2011). UV-resistant Acinetobacter sp. isolates from Andean wetlands display high catalase activity. FEMS Microbiol. Lett. 317 181–189. 10.1111/j.1574-6968.2011.02231.x [DOI] [PubMed] [Google Scholar]
- Dib J., Motok J., Zenoff V., Ordonez O., Farias M. E. (2008). Occurrence of resistance to antibiotics, UV-B, and arsenic in bacteria isolated from extreme environments in high-altitude (above 4400 m) andean wetlands. Curr. Microbiol. 56 510–517. 10.1007/s00284-008-9103-2 [DOI] [PubMed] [Google Scholar]
- Dib J., Weiss A., Neumann A., Ordoñez O., Estévez M., Farías M. E. (2009). Isolation of bacteria from remote high altitude Andean lakes able to grow in the presence of antibiotics. Recent Pat. Antiinfect. Drug Discov. 4 66–76. 10.2174/157489109787236300 [DOI] [PubMed] [Google Scholar]
- Dorador C., Busekow A., Vila I., Imhoff J., Witzel K. P. (2008a). Molecular analysis of enrichment cultures of ammonia oxidizers from the Salar de Huasco, a high altitude saline wetland in northern Chile. Extremophiles 12 405–414. 10.1007/s00792-008-0146-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorador C., Meneses D., Urtuvia V., Demergasso C., Vila I., Witzel K. P., et al. (2009). Diversity of Bacteroidetes in high altitude saline evaporitic basins in northern Chile. J. Geophys. Res. 114 G00D05. 10.1029/2008JG000837 [DOI] [Google Scholar]
- Dorador C., Remonsellez F., Vila I., Imhoff J. F., Witzel K. P. (2010). Unique clusters of Archaea in Salar de Huasco, an athalassohaline evaporitic basin of the Chilean Altiplano. FEMS Microbiol. Ecol. 73 291–302. 10.1111/j.1574-6941.2010.00891.x [DOI] [PubMed] [Google Scholar]
- Dorador C., Vila I., Imhoff J. F., Witzel K. P. (2008b). Cyanobacterial diversity in Salar de Huasco, a high altitude saline wetland in northern Chile: an example of geographical dispersion? FEMS Microbiol. Ecol. 64 419–432. 10.1111/j.1574-6941.2008.00483.x [DOI] [PubMed] [Google Scholar]
- Dorador C., Vila I., Witzel K. P., Imhoff J. F. (2013). Bacterial and archaeal diversity in high altitude wetlands of the Chilean Altiplano. Fundam. Appl. Limnol. 182 135–159. 10.1127/1863-9135/2013/0393 [DOI] [Google Scholar]
- Duché O., Trémoulet F., Glaser P., Labadie J. (2002). Salt stress proteins induced in Listeria monocytogenes. Appl. Environ. Microbiol. 68 1491–1498. 10.1128/AEM.68.4.1491-1498.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eggington J., Haruta N., Wood E., Cox M. (2004). The single-stranded DNA-binding protein of Deinococcus radiodurans. BMC Microbiol. 4:2 10.1186/1471-2180-4-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farias M., Fernandez-Zenoff V., Flores R., Ordonez O., Estevez C. (2009). Impact of solar radiation on bacterioplankton in Laguna Vilama, a hypersaline Andean lake (4650 m). J. Geophys. Res. 114 G00D04 10.1029/2008JG000784 [DOI] [Google Scholar]
- Farías M. E., Rascovan N., Toneatti D. M., Albarracín V. H., Flores M. R., Poiré D. G., et al. (2013). The discovery of stromatolites developing at 3570 m above sea level in a high-altitude volcanic lake Socompa, Argentinean Andes.PLoS ONE 8:e53497 10.1371/journal.pone.0053497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Zenoff V., Sineriz F., Farias M. E. (2006). Diverse responses to UV-B radiation and repair mechanisms of bacteria isolated from high-altitude aquatic environments. Appl. Environ. Microbiol. 72 7857–7863. 10.1128/AEM.01333-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flores M., Ordoñez O., Farías M. E. (2009). Isolation of UV-B resistant bacteria from two high altitude Andean lakes (4,400 m) with saline and non saline conditions. J. Gen. Appl. Microbiol. 55 447–458. 10.2323/jgam.55.447 [DOI] [PubMed] [Google Scholar]
- Fourment M., Holmes E. C. (2016). Seqotron: a user-friendly sequence editor for Mac OS X. BMC Res. Notes 9:106 10.1186/s13104-016-1927-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goris J., Konstantinidis K., Klappenbach J., Coenye T., Vandamme P., Tiedje J. M. (2007). DNA-DNA hybridization values and their relationship to whole-genome sequence similarities. Int. J. Syst. Evol. Microbiol. 57 81–91. 10.1099/ijs.0.64483-0 [DOI] [PubMed] [Google Scholar]
- Hara I., Ichise N., Kojima K., Kondo H., Ohgiya S., Matsuyama H., et al. (2007). Relationship between the size of the bottleneck 15 angstrom from iron in the main channel and the reactivity of catalase corresponding to the molecular size of substrates. Biochemistry 46 11–22. 10.1021/bi061519w [DOI] [PubMed] [Google Scholar]
- Hernández K., Yannicelli B., Olsen L., Dorador C., Menschel E., Molina V., et al. (2016). Microbial activity response to solar radiation across contrasting environmental conditions in Salar de Huasco, Northern Chilean Altiplano. Front. Microbiol. 7:1857 10.3389/fmicb.2016.01857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y., Yan C., Hsu C., Chen Q., Niu K., Komatsoulis G., et al. (2015). OmicCircos: a simple-to-use R package for the circular visualization of multidimensional omics data. Cancer Inform. 13 13–20. 10.4137/CIN.S13495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hugenholtz P., Skarshewski A., Parks D. (2016). Genome-based microbial taxonomy coming of age. Cold Spring Harb. Perspect. Biol. 8:a018085 10.1101/cshperspect.a018085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang B. Y., Kim J. H., Kim J., Kim B. G. (2005). Screening of Exigubacterium acetylicum from soil samples showing enantioselective and alkalitolerant esterase activity. Biotechnol. Bioprocess Eng. 10 367–371. [Google Scholar]
- Hyson P., Shapiro J., Wien M. W. (2015). Draft genome sequence of Exiguobacterium sp. strain BMC-KP, an environmental isolate from Bryn Mawr, Pennsylvania. Genome Announc. 3:e01164-15. 10.1128/genomeA.01164-1115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang X., Xue Y., Wang L., Yu B., Ma Y. (2013). Genome sequence of a novel polymer-grade l-lactate-producing alkaliphile, Exiguobacterium sp. strain 8-11-1. Genome Announc. 1:e00616-13 10.1128/genomeA.00616-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones P., Binns D., Chang H., Fraser M., Li W., McAnulla C., et al. (2014). InterProScan 5: genome-scale protein function classification. Bioinformatics 30 1236–1240. 10.1093/bioinformatics/btu031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasana R. C., Yadav S. K. (2007). Isolation of a psychrotrophic Exiguobacterium sp. SKPB5 (MTCC 7803) and characterization of its alkaline protease. Curr. Microbiol. 54 224–229. 10.1007/s00284-006-0402-1 [DOI] [PubMed] [Google Scholar]
- Katoh K., Standley D. M. (2013). MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 30 772–780. 10.1093/molbev/mst010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearse M., Moir R., Wilson A., Stones-Havas S., Cheung M., Sturrock S., et al. (2012). Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28 1647–1649. 10.1093/bioinformatics/bts199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolde R. (2015). pheatmap: Pretty Heatmaps. R Package Version 1.0.8. Available at: https://CRAN.R-project.org/package=pheatmap [Google Scholar]
- Konstantinidis K. T., Tiedje J. M. (2005a). Towards a genome-based taxonomy for prokaryotes. J. Bacteriol. 187 6258–6264. 10.1128/JB.187.18.6258-6264.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konstantinidis K. T., Tiedje J. M. (2005b). Genomic insights that advance the species definition for prokaryotes. Proc. Natl. Acad. Sci. U.S.A. 102 2567–2572. 10.1073/pnas.0409727102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruger M., Bertin P. N., Heipieper H., Arsene-Ploetze F. (2013). Bacterial metabolism of environmental arsenic—mechanisms and biotechnological applications. Appl. Microbiol. Biotechnol. 97 3827–3841. 10.1007/s00253-013-4838-5 [DOI] [PubMed] [Google Scholar]
- Kumar A., Singh V., Kumar R. (2006). “Characterization of an alkaliphile, Exiguobacterium sp. and it’s application in bioremediation”, in Proceedings of the International Conference on Extremophiles Brest-France: 115. [Google Scholar]
- Kurth D., Belfiore C., Gorriti M., Cortez N., Farias M. E., Albarracín V. H. (2015). Genomic and proteomic evidences unravel the UV-resistome of the poly-extremophile Acinetobacter sp. Ver3. Front. Microbiol. 6:328 10.3389/fmicb.2015.00328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurtz S., Phillippy A., Delcher A., Smoot M., Shumway M., Antonescu C., et al. (2004). Versatile and open software for comparing large genomes. Genome Biol. 5:R12 10.1186/gb-2004-5-2-r12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez L., Pozo C., Rodelas B., Calvo C., Juarez B., Martinez-Toledo M., et al. (2005). Identification of bacteria isolated from an oligotrophic lake with pesticide removal capacities. Ecotoxicology 14 299–312. 10.1007/s10646-003-6367-y [DOI] [PubMed] [Google Scholar]
- Lynch R., King A., Farías M. E., Sowell P., Vitry C., Schmidt S. K. (2012). The potential for microbial life in the highest-elevation (> 6000 masl) mineral soils of the Atacama region. J. Geophys. Res. Atmos. 117 G02028 10.1029/2012JG001961 [DOI] [Google Scholar]
- Marchler-Bauer A., Anderson J. B., DeWeese-Scott C., Fedorova N. D., Geer L. Y., He S., et al. (2003). CDD: a curated Entrez database of conserved domain alignments. Nucleic Acids Res. 31 383–387. 10.1093/nar/gkg087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Márquez-García M., Vila I., Hinojosa L. F., Mendez M. A., Carvajal J., Sabando M. C. (2009). Distribution and seasonal fluctuations in the aquatic biodiversity of the southern Altiplano. Limnologica 39 314–318. 10.1016/j.limno.2009.06.007 [DOI] [Google Scholar]
- Midha S., Bansal K., Sharma S., Kumar N., Patil P., Chaudhry V., et al. (2015). Genomic resource of rice seed associated bacteria. Front. Microbiol. 6:1551 10.3389/fmicb.2015.01551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell A., Chang H., Daugherty L., Fraser M., Hunter S., Lopez R., et al. (2014). The InterPro protein families database: the classification resource after 15 years. Nucleic Acids Res. 43 D213–D221. 10.1093/nar/gku1243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molina V., Hernández K., Dorador C., Eissler Y., Hengst M., Pérez V., et al. (2016). Bacterial active community cycling in response to solar radiation and their influence on nutrient changes in a high-altitude wetland. Front. Microbiol. 7:1823 10.3389/fmicb.2016.01823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers E., Sutton G., Delcher A., Dew I., Fasulo D., Flanigan M., et al. (2000). A whole-genome assembly of Drosophila. Science 287 2196–2204. 10.1126/science.287.5461.2196 [DOI] [PubMed] [Google Scholar]
- Nonaka K., Yoon K., Ogo S. (2014). Biochemical characterization of psychrophilic Mn-superoxide dismutase from newly isolated Exiguobacterium sp. OS-77. Extremophiles 18 363–373. 10.1007/s00792-013-0621-x [DOI] [PubMed] [Google Scholar]
- Norais C., Chitteni-Pattu S., Wood E., Inman R., Cox M. (2009). DdrB protein, an alternative Deinococcus radiodurans SSB induced by ionizing radiation. J. Biol. Chem. 284 21402–21411. 10.1074/jbc.M109.010454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okeke B. C. (2008). Bioremoval of hexavalent chromium from water by a salt tolerant bacterium, Exiguobacterium sp. GS1. J. Ind. Microbiol. Biotechnol. 35 1571–1579. 10.1007/s10295-008-0399-5 [DOI] [PubMed] [Google Scholar]
- Ordoñez O., Flores M., Dib J., Paz A., Farias M. E. (2009). Extremophile culture collection from Andean lakes: extreme pristine environments that host a wide diversity of microorganisms with tolerance to UV radiation. Microb. Ecol. 58 461–473. 10.1007/s00248-009-9527-7 [DOI] [PubMed] [Google Scholar]
- Ordoñez O., Lanzarotti E., Kurth D., Cortez N., Farias M. E., Turjanski A. G. (2015). Genome comparison of two Exiguobacterium strains from high altitudeandean lakes with different arsenic resistance: identification and 3Dmodeling of the Acr3 efflux pump. Front. Environ. Sci. 3:50 10.3389/fenvs.2015.00050 [DOI] [Google Scholar]
- Ordoñez O., Lanzarotti E., Kurth D., Gorriti M., Revale S., Cortez N., et al. (2013). Draft genome sequence of the polyextremophilic Exiguobacterium sp. strain S17, isolated from hyperarsenic lakes in the Argentinian Puna. Genome Announc. 1:e00480-13 10.1128/genomeA.00480-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Páez-Espino D., Tamames J., de Lorenzo V., Cánovas D. (2009). Microbial responses to environmental arsenic. Biometals 22 117–130. 10.1007/s10534-008-9195-y [DOI] [PubMed] [Google Scholar]
- Pattanapipitpaisal P., Mabbett A., Finlay J., Beswick A., Paterson-Beedle M., Essa A., et al. (2002). Reduction of Cr (VI) and bioaccumulation of chromium by gram positive and gram negative microorganisms not previously exposed to Cr-stress. Environ. Technol. 23 731–745. 10.1080/09593332308618367 [DOI] [PubMed] [Google Scholar]
- Petrova M., Mindlin S., Gorlenko Z., Kalyaeva E., Soina V., Bogdanova E. S. (2002). Mercury-resistant bacteria from permafrost sediments and prospects for their use in comparative studies of mercury resistance determinants. Russ. J. Genet. 38 1330–1334. [PubMed] [Google Scholar]
- Pfaffl M. W. (2001). A new mathematical model for relative quantification in real-time RT–PCR. Nucleic Acids Res. 29:e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin Q., Xie B., Zhang X., Chen X., Zhou B., Zhou J., et al. (2014). A proposed genus boundary for the prokaryotes based on genomic insights. J. Bacteriol. 196 2210–2215. 10.1128/JB.01688-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebollar E. A., Avitia M., Eguiarte L., González-González A., Mora L., Bonilla-Rosso G. Y., et al. (2012). Water–sediment niche differentiation in ancient marine lineages of Exiguobacterium endemic to the Cuatro Cienegas Basin. Environ. Microbiol. 14 2323–2333. 10.1111/j.1462-2920.2012.02784.x [DOI] [PubMed] [Google Scholar]
- Richter M., Rosselló-Mora R. (2009). Shifting the genomic gold standard for the prokaryotic species definition. Proc. Natl. Acad. Sci. U.S.A. 106 19126–19131. 10.1073/pnas.0906412106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigues D., Goris J., Vishnivetskaya T., Gilichinsky D., Thomashow M. F., Tiedje J. M. (2006). Characterization of Exiguobacterium isolates from the Siberian permafrost. Description of Exiguobacterium sibiricum sp. nov. Extremophiles 10 285–294. 10.1007/s00792-005-0497-5 [DOI] [PubMed] [Google Scholar]
- Rothschild L. J., Mancinelli R. L. (2001). Life in extreme environments. Nature 409 1092–1101. 10.1038/35059215 [DOI] [PubMed] [Google Scholar]
- Rout S., Rai A., Humphreys P. N. (2015). Draft genome sequence of alkaliphilic Exiguobacterium sp. strain HUD, isolated from a polymicrobial consortia. Genome Announc. 3:e01451-14. 10.1128/genomeA.01451-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarangi A., Krishnan C. (2008). Comparison of in vitro Cr (VI) reduction by CFEs of chromate resistant bacteria isolated from chromate contaminated soil. Bioresour. Technol. 99 4130–4137. 10.1016/j.biortech.2007.08.059 [DOI] [PubMed] [Google Scholar]
- Seemann T. (2014). Prokka: rapid prokaryotic genome annotation. Bioinformatics 30 2068–2069. 10.1093/bioinformatics/btu153 [DOI] [PubMed] [Google Scholar]
- Semova I., Carten J. D., Stombaugh J., Mackey L., Knight R., Farber S., et al. (2012). Microbiota regulate intestinal absorption and metabolism of fatty acids in the zebrafish. Cell Host Microbe 12 277–288. 10.1016/j.chom.2012.08.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seufferheld M. J., Alvarez H. M., Farias M. E. (2008). Role of polyphosphates in microbial adaptation to extreme environments. Appl. Environ. Microbiol. 74 5867–5874. 10.1128/AEM.00501-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slyemi D., Bonnefoy V. (2012). How prokaryotes deal with arsenic. Environ. Microbiol. Rep. 4 571–586. 10.1111/j.1758-2229.2011.00300.x [DOI] [PubMed] [Google Scholar]
- Suga S., Koyama N. (2000). Purification and properties of a novel azide-sensitive ATPase of Exiguobacterium aurantiacum. Arch. Microbiol. 173 200–205. 10.1007/s002039900129 [DOI] [PubMed] [Google Scholar]
- Sukumaran J., Holder M. (2010). DendroPy: a python library for phylogenetic computing. Bioinformatics 26 1569–1571. 10.1093/bioinformatics/btq228 [DOI] [PubMed] [Google Scholar]
- Takebe F., Hara I., Matsuyama H., Yumoto I. (2007). Effect of H2O2 under low- and high-aeration-level conditions on growth and catalase activity in Exiguobacterium oxidotolerans T-2-2T. J. Biosci. Bioeng. 104 464–469. 10.1263/jbb.104.464 [DOI] [PubMed] [Google Scholar]
- Tang J., Zhang Y., Meng H., Xue Z., Ma J. (2013). Complete genome sequence of Exiguobacterium sp. strain MH3, isolated from rhizosphere of Lemna minor. Genome Announc. 1:e01059-13 10.1128/genomeA.01059-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatusova T., DiCuccio M., Badretdin A., Chetvernin V., Nawrocki E., Zaslavsky L., et al. (2016). NCBI prokaryotic genome annotation pipeline. Nucleic Acids Res. 1 1–11. 10.1093/nar/gkw569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Gene Ontology Consortium (2015). Gene ontology consortium: going forward. Nucleic acids Res. 43 D1049–D1056. 10.1093/nar/gku1179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Usuda Y., Kawasaki H., Shimaoka M., Utagawa T. (1998). Molecular characterization of guanosine kinase gene from a facultative alkalophile, Exiguobacterium aurantiacum ATCC 35652. Biochim. Biophys. Acta 1442 373–379. 10.1016/s0167-4781(98)00166-3 [DOI] [PubMed] [Google Scholar]
- Varghese N., Mukherjee S., Ivanova N., Konstantinidis K., Mavrommatis K., Kyrpides N. C., et al. (2015). Microbial species delineation using whole genome sequences. Nucleic Acids Res. 43 6761–6771. 10.1093/nar/gkv657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vishnivetskaya T. A., Chauhan A., Layton A., Pfiffner S., Huntemann M., Copeland A., et al. (2014). Draft genome sequences of 10 strains of the genus Exiguobacterium. Genome Announc. 2:e01058-14 10.1128/genomeA.01058-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vishnivetskaya T. A., Kathariou S. (2005). Putative transposases conserved in Exiguobacterium isolates from ancient Siberian permafrost and from contemporary surface habitats. Appl. Environ. Microbiol. 71 6954–6962. 10.1128/AEM.71.11.6954-6962.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vishnivetskaya T. A., Kathariou S., Tiedje J. M. (2009). The Exiguobacterium genus: biodiversity and biogeography. Extremophiles 13 541–555. 10.1007/s00792-009-0243-5 [DOI] [PubMed] [Google Scholar]
- Vishnivetskaya T. A., Lucas S., Copeland A., Lapidus A., Glavina del Rio T., Dalin E., et al. (2011). Complete genome sequence of the Thermophilic Bacterium Exiguobacterium sp. AT1b. J. Bacteriol. 193 2880–2881. 10.1128/JB.00303-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vishnivetskaya T. A., Siletzky R., Jefferies N., Tiedje J. M., Kathariou S. (2007). Effect of low temperature and culture media on the growth and freeze-thawing tolerance of Exiguobacterium strains. Cryobiology 54 234–240. 10.1016/j.crybiol.2007.01.008 [DOI] [PubMed] [Google Scholar]
- Wayne L., Brenner D., Colwell R., Grimont P., Kandler O., Krichevsky M., et al. (1987). Report of the ad hoc committee on reconciliation of approaches to bacterial systematics. Int. J. Syst. Bacteriol. 37 463–464. 10.1099/00207713-37-4-463 [DOI] [Google Scholar]
- White R., Grassa C., Suttle C. A. (2013). Draft genome sequence of Exiguobacterium pavilionensis strain RW-2, with wide thermal, salinity, and pH tolerance, isolated from modern freshwater microbialites. Genome Announc. 1:e00597-13 10.1128/genomeA.00597-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickham H. (2009). ggplot2: Elegant Graphics for Data Analysis. New York, NY: Springer-Verlag. [Google Scholar]
- Wu D., Hugenholtz P., Mavromatis K., Pukall R., Dalin E., Ivanova N., et al. (2009). A phylogeny-driven genomic encyclopaedia of Bacteria and Archaea. Nature 462 1056–1060. 10.1038/nature08656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu M., Scott A. J. (2012). Phylogenomic analysis of bacterial and archaeal sequences with AMPHORA2. Bioinformatics 28 1033–1034. 10.1093/bioinformatics/bts079 [DOI] [PubMed] [Google Scholar]
- Yang J., Yan R., Roy A., Xu D., Poisson J., Zhang Y. (2015). The I-TASSER Suite: protein structure and function prediction. Nat. Methods 12 7–8. 10.1038/nmeth.3213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu G., Smith D., Zhu H., Guan Y., Lam T. (2016). ggtree: an R package for visualization and annotation of phylogenetic trees with their covariates and other associated data. Methods Ecol. Evol. 8 28–36. 10.1111/2041-210X.12628 [DOI] [Google Scholar]
- Yumoto I., Hishinuma-Narisawa M., Hirota K., Shingyo T., Takebe F., Nodasaka Y., et al. (2004). Exiguobacterium oxidotolerans sp. nov., a novel alkaliphile exhibiting high catalase activity. Int. J. Syst. Evol. Microbiol. 54 2013–2017. 10.1099/ijs.0.63129-0 [DOI] [PubMed] [Google Scholar]
- Zenoff V., Heredia J., Ferrero M., Sineriz F., Farias M. (2006). Diverse UV-B resistance of culturable bacterial community from high-altitude wetland water. Curr. Microbiol. 52 359–362. 10.1007/s00284-005-0241-5 [DOI] [PubMed] [Google Scholar]
- Zhang D., Liu Y., Huo Y., Xu X., Li X. (2015). Draft genome sequence of Thermophilic Exiguobacterium sp. strain JLM-2, isolated from deep-sea ferromanganese nodules. Genome Announc. 3:e00794-15 10.1128/genomeA.00794-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


